Improved Pullulan Production and Process Optimization Using Novel GA–ANN and GA–ANFIS Hybrid Statistical Tools
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth and Culture of Aureobasidium Pullulans
2.2. Optimization of Fermentation Variables
2.2.1. Process Optimization by Conformist Approach
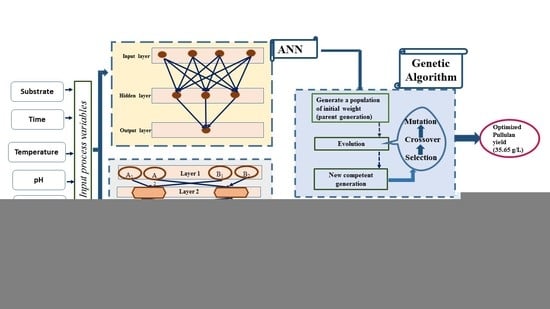
2.2.2. Development of Artificial Neural Networking (ANN) Model for Optimization of Process Variables
2.2.3. Implementing the GA–ANN Model to Optimize the Process Parameters for Pullulan Yield
2.2.4. Development of ANFIS Model for Experimental Data Training
2.2.5. Optimization of Process Parameters for Pullulan Production Using the GA–ANFIS Model
2.3. Extraction and Estimation of Pullulan Yield
3. Results and Discussion
3.1. Optimization of Various Production Parameters Using the OVAT Approach
3.1.1. Substrate Optimization by the OVAT Approach
3.1.2. Optimization of Temperature by the OVAT Approach
3.1.3. Optimization of pH by the OVAT Approach
3.1.4. Optimization of Duration of Fermentation (Time) by the OVAT Approach
3.1.5. Optimization of Agitation Speed by the OVAT Approach
3.2. Optimization of Fermentation Process Variable and Pullulan Yield by Hybrid Learning Tools
3.2.1. ANN Model for Training of Experimental Data
3.2.2. GA–ANN Model Executed to Optimize the Process Parameters for Pullulan Weight
3.2.3. ANFIS Model for the Experimental Data Training
3.2.4. GA–ANFIS Model for Optimization of Process Parameters and Pullulan Weight
3.2.5. Comparative Analysis of the Artificial Intelligence and Simulation Approaches Employed (GA–ANN and GA–ANFIS)
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lin, Y.; Zhang, Z.; Thibault, J. Aureobasidium pullulans batch cultivations based on a factorial design for improving the production and molecular weight of exopolysaccharides. Process Biochem. 2007, 42, 820–827. [Google Scholar] [CrossRef]
- Dubey, K.K. Insights of Microbial Pullulan Production: A Bioprocess Engineer Assessment. Curr. Biotechnol. 2018, 7, 262–272. [Google Scholar]
- Wang, D.; Yu, X.; Gongyuan, W. Pullulan production and physiological characteristics of Aureobasidium pullulans under acid stress. Appl. Microbiol. Biotechnol. 2013, 97, 8069–8077. [Google Scholar] [CrossRef]
- Deshpande, M.S.; Rale, V.B.; Lynch, J.M. Aureobasidium pullulans in applied microbiology: A status report. Enzym. Microb. Technol. 1992, 14, 514–527. [Google Scholar] [CrossRef]
- Mishra, B.; Vuppu, S. A study on downstream processing for the production of pullulan by Aureobasidium pullulans-SB-01 from the fermentation broth. Res. J. Recent Sci. 2013, 2277, 2502. [Google Scholar]
- Reeslev, M.; Jensen, B. Influence of Zn2+ and Fe3+ on polysaccharide production and mycelium/yeast dimorphism of Aureobasidium pullulans in batch cultivations. Appl. Microbiol. Biotechnol. 1995, 42, 910–915. [Google Scholar] [CrossRef]
- Shingel, K.I. Current knowledge on biosynthesis, biological activity, and chemical modification of the exopolysaccharide, pullulan. Carbohydr. Res. 2004, 339, 447–460. [Google Scholar] [CrossRef]
- Lin, Y.; Thibault, J. Pullulan fermentation using a prototype rotational reciprocating plate impeller. Bioprocess Biosyst. Eng. 2013, 36, 603–611. [Google Scholar] [CrossRef]
- Moscovici, M.; Ionescu, C.; Oniscu, C.; Fotea, O.; Hanganu, L.D. Exopolysaccharide biosynthesis by a fast-producing strain of Aureobasidium pullulans. Biotechnol. Lett. 1993, 15, 1167–1172. [Google Scholar] [CrossRef]
- Wiley, B.; Ball, D.; Arcidiacono, S.; Sousa, S.; Mayer, J.; Kaplan, D. Control of molecular weight distribution of the biopolymer pullulan produced byAureobasidium pullulans. J. Environ. Polym. Degrad. 1993, 1, 3–9. [Google Scholar] [CrossRef]
- Wu, S.; Jin, Z.; Kim, J.M.; Tong, Q.; Chen, H. Downstream processing of pullulan from fermentation broth. Carbohydr. Polym. 2009, 77, 750–753. [Google Scholar] [CrossRef]
- Abdel Hafez, A.; Abdelhady, H.M.; Sharaf, M.; El-Tayeb, T. Bioconversion of various industrial by–products and agricultural wastes into pullulan. J. Appl. Sci. Res. 2007, 3, 1416–1425. [Google Scholar]
- Choudhury, A.R.; Sharma, N.; Prasad, G. Deoiledjatropha seed cake is a useful nutrient for pullulan production. Microb. Cell Factories 2012, 11, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, A.; Prasad, G.; Choudhury, A.R. Cost effective production of pullulan from agri-industrial residues using response surface methodology. Int. J. Biol. Macromol. 2014, 64, 252–256. [Google Scholar] [CrossRef]
- Kana, E.B.G.; Olokeb, J.K.; Lateefb, A.; Oyebanjib, A. Comparative evaluation of artificial neural network coupled genetic algorithm and response surface methodology for modeling and optimization of citric acid production by Aspergillus niger MCBN297. Chem. Eng. 2012, 27, 397–402. [Google Scholar]
- Uzuner, S.; Cekmecelioglu, D. Comparison of artificial neural networks (ANN) and adaptive neuro-fuzzy inference system (ANFIS) models in simulating polygalacturonase production. BioResources 2016, 11, 8676–8685. [Google Scholar] [CrossRef] [Green Version]
- Valdez-Castro, L.; Baruch, I.; Barrera-Cortes, J. Neural networks applied to the prediction of fed-batch fermentation kinetics of Bacillus thuringiensis. Bioprocess Biosyst. Eng. 2003, 25, 229–233. [Google Scholar] [CrossRef]
- Haider, M.; Pakshirajan, K.; Singh, A.; Chaudhry, S. Artificial neural network-genetic algorithm approach to optimize media constituents for enhancing lipase production by a soil microorganism. Appl. Biochem. Biotechnol. 2008, 144, 225–235. [Google Scholar] [CrossRef]
- Nagata, Y.; Chu, K.H. Optimization of a fermentation medium using neural networks and genetic algorithms. Biotechnol. Lett. 2003, 25, 1837–1842. [Google Scholar] [CrossRef]
- Jang, J.-S. ANFIS: Adaptive-network-based fuzzy inference system. IEEE Trans. Syst. ManCybern. 1993, 23, 665–685. [Google Scholar] [CrossRef]
- Baş, D.; Boyacı, İ.H. Modeling and optimization II: Comparison of estimation capabilities of response surface methodology with artificial neural networks in a biochemical reaction. J. Food Eng. 2007, 78, 846–854. [Google Scholar] [CrossRef]
- Dutta, J.R.; Dutta, P.K.; Banerjee, R. Optimization of culture parameters for extracellular protease production from a newly isolated Pseudomonas sp. using response surface and artificial neural network models. Process Biochem. 2004, 39, 2193–2198. [Google Scholar] [CrossRef]
- Chiranjeevi, P.V.; Pandian, M.R.; Thadikamala, S. Integration of artificial neural network modeling and genetic algorithm approach for enrichment of laccase production in solid state fermentation by Pleurotus ostreatus. BioResources 2014, 9, 2459–2470. [Google Scholar] [CrossRef] [Green Version]
- Rekha, V.; Ghosh, M.; Adapa, V.; Oh, S.-J.; Pulicherla, K.; Sambasiva Rao, K. Optimization of polygalacturonase production from a newly isolated Thalassospira frigidphilosprofundus to use in pectin hydrolysis: Statistical approach. Biomed Res. Int. 2013, 2013, 750187. [Google Scholar] [CrossRef] [PubMed]
- Dhankhar, R.; Kumar, A.; Kumar, S.; Chhabra, D.; Shukla, P.; Gulati, P. Multilevel algorithms and evolutionary hybrid tools for enhanced production of arginine deiminase from Pseudomonas furukawaii RS3. Bioresour. Technol. 2019, 290, 121789. [Google Scholar] [CrossRef] [PubMed]
- Badhwar, P.; Kumar, P.; Dubey, K.K. Development of aqueous two-phase systems comprising poly ethylene glycol and dextran for purification of pullulan: Phase diagrams and fiscal analysis. Eng. Life Sci. 2018, 18, 524–531. [Google Scholar] [CrossRef] [Green Version]
- Badhwar, P.; Kumar, P.; Dubey, K.K. Extractive fermentation for process integration and amplified pullulan production by A. pullulans in Aqueous two phase systems. Sci. Rep. 2019, 9, 32. [Google Scholar] [CrossRef] [Green Version]
- West, T.; Strohfus, B. Polysaccharide production by a reduced pigmentation mutant of Aureobasidium pullulans NYS-1. Lett. Appl. Microbiol. 2001, 33, 169–172. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y.; Zhao, S.; Zhou, Q.; Xin, X.; Chen, L. Statistical optimization of medium for pullulan production by aureobasidium pullulans NCPS2016 using fructose and soybean meal hydrolysates. Molecules 2018, 23, 1334. [Google Scholar] [CrossRef] [Green Version]
- Cheng, K.-C.; Demirci, A.; Catchmark, J.M. Evaluation of medium composition and fermentation parameters on pullulan production by Aureobasidium pullulans. Food Sci. Technol. Int. 2011, 17, 99–109. [Google Scholar] [CrossRef]
- Sheng, L.; Tong, Q.; Ma, M. Why sucrose is the most suitable substrate for pullulan fermentation by Aureobasidium pullulans CGMCC1234? Enzym. Microb. Technol. 2016, 92, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.C.; Kim, Y.H.; Lee, H.S.; Kim, Y.N.; Byun, S.M. Production of pullulan by a fed-batch fermentation. Biotechnol. Lett. 1987, 9, 621–624. [Google Scholar] [CrossRef]
- Lazaridou, A.; Biliaderis, C.G.; Roukas, T.; Izydorczyk, M. Production and characterization of pullulan from beet molasses using a nonpigmented strain of Aureobasidium pullulans in batch culture. Appl. Biochem. Biotechnol. 2002, 97, 1–22. [Google Scholar] [CrossRef]
- McNeil, B.; Kristiansen, B. Temperature effects on polysaccharide formation by Aureobasidium pullulans in stirred tanks. Enzym. Microb. Technol. 1990, 12, 521–526. [Google Scholar] [CrossRef]
- Ürküt, Z.; Daǧbaǧlı, S.; Göksungur, Y. Optimization of pullulan production using Ca-alginate-immobilized Aureobasidium pullulans by response surface methodology. J. Chem. Technol. Biotechnol Int. Res. ProcessEnviron. Clean Technol. 2007, 82, 837–846. [Google Scholar]
- Alzoubi, I.; Delavar, M.R.; Mirzaei, F.; Nadjar Arrabi, B. Comparing ANFIS and integrating algorithm models (ICA-ANN, PSO-ANN, and GA–ANN) for prediction of energy consumption for irrigation land leveling. Geosystem Eng. 2018, 21, 81–94. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, A.; Chhabra, D.; Shukla, P. Improved biobleaching of mixed hardwood pulp and process optimization using novel GA–ANN and GA–ANFIS hybrid statistical tools. Bioresour. Technol. 2019, 271, 274–282. [Google Scholar] [CrossRef]




| Sr. No | Training Algorithm | Code Name Used | Training: R | Validation: R | Test: R | All: R |
|---|---|---|---|---|---|---|
| 1 | Levenberg–Marquardt (LM) | trainlm1 | 0.97759 | 0.94728 | 0.92455 | 0.94799 |
| 2 | Levenberg–Marquardt (LM) | trainlm2 | 0.99775 | 0.95739 | 0.89542 | 0.95895 |
| Sr. No. | ANN Training Algorithm | Optimized GA Value | Pullulan Yield (g/L) (Best Value) | Pullulan Yield (g/L) (Mean Value) | ||||
|---|---|---|---|---|---|---|---|---|
| Substrate ((g/L) | Time (h) | Temperature (°C) | pH | Agitation Speed (rpm) | ||||
| 1 | trainlm1 | 48.9 | 172.62 | 26.89 | 6.56 | 224.85 | 39.4918 | 39.3691 |
| 2 | trainlm2 | 44.9 | 172.33 | 27.85 | 6.99 | 210.56 | 35.5672 | 35.5552 |
| Sr. No. | MF Type | Epoch 3: Error | Method Adopted to Generate FIS | Train FIS Optimized Method | Optimized GA Value | Pullulan Yield (g/L) (Best Value) | Pullulan Yield (g/L) (Mean Value) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Substrate (g/L) | Time (h) | Temp. (◦C) | pH | Agitation Speed (rpm) | ||||||||
| 1 | Constant | gaussmf | 6.1055 × 10−5 | Grid-Partition | Hybrid | 49.94 | 182.39 | 27.41 | 6.99 | 190.08 | 36.0788 | 36.0425 |
| 2 | Constant | trimf | 4.7389 × 10−5 | Grid-Partition | Hybrid | 49.7 | 180.04 | 27.45 | 6.99 | 190.29 | 35.8462 | 35.1299 |
| 3 | Constant | gbellmf | 0.00011749 | Grid-Partition | Hybrid | 49.94 | 181.51 | 27.39 | 6.98 | 190.07 | 35.8332 | 35.6952 |
| 4 | Constant | gauss2mf | 0.0002537 | Grid-Partition | Hybrid | 49.902 | 179.99 | 27.46 | 6.95 | 190.01 | 35.7434 | 35.7379 |
| 5 | Constant | dsigmf | 0.00032176 | Grid-Partition | Hybrid | 49.92 | 179.42 | 27.52 | 6.98 | 190.17 | 35.6809 | 35.6671 |
| Sr. No | Optimization Tool/Method | Optimized Process Parameters | Predicted Pullulan Yield (g/L) | Experimental Pullulan Yield (g/L) | Percentage of Accuracy | ||||
|---|---|---|---|---|---|---|---|---|---|
| Substrate (g/L) | Time (h) | Temp (°C) | pH | Agitation Speed (rpm) | |||||
| 1 | Laboratory Fermentation | 45.00 | 180 | 27.5 | 7.5 | 200 | - | 35.55 | - |
| 2 | GA–ANN | 48.90 | 172.62 | 26.89 | 6.56 | 224.85 | 39.4918 | 37.642 ± 0.521 | 95.32 |
| 3 | GA–ANFIS | 49.94 | 182.39 | 27.41 | 6.99 | 190.08 | 36.0788 | 35.652 ± 0.348 | 98.82 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badhwar, P.; Kumar, A.; Yadav, A.; Kumar, P.; Siwach, R.; Chhabra, D.; Dubey, K.K. Improved Pullulan Production and Process Optimization Using Novel GA–ANN and GA–ANFIS Hybrid Statistical Tools. Biomolecules 2020, 10, 124. https://doi.org/10.3390/biom10010124
Badhwar P, Kumar A, Yadav A, Kumar P, Siwach R, Chhabra D, Dubey KK. Improved Pullulan Production and Process Optimization Using Novel GA–ANN and GA–ANFIS Hybrid Statistical Tools. Biomolecules. 2020; 10(1):124. https://doi.org/10.3390/biom10010124
Chicago/Turabian StyleBadhwar, Parul, Ashwani Kumar, Ankush Yadav, Punit Kumar, Ritu Siwach, Deepak Chhabra, and Kashyap Kumar Dubey. 2020. "Improved Pullulan Production and Process Optimization Using Novel GA–ANN and GA–ANFIS Hybrid Statistical Tools" Biomolecules 10, no. 1: 124. https://doi.org/10.3390/biom10010124