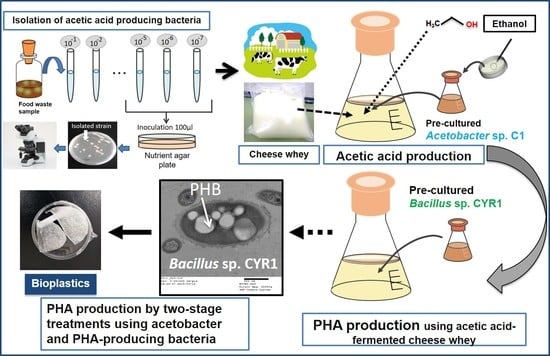
Two-Stage Polyhydroxyalkanoates (PHA) Production from Cheese Whey Using Acetobacter pasteurianus C1 and Bacillus sp. CYR1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biocatalyst
2.2. Isolation of Acetic-Acid-Producing Bacteria and Optimum Conditions for Acetic Acid Production
2.3. Acetic Acid Production with CW
2.4. Lactose Utilization Experiments
2.5. PHA Production without Pre-Treatment of CW and Quantification
2.6. Effect of Pre-Treatment of CW for the Production of PHA
2.7. PHB Identifivation
2.8. Analysis
3. Results and Discussion
3.1. Acetobacter Pasteurianus C1 Strain
3.2. Optimum Conditions for the Production of Acetic Acid
| Strain | Acetic Acid (g/L) | pH | Temperature (°C) | Ethanol (g/L) | Rotation Speed (rpm) | References |
|---|---|---|---|---|---|---|
| Acetobacter pasteurianus CWBI-B419 | 16 | n.a | 38 | 19.4 | 130 | [21] |
| Acetobacter pasteurianus MSU10 | 25 | n.a | 37 | 38.82–46.58 | 200 | [22] |
| Acetobacter pasteurianus SKU1108 | 35 | n.a | 37 | 38.82–46.58 | 200 | [22] |
| Acetobacter pasteurianus SL13E-2 | 40 | n.a | 37 | 46.58 | 200 | [23] |
| Acetobacter pasteurianus SL13E-3 | 40 | n.a | 37 | 46.58 | 200 | [23] |
| Acetobacter pasteurianus SL13E-4 | 40 | n.a | 37 | 46.58 | 200 | [23] |
| Acetobacter malorum | 40 | n.a | 37 | n.a | n.a | [24] |
| Acetobacter rancens SKU 1102 | 43.0 | n.a | 38 | 31.05 | 220 | [27] |
| Acetobacter pasteurianus SKYAA25 | 52.5 g/100 g of dry substrate | 5.5 | 37 | 62.10 | 180 | [28] |
| Acetobacter pasteurianus AAB4 | 42 | n.a | 37 | 77.63 | 180 | [29] |
| Acetobacter pasteurianus CICC 20001 | 48.24 | n.a | 32 | 35.70 | 180 | [30] |
| Acetobacter pasteurianus C1 | 37.02 | 4–5 | 30 | 54.34 | 180 | This study |
3.3. Lactose Utilization
3.4. PHA Production with Two-Stage Methodology
3.4.1. First Stage—Acetic Acid Production
3.4.2. Second Stage—PHA Production
Effect of Nitrogen Concentration on PHA Production
PHA Production with Pretreated CW
3.5. Identification of PHB Produced from the Two-Stage Process
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khanna, S.; Srivastava, A.K. Recent advances in microbial polyhydroxyalkanoates. Process. Biochem. 2005, 40, 607–619. [Google Scholar] [CrossRef]
- Berezina, N.; Yada, B.; Lefebvre, R. From organic pollutants to bioplastics: Insights into the bioremediation of aromatic compounds by Cupriavidus necator. New Biotechnol. 2015, 32, 47–53. [Google Scholar] [CrossRef]
- Castilho, L.R.; Mitchell, D.A.; Freire, D.M.G. Production of polyhydroxyalkanoates (PHAs) from waste materials and by-products by submerged and solid-state fermentation. Bioresour. Technol. 2009, 100, 5996–6009. [Google Scholar] [CrossRef] [PubMed]
- Valta, K.; Damala, P.; Angeli, E.; Antonopoulou, G.; Malamis, D.; Haralambous, K.J. Current treatment technologies of cheese whey and wastewater by Greek cheese manufacturing units and potential valorization opportunities. Waste Biomass Valoriz. 2017, 8, 1649–1663. [Google Scholar] [CrossRef]
- Koller, M.; Salerno, A.; Muhr, A.; Reiterer, A.; Chiellini, E.; Casella, S.; Horvat, P.; Braunegg, P.H.A.G. Whey Lactose as a Raw Material for Microbial Production of Biodegradable Polyesters. In Poliester; Saleh, H.E.-D.M., Ed.; IntechOpen: London, UK, 2013. [Google Scholar] [CrossRef] [Green Version]
- Pescuma, M.; de Valdez, G.F.; Mozzi, F. Whey-derived valuable products obtained by microbial fermentation. Appl. Microbiol. Biotechnol. 2015, 99, 6183–6196. [Google Scholar] [CrossRef] [PubMed]
- Berwig, K.H.; Baldasso, C.; Dettmer, A. Production and characterization of poly(3-hydroxybutyrate) generated by Al-caligenes latus using lactose and whey after acid protein precipitation process. Bioresour. Technol. 2016, 218, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Khatami, K.; Perez-Zabaleta, M.; Owusu-Agyeman, I.; Cetecioglu, Z. Waste to bioplastics: How close are we to sustainable polyhydroxyalkanoates production? Waste Manag. 2021, 119, 374–388. [Google Scholar] [CrossRef] [PubMed]
- Amaro, T.M.M.M.; Rosa, D.; Comi, G.; Iacumin, L. Prospects for the Use of Whey for Polyhydroxyalkanoate (PHA) Production. Front. Microbiol. 2019, 10, 992. [Google Scholar] [CrossRef]
- Reddy, M.V.; Mawatari, Y.; Onodera, R.; Nakamura, Y.; Yajima, Y.; Chang, Y.-C. Bacterial conversion of waste into polyhydroxybutyrate (PHB): A new approach of bio-circular economy for treating waste and energy generation. Bioresour. Technol. Rep. 2019, 7, 100246. [Google Scholar] [CrossRef]
- Reddy, M.V.; Watanabe, A.; Onodera, R.; Mawatari, Y.; Tsukiori, Y.; Watanabe, A.; Kudou, M.; Chang, Y.-C. Polyhydroxyalkanoates (PHA) production using single or mixture of fatty acids with Bacillus sp. CYR1: Identification of PHA synthesis genes. Bioresour. Technol. Rep. 2020, 11, 100483. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Ikeutsu, K.; Toyama, T.; Choi, D.; Kikuchi, S. Isolation and characterization of tetrachloroethylene- and cis-1,2-dichloroethylene-dechlorinating propionibacteria. J. Ind. Microbiol. Biotechnol. 2011, 38, 1667–1677. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.C.; Takada, K.; Choi, D.B.; Toyama, T.; Sawada, K.; Kikuchi, S. Isolation of biphenyl and polychlorinated bi-phenyl-degrading bacteria and their degradation pathway. Appl. Biochem. Biotechnol. 2013, 170, 381–398. [Google Scholar] [CrossRef]
- Saichana, N.; Matsushita, K.; Adachi, O.; Frébort, I.; Frebortova, J. Acetic acid bacteria: A group of bacteria with versatile biotechnological applications. Biotechnol. Adv. 2015, 33, 1260–1271. [Google Scholar] [CrossRef]
- Law, J.H.; Slepecky, A.S. Assay of poly-β-hydroxybutyric acid. J. Bacteriol. 1961, 82, 33–36. [Google Scholar] [CrossRef] [Green Version]
- Obruca, S.; Marova, I.; Melusova, S.; Mravcova, L. Production of polyhydroxyalkanoates from cheese whey employing Bacillus megaterium CCM 2037. Ann. Microbiol. 2011, 61, 947–953. [Google Scholar] [CrossRef]
- Yellore, V.; Desai, A. Production of poly-3-hydroxybutyrate from lactose and whey by Methylobacterium sp. ZP24. Lett. Appl. Microbiol. 1998, 26, 391–394. [Google Scholar] [CrossRef]
- Sengun, I.Y.; Karabiyikli, S. Importance of acetic acid bacteria in food industry. Food Control. 2011, 22, 647–656. [Google Scholar] [CrossRef]
- Pallach, M.; di Lorenzo, F.; Facchini, F.A.; Gully, D.; Giraud, E.; Peri, F.; Duda, K.A.; Molinaro, A.; Silipo, A. Structure and inflammatory activity of the LPS isolated from Acetobacter pasteurianus CIP103108. Int. J. Biol. Macromol. 2018, 119, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Gullo, M.; Verzelloni, E.; Canonico, M. Aerobic submerged fermentation by acetic acid bacteria for vinegar production: Process and biotechnological aspects. Process. Biochem. 2014, 49, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- Ndoye, B.; Lebecque, S.; Dubois-Dauphin, R.; Tounkara, L.; Guiro, A.-T.; Kere, C.; Diawara, B.; Thonart, P. Thermoresistant properties of acetic acids bacteria isolated from tropical products of Sub-Saharan Africa and destined to industrial vinegar. Enzym. Microb. Technol. 2006, 39, 916–923. [Google Scholar] [CrossRef]
- Kanchanarach, W.; Theeragool, G.; Yakushi, T.; Toyama, H.; Osao Adachi, O.; Matsushita, K. Characterization of thermotolerant Acetobacter pasteurianus strains and their quinoprotein alcohol dehydrogenases. Appl. Microbiol. Biotechnol. 2010, 85, 741–751. [Google Scholar] [CrossRef]
- Perumpuli, P.A.B.N.; Watanabe, T.; Toyama, H. Identification and characterization of thermotolerant acetic acid bacteria strains isolated from coconut water vinegar in Sri Lanka. Biosci. Biotechnol. Biochem. 2014, 78, 533–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Es-Sbata, I.; Lakhlifi, T.; Yatim, M.; El-Abid, H.; Belhaj, A.; Hafidi, M.; Zouhair, R. Screening and molecular characterization of new thermo- and ethanol-tolerant Acetobacter malorum strains isolated from two biomes Moroccan cactus fruits. Biotechnol. Appl. Biochem. 2021, 68, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Kadere, T.T.; Miyamoto, T.; Oniang‘o, R.K.; Kutima, P.M.; Njoroge, S.M. Isolation and identification of the genera Acetobacter and Gluconobacter in coconut toddy (mnazi). Afr. J. Biotechnol. 2008, 7, 2963–2971. [Google Scholar]
- Iino, T.; Suzuki, R.; Tanaka, N.; Kosako, Y.; Ohkuma, M.; Komagata, K.; Uchimura, T. Gluconacetobacter kakiaceti sp. nov., an acetic acid bacterium isolated from a traditional Japanese fruit vinegar. Int. J. Syst. Evol. Microbiol. 2012, 62, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Saeki, A.; Theeragool, G.; Matsushita, K.; Toyama, H.; Lotong, N.; Adachi, O. Development of thermotolerant acetic acid bacteria useful for vinegar fermentation at higher temperaturest. Biosci. Biotech. Biochem. 1997, 61, 138–145. [Google Scholar] [CrossRef]
- Vashisht, A.; Thakur, K.; Kauldhar, B.S.; Kumar, V.; Yadav, S.K. Waste valorization: Identification of an ethanol tolerant bacterium Acetobacter pasteurianus SKYAA25 for acetic acid production from apple pomace. Sci. Total. Environ. 2019, 690, 956–964. [Google Scholar] [CrossRef]
- Chen, Y.; Bai, Y.; Li, D.; Wang, C.; Xu, N.; Hu, Y. Screening and characterization of ethanol-tolerant and thermotolerant acetic acid bacteria from Chinese vinegar Pei. World J. Microbiol. Biotechnol. 2016, 32, 14. [Google Scholar] [CrossRef]
- Wu, X.; Yao, H.; Liu, Q.; Zheng, Z.; Cao, L.; Mu, D.; Wang, H.; Jiang, S.; Li, X. Producing acetic acid of Acetobacter pas-teurianus by fermentation characteristics and metabolic flux analysis. Appl. Biochem. Biotechnol. 2018, 186, 217–232. [Google Scholar] [CrossRef]
- Fukaya, M.; Tayama, K.; Tamaki, T.; Tagami, H.; Okumura, H.; Kawamura, Y.; Beppu, T. Cloning of the membrane-bound aldehyde dehydrogenase gene of Acetobacterpolyoxogenes and improvement of acetic acid production by use of the clonedgene. Appl. Environ. Microbiol. 1989, 55, 171–176. [Google Scholar] [CrossRef] [Green Version]
- Qi, Z.; Wang, W.; Yang, H.; Xia, X.; Yu, X. Mutation of Acetobacter pasteurianus by UV irradiation under acidic stress for high-acidity vinegar fermentation. Int. J. Food Sci. Technol. 2013, 49, 468–476. [Google Scholar] [CrossRef]
- Lustrato, G.; Salimei, E.; Alfano, G.; Belli, C.; Fantuz, F.; Grazia, L.; Ranalli, G. Cheese whey recycling in traditional dairy food chain: Effects of vinegar from whey in dairy cow nutrition. Acetic Acid Bact. 2013, 2, e8. [Google Scholar] [CrossRef]
- Veeravalli, S.S.; Mathews, A.P. Exploitation of acid-tolerant microbial species for the utilization of low-cost whey in the production of acetic acid and propylene glycol. Appl. Microbiol. Biotechnol. 2018, 102, 8023–8033. [Google Scholar] [CrossRef] [PubMed]
- Rai, P.; Singh, S.P.; Asthana, R.K. Biohydrogen Production from Cheese Whey Wastewater in a Two-Step Anaerobic Process. Appl. Biochem. Biotechnol. 2012, 167, 1540–1549. [Google Scholar] [CrossRef]
- Pandey, A.; Srivastava, S.; Rai, P.; Duke, M. Cheese whey to biohydrogen and useful organic acids: A non-pathogenic microbial treatment by L. acidophilus. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Ricciardi, A.; Zotta, T.; Ianniello, R.G.; Boscaino, F.; Matera, A.; Parente, E. Effect of Respiratory Growth on the Metabolite Production and Stress Robustness of Lactobacillus casei N87 Cultivated in Cheese Whey Permeate Medium. Front. Microbiol. 2019, 10, 851. [Google Scholar] [CrossRef]
- Koller, M.; Hesse, P.; Salerno, A.; Reiterer, A.; Braunegg, G. A viable antibiotic strategy against microbial contamination in biotechnological production of polyhydroxyalkanoates from surplus whey. Biomass- Bioenergy 2011, 35, 748–753. [Google Scholar] [CrossRef]
- Wong, T.Y.; Pei, H.; Bancroft, K.; Wang, F.; Lee, S.Y. Poly(3-hydroxybutyrate) production with high polymer content by fed-batch culture of Alcaligenes latus under nitrogen limitation. Appl. Environ. Microbiol. 1997, 63, 3703–3706. [Google Scholar] [CrossRef] [Green Version]
- Kessler, B.; Witholt, B. Factors involved in the regulatory network of polyhydroxyalkanoate metabolism. J. Biotechnol. 2001, 86, 97–104. [Google Scholar] [CrossRef]
- Reddy, M.V.; Yajima, Y.; Mawatari, Y.; Hoshino, T.; Chang, Y.-C. Degradation and conversion of toxic compounds into useful bioplastics by Cupriavidus sp. CY-1: Relative expression of the PhaC gene under phenol and nitrogen stress. Green Chem. 2015, 17, 4560–4569. [Google Scholar] [CrossRef] [Green Version]
- Nath, A.; Dixit, M.; Bandiya, A.; Chavda, S.; Desai, A.J. Enhanced PHB production and scale up studies using cheese whey in fed batch culture of Methylobacterium sp. ZP24. Bioresour. Technol. 2008, 99, 5749–5755. [Google Scholar] [CrossRef]
- Verlinden, R.A.J.; Hill, D.J.; Kenward, M.A.; Williams, C.D.; Radecka, I. Bacterial synthesis of biodegradable polyhy-droxyalkanoates. J. Appl. Microbiol. 2007, 102, 1437–1449. [Google Scholar] [CrossRef]
- Chinwetkitvanich, S.; Randall, C.W.; Panswad, T. Effects of phosphorus limitation and temperature on PHA production in activated sludge. Water Sci. Technol. 2004, 50, 135–143. [Google Scholar] [CrossRef]
- Shi, H.P.; Lee, C.M.; Ma, W.H. Influence of electron acceptor, carbon, nitrogen, and phosphorus on polyhy-droxyalkanoate (PHA) production by Brachymonas sp. P12. World J. Microbiol. Biotechnol. 2007, 23, 625–632. [Google Scholar] [CrossRef]
- Oliveira-Filho, E.; Silva, J.G.P.; de Macedo, M.A.; Taciro, M.K.; Gomez, J.G.C.; Silva, L.F. Investigating Nutrient Limitation Role on Improvement of Growth and Poly(3-Hydroxybutyrate) Accumulation by Burkholderia sacchari LMG 19450 From Xylose as the Sole Carbon Source. Front. Bioeng. Biotechnol. 2020, 7, 416. [Google Scholar] [CrossRef] [Green Version]
- Montiel-Jarillo, G.; Carrera, J.; Suárez-Ojeda, M.E. Enrichment of a mixed microbial culture for polyhydroxyalkanoates production: Effect of pH and N and P concentrations. Sci. Total. Environ. 2017, 583, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Sedlacek, P.; Slaninova, E.; Koller, M.; Nebesarova, J.; Marova, I.; Krzyzanek, V.; Obruca, S. PHA granules help bacterial cells to preserve cell integrity when exposed to sudden osmotic imbalances. New Biotechnol. 2019, 49, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Pernicova, I.; Novackova, I.; Sedlacek, P.; Kourilova, X.; Koller, M.; Obruca, S. Application of osmotic challenge for enrichment of microbial consortia in polyhydroxyalkanoates producing thermophilic and thermotolerant bacteria and their subsequent isolation. Int. J. Biol. Macromol. 2020, 144, 698–704. [Google Scholar] [CrossRef]
- Taïeb, H.M.; Garske, D.S.; Contzen, J.; Gossen, M.; Bertinetti, L.; Robinson, T.; Cipitria, A. Osmotic pressure modulates single cell cycle dynamics inducing reversible growth arrest and reactivation of human metastatic cells. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Colombo, B.; Sciarria, T.P.; Reis, M.A.; Scaglia, B.; Adani, F. Polyhydroxyalkanoates (PHAs) production from fermented cheese whey by using a mixed microbial culture. Bioresour. Technol. 2016, 218, 692–699. [Google Scholar] [CrossRef]
- Ryan, M.P.; Walsh, G. The biotechnological potential of whey. Rev. Environ. Sci. Bio/Technol. 2016, 15, 479–498. [Google Scholar] [CrossRef] [Green Version]
- Chandrapala, J.; Duke, M.C.; Gray, S.R.; Zisu, B.; Weeks, M.; Palmer, M.; Vasiljevic, T. Properties of acid whey as a function of pH and temperature. J. Dairy Sci. 2015, 98, 4352–4363. [Google Scholar] [CrossRef] [PubMed]
- Ahn, W.S.; Park, S.J.; Lee, S.Y. Production of Poly(3-Hydroxybutyrate) by Fed-Batch Culture of Recombinant Escherichia coli with a Highly Concentrated Whey Solution. Appl. Environ. Microbiol. 2000, 66, 3624–3627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, W.S.; Park, S.J.; Lee, S.Y. Production of poly(3-hydroxybutyrate) from whey by cell recycle fed-batch culture of re-combinant Escherichia coli. Biotechnol. Lett. 2001, 23, 235–240. [Google Scholar] [CrossRef]
- Koller, M. Recycling of Waste Streams of the Biotechnological Poly(hydroxyalkanoate) Production byHaloferax mediterraneion Whey. Int. J. Polym. Sci. 2015, 2015, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Povolo, S.; Toffano, P.; Basaglia, M.; Casella, S. Polyhydroxyalkanoates production by engineered Cupriavidus necator from waste material containing lactose. Bioresour. Technol. 2010, 101, 7902–7907. [Google Scholar] [CrossRef]
- Yee, L.-N.; Mumtaz, T.; Mohammadi, M.; Phang, L.-Y.; Ando, Y.; Raha, A.R.; Sudesh, K.; Ariffin, H.; Hassan, M.A.; Zakaria, M.R. Polyhydroxyalkanoate Synthesis by Recombinant Escherichia coli JM109 Expressing PHA Biosynthesis Genes from Comamonas sp. EB172. J. Microb. Biochem. Technol. 2012, 4, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Li, W.; Zhang, Z.-Z.; Tan, T.-W.; Li, Z.-J. Metabolic engineering of Escherichia coli for the synthesis of polyhydroxyalkanoates using acetate as a main carbon source. Microb. Cell Factories 2018, 17, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Zotta, T.; Solieri, L.; Iacumin, L.; Picozzi, C.; Gullo, M. Valorization of cheese whey using microbial fermentations. Appl. Microbiol. Biotechnol. 2020, 104, 2749–2764. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Reddy, M.V.; Choi, D. Cometabolic degradation of toxic trichloroethene or cis-1,2-dichloroethene with phenol and production of poly-β-hydroxybutyrate (PHB). Green Chem. 2021, 23, 2729–2737. [Google Scholar] [CrossRef]
- Reddy, M.V.; Onodera, R.; Chang, Y.-C. Degradation of aromatic compounds and their conversion into useful energy by bacteria. In Microbial Biodegradation of Xenobiotic Compounds; Chang, Y.-C., Ed.; CRC Press: Boca Raton, FL, USA, 2020; pp. 1–23. [Google Scholar]
- Baikar, V.; Rane, A.; Deopurkar, R. Characterization of Polyhydroxyalkanoate Produced by Bacillus megaterium VB89 Isolated from Nisargruna Biogas Plant. Appl. Biochem. Biotechnol. 2017, 183, 241–253. [Google Scholar] [CrossRef]
- Hassan, M.; Bakhiet, E.; Ali, S.; Hussien, H. Production and characterization of polyhydroxybutyrate (PHB) produced by Bacillus sp. isolated from Egypt. J. Appl. Pharm. Sci. 2016, 6, 46–51. [Google Scholar] [CrossRef] [Green Version]
- Koller, M.; Braunegg, G. Potential and Prospects of Continuous Polyhydroxyalkanoate (PHA) Production. Bioengineering 2015, 2, 94–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koller, M.; Hesse, P.; Bona, R.; Kutschera, C.; Atlić, A.; Braunegg, G. Potential of Various Archae- and Eubacterial Strains as Industrial Polyhydroxyalkanoate Producers from Whey. Macromol. Biosci. 2007, 7, 218–226. [Google Scholar] [CrossRef]
- Reddy, M.V.; Mawatari, Y.; Yajima, Y.; Seki, C.; Hoshino, T.; Chang, Y.C. Chang. Poly-3-hydroxybutyrate (PHB) pro-duction from alkylphenols, mono and poly-aromatic hydrocarbons using Bacillus sp. CYR1: A new strategy for wealth from waste. Bioresour. Technol. 2015, 192, 711–717. [Google Scholar] [CrossRef] [Green Version]
- Sirohi, R.; Pandey, J.P.; Gaur, V.K.; Gnansounou, E.; Sindhu, R. Critical overview of biomass feedstocks as sustainable substrates for the production of polyhydroxybutyrate (PHB). Bioresour. Technol. 2020, 311, 123536. [Google Scholar] [CrossRef]
- Bomrungnok, W.; Arai, T.; Yoshihashi, T.; Sudesh, K.; Hatta, T.; Kosugi, A. Direct production of polyhydroxybutyrate from waste starch by newly-isolated Bacillus aryabhattai T34-N4. Environ. Technol. 2020, 41, 3318–3328. [Google Scholar] [CrossRef]
- Porwal, S.; Kumar, T.; Lal, S.; Rani, A.; Kumar, S.; Cheema, S.; Purohit, H.J.; Sharma, R.; Patel, S.K.S.; Kalia, V.C. Hydrogen and polyhydroxybutyrate producing abilities of microbes from diverse habitats by dark fermentative process. Bioresour. Technol. 2008, 99, 5444–5451. [Google Scholar] [CrossRef]
- Hassan, M.A.; Bakhiet, E.K.; Hussein, H.R.; Ali, S.G. Statistical optimization studies for polyhydroxybutyrate (PHB) production by novel Bacillus subtilis using agricultural and industrial wastes. Int. J. Environ. Sci. Technol. 2019, 16, 3497–3512. [Google Scholar] [CrossRef]
- Penkhrue, W.; Jendrossek, D.; Khanongnuch, C.; Pathom-Aree, W.; Aizawa, T.; Behrens, R.L.; Lumyong, S. Response surface method for polyhydroxybutyrate (PHB) bioplastic accumulation in Bacillus drentensis BP17 using pineapple peel. PLoS ONE 2020, 15, e0230443. [Google Scholar] [CrossRef] [Green Version]
- Güngörmedi, G.; Demirbilek, M.; Mutlu, M.B.; Denkbaş, E.B.; Çabuk, A. Polyhydroxybutyrate and hydroxyvalerate production by Bacillus megaterium strain A1 isolated from hydrocarbon-contaminated soil. J. Appl. Polym. Sci. 2014, 131, 1–8. [Google Scholar] [CrossRef]
- Dave, H.; Ramakrishna, C.; Desai, J. Production of polyhydroxybutyrate by petrochemical activated sludge and Bacillus sp. IPCB-403. Indian J. Exp. Biol. 1996, 34, 216–219. [Google Scholar]
- Rodriguez-Contreras, A.; Koller, M.; de Sousa Dias, M.M.; Calafell, M.; Braunegg, G.; Marqués-Calvo, M.S. Novel poly [(R)-3-hydroxybutyrate]-producing bacterium isolated from a Bolivian hypersaline lake. Food Technol. Biotechnol. 2013, 51, 123–130. [Google Scholar]
- Sindhu, R.; Silviya, N.; Binod, P.; Pandey, A. Pentose-rich hydrolysate from acid pretreated rice straw as a carbon source for the production of poly-3-hydroxybutyrate. Biochem. Eng. J. 2013, 78, 67–72. [Google Scholar] [CrossRef]
- Sindhu, R.; Gnansounou, E.; Binod, P.; Pandey, A. Bioconversion of sugarcane crop residue for value added products—An overview. Renew. Energy 2016, 98, 203–215. [Google Scholar] [CrossRef]
- Halami, P.M. Production of polyhydroxyalkanoate from starch by the native isolate Bacillus cereus CFR06. World J. Microbiol. Biotechnol. 2008, 24, 805–812. [Google Scholar] [CrossRef]
- Yustinah; Hidayat, N.; Alamsyah, R.; Roslan, A.M.; Hermansyah, H.; Gozan, M. Production of polyhydroxybutyrate from oil palm empty fruit bunch (OPEFB) hydrolysates by Bacillus cereus suaeda B-001. Biocatal. Agric. Biotechnol. 2019, 18, 101019. [Google Scholar] [CrossRef]
- Khattab, A.M.; Esmael, M.E.; Farrag, A.A.; Ibrahim, M.I. Structural assessment of the bioplastic (poly-3-hydroxybutyrate) produced by Bacillus flexus Azu-A2 through cheese whey valorization. Int. J. Biol. Macromol. 2021, 190, 319–332. [Google Scholar] [CrossRef]
- Li, J.; Yang, Z.; Zhang, K.; Liu, M.; Liu, D.; Yan, X.; Si, M.; Shi, Y. Valorizing waste liquor from dilute acid pretreatment of lignocellulosic biomass by Bacillus megaterium B-10. Ind. Crop. Prod. 2021, 161, 113160. [Google Scholar] [CrossRef]
- Silambarasan, S.; Logeswari, P.; Sivaramakrishnan, R.; Pugazhendhi, A.; Kamaraj, B.; Ruiz, A.; Ramadoss, G.; Cornejo, P. Polyhydroxybutyrate production from ultrasound-aided alkaline pretreated finger millet straw using Bacillus megaterium strain CAM12. Bioresour. Technol. 2021, 325, 124632. [Google Scholar] [CrossRef]
- Susithra, K.; Narayanan, K.B.; Ramesh, U.; Raja, C.E.; Premkumar, G.; Varatharaju, G.; Vijayakumar, A.; Kannan, M.; Rajarathinam, K. Statistical Optimization of Poly-β-Hydroxybutyrate Biosynthesis Using the Spent Mushroom Substrate by Bacillus tequilensis PSR-2. Waste Biomass Valoriz. 2021, 1–17. [Google Scholar] [CrossRef]
- Suryawanshi, S.S.; Sarje, S.S.; Loni, P.C.; Bhujbal, S.; Kamble, P.P. Bioconversion of Sugarcane Molasses into Bioplastic (Polyhydroxybutyrate) using Bacillus cereus 2156 under Statistically Optimized Culture Conditions. Anal. Chem. Lett. 2020, 10, 80–92. [Google Scholar] [CrossRef]
- Getachew, A.; Woldesenbet, F. Production of biodegradable plastic by polyhydroxybutyrate (PHB) accumulating bacteria using low cost agricultural waste material. BMC Res. Notes 2016, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sheekh, M.M.; el-Abd, M.A.; el-Diwany, A.I.; Ismail, A.M.S.; Mohsen, S.; Omar, T.H. Poly-3-hydroxybutyrate (PHB) production by Bacillus flexus ME-77 using some industrial wastes. Rend. Fis. Acc. Lincei 2015, 26, 109–119. [Google Scholar] [CrossRef]
- Odeniyi, O.A.; Adeola, O.J. Production and characterization of polyhydroxyalkanoic acid from Bacillus thuringiensis using different carbon substrates. Int. J. Biol. Macromol. 2017, 104, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Werlang, E.B.; Moraes, L.B.; Muller, M.V.G.; Julich, J.; Corbellini, V.A.; Neves, F.D.F.; de Souza, D.; Benitez, L.B.; de Cassia de Souza Schneider, R. Polyhydroxybutyrate (PHB) Production via bioconversion using Bacillus pumilus in liquid phase cultivation of the biomass of Arthrospira platensis hydrolysate as a carbon source. Waste Biomass Valoriz. 2021, 12, 3245–3255. [Google Scholar] [CrossRef]














| Culture Condition | 20-Fold | 10-Fold | 4-Fold | 2-Fold |
|---|---|---|---|---|
| Initial concentration of acetic acid (g/L) | 3.8 | 5.7 | 12.3 | 24.4 |
| Final concentration of acetic acid (g/L) | 2.6 | 2.9 | 10.6 | 22.2 |
| The utilized acetic acid (g/L) | 1.2 | 2.8 | 1.7 | 2.2 |
| The utilized acetic acid (%) | 31.6 | 49.1 | 13.8 | 9.0 |
| Culture Condition | 20-Fold (Acetic Acid; 3.8 g/L) | 10-Fold (Acetic Acid; 5.7 g/L) | 4-Fold (Acetic Acid; 12.3 g/L) | 2-Fold (Acetic Acid; 24.4 g/L) |
|---|---|---|---|---|
| CDW (mg) | 151.6 | 319.4 | 638.2 | 831.7 |
| PHA production (mg/L) | 43.6 | 240.6 | 126.2 | 77.2 |
| PHA production (%CDW) | 28.8 | 75.4 | 19.8 | 9.2 |
| Yield of PHA from acetic acid (%) | 3.6 | 8.6 | 7.4 | 3.5 |
| PHA volumetric productivity (mg PHA/ L/hour) | 1.21 | 4 | 2.1 | 1.28 |
| Media (Substrates) | Initial Acetic Acid Concentration (g/L) | Extracted PHA (mg/L) | |
|---|---|---|---|
| Acetic acid fermented from CW | 2-fold | 24.4 | 77.2 |
| 4-fold | 12.3 | 126.1 | |
| 10-fold | 5.7 | 240.6 | |
| 20-fold | 3.8 | 43.6 | |
| One step production of PHA using raw CW | 30-fold | not detected | 46.6 |
| One step production of PHA using mineral medium containing pure acetic acid | 30 g/L | 30.0 | 8.6 |
| 20 g/L | 20.0 | 231.0 | |
| 10 g/L | 10.0 | 287.0 | |
| 5 g/L | 5.0 | 146.6 | |
| Culture Condition | 20-Fold (Acetic Acid; 2.3 g/L) | 10-Fold (Acetic Acid; 4.5 g/L) | 4-Fold (Acetic Acid; 11.3 g/L) |
|---|---|---|---|
| CDW (mg) | 155.2 | 261.4 | 1227.2 |
| PHA production (mg/L) | 58.0 | 96.6 | 411.2 |
| PHA production (%CDW) | 37.4 | 37.0 | 33.5 |
| Strain | Carbon Source | PHAs Monomer | PHB Conc. (g/L) | PHB Content. (wt%) | References |
|---|---|---|---|---|---|
| Bacillus megaterium | lactose | PHB | 0.47 | 26.0 | [5] |
| Bacillus megaterium VB89 | slurry from Nisargruna biogas plant | PHB | 0.67 | 36.17 | [63] |
| Bacillus sp. N-2 | glucose | PHB | 0.17 | 20.0 | [64] |
| Bacillus sp. CYR1 | phenol | PHB | 0.51 | 51.0 | [67] |
| Bacillus sp. CYR1 | naphthalene | PHB | 0.42 | 42.0 | [67] |
| Bacillus sp. CYR1 | 4-chlorophenol | PHB | 0.32 | 32.0 | [67] |
| Bacillus sp. CYR1 | 4-nonylphenol | PHB | 0.29 | 29.0 | [67] |
| Bacillus aryabhattai T34-N4 | oil palm trunk starch | PHB | 0.33 | 17.0 | [69] |
| Bacillus cereus EGU3 | contaminated food | PHB | 0.5 | 66.6 | [70] |
| Bacillus subtilis | rice bran | PHB | 0.81 | 62.6 | [71] |
| Bacillus drentensis BP17 | pineapple peel | PHB | 5.55 | n.a | [72] |
| Bacillus megaterium strain A1 | molasses | PHB | 14.6 | 48.7 | [73] |
| Bacillus megaterium S29 | glucose | PHB | n.a | ≧70 | [74] |
| Bacillus sp. IPCB-403 | activated sludges | PHB | n.a | ≧70 | [75] |
| Bacillus firmus NII 0830 | rice straw | PHB | 1.9 | 89.0 | [76,77] |
| Bacillus cereus CFR06 | starch-based materials | PHB | 0.48 | 48.0 | [78] |
| Bacillus cereus suaeda B-001 | oil palm empty fruit bunch hydrolysates | PHB | 0.47 | 55.4 | [79] |
| Bacillus flexus Azu-A2 | cheese whey | PHB | 0.95 | 20.96 | [80] |
| Bacillus megaterium B-10 | dilute acid pretreatment liquor of lignocellulose from rice straw | PHB | 1.496 | 32.56 | [81] |
| Bacillus megaterium strain CAM12 | finger millet straw hydrolysates | PHB | 8.31 | 51.0 | [82] |
| Bacillus tequilensis PSR-2 | glucose | PHB | 2.8 | 49.12 | [83] |
| Bacillus tequilensis PSR-2 | an alkali-pretreated spent mushroom substrate of sugarcane bagasse | PHB | 12.4 | n.a | [83] |
| B. cereus 2156 | sugarcane molasses | PHB | 2.2 | n.a | [84] |
| Bacillus sp. | sugarcane bagasse hydrolysates | PHB | 5.0 | 56.0 | [85] |
| Bacillus flexus ME-77 | sugarcane molasses | PHB | 4.5 | 15.0 | [86] |
| Bacillus thuringiensis SBC4 | glucose and corn cob | PHB | 0.4 | 43.95 | [87] |
| Bacillus pumilus | hydrolysate of the Arthrospira platensis biomass | PHB | n.a | 0.3 | [88] |
| Bacillus sp. CYR1 | cheese whey | PHB | 0.41 | 33.5 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.-C.; Reddy, M.V.; Imura, K.; Onodera, R.; Kamada, N.; Sano, Y. Two-Stage Polyhydroxyalkanoates (PHA) Production from Cheese Whey Using Acetobacter pasteurianus C1 and Bacillus sp. CYR1. Bioengineering 2021, 8, 157. https://doi.org/10.3390/bioengineering8110157
Chang Y-C, Reddy MV, Imura K, Onodera R, Kamada N, Sano Y. Two-Stage Polyhydroxyalkanoates (PHA) Production from Cheese Whey Using Acetobacter pasteurianus C1 and Bacillus sp. CYR1. Bioengineering. 2021; 8(11):157. https://doi.org/10.3390/bioengineering8110157
Chicago/Turabian StyleChang, Young-Cheol, Motakatla Venkateswar Reddy, Kazuma Imura, Rui Onodera, Natsumi Kamada, and Yuki Sano. 2021. "Two-Stage Polyhydroxyalkanoates (PHA) Production from Cheese Whey Using Acetobacter pasteurianus C1 and Bacillus sp. CYR1" Bioengineering 8, no. 11: 157. https://doi.org/10.3390/bioengineering8110157