Black Heart Detection in White Radish by Hyperspectral Transmittance Imaging Combined with Chemometric Analysis and a Successive Projections Algorithm
Abstract
:1. Introduction
2. Materials and Methods
2.1. White Radish Samples
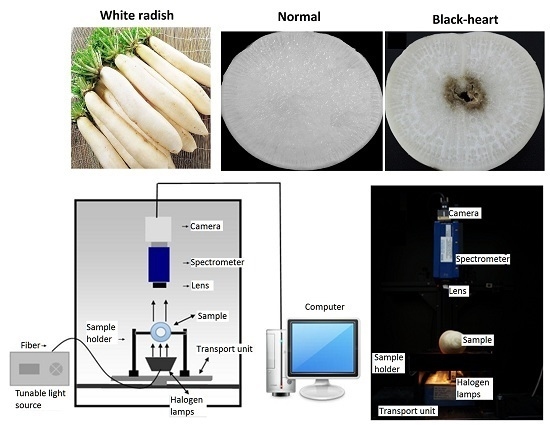
2.2. Hyperspectral Imaging System
2.3. Image Calibration and Roi Identification
2.4. Wavelength Selection
2.5. Spectra Processing and Chemometric Model Development
3. Results and Discussion
3.1. Mean Spectra for Normal and Defective White Radishes
3.2. Spectra Preprocessing Method Selection
3.3. Optimum Wavelength Selection
3.4. Classification Results between Normal and Defective White Radishes
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hu, X.; Li, N.; He, Z. Analysis of the current situation and prospect of radish industry development in China. Agric. Outlook 2012, 10, 35–37. [Google Scholar]
- Li, Y.; Li, H.; Zhang, X.; Li, W.; Wang, Y. Cause and control measures of piquancy, bitterness, black heart and skin in radish. Northern Hortic. 2015, 11, 53–54. [Google Scholar]
- Elbatawi, I.E. An acoustic impact method to detect hollow heart of potato tubers. Biosyst. Eng. 2008, 100, 206–213. [Google Scholar] [CrossRef]
- Ying, Y.; Liu, Y. Nondestructive measurement of internal quality in pear using genetic algorithms and FT-NIR spectroscopy. J. Food Eng. 2008, 84, 206–213. [Google Scholar] [CrossRef]
- Mathanker, S.K.; Weckler, P.R.; Bowser, T.J. X-ray applications in food and agriculture: A review. Trans. ASABE 2013, 56, 1227–1239. [Google Scholar]
- Pu, H.; Liu, D.; Wang, L.; Sun, D. Soluble solids content and ph prediction and maturity discrimination of lychee fruits using visible and near infrared hyperspectral imaging. Food Anal. Method 2016, 9, 235–244. [Google Scholar] [CrossRef]
- Han, D.; Tu, R.; Lu, C.; Liu, X.; Wen, Z. Nondestructive detection of brown core in the Chinese pear ‘Yali’ by transmission visible–NIR spectroscopy. Food Control 2006, 17, 604–608. [Google Scholar] [CrossRef]
- Clark, C.J.; McGlone, V.A.; Jordan, R.B. Detection of Brownheart in ‘Braeburn’ apple by transmission NIR spectroscopy. Postharvest Biol. Technol. 2003, 28, 87–96. [Google Scholar] [CrossRef]
- McGlone, V.A.; Martinsen, P.J.; Clark, C.J.; Jordan, R.B. On-line detection of Brownheart in Braeburn apples using near infrared transmission measurements. Postharvest Biol. Technol. 2005, 37, 142–151. [Google Scholar] [CrossRef]
- Zerbini, P.E.; Grassi, M.; Cubeddu, R.; Pifferi, A.; Torricelli, A. Nondestructive detection of brown heart in pears by time-resolved reflectance spectroscopy. Postharvest Biol. Technol. 2002, 25, 87–97. [Google Scholar] [CrossRef]
- Teerachaichayut, S.; Kil, K.Y.; Terdwongworakul, A.; Thanapase, W.; Nakanishi, Y. Non-destructive prediction of translucent flesh disorder in intact mangosteen by short wavelength near infrared spectroscopy. Postharvest Biol. Technol. 2007, 43, 202–206. [Google Scholar] [CrossRef]
- Takizawa, K.; Nakano, K.; Ohashi, S.; Yoshizawa, H.; Wang, J.; Sasaki, Y. Development of nondestructive technique for detecting internal defects in Japanese radishes. J. Food Eng. 2014, 126, 43–47. [Google Scholar] [CrossRef]
- Valero, C.; Barreiro, P.; Ruiz-altisent, M.; Cubeddu, R.; Pifferi, A.; Taroni, P.; Torricelli, A.; Valentini, G.; Johnson, D.; Dover, C. Mealiness detection in apples using time resolved reflectance spectroscopy. J. Texture Stud. 2005, 36, 439–458. [Google Scholar] [CrossRef] [Green Version]
- Pan, L.; Zhu, Q.; Lu, R.; McGrath, J.M. Determination of sucrose content in sugar beet by portable visible and near-infrared spectroscopy. Food Chem. 2015, 167, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Ariana, D.P.; Lu, R. Hyperspectral waveband selection for internal defect detection of pickling cucumbers and whole pickles. Comput. Electron. Agric. 2010, 74, 137–144. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; ElMasry, G.; Sun, D.; Allen, P. Application of NIR hyperspectral imaging for discrimination of lamb muscles. J. Food Eng. 2011, 104, 332–340. [Google Scholar] [CrossRef]
- Pu, Y.; Feng, Y.; Sun, D. Recent progress of hyperspectral imaging on quality and safety inspection of fruits and vegetables: a review. Compr. Rev. Food Sci. 2015, 14, 176–188. [Google Scholar] [CrossRef]
- Sanz, J.A.; Fernandes, A.M.; Barrenechea, E.; Silva, S.; Santos, V.; Gonçalves, N.; Paternain, D.; Jurio, A.; Melo-Pinto, P. Lamb muscle discrimination using hyperspectral imaging: Comparison of various machine learning algorithms. J. Food Eng. 2016, 174, 92–100. [Google Scholar] [CrossRef]
- Lorente, D.; Aleixos, N.; Gómez-Sanchis, J.; Cubero, S.; García-Navarrete, O.L.; Blasco, J. Recent advances and applications of hyperspectral imaging for fruit and vegetable quality assessment. Food Bioprocess Technol. 2012, 5, 1121–1142. [Google Scholar] [CrossRef]
- Cen, H.; Lu, R.; Ariana, D.P.; Mendoza, F. Hyperspectral imaging-based classification and wavebands selection for internal defect detection of pickling cucumbers. Food Bioprocess Technol. 2014, 7, 1689–1700. [Google Scholar] [CrossRef]
- Ariana, D.P.; Lu, R. Evaluation of internal defect and surface color of whole pickles using hyperspectral imaging. J. Food Eng. 2010, 96, 583–590. [Google Scholar] [CrossRef]
- Pan, L.; Zhang, Q.; Zhang, W.; Sun, Y.; Hu, P.; Tu, K. Detection of cold injury in peaches by hyperspectral reflectance imaging and artificial neural network. Food Chem. 2016, 192, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Lu, R. Apple mealiness detection using hyperspectral scattering technique. Postharvest Biol. Technol. 2010, 58, 168–175. [Google Scholar] [CrossRef]
- Fan, S.; Huang, W.; Guo, Z.; Zhang, B.; Zhao, C. Prediction of soluble solids content and firmness of pears using hyperspectral reflectance imaging. Food Anal. Method 2015, 8, 1936–1946. [Google Scholar] [CrossRef]
- Leiva-Valenzuela, G.A.; Lu, R.; Aguilera, J.E.M. Prediction of firmness and soluble solids content of blueberries using hyperspectral reflectance imaging. J. Food Eng. 2012, 115, 91–98. [Google Scholar] [CrossRef]
- Leiva-Valenzuela, G.A.; Lu, R.; Aguilera, J.M. Assessment of internal quality of blueberries using hyperspectral transmittance and reflectance images with whole spectra or selected wavelengths. Innov. Food Sci. Emerg. 2014, 24, 2–13. [Google Scholar] [CrossRef]
- Chao, K.; Yang, C.; Kim, M.S. Line-scan spectral imaging system for online poultry carcass inspection. J. Food Process. Eng. 2011, 34, 125–143. [Google Scholar] [CrossRef]
- Cen, H.; Lu, R.; Zhu, Q.; Mendoza, F. Nondestructive detection of chilling injury in cucumber fruit using hyperspectral imaging with feature selection and supervised classification. Postharvest Biol. Technol. 2016, 111, 352–361. [Google Scholar] [CrossRef]
- Meenu, G.; Amit, V.; Narender, B. Black rot-A devastating disease of crucifers: A review. Agric. Rev. 2013, 34, 269–278. [Google Scholar]
- Araújo, M.C.U.; Saldanha, T.C.B.; Galvão, R.K.H.; Yoneyama, T.; Chame, H.C.; Visani, V. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemom. Intell. Lab. Syst. 2001, 57, 65–73. [Google Scholar] [CrossRef]
- Soares, S.F.C.; Gomes, A.A.; Araujo, M.C.U.; Filho, A.R.G.; Galvão, R.K.H. The successive projections algorithm. TrAC Trends Anal. Chem. 2013, 42, 84–98. [Google Scholar] [CrossRef]
- Galvão, R.K.H.; Araújo, M.C.U.; Fragoso, W.D.; Silva, E.C.; José, G.E.; Soares, S.F.C.; Paiva, H.M. A variable elimination method to improve the parsimony of MLR models using the successive projections algorithm. Chemom. Intell. Lab. Syst. 2008, 92, 83–91. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; ElMasry, G.; Sun, D.; Allen, P. Non-destructive assessment of instrumental and sensory tenderness of lamb meat using NIR hyperspectral imaging. Food Chem. 2013, 141, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, J.; Fan, S.; Huang, W.; Zhao, C.; Liu, C.; Huang, D. Hyperspectral imaging combined with multivariate analysis and band math for detection of common defects on peaches (Prunus persica). Comput. Electron. Agric. 2015, 114, 14–24. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Pan, L.; Zhan, G.; Tu, K.; Tu, S.; Liu, P. Eggshell crack detection based on computer vision and acoustic response by means of back propagation artificial neural network. Eur. Food Res. Technol. 2011, 233, 457–463. [Google Scholar] [CrossRef]
- Vapnik, V.N. An overview of statistical learning theory. IEEE Trans. Neural Netw. 1999, 10, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Galvao, R.; Araujo, M.; Jose, G.; Pontes, M.; Silva, E.; Saldanha, T. A method for calibration and validation subset partitioning. Talanta 2005, 67, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Dachoupakan Sirisomboon, C.; Putthang, R.; Sirisomboon, P. Application of near infrared spectroscopy to detect aflatoxigenic fungal contamination in rice. Food Control 2013, 33, 207–214. [Google Scholar] [CrossRef]




| Pretreatment Methods | NP | SGFD | D | SGS | MSC | SNV | A | MC | SNV+MC |
|---|---|---|---|---|---|---|---|---|---|
| LVs | 4 | 4 | 4 | 3 | 3 | 4 | 4 | 8 | 5 |
| Calibration (%) | 97.5 | 98.1 | 98.1 | 96.6 | 98.1 | 99.1 | 98.1 | 98.8 | 100 |
| Prediction (%) | 95.6 | 96.1 | 96.7 | 95.6 | 96.7 | 98.9 | 96.7 | 98.3 | 99.4 |
| Wavelengths | 545 nm | 564 nm | 611 nm | 629 nm | 642 nm | 655 nm | 724 nm | 730 nm | 738 nm | 809 nm |
|---|---|---|---|---|---|---|---|---|---|---|
| 545 nm | 1.000 | - | - | - | - | - | - | - | - | - |
| 564 nm | 0.998 **,1 | 1.000 | - | - | - | - | - | - | - | - |
| 611 nm | 0.988 ** | 0.994 ** | 1.000 | - | - | - | - | - | - | - |
| 629 nm | 0.987 ** | 0.993 ** | 1.000 ** | 1.000 | - | - | - | - | - | - |
| 642 nm | 0.984 ** | 0.992 ** | 0.999 ** | 1.000 ** | 1.000 | - | - | - | - | - |
| 655 nm | 0.980 ** | 0.989 ** | 0.998 ** | 0.999 ** | 1.000 ** | 1.000 | - | - | - | - |
| 724 nm | 0.943 ** | 0.951 ** | 0.963 ** | 0.966 ** | 0.969 ** | 0.974 ** | 1.000 | - | - | - |
| 730 nm | 0.928 ** | 0.934 ** | 0.945 ** | 0.949 ** | 0.953 ** | 0.958 ** | 0.998 ** | 1.000 | - | - |
| 738 nm | 0.911 ** | 0.916 ** | 0.926 ** | 0.931 ** | 0.935 ** | 0.941 ** | 0.992 ** | 0.998 ** | 1.000 | - |
| 809 nm | 0.843 ** | 0.846 ** | 0.854 ** | 0.860 ** | 0.866 ** | 0.874 ** | 0.957 ** | 0.974 ** | 0.984 ** | 1.000 |
| Model | Wavelengths Number | Parameters | Class | Calibration (%) | Prediction (%) | ||
|---|---|---|---|---|---|---|---|
| Accuracy | Overall | Accuracy | Overall | ||||
| PLS-DA 1 | 420 | LVs: 5 | Normal | 100 | 100 | 98.9 | 99.4 |
| Black heart | 100 | 100 | |||||
| 10 | LVs: 7 | Normal | 100 | 99.7 | 100 | 99.4 | |
| Black heart | 99.4 | 98.9 | |||||
| 4 | LVs: 1 | Normal | 98.7 | 97.2 | 98.9 | 96.1 | |
| Black heart | 95.7 | 93.5 | |||||
| SVM | 420 | KT: RBF; C: 100; γ: 0.031623 | Normal | 100 | 100 | 100 | 98.9 |
| Black heart | 100 | 97.8 | |||||
| 10 | KT: RBF; C: 100; γ: 3.1623 | Normal | 100 | 100 | 98.9 | 98.9 | |
| Black heart | 100 | 98.9 | |||||
| 4 | KT: RBF; C: 100; γ: 0.0001 | Normal | 98.7 | 97.2 | 97.7 | 95.6 | |
| Black heart | 95.7 | 93.5 | |||||
| ANN | 420 | HLAF: tangent; OLAF: softmax; HLN: 15 | Normal | 100 | 100 | 100 | 100 |
| Black heart | 100 | 100 | |||||
| 10 | HLAF: Hyperbolic tangent; OLAF: softmax; HLN: 6 | Normal | 100 | 100 | 100 | 100 | |
| Black heart | 100 | 100 | |||||
| 4 | HLAF: Hyperbolic tangent; OLAF: softmax; HLN: 3 | Normal | 99.4 | 98.8 | 98.9 | 97.2 | |
| Black heart | 98.1 | 95.7 | |||||
| FLDA | 420 | - | Normal | - | - | - | - |
| Black heart | - | - | |||||
| 10 | - | Normal | 99.4 | 99.7 | 100 | 99.4 | |
| Black heart | 100 | 98.9 | |||||
| 4 | - | Normal | 99.4 | 98.4 | 100 | 97.8 | |
| Black heart | 97.5 | 95.7 | |||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, D.; Song, L.; Sun, Y.; Hu, P.; Tu, K.; Pan, L.; Yang, H.; Huang, M. Black Heart Detection in White Radish by Hyperspectral Transmittance Imaging Combined with Chemometric Analysis and a Successive Projections Algorithm. Appl. Sci. 2016, 6, 249. https://doi.org/10.3390/app6090249
Song D, Song L, Sun Y, Hu P, Tu K, Pan L, Yang H, Huang M. Black Heart Detection in White Radish by Hyperspectral Transmittance Imaging Combined with Chemometric Analysis and a Successive Projections Algorithm. Applied Sciences. 2016; 6(9):249. https://doi.org/10.3390/app6090249
Chicago/Turabian StyleSong, Dajie, Lijun Song, Ye Sun, Pengcheng Hu, Kang Tu, Leiqing Pan, Hongwei Yang, and Min Huang. 2016. "Black Heart Detection in White Radish by Hyperspectral Transmittance Imaging Combined with Chemometric Analysis and a Successive Projections Algorithm" Applied Sciences 6, no. 9: 249. https://doi.org/10.3390/app6090249