1. Introduction
S-nitrosoglutathione (GSNO) is formed in response to nitric oxide (NO) fluxes by the reaction of oxidized forms of NO (e.g., N
2O
3) with the free thiol of glutathione. GSNO can in turn modulate protein
S-nitrosylation via transnitrosylation—that is, the transfer of a NO
+ moiety to protein thiols. A large body of evidence indicates that the under or over
S-nitrosylation of proteins has a great impact on many pathologies [
1].
S-nitrosoglutathione reductase (GSNOR), coded by the alcohol dehydrogenase ADH5 gene in humans, was first identified by Koivusalo et al. [
2] as a NAD
+-dependent aldehyde dehydrogenase. In 1998, Jensen et al. [
3] reported that ADH5 could also catalyze the NADH-coupled reduction of
S-nitrosoglutathione (GSNO) to N-hydroxysulphenamido glutathione (Equation (1)). In the presence of glutathione (GSH), N-hydroxysulphenamido glutathione is converted to hydroxylamine and glutathione disulfide (GSSG) (Equation (2)).
Additional studies indicated that ADH5 and its GSNO metabolic activity are conserved from prokaryotes to eukaryotes [
4]. ADH5, which is expressed in all human tissues [
5,
6], was renamed GSNOR by researchers in the
S-nitrosothiol (SNO) signaling field.
GSNO metabolic activity is not specific to GSNOR. Super oxide dismutase [
7], glutathione peroxidase [
8], protein disulfide isomerase [
9], thioredoxin [
10], and carbonyl reductase [
11] can all metabolize GSNO. However, only GSNOR and carbonyl reductase can reduce NO
+ to NH
2OH, which cannot be converted back to NO
x via biological systems. In addition, the GSNO metabolic activity of only thioredoxin and GSNOR have been demonstrated to be physiologically relevant [
12].
GSNOR (ADH5) is generally found to be elevated in respiratory, cardiovascular, smooth muscle, and autoimmune disease states. Many studies indicate that the lowering of GSNOR (AHD5) has beneficial effects. For example, the deletion of GSNOR promotes bronchodilation and protection against asthma in the lungs [
13], improves post-cardiac arrest resuscitation [
14], increases cell senescence in mouse and human models [
15], protects against autoimmune disease [
16], improves skeletal muscle resistance and fatigue resistance [
17], and increases cardiomyocyte proliferation [
18] and neuronal differentiation [
19].
Interest in GSNOR as a therapeutic target is growing. Several drugs that inhibit GSNOR were developed for treating asthma, cystic fibrosis, and chronic obstructive pulmonary disease (COPD) [
14,
15,
16]. More recently, the demonstration of a direct role for GSNOR in dysfunctional relaxation in preterm labor has sparked interest in GSNOR inhibitors as potential new tocolytic drugs.
Clearly, controlling the activity of GSNOR is of ongoing pharmacological interest. The current study could potentially provide this opportunity, as it not only presents evidence for an allosteric GSNO binding site, but it also shows that the activity of GSNOR is very sensitive to perturbations at this site.
2. Materials and Methods
2.1. Materials
All reagents were purchased from Sigma-Aldrich (Oakville, ON, Canada) unless otherwise stated in the methods section.
All solutions were prepared with MilliQ water (Advantage A10 Water Purification System, Millipore Sigma, Etobicoke, ON, Canada).
2.2. Methods
2.2.1. GSNOR WT Cloning, Mutagenesis, and Protein Isolation
GSNOR (ADH5) Sub-Cloning
Human GSNOR (ADH5) was purchased from Origene (SC119755) and subcloned into bacterial expression vector pET28b using Cold Fusion Cloning Kit, SYMC010A1 (MJS BioLynx Inc., Brockville, ON, Canada). The destination vector pET28b was linearized via digestion with restriction enzymes NdeI and XhoI, PCR amplified and purified by electrophoresis on agarose gels.
The primers used for GSNOR (ADH5-ScSFA1) subcloning were:
Forward 5′– GTGCCGCGCGGCAGCCATATGGCGAACGAGGTTATCAAG –3′
Reverse 5′– GTGGTGGTGGTGGTGCTCGAGAATCTTTACAACAGTTCGAATG –3′.
The ligated plasmid DNA was transformed directly into chemically competent E. coli. Colonies were screened using diagnostic restriction enzyme digest and by partial sequencing (Robarts Research, London, ON, Canada).
GSNOR Site-Directed Mutagenesis
Site-directed mutagenesis of the residues at the postulated GSNO binding site was outsourced to Genscript. The following primers (NP_000662) for PCR were designed and sent to Genscript:
- Lys188Ala Forward
5′ ACT GCC GCG TTG GAG CCT 3′
- Lys188Ala Reverse
5′ GTT CAC AGC AGC ACC ATA ACC GGT 3′
- Lys323Ala Forward
5′ GCC TTT GGA GGA TGG GCG AGT GTA GAA AGT GTC 3′
- Lys323Ala Reverse
5′ AGT GCC TTT CCA TGT GCG ACC 3′
The resultant PCR product was treated with a kinase, ligase, and Dpn1 enzyme mix according to the manufacturer’s protocol, and used to transform chemically competent DH5 E. coli (New England BioLabs C2987, Whitby, ON, Canada). Individual colonies from the transformation plates were selected, and each colony was inoculated into 3 mL of LB medium (with 50 µg/mL kanamycin) for overnight growth at 37 °C. Then, plasmid DNA was isolated from these cultures using a standard bacterial miniprep procedure (Qiagen, Montreal, PQ, Canada).
2.2.2. GSNOR Expression and Purification
A single transformed colony was inoculated into 25 mL of 2× YT medium containing 50 µg/mL kanamycin and cultured overnight at 37 °C with shaking. The starter culture was poured into 1 L of 2× YT medium containing 50 µg/mL kanamycin and incubated at 37 °C until an OD of approximately 0.6 was attained at which time GSNOR (wt and mutant) expression was induced by the addition of Image result for IPTG.
Isopropyl β-d-1-thiogalactopyranoside (IPTG) (0.4 mM). The induced culture was grown for an additional 24 h at room temperature with shaking, and the cells were harvested by centrifugation (4000× g, 30 min, 4 °C). The bacterial cell pellet was resuspended in lysis buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 15 mM imidazole, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.5% Triton X-100, 50 µg/mL DNase I, and 100 µg/mL lysozyme). The suspension was incubated on ice for 30 min and pulse sonicated (30 cycles of 20 s on/20 s off) (Fisher Scientific Model FB120, Ottawa, ON, Canada). Then, the lysate was centrifuged (11,000× g, 30 min, 4 °C), and recombinant GSNOR in the supernatant was purified by HIS-Select® Nickel Affinity Gel chromatography (Sigma-Aldrich). The buffers employed were: (i) wash buffer, 50 mM Tris-HCl pH 8, 150 mM NaCl, and 50 mM imidazole; (ii) elution buffer, 50 mM Tris-HCl pH 8, 150 mM NaCl, and 300 mM imidazole. The eluted protein was buffer exchanged into storage solution (58 mM Na2HPO4, 17 mM NaH2PO4, 68 mM NaCl, 15% glycerol) using an Amicon centrifugal filter (Millipore Sigma), divided into 100-µL aliquots, and frozen at −80 °C.
GSNOR Kinetics
S-nitrosoglutathione (GSNO) was synthesized according to the method of Hart [
17]. The concentrations of GSNO and NADH were determined from the absorbance of their respective solutions using their extinction coefficients: GSNO, 922 M
−1 cm
−1 at 335 nm; NADH, 6220 M
−1 cm
−1 at 340 nm.
The kinetic studies were performed under steady-state conditions where constant concentrations of the enzyme (approximately 20 nM) and the cofactor [NADH] (80 μM) were used with varying [GSNO] (2 μM to 200 μM). The reaction was initiated by the addition of GSNOR. Due to the partial overlap of the absorption maxima of NADH and GSNO, the decrease at 340 nm due to GSNOR activity is 85% from NADH oxidation and 14% from GSNO reduction across the entire substrate range employed. The absorbance at 340 nm was monitored (Agilent 8453 UV/Vis Spectrophotometer, Mississauga, ON, Canada) for 180 s, and initial rates were extracted from the kinetic data and fitted to the Hill–Langmuir equation [
18,
19] (Equation (3)) where
n is the Hill coefficient.
The kinetic studies were performed as eight replicate experiments in triplicate.
2.3. Computational Methods
Docking and Molecular Dynamics (MD) Simulation
The Molecular Operating Environment (MOE) program was used to prepare all systems for docking and molecular dynamics (MD) simulations [
20]. For the chemical models, an experimental X-ray crystal structure of suitably high resolution (1.9 Å) of
S-nitrosoglutathione reductase (GSNOR) with a bound reversible inhibitor (PDB ID: 3QJ5) [
21] was used as the initial template structure. The bound inhibitor was deleted from the structure. The protonation states of all residues were assigned using the Propka tool as available in the MOE program. All crystallographic waters were removed. Subsequently, the substrate
S-nitrosoglutathione (GSNO) was docked in the whole protein using the virtual screening formulism. To evaluate the top poses, the London dG scoring function was used with the AMBER12:EHT force field refinement keeping the top 10 scores [
20]. Then, molecular dynamics simulations were performed on the top scoring structure (i.e., predicted most preferred bound complex) obtained. The MD simulations were run using the NAMD program using its default settings as provided through MOE [
22]. Each system was resolvated to a water depth of 6 Å away from any residue. The solvated structure generated was optimized using the AMBER12: EHT molecular mechanics force field until the root-mean-square gradient fell below 0.01 kcal/mol Å
−1. In order to allow for thermal relaxation and equilibration, the minimized structures were each annealed over 100 ps, during which the temperature was raised from 0 to 300 k at constant pressure. Then, this was followed for each equilibrated complex by a 3-ns production run MD simulation, in which all the atoms were free to move, with a time step of 2 fs under constant pressure and temperature. It is noted that the default settings include use of the Particle mesh Ewald (PME) method for calculating Coulombic interactions, a cut-off for nonbonded interactions of 8–10 Å, and tether ranges of 0 to 100 Å applied to heavy atoms. For each production simulation, cluster analyses were performed on the conformations obtained (for instance, based on the root-mean-square deviation (rmsd) of the protein backbone, and the bound GSNO and putative binding site residues) and a representative structure was selected from the most populated cluster for further analysis.
2.4. MS-MS for GSNOR Peptide Identification
Tandem mass spectrometry (MS-MS) was performed on a Waters, Synapt G1 for characterization of the peptides fragmented from the pepsin column. By acquiring data in MS-TOF mode, a precursor ion can be chosen to undergo further fragmentation by collision-induced dissociation (CID) The collision energy applied was customized for each precursor ion to obtain an ideal fragmentation pattern. The fingerprint spectra were collected within a mass range of 100–2000 m/z.
A theoretical pepsin digest was performed using the FindPept tool on the ExPASy Proteomics server (Swiss Institute of Bioinformatics, Basel, Switzerland). Search parameters were set to pepsin (porcine A) at pH > 2 with a mass tolerance of ± 0.5 Da. The possible identities of the parent ion were chosen from the resulting list. Each possible parent ion was theoretically fragmented by the spectra-viewing software mMass and compared with the fingerprint. The parameters used included searching for the loss of –H2O and –NH3, as well as identifying y, a, b, int-a, and int-b fragment ions.
2.5. HDX-MS
Hydrogen–deuterium (D) exchange (HDX) MS was made possible by outfitting the Synapt G1 with the custom TRESI apparatus as described by Rob et al. [
23]. The reagents utilized include 5% (
v/v) acetic acid, GSNOR in 200 mM ammonium acetate, and deuterium oxide (D
2O) (99.9% purity of LC-MS grade). These reagents are pumped through a polyamide-coated glass capillary with an outer diameter (o.d.) of 109.2 μm using Harvard Apparatus Pump 11 Elite infusion syringe pumps (Holliston, MA, USA). Protein and D
2O were pumped at a rate of 2 μL/min with 0.5-mL syringes (Hamilton 700 78 Series Gastight Syringe Cole-Parmer Montreal, PQ, Canada), while acid was pumped at a rate of 16 μL/min with a 5-mL or 2.5-mL syringe (Hamilton 1000 Series Gastight Syringe).
The 109.2-μm o.d. glass capillary-containing protein is encased in a metal capillary with an inner diameter (i.d.) of 132.6 μm. A 2-mm notch was made, and the end of the glass capillary was sealed prior to each experiment using a VersaLaser™ [
24]. This allowed for efficient kinetic mixing before the exchange reaction was quenched by the acid and sent to the PMMA chip containing the pepsin agarose beads for MS1 fragmentation. The duration of the D
2O–protein exchange reaction is controlled by varying the length of the capillary past the protein–D
2O mixing point. In this study, 5-mm and 10-mm long capillaries were used post-mixing, corresponding to 2.07 s (approximately 2 s) and 4.14 s (approximately 4 s), respectively.
Data was collected in IMS mode in the 400–1500 m/z range. The experimental deuterium uptake of each peptide obtained was calculated using a custom-built software program (DJW, unpublished results).
Each set of data was collected on the same day, including six sets of 3 × 5 min spectra acquisitions of protein GSNOR without deuterium exchange, 2-s exchange, and 4-s exchange. This was followed by two sets of 3 × 5 min spectra acquisitions of protein GSNOR in the presence of a 20-fold molar excess of GSNO with deuterium exchange: 2-s exchange, and 4-s exchange. Concentrations of GSNOR and GSNO were calculated by Bradford assay and the GSNO extinction coefficient (λmax = 335 nm, εM = 920 M−1 cm−1), respectively, to confirm a 20× stoichiometric addition of GSNO. Integrity of the notch was confirmed to be maintained at the end of the experiment.
4. Discussion
The postulated physiological role of GSNOR is to irreversibly remove nitric oxide equivalents, thereby attenuating protein S-nitrosylation and blocking SNO-signaling pathways.
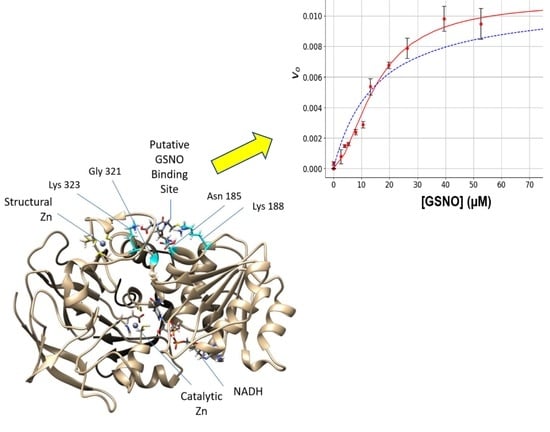
In this study, we observed that under steady-state kinetic conditions, the catalysis of GSNO reduction by GSNOR did not follow simple hyperbolic behavior i.e., the initial rate versus GSNO plots were sigmoidal (
Figure 1). These data were well accommodated by the Hill–Langmuir algorithm for allosteric kinetic behavior (Equation (3)) with an estimate Hill coefficient of approximately 1.5, which indicates positive cooperativity; that is, as the GSNO concentrations increase, the enzyme becomes activated. To the best of our knowledge, GSNOR allostery has not been reported previously. Allostery is manifested in multimeric proteins such as hemoglobin where allosteric behavior was first observed and characterized. In hemoglobin, as O
2 tension increases, one of its four subunits undergoes a conformational change that is translated to the other subunits, lowering their affinity to O
2. GSNOR exists as a homodimer; therefore, it possesses the minimum structural requirement for allosteric behavior. The kinetic behavior observed here with GSNOR can be explained by its substrate GSNO binding to an allosteric site on the enzyme and changing its conformation of its subunit(s) to activate the enzyme at larger levels of GSNO. The substrate-induced activation of GSNOR is logical from the physiological point of view, as GSNOR would work more efficiently to remove GSNO upon the sensing of a larger GSNO-flux.
We next probed the GSNOR structure for potential GSNO allosteric domains via dynamics and docking studies utilizing the GSNOR crystal structure PDB ID: 3QJ5 [
21] as the template structure. The computational investigations revealed a stable GSNO complex with a tetrad of residues Asn
185, Lys
188, Gly
321, and Lys
323 (
Figure 2). In fact, this postulated GSNO allosteric site yielded a more stable GSNO domain complex in comparison to that observed for the active site GSNO-binding domain (
Table S1).
The existence of the GSNO allosteric domain was tested with HDX-MS. In these experiments, D-exchange was performed on the protein in the absence and presence of GSNO. In theory, if GSNO bound at or near the residues comprising the GSNO-binding domain, then these residues could be identified as they would have less exposure to solvent and as a result yield lower D-exchange endpoints. The HDX-MS was performed at two exchange endpoints, 2 s and 4 s. The 2-s data corroborated the dynamics and docking studies as the postulated GSNO allosteric site residues Lys
188, Gly
321, and Lys
323, as well as the active site residues, showed decreased D-exchange endpoints (
Table S2,
Figure 3). At the 4-s D-exchange, the postulated allosteric site residue Lys
188 decreased further, whereas Gly
321 and Lys
323 as well as the active site residues yielded increased D-exchange endpoints, indicating more solvent exposure. We interpret these data as evidence that GSNO binding to the allosteric site ‘primes’ the enzyme for catalysis by lowering one or more activation barriers for the conformational transitions associated with catalysis.
As further confirmatory evidence for the GSNO allosteric site, we employed site-directed mutagenesis to change the postulated allosteric site lysines to alanines (K188A and K323A). Interestingly, K188A totally eliminated the +ve allosteric kinetic behavior as the intial rate (
vo) versus GSNO plots no longer displayed sigmoidal behavior (
Figure 4). The alteration of the other lysine (K323A) drastically lowered the catalytic activity of the enzyme, once again indicating that any changes in this region of the protein can drastically affect GSNOR structure and function. Double mutation of the lysines (K188A/K323A) again essentially inactivated the enzyme.
The HXD and the mutagenesis studies strongly suggest that GSNOR contains a second allosteric binding site for GSNOR.