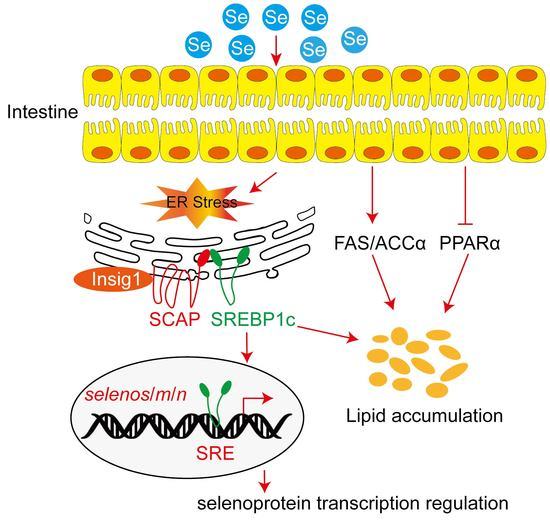
Dietary Marginal and Excess Selenium Increased Triglycerides Deposition, Induced Endoplasmic Reticulum Stress and Differentially Influenced Selenoproteins Expression in the Anterior and Middle Intestines of Yellow Catfish Pelteobagrus fulvidraco
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expt 1: Diets, Animals, and Management
2.2. Expt 2: The Analysis for the Regions of Selenos, Selenom, and Selenon Promoters
2.3. Sample Analysis
2.3.1. Histochemical Observations
2.3.2. Determination of TGs Concentrations and Se Contents in AI and MI
2.3.3. Enzyme Activity and Real-Time Quantitative PCR (qPCR)
2.3.4. Western Blot Analysis
2.3.5. Immunofluorescence Analysis
2.3.6. Assays for Luciferase Activities
2.3.7. EMSA
2.3.8. Statistical Analysis
3. Results
3.1. Experiment 1: In Vivo Study
3.1.1. Growth Performance, Feed Intake (FI), Morphological Parameters, and GPX Activity in Plasma and Liver
3.1.2. Oil Red O Staining, TGs Concentrations, and Se Contents
3.1.3. Enzymatic Activities
3.1.4. The Expression of Genes and Proteins Associated with Lipid Metabolism, ER Stress, ER Ca2+ Channels, and Selenogenome
3.1.5. Correlation Between the mRNA Levels of ER Stress Genes and Lipogenic Genes
3.2. Experiment 2: In Vitro Study
Characterization of Se-Responsive Element in Selenos, Selenom and Selenon Promoter
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa-Wong, A.N.; Berry, M.J.; Seale, L.A. Selenium and Metabolic Disorders: An Emphasis on Type 2 Diabetes Risk. Nutrients 2016, 8, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.C.; Zhao, H.; Li, J.G.; Xia, X.J.; Wang, K.N.; Zhang, Y.J.; Liu, Y.; Zhao, Y.; Lei, X.G. Selenoprotein gene expression in thyroid and pituitary of young pigs is not affected by dietary selenium deficiency or excess. J. Nutr. 2009, 139, 1061–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, X.; Pepper, M.P.; Vatamaniuk, M.Z.; Roneker, C.A.; Li, L.; Lei, X.G. Dietary selenium deficiency partially rescues type 2 diabetes-like phenotypes of glutathione peroxidase-1-overexpressing male mice. J. Nutr. 2012, 142, 1975–1982. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Kim, J.; Lei, X.G. High dietary fat and selenium concentrations exert tissue- and glutathione peroxidase 1-dependent impacts on lipid metabolism of young-adult mice. J. Nutr. 2020, 150, 1738–1748. [Google Scholar] [CrossRef]
- Akahoshi, N.; Anan, Y.; Hashimoto, Y.; Tokoro, N.; Mizuno, R.; Hayashi, S.; Yamamoto, S.; Shimada, K.; Kamata, S.; Ishii, I. Dietary selenium deficiency or selenomethionine excess drastically alters organ selenium contents without altering the expression of most selenoproteins in mice. J. Nutr. Biochem. 2019, 69, 120–129. [Google Scholar] [CrossRef]
- Huang, J.Q.; Li, D.L.; Zhao, H.; Sun, L.H.; Xia, X.J.; Wang, K.N.; Luo, X.; Lei, X.G. The selenium deficiency disease exudative diathesis in chicks is associated with downregulation of seven common selenoprotein genes in liver and muscle. J. Nutr. 2011, 141, 1605–1610. [Google Scholar] [CrossRef]
- Zhao, Z.; Barcus, M.; Kim, J.; Lum, K.L.; Mills, C.; Lei, X.G. High dietary selenium intake alters lipid metabolism and protein synthesis in liver and muscle of pigs. J. Nutr. 2016, 146, 1625–1633. [Google Scholar] [CrossRef]
- Cao, L.; Zhang, L.; Zeng, H.; Wu, R.T.Y.; Wu, T.L.; Cheng, W.H. Analyses of selenotranscriptomes and selenium concentrations in response to dietary selenium deficiency and age reveal common and distinct patterns by tissue and sex in telomere-dysfunctional mice. J. Nutr. 2017, 147, 1858–1866. [Google Scholar] [CrossRef] [Green Version]
- Ruhr, I.M.; Takei, Y.; Grosell, M. The role of the rectum in osmoregulation and the potential effect of renoguanylin on SLC26a6 transport in the gulf toadfish (Opsanus beta). Am. J. Physiol. 2016, 311, 179–191. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.H.; Hogstrand, C.; Luo, Z.; Zhang, D.G.; Ling, S.C.; Wu, K. Dietary zinc addition influenced zinc and lipid deposition in the fore- and mid-intestine of juvenile yellow catfish Pelteobagrus fulvidraco. Br. J. Nutr. 2017, 118, 570–579. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.H.; Luo, Z.; Hogstrand, C.; Wu, K.; Ling, S.C. SREBP1, PPARG and AMPK pathways mediated the Cu-induced change in intestinal lipogenesis and lipid transport of yellow catfish Pelteobagrus fulvidraco. Food Chem. 2018, 269, 595–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shchedrina, V.A.; Zhang, Y.; Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Structure-function relations, physiological roles, and evolution of mammalian ER-resident selenoproteins. Antioxid. Redox Signal. 2010, 12, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Addinsall, A.B.; Wright, C.R.; Andrikopoulos, S.; Poel, C.V.D.; Stupka, N. Emerging roles of endoplasmic reticulum-resident selenoproteins in the regulation of cellular stress responses and the implications for metabolic disease. Biochem. J. 2018, 475, 1037–1057. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitts, M.W.; Hoffmann, P.R. Endoplasmic reticulum-resident selenoproteins as regulators of calcium signaling and homeostasis. Cell Calcium 2018, 70, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.F.; Luo, Z.; Zhang, L.H.; Hogstrand, C.; Pan, Y.X. Endoplasmic reticulum stress and disturbed calcium homeostasis are involved in copper-induced alteration in hepatic lipid metabolism in yellow catfish Pelteobagrus fulvidraco. Chemosphere 2016, 144, 2443–2453. [Google Scholar] [CrossRef]
- Xu, Y.H.; Xu, Y.C.; Hogstrand, C.; Zhao, T.; Wu, L.X.; Zhuo, M.Q.; Luo, Z. Waterborne copper exposure up-regulated lipid deposition through the methylation of GRP78 and PGC1α of grass carp Ctenopharyngodon idella. Ecotoxicol. Environ. Saf. 2020, 205, 111089. [Google Scholar] [CrossRef] [PubMed]
- Rawson, R.B.; Zelenski, N.G.; Nijhawan, D.; Ye, J.; Sakai, J.; Hasan, M.T.; Chang, T.Y.; Brown, M.S.; Goldstein, J.L. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1997, 1, 47–57. [Google Scholar] [CrossRef]
- Tajima-Shirasaki, N.; Ishii, K.A.; Takayama, H.; Shirasaki, T.; Iwama, H.; Chikamoto, K.; Saito, Y.; Iwasaki, Y.; Teraguchi, A.; Lan, F.; et al. Eicosapentaenoic acid down-regulates expression of the selenoprotein P gene by inhibiting SREBP-1c protein independently of the AMP-activated protein kinase pathway in H4IIEC3 hepatocytes. J. Biol. Chem. 2017, 292, 10791–10800. [Google Scholar] [CrossRef] [Green Version]
- Mariotti, M.; Ridge, P.G.; Zhang, Y.; Lobanov, A.V.; Pringle, T.H.; Guigo, R.; Hatfield, D.L.; Gladyshev, V.N. Composition and evolution of the vertebrate and mammalian selenoproteomes. PLoS ONE 2012, 7, e33066. [Google Scholar] [CrossRef]
- Gong, G.R.; Dan, C.; Xiao, S.J.; Guo, W.J.; Huang, P.P.; Xiong, Y.; Wu, J.J.; He, Y.; Zhang, J.C.; Li, X.H.; et al. Chromosomal-level assembly of yellow catfish genome using third-generation DNA sequencing and Hi-C analysis. Gigascience 2018, 7, giy120. [Google Scholar] [CrossRef]
- Xu, X.J.; Zhang, D.G.; Zhao, T.; Xu, Y.H.; Luo, Z. Characterization and expression analysis of seven selenoprotein genes in yellow catfish Pelteobagrus fulvidraco to dietary selenium levels. J. Trace Elem. Med. Biol. 2020, 62, 126600. [Google Scholar] [CrossRef] [PubMed]
- Men, L.; Yao, J.; Yu, S.; Li, Y.; Cui, S.; Jin, S.; Zhang, G.; Ren, D.; Du, J. Selenoprotein S regulates adipogenesis through IRE1alpha-XBP1 pathway. J. Endocrinol. 2020, 244, 431–443. [Google Scholar] [CrossRef]
- Pitts, M.W.; Reeves, M.A.; Hashimoto, A.C.; Ogawa, A.; Kremer, P.; Seale, L.A.; Berry, M.J. Deletion of selenoprotein M leads to obesity without cognitive deficits. J. Biol. Chem. 2013, 288, 26121–26134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varone, E.; Pozzer, D.; Di Modica, S.; Chernorudskiy, A.; Nogara, L.; Baraldo, M.; Cinquanta, M.; Fumagalli, S.; Villar-Quiles, R.N.; De Simoni, M.G.; et al. SELENON (SEPN1) protects skeletal muscle from saturated fatty acid-induced ER stress and insulin resistance. Redox Biol. 2019, 24, 101176. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.R.; Huang, Y.H.; Wang, G.X.; Wu, Y.X.; Xian, J.A.; Wang, A.L.; Cao, J.M. Deficient and excess dietary selenium levels affect growth performance, blood cells apoptosis and liver HSP70 expression in juvenile yellow catfish Pelteobagrus fulvidraco. Fish Physiol. Biochem. 2016, 42, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.C.; Wu, K.; Zhang, D.G.; Luo, Z. Endoplasmic reticulum stress-mediated autophagy and apoptosis alleviate dietary fat-induced triglyceride accumulation in the intestine and in isolated intestinal epithelial cells of yellow catfish. J. Nutr. 2019, 149, 1732–1741. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.C.; Luo, Z.; Hogstrand, C.; Xu, Y.H.; Wu, L.X.; Chen, G.H.; Pan, Y.X.; Song, Y.F. Zinc reduces hepatic lipid deposition and activates lipophagy via Zn2+/MTF-1/PPARα and Ca2+/CaMKKβ/AMPK pathways. FASEB J. 2018, 32, 6666–6680. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Wu, K.; Hogstrand, C.; Xu, Y.H.; Chen, G.H.; Wei, C.C.; Luo, Z. Lipophagy mediated carbohydrate-induced changes of lipid metabolism via oxidative stress, endoplasmic reticulum (ER) stress and ChREBP/PPARγ pathways. Cell Mol. Life Sci. 2020, 77, 1987–2003. [Google Scholar] [CrossRef] [PubMed]
- Briens, M.; Mercier, Y.; Rouffineau, F.; Mercerand, F.; Geraert, P.A. 2-Hydroxy-4-methylselenobutanoic acid induces additional tissue selenium enrichment in broiler chickens compared with other selenium sources. Poul. Sci. 2014, 93, 85–93. [Google Scholar] [CrossRef]
- Poston, H.A.; Combs, G.F., Jr.; Leibovitz, L. Vitamin E and selenium interrelations in the diet of Atlantic salmon (Salmo salar): Gross, histological and biochemical deficiency signs. J. Nutr. 1976, 106, 892–904. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.G.; Pirie, B.J.; Adron, J.W.; Cowey, C.B. Some effects of selenium deficiency on glutathione peroxidase (EC 1.11.1.9) activity and tissue pathology in rainbow trout Salmo gairdneri. Br. J. Nutr. 1986, 55, 305–311. [Google Scholar] [CrossRef] [Green Version]
- Sunde, R.A.; Li, J.L.; Taylor, R.M. Insights for Setting of Nutrient Requirements, Gleaned by Comparison of Selenium Status Biomarkers in Turkeys and Chickens versus Rats, Mice, and Lambs. Adv. Nutr. 2016, 7, 1129–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Tang, J.; Xu, J.; Jia, G.; Liu, G.; Chen, X.; Cai, J.; Shang, H.; Zhao, H. Supranutritional dietary selenium induced hyperinsulinemia and dyslipidemia via affected expression of selenoprotein genes and insulin signal-related genes in broiler. RSC. Adv. 2016, 6, 4990–4998. [Google Scholar] [CrossRef]
- Zhao, L.; Feng, Y.; Deng, J.; Zhang, N.Y.; Zhang, W.P.; Liu, X.L.; Rajput, S.A.; Qi, D.S.; Sun, L.H. Selenium deficiency aggravates aflatoxin B1-induced immunotoxicity in chick spleen by regulating 6 selenoprotein genes and redox/inflammation/apoptotic signaling. J. Nutr. 2019, 149, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chandler, J.D.; Orr, M.L.; Hao, L.; Liu, K.; Uppal, K.; Go, Y.M.; Jones, D.P. Selenium supplementation alters hepatic energy and fatty acid metabolism in mice. J. Nutr. 2018, 148, 675–684. [Google Scholar] [CrossRef]
- Zhang, K.; Han, Y.; Zhao, Q.; Zhan, T.; Li, Y.; Sun, W.; Li, S.; Sun, D.; Si, X.; Yu, X.; et al. Targeted metabolomics analysis reveals that dietary supranutritional selenium regulates sugar and acylcarnitine metabolism homeostasis in pig liver. J. Nutr. 2020, 150, 704–711. [Google Scholar] [CrossRef]
- Ruiz-Jarabo, I.; Gregorio, S.F.; Gaetano, P.; Trischitta, F.; Fuentes, J. High rates of intestinal bicarbonate secretion in seawater tilapia (Oreochromis mossambicus). Comp. Biochem. Physiol. 2017, 207, 57–64. [Google Scholar] [CrossRef]
- Ling, S.C.; Luo, Z.; Chen, G.H.; Zhang, D.G.; Liu, X. Waterborne Zn influenced Zn uptake and lipid metabolism in two intestinal regions of juvenile goby Synechogobius hasta. Ecotoxicol. Environ. Saf. 2018, 148, 578–584. [Google Scholar] [CrossRef]
- Rutkowski, D.T.; Wu, J.; Back, S.H.; Callaghan, M.U.; Ferris, S.P.; Iqbal, J.; Clark, R.; Miao, H.; Hassler, J.R.; Fornek, J.; et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell 2008, 15, 829–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luciani, D.S.; Gwiazda, K.S.; Yang, T.L.; Kalynyak, T.B.; Bychkivska, Y.; Frey, M.H.; Jeffrey, K.D.; Sampaio, A.V.; Underhill, T.M.; Johnson, J.D. Roles of IP3R and RyR Ca2+ channels in endoplasmic reticulum stress and beta-cell death. Diabetes 2009, 58, 422–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wullschleger, M.; Blanch, J.; Egger, M. Functional local crosstalk of inositol 1,4,5-trisphosphate receptor- and ryanodine receptor-dependent Ca2+ release in atrial cardiomyocytes. Cardiovasc. Res. 2017, 113, 542–552. [Google Scholar] [CrossRef] [Green Version]
- Bagur, R.; Hajnoczky, G. Intracellular Ca2+ sensing: Its role in calcium homeostasis and signaling. Mol. Cell 2017, 66, 780–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kammoun, H.L.; Chabanon, H.; Hainault, I.; Luquet, S.; Magnan, C.; Koike, T.; Ferre, P.; Foufelle, F. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J. Clin. Investig. 2009, 119, 1201–1215. [Google Scholar] [CrossRef]
- Lee, J.N.; Ye, J. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J. Biol. Chem. 2004, 279, 45257–45265. [Google Scholar] [CrossRef] [Green Version]
- Bobrovnikova-Marjon, E.; Hatzivassiliou, G.; Grigoriadou, C.; Romero, M.; Cavener, D.R.; Thompson, C.B.; Diehl, J.A. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc. Natl. Acad. Sci. USA 2008, 105, 16314–16319. [Google Scholar] [CrossRef] [Green Version]
- Sunde, R.A.; Raines, A.M.; Barnes, K.M.; Evenson, J.K. Selenium status highly regulates selenoprotein mRNA levels for only a subset of the selenoproteins in the selenoproteome. Biosci. Rep. 2009, 29, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhao, H.; Zhang, Q.; Tang, J.; Li, K.; Xia, X.J.; Wang, K.N.; Li, K.; Lei, X.G. Prolonged dietary selenium deficiency or excess does not globally affect selenoprotein gene expression and/or protein production in various tissues of pigs. J. Nutr. 2012, 142, 1410–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burk, R.F.; Hill, K.E. Regulation of selenium metabolism and transport. Annu. Rev. Nutr. 2015, 35, 109–134. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.G.; Evenson, J.K.; Thompson, K.M.; Sunde, R.A. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J. Nutr. 1995, 125, 1438–1446. [Google Scholar] [PubMed]
- Campo-Sabariz, J.; Moral-Anter, D.; Brufau, M.T.; Briens, M.; Pinloche, E.; Ferrer, R.; Martin-Venegas, R. 2-hydroxy-(4-methylseleno) butanoic acid is used by intestinal Caco-2 cells as a source of selenium and protects against oxidative stress. J. Nutr. 2019, 149, 2191–2198. [Google Scholar] [CrossRef]
- Speckmann, B.; Bidmon, H.J.; Borchardt, A.; Sies, H.; Steinbrenner, H. Intestinal selenoprotein P in epithelial cells and in plasma cells. Arch. Biochem. Biophys. 2014, 541, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Kipp, A.P.; Muller, M.F.; Goken, E.M.; Deubel, S.; Brigelius-Flohe, R. The selenoproteins GPx2, TrxR2 and TrxR3 are regulated by Wnt signaling in the intestinal epithelium. Biochim. Biophys. Acta. 2012, 820, 588–596. [Google Scholar]
- Maseko, T.; Howell, K.; Dunshea, F.R.; Ng, K. Selenium-enriched Agaricus bisporus increases expression and activity of glutathione peroxidase-1 and expression of glutathione peroxidase-2 in rat colon. Food Chem. 2014, 46, 27–33. [Google Scholar] [CrossRef] [PubMed]
- De Spirt, S.; Eckers, A.; Wehrend, C.; Micoogullari, M.; Sies, H.; Stahl, W.; Steinbrenner, H. Interplay between the chalcone cardamonin and selenium in the biosynthesis of Nrf2-regulated antioxidant enzymes in intestinal Caco-2 cells. Free Radic. Biol. Med. 2016, 91, 164–171. [Google Scholar] [CrossRef]
- Sun, L.H.; Zhang, N.Y.; Zhu, M.K.; Zhao, L.; Zhou, J.C.; Qi, D.S. Prevention of aflatoxin B1 hepatoxicity by dietary selenium is associated with inhibition of cytochrome P450 isozymes and up-regulation of 6 selenoprotein genes in chick liver. J. Nutr. 2016, 146, 655–661. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, L.H.; Huang, J.Q.; Briens, M.; Qi, D.S.; Xu, S.W.; Lei, X.G. A novel organic selenium compound exerts unique regulation of selenium speciation, selenogenome, and selenoproteins in broiler chicks. J. Nutr. 2017, 147, 789–797. [Google Scholar] [CrossRef] [Green Version]
- Chernorudskiy, A.; Varone, E.; Colombo, S.F.; Fumagalli, S.; Cagnotto, A.; Cattaneo, A.; Briens, M.; Baltzinger, M.; Kuhn, L.; Bachi, A.; et al. Selenoprotein N is an endoplasmic reticulum calcium sensor that links luminal calcium levels to a redox activity. Proc. Natl. Acad. Sci. USA 2020, 117, 21288–21298. [Google Scholar] [CrossRef]
- Olsson, M.; Olsson, B.; Jacobson, P.; Thelle, D.S.; Bjorkegren, J.; Walley, A.; Froguel, P.; Carlsson, L.M.; Sjoholm, K. Experssion of the selenoprotein S (SELS) gene in subcutaneous adipose tissue and SELS genotype are associated with metabolic risk factors. Metabolism 2011, 60, 114–120. [Google Scholar] [CrossRef]
- Takayama, H.; Misu, H.; Iwama, H.; Chikamoto, K.; Saito, Y.; Murao, K.; Teraguchi, A.; Lan, F.; Kikuchi, A.; Saito, R.; et al. Metformin suppresses expression of the selenoprotein P gene via an AMP-activated kinase (AMPK)/FoxO3a pathway in H4IIEC3 hepatocytes. J. Biol. Chem. 2014, 289, 335–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, B.; Huang, Y.; Zou, C.; Wu, Y.; Huang, Y.; Ni, J.; Tian, J. Transcriptional regulation of selenoprotein F by heat shock factor 1 during selenium supplementation and stress response. Cells 2019, 8, 479. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| Se Supplementation | |||
|---|---|---|---|
| 0.03 mg/kg | 0.25 mg/kg | 6.39 mg/kg | |
| IBW, g/fish | 8.34 ± 0.03 | 8.26 ± 0.03 | 8.21 ± 0.05 |
| FBW, g/fish | 21.6 ± 0.72 b | 25.6 ± 0.71 a | 21.3 ± 0.32 b |
| WG 1, % | 160 ± 14.1 b | 210 ± 8.0 a | 159 ± 4.1 b |
| FCR 2 | 1.77 ± 0.03 a | 1.50 ± 0.04 b | 1.54 ± 0.03b |
| FI, g/fish | 23.5 ± 2.1 | 25.9 ± 1.3 | 20.15 ± 3.2 |
| Survival, % | 100.0 ± 0.00 | 100.0 ± 0.00 | 100.0 ± 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.-G.; Zhao, T.; Xu, X.-J.; Lv, W.-H.; Luo, Z. Dietary Marginal and Excess Selenium Increased Triglycerides Deposition, Induced Endoplasmic Reticulum Stress and Differentially Influenced Selenoproteins Expression in the Anterior and Middle Intestines of Yellow Catfish Pelteobagrus fulvidraco. Antioxidants 2021, 10, 535. https://doi.org/10.3390/antiox10040535
Zhang D-G, Zhao T, Xu X-J, Lv W-H, Luo Z. Dietary Marginal and Excess Selenium Increased Triglycerides Deposition, Induced Endoplasmic Reticulum Stress and Differentially Influenced Selenoproteins Expression in the Anterior and Middle Intestines of Yellow Catfish Pelteobagrus fulvidraco. Antioxidants. 2021; 10(4):535. https://doi.org/10.3390/antiox10040535
Chicago/Turabian StyleZhang, Dian-Guang, Tao Zhao, Xiao-Jian Xu, Wu-Hong Lv, and Zhi Luo. 2021. "Dietary Marginal and Excess Selenium Increased Triglycerides Deposition, Induced Endoplasmic Reticulum Stress and Differentially Influenced Selenoproteins Expression in the Anterior and Middle Intestines of Yellow Catfish Pelteobagrus fulvidraco" Antioxidants 10, no. 4: 535. https://doi.org/10.3390/antiox10040535