1
Bioinformatics Graduate Program, University of British Columbia, Vancouver, BC, Canada
2
Centre for High-Throughput Biology, University of British Columbia, Vancouver, BC, Canada
3
Department of Psychiatry, University of British Columbia, Vancouver, BC, Canada
The ability to computationally extract mentions of neuroanatomical regions from the literature would assist linking to other entities within and outside of an article. Examples include extracting reports of connectivity or region-specific gene expression. To facilitate text mining of neuroscience literature we have created a corpus of manually annotated brain region mentions. The corpus contains 1,377 abstracts with 18,242 brain region annotations. Interannotator agreement was evaluated for a subset of the documents, and was 90.7% and 96.7% for strict and lenient matching respectively. We observed a large vocabulary of over 6,000 unique brain region terms and 17,000 words. For automatic extraction of brain region mentions we evaluated simple dictionary methods and complex natural language processing techniques. The dictionary methods based on neuroanatomical lexicons recalled 36% of the mentions with 57% precision. The best performance was achieved using a conditional random field (CRF) with a rich feature set. Features were based on morphological, lexical, syntactic and contextual information. The CRF recalled 76% of mentions at 81% precision, by counting partial matches recall and precision increase to 86% and 92% respectively. We suspect a large amount of error is due to coordinating conjunctions, previously unseen words and brain regions of less commonly studied organisms. We found context windows, lemmatization and abbreviation expansion to be the most informative techniques. The corpus is freely available at http://www.chibi.ubc.ca/WhiteText/
.
Bioinformatics has proven the value of databasing and formalizing knowledge. Traditionally much of the focus is on molecular biology but neuroscience researchers are taking note (French and Pavlidis, 2007
). One means of building, or at least seeding, knowledge bases is text mining, or the automated extraction and formalization of information from free text sources such as the biomedical literature. There has been much interest in applying text mining to extracting information about genes and proteins. In the BioCreative 2 challenge, 44 teams competed to extract, resolve and link protein and gene mentions (Krallinger et al., 2008
), and the methods work well enough to be of practical importance in creating databases (Leitner et al., 2008
). There has been less work on how to apply such techniques to domain-specific knowledge in neuroscience.
One entity of interest in the neuroscience literature is mentions of neuroanatomical regions (which we call brain regions for short). By analogy to the task of extracting gene mentions, the ability to computationally extract mentions of brain regions would be of potential value in building neurobiological knowledge bases. This is because many neurobiological studies only make sense in the context of the specific brain regions studied. Furthermore anatomical or functional connections between regions are commonly described. Computationally extracting these locations would allow faster organization and mining of neuroscience data.
We hypothesize that many of the methods and approaches developed for extraction of information about genes can be applied to extraction of information about brain areas. This is an attractive approach because many of the challenges in analyzing text for information about genes are also faced in trying to mine information about brain regions. These challenges include abbreviations, synonyms, lexical variation and ambiguity. For example, the gene “carbonic anhydrase 1” has synonyms including “carbonate dehydratase I”, “Car1”, and “CA-I”. Its official symbol, CA1, is ambiguious in that it also matches a drug (the abbreviated form of coumermycin A1) and a brain region (the CA1 field of the hippocampus). Similarly brain regions have a variety of names and abbreviations, and can be confused with other types of entities. Approaches have been developed for addressing these problems for genes, so it seems reasonable to expect that the lessons learned will apply at least partly to other domains. However, before these approaches can be applied to brain regions, a “gold standard” corpus is needed. Such a corpus is needed both as training data for algorithms and for evaluation of methods. To our knowledge, no such resource exists for neuroscience text mining.
Past efforts in neuroscience text mining provided limited ability to retrieve brain region mentions, by looking for exact matches of brain region names from small lists (Crasto et al., 2003
, 2007
; Muller et al., 2008
). This limits the recall to a small number of (usually broad or large) brain regions. The most extensive effort is “Textpresso for Neuroscience”, with a list of 4,800 brain region terms (Muller et al., 2008
). Unfortunately evaluations of these tools are lacking, as the methods were not checked against a gold standard set of annotated abstracts, leaving accuracy in question. The Neuroscholar project was the first to explore advanced natural language processing methods to extraction of neuroscience data (Burns et al., 2007
). Focusing on neuroanatomical connectivity, Burns et al. sought to extract and annotate detailed statements from full-text articles. Their goal was extraction of relatively detailed experimental parameters and descriptions of results. They manually annotated 1,047 sentences from 21 articles. Text spans were tagged with five different labels including two that represented brain regions. These annotations provided the test and training examples for a CRF that was able to produce the same tags at an overall 79% F-Measure (performance for brain-region recognition alone was not reported). Although it was a small dataset they found the CRF could be joined with manual curation to increase annotation rate by 255%.
The goals of the current work are two-fold. First, we provide a reasonably large corpus of article abstracts manually annotated for brain region mentions. Second, we develop and evaluate methods for extraction of brain region mentions from text, using the corpus. We also describe preliminary attempts to normalize the mentions to brain region terms from a common neuroanatomical database. This sets the stage for further efforts at improving and applying text-mining methods to neuroanatomical questions.
Corpus Creation
Articles for the corpus were initially selected manually but later an automated procedure was employed. The first 119 articles in the corpus were selected with the help of PubMed
1
searches using keywords such as “afferent” and “efferent”. The process was then automated to increase speed of curation and reduce bias in selection. The automated procedure picks random articles from the Journal of Comparative Neurology. There was no limitation placed on the topic organism (rat and cat were most common but insects were the topics of some abstracts). We also experimented with other search strategies, for example the MeSH keyword of “Neural Pathways”. The Journal of Comparative Neurology was chosen to maximize the number of abstracts that included brain region mentions. It has also been used in previous work (Burns et al., 2007
). A total of 1,377 abstracts were used.
The selected abstracts were retrieved in MEDLINE XML format for preprocessing. For each abstract the PubMed identifier, title and abstract were stored. The abstract text was then processed by the Schwartz and Hearst (2003)
abbreviation expansion algorithm. This identifies the short and long forms of abbreviations in the abstract with high accuracy. All short forms of the abbreviation are replaced with the long form followed by its short form in parentheses. Thirty-two abstracts (2.3%) were reloaded without expansion due to encoding errors. The abbreviation expansion changes are expressed in the XML markup and can be reversed. Finally, annotators are provided the abstract and title for annotation. Figure 1
shows an example of a final annotated abstract with abbreviations expanded. The General Architecture for Text Engineering (GATE)
2
was used to create, compare and visualize the document annotations. Additionally, GATE provided a helpful interface and API for managing the document collections.
Figure 1. A representative annotated abstract with several expanded abbreviations (Gabbott et al., 2005
).
Manual Annotation Guidelines
The annotators were presented with the title and abstract text in the GATE interactive document display. Using the computer mouse, regions of text were selected and then “tagged” as representing a brain region mention. One annotator (the “primary” annotator, SL) annotated all abstracts. A secondary annotator (LX) re-annotated a random subset of abstracts annotated by the primary annotator (to allow estimation of the human component in annotation accuracy). The annotators used their own knowledge of neuroanatomy, supplemented by online resources such as medical dictionaries, neuroanatomical atlases and BrainInfo
3
. An initial set of guidelines were developed prior to the annotation starting; these guidelines were amended in response to the outcome of periodic discussion of problems and manual review of the corpus.
Brain (and spinal cord) regions were the primary targets of our manual annotation efforts. We annotated all mentions of brain regions in both the abstracts and titles according to our guidelines. Although we annotated all brain region mentions, our guidelines are influenced by our interest in mentions that describe higher-level features such as neuroanatomical connections.
A key set of guidelines involves the level of detail. In particular, we did not attempt to annotate details such as specific cortical layers, in part because they cover the whole cortex but also because these were judged to present an additional challenge that would be a topic of future work. Conversely, very broad mentions of “systems” were not annotated (e.g. “orexin/hypocretin system” or “vestibular system”). However, mentions such as “cortex” were captured. Further, mentions of white matter tracts or fasciculi were not annotated. Annotations also included text that modified the mention. An example is “motor related areas of the hippocampus”. We annotated the adjective forms of brain regions, for example “thamalic” or “cortical”. We also annotated parts that were identified by a number (primarily this applied to Brodmann areas or cortical regions such as V1). Brain region mentions were not extended to include organism name, so “rat hippocampus” would always be annotated only as “hippocampus”. We annotated text segments that referred to a specific region but might not be resolvable without more context. For example, in an abstract about the cerebellum we might find mentions of “medial zone”. As a fragment, “medial zone” cannot be assigned to a specific region.
One particular problem area is conjunctions or coordination ellipses. Examples are “dorsal and ventral cortex” or “lower thoracic and lumbosacral segments”. The difficulty is determining whether these should be broken up into two brain region mentions or treated together. Past annotation efforts have recognized this difficulty (Tanabe et al., 2005
). Unlike abbreviations there is no reliable method to automatically expand such expressions (Buyko et al., 2007
). In the corpus, annotation of conjunctions varies except in the abstracts annotated by both annotators where consistency was enforced. To achieve this, the whole conjunction was annotated if the contained brain region names have been shortened.
Dictionary Matching
To test dictionary matching approaches we created term lists from neuroanatomical nomenclature sources. Although several lexicons exist we focused on Neuronames, the largest source of brain region names (Bowden and Dubach, 2003
). We extracted terms from both Nomenclatures of Canonical Mouse and Rat Brain Atlases and the Ontology of Human and Macaque Neuroanatomy. From the later, a total of 6,462 terms were extracted from the primary names, synonyms, ancillary structures and Latin terms. We additionally extracted 1,900 terms from the Nomenclatures of Canonical Mouse and Rat Brain Atlases that organizes terms from mouse (Hof et al., 2000
; Paxinos and Franklin, 2001
; Dong, 2007
) and rat brain atlases (Swanson, 1999
). Since we expand abbreviations within the abstracts we excluded abbreviations contained in Neuronames.
To match the Neuronames terms to the document text we used a GATE Gazetteer. Bracketed text in Neuronames terms were removed before matching. We set the Gazetteer to use case insensitive exact string matching. Resulting annotations were joined to remove overlapping matches.
To compare our method to that used by “Textpresso for Neuroscience” we used its lexicon files
4
, with case sensitive exact matching. To further replicate conditions used by Textpresso we reverted the expanded abbreviations in the abstracts and did not filter abbreviation terms from the lexicon.
Conditional Random Field
For automated annotation of brain region mentions, we applied a linear chain conditional random field (CRF) using the Mallet software toolkit (Lafferty et al., 2001
; McCallum, 2002
). A linear chain CRF is similar to a hidden Markov model (HMM). Like an HMM, a CRF is a method for sequence processing that takes a series of symbols (in our case, words) as input and provides as output the predicted state (in our case, whether the symbol is part of a brain region mention or not). Unlike HMM’s, in which state probabilities are conditioned only on the state of the previous token, CRF state probabilities are computed by conditioning on the entire input sequence. Therefore, it cannot compare the probabilities of labellings across sentences. In return CRF models allow token descriptions (features) with complex dependencies. For example, HMM’s use current token type but a CRF feature design can examine the previous and next two tokens.
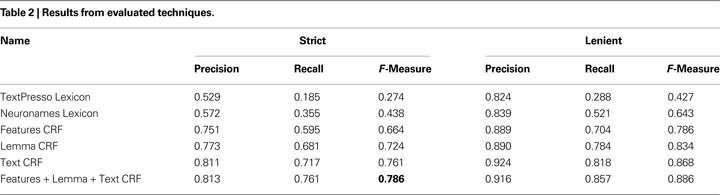
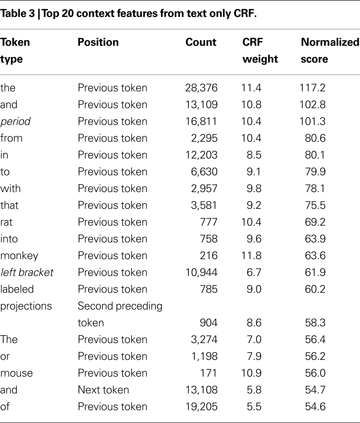
To start, the CRF model must be trained, by computing features for tokens with known label sequences (training set). In our case each feature has a Boolean value (details on the features are given in the next section). For example a feature named “text = red” is true if the current token is “red”. These features combined with the state transitions form feature functions. The feature functions are then given weights, so that a specific feature can influence the likelihood of specific state transition. The weights are learned from the known state sequences using an optimization procedure. For example, in Table 3
we can see that the probability of the label sequence changing from outside of a brain region to inside is increased when the preceding token is “the”. For test sequences or sentences, probabilities of state sequences are computed. The most probable state sequence then forms the predicted brain region mention spans. For further detail we point our readers to a more complete introduction of CRFs (Wallach, 2004
).
The GATE software was used to segment the abstracts into sentences and tokens. For Mallet we used default CRF settings from the SimpleTagger class except Gaussian variance was set to 1.
Features
As mentioned, all of the features we used were binary. Thus the representation of each token was a long binary vector representing, for each feature, whether it was present for the given token. The simplest feature is the token itself, generated for every word/token in the corpus. We tested orthographic features, for example an uppercase first letter or presence of a numerical digit. The part of speech tag and lemma of the word were also computed and tested. Like the text features, the lemmas of every word become a feature that is set to true if a word’s canonical form matches that lemma. To determine lemmas and tags we employed a model for the TreeTagger software (Schmid, 1994
) that was extensively trained on the GENIA biomedical corpus (Kim et al., 2003
) for STRING-IE (Saric et al., 2006
).
The token is compared to several term lists and lexical resources. For complete matching a word and neighbouring words must exactly match a brain region name in one of many neuroanatomical lexicons. Further we segmented the brain region names into word n-grams. For example “ventral anterior nucleus” is fragmented into the 2-grams of “ventral anterior” and “anterior nucleus”. The tokens are then matched against these n-grams allowing relaxed matches to the lexicons. We further employed word lists for neuroanatomical terms describing boundaries or regions (e.g. bank, sulci, surface, area), neuroanatomical directions (e.g. dorsal, superior), root neuroscience terms (e.g. chiasm, raphe, striated) and stop words (e.g. on, this, is). Root neuroscience terms were extracted from Dr. Eric Chudler’s resource for neuroanatomical, neurophysiological and neuropsychological terminology
5
. We used the stop word list from the Snowball small string processing language software
6
. We also added regular expression features that match common templates, for example Brodmann’s areas and spinal vertebrae. Finally, we employed window features that add context information to the current words feature set. This is done by encoding features from previous and following words into the current word’s set.
To rank the context features we averaged feature weights from eight cross-validation folds. The weights are from CRFs using only the text feature with a context window of two tokens on each side. We show the top weights for the state transition of outside a brain region mention into inside one, which occurs at the first word of a brain region mention. We filtered out the direct features from the current word to leave only the weights and rankings of features derived from the neighbouring words. Next we calculated a normalized score by multiplying the weight by the natural logarithm of its frequency.
Experiment Setup
Manual feature design and initial tests were performed using eight-fold cross-validation on the 1,146 abstracts annotated only by the primary annotator. Annotations from both curators were merged by a logical OR operation at the character level (if an annotator marked that character as a brain region then it was kept). Sentences of an abstract were not split between training and testing sets. Each sentence became an input instance for the CRF. Final results were generated on the same eight-fold cross-validation across all abstracts.
Resolution
To separate this task from recognition of brain region mentions we attempted to resolve manually annotated brain region mentions. The target term set are the same Neuroname entries previously described for dictionary matching. The first method ignored case and removed text surrounded by brackets. The “bag of words” method is similar except word order is ignored. The lexicon entries and mentions are tokenized with “of” and “the” removed. Matches are then found by exact matching of the token sets or bags of words. For example “ventral posterolateral nucleus, caudal part” matches “caudal part of ventral posterolateral nucleus”. Because both of these methods are very strict we did not evaluate the results for accuracy and instead provide coverage.
Evaluation
We used standard evaluation measures that ignore true negatives and operate at the annotation level instead of the token. Precision is defined as the proportion of predictions matching the annotated spans with recall being the proportion of annotated spans that match a prediction. F-measure is the harmonic mean of precision and recall. In the strict case annotation spans must match exactly. Lenient measures are computed by counting partially overlapping spans as matches.
In total 1,377 abstracts were annotated by the primary curator. A second curator annotated 231 of those abstracts for agreement evaluation. The average number of brain region annotations per abstract from the primary curator was 13.2 and 14.6 for the second. Interannotator agreement was 90.7% (F-measure), increasing to 96.7% for the lenient measure. Table 1
displays the top 40 occurring mentions and their frequencies in the corpus.
The GATE tokenizer split the corpus into 17,247 sentences then 461,552 tokens with 46,340 labelled as brain regions. On average each brain region is 2.3 tokens in length. We observed a large vocabulary of 17,901 token types.
Lexicon-based methods directed from neuroanatomical atlases performed poorly on the dataset, reaching 43.8% F-measure (precision = 57.2%, recall = 35.5%). We expected a higher level of precision; we believe variances in applying the annotation guidelines account for some of the false positives. Neuronames contains terms for layers, systems and tracts all of which we did not annotate. In addition, TextPresso contains abbreviations which possibly cause additional false positives.
The next best performance of 66.4% F-Measure was attained by a CRF using 625 features we derived primarily from neuroanatomical lexicons. The lemma and text based CRF’s demonstrate the effect of the context window. These classifiers only look at the token type, or word. Without the window features the text based CRF achieves 66.7% F-measure. Adding information about the previous and next two words increases F-Measure to 76.1%. By combining any two of the designed, lemma and text feature sets the CRF reaches F-measures in the range 76–78%. Combining the text and lemma features only slightly improves on text alone suggesting the features are very similar. By combing all three feature sets, the F-Measure peaks at 78.6%, with most of the gain from recall. This CRF that combined all features perfectly predicted all brain region mentions for 174 abstracts that had on average 6.8 brain region mentions per abstract.
We were unable to clearly determine which of our designed features contributed most to the final performance. This is due to the high dependency between the designed features and the simple text features. Furthermore, F-Measure varies by about 1% across different cross-validation splits, so improvements of less than 1% are not significant.
Throughout Table 2
the recall rate is below precision. This suggests many novel brain regions are left unrecognized, also known as out-of-vocabulary error. Indeed, we find that on average 19.3% of text features are observed in the test folds but not in the training folds. To test the impact of this effect, we repeated the experiment but allowing the sentences of an abstract to be spread across training and testing sets. This decreases unseen words to 10.4% because new terms are often mentioned many times throughout an abstract. At this sentence level performance improves; F-measure reaches 0.813 with the gain in recall twice that of precision. This suggests that, not surprisingly, performance can be improved simply by having more diverse training data.
We found some of the poorly classified examples were studies of brain regions from insects or other organisms underrepresented in the corpus. These abstracts tended to lack relevant training samples, and the regions they mention are not contained in the brain region lexicons we collected, resulting in very poor recall. To examine this effect in more detail, we used a subset of abstracts for which we annotated the organism of study. This subset was further reduced to those studying monkey, cat, rat and mouse brains. A full featured CRF trained on this set of 214 common organism abstracts demonstrates much higher performance than a CRF trained on a random subset of the same size. This is demonstrated primarily by recall which increases to 75.7% from 67.6%, combined with a small increase in precision we find F-Measure increases to 77.8% from 72.5%. In terms of unseen features, the random set has 20.2% compared to 17.6% for the common organism set. This suggests that both sets have a similar out-of-vocabulary error.
We began by assuming that expanding abbreviations to the full forms would increase performance. As a test of this assumption, we reverted the expanded abbreviations back to the original, resulting in an F-Measure decrease of only 2.1 (to 76.5%). If we include the Neuronames abbreviation terms as an added feature this difference is reduced to 1.4.
We observed that coordinating conjunctions (see Materials and Methods) cause a significant amount of error. Examples are “middle and caudal amydgala” or “hippocampus and amydgala”. Five percent of annotations have a similar form with 893 annotations in 403 of the abstracts containing “and”, “or”, comma, semicolon, or a slash. By removing these abstracts we remove annotations that span conjunctions, the remaining abstracts still have conjunctions but each part is annotated separately. By training and testing the CRF on the reduced set of 974 the F-Measure increases to 79.9. This is significant compared to 76.5% reached by a CRF trained on a random set of the same size. With these consistently annotated conjunctions the strict precision gains the most, while lenient precision is almost unchanged. This suggests both datasets produce similar predictions but consistent annotations produce more precise spans.
Table 3
presents the context feature weights derived from a text only conditional random field. The window size ranged from the two preceding and following tokens. Although we only display the top 20, this CRF has over 300,000 weights for 17,901 token types times 5 token locations across four state changes. As expected common prepositions or adpositions are the most informative. Interestingly, “rat” and “monkey” have top scores. It seems the CRF learned that an organism name often precedes a brain region mention. Another entry is “projections” that is informative when seen two words before the current token. This connectivity related word makes sense given the high number of tract tracing experiments in the Journal of Comparative Neurology.
We found several techniques frequently used in general and biomedical named entity recognition research did not improve performance. Guided by work on gene name extraction we experimented with bidirectional parsing and beginning-inside-outside labels (Hsu et al., 2008
). We processed the text using MMTx and extracted rich semantic features (Aronson, 2006
). We tested feature induction (McCallum, 2003
), an extension of the CRF framework. To treat the abstract as a whole we tested treating each abstract as a sequence instead of its sentences and also carried the features from the first mention of a word to all the following. The large vocabulary suggested semi-supervised learning may help; we tested a self training approach using an additional set of 3,881 unlabelled abstracts. Unfortunately, all of these methods failed to produce a significant increase in performance when compared to our best results.
We tested the two simple normalization procedures for resolving a brain region mention to its term in the Neuronames vocabulary. The first approach of direct matching covers 33.1% of annotations. This mirrors the recall of the lexicon based method for recognition. By ignoring the number of times a mention occurs we find that 11.4% of the 6,146 unique text mentions are matched to a term. The “bag of words” method that disregards the word order matched slightly more regions at 34.6% with 13.0% unique term matches. The bag of words method reduced the total number of Neuronames entries by 96 to 6,366 suggesting that terms maintain their uniqueness when word order is ignored. Because of the strict constraints enforced by these methods we believe that almost all of the term matches are correct.
We have provided the first corpus of manually annotated brain region mentions in biomedical abstracts. The corpus is large enough to allow statistical models to learn the nomenclature. This is demonstrated by the text-based CRF which reached a 76.1% F-Measure without outside resources. We found context windows, lemmatization and abbreviation expansion to be the most informative features for CRF labelling. A CRF using all the features provided the best performance of 78.6% F-Measure.
Compared to more advanced techniques, the dictionary approach based on neuroanatomical lexicons performed poorly. However, it has the advantage of speed and easier resolution to standardized names. Furthermore, features derived from these lexicons provide valuable information to the CRF models.
We demonstrated that significant amounts of error are due to coordinating conjunctions, previously unseen words and brain regions of less commonly studied organisms. The poor performance of the lexicon combined with recall values consistently below precision suggest that lexical resources for neuroscience need to be improved. Current resources are based primary on neuroanatomical atlases of a few organisms. With open initiatives like NeuroLex we hope richer resources will be generated by a broader audience
7
.
We performed a preliminary examination of normalization of mentions to standardized identifiers. This task is more difficult than mention extraction alone, as demonstrated by our baseline methods covering just over one-third of mentions. One reason for the difficulty of the normalization task is that researchers do not use standardized nomenclatures for brain regions in their papers. This is a recognized problem for resolving gene mentions (where aliases are common) which has been ameliorated to some extent by efforts by nomenclature standardization committees (Wain et al., 2004
). Such efforts would be of obvious value in neuroscience (Bug et al., 2008
). When combined with organism identification it grows in difficulty.
Leon French and Paul Pavlidis designed the study. Leon French implemented the software and performed all analyses. Suzanne Lane and Lydia Xu performed evaluations. Leon French and Paul Pavlidis wrote the manuscript. Paul Pavlidis provided project management and oversight.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
LF is supported by Natural Sciences and Engineering Research Council of Canada. PP is supported by a career award from the Michael Smith Foundation for Health Research, a CIHR New Investigator award, and a Human Brain Project grant from the National Institutes of Health (GM076990).
Aronson, A. R. (2006). MetaMap: Mapping Text to the UMLS Metathesaurus. Available at: http://skr.nlm.nih.gov/papers/references/metamap06.pdf
.
Leitner, F., Krallinger, M., Rodriguez-Penagos, C., Hakenberg, J., Plake, C., Kuo, C. J., Hsu, C. N., Tsai, R. T., Hung, H. C., Lau, W. W., Johnson, C. A., Saetre, R., Yoshida, K., Chen, Y. H., Kim, S., Shin, S. Y., Zhang, B. T., Baumgartner, W. A., Jr., Hunter, L., Haddow, B., Matthews, M., Wang, X., Ruch, P., Ehrler, F., Ozgur, A., Erkan, G., Radev, D. R., Krauthammer, M., Luong, T., Hoffmann, R., Sander, C., and Valencia, A. (2008). Introducing meta-services for biomedical information extraction. Genome Biol. 9(Suppl. 2), S6.
McCallum, A. (2002). MALLET: A Machine Learning for Language Toolkit. Available at: http://mallet.cs.umass.edu
.