Liver transcriptome and physiological analyses preliminarily revealed the adaptation mechanisms of Amur grayling (Thymallus arcticus grubei, Dybowski, 1869) fry for dietary lipid nutrition
- 1Key Laboratory of Aquatic Animal Diseases and Immune Technology of Heilongjiang Province, Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Harbin, China
- 2Tianjin Key Lab of Aqua-Ecology and Aquaculture, College of Fisheries, Tianjin Agricultural University, Tianjin, China
- 3Supervision, Inspection and Testing Center for Fishery Environment and Aquatic Products (Harbin), Ministry of Agriculture and Rural Affairs, Harbin, China
- 4Heilongjiang Aquatic Animal Resource Conservation Center, Harbin, China
The Amur grayling (Thymallus arcticus grubei Dybowski, 1869), a species of potentially economic and research value, is renowned for its tender meat, exquisite flavor, and high nutritional contents. This study was conducted to investigate the physiological adaptation mechanisms to dietary lipids in Amur grayling fry (with average initial weight 4.64±0.03 g). This study involved a 56-day feeding trial with diets containing varying lipid levels (9.07%, 12.17%, 15.26%, 18.09%, 21.16%, and 24.07%, designated as GL1 through GL6, respectively) to explore the impact of dietary lipids on growth performance, intestinal digestion, liver antioxidative function, and transcriptomic profiles. Results showed that The group receiving 18% dietary lipid exhibited a markedly higher weight gain rate (WGR) and specific growth rate compared to other groups, alongside a reduced feed conversion ratio (FCR), except in comparison to the 15% lipid group. Activities of lipase in pancreatic secretion and amylase in stomach mucosa peaked in the 18% lipid treatment group, indicating enhanced digestive efficiency. The liver of fish in this group also showed increased activities of antioxidative enzymes and higher levels of glutathione and total antioxidative capacity, along with reduced malondialdehyde content compared to the 9% and 24% lipid treatments. Additionally, serum high-density lipoprotein cholesterol levels were highest in the 18% group. Transcriptomic analysis revealed four significant metabolic pathways affected: Cholesterol metabolism, Fat digestion and absorption, PPAR signaling, and Fatty acid degradation, involving key genes such as Lipase, Lipoprotein lipase, Fatty acid-binding protein, and Carnitine palmitoyltransferase I. These findings suggest that the liver of Amur grayling employs adaptive mechanisms to manage excessive dietary lipids. Quadratic regression analysis determined the optimal dietary lipid levels to be 16.62% and 16.52%, based on WGR and FCR, respectively. The optimal dietary lipid level for juvenile Amur grayling appears to be around 18%, as evidenced by improved growth performance, digestive function, balanced serum lipid profile, and enhanced liver antioxidative capacity. Exceeding this lipid threshold triggers both adaptive and potentially detrimental liver responses.
Introduction
The Amur grayling (Thymallus arcticus grubei, Dybowski, 1869), a member of the Salmoniformes, Salmonidae, and Thymallus families, is mainly distributed in Eurasia and the tributaries of Heilongjiang River basin in China. It is a rare cold-water fish unique to the Heilongjiang River system. This species is esteemed for its tender meat, appealing taste, and high nutritional value (1). Recent declines in the wild population, primarily due to overfishing, environmental degradation, and habitat destruction, have led to the Amur grayling being classified as a vulnerable species in China (2). Despite advances in artificial breeding (3), seed cultivation (4), and germplasm identification (5) in China, comprehensive studies on the nutritional needs of Amur grayling remain unreported.
Identifying optimal dietary protein and fat levels is fundamental in the nutrition research of cold-water fish (6). Research by Gu et al. (7) indicated that the crude protein content in Amur grayling muscle is 17.93%, lower than that in rainbow trout (Oncorhynchus mykiss) (8), Oncorhynchus mosou (9), and taimen (Hucho taimen) (10), suggesting a relatively higher protein requirement for Amur grayling compared to lipid. The persistent escalation in prices for premium animal and plant protein sources (e.g., fishmeal and soybean meal) has increased aquaculture costs significantly (11). Leveraging the protein-sparing effect of lipids offers a viable solution to this challenge (12). Additionally, dietary lipids serve not only as a source of energy and essential fatty acids for Amur grayling but also play a role in vitamin and hormone synthesis and in the absorption of fat-soluble vitamins (13). Thus, it is critical to delineate the nutritional and physiological adaptive ranges, as well as the adaptive mechanisms, of Amur grayling to dietary lipid supplementation to support sustainable and healthy aquaculture practices.
RNA sequencing (RNA-Seq), or transcriptome sequencing, entails the sequencing of all RNA types within a cell or tissue at a given developmental stage or under specific conditions, encompassing both mRNA and non-coding RNA (14). This technology has been extensively applied across various fish species, including largemouth bass (Micropterus salmoides) (15) and Brachymystax lenok (16), to elucidate physiological regulatory mechanisms at the molecular level in response to varying dietary lipid levels. For fish, the liver is a major site where lipolysis and lipogenesis occur, which plays essential roles in the synthesis and utilization of lipids. Many key enzymes and transcriptional factors are involved in the processes of lipolysis and lipogenesis, including fatty acid desaturas 2 (FADS2), Δ6 desaturase (d6D), fatty acid elongases 2 and 5 (ELOVL2 and ELOVL5), carnitine palmitoyltransferase I (CPT I), lipoprotein lipase (LPL), fatty acid synthetase (FAS), glucose 6-phosphate dehydrogenase (G6pd), PPARs, etc. (17). Li e al. (18) found that the ability of triploid rainbow trout to synthesize polyunsaturated fatty acids (PUFAs) is weaker than that of diploids depending on comparative metabolomics analysis and quantitative PCR (qPCR) verification of the liver. Hence, studies on the response of the liver to different dietary lipid levels could help us to understand the adaptation mechanism of Amur grayling to dietary lipid nutrition.
Building on this foundation, our study focuses on the regulatory mechanisms of lipid nutrition in Amur grayling. The objective is to investigate the physiological adaptation mechanisms to dietary lipids in Amur grayling fry by examining liver and intestinal digestive physiology and hepatic antioxidant capacity and conducting comprehensive hepatic transcriptome sequencing analysis. Ultimately, this research aims to provide a theoretical framework for the development of scientifically formulated compound feeds tailored to the nutritional needs of Amur grayling.
Materials and methods
Ethics statement
All animal procedures employed in this study adhered to the Heilongjiang River Fisheries Research Institute’s guidelines for the care and use of laboratory animals. Ethical review and approval were obtained from the Committee for the Welfare and Ethics of Laboratory Animals at the Heilongjiang River Fisheries Research Institute (CAFS), ensuring compliance with established ethical standards.
Experimental diet preparation
According to Cui’s (3) introduction of artificial breeding technology of Amur grayling fry, the crude protein content of the feed was 46–49%. Based on this, the protein level of the experimental diets in our study was designed to be approximately 47.5%. Fishmeal, chicken powder, cottonseed protein concentrate, and soybean protein concentrate were used as the main protein sources. Referring to a description of the range of dietary lipid requirements for cold water fish (such as rainbow trout, sturgeon, taimen, etc.) by Wang and Liu (13), the experimental diets were designed to contain six lipid levels, and the analyzed lipid concentrations were 9.07, 12.17, 15.26, 18.09, 21.16, and 24.07% (measured using the Soxhlet method) and were named as GL1, GL2, GL3, GL4, GL5, and GL6, respectively. Equal amounts of fish oil and soybean oil were used as sources of lipids, and the graded concentrations of 2.4, 3.9, 5.4, 6.9, 8.4, and 9.9% dry matter were designed to realize the increasing dietary lipid level. Corn starch is the only source of carbohydrates, the contents of which are gradually decreased with increased lipid contents. The proximate composition of experimental diets is presented in Table 1. Feed processing operations were consistent with a previous study (21). The diets were air-dried to 10% moisture at 60°C, sealed, and kept at −20°C until used.
Fish rearing and experimental conditions
The Amur grayling fry was provided by the Bohai Cold-Water Fish Experimental Station of Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences. Amur grayling fry were reared in 120 L recirculating aquaculture system (RAS) aquaria (38 cm × 39 cm × 80 cm) for 2 weeks before the feeding trial to adapt to the environment. A total of 450 Amur grayling fry (initial body weight 4.64 ± 0.03 g) were randomly distributed into 18 aquaria, with 25 Amur grayling fry in each aquarium. Each aquarium was randomly assigned to one of the six experimental diets, resulting in three replicates for each diet. They were artificially fed 2–3% of body mass three times daily (8:00, 13:00, and 17:00 h) based on the description of Cui (3) and Feng et al. (22) and re-weighted to adjust their daily feeding amount at fortnightly intervals. During the 8-week breeding trial, approximately one-third of water was exchanged to maintain the favorable water quality indices every third day. The water quality parameters during the experiment were as follows: water temperature was 13 ± 0.5°C, total ammonia was approximately 0.20 mg/L, and dissolved oxygen ranged from 5.8 to 6.2 mg/L.
Sample collection
Amur grayling were weighed and counted after the 8-week breeding trial to calculate weight gain rate (WGR), specific growth rate (SGR), feed conversion rate (FCR), and survival rate. Depending on the operation procedure described by Feng et al. (22) and Zhang et al. (23), experimental fish were sacrificed on ice using 100 mg/L of MS-222 (tricaine methanesulfonate; Sigma, St. Louis, MO, United States) and dissected immediately to collect the tissue samples. MS-222 (Sigma Chemical E10521, St Louis, MO, United States) used in the experiment was solubilized in deionized water and buffered with sodium bicarbonate at a ratio of 1:1 (sodium bicarbonate:tricaine methanesulfonate powder), providing a solution with a final MS-222 concentration of 10 mg/mL (pH 7.0) from which aliquots were taken and used in anesthetization for fish (24). The mixed liver and intestine samples from three Amur grayling in each replicate were taken for enzyme activity analysis. Additionally, the mixed liver samples from the other three Amur grayling in each replicate were quickly removed and put into a 1.5-mL centrifuge tube, which was then immediately frozen with liquid nitrogen to prevent RNA degradation and stored at −80°C for gene expression analysis. Moreover, according to the growth performance, the obtained liver samples from three other Amur grayling in GL1, GL3, and G6 groups were used for transcriptome analysis.
Detection of enzyme activity
Each of the liver, stomach, and intestine in different groups was accurately weighted (0.2 g) and homogenized in 0.9 mL of 0.9% saline with a low-temperature homogenizer and high-speed refrigerated centrifuge according to our previous study (21).
An assay kit produced by Nanjing Jiancheng Bioengineering Institute was used to assess the digestive ability of the stomach and intestine, including lipase (Cat. No. A054-2-1) and amylase (Cat. No. C016-1-1). Another assay kit produced by Nanjing Jiancheng Bioengineering Institute was used to assess the antioxidant ability in liver and intestine by measuring the activities of catalase (Cat. No. A007-2-1) and peroxidase (POD, Cat. No. A084-3-1), as well as the contents of glutathione (GSH, Cat. No. A006-2-1), malondialdehyde (MDA), and total antioxidant capacity (T-AOC, Cat. No. A015-2-1). The double antibody sandwich method was used for the activities of protease in the stomach and intestine, as well as superoxide dismutase (SOD) in the liver and intestine, using an enzyme-linked immunosorbent assay (ELISA) kit produced by Shanghai Enzyme-Linked Biotechnology Co., LTD, as described in our previous study (21). The activities of glutamic oxalacetic transaminase (GOT) and glutamic-pyruvic transaminase (GPT) in the liver were detected using commercial kits from Nanjing Jiancheng Bioengineering Institute (Corresponding commodity serial numbers: Cat. No C0013-2-1, C010-2-1). The protein concentration of the tissue homogenate determined by coomassie blue staining was used to calculate relevant enzyme activity indicators.
Detection of serum biochemical parameters
The contents of triglyceride (TG), total cholesterol (TCHO), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C) in the serum were detected using commercial kits from Nanjing Jiancheng Bioengineering Institute (Corresponding commodity serial numbers: Cat. No C009-2-1, A110-1-1, A111-1-1, A112-1-1, A113-1-1).
Transcriptome profiling analysis of liver
Liver total RNA from Amur grayling was extracted with an RNAiso Plus Kit (Cat. No. 9109, TaKaRa, Co. Ltd., Dalian, China). The RNA integrity and quantity were assessed using an Agilent 2,100 Bioanalyzer (Agilent, Santa Clara, CA). Library construction and sequencing were carried out by Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China). Three samples were collected in each library with an equal amount of purified RNA. Each library was sequenced using 2 × 125 bp pair-end (PE) mode in the Illumina HiSeq 6,000 sequencing platform.
Raw paired-end reads were trimmed using SeqPrep software1 and quality controlled using Sickle software2 with default parameters. Clean reads were then used for de novo assembly with Trinity software.3 The web program BLAST was used to annotate unigenes with the Nt database to obtain complete gene function information. All the assembled sequences were assigned to the NCBI protein non-redundant (NR), the Swiss-Prot protein database, Clusters of Orthologous Groups of proteins (COG), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using BLASTX (E-value cutoff of 1e-5) for functional annotations (25).
Bowtie2 software in RSEM was used to compare the clean reads of each sample with the transcriptome spliced by Trinity to obtain the number of read counts on each gene. Then, the read count data were standardized to analyze the gene expression level. DEG-seq provided a statistical method for determining differentially expressed genes (DEGs), and DEG-seq was used for differential analysis. To reduce the number of false positives, the screening threshold was set to q-value < 0.01 and |log2 Fold Change| > 2.
Data processing and analysis
To assess the growth performance of the common carp, the weight gain rate (WGR), specific growth rate (SGR), protein efficiency ratio (PER), and feed conversion ratio (FCR) were computed in terms of the following formula:
The sum of feed input of 2–3% body weight for 56 days per aquarium was deemed as total feed giver, which was used for calculating the FCR and PER. Data analyses were carried out using SPSS 23.0 software (SPSS Inc., Chicago, IL, United States). The results were subjected to a one-way analysis of variance (ANOVA), followed by Duncan’s test to delineate significance among fish groups. Moreover, quadratic regression analyses were applied to determine the optimum lipid level based on WGR and FCR according to their changing trend (26). Experimental results were presented as mean values ± standard deviations. Statistically significant levels were assigned at p < 0.05 unless otherwise stated.
Results and analysis
Growth parameters and feed utilization
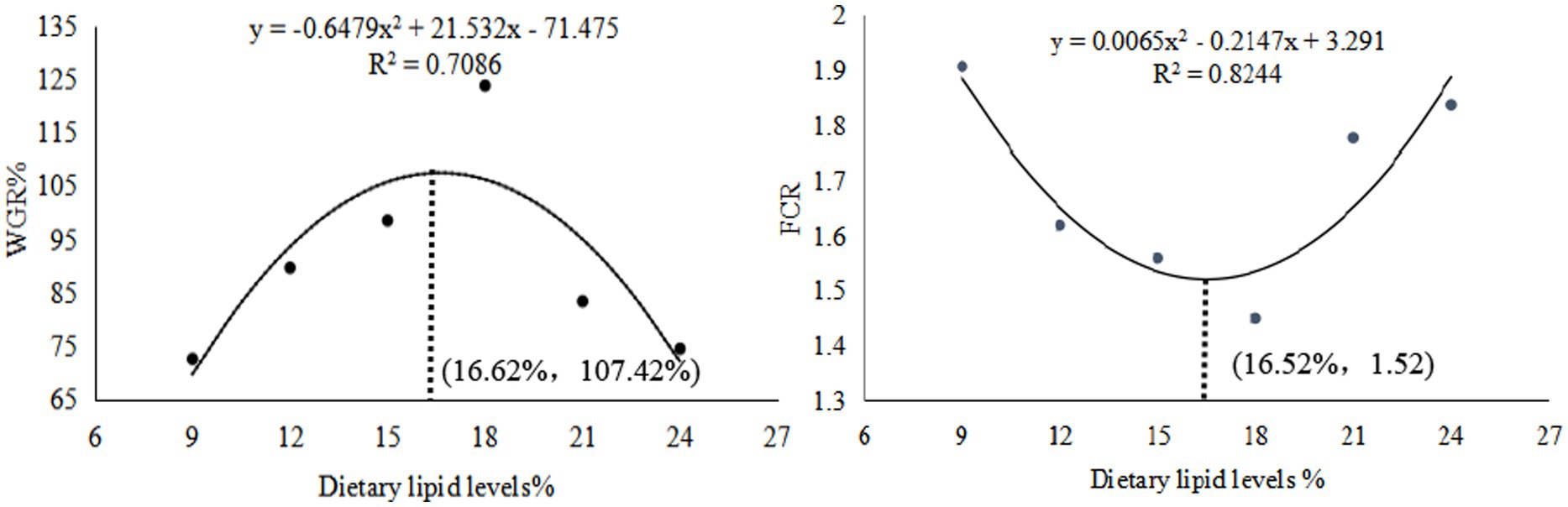
The weight gain ratio (WGR), specific growth rate (SGR), and protein efficiency ratio (PER) exhibited an initial increase followed by a decrease, peaking in the GL4 group. Specifically, the Amur grayling in the 18% lipid group demonstrated a significantly higher WGR and SGR than other groups (p < 0.05). Conversely, the feed conversion ratio (FCR) displayed an inverse trend, being significantly lower in the GL4 group compared to all others except the GL3 group (p < 0.05) (Table 2). Figure 1 presents the quadratic regression analysis. Using WGR as the metric, the quadratic regression equation was y1 = −0.6479x2 + 21.532x−71.475 (R2 = 0.7086), indicating an optimal dietary lipid level of 16.62%. Similarly, based on FCR, the quadratic regression equation was y2 = 0.0065x2–0.2147x + 3.291 (R2 = 0.8244), suggesting an optimal lipid level of 16.52%.
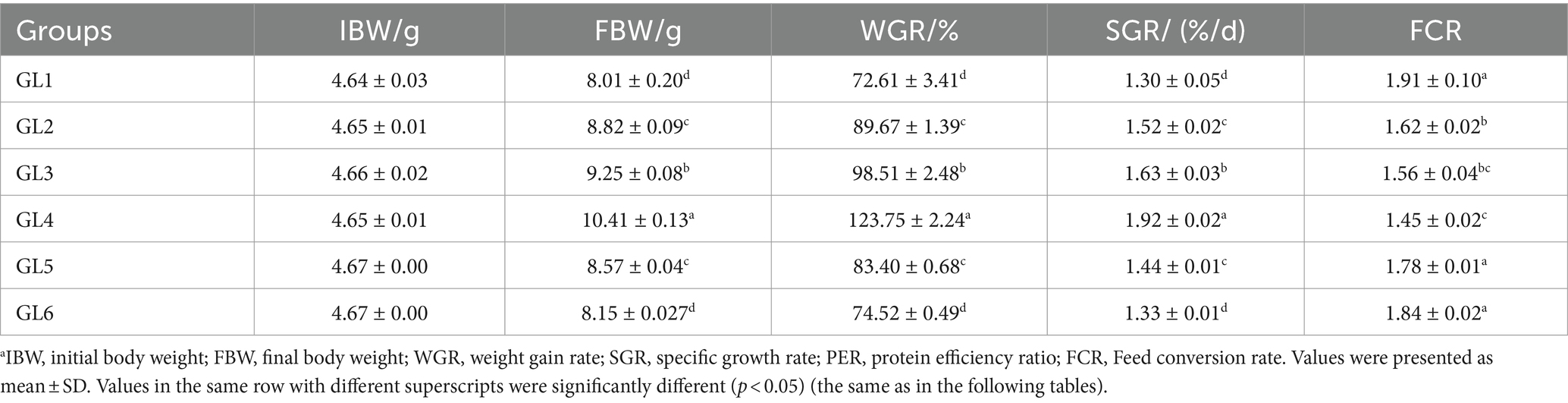
Table 2. Growth performance and feed utilization of Amur grayling fed the experimental diets for 56 days.a
Digestive enzyme activity in the stomach
Specifically, the activities of lipase in pancreatic secretion (LPS) and amylase in stomach mucosa (AMS) followed a pattern of initial increase followed by a decrease, peaking in the GL4 group. Notably, the AMS activity in the 18% group (GL4) exhibited significant increases compared to the GL1, GL5, and GL6 groups (p < 0.05) (Table 3).
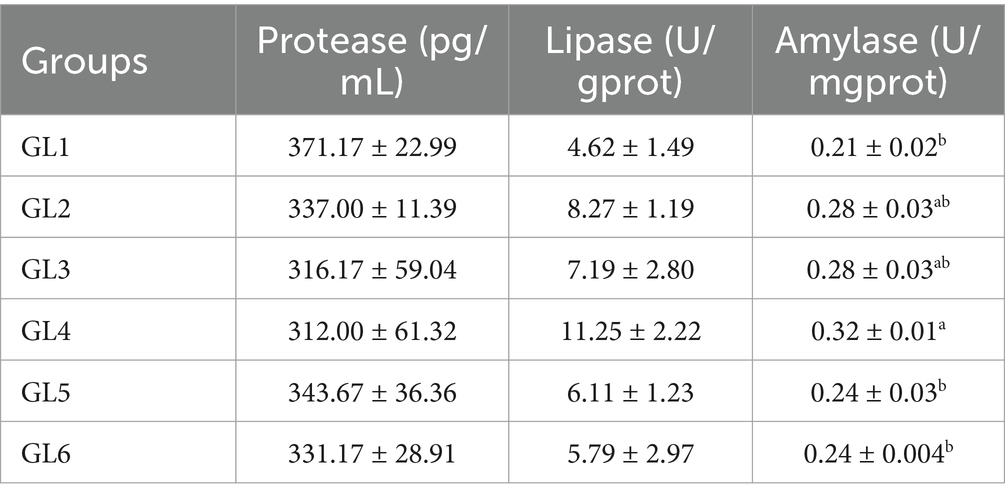
Table 3. Digestive enzyme activity in the stomach of Amur grayling fed the experimental diets for 56 days.
Digestive enzyme activity in the intestine
Data presented in Table 4 show that the activities of intestinal protease, lipase in pancreatic secretion (LPS), and amylase (AMS) in stomach mucosa demonstrated a trend of initial increase and subsequent decrease correlating with rising dietary lipid levels. All three enzymes reached their peak activities in the GL4 group. Specifically, the GL4 group exhibited significantly higher protease activity compared to the GL1, GL2, and GL6 groups and markedly higher AMS activities relative to all groups except GL3 (p < 0.05).
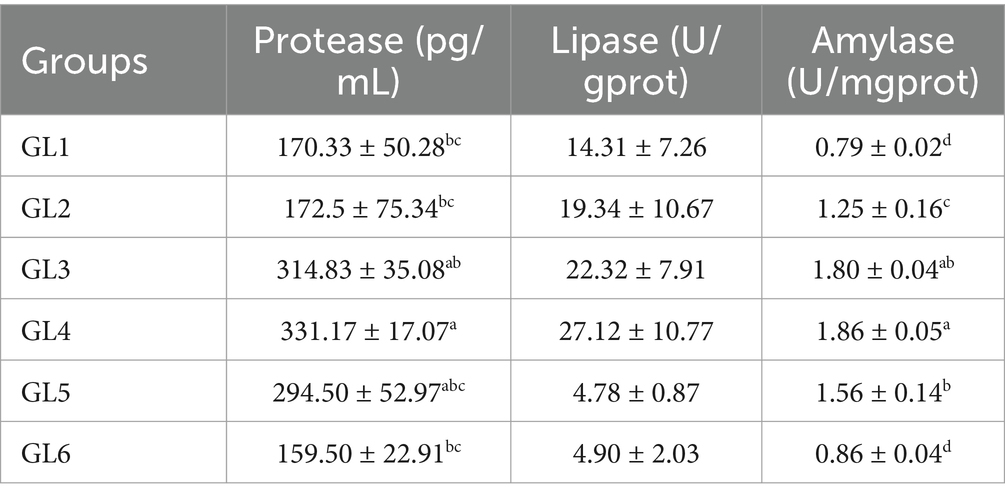
Table 4. Digestive enzyme activities in the intestine of Amur grayling fed the experimental diets for 56 days.
Got and GPT activities related to liver function
Table 5 illustrates that the activities of glutamic-oxaloacetic transaminase (GOT) and glutamic-pyruvic transaminase (GPT) in the liver initially increased and then decreased with an increase in dietary lipid level. Notably, the GL4 group exhibited significantly higher GOT activity than all other groups, except for the GL3 group, and demonstrated higher GPT activity than all other groups (p < 0.05).
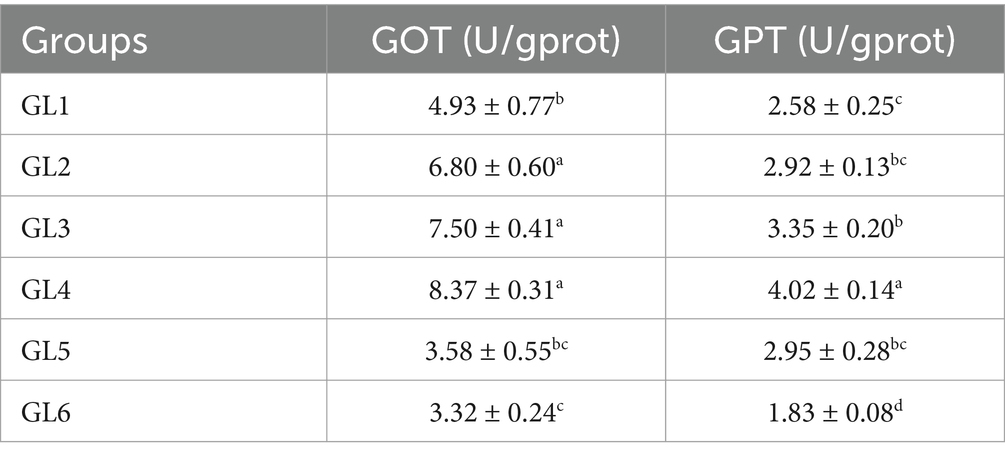
Table 5. GOT and GPT activities related to the liver function of Amur grayling fed the experimental diets for 56 days.
Antioxidant parameters in the liver
Table 6 demonstrates the variation in antioxidant parameters in the liver, indicating that the activities of superoxide dismutase (SOD), catalase (CAT), peroxidase (POD), glutathione (GSH), and total antioxidant capacity (T-AOC) initially increased and then decreased, reaching peak values in the GL4 group. Specifically, the GL4 group exhibited significantly higher SOD activity and GSH content than all groups except GL3, increased CAT activity relative to the GL1 and GL6 groups, and elevated T-AOC content in comparison to the GL1 and GL2 groups (p < 0.05). Additionally, this group showed notably lower malondialdehyde (MDA) content than the other groups (p < 0.05).
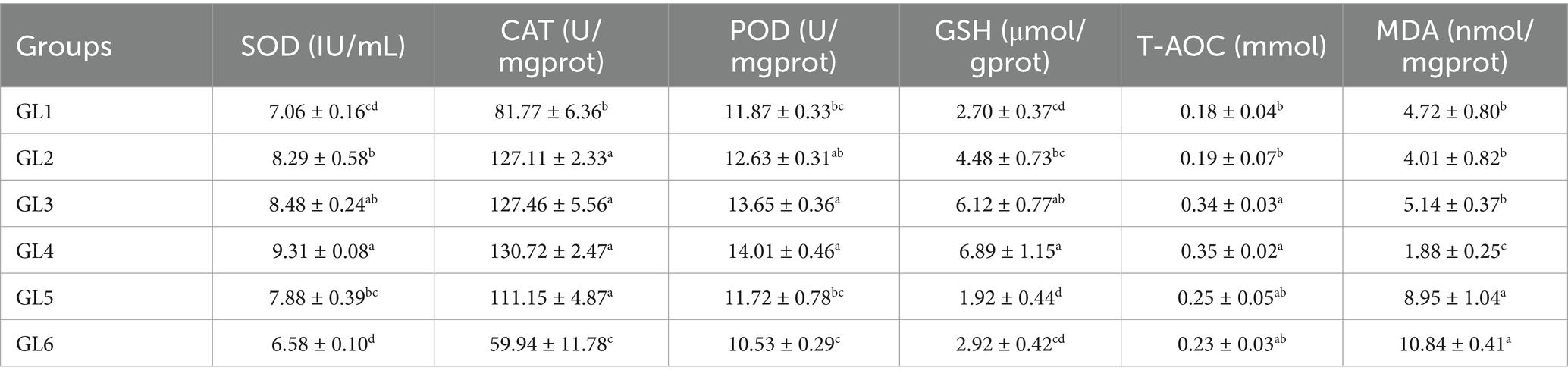
Table 6. Antioxidant parameters in the liver of Amur grayling fed the experimental diets for 56 days.
Lipid composition in serum
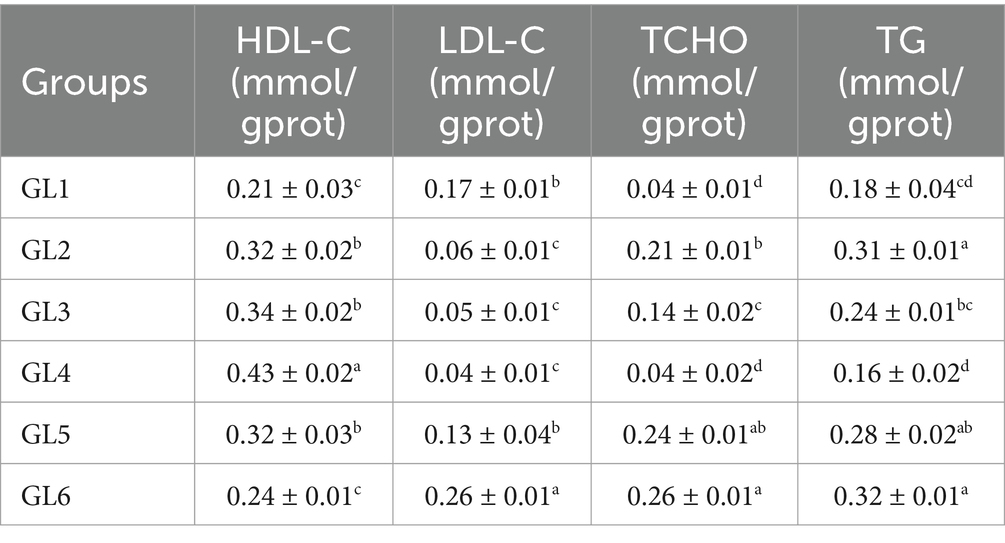
High-density lipoprotein cholesterol (HDL-C) levels initially decreased and then increased, reaching their highest values in the GL4 group. This group exhibited significantly higher HDL-C content compared to the other groups and elevated triglyceride (TG) content, except when compared to the GL2 and GL3 groups (p < 0.05). Conversely, low-density lipoprotein cholesterol (LDL-C), total cholesterol (TCHO), and TG displayed the reverse trend. Notably, the GL4 group had significantly lower LDL-C content than other groups, except GL2 and GL3, and lower T-CHO and TG contents compared to all groups except GL1 (p < 0.05) (Table 7).
Overall, the optimal dietary lipid level for Amur grayling is identified as 18%. Levels exceeding 18% or as low as 9% significantly impede the growth, digestibility, and antioxidant performance of Amur grayling. Based on these findings, the 9% (GL1), 18% (GL4), and 24% (GL6) groups were selected for further liver transcriptome response analysis.
Transcriptome profiling analysis of liver
Illumina sequencing and de novo assembly
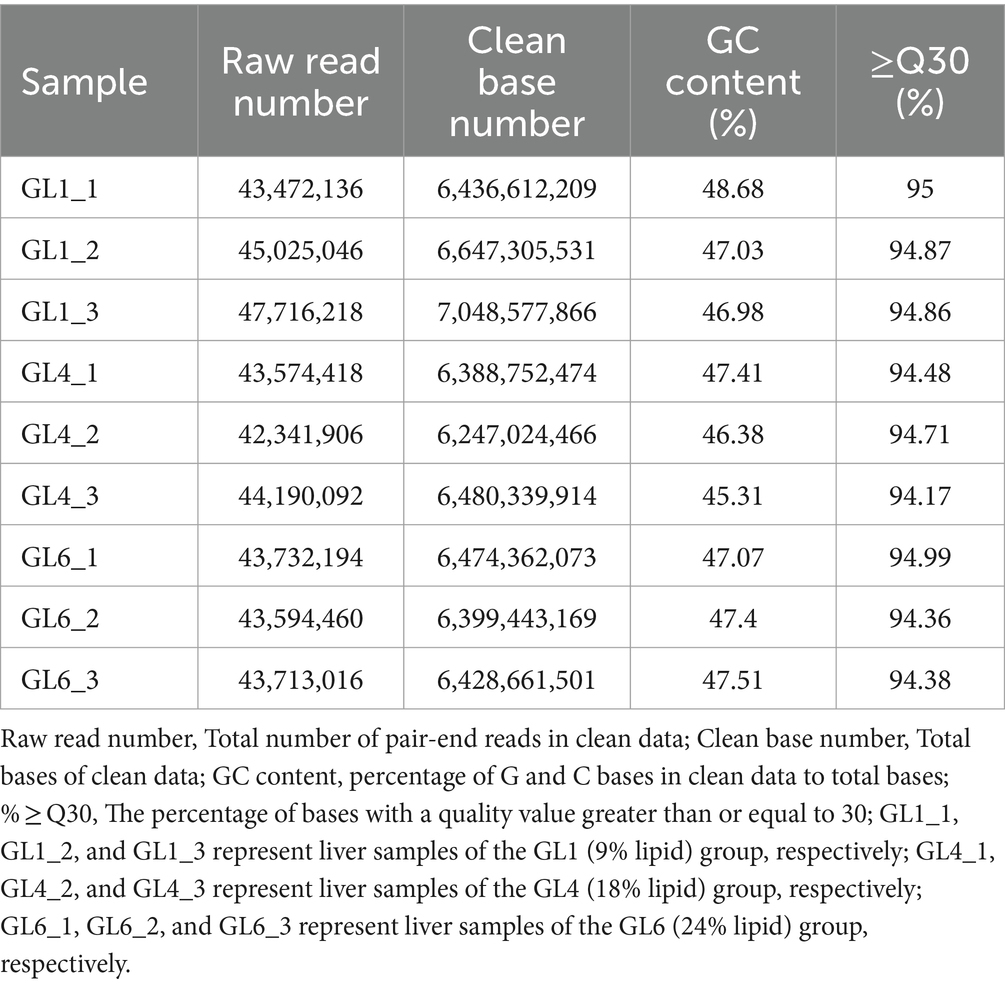
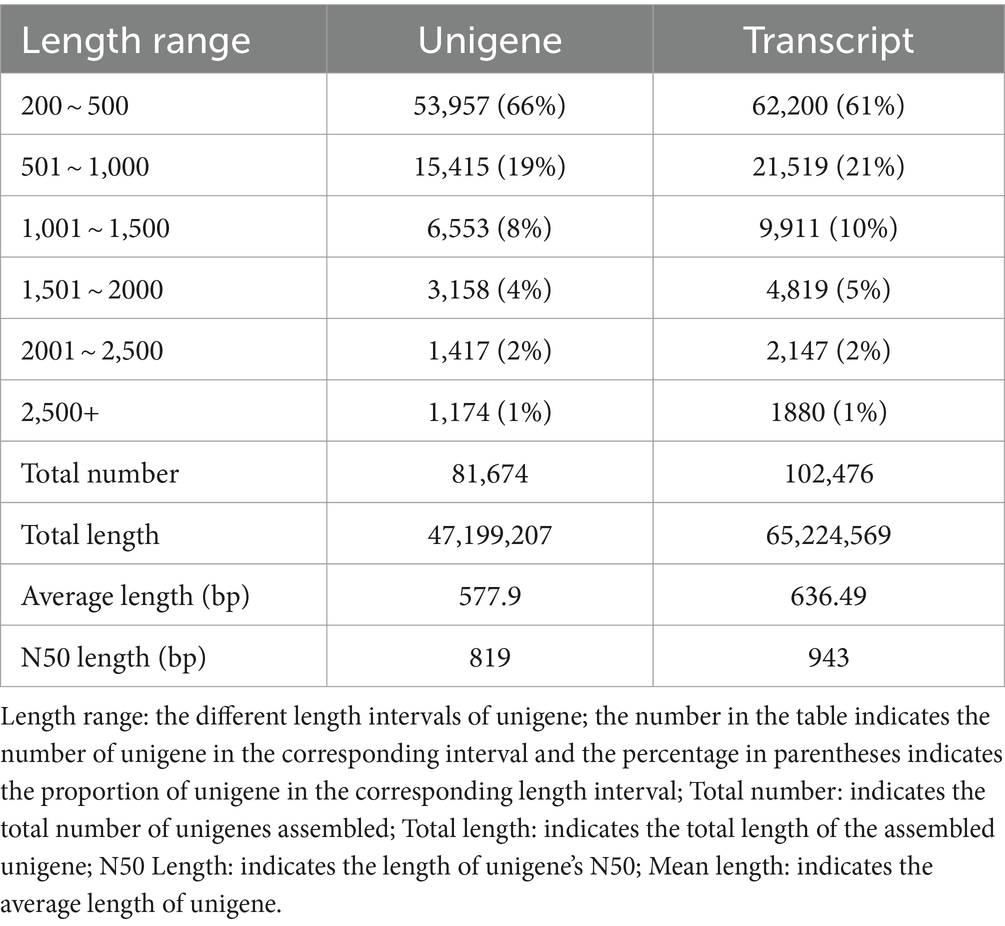
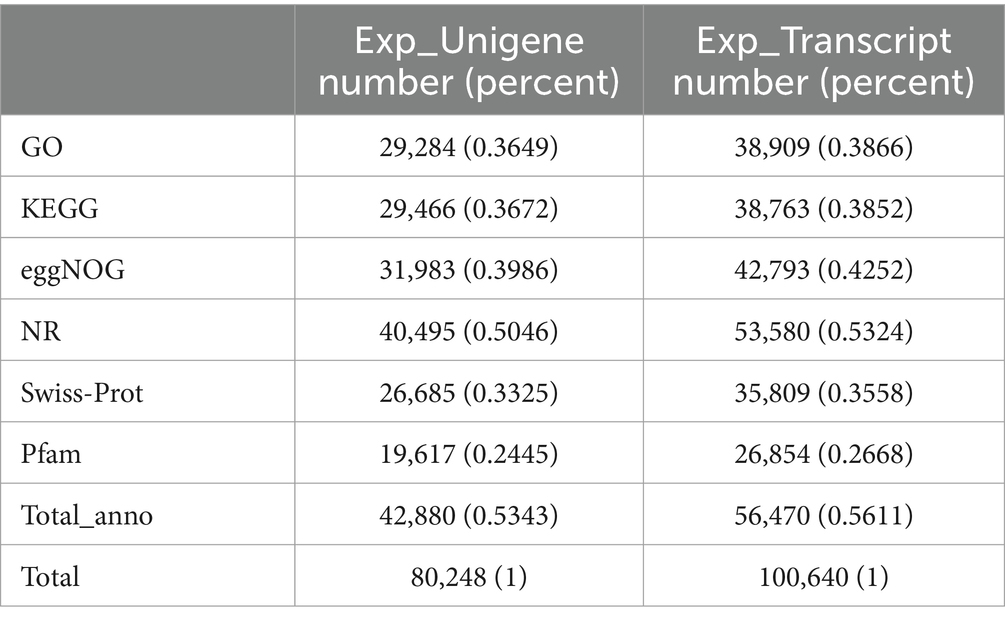
Transcriptome sequencing yielded a total of 58.55 Gb of clean data, with each sample contributing approximately 6.25 Gb. The percentage of Q30 bases, an indicator of sequencing quality, exceeded 94.17% (Table 8). From the assembled clean data, a total of 81,674 unigenes were identified, among which 12,302 unigenes were over 1 kb in length (Table 9).
Functional annotation and classification
Functional annotation of the 80,248 unigenes was conducted using various databases, including NR, Swiss-Prot, Pfam, eggNOG, GO, and KEGG (Table 10). Of these, 29,284 unigenes (representing 36.5% of the total) were annotated in the Gene Ontology (GO) database, encompassing three main categories: biological process, cellular component, and molecular function (Figure 2). Within the biological process category, the predominant sub-categories were cellular processes, metabolic processes, and biological regulation. The cellular component category was primarily characterized by sub-categories such as cell part, membrane, and organelle. In the molecular function category, binding and catalytic activities emerged as the principal sub-categories.
Identification of DEGs, KEGG enrichment, and function analysis
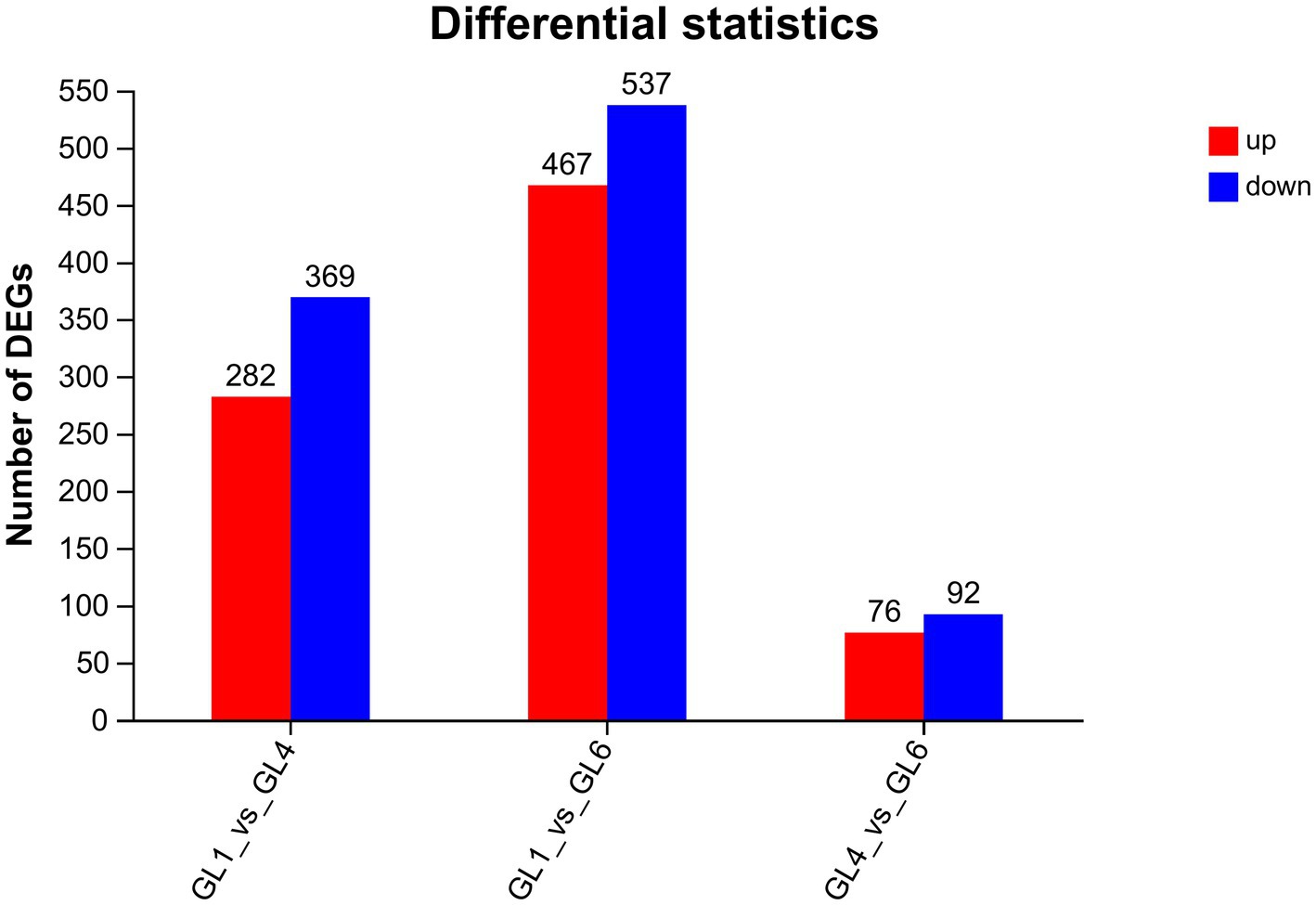
The GL1 group was designed as the matched group to detect the molecular mechanisms of liver function caused by increasing dietary lipid levels. Based on the threshold of |log2(Fold change)| > 1 and FDR < 0.05, a total of 678, 1,004, and 168 DEGs were identified in the GL1 vs. GL4, GL1 vs. GL6, and GL4 vs. GL6 comparisons, respectively (Figure 3). Moreover, Figure 3 revealed that an increment in downregulated genes was observed with increasing dietary lipid levels.
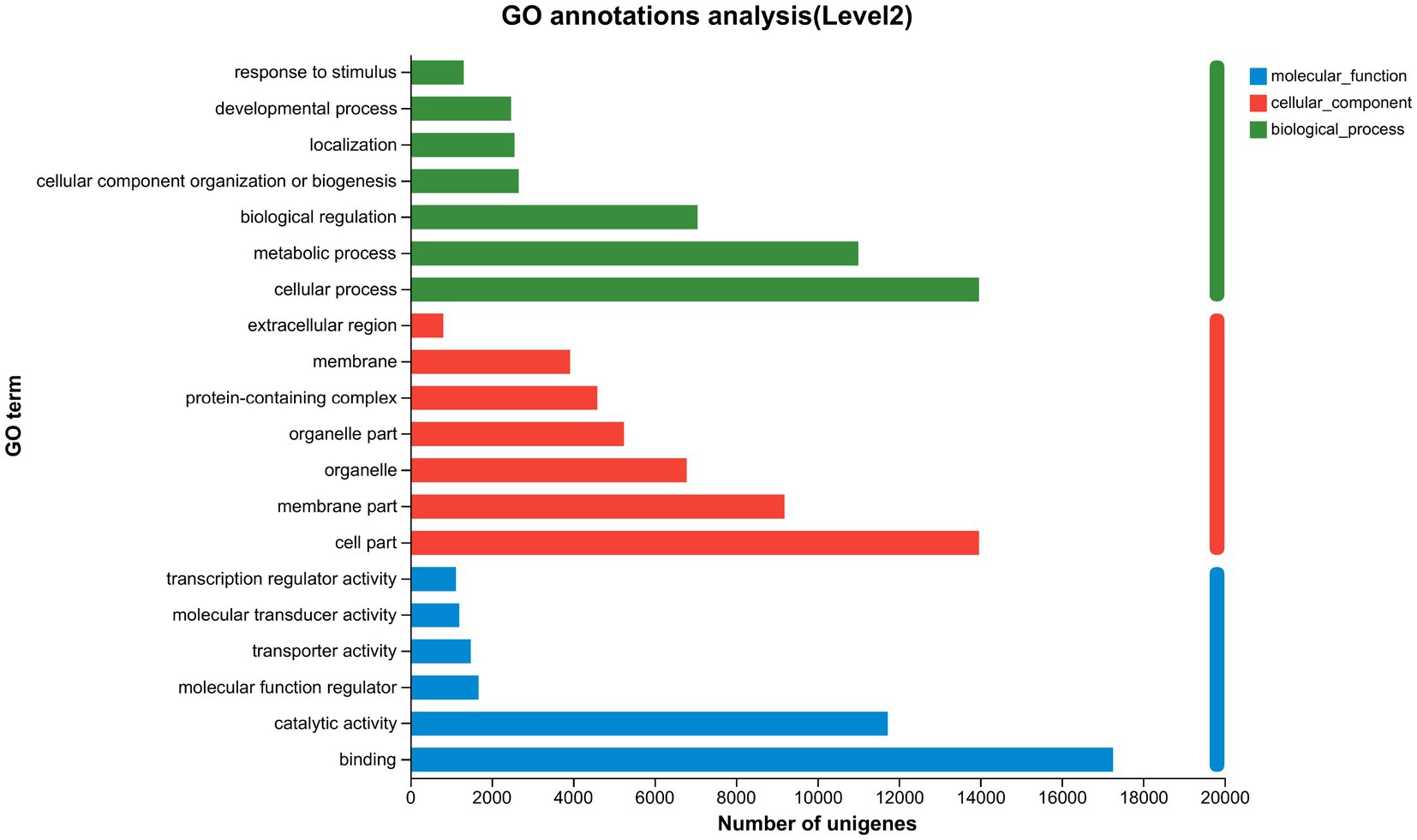
Figure 3. Numbers of DEGs in GL1 vs. GL4, GL1 vs. GL6, and GL4 vs. GL6 comparisons. Red and blue indicate upregulated and downregulated DEGs, respectively.
The top 20 pathways of differentially expressed genes (DEGs), ranked by Q-value based on KEGG enrichment analyses, are displayed in Figures 4A–C. Notably, in the GL1 vs. GL4 comparison, the most significant pathways were protein processing in the endoplasmic reticulum, ribosome, and protein. Additionally, several pathways linked to lipid metabolism were prominently enriched in GL1 vs. GL6 and GL4 vs. GL6 comparisons. These include cholesterol metabolism, fat digestion and absorption, and PPAR signaling pathways. Steroid biosynthesis and fatty acid degradation pathways were particularly noted in GL1 vs. GL6, while non-alcoholic fatty liver disease was significant in GL4 vs. GL6.
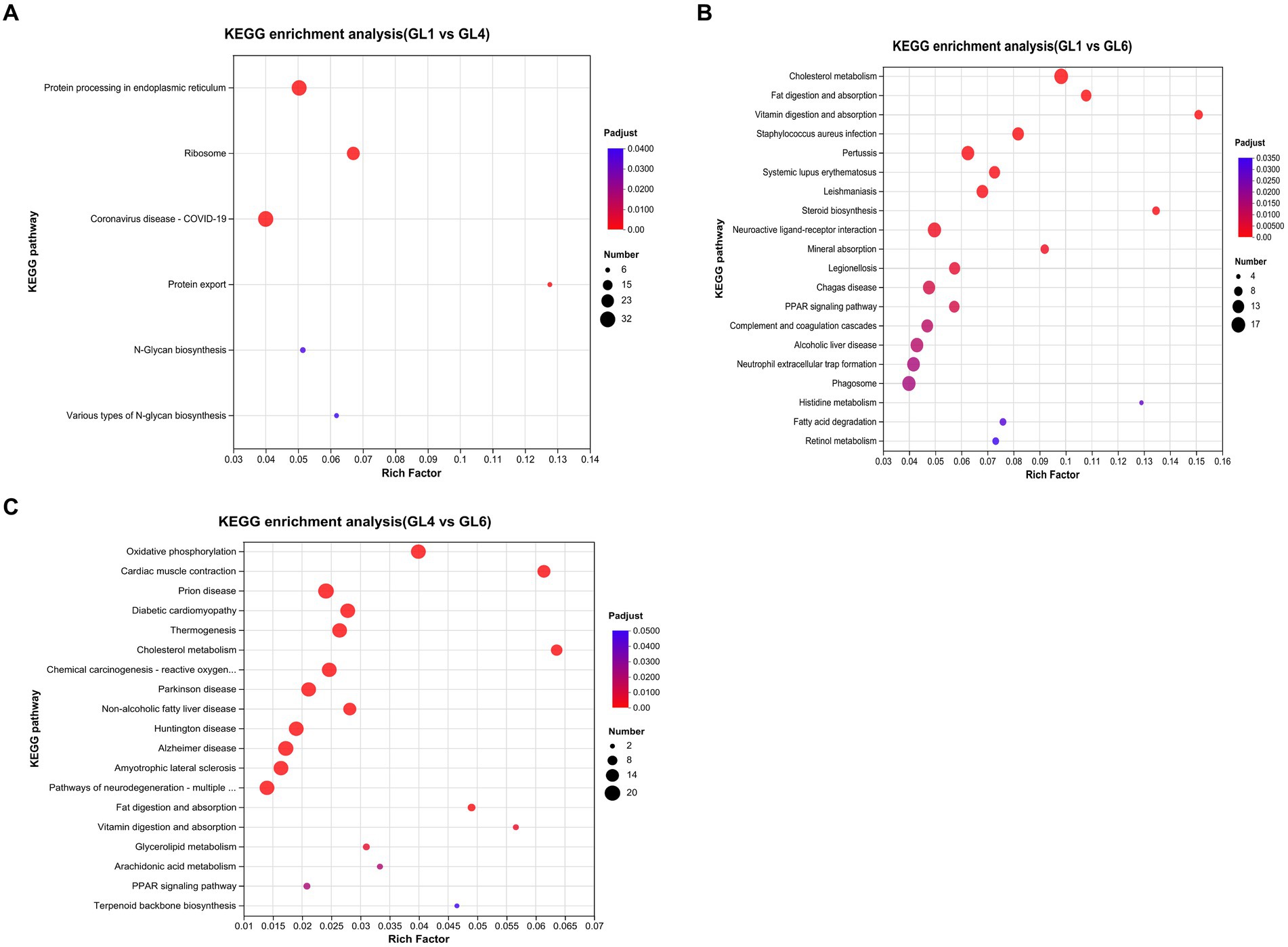
Figure 4. The top 20 enriched KEGG pathways in GL1 vs. GL4 (A), GL1 vs. GL6 (B), and GL4 vs. GL6 (C) comparisons. The x-axis shows the value of the gene ratio (number of DEGs enriched in the pathway/number of all genes in the annotation gene set) for each pathway. The y-axis indicates the specific KEGG pathway. The gene numbers and ranges of the Q-values are represented by the size and color of the dots, respectively.
Figure 5 illustrates these enriched pathways and their key DEGs for clearer understanding. RNA-seq analysis revealed that lipoprotein lipase (LPL) in the cholesterol metabolism pathway, lipase in the fat digestion and absorption pathway, and fatty acid-binding protein in the PPAR signaling pathway were downregulated with increased dietary lipid levels, with lipase expression being significantly higher in the GL1 group than in the GL6 group (p < 0.05). In contrast, carnitine palmitoyltransferase I (CPT1) in the fatty acid degradation pathway was upregulated with increased dietary lipids, though no significant differences were observed among the three groups (Figure 6). The potential functional roles of these DEGs in lipid metabolism in Amur grayling will be discussed in detail.
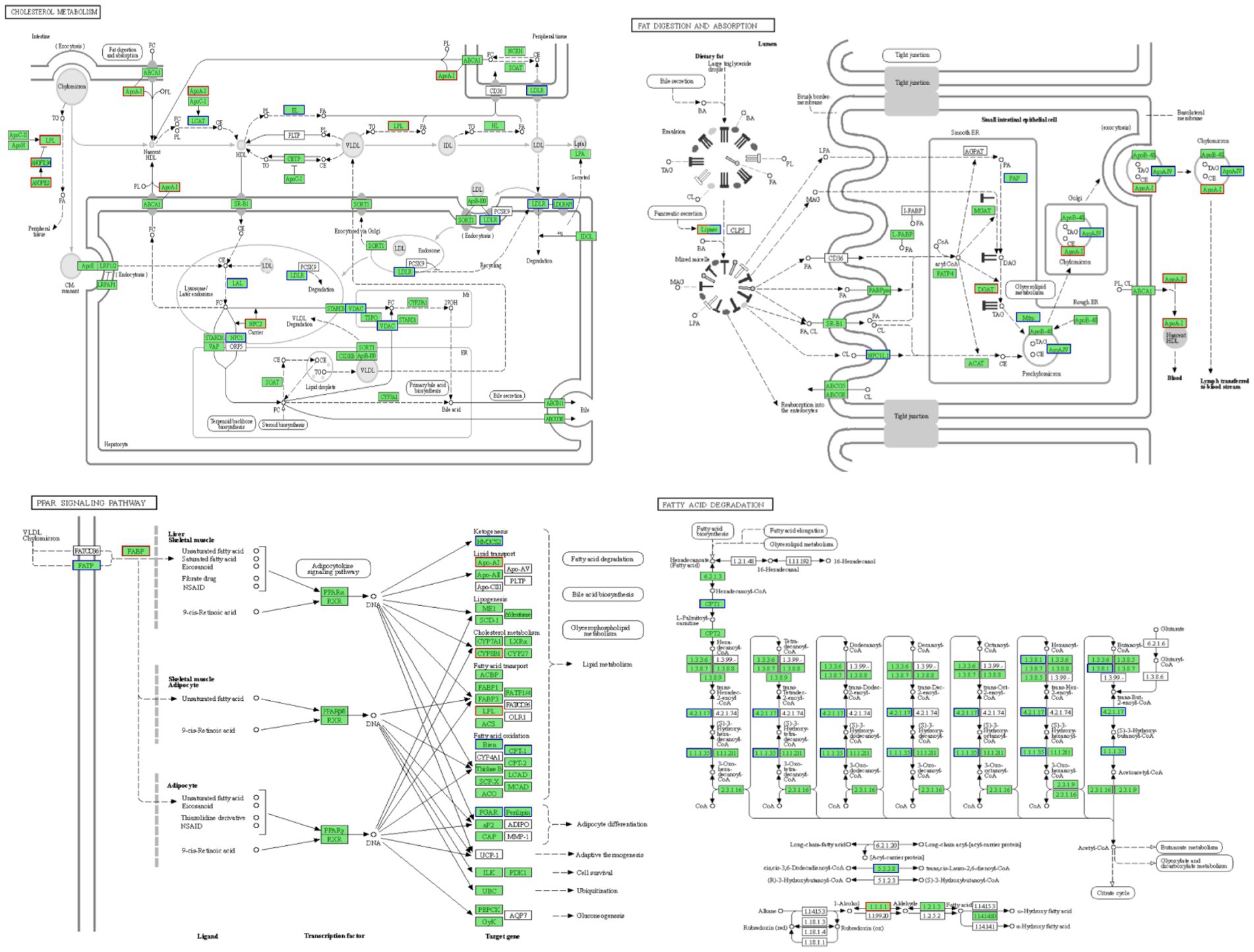
Figure 5. Four enriched KEGG pathways in the GL1 vs. GL6 and GL4 vs. GL6 comparisons and their involved genes.
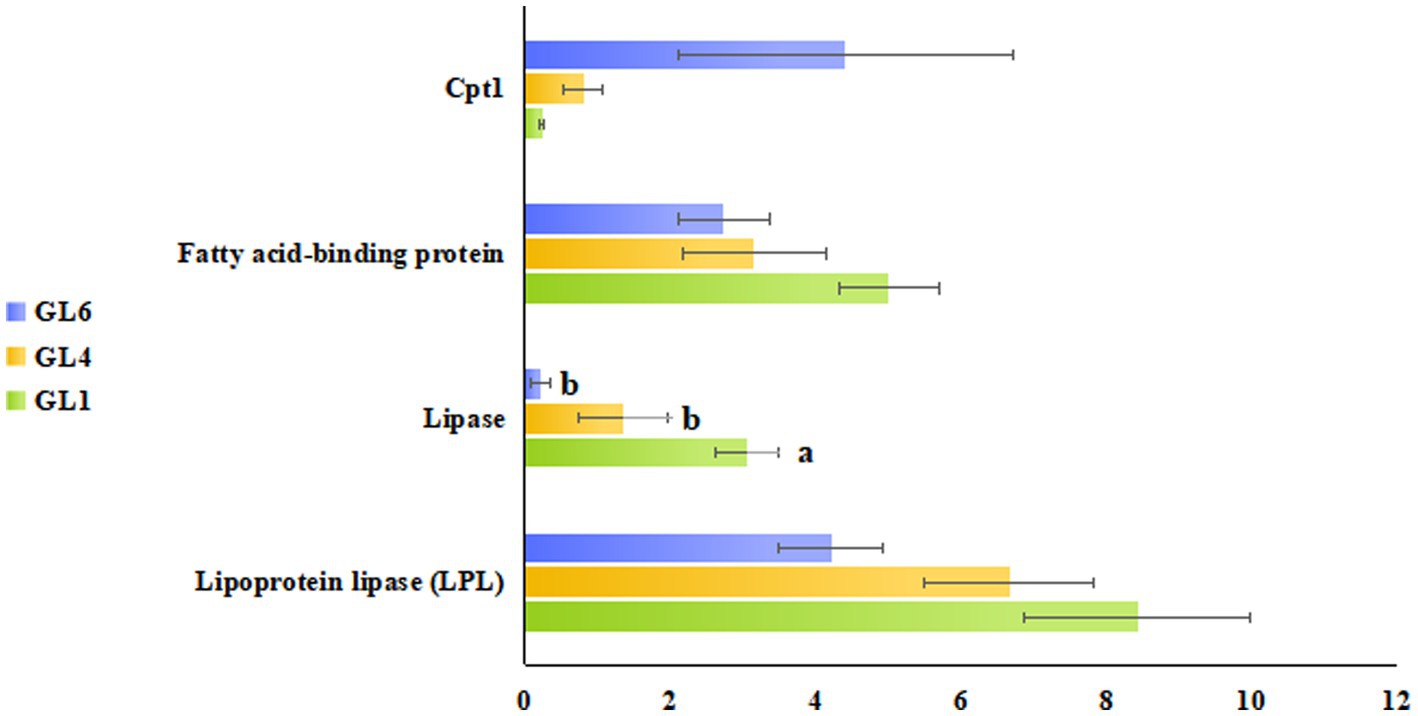
Figure 6. RNA-seq identification of lipoprotein lipase (LPL) in the cholesterol metabolism pathway, lipase in fat digestion and absorption pathway, fatty acid-binding protein in the PPAR signaling pathway, and Cpt1 in the fatty acid degradation pathway. Data were expressed as mean ± standard error (means ± SD) with three biological replicates.
Discussion
Compared to warm-water fish, cold-water fish exhibit a superior ability to utilize dietary lipids. For instance, salmon and trout can thrive with dietary lipid levels as high as 35%, while sturgeon optimal lipid levels range between 5 and 12% (13). Our study reveals that Amur grayling shows optimal performance with 18% dietary lipid, indicating efficient lipid utilization characteristic of cold-water fish. We observed that dietary lipid levels ranging from 12 to 21% positively influenced the WGR, SGR, and FCR of Amur grayling, with 18% proving to be the most effective. This pattern aligns with the study of common carp (Cyprinus carpio), suggesting that growth performance in fish presented the trend of initially increasing and then decreasing as dietary lipid levels rise when the designed lipid gradient is reasonable and differences between groups are adequately distinct (27). Within an appropriate physiological range, dietary lipids can significantly enhance growth, but beyond this range, they may become unsuitable and exert inhibitory effects. However, findings on lipid nutritional requirements in cold-water fish are not universally consistent. As Liu et al. discovered during their research, rainbow trout (Oncorhynchus mykiss) in cage culture with flowing water showed a positive correlation between growth parameters and increasing dietary lipid levels, identifying a minimum suitable lipid requirement of 26.06% (28). This suggests metabolic differences in lipid processing among various cold-water fish species, underscoring the need for further research to understand these variations.
Digestive capacity and nutrient absorption efficiency are key determinants of feed utilization and fish growth. Among digestive enzymes, protease, lipase, and amylase are critical for these processes (29). Enzyme activity is a crucial indicator of the digestive system’s physiological function, influencing nutrient digestion and absorption and ultimately affecting growth, development, and reproduction in fish (21). The Amur grayling possesses a comprehensive digestive system, including the esophagus, pyloric cecum, stomach, and intestine. Zhao and Li (29) observed that wild Amur grayling have higher trypsin and amylase activities in the intestine and pyloric cecum and elevated lipase activities in the pyloric cecum and stomach, which is consistent with their carnivorous diet. Our study revealed both similarities and notable differences in digestive enzyme activities between the stomach and intestine in response to increasing dietary lipid levels. The activities of lipase in pancreatic secretion and amylase in stomach mucosa peaked at 18% lipid treatment in both organs. Interestingly, we noted a divergence in the activity patterns of stomach proteases, which were restrained with higher lipid levels, and intestinal proteases, peaking at 18% lipid treatment. These findings suggest a gradual increase in the activities of digestive enzymes related to energy substances with higher dietary lipid content in Amur grayling fry. Importantly, the increased activity of these enzymes appears to enhance intestinal protease activity, reinforcing the protein-sparing effect of lipids. Similar observations have been reported in studies on paddlefish (Polyodon spathula) (30), Pengze crucian carp (Carassius auratus var. Pengze) (31), and common carp (27).
GPT and GOT existing in liver cells are important indicators to reflect whether the liver function is normal, as well as important enzymes to reflect the amino acid metabolism (32). When liver cells are damaged by foreign substances, a large amount of GPT and GOT penetrates the blood due to the increasing permeability of the cell membrane, resulting in increased activity of GPT and GOT in the blood (33). In the present research, Amur grayling fry in the 18% dietary lipid treatment gained higher GOT and GPT activities, indicating that the corresponding serum of this treatment has lower GOT and GPT activities. This result may indicate that under the conditions of appropriate lipid levels, the liver function of Amur grayling could not be damaged but could be improved, which was in line with the study on paddlefish (30) and Procypris merus (34).
High lipid levels typically reduce antioxidant capacity in fish, as observed in large yellow croaker (Larimichthys crocea) (35) and rainbow trout (36), characterized by disruptions in both enzymatic and non-enzymatic antioxidant systems responding to reactive oxygen species (ROS) generated during nutrient metabolism (37). Studies on largemouth bass (Micropterus salmoides) (38) and GIFT tilapia (Oreochromis niloticus) (39) found that the optimal lipid levels (11.05 and 9.35%) led to increases in SOD and GSH-Px activities and a decrease in MDA content in the liver. Analogously, our results showed that at 18% dietary lipid, the liver exhibited increased activities of SOD, CAT, POD, GSH, and T-AOC content and decreased MDA content compared to the 9 and 24% treatments. This indicates that an optimal lipid diet boosts the fish’s antioxidant capacity, facilitating a balance between ROS production and elimination, thereby mitigating oxidative stress. Hence, it can be hypothesized that the dietary lipid tolerance threshold for Amur grayling fry, based on liver antioxidant capacity, is approximately 18%.
Blood lipids play essential roles in maintaining body temperature, transporting foreign substances, preserving acid–base balance, participating in cell metabolism, nourishing cells, and supporting immunity. Serum TCHO and TG are indicators of fish nutritional status, with elevated levels reflecting active endogenous lipid transport as a response to dietary lipid intake (40). HDL acts as a cholesterol transporter in the blood, forming HDL-C upon cholesterol binding, while low-density lipoprotein (LDL) performs the opposite function, forming LDL-C (41). In our study, serum LPL-C, TCHO, and TG content significantly decreased with dietary lipid levels from 12 to 18%, in contrast to other treatments. Conversely, serum HDL-C peaked at the 18% dietary lipid level. The above results indicated that a more active endogenous lipid transport would be triggered in response to the appropriate dietary lipid level, which is similar to previous studies of Atlantic salmon (Salmo salar, L.) (42) and Acipenser gueldenstaedti (43). Additionally, Our findings suggest that in Amur grayling, excessive dietary lipids (beyond 18%) impair liver function, reducing the capacity to synthesize and secrete proteins necessary for cholesterol transport. This results in increased serum TC content and stable or declining HDL-C. A similar result was observed in the study on yellow drum (Nibea albiflora) (44). The specific underlying mechanisms warrant further investigation.
To elucidate the lipid metabolism regulatory mechanisms in Amur grayling, transcriptome sequencing was performed on liver samples from the 9, 18, and 24% dietary lipid groups. The liver, as the hub of lipid metabolism and the primary site for lipolysis and lipogenesis (45), is highly responsive to dietary lipids (46). Key enzymes and transcription factors play pivotal roles in lipid production and breakdown (47). Our study identified differential expression of key enzymes in pathways such as cholesterol metabolism, fat digestion and absorption, PPAR signaling, and fatty acid degradation, reflecting liver metabolic function alterations. We focused on the trends and functions of these enzymes to understand lipid metabolism regulation. Lipase, a member of the carboxyl ester hydrolase family, hydrolyzes triglycerides (TG) into glycerol and fatty acids. Lipoprotein lipase (LPL), within this family, regulates TG breakdown (48). Fatty acid-binding proteins bind free fatty acids and other hydrophobic ligands, facilitating their transport to organelles for processes such as fatty acid oxidation and TG and phospholipid synthesis (49). This study observed significant downregulation of lipase and LPL at 24% dietary lipid, suggesting inhibited lipoprotein hydrolysis and lipid absorption, and a reduction in fatty acid-binding protein levels, indicating decreased TG synthesis. This gene expression differentiation and metabolic pathway alteration might result from a negative feedback mechanism, preventing cytotoxicity from excessive lipid accumulation (50). Once ample lipid is stored in Amur grayling’s liver or other tissues in high-lipid diet groups, lipid intake and synthesis are curbed via metabolic pathways. These findings align with studies on large yellow croaker, where high lipid levels (18%) led to reduced LPL and related gene expressions (19). Intriguingly, the Amur grayling liver also appeared to engage a positive feedback mechanism to metabolize excess lipids, especially TG. Fatty acid β-oxidation, crucial for maintaining liver TG balance, involves CPT1 as a rate-limiting enzyme (51). The transcription level of CPT1 increased in the liver at 24% lipid, potentially counteracting oxidative stress and liver damage from lipid accumulation through β-oxidation, preventing lipid droplet accumulation and lipotoxicity (52). This indicates Amur grayling’s attempt to balance lipid intake and breakdown, although the equilibrium was not fully achieved. Future studies are warranted to further elucidate Amur grayling’s lipid metabolism regulation through genetic or nutritional interventions.
Conclusion
This study is the first to investigate the lipid requirements and lipid metabolism mechanisms in Amur grayling, an economically valuable species. Our findings suggest that a dietary lipid level of 18% fulfills the lipid requirements for Amur grayling, leading to enhanced growth performance, improved intestinal digestion, optimized serum lipid profile, and augmented liver antioxidant capacity. Transcriptome analysis revealed four differentially expressed genes (DEGs)-lipase, LPL, fatty acid-binding protein, and CPT1, suggesting that the Amur grayling liver employs both positive and negative regulatory mechanisms in response to excessive dietary lipid. Quadratic regression analysis determined the optimal dietary lipid levels to be 16.62 and 16.52%, based on WGR and FCR, respectively. These outcomes offer valuable theoretical insights for developing specialized feeds for Amur grayling and provide an in-depth understanding of its lipid metabolism mechanisms.
Limitations and prospects
First, the observed growth performance in our feeding trial highlighted a significant issue of slow growth in Amur grayling fry, attributable to factors such as breeding environment, feed composition, and feeding practices. We believe the most critical factor is the lack of comprehensive understanding of Amur grayling’s nutritional requirements. Enhancing growth rates through nutritional interventions is poised to become a focal point of future research.
Verifying gene expression using qPCR is crucial for elucidating Amur grayling’s adaptive mechanisms to dietary lipid nutrition. However, this study faced two main limitations. First, the scarcity of research on Amur grayling has resulted in limited availability of related gene sequences, an issue we aim to address in future studies. Second, given this constraint, we conducted liver transcriptome analysis using Eukaryotic reference-free transcriptome cloud analysis. This approach underscores our study’s objective to uncover the liver’s key adaptation mechanisms to dietary lipids and the genes involved in essential signaling pathways, utilizing cutting-edge transcriptome technology. This lays the groundwork for cloning-related genes in future investigations.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation. The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database, accession number PRJNA1089361. This data can be found in http://www.ncbi.nlm.nih.gov/bioproject/1089361.
Ethics statement
The animal studies were approved by the Committee for the Welfare and Ethics of Laboratory Animals of Heilongjiang River Fisheries Research Institute (CAFS). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
ZF: Writing – original draft, Project administration, Funding acquisition. KM: Methodology, Writing – review & editing. YW: Writing – review & editing, Formal analysis, Data curation. LW: Conceptualization, Supervision, Writing – review & editing, Resources. YZ: Writing – review & editing, Funding acquisition, Project administration. CL: Writing – review & editing, Investigation. JiaL: Writing – review & editing, Formal analysis, Data curation. DW: Resources, Writing – original draft. JinL: Resources, Writing – original draft. ZL: Investigation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was financially supported by the Central Public-interest Scientific Institution Basal Research Fund, HRFRI (HSY202205M) and the Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 2023TD60).
Acknowledgments
The authors thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Zhang, YZ, Fu, LW, Li, MY, Li, YY, and Liu, YG. A review: research progress on taxonomy and molecular phylogeny of genus Thymallus, grayling. Chin J Fish. (2020) 33:19–23. doi: 10.19720/j.cnki.issn.1005-3832.2020.03.0019.05
2. Liu, RY. The status of marine biodiversity and resources in China from the red list of species of China. Chin Nat. (2004) 6:15–7.
3. Cui, HX. Biological characteristics and artificial breeding technology of Thymallus arcticus grubei. North Chin Fish. (2021) 40:51–3.
4. Li, HL, Zhang, LL, Hu, S, Mou, ZB, Xu, GF, and Han, T. Breeding and disease control of fennel fry in Heilongjiang Province. North Chin Fish. (2009) 6:28–31.
5. Ma, B, Sun, JX, and Jiang, ZF. Taxonomic status of three fish species in Thymallus from upper Heilongjiang River based on mitochondrial control region sequence variation. J Fish Sci China. (2011) 18:314–21. doi: 10.3724/SP.J.1118.2011.00314
6. Yin, HF, Du, G, Han, Y, and Kong, LY. Nutrient requirements of protein and amino acid in cold water fishes. Feed Industry. (2005) 2005:24–8.
7. Gu, W, Wang, BQ, Zhao, JW, Yin, HB, and Wang, CA. Nutritional composition in muscles of cutivated Amur grayling (Thymallus grubii). J Northeast Agric Univ. (2009) 40:86–90. doi: 10.19720/j.cnki.issn.1005-9369.2009.11.019
8. Zhang, CJ, Liu, Z, and Wang, SY. Analysis of meat content and muscle nutrients of rainbow trout. J Hydrol. (2006) 26:83–5. doi: 10.15928/j.1674-3075.2006.04.039
9. Yin, HB, Sun, ZW, Shun, SX, and Wang, ZM. Analysis of muscle nutrition composition for Oncorhynchus masou. Acta Hydrobiol Sin. (2004) 28:577–80.
10. Jiang, ZF, Liu, Y, Li, YF, Bai, QL, and Jia, ZC. Analysis of biochemical components and evaluation of nutritional quality of wild and artificial Hucho taimen. J Northeast For Univ. (2005) 33:34–6.
11. Boyd, CE, D'Abramo, LR, Glencross, BD, Huyben, DC, Juarez, LM, Lockwood, GS, et al. Achieving sustainable aquaculture: historical and current perspectives and future needs and challenges. J World Aquacult Soc. (2021) 51:578–633. doi: 10.1111/jwas.12714
12. Franccis, DS, and Turchini, GM. Retro-engineering the protein sparing effect to preserve n-3 LC-PUFA from catabolism and optimise fish oil utilisation: a preliminary case study on juvenile Atlantic salmon. Aquaculture. (2017) 468:184–92. doi: 10.1016/j.aquaculture.2016.10.013
13. Wang, CA, and Liu, HB. Cold water fish feed and feeding application technology. Harbin: Heilongjiang Science & Technology Press (2020).
14. Wong, LL, Razali, SA, Mat Deris, Z, Danish-Daniel, M, Tan, MP, Mohd Nor, SA, et al. Application of second-generation sequencing (SGS) and third generation sequencing (TGS) in aquaculture breeding program. Aquaculture. (2002) 548:737633. doi: 10.1016/j.aquaculture.2021.737633
15. Peng, XH. The physiological adaptation mechanisms of Micropterus salmoides for high-lipid diet. Chongqing: Southwest University (2017).
16. Wu, ZX. Effects of exogenous lipase on growth and lipid metabolism of Brachymystax lenok under high fat stress and its mechanism. Tongliao: Inner Mongolia Minzu University (2022).
17. Cheng, HL, Xia, DQ, and Wu, TT. Fatty liver and regulation of lipids Metabolismin fish. Chin J Animal Nut. (2006) 18:294–8.
18. Li, CH, Chen, ZX, Qin, DL, Xu, GF, Gao, L, Hao, QR, et al. Comparative metabolomics analysis based on LC-MS method reveals regulatory difference in lipid metabolism and fatty acids metabolism between diploid and triploid rainbow trout (Oncorhynchus mykiss). Aquaculture Rep. (2024) 35:101933. doi: 10.1016/j.aqrep.2024.101933
19. Association of Official Analytical Chemists. Official methods of analysis. 15th ed. Arlington, VA: Association of Official Analytical Chemists Inc (2005).
20. AOAC- Association of Official Analytical Chemists. Official Methods of Analysis of the Association of Official Analytical Chemists International. 16th edition. Arlington, Virginia, USA: AOAC (2006).
21. Fan, Z, Wu, D, Li, CH, Li, JN, Liu, QW, and Wang, LS. Effects of bitter melon Momordica charantia saponins on growth, digestion and hepatopancreas health of juvenile Songpu mirror carp (Cyprinus carpio Songpu) fed high-carbohydrate diets. J. Fish. China. (2023) 36:29–37. doi: 10.16378/j.cnki.1005.3832(2023)03.0029.09
22. Feng, CJ, Liu, XF, Zhang, Y, Lv, WH, Han, SC, Zhang, YQ, et al. Histological structure and transcriptome characteristics in liver of juvenile Amur grayling (Thymallus arcticus grubei) under high temperature stress. J. Dalian Ocean Univ. (2023) 38:603–14. doi: 10.16535/j.cnki.dlhyxb.2022-237
23. Zhang, ZM, Liu, DC, Ding, HP, and Jiang, ZQ. Effects of MS-222 and clove oil on blood biochemical parameters and immune indices of koi carp Cyprinus carpio. Fish Sci. (2015) 2015:71–6.
24. Barbas, LAL, Pereira-Cardona, PM, Maltez, LC, Garcia, LO, Monserrat, JM, and Sampaio, LA. Anaesthesia and transport of juvenile tambaqui Colossoma macropomum (Cuvier, 1818) with tricaine methane-sulphonate: implications of secondary and oxidative stress responses. J Appl Ichthyol. (2017) 33:720–30. doi: 10.1111/jai.13382
25. Yan, J. Effects of dietary lipid levels and types of fatty acids on lipid deposition in large yellow croaker (Laarmichtins crocea). Qingdao: Ocean University of China (2015).
26. Liu, M, and Zhang, H. Research progress on the digestive enzyme of the fish. Fish Econ Res. (2008) 6:6–10.
27. Fan, Z, Li, JN, Zhang, YY, Wu, D, Zheng, XH, Wang, CA, et al. Excessive dietary lipid affecting growth performance, feed utilization, lipid deposition, and hepatopancreas lipometabolism of large-sized common carp (Cyprinus carpio). Front Nutr. (2021) 8:694426. doi: 10.3389/fnut.2021.694426
28. Liu, ZL, Zhao, W, Hu, WS, Zhu, B, Xie, JJ, Liu, YJ, et al. Lipid metabolism, growth performance, antioxidant ability and intestinal morphology of rainbow trout (Oncorhynchus mykiss) under cage culture with flowing water were affected by dietary lipid levels. Aquaculture Rep. (2021) 19:100593. doi: 10.1016/j.aqrep.2021.100593
29. Zhao, JW, and Li, X. Comparitive study on the distribution of digestive enzymes in wild and cultured Thymallus grubii. J Anhui Agric Univ. (2012) 39:26–30. doi: 10.13610/j.cnki.1672-352x.2012.01.030
30. Liu, YY, Yu, HB, Wu, WY, Zhong, MZ, Xing, JX, Zhou, JS, et al. Effects of dietary lipid levels on growth, body composition, digestive enzyme activities, serum biochemical indexes and antioxidant performance of Polyodon spathula. J Fish China. (2018) 42:1940–56. doi: 10.11964/jfc.20180111138
31. Fu, HY, Wan, GY, Fu, YL, Rao, Y, Zhang, GF, Chen, WJ, et al. Effect of dietary lipid levels on immune, antioxidant function and digestive enzyme activities of juvenile Pengze crucian carp (Carassius auratus var. Pengze). Feed Res. (2020) 43:14–7. doi: 10.13557/j.cnki.issn1002-2813.2020.01.005
32. Jia, R, Cao, L, Xu, P, Jeney, G, and Yin, GJ. In vitro and in vivo hepatoprotective and antioxidant effects of astragalus polysaccharides against carbon tetrachloride-induced hepatocyte damage in common carp (Cyprinus carpio). Fish Physiol Biochem. (2012) 38:871–81. doi: 10.1007/s10695-011-9575-z
33. Sun, JH, Fan, Z, Zhang, MJ, Cheng, BQ, Bai, DQ, Qiao, XT, et al. Effect of dietary protein level on hepatic function and antioxidant capacity of juvenile common carps (Cyprinus carpio). South China Fish Sci. (2017) 13:113–9. doi: 10.3969/j.issn.2095-0780.2017.03.015
34. Xie, YT, Mo, WY, Chen, B, Cao, JM, Li, CY, and Wang, GX. Effects of crude crude fat level on growth, body composition, serum enzyme activity and antioxidant capacity of Procypris merus. Guangdong J Anim Vet Sci. (2023) 48:1–14. doi: 10.19978/j.cnki.xmsy.2023.01.01
35. Ding, T, Xu, N, Liu, YT, Du, JL, Xiang, XJ, Xu, D, et al. Effect of dietary bile acid (BA) on the growth performance, body composition, antioxidant responses and expression of lipid metabolism-related genes of juvenile large yellow croaker (Larimichthys crocea) fed high-lipid diets. Aquaculture. (2019) 518:734768. doi: 10.1016/j.aquaculture.2019.734768
36. Zhao, W, Yao, R, Wei, HL, Guo, YC, Chen, AQ, Chen, BY, et al. Astaxanthin, bile acid and chlorogenic acid attenuated the negative effects of high-fat diet on the growth, lipid deposition, and liver health of Oncorhynchus mykiss. Aquaculture. (2023) 567:739255. doi: 10.1016/j.aquaculture.2023.739255
37. Zhou, JH, Liu, N, and Yang, Y. Keap1-Nrf2/are signaling pathway and its antioxidant regulation in teleost fish: a review. Chin J Fish. (2022) 35:117–24. doi: 10.16378/j.cnki.1005-3832.2022.04.0117.08
38. Zhu, TT, Jin, M, Sun, P, Li, CC, Ma, HN, Luo, JX, et al. Effects of dietary lipid level on morphology indexes, tissue fatty acid composition, serum biochemical indexes and liver antioxidant indexes of largemouth bass (Micropterus salmoides). Chin J Anim Nut. (2018) 30:126–37. doi: 10.3969/j.issn.1006-267x.2018.01.017
39. Liu, YQ. Effect of different dietary lipid levels on the growth performance, antioxidantion, immunity, fatty acid metabolism and related gene expression in juvenile GIFT tilapia. Nanning: Guangxi University (2021).
40. Chen, HJ, and Luo, DJ. Application of haematology parameters for health management in fish farms. Rev Aquac. (2023) 15:704–37. doi: 10.1111/raq.12753
41. Liao, RF, and Tang, CK. Basis and clinic of cholesterol reverse transport. Beijing: Science Press (2009).
42. Hamre, K, Christianse, R, Waagbø, R, Maage, A, Torstensen, BE, Lygren, B, et al. Antioxidant vitamins, minerals and lipid levels in diets for Atlantic salmon (Salmo salar, L.): effects on growth performance and fillet quality. Aquac Nutr. (2004) 10:113–23. doi: 10.1111/j.1365-2095.2003.00288.x
43. Zhu, TT, Li, Q, Zhu, HY, Wang, HW, Li, EC, and Chen, LQ. Effects of dietary lipid level on growth performance, blood biochemical index and antioxidant status of juvenile Acipenser guelden staedti. Marine Fish. (2017) 39:58–67. doi: 10.13233/j.cnki.mar.fish.2017.01.007
44. Wang, LG, Lu, Q, Luo, SY, Zhan, W, Chen, RY, Lou, B, et al. Effect of dietary lipid on growth performance, body composition, plasma biochemical parameters and liver fatty acids content of juvenile yellow drum Nibea albiflora. Aquacult Rep. (2016) 4:10–6. doi: 10.1016/j.aqrep.2016.05.002
45. Pierron, F, Baudrimont, M, Bossy, A, Bourdineaud, J, Brethes, D, Elie, P, et al. Impairment of lipid storage by cadmium in the European eel (Anguilla anguilla). Aquat Toxicol. (2007) 81:304–11. doi: 10.1016/j.aquatox.2006.12.014
46. Rashidian, G, Gorji, SB, Farsani, MN, Prokić, MD, and Faggio, C. The oak (Quercus brantii) acorn as a growth promotor for rainbow trout (Oncorhynchus mykiss): growth performance, body composition, liver enzymes activity and blood biochemical parameters. Nat Prod Res. (2018) 34:2413–23. doi: 10.1080/14786419.2018.1538994
47. Zheng, JL, Luo, Z, Zhu, QL, Tan, XY, Chen, QL, Sun, LD, et al. Molecular cloning and expression pattern of 11 genes involved in lipid metabolism in yellow catfish Pelteobagrus fulvidraco. Gene. (2013) 531:53–63. doi: 10.1016/j.gene.2013.08.028
48. Young, SG, Fong, LG, Beigneux, AP, Allan, CM, He, C, Jiang, H, et al. GPIHBP1 and lipoprotein lipase, partners in plasma triglyceride metabolism. Cell Metab. (2019) 30:51–65. doi: 10.1016/j.cmet.2019.05.023
49. Sui, XL, and Zhang, YR. Research progress of fatty acid binding proteins fish: a review. Fish Sci. (2019) 28:563–74. doi: 10.16378/j.cnki.1003-1111.2019.04.019
50. Bonen, A, Parolin, ML, Steinberg, GR, Cales Escandon, J, Tandon, NN, Glatz, JF, et al. Triacylglycerol accumulation in human obesity and type 2 diabetes is associated with increased rates of skeletal muscle fatty acid transport and increased sarcolemmal FAT/CD36. FASEB J. (2004) 18:1144–6. doi: 10.1096/fj.03-1065fje
51. Zhang, Y, Jin, Y, Guo, YY, and Yao, D. Research progress on biological function of carnitine palmitoyl transferase l and its influence on meat quality. Chin J Anim Nut. (2021) 33:4315–22. doi: 10.3969/j.isn.1006-267x.2021.08.012
52. Bai, JH, Li, CY, Tang, ZT, Wu, C, and Wei, ZH. Comparative study of carbohydrate levels on growth, oxidative stress and glucolipid metabolism of hybrid fish between Megalobrama amblycephala (♀) × Culter alburnus (♂) and Culter alburnus. Reproduct Breed. (2023) 3:131–42. doi: 10.1016/j.repbre.2023.07.002
Keywords: Amur grayling, dietary lipid, growth performance, liver lipid metabolism, feedback regulation mechanism
Citation: Fan Z, Ma K, Wang Y, Wang L, Zhang Y, Li C, Li J, Wu D, Li J and Li Z (2024) Liver transcriptome and physiological analyses preliminarily revealed the adaptation mechanisms of Amur grayling (Thymallus arcticus grubei, Dybowski, 1869) fry for dietary lipid nutrition. Front. Vet. Sci. 11:1369845. doi: 10.3389/fvets.2024.1369845
Edited by:
Ana Couto, University of Porto, PortugalReviewed by:
Seyed Pezhman Hosseini Shekarabi, Agricultural Research, Education and Extension Organization (AREEO), IranKenneth Prudence Abasubong, University of South Bohemia, Czechia
Copyright © 2024 Fan, Ma, Wang, Wang, Zhang, Li, Li, Wu, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongquan Zhang, zhangyongquan@hrfri.ac.cn; Liansheng Wang, wangliansheng@hrfri.ac.cn
 Ze Fan
Ze Fan Kai Ma1
Kai Ma1  Di Wu
Di Wu