RAP44 phage integrase-guided 50K genomic island integration in Riemerella anatipestifer
- 1College of Veterinary Medicine, Shanxi Agricultural University, Jinzhong, China
- 2Institute of Animal Science and Veterinary Medicine, Shandong Academy of Agricultural Sciences, Jinan, China
Bacteriophages are viruses that infect bacteria. Bacteria and bacteriophages have been fighting for survival. Over time, the evolution of both populations has been affected. Pathogenic Flavobacteriaceae species including Riemerella anatipestifer mainly infects ducklings, geese, and turkeys. However, it does not infect humans, rats, or other mammals, and is a suitable and safe research object in the laboratory. Our previous study showed that there is a 10K genomic island in R. anatipestiferIn this study, we found another integrated 50K genomic islands and focused on the relationship between R. anatipestifer genomic islands and the RAP44 phage genome. The phage RAP44 genome was integrated into R. anatipestifer chromosome, and an evolutionary relationship was evident between them in our comparative analysis. Furthermore, the integrated defective RAP44 phage sequence had the function of integration, excision, and cyclization automatically. Integrases are important integration elements. The integrative function of integrase was verified in R. anatipestifer. The integrase with the attP site can be integrated stably at the attB locus of the R. anatipestifer genome. A recombinant strain can stably inherit and express the exogenous gene. By studying the integration between host bacterium and phage, we have provided evidence for the evolution of the genomes in R. anatipestifer.
Introduction
Bacteriophages are viruses that infect bacterial cells. However, bacteria and bacteriophages are fighting for survival; over time, the evolution of both populations has been affected (1–3). It is possible to divide bacteriophages into temperate phages (prophages) and virulent phages (lytic bacteriophages) based on the replication process and survival status of the host bacteria.
Prophages are the main contributors to bacterial genetic diversity (4), and most vertically inherited prophage sequences are highly conserved. A large part of bacterial genome sequences is derived from prophages, and the domestication of prophages can promote the adaptability of bacteria. Approximately, 300 complete and defective prevention fragments were found in the intestinal bacterial genome. The sizes of these fragments showed a bimodal distribution; one peak represented a fully functioning prophage, ranging in size from 30 to 70 kb, and the other peak represented a defective prophage, ranging in size from 5 to 30 kb (4). Prophages are nucleic acids containing mild phages integrated into the host genome. Fully functioning prophages have excision, virion formation, lysis, or infective abilities. Prophages lacking these functions are referred to as defective prophages. Defective prophages can be beneficial to bacteria by protecting them from further bacteriophage infection and improving their antibiotic tolerance. Bacteria can also benefit from fully functional prophages. In Listeria monocytogenes, prophage excision promotes the expression of functional genes and enhances phagosomal escape and bacterial virulence (5). The prophage inserted into the bacterial chromosome has two outcomes: one is through a replicating lytic life cycle, and the other is that the prophage genome may become part of the bacterial chromosome through progressive mutations of critical genes or sites.
Prophages significantly influence the phenotype and pathogenicity of bacteria. For example, the integration of prophages results in the interruption or ectopic coding of host genes, leading to changes in the host phenotype. Similarly, the insertion of prophages introduces new encoded genes to enhance bacterial adaptability to the environment and pathogenicity to the host. The lysogenic conversion of prophages was also regulated by new bacterial phenotype. Therefore, prophages may be the main factors in horizontal gene transfer (HGT), evolution, and genetic diversity in bacteria (6). Penadés et al. recently reported a new family of pathogenic islands, known as phage-induced chromosomal islands. These islands are ubiquitous in bacteria and play an important role in the HGT, adaptability, and virulence of bacteria (7, 8). Staphylococcus aureus pathogenicity islands (SaPIs), also known as bacteriophage satellites, represent this new pathogenic island family. SaPIs are closely related to prophages because they exist throughout the life cycle of phages. SaPIs encode integrases (int) (9) and exonucleases (xis) (10) and can be removed from bacterial genomes to form large plasmid-like closed-loop DNA (11). Closed-loop DNA can then be packaged into infectious bacteriophage particles. These infectious particles are released by bacterial lysis and can adsorb and import SaPI DNA into the bacteria. The HGT can be accomplished by integrase-guided insertion into host chromosomes or replication of plasmids.
Almost half of the sequenced bacterial genomes contain complete prophages (12). There is a prophage named PP3 in the clinical isolates of Pseudomonas aeruginosa PAO1. PP3 can spontaneously be removed from the PAO1 chromosome at a frequency of approximately 25%. However, no phage plaque was formed in P. aeruginosa by induction, and no phage particles were detected in the supernatant; therefore, it was defined as a defective prophage. PP3 can be transferred to other P. aeruginosa strains. The first step after the transfer is integrating DNA into the bacterial chromosome. PP3 can be integrated into the same position as PAO1 and maintain circulatory capacity for integration and excision (13).
The pathogenic Flavobacteriaceae species including Riemerella anatipestifer mainly infects ducklings, geese, and turkeys. It does not infect humans, rats, or other mammals and is a suitable and safe research object in the laboratory (14). A 10K genomic island of R. anatipestifer was previously reported (15). RAP44, a strong bacteriophage of R. anatipestifer belonging to the Siphoviridae family of tailed phages, was isolated from the feces of healthy Chinese Muscovy ducks. The complete genome consists of 49,329 nucleotides of linear double-stranded DNA molecules with 80 open-reading frames. This study revealed the relationship between the newly discovered R. anatipestifer 50K genomic island and the R. anatipestifer phage RAP44. In our research, we focused on the relationship between bacterial genomic islands and phage genomes, and through comparative analysis, we found an evident evolutionary relationship between them. This is significant because phage integration promotes bacterial evolution and genomic diversity. We also investigated integrase that plays an essential role in integrating the phage genome into bacterial chromosomes. By studying the integration between host bacteria and phages, we provided new insights into the evolution of the genomes of R. anatipestifer.
Materials and methods
Bacterial strains, plasmids, and growth conditions
Riemerella anatipestifer ATCC11845 was purchased from the Microbial Preservation Center (Guangzhou, Guangdong), whereas R. anatipestifer RA-YM was obtained from a laboratory collection (16). The bacterial strains and plasmids used in this study are listed in Table 1. R. anatipestifer and Escherichia coli X7213 culture conditions have been previously described (17). When needed, spectinomycin and chloramphenicol antibiotics were used at a final concentration of 100 μg/ml.
Genomic island prediction in R. anatipestifer ATCC 11845
Our previous studies identified a 10K genomic island integrase function using IslandViewer 4. This study used IslandViewer 4 and the Phage Search Tool to predict genomic islands in the R. anatipestifer ATCC 11845 genome (PHAST, http://phast.wishartlab.com/) (18). Six genomic islands were predicted in R. anatipestifer ATCC 11845. We obtained the whole-genome data of R. anatipestifer from NCBI and used the Artemis Comparison Tool (ACT) to compare differences between prophage sequences and R. anatipestifer genomes (19).
The 50K genomic island in R. anatipestifer
We conducted a comparative genomic analysis of the R. anatipestifer 50K genomic islands and the RAP44 genome. The ACT was used to compare the differences in the 50K genomic island of R. anatipestifer. Nucleotide sequence alignment was used to construct a phylogenetic tree using the neighbor-joining (NJ) method and was further analyzed using MEGA v6.06 software (17).
The primers used in this study are listed in Table 2. The existence of the 50K genomic island in R. anatipestifer ATCC11845, R. anatipestifer RA-YM, and 20 clinical isolates was determined by polymerase chain reaction (PCR) amplification of the integrase marker gene using the P1/P2 primer pair. Amplification was performed using 2× MonAmpTM Taq Mix (Code No. RN03001M). We performed pre-denaturation at 95°C for 5 min, denaturation at 95°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 90 s (extension time selected according to fragment size, 1,000 bp/min). The detailed method has been described in our previous article (15). The PCR products were separated and detected by 0.8% agarose gel electrophoresis. The integrase homologous amino acid sequences were identified by searching the GenBank database using BLASTX protein homology/analogy recognition (20).
50K genomic island integration and excision
Genomic island integration and excision were also determined using PCR amplification. P3/P4 and P5/P6 primer pairs were used for detecting the excision of the 50K genomic island. P3/P5 and P4/P6 primer pairs were used for detecting the integration of the 50K genomic island. The PCR products were separated and detected by 0.8% agarose gel electrophoresis. An integration and excision model of the 50K genomic island was predicted.
Suicide recombinant vector integration into R. anatipestifer
Integration of the suicide recombinant vector into R. anatipestifer was performed according to our previously described method (17). R. anatipestifer no. 1 was selected as the receptor strain because it did not have the 50K genomic island but had a 19-bp attachment site attB (Supplementary Table S1). Integrase and the attP sites were amplified using the P7/P8 primer pair. A pRE112-Spec-int integration vector was constructed, and the detailed method is described in a published article (15). The pRE112-Spec-int plasmid was transformed into donor strain E. coli X7213. The R. anatipestifer integration strain was confirmed by PCR, and the integration and excision functions of the recombinant strains were verified. The genetic stability of the recombinant strain was detected. The recombinant bacteria regrew in the TSA pallet with Spec antibiotic after 20 passages without antibiotics.
Results
Comparative analysis of the 50K genomic island and RAP44 phage
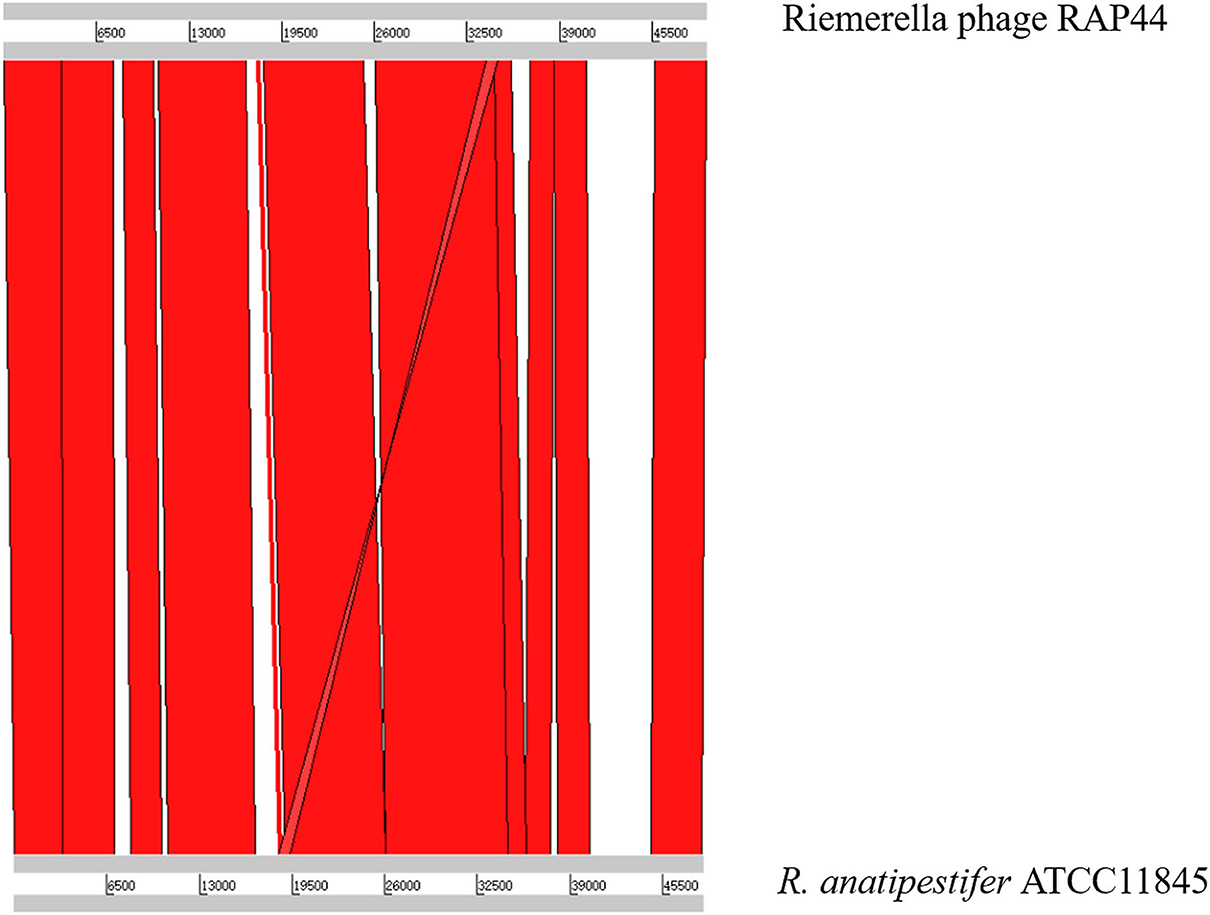
We selected the R. anatipestifer ATCC 11845 standard strain as our research target. The whole genome of R. anatipestifer ATCC 11845 has been published. IslandViewer 4 was used to predict bacterial GIs. There were six predicted genomic islands in R. anatipestifer ATCC 11845 (Supplementary Table S2). Among them, the 10K genomic island was reported in our previous articles. PHAST was used to predict prophages in R. anatipestifer ATCC 11845. There was a 50K intact prophage predicted by PHAST. The 50K prophage was also predicted to be a genomic island; thus, it was called the 50K genomic island and was highly homologous to the R. anatipestifer RAP44 phage. The 50K genomic island-coded protein predicted by PHAST is shown in Figure 1. Most proteins were RAP44 phage-related proteins. The results showed that the phage genomes were integrated into the bacterial genome. We also compared the differences between the 50K genomic island and the RAP44 phage. Large deletions and insertions were also identified when the phage genome and genomic island were analyzed using ACT (Figure 2).
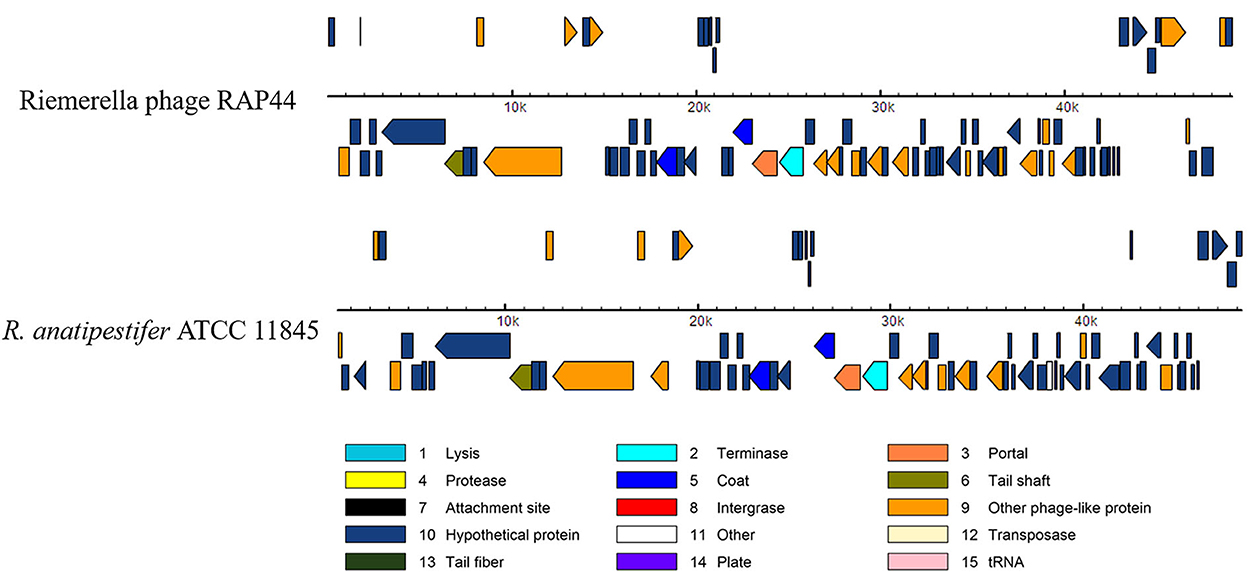
Figure 1. The 50K genomic island proteins related to RAP44 phage. PHAST was used to analyze 50K genomic island, and different phage proteins are shown.
Analysis of the 50K genomic island structure in R. anatipestifer
Whole-genome comparisons revealed that the RAP44 phage genome was integrated into the chromosome and formed a 50K genomic island in R. anatipestifer. Supplementary Table S3 shows the results of the homology comparison of the RAP44 and 50K genomic island genes. The replication proteins and other proteins of RAP44 were deleted, and several exogenous genes were inserted into R. anatipestifer. We performed a comparative genomic analysis between R. anatipestifer genomes and RAP44 phage genomes. Nucleotide sequence alignment was used to construct a phylogenetic tree using the NJ algorithm (Figure 3A). ACT was used to compare the differences between the 50K genomic island and RAP44 (Figure 3B). The prophages and genomic islands sequences had some common characteristics: (1) direct repeat sequences attL and attR were located on the two flanks of the genomic island and (2) integrase was located on the medial side of direct repeat sequences. The 50K genomic island was located downstream of transfer-ribonucleic acid (tRNA), and 19-bp repeat sequences (TCCCTCTCTCTCCGCAAAA) were present in the two flanks. A 50K genomic island simulation diagram was constructed (Figure 3C).
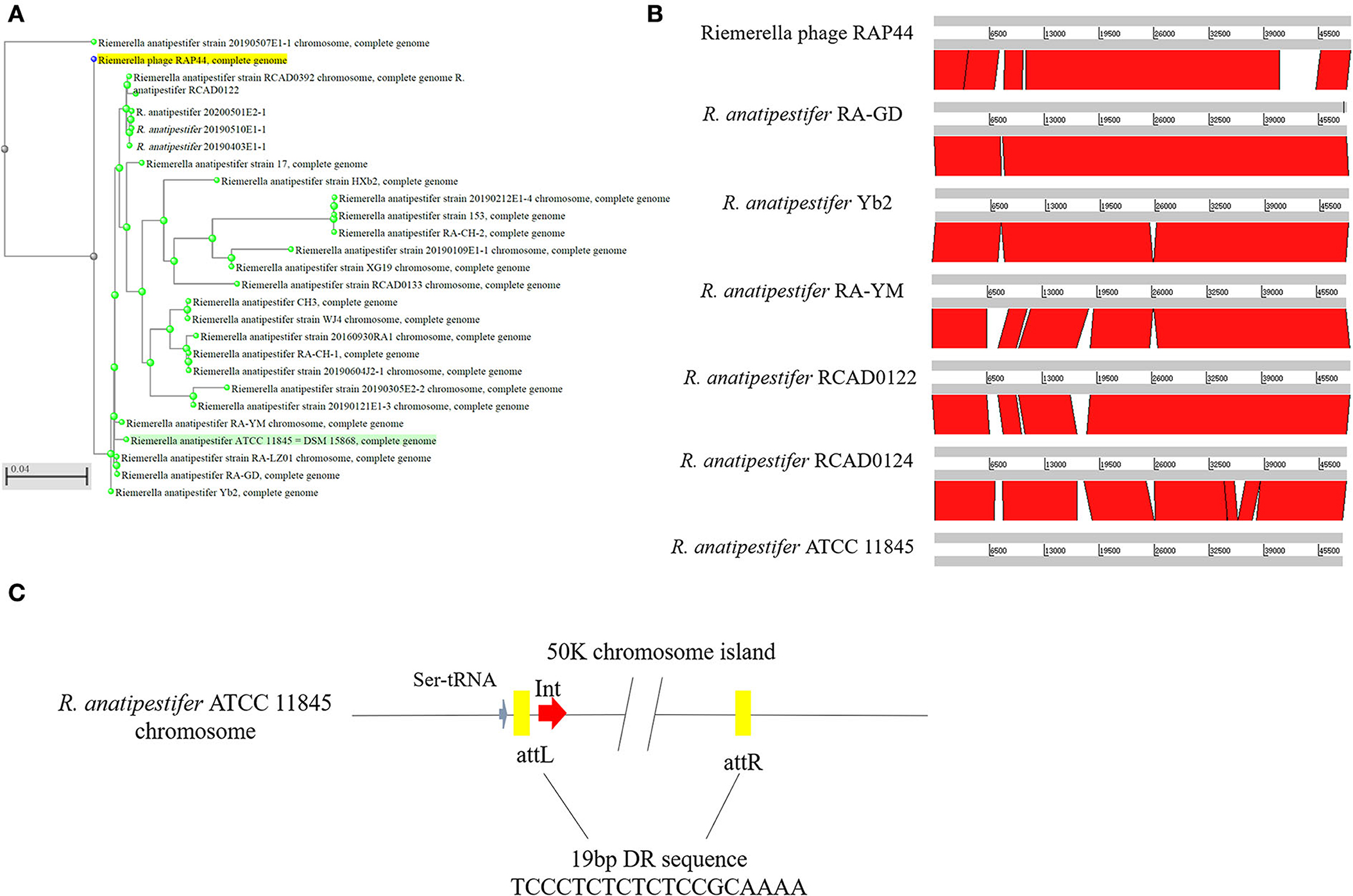
Figure 3. Evolution and components of 50K genomic island in R. anatipestifer. (A) Comparative genomic analysis of R. anatipestifer genome 50K genomic island and RAP44 genome. The nucleotide sequences alignment was used for the construction of a phylogenetic tree using the neighbor-joining (NJ). (B) The ACT for comparing the difference in 50K genomic islands from six R. anatipestifer strains. (C) The 50K genomic island simulation diagram. 50K genomic island located downstream of tRNA, and 19-bp repeat sequences (TCCCTCTCTCTCCGCAAAA) in the two flanks.
Conservation analysis of 50K genomic island integrase
The prophage was preferentially integrated into the tRNA gene through its encoded integrase, so its attachment site was tRNA, which was highly conserved and contributed to the genetic variation of bacteria. Integrase had high specificity and was highly efficient. Research has shown that prophage integrase typing is a useful indicator of genetic diversity in Salmonella enterica (21). RAP44 encodes a lambda repressor-like DNA-binding domain protein sequence, which is the same as that of R. anatipestifer tyrosine-type recombinase/integrase. In this study, the integrase gene was detected with the P1/P2 primer pairs in R. anatipestifer ATCC 11845, RA-YM, and 20 clinically isolated R. anatipestifer strains (Figure 4A). The results indicated ATCC 11845, RA-YM, and clinically isolated strain no. 9 with integrase gene (Figure 4B). Sequence alignment analysis showed that the integrase was a tyrosine integrase and was highly conserved in R. anatipestifer (Figure 4C). Three-dimensional structures of integrase protein (protein ID: YP_007003674.1) were predicted and modeled from the Phyre2 database. We found that it was similar to tyrosine recombinase Cre (protein ID: AAQ13978.1) (Figure 4D).
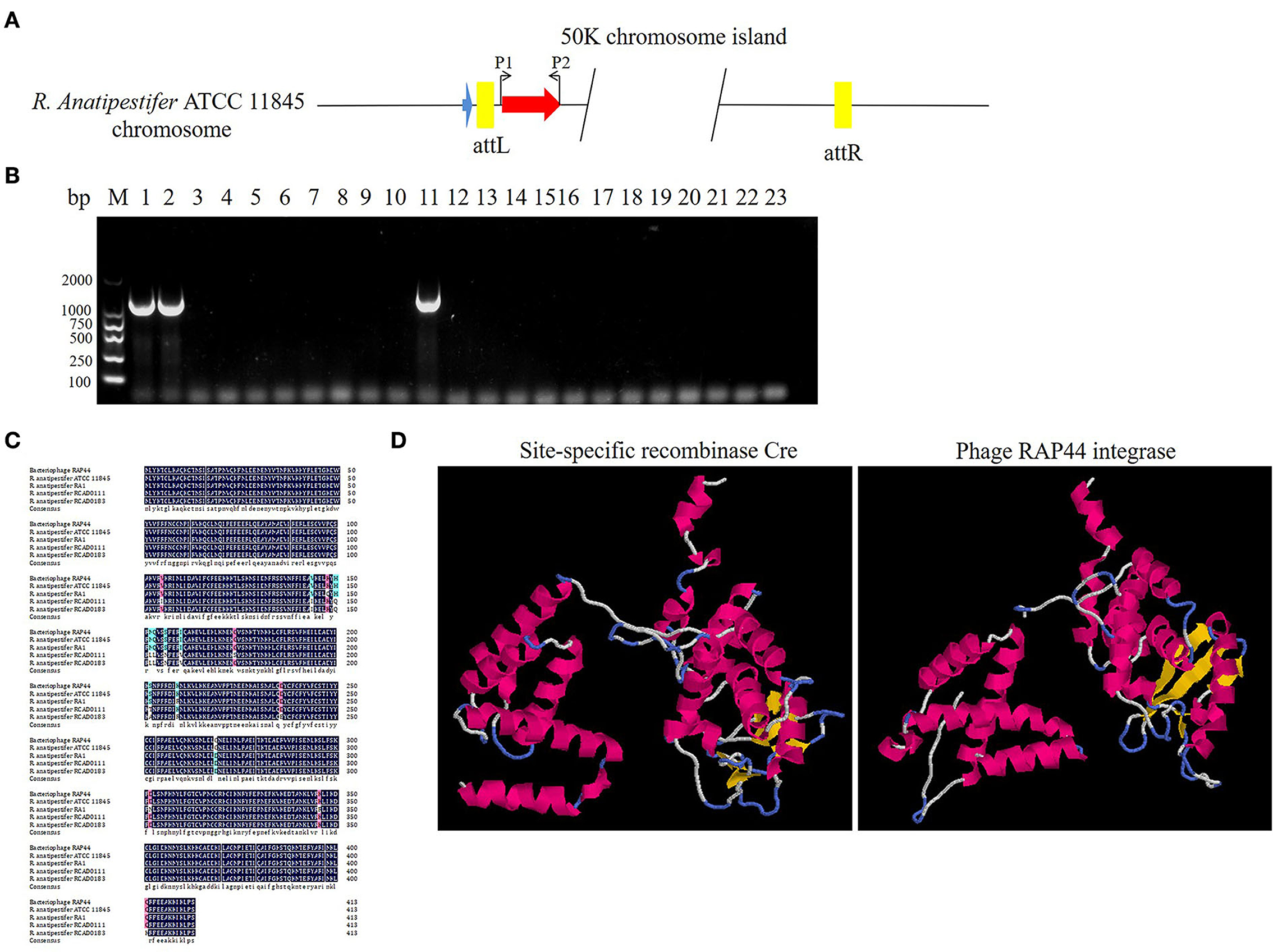
Figure 4. Analysis of RAP44 integrase and genomic island in R. anatipestifer. (A) Integrase gene with P1/P2 primer pairs was detected in R. anatipestifer ATCC 11845, RA-YM, and 20 R. anatipestifer clinical isolated strains. (B) Detecting of integrase in R. anatipestifer clinical isolated strain with primer pair P1/P2. Lane 1: R. anatipestifer ATCC11845 strain; lane 2, R. anatipestifer YM strain; lanes 3–22: 20 clinical isolated strains no. 1–20; lane 23: negative control; ATCC 11845, RA-YM, and clinical isolated strain no. 9 with the integrase. (C) Sequence alignment analysis shows that the integrase was tyrosine integrase and highly conserved in R. anatipestifer. (D) Phyre2 was applied to predict the integrase protein 3D structure. We found that the integrase protein structure is similar to tyrosine recombinase Cre.
50K genomic island integration and excision
The pathogenic island can be excised from the chromosome to form an unstable plasmid, replicating in S. aureus using its replicon (11). Three pathogenic islands of Vibrio cholerae (SPI-2, VSP-I, and VSP-II) can also be excised from the chromosome, forming circular intermediates (22). In this study, the primer pairs P3/P4 and P5/P6 were used to detect 50K genomic island excision. The primer pairs P3/P5 and P4/P6 were used to detect genomic island integration in R. anatipestifer ATCC 11845, RA-YM, and clinically isolated strain no. 1. R. anatipestifer ATCC 11845 and the RA-YM genomic islands had integration and excision functions (Figure 5A). This genomic island can be spontaneously excised and circularized from the R. anatipestifer chromosome. Figure 5B shows a schematic of the integration and excision models. Our research indicates that the 50K genomic island has a mobile function. The R. anatipestifer with this genomic island had chromosomal heterogeneity.
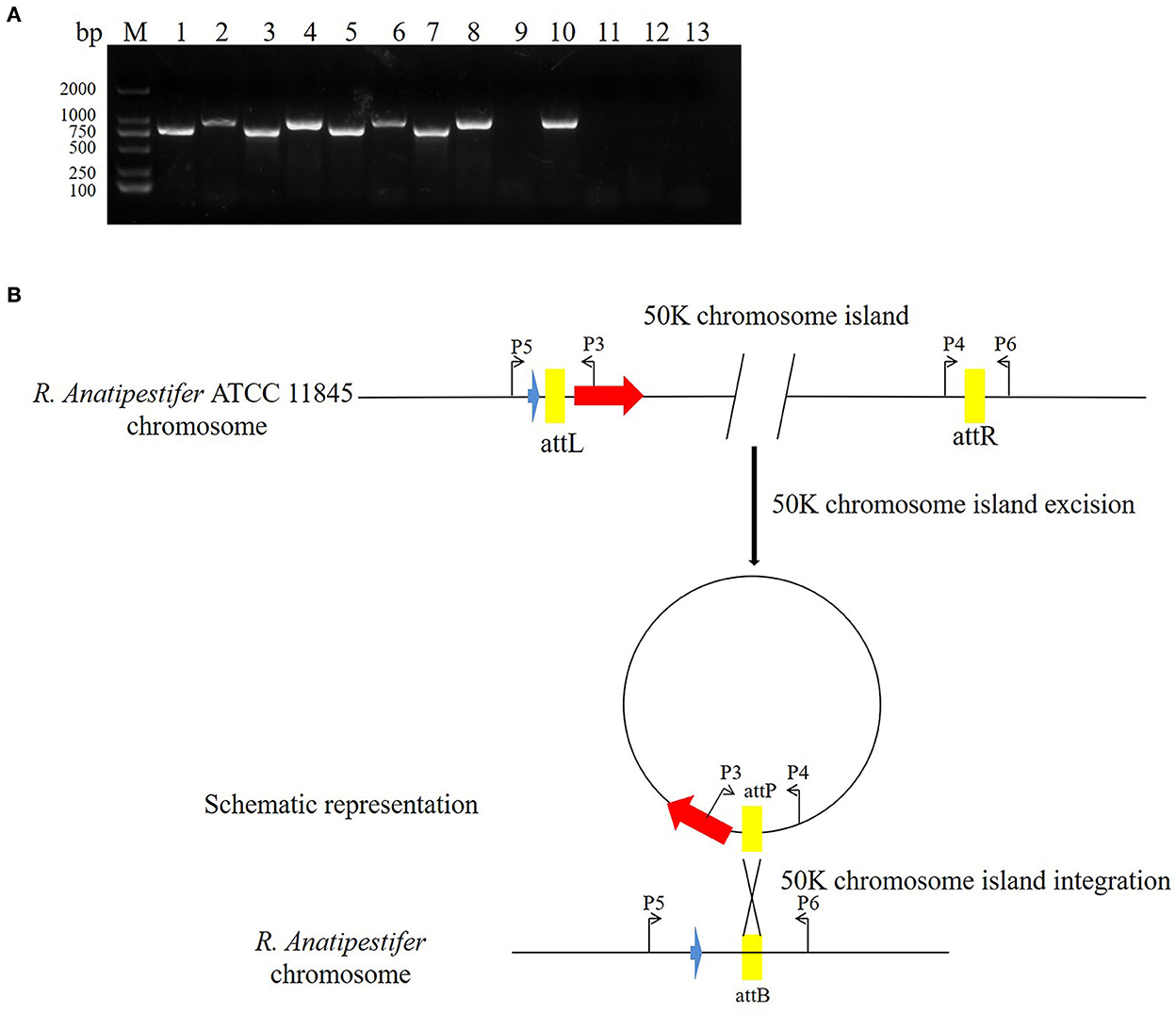
Figure 5. The 50K genomic island integration and excision. (A) The primer pairs P3/P4, P5/P6 were used for detecting genomic island excision, and P3/P5, P4/P6 were used for detecting genomic island integration in R. anatipestifer ATCC 11845, RA-YM, and clinical isolated strain no. 1. M: DL2000 DNA marker; lane 1: DNA fragment was amplified with P3/P4 primer pair from R. anatipestifer ATCC11845; lane 2: DNA fragment was amplified with P5/P6 primer pair from R. anatipestifer ATCC11845; lane 3: DNA fragment was amplified with P4/P6 primer pair from R. anatipestifer ATCC11845; lane 4: DNA fragment was amplified with P3/P5 primer pair from R. anatipestifer ATCC11845; lanes 5–8: DNA fragment was amplified with same primer from R. anatipestifer YM; lanes 9–12: DNA fragment was amplified with same primer from R. anatipestifer clinical isolated strain no. 1; lane 13: negative control; R. anatipestifer ATCC 11845 and RA-YM genomic islands had the integration and excision function. (B) Schematic representation of the integration and excision model.
50K genomic island integrase-mediated integration in R. anatipestifer
Here, we studied the integration mechanism of the genomic islands in R. anatipestifer. Integrase with the promoter and attP site was cloned with P7/P8 primer pairs and then connected with suicide vector pRE112-Spec (Figure 6A). The recombinant vector was named pRE112-Spec-int. The construction diagram of the integration vector is shown in Figure 6B. The recombinant suicide vector was transferred from the E. coli X7213 strain to the clinically isolated strain no. 1 via conjugal transfer. The recombinant strain was selected by Spec antibiotic plate and was called RAint. Integration and excision were detected by PCR of the recombinant strain. The results indicated that the plasmid was integrated into R. anatipestifer, and the insert site was consistent with R. anatipestifer ATCC 11845 and RA-YM. The recombinant strain only had the function of integration and lost its excision function (Figure 6C). Integrase-mediated integration is genetically stable. The recombinant bacteria after 20 passages in tryptone soybean agar pallet without antibiotics also have stable expression Spec-resistance cassette (Figure 6D). The recombinant strain was analyzed using high-throughput sequencing, and the insertion site and sequence were confirmed (Supplementary material). Our results indicate that the integrase from 50K genomic island mediated integration and stable protein expression in R. anatipestifer.
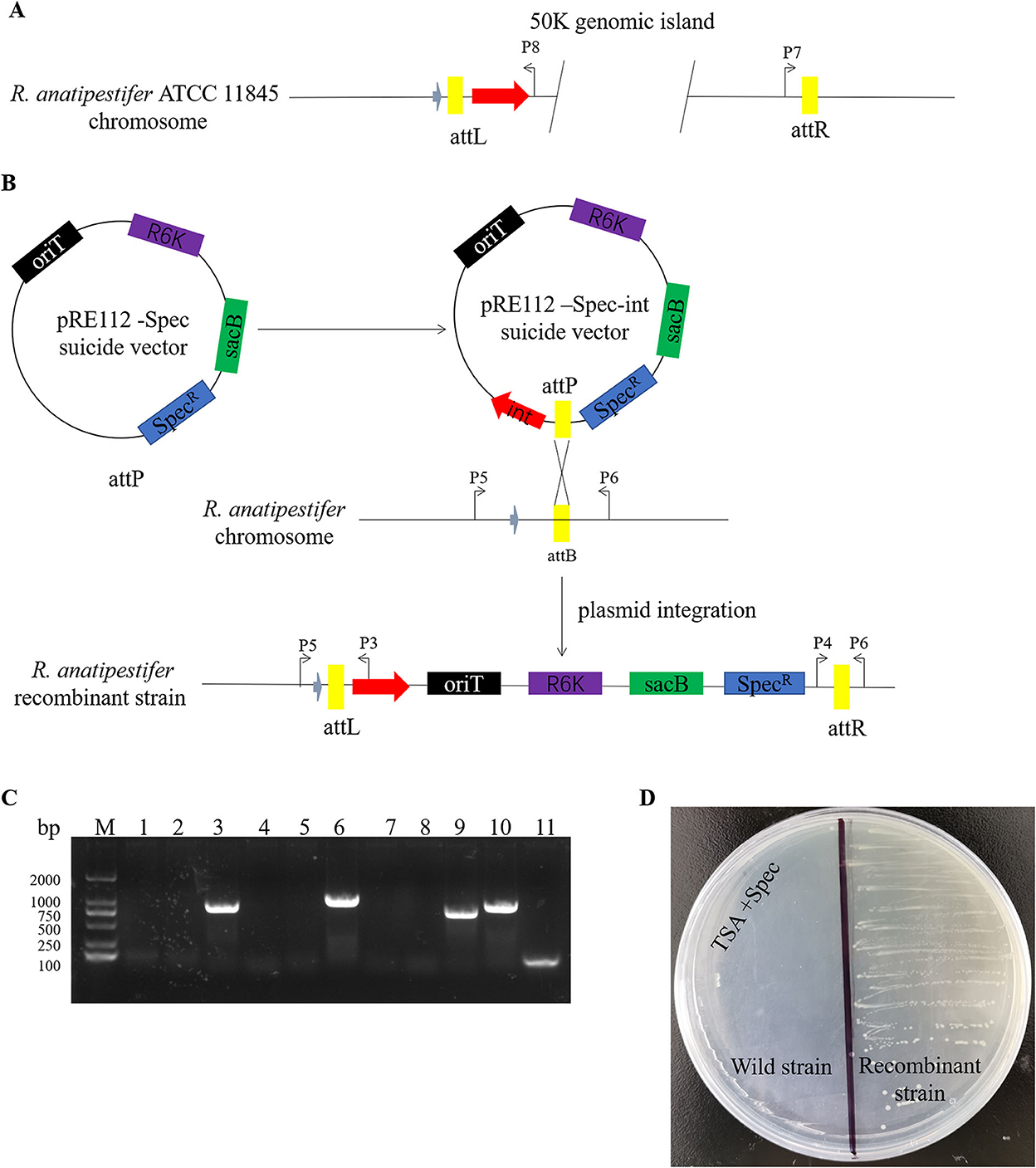
Figure 6. Identification of an integrase from phage RAP44-mediated integration in R. anatipestifer. (A) The integase with promoter and attP site was cloned with P7/P8 primer pairs. (B) The DNA fragments were cloned into suicide vector pRE112-Spec, the recombinant vector named pRE112-Spec-int. The construction diagram of integration vector is shown. (C) The recombinant strain was selected using Spec antibiotic plate and called R. anatipestifer RAint. Integration and excision were detected by PCR in the recombinant strain. M: DL2000 DNA marker; lane 1: Spec gene fragment was amplified with Spec F/R primer pair from R. anatipestifer clinical isolated strain no. 1; lane 2: DNA fragment was amplified with P3/P4 primer pair from R. anatipestifer clinical isolated strain no. 1; lane 3: DNA fragment was amplified with P5/P6 primer pair from R. anatipestifer clinical isolated strain no. 1; lane 4: DNA fragment was amplified with P4/P6 primer pair from R. anatipestifer clinical isolated strain no. 1; lane 5: DNA fragment was amplified with P3/P5 primer pair from R. anatipestifer clinical isolated strain no. 1; lanes 6–10: DNA fragment was amplified with same primer from recombinant strain; lane 11: negative control. Plasmid was integrated into R. anatipestifer, and the insert site was consistent with the R. anatipestifer ATCC 11845 and RA-YM. The recombinant strain only had the function of integration and loss of the function of excision. (D) Integrase-mediated integration has genetic stability. The well expression of Spec resistance cassette in recombinant bacteria after 20 passages in TSA pallet without antibiotic.
Discussion
With super bacteria's increasing virulence and drug resistance, it is particularly important to understand the causes that drive bacterial evolution. Phages are vital gene-transfer particles. Phage transduction is considered the main cause of HGT between bacteria. HGT is the primary method for bacteria to acquire virulence and drug-resistance genes, which is of great significance in medicine (23). There are three modes of HGT, namely, transformation, conjugation, and transduction. Phage genomes are integrated into bacterial chromosomes with replication of the host chromosome-integrated phage genome passive replication. Phage transduction is the main driving force of microbial evolution (24).
In this study, the R. anatipestifer genomic islands and prophages were predicted using IslandViewer 4 and PHAST, and a 50K genomic island was identified (Figure 1). The 50K genomic island sequence alignment indicated that most sequences were consistent with the R. anatipestifer phage RAP44 (Figure 2). Phage RAP44 was the first virulent phage to be reported in 2012. RAP44 phage with double-stranded DNA and 80 coding sequences were identified (25). Further comparative genomic analysis revealed significant differences between 50K genomic islands of different R. anatipestifer strains and RAP44 phage genome (Figures 3A,B). The inserted phage genes resulted in host evolution, whereas the deleted phage genes promoted the loss of phage function. The deletion of the RAP44 phage-related genes and the insertion of foreign genes made it impossible to form complete phage particles. Insertion of the phage genome promoted the evolution of R. anatipestifer.
We analyzed the locus of the 50K genomic island and found that this genomic island is located downstream of tRNA, and the integrase is located downstream of tRNA. The 19 bp repeats sequence is located at the flanks of the genomic island. According to the above analysis, we confirmed the basic molecular characteristics of the genomic island (Figure 3C). We used integrase as the marker gene to identify the genomic island of clinically isolated strains and found that only one clinically isolated strain contains the 50K genomic island (Figure 4B). Phages are the most abundant organisms on earth and play a critical role in microbial evolution. They are the agents of HGT that form a vast network of biology that connects all bacterial genomes. Bacteria often require an exchange of genetic material to adapt quickly to the environment (26). Bacteriophages shape bacterial evolution in various ways; they increase bacterial diversity through the selective capture of species (27), promote HGT (28), and serve as a repository of genetic innovations (29). Phages are thought to be the primary source of genetic material for bacteria to produce new genes (30). We also found that other bacteria have similar genomic islands. Enterobacter phase mEp460 and Pseudomonas phase D3 were predicted as genomic islands in Gram-negative bacteria E. coli and P. aeruginosa, respectively. Staphylococcus phage IME1364_01, Listeria phase B025, and Lactobacillus phase phiJB were predicted as genomic islands in Gram-positive bacteria S. aureus, L. monocytogenes, and Lactobacillus delbrueckii, respectively. Therefore, the integration of phage genome-mediated HGT is ubiquitous in bacteria.
Prophages are parasitic bacterial viruses that can integrate into the bacterial genome by integrase, replicating on the host chromosome. Bacteria that integrate phage genes can produce additional virulence factors that enhance bacterial virulence and the ability to adapt and survive in various environments (31). HGT enhances the frequency of exchange between strains (32). Both exist in integrative and excisional forms of prophages in bacteria. Most of these exist in an integrative form (33). In addition to phages, pathogenic islands function in integrating and excising chromosomes. There are differences in the integrase amino acid sequence of the 50K genomic island and RAP44 in several sites (Figure 4C). We found that the 3D structure of the 50K genomic island integrase may be similar to lambda integrase Cre (Figure 4D). In the λ bacteriophage, λ integrase catalyzes the integration between attP and attB sites, forming attL and attR sites. SOS or mitomycin C can induce the excision–replication–packaging cycle of SaPIs. Almost all SaPIs have attL and attR direct repeats on both sides, which encode integrases and insert them in the same direction at specific chromosomal locations. Here, we described a 50K genomic island, a phage cluster, in R. anatipestifer ATCC 11845 and RA-YM. This genomic island can be spontaneously excised and circularized from the R. anatipestifer chromosome (Figure 5). Here, the SOS reaction was used to induce prophage of R. anatipestifer ATCC 11845 and RA-YM, and no phage particles were detected in the supernatant (data not shown). Prophage Spn1 of Streptococcus pneumoniae and PP3 of P. aeruginosa had similar characteristics.
Phage-derived integrases play an important role in gene expression in microorganisms, cells, plants, and animals. Stabilization of chromosomal integration of large DNA fragments is mediated by phage integrase in Clostridium ljungdahlii (34). A recent study showed that the BxB1-integrase system guided stable exogenous gene expression in Pseudomonas (35). In addition, λ integrase mediates a seamless vector transgenesis platform for therapeutic protein expression (36). Furthermore, a novel approach to plastid transformation uses phiC31 phage integrase (37). Cre/lox-mediated genome modifications have been widely used in mouse functional genomic research (38). We constructed the recombinant suicide plasmid containing integrase and attP site. The recombinant suicide plasmid had been integrated into clinical isolates of R. anatipestifer through conjugation transfer. The results indicated that integrase only possesses integrase activity, not possess exonuclease activity (Figure 6C). The recombinant strain obtained by resistance screening, and the foreign resistance gene were stable expression (Figure 6D). Integrase can catalyze the removal and reconnection of attL and attR loci. In addition to integrase, integration and excision reactions require the integration of host factors, and excision requires λ excisionase. The excision function of this genomic island may be mediated by exonuclease, and we will further explore the molecular mechanism of the excision function of this genomic island.
Phage-derived integrases can be used in various hosts. Clostridium phages reported in infections include C. difficile and pathogenic C. perfringens. The phage attachment site attP and bacterial attachment site attB have also been identified (39, 40). Moreover, the integrase systems of Clostridium phages have shown universality in different bacteria (41). They are functional and can be used as genetic manipulation tools for Clostridium. Therefore, further research is required as these integrase systems are expected to be widely used as genetic tools. We identified phage-derived integrases and analyzed their integrative characteristics of R. anatipestifer. Based on this, we propose a straightforward method for establishing stable expression of exogenous genes in R. anatipestifer.
In this study, by comparing the genomic islands with the whole-genome sequences of the RAP44 phage, we found a 50K genomic island originating from the phages. The inserted phage lost its characteristics through deletion and insertion, and the RAP44 phage genome was a part of the bacteria. Furthermore, the genomic islands contain numerous integrated mobile genetic elements. In R. anatipestifer ATCC 11845, the 50K genomic island was integrated, excised, and cyclized automatically. Integrases are essential elements of integration, and the integrative function of integrase was verified in R. anatipestifer; the integrase with the attP site could be stably integrated at the attB locus of the R. anatipestifer genome. Recombinant strains can stably inherit and express exogenous genes. We identified phage-derived integrase and analyzed their integrative characteristics. Based on this, we propose evidence for the evolution of R. anatipestifer genomes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
YY and YW: funding acquisition, supervision, validation, writing (review and editing), and resources. YW: investigation, visualization, and writing—original draft. JD, JR, LL, and JL: methodology. SN, YL, YZ, SG, FY, HM, and W-xT: project administration. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the National Natural Science Foundation of China (No. 32202808), Shanxi Province Excellent Doctoral Work Award-Scientific Research Project Genomic Island Integration in Riemerella anatipestifer (Nos. SXBYKY2021007 and SXBYKY2021043), Start-up Fund for doctoral research of Shanxi Agricultural University (Nos. 2020BQ63 and 2021BQ08), Shanxi Province Colleges and Universities Science and Technology Innovation Project (Nos. 2021L159 and 2021L132), Shanxi 1331 Project (Grant No. 20211331-13), Key Research Development Program of Shandong Province (No. 2019JZZY010719), and Shanxi Agriculture Research System (2022-07).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.961354/full#supplementary-material
References
1. Dy RL, Richter C, Salmond GP, Fineran PC. Remarkable Mechanisms in Microbes to Resist Phage Infections. Annu Rev Virol. (2014) 1:307–31. doi: 10.1146/annurev-virology-031413-085500
2. Samson JE, Magadan AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol. (2013) 11:675–87. doi: 10.1038/nrmicro3096
3. Koskella B, Brockhurst MA. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. (2014) 38:916–31. doi: 10.1111/1574-6976.12072
4. Bobay LM, Touchon M, Rocha EP. Pervasive domestication of defective prophages by bacteria. Proc Natl Acad Sci U S A. (2014) 111:12127–32. doi: 10.1073/pnas.1405336111
5. Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell. (2012) 150:792–802. doi: 10.1016/j.cell.2012.06.036
6. Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. (2004) 68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004
7. Martinez-Rubio R, Quiles-Puchalt N, Marti M, Humphrey S, Ram G, Smyth D, et al. Phage-inducible islands in the Gram-positive cocci. ISME J. (2017) 11:1029–42. doi: 10.1038/ismej.2016.163
8. Fillol-Salom A, Martinez-Rubio R, Abdulrahman RF, Chen J, Davies R, Penades JR, et al. Phage-inducible chromosomal islands are ubiquitous within the bacterial universe. ISME J. (2018) 12:2114–28. doi: 10.1038/s41396-018-0156-3
9. Ubeda C, Tormo MA, Cucarella C, Trotonda P, Foster TJ, Lasa I, et al. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol Microbiol. (2003) 49:193–210. doi: 10.1046/j.1365-2958.2003.03577.x
10. Mir-Sanchis I, Martinez-Rubio R, Marti M, Chen J, Lasa I, Novick RP, et al. Control of Staphylococcus aureus pathogenicity island excision. Mol Microbiol. (2012) 85:833–45. doi: 10.1111/j.1365-2958.2012.08145.x
11. Ubeda C, Barry P, Penades JR, Novick RP. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proc Natl Acad Sci U S A. (2007) 104:14182–8. doi: 10.1073/pnas.0705994104
12. Touchon M, Bernheim A, Rocha EP. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. (2016) 10:2744–54. doi: 10.1038/ismej.2016.47
13. Li G, Lu S, Shen M, Le S, Shen W, Tan Y, et al. Characterization and interstrain transfer of prophage pp3 of Pseudomonas aeruginosa. PLoS ONE. (2017) 12:e0174429. doi: 10.1371/journal.pone.0174429
14. Leavitt S, Ayroud M. Riemerella anatipestifer infection of domestic ducklings. Can Vet J. (1997) 38:113.
15. Wang Y, Zhang Y, Cui Y, Sun Z, Zhou Z, Hu S, et al. Identification of an integrase that responsible for precise integration and excision of Riemerella anatipestifer genomic island. Front Microbiol. (2019) 10:2099. doi: 10.3389/fmicb.2019.02099
16. Zhou Z, Peng X, Xiao Y, Wang X, Guo Z, Zhu L, et al. Genome sequence of poultry pathogen Riemerella anatipestifer strain RA-YM. J Bacteriol. (2011) 193:1284–5. doi: 10.1128/JB.01445-10
17. Wang Y, Lu T, Yin X, Zhou Z, Li S, Liu M, et al. A novel RAYM_RS09735/RAYM_RS09740 two-component signaling system regulates gene expression and virulence in Riemerella anatipestifer. Front Microbiol. (2017) 8:688. doi: 10.3389/fmicb.2017.00688
18. Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. (2011) 39:347–52. doi: 10.1093/nar/gkr485
19. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J, et al. ACT: the artemis comparison tool. Bioinformatics. (2005) 21:3422–3. doi: 10.1093/bioinformatics/bti553
20. Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. (2017) 45:200–3. doi: 10.1093/nar/gkw1129
21. Colavecchio A, D'Souza Y, Tompkins E, Jeukens J, Freschi L, Emond-Rheault JG, et al. Prophage integrase typing is a useful indicator of genomic diversity in Salmonella enterica. Front Microbiol. (2017) 8:1283. doi: 10.3389/fmicb.2017.01283
22. Murphy RA, Boyd EF. Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J Bacteriol. (2008) 190:636–47. doi: 10.1128/JB.00562-07
23. Touchon M, Moura J, de Sousa A, Rocha EP. Embracing the enemy: the diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr Opin Microbiol. (2017) 38:66–73. doi: 10.1016/j.mib.2017.04.010
24. Chen J, Quiles-Puchalt N, Chiang YN, Bacigalupe R, Fillol-Salom A, Chee MSJ, et al. Genome hypermobility by lateral transduction. Science. (2018) 362:207–12. doi: 10.1126/science.aat5867
25. Cheng LF, Chen HM, Zheng T, Fu GH, Shi SH, Wan CH, et al. Complete genomic sequence of the virulent bacteriophage RAP44 of Riemerella anatipestifer. Avian Dis. (2012) 56:321–7. doi: 10.1637/9770-050411-Reg.1
26. Chiang YN, Penadés JR, Chen J. Genetic transduction by phages and chromosomal islands: The new and noncanonical. PLoS Pathog. (2019) 15:e1007878. doi: 10.1371/journal.ppat.1007878
27. Koonin EV. Viruses and mobile elements as drivers of evolutionary transitions. Philos Trans R Soc Lond B Biol Sci. (2016) 371:20150442. doi: 10.1098/rstb.2015.0442
28. Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. Phage as agents of lateral gene transfer. Curr Opin Microbiol. (2003) 6:417–24. doi: 10.1016/S1369-5274(03)00086-9
29. Cortez D, Forterre P, Gribaldo S. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol. (2009) 10:R65. doi: 10.1186/gb-2009-10-6-r65
30. Daubin V, Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. (2004) 14:1036–42. doi: 10.1101/gr.2231904
31. Canchaya C, Fournous G, Brüssow H. The impact of prophages on bacterial chromosomes. Mol Microbiol. (2004) 53:9–18. doi: 10.1111/j.1365-2958.2004.04113.x
32. Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. (2009) 323:139–41. doi: 10.1126/science.1164783
33. Denou E, Pridmore RD, Ventura M, Pittet AC, Zwahlen MC, Berger B, et al. The role of prophage for genome diversification within a clonal lineage of Lactobacillus johnsonii: characterization of the defective prophage LJ771. J Bacteriol. (2008) 190:5806–13. doi: 10.1128/JB.01802-07
34. Huang H, Chai C, Yang S, Jiang W, Gu Y. Phage serine integrase-mediated genome engineering for efficient expression of chemical biosynthetic pathway in gas-fermenting Clostridium ljungdahlii. Metab Eng. (2019) 52:293–302. doi: 10.1016/j.ymben.2019.01.005
35. Otto M, Wynands B, Drepper T, Jaeger KE, Thies S, Loeschcke A, et al. Targeting 16S rDNA for Stable Recombinant Gene Expression in Pseudomonas. ACS Synth Biol. (2019) 8:1901–12. doi: 10.1021/acssynbio.9b00195
36. Makhija H, Roy S, Hoon S, Ghadessy FJ, Wong D, Jaiswal R, et al. A novel λ integrase-mediated seamless vector transgenesis platform for therapeutic protein expression. Nucleic Acids Res. (2018) 46:e99. doi: 10.1093/nar/gky500
37. Lutz KA, Corneille S, Azhagiri AK, Svab Z, Maliga P. A novel approach to plastid transformation utilizes the phiC31 phage integrase. Plant J. (2004) 37:906–13. doi: 10.1111/j.1365-313X.2004.02015.x
38. Thomson JG, Rucker EB, Piedrahita JA. Mutational analysis of loxP sites for efficient Cre-mediated insertion into genomic DNA. Genesis. (2003) 36:162–7. doi: 10.1002/gene.10211
39. Goh S, Ong PF, Song KP, Riley TV, Chang BJ. The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology (Reading). (2007) 153:676–85. doi: 10.1099/mic.0.2006/002436-0
40. Williams R, Meader E, Mayer M, Narbad A, Roberts AP, Mullany P, et al. Determination of the attP and attB sites of phage CD27 from Clostridium difficile NCTC 12727. J Med Microbiol. (2013) 62:1439–43. doi: 10.1099/jmm.0.058651-0
Keywords: Riemerella anatipestifer, RAP44 phage, integrase, genomic island, integration
Citation: Wang Y, Deng J, Ren J, Liang L, Li J, Niu S, Wu X, Zhao Y, Gao S, Yan F, Liu Y, Ma H, Tian W-x and Yan Y (2022) RAP44 phage integrase-guided 50K genomic island integration in Riemerella anatipestifer. Front. Vet. Sci. 9:961354. doi: 10.3389/fvets.2022.961354
Received: 04 June 2022; Accepted: 08 November 2022;
Published: 29 November 2022.
Edited by:
Thi Thu Hao Van, RMIT University, AustraliaReviewed by:
Qihui Luo, Sichuan Agricultural University, ChinaMarja-Liisa Hänninen, University of Helsinki, Finland
Rafael Antonio Casarin Penha Filho, São Paulo State University, Brazil
Copyright © 2022 Wang, Deng, Ren, Liang, Li, Niu, Wu, Zhao, Gao, Yan, Liu, Ma, Tian and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Yan, yanyi@sxau.edu.cn
 Ying Wang
Ying Wang Jianfeng Deng1
Jianfeng Deng1  Jianle Ren
Jianle Ren Libin Liang
Libin Liang Wen-xia Tian
Wen-xia Tian Yi Yan
Yi Yan