Molecular Survey on Kobuviruses in Domestic and Wild Ungulates From Northwestern Italian Alps
- 1Faculty of Veterinary Medicine, Università degli Studi di Teramo, Teramo, Italy
- 2Istituto Zooprofilattico Sperimentale del Piemonte, Liguria e Valle d'Aosta, Centro di Referenza Nazionale per le Malattie degli Animali Selvatici (CeRMAS), Aosta, Italy
- 3Department of Veterinary Medicine, Università Aldo Moro di Bari, Valenzano, Italy
Since the first identification in 1989 in humans, kobuviruses (KoVs) have been identified from a wide range of animal species including carnivores, rodents, birds, ungulates, rabbits, and bats. Several studies have described the identification of genetically related KoVs in the fecal virome of domestic and wild animals suggesting a mutual exchange of viruses. By screening a total of 231 fecal samples from wild and domestic ungulates, KoVs RNA was detected in wild boars (3.2%; 2/63), chamois (4.6%; 2/43), and goats (2.6%; 2/77). On phylogenetic analysis of the partial RdRp sequence, the wild boar strains clustered within the species Aichivirus C whilst the strains identified in domestic and wild ruminants grouped into the species Aichivirus B. The complete VP1 gene was obtained for chamois and goat KoVs. Interestingly, upon phylogenetic analysis the strains grouped together with a KoV of ovine origin within a distinct genetic type (B3) of the species Aichivirus B.
Introduction
Kobuviruses (KoVs) are small (~30–32 nm), icosahedral, non-enveloped viruses with a single stranded positive sense RNA genome of 8.2–8.4 kb in length, classified in the genus Kobuvirus within the family Picornaviridae (1). The viral RNA, polyadenylated at the 3′-end and covalently linked to a virus-encoded protein (VPg) at its 5′-end, consists of a 5′ untranslated region (UTR) of 646–717 nucleotides (nt), an open reading frame (ORF) of 7,308–7,467 nt and a 3′ UTR of 241–244 nt. The unique ORF encodes a single large polyprotein that is post-translationally cleaved into three distinct functional P regions (P1–P3) with P1 encoding the viral capsid proteins (VP0, VP3, and VP1) and P2 and P3 encoding proteins involved in protease processing and genome replication (2).
KoVs were first recognized in 1989 as the cause of oyster-associated non-bacterial gastroenteritis in humans in Aichi Prefecture, Japan (3). Since then, an increasing number of novel KoVs have been repeatedly identified from a large diversity of animal species, including ungulates, carnivores, rodents, birds, rabbits, and bats (4–21). Based on the phylogenetic analysis of the complete nt sequence of the VP1 encoding gene (15), the genus Kobuvirus is currently classified into six established species, Aichivirus A to F, and 20 genetic types [https://talk.ictvonline.org]. Aichivirus A includes a total of ten genetic types (Aichivirus A1–A10) identified in humans (A1) (3), canids (A2) (10, 11), rodents (A3, A6–A10) (14–17), domestic cats (A4) (12), and birds (A5) (18); Aichivirus B comprises KoVs detected in cattle (B1) (4), ferret (B2) (13), and sheep (B3) (6); within the Aichivirus C are classified KoVs detected in swine (C1) (5) and in goats (C2) (8); Aichivirus D includes newly discovered KoVs detected in Japanese black cattle (9), designed Kagovirus 1 and 2 (D1 and D2); the species Aichivirus E and F contains KoVs identified, respectively, in rabbits (E1) (19) and bats (F1 and F2) (21). Although information on the epidemiology of KoVs is still limited, cross transmission of these viruses has been revealed at the level of host order, family and species, with well-founded suspicious that wildlife ecosystem may constitute a large reservoir. This suggestion is supported by the recurrent identification of genetically related strains, practically indistinguishable from each other, in the fecal virome of domestic mammals and in their wildlife counterparts, such as Aichivirus A-2 (formerly canine kobuvirus) in dogs and wild canids (22–24), Aichivirus C-1 (formerly porcine kobuvirus) in pigs and wild boars (25, 26), Aichivirus B-1 (formerly bovine kobuvirus) and Aichivirus C-2 (formerly caprine kobuvirus) in domestic and wild ruminants (27). A molecular survey performed in the Serengeti National Park in Tanzania (23) has documented the circulation of highly genetically related strains between domestic and wild carnivores. In Serbia, porcine kobuvirus sequences identified from wild boars and domestic pigs revealed an amino acid diversity of 1%, emphasizing the role of wild boars as potential reservoirs for domestic pigs (26). In Italy, despite the identification of genetically similar bovine and caprine KoV strains in domestic ruminants and roe deer (27), systematic studies in a multi-host landscape exploring the role of different domestic and wild species in spreading and maintaining KoVs are still lacking. Herewith, we report the results of a molecular survey conducted in a defined geographical setting (Northwestern Italian Alps) where wild ungulates are abundant and occasionally enter in contact with livestock animals (seasonal grazing of goats and sheep).
Materials and Methods
Study Area and Sampling
Sampling covered the Region of Valle d'Aosta, in Northwestern Italy, with an overall geographic area of 3,261 km2. It is an alpine area characterized by the concomitant presence of domestic ruminant semi-rural farms and abundant ungulates wildlife. In this setting, the population of wild ungulates, composed primarily by wild boar (Sus scrofa), red deer (Cervus elaphus), and chamois (Rupicapra rupicapra), may occasionally enter in contact with domestic small ruminants through the use of pastures during seasonal transhumance or in the surrounding of backyard farms. Between September 2017 and December 2019, using a convenience sampling strategy, a total of 231 fecal samples was obtained from domestic and wild ungulates. Briefly, 95 fecal specimens were collected from 77 goats and 18 sheep, respectively, in 14 caprine and 3 ovine small farms from 16 municipalities in the Valle d'Aosta region (Figure 1A), consisting of 4–10 animals. All animals were female and clinically healthy at the time of sampling. On the basis of the age, 25 goats and 6 sheep were from the defined age group <3 years, 22 goats and 3 sheep from 3 to 4 years group, 16 goats and 9 sheep were from the age group >4 years. Age was unknown for 15 goats. Among wild animals, a total of 136 stool specimens was collected from 63 wild boars, 30 red deer, and 43 chamois sampled during the regular hunting season in Valle d'Aosta Region and submitted to the National Reference Center for Wild Animal Diseases (Italy). Age data were not available for many of the wild species investigated.
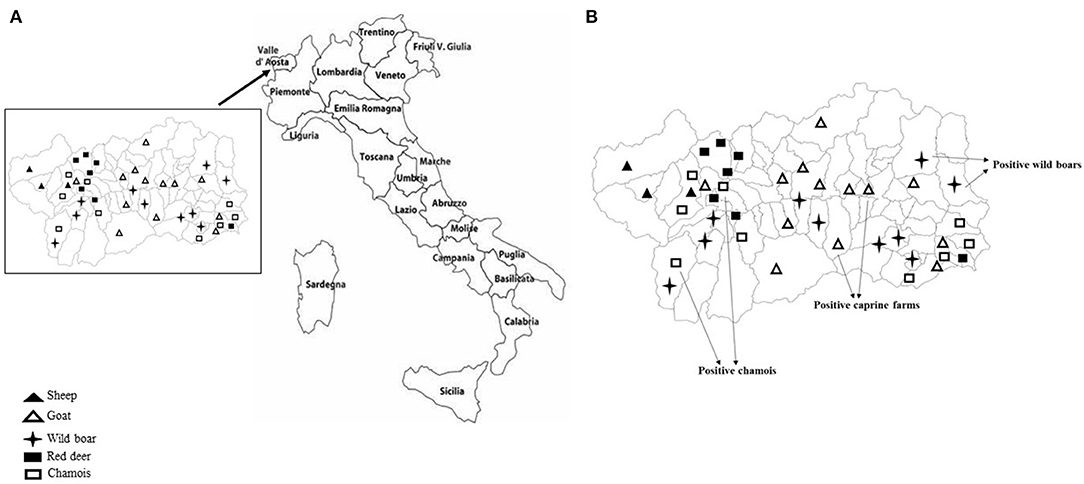
Figure 1. (A) Mapping of sheep and goat farms investigated and areas in which wild ungulates were sampled. (B) Sites where positive goats and wild ungulates were sampled.
Molecular Analyses
Total RNA was extracted from 200 μl of 10% (wt/vol) fecal suspension using TRIzol LS (Invitrogen, Ltd, Paisley, UK) procedure. KoVs RNA was detected by RT-PCR using broadly reactive primer pair, UNIV-kobu-F/UNIV-kobu-R (28) designed to amplify all members of the genus Kobuvirus and targeting a region of 217-bp of the viral RNA-dependent RNA polymerase complex (RdRp). In order to further investigate the genetic heterogeneity of the strains detected, all the samples yielding amplicons of the expected sizes were screened using specific primer sets (Table 1) designed by multiple alignment of sequences currently available in GenBank, able to amplify the complete VP1 encoding gene. All the positive amplicons were purified with the QIAquick Gel Extraction Kit (Qiagen, Milan, Italy) and sequenced by using BigDye Terminator Cycle chemistry (Applied Biosystems, Foster City, California, US). Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov) and FASTA (http://www.ebi.ac.uk/fasta33) with default values were used to find homologous hits. Multiple alignments were performed using the commercially available Geneious software package version 9.1.6 (Geneious software package vers. 9, Biomatters, New Zealand, https://www.geneious.com/ Biomatters). Phylogenetic tree (neighbor joining and Kimura 2-parameter model) with bootstrap analysis (1,000 replicates) were constructed by using the MEGA software package, version X (29).
Results
KoVs RNA was detected in a total of 6 fecal samples with an overall prevalence of 2.6% (6/231). Among the five ungulate species investigated in this study, viral RNA was found in wild boars (3.2%; 2/63), chamois (4.6%; 2/43), and goats (2.6%; 2/77), whilst all the samples collected from red deer and sheep resulted negative (Figure 1B). The two positive goats were aged 1 and 5 years old, respectively.
However, none significant association was found between the detection of KoVs RNA and age. Also, on the basis of the positivities obtained, a geographical clustering between wild and domestic animals was not observed.
Partial RdRp sequences were determined from the KoV positive samples. By sequence analysis, two wild boar strains, WB-15/ITA and WB-28/ITA (GenBank accession numbers: MW307937-8), shared 98.6% nt identity to each other and displayed a closed relatedness (94.0–96.1% nt identity) to porcine KoV sequences detected from swine enteric samples with or without diarrhea (30–35), whilst identities to KoVs identified in wild boar fecal samples (25, 26) ranged from 91.7 to 93.1%. On phylogenetic analysis, strains WB-15/ITA and WB-28/ITA clustered together with pigs and wild boar KoVs within the species Aichivirus C (Figure 2). Despite several attempts, additional genetic information was not obtained. The chamois KoV strains Chamois-8/ITA and Chamois-22/ITA (accessions MW307939-40) and the goat strains Goat-23/ITA and Goat-24/ITA (accessions MW307941-2) displayed the highest sequence identity (97.0–98.2% nt) to KoVs detected in calves and goats in previous surveys performed in an Italian study in Abruzzo, Southern Italy (36, 37). In the phylogenetic tree, the chamois, and goat KoV strains segregated within the species Aichivirus B, falling into two distinct clades. Strains Chamois-8/ITA and Chamois-22/ITA grouped with bovine KoVs, whilst strains Goat-23/ITA and Goat-24/ITA grouped with KoVs previously identified in goats and sheep (6, 37) (Figure 2). Out of the four KoV strains detected in ruminants, the complete VP1 encoding gene was obtained from three samples collected, respectively, from the two goats (Goat-23/ITA and Goat-24/ITA, MW307943-4) and a chamois (Chamois-8/ITA, MW307945). A selection of VP1 encoding gene representative of the Aichivirus B species was retrieved from GenBank. Based on the inspection of tree and according to the distance matrix, three genetic type groups could be distinguished (Figure 3). A large group included only viruses of bovine origin (Aichivirus B-1). The mean nt identity within this group was 88.7%. A second group, sharing more than 85.5% nt identity (mean identity 86.4%) comprised KoVs detected from ferrets (Aichivirus B-2). Group 3 included a strain of ovine origin, TB3/HUN/2009 (6), and the three KoVs found in goats and chamois in this study (Aichivirus B-3). Identity among these VP1 sequences was higher than 90% nt (mean identity 93.2%).
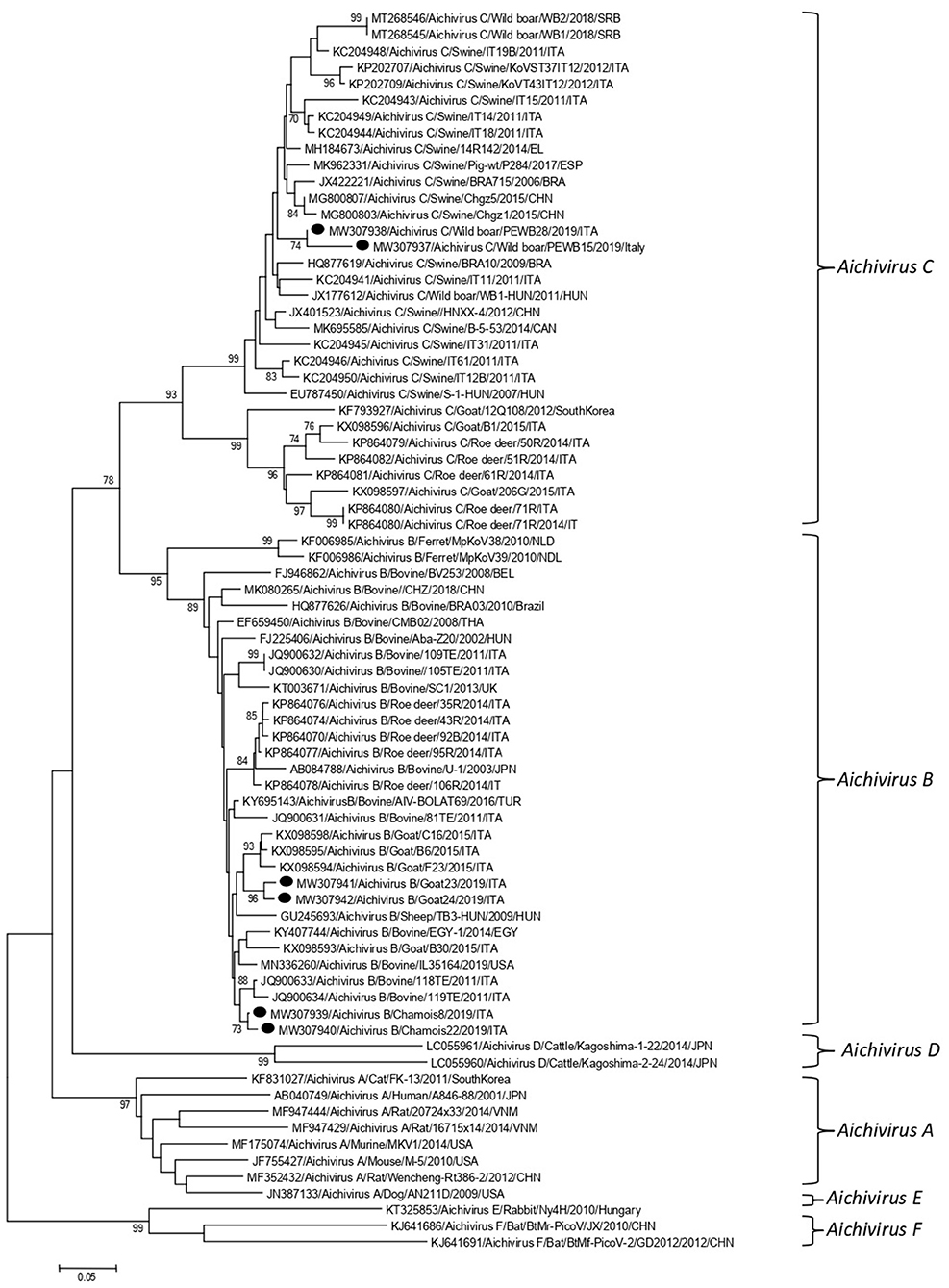
Figure 2. Phylogenetic tree based on the nt sequence of the partial RdRp gene of the KoV strains detected in this study. A selection of 70 sequences representative of the genus Kobuvirus was retrieved from GenBank. The tree was generated using the neighbor joining method and kimura 2-parameter model supplying statistical support with bootstrapping of 1,000 replicates. Labels indicate KoVs identified in this survey.
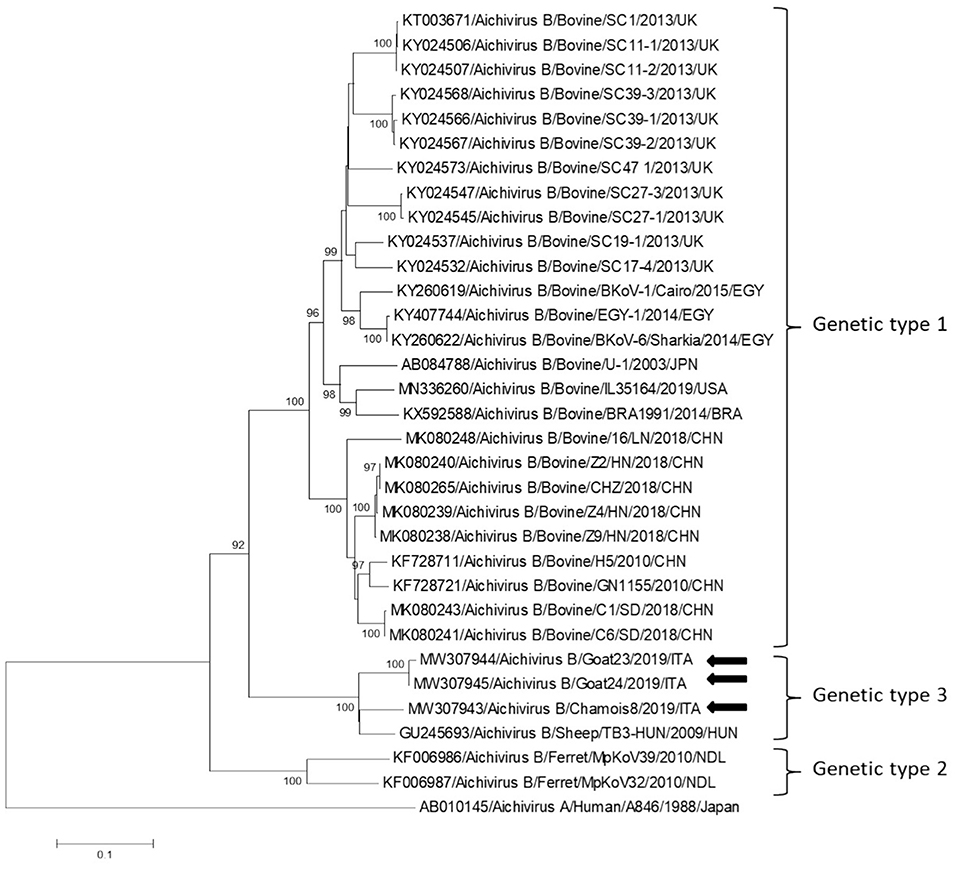
Figure 3. Phylogenetic tree based on the nt sequence of the VP1 encoding gene constructed with a selection of strains representative of the species Aichivirus B. Tree was generated using the neighbor-joining method with kimura 2-parameter model and supplying statistical support with bootstrapping of 1,000 replicates. The arrows indicate the KoV strains detected in this study. Human aichivirus A strain A8461 (GenBank accession no. AB010145) was used as outgroup.
Discussion
In this study we monitored circulation of KoVs among wild boars, goats and chamois in a restricted Italian geographical area. Identification of these viruses in wild boars and goats has been already described elsewhere (7, 8, 25, 26, 38, 39), whilst KoV infection has never been documented thus far in chamois. Viruses virtually identical to porcine KoVs (species Aichivirus C-1) were first identified in wild boars in Hungary (25) with a prevalence of 100% (10/10) and subsequently in Serbia with a rate of 6.0% (6/100) (26). Although the detection rate obtained in our survey was lower (3.2%, 2/63), our results indicate that circulation of porcine KoVs among wild boars is not uncommon and it is not limited to some settings (25, 26). In our investigation, by screening fecal samples from goat and chamois, only aichivirus B-like strains were identified, with the highest prevalence rate in chamois (4.6%, 2/43). Interestingly, all the four KoVs resulted genetically less related (92.1–95.5%) in the partial RdRp region to aichivirus B strains previously identified in roe deer from Valle d'Aosta Region (27) than to viruses detected in goats in Southern Italy (97.0–98.2%) (37). On analysis of the complete VP1 gene, a more definitive characterization of strains Goat-23/ITA, Goat-24/ITA, and Chamois-8/ITA was obtained, as all the strains grouped together with a KoV of ovine origin (TB3-HUN) in a separate genetic type (B3), distinct from viruses identified in calves (B1) and ferrets (B2). Accordingly, a definitive classification of KoV genetic types necessarily relies on sequence analysis of the full-length VP1 gene. The identification of KoVs in chamois, while expanding the host range of these viruses, reinforce the hypothesis that wildlife may represent an important reservoir of KoVs for livestock animals. However, despite the findings of this study confirmed a mild circulation of KoVs in wild and in domestic animals, a bi-directional flow was not revealed at least in the context analyzed. Further studies involving larger animal populations in other geographic areas where wild ungulates are in contact with domestics could be useful to obtain a more complete picture of the ecology of these viruses. In our analysis, all of the tested animals were apparently healthy at time of sampling. The role of KoVs in the etiology of enteritis in animals is still controversial (36, 40, 41). In previous studies (36), Aichivirus B-3 has been found at higher positivity rates in goats with enteritis (6.5%, 3/46) than in goats without enteritis (5.4%, 5/93), although the difference was not statistically significant. The possible role of Aichivirus B-1 as enteric pathogen involved in neonatal calf diarrhea has been hypothesed (41). Furthermore, in many cases, KoVs have been reported as the sole enteric pathogen detected in diarrheic pigs (42, 43), dogs (44), and cats (45, 46). Structured surveillance studies could help understand the overall impact of KoVs on livestock and wild animals in terms of health and production.
Data Availability Statement
The data that supports the findings of this study are openly available in the GenBank database at https://www.ncbi.nlm.nih.gov/nucleotide/ under accession numbers MW307937-42 (BankIt2404995).
Ethics Statement
For this study ethical statement was not required (Decreto Legislativo 4 March 2014, n. 26). Faecal samples from wild ruminants were collected during the routinely monitoring activities performed by the National Reference Centre for Wild Animal Diseases (CeRMAS – IZS PLV), whilst samples from sheep and goats were collected only for veterinary diagnostic purposes.
Author Contributions
BDM, FDP, and SR: conceptualization. BDM, FDP, AP, PF, VS, and IM: methodology. AP, SR, PF, VS, FDP, IM, and RO: investigation. BDM, FDP, VM, and FM: writing—original draft preparation. BDM, SR, and RO: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Italian Ministry of Health (Progetto di Ricerca Corrente 2018 IZS PLV 12/18 RC Sorveglianza epidemiologica sulla circolazione in Italia Nord-Occidentale di virus enterici a carattere zoonosico emergenti e riemergenti nell'interfaccia domestico-selvatico).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Marco Ragionieri (Azienda USL della Valle d'Aosta, Quart, Italy) for collection of biological samples and Aosta Valley Forestry Corp (CFVdA) for their technical assistance.
References
1. Knowles NJ, Hovi T, Hyypiä T, King AMQ, Lindberg AM, Pallansch MA, et al. Picornaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Classification and Nomenclature of Viruses: 9th Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier (2012). p. 855–80.
2. Racaniello VR. Picornaviridae: The viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Strauss SE, editors. Fields Virology, 5th Edn. Philadelphia, PA: Lippincott Williams & Wilkins (2007). p. 795–838.
3. Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, et al. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J Infect Dis. (1991) 164:954–7. doi: 10.1093/infdis/164.5.954
4. Yamashita T, Ito M, Kabashima Y, Tsuzuki H, Fujiura A, Sakae K. Isolation and characterization of a new species of kobuvirus associated with cattle. J Gen Virol. (2003) 84:3069–77. doi: 10.1099/vir.0.19266-0
5. Reuter G, Boldizsár A, Kiss I, Pankovics P. Candidate new species of Kobuvirus in porcine hosts. Emerg Infect Dis. (2008) 14:196870. doi: 10.3201/eid1412.080797
6. Reuter G, Boros A, Pankovics P, Egyed L. Kobuvirus in domestic sheep, Hungary. Emerg Infect Dis. (2010) 16:869–70. doi: 10.3201/eid1605.091934
7. Lee MH, Jeoung HY, Lim JA, Song JY, Song DS, An DJ. Kobuvirus in South Korean black goats. Virus Genes. (2012) 45:186–9. doi: 10.1007/s11262-012-0745-6
8. Oem JK, Lee MH, Lee KK, An DJ. Novel Kobuvirus species identified from black goat with diarrhea. Vet Microbiol. (2014) 172:563–7. doi: 10.1016/j.vetmic.2014.06.009
9. Otomaru K, Naoi Y, Haga K, Omatsu T, Uto T, Koizumi M, et al. Detection of novel kobu-like viruses in Japanese black cattle in Japan. J Vet Med Sci. (2016) 78:321–4. doi: 10.1292/jvms.15-0447
10. Kapoor A, Simmonds P, Dubovi EJ, Qaisar N, Henriquez JA, Medina J, et al. Characterization of a canine homolog of human Aichivirus. J Virol. (2011) 85:11520–5. doi: 10.1128/JVI.05317-11
11. Li L, Pesavento PA, Shan T, Leutenegger CM, Wang C, Delwart E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J Gen Virol. (2011) 92:2534–41. doi: 10.1099/vir.0.034611-0
12. Chung JY, Kim SH, Kim YH, Lee MH, Lee KK, Oem JK. Detection and genetic characterization of feline kobuviruses. Virus Genes. (2013) 47:559–62. doi: 10.1007/s11262-013-0953-8
13. Smits SL, Raj VS, Oduber MD, Schapendonk CM, Bodewes R, Provacia L, et al. Metagenomic analysis of the ferret fecal viral flora. PloS One. (2013) 8:e71595. doi: 10.1371/journal.pone.0071595
14. Phan TG, Kapusinszky B, Wang C, Rose RK, Lipton HL, Delwart E. The fecal viral flora of wild rodents. PLoS Pathog. (2011) 9:e1002218. doi: 10.1371/journal.ppat.1002218
15. Lu L, Van Dung N, Ivens A, Bogaardt C, O'Toole A, Bryant JE, et al. Genetic diversity and cross-species transmission of Kobuviruses in Vietnam. Virus Evol. (2018) 4:vey002. doi: 10.1093/ve/vey002
16. Wu Z, Lu L, Du J, Yang L, Ren X, Liu B, et al. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome. (2018) 6:178. doi: 10.1186/s40168-018-0554-9
17. Williams SH, Che X, Garcia JA, Klena JD, Lee B, Muller D, et al. Viral diversity of house mice in New York City. mBio. (2018) 9:e01354–17. doi: 10.1128/mBio.01354-17
18. Pankovics P, Boros Á, Kiss T, Reuter G. Identification and complete genome analysis of Kobuvirus in faecal samples of European roller (Coracias garrulus): for the first time in a bird. Arch Virol. (2015) 160:345–51. doi: 10.1007/s00705-014-2228-7
19. Pankovics P, Boros Á, Bíró H, Horváth KB, Phan TG, Delwart E, et al. Novel picornavirus in domestic rabbits (Oryctolagus cuniculus var. domestica). Infect Genet Evol. (2016) 37:117–22. doi: 10.1016/j.meegid.2015.11.012
20. Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH, et al. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol. (2010) 84:6955–65. doi: 10.1128/JVI.00501-10
21. Wu Z, Yang L, Ren X, He G, Zhang J, Yang J, et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. (2016) 10:609–20. doi: 10.1038/ismej.2015.138
22. Di Martino B, Di Profio F, Melegari I, Robetto S, Di Felice E, Orusa R, et al. Molecular evidence of kobuviruses in free-ranging red foxes (Vulpes vulpes). Arch Virol. (2014) 159:1803–6. doi: 10.1007/s00705-014-1975-9
23. Olarte-Castillo XA, Heeger F, Mazzoni CJ, Greenwood AD, Fyumagwa R, Moehlman PD, et al. Molecular characterization of canine kobuvirus in wild carnivores and the domestic dog in Africa. Virology. (2015) 477:89–97. doi: 10.1016/j.virol.2015.01.010
24. Melegari I, Sarchese V, Di Profio F, Robetto S, Carella E, Bermudez Sanchez S, et al. First molecular identification of kobuviruses in wolves (Canis lupus) in Italy. Arch Virol. (2018) 163:509–13. doi: 10.1007/s00705-017-3637-1
25. Reuter G, Nemes C, Boros A, Kapusinszky B, Delwart E, Pankovics P. Porcine kobuvirus in wild boars (Sus scrofa). Arch Virol. (2013) 158:281–2. doi: 10.1007/s00705-012-1456-y
26. Milićević V, Kureljušić B, Maksimović-Zorić J, Savić B, Spalević L, Žutić J. Molecular detection and characterization of Porcine Kobuvirus in domestic pigs and wild boars in Serbia. Res Vet Sci. (2020) 132:404–6. doi: 10.1016/j.rvsc.2020.07.028
27. Di Martino B, Di Profio F, Melegari I, Di Felice E, Robetto S, Guidetti C, et al. Molecular detection of kobuviruses in European roe deer (Capreolus capreolus) in Italy. Arch Virol. (2015) 160:2083–6. doi: 10.1007/s00705-015-2464-5
28. Reuter G, Boldizsár A, Pankovics P. Complete nucleotide and amino acid sequences and genetic organization of Porcine Kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch Virol. (2009) 154:101–8. doi: 10.1007/s00705-008-0288-2
29. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
30. Cao W, Zheng H, Zhang K, Jin Y, Lv L, Yang F, et al. Complete genome sequence of the porcine kobuvirus variant CH/HNXX-4/2012. J Virol. (2012) 86:11947. doi: 10.1128/JVI.02137-12
31. Di Profio F, Ceci C, Di Felice E, Marsilio F, Di Martino B. Molecular detection of porcine kobuviruses in Italian swine. Res Vet Sci. (2013) 95:782–5. doi: 10.1016/j.rvsc.2013.06.020
32. Ribeiro J, de Arruda Leme R, Alfieri AF, Alfieri AA. High frequency of Aichivirus C (Porcine Kobuvirus) infection in piglets from different geographic regions of Brazil. Trop Anim Health Prod. (2013) 45:1757–62. doi: 10.1007/s11250-013-0428-x
33. Theuns S, Vanmechelen B, Bernaert Q, Deboutte W, Vandenhole M, Beller L, et al. Nanopore sequencing as a revolutionary diagnostic tool for porcine viral enteric disease complexes identifies Porcine Kobuvirus as an important enteric virus. Sci Rep. (2018) 8:9830. doi: 10.1038/s41598-018-28180-9
34. Cortey M, Díaz I, Vidal A, Martín-Valls G, Franzo G, Gómez de Nova PJ, et al. High levels of unreported intraspecific diversity among RNA viruses in faeces of neonatal piglets with diarrhoea. BMC Vet Res. (2019) 15:441. doi: 10.1186/s12917-019-2204-2
35. Nantel-Fortier N, Lachapelle V, Letellier A, L'Homme Y, Brassard J. Kobuvirus shedding dynamics in a swine production system and their association with diarrhea. Vet Microbiol. (2019) 235:319–26. doi: 10.1016/j.vetmic.2019.07.023
36. Di Martino B, Di Profio F, Di Felice E, Ceci C, Pistilli MG, Marsilio F. Molecular detection of bovine kobuviruses in Italy. Arch Virol. (2012) 157:2393–6. doi: 10.1007/s00705-012-1439-z
37. Melegari I, Di Profio F, Sarchese V, Martella V, Marsilio F, Di Martino B. First molecular evidence of kobuviruses in goats in Italy. Arch Virol. (2016) 161:3245–8. doi: 10.1007/s00705-016-3017-2
38. Abi KM, Zhang Q, Jing ZZ, Tang C. First detection and molecular characteristics of caprine kobuvirus in goats in China. Infect Genet Evol. (2020) 85:104566. doi: 10.1016/j.meegid.2020.104566
39. Sobhy NM, Armién AG, Wünschmann A, Muldoon D, Goyal SM, Mor SK. Detection and molecular characterization of kobuvirus from diarrheic goats in Minnesota. J Vet Diagn Invest. (2020) 32:873–9. doi: 10.1177/1040638720949475
40. Reuter G, Boros A, Pankovics P. Kobuviruses - a comprehensive review. Rev Med Virol. (2011) 21:32–41. doi: 10.1002/rmv.677
41. Hao L, Chen C, Bailey K, Wang L. Bovine kobuvirus-a comprehensive review. Transbound Emerg Dis. (2020). doi: 10.1111/tbed.13909. [Epub ahead of print].
42. Park SJ, Kim HK, Moon HJ, Song DS, Rho SM, Han JY, et al. Molecular detection of porcine kobuviruses in pigs in Korea and their association with diarrhea. Arch Virol. (2010) 155:1803–11. doi: 10.1007/s00705-010-0774-1
43. Almeida PR, Lorenzetti E, Cruz RS, Watanabe TT, Zlotowski P, Alfieri AA, et al. Diarrhea caused by rotavirus A, B, and C in suckling piglets from southern Brazil: molecular detection and histologic and immunohistochemical characterization. J Vet Diagn Invest. (2018) 30:370–6. doi: 10.1177/1040638718756050
44. Di Martino B, Di Felice E, Ceci C, Di Profio F, Marsilio F. Canine kobuviruses in diarrhoeic dogs in Italy. Vet Microbiol. (2013) 166:246–9. doi: 10.1016/j.vetmic.2013.05.007
45. Di Martino B, Di Profio F, Melegari I, Marsilio F, Martella V. Detection of feline kobuviruses in diarrhoeic cats, Italy. Vet Microbiol. (2015) 176:186–9. doi: 10.1016/j.vetmic.2015.01.012
Keywords: kobuvirus, wild boar, goat, chamoise, Aichivirus B, Aichivirus C
Citation: Di Martino B, Di Profio F, Robetto S, Fruci P, Sarchese V, Palombieri A, Melegari I, Orusa R, Martella V and Marsilio F (2021) Molecular Survey on Kobuviruses in Domestic and Wild Ungulates From Northwestern Italian Alps. Front. Vet. Sci. 8:679337. doi: 10.3389/fvets.2021.679337
Received: 11 March 2021; Accepted: 17 May 2021;
Published: 14 June 2021.
Edited by:
Jesus Hernandez, Consejo Nacional de Ciencia y Tecnología (CONACYT), MexicoReviewed by:
Muhammad Zubair Shabbir, University of Veterinary and Animal Sciences, PakistanAbdelaziz Ed-Dra, Zhejiang University, China
Copyright © 2021 Di Martino, Di Profio, Robetto, Fruci, Sarchese, Palombieri, Melegari, Orusa, Martella and Marsilio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara Di Martino, bdimartino@unite.it
 Barbara Di Martino
Barbara Di Martino Federica Di Profio
Federica Di Profio Serena Robetto2
Serena Robetto2  Vittorio Sarchese
Vittorio Sarchese Andrea Palombieri
Andrea Palombieri Vito Martella
Vito Martella Fulvio Marsilio
Fulvio Marsilio