Salmonella Excludes Salmonella in Poultry: Confirming an Old Paradigm Using Conventional and Barcode-Tagging Approaches
- 1Department of Poultry Science, University of Arkansas, Fayetteville, AR , United States
- 2Department of Veterinary Pathobiology, Texas A&M University, College Station, TX, United States
- 3Facultad de Medicina Veterinaria y Zootecnia, Universidad Nacional Autónoma de México, Mexico City, Mexico
- 4Cell and Molecular Biology Program, University of Arkansas, Fayetteville, AR, United States
- 5Center of Food Safety, Department of Food Science, University of Arkansas, Fayetteville, AR, United States
Salmonella is one of the major foodborne bacterial pathogens, and the consumption of contaminated chicken meats isa primary route of Salmonella transmission into human food chains. However, the mechanism of Salmonella transmission within the chicken flock is not fully understood, including competition among Salmonella strains during chicken infection. The purpose of the present study was to evaluate the competitive exclusion (CE) between different or same Salmonella species consecutively challenged through the oral route. Two different approaches were used to evaluate the CE effect, including tracking Salmonella colonization by wild-type strains with difference in natural antibiotic resistance or DNA barcode-tagged isogenic strains. When day-of-hatch chicks were administered by wild-type S. Typhimurium (ST) on day 1, followed by infection on day 2 by S. Enteritidis (SE) or vice versa, most of the birds were colonized only by the first strains administered (82% by ST or 83% by SE). When similar experiments were performed using two different isogenic barcode-tagged SE strains, Illumina sequencing analysis of the barcode region showed that the first barcode-tagged strains administered were dominant strains, ranging from 92 to 99% of the Salmonella recovered from ceca. These results provide quantitative evidence supporting the CE theory that oral administration of Salmonella will produce predominant inhibition over the subsequent colonization of ceca by the following administration one day later by different or same Salmonella species. We also showed that the use of barcode-tagged isogenic strains in combination with deep profiling of barcodes by Illumina sequencing can serve as a quantitative method for studying complex dynamics of Salmonella infection, transmission and colonization in poultry.
Introduction
Due to the common foodborne illness cases caused by Salmonella, prevention of Salmonella colonization in the gastrointestinal tract (GIT) of chickens is necessary. Because normal gut microbiota are not fully developed until 3–6 weeks of age, chicks are particularly vulnerable and susceptible to infection by Salmonella (1, 2). The live attenuated Salmonella vaccine strain has been identified as an effective approach for controlling gut colonization by pathogenic Salmonella. These vaccine strains are believed to reduce the ability of Salmonella to colonize in the chickens primarily via stimulating cell-mediated immunity (3, 4). However, another possibility is that vaccination may do so via modulating the diversity and structure of gut microbiome in the chicken (5). Crhanova et al. (6) also suggested that attenuated Salmonella vaccines are able to modify the chicken gut microbiota, enhance the maturity of gut immune system, and subsequently increase resistance to infection by pathogenic strains. However, these may not be the only mechanisms by which a decrease in Salmonella colonization occurs in the chickens. As compared to stimulation of the immune system and modulation of gut microbiota, which takes a longer time, competitive exclusion (CE) mechanism might be acting for immediate effect in suppressing other Salmonella strains (6). Methner et al. (7) proposed that vaccination of chicks at day one post-hatch ensures colonization by the live attenuated Salmonella vaccine strain, which produces an inhibitory effect and stimulates the development of an immunological response to the following infection (7).
Barrow et al. (8) also demonstrated that oral administration of live strains of virulent Salmonella to day-old chicks produced inhibition in the subsequent cecal colonization by Salmonella Typhimurium (ST) strain administered one day later. Interestingly, closely related enterobacteria were unable to induce the same effect (8). Rabsch et al. (9), used mathematical models that combined epidemiology and population biology to postulate a theory that Salmonella Enteritidis (SE) occupied the ecological niche vacated by eradication of Salmonella Gallinarum (SG) and Salmonella Pullorum (SP) from poultry (9). The theory suggests that SG was able to competitively exclude SE from poultry flocks in the early 20th century, and the elimination of SG and SP in poultry led to an epidemic increase of SE in poultry and human infections in the 1980’s. Protection against colonization by SE, but not ST, through immunization of chickens with SG was possible because both SG and SE possess the same immune-dominant O-antigen on their cell surfaces (10–12). Holt and Gast (13) reported that prior infection of hens with ST or Salmonella Muenchen (SM) reduced the infection by SE, which indicated that cross-serovar protection occurred among ST, SM and SE (13). Growth inhibition by different serovars may also occur in a serovar-specific manner through inhibitory metabolites. For example, in an in vitro experiment conducted by Calo et al. (14), decreased growth of ST occurred in spent media originating from S. Heidelberg growth cultures, but not from other serovars (14).
CE is a theory in ecology, which states that two closely related species that compete for the same resources cannot stably occupy the same ecological niche. This CE concept has been explored extensively as an effective strategy to control Salmonella in poultry since the landmark research by Nurmi and Rantala (15). To better describe CE phenomenon that may exist between different or same Salmonella serovars accurately, it is important to employ a quantitatively approach. For evaluation of CE theory, strains carrying different antibiotic markers have been used widely to differentiate two different strains from tissues or organs with mixed infections. However, these marker strains may not behave exactly in the same manner as the corresponding wild type strains due to the phenotypic changes caused by antibiotic markers (16). Hence, it is critical to use the isogenic strains phenotypically identical for studying CE to decrease the bias from different infection strains. We have constructed a set of isogenic SE barcode-tagged strains in which unique DNA barcodes were inserted in a functionally neutral locus in the genome of SE and the resulting strains can be used to quantitatively track the colonization by the respective strains by profiling the barcode-regions using Illumina sequencing method (16). The main advantage of using these barcode-tagged strains over previously used marker strains is that each strain can be tracked quantitatively within the entire population of barcode-tagged strains at high accuracy. This similar strategy has been used in several studies to allow quantitative profiling among multiple barcode-tagged strains as well as discrimination from the environmental bacteria or viruses without altering phenotypes or behaviors during infection, colonization and dissemination (16–20). The purpose of the present study was to evaluate the CE responses of Salmonella consecutively challenged in chickens using both conventional and barcode-tagging approaches.
Materials and Methods
Experiment 1: Recovery of Salmonella After Consecutive Salmonella Serovar Challenges in Day-Old Lenghorn Chicks (Trials 1 and 2)
Salmonella cultures. A highly invasive poultry isolate of SE was obtained from the USDA National Veterinary Services Laboratory (Ames, IA 50011). A spontaneous mutant that is resistance to Nalidixic acid (NA; Sigma, St. Louis, MO) was selected and used in this study. This strain was also found to be naturally sensitive to gentamicin (GM10) on antibiotic sensitivity discs. A spontaneous mutant of an invasive isolate of ST (ATCC13311) that is resistance to NA was also selected and used in this study. This ST was found to be resistant to GM10. Both strains of SE and ST were inherently resistance to Novobiocin (NO). In Experiment 1, we conveniently used the difference in the sensitivity to gentamicin between SE (GM10S) and ST (GM10R) to differentiate these two strains. For the present studies, 100 µL of SE or ST from a frozen stock was added to 10 mL of tryptic soy broth (TSB, Becton Dickinson, Sparks, MD) and incubated at 37°C for 8 h. This was followed by 3 passages every 8 h into fresh TSB. After incubation, bacterial cells were washed 3 times in sterile 0.9% saline by centrifugation (1,864 × g, 4°C, 15 min), and quantified with a spectrophotometer (Spectronic 20D+, Thermo Scientific, Waltham, MA) at 625 nm using an established standard curve. The cell suspensions were then diluted in sterile 0.9% saline as per required concentrations for the corresponding trials. Concentrations of SE or ST were also determined retrospectively by plating appropriate serial dilutions onto brilliant green agar (BGA, Sigma) supplemented with NO (25 µg/mL) and NA (20 µg/mL) for enumeration of actual CFU/mL used for the challenge studies.
Experimental birds. Naïve, day-of hatch, single comb white Leghorn male chicks obtained from a local hatchery were randomly placed in cages (n = 30 for each Control group or n = 60 for Treatment group for Trial 1; n = 20 for each Control group or n = 40 for Treatment group for Trial 2) within electrically heated starter batteries. The cages were located within a modern biological hazard isolation unit on the research farm of the College of Veterinary Medicine, Texas A & M University (College Station, TX). Chicks were provided ad libitum access to water and a balanced unmedicated corn-soybean diet meeting the nutrition requirements of poultry recommended by the NRC (1994). Adequate body temperature was maintained using heat lamps placed within the isolators. This study was carried out in accordance with the Guideline for the Care and Use of Agricultural Animals in Teaching and Research (Federation of Animal Science Societies), and the protocol was approved by the Institutional Animal Care and Use (IACUC) Committee at Texas A&M University. To check the presence of Salmonella, a subset of chicks for each trial were humanely killed, transported and sampled upon arrival at the laboratory. Whole ceca-cecal tonsils (CCT), liver, spleen and yolk sac were aseptically removed from these neonatal chicks, incised, and cultured in 10 mL of tetrathionate enrichment broth (TEB) (Tet, Becton Dickinson, Sparks, MD) and incubated overnight at 37°C. The samples were confirmed negative for Salmonella by plating them onto selective BGA plates.
Experimental design For Trial 1, chicks in Treatment group were ora lly gavaged with 104 CFU ST on day 1, and consecutively with 105 CFU SE on day 2. For the control groups, two groups of chicks were orally challenged only with 104 CFU ST on day 1 (ST control) or 105 CFU SE on day 2 (SE control) (see Tables 1 and 2). On day 3, chickens were euthanized and cultured for recovery of Salmonella in CCT. To determine the incidence and frequency of SE and ST in CCT, whole CCT was enriched in TEB and incubated for 24 h at 37°C. Samples were taken from the enriched broth and subcultured on BGA plates containing 20 µg/mL NA and 25 µg/mL NO for approximately 18 h at 37°C. For the control groups, approximately 10 isolated colonies per bird were taken from each BGA and streaked as separate lines onto Mueller-Hinton plates 19. For the treatment group, approximately 20 isolated colonies per bird were streaked in the same manner onto Mueller-Hinton plates. An antibiotic disc of GM10 was placed on each streaked line of colony inoculation and the results tabulated 24 h later as either Resistant (indicating ST) or Sensitive (indicating SE). From Experiment 1, total 20 isolates were randomly selected and serogrouped using commercially available antisera to verify the accuracy of serovar identification based on sensitivity or resistance to GM10. Trial 2 was performed in the same manner as Trial 1 except that SE was used for the first challenge (104 CFU) on day one, which was followed by ST challenge (105 CFU) on day 2 (see Tables 1 and 2).
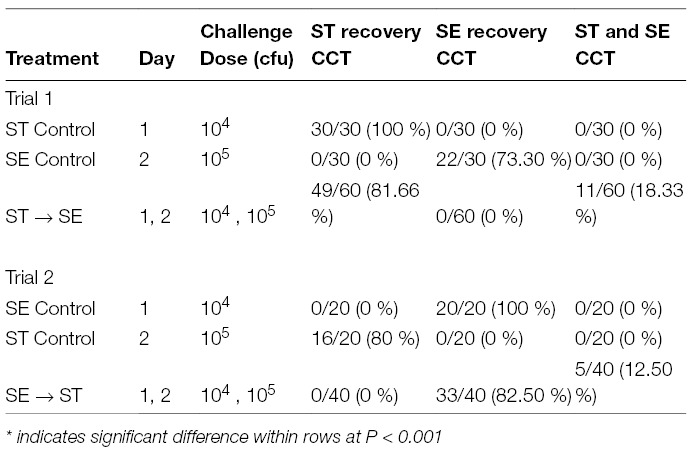
Table 1. Recovery of Salmonella in whole ceca-cecal tonsils (CCT) after consecutive Salmonella serovar challenges in day-old Lenghorn chicks in Trial 1 and Trial 2 of Experiment 1. Data is expressed as number of Salmonella (ST, SE or both) culture positive birds/ total number birds tested (%).
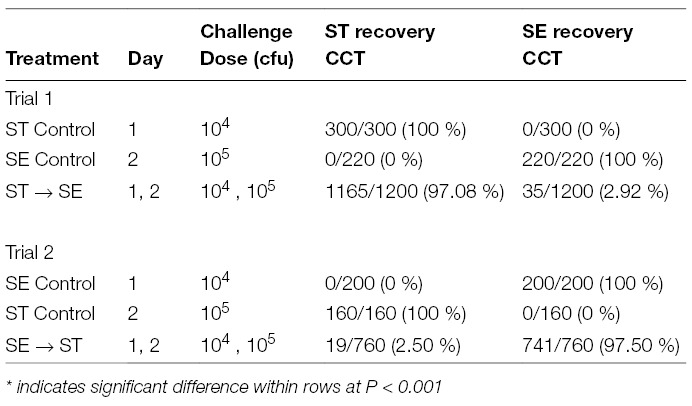
Table 2. Salmonella isolates recovered and serotyped in whole ceca-cecal tonsils (CCT) after consecutive Salmonella serovar challenges in day-old Lenghorn chicks in Trial 1 and Trial 2 of Experiment 1. Data is expressed as number of ST or SE isolates/ total number Salmonella isolates tested (%).
Experiment 2: Recovery of Barcode-Tagged Isogenic SE Strains After Consecutive Challenges in Day-Old Broiler Chicks
Construction of barcode-tagged SE strains. The method for construction of barcode-tagged strains was described previously (16). Briefly, SE 13A strain containing pKD46 that expresses the Red recombinase system was used for construction of barcode-tagged strains via electroporation. Overlapping extension PCR was used to join the three PCR products corresponding to upstream fragment (of the insertion site) plus a 6 nt random barcode, Km resistance gene and downstream fragment (of the insertion site). After electroporation, the mutants carrying the barcode sequence along with the kanamycin resistance gene inserted into a functionally neutral intergenic region between SEN1521 and SEN1522 were selected and used in this study as previously described in details (16).
Bacterial strains and culture condition. Two SE barcode-tagged isogenic strains (hereafter, BC1 and BC2) were incubated in Luria-Bertani (LB) broth supplemented with kanamycin (50 µg/mL) overnight at 37°C, and were harvested by centrifugation at 4°C. The cell pellet was washed three times and resuspended in distilled 0.9% saline. A suspension of 108 CFU/mL was obtained by using a spectrophotometer to adjust OD625 = 0.147. The cell suspensions were subsequently diluted to 105 CFU/mL for chick infection.
Experimental birds. Day-of-hatch, male broiler chicks obtained from Cobb-Vantress (Siloam Springs, AR) were placed in floor pens with a controlled age-appropriate environment. Chicks were provided ad libitum access to water and a balanced unmedicated corn-soybean diet meeting the nutrition requirements of poultry recommended by the NRC (1994). Adequate body temperature was maintained using heat lamps placed within the isolators. All animal handling procedures were in compliance with the Guideline for the Care and Use of Agricultural Animals in Teaching and Research (Federation of Animal Science Societies), and the experimental protocol was approved by IACUC Committee at the University of Arkansas. Twelve chickens for each trial were euthanized and sampled upon arrival at the laboratory to confirm that the chicks were Salmonella-negative as described in Experiment 1.
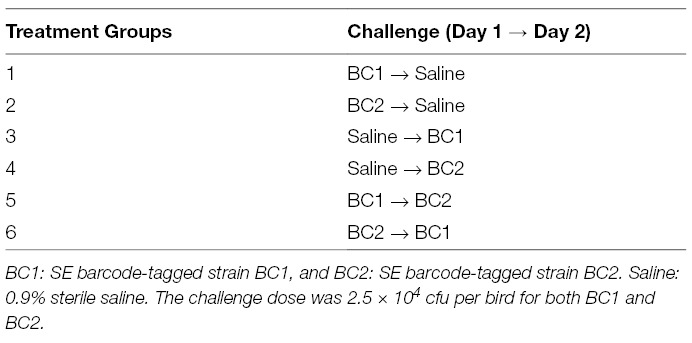
Experimental design. This experiment was set up to confirm if oral administration with one SE barcode-tagged strain inhibits the colonization by the other SE barcode-tagged strain in the ceca of chickens. A total of 90 day-of-hatch broiler chicks were randomly separated into six groups (n = 15 chicks/group). Description of the 6 treatment groups are shown in Table 3. On day three, 12 chickens from each group were euthanized. CCT and liver/spleen were collected, macerated, and suspended in 0.9% saline in 1:4 ratio in sterile bags. One mL of suspension from each bag was collected for genomic DNA isolation, and 100 µL of suspension from each bag was used for serial dilution and enumeration of CFU using BGA agar plates. Two-fold volume of tetrathionate broth (TET) was added into the remaining suspension for enrichment and detection of the positive/negative presence of Salmonella in each sample.
Statistical Analysis
The data expressed as positive/total chicks in % and the % recovery of ST or SE were compared using the chi-squared test of independence to determine the significance (P ≤ 0.001) (21). Barcode % recovery data within experimental groups were subjected to one way ANOVA (SAS Institute, 2002). Barcode % recoveries were expressed as means and considered significant at P ≤ 0.001.
Results
Experiment 1: Recovery of Salmonella After Consecutive Salmonella Serovar Challenges in Day-Old Lenghorn Chicks (Trials 1 and 2)
The recovery results of Salmonella in CCT from consecutive challenges with different Salmonella serovars in day-old Leghorn chicks in Trial 1 and Trial 2 are shown in Table 1. This table shows the number of Salmonella culture positive birds (ST positive, SE positive, or ST & SE positive) per total number birds tested. For both ST control and SE control groups in both trials, only the serovars that were used for oral administration were exclusively recovered from CCT. However, when ST was administered on day one at 104 CFU, followed by the consecutive challenge of SE twenty-four hours later at 105 CFU (ST→SE group in Trial 1), the birds were predominantly colonized by ST alone (81.66%; P < 0.001). On the contrary, in no case was SE alone recovered from any bird. Those colony isolates of SE came only from birds with mixed infections of ST and SE. In addition, when the percentage of ST or SE in all tested Salmonella colony isolates were determined within each group, ST and SE recovery were 97.08 and 2.92%, respectively, in the ST→SE group. On the contrary, it was 100% ST or 100% SE in the respective control groups (Table 2).
Similar results were observed in Trial 2, when chickens were challenged with SE on day one at 104 CFU followed by the consecutive oral challenge of ST on day two at 105 CFU (SE→ST group). In Trial 2, 82.5% of the birds were positive for SE only, the first serovar administered (Table 1). In the same SE→ST group, colonies isolated as SE from CCT were 97.5% as compared to ST colonies at 2.5% (Table 2).
Experiment 2: Recovery of Barcode-Tagged Isogenic SE Strains After Consecutive Challenges in Day-Old Broiler Chicks
The results of the percentage Salmonella barcode strains recovered from cecal samples enumerated from Illumina sequence data in day-old broiler chicks in Experiment 2 are summarized in Figure 1 (cecal samples) and Figure 2 (liver/spleen samples). A total of 3,138,578 sequence reads of 167 bp was obtained from the MiSeq sequencing run. The sequence reads were binned into different files according to the combinatorial index sequences corresponding to the samples from the six treatment groups (Table 3). We demanded perfect matches to the 6-nt six barcode sequences, discarding any reads without perfectly matching barcodes. The read numbers reflect only relative frequency of each barcode-tagged strain in a given sample. Therefore, the original read numbers were converted to the percentage of each barcode-tagged strain in each sample. The result from CCT indicated that oral gavage of BC1 on day one followed by saline on day two in Group 1 resulted in 100% BC1 recovery (Figure 1). Similarly, in Group 2, administration of BC2 on day one followed by saline on day two in Group 2 resulted in 84% BC2 recovery. However, oral gavage of BC1 on day one and BC2 on day two in Group 5 resulted in 99% BC1 recovery while only 1% of BC2 was recovered. The opposite effect was observed when BC2 was administered first with 92% recovery of BC2 and only 8% of BC1 recovered from CCT in Group 6. However, both BC1 and BC2 strains were isolated from ceca in the chicks from Group 3 and 4, even though only one single SE barcode-tagged strain was introduced on day two (Figure 1). The level of recovered Salmonella from CCT in Groups 1, 2, 5, and 6 (6.55 ± 0.22 CFU/mla; 5.57 ± 0.85 a; 4.87 ± 0.87 ab; 3.31 ± 1.03 bc, respectively) was higher than Group 3 and 4 (0.42 ± 0.42 CFU/mld; 1.73 ± 0.64 dc, respectively). The enrichment result from CCT exhibited a similar tendency: the percentage of Salmonella positive chicks from Groups 1, 2, 5, and 6 (100%, 83.33%, 75 and 50%, respectively) was higher than Group 3 and 4 (33.33% and 58.33%, respectively).
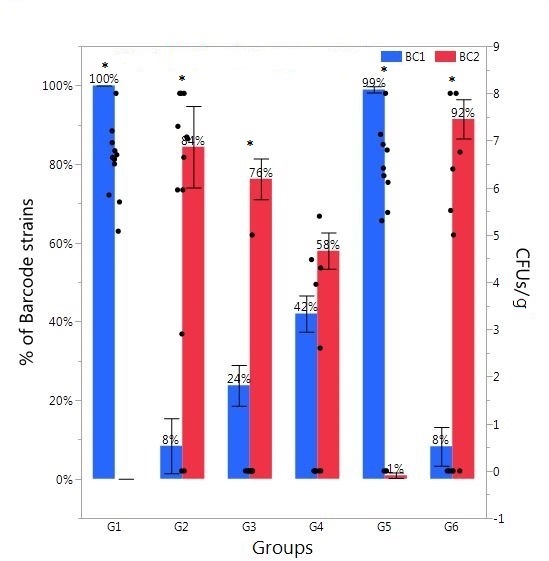
Figure 1. Relative abundance of Salmonella barcode-tagged strains (BC1 and BC2) in cecal samples in Experiment 2. The relative abundance of each barcode-tagged strain (BC1 in blue bars, and BC2 in red bars) in % (left Y axis) was determined from MiSeq data. Black dots (right Y axis) indicate CFU/g of cecal contents as determined by plating of serial dilutions. The black dots corresponding to 0 CFU/g indicate the samples in which Salmonella level was lower than detection limit. Day-of-hatch chickens (n = 12/group) were orally gavaged in Group 1 (G1) with BC1 (Day 1) and saline (Day 2); in G2 with BC2 (Day 1) and saline (Day 2); in G3 with saline (Day 1) and BC1 (Day 2); in G4, with saline (Day 1) and BC2 (Day 2), in G5 with BC1(Day 1) and BC2 (Day 2), and in G6 with BC2 (Day 1) and BC1 (Day 2). For all oral gavage with either BC1 or BC2, each chick received 2.5 × 104 CFU. Asterisk (*) on the top of the bars indicates significant difference between BC1 and BC2 within the same group at P < 0.01.
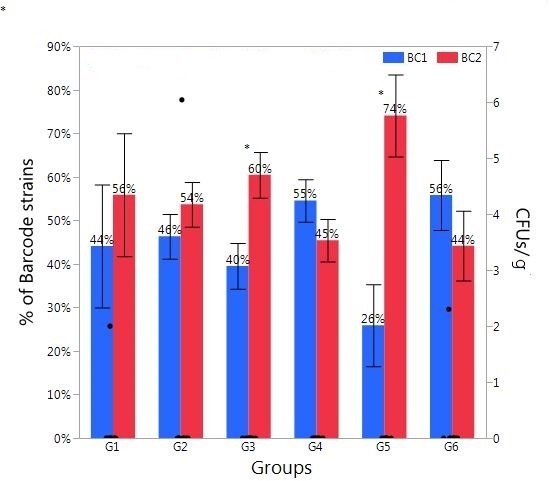
Figure 2. Relative abundance of Salmonella barcode-tagged strains (BC1 and BC2) in liver/spleen samples in Experiment 2. The relative abundance of each barcode-tagged strain (BC1 in blue bars, and BC2 in red bars) in % (left Y axis) was determined from MiSeq data. Black dots (right Y axis) indicate CFU/g of liver/spleen samples as determined by plating of serial dilutions. The black dots corresponding to 0 CFU/g indicate the samples in which Salmonella level was lower than detection limit. The treatment groups G1-G6 are the same to those in Figure 1. Asterisk (*) on the top of the bars indicates significant difference between BC1 and BC2 within the same group at P < 0.01.
Discussion
According to Disease Outbreak Surveillance System from 1998 to 2012, poultry was associated with 279 (25%) out of 1114 outbreaks in which the implicated source could be traced back to one food category, accounting for the highest number of outbreaks, illnesses, and hospitalizations. Among those 149 poultry-associated outbreaks were caused by a confirmed pathogen, and Salmonella enterica (43%) was the most common pathogens (22). Hence, understanding the mechanisms of infection of Salmonella in poultry is critical in order to find alternative methods to antibiotics that can eliminate or reduce this pathogen from poultry and poultry products.
Several investigators have demonstrated a significant protection from the second Salmonella challenge with sequential administration of Salmonella serovars in mice (23) or chicks (24–26). Early studies indicated that prior intravenous infection with ST could protect mice from the intravenous challenge of another Salmonella serovar (27, 28). Similar investigations have demonstrated that intravenous challenge of SG caused protection against subsequent intravenous challenge with SE (29). In their study, Collins et al. (29) demonstrated that SG was able to persist in the tissues therefore protecting against SE challenge. However, SP, an antigenically similar organism, was unable to establish within the tissues which apparently allowed SE to colonize. In addition, live attenuated Salmonella vaccines provided protection from subsequent Salmonella challenges within 4 weeks of vaccination (25, 30, 31). Similarly, the results of the present study confirm those by Barrow et al. (8) in which day-old chicks which received sequential Salmonella challenge resulted in an almost exclusive infection by the first challenge strain within 24 h. Hence, regardless of route of challenge, experimental animal or time intervals between challenges, sequential Salmonella challenges allow chickens to become refractory to the second Salmonella serovar administered. Our results also suggest that Salmonella-infected chicks become refractory to a second challenge serovar within 24 h, confirming previous reports of rapid induction of resistance to consecutive Salmonella challenge.
Several investigators have evaluated Salmonella transmission in commercial poultry flocks using conventional bacteriology and serological methods (32–34). These studies have helped in gaining an understanding on the impact of different phage types or housing systems on the frequency of horizontal transmission. However, comprehensive elucidation of the transmission and pathogenesis mechanisms involving interactions among multiple serovars cannot be delineated using the traditional culture methods. In the present study, two barcode-tagged SE strains were used to investigate transmission dynamics of Salmonella in chickens quantitatively after consecutive challenges. These strains have served as an initial conceptual proof to quantitatively track the Salmonella transmission routes from environment to flock, since they carry distinct barcode tags that allow them to be identified unambiguously and quantified accurately by Illumina sequencing of the barcode regions (16).
In summary, utilizing isogenic barcode-tagged strains, the population structure can be quantified to evaluate the patterns of SE infection and dissemination in chickens, and determine whether infection of neonatal chicks with one Salmonella strain excludes the infection by a second strain. Our Illumina sequence data indicated that any BC strain used for infection on day one became the predominant strain, whether there was a second infection by the other BC strain (Group 5 and 6 in Figure 1) or not (Group 1 and 2 in Figure 1). Unexpectedly, there was a mixed population of two BC strains when the first infection by BC strain was delayed to day 2 (Group 3 and 4 in Figure 1). In case of Group 3 and 4, the chicks were infected only by one BC strain (BC1 and BC2, respectively) on day 2, but both BC strains could be recovered at significant levels. It is important to note that the level of recovered Salmonella from CCT was significantly lower in Group 3 (0.42 ± 0.42 CFU/ml) and 4 (1.73 ± 0.64 CFU/ml) as compared to other groups (≥3.31 ± 1.03). The result indicates the possible aerosol transmission may exist from other groups in the same isolation room through the respiratory tract (35).
Based on the dilution, counting and enrichment method, the Salmonella level was too low to be detected for most liver/spleen samples (data not shown). However, both Salmonella BC strains could be detected in most of the liver/spleen samples by using PCR and sequencing (Figure 2). This result indicated that this strategy is more sensitive than the traditional culture method. Surprisingly, all groups had significant levels of both BC1 (≥26%) and BC2 (≥44%) (Figure 2), whether it was infected by one or two BC strains. This also provides additional evidence for the aerosol transmission through the respiratory tract and it indicates that dissemination into internal organs (liver and spleen) might be more efficient than dissemination into ceca after infection through the respiratory tract. There is a possibility that once Salmonella infection appears in the tracheal route, they may migrate into different body sites more rapidly with higher efficiency than infection through the oral route. Salmonella can replicate in the respiratory macrophages and transport from the lungs to the secondary lymphoid organs, such as spleen, and spread systemically to liver and potentially to cecal tonsil later (36, 37).
Alternative explanations for the result presented in Figure 2 include inaccurate differentiation between BC1 and BC2 or instability of barcode-tags. However, we used 2-step PCR to amplify barcode regions using 2 sets of primers, enhancing specificity of amplification and ensuring barcode tags recovered are from the originally inserted chromosomal locus. When the sequence reads were processed, we demanded perfect matches to 6nt barcode sequences, discarding any reads with no perfect matches. For these reasons, we argue that the observed barcode profiles accurately reflect relative abundance of BC1 and BC2 in the given samples. It is also important to note that we cannot exclude the possibility that the barcode sequences could have been amplified from dead cells or DNA, based on the fact that most liver/spleen samples were Salmonella-negative on enrichment.
In order to better comprehend the implications of CE and intratracheal infection of Salmonella in commercial poultry, larger scale experiments are necessary to assess additional environmental and host factors. Nevertheless, the current experiment further confirmed that the use of barcode-tagged strains is an original and an effective method to understand the dynamics of Salmonella infection, which provides valuable opportunities to develop and improve effective measures to control Salmonella in poultry flocks. Currently, studies to evaluate and confirm our previous work (35) that demonstrated the importance of airborne transmission of Salmonella via an intratracheal route versus oral infection are conducted using these SE barcode-tagged strains.
Ethics Statement
All animal handling procedures were in compliance with Institutional Animal Care and Use Committees at Texas A & M University, and University of Arkansas.
Author Contributions
GT, BH, and YK designed the experiments. YY and PR conducted experiments and analyzed the data. YY, GT, JL, XH, and SR wrote the manuscript. YY and YMK revised the manuscript.
Funding
This study was conducted by the funding support from Arkansas Biosciences Institute (ABI).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Barnes EM, Mead GC, Barnum DA, Harry EG, Ranjitkar S, Lawley B. The intestinal flora of the chicken in the period 2 to 6 weeks of age, with particular reference to the anaerobic bacteria. Br Poult Sci (1972) 13(3):311–26. doi: 10.1080/00071667208415953
2. Ranjitkar S, Lawley B, Tannock G, Engberg RM. Bacterial succession in the broiler gastrointestinal tract. Appl Environ Microbiol (2016) 82(8):2399–410. doi: 10.1128/AEM.02549-15
3. Lillehoj EP, Yun CH, Lillehoj HS. Vaccines against the avian enteropathogens Eimeria, Cryptosporidium and Salmonella. Anim Health Res Rev (2000) 1(1):47–65. doi: 10.1017/S1466252300000050
4. Babu U, Dalloul RA, Okamura M, Lillehoj HS, Xie H, Raybourne RB, et al. Salmonella enteritidis clearance and immune responses in chickens following Salmonella vaccination and challenge. Vet Immunol Immunopathol (2004) 101(3-4):251–7. doi: 10.1016/j.vetimm.2004.05.002
5. Park SH, Kim SA, Rubinelli PM, Roto SM, Ricke SC. Microbial compositional changes in broiler chicken cecal contents from birds challenged with different Salmonella vaccine candidate strains. Vaccine (2017) 35(24):3204–8. doi: 10.1016/j.vaccine.2017.04.073
6. Crhanova M, Hradecka H, Faldynova M, Matulova M, Havlickova H, Sisak F, et al. Immune response of chicken gut to natural colonization by gut microflora and to Salmonella enterica serovar enteritidis infection. Infect Immun (2011) 79(7):2755–63. doi: 10.1128/IAI.01375-10
7. Methner U, Barrow PA, Berndt A, Steinbach G. Combination of vaccination and competitive exclusion to prevent Salmonella colonization in chickens: experimental studies. Int J Food Microbiol (1999) 49(1-2):35–42. doi: 10.1016/S0168-1605(99)00051-3
8. Barrow PA, Tucker JF, Simpson JM. Inhibition of colonization of the chicken alimentary tract with Salmonella typhimurium gram-negative facultatively anaerobic bacteria. Epidemiol Infect (1987) 98(3):311–22. doi: 10.1017/S0950268800062063
9. Rabsch W, Hargis BM, Tsolis RM, Kingsley RA, Hinz KH, Tschäpe H, et al. Competitive exclusion of Salmonella enteritidis by Salmonella gallinarum in poultry. Emerging Infect Dis (2000) 6(5):443–8. doi: 10.3201/eid0605.000501
10. Silva EN, Snoeyenbos GH, Weinack OM, Smyser CF. Studies on the use of 9R strain of Salmonella gallinarum as a vaccine in chickens. Avian Dis (1981) 25(1):38–52. doi: 10.2307/1589825
11. Barrow PA, Berchieri A, Al-Haddad O. Serological response of chickens to infection with Salmonella gallinarum-S. pullorum detected by enzyme-linked immunosorbent assay. Avian Dis (1992) 36(2):227–36. doi: 10.2307/1591495
12. Nassar TJ, Al-Nakhli HM, Al-Ogaily ZH. Use of live and inactivated Salmonella enteritidis phage type 4 vaccines to immunise laying hens against experimental infection. Rev - Off Int Epizoot (1994) 13(3):855–67. doi: 10.20506/rst.13.3.804
13. Holt PS, Gast RK. Effects of prior coinfection with different Salmonella serovars on the progression of a Salmonella enterica serovar enteritidis infection in hens undergoing induced molt. Avian Dis (2004) 48(1):160–6. doi: 10.1637/7101
14. Calo JR, Park SH, Baker CA, Ricke SC. Specificity of Salmonella Typhimurium strain (ATCC 14028) growth responses to Salmonella serovar-generated spent media. J Environ Sci Health B (2015) 50(6):422–9. doi: 10.1080/03601234.2015.1011962
15. Nurmi E, Rantala M. New aspects of Salmonella infection in broiler production. Nature (1973) 241(5386):210–1. doi: 10.1038/241210a0
16. Yang Y, Ricke SC, Tellez G, Kwon YM. Quantitative tracking of Salmonella enteritidis transmission routes using barcode-tagged isogenic strains in chickens: proof-of-concept study. Front Vet Sci (2017) 4:15. doi: 10.3389/fvets.2017.00015
17. Troy EB, Lin T, Gao L, Lazinski DW, Camilli A, Norris SJ, et al. Understanding barriers to Borrelia burgdorferi dissemination during infection using massively parallel sequencing. Infect Immun (2013) 81(7):2347–57. doi: 10.1128/IAI.00266-13
18. Varble A, Albrecht RA, Backes S, Crumiller M, Bouvier NM, Sachs D, et al. Influenza A virus transmission bottlenecks are defined by infection route and recipient host. Cell Host Microbe (2014) 16(5):691–700. doi: 10.1016/j.chom.2014.09.020
19. Lam LH, Monack DM. Intraspecies competition for niches in the distal gut dictate transmission during persistent Salmonella infection. PLoS Pathog (2014) 10(12):e1004527. doi: 10.1371/journal.ppat.1004527
20. Zhang T, Abel S, Abel Zur Wiesch P, Sasabe J, Davis BM, Higgins DE, et al. Deciphering the landscape of host barriers to Listeria monocytogenes infection. Proc Natl Acad Sci USA (2017) 114(24):6334–9. doi: 10.1073/pnas.1702077114
22. Chai SJ, Cole D, Nisler A, Mahon BE. Poultry: the most common food in outbreaks with known pathogens, United States, 1998-2012. Epidemiol Infect (2017) 145(2):316–25. doi: 10.1017/S0950268816002375
23. Collins FM. Cross-protection against Salmonella enteritidis infection in mice. J Bacteriol (1968) 95(4):1343–9.
24. Barrow PA, Hassan JO, Berchieri A. Reduction in faecal excretion of Salmonella typhimurium strain F98 in chickens vaccinated with live and killed S. typhimurium organisms. Epidemiol Infect (1990) 104(3):413–26. doi: 10.1017/S0950268800047439
25. Berchieri A, Barrow PA. Further studies on the inhibition of colonization of the chicken alimentary tract with Salmonella typhimurium by pre-colonization with an avirulent mutant. Epidemiol Infect (1990) 104(3):427–41. doi: 10.1017/S0950268800047440
26. Barrow PA, Page K. Inhibition of colonisation of the alimentary tract in young chickens with Campylobacter jejuni by pre-colonisation with strains of C. jejuni. FEMS Microbiol Lett (2000) 182(1):87–91. doi: 10.1111/j.1574-6968.2000.tb08879.x
27. Howard JG. Resistance to infection with Salmonella paratyphi C in mice parasitized with a relatively avirulent strain of Salmonella typhimurium. Nature (1961) 191:87–8. doi: 10.1038/191087a0
28. Collins FM. Cross-protection against Salmonella enteritidis infection in mice. J Bacteriol (1968) 95(4):1343–9.
29. Collins FM, Mackaness GB, Blanden RV. Infection-immunity in experimental salmonellosis. J Exp Med (1966) 124(4):601–19. doi: 10.1084/jem.124.4.601
30. Cooper GL, Venables LM, Nicholas RA, Cullen GA, Hormaeche CE. Vaccination of chickens with chicken-derived Salmonella enteritidis phage type 4 aroA live oral Salmonella vaccines. Vaccine (1992) 10(4):247–54. doi: 10.1016/0264-410X(92)90160-L
31. Dougan G, John V, Palmer S, Mastroeni P. Immunity to salmonellosis. Immunol Rev (2011) 240(1):196–210. doi: 10.1111/j.1600-065X.2010.00999.x
32. Grant AJ, Restif O, Mckinley TJ, Sheppard M, Maskell DJ, Mastroeni P. Modelling within-host spatiotemporal dynamics of invasive bacterial disease. PLoS Biol (2008) 6(4):e74. doi: 10.1371/journal.pbio.0060074
33. Thomas E, Bouma A, Klinkenberg D. A comparison of transmission characteristics of Salmonella enterica serovar Enteritidis between pair-housed and group-housed laying hens. Vet. Res. (2011) 42:40. doi: 10.1186/1297-9716-42-40
34. Gast RK, Guraya R, Jones DR, Anderson KE. Horizontal transmission of Salmonella Enteritidis in experimentally infected laying hens housed in conventional or enriched cages. Poult Sci (2014) 93(12):3145–51. doi: 10.3382/ps.2014-04237
35. Kallapura G, Morgan MJ, Pumford NR, Bielke LR, Wolfenden AD, Faulkner OB, et al. Evaluation of the respiratory route as a viable portal of entry for Salmonella in poultry via intratracheal challenge of Salmonella Enteritidis and Salmonella Typhimurium. Poult Sci (2014c) 93(2):340–6. doi: 10.3382/ps.2013-03602
36. Kallapura G, Botero A, Layton S, Bielke LR, Latorre JD, Menconi A, et al. Evaluation of recovery of Salmonella from trachea and ceca in commercial poultry. J Appl Poult Res (2014a) 23(1):132–6. doi: 10.3382/japr.2013-00854
Keywords: Salmonella, poultry, competitive exclusion, barcode-tagged isogenic strain, intestinal colonization
Citation: Yang Y, Tellez G, Latorre JD, Ray PM, Hernandez X, Hargis BM, Ricke SC and Kwon YM (2018). Salmonella Excludes Salmonella in Poultry: Confirming an Old Paradigm Using Conventional and Barcode-Tagging Approaches. Front. Vet. Sci. 5:101. doi: 10.3389/fvets.2018.00101
Received: 06 February 2018; Accepted: 23 April 2018;
Published: 16 May 2018
Edited by:
Joy Scaria, South Dakota State University, United StatesReviewed by:
Raghavendra G Amachawadi, Kansas State University, United StatesShankar Thangamani, Midwestern University, United States
Orhan Sahin, Iowa State University, United States
Copyright © 2018 Yang, Tellez, Latorre, Ray, Hernandez, Hargis, Ricke and Kwon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven C. Ricke, sricke@uark.edu
 Yichao Yang
Yichao Yang Guillermo Tellez
Guillermo Tellez Juan D. Latorre
Juan D. Latorre Pamela M. Ray2
Pamela M. Ray2  Xochitl Hernandez
Xochitl Hernandez Billy M. Hargis
Billy M. Hargis Steven C. Ricke
Steven C. Ricke Young Min Kwon
Young Min Kwon