- 1School of Rehabilitation Sciences, Southern Medical University, Guangzhou, China
- 2First Clinical Medical College, Guangzhou University of Chinese Medicine, Guangzhou, China
- 3Medical College of Acupuncture-Moxibustion and Rehabilitation, Guangzhou University of Chinese Medicine, Guangzhou, China
Objective: To determine the diagnostic accuracy of serological tests for coronavirus disease-2019 (COVID-19).
Methods: PubMed, Embase and the Cochrane Library were searched from January 1 2020 to September 2 2022. We included studies that measured the sensitivity, specificity or both qualities of a COVID-19 serological test and a reference standard of a viral culture or reverse transcriptase polymerase chain reaction (RT–PCR). The risk of bias was assessed by using quality assessment of diagnostic accuracy studies 2 (QUADAS-2). The primary outcomes included overall sensitivity and specificity, as stratified by the methods of serological testing [enzyme-linked immunosorbent assays (ELISAs), lateral flow immunoassays (LFIAs) or chemiluminescent immunoassays (CLIAs)] and immunoglobulin classes (IgG, IgM, or both). Secondary outcomes were stratum-specific sensitivity and specificity within the subgroups, as defined by study or participant characteristics, which included the time from the onset of symptoms, testing via commercial kits or an in-house assay, antigen target, clinical setting, serological kit as the index test and the type of specimen for the RT–PCR reference test.
Results: Eight thousand seven hundred and eighty-five references were identified and 169 studies included. Overall, we judged the risk of bias to be high in 47.9 % (81/169) of the studies, and a low risk of applicability concerns was found in 100% (169/169) of the studies. For each method of testing, the pooled sensitivity of the ELISAs ranged from 81 to 82%, with sensitivities ranging from 69 to 70% for the LFIAs and 77% to 79% for the CLIAs. Among the evaluated tests, IgG (80–81%)-based tests exhibited better sensitivities than IgM-based tests (66–68%). IgG/IgM-based CLIA had the highest sensitivity [87% (86–88%)]. All of the tests displayed high specificity (97–98%). Heterogeneity was observed in all of the analyses. The detection of nucleocapsid protein (77–80%) as the antigen target was found to offer higher sensitivity results than surface protein detection (66–68%). Sensitivity was higher in the in-house assays (78–79%) than in the commercial kits (47–48%).
Conclusion: Among the evaluated tests, ELISA and CLIA tests performed better in terms of sensitivity than did the LFIA. IgG-based tests had higher sensitivity than IgM-based tests, and combined IgG/IgM test-based CLIA tests had the best overall diagnostic test accuracy. The type of sample, serological kit and timing of use of the specific tests were associated with the diagnostic accuracy. Due to the limitations of the serological tests, other techniques should be quickly approved to provide guidance for the correct diagnosis of COVID-19.
Introduction
Coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has affected 219 countries and territories, with 614,385,693 confirmed cases; additionally, 6,522,600 deaths have been reported by the World Health Organization last update 30 September 2022. Accurate and rapid diagnostic tests are critical in achieving the global control of COVID-19. There are two main diagnostic tests for COVID-19: molecular tests that detect viral RNA, and serological tests that detect anti-SARS-CoV-2 immunoglobulin (1). Reverse transcription polymerase chain reaction (RT–PCR) is the gold standard diagnostic test recommended by the current guidelines (2). However, RT–PCR exhibits its own limitations, including inappropriate specimen collection techniques, viral load time since the time of exposure (3) and the source of the specimen, which can contribute to false-negative test results (4). The rates of false-positive RT–PCR performance on the day of the onset of symptoms are 100% but decrease to 38% 5 days later (5). Serological testing is a blood test that can detect specific antibodies against COVID-19, including immunoglobulin M (IgM), IgG and IgA antibodies. Serological tests have been developed as supplementary diagnostic methods, as they can take several days or weeks to develop antibodies after viral exposure; therefore, they can provide information about recent or prior infections (1). As such, serological tests can be used as surveillance tools to better understand the overall infection rate in different regions and populations wherein quantitative PCR assays are not available or are delayed (6). Given the importance of serological tests in combating COVID-19, systematic reviews and meta-analyses that aim to summarize the accuracy parameters of serological tests and to investigate whether they are sufficiently specific or sensitive to achieve their role in practice are urgently needed.
Although some studies have compared pooled sensitivities and specificities of serological test methods, as well as identifying study and patient characteristics (7–10), high-quality evidence supporting the use of antibody tests for COVID-19 in practice is missing, due to a fast-growing field; additionally, ongoing updates of this systematic review will be implemented (11). Therefore, we conducted a systematic review and meta-analysis to assess the diagnostic accuracy of serological tests for COVID-19 infection. We aimed to understand the global serological tests of coronavirus with maps and updates on the overall sensitivity and specificity. To reduce variability in the estimates and to enhance generalizability, both sensitivity and specificity were stratified by clinical setting (outpatient vs. inpatient), antigen target, serological kit as the index test and the number of days that elapsed since the onset of symptoms. Analyses on the sensitivity and specificity of the different testing methods were performed to provide scientific guidance for the design and evaluation of vaccines and therapeutic antibodies in the future (1).
Methods
Search strategy
This meta-analysis was conducted according to the Preferred Reporting Project for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (12) and recommends best practices (13). We searched the PubMed, Embase and the Cochrane Library. The search terms used were (SARS-CoV-2 OR Coronavirus disease 2019 OR COVID-19) AND (IgM OR IgG). The searches ends September 2, 2022, with no restrictions on language. The detailed search strategy is in Supplementary material.
Types of studies
We included studies that met the following criteria. (1) Eligible studies, including randomized trials, cohort studies, or case-control studies, and case series reporting sensitivity, specificity, or both qualities of serological testing for COVID-19. (2) Studies evaluating any test that detects antibodies to SARS-CoV-2, including laboratory-based methods and tests designed for use in field therapy. Test methods include: laboratory-based enzyme-linked immunosorbent assay (ELISA) and chemiluminescence immunoassay (CLIA). Rapid diagnostic tests use lateral flow assays (LFIA), including colloidal gold or fluorescently labeled immunochromatographic assays (CGIA or FIA). (3) Serological diagnostic tests not limited to any antibodies, antigens or test methods.
The exclusion criteria were as follows: (1) case reports, review articles and editorials; (2) studies that focus on ineligible populations, such as vaccinated patients and people not infected with the coronavirus.
Three different researchers independently screened literature, extracted data and validated the results. If there is an objection, resolve it by discussion or negotiation with a third researcher.
Participants
We included studies that recruited people with suspicion of current or previous SARS-CoV-2 infection confirmed by NAT (such as RT PCR or sequencing) or NAT in combination with clinical outcomes.
Outcomes
The primary outcomes included overall sensitivity and specificity, stratified by serological tests (ELISA, LFIA, and CLIA) and immunoglobulin class (IgG, IgM, or both). Secondary outcomes include layer-specific sensitivity and specificity within subgroups, defined by study or participant characteristics.
Data extraction and bias assessment
The following data were independently extracted by 2 professional researchers: general study details (authors, year of publication, country of origin, study design, sample size, reagent company, time from symptom onset to index test and clinical setting and whether testing was performed via commercial kits or an in-house assay), methods, characteristics and diagnostic test results [true positive (TP), true negative (TN), false-positive (FP), false negative (FN), sensitivity, specificity and accuracy] (7).
Two researchers independently assessed the risk of bias for each study using the Cochrane Collaboration recommended Diagnostic Precision Study Quality Assessment Tool (QUADAS-2) (14). Quadas-2 is a quality assessment tool developed specifically for the systematic evaluation of accuracy studies, covering the following four key areas: patient selection, index test, reference standard and flow and timing. Additionally, each area was divided into low risk, high risk and unclear risk. The tool classifies evidence from observational studies into “low risk of bias,” “unclear” and “high risk of bias” level. If at least 50% of the fields are classified as low bias risk, the overall risk of bias for individual studies is classified as low bias risk; Otherwise, a higher risk of bias is defined (15).
Statistical analysis
Sensitivity and specificity of calculated estimates for each individual study (based on 2 × 2 contingency tables). All of the results are reported with 95% confidence intervals (CIs). Data are summarized as paired Forest plots. Since different studies have different cutoff values, a two-variable random effects model was used for meta-analysis. A summary receiver operating characteristic (ROC) curve based on TP and FP rates was established to describe the relationship between detection sensitivity and specificity. The area under the curve (AUC) is close to 1, indicating that the test has good diagnostic performance. All of the analyses were performed using Meta-Disc version 1.4.7. Random effects logistic regression model was used to compare the diagnostic accuracy of different antibodies, different antibody detection methods and different antigens. The heterogeneity of the study was determined by summary ROC curves with 95% prediction regions, estimated using bivariate meta-analysis with a test level random effect only, and forest plots. As our models were bivariate, we did not use the I2 statistic.
In the subgroup analysis, to assess pre-specified variables as potential determinants of diagnostic accuracy, we collected samples at times associated with symptom onset (at week 1, week 2, at week 3, or after week 3); Depending on the antigen target [surface protein (S), nucleocapsid protein (N), surface and nucleocapsid protein], the test is performed using a commercial kit or an internal test; Clinical institutions (inpatient, outpatient, inpatient, outpatient only); Serological kits as indicative tests (using commercial kits or in-house tests); and the type of specimen used for RT PCR reference testing (nasopharyngeal or sputum, saliva or oral, throat and pharyngeal). In these analyses, we pooled data according to the test method (ELISA, LFIA and CLIA) and immunoglobulin class (IgM, IgG or both).
Patient and public involvement
Patients were not involved in the formulation of study questions or outcome measurements, the conduct of the study or the preparation of the manuscript (7).
Results
Description of included studies
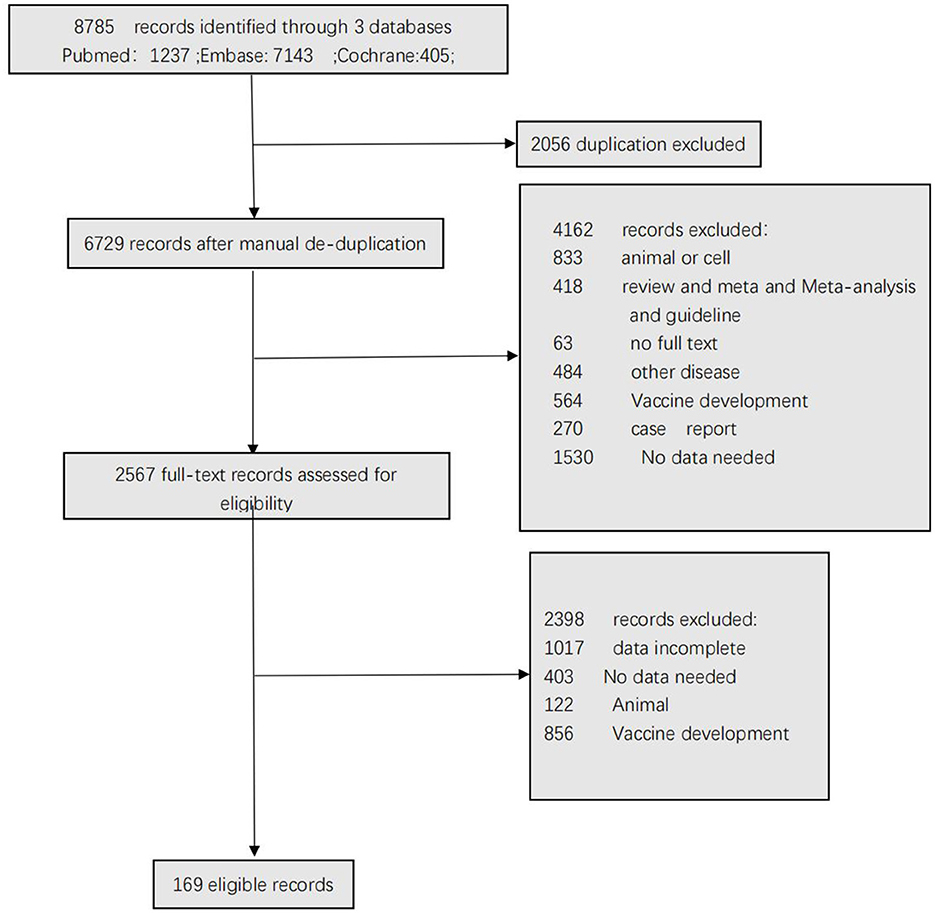
Figure 1 shows the selection of the studies. A total of 8,785 articles were identified after the removal of duplicate articles. Of these articles, 2,056 articles were excluded during the screening phase (title and abstract reading), with 6,560 records being fully appraised. Finally, 169 articles met the inclusion criteria.
Participant characteristics
Table 1 summarizes the studies by test method; the sum of the number of studies exceeded 169 because some studies evaluated more than one method. For example, a study that assessed 2 LFIAs and 3 ELISAs on the same set of patients would contribute 5 study arms. Twenty percent (33/169) of the studies were from the United States, Fifteen percent (26/169) of the studies were from China and the remainder of the studies were from Italy (12/169), Germany (9/169), Belgium (8/169), France (7/169), Japan (6/169), UK (6/169), Australia (5/169), Spain (5/169), Switzerland (5/169), Brazil (4/169), Saudi Arabia (4/169), Singapore (4/169), Austria (3/169), Sweden (3/169), Canada (2/169), Ecuador (2/169), Liechtenstein (2/169), Netherlands (2/169), Thailand (2/169), Bangladesh (1/169), Chile (1/169), Colombia (1/169), Croatia (1/169), Finland (1/169), Greece (1/169), India (1/169), Iran (1/169), Israel (1/169), Kenya (1/169), Korea (1/169), Mexico (1/169), New Zealand (1/169), Nigeria (1/169), Qatar (1/169), Serbia (1/169), South Africa (1/169), Uganda (1/169), and United Arab Emirates (1/169). Three SARS-CoV-2 antigens, including surface protein (S), nucleocapsid protein (N) and envelope protein (E), were used either together or separately in the studies that were included in the review. The spike protein was used as the antigen in 31 study arms, and the nucleocapsid protein was used in 21 study arms. Fifty-two study arms separately used both S and N as antigens. In 19 study arms, S and N antigens (S-N) were used together as the antigen. In 17 study arms, N and E antigens (N-E) were used together as the antigen. The sample was collected from inpatients in 48 articles and in 11 articles regarding outpatients. Fifty-nine study arms were separately comprised of outpatients and inpatients. In 42 study arms, samples were collected from inpatients and outpatients together. Most of the serological assay test kits were commercial (n = 173 study arms), and 22 study arms involved in-house assays. When regarding the type of specimen used for the RT–PCR reference test, 70 study arms involved nasopharyngeal samples, and 47 study arms involved sputum, saliva or oral, throat and pharyngeal samples. Table 1 reports the characteristics of each individual study.
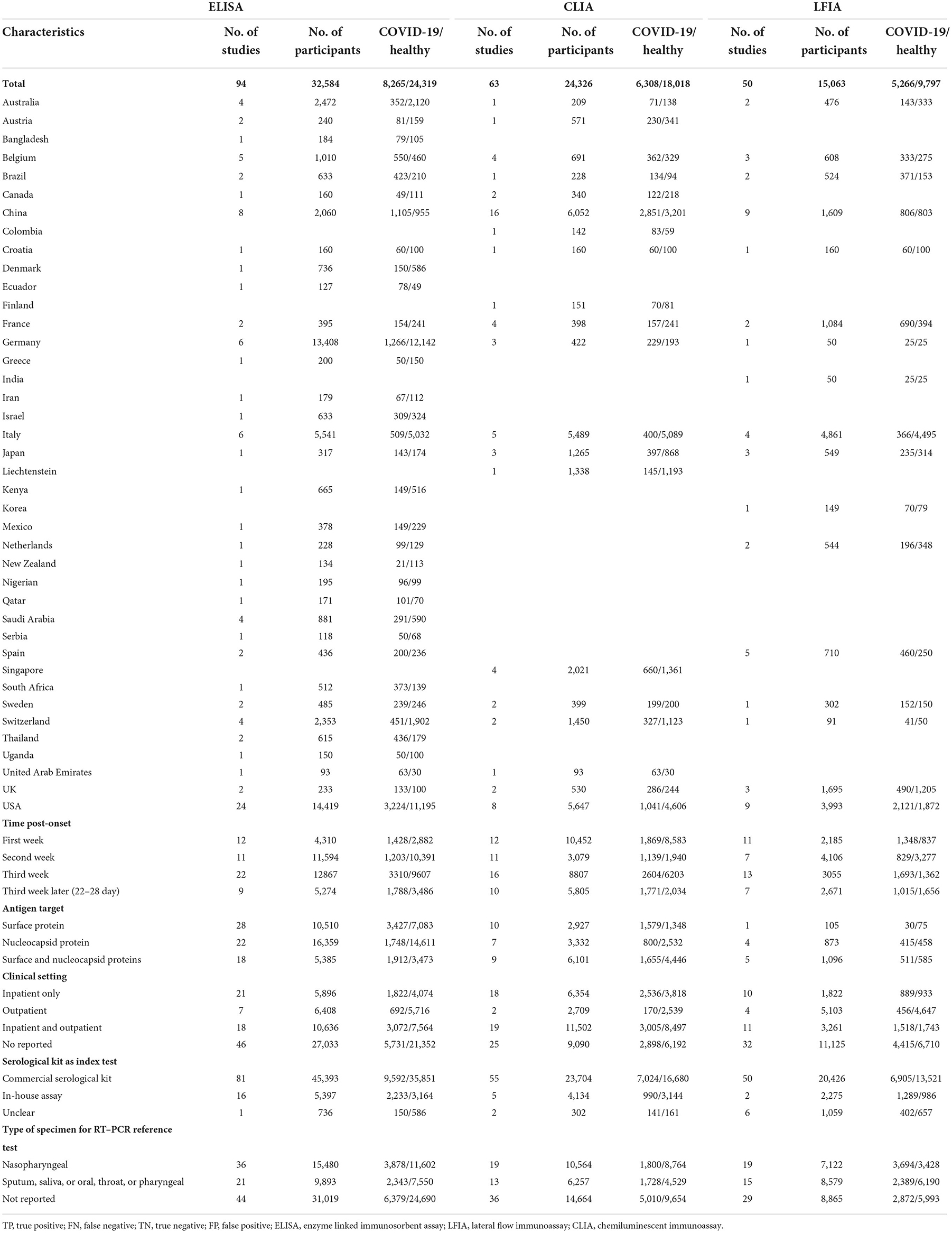
Table 1. Summary of characteristics of included studies, stratified by method of serological testing.
Methodological qualities of the included studies
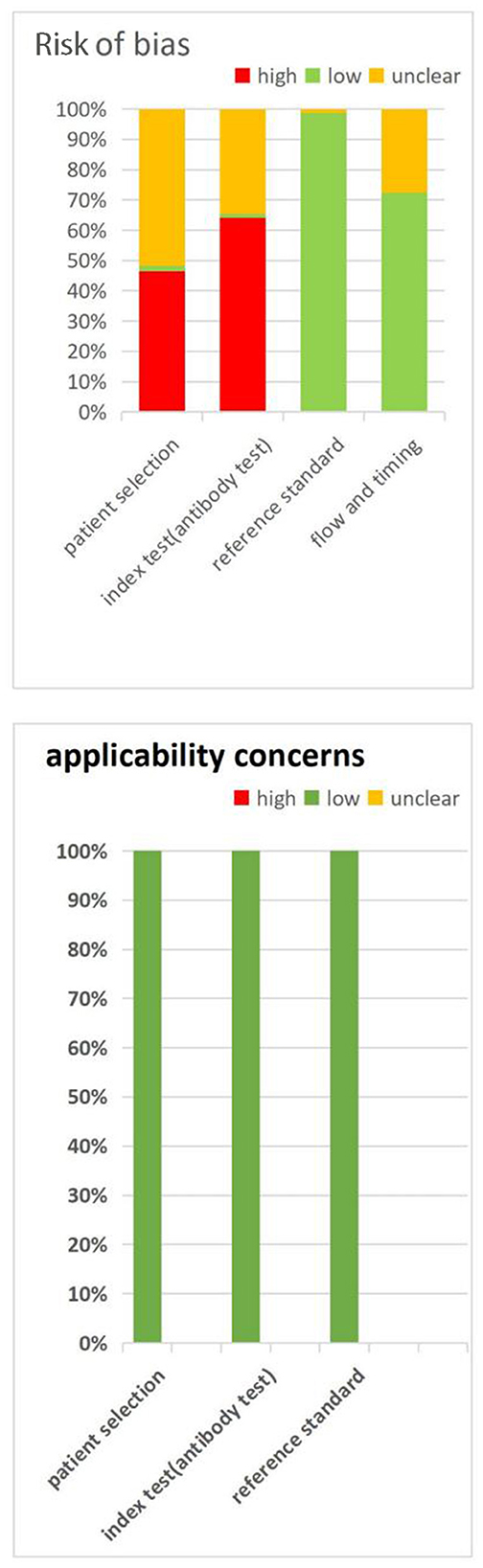
Figure 2 summarizes the QUADA-2 assessment, and Supplementary Table S1 provides details for each study QUADAS-2 evaluations. For the patient selection domain, a high or unclear risk of bias was observed in 98% (166/169) of the QUADAS-2 assessments, with the risks of bias mostly related to a case-control design and not due to conductive or random sampling. For the index test domain, 99% (167/169) of the assessments demonstrated a high or unclear risk of bias because it was not clear whether the serological test was interpreted blindly to the reference standard or whether the cut-off values for classifying the results were positive or negative. For the reference standard domain, 99% (167/169) of the assessments concluded a low risk of bias because the RT–PCR test is currently the best diagnostic method for use in novel coronavirus patients and is evaluated without knowing the results of the novel coronavirus serum test. The risk of bias from flow and timing was high or unclear in 27.2% (46/169) of the assessments, which was due to an appropriate time interval between the new coronavirus serum test that we investigated and the gold standard RT–PCR test. All of the patients underwent the same gold standard test, and most of the researched cases were included in the analysis.
Overall sensitivity
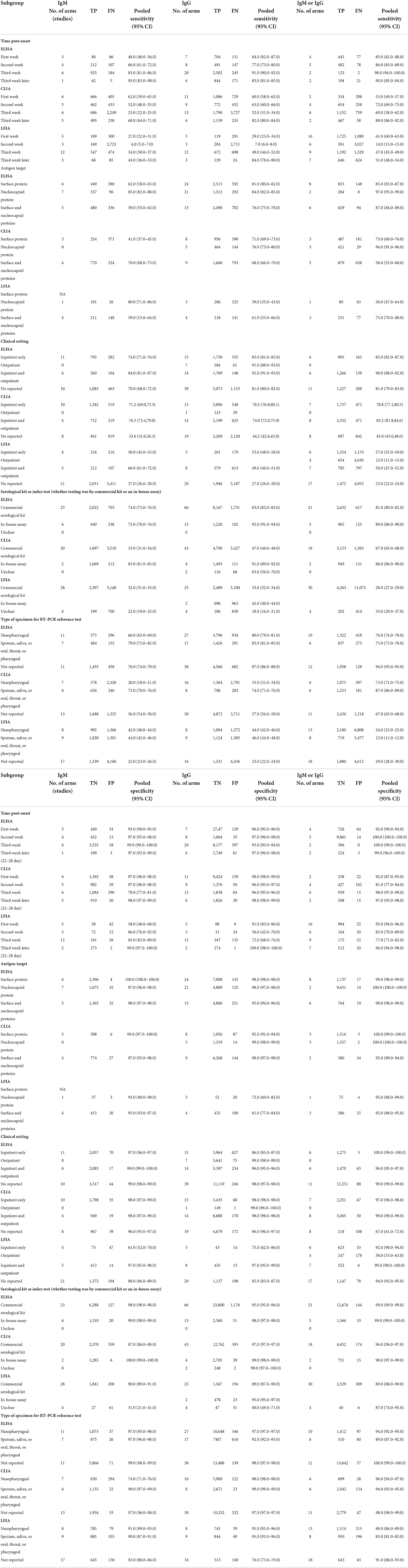
Table 2 reports on the sensitivity that was stratified by test type and immunoglobulin class. Within each test method (CLIA, ELISA, and LFIA), point estimates were similar between the different types of immunoglobulins, and the confidence intervals overlapped. Within each class of immunoglobulin, the sensitivity was lowest for the LFIA method. The pooled sensitivity of the ELISAs measuring IgM was 71% (95% CI: 70–73%), with IgG being 84% (95% CI: 83–84%) and IgM or IgG being 84% (95% CI: 83–85%). The pooled sensitivity of the LFIAs measuring IgM was 65% (95% CI: 64–67%), with IgG being 73% (95% CI: 71–74%) and IgM/IgG being 69% (95% CI: 68–71%). The pooled sensitivity of the CLIAs measuring IgM was 70% (95% CI: 69–72%), with IgG being 80% (95% CI: 79–81%) and IgM/IgG being 87% (95% CI: 86–88%). For all of the test methods and immunoglobulin classes, visual inspections of the summary ROC curves (Supplementary Figure S1) and of the forest plots (Supplementary Figure S2) exhibited significant heterogeneity.
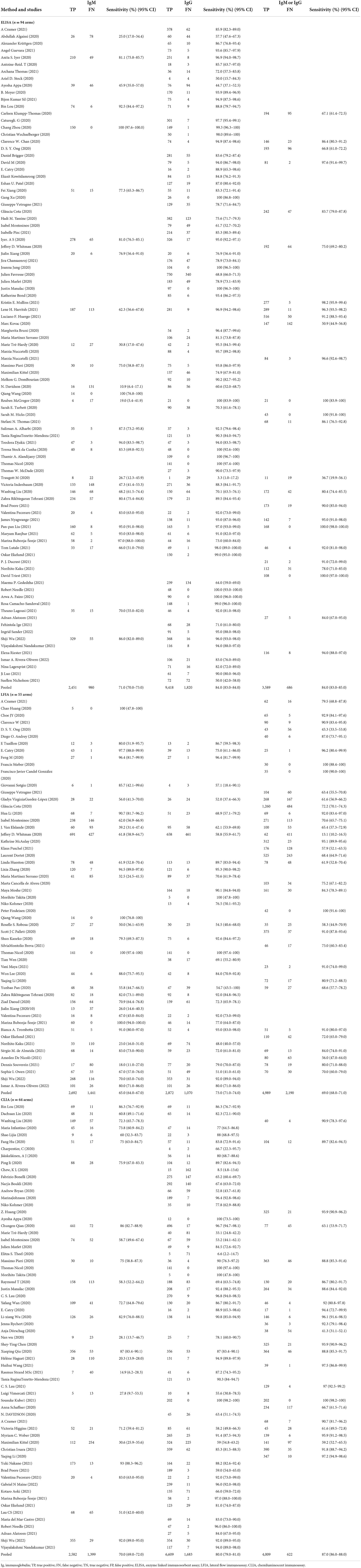
Table 2. Individual and pooled sensitivity by serological test method and immunoglobulin class detected.
Overall specificity
Table 3 describes the within study and pooled specificities, as stratified by test type and immunoglobulin class. The pooled specificity of the ELISAs measuring IgM was 98% (95% CI: 98–99%), with IgG being 96% (95% CI: 95–96%), and IgM or IgG being 99% (95% CI: 99–99%). The pooled specificity of the LFIAs measuring IgM was 96% (95% CI: 96–97%), with IgG being 97.0% (95% CI: 96–97%) and IgM or IgG being 97% (95% CI: 97–98%). The pooled specificity of the CLIAs measuring IgM was 94% (95% CI: 93–95%), with IgG being 98% (95% CI: 98–99%) and IgM or IgG being 97% (95% CI: 97–97%). Within each class of immunoglobulin, the specificity was the lowest for the IgM-based CLIA tests. All of the tests displayed high specificity (ranging from 93.0 to 99.0%). For all of the test methods and immunoglobulin classes, visual inspections of the summary ROC curves (Supplementary Figure S1) and of the forest plots (Supplementary Figure S3) showed meta-analytical estimates of specificity (with a value of 95%) by using the serological test method and antibody class.
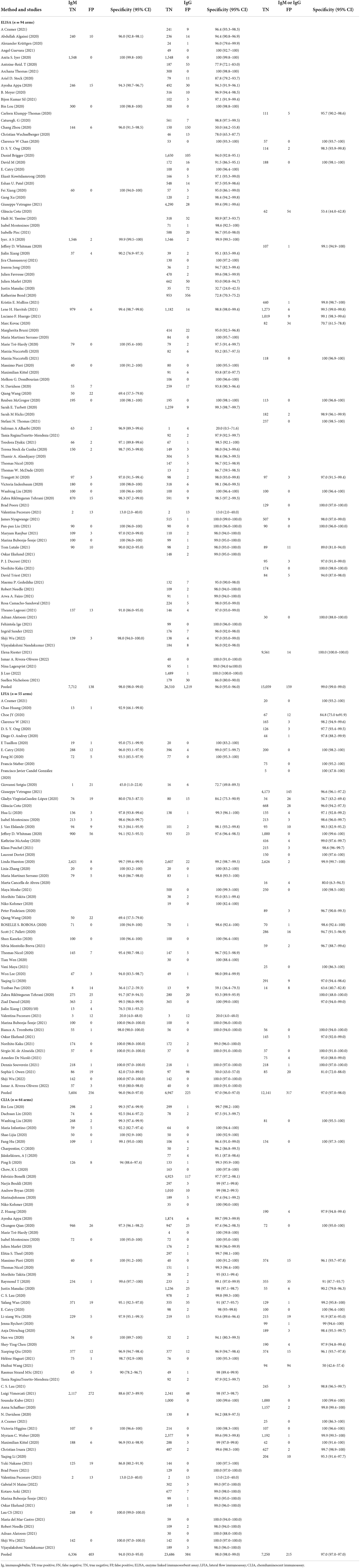
Table 3. Individual and pooled specificity by serological test method and immunoglobulin class detected.
Sensitivity and specificity by potential sources of heterogeneity
Table 4 reports the stratified meta-analyses for evaluating potential sources of heterogeneity in sensitivity and specificity. Heterogeneity was observed in all of the analyses.
Subgroup analysis of the timing of sample collection in relation to symptom onset
The average sensitivity across all of the included studies for ELISA-tested IgG, IgM and IgG/IgM showed low sensitivity during the first week after the onset of symptoms, after which they increased in the second week and reached their highest values beyond 3 weeks. For the ELISAs, sensitivity estimates were higher in the third week or later after symptom onset (ranging from 83.0 to 90.0%). In contrast, for the CLIAs, pooled sensitivity was lower in the third week (<30%); for the LFIAs, pooled sensitivity was lower in the second week (<10%) after symptom onset. Very few studies have evaluated tests beyond 35 days to estimate accuracy. Data on specificity, as stratified by timing, showed that the pooled data were highest in the second week. Specificity was higher at least 2 weeks after symptom onset (ranging from 98.0 to 98.0%) than within the first week (ranging from 96.0 to 97.0%). For the ELISA test method, the pooled specificity of 99% (ranging from 99 to 100%) was high when the measured time post-onset was in the second week. For the CLIA and FLIA test methods, the pooled specificity was high when measured time post-onset was in the third week later (ranging from 97 to 99%) (Table 4).
Subgroup analysis of test technology type
Point estimates for the pooled sensitivity and specificity were higher when the N protein was used. A subgroup meta-analysis showed that tests using the N antigen (ranging from 77 to 80%) were more sensitive than with the use of S protein (ranging from 66.0 to 68.0.0%) antigen tests. Moreover, IgG-based serological assays that used the N antigen were more sensitive than IgG-based serological assays that used the S antigen. For the ELISAs, specificity was higher when the nucleocapsid protein was used; however, this was not the case for the LFIAs or CLIAs. For the CLIAs, specificity and sensitivity were higher from reported studies that used the nucleocapsid proteins (ranging from 99 to 100%) (Table 4).
Subgroup analysis of setting (outpatient vs. inpatient)
For the ELISAs, point estimates for pooled sensitivity were higher when estimates at the sample level for both inpatients and outpatients were included, in which case the sensitivity was 90% (ranging from 89 to 91%). For the LFIAs, pooled specificity was higher when estimates at the sample level for both inpatient and outpatient were included, in which case the specificity was 98% (ranging from 97 to 98%). Among the three test methods, point estimates for pooled sensitivity and specificity were higher when estimates at the sample level included both inpatients and outpatients (Table 4).
Subgroup analysis of serological kits as index tests (whether testing was performed by using commercial kits or an in-house assay)
Both in-house and commercial kits are the preferred molecular tests being used worldwide in the COVID-19 diagnosis. We compared pooled sensitivity and specificity across subgroup according to serological kit as index test (whether testing was by commercial kit or an in-house assay). For all three of the test methods, point estimates of sensitivity and specificity were higher for in-house assays vs. commercial kits. The pooled sensitivity was higher for in-house assays (ranging from 78 to 79%) than for commercial kits (ranging from 47 to 48%). The pooled specificity was higher for in-house assays (ranging from 98 to 99%) than for commercial kits (ranging from 96 to 96%) (Table 4).
Subgroup analysis of the type of specimen used for the RT–PCR reference test
For the ELISA and CLIA test methods, pooled specificity and sensitivity were high when the types of specimens that were used for the RT–PCR were sputum, saliva, oral, throat or pharyngeal samples. However, when the sample was nasopharyngeal, the pooled sensitivity and specificity were high, as indicated by the LFIA test method (Table 4).
The accuracy of serological tests world map for COVID-19
We pooled the sensitivity and specificity of COVID-19 serological tests that are used worldwide. For the ELISA, pooled sensitivity was higher in Canada (100%) than in other areas; For the CLIA, pooled sensitivity was higher in Croatia (97%) than in other areas (Supplementary Figures S5–S7). Among these three serological tests, ELISA exhibited higher sensitivity (ranging from 50 to 100%) and higher specificity (ranging from ≥73–100%). For the CLIA, Italy, Switzerland and Singapore had lower sensitivities (< 30%) (Supplementary Table S3). Among the three test methods, point estimates for pooled specificity were higher in Latin America (ranging from 99.0 to 100%) (Supplementary Figures S4–S9, Supplementary Table S3).
Discussion
In this systematic review and meta-analysis, we found that ELISA and CLIA methods performed better in terms of sensitivity than the LFIA method, thus indicating that viral infections can lead to false-positive results for the LIFA method. For each test method, the type of immunoglobulin being measured (IgM, IgG or both) was associated with diagnostic accuracy, and sensitivities were consistently higher with IgG than with IgM. Moreover, IgG-IgM-based CLIA tests exhibited the best overall diagnostic test accuracy. Moreover, pooled specificities of each test method were high. Pooled sensitivities and specificities were higher with in-house assays vs. commercial kits and in the third week or later, compared with the first and second weeks after symptom onset. Additionally, point estimates for pooled sensitivity and specificity were higher when estimates at the sample level were both inpatient and outpatient; therefore, serological tests are able to detect lower antibody levels that are likely observed with milder and asymptomatic COVID-19 disease.
Research implications
1. For all three of the methods, the LFIA method had lower sensitivity than the ELISA or CLIA methods for IgM (similar data were available for IgG and IgM/IgG). For the LFIAs, pooled sensitivity was lowest in the second week of symptom onset and highest in the first week. These observations can provide recommendations to the World Health Organization for improving test accuracy when using LFIA serological tests. Given the poor performances of the current LFIA devices (7, 16), LFIA tests for COVID-19 in the second week of symptom onset (with an average sensitivity of 9%) will be falsely identified as not being positive for infection. In addition, sensitivity estimates are likely to increase in the first week, compared with other time points of sample collection. Our time-stratified analyses suggest that LFIA seems to be a better choice (in terms of sensitivity) at the first week of sample collection, in relation to symptom onset.
2. For all three of the test methods, pooled sensitivities and specificities were higher with in-house assays vs. commercial kits. These findings are expected, given that the pooled sensitivities were lower with the commercial kits than with in-house assays (7). Point estimates of pooled sensitivity were lower for commercial kits vs. in-house assays, for all three methods, with the strongest difference seen for LFIAs, where the sensitivity of commercial kits was 28.0% sensitivity and 89.0% specificity with IgM or IgG. For commercial kits based LFIA, the sensitivity was found to be below 50% and higher quality clinical studies assessing the diagnostic accuracy of commercial kits based LFIA are urgently needed.
3. Sensitivity varied with the time since the onset of symptoms and technology test methods. Our findings should give pause to governments that are contemplating the use of serological tests. For example, if LFIAs are applied to a population in the second week after the onset of symptoms, the average sensitivity of the test may be 9%; thus, only 9 patients out of 100 true positive patients can be detected. Serological tests are likely to have a useful role in detecting previous COVID-19 infections if they are used at 15 or more days after the onset of symptoms, except with estimated pooled specificities using CLIAs and LFIAs test methods, which are more suitable for use at 7 days after the onset of symptoms. Overall, the type of sample should be collected with consideration of the timing of the infection. It is necessary to perform the correct test at the correct time in the sample collection process, in order to avoid misdiagnoses of asymptomatic patients who are negative for serological tests.
4. Sensitivity has mainly been evaluated in hospitalized patients (7, 10); therefore, it is unclear whether the tests are able to detect lower antibody levels that are likely observed with milder and asymptomatic COVID-19 disease. Few studies have solely evaluated outpatient sensitivity accuracy. Point estimates for pooled sensitivity and specificity were higher when estimates at the sample level included both inpatients and outpatients. Our findings support the use of serological tests that are applied to people with mild symptoms who were not hospitalized, thus reducing variability in the estimates and enhancing generalizability.
5. There was little clear evidence of differences in specificity between the technology types. Specifically, all of the tests displayed high specificities. Within each class of immunoglobulin, specificity was lowest for the IgM-based CLIA tests.
6. Generally, IgG-based serological tests demonstrated a better choice in terms of sensitivity than IgM-based serological assays in each respective test method. IgG-based tests may be a safer choice at this stage of the pandemic. Low IgM antibody concentrations could potentially be explained by the fact that, immediately after a person is infected, antibodies may not have been developed yet; additionally, if it is too late after a person has been infected, IgM antibodies may have decreased or disappeared (17). The nucleocapsid protein and surface protein were used for detecting IgM and IgG antibodies, and their diagnostic feasibilities were evaluated. A subgroup meta-analysis showed that nucleocapsid antigen-based IgG serological assays are more sensitive than S antigen-based IgG serological assays that use the S antigen, thus indicating that combined IgG/IgM test antigen target nucleocapsid protein-based CLIA tests have the best overall diagnostic test accuracy.
Comparison to previous studies
The sensitivities of all of the serological assays varied widely across the studies. Similar to other meta-analyses (7, 16, 18), the LFIA method had lower sensitivities than the CLIA and ELISA methods within each antibody class. CLIA and ELISA may be a safer choice at this stage of the pandemic. In addition, similar to other meta-analyses (17), IgM-based serological assays had the lowest sensitivities, compared with IgG-based serological tests, in each respective test method. From this study, we showed that IgG-IgM-based CLIA tests had a higher pooled sensitivity than the ELISA and LFIA tests. Moreover, it must be noted that a meta-analysis by Vengesai et al. (16) found that IgG-IgM-based ELISA tests have the best overall diagnostic test accuracy; however, in that review, they did not estimate the pooled sensitivity of IgG-IgM-based CLIA, due to the limited number of studies.
Few studies have evaluated tests beyond 35 days to estimate accuracy. For ELISAs, sensitivity estimates were higher in the third week or later after the onset of symptoms (ranging from 88 to 90%). In contrast, for the CLIAs, pooled sensitivity was lower in the third week (< 35%); For LFIAs, pooled sensitivity was lower in the second week (< 10%) after symptom onset. These findings differ from those of previous studies, in which sensitivity estimates were lowest in the first week of symptom onset and highest in the third week or later (7, 10). These observations argue against the use of serological tests for COVID-19 that exhibit higher sensitivity when performed later during the course of the disease.
A subgroup meta-analysis showed that tests using the nucleocapsid antigen were more sensitive than surface antigen tests in each immunoglobulin (IgM, IgG or both) test method. The pooled sensitivity results are in agreement with other meta-analyses that demonstrated that IgG-based serological assays that use the N antigen are more sensitive than IgG-based serological assays that use the S antigen (17). However, it must be noted that a meta-analysis by Liu et al. (19) showed that the S antigen is more sensitive than IgM-based serological assays that used N antigen tests. Thus, there is a need for more research concerning a higher sensitivity and earlier immune response to the nucleocapsid antigen.
Strengths and limitations of this review
Our review had several strengths. For example, our review involved two independent reviewers who systematically assessed potential sources of bias. Additionally, the entire search strategy and data analysis process were relatively standardized. Moreover, we included 134 published articles on SARS-CoV-2 infections that were defined by RT–PCR because a considerable amount of new research is being published in this field. The advantages of large studies and large sample sizes allow researchers to magnify the bias associated with error, which can result from sampling or study design. Another strength of our review was that the study was conducted using in-depth subgroup meta-analyses to evaluate potential sources of heterogeneity in sensitivity and specificity, which reduces variability in the estimates and enhances diagnostic accuracy.
Our study also had some limitations. For example, we did not pool sensitivity and specificity for measurements of IgA or total immunoglobulin levels, due to small numbers. Another limitation was that we did not search for studies from individuals who were not suspected of having COVID-19 or specimens from individuals with COVID-19 symptoms and a negative RT–PCR result for SARS-CoV-2.
Conclusions
Seroconversion occurred after 7 days in 50% of patients (and by day 14 in all of the patients), but this was not followed by a rapid decline in viral load (20). There is an urgent need for an effective and accurate diagnostic method to limit the spread of the COVID-19 infection. At present, rapid antigen or antibody tests, immunoenzymatic serological tests and molecular tests based on RT–PCR are the most widely used and validated techniques worldwide (21). We have found major weaknesses in the evidence base for serological tests for COVID-19. It is necessary to take into account not only the right test method (ELISAs, LFIAs, or CLIAs) but also the correct time from the onset of symptoms and from the correct biological sample for a successful outcome of the diagnostic test. Due to the limitations of serological tests, other techniques, including isothermal nucleic acid amplification techniques, clusters of regularly interspaced short palindromic repeats/Cas (CRISPR/Cas)-based approaches or digital PCR methods, should be quickly approved to provide guidance for a correct diagnosis of COVID-19.
Author contributions
XZ and RD: drafting and revision of the manuscript for content, including medical writing for content, analysis or interpretation of data, and major role in the acquisition of data. FG, XW, YD, RC, and ML: major role in the acquisition of data. CT and LL: study concept or design. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (NSFC)/82205245 to XZ, NSFC/82174479 to CT, and NSFC/82174527 and the special project of Lingnan Modernization of Traditional Chinese Medicine in 2019 Guangdong Provincial R&D Program (No. 2020B1111100008) to LL.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2022.923525/full#supplementary-material
Supplementary Figure S1. Visual inspection of summary ROC curves by test method and antibody class.
Supplementary Figure S2. Meta-analytical estimates of sensitivity (with 95%) by serological test method and antibody class.
Supplementary Figure S3. Meta-analytical estimates of specificity (with 95%) by serological test method and antibody class.
Supplementary Figure S4. Sensitivity of ELISA around the world.
Supplementary Figure S5. Sensitivity of CLIA around the world.
Supplementary Figure S6. Sensitivity of LFIA around the world.
Supplementary Figure S7. Specificity of ELISA around the world.
Supplementary Figure S8. Specificity of CLIA around the world.
Supplementary Figure S9. Specificity of LFIA around the world.
Supplementary Table S1. The included study individual QUADAS-2 evaluations.
Supplementary Table S2. Report the PubMed ID for each included study.
Supplementary Table S3. The accuracy of serological tests for COVID-19 around the world.
References
1. Sidiq Z, Hanif M, Dwivedi KK, Chopra KK. Benefits and limitations of serological assays in COVID-19 infection. Indian J Tuberc. (2020) 67:S163–66. doi: 10.1016/j.ijtb.2020.07.034
2. Pau AK, Aberg J, Baker J, Belperio PS, Coopersmith C, Crew P, et al. Convalescent plasma for the treatment of COVID-19: perspectives of the national institutes of health COVID-19 treatment guidelines panel. Ann Intern Med. (2021) 174:93–5. doi: 10.7326/M20-6448
3. Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. (2020) 382:1177–79. doi: 10.1056/NEJMc2001737
4. Winichakoon P, Chaiwarith R, Liwsrisakun C, Salee P, Goonna A, Limsukon A, et al. Negative nasopharyngeal and oropharyngeal swabs do not rule out COVID-19. J Clin Microbiol. (2020) 58:e00297-20. doi: 10.1128/JCM.00297-20
5. Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med. (2020) 173:262–67. doi: 10.7326/M20-1495
6. Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. (2020) 71:778–85. doi: 10.1093/cid/ciaa310
7. Bastos ML, Tavaziva G, Abidi SK, Campbell JR, Haraoui L-P, Johnston JC, et al. Diagnostic accuracy of serological tests for COVID-19: systematic review and meta-analysis. BMJ. (2020) 370:m2516. doi: 10.1136/bmj.m2516
8. Chen M, Qin R, Jiang M, Yang Z, Wen W, Li J. Clinical applications of detecting IgG, IgM or IgA antibody for the diagnosis of COVID-19: A meta-analysis and systematic review. Int J Infect Dis. (2021) 104:415–22. doi: 10.1016/j.ijid.2021.01.016
9. Boger B, Fachi MM, Vilhena RO, Cobre AF, Tonin FS, Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am J Infect Control. (2021) 49:21–9. doi: 10.1016/j.ajic.2020.07.011
10. Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. (2020) 6:CD013652. doi: 10.1002/14651858.CD013652
11. Vandenberg O, Martiny D, Rochas O, van Belkum A, Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat Rev Microbiol. (2021) 19:171–83. doi: 10.1038/s41579-020-00461-z
12. Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JPA, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. (2009) 339:b2700. doi: 10.1136/bmj.b2700
13. Forero DA, Lopez-Leon S, González-Giraldo Y, Bagos PG. Ten simple rules for carrying out and writing meta-analyses. PLoS Comput Biol. (2019) 15:e1006922. doi: 10.1371/journal.pcbi.1006922
14. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deels JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. (2011) 155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009
15. Savović J, Weeks L, Sterne JAC, Turner L, Altman DG, Moher D, et al. Evaluation of the Cochrane Collaboration's tool for assessing the risk of bias in randomized trials: focus groups, online survey, proposed recommendations and their implementation. Syst Rev. (2014) 3:37. doi: 10.1186/2046-4053-3-37
16. Vengesai A, Midzi H, Kasambala M, Mutandadzi H, Mduluza-Jokonya TL, Rusakaniko S, et al. A systematic and meta-analysis review on the diagnostic accuracy of antibodies in the serological diagnosis of COVID-19. Syst Rev. (2021) 10:155. doi: 10.1186/s13643-021-01689-3
17. Kontou PI, Braliou GG, Dimou NL, Nikolopoulos G, Bagos PG. Antibody Tests in detecting SARS-CoV-2 infection: a meta-analysis. Diagnostics. (2020) 10:319. doi: 10.3390/diagnostics10050319
18. Berger RS, Mandel EB, Hayes TJ, Grimwood RR. Minocycline staining of the oral cavity. J Am Acad Dermatol. (1989) 21:1300–1. doi: 10.1016/S0190-9622(89)80309-3
19. Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. (2020) 58:e00461-20. doi: 10.1128/JCM.00461-20
20. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. (2020) 581:465–69. doi: 10.1038/s41586-020-2196-x
Keywords: serological tests, COVID-19, systematic review, meta-analysis, RT–PCR
Citation: Zheng X, Duan Rh, Gong F, Wei X, Dong Y, Chen R, yue Liang M, Tang C and Lu L (2022) Accuracy of serological tests for COVID-19: A systematic review and meta-analysis. Front. Public Health 10:923525. doi: 10.3389/fpubh.2022.923525
Received: 20 April 2022; Accepted: 15 November 2022;
Published: 16 December 2022.
Edited by:
Jayanthi S. Shastri, Topiwala National Medical College and BYL Nair Charitable Hospital, IndiaReviewed by:
Citra Fragrantia Theodorea, University of Indonesia, IndonesiaZhen Qin, University of Toronto, Canada
Copyright © 2022 Zheng, Duan, Gong, Wei, Dong, Chen, yue Liang, Tang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunzhi Tang, jordan664@163.com; Liming Lu, lulimingleon@126.com
†These authors have contributed equally to this work
 Xiaoyan Zheng
Xiaoyan Zheng Rui hua Duan
Rui hua Duan Fen Gong
Fen Gong Xiaojing Wei
Xiaojing Wei Yu Dong
Yu Dong Rouhao Chen
Rouhao Chen Ming yue Liang
Ming yue Liang Chunzhi Tang
Chunzhi Tang Liming Lu
Liming Lu