- 1Department of Epidemiology and Health Statistics, Xiangya School of Public Health, Central South University, Changsha, China
- 2Department of Mathematics and Statistics, Mzuzu University, Mzuzu, Malawi
- 3OMNI Research Group, Ottawa Hospital Research Institute, Ottawa, ON, Canada
- 4Department of Obstetrics and Gynaecology, University of Ottawa Faculty of Medicine, Ottawa, ON, Canada
- 5School of Epidemiology and Public Health, University of Ottawa Faculty of Medicine, Ottawa, ON, Canada
- 6School of Resource and Environmental Sciences, Wuhan University, Wuhan, China
- 7International Institute of Spatial Lifecourse Health (ISLE), Wuhan University, Wuhan, China
Background: The 2019 novel coronavirus (COVID-19) pandemic remains rampant in many countries/regions. Improving the positive detection rate of COVID-19 infection is an important measure for control and prevention of this pandemic. This meta-analysis aims to systematically summarize the current characteristics of the auxiliary screening methods by serology for COVID-19 infection in real world.
Methods: Web of Science, Cochrane Library, Embase, PubMed, CNKI, and Wangfang databases were searched for relevant articles published prior to May 1st, 2022. Data on specificity, sensitivity, positive/negative likelihood ratio, area under curve (AUC), and diagnostic odds ratio (dOR) were calculated purposefully.
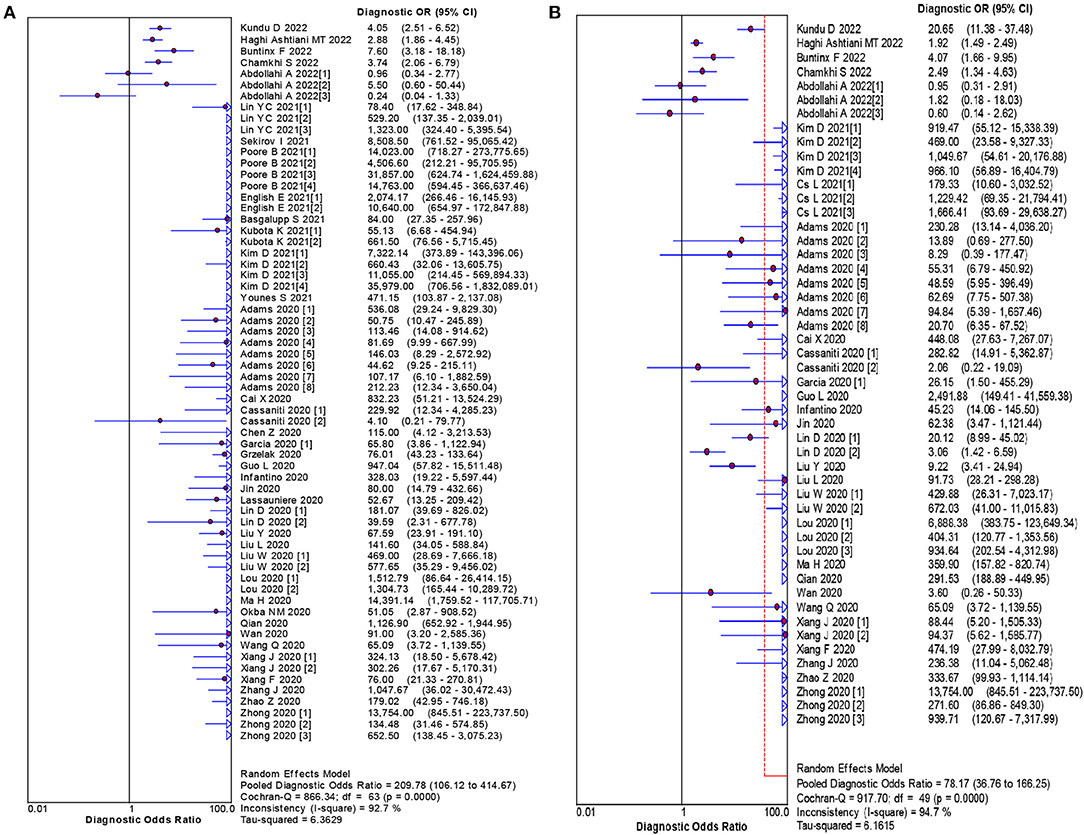
Results: Sixty-two studies were included with 35,775 participants in the meta-analysis. Among these studies, the pooled estimates for area under the summary receiver operator characteristic of IgG and IgM to predicting COVID-19 diagnosis were 0.974 and 0.928, respectively. The IgG dOR was 209.78 (95% CI: 106.12 to 414.67). The IgM dOR was 78.17 (95% CI: 36.76 to 166.25).
Conclusion: Our findings support serum-specific antibody detection may be the main auxiliary screening methods for COVID-19 infection in real world.
Introduction
Three unprecedented outbreaks of human coronavirus (HCoV) at the beginning of the 21st century, indicated coronavirus as a major public health problem worldwide (1, 2). Less than a decade after the last human disease outbreak, caused by the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in 2012, a new outbreak of the severe acute respiratory syndrome, caused by coronavirus 2 (SARS-CoV-2), is spreading around the world (2). This pandemic is now defined as the 2019 novel coronavirus (COVID-19). The virus was primarily spread by COVID-19 infected individuals. Therefore, the main way to control the spread of COVID-19 disease is to isolate the source of infection. In this regard, the differential screening of COVID-19 highlights the necessity for readily available, accurate and fast screening testing methods (3).
The current gold standard for etiological diagnosis of COVID-19 infection is real time reverse transcription polymerase chain reaction (RT-PCR) in respiratory specimens (4). However, some studies have shown some problems in using this method to detect COVID-19 infection, such as low sensitivity and specificity (5). These studies suggested that causes of these problems may include performing screening outside the diagnostic window, virus mutation and recombination, using insufficiently validated tests, and instrument failure (5). Therefore, more and more studies are taking cross-disciplinary methods to better understand screening and diagnosis of COVID-19 infection in order to improve diagnostic accuracy, for example, by combining clinical evidence, and COVID-19 antibody test results, and interpreting RT-PCR results on the basis of epidemiological and clinical evidence (6). However, none of the current meta-analysis have systematically summarized these strategies (7–9). Although the association between antibodies and SARS-CoV-2 infection has been discussed in some recently published meta-analyses, the studies included in these studies are relatively limited, ranging from 14 to 29 (10–12).
This study aims to (1) systematically summarize the sensitivity and specificity of the major screening methods for the COVID-19, (2) analyze the possible causes of false negatives or false positives as regards the efficacy of the screening methods, and (3) explore how to further improve the sensitivity and specificity of the screening methods. The study will also discuss epidemiological methods that could be used to further help identify more patients with latent infections by investigating their exposure history. It was speculated that, in a public health emergency, a combination of multidisciplinary approaches may be of some help in the control and prevention of COVID-19.
Methods
Search Strategy and Selection Criteria
Standard procedures for meta-analyses according to the Cochrane Handbook; and the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines were used to conduct this study (13). Thus, using these procedures, two independent reviewers (XP and AK) systematically searched in the electronic databases, Web of Science, Cochrane Library, Embase, PubMed, CNKI, and Wangfang, for relevant studies published in May 1st, 2022, or earlier. Then, from the search outcomes, they selected eligible studies according to the purpose of this study, using predefined selection criteria. Noteworthy, China has accumulated valuable experience in screening and diagnosing COVID-19, but very much literature related to China on this subject has been published in Chinese language. Therefore, to avoid publication bias, gray literature and studies published in Chinese were included in this study. That is, studies published in English or Chinese were included in this study. In order to retrieve as much literature as possible, the search strategy, among other important terms, included the professional name, COVID-19, and its variant names. Based on both historical and current COVID-19 names, Boolean operators, truncation and wildcards were used appropriately to include all other variant names for COVID-19. The complete search strategy is shown in the Supplementary Materials (Appendix 1). Experienced librarians designed the search strategy and adjusted it to meet the requirements of each of the databases specified above. For example, a search strategy in the database of PubMed was structured as follows using keywords (search terms): (((“COVID-19”[Title/Abstract] OR “2019 novel coronavirus infection”[Title/Abstract] OR “COVID19”[Title/Abstract] OR “coronavirus disease 2019”[Title/Abstract] OR “coronavirus disease-19”[Title/Abstract] OR “2019-nCoV disease”[Title/Abstract] OR “2019 novel coronavirus disease”[Title/Abstract] OR “2019-nCoV infection”[Title/Abstract] OR “coronavirus disease 2019 virus”[Title/Abstract] OR “SARS-CoV-2”[Title/Abstract] OR “SARS2”[Title/Abstract] OR “2019-nCoV”[Title/Abstract] OR “2019 novel coronavirus”[Title/Abstract] OR “severe acute respiratory syndrome coronavirus 2”[Title/Abstract] OR “COVID Asymptomatic Infections”[Title/Abstract]) AND (“Nucleic Acid Detection”[Title/Abstract] OR “Nucleic Acid Probes”[Title/Abstract] OR “Molecular Probes”[Title/Abstract] OR “Nucleic Acid Probes”[Title/Abstract] OR “Reagent Kits, Diagnostic”[Title/Abstract] OR “Reagent Strips”[Title/Abstract] OR “polymerase chain reaction”[Title/Abstract] OR “PCR*”[Title/Abstract])) AND (“Serology”[Title/Abstract] OR “Antibody”[Title/Abstract])).
Study Selection
Selection of studies for this meta-analysis was based on the following inclusion criteria: (1) diagnostic and screening studies; (2) studies reported methods for diagnosing COVID-19; and (3) original studies. Studies were excluded based on the following criteria: (1) conference papers, case reports, letters, or reviews; and (2) studies not on humans.
Data Extraction
Two reviewers (SW and AK) translated the Chinese articles into English, and entered all the data and removed duplicates in EndNote (version x9.1), Then used custom data extraction tool, EpiData (version 3.0) grids, and extracted (14, 15). The key characteristics of interest were extracted from each eligible study in the EpiData (version 3.0) grids, including first author, year of publication, study area, number of subjects, sensitivity, and specificity. Any differences between the two reviewers were resolved by consensus involving the third reviewer (AL).
Quality Evaluation
The Quality Assessment of Diagnostic Accuracy Studies-2 checklist (QUADAS-2) was used to assess the quality of the eligible studies. The selected studies were grouped based on their score into high (6–7 points), moderate (4–5 points), and low (0–3 points) quality categories (16, 17).
Statistical Analysis
In this study, meta-analyses were carried out using MetaDiSc (version 1.4) and R software (version R i386 3.4.2). For the diagnostic meta-analysis, the number of subjects with a true-positive (TP), false-positive (FP), true-negative (TN), and false-negative (FN) values for each study unit was extracted to calculate the pooled sensitivity [TP/(TP+FN)], specificity [TN/(TN+FP)], positive likelihood ratio (PLR) [(sensitivity/(1–sensitivity)], negative likelihood ratio (NLR), [(1–specificity)/specificity)], diagnostic odds ratio (dOR) [PLR/NLR], and their corresponding 95% confidence intervals (CIs) using a bivariate random-effect meta-analysis model (18). The summary receiver operator characteristic (SROC) curve was plotted, and the area under the SROC curve (AUC) was calculated to evaluate the pooled diagnostic performance of IgG and IgM for predicting COVID-19 diagnosis (19, 20). Heterogeneity between enrolled studies was quantified by the I2 statistic and assessed by the Cochran's Q-statistic. I2 = 0% indicated no heterogeneity, and I2 = 100% indicated maximal heterogeneity (21, 22). Publication bias was assessed using the Deeks' funnel plot asymmetry test, when the number of studies reporting meta-analysis results was 10 or more. Finally, in all analyses, the level of significance for the effect size estimation was set at 5%, and all tests were two-sided.
Results
Characteristics of Eligible Studies
The search strategy retrieved a total of 7,339 studies, of which 2,631 from Web of Science, 261 from Cochrane library, 2,293 from Embase, 1,353 were from PubMed, 503 from CNKI and 298 from Wanfang. Following a full review of 658 of these studies, 62 met the inclusion criteria for this meta-analysis. The PRISMA flowchart in Figure 1 shows the number of studies and the selection process.
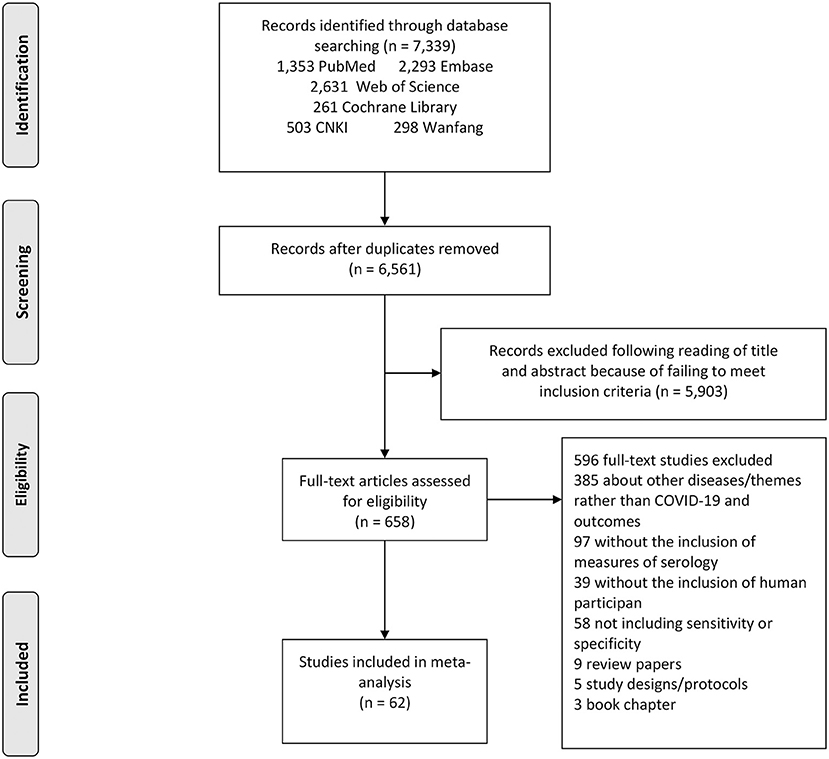
Figure 1. Study selection flow chart. A Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow chart demonstrating the selection process of articles included in the analysis as well as in the qualitative summary.
Characteristics of Eligible Studies
Sixty-two studies were included with 35,775 participants in our meta-analysis. Characteristics of the eligible studies are shown in Appendix 2. Further, focusing on test method, 62 studies reported on antibody tests. The QUADAS-2 score of these studies varied between 3 and 6, with 27 studies of high quality and 35 of moderate quality. There are 44 studies from Asia, 12 from Europe, 4 from the Americas and 2 from Africa.
Main Outcomes
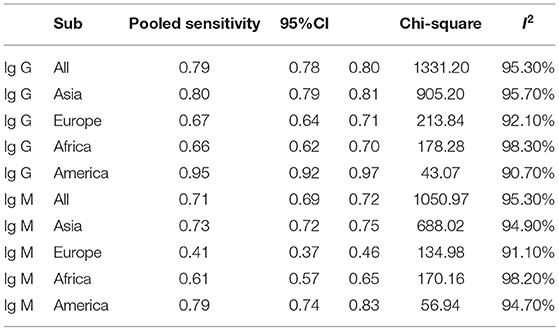
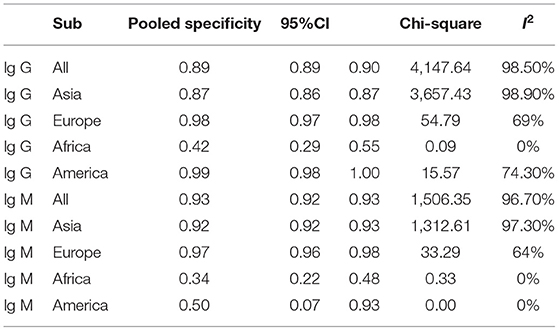
The pooled estimates for sensitivity and specificity of IgG to predicting COVID-19 diagnosis were 0.79 (95% CI: 0.78–0.80) and 0.89 (95% CI: 0.89–0.90), respectively, (Figure 2), corresponding to a PLR of 37.47 (95% CI: 14.59–96.26) (Appendix 3) and an NLR of 0.20 (95% CI: 0.16–0.27) (Appendix 4). The overall AUC was 0.97 (Figure 3) and the dOR was 209.78 (95% CI: 106.12–414.67) (Figure 4).
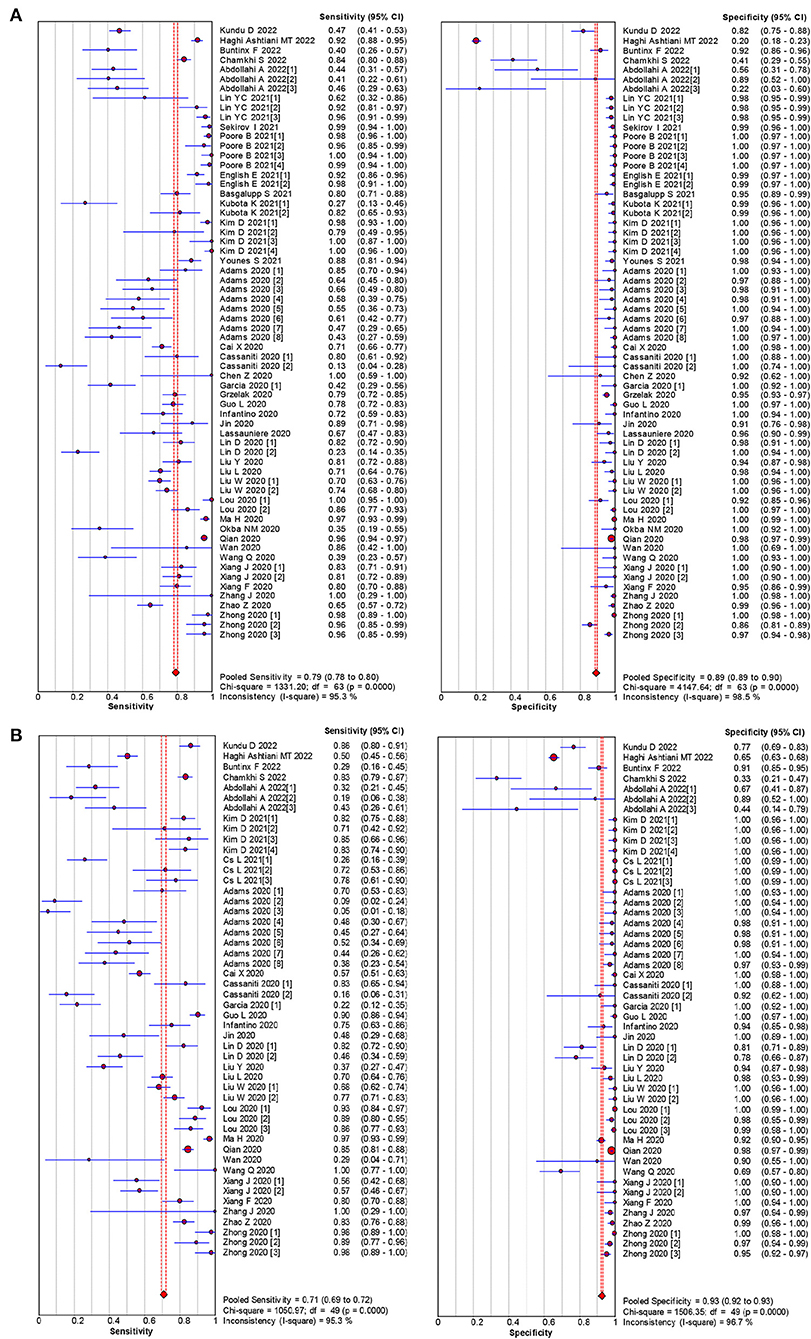
Figure 2. Forest plot of sensitivities and specificities IgG (A) and IgM (B) for predicting COVID-19 diagnosis.
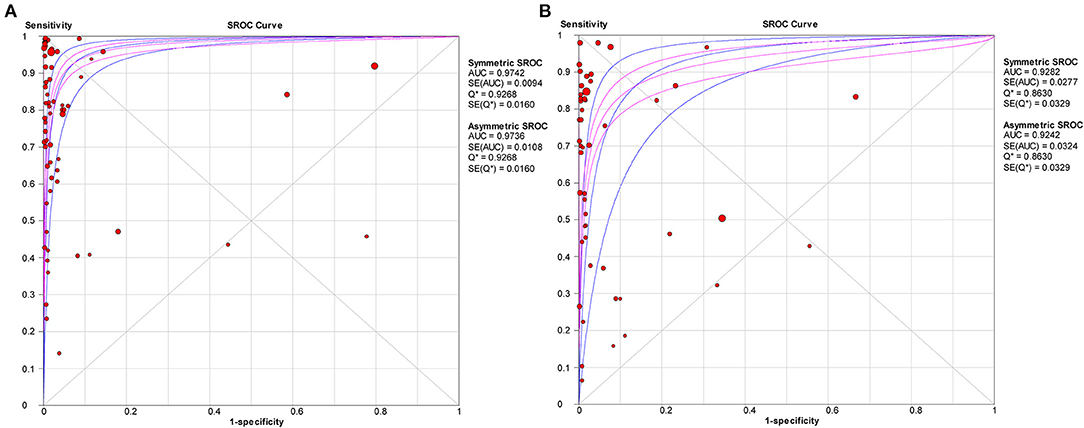
Figure 3. SROC curve with pooled estimates of sensitivity, specificity, and the AUC for all included studies IgG (A) and IgM (B) for predicting COVID-19 diagnosis. AUC, area under the SROC curve; SROC, summary receiver operator characteristic.
The pooled estimates for sensitivity and specificity of IgM to predicting COVID-19 diagnosis were 0.71 (95% CI: 0.69–0.72) and 0.93 (95% CI: 0.92–0.93), respectively, (Figure 2), corresponding to a PLR of 20.40 (95% CI: 11.71–35.55) (Appendix 5) and an NLR of 0.33 (95% CI: 0.25–0.42) (Appendix 6). The overall AUC was AUC was 0.93 (Figure 3) and the dOR was 78.17 (95% CI: 36.76–166.25) (Figure 4).
Subgroup analysis showed that there were differences among different regions. The studies in Asia with higher sensitivity of IgG and IgM to predicting COVID-19 diagnosis were 0.80 (95% CI: 0.79–0.81) and 0.73 (95% CI: 0.72–0.75), respectively (Table 1). However, the studies in Europe with lower sensitivity of IgG and IgM to predicting COVID-19 diagnosis were 0.67 (95% CI: 0.64–0.71) and 0.41 (95% CI: 0.37–0.46), respectively (Table 1). The studies in Europe with higher specificity of IgG and IgM to predicting COVID-19 diagnosis were 0.98 (95% CI: 0.97–0.98) and 0.97 (95% CI: 0.96–0.98), respectively, (Table 2). However, the studies in Africa with lower specificity of IgG and IgM to predicting COVID-19 diagnosis were 0.42 (95% CI: 0.29–0.55) and 0.34 (95% CI: 0.22–0.48), respectively, (Table 2). The Deeks' funnel plot for IgG and IgM was symmetrical (Figure 5), and the Deeks' test did not reject the hypothesis that there was no publication bias for IgG (t = −1.97, p = 0.052) and IgM (t = −1.88, p = 0.064).
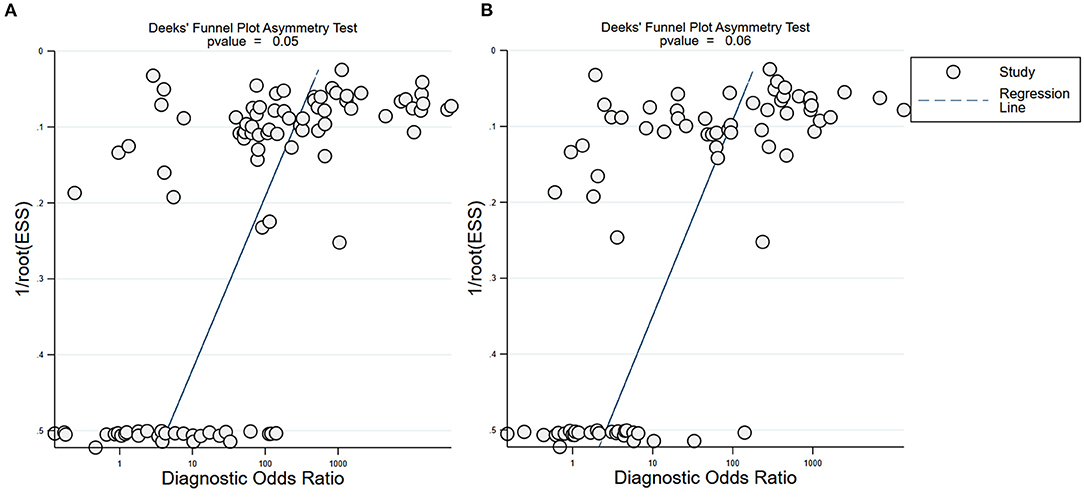
Figure 5. Deeks' funnel plot for IgG and IgM. Deeks' funnel plot to assess publication bias for IgG (A) and IgM (B). Plots show study size as a function of effect size for studies included in the meta-analysis. The dots represent each study.
Discussion
Although the association between antibodies and SARS-CoV-2 infection has been discussed in some recently published meta-analyses, the studies included in these studies are relatively limited, ranging from 14 to 29 (10–12). This is the maximum sample size, comprehensive, and updated review of the latest advances in the diagnosis of COVID-19 (7, 9, 23). Sixty-two studies were included with 35,775 participants in our meta-analysis. In view of the relatively strong infectivity of COVID-19, early detection, reporting, isolation and treatment are of great significance for the prevention and control of the spread of the infection. Following their encouraging accuracy as shown in the reviewed studies, RT-PCR nucleic acid and serum-specific antibody detection are recommended as the main screening criteria (24). At the same time, different methods of cross-test verification may further improve the sensitivity and specificity, such as serum-specific antibody detection, hence this may further reduce false negatives and false positives, and increase the accuracy of screening.
Furthermore, laboratory confirmed positive cases should meet one of the following two conditions. First, the two targets of COVID-19 (ORF1ab, N or E) in the same specimen should all be positive in real-time fluorescence RT-PCR (25). For example, if one target tests positive, it is necessary to re-sample and re-test it. If it still tests positive after re-sampling, then it is positive. Second, both samples should be positive for a single target by the real-time fluorescent RT-PCR, or both samples of the same type should be positive for a single target in 2 sampling tests, which could be judged as positive. In addition, the results of the reviewed studies suggest that N gene or E gene test could be used as a screening tool in the routine workflow, whereas RdRp gene (ORF1ab) could be used as a diagnostic tool, or it could be directly detected separately (26).
Current studies have shown that nucleic acid detection technology has the characteristics of early diagnosis, high sensitivity and specificity (27). However, in practice, this technology often yields false negatives. Therefore, the potential methods to reduce false negative results from the aspects of sample collection time, sample collection site and nucleic acid extraction process are worth discussing (7). First, mutations in the primers design area of the viral RNA may result in false negative test results. Thus, based on the preceding possibility, we recommend the following: collecting nasopharyngeal swabs within 3–7 days of onset (28), testing sputum samples in patients with negative RT-PCR results from pharyngeal swabs, and suspect or confirm a high probability of infection (29). However, most patients have been sick for more than 1 week at the time of treatment. In this regard, we recommend combining these with serological antibody detection (24, 30).
Additionally, due to the differences in the incubation period of individuals, we recommend multiple nucleic acid tests for clinically symptomatic patients in order to improve the detection rate (31). Alveolar lavage fluid sampling is not recommended because of the risk of trauma and cross-infection. According to the accurate selection of appropriate detection methods at different infection periods, we recommend nucleic acid detection immediately after showing clinical symptoms, and serological detection 7 days after having shown clinical symptoms (Figure 6).
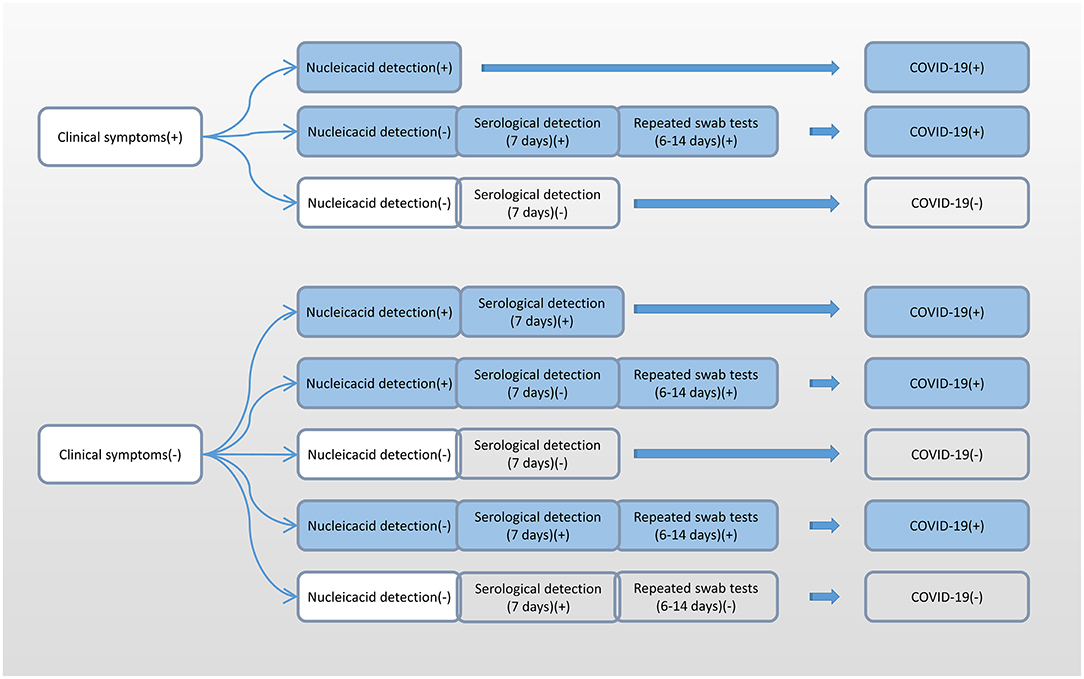
Figure 6. Flow chart of the selection of different detection methods for SARS-CoV-2 at different infection periods. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Moreover, studies have shown that both IgG and IgM positive rates increase after COVID-19 infection (24). Serum specific antibody can be used as an important indicator to evaluate COVID-19 infection, and IgM can effectively diagnose and exclude COVID-19 nucleic acid negative patients (24). Therefore, dynamic monitoring of viral IgM or IgG can be considered as a complementary means for nucleic acid detection (32). The flow chart of serum-specific antibody binding RT-PCR nucleic acid detection is summarized in Figure 7. Studies have shown that if nucleic acid is positive, then IgG and IgM are all negative, while if nucleic acid and IgM are positive, but IgG is negative, then patients are in the early stage of infection (32). In addition, patients with nucleic acid and IgG positive, but IgM negative, may be in the middle or late stage of COVID-19 infection or recurrent infection. Also, patients with nucleic acid positive, IgM positive, and IgG positive may be in the active period of the infection (33). Further, nucleic acid negative, IgM positive, and IgG negative results most likely suggest acute stage of the COVID-19 infection (34). Besides, nucleic acid negative, IgM negative, and IgG positive suggest that the patient may have been previously infected with COVID-19 (24).
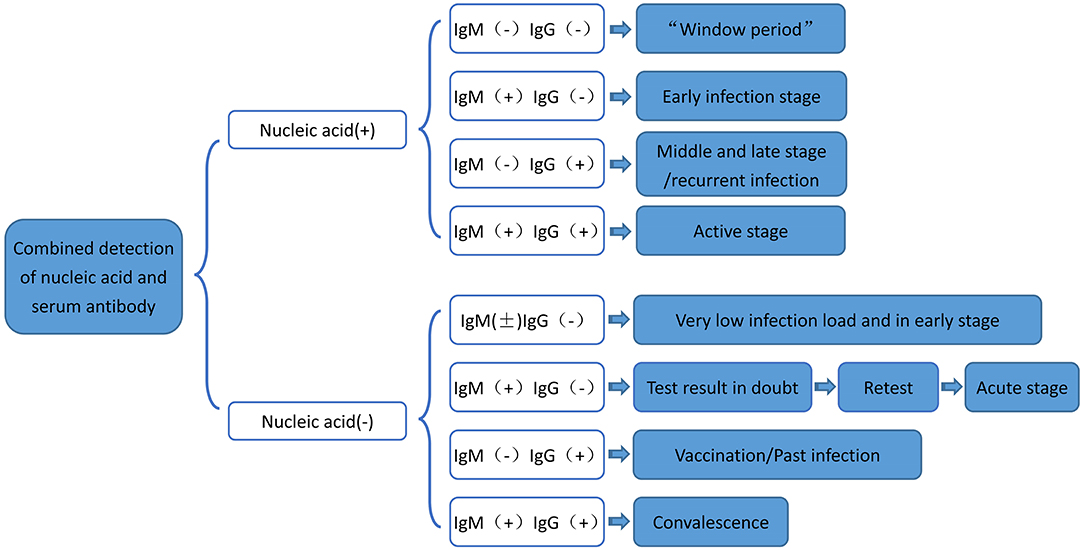
Figure 7. Flow chart of serum-specific antibody binding RT-PCR nucleic acid detection. RT-PCR, Real-time fluorescence quantification reverse transcriptase; COVID-19, 2019 novel coronavirus.
Subgroup analysis showed that there were differences among different regions. The studies in Asia with higher sensitivity of IgG and IgM to predicting COVID-19 diagnosis. However, the studies in Europe with lower sensitivity of IgG and IgM to predicting COVID-19 diagnosis. The studies in Europe with higher specificity of IgG and IgM to predicting COVID-19 diagnosis. However, the studies in Africa with lower specificity of IgG and IgM to predicting COVID-19 diagnosis. These results may be related to the intensity of immune response between different races to produce antibodies against the SARS-CoV-2 infection (35, 36). Previous study has shown that Hispanics and Blacks had higher rates of SARS-CoV-2 antibodies than Whites, indicating that SARS-CoV-2 spread disproportionately in racial and ethnic minorities during the COVID-19 pandemic (37, 38). Future studies should further explore the possible molecular mechanisms (39, 40).
The biggest advantage of the serum antibody kit is that the sample collection is easy, the sample processing and the experimental operation is simple, the test time is short, hence the speed is fast, and it is suitable for large-sample screening. However, antibodies also have the limitation of false negatives (33, 34), particularly resulting from (1) the type of preservatives and anticoagulants used; (2) improper specimen preservation; (3) laboratory operator error or improper preservation of the kit; and (4) too fast chromatography. In relation to (1), treating specimens with preservative sodium azide and anticoagulant EDTA can inhibit the activity of horseradish peroxidase (HRP), resulting in false negative enzyme linked immunosorbent assay (ELISA). As regards (2), when the specimen is kept in the refrigerator for too long, the IgG in the serum polymerizes into a polymer, and the immune activity of the antibody is weakened. Considering (3), if the reagent is not balanced to room temperature before use, the reagent dosage is insufficient, and the incubation time or temperature is insufficient; then the reagent is not stored as required, as a result it becomes contaminated or ineffective. Finally, taking (4), if the antigen antibody complex has not yet bound with the antibody on the detection line, then it is more likely that this occurs out of the detection line, resulting in false negatives. In practice, these problems should be avoided so that false negative results should be minimized.
Conclusions
Multidisciplinary cooperation can improve the diagnostic efficiency of COVID-19. We recommend the use of RT-PCR nucleic acid and serum-specific antibody detection as the main screening criteria. Through standard sampling, sample processing, combined accurate selection of appropriate detection methods in different infection stages of laboratory methods, sensitivity of detection can be improved.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
XP, PJ, and, AL contributed to the study design, while YF and AK contributed to the data collection. Interpretation of results was performed by AL and HL, whereas XP, YC, AL, and SW drafted the manuscript and edited the language. All authors participated in the critical revisions and approved the final version of the manuscript.
Funding
This study was supported by the International Institute of Spatial Lifecourse Health (ISLE), Wuhan University Specific Fund for Major School-level Internationalization Initiatives (WHU-GJZDZX-PT07), Hunan Provincial Key Laboratory of Clinical Epidemiology, and the Hunan Provincial Key Research and Development Program (2018SK2065), China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Central South University Library for his assistance during the literature search. Furthermore, we sincerely appreciate the hard work of the distinguished editors and reviewers, which improved the clarity of this article.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2022.819841/full#supplementary-material
References
1. The Lancet Infectious Disease. COVID-19, a pandemic or not? Lancet. Infect Dis. (2020) 20:383. doi: 10.1016/S1473-3099(20)30180-8
2. Ammad Ud Din M, Boppana LKT. An update on the 2019-nCoV outbreak. Am J Infect Control. (2020) 48:713. doi: 10.1016/j.ajic.2020.01.023
3. Pan Y, Li X, Yang G, Fan J, Tang Y, Zhao J, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect. (2020) 81:e28–32. doi: 10.1016/j.jinf.2020.03.051
4. Tang A, Tong Z-D, Wang H-L, Dai Y-X, Li K-F, Liu J-N, et al. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic Child, China. Emerg Infect Dis. (2020) 26:1337–9. doi: 10.3201/eid2606.20.0301
5. Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. (2020) 505:172–5. doi: 10.1016/j.cca.2020.03.009
6. Yang W, Yan F. Patients with RT-PCR confirmed COVID-19 and normal chest CT. Radiology. (2020) 295:E3. doi: 10.1148/radiol.2020200702
7. Singhal T. A review of coronavirus disease-2019 (COVID-19). Indian J Pediatr. (2020) 87:281–6. doi: 10.1007/s12098-020-03263-6
8. Salehi S, Abedi A, Balakrishnan S, Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR Am J Roentgenol. (2020) 215:87–93. doi: 10.2214/AJR.20.23034
9. Rodriguez-Morales AJ, Cardona-Ospina JA, Gutierrez-Ocampo E, Villamizar-Pena R, Holguin-Rivera Y, Escalera-Antezana JP, et al. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis. (2020) 34:101623. doi: 10.1016/j.tmaid.2020.101623
10. Zhang JJY, Lee KS, Ong CW, Chan MY, Ang LW, Leo YS, et al. Diagnostic performance of COVID-19 serological assays during early infection: a systematic review and meta-analysis of 11 516 samples. Influenza Other Respir Viruses. (2021) 15:529–38. doi: 10.1111/irv.12841
11. Vengesai A, Midzi H, Kasambala M, Mutandadzi H, Mduluza-Jokonya TL, Rusakaniko S, et al. A systematic and meta-analysis review on the diagnostic accuracy of antibodies in the serological diagnosis of COVID-19. Syst Rev. (2021) 10:155. doi: 10.1186/s13643-021-01689-3
12. Boger B, Fachi MM, Vilhena RO, Cobre AF, Tonin FS, Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am J Infect Control. (2021) 49:21–9. doi: 10.1016/j.ajic.2020.07.011
13. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. (2009) 339:b2535. doi: 10.1136/bmj.b2535
14. Pan X, Chiwanda Kaminga A, Liu A, Wen SW, Chen J, Luo J. Chemokines in non-alcoholic fatty liver disease: a systematic review and network meta-analysis. Front Immunol. (2020) 11:1802. doi: 10.3389/fimmu.2020.01802
15. Pan X, Kaminga AC, Chen J, Luo M, Luo J. Fetuin-A and Fetuin-B in non-alcoholic fatty liver disease: a meta-analysis and meta-regression. Int J Environ Res Public Health. (2020) 17:2735. doi: 10.3390/ijerph17082735
16. Pan X, Kaminga AC, Wen SW, Wu X, Acheampong K, Liu A. Dopamine and dopamine receptors in alzheimer's disease: a systematic review and network meta-analysis. Front Aging Neurosci. (2019) 11:175. doi: 10.3389/fnagi.2019.00175
17. Pan X, Wu X, Kaminga AC, Wen SW, Liu A. Dehydroepiandrosterone and dehydroepiandrosterone sulfate in alzheimer's disease: a systematic review and meta-analysis. Front Aging Neurosci. (2019) 11:61. doi: 10.3389/fnagi.2019.00061
18. Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-diSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol. (2006) 6:31. doi: 10.1186/1471-2288-6-31
19. Westwood ME, Whiting PF, Kleijnen J. How does study quality affect the results of a diagnostic meta-analysis? BMC Med Res Methodol. (2005) 5:20. doi: 10.1186/1471-2288-5-20
20. Pan X, Kaminga AC, Wen SW, Liu A. Catecholamines in post-traumatic stress disorder: a systematic review and meta-analysis. Front Mol Neurosci. (2018) 11:450. doi: 10.3389/fnmol.2018.00450
21. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. (2003) 327:557–60. doi: 10.1136/bmj.327.7414.557
22. Pan X, Wang Z, Wu X, Wen SW, Liu A. Salivary cortisol in post-traumatic stress disorder: a systematic review and meta-analysis. BMC Psychiatry. (2018) 18:324. doi: 10.1186/s12888-018-1910-9
23. Adhikari SP, Meng S, Wu Y-J, Mao Y-P, Ye R-X, Wang Q-Z, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. (2020) 9:29. doi: 10.1186/s40249-020-00646-x
24. Liang Y, Si Liu mS, Wu SM. Diagnostic value of novel coronavirus antibody in the diagnosis of new coronavirus pneumonia. J Wuhan Univ. (2020) 33:21–4.
25. Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. (2020) 66:549–55. doi: 10.1093/clinchem/hvaa029
26. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. (2020) 25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045
27. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. (2020) 581:465–9. doi: 10.1101/2020.03.05.20030502
28. Liu YB, Liu T, Cui Y, Wang B, Luo FM. Comparative study of novel coronavirus nucleic acid screening methods using two methods of nasal swabs and throat swabs. Chin J Respir Crit Care. (2020) 23:1–3.
29. Chen W, Chun YZ, Zhu Y, Yan HZ, You LB, Wu BS, et al. Chinese new novel coronavirus infection: comparison of virus nucleic acids in throat swabs and sputum specimens. Chin J Zoonosis. (2020) 20:211–7.
30. Chen J, Qi T, Liu L, Ling Y, Qian Z, Li T, et al. Clinical progression of patients with COVID-19 in Shanghai, China. J Infect. (2020) 80:e1–6. doi: 10.1016/j.jinf.2020.03.004
31. Shi S, Nie B, Guo Rowley Y, Zhang LL, Wen QL. Novel coronavirus pneumonia cases with virus nucleic acid detection results of various biological samples. West Chin Med J. (2020) 35:132–6.
32. Shanmugaraj B, Siriwattananon K, Wangkanont K, Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19). Asian Pac J Allergy Immunol. (2020) 38:10–8. doi: 10.12932/AP-200220-0773
33. Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. (2020) 9:382–5. doi: 10.1080/22221751.2020.1729069
34. Zheng M, Song L. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell Mol Immunol. (2020) 17:536–8. doi: 10.1038/s41423-020-0385-z
35. Pressman A, Lockhart SH, Wilcox J, Smits K, Etzell J, Albeiroti S, et al. COVID-19 in pregnancy by race and ethnicity: implications for development of a vaccination strategy. Womens Health. (2021) 17:17455065211063300. doi: 10.1177/17455065211063300
36. Coyer L, Boyd A, Schinkel J, Agyemang C, Galenkamp H, Koopman ADM, et al. SARS-CoV-2 antibody prevalence and correlates of six ethnic groups living in Amsterdam, the Netherlands: a population-based cross-sectional study, June-October 2020. BMJ Open. (2022) 12:e052752. doi: 10.1136/bmjopen-2021-052752
37. Kennedy JL, Forrest JC, Young SG, Amick B, Williams M, James L, et al. Temporal variations in seroprevalence of severe acute respiratory syndrome coronavirus 2 infections by race and ethnicity in arkansas. Open Forum Infect Dis. (2022) 9:ofac154. doi: 10.1093/ofid/ofac154
38. Coyer L, Boyd A, Schinkel J, Agyemang C, Galenkamp H, Koopman ADM, et al. Differences in SARS-CoV-2 infections during the first and second wave of SARS-CoV-2 between six ethnic groups in Amsterdam, the Netherlands: a population-based longitudinal serological study. Lancet Reg Health Eur. (2022) 13:100284. doi: 10.1016/j.lanepe.2021.100284
39. Yang S, Pan X, Yuan D, Zeng P, Jia P. Cross-disciplinary approaches to assist with nucleic acid testing for SARS-CoV-2. Appl Microbiol Biotechnol. (2020) 105:6291–9. doi: 10.1007/s00253-021-11498-2
Keywords: COVID-19, serum specific antibody, novel coronavirus, meta-analysis, nucleic acid detection
Citation: Pan X, Kaminga AC, Chen Y, Liu H, Wen SW, Fang Y, Jia P and Liu A (2022) Auxiliary Screening COVID-19 by Serology. Front. Public Health 10:819841. doi: 10.3389/fpubh.2022.819841
Received: 22 November 2021; Accepted: 21 June 2022;
Published: 02 August 2022.
Edited by:
Michaela Cellina, ASST Fatebenefratelli Sacco, ItalyCopyright © 2022 Pan, Kaminga, Chen, Liu, Wen, Fang, Jia and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Jia, jiapengff@hotmail.com; Aizhong Liu, lazroy@live.cn
 Xiongfeng Pan
Xiongfeng Pan Atipatsa C. Kaminga
Atipatsa C. Kaminga Yuyao Chen1
Yuyao Chen1 Shi Wu Wen
Shi Wu Wen Peng Jia
Peng Jia Aizhong Liu
Aizhong Liu