- 1College of Life Science and Oceanography, Shenzhen University, Shenzhen, China
- 2Department of Dermatology, Peking University Shenzhen Hospital, Shenzhen, China
- 3Department of Child Healthcare, Maternal and Child Health Hospital of Baoan, Shenzhen, China
Background: Autism spectrum disorder (ASD) is a complex neurodevelopmental disorder that affects millions of people worldwide. However, there are currently no reliable biomarkers for ASD diagnosis.
Materials and Methods: The strategy of computational prediction combined with experimental verification was used to identify blood protein biomarkers for ASD. First, brain tissue–based transcriptome data of ASD were collected from Gene Expression Omnibus database and analyzed to find ASD-related genes by bioinformatics method of significance analysis of microarrays. Then, a prediction program of blood-secretory proteins was applied on these genes to predict ASD-related proteins in blood. Furthermore, ELISA was used to verify these proteins in plasma samples of ASD patients.
Results: A total of 364 genes were identified differentially expressed in brain tissue of ASD, among which 59 genes were predicted to encode ASD-related blood-secretory proteins. After functional analysis and literature survey, six proteins were chosen for experimental verification and five were successfully validated. Receiver operating characteristic curve analyses showed that the area under the curve of SLC25A12, LIMK1, and RARS was larger than 0.85, indicating that they are more powerful in discriminating ASD cases from controls.
Conclusion: SLC25A12, LIMK1, and RARS might serve as new potential blood protein biomarkers for ASD. Our findings provide new insights into the pathogenesis and diagnosis of ASD.
Introduction
Autism spectrum disorder (ASD) is a complex neurodevelopment disorder characterized by impairments in social interaction and communication, as well as expression of restricted interests and repetitive behavior (1). These symptoms would be presented during the first 3 years of life. Boys are with four to five times higher risk of autism than girls (2). According to reports of 2015, ~24.8 million people worldwide were affected by autism (3). In developed countries, the proportion of children with autism increased from 0.67% in 2000 to 1.5% in 2017 (4, 5). Obviously, the number of patients with ASD is increasing year by year.
Genetic and environmental factors are generally acknowledged as important contributors to the pathogenesis of ASD (6). However, the exact pathological mechanism remains uncertain and there are no effective treatments for ASD. Studies show that early intervention with behavioral therapy at an early stage can improve patient social communication and reduce anxiety and aggression. Thus, it is critical to detect ASD at an early stage (7, 8). Currently, the clinical diagnosis of ASD is based on the fifth edition of the Statistical Manual of Mental Disorder (DSM-V) (9), which may lead to exclusion of autistic individuals with mild form. Therefore, there is a need to find useful and reliable biomarkers to assist the diagnosis of autism.
Blood is a potential source for disease biomarker discovery because it contains large numbers of proteins associated with the physiology or pathology of disease. Several studies have been performed to search for blood biomarkers of ASD. Smith et al. (10) found that the combination of glutamine, glycine, and ornithine amino acid dysregulation identified a dysregulation in amino acid/branched-chain amino acid metabolism with good specificity and positive predictive value in the ASD subject cohort. Momeni et al. (11) found three differentially expressed peptides in the heparin plasma of children with ASD. Ngounou Wetie et al. (12) reported that apolipoproteins (Apos) ApoA1, ApoA4, and serum paraoxanase/arylesterase 1 (PON1) were increased in the sera of children with ASD. Wu et al. (13) proposed a movement biomarker to characterize the neurodevelopment level, which could differentiate ASD subjects from typically developing individuals. Howsmon et al. (14) developed multivariate statistical models to distinguish children with ASD from controls based on the metabolic abnormalities. Oztan et al. (15) employed a multidimensional neuropeptide analysis and found low blood neuropeptide receptor might act as promising biomarker of disease presence and symptom severity in ASD. Recently, we found a protein pattern that could distinguish the plasma samples of autistic children from healthy controls (16). In addition, we identified 41 proteins as differentially expressed proteins in the peripheral blood mononuclear cells of autistic children (17); three of them, i.e., complement C3 (C3), calreticulin (CALR), and alpha-1-antitrypsin (SERPINA1), are common differential proteins in the plasma (16). Despite these advances, there are still no diagnostic biomarkers available for ASD nowadays.
It is generally known that the blood–brain barrier (BBB) plays an important role in the defense of the central nervous system by limiting harmful solutes, macromolecules, and cells circulating from the bloodstream into the brain. However, several studies have shown that dysfunctions of BBB had a relationship with pathogenesis of neurological diseases including ASD (18–21), suggesting that some ASD-related proteins might be secreted from brain into blood as potential biomarkers. In addition, Cui et al. (22) developed a computational method to predict whether a protein could be secreted from tissue into blood with a high accuracy. Therefore, it would be possible to apply this program on the proteins encoded by the ASD-related genes to predict some potential ASD-related proteins in blood.
In this study, we identified blood protein biomarkers for ASD through computational prediction combined with experimental verification. First, we identified ASD-related genes by analyzing brain tissue–based gene expression data of autistic patients and healthy controls collected from a public database. Then, we predicted whether the protein products of these genes could be secreted into blood as ASD-related proteins. Further, we made bioinformatics analysis and literature survey on these proteins, and then selected some ASD-related proteins for verification in plasma of children with ASD by ELISA analysis.
Materials and Methods
Collection of Brain Tissue–Based Gene Expression Data of ASD Patients
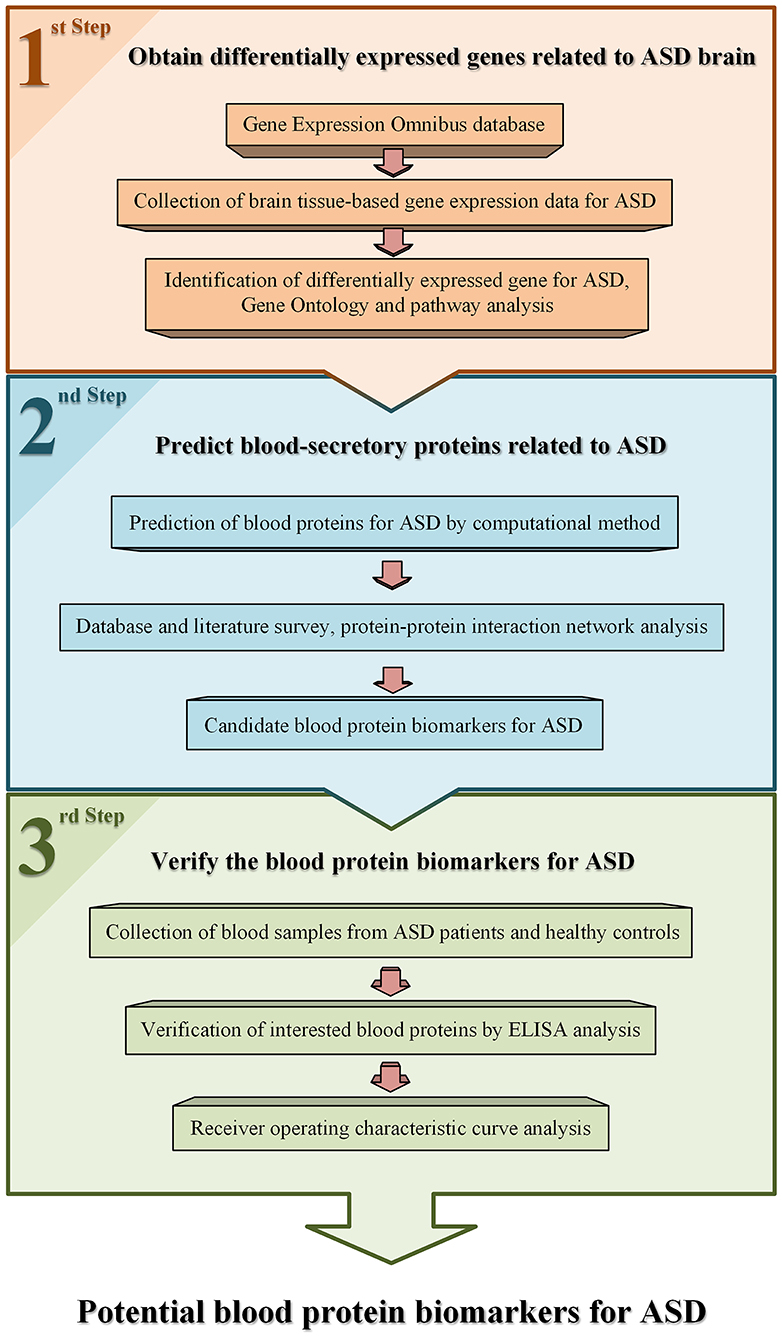
The workflow used in this study is shown in Figure 1. Gene expression data of brain tissues from patients with ASD and healthy controls were collected from public database Gene Expression Omnibus (GEO) (23). One dataset, GSE28521 (24), was selected for data analysis according to the following criteria. First, it contains both brain tissue samples of ASD patients and healthy controls. Second, the number of samples for ASD and controls are larger than 10, respectively. There are 79 brain tissue samples obtained from 19 autistic patients and 17 healthy controls in this dataset. Among these samples, 10, 16, and 13 samples are from cerebellum, frontal cortex, and temporal cortex of autism cases, and 11, 16, and 13 samples are from corresponding tissues of controls, respectively. The average age of the patients and controls were 24 (ranged from 2 to 56) and 34.6 (ranged from 16 to 56), respectively. The ratio of male to female was about 14:5 and 16:1 in the patients and controls. Detailed information of these samples in this dataset can be accessed from the GEO database.
Identification of Differentially Expressed Genes for ASD
Generally, different tissues have different gene expression patterns. We investigated the differentially expressed genes of cerebellum, frontal cortex, and temporal cortex for ASD patients, respectively. A computational method, significance analysis of microarrays (SAM) (25), was employed to identify differentially expressed genes for ASD. A statistic delta was calculated for each gene in SAM, measuring how strong the relationship between gene expression and a response variable. R package “siggenes” was used to implement SAM analysis. To obtain the appropriate number of differentially expressed genes of ASD, delta was 1.2 and the false discovery rate (FDR) was 0.05 as cutoff.
To understand the functions of the differentially expressed genes, Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) (26) was used to conduct Gene Ontology (GO) annotation and pathway analysis on these genes. In addition, functional interaction network analysis was performed using ClueGO cytoscape plugin [GlueGO v2.5.7; (27)].
Prediction of ASD-Related Proteins in Blood
All differentially expressed genes of ASD were analyzed to determine whether their protein products could be secreted into blood by using a prediction program developed by Cui et al. (22). The main idea of the program is described as follows. Human proteins known to be blood secretory or not were collected from the published data to constitute the positive and negative training data, respectively. A list of protein features including sequence, structure, and chemical and physical properties was examined, and core features were selected according to their abilities in distinguishing the positive data from the negative. Based on the core features and training data, a prediction program for blood-secretory proteins was constructed by using support vector machine (SVM) (28) method.
In addition, to further determine whether these predicted proteins associated with ASD and presented in blood, we compared their genes with autism-associated gene database AutismKB (http://db.cbi.pku.edu.cn/autismkb_v2/) (29), and the proteins with plasma protein database (PPD, http://www.plasmaproteomedatabase.org/) (30), respectively. Moreover, protein–protein interaction (PPI) network analysis was conducted by using Lens for Enrichment and Network Studies of Proteins (LENS, http://severus.dbmi.pitt.edu/LENS/) (31) and Search Tool for the Retrieval of Interacting Genes/Proteins (String database, http://string-db.org/) (32).
Verification of Potential Blood Protein Biomarkers for ASD by Using ELISA
After the aforementioned prediction of blood-secretory proteins associated with ASD, we selected some potential protein biomarkers for ASD to validate according to the following criteria. First, we ranked these proteins according to the likelihood of protein secretion into blood derived from the prediction program. Then we compared their genes with autism-associated gene database AutismKB and the proteins with plasma protein database. Further, we made functional analysis and literature survey on these proteins. Based on this criterion, we selected six proteins for verification by ELISA. ELISA analysis was conducted on blood samples of children with ASD and healthy controls. The research protocol of this study was permitted by the Human Research Ethics Committee of Shenzhen University and performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. A total of 40 subjects were recruited from Maternal and Child Health Hospital of Baoan between September 2017 and September 2018, including 20 children with ASD, and 20 age- and gender-matched healthy controls. The written consents were obtained from the caregivers of the participating children before this experiment. These patients were all diagnosed by a child neuropsychiatrist based on the criteria defined in the DSM-V (33). The male to female ratio was 4:1. The average age was 4.7 for patients and 4.5 for controls. The control cases had no known neurological disorders. There were no significant differences in weight, height, or body mass index (BMI) between the autistic children and healthy subjects. Blood samples (5 ml) were collected with EDTA-coated plastic tubes in the morning and then centrifuged at 3,000 × g for 10 min at room temperature. The supernatants were divided into aliquots and stored at −80°C until further analysis.
For ELISA analysis, the protein concentration was measured by a commercial ELISA kit (Uscn Life Science, Wuhan, China) according to the manufacturer's instructions, and then normalized by the total protein concentration determined by bicinchoninic acid (BCA) protein assay kit (Beyotime, Jiangsu, China). G-test (34) was used to detect the outliers in the normalized data. GraphPad Prism 5 software (GraphPad Software, San Diego, California) was applied to make statistical analyses on the concentrations of protein in ASD patients vs. healthy controls by using t-test with p-value < 0.05 as cutoff.
Results
Identification of Differentially Expressed Genes in Brain Tissues of ASD
There were 283 probes (3 up-regulated, 280 down-regulated) and 142 probes (3 up-regulated and 139 down-regulated) identified differentially expressed with FDR <0.05 as cutoff in the frontal cortex and temporal cortex of ASD, respectively. There were no differentially expressed probes found in the cerebellum of ASD. After combining the up- and down-regulated probes identified in the frontal and temporal cortex of ASD, six probes (corresponding to six genes) and 373 probes (corresponding to 358 genes) were differentially up- and down-regulated in cortex of ASD, respectively (Supplementary Tables 1, 2).
To assess the functions of these differentially expressed genes, GO annotation and pathway analyses were conducted by using DAVID database. A total of 72 GO terms and 18 pathways were significantly enriched by these genes with p-value <0.05 as threshold (Supplementary Tables 3–6). The enriched GO terms include 31 biological processes (BP), 19 cellular components (CC), and 22 molecular functions (MF). The top 10 enriched terms of BP, CC, and MF are shown in Figures 2A–C. BP analysis showed that they were involved in tricarboxylic acid (TCA) cycle, neurotransmitter transport, ATP hydrolysis coupled proton transport, synaptic vesicle exocytosis, canonical glycolysis, exocytosis, response to calcium ion, glycolytic process, actin and cytoskeleton organization, etc. CC analysis showed that they were associated with mitochondrion, myelin sheath, synaptic vesicle membrane, mitochondrial matrix, neurofilament, postsynaptic density, and synaptic vesicle. MF included ATP binding, calcium ion binding, syntaxin binding, etc. In addition, pathway analysis showed that some metabolic pathways were enriched, including carbon metabolism, TCA cycle, and oxidative phosphorylation (Figure 2D). Interestingly, pathways of Alzheimer's disease (AD), Huntington disease (HD), and amyotrophic lateral sclerosis (ALS) were also enriched by these genes. Similarly, the enrichment of BP and CC on these genes by ClueGO cytoscape plugin showed that they were mainly associated with glucose catabolic process, proton-transporting two-sector ATPase complex, catalytic domain, Schaffer collateral-CA1 synapse, and vesicle-mediated transport in synapse (Figure 3A, Supplementary Table 7). Pathway analysis showed that they were mainly involved in the citric acid (TCA) cycle and respiratory electron transport, mitochondrial protein import, and glycolysis and gluconeogenesis (Figure 3B, Supplementary Table 8).
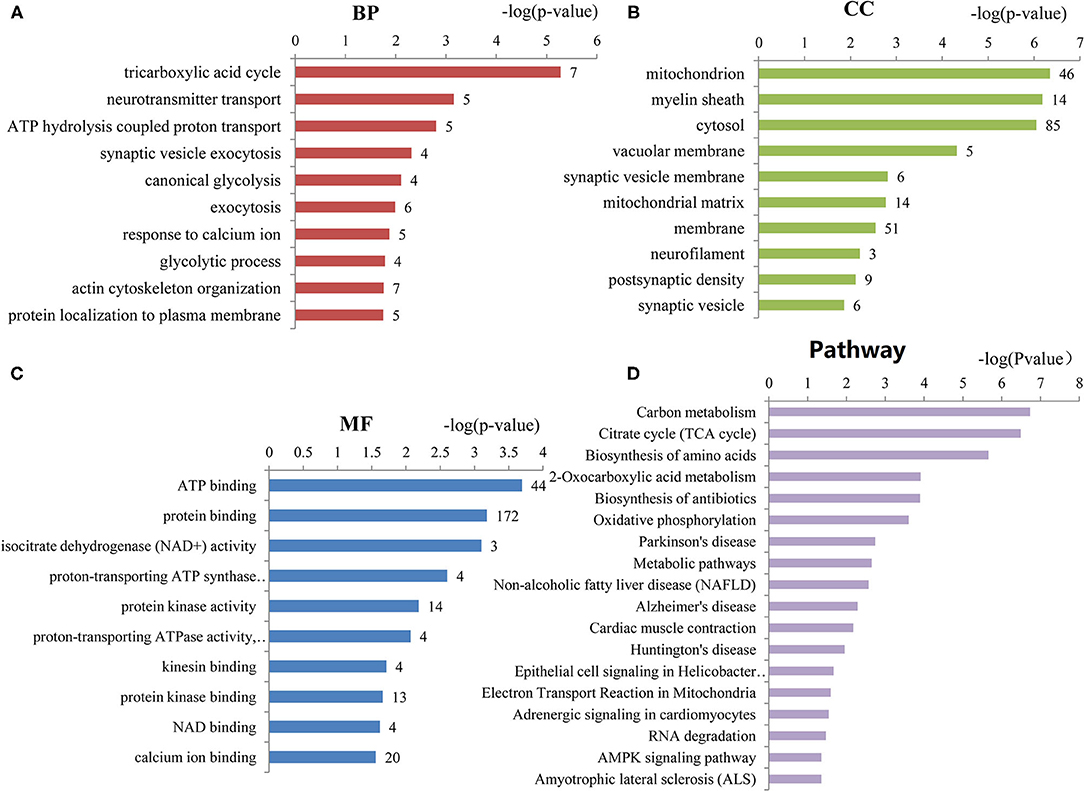
Figure 2. The top 10 enriched GO terms of biological processes, cellular components, and molecular functions. (A) Biological processes (BP) enriched by the differentially expressed genes of ASD. (B) Cellular components (CC) enriched by the differentially expressed genes of ASD. (C) Molecular functions (MF) enriched by the differentially expressed genes of ASD. The number of proteins associated with each category is presented at the end of each bar. (D) The pathways enriched by the differentially expressed genes of ASD. The number of proteins enriched in each pathway is at the end of each bar.
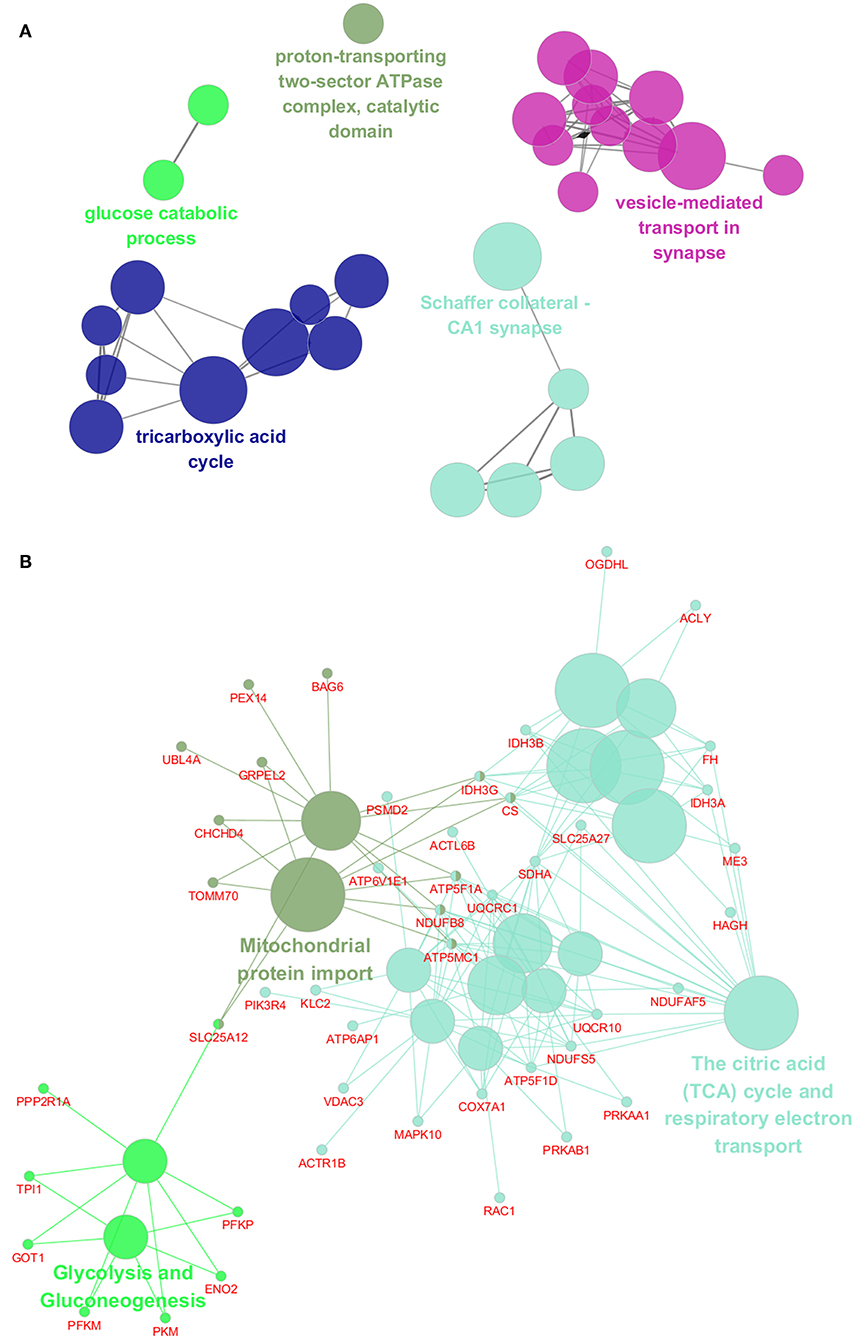
Figure 3. Functional interaction network analysis of the differentially expressed genes. (A) The differentially expressed genes were mapped to the GO categories, including biological processes and cellular components by using ClueGO cytoscape plugin. (B) The differentially expressed genes were mapped to the KEGG pathway, REACTOME pathway, and Wiki pathway by using ClueGO cytoscape plugin.
Prediction of ASD-Related Proteins in Blood
Based on the differentially expressed genes of ASD, we applied a computational program developed by Cui et al. (22) on these genes to predict whether their protein products could be secreted from tissue into blood. Subsequently, 59 proteins encoded by down-regulated genes were predicted to be blood-secretory proteins, suggesting that they might act as ASD-related proteins in blood (Supplementary Table 9).
To examine whether the predicted blood-secretory proteins were present in blood, we compared these proteins with proteins in PPD (30) and found that 43 proteins were in common (Figure 4A, Supplementary Table 9). To further determine whether these predicted proteins were associated with ASD, we compared their corresponding genes with autism-related genes listed in the AutismKB (29) and found that 55 genes were reported associated with ASD except TTC13, RARS, THOC5, and ATPAF1 (Figure 4B, Supplementary Table 9). There were 40 overlapping blood-secretory proteins between PPD and AutismKB database (Figure 4C, Supplementary Table 9). After literature survey on the four genes, we found that RARS and THOC5 had been reported related to brain development (35, 36).
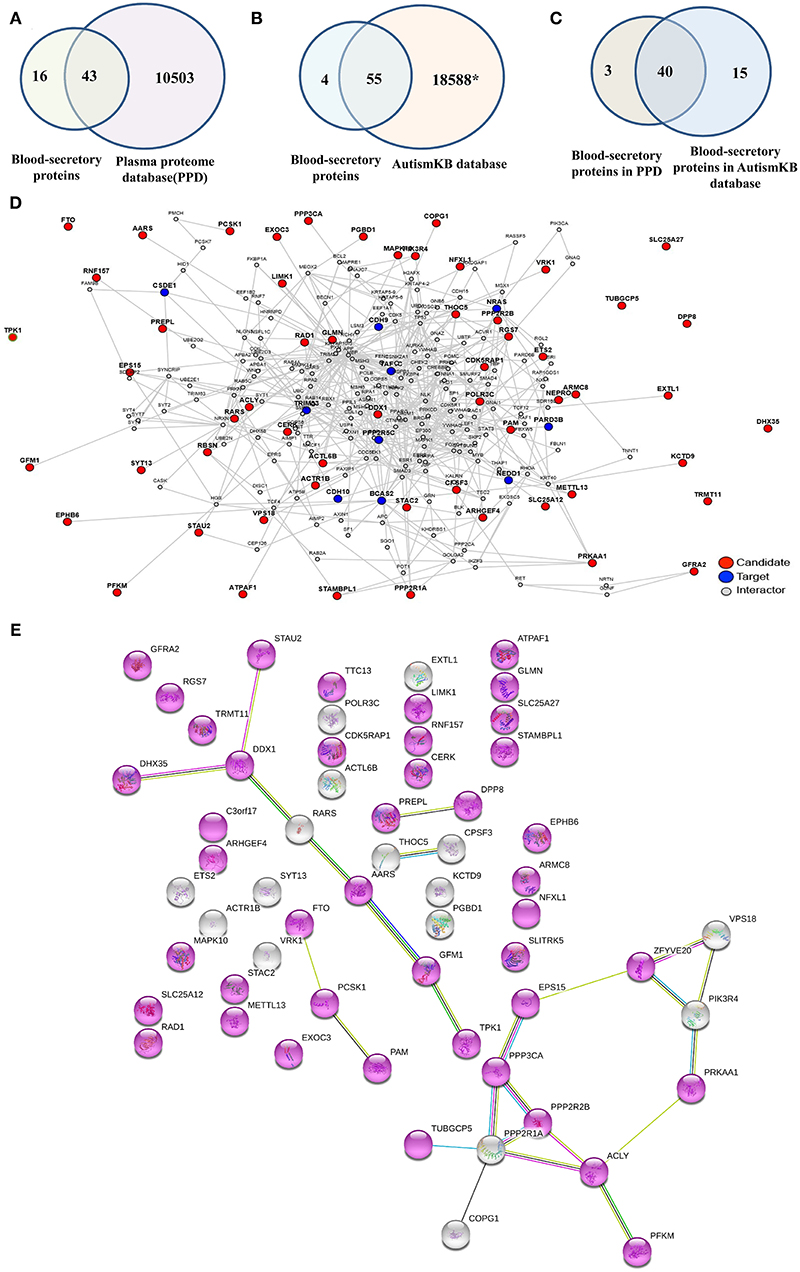
Figure 4. Database survey and protein–protein interaction network analysis on the 59 blood-secretory proteins. (A) Compared with plasma protein database. (B) Compared with ASD-related database (AutismKB). *The AutismKB database contains 1,379 genes, 5,420 copy number variants (CNVs)/structural variations (SVs), 11,669 single-nucleotide variations (SNVs)/insertions and deletions (InDels), and 172 linkage regions associated with ASD. (C) The blood-secretory proteins overlapped in plasma protein database and AutismKB database. (D) The protein–protein interaction network of these 59 blood-secretory proteins. The predicted blood-secretory proteins are shown as red nodes and autism pathology-related proteins are shown as blue nodes. (E) UniProt keywords were enriched by using String database. Gene with a node color of purplish red, whose Uniprot keyword is alternative splicing.
In order to understand how these predicted proteins were involved in the pathogenesis of ASD, we conducted a PPI network analysis on these proteins by using a web tool of LENS. Figure 4D shows the network, which was constructed by 59 blood-secretory proteins (red nodes) input as candidate proteins and 15 proteins known related to autism pathology (blue nodes) provided as targeted proteins. From the network, we found that most of these blood-secretory proteins were connected with the targeted proteins except SLITRK5, TPK1, SLC25A27, DHX35, DPP8, TTC13, TUBGCP5, FTO, and TRMT11. Interestingly, String database analysis showed that 43 proteins might be associated with alternative splicing (Figure 4E).
Verification of the Potential Protein Biomarkers for ASD by ELISA
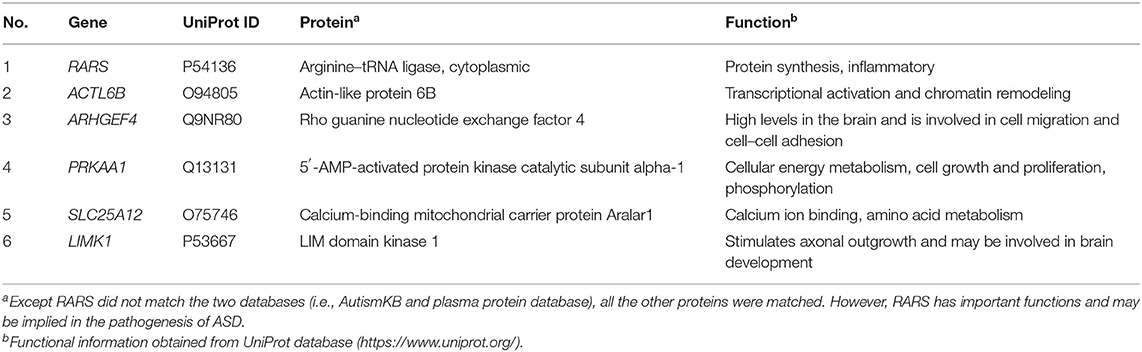
Based on the possibilities of proteins secreting into blood and their functions, six proteins were selected for validation in blood samples of children with ASD and healthy controls, including arginine-tRNA ligase, cytoplasmic (RARS), actin-like protein 6B (ACTL6B), 5′-AMP-activated protein kinase catalytic subunit alpha-1 (PRKAA1), calcium-binding mitochondrial carrier protein Aralar1 (SLC25A12), LIM domain kinase 1 (LIMK1), and rho guanine nucleotide exchange factor 4 (ARHGEF4) (Table 1). As shown in Figure 5, five proteins, RARS, ACTL6B, PRKAA1, SLC25A12, and LIMK1, were significantly down-regulated in plasma samples of ASD, which were consistent with the expression changes of their corresponding genes mentioned previously. Even though the expression level of ARHGEF4 was not significantly down-regulated in autism samples, it was still expressed lower in autism samples compared with controls.
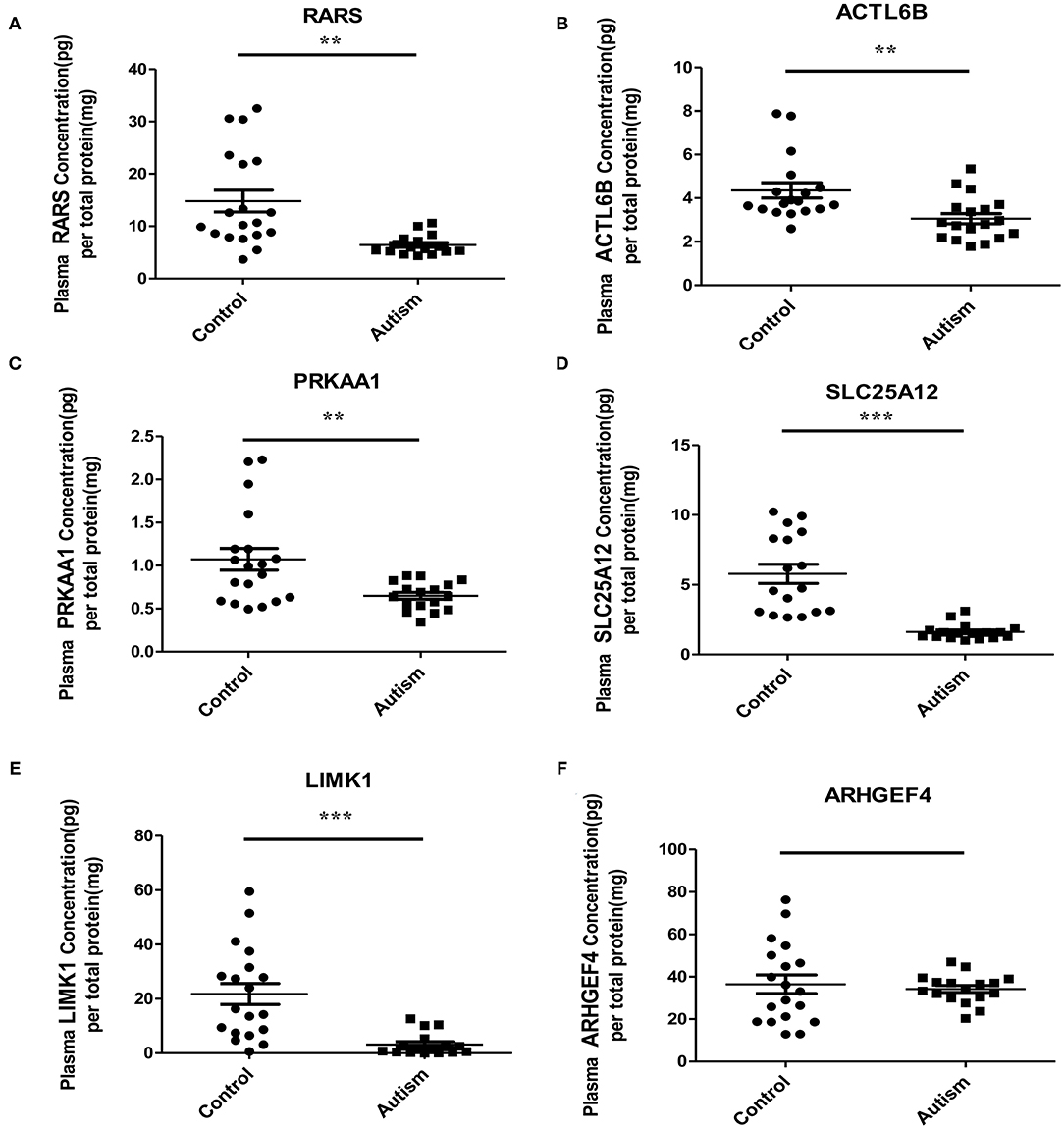
Figure 5. Verification of the potential blood protein biomarkers for ASD by ELISA. **p < 0.005; ***p < 0.0005.
To evaluate the performance of these five proteins in distinguishing samples of ASD from healthy controls, receiver operating characteristic (ROC) curve analyses were carried out on protein concentrations measured by ELISA. Figure 6 shows that SLC25A12 has the most discriminative ability with the area under curve (AUC) of 0.976 (sensitivity 100%, specificity 88.2%), and the AUCs of LIMK1 and RARS are 0.898 (sensitivity 94.7%, specificity 75.0%) and 0.862 (sensitivity 84.2%, specificity 81.2%), respectively. The remaining two proteins ACTL6B and PRKAA1 are with AUCs of 0.793 and 0.768, respectively.
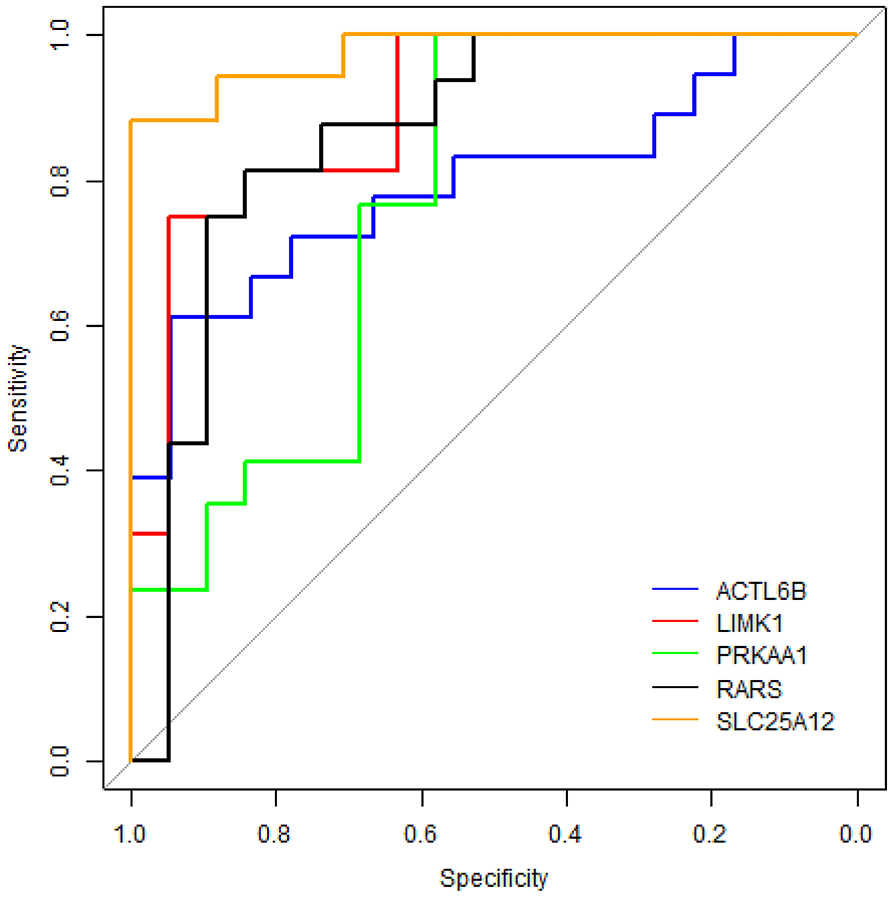
Figure 6. ROC curve analyses on five differentially expressed proteins in plasma samples of ASD patients. The blue line represents protein ACTL6B, the red line is LIMK1, the green line is PRKAA1, the black line is RARS, and the orange line is SLC25A12.
Discussion
ASD is a neurodevelopment disorder that has affected the health of millions of people. However, the pathogenesis of ASD is poorly understood and there are no reliable diagnostic biomarkers currently. In this study, we identified the potential blood protein biomarkers for ASD by a new strategy of computational prediction in conjunction with experimental validation, which could provide a more effective and specific way for biomarker discovery in blood (37).
First of all, 364 differentially expressed genes were identified for ASD based on transcriptome analysis. Functional enrichment analysis showed that these genes were mainly involved in BPs of TCA cycle, neurotransmitter transport, and synaptic vesicle exocytosis; CCs of mitochondrion, myelin sheath, and synaptic vesicle membrane; actin and neurofilament cytoskeleton organization and synapse; MFs of ATP binding, calcium ion binding, and syntaxin binding; and pathways of metabolic, nervous system diseases, and alternative splicing, which are all known to be associated with the pathophysiology of ASD.
From the aforementioned functional analysis, it could be speculated that the mitochondria, myelin sheath, synapses, and cytoskeleton of neurofilaments are impaired in the brains of children with ASD. Previous studies have reported that mitochondrial dysfunction seemed to be the most prevalent metabolic disease associated with ASD (38, 39). Mitochondrial dysfunction could lead to metabolic changes. Here, the metabolic abnormalities include carbon metabolism, TCA cycle, oxidative phosphorylation, glycolysis, and gluconeogenesis, which are consistent with previous published research (40–42). In addition, changes in myelin sheath, and actin and neurofilament cytoskeleton have been reported associated with ASD (43–47). In agreement with previous research (24, 44, 45, 47–51), the genes associated with pre-synaptic and post-synaptic proteins, synaptic vesicles, and neurotransmitter transport were observed as significantly changed in ASD subjects vs. controls. Furthermore, it has been reported that differential alternative splicing was observed in ASD brains and blood (24, 52, 53), and the unfolded protein response and altered endoplasmic reticulum (ER) stress have also been reported to be associated with ASD (17, 54, 55). It should be a concern that these factors are interrelated with each other. Dysfunction of mitochondria might cause impairment of synaptic function, and both of them are related to neurological diseases such as AD, schizophrenia, and so on (42, 56). Alternative splicing has been reported to be related to the expression of synaptic-related genes in ASD (57). Mutations linked to ASDs in synaptic proteins such as NLGN3, CASPR2, and CADM1 might lead to ER stress conditions (58).
After functional analysis and literature survey, six proteins were selected for verified in plasma samples of ASD, and five were successfully verified, including RARS, ACTL6B, PRKAA1, SLC25A12, and LIMK1. Among them, RARS acts as an enzyme essential for RNA translation and plays an important role in myelination (35). ACTL6B was identified as a candidate risk gene for ASD with functions of neuron-specific chromatin remodeling and neurodevelopment (59, 60). PRKAA1, a catalytic subunit of protein kinase A (PKA), plays a key role in regulating cellular energy metabolism. It was found that regression in ASD might be associated with decreased PKA-mediated phosphorylation of proteins and abnormalities in cellular signaling (61). PRKAA1 has also been reported in several studies related to autism and/or ASD including linkage studies (62–64), NGS de novo mutation studies (65), and genome-wide association studies (66). SLC25A12 has been proposed as a candidate gene for ASD due to its important role in mitochondrial function and ATP synthesis (67). Some research showed that single nucleotide polymorphism in SLC25A12 might significantly contribute to ASD risk (68, 69). Increased evidence suggests that it may play a critical role in the pathogenesis of ASD (69–72). In particular, it has been reported that SLC25A12 is associated with autism of the Han Chinese in Taiwan (70). Meanwhile, it is worth noting that SLC25A12 is with the highest “evidence score” in the AutismKB, indicating that it is closely associated with ASD. Moreover, LIMK1 stimulates axonal outgrowth and involves in neurodevelopment and synaptic plasticity (73, 74). It has been reported to be related to ASD (24, 63). Furthermore, ARHGEF has been reported to be associated with copy number variants (CNVs) in children with ASD (75–77). Here, although it has no significant difference between the cases and the control group, the trend was in line with expectations and a larger sample size might be needed to verify the change.
ROC curve analyses showed that the AUCs of SLC25A12, LIMK1, and RARS were larger than 0.85, indicating that they are more powerful in distinguishing samples of ASD from healthy controls and might serve as new potential protein biomarkers for ASD in blood. As far as we know, this is the first study to investigate blood protein biomarkers for ASD through such a strategy. Proteins SLC25A12, LIMK1, and RARS were first reported here as new potential blood protein biomarkers for ASD. Clearly, these findings are needed to be confirmed on large number of samples.
In conclusion, the combination of computational prediction and experimental validation was used to identify blood protein biomarkers for ASD. A total of 364 differentially expressed genes were found in ASD, out of which 59 genes were predicted that their protein products could be secreted into blood as candidate ASD-related blood proteins. After functional analysis and literature survey, six proteins were selected for experimental validation and five were successfully verified in the plasma samples of ASD. ROC analysis showed that SLC25A12, LIMK1, and RARS are more powerful in differentiating ASD samples from controls and might serve as new potential protein biomarkers for ASD in blood.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Human Research Ethics Committee of Shenzhen University. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
LS conceived and designed the study and revised the article. FY performed statistical analysis, data interpretation, and drafted the first article. KZ and XL contributed to experimental validation and data analysis. CF and YG contributed to acquisition of blood samples and performed correlation analysis. JN coordinated the study design and article revision. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 31870825), the Shenzhen Science and Technology Innovation Commission (Grant Nos. JCYJ20170818102400688 and JCYJ20170412110026229), and Shenzhen-Hong Kong Institute of Brain Science–Shenzhen Fundamental Research Institutions (Grant No. 2019SHIBS0003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Yan Zhang for some advices on data analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2021.554621/full#supplementary-material
Abbreviations
ASD, autism spectrum disorder; BBB, blood–brain barrier; GEO, gene expression omnibus; SAM, significance analysis of microarrays; FDR, false discovery rate; DAVID, database for annotation, visualization and integrated discovery; GO, gene ontology; SVM, support vector machine; PPD, plasma protein database; PPI, protein–protein interaction; LENS, lens for enrichment and network studies of proteins; BP, biological process; CC, cellular component; MF, molecular function; HD, Huntington disease; ALS, amyotrophic lateral sclerosis; ROC, receiver operating characteristic; AUC, area under curve.
References
1. Black DW, Grant JE, American Psychiatric Association. DSM-5 Guidebook: The Essential Companion to the Diagnostic and Statistical Manual of Mental Disorders. 5th ed. Washington, DC: American Psychiatric Publishing (2014).
2. Werling DM, Geschwind DH. Sex differences in autism spectrum disorders. Curr. Opin. Neurol. (2013) 26:146–53. doi: 10.1097/WCO.0b013e32835ee548
3. G.D.a.I.I.a.P Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the global burden of disease study 2015. Lancet. (2016) 388:1545–602. doi: 10.1016/S0140-6736(16)31678-6
4. Lyall K, Croen L, Daniels J, Fallin MD, Ladd-Acosta C, Lee BK, et al. The changing epidemiology of autism spectrum disorders. Annu. Rev. Public Health. (2017) 38:81–102. doi: 10.1146/annurev-publhealth-031816-044318
5. Xu G, Strathearn L, Liu B, Bao W. Prevalence of autism spectrum disorder among US children and adolescents, 2014-2016. JAMA. (2018) 319:505. doi: 10.1001/jama.2018.0001
6. Chaste P, Leboyer M. Autism risk factors: genes, environment, and gene-environment interactions. Dialogues Clin. Neurosci. (2012) 14:281–92. doi: 10.31887/DCNS.2012.14.3/pchaste
7. Dawson G, Burner K. Behavioral interventions in children and adolescents with autism spectrum disorder: a review of recent findings. Curr. Opin. Pediatr. (2011) 23:616–20. doi: 10.1097/MOP.0b013e32834cf082
8. Lai MC, Lombardo MV, Baron-Cohen S. Autism. Lancet. (2014) 383:896–910. doi: 10.1016/S0140-6736(13)61539-1
9. Kocsis RN. Diagnostic and statistical manual of mental disorders: fifth edition (DSM-5). Int. J. Offender Ther. Comp. Criminol. (2013) 57:1546–8. doi: 10.1177/0306624X13511040
10. Smith AM, King JJ, West PR, Ludwig MA, Donley ELR, Burrier RE, et al. Amino acid dysregulation metabotypes: potential biomarkers for diagnosis and individualized treatment for subtypes of autism spectrum disorder. Biol. Psychiatry. (2019) 85:345–54. doi: 10.1016/j.biopsych.2018.08.016
11. Momeni N, Bergquist J, Brudin L, Behnia F, Sivberg B, Joghataei MT, et al. A novel blood-based biomarker for detection of autism spectrum disorders. Transl. Psychiatry. (2012) 2:e91. doi: 10.1038/tp.2012.19
12. Ngounou Wetie AG, Wormwood K, Thome J, Dudley E, Taurines R, Gerlach M, et al. A pilot proteomic study of protein markers in autism spectrum disorder. Electrophoresis. (2014) 35:2046–54. doi: 10.1002/elps.201300370
13. Wu D, Jose JV, Nurnberger JI, Torres EB. A biomarker characterizing neurodevelopment with applications in autism. Sci. Rep. (2018) 8:614. doi: 10.1038/s41598-017-18902-w
14. Howsmon DP, Kruger U, Melnyk S, James SJ, Hahn J. Classification and adaptive behavior prediction of children with autism spectrum disorder based upon multivariate data analysis of markers of oxidative stress and DNA methylation. PLoS Comput. Biol. (2017) 13:e1005385. doi: 10.1371/journal.pcbi.1005385
15. Oztan O, Jackson LP, Libove RA, Sumiyoshi RD, Phillips JM, Garner JP, et al. Biomarker discovery for disease status and symptom severity in children with autism. Psychoneuroendocrinology. (2018) 89:39–45. doi: 10.1016/j.psyneuen.2017.12.022
16. Shen L, Zhang K, Feng C, Chen Y, Li S, Iqbal J, et al. iTRAQ-based proteomic analysis reveals protein profile in plasma from children with autism. Proteomics Clin. Appl. (2018) 12:e1700085. doi: 10.1002/prca.201700085
17. Shen L, Feng C, Zhang K, Chen Y, Gao Y, Ke J, et al. Proteomics study of peripheral blood mononuclear cells (PBMCs) in autistic children. Front. Cell. Neurosci. (2019) 13:105. doi: 10.3389/fncel.2019.00105
18. Theoharides TC, Doyle R. Autism, gut-blood-brain barrier, and mast cells. J. Clin. Psychopharmacol. (2008) 28:479–83. doi: 10.1097/JCP.0b013e3181845f48
19. Theoharides TC, Doyle R, Francis K, Conti P, Kalogeromitros D. Novel therapeutic targets for autism. Trends. Pharmacol. Sci. (2008) 29:375–82. doi: 10.1016/j.tips.2008.06.002
20. Theoharides TC, Zhang B. Neuro-inflammation, blood-brain barrier, seizures and autism. J Neuroinflammation. (2011) 8:168. doi: 10.1186/1742-2094-8-168
21. Fiorentino M, Sapone A, Senger S, Camhi SS, Kadzielski SM, Buie TM, et al. Blood-brain barrier and intestinal epithelial barrier alterations in autism spectrum disorders. Mol. Autism. (2016) 7:49. doi: 10.1186/s13229-016-0110-z
22. Cui J, Liu Q, Puett D, Xu Y. Computational prediction of human proteins that can be secreted into the bloodstream. Bioinformatics. (2008) 24:2370–5. doi: 10.1093/bioinformatics/btn418
23. Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. (2002) 30:207–10. doi: 10.1093/nar/30.1.207
24. Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. (2011) 474:380–4. doi: 10.1038/nature10110
25. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. (2001) 98:5116–21. doi: 10.1073/pnas.091062498
26. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. (2009) 37:1–13. doi: 10.1093/nar/gkn923
27. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. (2009) 25:10913. doi: 10.1093/bioinformatics/btp101
28. Souza BF, Carvalho AP. Gene selection based on multi-class support vector machines and genetic algorithms. Genet. Mol. Res. (2005) 4:599–607.
29. Yang C, Li J, Wu Q, Yang X, Huang AY, Zhang J, et al. AutismKB 2.0: a knowledgebase for the genetic evidence of autism spectrum disorder. Database. (2018) 2018:bay106. doi: 10.1093/database/bay106
30. Nanjappa V, Thomas JK, Marimuthu A, Muthusamy B, Radhakrishnan A, Sharma R, et al. Plasma proteome database as a resource for proteomics research: 2014 update. Nucleic Acids Res. (2014) 42:D959–65. doi: 10.1093/nar/gkt1251
31. Handen A, Ganapathiraju MK. LENS: web-based lens for enrichment and network studies of human proteins. BMC Med. Genomics. (2015) 8(Suppl. 4):S2. doi: 10.1186/1755-8794-8-S4-S2
32. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. (2019) 47:D607–13. doi: 10.1093/nar/gky1131
33. Francesmonneris APH, First M. Diagnostic and Statistical Manual of Mental Disorders: DSM-V. Washington, DC: American Psychiatric Association (2013).
34. McDonald JH. G-test of goodness-of-fit. In: Handbook of Biological Statistics. 3rd ed. Baltimore, MD: Sparky House Publishing (2014).
35. Wolf NI, Salomons GS, Rodenburg RJ, Pouwels PJ, Schieving JH, Derks TG, et al. Mutations in RARS cause hypomyelination. Ann. Neurol. (2014) 76:134–9. doi: 10.1002/ana.24167
36. Maeder CI, Kim JI, Liang X, Kaganovsky K, Shen A, Li Q, et al. The THO complex coordinates transcripts for synapse development and dopamine neuron survival. Cell. (2018) 174:1436–49.e1420. doi: 10.1016/j.cell.2018.07.046
37. Yao F, Zhang K, Zhang Y, Guo Y, Li A, Xiao S, et al. Identification of blood biomarkers for Alzheimer's disease through computational prediction and experimental validation. Front. Neurol. (2018) 9:1158. doi: 10.3389/fneur.2018.01158
38. Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol. Psychiatry. (2012) 17:290–314. doi: 10.1038/mp.2010.136
39. Frye RE. Metabolic and mitochondrial disorders associated with epilepsy in children with autism spectrum disorder. Epilepsy Behav. (2015) 47:147–57. doi: 10.1016/j.yebeh.2014.08.134
40. Frye RE, Melnyk S, Macfabe DF. Unique acyl-carnitine profiles are potential biomarkers for acquired mitochondrial disease in autism spectrum disorder. Transl. Psychiatry. (2013) 3:e220. doi: 10.1038/tp.2012.143
41. Cheng N, Rho JM, Masino SA. Metabolic dysfunction underlying autism spectrum disorder and potential treatment approaches. Front. Mol. Neurosci. (2017) 10:34. doi: 10.3389/fnmol.2017.00034
42. Kim Y, Vadodaria KC, Lenkei Z, Kato T, Gage FH, Marchetto MC, et al. Mitochondria, metabolism, and redox mechanisms in psychiatric disorders. Antioxid Redox Signal. (2019) 31:275–317. doi: 10.1089/ars.2018.7606
43. Zikopoulos B, Barbas H. Changes in prefrontal axons may disrupt the network in autism. J. Neurosci. (2010) 30:14595–609. doi: 10.1523/JNEUROSCI.2257-10.2010
44. Lewis TLJr, Courchet J, Polleux F. Cell biology in neuroscience: cellular and molecular mechanisms underlying axon formation, growth, and branching. J. Cell Biol. (2013) 202:837–48. doi: 10.1083/jcb.201305098
45. Krishnan A, Zhang R, Yao V, Theesfeld CL, Wong AK, Tadych A, et al. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat. Neurosci. (2016) 19:1454–62. doi: 10.1038/nn.4353
46. Wei H, Ma Y, Liu J, Ding C, Hu F, Yu L. Proteomic analysis of cortical brain tissue from the BTBR mouse model of autism: evidence for changes in STOP and myelin-related proteins. Neuroscience. (2016) 312:26–34. doi: 10.1016/j.neuroscience.2015.11.003
47. Chang Q, Yang H, Wang M, Wei H, Hu F. Role of microtubule-associated protein in autism spectrum disorder. Neurosci. Bull. (2018) 34:1119–26. doi: 10.1007/s12264-018-0246-2
48. Kwong WH, Chan WY, Lee KK, Fan M, Yew DT. Neurotransmitters, neuropeptides and calcium binding proteins in developing human cerebellum: a review. Histochem. J. (2000) 32:521–34. doi: 10.1023/A:1004197210189
49. Berger RH, Miller AL, Seifer R, Cares SR, LeBourgeois MK. Acute sleep restriction effects on emotion responses in 30- to 36-month-old children. J. Sleep Res. (2012) 21:235–46. doi: 10.1111/j.1365-2869.2011.00962.x
50. Geoffray MM, Nicolas A, Speranza M, Georgieff N. Are circadian rhythms new pathways to understand autism spectrum disorder? J. Physiol. (2016) 110:434–8. doi: 10.1016/j.jphysparis.2017.06.002
51. Miyamoto H, Shimohata A, Abe M, Abe T, Mazaki E, Amano K, et al. Potentiation of excitatory synaptic transmission ameliorates aggression in mice with Stxbp1 haploinsufficiency. Hum. Mol. Genet. (2017) 26:4961–74. doi: 10.1093/hmg/ddx379
52. Stamova BS, Tian Y, Nordahl CW, Shen MD, Rogers S, Amaral DG, et al. Evidence for differential alternative splicing in blood of young boys with autism spectrum disorders. Mol. Autism. (2013) 4:30. doi: 10.1186/2040-2392-4-30
53. Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. (2016) 540:423–7. doi: 10.1038/nature20612
54. Crider A, Ahmed AO, Pillai A. Altered expression of endoplasmic reticulum stress-related genes in the middle frontal cortex of subjects with autism spectrum disorder. Mol. Neuropsychiatry. (2017) 3:85–91. doi: 10.1159/000477212
55. Kawada K, Mimori S. Implication of endoplasmic reticulum stress in autism spectrum disorder. Neurochem. Res. (2018) 43:147–52. doi: 10.1007/s11064-017-2370-1
56. Hollis F, Kanellopoulos AK, Bagni C. Mitochondrial dysfunction in autism spectrum disorder: clinical features and perspectives. Curr. Opin. Neurobiol. (2017) 45:178–87. doi: 10.1016/j.conb.2017.05.018
57. Smith RM, Sadee W. Synaptic signaling and aberrant RNA splicing in autism spectrum disorders. Front. Synaptic Neurosci. (2011) 3:1. doi: 10.3389/fnsyn.2011.00001
58. Trobiani L, Favaloro FL, Di Castro MA, Di Mattia M, Cariello M, Miranda E, et al. UPR activation specifically modulates glutamate neurotransmission in the cerebellum of a mouse model of autism. Neurobiol. Dis. (2018) 120:139–50. doi: 10.1016/j.nbd.2018.08.026
59. Vogel-Ciernia A, Wood MA. Neuron-specific chromatin remodeling: a missing link in epigenetic mechanisms underlying synaptic plasticity, memory, and intellectual disability disorders. Neuropharmacology. (2014) 80:18–27. doi: 10.1016/j.neuropharm.2013.10.002
60. Krupp DR, Barnard RA, Duffourd Y, Evans SA, Mulqueen RM, Bernier R, et al. Exonic mosaic mutations contribute risk for autism spectrum disorder. Am. J. Hum. Genet. (2017) 101:369–90. doi: 10.1016/j.ajhg.2017.07.016
61. Ji L, Chauhan V, Flory MJ, Chauhan A. Brain region-specific decrease in the activity and expression of protein kinase A in the frontal cortex of regressive autism. PLoS ONE. (2011) 6:e23751. doi: 10.1371/journal.pone.0023751
62. Liu J, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind D, et al. A genomewide screen for autism susceptibility loci. Am. J. Hum. Genet. (2001) 69:327–40. doi: 10.1086/321980
63. Buxbaum JD, Silverman J, Keddache M, Smith CJ, Hollander E, Ramoz N, et al. Linkage analysis for autism in a subset families with obsessive-compulsive behaviors: evidence for an autism susceptibility gene on chromosome 1 and further support for susceptibility genes on chromosome 6 and 19. Mol. Psychiatry. (2004) 9:144–50. doi: 10.1038/sj.mp.4001465
64. Ylisaukko-oja T, Alarcon M, Cantor RM, Auranen M, Vanhala R, Kempas E, et al. Search for autism loci by combined analysis of autism genetic resource exchange and finnish families. Ann. Neurol. (2006) 59:145–55. doi: 10.1002/ana.20722
65. Cho SC, Yoo HJ, Park M, Cho IH, Kim BN, Kim JW, et al. Genome-wide association scan of korean autism spectrum disorders with language delay: a preliminary study. Psychiatry Invest. (2011) 8:61–6. doi: 10.4306/pi.2011.8.1.61
66. Dong S, Walker MF, Carriero NJ, DiCola M, Willsey AJ, Ye AY, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. (2014) 9:16–23. doi: 10.1016/j.celrep.2014.08.068
67. Napolioni V, Persico AM, Porcelli V, Palmieri L. The mitochondrial aspartate/glutamate carrier AGC1 and calcium homeostasis: physiological links and abnormalities in autism. Mol. Neurobiol. (2011) 44:83–92. doi: 10.1007/s12035-011-8192-2
68. Turunen JA, Rehnstrom K, Kilpinen H, Kuokkanen M, Kempas E, Ylisaukko-Oja T. Mitochondrial aspartate/glutamate carrier SLC25A12 gene is associated with autism. Autism Res. (2008) 1:189–92. doi: 10.1002/aur.25
69. Kim SJ, Silva RM, Flores CG, Jacob S, Guter S, Valcante G, et al. A quantitative association study of SLC25A12 and restricted repetitive behavior traits in autism spectrum disorders. Mol. Autism. (2011) 2:8. doi: 10.1186/2040-2392-2-8
70. Chien WH, Wu YY, Gau SS, Huang YS, Soong WT, Chiu YN, et al. Association study of the SLC25A12 gene and autism in Han Chinese in Taiwan. Prog. Neuropsychopharmacol. Biol. Psychiatry. (2010) 34:189–92. doi: 10.1016/j.pnpbp.2009.11.004
71. Carayol J, Schellenberg GD, Dombroski B, Genin E, Rousseau F, Dawson G. Autism risk assessment in siblings of affected children using sex-specific genetic scores. Mol. Autism. (2011) 2:17. doi: 10.1186/2040-2392-2-17
72. Jiao Y, Chen R, Ke X, Cheng L, Chu K, Lu Z, et al. Single nucleotide polymorphisms predict symptom severity of autism spectrum disorder. J. Autism Dev. Disord. (2012) 42:971–83. doi: 10.1007/s10803-011-1327-5
73. Dong Q, Ji YS, Cai C, Chen ZY. LIM kinase 1 (LIMK1) interacts with tropomyosin-related kinase B (TrkB) and Mediates brain-derived neurotrophic factor (BDNF)-induced axonal elongation. J. Biol. Chem. (2012) 287:41720–31. doi: 10.1074/jbc.M112.405415
74. Cuberos H, Vallee B, Vourc'h P, Tastet J, Andres CR, Benedetti H. Roles of LIM kinases in central nervous system function and dysfunction. FEBS Lett. (2015) 589:3795–806. doi: 10.1016/j.febslet.2015.10.032
75. van der Zwaag B, Franke L, Poot M, Hochstenbach R, Spierenburg HA, Vorstman JA, et al. Gene-network analysis identifies susceptibility genes related to glycobiology in autism. PLoS ONE. (2009) 4:e5324. doi: 10.1371/journal.pone.0005324
76. Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. (2010) 466:368–72. doi: 10.1038/nature09146
Keywords: autism spectrum disorder, blood, protein, biomarker, computational, experimental
Citation: Yao F, Zhang K, Feng C, Gao Y, Shen L, Liu X and Ni J (2021) Protein Biomarkers of Autism Spectrum Disorder Identified by Computational and Experimental Methods. Front. Psychiatry 12:554621. doi: 10.3389/fpsyt.2021.554621
Received: 29 April 2020; Accepted: 19 January 2021;
Published: 25 February 2021.
Edited by:
Raz Gross, Sheba Medical Center, IsraelReviewed by:
Idan Menashe, Ben-Gurion University of the Negev, IsraelSalvatore Chirumbolo, University of Verona, Italy
A. Gulhan Ercan-Sencicek, Masonic Medical Research Institute (MMRI), United States
Copyright © 2021 Yao, Zhang, Feng, Gao, Shen, Liu and Ni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liming Shen, slm@szu.edu.cn
 Fang Yao
Fang Yao Kaoyuan Zhang
Kaoyuan Zhang Chengyun Feng
Chengyun Feng Yan Gao3
Yan Gao3 Liming Shen
Liming Shen