- 1National Tobacco Genetic Engineering Research Center, Yunnan Academy of Tobacco Agricultural Sciences, Kunming, Yunnan, China
- 2College of Resources and Environmental Science, Yunnan Agricultural University, Kunming, Yunnan, China
- 3Plant Functional Component Research Center, Tobacco Research Institute of Chinese Academy of Agricultural Sciences, Qingdao, Shandong, China
- 4Key Laboratory of Economic Plants and Biotechnology, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, China
- 5State Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
The pyridine alkaloid nicotine acts as one of best-studied plant resistant traits in tobacco. Previous research has shown that NtERF199 and NtERF189, acting as master regulators within the NIC1 and NIC2 locus, quantitatively contribute to nicotine accumulation levels in N. tabacum. Genome editing-created Nic1(Nterf199) and Nic2 (Nterf189) double mutant provides an ideal platform for precisely dissecting the defensive role of nicotine and the connection between the nicotine biosynthetic pathway with other putative metabolic networks. Taking this advantage, we performed a comparative transcriptomic analysis to reevaluate the potential physiological and metabolic changes in response to nicotine synthesis defect by comparing the nic1nic2 and NIC1NIC2 plants. Our findings revealed that nicotine reduction could systematically diminishes the expression intensities of genes associated with stimulus perception, signal transduction and regulation, as well as secondary metabolic flux. Consequently, this global expression reduction might compromise tobacco adaptions to environmental fitness, herbivore resistances, and plant growth and development. The up-regulation of a novel set of stress-responsive and metabolic pathway genes might signify a newly established metabolic reprogramming to tradeoff the detrimental effect of nicotine loss. These results offer additional compelling evidence regarding nicotine’s critical defensive role in nature and highlights the tight link between nicotine biosynthesis and gene expression levels of quantitative resistance-related genes for better environmental adaptation.
Introduction
Plant secondary metabolites play a pivotal role as essential bioactive compounds in defending against predators and optimizing plant environmental fitness (Steppuhn et al., 2004). Among these bioactive compounds, alkaloids constitute a class of alkaline organic substances characterized by a cyclic nitrogenous nucleus and are known for their functions in defending against herbivores and pathogen attacks (Tso, 1990; Ashihara et al., 2010). In Nicotiana species, 3-pyridyl derivative alkaloids are in vivo synthesized and amounts vary in organs with distinct major alkaloid profiles, such as nicotine, nornicotine, and anabasine (Sisson and Saunders, 1982; Saitoh et al., 1985). In cultivated tobacco (Nicotiana tabacum), nicotine is the predominant alkaloid and its content in tobacco leaves is recognized as an essential standard for commercial use (Davis and Nielsen, 1999). Pharmaceutically, nicotine binds to neuronal nicotinic acetylcholine receptor subtypes throughout the nervous systems and stimulates midbrain dopamine neuron firing rates and phasic burst firing, and results in neuroadaptations (Tobacco TCPGT, 2008; Wittenberg et al., 2020). Therefore, nicotine serves as the primary addictive substance in combusted tobacco products and e-cigarettes. For public health concerns and the goal of reducing addiction, both the World Health Organization (2015) and the Food and Drug Administration of the United States (2018) have considered mandating standards to minimize nicotine levels in combustible cigarettes to nonaddictive levels (Lewis et al., 2020). The recommended threshold of non-addiction nicotine in the tobacco filler is set below 0.04% (World Health Organization, 2015).
In N. tabacum, nicotine is exclusively synthesized in root tissues and subsequently translocated to aerial organs via the xylem system, and accumulates in leaf cells (Dawson, 1941; Thurston et al., 1966). The nicotine molecule is characterized by the presence of a pyrrolidine ring and a pyridine ring. The nicotine content in commercial tobacco cultivars typically ranges between 2% and 4% of the total dry weight basis (Saitoh et al., 1985) and exhibits significant variability influenced by genetic factors, environmental conditions, and agronomic practices (Tso, 1990; Bush et al., 1993). Genetic studies have revealed that alkaloid levels in cultivated tobacco are primarily regulated by two loci, NIC1 and NIC2 (Legg et al., 1969 and Legg and Collins, 1971). Among these loci, NIC1 exerts a 2.4 times stronger effect in determining nicotine and associated alkaloid levels when compared to NIC2 (Legg and Collins, 1971). Both NIC1 and NIC2 loci consist of tandem sets of homologous ERF transcription factors located on chromosomes 7 and 19 within the tobacco genome (Shoji et al., 2010; Kajikawa et al., 2017; Sui et al., 2020). The recessive alleles of Nic1 and Nic2 in nicotine mutants have been attributed to chromosomal deletions (Shoji et al., 2010; Kajikawa et al., 2017) and potentially chromosomal deletion-induced structural variations (Sui et al., 2020; Shoji et al., 2022). Two homologous genes, NtERF199 and NtERF189, which act as master transcriptional activators within the NIC1 and NIC2 ERF clusters, have been found to directly bind to the GCC-box element in the promoters of pathway structural genes, thereby coordinately activating their expression (Shoji et al., 2010; Sui et al., 2020). Furthermore, CRISPR/Cas9-based double mutants have exhibited a low-nicotine phenotype through the simultaneous genome editing of NtERF199 and NtERF189 (referred to as Nic1-2 and Nic2-2; Hayashi et al., 2020). Natural mutations in Nic1 and Nic2 have been associated with pleiotropic effects on leaf yields, chlorophyll content, and harvest timing (Chaplin and Weeks, 1976). However, near-isogenic Nic1 and Nic2 lines generated through traditional breeding methods tend to introduce additional variations caused by chromosomal structural changes which are tightly linked to target genes (Shoji et al., 2010; Kajikawa et al., 2017; Sui et al., 2020). This fact inevitably impedes the precise pinpointing of the impact of the defensive resistance conferred by nicotine. In contrast, the Nic1-2 and Nic2-2 double mutants exhibited no abnormalities in growth and development under experimental conditions (Hayashi et al., 2020; Shoji et al., 2023). Therefore, these double mutants provide an ideal system to evaluate the putative defense trait of nicotine in nature. Since they differ only in the loss of function of NtERF199 and NtERF189 while otherwise remaining genetically identical (Bergelson and Purrington, 1996; Steppuhn et al., 2004). Taking this advantage, it is interesting to investigate the correlation between plant innate immunity system and the defensive trait of nicotine in tobacco.
In the present study, we conducted a comprehensive comparative transcriptome analysis between wild-type tobacco (NIC1NIC2) and low-nicotine lines (Nic1-2Nic2-2) that share an identical genomic background. This transcriptomic profiling spanned the entire spectrum of tobacco growth and encompassed early stages in response to topping. The transcriptome analysis provided compelling evidence that nicotine reduction leads to differentially transcriptomic and metabolic alterations at each of the investigated growth stages. The differentially expressed genes identified are intricately linked to various aspects of the plant innate immunity system, including environmental stimulus perception, signal transduction cascade, defensive response, transcriptional regulation, as well as secondary metabolite biosynthesis. In summary, our findings demonstrate that loss-of-nicotine leads to a comprehensive reduction in the expression of a diverse array of genes associated with environmental adaptability, herbivore resistance, and various facets of plant growth and development. Notably, we also observed the up-regulation of a novel set of stress-responsive and metabolic pathway genes which might signify a newly metabolic reprogramming strategy to trade-off the detrimental effects of nicotine loss and compensate overall environmental fitness.
Materials and methods
Experimental materials and RNA extraction
Flue-cured tobacco variety Yunyan87 (WT) and Nic1-2Nic2-2 lines (Hayashi et al., 2020) with identical genomic background were cultivated under standard greenhouse condition at the Yanhe Tobacco Experimental Station in Yuxi, Yunnan province. Seedlings were transplant into plastic pots (33 cm × 27 cm) with cultural media and managed following routine greenhouse practices. To comprehensively capture the transcriptomic differences throughout the different representative growth periods between WT and Nic1-2Nic2-2, three vegetative growth stages were selected, including the seedling stage (SD), rosette stage (RS), and before topping stage (BT). In the SD, the typical seedlings usually have thick leaves with relatively large leaf area and enhanced photosynthetic capability. The plantlet also has newly established root system and transport tissue with strong absorption capabilities. The RS refers to the period from seedling transplant to a tobacco plant with 30~35cm in height and 12-16 unfolded leaves. These leaves horizontally developed with the width as twice as the height of the plant. This stage signifies the peak of root growth after tobacco transplant and the underground growth outweighs above-ground growth, with a substantial development of lateral and fine roots. The BT stage extends from plant rosette to the emergence of flower buds and is characterized by vigorous vegetative growth accompanied by flower bud differentiation. During this period, the lateral and fine roots continue to grow, and adventitious roots are formed abundantly. The stem elongates rapidly and the leaves are enlarged and overlapped, spreading out in a trumpet shape. Importantly, the emergence of the flower buds in a tobacco plant signals a transition from a vegetative growth stage to a reproductive growth stage. Flower buds must be removed by topping practice to allow the plant to reach its full yield and improved quality at harvest. After topping (T) practices, different plant phytohormone signaling cascades could be triggered within tobacco plant, which in turn activates their downstream gene expression and initiates the accumulation of diverse array of secondary metabolites. Therefore, leaves from the two groups were also collected at different time points (0.5 h, 1 h, 2 h, 4 h, and 6 h) after topping (T05, T1, T2, T4, and T6) for transcriptomic sequencing and/or gene expression validation. Both WT and Nic1-2Nic2-2 plants with similar physiological status were selected for sampling. Middle leaves were gently excised using a surgical scissor and frozen immediately in liquid nitrogen and stored in -80°C for RNA extraction. For RNA extraction, the frozen leaf tissues of all seven sampling stages were ground into powder, and total RNA were extracted using RNeasy Plant Mini Kit (Qiagen, USA) following the manufacturer’s guidelines.
Library preparation and transcriptome sequencing
For RNA sequencing library construction, 2 µg of total RNA was employed. The quality of these RNA samples was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Samples with RNA integrity number (RIN) ≥ 8 were proceeded for library preparation. The library construction was performed using TruSeq® RNA Sample Preparation Kit (Illumina, USA) according to the manufacturer’s protocol. Sequencing reaction was conducted on both the WT control and Nic1-2Nic2-2 lines with pair-end run (150bp × 2). The amplified libraries were subsequently sequenced on the MGISEQ-2000 sequencing platform, generating a total of 8 GB of 150-bp paired-end reads per sample. Demultiplexing, adapter and barcode sequences trimming were performed on Trimmomatic v0.32 (Bolger et al., 2014). The quality of the data was verified, and sequencing reads were provided in FASTq format. The transcriptome data has been deposited in the Genome Sequence Archive (Chen et al., 2021) in National Genomics Data Center (CNCB-NGDC Members and Partners, 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Science (GSA: CRA014456) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
Read assembling and gene expression quantification
The raw paired-end reads were cleaned through removing adaptor sequences, poly-N, and low-quality sequences (i.e. the percentage of bases of quality value≤ 5 exceeds 50% in the read). In addition, short sequences (< 50bp) were also removed using a custom Perl program. All filtered reads were aligned to the tobacco reference genome (Nitab-v4.5_genome_Scf_Edwards2017.fasta.gz) accessible from Sol Genomics Network (Fernandez-Pozo et al., 2015) using HISAT v2.2.1 (Kim et al., 2019). Gene expression levels were quantified as FPKM (fragments per kilobase of exon per million reads fragments mapped reads) value using Cufflinks software version 1.2.1 (Trapnell et al., 2012). Differential expressed genes (DEGs) were identified using the DESeq R package (v1.18.0) and edgeR package (v 3.24.3) (Robinson et al., 2010). The criteria for identifying DEGs were set at a |log2 fold change (FC)| threshold of ≥ 1 and a corrected p-value (q-value) < 0.05, as previously described (Hu et al., 2021). Pair-wise Pearson correlation coefficients for each transcript were calculated using the RPKM. The Matrix distances for the expression heat-map were computed with Pearson correlations of gene expression values (RPKM) by heatmap.2 function of gplots (version 3.0.1) within the Bioconductor package in R (version 3.2.2) (Wickham, 2009).
Functional annotation and gene ontology
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted to identity the enrichment of DEGs in GO terms and metabolic pathways, respectively. A threshold of a corrected p-value (q-value) of ≤ 0.05 was applied to define significantly enriched GO terms and KEGG pathways. Functional analysis of these DEGs was performed in the KEGG Automatic Annotation Server (KAAS) (http://www.genome.jp/tools/kaas/). The KEGG Orthology (KO) terms assigned to the transcripts and categorized them into KEGG pathways with default parameters. Genes identified by DEG analysis were subjected to a GO enrichment analysis using clusterProfiler (version 4.4.2) package (Yu et al., 2012a).
Quantitative real-time PCR analysis
The qRT-PCR experiments were conducted in a 20 ml reaction volume using the LC480 system (Roche, USA) with three biological replicates. The tobacco Actin gene (accession No.: X63603) was used as an internal standard for normalization, and SYBR Green served as the fluorochrome. A three-step PCR process was performed with a pre-denaturation at 95°C for 30 s, followed by 45 cycles of denaturation at 95°C for 15 s, annealing at the optimal temperature of each primer pair for 15 s, and elongation at 72°C for 20 s, and finally for melting point curve analysis (95°C for 5 s, 70°C for 1 min and 95°C for 15 s) to test amplicon specificity. Relative quantification of gene expression levels was calculated using 2-△△Ct method. Sequence information of the qRT-PCR primers are listed in Supplementary Table 8.
Chemical quantification
In total of 2.0 kilograms of flue-cured middle leaves from both wild-type and Nic1-2Nic2-2 transgenic tobacco plants were collected for chemical measurements. The contents of six carbohydrate compounds (total sugar, reducing sugar, fructose, starch, glucose, and saccharose) and total nitrogen from the two groups were measured according to Tobacco and Tobacco Products-Determination YC/T 159-2019, YC/T 216-2013, YC/T 251-2008, and YC/T 161-2002, respectively. The contents of total amino acid and carotenoid in each leaf sample were quantified using ninhydrin color reaction and colorimetry as previously described (Wei et al., 2006; Li et al., 2010).
Myzus persicae performance and feeding preference
2-weeks-old seedlings were transplanted individually into plastic pots (10 cm × 7.5 cm) with cultural media at 25°C under 16h artificial light in the growth chamber (Percival Scientific, USA). For analysis of feeding preference, newly enclosed M. persicae larvae were placed on the newly developing leaf of 4-week-old WT and Nic1-2Nic2-2 seedling plants (5 and 5) and allowed to feed for 15 days. The feeding preference of M. persicae was determined by the larval mass on the surface of the seedling leaves.
Agrobacterium infiltration
For transient expression assays, plasmids encoding the TMV-Cg CP protein was introduced into Agrobacterium strain EHA105. The bacteria were grown overnight at 25°C and diluted to an 0.5 A600 density with a 1-mL needleless syringe before infiltration into 4-week-old WT and Nic1-2Nic2-2 seedling (at the 5 to 8 leaf stage) leaves immediately above the cotyledon (Li et al., 2023). Infiltrated seedling plants were grown in cultural media in an environmentally controlled growth chamber (Percival Scientific, USA). At least 3 seedling plants were used for each experiment and all experiments were repeated three times.
Statistical analysis
All data are presented as the mean ± standard deviation (SD). Statistical analyses were conducted using a one-way ANOVA model, followed by Student’s t-tests for multiple group comparisons. Significance was determined with unpaired p-values ≤ 0.05 were considered statistically significant.
Results
RNA−seq analysis reveals differential gene expression between WT plants and Nic1-2Nic2-2 lines
Transcriptomic analysis was conducted to comprehensively assess the impact of NtERF189/199 knockdown on gene expression across three vegetative growing stages and four after-topping time stages. A total of 42 samples, consisting of three biological replicates for both the WT control and Nic1-2Nic2-2 lines, were utilized for library preparation and subsequent RNA sequencing. Initial filtering of low-quality sequencing reads was performed using FastQC (Andrews, 2010), resulting in a total of 1,582 million (M) clean reads for the WT samples and 1,600 million (M) for the Nic1-2Nic2-2 samples. On average, each sequencing library contained more than 56 M reads with high-quality base scores (Supplementary Table 1). All clean reads from each library were successfully mapped to the tobacco reference genome sequence (Edwards et al., 2017) with genome mapping rates ranging from 91.19% to 96.99% and over 88% of the clean reads mapped to unique genomic positions (Supplementary Table 2).
Nicotine deficient significantly changes gene expression patterns in the Nic1-2Nic2-2 leaves at different growth stages and response to topping
To assess the impact of NtERF189/199 knockdown on tobacco physiological and metabolic changes, a comprehensive transcriptomic analysis was carried out to identify potential gene expression variations. Differential expression analysis was performed to identify DEGs and the quantification of gene expression levels were were quantified using FPKM values. High correlation coefficients (approximately 0.93) were observed across different biological replicates (Supplementary Tables 3, 4). Differentially expressed genes (DEGs) were identified based on FPKM with a threshold set at |log2 fold-change (FC)| ≥ 1 and a false discovery rate (FDR) < 0.05. The knockdown of NtERF189/199, in comparison to the WT control, led to significant changes in gene expression patterns during both the vegetative growth stages and after topping practices. In detail, 110 DEGs (55 upregulated, 54 downregulated) were identified at the SD stage, 136 DEGs (58 upregulated, 78 downregulated) at the RS stage, and 673 DEGs (117 upregulated, 556 downregulated) at the BT stage (Figure 1). Physical topping also resulted in distinct expression patterns between the WT control and Nic1-2Nic2-2 transcriptomes at four time points. Specifically, 880 DEGs (737 upregulated, 143 downregulated) were detected at the T0.5 stage, 68 DEGs (33 upregulated, 35 down-regulated) at T1 stage, 749 DEGs (175 upregulated, 574 down-regulated) at T2 stage, and 140 DEGs (45 upregulated, 95 down-regulated) at T4 stage (Figure 1; Supplementary Table 5). The relationships among the DEGs identified within the three vegetative stages and four after topping stages were illustrated separately using Venn diagrams. As illustrated, the Venn diagram revealed that only 46 DEGs were shared within at least two vegetative growth stages (Figure 2A). Similarly, there were 149 DEGs shared by at least two stages among the before and after topping stages (Figure 2B). Among the total of 195 DEGs, the downregulated genes were predominantly associated with plant-pathogen interaction and the mitogen-activated protein kinase (MAPK) signaling pathway. In contrast, genes related to starch and sucrose metabolism, amino acid metabolism, heat shock protein (HSP) 20 family genes, and carotenoid biosynthesis were upregulated in Nic1-2Nic2-2 lines compared to the WT control. The differentially expressed genes for each growth stage are listed in Supplementary Table 5.
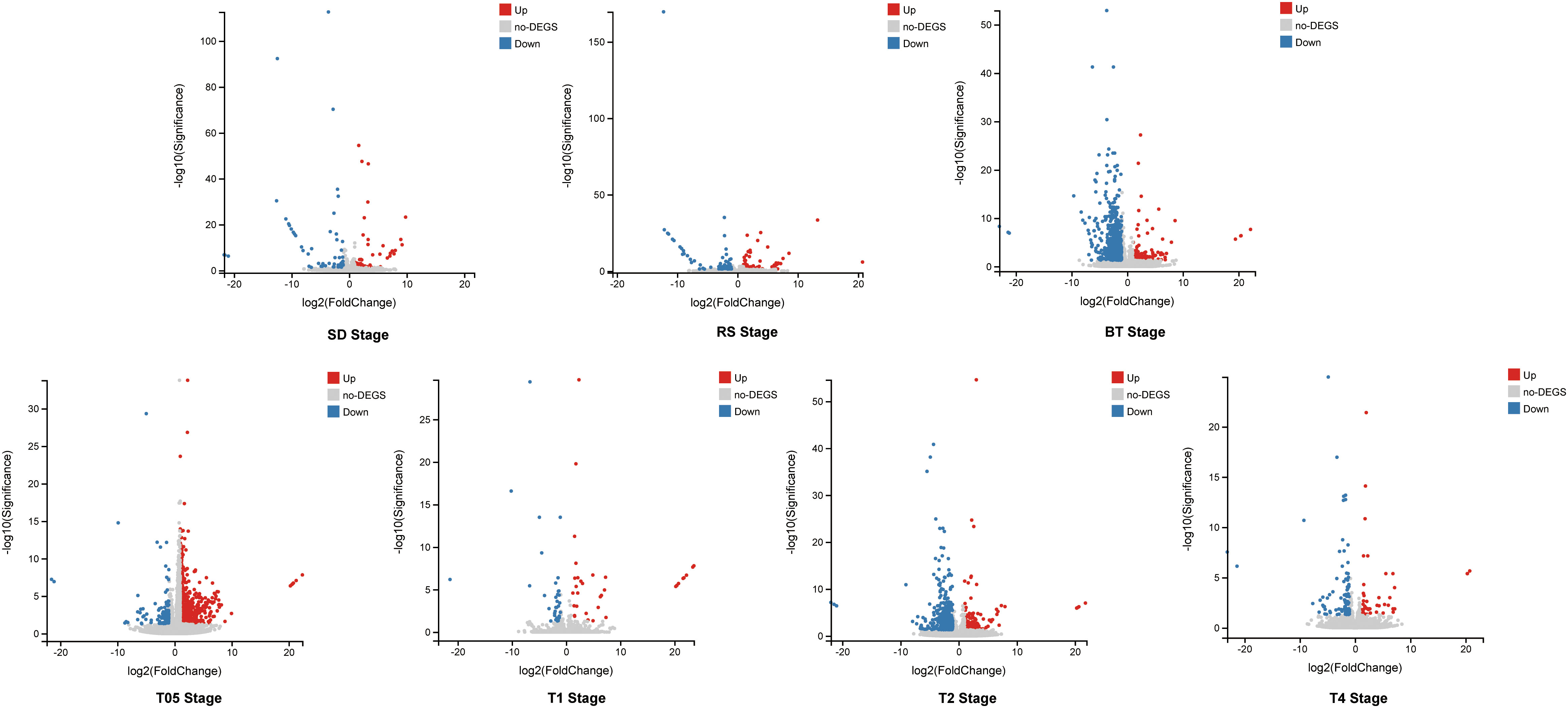
Figure 1 Differential expression analysis of leaf transcriptomes from wild-type and Nic1-2Nic2-2 transgenic tobaccos at different growing stages. Volcano plot identified DEGs in the transcriptomes of seedling stage (SD), rosette stage (RS), before topping stage (BT), and after topping stages (T05, T1, T2, T4), respectively. The value of |log2 fold changes (FC)|≥1 and an corrected p-value (q-value) ≤ 0.05 was used as criteria to select significantly expressed gene. The red dots denote the significantly up-regulated DEGs, and the blue dots denote down-regulated DEGs, and the gray dots denote genes are not significantly expressed.
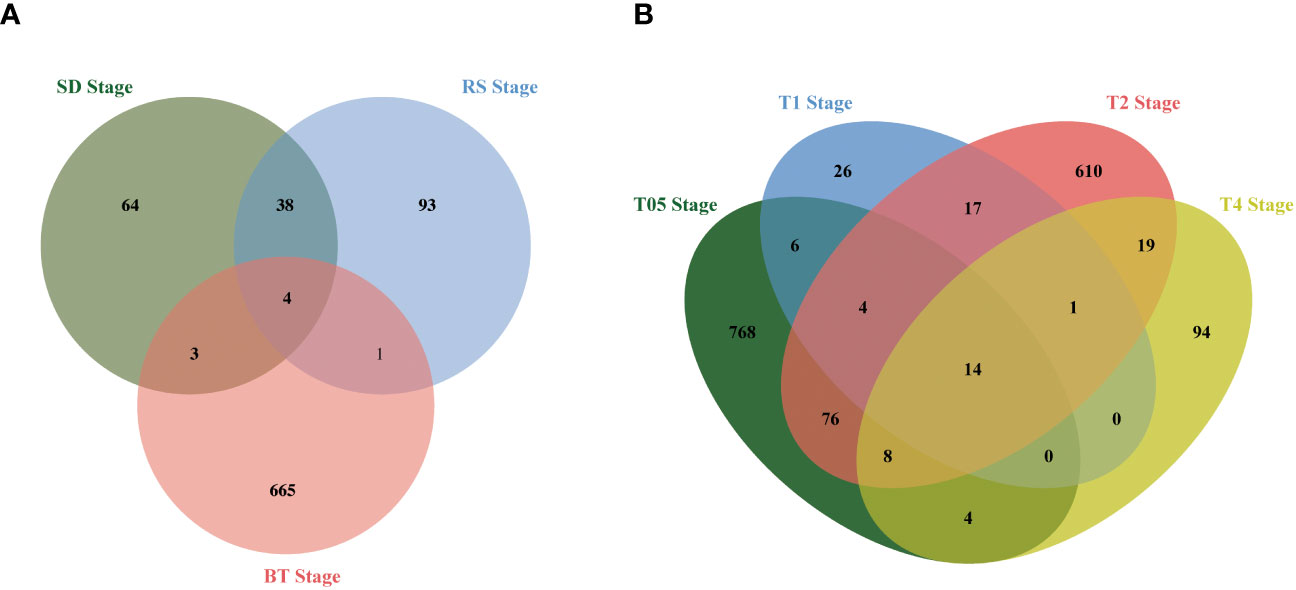
Figure 2 Venn diagram analysis of the DEGs at different growing stages. (A) Venn diagram showing the overlap of DEGs among the transcriptomes from the SD stage, RS stage, BT stage, respectively. Compared with the EV control, double knock off of NtERF189 and NtERF199 resulted in 109, 136, and 673 DEGs in these three developmental stage transcriptomes. (B) Venn diagram showing the overlap of DEGs after topping four time points. Compared with the wild-type control, knocking down of NtERF189 and NtERF199 resulted in 880, 68, 749 and 140 DEGs at after topping four-time course points.
Gene ontology enrichment analysis highlights the critical role of nicotine biosynthesis in maintaining typical responses to stress stimuli, secondary metabolite biosynthesis, and regulatory processes
To assess the impact of NtERF189/199 knockdown on the physiological status of tobacco, we conducted gene ontology (GO) enrichment analysis to further explore the DEGs across various growth stages. Significantly enriched GO terms for DEGs were determined with a Q-value threshold of ≤ 0.05. Specifically, three key growth stages (BT, T0.5 and T2) were selected for GO enrichment analysis as these results hold particular significance in addressing our research objectives (Figure 3). The DEGs were categorized into three ontologies: biological processes, cellular components, and molecular functions.
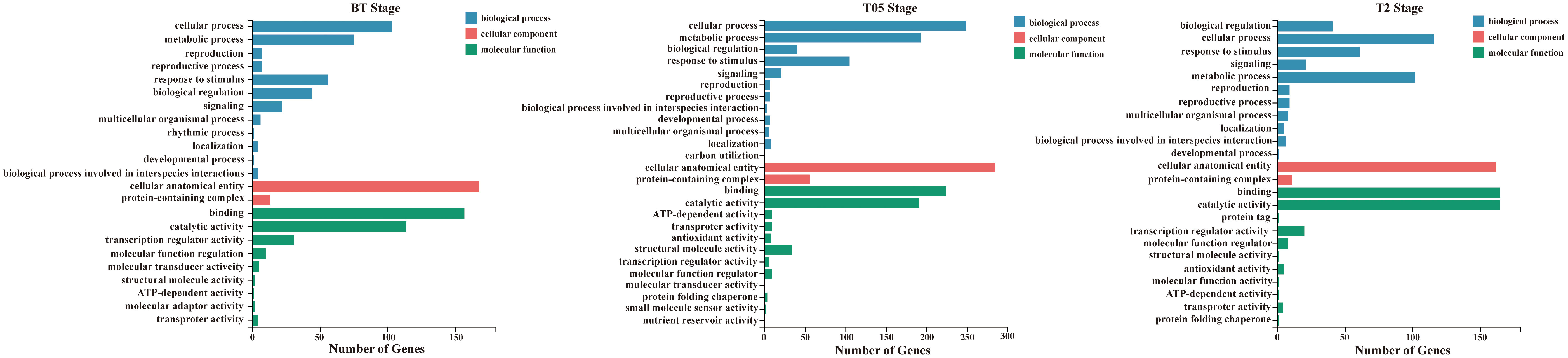
Figure 3 Gene ontology (GO) analysis of three selected growing stage (BT, T05, and T2) transcriptomes. GO term enrichment analysis. Significantly enriched GO terms were selected based on an FDR<0.05. GO terms of the categories of biological processes, cellular components, and molecular functions are depicted in blue, red, and green, respectively.
In the BT stage, genes that were upregulated in response to the knockdown of NtERF189/199 were significantly enriched in several GO terms, including glutathione metabolic process (GO:0006749), response to temperature stimulus (GO:0009266), diterpenoid biosynthetic process (GO:0016102), and glycogen metabolic process (GO:0005977). For example, the GO term response to temperature stimulus encompassed genes from the heat shock protein 20 family, which are known for their role in responding to a variety of abiotic and biotic stresses. On the contrary, downregulated genes in the BT stage were primarily associated with GO terms related to calcium ion binding (GO:0005509), xyloglucan metabolic process (GO:0010411), ubiquitin-protein transferase activity (GO:0004842), protein serine/threonine kinase activity (GO:0004674), and sequence-specific DNA binding (GO:0043565) (Figure 4). In the initial plant immunity steps, specific cytosolic Ca2+ rises in the cytosol and in organelles, then these Ca2+ specifically binds to a plethora of sensors including calmodulin (CaM), CaM-like proteins (CML), calcium-dependent protein kinases (CDPK), which, in turn, activate target proteins either through direct binding or phosphorylation (Aldon et al., 2018). Additionally, the GO terms sequence-specific DNA binding includes different stress-induced TF family genes, such as ERF, WRKY, jasmonate ZIM domain (JAZ) proteins and GRAS. These genes were mostly downregulated by knocking down of NtERF189/199 in the BT transcriptomes.
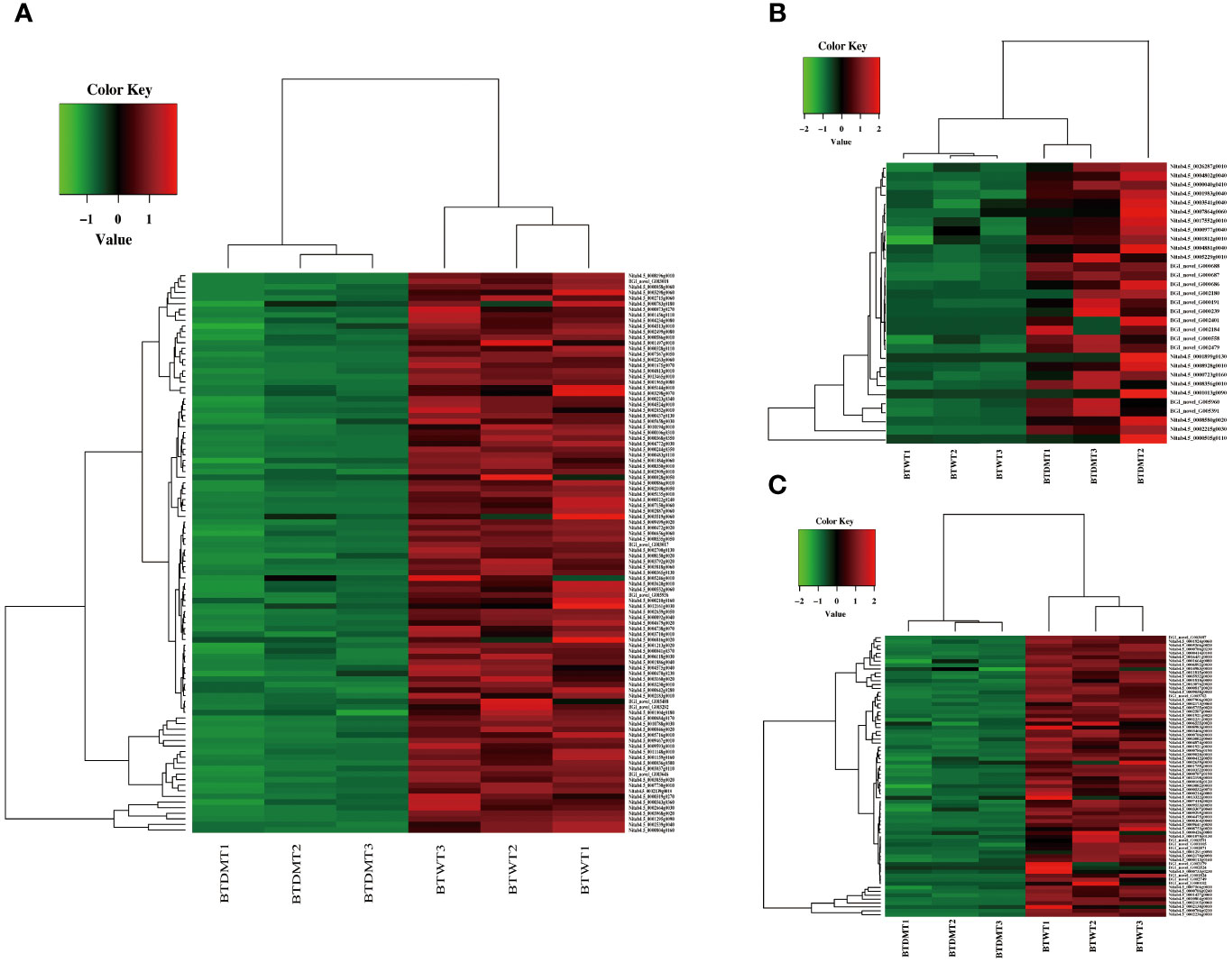
Figure 4 Differential gene expression analysis and hierarchical cluster analysis of the BT stage transcriptomes. (A) Heatmap analysis of DEGs based on the KO terms in plant-pathogen interaction, plant MAPK signaling pathway, and plant hormone signal transduction. (B) Hierarchical cluster analysis of the upregulated DEGs which were the KO terms related to secondary metabolite biosynthesis in the BT stage transcriptomes. (C) Hierarchical cluster analysis of the downregulated DEGs which were KO terms related to secondary metabolite biosynthesis in the BT stage transcriptomes. The red color denotes the highly expressed up-regulated DEGs, and the green color denotes down-regulated DEGs with lower expression levels. The gradation from red to green represents the transition from large to small values of a FPKM normalized log2 transformed counts. The sample names are indicated below each column. DEGs are defined by different colors, with the normalized expression levels employing a color gradient from low (green) to high (red).
In the T0.5 stage, there were more upregulated DEGs compared to downregulated genes (353 upregulated and 63 downregulated). These upregulated DEGs were enriched in GO terms associated with catalytic activity (GO:0003824), protein binding (GO:0005515), and ion binding (GO:0043167). These terms included genes belonging to ribosomal proteins, chlorophyll A-B binding proteins, cytochrome P450, photosystem-related proteins, and heat shock proteins (HSPs) (Supplementary Table 6). Notably, the presence of a significant number of DEGs related to HSPs in both BT and T0.5 transcriptomes suggests their essential role in response to nicotine deficiency. HSPs play vital roles in accumulating resistance proteins and coordinating defense signaling cascades in plants (Park and Seo, 2015). Additionally, HSPs act as crucial components in both prokaryotic and eukaryotic organisms by preventing protein aggregation during abiotic stresses (Muthusamy et al., 2017; Hu et al., 2022). HSPs can be categorized into different groups based on their molecular weights, including large HSPs, HSP90, HSP70, HSP60, HSP20, and small HSPs (Hu et al., 2022). These chaperone proteins help maintain cellular proteostasis, facilitate protein synthesis, folding, degradation, and signal transduction. Expression of HSP20s in plants is known to be induced by various stressors, including heat, drought, osmotic stress, phytohormones, and oxidative stresses (Guo et al., 2020). To be specific, a number of HSP genes belonging to HSP20, HSP70, and HSP90 classes were significantly upregulated in response to topping in low nicotine tobacco. The increased expression of HSP genes in low-nicotine tobacco may represent a strategy to enhance overall stress tolerance and compensate for the loss of pyridine alkaloids in tobacco. Furthermore, the upregulation of genes related to ribosomes in response to nicotine deficiency suggests their role in translational regulation during the protein synthesis process. In fact, ribosomes play a housekeeping role in translational regulation during protein synthesis process (Horiguchi et al., 2012). The 30S ribosomal subunit works with the 50S subunit to ensure genetic message translation accuracy and move the tRNAs to associated mRNA for translocation (Carter et al., 2000). Accumulating evidence suggested that loss of ribosomal proteins result in a reduced growth rate and developmental defects in plants. For instance, ribosomal protein L27 (RPL27), RPL11, and ribosomal protein S1(RPS1) are known regulatory genes for growth and patterning, photosynthesis, heat stress-responsive gene expression in Arabidopsis (Pesaresi et al., 2001; Szakonyi and Byrne, 2011; Yu et al., 2012b). The upregulation of Nicotiana homologous genes related to ribosomal proteins in low-nicotine tobacco further supports their importance in plant defesive responses. Moreover, the genes related to carbohydrate metabolism (GO:0005975), including galactinol synthase (GolS), glucan endo-1,3-beta-glucosidase, and inositol-3-phosphate synthase, which functions in responding to abiotic and biotic stresses, fungal pathogen resistance, and salt tolerance, are upregulated in low nicotine condition (Supplementary Table 6).
In the T2 stage, we observed that most of the DEGs were downregulated (175 upregulated and 574 downregulated) when compared to wild-type plants. The downregulated genes in this stage were associated with GO terms related to metabolic processes (GO:0008152) and biological regulation (GO:0065007). They also had molecular functions related to calcium ion binding (GO:0005509), heterocyclic compound binding (GO:1901363), nitrogen compound transport (GO:0071705), DNA-binding transcription factor activity (GO:0003700), carotenoid dioxygenase activity (GO:0010436), and ubiquitin-protein transferase activity (GO:0004842) (Supplementary Table 6). These downregulated genes belonged to various gene families, including calmodulin-binding proteins, cytochrome P450 enzymes, exocyst complex component proteins, plant transcription factor genes (ERF, MYB, Zinc-finger, NAC, GRAS, and bHLH), epoxycarotenoid dioxygenase genes, and U-box domain-containing proteins. Notably, the plant U-box proteins (PUBs) were among the downregulated genes. PUBs are closely associated with protein kinases involved in stress signaling and development and paired with E2-ubiquitin-conjugating enzymes to participate in signal hubs and mark substrates for degradation (Azevedo et al., 2001; Trenner et al., 2022). The OsPUB protein spotted leaf11 (spl11) mutant showed enhanced rice blast and bacterial blight resistance (Zeng et al., 2004). Expression of PUB22, PUB23, and PUB24 was induced by pathogen infections and pub22/pub23/pub24 triple mutant displayed enhanced activation of PAMP-triggered immunity as well as increased resistance against pathogens in Arabidopsis (Trujillo et al., 2008). PUB22 and PUB23 also showed to interact with ABA receptor pyrabactin resistance-like 9 (PYL9) to facilitate its own degradation and increase plant drought tolerance in Arabidopsis (Zhao et al., 2017). In addition to PUBs, the downregulated DEGs included members of the cytochrome P450 family, which are involved in various aspects of secondary metabolite biosynthesis and phytohormone signaling (GO:0009695) (Pandian et al., 2020). For instance, the expression of CYP450 proteins such as TRANSPARENT TESTA 7 (TT7) and allene oxide synthase (AOS) was suppressed. These genes are well-known structural genes involved in flavonoid biosynthesis and alpha-linolenic acid metabolism in Arabidopsis. Furthermore, this study also suggests that nicotine reduction in tobacco has a significant impact on the expression of various receptor serine/threonine kinase (RSTK) genes, which are a subclass of receptor-like kinase (RLK) proteins (Figure 5; Supplementary Table 6). Plant receptor-like kinase (RLK) proteins interact with diverse group of proteins to determine combinatorial variations in signal response specificity in the process of pathogen recognition, plant defense mechanism activation (Afzal et al., 2008). Receptor protein kinase can be divided into three subclasses according to different substrate specificities among the kinase domains, including receptor tyrosine kinase (RTK), receptor serine/threonine kinase (RSTK), and receptor histidine kinase (RHK), respectively (Becraft, 2002). RSTK genes, known as the receptor-like kinase, are involved in cell growth and development (Li and Chory, 1997; Jinn et al., 2000) as well as plant-pathogen interaction and defense responses (Song et al., 1995; Gómez-Gómez and Boller, 2000) in plants. For instance, three CLAVATA-like genes CLV1/CLV2/CLV3 are involved in meristem-size homeostasis and loss of function of each of them result in a shoot meristem enlargement phenotype in Arabidopsis (Clark et al., 1997). In addition, the receptor kinase FLS2 interacts with bacterial elicitor to activate its kinase domain which in turns to initiate downstream signaling for defense response in Arabidopsis (Haffani et al., 2004; Chinchilla et al., 2006). These results indicates that NtERF189/199 appear to play central roles in play central roles in enhancing plant defense signaling cascades and, in turn, activating the biosynthesis of specialized metabolites in tobacco.
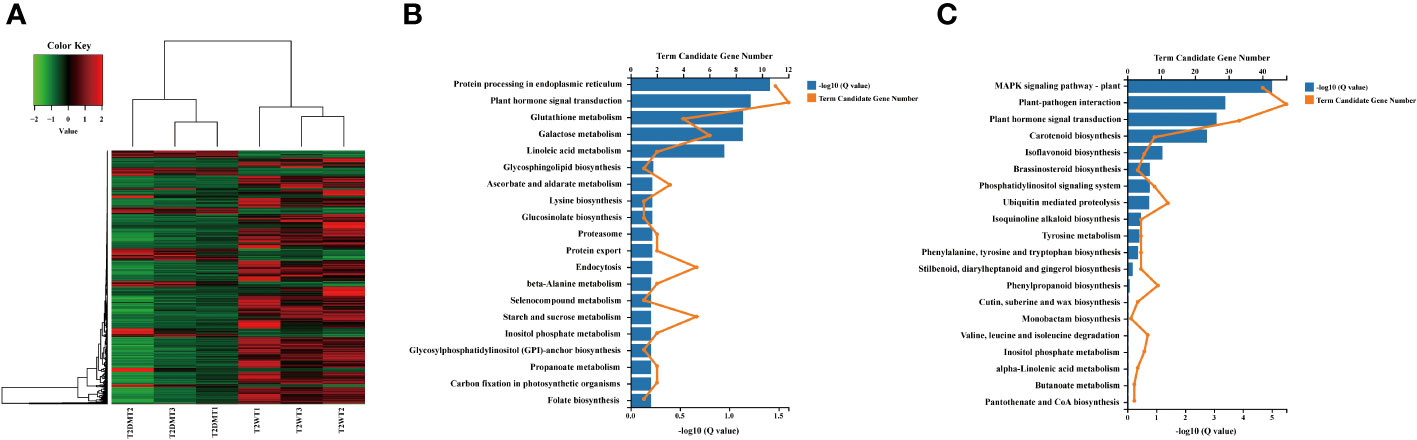
Figure 5 Hierarchical cluster analysis and the KEGG enrichment of the DEGs identified in the T2 stage transcriptomes. (A) Heatmap analysis of DEGs based on the KO terms in secondary metabolite biosynthesis in the T2 transcriptomes. The red color denotes the highly expressed up-regulated DEGs, and the green color denotes down-regulated DEGs with lower expression levels. The gradation from red to green represents the transition from large to small values of a FPKM normalized log2 transformed counts. The sample names are indicated below each column. DEGs are defined by different colors, with the normalized expression levels employing a color gradient from low (green) to high (red). (B) KEGG enrichment analysis of the upregulated DEGs identified in the T2 transcriptomes. (C) KEGG enrichment analysis of the down-regulated DEGs identified in the T2 transcriptomes. The blue column represents the degree of significance of the enrichment of DEGs in a pathway is indicated by -log10 (Q value). The x-axis represents the number of DEGs annotated to a particular pathway.
Collectively, these findings suggest that the expression intensity of a wide range of genes associated with various biological processes, such as biotic and abiotic stress responses, secondary metabolite biosynthetic pathways, and transcriptional regulations, were significantly decreased in response to knocking down NtERF189/199 when compared to the wild-type control. This reduction in gene expression in these crucial pathways reflects the broader impact of nicotine reduction on the plant’s overall physiology and defense mechanisms. In contrast, genes involved in heat shock responses, carbohydrate metabolism, ribosomal proteins, and plant U-box proteins were upregulated and are likely compensatory mechanisms that help offset the negative effects of nicotine synthesis defects in low nicotine tobacco. The study’s results provide a global perspective on why nicotine reduction in tobacco can render Nicotiana species more vulnerable to a wide range of biotic and abiotic stresses, more attractive targets for herbivore attacks compared to wild-type plants. A complete list of GO terms associated with three different growing stages were summarized in Supplementary Table 6.
KEGG pathway enrichment and functional classification
KEGG pathway enrichment analysis was performed to assign biological functions to these DEGs within known biological pathways in response to alkaloid reduction in tobacco, with particular emphasis on the T2 stage. This stage is of particular interest due to its potential implications for the differential synthesis of metabolites in wild-type and ultra-low nicotine tobacco plants. The enriched specialized pathways were separately categorized into two groups based on their expression status (|log2-fold change| ≥1) to visualize affected pathways and predict differentially accumulated metabolites (Figure 6). In the nicotine-deficient condition, most of upregulated DEGs were significantly enriched in the classes of galactose metabolism (sly04075, 24.00%), amino sugar and nucleotide sugar metabolism (sly00940, 19.00%), carbon metabolism (16.00%), and starch and sucrose metabolism (sly00592, 10.00%). In detail, genes encoding enzymes in raffinose family oligosaccharides, e.g. alkaline alpha galactosidase (Nitab4.5_0002649g0010 and Nitab4.5_0009705g0010) and galactinol synthase (Nitab4.5_0001013g0090), were upregulated in response to the lack of alkaloid accumulation (Figure 6A; Supplementary Table 7). Additionally, genes related to glycoside hydrolase (glucan endo-1,3-beta-glucosidase 5), glycosyl hydrolase (1,4-alpha-glucan branching enzyme II) and glycolysis (triosephosphate isomerase and glyceraldehyde-3-phosphare dehydrogenase) were also significantly induced (Figure 6A; Supplementary Table 7). In addition, alkaloid reduction in tobacco also triggered the activation of pathways leading to the biosynthesis of other specialized metabolites, such as betalains (Nitab4.5_0002352g0120), indole alkaloids (Nitab4.5_0010352g0040 and Nitab4.5_0001048g0050), isoquinoline alkaloids (Nitab4.5_0010352g0010), phenylpropanoids (Nitab4.5_0016891g0010), and sesquiterpenoids and triterpenoids (Nitab4.5_0000040g0410) (Figure 6B; Supplementary Table 7). Conversely, genes related to certain biosynthetic pathways were downregulated in the ultra-low nicotine condition, such as the biosynthesis of amino acids (valine, leucine, isoleucine, cysteine, methionine, and arginine), ascorbate and aldarate metabolism, and carotenoid biosynthesis. For example, genes related to amino acid biosynthesis such as arginine decarboxylase (Nitab4.5_0005266g0020), aminotransferases (Nitab4.5_0002231g0020), tryptophan synthase (Nitab4.5_0002155g0050) and prephenate dehydratase (Nitab4.5_0008983g0010) were downregulated (Figure 6B; Supplementary Table 7). Also, KEGG enrichment analyses indicated the DEGs were significantly downregulated in the various pathways like carotenoid biosynthesis (Nitab4.5_0001070g0130 and Nitab4.5_0000192g0200), biosynthesis of cofactors (Nitab4.5_0010352g0010), brassinosteroid biosynthesis (Nitab4.5_0000364g0040 and Nitab4.5_0002898g0020), and pentose and glucuronate interconversions (Nitab4.5_0000741g0040 and Nitab4.5_0005235g0040) (Figure 6B; Supplementary Table 7). Taken together, these results indicate that the knockdown of NtERF189/199, two dominant activators for nicotine synthesis, has a substantial impact on the metabolic reprogramming and metabolite accumulation. The plant appears to compensate for nicotine deficiency by modulating the biosynthesis of other specialized metabolites.
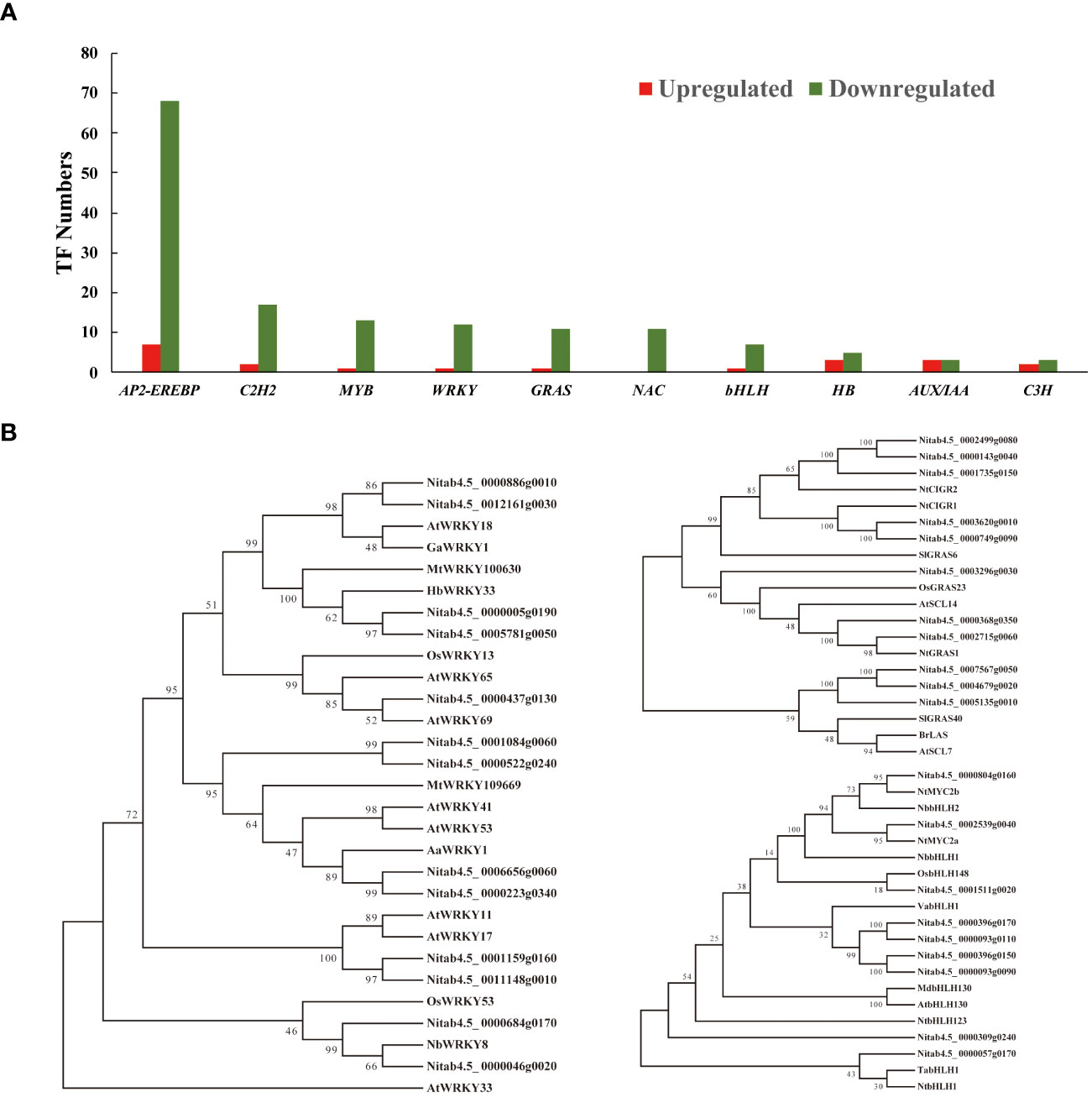
Figure 6 The transcription factor statistics and phylogenetic analysis. (A) The statistical analysis of the transcriptional regulators identified in all transcriptomes. (B) Amino acid sequences of three TF family genes (WRKY, GRAS, and bHLH) were selected to construct individual phylogenetic trees using MEGA-X software with the neighbor-joining method (with 2000 replications). Bootstrap values are indicated at branch nodes.
Nicotine reduction suppresses TF expression intensity involved in disease resistances, environmental stress tolerances and secondary metabolism
To investigate the wide-ranging impact of nicotine synthesis deficiency on transcriptional regulators, we conducted a systematic analysis across our transcriptomes. Our analysis revealed key TFs representing diverse families, including AP2-EREBP, C2H2, MYB, WRKY, GRAS, NAC, and bHLH, which play pivotal roles in modulating both defense responses and secondary metabolism as either positive or negative regulators (Figure 6A). Among these TF families, the AP2-EREBP family is most noticeable, encompassing a substantial number of ERFs. Previous studies have identified at least 239 AP2/ERF TFs in tobacco genome can be classified into ten subgroups, from group I to X (Rushton et al., 2008). AP2/ERF TFs comprise a diverse and influential group of factors that play essential roles in regulating various biological and physiological processes in plants (Licausi et al., 2013). These processes include plant morphogenesis, phytohormone signaling, responsiveness to environmental stimuli, and the biosynthesis of specialized metabolites (Feng et al., 2020). Our analysis identified a total of 75 AP2/ERF TFs (7 upregulated and 68 downregulated) displaying differential expression patterns, which could be further categorized into groups III, VII, IX, and X, based on phylogenetic analysis. Notably, several ERF TFs within this family have been recognized for their involvement in regulating a wide array of responses to biotic and abiotic stresses in tobacco. This group includes previously documented members such as NtERF1, NtERF5, NtERF172, ACRE111a/111b, ACRE1, and EREBP2/3/5/6 (Supplementary Table 5). EREBP proteins, functioning as activators or repressors, are known to specifically bind to the ethylene-responsive element (GCC-box) and regulate ethylene-responsive gene transcription (Ohme-Takagi and Shinshi, 1995; Ohta et al., 2001). For instance, NtERF5 has demonstrated enhanced resistance to tobacco mosaic virus (TMV), accompanied by reduced hypersensitive-response lesions, suggesting a role in controlling viral propagation (Fischer and Dröge-Laser, 2004). Additionally, NtERF172 has been identified as a positive regulator of tobacco drought resistance, achieved by direct interaction with the catalase NtCAT gene promoter to enhance CAT-mediated H2O2 homeostasis in tobacco (Zhao et al., 2020). Furthermore, the Tobacco Avr9/Cf-9 rapidly elicited (ACRE) genes, induced by elicitors and wounding, encode putative signaling components that initiate the defense response (Rowland et al., 2005).
Furthermore, the impact of alkaloid reduction extended to the downregulation of WRKY TFs, which are extensively recognized for their pivotal roles in plant innate immunity, serving as responsive genes to pathogen infections, elicitors, and phytohormone inductions. WRKY TFs contribute significantly to regulating both biotic and abiotic stress tolerance and play a substantial role in specialized metabolism in plants (Schluttenhofer and Yuan, 2015). Our transcriptomic analysis revealed a considerable decrease in the expression levels of these differentially expressed WRKY TFs in comparison to the control. Phylogenetic analysis suggested that these WRKY protein orthologs have functions in defense responses and secondary metabolite biosynthesis in various species, including Arabidopsis, wheat, Oryza sativa, and N. benthamiana. Notably, we identified two orthologous genes (Nitab4.5_0000046g0020 and Nitab4.5_0000684g0170) clustered with NbWRKY8 (Figure 6B). Previously, phosphorylation of NbWRKY8 induce the expression of defense-related genes, specifically 3-hydroxy-3-methylglutaryl CoA reductase 2 and NADP-malic enzyme expression. VIGS-based silencing of WRKY8 leads to increased disease susceptibility to Phytophthora and Colletotrichum pathogens (Ishihama et al., 2011). Furthermore, AaWRKY1, which cladding with two tobacco WRKY genes (Nitab4.5_0000665g0060 and Nitab4.5_0000223g0340), serves as a positive regulator of jasmonic acid (JA)-mediated signaling. It enhances artemisinin biosynthesis by trans-activating the expression of the amorpha-4,11-diene synthase gene in Artemisia annua (Ma et al., 2009). Additionally, overexpression of OsWRKY13 has been demonstrated to activate the expression of salicylic acid (SA) synthetic genes while simultaneously repressing JA signaling-responsive genes, thus increasing resistance to bacterial blight and fungal blast in rice (Qiu et al., 2007). Previous studies have shown that HbWRKY3, which is JA-inducible, plays a significant role in regulating rubber biosynthesis in Hevea brasiliensis (Li et al., 2014). Similarly, Medicago truncatula WRKY100630 is known to activate the biosynthesis of phenolic compounds and lignin accumulation in transgenic tobacco (Naoumkina et al., 2008). Notably, two NtWRKY proteins (Nitab4.5_0000886g0010 and Nitab4.5_0012161g0030) are affiliated with these two WRKY TFs, further suggesting their role as positive regulators in JA signaling and lignin biosynthesis (Figure 6B).
Significantly, we also observed the downregulation of several GRAS family TFs in the transcriptomes of low-nicotine tobacco (Figure 6B). GRAS family genes are known for their multifaceted roles in plant growth, development, and stress tolerance (Waseem et al., 2022). Notably, we identified five tobacco GRAS TFs (Nitab4.5_0002499g0080, Nitab4.5_0000143g0040, Nitab4.5_0001735g0150, Nitab4.5_0003620g0010, and Nitab4.5_0000749g0090) that are phylogenetically related to their orthologous genes in rice and tomato, which are implicated in responses to both biotic and abiotic stress. In rice, CIGR1 and CIGR2, members of the GRAS gene family, are significantly induced by exogenous gibberellins in a dose-dependent manner, and their expression is regulated by phosphorylation cascades (Day et al., 2004). Specifically, GIGR2 acts as a transcriptional activator of the B-type heat shock protein OsHsf23, enhancing disease resistance to rice blast fungus infection (Tanabe et al., 2016). Similarly, in tomato, the transcripts of SlGRAS6 and its orthologue preferentially accumulate in response to avirulent bacteria and fungal elicitors. Suppression of SlGRAS6 through VIGS-mediated gene silencing results in impaired drought tolerance (Mayrose et al., 2006). The downregulation of three tobacco GRAS family genes (Nitab4.5_0003296g0030, Nitab4.5_0000368g0350, and Nitab4.5_0002715g0060) in conjunction with the low-nicotine phenotype aligns with previous findings indicating that low nicotine tobacco exhibits reduced disease tolerance and stress resistance in tobacco (Steppuhn et al., 2004). Furthermore, overexpression of stress-induced OsGRAS23 has been demonstrated to confer improved drought resistance and oxidative stress tolerance in rice plants (Xu et al., 2015). Tobacco NtGRAS1 is another example, as it is strongly induced by various environmental stresses and correlates with increased intracellular reactive oxygen species (ROS) levels (Czikkel and Maxwell, 2007). Similarly, GRAS family TFs, such as SlGRAS40, BrLAS, and AtSCL7, have been identified as positive regulators of drought and salinity tolerance in tomato, Brassica, and Arabidopsis (Ma et al., 2010; Liu et al., 2017; Li et al., 2018). The expression of their tobacco orthologues (Nitab4.5_0007567g0050, Nitab4.5_0004679g0020, and Nitab4.5_0005135g0010) in our transcriptome was also notably decreased (Figure 6B).
The bHLH TF family is known for its diverse and vital roles in various biological processes within plants. Notably, a bHLH TF, NtMYC2a, has been identified as a master regulator in the JA (jasmonic acid) signaling transduction cascade, functioning upstream to regulate nicotine synthesis through ERF clusters (Shoji et al., 2010; Sui et al., 2020; Hou et al., 2023). In our study, the homologs of NtMYC2a/b (Nitab4.5_0002539g0040 and Nitab4.5_0000804g0160) were also found to be downregulated in low-nicotine tobacco leaves upon the knockdown of NtERF189 and NtERF199. In wheat, the transcript level of the TabHLH1 gene is induced by different osmotic stresses, phosphate, and nitrogen deprivation. Overexpressing TabHLH1 in transgenic tobacco results in enhanced drought resistance and salinity tolerance, presumably through its modulation of the ABA signaling pathway, nutrient transporter genes, and ROS homeostasis (Yang et al., 2016). Another example is NtbHLH1, which functions as a transcription activator and responds to iron deficiency by activating the expression of iron deficiency-related genes (Li et al., 2020). In our study, the expression of orthologues of TabHLH1 and NtbHLH1 in tobacco (Nitab4.5_0000057g0170) was also found to be repressed along with the decline in nicotine levels in tobacco leaves. Furthermore, the stress-inducible gene OsbHLH148, a homolog of the tobacco bHLH TF (Nitab4.5_0001511g0020), has been demonstrated to respond to environmental stresses and phytochrome inductions. It interacts with the OsJAZ1 protein to initiate jasmonate-regulated gene expression, contributing to drought tolerance (Seo et al., 2011). Collectively, these findings suggest that NtERF189 and NtERF199 not only play a role in nicotine biosynthesis and accumulation for insect resistance but also interconnecting with the regulatory mechanisms of ERF, WRKY, GRAS, and bHLH TFs. These TFs exhibit functional versatility, responding to a wide range of biotic and abiotic stresses in plants.
Transcriptomic expression level validation and biotic stress tolerance assays
Based on the KEGG results obtained by RNA-seq analysis, we validated the expression levels of genes that belong to different TF families by using qRT-PCR analysis. As expected, the expression patterns of these selected TF genes analyzed were consistent with the transcriptomic data (Figure 7A). For example, expression of these ERF genes and their homologs (Nitab4.5_0004445g0080 and Nitab4.5_0001393g0040; Nitab4.5_0002236g0040 and Nitab4.5_0002211g0060; Nitab4.5_0002211g0120) was significantly downregulated at different growing stages in the Nic1-2Nic2-2 transgenic lines, compared with the WT control (Figure 7). In addition, we also measured the expression levels of one bHLH gene (Nitab4.5_0000804g0160) and one C2H2 gene (Nitab4.5_0002105g0060). Similarly, expression of these two transcripts was also significantly reduced in several growing stages by the loss-of-function NtERF189 and NtERF199 (Figure 7). Moreover, we also checked expression of other TF family genes, including two C3H homologous genes (Nitab4.5_00044629g0020 and Nitab4.5_0013137g0020), AUX/IAA homologous genes (Nitab4.5_0003159g0030 and Nitab4.5_0003885g0020), and MYB homologous genes (Nitab4.5_0001373g0030/Nitab4.5_0002578g0010), and these genes were also significantly decreased in the loss-of-nicotine transcriptomes. Furthermore, transient expression of TMV-Cg CP in WT and Nic1-2 Nic2-2 were carried out. The TMV-Cg CP was transiently expressed in N. tabacum leaves using agroinfiltration. As the N. tabacum contains N’ gene, which is resistance to TMV-Cg, the transient expression of TMV-Cg CP could induce hypersensitive response (HR) symptoms in infiltrated leaves. As illustrated, at 24 hours post infiltration (hpi), the WT leaves exhibited initial HR symptoms, while the Nic1-2 Nic2-2 plants has no HR symptoms. At 72 hpi, the WT leaves exhibited solid HR symptoms, while the Nic1-2 Nic2-2 plants has only mild HR symptoms (Figure 7B). The HR implies that the N. tabacum plants are resistant to TMV-Cg, whereas the severity of HR indicates the resistance level of WT and Nic1-2 Nic2-2 plants. The postponed and milder HR symptom in Nic1-2 Nic2-2 plants indicated that the resistance level was down-regulated in these nicotine deficient plants compared with WT. Similarly, the aphid feeding preference assay demonstrated that the low nicotine seedlings (Nic1-2Nic2-2) are more susceptible to Myzus persicae and attract significantly higher number of aphid than did WT plants after feeding with aphid larvae for 15 days (Figure 7C). Taken together, these results confirmed the reliability of our transcriptome data and demonstrate that nicotine resistance trait is tightly linked to control the expression levels of various abiotic and biotic stresses related and quantitative resistance related genes in vivo to increase its own environmental fitness.
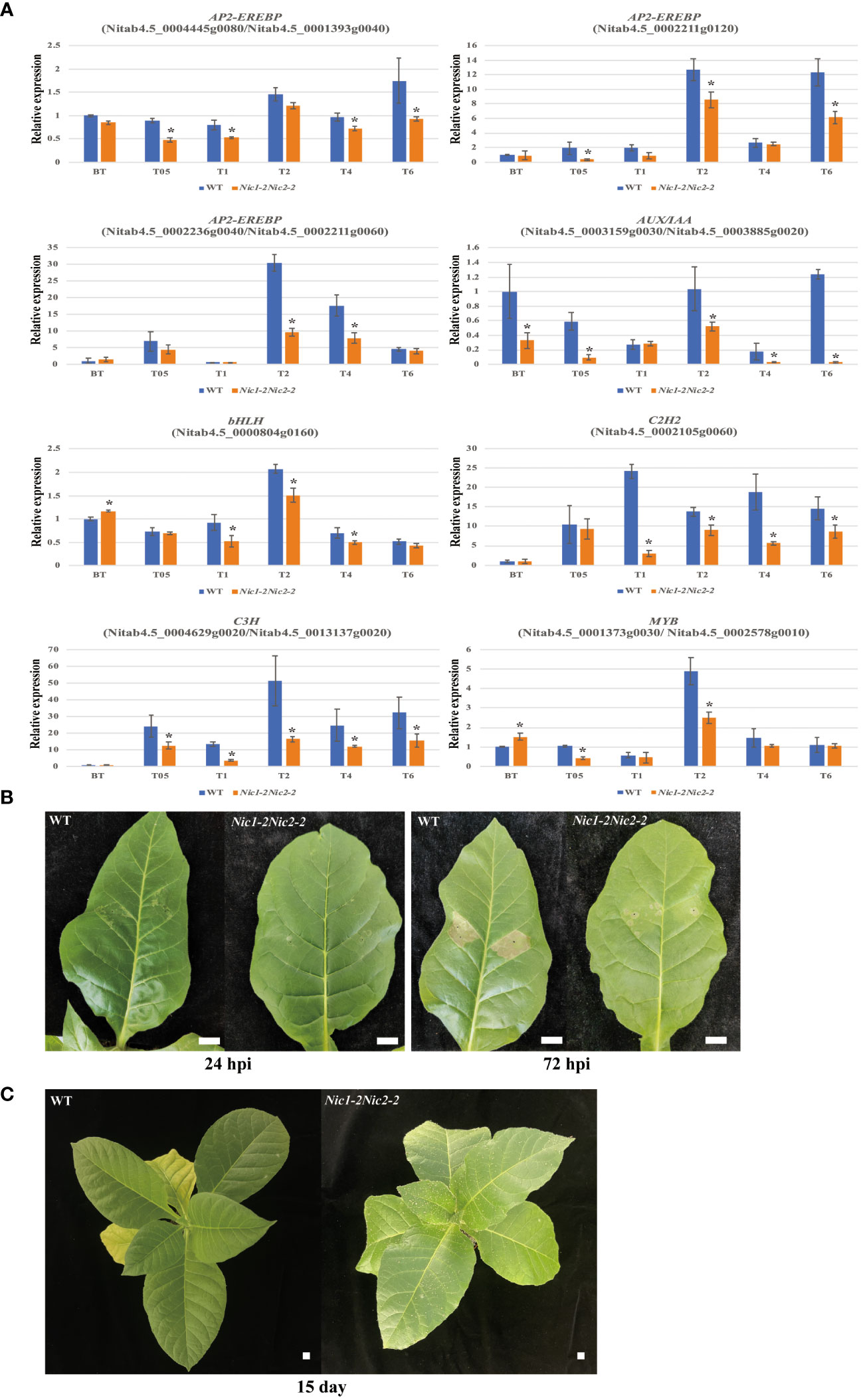
Figure 7 Verification of the expression level of stress-related genes and biotic stress tolerances in low nicotine tobacco plants. (A) qRT-PCR validation of the downregulated DEGs related to transcriptional regulation in low nicotine plants. The relative expression levels of 8 candidate genes and their homologous genes (AP2-EREBP, AUX/IAA, bHLH, C2H2, C3H and MYB) in the leaves of the Nic1-2Nic2-2 lines. The actin gene (accession No.: X63603) was used as an internal control. Data represent means ± standard deviation (SD) of three replicates. *P<0.05(n=3), by Student’s t-test. (B) Transient expression of TMV-Cg CP in wild type (WT) and low nicotine plant (Nic1-2 Nic2-2). The TMV-Cg CP was transiently expressed in N. tabacum leaves using agroinfiltration. As the N. tabacum contains N’ gene, which is resistance to TMV-Cg, the transient expression of TMV-Cg CP could induce hypersensitive response (HR) symptoms in infiltrated leaves. At 24 hours post infiltration (hpi), the WT leaves exhibited initial HR symptoms, while the Nic1-2Nic2-2 plants has no HR symptoms. At 72 hours post infiltration (hpi), the WT leaves exhibited solid HR symptoms, while the Nic1-2Nic2-2 plants has only mild HR symptoms. (C) The feeding preference of tobacco aphid to WT and Nic1-2Nic2-2 plants. The same amount of aphid larva was placed on N. tabacum seedling leaves. After fed for 15 days, the nicotine-deficient seedlings (Nic1-2Nic2-2) are more susceptible to Myzus persicae and attract significantly higher number of aphid than did WT plants. Bar = 1 cm.
Discussion
Alkaloids represent a diverse group of nitrogen-containing compounds produced in various plant species, and they have been attributed multiple functions, including serving as nitrogen reserves, aiding in detoxification processes, regulating plant growth, and acting as a defense mechanism against insects and predatory herbivores (Bush et al., 1993; Ali et al., 2019). Nicotine, a well-documented defensive alkaloid in Nicotiana species, is also employed as an insecticide in agricultural applications (Shepard, 1951; Schmeltz, 1971; Baldwin, 1996; Baldwin et al., 1997; Baldwin, 1998, Baldwin, 2001; Voelckel et al., 2001). Herbivore attack triggers a de novo nicotine biosynthesis through induce jasmonic acid signaling transduction cascade achieved by trans-activating multiple transcriptional regulators (Kazan and Manners, 2008, Kazan and Manners, 2013; Shoji and Hashimoto, 2011a; Shoji and Hashimoto, 2011b; Shoji et al., 2008; Shoji and Hashimoto, 2013, Shoji and Hashimoto, 2015, Shoji et al., 2010; Zhang et al., 2012; Sears et al., 2014; Sui et al., 2019; Sui et al., 2020; Qin et al., 2021; Godbole et al., 2022).
Historically, the pursuit of breeding low-nicotine tobacco dates back to the 1940s with the primary objective was to mitigate the throat irritation caused by nicotine during cigar inhalation (Valleau, 1949). The low alkaloid trait from Cuba cigar varieties were then incorporated into Burley and flue-cured tobaccos using traditional breeding methods (Legg et al., 1969; Chaplin, 1975). Genetic studies indicate that the level of total alkaloids in tobacco is primarily regulated by two independent loci, denoted as A and B (also referred to as NIC1 and NIC2, respectively) (Legg et al., 1969; Legg and Collins, 1971). Consequently, different nearly isogenic lines (NILs) have been developed, carrying recessive alleles at the Nic1 and/or Nic2 loci, resulting in four distinct levels of alkaloids in tobacco, namely high alkaloid, high-intermediate, low-intermediate, and low alkaloid (Chaplin, 1975; Chaplin and Burk, 1984, and Chaplin, 1986; Legg et al., 1970; Nielsen et al., 1988). However, the allelic variability in nicotine content was also found to have pleiotropic effect on tobacco agronomic traits, such as yield, leaf maturation, reducing sugar, polyamines levels, mesophyll cells and chlorophyll contents (Chaplin and Weeks, 1976; Nölke et al., 2018).
Recent advancements in tobacco genomics have facilitated a more comprehensive understanding of the physical locations and genetic compositions of the NIC1 and NIC2 loci within the tobacco genome (Shoji et al., 2010; Kajikawa et al., 2017; Sui et al., 2019; Sui et al., 2020). It has been established those recessive alleles of Nic1 and Nic2 in natural nicotine mutants result from chromosomal deletions (Shoji et al., 2010; Kajikawa et al., 2017) and chromosomal deletion-induced structural variations (Sui et al., 2020; Shoji et al., 2010). However, it’s important to note that allelic variations arising from naturally occurring mutations in these nearly isogenic lines (NILs), which were generated through repeated backcrossing, can introduce confounding variations. These variations may encompass not only the regulatory genes themselves but also genomic structural changes that could potentially be linked to both agronomic traits and the physiological alterations related to nicotine. A supporting observation in this scenario is that Nic1-2 and Nic2-2 double mutants did not exhibit any discernible abnormalities in terms of growth and development during laboratory trials (Hayashi et al., 2020).
For precise estimation of specific defense traits in plants, it is ideal to work with plant mutants that differ only in a single gene controlling the expression of the target trait while all other characteristics are identical (Bergelson and Purrington, 1996; Steppuhn et al., 2004). To reduce nicotine and other alkaloid levels, many attempts have been employed, including the reduction of expression levels or specific mutations of nicotine synthesis structural genes and their transcriptional regulators (such as NtQPT, NtPMTs, NtBBLs, NtA622s, NtMPO) using either RNA interference or CRISPR/Cas9-based editing technologies (Conkling et al., 2001; Chintapakorn and Hamill, 2003; Steppuhn et al., 2004; Wang et al., 2009; Lewis et al., 2015; Lewis, 2019; Hayashi et al., 2020; Sui et al., 2020; Burner et al., 2022). For example, silencing the expression of PMT in Nicotiana attenuata has shown that transgenic plants become more attractive to herbivore attacks by nicotine-adapted herbivores compared to wild-type plants (Steppuhn et al., 2004). In line with this rationale, the creation of nicotine mutants by simultaneously loss-of-function of NtERF199 and NtERF189, resulting in reduced biotic and abiotic stress tolerance, precisely meets the requirements for studying nicotine’s role in plant defense resistance in tobacco (Figure 7). Therefore, it is significance to provide a comprehensive transcriptomic overview of the transcriptional and metabolic differences between Nic1-2Nic2-2 mutants and wild-type plants.
In this study, we conducted a comparative transcriptomic analysis to provide additional evidence supporting the notion that the nicotine synthesis regulon closely coordinates the intensity of gene expression related to various aspects of stimulus perceptions, signal transduction, and defensive response in plant innate immunity system. The individual sequencing library generated more than 56 M reads with more than 90% mapping rates on the reference genome suggests the high quality and sequencing depth of the library (Supplementary Tables 1, 2). The differential gene expression patterns in each of the investigated stages, ranging from 68 to 880, exhibited distinct transcriptional patterns between the Nic1-2Nic2-2 mutants and the wild-type plants, indicating that nicotine deficiency significantly altered the gene expression and metabolic consequence in tobacco. Notably, three stages (BT, T05, and T2) exhibited a higher number of DEGs in the Nic1-2Nic2-2 mutants compared to the wild-type control (Figure 1).
Nicotine, a well-known pyridine alkaloid, serves as a prominent plant resistance trait in Nicotiana species. Loss of function in nicotine production renders N. attenuata more attractive to tobacco-chewing insects (Steppuhn et al., 2004). Our Gene Ontology (GO) enrichment analysis has shown that the biological processes associated with these GO terms related to cellular processes (GO:0009987), metabolic processes (GO:0008152), responses to stimuli (GO:0050896), biological regulation (GO:0065007), and signaling (GO:0023052) are significantly altered (Figure 3). The downregulation of DEGs associated with calcium ion binding (GO:0005509), protein serine/threonine kinase activity (GO:0004674), and ubiquitin-protein transferase activity (GO:0004842) suggests that plant pathogen perception within the innate immune system, to some extent, is compromised in low-nicotine plants. In signal transduction process, the expression levels of DEGs related to the MAP kinase cascade and sequence-specific DNA binding (GO:0043565), which includes members of the ERF, WRKY, bHLH, and GRAS TF families, are significantly reduced. This reduction implies a limited capacity for downstream signal transduction to induce sufficient critical defense and metabolic processes, such as the production of reactive oxygen species and the biosynthesis of various specialized metabolites. Indeed, several GO terms related to metabolic processes (GO:0008152) also exhibit significant weakening. These terms include xyloglucan metabolic process (GO:0010411), heterocyclic compound binding (GO:1901363), nitrogen compound transport (GO:0071705), and carotenoid dioxygenase activity (GO:0010436) (Supplementary Table 6). Different from WT plants, low-nicotine tobacco appears to reconfigure its innate immune system through the activation of other specific GO processes. These include temperature stimulus (GO:0009266), glycogen metabolic processes (GO:0005977), catalytic activity (GO:0003824), carbohydrate metabolism (GO:0005975), and diterpenoid biosynthetic processes (GO:0016102), evident by the upregulation of key genes belonging to various families, such as heat shock proteins, ribosomal proteins, galactinol synthases, glucan endo-1,3-beta-glucosidases, and alpha-farnesene synthases. Furthermore, this unique downstream signaling cascade under nicotine-deficient conditions has the potential to uncover a new subset of TF family genes with functions related to enhancing plant-pathogen resistance. Typically, the contributions of these genes are probably overlooked in wild-type tobacco (Supplementary Table 6).
Carbon-nitrogen interactions between the shoots and roots, transported by the xylem and phloem, play a crucial role in secondary metabolism in tobacco. The reduction of photosynthetic carbon metabolism leads to a decreased nitrate assimilation, amino acid levels, as well as nicotine accumulation (Matt et al., 2002). Additionally, topping also induces significant changes in carbohydrate metabolism (e.g., starch metabolism, glycolysis/gluconeogenesis) and signal transduction in tobacco axillary shoot growth (Wang et al., 2018). In our case, we found a defect in nicotine synthesis leads to a profound shift in the balance between carbon -nitrogen metabolism and secondary metabolic consequences in nicotine deficient condition (Figure 8). Notably, we identified a substantial increase in KEGG pathways related to galactose metabolism (sly04075), amino sugar and nucleotide sugar metabolism (sly00940), carbon metabolism (ko01200), and starch and sucrose metabolism (sly00592) (Figure 6). These findings are also supported by the significant increase in six carbohydrate compounds as well as decrease in total nitrogen and total amino acid contents in low-nicotine leaves (Figures 8A, B). Interestingly, this alteration in carbon and nitrogen metabolism appears to activate a range of plant specialized metabolic pathways, likely driven by these newly identified TFs, including betalain biosynthesis, indole alkaloid biosynthesis, isoquinoline alkaloid biosynthesis, phenylpropanoid metabolism, and sequiterpenoid and triterpenoid biosynthesis (Figure 5). Conversely, the knockdown of NtERF189/199 in N. tabacum appears to result in decreased nitrogen absorption and assimilation, as evidenced by reduced total nitrogen accumulation and the downregulation of amino acid metabolism pathways, such as valine, leucine, and isoleucine biosynthesis, ascorbate and aldarate metabolism, cysteine and methionine metabolism, and arginine and proline metabolism (Figure 6, Supplementary Table 7). Taken together, these results support the notion that the inhibition of nicotine synthesis, a nitrogen-rich secondary metabolite in tobacco leaves, triggers a broad activation of photosynthetic carbon metabolism and a reduction in nitrogen related metabolism, leading to significant changes in metabolic outcomes.
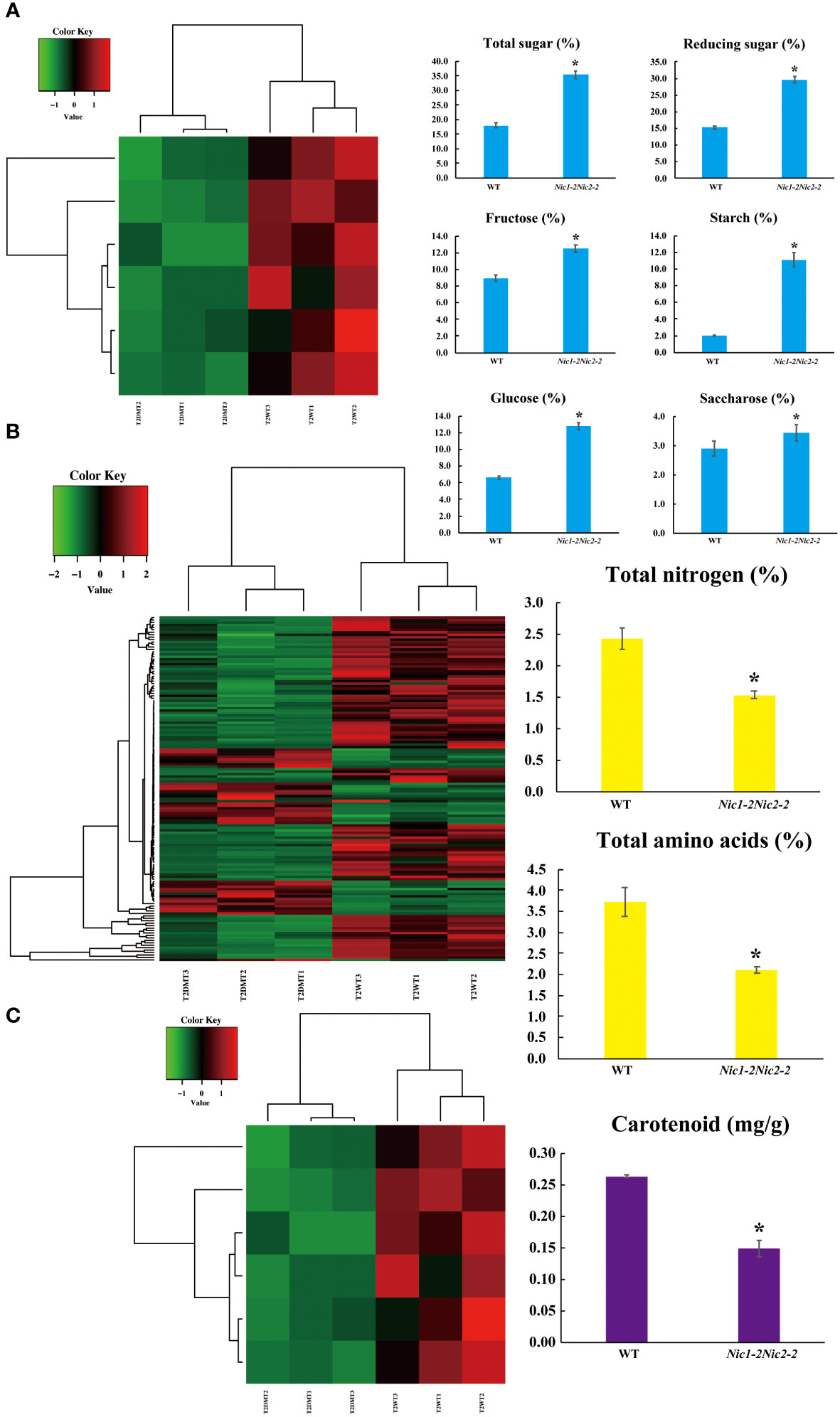
Figure 8 Correlation analysis between the DEGs identified and selected metabolites in the T2 stage transcriptomes. (A) Heatmap analysis of DEGs based on the GO terms related to carbohydrate metabolic process in the T2 transcriptomes. (B) Heatmap analysis of DEGs based on the GO terms involve in nitrogen related metabolic process and/or cellular amino acid metabolic process in the T2 transcriptomes. (C) Heatmap analysis of DEGs based on the GO terms related to carotenoid dioxygenase activity in the T2 transcriptomes. The red color denotes the highly expressed up-regulated DEGs, and the green color denotes down-regulated DEGs with lower expression levels. The gradation from red to green represents the transition from large to small values of a FPKM normalized log2 transformed counts. The sample names are indicated below each column. DEGs are defined by different colors, with the normalized expression levels employing a color gradient from low (green) to high (red). Metabolic profile of six carbohydrate compounds (blue color), total nitrogen and total amino acid (yellow color), and carotenoid (purple color) in the flue-cured leaves of the wild-type control and Nic1-2Nic2-2 transgenic plants. Data represent means ± standard deviation (SD) of three replicates. *P<0.05(n=3), by Student’s t-test.
Nicotine is recognized as a qualitative resistance trait with a substantial impact on defending against pathogen attacks and herbivores. In contrast to predominant loci, which have large effects on resistance, studies on quantitative resistance, associated with small-effect loci responding to pathogen attacks, have been focused on specific defense-related outputs, such as reinforcing the cell wall or biosynthesis of defense compounds (Corwin and Kliebenstein, 2017). The successful identification of most causal genes for quantitative resistance often relies on our understanding of the known components of the plant innate immune system, which can be divided into three stages: pathogen perception, signal transduction, and defense response. In line with this model, our transcriptomic analysis revealed that a substantial number of genes associated with these three aspects are downregulated in the absence of nicotine. For instance, many causal genes involved in the pathogen perception process, such as calmodulin and calmodulin-like proteins, receptor-like protein kinase proteins, and plant U-box proteins, have been identified. The second-largest group of causal loci includes potential transcription factors and secondary metabolite enzymes, which are associated with signal transduction. This group encompasses ERF, bHLH, WRKY, GRAS, as well as genes related to downstream defense mechanisms, such as xyloglucan endotransglucosylase hydrolases, plant polyphenol oxidase, and 9-cis-epoxycarotenoid dioxygenase (Figures 4–6). Many of these identified genes have homologous counterparts in other plant species that function in response to both biotic and abiotic stresses. This observation further supports the idea that these identified genes likely include many causal genes associated with small-effect quantitative resistance in tobacco. The collective downregulation of these genes in the absence of nicotine suggests a compromised defense response in the low-nicotine condition, making the plant more vulnerable to various stressors.
In this study, we have conducted a comprehensive comparative transcriptomic analysis to explore the biological consequences of the loss of nicotine biosynthesis in tobacco across various growth periods. The results presented here contribute to a broader understanding of the regulatory role of nicotine biosynthesis in the entire primary and secondary metabolic networks. Furthermore, our findings shed light on the tight link between qualitative resistance (nicotine biosynthesis) and quantitative resistances in tobacco. Specifically, we have demonstrated that the disruption of nicotine synthesis leads to a significant decrease in the expression of genes involved in responding to diverse abiotic and biotic stresses, as well as herbivore resistance. The candidate genes we have identified, which play roles in the plant innate immunity system, hold the potential to serve as causal genes for quantitative resistance. Their molecular functions will be a subject of further investigation in future studies. This research not only deepens our understanding of the intricate interplay between nicotine biosynthesis and plant defense mechanisms but also opens up new avenues for exploring the molecular basis of quantitative resistance in tobacco.
Data availability statement
The data presented in this study are deposited in the National Genomics Data Center (NGDC) repository, accession number CRA014456 (https://bigd.big.ac.cn/gsa/browse/CRA014456).
Author contributions
XS: Data curation, Funding acquisition, Writing – review & editing. ZS: Data curation, Methodology, Writing – original draft, Writing – review & editing. RW: Data curation, Writing – original draft. HZ: Data curation, Writing – review & editing. ZT: Data curation, Writing – review & editing. CY: Data curation, Writing – review & editing. YL: Data curation, Writing – review & editing. CH: Data curation, Writing – review & editing. LZ: Data curation, Writing – review & editing. YW: Methodology, Writing – review & editing. YD: Methodology, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Projects of YATAS (Contract No. 2022530000241012 and 110202103013), and the National Natural Science Foundation of China (Grant No. 31860069) to XS.
Acknowledgments
The authors thank the Beijing Genomics Institution (Beijing, China) for carrying out RNA-seq service for this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1338169/full#supplementary-material
References
Afzal, A. J., Wood, A. J., Lightfoot, D. A. (2008). Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol. Plant-Microbe Interactions. 21, 507–517. doi: 10.1094/MPMI-21-5-0507
Aldon, D., Mbengue, M., Mazars, C., Galaud, J. P. (2018). Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 19, 665. doi: 10.3390/ijms19030665
Ali, A. H., Abdelrahman, M., El-Sayed, M. A. (2019). Alkaloid role in plant defense response to growth and stress. Bioactive molecules Plant defense: Signaling Growth Stress. (Switzerland: Springer), 145–158. doi: 10.1007/978-3-030-27165-7_9
Andrews, S. (2010). “FastQC: a quality control tool for high throughput sequence data,” in Babraham Bioinformatics (Babraham Institute, Cambridge, United Kingdom).
Ashihara, H., Crozier, A., Komamine, A. (2010). Plant metabolism and biotechnology (Chichester, West Sussex: John Wiley & Sons).
Azevedo, C., Santos-Rosa, M. J., Shirasu, K. (2001). The U-box protein family in plants. Trends Plant Sci. 6, 354–358. doi: 10.1016/S1360-1385(01)01960-4
Baldwin, I. T. (1996). Methyl jasmonate-induced nicotine production in Nicotiana attenuata: inducing defenses in the field without wounding. Entomol. Exp. Appl. 80, 213-220.
Baldwin, I. T. (1998). Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc. Natl. Acad. Sci. U S A. 95, 8113–8118. doi: 10.1073/pnas.95.14.8113
Baldwin, I. T. (2001). An ecologically motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiol. 127, 1449–1458. doi: 10.1104/pp.010762
Baldwin, I. T., Zhang, Z. P., Diab, N., Ohnmeiss, T. E., McCloud, E. S., Lynds, G. Y., et al. (1997). Quantification, correlations and manipulations of wound-induced changes in jasmonic acid and nicotine in Nicotiana sylvestris. Planta. 201, 397–404. doi: 10.1007/s004250050082
Becraft, P. W. (2002). Receptor kinase signaling in plant development. Annu. Rev. Cell Dev. Biol. 18, 163–192. doi: 10.1146/annurev.cellbio.18.012502.083431
Bergelson, J., Purrington, C. B. (1996). Surveying patterns in the cost of resistance in plants. Am. Naturalist. 148, 536–558. doi: 10.1086/285938
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burner, N., McCauley, A., Pramod, S., Frederick, J., Steede, T., Kernodle, S. P., et al. (2022). Analyses of diverse low alkaloid tobacco germplasm identify naturally occurring nucleotide variability contributing to reduced leaf nicotine accumulation. Mol. Breed. 42, 4. doi: 10.1007/s11032-021-01274-5
Bush, L., Fannin, F., Chelvarajan, R., Burton, H. (1993). Biosynthesis and metabolism of nicotine and related alkaloids. (Wiltshire: Springer), 1–30. doi: 10.1007/978-94-011-2110-1
Carter, A. P., Clemons, W. M., Brodersen, D. E., Morgan-Warren, R. J., Wimberly, B. T., Ramakrishnan, V. (2000). Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 407, 340–348. doi: 10.1038/35030019
Chaplin, J. F. (1975). Registration of LAFC 53 tobacco germplasm. Crop Sci. 15, 282. doi: 10.2135/cropsci1975.0011183X001500020048x
Chaplin, J. F. (1986). Registration of MAFC 5 tobacco germplasm. Crop Sci. 26, 214. doi: 10.2135/cropsci1986.0011183X002600010081x
Chaplin, J. F., Burk, L. (1984). Registration of LMAFC 34 tobacco germplasm. Crop Sci. 24, 1220. doi: 10.2135/cropsci1984.0011183X002400060067x
Chaplin, J. F., Weeks, W. (1976). Association between percent total alkaloids and other traits in flue-cured tobacco 1. Crop Sci. 16, 416–418. doi: 10.2135/cropsci1976.0011183X001600030026x
Chen, T., Chen, X., Zhang, S., Zhu, J., Tang, B., Wang, A., et al. (2021). The Genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinf. 19, 578–583. doi: 10.1016/j.gpb.2021.08.001
Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476. doi: 10.1105/tpc.105.036574
Chintapakorn, Y., Hamill, J. D. (2003). Antisense-mediated down-regulation of putrescine N-methyltransferase activity in transgenic Nicotiana tabacum L. can lead to elevated levels of anatabine at the expense of nicotine. Plant Mol. Biol. 53, 87–105. doi: 10.1023/B:PLAN.0000009268.45851.95
Clark, S. E., Williams, R. W., Meyerowitz, E. M. (1997). The CLAVATA1gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 89, 575–585. doi: 10.1016/S0092-8674(00)80239-1
CNCB-NGDC Members and Partners (2022). Database resources of the national genomics data center, China national center for bioinformation in 2022. Nucleic Acids Res. 50, 27–38. doi: 10.1093/nar/gkab951
Conkling, M. A., Mendu, N., Song, W. (2001). Regulation of quinolate phosphoribosyl transferase expression. WO1998/056923 A1.
Corwin, J. A., Kliebenstein, D. J. (2017). Quantitative resistance: more than just perception of a pathogen. Plant Cell. 29, 655–665. doi: 10.1105/tpc.16.00915
Czikkel, B. E., Maxwell, D. P. (2007). NtGRAS1, a novel stress-induced member of the GRAS family in tobacco, localizes to the nucleus. J. Plant Physiol. 164, 1220–1230. doi: 10.1016/j.jplph.2006.07.010
Davis, D. L., Nielsen, M. T. (1999). Tobacco: production, chemistry and technology. (Wiley-Blackwell, Massachusetts).
Dawson, R. F. (1941). The localization of the nicotine synthetic mechanism in the tobacco plant. Science. 94, 396–397. doi: 10.1126/science.94.2443.396
Day, R. B., Tanabe, S., Koshioka, M., Mitsui, T., Itoh, H., Ueguchi-Tanaka, M., et al. (2004). Two rice GRAS family genes responsive to N-acetylchitooligosaccharide elicitor are induced by phytoactive gibberellins: evidence for cross-talk between elicitor and gibberellin signaling in rice cells. Plant Mol. Biol. 54, 261–272. doi: 10.1023/B:PLAN.0000028792.72343.ee
Edwards, K. D., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A. D., Bombarely, A., et al. (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 18, 1–14. doi: 10.1186/s12864-017-3791-6
Feng, K., Hou, X. L., Xing, G. M., Liu, J. X., Duan, A. Q., Xu, Z. S., et al. (2020). Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 40, 750–776. doi: 10.1080/07388551.2020.1768509
Fernandez-Pozo, N., Menda, N., Edwards, J. D., Saha, S., Tecle, I. Y., Strickler, S. R., et al. (2015). The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res. 43, D1036–D1041. doi: 10.1093/nar/gku1195
Fischer, U., Dröge-Laser, W. (2004). Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Mol. Plant-Microbe Interactions. 17, 1162–1171. doi: 10.1094/MPMI.2004.17.10.1162
Food and Drug Administration of the United States. (2018). Tobacco product standard for nicotine level of combusted cigarettes. Fed. Reg. 83, 11818–11843. doi: 10.2105/AJPH.2019.305067
Godbole, R. C., Pable, A. A., Singh, S., Barvkar, V. T. (2022). Interplay of transcription factors orchestrating the biosynthesis of plant alkaloids. 3 Biotech. 12, 250. doi: 10.1007/s13205-022-03316-x
Gómez-Gómez, L., Boller, T. (2000). FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell. 5, 1003–1011. doi: 10.1016/S1097-2765(00)80265-8
Guo, L. M., Li, J., He, J., Liu, H., Zhang, H. M. (2020). A class I cytosolic HSP20 of rice enhances heat and salt tolerance in different organisms. Sci. Rep. 10, 1383. doi: 10.1038/s41598-020-58395-8
Haffani, Y. Z., Silva, N. F., Goring, D. R. (2004). Receptor kinase signalling in plants. Can. J. botany. 82, 1–15. doi: 10.1139/b03-126
Hayashi, S., Watanabe, M., Kobayashi, M., Tohge, T., Hashimoto, T., Shoji, T. (2020). Genetic manipulation of transcriptional regulators alters nicotine biosynthesis in tobacco. Plant Cell Physiol. 61, 1041–1053. doi: 10.1093/pcp/pcaa036
Horiguchi, G., Van Lijsebettens, M., Candela, H., Micol, J. L., Tsukaya, H. (2012). Ribosomes and translation in plant developmental control. Plant Sci. 191, 24–34. doi: 10.1016/j.plantsci.2012.04.008
Hou, X., Singh, S. K., Werkman, J. R., Liu, Y., Yuan, Q., Wu, X., et al. (2023). Partial desensitization of MYC2 transcription factor alters the interaction with jasmonate signaling components and affects specialized metabolism. Int. J. Biol. Macromol. 252, 126472. doi: 10.1016/j.ijbiomac.2023.126472
Hu, C., Yang, J., Qi, Z., Wu, H., Wang, B., Zou, F., et al. (2022). Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm. 3, e161. doi: 10.1002/mco2.161
Hu, M., Zhang, H., Wang, B., Song, Z., Gao, Y., Yuan, C., et al. (2021). Transcriptomic analysis provides insights into the AUXIN RESPONSE FACTOR 6-mediated repression of nicotine biosynthesis in tobacco (Nicotiana tabacum L.). Plant Mol. Biol. 107, 21–36. doi: 10.1007/s11103-021-01175-3
Ishihama, N., Yamada, R., Yoshioka, M., Katou, S., Yoshioka, H. (2011). Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell. 23, 1153–1170. doi: 10.1105/tpc.110.081794
Jinn, T. L., Stone, J. M., Walker, J. C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108–117.
Kajikawa, M., Sierro, N., Kawaguchi, H., Bakaher, N., Ivanov, N. V., Hashimoto, T., et al. (2017). Genomic insights into the evolution of the nicotine biosynthesis pathway in tobacco. Plant Physiol. 174, 999–1011. doi: 10.1104/pp.17.00070
Kazan, K., Manners, J. M. (2008). Jasmonate signaling: toward an integrated view. Plant Physiol. 146, 1459–1468. doi: 10.1104/pp.107.115717
Kazan, K., Manners, J. M. (2013). MYC2: the master in action. Mol. Plant 6, 686–703. doi: 10.1093/mp/sss128
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Legg, P. D., Chaplin, J. F., Collins, G. B. (1969). Inheritance of percent total alkaloids in Nicotiana tabacum L. Populations derived from crosses of low alkaloid lines with burley and flue-cured varieties. J. Hered. 60, 213–217. doi: 10.1093/oxfordjournals.jhered.a107974
Legg, P. D., Collins, G. B., Litton, C. C. (1970). Registration of LA Burley 21 tobacco germplasm. Crop Sci. 10, 212. doi: 10.2135/cropsci1970.0011183x001000020041x
Legg, P. D., Collins, G. B. (1971). Inheritance of per cent total alkaloids in Nicotiana tabacum L. II. Genetic effects of two loci in Burley 21× LA Burley 21 populations. Can. J. Genet. Cytol. 13, 287–291. doi: 10.1139/g71-047
Lewis, R. S. (2019). Potential mandated lowering of nicotine levels in cigarettes: a plant perspective. Nicotine Tobacco Res. 21, 991–995. doi: 10.1093/ntr/nty022
Lewis, R. S., Drake-Stowe, K. E., Heim, C., Steede, T., Smith, W., Dewey, R. E. (2020). Genetic and agronomic analysis of tobacco genotypes exhibiting reduced nicotine accumulation due to induced mutations in berberine bridge like (BBL) genes. Front. Plant science. 11, 368. doi: 10.3389/fpls.2020.00368
Lewis, R. S., Lopez, H. O., Bowen, S. W., Andres, K. R., Steede, W. T., Dewey, R. E. (2015). Transgenic and mutation-based suppression of a berberine bridge enzyme-like (BBL) gene family reduces alkaloid content in field-grown tobacco. PLoS One 10, e0117273. doi: 10.1371/journal.pone.0117273
Li, J., Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 90, 929–938. doi: 10.1016/S0092-8674(00)80357-8
Li, H. L., Guo, D., Yang, Z. P., Tang, X., Peng, S. Q. (2014). Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomics. 104, 14–23. doi: 10.1016/j.ygeno.2014.04.004
Li, P., Zhang, B., Su, T. B., Li, P. R., Xin, X. Y., Wang, W. H., et al. (2018). BrLAS, a GRAS transcription factor from Brassica rapa, is involved in drought stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 9, 1792. doi: 10.3389/fpls.2018.01792
Li, Y. L., Huang, C. J., Liu, Y., Zeng, J. M., Yu, H. Q., Tong, Z. J., et al. (2023). CRISPR/Cas9-mediated seamless gene replacement in protoplasts expands the resistance spectrum to TMV-U1 strain in regenerated Nicotiana tabacum. Plant Biotechnol. J. 21, 2641–2653. doi: 10.1111/pbi.14159
Li, Y. Y., Sui, X. Y., Yang, J. S., Xiang, X. H., Li, Z. Q., Wang, Y. Y., et al. (2020). A novel bHLH transcription factor, NtbHLH1, modulates iron homeostasis in tobacco (Nicotiana tabacum L.). Biochem. Biophys. Res. Commun. 522, 233–239. doi: 10.1016/j.bbrc.2019.11.063
Li, H. W., Wang, L., Zhou, J. Y., Dong, G. F., Zhang, Q. (2010). Determination on amino acids contents in tobacco by ninhydrin color reaction. J. Anhui Agric. Sci. 38, 14926–14928. doi: 10.13989/j.cnki.0517-6611.2010.27.132
Licausi, F., Ohme-Takagi, M., Perata, P. (2013). APETALA 2/Ethylene Responsive Factor (AP 2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liu, Y., Huang, W., Xian, Z., Hu, N., Lin, D., Ren, H., et al. (2017). Overexpression of SlGRAS40 in tomato enhances tolerance to abiotic stresses and influences auxin and gibberellin signaling. Front. Plant Science. 8, 1659. doi: 10.3389/fpls.2017.01659
Ma, H. S., Liang, D., Shuai, P., Xia, X. L., Yin, W. L. (2010). The salt-and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 61, 4011–4019. doi: 10.1093/jxb/erq217
Ma, D., Pu, G., Lei, C., Ma, L., Wang, H., Guo, Y., et al. (2009). Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 50, 2146–2161. doi: 10.1093/pcp/pcp149
Matt, P., Krapp, A., Haake, V., Mock, H. P., Stitt, M. (2002). Decreased Rubisco activity leads to dramatic changes of nitrate metabolism, amino acid metabolism and the levels of phenylpropanoids and nicotine in tobacco antisense RBCS transformants. Plant J. 30, 663–677. doi: 10.1046/j.1365-313X.2002.01323.x
Mayrose, M., Ekengren, S. K., Melech-Bonfil, S., Martin, G. B., Sessa, G. (2006). A novel link between tomato GRAS genes, plant disease resistance and mechanical stress response. Mol. Plant Pathol. 7, 593–604. doi: 10.1111/j.1364-3703.2006.00364.x
Muthusamy, S. K., Dalal, M., Chinnusamy, V., Bansal, K. C. (2017). Genome-wide identification and analysis of biotic and abiotic stress regulation of small heat shock protein (HSP20) family genes in bread wheat. J. Plant Physiol. 211, 100–113. doi: 10.1016/j.jplph.2017.01.004
Naoumkina, M. A., He, X., Dixon, R. A. (2008). Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol. 8, 1–14. doi: 10.1186/1471-2229-8-132
Nielsen, M. T., Legg, P. D., Collins, G. B. (1988). Registration of HI and LI burley 21 tobacco germplasms. Crop Sci. 28, 206–207. doi: 10.2135/cropsci1988.206b.rgp
Nölke, G., Volke, D., Chudobová, I., Houdelet, M., Lusso, M., Frederick, J., et al. (2018). Polyamines delay leaf maturation in low-alkaloid tobacco varieties. Plant direct 2, e00077. doi: 10.1002/pld3.77
Ohme-Takagi, M., Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 7, 173–182. doi: 10.1105/tpc.7.2.173
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 13, 1959–1968. doi: 10.1105/TPC.010127
Organization W.H (2015). Advisory note: global nicotine reduction strategy (WHO Study Group on Tobacco Product Regulation).
Pandian, B. A., Sathishraj, R., Djanaguiraman, M., Prasad, P. V., Jugulam, M. (2020). Role of cytochrome P450 enzymes in plant stress response. Antioxidants. 9, 454. doi: 10.3390/antiox9050454
Park, C. J., Seo, Y. S. (2015). Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol. J. 31, 323. doi: 10.5423/PPJ.RW.08.2015.0150
Pesaresi, P., Varotto, C., Meurer, J., Jahns, P., Salamini, F., Leister, D. (2001). Knock-out of the plastid ribosomal protein L11 in Arabidopsis: effects on mRNA translation and photosynthesis. Plant J. 27, 179–189. doi: 10.1046/j.1365-313x.2001.01076.x
Qin, Q., Humphry, M., Gilles, T., Fisher, A., Patra, B., Singh, S. K., et al. (2021). NIC1 cloning and gene editing generates low-nicotine tobacco plants. Plant Biotechnol. J. 19, 2150. doi: 10.1111/pbi.13694
Qiu, D., Xiao, J., Ding, X., Xiong, M., Cai, M., Cao, Y., et al. (2007). OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate-and jasmonate-dependent signaling. Mol. Plant-Microbe interactions. 20, 492–499. doi: 10.1094/MPMI-20-5-0492
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rowland, O., Ludwig, A. A., Merrick, C. J., Baillieul, F., Tracy, F. E., Durrant, W. E., et al. (2005). Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9–dependent disease resistance in tomato. Plant Cell. 17, 295–310. doi: 10.1105/tpc.104.026013
Rushton, P. J., Bokowiec, M. T., Han, S., Zhang, H., Brannock, J. F., Chen, X., et al. (2008). Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 147, 280–295. doi: 10.1104/pp.107.114041
Saitoh, F., Noma, M., Kawashima, N. (1985). The alkaloid contents of sixty Nicotiana species. Phytochemistry. 24, 477–480. doi: 10.1016/S0031-9422(00)80751-7
Schluttenhofer, C., Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. doi: 10.1104/pp.114.251769
Schmeltz, I. (1971). Nicotine and other tobacco alkaloids. Naturally occurring insecticides (New York: Marcel Dekker, Inc), 99-136.
Sears, M. T., Zhang, H., Rushton, P. J., Wu, M., Han, S., Spano, A. J., et al. (2014). NtERF32: a non-NIC2 locus AP2/ERF transcription factor required in jasmonate-inducible nicotine biosynthesis in tobacco. Plant Mol. Biol. 84, 49–66. doi: 10.1007/s11103-013-0116-2
Seo, J. S., Joo, J., Kim, M. J., Kim, Y. K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65, 907–921. doi: 10.1111/j.1365-313X.2010.04477.x
Shepard, H. H. (1951). Plant products: nicotine and other alkaloids. The chemistry and action of insecticides. (New York: McGraw-Hill).
Shoji, T., Kajikawa, M., Hashimoto, T. (2010). Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell. 22, 3390–3409.
Shoji, T., Hashimoto, T. (2011a). Recruitment of a duplicated primary metabolism gene into the nicotine biosynthesis regulon in tobacco. Plant J. 67, 949–959. doi: 10.1111/j.1365-313X.2011.04647.x
Shoji, T., Hashimoto, T. (2011b). Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol. 52, 1117–1130. doi: 10.1093/pcp/pcr063
Shoji, T., Hashimoto, T. (2013). Smoking out the masters: transcriptional regulators for nicotine biosynthesis in tobacco. Plant Biotechnol. 30, 217–224. doi: 10.5511/plantbiotechnology.13.0221a
Shoji, T., Hashimoto, T. (2015). Stress-induced expression of NICOTINE2-locus genes and their homologs encoding Ethylene Response Factor transcription factors in tobacco. Phytochemistry. 113, 41–49. doi: 10.1016/j.phytochem.2014.05.017
Shoji, T., Hashimoto, T., Saito, K. (2023). Genetic regulation and manipulation of nicotine biosynthesis in tobacco: Strategies to eliminate addictive alkaloids. J. Exp. Bot., erad341. doi: 10.1093/jxb/erad341
Shoji, T., Kajikawa, M., Hashimoto, T. (2010). Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell. 22, 3390–3409. doi: 10.1105/tpc.110.078543
Shoji, T., Moriyama, K., Sierro, N., Ouadi, S., Ivanov, N. V., Hashimoto, T., et al. (2022). Natural and induced variations in transcriptional regulator genes result in low-nicotine phenotypes in tobacco. Plant J. 111, 1768–1779. doi: 10.1111/tpj.15923
Shoji, T., Ogawa, T., Hashimoto, T. (2008). Jasmonate-induced nicotine formation in tobacco is mediated by tobacco COI1 and JAZ genes. Plant Cell Physiol. 49, 1003–1012. doi: 10.1093/pcp/pcn077
Sisson, V., Saunders, J. (1982). Alkaloid composition of the USDA tobacco (Nicotiana tabacum L.) introduction collection. Tob. Sci. 16, 117-120.
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Steppuhn, A., Gase, K., Krock, B., Halitschke, R., Baldwin, I. T. (2004). Nicotine's defensive function in nature. PLoS Biol. 2, e217. doi: 10.1371/journal.pbio.0020217
Sui, X., Xie, H., Tong, Z., Zhang, H., Song, Z., Gao, Y., et al. (2020). Unravel the mystery of NIC1-locus on nicotine biosynthesis regulation in tobacco. bioRxiv. doi: 10.1101/2020.07.04.187922
Sui, X., Zhang, H., Song, Z., Gao, Y., Li, W., Li, M., et al. (2019). Ethylene response factor NtERF91 positively regulates alkaloid accumulations in tobacco (Nicotiana tabacum L.). Biochem. Biophys. Res. Commun. 517, 164–171. doi: 10.1016/j.bbrc.2019.07.037
Szakonyi, D., Byrne, M. E. (2011). Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J. 65, 269–281. doi: 10.1111/j.1365-313X.2010.04422.x
Tanabe, S., Onodera, H., Hara, N., Ishii-Minami, N., Day, B., Fujisawa, Y., et al. (2016). The elicitor-responsive gene for a GRAS family protein, CIGR2, suppresses cell death in rice inoculated with rice blast fungus via activation of a heat shock transcription factor, OsHsf23. Bioscience Biotechnology Biochem. 80, 145–151. doi: 10.1080/09168451.2015.1075866
Thurston, R., Smith, W. T., Cooper, B. P. (1966). Alkaloid secretion by trichomes of Nicotiana species and resistance to aphids. Entomol Exp. Appl. 9, 428–432. doi: 10.1111/j.1570-7458.1966.tb01003.x
Tobacco TCPGT (2008). A clinical practice guideline for treating tobacco use and dependence: 2008 update: a US public health service report. Am. J. Prev. Med. 35, 158–176. doi: 10.1016/j.amepre.2008.04.009
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Trenner, J., Monaghan, J., Saeed, B., Quint, M., Shabek, N., Trujillo, M. (2022). Evolution and functions of plant U-box proteins: from protein quality control to signaling. Annu. Rev. Plant Biol. 73, 93–121. doi: 10.1146/annurev-arplant-102720-012310
Trujillo, M., Ichimura, K., Casais, C., Shirasu, K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18, 1396–1401. doi: 10.1016/j.cub.2008.07.085
Voelckel, C., Krügel, T., Gase, K., Heidrich, N., Van Dam, N. M., Winz, R., et al. (2001). Anti-sense expression of putrescine N-methyltransferase confirms defensive role of nicotine in Nicotiana sylvestris against Manduca sexta. Chemoecology. 11, 121–126. doi: 10.1007/PL00001841
Wang, W. F., Chen, P., Lv, J., Chen, L., Sun, Y. H. (2018). Transcriptomic analysis of topping-induced axillary shoot outgrowth in Nicotiana tabacum. Gene. 646, 169–180. doi: 10.1016/j.gene.2017.12.053
Wang, P., Zeng, J., Liang, Z., Miao, Z., Sun, X., Tang, K. (2009). Silencing of PMT expression caused a surge of anatabine accumulation in tobacco. Mol. Biol. Rep. 36, 2285–2289. doi: 10.1007/s11033-009-9446-1
Waseem, M., Nkurikiyimfura, O., Niyitanga, S., Jakada, B. H., Shaheen, I., Aslam, M. M. (2022). GRAS transcription factors emerging regulator in plants growth, development, and multiple stresses. Mol. Biol. Rep. 49, 9673–9685. doi: 10.1007/s11033-022-07425-x
Wei, F. J., Wang, F., Liu, G. S., Li, Y. J., Guo, Q. Y. (2006). Research on metabolism of flue-cured tobacco leaf carotenoids during its growth development. J. Henan Agric. Univ. 40, 588–591. doi: 10.3969/j.issn.1000-2340.2006.06.004
Wickham, H. (2009). Getting started with qplot. ggplot2: elegant graphics for data analysis (Maryland: Springer). 9–26.
Wittenberg, R. E., Wolfman, S. L., De Biasi, M., Dani, J. A. (2020). Nicotinic acetylcholine receptors and nicotine addiction: A brief introduction. Neuropharmacology. 177, 108256. doi: 10.1016/j.neuropharm.2020.108256
World Health Organization. (2015). Advisory Note: Global Nicotine Reduction Strategy: WHO Study Group on Tobacco Product Regulation (Geneva: WHO Press).
Xu, K., Chen, S., Li, T., Ma, X., Liang, X., Ding, X., et al. (2015). OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 15, 1–13. doi: 10.1186/s12870-015-0532-3
Yang, T., Yao, S., Hao, L., Zhao, Y., Lu, W., Xiao, K. (2016). Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway. Plant Cell Rep. 35, 2309–2323. doi: 10.1007/s00299-016-2036-5
Yu, G., Wang, L. G., Han, Y., He, Q. Y. (2012a). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics: J. Integr. Biol. 16, 284–287. doi: 10.1089/omi.2011.0118
Yu, H. D., Yang, X. F., Chen, S. T., Wang, Y. T., Li, J. K., Shen, Q., et al. (2012b). Downregulation of chloroplast RPS1 negatively modulates nuclear heat-responsive expression of HsfA2 and its target genes in Arabidopsis. PLoS Genet. 8, e1002669. doi: 10.1371/journal.pgen.1002669
Zeng, L. R., Qu, S., Bordeos, A., Yang, C., Baraoidan, M., Yan, H., et al. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell. 16, 2795–2808. doi: 10.1105/tpc.104.025171
Zhang, H. B., Bokowiec, M. T., Rushton, P. J., Han, S. C., Timko, M. P. (2012). Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol. Plant 5, 73–84. doi: 10.1093/mp/ssr056
Zhao, Q., Hu, R. S., Liu, D., Liu, X., Wang, J., Xiang, X. H., et al. (2020). The AP2 transcription factor NtERF172 confers drought resistance by modifying NtCAT. Plant Biotechnol. J. 18, 2444–2455. doi: 10.1111/pbi.13419
Keywords: ethylene response factor, transcriptomic analysis, nicotine biosynthesis, defensive chemical, environmental fitness, quantitative resistance, Nicotiana tabacum
Citation: Song Z, Wang R, Zhang H, Tong Z, Yuan C, Li Y, Huang C, Zhao L, Wang Y, Di Y and Sui X (2024) Comparative transcriptome analysis reveals nicotine metabolism is a critical component for enhancing stress response intensity of innate immunity system in tobacco. Front. Plant Sci. 15:1338169. doi: 10.3389/fpls.2024.1338169
Received: 14 November 2023; Accepted: 05 March 2024;
Published: 21 March 2024.
Edited by:
Ute Vothknecht, University of Bonn, GermanyReviewed by:
Panagiotis Madesis, University of Thessaly, GreeceChristell Van Der Vyver, Stellenbosch University, South Africa
Copyright © 2024 Song, Wang, Zhang, Tong, Yuan, Li, Huang, Zhao, Wang, Di and Sui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueyi Sui, xueyisui2017@hotmail.com
†These authors have contributed equally to this work and share first authorship
 Zhongbang Song
Zhongbang Song Ruixue Wang1,2†
Ruixue Wang1,2† Hongbo Zhang
Hongbo Zhang Zhijun Tong
Zhijun Tong Cheng Yuan
Cheng Yuan Yong Li
Yong Li Changjun Huang
Changjun Huang Lu Zhao
Lu Zhao Yingtong Di
Yingtong Di Xueyi Sui
Xueyi Sui