- 1Department of Bioengineering and Technology, Gauhati University, Guwahati, Assam, India
- 2Symbiosis School of Biological Sciences, Symbiosis International (Deemed University), Pune, India
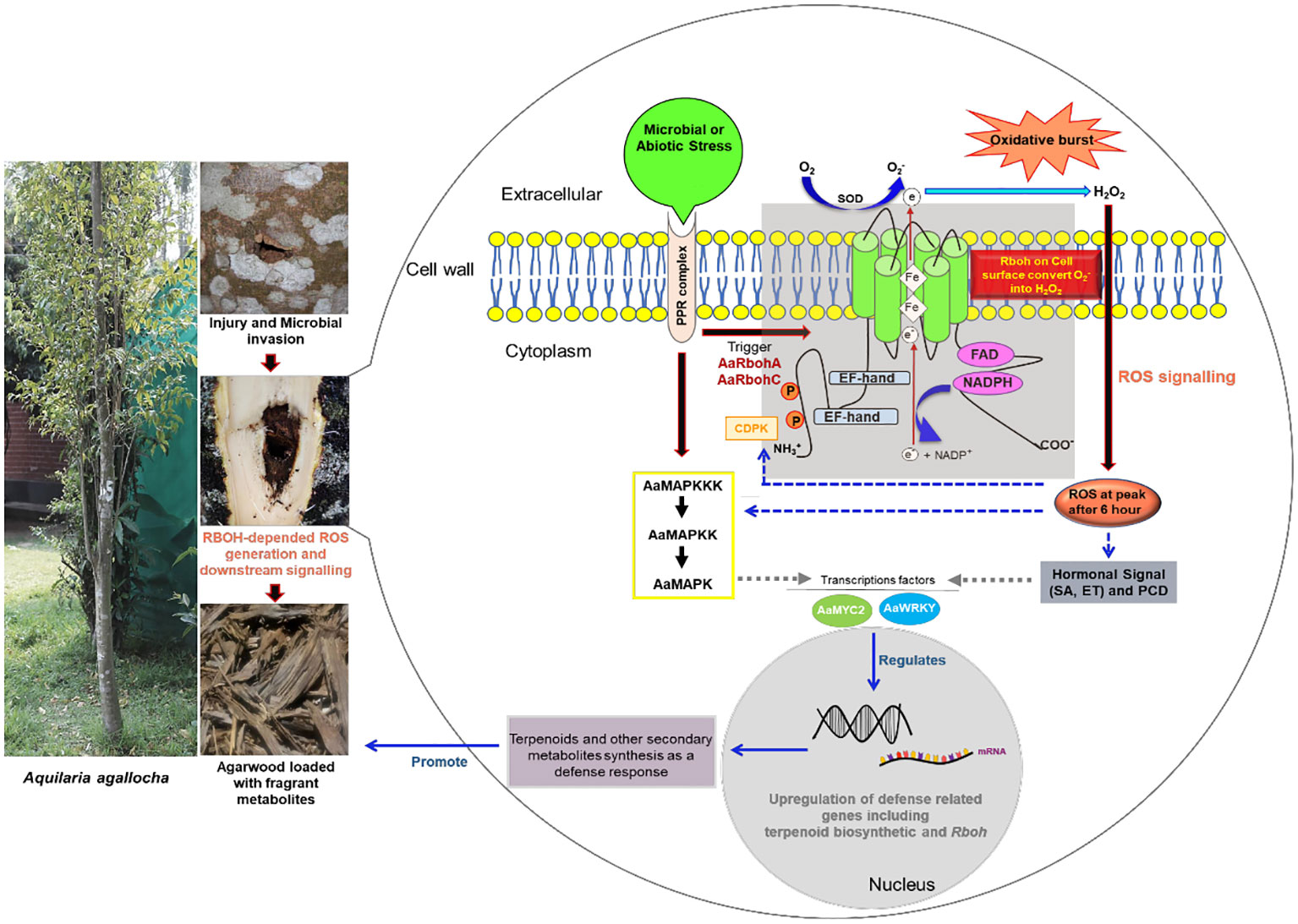
Respiratory burst oxidase homolog (Rboh) generates reactive oxygen species (ROS) as a defense response during biotic and abiotic stress. In Aquilaria plants, wounding and fungal infection result in biosynthesis and deposition of secondary metabolites as defense responses, which later form constituents of fragrant resinous agarwood. During injury and fungal invasion, Aquilaria tree generates ROS species via the Rboh enzymes. Despite the implication of Rboh genes in agarwood formation, no comprehensive genomic-level study of the Rboh gene family in Aquilaria is present. A systematic illustration of their role during stress and involvement in initiating signal cascades for agarwood metabolite biosynthesis is missing. In this study, 14 Rboh genes were retrieved from genomes of two Aquilaria species, A. agallocha and A. sinensis, and were classified into five groups. The promoter regions of the genes had abundant of stress-responsive elements. Protein–protein network and in silico expression analysis suggested their functional association with MAPK proteins and transcription factors such as WRKY and MYC2. The study further explored the expression profiles of Rboh genes and found them to be differentially regulated in stress-induced callus and stem tissue, suggesting their involvement in ROS generation during stress in Aquilaria. Overall, the study provides in-depth insight into two Rboh genes, AaRbohC and AaRbohA, highlighting their role in defense against fungal and abiotic stress, and likely during initiation of agarwood formation through modulation of genes involved in secondary metabolites biosynthesis. The findings presented here offer valuable information about Rboh family members, which can be leveraged for further investigations into ROS-mediated regulation of agarwood formation in Aquilaria species.
1 Introduction
The evolution of plants as sessile organisms presents a unique set of challenges. They are constantly exposed to various environmental stresses, both biotic (infection by bacteria, fungi, nematodes, etc.) and abiotic (drought, heavy metals, radiation, salinity, etc.), which can profoundly impact plant growth and yield (Wang et al., 2020; Mahalingam et al., 2021). Plant cells resist or respond to such stresses by promoting accumulation of reactive oxygen species (ROS). These species regulate almost all biological processes associated with developmental stages, stresses, and immunity responses (Castro et al., 2021). Although, ROS were once considered toxic by-products inside the cells because of cellular aerobic metabolism (Inupakutika et al., 2016). Recent studies affirm that they act as crucial signaling molecules to activate signal transduction cascades related to stress responses (Hu et al., 2018; Hawamda et al., 2020). However, beyond a specific threshold level, accumulation of ROS species can cause abnormal and irreparable metabolic changes and cell damage (Liu and He, 2016; Cheng et al., 2020; Wang et al., 2020). In plants, hydrogen peroxide (H2O2) is primary ROS species produced during C2 cycle in peroxisome (photorespiration) (Liu and He, 2016). In addition, as a by-product of photosynthesis and respiration, the photosynthesizing chloroplast and respiring mitochondria produce superoxide and hydrogen peroxide (Navathe et al., 2019). Superoxide anion (O−2) first generated from apoplastic molecular oxygen (O2) by the enzyme respiratory burst oxidase homolog protein (Rboh), which is next conFIGverted to H2O2 through superoxide dismutation reaction by the enzyme superoxide dismutase (Navathe et al., 2019). The different Rboh isoforms, also known as Nicotinamide Adenine Dinucleotide Phosphate Hydrogen (NADPH) oxidase in plasma membrane, transfer electrons from cytosolic NADPH/Nicotinamide Adenine Dinucleotide (NAD) + Hydrogen (H) (NADH) to apoplastic oxygen, producing the ROS in the cells (Yu et al., 2020). Plant Rboh is an intrinsic protein with six conserved transmembrane helices containing two basic helix-loop-helix calcium-binding structural domains (EF-hands) that are directly controlled by Ca2+ ions (Yu et al., 2020). Plant Rboh proteins share structural and functional domains with the mammalian homolog catalytic unit gp91phox, with the exception of an extended N-terminal sequence (Cheng et al., 2013; Cheng et al., 2019). The extended N-terminal region of plant Rboh contains two potential calcium-binding sites regulated by Ca2+ ion. The hydrophilic C-terminal domain has cytosolic-facing flavin adenine dinucleotide (FAD) and NADPH-binding sites. At the apoplast, heme groups are necessary for electron transport across the membrane to oxygen (O2, the electron acceptor) through FAD (Mahalingam et al., 2021).
Rboh of plants are a small multigene protein family (Marino et al., 2012). To date, genes encoding the Rboh proteins have been investigated and delineated in several plant species, namely, Citrus sinensis (Zhang et al., 2022); Capsicum annuum (Zhang et al., 2021); Hordeum vulgare (Lightfoot et al., 2008; Mahalingam et al., 2021); Nicotiana tobacum (Yu et al., 2020); Prunus avium, Prunus dulcis, Malus domestica Rubus occidentalis, Fragaria vesca, and Rosa chinensis (Cheng et al., 2020); Tritium aestivum (Hu et al., 2018; Navathe et al., 2019); Glycine max (Liu et al., 2019); Oryza sativa (Wong et al., 2007; Yamauchi et al., 2017); Malus domestica (Cepauskas et al., 2015); Vitis vinifera (Cheng et al., 2013); Medicago truncatula (Marino et al., 2011); Zea mays (Lin et al., 2009); Arabidopsis thaliana (Torres and Dangl, 2005; Torres et al.,1998); and Lycopersicon esculentum (Sagi and Fluhr, 2001). The rice OsRbohA was the first Rboh protein identified in plants (Navathe et al., 2019). The model plant Arabidopsis thaliana genome has 10 numbers of AtRboh genes, and, as per GeneVestigator microarray datasets (Zimmermann et al., 2004), of these, AtRbohH and AtRbohJ are involved with the growth of tip of pollen tube, whereas AtRbohA, AtRbohB, AtRbohC, AtRbohG, AtRbohE, and AtRbohI are expressed in root tissues; AtRbohD and AtRbohF are expressed across all A. thaliana tissues (Hawamda et al., 2020). On the other hand, AtRbohB was responsible for seed ripening and root hair formation, whereas AtRbohC controlled growth of the root hair cell (Navathe et al., 2019). Expression of AtRbohD and AtRbohE was also induced by plant hormone jasmonic acid, indicating their role in stress response and signaling (Maruta et al., 2011). AtRbohE was also reported to regulate the tapetal programmed cell death (PCD) and pollen formation in wheat (Hu et al., 2018).
In Aquilaria plants, as a result of biotic and abiotic stress, heartwood of the tree transforms into worthy resinous dark wood known as agarwood (Das et al., 2021). In general, Aquilaria species are diploid in nature. The genome size of A. sinensis is 726.5 Mb (Ding et al., 2020) and of A. agallocha is 736 Mb (Chen et al., 2014). Agarwood is well-known around the world for its usage as a primary ingredient in perfume, incense, and medicine (Monggoot et al., 2017). It has been traded and utilized for centuries to create perfume, which is still employed in religious and cultural ceremonies (López-Sampson and Page, 2018; Barden et al., 2000). Global agarwood prices can range from US$ 20 to US$ 6,000/kg for wood chips, depending on quality, or US$ 10,000/kg for the actual wood (Abdin, 2014). Agarwood essential oil can also fetch up to US$ 30,000/kg. According to estimates, the agarwood market in the world is worth between $6 and $8 billion annually (Tan et al., 2019). Among all the species of this genus, A. agallocha and A. sinensis, are known for producing high quality agarwood (Kristanti et al., 2018). When the tree is physiologically triggered by physical wound, followed by insect invasion or microbial infection, it activates defense-related signal transduction pathways, leading to accumulation of fragrant metabolites (Liu et al., 2013; Mohamed et al., 2014; Tan et al., 2019). Sesquiterpenes and 2-(2-phenylethyl)chromones are the two prominent chemical types found to be deposited in agarwood (Naef, 2011; Yang et al., 2021; Zhang et al., 2013). In addition, H2O2 burst (ROS production) is known to occur in wounded Aquilaria trees, leading to deposition of resins loaded with these metabolites (Zhang et al., 2013). Also, in plants, H2O2 is known to play a role in the regulation of the biogenesis of the secondary metabolites, viz., capsodiol, phenolics, and β-coumaroyl octopamine in tobacco, carrot, and potato, respectively (Matsuda et al., 2001). Previous studies have established role of Rboh gene families in ROS molecules accumulations after microbial invasion in plants (Morales et al., 2016; Chang et al., 2020; Pacheco-Trejo et al., 2022). Because agarwood resin formation in Aquilaria tree is an outcome of microbe-mediated stress, it leads us to hypothesize that, during microbial infections, Aquilaria trees accumulate ROS through the action of Rboh proteins. The ROS produced initiate a cascade of biochemical reactions that activate the defense-related signal transduction processes, eventually activating secondary metabolite biosynthetic genes for defense responses (Xu et al., 2016; Tan et al., 2019; Das et al., 2021).
To the best of our knowledge, a comprehensive genome-level illustration of the Rboh family members and their role during stress, and relation with downstream cascades leading to secondary metabolite biosynthesis is missing in A. agallocha. Therefore, this study aims to systemically identify, characterize, and analyze the expression of the Rboh genes in stress-induced tissues, which will likely identify the key members involved in ROS generation. In addition, findings in this current study will lay the foundation for understanding the molecular basis and regulatory mechanisms of Aquilaria species Rboh genes and their possible involvement in secondary metabolite biosynthesis and agarwood resin deposition.
2 Materials and methods
2.1 Sequence retrieval and identification of Rboh genes
The genomic sequence data of A. agallocha and A. sinensis were collected from previous annotation projects (Das et al., 2021 and Ding et al., 2020). Following that the sequence alignment of respiratory burst NADPH oxidase (PF08414), ferric reductase-like transmembrane component (PF01794), FAD-binding (PF08022), and ferric reductase NAD (PF08030) were obtained from the Pfam database and were used to build hidden Markov model (HMM) profile utilizing hmmbuild in the software HMMER 3.3.2 (Potter et al., 2018). Subsequently, the hmmsearch program was utilized to identify the putative AaRbohs and AsRbohs proteins, and the redundant protein sequences were discarded. All the putative sequences were further confirmed through Pfam database (http://pfam.xfam.org/) and SMART database (http://smart.embl-heidelberg.de/) for the presence of conserved NADPH_Ox (PF08414) domain (Cheng et al., 2020; Zhang et al., 2023). The physiological properties such as molecular weight (kDa), isoelectric point (pI), instability index (II), aliphatic index Ai), and the grand average of hydropathicity (GRAVY) were calculated by using the ExPASy-ProtParam tool (http://web.expasy.org/protparam/). The subcellular location of the Rboh proteins was predicted using online web server CELLO version 2.5 (http://cello.life.nctu.edu.tw/).
2.2 Multiple sequence alignments and phylogenetic analysis
Sequences of Rboh proteins of Solanum tuberosum, Arabidopsis thaliana, Hordeum vulgare, Oryza sativa, and Glycine max were downloaded from UniProtKB and aligned with predicted AaRboh and AsRboh protein sequences in the MEGA-X program (https://www.megasoftware.net/). The sequence alignment was presented with ESPrit 2.2-ENDscript 1.0 (Robert and Guoet, 2014). A phylogenetic tree was constructed on the basis of the alignment in the MEGA-X program using neighbour-joining method with parameter set as P distance model and 1,000 bootstrap replicates (Kumar et al., 2016).
2.3 Gene structure, cis-acting elements analysis
The intron–exon structure of individual Rboh genes was predicted utilizing genomic DNA and complete coding sequence (CDS) in Gene structure Display Server GSDS v2to.0 (http://gsds.cbi.pku.edu.cn) (Hu et al., 2015). The cis-acting regulatory elements were identified in 2.4 kb upstream of each gene using PlantCARE database.
2.4 Conserve motif in the proteins and homology modeling
Conserved motifs in the Rboh genes were predicted utilizing the MEME suite (http://meme-suite.org/). The analysis parameters were configured to identify the top 10 conserved motifs, whereas the remaining settings were kept at their default (Bailey et al., 2009) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). Homology modeling was employed to determine the 3D structures of the Rboh proteins using Swiss Model web server (https://swissmodel.expasy.org/). Structure assessment of the modeled structures was done considering the Ramachandran Plot and Stereochemistry (MolProbity score) and Clash score parameters (https://swissmodel.expasy.org/assess). Geometric and energetic validation of the structures was done using ERRAT server of SAVES v6.0 (https://saves.mbi.ucla.edu/).
2.5 Synteny and duplication analyses
To identify synteny blocks within the two Aquilaria genomes and with other plants (A. thaliana and S. tuberosum), blastp and Quick MCScanX Wrapper were employed and later visualized with the Dual Synteny plotter in TBtools (Chen et al., 2020). Duplicated genes were identified using DupGen-finder software (https://github.com/qiao-xin/DupGen_finder) using A. thaliana as outgroup and subsequently classified into duplication type following default parameters as per the user manual (Qiao et al., 2019). The non-synonymous substitution rate (Ka), synonymous substitution rate (Ks), and Ka/Ks ratio were calculated with TBtools (Wang et al., 2010). Divergence duration for duplication of paralogs pairs of the gene was calculated as per the formula T = Ks/2λ (where λ indicates the clock-of-like rate of 6.96 synonymous substitutions per 10−9 years) (Lopez-Ortiz et al., 2019).
2.6 Functional predictions and protein–protein interactions
Probable functions of Rboh proteins were predicted on the basis of assignment of Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations with e-value cutoff < 10−5. Regulatory network and their functional partners were identified through STRING v11.5 program with the following terms: databases, experimental evidences, gene neighborhood, gene co-occurrence gene fusion, co-expression, textminig, co-expression, and protein homology parameters utilizing Arabidopsis homologous proteins as reference.
2.7 The expression patterns of the AaRboh and AsRboh genes
To study transcript abundance of Rboh genes, RNA-seq data were downloaded from NCBI-SRA website (https://www.ncbi.nlm.nih.gov/sra). The transcript abundance of AaRboh genes in agarwood (SRX4149019-SRX4149021) and healthy (SRX4184708-SRX4184710) wood tissues were calculated and compared. Similarly, transcript abundance of AsRboh genes in the different tissues/organs, viz., aril (SRX6871071 and SRX6871068), seed (SRX6871057 and SRX6871070), flower (SRX6871060 and SRX6871063), bud (SRX6871059 and SRX6871062), leaf (SRX6871066 and SRX6871058), salinity stressed callus (SRX1495981 and SRX1495736), flower (SRX6871059 and SRX6871062), and wounded stem (SRX6871056 and SRX6871064), was accessed. First, the short reads were aligned to the genome using HISAT2 (Kim et al., 2015), following the reads were assembled and quantified using StringTie (Kovaka et al., 2019). Differentially expressed genes were then identified using DESeq2 software (Love et al., 2014).
2.8 Plant material, growth, and treatment
A. agallocha calli were induced from leaves on Murashige–Skoog (MS) medium supplemented with dichlorophenoxyacetic acid (6 mg/L) and kinetin (2 mg/L). The calli were transferred to fresh MS medium every month until the formation of friable calli. To induce stress, the calli were put into an MS medium containing 10 mM H2O2 and exposed to 5 mM dimethylthiourea (DMTU; an H2O2 scavenger) and combination of H2O2 with DMTU separately (Wang et al., 2018). Calli, without any treatment, were considered as control. After treatment, the samples were harvested at 0 h, 1 h, 2 h, 6 h, 12 h, 24 h, and 48 h.
Healthy saplings of A. agallocha maintained in pots at the Bioengineering and Technology Department of Gauhati University were selected for stress treatments as per standard methodology (Lv et al., 2019). The lateral stems were cut with scissors, and 1 cm from the apical end of the cut lateral stems was immersed separately in distilled H2O, H2O2, and DMTU solutions for stress treatments. The healthy Aquilaria lateral stems were taken as a control. After that, the portions immersed in treatment solutions were discarded, and the remaining treated stems (approximately 2 cm) were exposed to air for sample harvesting. Samples were harvested after 0 h, 1 h, 2 h, 6 h, 12 h, 24 h, and 48 h of air exposure. Treatment of seedlings after cutting refers to physical wounding. Wood samples (resin-embedded infected wood and healthy wood of A. agallocha) from Hoollongapar Gibbon Sanctuary in Jorhat, Assam, India, were collected following the methodology described by Islam et al. (2020) to analyze the AaRboh transcripts abundance. All sample sets were rapidly immersed in the liquid nitrogen and stored at −80°C until experiments were done.
2.9 RNA extraction and real-time reverse transcription PCR analysis
Total RNA from stem tissues was extracted following the RNA extraction method outlined by Islam and Banu (2019) and from callus tissues using the RNeasy Plant Mini Kit (Qiagen). The quality and concentration of extracted RNA were assessed with 1% agarose gel electrophoresis and estimated with the Multiskan Sky Microplate Spectrophotometer (Thermo Fisher Scientific, USA), respectively. One micrograms of RNA was used to synthesize the first strand of cDNA with SuperScript III Reverse Transcriptase (Thermofisher). The qRT-PCR was carried out using a QuantStudio™ 3 real-time PCR system (Applied Biosystems, USA) and the PowerUp SYBR Green Master Mix (Applied Biosystems). Seven AaRbohs gene-specific primer pairs were designed with PrimerQuest software of IDT (https://sg.idtdna.com/pages/tools/primerquest/) and are enlisted at Supplementary Table 1. The standardized GAPDH primer was utilized as the internal control (Islam et al., 2020). For each biological replicate, the analyses were performed with three technical replicates, each containing 20 µl of reaction volume in optical stripes, the temperature pattern of 95°C for 1 min, followed by 40 cycles at 95°C for 10 s and 60°C for 30 s, was followed as thermal cycler profile. Fold change in the gene expression was measured by the 2ΔΔCt method (Ding et al., 2021).
2.10 ROS determination of treated plant materials
The endogenous ROS production was determined according to Wang et al. (2018) with minor modifications. Plant samples (3 g of fresh weight) were subjected to individual and combined treatments with H2O2 and DMTU. The treated samples were homogenized in 3 mL of pre-cooled acetone using a mortar and pestle on ice. Obtained mixtures were centrifuged at 3,000 rpm for 10 min at 4°C. The supernatant obtained (0.1 mL) was quickly mixed with 0.1 mL of 5% TiSO4 and 0.2 mL of NH4OH was added to it. The resulting mixtures were centrifuged at 3,000 rpm for 10 min at 4°C to separate the titanium–hydroperoxide complex precipitate, and the supernatants were discarded. After three washes with pre-cooled acetone, the precipitates were dissolved in 2 mL of H2SO4 (2 mol/L). Absorbance of solution at 415 nm was monitored to quantify H2O2 content. Obtained absorbances were compared with the calibration curve derived from known concentrations of H2O2 (30%) (Wang et al., 2018; Zhang et al., 2021).
2.11 Statistical analysis
Three each biological and technical replicate were used for each control and treatment samples. T-test was performed to validate the expression differences of the Rboh genes in the different treated conditions. The P-value cutoff ≤ 0.05 was considered to be statistically significant test result. The methodology followed in the current study was respresented as Supplementary Figure 1.
3 Results
3.1 Identification of Rboh Genes in A. agallocha and A. sinensis
Seven genes each from A. agallocha (AaRboh) and A. sinensis (AsRboh) were identified and characterized (Supplementary Figure 2). The AsRboh genes were distributed on to four chromosomes (Chr02, Chr04, Chr06, and Chr07) and AaRbohs on seven scaffolds (KK901300.1, KK899295.1, KK902390.1, KK900302.1, KK899913.1, KK900079.1, and KK899008.1). The length of the protein varied, i.e., the shortest and the longest Rboh belonged to A. agallocha. For example, AaRbohJ had 663 amino acids (shortest), and AaRbohA had 946 amino acids (longest) (Table 1). Molecular weights varied from 75.72 kDa (AaRbohJ) to 107.77 kDa (AsRbohA) and pI from 8.93 (AsRbohC2) to 9.45 (AaRbohA). GRAVY values, representing the grand average of hydropathicity of the Rboh proteins were found to be < 0, which indicated their hydrophilic nature.
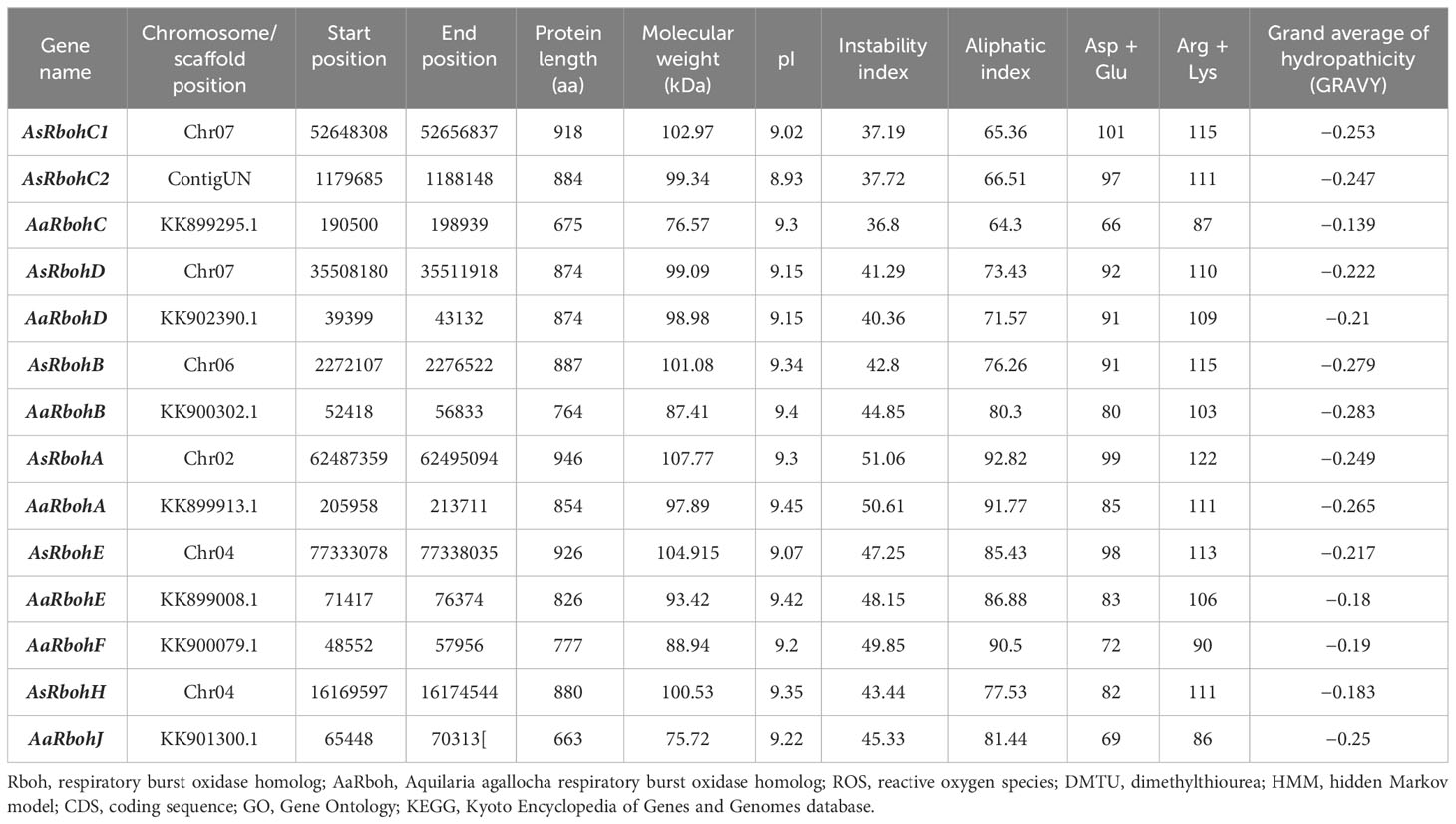
Table 1 Details of A. agallocha Rboh (AaRboh) and A. sinensis Rboh (AsRboh) identified in the genomes and properties of their deduced proteins.
3.2 Multiple sequence alignment and phylogenetic analysis
To determine the phylogenetic positions, a tree was build using 14 Aquilaria Rboh and 5, 12, 16, and 16 Rboh proteins of S. tuberosum, A. thaliana, H. vulgare, and Glycine max, respectively. Interestingly, RbohA-E were found in both the species, whereas RbohF and RbohJ were found in A. agallocha and RbohH in A. sinensis, respectively. In addition, A. sinensis had two members of RbohC, whereas A. agallocha had only one. The members of six plant species were majorly grouped into five major clades (Figure 1). The highest numbers of Rboh members were present in Group 1 (19), followed by Group 3 (17), Group 2 (15), Group 4 (12), and Group 5 (11). Group 1 composed of two AaRboh (AaRbohD and AaRbohC) and three AsRboh (AsRbohD, AsRbohC1, and AsRbohC2) proteins. Whereas, Group 2 composed of one each of AaRboh (AaRbohB) and AsRboh (AsRbohB). Similarly, Group 3 had one each of AaRboh (AaRbohA) and AsRboh (AsRbohA). Group 4 included two AaRboh proteins (AaRbohE and AaRbohF), one AsRboh (AsRbohE), one AaRboh (AaRbohJ), and one AsRboh (AaRbohH). All five groups contained at least one member from each of the six plant species, including A. agallocha and A. sinensis.
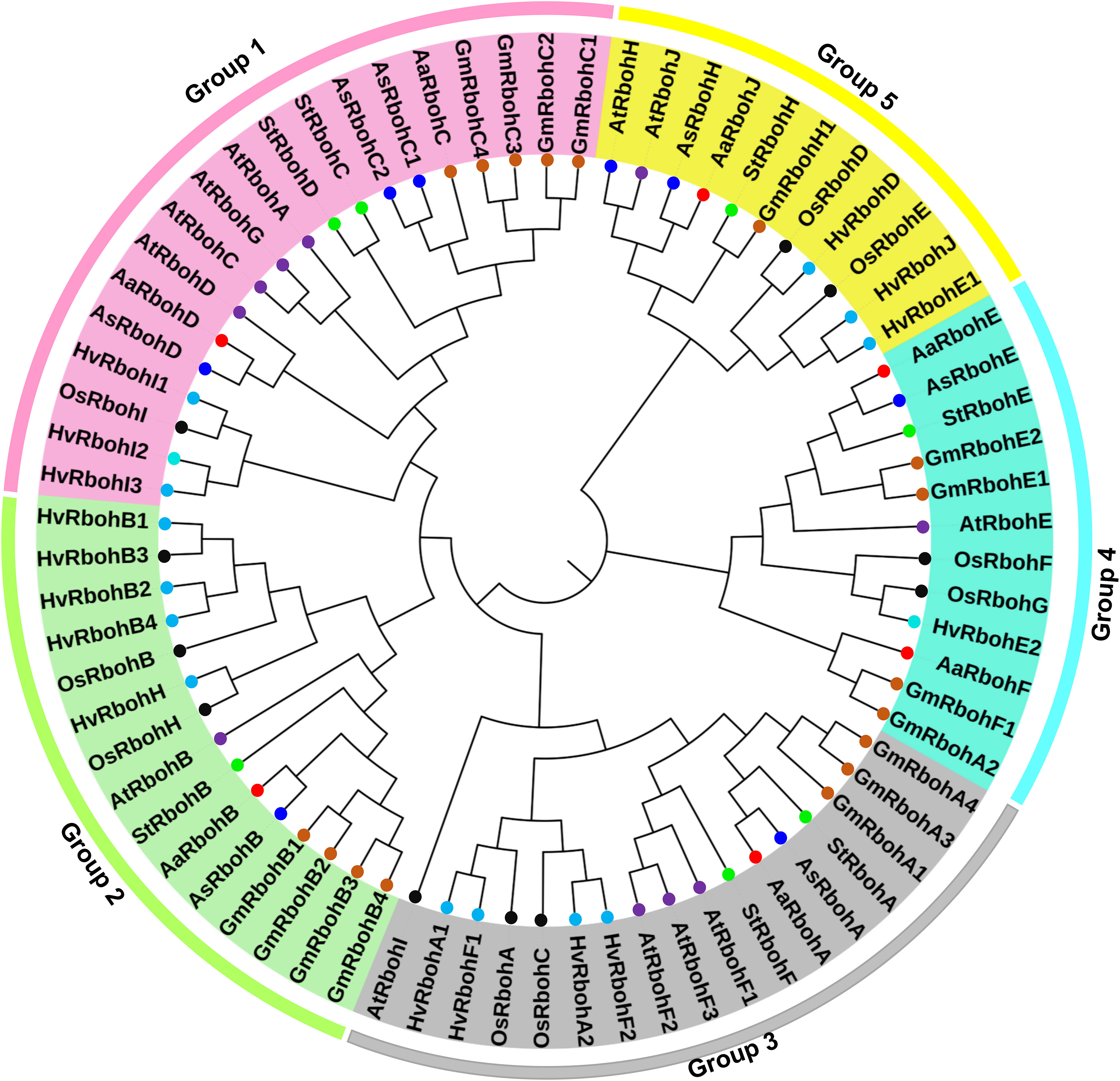
Figure 1 Phylogenetic relationship between the Rboh proteins of (A) agallocha, (A) sinensis, (A) thaliana, (G) max, (H) vulgare, and S. tuberosum. Molecular phylogenetic tree constructed using MEGA-X with NJ method–based P distance substitutions model. Bootstrap values used to assess the tree. Five group are shown as Groups 1–5 with different colors.
3.3 Subcellular location and gene structure
The results of subcellular location prediction indicated that all Rboh proteins are localized to the plasma membrane. Furthermore, the structural organization of exon–intron sequences in the clustered genes displayed notable similarities, suggesting a close evolutionary relationship among them. They exhibited varying numbers of exons, ranging from eight (AaRbohD and AsRbohD) to 15 (AaRbohE). Most Rboh genes, on the other hand, contained either 12 (AaRbohB, AaRbohA, and AsRbohC1) or 14 (AaRbohF, AsRbohB, AsRbohC2, AsRbohA, and AsRbohE) exons (Figure 2). In terms of intron composition, the members showed variability in both the number and types of introns, where AaRbohE had the maximum intron. Phase 0 introns were the most abundant, totaling 81, followed by phase 2 introns being 42 and phase 1 introns being 31. The presence of phase 0 introns ranged from two to nine in each member, whereas phase 1 introns varied from one to three and phase 2 introns varied from two to three, with the exception of AsRbohJ (Figure 2).
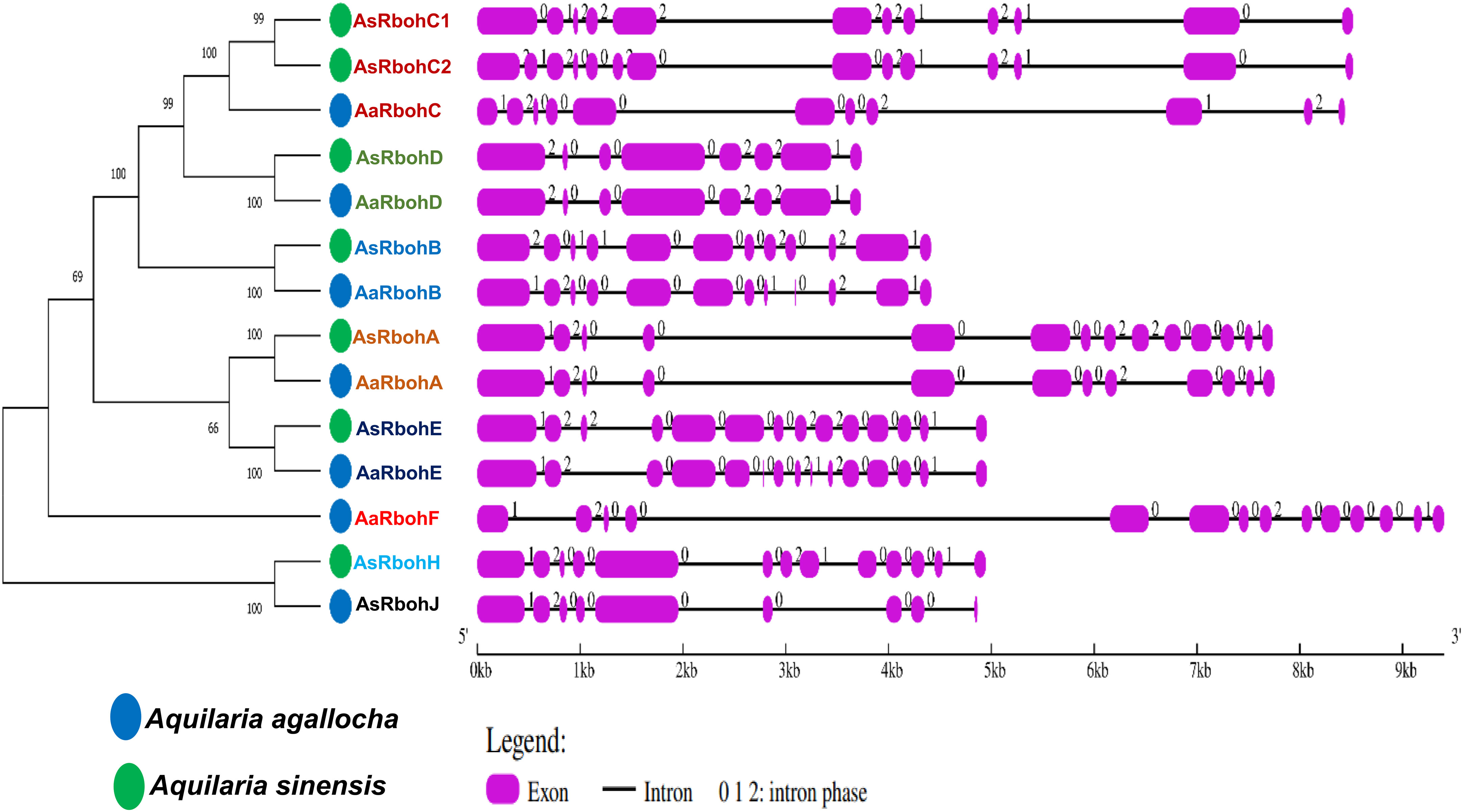
Figure 2 Schematic representation of structures of 14 putative Rboh genes in two Aquilaria species. The exons and introns are indicated with pink rectangles and black color lines, respectively, on the right of phylogenetic tree. The numbers (0, 1, and 2) on the gene structures indicate the intron phases.
3.4 Cis-acting elements of the putative Rboh promoter
A total of 56 types of cis-acting elements were identified on the 2.4-kb region upstream of the translational start site of each Rboh genes (Supplementary Table 2). These elements were categorized into four major functional groups: hormone regulation, stress response, and metabolism-responsive and development-related cis-acting elements. Stress and defense responsiveness cis-acting elements were ARE (cis-acting elements for anaerobiosis), MBS (drought response), LTR (low-temperature responsive cis-acting element), TC-rich repeats (defense), and WUN motif (wound stress) (Figure 3). In addition, six types of plant hormone regulatory elements were salicylic acid response element (TCA-element and SARE); Gibberellin response element (TATC-box, P-box, and GARE); Auxin response element (AuxRR-core and TGA-element); Ethylene response element (ERE); Abscisic acid (ABA) response element (ABRE); and methyl jasmonate response elements (TGACG-motif and CGTAC-motif) (Figure 3A). The promoters also had cell differentiation and developmental processes elements such as RY-elements and CAT box cis-elements. Interestingly, the numbers of defense and stress responsiveness elements were observed in all Rbohs gene’s promoter in highest numbers, ranging from 6 (AaRbohE) to 14 (AaRbohA) (Figure 3B).
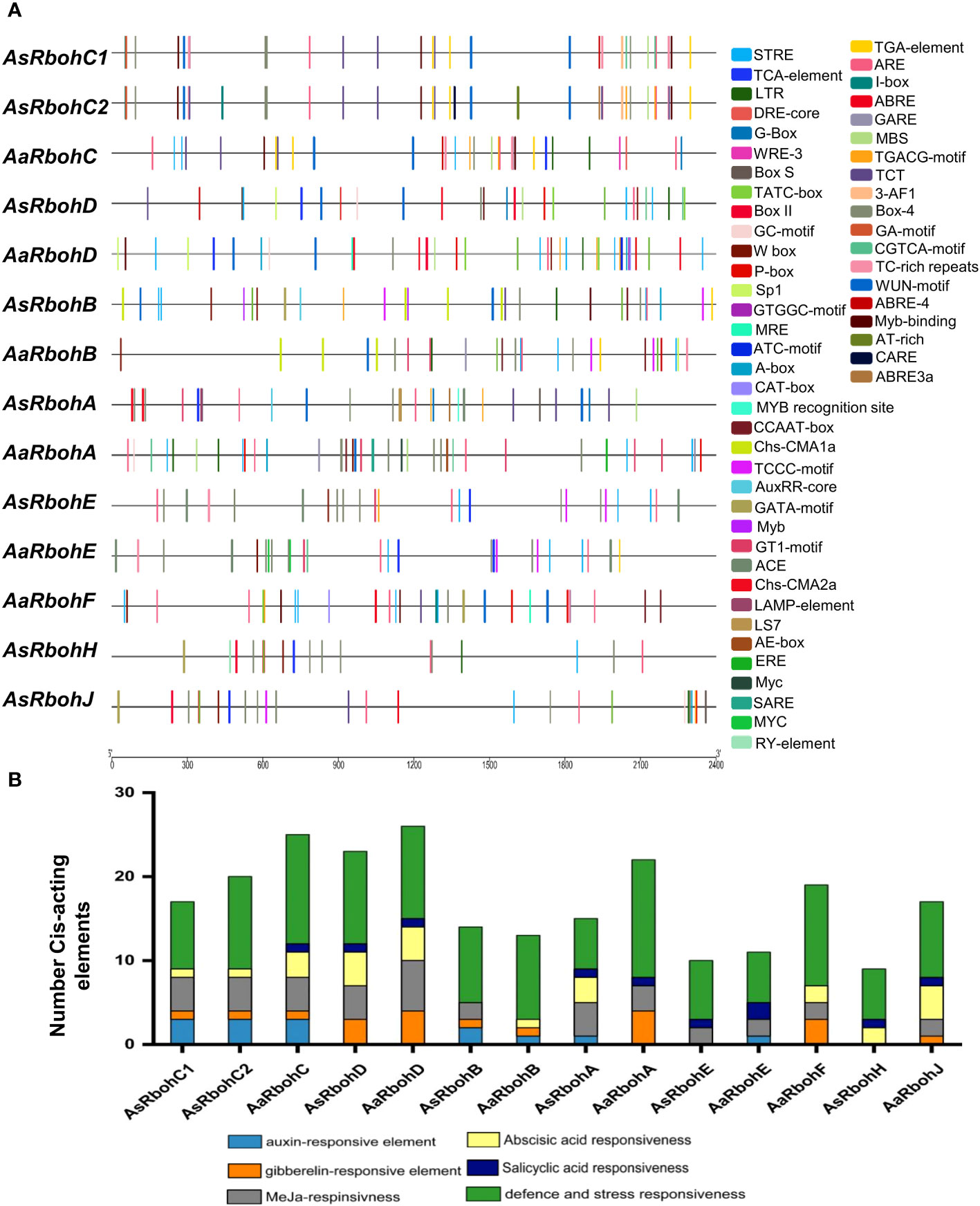
Figure 3 Distribution of major cis-acting elements in the promoter of AaRboh and AsRboh genes. (A) Cis-acting regulatory elements predicted in the 2.4-kb upstream regions of AaRboh and AsRboh genes, indicating with different color rectangular boxes. (B) The number of hormone responsiveness and defense-related cis-acting regulatory elements of AaRboh and AsRboh genes.
3.5 Gene location, synteny block analysis, and Ka/Ks calculation
The seven Rboh genes of A. agallocha were distributed in seven different scaffolds (Figure 4A), and AsRboh genes were distributed in Chromosome 2 (AsRbohA), Chromosome 4 (AsRbohE), Chromosome 6 (AsRbohB), Chromosome 7 (AsRbohC1 and AsRbohD), and ContigUN (AsRbohC2) (Figure 4B). The syntenic analysis unveiled a collinear relationship between five AaRboh genes, namely, AaRbohA, AaRbohB, AaRbohC, AaRbohF, and AaRbohE in A. agallocha and their counterparts in A. sinensis (Figure 4C). AaRbohE was found to be associated with two syntenic gene pairs in A. sinensis. The AaRbohD and AaRbohJ genes exhibited no collinear relationships with genes in A. sinensis. Furthermore, AaRbohA and AaRbohC displayed a collinear relationship with genes in A. thaliana, whereas AaRbohE exhibited synteny with S. tuberosum. The member AsRbohC2 exhibited synteny with genes present in both S. tuberosum and A. thaliana. The syntenic blocks with their genome location were summarized in Supplementary Table 3. Duplication analysis indicated that, in A. sinensis, one gene pair AsRbohC1 and AsRbohC2 undergone segmental duplication, whereas AsRbohD emerged from the parental gene AsRbohC1 through transposed duplication (TRD). Interestingly, in A. agallocha, three pairs of TRD genes were detected, where AaRbohB, AaRbohC, and AaRbohE duplicated from the parent gene AaRbohA. The Ka/Ks ratio of the duplicated gene pair was found to be < 1 (Supplementary Table 4). The divergent time of the duplicated members ranged from 1.4 to 227.85 million years ago (MYA).
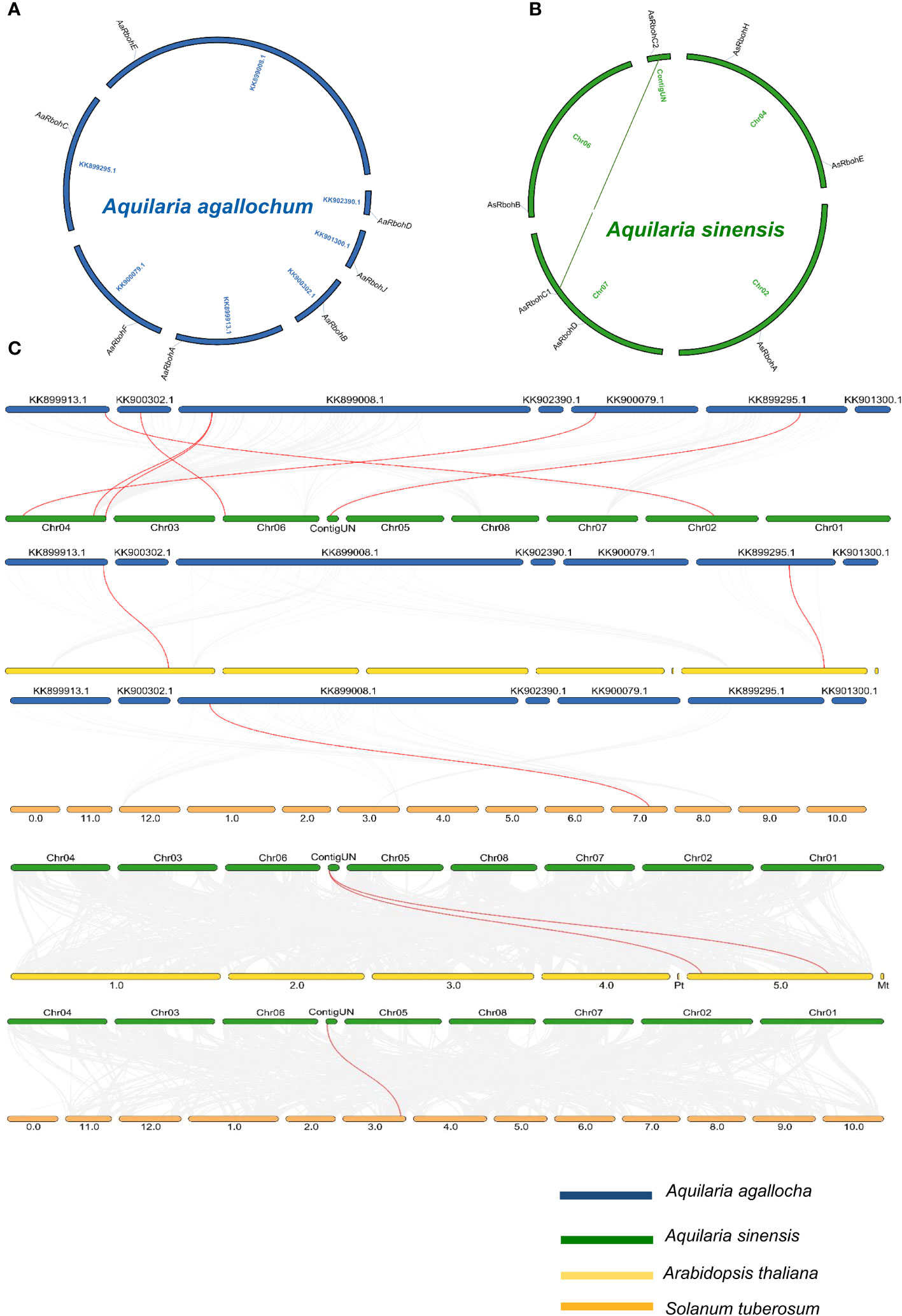
Figure 4 Overview of evolutionary relationship of Rboh of A. agallocha, A. sinensis, A. thaliana, and S. tuberosum. (A) Synteny analysis of AaRboh. (B) Synteny analysis of AsRboh and AsRboh (green line shows duplicate genes). (C) Synteny analysis among A. agallocha and A. sinensis; A. agallocha and A. thaliana; A. agallocha and S. tuberosum; A. sinensis and A. thaliana; and A. sinensis and S. tuberosum. Gray lines represent all collinearity blocks, whereas red lines show orthologous gene pairs among two species.
3.6 Amino acid sequence and characteristic domain analysis of Rboh proteins
MEME suite tool identified 10 consensus motifs in the Rboh proteins based on degree of conservation amino acid residues. The motifs, viz., motif 1 (except AaRbohJ), motif 2, motif-3 (except AaRbohE), motif 4, motif 5, motif 6, motif 7, and motif 8, existed in all Rboh proteins (Figure 5A). Whereas, both motif 9 and motif 10 were missing in Aquilaria RbohB, RbohC, and RbohJ. The four conserved motifs typically found in Rboh proteins existed in the Aquilaria Rboh members (Figure 5B). The most conserved amino acid within these motifs were represented by higher bits size (Figure 5C). Multiple sequence alignment revealed the presence of characteristics conserved domains, i.e., NADPH oxidase (PF08414), EF hand, Ferric reductase (PF01794), FAD binding (PF08022), and NAD binding (PF08030) (Figure 6). However, NAD-binding domain was missing in AaRbohB, AaRbohF, and AaRbohJ, and Ca2+-binding EF-hand domain in AaRbohF. The Rboh protein’s motif analysis revealed that motif 7, motif 4, motif 2, motif 5, motif 8, and motif 3 are parts of the NADPH oxidase (PF08414); motif 8 and motif 3 are part of the transmembrane helix; motif 9 is a part of the FAD binding PF08022); and motif 10, motif 6, and motif 1 are parts of the NAD binding (PF08030) (Supplementary Table 5). Overall, few motifs in the Rboh members (except RbohA and RbohC) of A agallocha were missing.
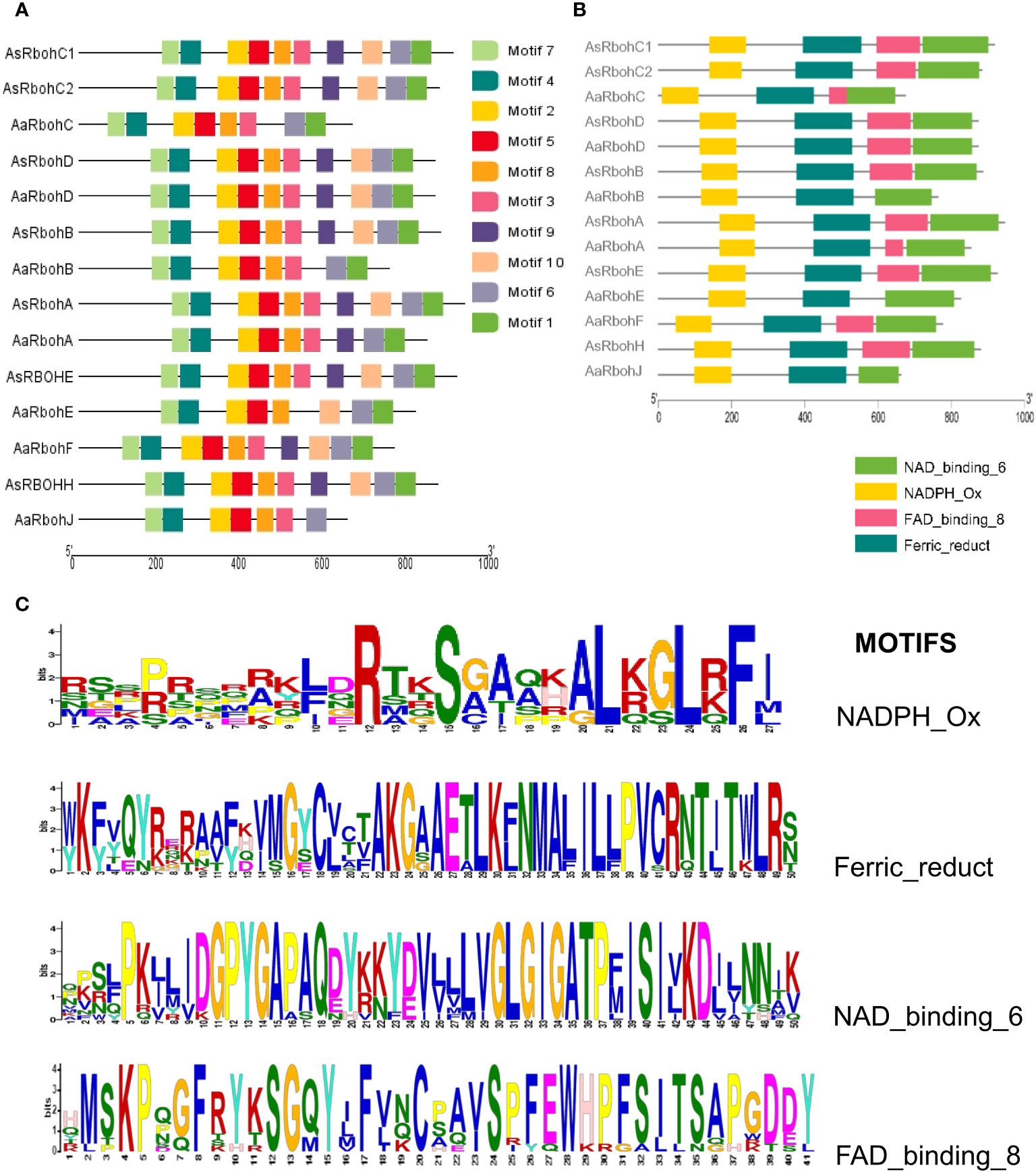
Figure 5 Conserved motifs distribution of AaRboh and AsRboh protein sequences. (A) Ten types of conserved motifs of AsRboh and AaRboh. (B) Four particular characteristics of motif of AsRboh and AaRboh. (C) Sequence logos of the NADPH_Ox, Ferric_reduct, FAD_binding_8, and NAD_binding_6 motif.

Figure 6 Multiple protein sequence alignment and domain structure of Rboh proteins of A. agallocha and A. sinensis. Highly conserved amino acids indicate with red shading, and low amino acid levels represent with lighter shading. The NADPH_ox (PF08414), EF-hand domain, Ferric_reduct (PF01794), FAD_binding_8(PF08022), and NAD_binding_6 (PF08030) were indicated with blue, black, violet, brown, and yellow color, respectively.
3.7 Secondary and tertiary structures
The secondary structures (SSs) of the Rboh proteins consisted of α-helix, coils, turns, and β-sheet (Supplementary Figure 3). Among all, α-helices were seen to be most dominant. The conserved motifs were identified in the models, where NADPH_Ox, EF-hand, and Ferric-reduct appeared as α-helix. More than one type of SS was found in a few motifs. For example, NAD_binding_6 consisted of both α-helices and coils, and FAD_binding_8 composed of a β-strand and coils. Note that the results of assessment parameters of the tertiary structures suggested a good quality of the models. For instance, the Mol Probity and Clash scores ranged from 0.96 to 1.98 and from 0.39 to 4.69. In addition, all the structures were Ramachandran favored with % above 92. The quality factor of the models calculated using ERRAT ranged from 84.13 to 96.5, indicating an acceptable quality of the constructed models.
3.8 Functional analysis and protein–protein interaction
GO terms were assigned to the Aquilaria Rboh members, and their participation in biological processes (BP), molecular function (MF), and cellular component (CC) were elucidated (Supplementary Table 6). The string network model that consisted of 26 nodes and 121 edges (P = 1.0e−16) helped identify their functional partners (Figure 7). In addition, functional information pulled from KEGG database revealed their involvement in signal transduction pathways [mitogen-activated protein kinase (MAPK) signaling], plant–pathogen interaction, and plant hormone. In plant–pathogen interaction, AaRbohA, AaRbohB, AaRbohC, AaRbohD, and AaRbohE were directly involved and interacted with their functional partners, namely, MPK3, CPK28, CDPK1, WRKY33, and EFR. Similarly, the five Rboh proteins mentioned above were involved in MAPK signaling and interacted with their partners MKP2, MPK3, WRKY3, ABI1, ABI2, and OST1. AaRbohA and AaRbohD were possibly involved in hormone transduction and interacted with BRI1, ABI1, ABI2, OST1, ABF2, HAB1, and PP2CA (Figure 7).
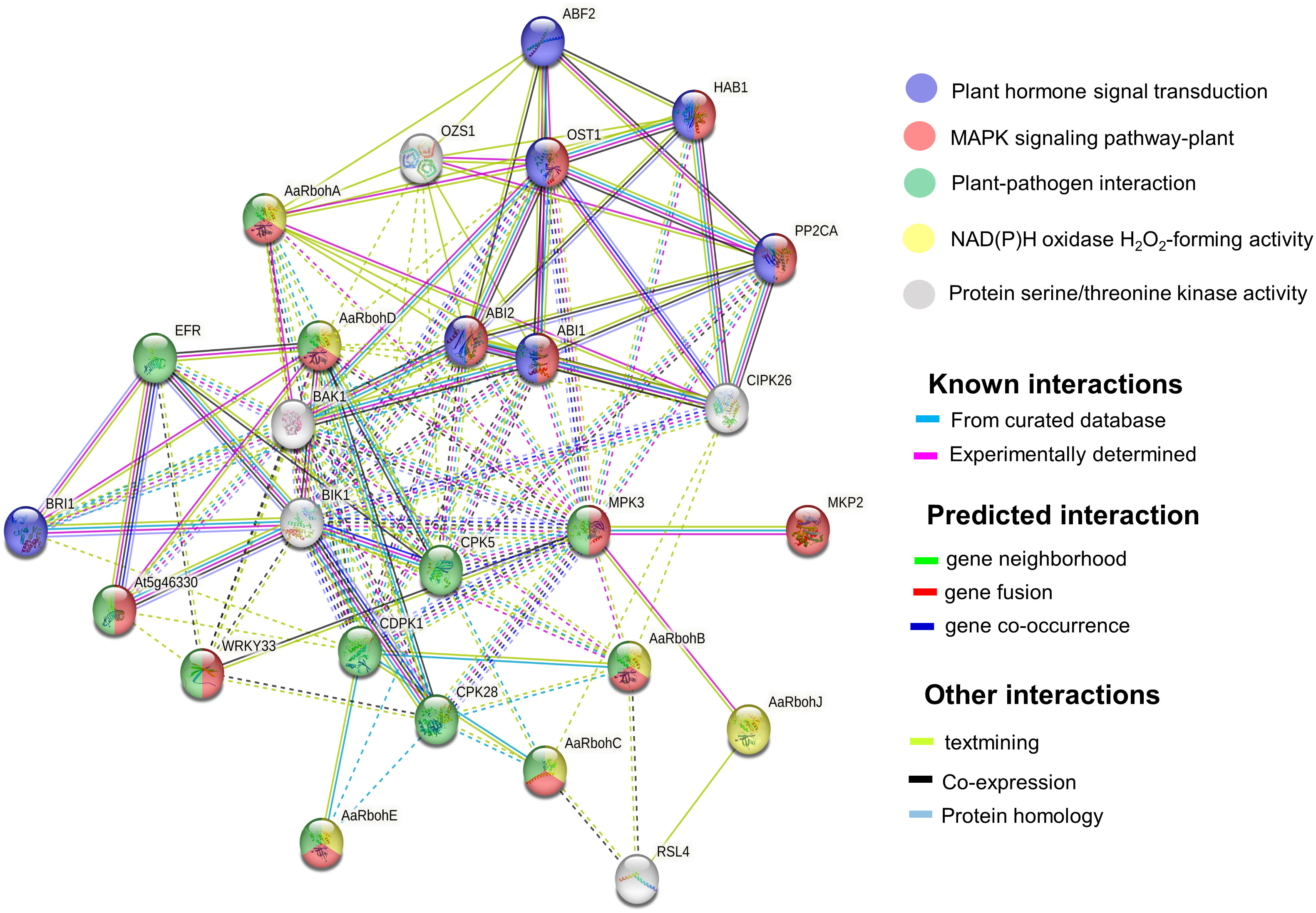
Figure 7 Protein interaction network of AaRboh in A. agallocha based on Arabidopsis orthologs. The potential AaRboh with their functional partners [MPK3 (mitogen-activated protein kinase 3), CPK28 (calcium-dependent protein kinase 28), CDPK1, WRKY33 (WRKY transcription factor 33), ERF (EF-TU receptor), BRI1 (brassinosteroid-insensitive 1), ABI1(abscisic acid–insensitive 1), ABI2, OST1(open stomata 1), ABF2 (abscisic acid–responsive element–binding factor 2), HAB (hypersensitive to ABA1), and PP2CA (protein phosphatase 2CA)] in each enriched pathway are displayed in a network model of proteins where the lines of various colors indicate the type of interactions between the potential AaRboh and their functional partners. The solid and dotted lines represent connections within the same and different clusters.
3.9 In silico expression analysis of the Rboh genes and their functional partners in A. agallocha and A. sinensis tissues
Quantification of transcripts accumulation of the Rboh genes in A. agallocha showed differential upregulation of AaRbohA (0.7 log2FC) and AaRbohC (1.5 log2FC) in agarwood tissue. In contrast, AaRbohB, AaRbohE, and AaRbohF significantly downregulated by 1.3, 0.9, and 0.3 log2FC, respectively. At the same time, AaRbohD and AaRbohJ showed no change in expression (Figure 8A). The genes that act as transcription factors (MYC2 and WRKY), in MAPK signaling cascade (MAPK, MAPKK, and MAPKKK) and in terpene backbone biosynthesis (DXS, HMGR, MVK, GGPS, and FPS), were significantly upregulated in agarwood tissue as shown in Figures 8B–D; Supplementary Table 7. In addition, expression of AsRboh in RNA-seq data of different tissues was estimated using aril tissue as control. Interestingly, AsRbohA was found to be significantly upregulated in all the tissues including wounded stem, callus, leaf, flower, and seed in the range of 4–8 log2FC (Figure 8E), whereas AaRbohC1 and AaRbohC2 upregulated only in wounded stem, callus, and flower in the range of 2–4 log2FC. Similarly, AsRbohB was comparatively higher in wounded stem (9 log2FC) and callus (15 log2FC), and AsRbohE in wounded stem (7.1 log2FC) and callus (7.3 log2FC). However, expression of AsRbohD and AsRbohH in the different tissues was either insignificant or had no difference (Supplementary Table 8).
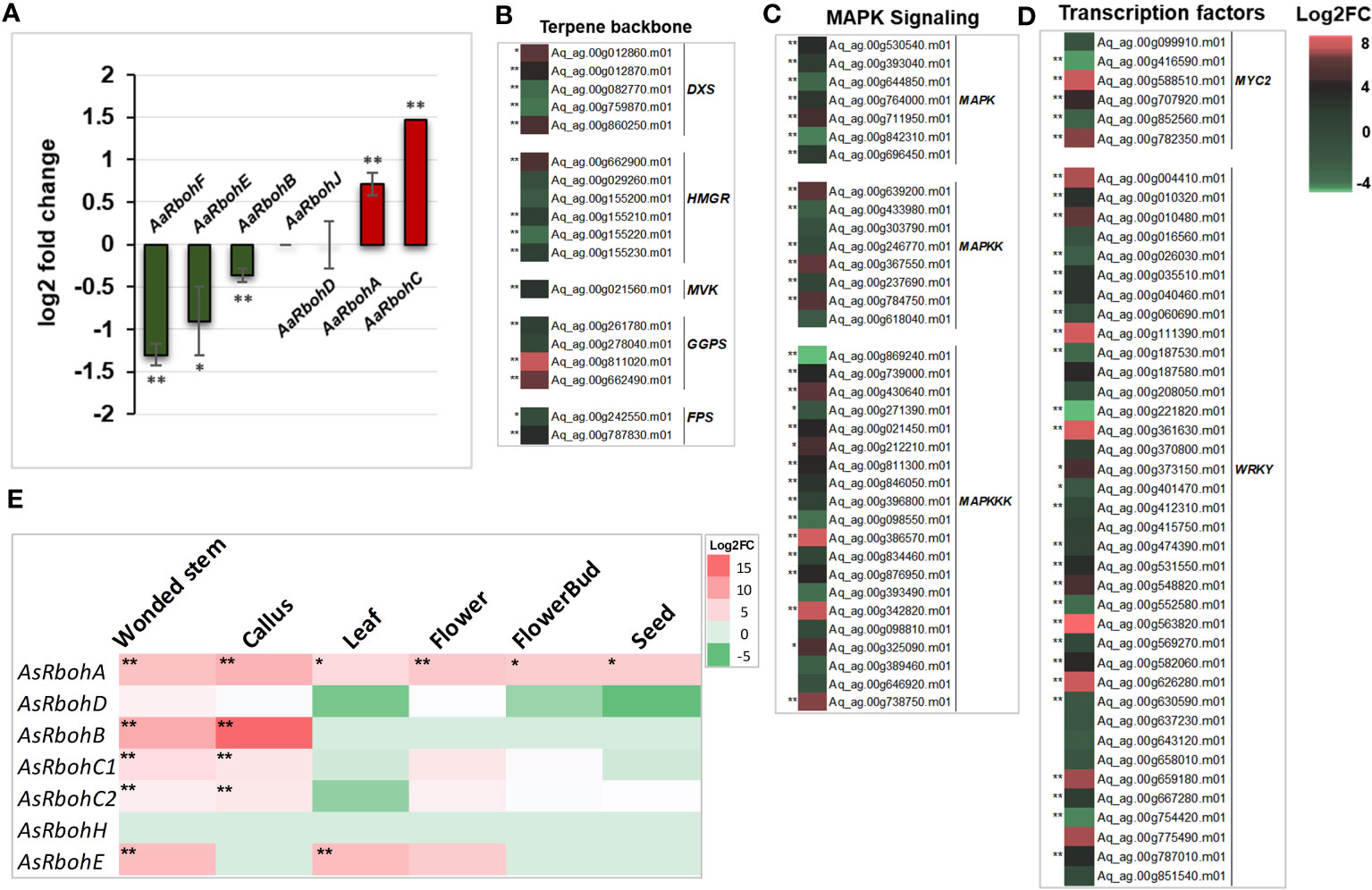
Figure 8 Expression profile of AaRboh and AsRboh genes of different types tissues of A. agallocha and A. sinensis. (A) AaRboh gene expression patterns in agarwood tissue. X-axis represents the AaRboh members, and Y-axis represents the log2 fold change value. (B) Expression patterns of genes involved in terpenoid biosynthesis genes where DXS indicates 1-deoxy-D-xylose-5-phosphate synthase, HMGR indicates 3-Hydroxy-3-methylglutaryl-coenzyme A reductase, MVK indicates mevalonate kinase, GGPS indicates geranylgeranyl diphosphate synthase, and FPS indicates farnesyl pyrophosphate synthase.(C) Expression patterns of mitogen-activated protein kinase (MAPK) signaling cascades genes. (D) Expression pattern of transcription factors. (E) AsRboh gene expression patterns in the six different tissues compared to aril tissue. * indicates p-value less than 0.05, and ** indicates p-value less than 0.01.
3.10 Validation of the expression of AaRboh genes in H2O2-treated callus and stem
To evaluate the impact of hydrogen peroxide (H2O2) on the transcript levels of AaRboh genes, calli tissues were subjected to treatments involving H2O2, DMTU (a ROS scavenger), and combination of them (H2O2 + DMTU). In calli, the exposure to H2O2 resulted in the upregulation of AaRboh genes. Specifically, the expression of AaRbohA peaked at 2 h, showing a 4.15-fold increase. Similarly, the expression of AaRbohB, AaRbohC, and AaRbohE reached their peaks at 6 h, exhibiting 6.07-fold, 24.40-fold, and 5.26-fold increases, respectively. It is worth noting that there was a subsequent decline in the expression of these genes from 6 h to 48 h (Figure 9). When subjected to a combination of H2O2 and DMTU, the expression of these genes also increased, although not to the same extent as when induced by H2O2 alone. In contrast, treatment with DMTU alone resulted in lower expression compared to the control. Interestingly, there were no significant variations in the expression levels of the three genes, AaRbohD, AaRBohF, and AaRbohJ, when compared to the control across the different time periods.
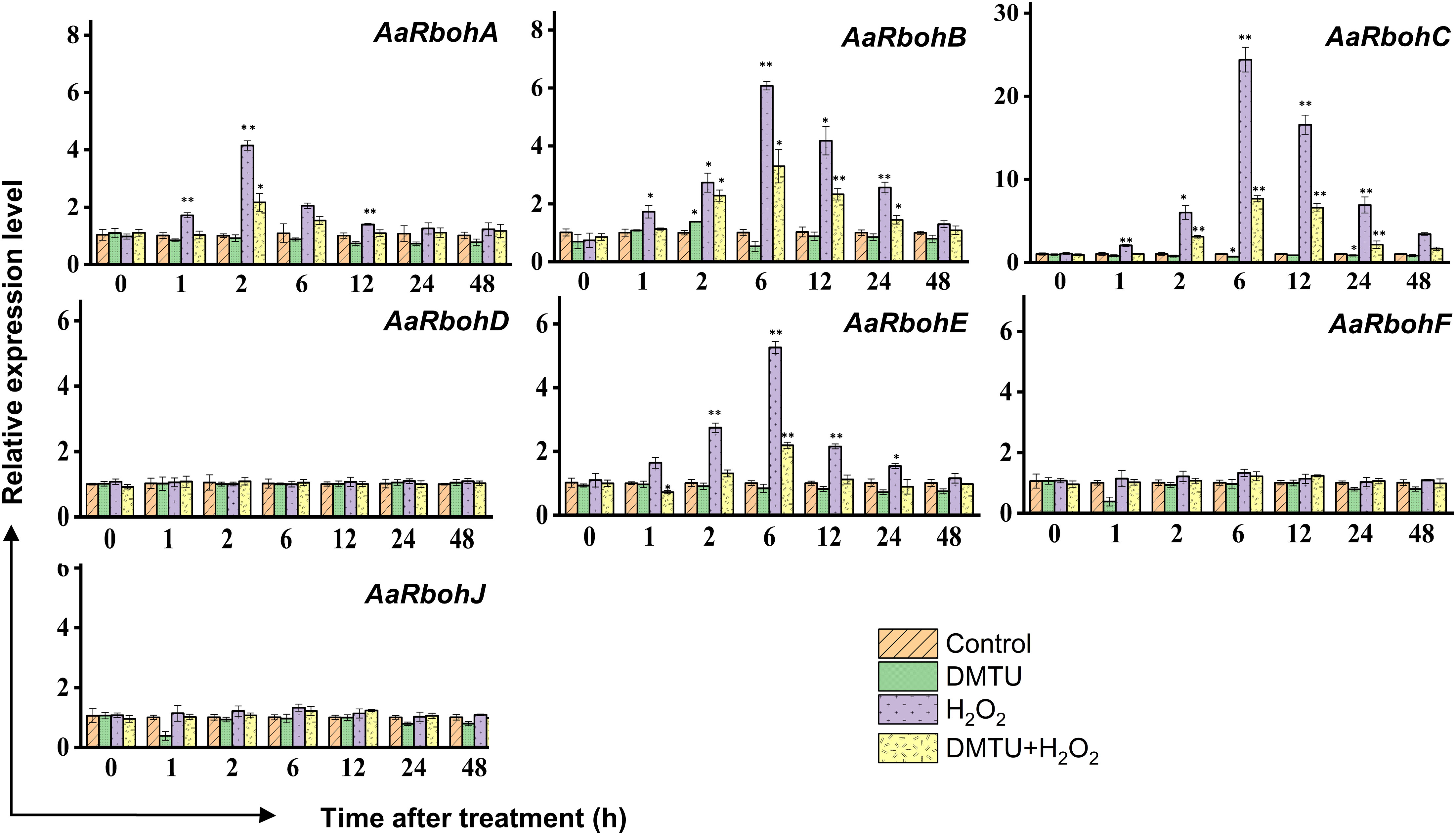
Figure 9 Relative expression levels of AaRboh genes in treated calli of A. agallocha. Relative transcripts abundance of seven AaRboh genes were measured in calli tissue transferred to MS media with H2O2, H2O2 + DMTU, DMTU, respectively, and calli without any treatment considered as control condition and samples harvested at 0 h, 1 h, 2 h, 6 h, 12 h, 24 h, and 48 h. Transcript abundances were measured using A. agallocha GAPDH as internal control. Asterisk (*) denotes a significant difference compared with healthy samples at 0.05 or **P < 0.01 (Student’s t-test). Data represent means ± SE off three independent experiments.
In the wounded stem treated with H2O2, H2O, and DMTU, separately, the transcript levels of AaRbohA and AaRbohC experienced significant increase in H2O2-treated stem, reaching 6.82-fold and 6.05-fold, respectively, within the first hour (Figure 10). Subsequently, after 2 h, their expression levels returned to the initial baseline. However, at the 6-h time point, both genes exhibited a remarkable surge in expression, with AaRbohA and AaRbohC showing increase of 21.64-fold and 40.21-fold, respectively. This heightened expression subsided from 12 h and progressively declined during the 48 h of air exposure. In contrast, the treatment with water (H2O) resulted in a peak in the level of both genes, AaRbohC and AaRbohA, at 6 h, and their expression had not reverted to pre-treatment levels even after 48 h. However, when wounded stems were treated with DMTU, the expression of both genes decreased by about three-fold and four-fold compared to wounded stems treated with H2O2 (Figure 10). Meanwhile, AaRbohB, AaRbohD, AaRbohF, and AaRbohJ exhibited no significant deviations in their expression patterns compared to the control.
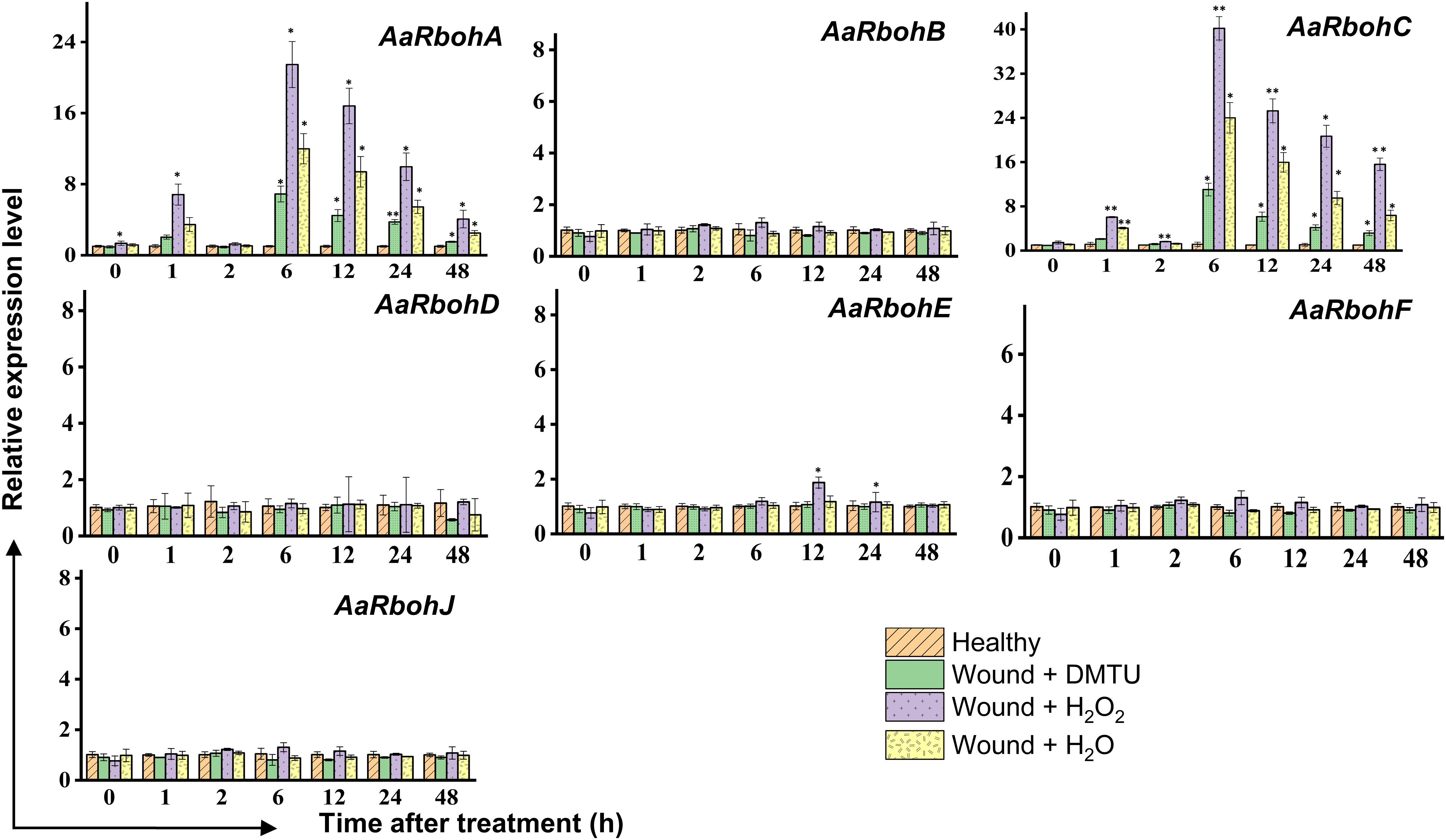
Figure 10 Relative expression levels of AaRboh genes in H2O2-treated stem of A. agallocha. The stems were cut, and the apical end of each cut stem was placed in distilled H2O, H2O2, DMTU, respectively, as appropriate. The pre-treating solution was thrown away after 2 h, and the stems were left exposed to air. The samples were taken at 0 h, 1 h, 2 h, 6 h, 12 h, 24 h, and 48 h following air exposure. The samples without any treatments are considered as healthy. Asterisks (*) denotes a significant difference compared with healthy samples at 0.05 or **P < 0.01 (Student’s t-test). Data represent means ± SE off three independent experiments.
3.11 ROS determination
The treatment with (H2O2) led to an increase in endogenous H2O2 content in both calli and actively growing wounded pruned stem tissues. In calli, a transient rise in H2O2 levels was observed at 6 h, reaching 2.96 μmol/g, after which it gradually decreased to 1.26 μmol/g by 48 h (Figure 11A). Moreover, treatment with DMTU alone resulted in a decrease in the accumulation of H2O2, which remained relatively constant throughout the study period. When H2O2 was applied in combination with DMTU, as expected, it led to the reduction in endogenous H2O2 production, which reached 0.92 μmol/g at 6 h. In the case of wounded stems treated with H2O2, the concentration of endogenous H2O2 experienced an initial peak at 1 h, reaching 2.12 μmol/g. Subsequently, it decreased to the baseline level at 2 h, followed by another increase. The maximum H2O2 production occurred during the second peak at 6 h, with an H2O2 concentration 31.16 times greater than the initial concentration, totalling 9.97 μmol/g. After 48 h of exposure to air, the H2O2 concentration decreased to 1.19 μmol/g. The elevated endogenous H2O2 levels were mitigated by DMTU application. Furthermore, the endogenous H2O2 content in wounded stems treated with H2O was lower than that in wounded stems treated with H2O2, but it was higher than that observed in the treatment with DMTU. Notably, there were no significant alteration in the endogenous H2O2 levels in healthy stems (Figure 11B).
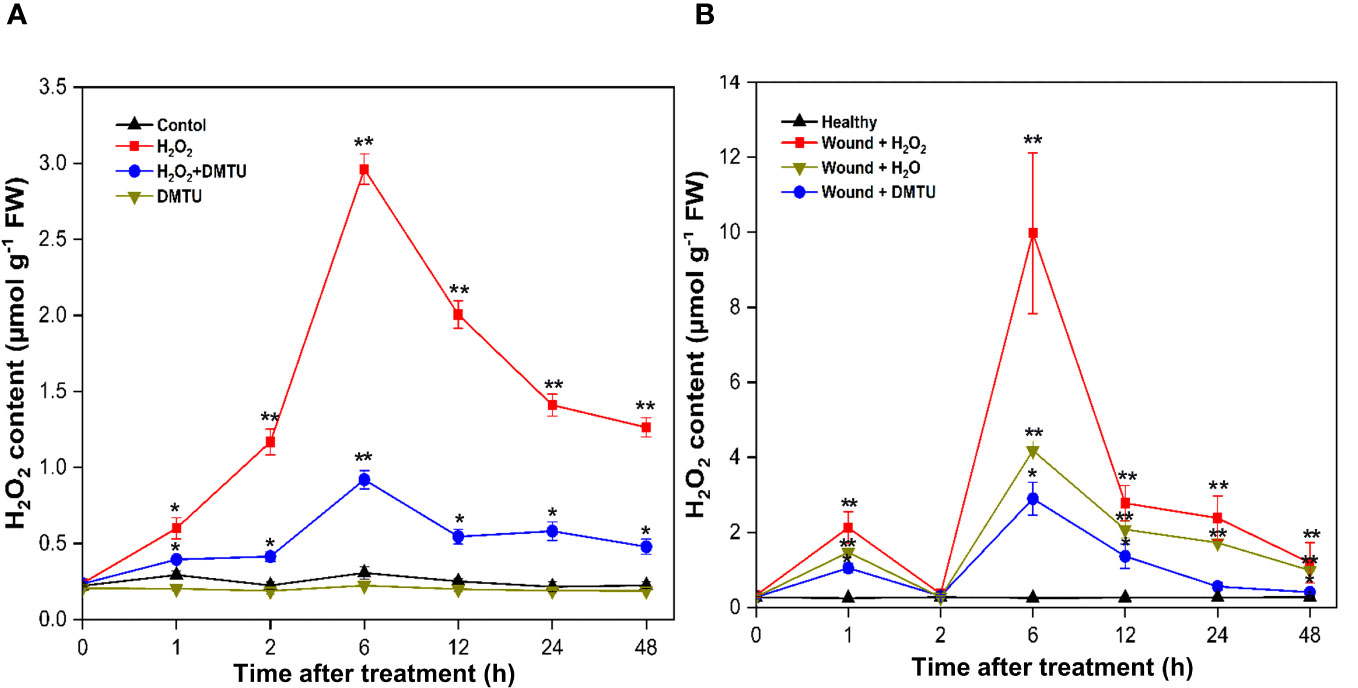
Figure 11 Endogenous H2O2 content in calli and stem of (A) agallocha. (A) Content of endogenous H2O2 in calli treated with H2O2, DMTU, and H2O2 + DMTU, respectively, for 0, 1 h, 2 h, 6 h, 12 h, 24 h, and 48 h. (B) Content of endogenous H2O2 in the 1-year-old stems after pruning, the cut ends were immersed in distilled H2O, H2O2, and DMTU. The pruned stems were exposed to air after 2 h, and the pretreating solution was discarded. The healthy condition indicates the samples without any treatment and served as control. Following air exposure, samples were collected at 0 h, 1 h, 2 h, 6 h, 12 h, 24 h, and 48 h. Asterisks (*) indicate a statistically significant difference from healthy samples at *P < 0.05 or **P < 0.01 (Student’s t-test). The data represent the means and standard deviations of three independent experiment.
3.12 Validation of the expression of AaRbohA and AaRbohC in naturally infected A. agallocha tree
The significantly higher AaRbohC and AaRbohA expression levels in both calli and stem tissues under various treatment conditions strongly suggest their involvement in stress responses. Their expression was assessed in naturally infected wood tissues to further investigate their role in response to stress. Both genes, AaRbohC and AaRbohA, exhibited substantial upregulation, with increases ranging from 22.61-folf to 76.94-fold, respectively, in the infected A. agallocha wood tissues compared with that in healthy wood tissues (Figure 12). This finding indicates the crucial role of these two members in stress responses and possibly during agarwood formation in A. agallocha.
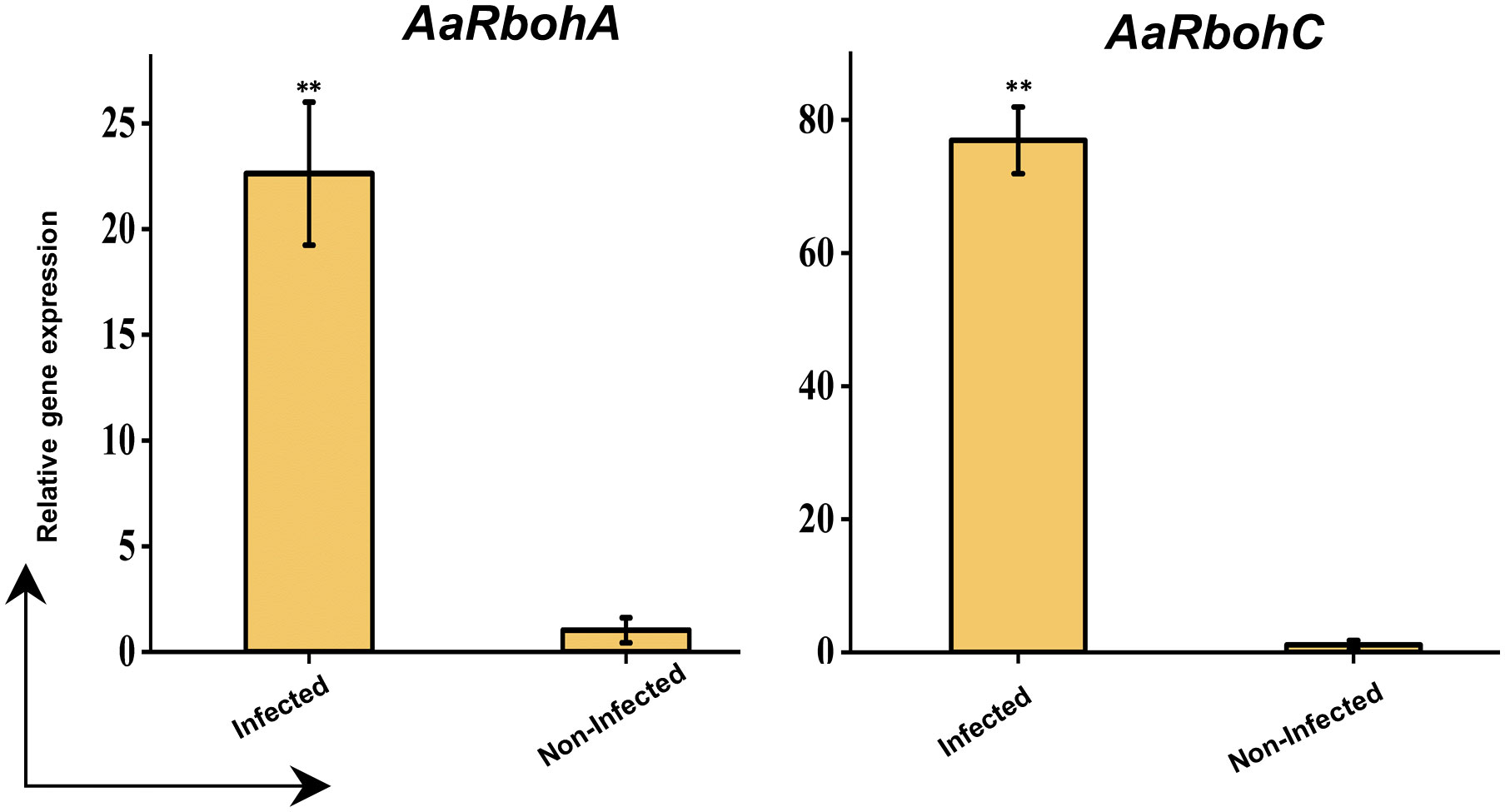
Figure 12 qRT-PCR analysis of two selected AaRboh genes. The −2ΔΔCT method was used to determine relative gene expression value. The house keeping gene GAPDH was used to normalized the data. The * symbol indicates transcript levels that differ statistically significantly based on the student t test, and the P-value (**P < 0.01). The mean SE of three technical replicates is used to calculate each expression value. The infected and non-infected plants from Hoollongapar Gibbon Sanctuary.
4 Discussion
In this study, a comprehensive examination of total 14 Rboh proteins was carried out in both Aquilaria species. Notably, an equivalent number of 7 Rboh proteins have been reported in the genomes of several other plant species, including Citrus sinensis (Zhang et al., 2022), Capsicum annuum (Zhang et al., 2021), Rubus occidentialis, Prunus dulsis (Cheng et al., 2020), Prunus persica (Cheng et al., 2019), Cucumis sativus (Li et al., 2019), Jatropha curcas, Ricinus communis (Zhao and Zou, 2019), Fragaria ananassa cv. Toyonaka (Zhang et al., 2018), and Vitis vinifera (Cheng et al., 2013) (Supplementary Figure 4). This intriguing consistency in the number of Rboh proteins underscores their importance across diverse plant species. However, certain differences within the members of both the Aquilaria species were observed. For instance, RbohF and RbohJ were identified only in A. agallocha, but not in A. sinensis, and vice-versa in the case of RbohH. Similarly, maximum intron, i.e., 15 was found in AaRbohE, whereas 14 in AsRbohE. The promoter of these genes composed of various cis-regulatory elements, a characteristic that affirms their involvement in stress responses, hormonal regulation, and developmental processes (Jakubowicz et al., 2010; Marino et al., 2012; Huang et al., 2021; Zhang et al., 2022). Thus, aligning with previous research carried out in O. sativa and A. thaliana, the presence of these elements within the putative Rboh genes of Aquilaria indicates notable similarities in their functions (Kaur et al., 2016). In addition, plant Rboh proteins are equipped with conserved domains that facilitate their vital functions, including ROS metabolism during stress conditions, the regulation of calcium ion (Ca2+) channels, and downstream signaling processes (Torres and Dangl, 2005; Yu et al., 2020). Recent studies have delved into their roles in stress responses across various angiosperms (Hu et al., 2018; Kaur and Pati, 2018; Chang et al., 2020; Zhang et al., 2022). The results of motif analysis and homology modeling indicate that the Aquilaria Rboh proteins possess the essential domain NADPH_ox, in addition to a transmembrane domain likely associated with ferric reductase activity and two calcium-binding structural motifs known as EF-hand motifs specifically in RbohA and RbohC. The presence of EF-hand suggests its crucial role in interaction with small GTPases (Herve et al., 2006). These structural features strongly suggest that the ROS generated by these Rboh proteins are integral components of the cellular signaling network. The oxidative burst, characterized by the production of hydrogen peroxide (H2O2), occurs as a result of the catalytic conversion of environmental oxygen into H2O2 through the NAD(P)H oxidase H2O2 forming activity of the putative Rboh proteins (Ben Rejeb et al., 2015; Wang et al., 2018). These conserved domains in the putative Rboh proteins imply their potential involvement in stress-induced ROS generation during exposure to various stressors by Aquilaria species. Interestingly, few motifs in the Rboh members (except RbohA and RbohC) of A agallocha were missing. Overall, the presence or absence of specific motifs and differences in certain characteristics in their gene or protein sequence may linked to functional divergence and conservation of Aquilaria Rboh members.
Gene duplication processes significantly influence protein families’ expansion and contraction to fulfill plants’ physiological requirements (Zhang et al., 2021). Duplication events have been identified as a major force behind the expansion of Rboh gene family across various plant species, including Brassica rapa (Li et al., 2019), Musa acuminate (Ying et al., 2020), Gossypium hirsutum (Wang et al., 2020), and A. thaliana (Zhou et al., 2020). Our current investigation identified a single instance of segmental duplication and TRD in A. sinensis. However, the TRD was found to be major force of expansion of Rboh family of A. agallocha. The result indicated that AaRbohA acted as the old parent copy and generated AaRbohB, AaRbohC, and AaRbohE in different divergent time. The presence of intron regions in these genes also indicated their possible generation by transposon-mediated event. Similar results have been reported in case of TRAM/LAG/CRN8 (TLC) genes in maize, where Whole genome duplication (WGD) and TRD contributed to expansion and diversification of the protein families (Si et al., 2019). A significant number of genes have also been shown to undergo TRD in A. thaliana (Wang et al., 2013). The Ka/Ks ratio of the duplicated gene pairs in this study was found to be <1, suggesting that these duplicated genes have undergone robust purifying selection during the evolutionary course. Furthermore, the reduction in the number of Rboh proteins in both Aquilaria genomes, compared to Arabidopsis Rboh family, implies that the loss of function events possibly transpired during the evolutionary course of the Aquilaria genomes. Alternatively, these gene losses could be attributed to functional redundancy (Espinosa-Cantú et al., 2015; Martin and Schnarrenberger, 1997).
KEGG pathway analysis and model interaction network predictions further substantiate the role of AaRboh and AsRboh genes in plant–pathogen interactions, hormone signaling, and MAPK pathway. Rboh proteins in H. vulgare, G. barbadense, Z. jujube, J. curcas, and M. sativa have been shown to perform function through generation of ROS, thereby conferring protection against invading plant pathogens (Trujillo et al., 2006; Zhao and Zhou, 2019; Cheng et al., 2020). But, a study comprehending on the ROS generation and associated Rboh proteins in the genomic-level is missing in Aquilaria plant. Hence, within the framework of this study, the in silico expression analysis has elucidated the differential upregulation of RbohA and RbohC in both plant species, as well as the elevation of RbohB expression in A. sinensis. To corroborate these findings, we validated their expression levels in H2O2-mediated stress-induced callus and stem tissues of A. agallocha using qRT-PCR, followed by quantifying the ensuing ROS generation. An increase in the expression levels of AaRbohA and AaRbohC significantly, coupled with the maximal accumulation of ROS in both callus and stem tissues following a 6-h exposure to H2O2, provides compelling evidence of their involvement in ROS production in response to stress stimuli. In addition, the elevated expression of AaRbohB and AaRbohE exclusively in callus tissue suggests their specialized role in ROS generation, particularly within the callus. However, in A. thaliana, RbohD and RbohF expressed in all tissue (Orman-Ligeza et al., 2016). But in this study, we have not detected any differential expression of these members. Nevertheless, in Solanum melongena, RbohB and RbohC were highly expressed in leaves (Du et al., 2023). The results of the expression study corroborates previous findings about accumulation of endogenous H2O2 under salt stress conditions in Aquilaria itself (Wang et al., 2018). In similar lines, in C. annum L., cold, drought, and salt stresses have been shown to trigger significant endogenous H2O2 production via Rboh enzymes (Zhang et al., 2021). Application of stress via methyl jasmonate was able to substantially upregulate expression of Rboh genes in A. sinensis calli (Xu et al., 2013); similar observation was reported in Nitraria tangutorum (Yang et al., 2012). In light of these findings, it is evident that, under stress conditions, Rboh genes, specifically AaRbohA and AaRbohC, are upregulated and play a pivotal role in ROS generation, which may be closely linked with stress responses and hormonal regulation in A. agallocha.
The generation of agarwood resin that is laden with a diverse array of fragrant metabolites when A. agallocha is subjected to injury and biotic stress is a well-established fact (Ahmead and Kulkarni, 2017; Gao et al., 2019; Huang et al., 2022). The mechanism initiated by infection and the engagement of the MAPK signaling pathway, coupled with the orchestration of defense responses through a network of hormonal biosynthesis and modulation of terpenoid pathway genes by transcription factors, such as WRKY and MYC2, have been postulated as fundamental components intricately involved in the biosynthesis and regulation of these aromatic metabolites in agarwood (Xu et al., 2013; Lv et al., 2019; Tan et al., 2019). Interestingly, recent research on similar line suggested plant–pathogen/microbial interaction as a factor leading to ROS-mediated activation of MAPK pathway (Pacheco-Trejo et al., 2022; Chuang et al., 2022). Thus, we were interested to quantify the transcript levels of AaRbohA and AaRbohC, aiming to explore whether a similar process is at play in the natural production of agarwood within A. agallocha trees. Intriguingly, the significant and distinct upregulation of both these genes compared to healthy tissue strongly suggests their pivotal role in generating ROS within the wood tissue. In the course of this study, we employed an in silico approach, unveiling the differential upregulation of genes associated with the MAPK signaling pathway, transcription factors, and the biosynthesis of terpene backbones. The same had also been validated and is in align with our few previous studies where we obtained differential regulation of genes encoding MAPK, WRKY, MYC2, and terpenoid biosynthesis through qRT-PCR in naturally infected wood (Islam et al., 2020; Das et al., 2021; Islam and Banu, 2021; Das et al., 2023). We have previously observed that naturally infected Aquilaria woods exhibited higher expression levels of the genes responsible for sesquiterpene biosynthesis, which include ADXPS, AHMGR, AFPS, ASS, DGS, and ADXPR (Islam et al., 2020). In the same way, in the infected agarwood, a higher expression of signaling genes (MK, WRKY1, and MAPK3), jasmonate biosynthesis genes (MYC2 and LOX), and sesquiterpenes genes (DSS, DGS, DXPS, FPS, SS4, and DGS1) was observed (Islam and Banu, 2021). An in silico investigation shows that the molecular mechanism behind the production of numerous types of aromatic chemicals is attributed by a AaTPS gene family (Das et al., 2021). The gene family AaCYPs, involved in sesquiterpenoids and phenylpropanoids biosynthesis, and these members were shown to be enhanced in methyl jasmonate–induced callus and infected Aquilaria trees (Das et al., 2023). In addition, in A. sinensis, upregulation of three sesquiterpene synthases (AsTPS10, AsTPS16, and AsTPS19) stem tissue has been linked to sesquiterpenes accumulation in the H2O2 pruned stem (Lv et al., 2019). These findings provide compelling evidence of a cascade of events, initiated by ROS-induced activation of the MAPK pathway, subsequently culminating in the hormonal regulation of terpenoid biosynthesis through TFs like WRKY and MYC2, contributing to the agarwood resin production in A. agallocha tree.
Overall, the findings of this study confirm that, under stress conditions in Aquilaria, Rboh genes play a pivotal role in ROS generation, subsequently leading to the upregulation of various genes responsible for the accelerated accumulation of specifically terpenoids and other secondary metabolites as part of the defense response mechanism (Zhang et al., 2013; Xu et al., 2016; Lv et al., 2019). The generated ROS molecules are likely to serve as signaling entities, modulating the genes involved in the biosynthesis fragrant resinous agarwood.
5 Conclusion
In summary, this study characterized seven Rboh genes in each Aquilaria species, delving into their structural and functional attributes. The comprehensive analyses of phylogenetic positions, exon–intron structures, and motif patterns highlight both divergence and conservation among Aquilaria Rboh members. Promoter analysis strongly indicates their active involvement in stress-related pathways. The study further suggests that Rboh genes are functionally linked with MAPK proteins and transcription factors, including WRKY and MYC2. The two members, viz., AaRbohA and AaRbohC, are likely to play a role in generating ROS and may have a significant impact on the signaling pathways associated with the biosynthesis of metabolites present in resinous agarwood. Although, the full intricate molecular mechanism underlying agarwood formation is still lacking. The functional characterization of this Rboh gene family is expected to expedite the understanding of the initiation of agarwood deposition in Aquilaria plants.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
KB: Conceptualization, Data curation, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. AD: Data curation, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. RA: Methodology, Software, Writing – original draft. SA: Data curation, Validation, Writing – original draft. RK: Data curation, Validation, Visualization, Writing – review & editing, Writing – original draft. SB: Conceptualization, Data curation, Investigation, Resources, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors are indebted to Gauhati University for providing the technical facility and space for carrying out experiments of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1326080/full#supplementary-material
Supplementary Figure 1 | Flowchart of the methodology obtained in the study.
Supplementary Figure 2 | Domain organization of 14 Rboh genes of A. agallocha and A. sinensis. The characteristics domains are displayed based upon results of putative Rboh SMART tool search. Blue rectangular boxes represent trans-membrane regions and small pink boxes represents low complexity regions.
References
Abdin, M. J. (2014). The agar wood industry: yet to utilize in Bangladesh. Int. J. Econ. Manage. Sci. 3, 163–166. doi: 10.2139/ssrn.2430055
Ahmead, D. T., Kulkarni, A. D. (2017). Sesquiterpenes and chromones of agarwood: a review. Malays J. Chem. 19 (1), 33–58.
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., Noble, W. S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Barden, A., Anak, N. A., Mulliken, T., Song, M. (2000). Heart of the matter: Agarwood use and trade and CITES implementation for Aquilaria malaccensis. Traffic Int., 17–18.
Ben Rejeb, K., Lefebvre-De Vos, D., Le Disquet, I., Leprince, A. S., Bordenave, M., Maldiney, R., et al. (2015). Hydrogen peroxide produced by NADPH oxidases increases proline accumulation during salt or mannitol stress in Arabidopsis thaliana. New Phytol. 208 (4), 1138–1148. doi: 10.1111/nph.13550
Castro, B., Citterico, M., Kimura, S., Stevens, D. M., Wrzaczek, M., Coaker, G. (2021). Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 7 (4), 403–412. doi: 10.1038/s41477-021-00887-0
Cepauskas, D., Miliute, I., Staniene, G., Gelvonauskiene, D., Stanys, V., Jesaitis, A. J., et al. (2015). Characterization of apple NADPH oxidase genes and their expression associated with oxidative stress in shoot culture in vitro. Plant Cell Tissue Organ Cult 124 (3), 621–633. doi: 10.1007/s11240-015-0920-2
Chang, Y., Li, B., Shi, Q., Geng, R., Geng, S., Liu, J., et al. (2020). Comprehensive analysis of respiratory burst oxidase homologs (Rboh) gene family and function of GbRboh5/18 on Verticillium wilt resistance. Gossypium barbadense. 11, 788. doi: 10.3389/fgene.2020.00788
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative toolkit developed for interactive analyses of big biological data. Nucleic Acids Res. 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C. H., Kuo, T. C., Yang, M. H., Chien, T. Y., Chu, M. J., Huang, L. C., et al. (2014). Identification of cucurbitacins and assembly of a draft genome for Aquilaria agallocha. BMC Genomics 15 (1), 578. doi: 10.1186/1471-2164-15-578
Cheng, C., Che, Q., Su, S., Liu, Y., Wang, Y., Xu, X. (2020). Genome-wide identification and characterization of Respiratory Burst Oxidase Homolog genes in six Rosaceae species and an analysis of their effects on adventitious rooting in apple. PloS One 15 (9), 1–8. doi: 10.1371/journal.pone.0239705
Cheng, C., Xu, X., Gao, M., Li, J., Guo, C., Song, J., et al. (2013). Genome-wide analysis of respiratory burst oxidase homologs in grape (Vitis vinifera L.). Int. J. Mol. Sci. 14 (12), 24169–24186. doi: 10.3390/ijms141224169
Cheng, X., Li, G., Manzoor, M. A., Wang, H., Abdullah, M., Su, X., et al. (2019). In Silico Genome-Wide Analysis of Respiratory Burst Oxidase Homolog (RBOH) family genes in five fruit-producing trees, and potential functional analysis on lignification of stone cells in chinese white pear. Cells 8 (6), 1–21. doi: 10.3390/cells8060520
Chuang, C. Y., Lin, S. T., Li, A. T., Li, S. H., Hsiao, C. Y., Lin, Y. H. (2022). Bacillus amyloliquefaciens PMB05 increases resistance to bacterial wilt by activating mitogen-activated protein kinase and reactive oxygen species pathway crosstalk in Arabidopsis thaliana. Phytopathology 112 (12), 2495–2502. doi: 10.1094/PHYTO-04-22-0134-R
Das, A., Begum, K., Akhtar, S., Ahmed, R., Kulkarni, R., Banu, S. (2021). Genome-wide detection and classification of terpene synthase genes in Aquilaria agallochum. Physiol. Mol. Biol. Plants 27 (8), 1711–1729. doi: 10.1007/s12298-021-01040-z
Das, A., Begum, K., Akhter, S., Ahmed, R., Tamuli, P., Kulkarni, R., et al. (2023). Genome-wide investigation of Cytochrome P450 superfamily of Aquilaria agallocha: association with terpenoids and phenylpropanoids biosynthesis. Int. J. Biol. Macromol. 234, 123758. doi: 10.1016/j.ijbiomac.2023.123758
Ding, B., Liu, T., Hu, C., Song, Y., Hao, R., Feng, X., et al. (2021). Comparative analysis of transcriptomic profiling to identify genes involved in the bulged surface of pear fruit (Pyrus bretschneideri Rehd. cv. Yuluxiangli). Physiol. Mol. Biol. Plants an Int. J. Funct. Plant Biol. 27 (1), 69–80. doi: 10.1007/s12298-021-00929-z
Ding, X., Mei, W., Lin, Q., Wang, H., Wang, J., Peng, S., et al. (2020). Genome sequence of the agarwood tree Aquilaria sinensis (Lour.) Spreng: the first chromosome-level draft genome in the Thymelaeceae family. GigaScience 9, 1–10. doi: 10.1093/gigascience/giaa013
Du, L., Jiang, Z., Zhou, Y., Shen, L., He, J., Xia, X., et al. (2023). Genome-wide identification and expression analysis of respiratory burst oxidase homolog (RBOH) gene family in eggplant (Solanum melongena L.) under abiotic and biotic stress. Genes 14 (9), 1665. doi: 10.3390/genes14091665
Espinosa-Cantú, A., Ascencio, D., Barona-Gómez, F., DeLuna, A.. (2015). Gene duplication and the evolution of moonlighting proteins. Front. Genet. 6, 227. doi: 10.3389/fgene.2015.00227
Gao, M., Han, X., Sun, Y., Chen, H., Yang, Y., Liu, Y., et al. (2019). Overview of sesquiterpenes and chromones of agarwood originating from four main species of the genus. Aquilaria. RSC Adv. 9 (8), 4113–4130. doi: 10.1039/C8RA09409H
Hawamda, A. I. M., Zahoor, A., Abbas, A., Ali, M. A., Bohlmann, H. (2020). The Arabidopsis RbohB encoded by At1g09090 is important for resistance against nematodes. Int. J. Mol. Sci. 21 (15), 1–20. doi: 10.3390/ijms21155556
Herve, C., Tonon, T., Collen, J., Corre, E., Boyen, C. (2006). NADPH oxidases in Eukaryotes: red algae provide new hints! Curr. Genet. 49 (3), 190–204. doi: 10.1007/s00294-005-0044-z
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, C. H., Wei, X. Y., Yuan, B., Yao, L. B., Ma, T. T., Zhang, P. P., et al. (2018). Genome-Wide identification and functional analysis of NADPH oxidase family genes in wheat during development and environmental stress responses. Front. Plant Sci. 9, 906. doi: 10.3389/fpls.2018.00906
Huang, S., Tang, Z., Zhao, R., Hong, Y., Zhu, S., Fan, R., et al. (2021). Genome-wide identification of cassava MeRboh genes and functional analysis in Arabidopsis. Plant Physiol. Biochem. 167, 296–308. doi: 10.1016/j.plaphy.2021.07.039
Huang, X. L., Zhou, Y. T., Yan, Y. M., Cheng, Y. X. (2022). Sesquiterpenoid-chromone heterohybrids from agarwood of Aquilaria sinensis as potent specific Smad3 phosphorylation inhibitors. J. Org Chem. 87 (12), 7643–7648. doi: 10.1021/acs.joc.2c00145
Inupakutika, M. A., Sengupta, S., Devireddy, A. R., Azad, R. K., Mittler, R. (2016). The evolution of reactive oxygen species metabolism. J. Exp. Bot. 67 (21), 5933–5943. doi: 10.1093/jxb/erw382
Islam, M. R., Banu, S. (2019). An improved cost-effective method of RNA extraction from Aquilaria malaccensis. Acta Sci. Agric. 3 (2), 30–38.
Islam, M. R., Banu., S. (2021). Transcript profiling leads to biomarker identification for agarwood resin-l oaded Aquilaria malaccensis. Trees 35, 2119–2132. doi: 10.1007/s00468-021-02180-1
Islam, M. R., Bhau, B. S., Banu, S. (2020). Gene expression analysis associated with agarwood formation in Aquilaria malaccensis. Plant Physiol. Rep. 25 (2), 304–314. doi: 10.1007/s40502-020-00505-9
Jakubowicz, M., Galganska, H., Nowak, W., Sadowski, J. (2010). Exogenously induced expression of ethylene biosynthesis, ethylene perception, phospholipase D, and Rboh-oxidase genes in broccoli seedlings. J. Exp. Bot. 61 (12), 3475–3491. doi: 10.1093/jxb/erq177
Kaur, N., Dhawan, M., Sharma, I., Pati, P. K. (2016). Interdependency of reactive oxygen species generating and scavenging system in salt sensitive and salt tolerant cultivars of rice. BMC Plant Biol. 16 (1), 131. doi: 10.1186/s12870-016-0824-2
Kaur, G., Pati, P. K. (2018). In silico insights on diverse interacting partners and phosphorylation sites of respiratory burst oxidase homolog (Rbohs) gene families from Arabidopsis and rice. BMC Plant Biol. 18 (1), 161. doi: 10.1186/s12870-018-1378-2
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/nmeth.3317
Kovaka, S., Zimin, A. V., Pertea, G. M., Razaghi, R., Salzberg, S. L., Pertea, M. (2019). Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20 (1), 278. doi: 10.1186/s13059-019-1910-1
Kristanti, A. N., Tanjung, M., Aminah, N. S. (2018). Review: Secondary metabolites of Aquilaria, a thymelaeaceae genus. Mini Rev. Org Chem. 15 (1), 36–55. doi: 10.2174/1570193X14666170721143041
Kumar, S., Stecher, G., Tamura, K. (2016). Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Peer, V. D., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30, 1, 1194–1202. doi: 10.1093/nar/30.1.325
Li, D., Wu, D., Li, S., Dai, Y., Cao, Y. (2019). Evolutionary and functional analysis of the plant-specific NADPH oxidase gene family in Brassica rapa L. R. Soc. Open Science. 6 (2), 181727. doi: 10.1098%2Frsos.181727
Lightfoot, D. J., Boettcher, A., Little, A., Shirley, N., Able, A. J. (2008). Identification and characterisation of barley (Hordeum vulgare) respiratory burst oxidase homologue family members. Funct. Plant Biol. 35 (5), 347–359. doi: 10.1071/FP08109
Lin, F., Zhang, Y., Jiang, M. Y. (2009). Alternative splicing and differential expression of two transcripts of nicotine adenine dinucleotide phosphate oxidase B gene from Zea mays. J. Integr. Plant Biol. 51 (3), 287–298. doi: 10.1111/j.1744-7909.2008.00808.x
Liu, Y., Chen, H., Yang, Y., Zhang, Z., Wei, J., Meng, H., et al. (2013). Whole-tree agarwood-inducing technique: an efficient novel technique for producing high-quality agarwood in cultivated. Aquilaria sinensis trees. Molecules 18 (3), 3086–3106. doi: 10.3390/molecules18033086
Liu, Y., He, C. (2016). Regulation of plant reactive oxygen species (ROS) in stress responses: learning from. AtRBOHD. Plant Cell Rep. 35 (5), 995–1007. doi: 10.1007/s00299-016-1950-x
Liu, J., Lu, H., Wan, Q., Qi, W., Shao, H. (2019). Genome-wide analysis and expression profiling of respiratory burst oxidase homologue gene family in Glycine max. Environ. Exp. Bot. 161, 344–356. doi: 10.1016/j.envexpbot.2018.07.015
Lopez-Ortiz, C., Dutta, S. K., Natarajan, P., Pena-Garcia, Y., Abburi, V., Saminathan, T., et al. (2019). Genome-wide identification and gene expression pattern of ABC transporter gene family in Capsicum spp. PloS One 14 (4), 1–23. doi: 10.1371/journal.pone.0215901
López-Sampson, A., Page, T. (2018). History of Use and Trade of Agarwood. Econ Bot. 72, 107–129. doi: 10.1007/s12231-018-9408-4
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Lv, F., Li, S., Feng, J., Liu, P., Gao, Z., Yang, Y., et al. (2019). Hydrogen peroxide burst triggers accumulation of jasmonates and salicylic acid inducing sesquiterpene biosynthesis in wounded Aquilaria sinesis. J. Plant Physiol. 234-235, 167–175. doi: 10.1016/j.jplph.2019.02.006
Mahalingam, R., Graham, D., Walling, J. G. (2021). The Barley (Hordeum vulgare ssp. vulgare) Respiratory Burst Oxidase Homolog (HvRBOH) gene family and their plausible role on malting quality. Front. Plant Sci. 12, 608541. doi: 10.3389/fpls.2021.608541
Marino, D., Andrio, E., Danchin, E. G., Oger, E., Gucciardo, S., Lambert, A., et al. (2011). A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning. New Phytol. 189 (2), 580–592. doi: 10.1111/j.1469-8137.2010.03509.x
Marino, D., Dunand, C., Puppo, A., Pauly, N. (2012). A burst of plant NADPH oxidases. Trends Plant Sci. 17 (1), 9–15. doi: 10.1016/j.tplants.2011.10.001
Martin, W., Schnarrenberger, C. (1997). The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: a case study of functional redundancy in ancient pathways through endosymbiosis. Curr. Genet. 32, 1–18. doi: 10.1007/s002940050241
Maruta, T., Inoue, T., Tamoi, M., Yabuta, Y., Yoshimura, K., Ishikawa, T., et al. (2011). Arabidopsis NADPH oxidases, AtrbohD and AtrbohF, are essential for jasmonic acid-induced expression of genes regulated by MYC2 transcription factor. Plant Sci. 180 (4), 655–660. doi: 10.1016/j.plantsci.2011.01.014
Matsuda, F., Miyagawa, H., Ueno, T. (2001). Involvement of reactive oxygen species in the induction of (S)-N-P coumaroyloctopamine accumulation by h-1,3-glucooligosaccharide elicitors in potato tuber tissues. J. Biosci. 56, 228–234. doi: 10.1515/znc-2001-3-410
Mohamed, R., Jong, P. L., Kamziah, A. K. (2014). Fungal inoculation induces agarwood in young Aquilaria malaccensis trees in the nursery. J. For Res. 25 (1), 201–204. doi: 10.1007/s11676-013-0395-0
Monggoot, S., Popluechai, S., Gentekaki, E., Pripdeevech, P. (2017). Fungal Endophytes: an alternative source for production of volatile compounds from agarwood oil of Aquilaria subintegra. Microb. Ecol. 74 (1), 54–61. doi: 10.1007/s00248-016-0908-4
Morales, J., Kadota, Y., Zipfel, C., Molina, A., Torres, M. A. (2016). The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 67 (6), 1663–1676. doi: 10.1093/jxb/erv558
Naef, R. (2011). The volatile and semi-volatile constituents of agarwood, the infected heartwood of Aquilaria species: a review. Flavour Fragr J. 26 (2), 73–87. doi: 10.1002/ffj.2034
Navathe, S., Singh, S., Singh, V. K., Chand, R., Mishra, V. K., Joshi, A. K. (2019). Genome-wide mining of respiratory burst homologs and its expression in response to biotic and abiotic stresses in Triticum aestivum. Genes Genom 41 (9), 1027–1043. doi: 10.1007/s13258-019-00821-x
Orman-Ligeza, B., Parizot, B., Rycke, D. R., Fernandez, A., Himschoot, E., Breusegem, V. F., et al. (2016). RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development 143 (18), 3328–3339. doi: 10.1242/dev.136465
Pacheco-Trejo, J., Aquino-Torres, E., Reyes-Santamaría, M. I., Islas-Pelcastre, M., Pérez-Ríos, S. R., Madariaga-Navarrete, A., et al. (2022). Plant defensive responses triggered by richoderma spp. as tools to face stressful conditions. Horticulturae. 8, 1181. doi: 10.3390/horticulturae8121181
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46 (W1), W200–W204. doi: 10.1093/nar/gky448
Qiao, X., Li, Q., Yin, H., Qi, K., Li, L., Wang, R., et al. (2019). Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 20, 38. doi: 10.1186/s13059-019-1650-2
Robert, X., Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42 (Web Server issue), W320–W324. doi: 10.1093/nar/gku316
Sagi, M., Fluhr, R. (2001). Superoxide production by plant homologues of the gp91(phox) NADPH oxidase modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol. Rep. 126 (3), 1281–1290. doi: 10.1104/pp.126.3.1281
Si, W., Hang, T., Guo, M., Chen, Z., Liang, Q., Gu, L., et al. (2019). Whole-Genome and transposed duplication contributes to the expansion and diversification of TLC Genes in Maize. Int. J. Mol. Sci. 20 (21), 5484. doi: 10.3390/ijms20215484
Tan, C. S., Isa, N. M., Ismail, I., Zainal, Z. (2019). Agarwood Induction: current developments and future perspectives. Front. Plant Sci. 10, 122. doi: 10.3389/fpls.2019.00122
Torres, M. A., Dangl, J. L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8 (4), 397–403. doi: 10.1016/j.pbi.2005.05.014
Torres, M. A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K. E., Jones, J. D. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14 (3), 365–370. doi: 10.1046/j.1365-313X.1998.00136.x
Trujillo, M., Altschmied, M., Schweizer, P., Kogel, K., Hückelhoven, R. (2006). Respiratory Burst Oxidase Homologue A of barley contributes to penetration by the powdery mildew fungus Blumeria graminis f. sp. hordei. J. Exp. Bot. 57 (14), 3781–3791. doi: 10.1093/jxb/erl191
Wang, W., Chen, D., Liu, D., Cheng, Y., Zhang, X., Song, L., et al. (2020). Comprehensive analysis of the Gossypium hirsutum L. respiratory burst oxidase homolog (Ghrboh) gene family. BMC Genomics 21 (1), 91. doi: 10.1186/s12864-020-6503-6
Wang, X., Dong, X., Feng, Y., Liu, X., Wang, J., Zhang, Z., et al. (2018). H2O2 and NADPH oxidases involve in regulation of 2-(2-phenylethyl)chromones accumulation during salt stress in Aquilaria sinensis calli. Plant Sci. 269, 1–11. doi: 10.1016/j.plantsci.2018.01.002
Wang, Y., Li, J., Paterson, H. A. (2013). MCScanX-transposed: detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 29 (11), 1458–1460. doi: 10.1093/bioinformatics/btt150
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteom Bioinform. 8 (1), 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wong, H. L., Pinontoan, R., Hayashi, K., Tabata, R., Yaeno, T., Hasegawa, K., et al. (2007). Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19 (12), 4022–4034. doi: 10.1007/978-1-4020-6635-1_32
Xu, Y. H., Liao, Y. C., Zhang, Z., Liu, J., Sun, P. W., Gao, Z. H., et al. (2016). Jasmonic acid is a crucial signal transducer in heat shock induced sesquiterpene formation in. Aquilaria sinensis. Sci. Rep. 6, 21843. doi: 10.1038/srep21843
Xu, Y., Zhang, Z., Wang, M., Wei, J., Chen, H., Gao, Z., et al. (2013). Identification of genes related to agarwood formation: transcriptome analysis of healthy and wounded tissues of Aquilaria sinensis. BMC Genomics 14, 227. doi: 10.1186/1471-2164-14-227
Yamauchi, T., Yoshioka, M., Fukazawa, A., Mori, H., Nishizawa, N. K., Tsutsumi, N., et al. (2017). An NADPH Oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 29 (4), 775–790. doi: 10.1105/tpc.16.00976
Yang, L., Yang, J. L., Dong, W. H., Wang, Y. L., Zeng, J., Yuan, J. Z., et al. (2021). The characteristic fragrant sesquiterpenes and 2-(2-Phenylethyl)chromones in wild and cultivated "Qi-Nan" agarwood. Molecules 26 (2), 1–12. doi: 10.3390/molecules26020436
Yang, Y. L., Zhang, Y. Y., Lu, J., Zhang, H., Liu, Y., Jiang, Y., et al. (2012). Exogenous H2O2 increased catalase and peroxidase activities and proline content in Nitraria tangutorum callus. Biol. Plantarum 56, 330–336. doi: 10.1007/s10535-012-0094-2
Ying, C. S., Tahir, N. F., Abu Bakar, N. S., How, T. C., N.F., M. Y., Baharum, N. A., et al. (2020). Genome-wide identification and expression analysis of banana Rboh genes in response to Fusarium oxysporum f. sp. cubense tropical race 4. Malaysian J. Biochem. Mol. Biol. 23, 46–52.
Yu, S., Kakar, K. U., Yang, Z., Nawaz, Z., Lin, S., Guo, Y., et al. (2020). Systematic study of the stress-responsive Rboh gene family in Nicotiana tabacum: Genome-wide identification, evolution and role in disease resistance. Genomics 112 (2), 1404–1418. doi: 10.1016/j.ygeno.2019.08.010
Zhang, Y., Li, Y., He, Y., Hu, W., Zhang, Y., Wang, X., et al. (2018). Identification of NADPH oxidase family members associated with cold stress in strawberry. FEBS Open Bio 8 (4), 593–605. doi: 10.1002/2211-5463.12393
Zhang, J., Xie, Y., Ali, B., Ahmed, W., Tang, Y., Li, H. (2021). Genome-wide identification, classification, evolutionary expansion and expression of Rboh Family genes in pepper (Capsicum annuum L.). Trop. Plant Biol. 14 (3), 251–266. doi: 10.1007/s12042-021-09286-3
Zhang, Y., Zhang, Y., Luo, L., Lu, C., Kong, W., Cheng, L., et al. (2022). Genome Wide identification of respiratory burst oxidase homolog (Rboh) genes in Citrus sinensis and functional analysis of CsRbohD in cold tolerance. Int. J. Mol. Sci. 23 (2), 1–13. doi: 10.3390/ijms23020648
Zhang, H., Wang, X., Yan, A., Deng, J., Xie, Y., Liu, S., et al. (2023). Evolutionary analysis of Respiratory Burst Oxidase Homolog (RBOH) genes in plants and characterization of ZmRBOHs. Int. J. Mol. Sci. 24 (4), 3858. doi: 10.3390/ijms24043858
Zhang, Z., Zhang, X., Yang, Y., Wei, J.-h., Meng, H., Gao, Z.-h., et al. (2013). Hydrogen peroxide induces vessel occlusions and stimulates sesquiterpenes accumulation in stems of Aquilaria sinensis. Plant Growth Regul. 72 (1), 81–87. doi: 10.1007/s10725-013-9838-z
Zhao, Y., Zou, Z. (2019). Genomics analysis of genes encoding respiratory burst oxidase homologs (RBOHs) in jatropha and the comparison with castor bean. PeerJ 7, 1–20. doi: 10.7717/peerj.7263
Zhou, X., Xiang, Y., Li, C., Yu, G. (2020). Modulatory role of reactive oxygen species in root development in model plant of Arabidopsis thaliana. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.485932
Keywords: Aquilaria, Rboh proteins, ROS generation, secondary metabolites, agarwood
Citation: Begum K, Das A, Ahmed R, Akhtar S, Kulkarni R and Banu S (2024) Genome-wide analysis of respiratory burst oxidase homolog (Rboh) genes in Aquilaria species and insight into ROS-mediated metabolites biosynthesis and resin deposition. Front. Plant Sci. 14:1326080. doi: 10.3389/fpls.2023.1326080
Received: 22 October 2023; Accepted: 18 December 2023;
Published: 09 February 2024.
Edited by:
Xupo Ding, Chinese Academy of Tropical Agricultural Sciences, ChinaReviewed by:
Baopeng Ding, Shanxi Datong University, ChinaFei Wei, Institute of Cotton Research (CAAS), China
Muhammad Tahir Ul Qamar, Government College University, Faisalabad, Pakistan
Xueyang Min, Yangzhou University, China
Copyright © 2024 Begum, Das, Ahmed, Akhtar, Kulkarni and Banu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sofia Banu, sofiabanu2@gmail.com
 Khaleda Begum
Khaleda Begum Ankur Das
Ankur Das Raja Ahmed
Raja Ahmed Suraiya Akhtar
Suraiya Akhtar Ram Kulkarni
Ram Kulkarni Sofia Banu
Sofia Banu