- 1Innovative Institute for Plant Health, Zhongkai University of Agriculture and Engineering, Guangzhou, China
- 2Innovative Center of Molecular Genetics and Evolution, College of Life Sciences, Guangzhou University, Guangzhou, Guangdong, China
- 3Research Center of Integrative Medicine, School of Basic Medical Sciences, Guangzhou University of Chinese Medicine, Guangzhou, Guangdong, China
The cultivated soybean (Glycine max (L.) Merrill) is domesticated from wild soybean (Glycine soja) and has heavier seeds with a higher oil content than the wild soybean. In this study, we identified a novel candidate gene associated with SW using a genome-wide association study (GWAS). The candidate gene GmWRI14-like was detected by GWAS analysis in three consecutive years. By constructing transgenic soybeans overexpressing the GmWRI14-like gene and gmwri14-like soybean mutants, we found that overexpression of GmWRI14-like increased the SW and increased total fatty acid content. We then used RNA-seq and qRT-PCR to identify the target genes directly or indirectly regulated by GmWRI14-like. Transgenic soyabeans overexpressing GmWRI14-like showed increased accumulation of GmCYP78A50 and GmCYP78A69 than non-transgenic soybean lines. Interestingly, we also found that GmWRI14-like proteins could interact with GmCYP78A69/GmCYP78A50 using yeast two-hybrid and bimolecular fluorescence complementation. Our results not only shed light on the genetic architecture of cultivated soybean SW, but also lays a theoretical foundation for improving the SW and oil content of soybeans.
1 Introduction
Seed weight (SW) is a complex quantitative trait controlled by genetic and environmental factors (Shimada et al., 2016; Goettel et al., 2022). Heavier seeds can provide sufficient energy for seed germination and ensure the competitiveness of seedlings during their growth process (Nakabayashi et al., 2010). At present, hundreds of genes that participate in SW regulation have been reported, which are related to the seed coat, endosperm development, and hormone regulation (Figure 1). GmGA20OX encodes a member of the gibberellin 20 oxidase superfamily, which affects seed development through regulation of gibberellin synthesis (Tudzynski et al., 2002; Carmen et al., 2007). The expression level of GmGA20OX is positively correlated with SW; overexpression of GmGA20OX increased SW in Arabidopsis (Hu et al., 2023). PP2C-1 encodes a phosphatase associated with the transcription factor GmBZR1, which is involved in brassinolide signaling (González-García et al., 2007; Singh et al., 2021), promoting the dephosphorylation of GmBZR1 and enhancing the size of integument cells. The overexpression of PP2C-1 can significantly increase the SW and seed size (Wang et al., 2018). Soybean cytochrome P450 (CYP450) positively regulates SW by controlling cell proliferation (Ohta et al., 2000; Guttikonda et al., 2010). Overexpression of the soybean CYP450 genes (GmCYP78A5 and GmCYP78A72) considerably increases soybean SW (Du et al., 2017).
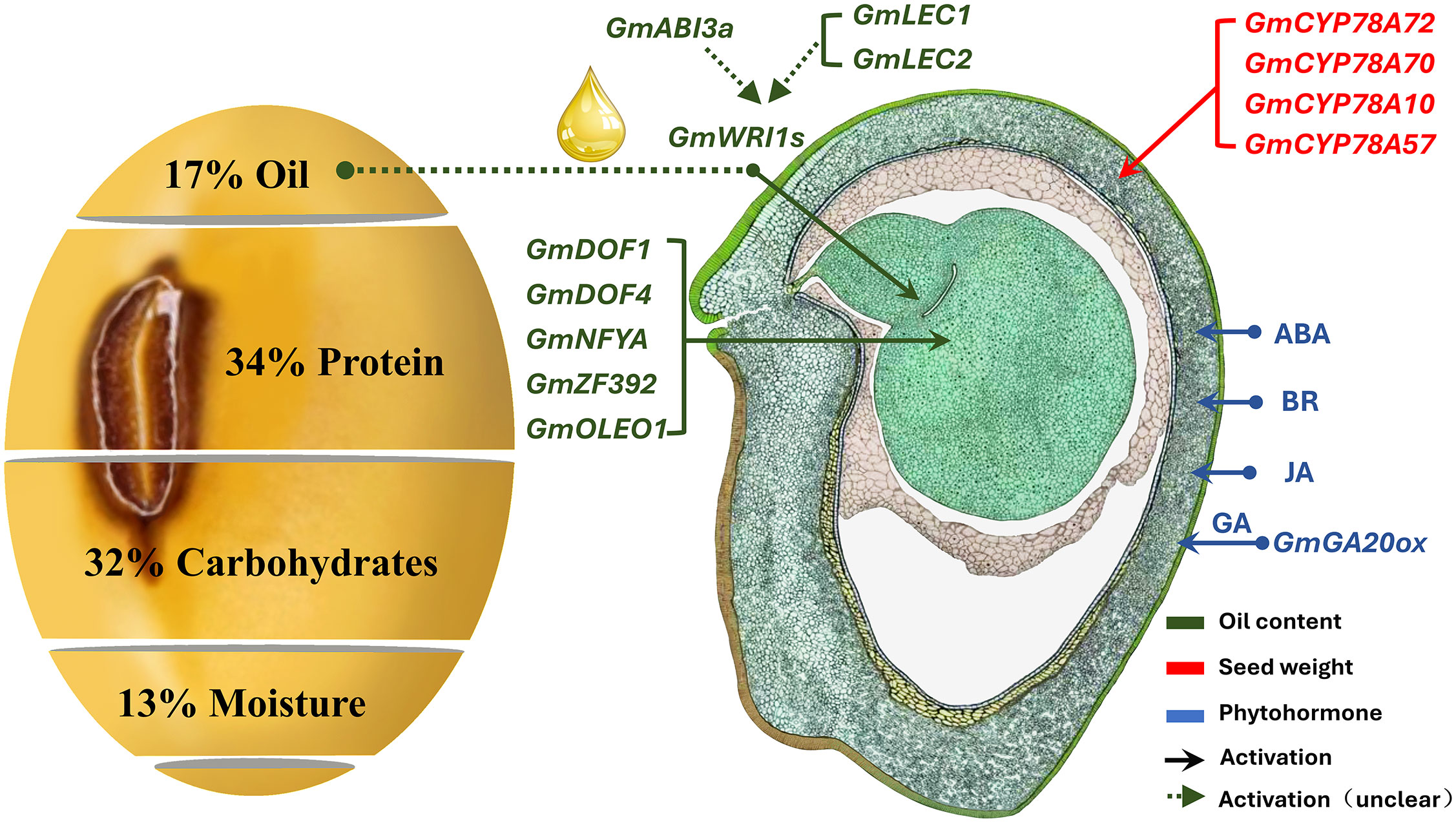
Figure 1 Schematic diagram of soybean seed weight (SW) regulation (Duan et al., 2023). The morphographic image was hand drawn by Sharon Lee Belkin (http://seedgenenetwork.net/soybean). The green box represents genes related to fatty acids in the process of SW regulation, and the red represents the process of seed development regulation.
In addition to the positive regulation of soybean SW by the aforementioned genes, certain genes also play a negative role in regulating SW. For example, a knockout of the cell wall invertase inhibitor gene (GIF1) results in a considerable increase in cell wall invertase activity and SW (Kim and Hans, 2004). The ubiquitin receptor, DA1, negatively regulates seed size and SW by limiting the cell proliferation cycle (Mora et al., 2021). Furthermore, the overexpression of DA1 interferes with mutations in rapeseed, leading to an increase in SW (Du et al., 2014). BIG SEEDS1 (BS1) encodes a plant-specific transcription regulatory factor, TIFY, which negatively regulates seed development by inhibiting cell proliferation (Yin et al., 2020). When soybean BS1 homologous genes (GmBA1 and GmBA2) were suppressed, SW increased. Many candidate genes related to SW have been identified; however, the low-density molecular makers mean that the candidate genes are less effective in crop improvement. In addition, the regulatory pathways of soybean SW have not yet formed a systematic molecular regulatory network.
The cultivated soybean (Glycine max (L.) Merrill) seeds contain approximately 17% oil (Figure 1). Soybean oil is stored mainly as fatty acids (FAs), triacylglycerols (TAGs), and tocopherols; there are five central FAs present in soybean seeds (Qin et al., 2021). The genes related to fatty acid synthesis, such as DREBL (Zhang et al., 2016), DGAT (Vyacheslav et al., 2010; Zhao et al., 2019), Dof (Zhang et al., 2014), and WRINKLED1 (Vogel et al., 2019), regulate the development and maturation process of seeds and affect SW via the redistribution of storage substances (Che et al., 2016; Chen et al., 2020). WRI1 is a core transcription factor that regulates key enzymes for fatty acid biosynthesis; however, we lack an understanding of the molecular mechanisms underlying the regulation of SW by WRI1. Furthermore, the genetic mechanism for the synergistic regulation of SW and FAs remains poorly understood (Jaromír et al., 2015; Lu et al., 2016b; Hacisalihoglu et al., 2018). SW is regulated by numerous genes; therefore, the genetic complexity of quantitative traits controlled by multiple genes and their interaction with the environment make it difficult for these candidate genes to be directly used to guide molecular marker-assisted breeding. The present study can serve as a good reference for future studies on high-quality soybean breeding.
2 Materials and methods
2.1 Phenotyping of soybean germplasms
The 1210 soybean accessions from China were collected to obtain accurate phenotypic data for the GWAS. In 2020–2022, the plant materials were planted in three regions of China (Gongzhuling, Yantai, and Guangzhou), which represent typical climates (subtropical, tropical, and temperate zones). Seeds of different soybean accessions were dried after harvest to measure the SW and fatty acid content. The NIRSTM DS 2500 (FOSS Inc., Hillerod, Denmark) was used to measure the oil content of the soybean seeds. Five FAs (linolenic acid, stearic acid, oleic acid, palmitic acid, and linoleic acid) were determined by matrix-assisted laser desorption/ionization time-of-flight imaging mass spectrometry (MALDI-TOF IMS) (Bruker Daltonics, Bremen, Germany). Each calculation was repeated thrice. The correlation coefficients were calculated using the SPSS software 20.0 (SPSS Inc., Chicago, IL, USA).
2.2 Genotyping of soybean germplasms
Genomic DNA was extracted from young leaves of the natural population consisting of 1210 soybean accessions using the cetyltrimethylammonium bromide (CTAB) method (Porebski et al., 1997). For each accession, 6 μg of DNA was used to construct a sequencing library according to the manufacturer’s instructions. The same genotyping method was performed as previously described (Qin et al., 2021). The restriction endonuclease combination was RsaI-HaeIII. Single nucleotide polymorphism (SNP) molecular markers were used for the GWAS analysis.
2.3 GWAS and identification of candidate genes
Based on the phenotypic data of 1210 soybean germplasms, the SNP markers with a minor allele frequency (MAF) > 0.06 obtained from genotyping were included in the GWAS to analyze the association between genotype and phenotype datasets. Haploview 4.2 software was used for Manhattan map construction. We used plink2 software to calculate the linkage disequilibrium (LD) decay distances. The candidate genes were within the LD distance (8.9 Kb). We used 3,212,756 high-quality SNPs (MAF > 0.06) to perform GWAS for SW in the 1210 accessions. The fastlmmc model were used for association analyses. The threshold value for GWAS was set to –log(p) > 6.20. The significantly associated regions were verified from the aligned sequencing reads against the Williams 82 genome (Glycine max Wm82.a2.v1) with SAMtools ver. 0.1.18 (Li and Durbin, 2009). For each statistically significant SNP of interest, the annotated genes within 150 Kb around the SNP were considered as candidate genes (based on Williams 82).
2.4 Plasmid construction and plant transformation
The GmWRI14-like gene from the soybean cultivar 010a (approval number: 2012010) was cloned into the BamHI-SacI site of the plasmid pTF101 named pTF101-GmWRI14-3×Flag, which was driven by the CaMV35S promoter. The target gene was terminated by the NOS terminator (Supplementary Figure A1). The recombinant plasmid was transformed into calli using the Agrobacterium tumefaciens strain (LBA4404). Transgenic soybean plants were generated as previously described (Holsters et al., 1978). Individual T0 plant lines were established in a greenhouse and three independent transgenic soybean lines with the same plasmid were harvested: GmWRI14-1, GmWRI14-2, and GmWRI14-3. Following a previous protocol (Bai et al., 2020), the novel CRISPR/Cas9 vector pGES201 was used for soybean genome editing. The Cas9 gene driven by the pM4 promoter (Supplementary Figure A2) and the Cas9 vector were gifted by Fujian Agriculture and Forestry University. The exon sequence of the GmWRI14-like gene in the soybean genome was determined using the CRISPR direct website (http://crispr.dbcls.jp/). We designed three target site sequences with a length of 20 bp (Supplementary Figure A2): WRI14-pM4-T1-F1/WRI14-pM4-T1-R1, WRI14-pM4-T2-F1/WRI14-pM4-T2-R1, and WRI14-pM4-T3-F1/WRI14-pM4-T3-R1. We simultaneously designed the CRISPR primers WRI14-CasF-Test/WRI14-CasR-Test. Three independent gmwri14-like soybean mutant lines containing the same plasmid were harvested: gmwri14-like-1, gmwri14-like-2, and gmwri14-like-3.
2.5 RNA extraction and sequencing
To explore the changes in differentially expressed genes (DEGs) and differential metabolic pathways in GmWRI14-like transgenic soybeans, RNA sequencing was performed on transgenic soybeans using a high-throughput Illumina HiSeq sequencing platform. Total RNA was extracted using the Eastep® Super total RNA extraction kit (TaKaRa, USA). RNA quality was assessed using the Nanodrop 2000c (Thermo Scientific, Hudson, NH, US). RNA-seq library preparation and sequencing were performed as previously described (Wang et al., 2014). The 2-ΔΔCt method was used to assess the fold change in gene expression (Livak and Schmittgen, 2001). To analyze the transduction pathways induced by the GmWRI14-like gene, the total RNA of each sample (GmWRI14-like transgenic soybeans, CK, gmwri14-like mutants) was isolated from the seeds for the RNA-sequencing; 2 g of RNA was obtained from each plant sample. The RNA-sequencing depth was 10 ×. The bioinformatics software (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) was used to improve data quality. In total, we obtained 41.66 Gb of clean data in this project. The statistical power of this experimental design, calculated in cluster Profiler Power, was equal to a q value ≤ 0.01. Gene Ontology (http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genome (KEGG) statistical analysis (http://www.genome.jp/kegg/) were performed to assess the function of the DEGs.
2.6 Quantitative reverse transcription-polymerase chain reaction
After 10 days of podding, RNA was extracted from the seeds of the soybean lines (GmWRI14-like transgenic soybeans, gmwri14-like mutants, and control soybean lines). We then conducted 24 h sampling daily at 10, 11, 12, 13, and 14 days after podding (GT0, GT4, GT8, GT12, GT16, GT20, and GT24 soybean seeds, respectively) to extract total RNA. qRT-PCR was used to compare the overexpression of GmWRI14-like soybeans on different days and at different times to explore the molecular regulatory network related to the GmWRI14-like, gmwri14-like mutants, and control soybean plants involving the DEGs. cDNA synthesis was performed using a reverse transcription kit EZBiosciences (Omega. USA). Real-time fluorescence analysis was performed using a LightCycler (Roche, Rotkreuz, Switzerland) according to the manufacturer’s instructions (Livak and Schmittgen, 2001). All primers used are listed in the additional files (Supplementary Table A1).
2.7 Yeast two-hybrid assays and bimolecular fluorescence complementation
We performed a Y2H assay by screening the cDNA library constructed from the seed aged 15 days. The full-length GmWRI14-like protein was used as a bait, and we identified its putative partner using Y2H library screening. Y2H assays were performed as described previously (Xiaofei et al., 2017). The BiFC assay was performed as previously described (Mou et al., 2021).
3 Results
3.1 Phenotypic variation of SW between 2020 and 2022
Over 3 years of measurements, the variation in SW ranged from 5.43 g to 27.11 g in 2020 (standard deviation (SD): 7.6), 17.88 g to 26.89 g in 2021 (SD: 3.7), and 2.21 g to 23.59 g in 2022 (SD: 6.7). The variation in oil content ranged from 3.35% to 25.12% in 2020 (SD: 7.9), 15.52% to 25.19% in 2021 (SD: 2.4), and 17.33% to 27.83% in 2022 (SD: 2.6). The distribution of the SW was continuous, and both the SW and oil content conformed to normal distributions (Figures 2A, B).
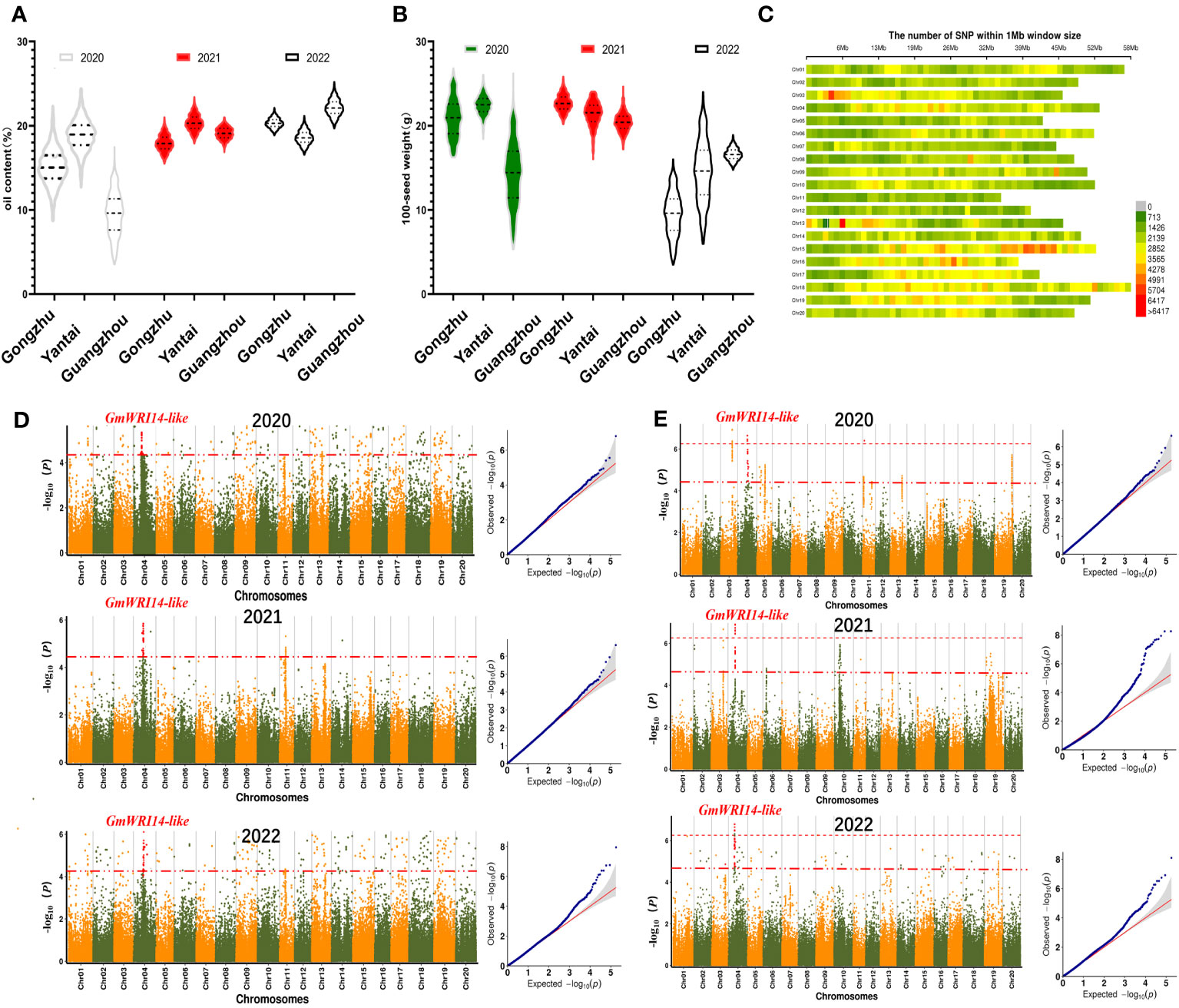
Figure 2 (A) Frequency distribution of oil content from the year 2020 to 2022. The 1210 soybean lines were grown in three different regions (Gongzhuling, Yantai, and Guangzhouling). Oil content conformed to normal distribution. In 2020, the oil content was the lowest in Gongzhuling (4.51–17.52%). (B) Frequency distribution of seed weight (SW) from the year 2020 to 2022. In 2022, the SW was the lowest in Guangzhou (3.85–14.26 g). (C) Distribution map of single nucleotide polymorphisms (SNPs) on different chromosomes. The abscissa is the length of the chromosomes. Each band represents one chromosome. The red area indicates that the number of SNPs exceeded 5704. The green area indicates that the number of SNPs was less than 1426. (D) From the year 2020 to 2022, GmWRI14-like was found to be a candidate gene for oil content. (E) From the year 2020 to 2022, genome-wide Manhattan plots of the associations of GmWRI14-like with SW.
3.2 A new candidate gene related to SW was discovered by GWAS
GWAS using the fastlmmc model was used to identify potential candidate genes associated with SW. The high-quality 3,212,756 SNP markers obtained by SLAF-Seq technology were used for GWAS for SW in the 1210 accessions. The SNP marker distributions are shown in Figure 2C. Manhattan maps were used to show the oil content (Figure 2D) and SW (Figure 2E) across different years; the threshold value was set at –log(p) > 6.20 (red). We identified one significant association loci for SW on chromosome 4 (Figure 2E), and we estimated the candidate region to be from 1.600 to 1.750 Mb (150 Kb) around the leading SNP in chromosome 4. This region contains 14 genes (Supplementary Table A2). According to Swissport annotation, these genes were annotated, such as gibberellin 20 oxidase, phosphatidylethanolamine-binding protein, and dof zinc finger protein, and had already been reported to affect SW (Wang et al., 2007; Lu et al., 2016a); therefore, we focused on Glyma.04G116500.1, which we labeled GmWRI14-like. It is speculated that GmWRI14-like belongs to the WRI1 family with two AP2/EREB domains (Supplementary Figure A3). The soybean line with the lowest SW (KH1088) harbored one SNP predicted to cause a gain in a stop codon, resulting in the premature termination of translation after 241 amino acids in the 710-amino acid GmWRI14-like protein.
3.3 Identification of the role of GmWRI14-like in controlling SW
Using a stable soybean genetic transformation system, transgenic soybean plants overexpressing GmWRI14-like were successfully obtained (Figure 3A). Overexpression of GmWRI14-like in soybeans significantly increased seed size and SW compared to the wild-type soyabeans (P<0.01), resulting in severe bending of soybean branches (Figure 3A). The gmwri14-like soybean mutant was obtained using CRISPR/Cas9 genome editing technology (Figure 3A). Compared with the non-transgenic soybean control, the seed size of the gmwri14-like soybean mutant was significantly reduced (P<0.01), and the seed epidermis of the gmwri14-like soybean mutant exhibited wrinkling (Figure 3B). Overexpression of GmWRI14-like resulted in a 15.32% increase in seed length (P<0.01), whereas the seed length of the gmwri14-like mutant was reduced by 23% (Figure 4F). Furthermore, the width of the GmWRI14-like seeds was 14.29% greater than that of the CK. GmWRI14-like-3 had the largest width, reaching 7.14 ± 0.03 mm, whereas the gmwri14-like mutant seeds showed a 18.23–23.57% decrease in width. The width of GmWRI14-like-3 reached 6.34 ± 0.16 mm (Figure 4G). The phenotype of GmWRI14-like transgenic soybeans showed a significant difference in the color of the seed coat from the different soybean lines (P<0.01). The non-transgenic soybean seed coat was yellow, whereas the seed coat color of GmWRI14-like soybeans tended to be white. These results suggested that the GmWRI14-like gene regulates the cytochrome gene expression related to the seed coat.
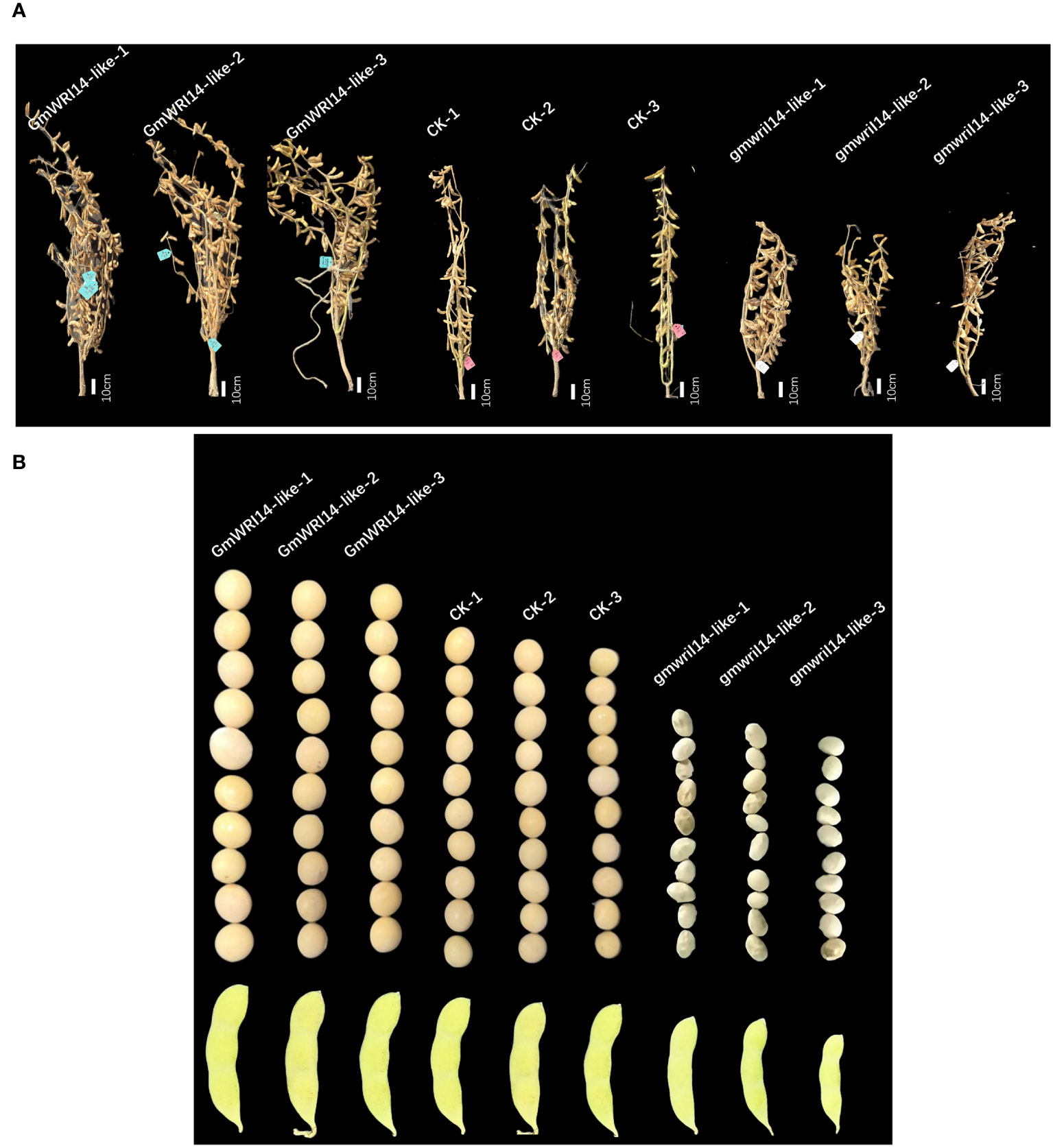
Figure 3 (A) GmWRI14-like overexpression soybean and gmwri1-like mutant phenotype. (B) The overexpression of the GmWRI14-like gene in soybeans resulted in an increase in seed size and SW, while the gmwri14-like mutant showed a decrease in SW and wrinkling. GmWRI14-like-1, 2, and 3: overexpressing GmWRI14-like lines, gmwri1-like-1, 2, and 3: different mutant lines.
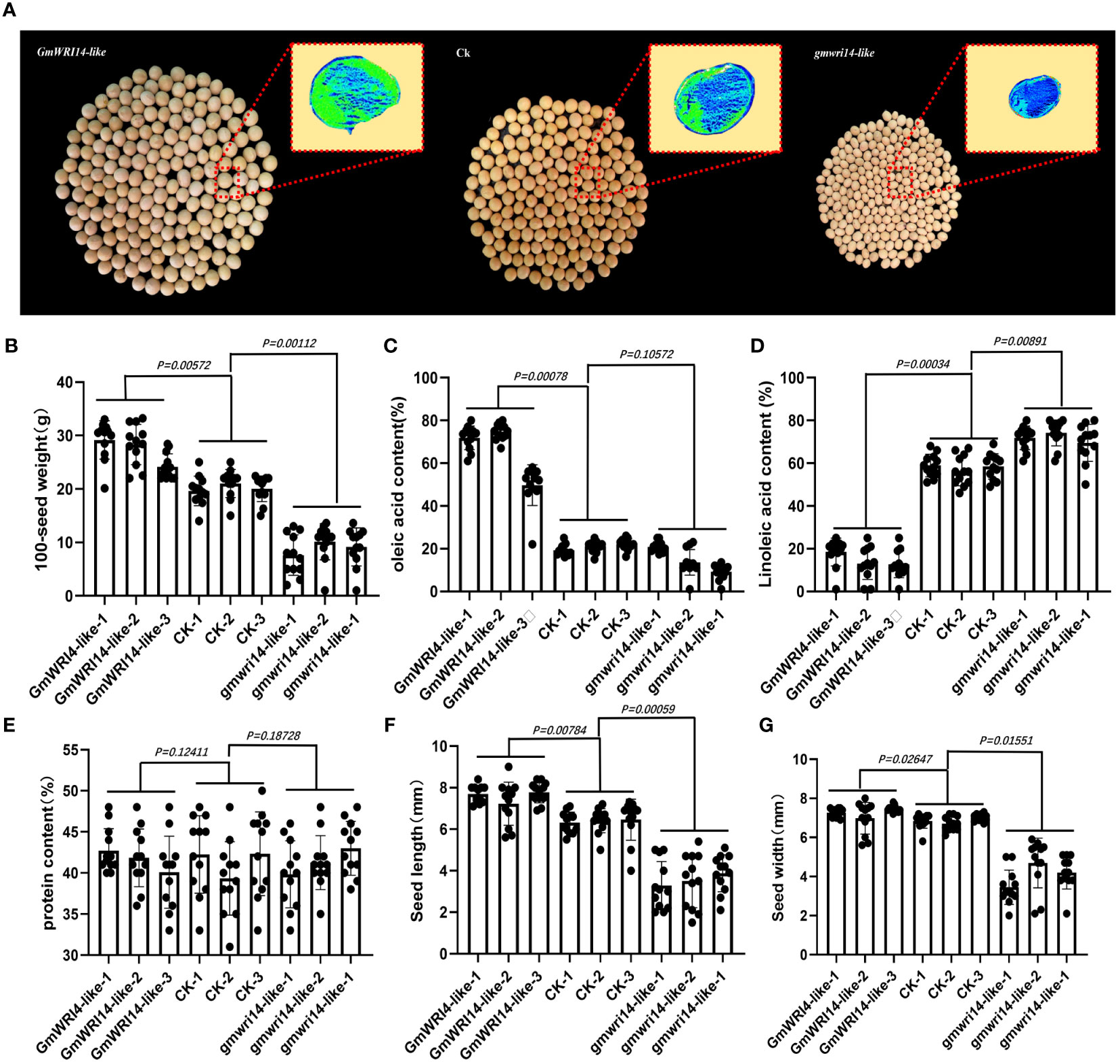
Figure 4 (A) Matrix-assisted laser desorption/ionization time-of-flight imaging mass spectrometry (MALDI-TOF IMS) analysis of the distribution of oil in the seeds. Survey results of 100 seed weights (SWs) (B), The green area represents a higher density of oil distribution in the seeds, with a unit of 1 mm. Survey results of 100 seed weights (SWs), oleic acid (C), linoleic acid (D), protein content (E), seed length (F), and seed width (G) in transgenic GmWRI14-like soybeans, non-transgenic soybean recipients, and gmwri14-like mutant soybeans.
3.4 Functional validation of GmWRI14-like by transgenic experiments
The oil content and distribution in gmwri14-like soybean mutants and overexpressing GmWRI14-like soybean seeds were detected by using MALDI-TOF IMS (Figure 4A). The GmWRI14-like overexpressing soybeans seeds showed a 16.14–19.92% increase in 100 seed weights (Figure 4B). The MALDI-TOF IMS results showed that, compared to control, the relative content of oleic acid in the gmwri1-like mutant significantly decreased from 22.12% to 15.14% (P<0.01), whereas the oleic acid content in the GmWRI14-like overexpressing soybeans increased from 22.12% to 74.31% (Figure 4C). The relative content of linoleic acid in the gmwri1-like mutant increased by 8.2% (P<0.01), whereas the linoleic acid content in GmWRI14-like soybeans decreased from 49.31% to 18.42% (Figure 4D). There was no significant difference in protein content among the GmWRI14-like soybean, gmwri14-like mutant, and non-transgenic soybean (Figure 4E). These results indicate that the GmWRI14-like gene does not affect the protein content in soybean seeds.
3.5 Transcriptome analysis of the GmWRI14-like overexpression soybean
RNA-seq was performed on the transgenic and control groups (three replicates), resulting in more than 5.1 × 107 clean reads; 2101 DEGs were screened through transcriptome analysis. The differential expression gene analysis (DEGVP) showed that two genes related to cytochrome metabolism were differentially expressed (more than 2.5 times), namely, GmCYP78A50 (Glyma.07G052300) and GmCYP78A69 (Glyma.16G021200) (Figure 5A). We analyzed the functions of the target genes and potential target genes using the KEGG pathway (Figure 5B), which showed that the DEGs were enriched in three signaling pathways related to seed development, including fatty acid biosynthesis (50.83%), fatty acid metabolism (36.37%), and cytochrome biosynthesis (19.17%) (Figure 5B).
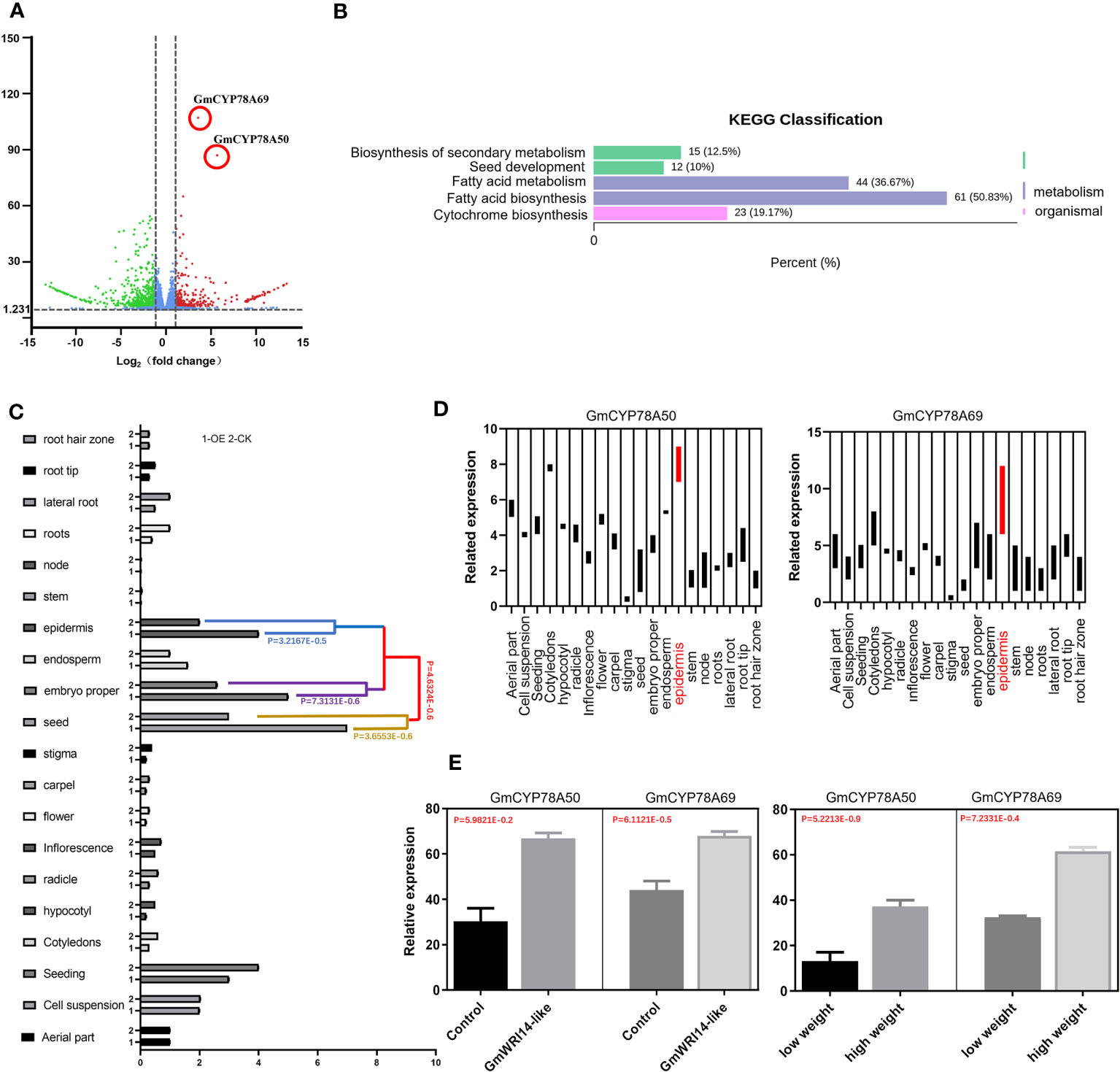
Figure 5 Differentially expressed genes (DEGs) related to seed weight (SW) in the GmWRI14-like transgenic soybeans. (A) DEGs of the GmWRI14-like transgenic soybean; the down-regulated differential genes (blue), and up-regulated genes (red). (B) KEGG pathway enrichment analysis. The differential genes were enriched in four signaling pathways, of which the signaling pathways with many enriched genes and related to seed development mainly included fatty acid biosynthesis (50.83%), fatty acid metabolism (36.37%), and cyto-chrome biosynthesis (19.17%). (C) GmWRI14-like expression in different tissues of the soybean. The GmWRI14-like expression increased in specific tissues (epidermis, embryo, and seed) of the GmWRI14-like transgenic soybean. (D) The GmCYP78A50 and GmCYP78A69 expression in different tissues of GmWRI14-like transgenic soybeans. (E) The GmCYP78A50 and GmCYP78A69 expression in the control soybean and GmWRI14-like transgenic soybean.
To explore the role of GmWRI14-like in more detail, we measured the expression level of the GmWRI14-like gene in different soybean tissues (root, stem, leaf, and seed) using qRT-PCR and found that the GmWRI14-like gene had the highest expression level in soybean seeds (Figure 5C). The qRT-PCR results showed that GmWRI14-like gene expression was highly tissue-specific in soybean plants (Figure 5C). We used the lectin gene (Gene ID: 100775957) as a reference gene and detected the expression levels of GmCYP78A50 and GmCYP78A69 in different transgenic soybean tissues using qRT-PCR, which suggested that the expression levels of GmCYP78A50 and GmCYP78A69 were the highest in the epidermis of transgenic soybean seeds (Figure 5D). GmCYP78A50 and GmCYP78A69 showed an increased expression level in the transgenic plants than in the control plants (Figure 5E). Based on the RNA-seq and qRT-PCR results, we speculate that the GmWRI14-like gene regulates the expression of GmCYP78A50 and GmCYP78A69 in soybean seeds.
3.6 GmWRI14-like interacts with GmCYP78A50 and GmCYP78A69
We found 140 unique cDNA clones using Y2H. Interestingly, among the interacting proteins, GmCYP78A50 (Glyma.07G052300) and GmCYP78A69 (Glyma.16G021200) were identified multiple times. We further confirmed the interaction between GmWRI14-like and GmCYP78A50/GmCYP78A69 by point-to-point Y2H. First, we used GmWRI14-like bait and GmCYP78A50/GmCYP78A69 as prey (i.e., fused with GAL4-BD and GAL4-AD, respectively) and confirmed the interaction through Y2H. As an additional control, we fused another coiled coil domain protein (structural maintenance of chromosome 5) with GAL4-AD and another AP2 transcription factor (dehydration response element binding protein 2A) with GAL4-8D and tested their interactions with WRI1-BD and GmCYP78A50/GmCYP78A69-AD, respectively. Only when GmWRI14-like-BD binds to GmCYP78A69/GmCYP78A50-AD clones can they grow on selective media, indicating that the interaction is specific (Figures 6A, B). In addition, the interaction between GmWRI14-like and GmCYP78A50/GmCYP78A69 was further confirmed by BiFC (Figure 6C). Therefore, these results show that the GmWRI14-like protein can interact with CYP78A50/GmCYP78A69.
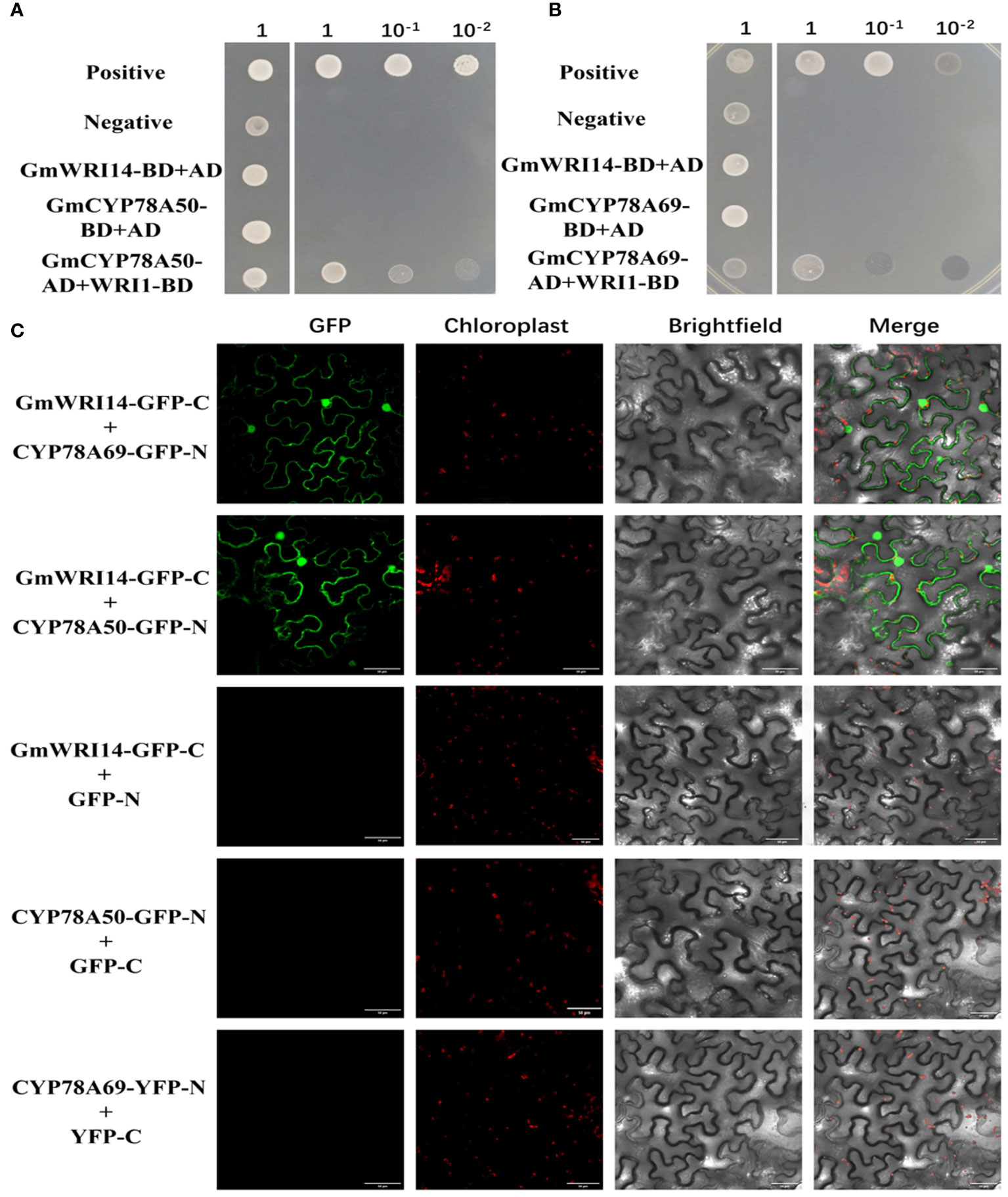
Figure 6 GmCYP78A69/GmCYP78A50 interacted with GmWRI114-like. (A) The interaction between GmCYP78A50 and GmWRI114-like was confirmed in the yeast two-hybrid (Y2H) assay. (B) The interaction between GmCYP78A69 and GmWRI114-like was confirmed in the yeast two-hybrid (Y2H) assay. Positive control: pGADT7-T + pGBKT7-53; negative control: pGADT7-T + pGBKT7-lam. GmWRI114-like-BD, GmWRI114-like fused to GAL4-BD; AD, GAL4-AD vectors without insertion. (C) The interaction between GmCYP78A69/GmCYP78A50 and GmWRI114-like was confirmed with bimolecular fluorescence complementation (BiFC).
4 Discussion
Cultivated soybeans (Glycine max L. Merr.) are domesticated from annual wild progenitors (Glycine soja) (Kuroda et al., 2009). There are significant differences in the SW and oil content between cultivated and wild soybeans (Dong et al., 2021). The composition and ratio of soybean FAs determine the quality of the soybean oil, which is mainly composed of five FAs (Rahman et al., 2003; Harris et al., 2008; Martin et al., 2008). SW and oil content are highly correlated agronomical traits; however, there is currently a lack of a molecular basis for the synergistic regulation of SW and oil content (Duan et al., 2022; Duan et al., 2023). The candidate genes that specifically regulate SW have no significant impact on seed oil content (Fasoula et al., 2004; Fasoula and Boerma, 2007; Ramya et al., 2010). Therefore, exploring new candidate genes associated with oil content and SW through GWAS would provide novel ideas for high-yield breeding (Surabhi et al., 2009).
Over recent decades, some genes have been shown to affect SW (Chen et al., 2020), including oleate dehydrogenase (FAD2), which is a key enzyme in linoleic acid synthesis (Pham et al., 2010). Soybean mutants with a low linoleic acid content were obtained through targeted editing of GmFAD2-1A, resulting in a 2.62% increase in yield and a significantly higher SW than that of the control group (Hoshino et al., 2010). Linoleic acid dehydrogenase (FAD3) catalyzes the conversion of linoleic acid to linolenic acid (Dominguez et al., 2010). The overexpression of GmFAD3 induces linolenic acid accumulation and jasmonic acid accumulation (Parida et al., 2017), which results in larger and heavier seeds (Bocianowski and Bartkowiak-Broda, 2012). DGAT is a rate-limiting enzyme in triacylglycerol synthesis, and the preference for the different types of DGAT selection could affect the SW (Charles et al., 2011). In a previous study, soybean GmDGAT2D was shown to prioritize the use of linoleic acid acyl chains as substrates; the linoleic acid content in OE-GmDGAT2D Arabidopsis was significantly higher than that of the control, whereas the thousand SW of OE-GmDGAT2D Arabidopsis decreased (Turchetto-Zolet et al., 2011). GmDGAT1A prioritizes the use of linolenic acid acyl chains as substrates, and GmDGAT1A reduces linoleic acid content in Arabidopsis, resulting in an increase in the thousand SW (Zhao et al., 2019).
The soybean transcription factors (TFs) Dof (GmDof4 and GmDofll) increase the oil content of Arabidopsis and the SW (Zhang et al., 2014). The soybean DREB transcription factor GmDREBL plays an important role in seed oil accumulation. Wang et al. (2007) overexpressed GmDREBL in Arabidopsis, which resulted in a significant increase in the SW (Wang et al., 2007).
Plant cytochrome P450 is a monooxygenase encoded by a super gene family (Bernard et al., 2004). This enzyme usually combines with the membrane system of organelles and participates in many metabolic processes, including various fatty acid conjugates, plant hormones, secondary metabolites, etc. (Fissithaler et al., 1999). Some studies have shown that the CYP83 family of genes CYP83A1 and CYP83B1 play important roles in the dynamic balance of auxin and the growth and development of Arabidopsis (Liu et al., 2012). The CYP78A5 gene in Arabidopsis affects cell proliferation and organ size. Soybean CYP78A10 and Arabidopsis CYP78A have similar functions, mainly in the regulation of seed size, SW, and pod number, suggesting that this gene may also regulate seed development (Bernard et al., 2004). Studies have also indicated that GmCYP78A72 regulates flower and seed development in soybeans; Arabidopsis thaliana lines overexpressing the GmCYP78A72 gene exhibit increased leaf, petal, seed, and carpel traits. The overexpression of GmCYP78A72 in soybeans can also cause soybean seeds to grow larger (Dai et al., 2016).
Although several TFs, such as WRI1, FUS3, and LEC1, play important roles in seed development (Liu et al., 2010), their mechanism is yet to be clarified, especially in soybean seeds (Duan et al., 2023). WRI1 is an important transcription factor that regulates the distribution of carbon elements to the synthesis of FAs in seeds (Baud et al., 2007; Vogel et al., 2019). WRI1 not only increases the FA content in the seed, but also alters the SW through cross-family interactions with multiple transcription, post-transcription, and post-translational regulators (Maeo et al., 2010; Dahee and Suh, 2015). Therefore, this gene has the potential to improve the plant yield. In the Arabidopsis mutant, Atwri1, the weight of 1000 seeds decreased, total seed oil decreased, and linoleic acid content significantly increased by 15% (Baud et al., 2007). After expressing the exogenous gene AtWRI1 in soybeans, the total oil content increased, and linoleic acid content decreased from 52.4% to 4.6% (Vogel et al., 2019). WRI1 can also directly or indirectly affect the activity of Dof11, ACBP, PDCT, and FAD2 by binding to AW-box cis-acting elements (Zhou et al., 2020). Similar results were reported in our research; we found that the overexpression of GmWRI14-like not only increased the FA content but also significantly increased the SW. However, the molecular mechanism of GmWRI14-like is unclear. RNA-seq and qRT-PCR results indicated that GmCYP78A50 and GmCYP78A69 expression is increased in the transgenic plants compared to the control plants. We further confirmed the interaction between GmWRI14-like and GmCYP78A50/GmCYP78A69 by point-to-point Y2H and BiFC. Taken together, our data indicate that GmWRI14-like not only regulates the expression level of GmCYP78A50 and GmCYP78A69, but also interacts with GmCYP78A50 and GmCYP78A69, which has not been reported in previous studies. According to our results, it is reasonable to speculate that the GmWRI14-like gene can directly or indirectly regulate GmCYP78A50 and GmCYP78A69 expression (depicted as “?” in Figure 7), and that GmCYP78A50 and GmCYP78A69 protein interaction with GmWRI14-like regulate its transcription in soybean seeds. Future studies should assess the precise function of GmCYP78A50 and GmCYP78A69 in soybean seeds, and their effect on GmWRI14-like transcription.
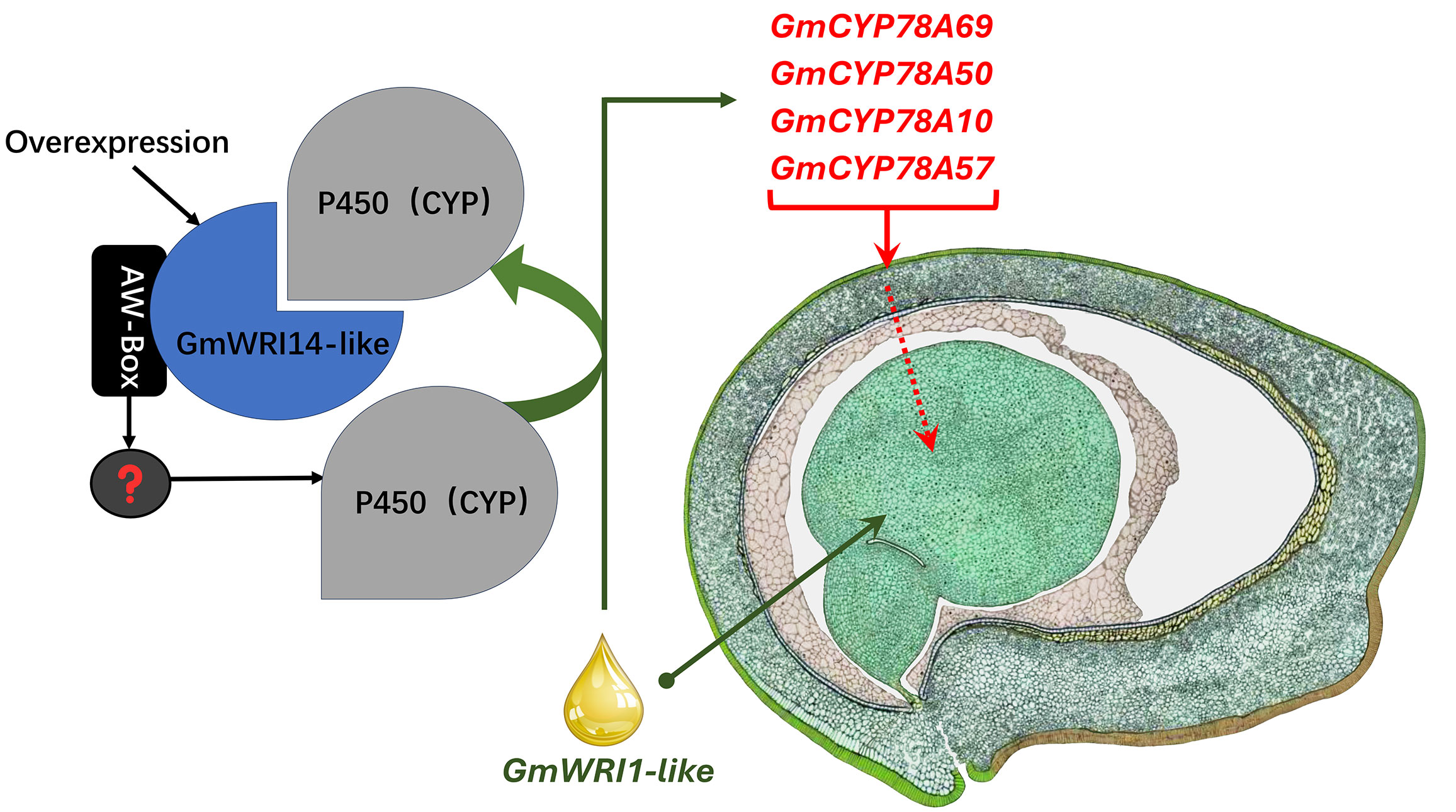
Figure 7 The model depicting transcriptional control by GmWRI14-like. The right image was hand drawn by Sharon Lee Belkin (http://seedgenenetwork.net/soybean).
5 Conclusions
In this study, we identified a novel candidate gene, GmWRI14-like, that is associated with soyabean SW using a GWAS. Overexpression of the GmWRI14-like gene not only increased the total FA content of the seeds, but also increased the SW. RNA-seq and qRT-PCR results indicated that the upregulation of GmCYP78A50 and GmCYP78A69 is triggered by the transcription factor GmWRI14-like. Furthermore, the protein interaction between GmCYP78A69/GmCYP78A50 and GmWRI14-like was confirmed by using Y2H and BiFC.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/ PRJNA608146 and PRJNA681350.
Author contributions
DQ: Data curation, Investigation, Software, Writing – original draft, Writing – review & editing. LD: Writing – original draft, Data curation. LJ: Conceptualization, Writing – original draft, Formal Analysis, Methodology. XL: Conceptualization, Investigation, Software, Writing – original draft. PC: Formal Analysis, Funding acquisition, Project administration, Validation, Writing – review & editing. BL: Writing – review & editing, Supervision. GY: Writing – original draft, Investigation, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The project was funded by GuangDong Basic and Applied Basic Research Foundation (2022A1515110617).
Acknowledgments
We are grateful for the generous grants from Guangzhou University and Guangzhou University of Chinese Medicine, and the University of Camerino for making this work possible. Guangzhou University provided part of the soybean materials, and Guangzhou University of Chinese Medicine and University of Camerino provided the bioinformatics analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1268511/full#supplementary-material
Supplementary Figure 3 | Multiple alignment. GmWRI14-like contained two (AP2/EREB) DNA-binding domains. The homology of the amino acid sequences between GmWRI14-like and AtWRI1 (Gene ID: 824599) is 62.34%.
Supplementary Table 1 | Primer sequence for qRT-PCR.
References
Bai, M., Yuan, J., Kuang, H., Gong, P., Li, S., Zhang, Z., et al. (2020). Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. Plant Biotechnol. J. 18, 721–731. doi: 10.1111/pbi.13239
Baud, S., Mendoza, S., To, A., Harscot, E., Lepiniec, L., Dubreucq, B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50, 825–838. doi: 10.1111/j.1365-313X.2007.03092.x
Bernard, M., Samuël, P., de, V., Sason, S. (2004). Mechanism of oxidation reactions catalyzed by cytochrome P450 enzymes. Chem. Rev. 89, 1020–1032. doi: 10.1021/cr020443g
Bocianowski, J., Bartkowiak-Broda, K. M. (2012). Determination of fatty acid composition in seed oil of rapeseed (Brassica napus L.). by mutated alleles of the FAD3 desaturase genes. J. Appl. Genet. 9, 202–215. doi: 10.1007/s13353-011-0062-0
Carmen, F., Francisco, R. T., Domingo, J. I., Laura, H., Ignacio, L., Ana, M., et al. (2007). Engineering of gibberellin levels in citrus by sense and antisense overexpression of a GA 20-oxidase gene modifies plant architecture. J. Exp. Bot. 58, 1407–1420. doi: 10.1385/criai:27:3:207
Charles, H., Joel, H., Ryan, S., Scot, G. (2011). DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J. Lipid Res. 42, 109–121. doi: 10.1194/jlr.M013003
Che, R., Tong, H., Shi, B., Liu, Y., Fang, S., Liu, D., et al. (2016). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants. 261 (4), 138–249. doi: 10.1038/nplants.2015.195
Chen, Y., Yan, Y., Wu, T. T., Zhang, G. L., Gou, J. Y. (2020). Cloning of wheat keto-acyl thiolase 2B reveals a role of jasmonic acid in grain weight determination. Nat. Commun. 11, 204–213. doi: 10.1038/s41467-020-20133-z
Dahee, A., Suh, M. C. (2015). Overexpression of Arabidopsis WRI1 enhanced seed mass and storage oil content in Camelina sativa. Plant Biotechnol. Rep. 9, 137–148. doi: 10.1007/s11816-015-0351-x
Dai, A., Jiang, N., Wei, H., Yang, S., Feng, X. (2016). Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 83, 201–221. doi: 10.1093/jxb/erw460
Dominguez, T., Hernandez, M. L., Pennycooke, J. C., Jimenez, P., Martinez-Rivas, J. M., Sanz, C., et al. (2010). Increasing-3 desaturase expression in tomato results in altered aroma profile and enhanced resistance to cold stress. Plant Physiol. 65, 604–616. doi: 10.1104/pp.110.154815
Dong, L., Fang, C., Cheng, Q., Su, T., Liu, B. (2021). Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Commun. 12 (1), 5445–5455. doi: 10.1038/s41467-021-25800-3
Du, J., Wang, S., He, C., Zhou, B., Ruan, Y. L., Shou, H. (2017). Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis. J. Exp. Bot. 12, 1010–1032. doi: 10.1093/jxb/erw460
Du, L., Li, N., Chen, L., Xu, Y., Li, Y. (2014). The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in arabidopsis. Plant Cell 54, 1087–1095. doi: 10.1105/tpc.114.122663
Duan, Z., Li, Q., Wang, H., He, X., Zhang, M. (2023). Genetic regulatory networks of soybean seed size, oil and protein contents. Front. Plant Sci. 14 (11). doi: 10.3389/fpls.2023.1160418
Duan, Z., Zhang, M., Zhang, Z., Liang, S., Fan, L., Yang, X., et al. (2022). Natural allelic variation of GmST05 controlling seed size and quality in soybean. Plant Biotechnol. J. 20 (9), 1807–1818. doi: 10.1111/pbi.13865
Fasoula, V. A., Boerma, H. R. (2007). Intra-cultivar variation for seed weight and other agronomic traits within three elite soybean cultivars. Crop Sci. 47, 367–373. doi: 10.2135/cropsci2005.09.0334
Fasoula, V. A., Harris, D. K., Roger, B. H. (2004). Validation and designation of quantitative trait loci for seed protein, seed oil, and seed weight from two soybean populations. Crop Sci. 44, 1218–1225. doi: 10.2135/cropsci2004.1218
Fissithaler, B., Popp, R., Harder, D. R., Fleming, I., Busse, R. (1999). Cytochrome P450 2C is an EDHF synthase in coronary arteries. Nature 76, 121–131. doi: 10.1016/S1050-1738(00)00065-7
Goettel, W., Zhang, H., Li, Y., Qiao, Z., Jiang, H., Hou, D., et al. (2022). POWR1 is a domestication gene pleiotropically regulating seed quality and yield in soybean. Nat. Commun. 13, 1340–1391. doi: 10.1038/s41467-022-30314-7
González-García, M., Rodríguez, D., Nicolás, C., Nicolás, G., Lorenzo, O. (2007). Transcriptomic and proteomic profiling of FsPP2C1-overexpressing Arabidopsis plants. Plant J. 21, 235–244. doi: 10.1002/pmic.200700861
Guttikonda, S. K., Trupti, J., Bisht, N. C., Chen, H., An, Y., Pandey, S., et al. (2010). Whole genome co-expression analysis of soybean cytochrome P450 genes identifies nodulation-specific P450 monooxygenases. BMC Plant Biol. 10, 243–243. doi: 10.1186/1471-2229-10-243
Hacisalihoglu, G., Burton, A. L., Gustin, J. L., Eker, S., Asikli, S., Heybet, E. H. (2018). Quantitative trait loci associated with soybean seed weight and composition under different phosphorus levels. J. Integr. Plant Biol. 60 (3), 232–241. doi: 10.1111/jipb.12612
Harris, W. S., Lemke, S. L., Hansen, S. N., Goldstein, D. A., Dirienzo, M. A., Su, H., et al. (2008). Stearidonic acid-enriched soybean oil increased the omega-3 index, an emerging cardiovascular risk marker. Lipids 43, 805–811. doi: 10.1007/s11745-008-3215-0
Holsters, M., Waele, D., Depicker, A., Messens, E., Montagu, M., Schell, J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163, 181–187. doi: 10.1007/bf00267408
Hoshino, T., Takagi, Y., Anai, T. (2010). Novel GmFAD2-1b mutant alleles created by reverse genetics induce marked elevation of oleic acid content in soybean seeds in combination with GmFAD2-1a mutant alleles. Breed. Sci. 60, 419–425. doi: 10.1270/jsbbs.60.419
Hu, Y., Liu, Y., Tao, J. J., Lu, L., Jiang, Z. H., Wei, J. J., et al. (2023). GmJAZ3 interacts with GmRR18a and GmMYC2a to regulate seed traits in soybean. J. Integr. Plant Biol. 12, 15–34. doi: 10.1111/jipb.13494
Jaromír, L., Alena, H., Táborský, L., Zora, K., Vladimír, P., Střalková, R. (2015). Evaluation of oil content and fatty acid composition in the seed of grapevine varieties. LWT - Food Sci. Technol. 23, 605–610. doi: 10.1016/j.lwt.2015.03.044
Kim, J. H., Hans, K. (2004). A Transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. United States America 75, 1266–1289. doi: 10.1073/pnas.0405450101
Kuroda, Y., Tomooka, N., Kaga, A., Wanigadeva, S., Vaughan, D. A. (2009). Genetic diversity of wild soybean (Glycine soja Sieb. et Zucc.). and Japanese cultivated soybeans [G. max (L.). Merr.] based on microsatellite (SSR). analysis and the selection of a core collection. Genet. Resour. Crop Evol. 56, 1045–1055. doi: 10.1007/s10722-009-9425-3
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25 (14), 1754–1760. doi: 10.1093/bioinformatics/btp324
Liu, J., Wei, H., Zhan, G., Wei, F., Wang, X., Liu, G., et al. (2010). Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 48, 9–15. doi: 10.1016/j.plaphy.2009.09.007
Liu, K., Liu, F., Zhang, N., Liu, S., Jiang, Y. (2012). Isolation and Expression of Glucosinolate Synthesis Genes CYP83A1 and CYP83B1 in Pak Choi (Brassica rapa L. ssp. chinensis var. communis (N. Tsen & S.H. Lee). Hanelt). Int. J. Mol. Sci. 51, 1021–1030. doi: 10.3390/ijms13055818
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method. Methods: A Companion to Methods Enzymol. 41, 25–33. doi: 10.1006/meth.2001.1262
Lu, X., Li, Q., Xiong, Q., Li, W., Bi, Y., Lai, Y., et al. (2016a). The transcriptomic signature of developing soybean seeds reveals the genetic basis of seed trait adaptation during domestication. Plant J. 86 (6), 530–544. doi: 10.1111/tpj.13181
Lu, X., Li, Q. T., Xiong, Q., Li, W., Zhang, J. S. (2016b). The transcriptomic signature of developing soybean seeds reveals genetic basis of seed trait adaptation during domestication. Plant J. 86 (6), 530–544. doi: 10.1111/tpj.13181
Maeo, K., Tokuda, T., Ayame, A., Mitsui, N., Nakamura, K. (2010). An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 60, 476–487. doi: 10.1111/j.1365-313X.2009.03967.x
Martin, C. A., Visentainer, J. V., Oliveira, A., Oliveira, C., Souza, N. (2008). Fatty acid contents of Brazilian soybean oils with emphasis on trans fatty acids. J. Braz. Chem. Soc. 19, 117–122. doi: 10.1590/S0103-50532008000100017
Mora, A., Weichert, H., Wirén, N., Frohberg, C., Weber, H. (2021). The da1 mutation in wheat increases grain size under ambient and elevated CO2 but not grain yield due to trade-off between grain size and grain number. Plant J. 964, 36–46. doi: 10.22541/au.160218279.96892332/v1
Mou, S., Meng, Q., Gao, F., Zhang, T., He, S. (2021). A cysteine-rich receptor-like protein kinase cackr5 modulates immune response against ralstonia solanacearum infection in pepper. BMC Plant Biol. 21 (1), 382–390. doi: 10.1186/s12870-021-03150-y
Nakabayashi, K., Okamoto, M., Koshiba, T., Kamiya, Y., Nambara, E. (2010). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 41, 697–709. doi: 10.1111/j.1365-313X.2005.02337.x
Ohta, H., Sueoka, H., Takamiya, K. I. (2000). Mechanism for the regulation of the gene expression of CYP93A gene family involved in isoflavone biosynthesis of soybean. Soy Protein Res. Japan 234, 1–5. doi: 10.1007/s11032-009-9305-8
Parida, S. K., Kujur, A., Bajaj, D., Das, S., Srivastava, R., Badoni, S., et al. (2017). Integrative genome-wide association studies (GWAS). to understand complex genetic architecture of quantitative traits in chickpea. Open Access Repository of ICRISAT 46, 181–190.
Pham, A. T., Lee, J. D., Shannon, J. G., Bilyeu, K. D. (2010). Mutant alleles of FAD2-1A and FAD2-1B combine to produce soybeans with high oleic acid seed oil trait. BMC Plant Biol. 10, 195–203. doi: 10.1186/1471-2229-10-195
Porebski, S., Bailey, L. G., Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. doi: 10.1007/BF02772108
Qin, D., Angela, P., Qi, Z., Cristina, M., Hui, L., Xiaoyi, L. (2021). Genome-wide association study identifies candidate genes related to the linoleic acid content in soybean seeds. Int. J. Mol. Sci. 23 (6), 530–544. doi: 10.3390/ijms23010454
Rahman, S. M., Anai, T., Kinoshita, T., Takagi, Y. (2003). A novel soybean germplasm with elevated saturated fatty acids. Crop Sci. 43, 527–531. doi: 10.2135/cropsci2003.0527
Ramya, P., Chaubal, A., Kulkarni, K., Gupta, L., Kadoo, N. (2010). QTL mapping of 1000-kernel weight, kernel length, and kernel width in bread wheat (Triticum aestivum L.). J. Appl. Genet. 85, 564–575. doi: 10.1007/BF03208872
Shimada, T., Takahashi, A., Shibata, M., Yagihashi, T. (2016). Effects of within plant variability in seed weight and tannin content on foraging behaviour of seed consumers. Funct. Ecol. 29, 56–63. doi: 10.1111/1365-2435.12464
Singh, M., Kaur, K., Sharma, A., Kaur, R., Singh, P. (2021). Genome-wide characterization of peptidyl-prolyl cis-trans isomerases in penicillium and their regulation by salt stress in a halotolerant P. Oxalicum. Sci. Rep. 11, 76–89. doi: 10.21203/rs.3.rs-223100/v1
Surabhi, G. K., Reddy, K. R., Singh, S. K. (2009). Photosynthesis, fluorescence, shoot biomass and seed weight responses of three cowpea (Vigna unguiculata (L.). Walp.). cultivars with contrasting sensitivity to UV-B radiation. Environ. Exp. Bot. 66, 160–171. doi: 10.1016/j.envexpbot.2009.02.004
Tudzynski, B., Mrria, C. R., Paul, G., Peter, H. (2002). The gibberellin 20-oxidase of Gibberella fujikuroi is a multifunctional monooxygenase. J. Biol. Chem. 277, 21246–21253. doi: 10.1074/jbc.M201651200
Turchetto-Zolet, A. C., Maraschin, F. S., Morais, G., Cagliari, A., Margis, R. (2011). Evolutionary view of acyl-CoA diacylglycerol acyltransferase (DGAT), a key enzyme in neutral lipid biosynthesis. BMC Evolution. Biol. 11, 263–272. doi: 10.1186/1471-2148-11-263
Vogel, P. A., Shen, B., Park, H., Nguyen, H., Hou, L., Changa, T., et al. (2019). Expression of the Arabidopsis WRINKLED 1 transcription factor leads to higher accumulation of palmitate in soybean seed. Plant Biotechnol. J. 51, 121–133. doi: 10.1111/pbi.13061
Vyacheslav, A., Nikolai, B., Natalia, P., Anita, B., Joseph, D., Sergei, S., et al. (2010). Tobacco as a production platform for biofuel: overexpression of arabidopsis dgat and lec2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 8 (3), 277–287. doi: 10.1111/j.1467-7652.2009.00458.x
Wang, L., Cao, C., Ma, Q., Zeng, Q., Haifeng, W. (2014). RNA-seq analyses of multiple meristems of soybean: novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 14 (1), 169–180. doi: 10.1186/1471-2229-14-169
Wang, K., He, J., Zhao, Y., Wu, T., Zhou, X., Ding, Y., et al. (2018). EAR1 negatively regulates ABA signaling by enhancing 2C protein phosphatase activity. Plant Cell 5, 876–883. doi: 10.1105/tpc.17.00875
Wang, H., Zhang, B., Hao, Y., Huang, J., Tian, A., Liao, Y. (2007). The soybean dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 52 (4), 716–729. doi: 10.1111/j.1365-313X.2007.03268.x
Xiaofei, C., Ruyi, X., Yinzi, L., Fangfang, L., Xueping, Z., Aiming, W. (2017). Sumoylation of turnip mosaic virus rna polymerase promotes viral infection by counteracting the host npr1-mediated immune response. Plant Cell 29 (3), 508–525. doi: 10.1105/tpc.16.00774
Yin, P., Ma, Q., Wang, H., Feng, D., Wang, X., Pei, Y., et al. (2020). SMALL LEAF AND BUSHY1 controls organ size and lateral branching by modulating the stability of BIG SEEDS1 in Medicago truncatula. New Phytol. 226 (11), 2103–2127. doi: 10.1111/nph.16449
Zhang, J., Hao, Q., Bai, L., Xu, J., Yin, W. (2014). Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea. Biotechnol. Biofuels 7, 56–73. doi: 10.1186/s13068-014-0128-4
Zhang, Y. Q., Lu, X., Zhao, F. Y., Li, Q., Niu, S., Wei, W., et al. (2016). Soybean GmDREBL increases lipid content in seeds of transgenic Arabidopsis. Sci. Rep. 6, 343–354. doi: 10.1038/srep34307
Zhao, J., Bi, R., Li, S., Zhou, D., Zhang, W. (2019). Genome-wide analysis and functional characterization of Acyl-CoA:diacylglycerol acyltransferase from soybean identify GmDGAT1A and 1B roles in oil synthesis in Arabidopsis seeds. J. Plant Physiol. 242, 1530–1541. doi: 10.1016/j.jplph.2019.153019
Keywords: soybean, seed weight, oil content, GWAS, candidate gene
Citation: Di Q, Dong L, Jiang L, Liu X, Cheng P, Liu B and Yu G (2023) Genome-wide association study and RNA-seq identifies GmWRI1-like transcription factor related to the seed weight in soybean. Front. Plant Sci. 14:1268511. doi: 10.3389/fpls.2023.1268511
Received: 28 July 2023; Accepted: 30 October 2023;
Published: 17 November 2023.
Edited by:
Yi-Hong Wang, University of Louisiana at Lafayette, United StatesReviewed by:
Hengyou Zhang, Chinese Academy of Sciences (CAS), ChinaShancen Zhao, Beijing Genomics Institute (BGI), China
Copyright © 2023 Di, Dong, Jiang, Liu, Cheng, Liu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qin Di, di.qin@unicam.it; Baohui Liu, liubh@gzhu.edu.cn; Guohui Yu, yuguohui189@163.com
 Qin Di
Qin Di Lidong Dong2
Lidong Dong2 Li Jiang
Li Jiang Xiaoyi Liu
Xiaoyi Liu