- 1Key Laboratory of Biology and Germplasm Enhancement of Horticultural Crops in South China, Ministry of Agriculture and Rural Areas, College of Horticulture, South China Agricultural University, Guangzhou, China
- 2Tropical Crops Genetic Resources Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
Background: Pineapple is the only commercially grown fruit crop in the Bromeliaceae family and has significant agricultural, industrial, economic, and ornamental value. GRF (growth-regulating factor) proteins are important transcription factors that have evolved in seed plants (embryophytes). They contain two conserved domains, QLQ (Gln, Leu, Gln) and WRC (Trp, Arg, Cys), and regulate multiple aspects of plant growth and stress response, including floral organ development, leaf growth, and hormone responses. The GRF family has been characterized in a number of plant species, but little is known about this family in pineapple and other bromeliads.
Main discoveries: We identified eight GRF transcription factor genes in pineapple, and phylogenetic analysis placed them into five subfamilies (I, III, IV, V, VI). Segmental duplication appeared to be the major contributor to expansion of the AcGRF family, and the family has undergone strong purifying selection during evolution. Relative to that of other gene families, the gene structure of the GRF family showed less conservation. Analysis of promoter cis-elements suggested that AcGRF genes are widely involved in plant growth and development. Transcriptome data and qRT-PCR results showed that, with the exception of AcGRF5, the AcGRFs were preferentially expressed in the early stage of floral organ development and AcGRF2 was strongly expressed in ovules. Gibberellin treatment significantly induced AcGRF7/8 expression, suggesting that these two genes may be involved in the molecular regulatory pathway by which gibberellin promotes pineapple fruit expansion.
Conclusion: AcGRF proteins appear to play a role in the regulation of floral organ development and the response to gibberellin. The information reported here provides a foundation for further study of the functions of AcGRF genes and the traits they regulate.
1 Introduction
Pineapple is one of the three most widely cultivated tropical fruit crops in the world and the only commercially grown member of the Bromeliaceae family (Lobo and Paull, 2017). Because of its excellent flavor and texture, it has been called the queen of fruits (Baruwa, 2013). Its unique shape, fiber content, and nutritional value give it an important place in medicine and industry (Sanewski et al., 2018). However, adverse environmental factors such as temperature extremes, as well as changes in hormone levels, can seriously affect pineapple growth and development, reducing yield and quality. Exploring the mechanisms that regulate flower and fruit development and stress responses is important for maintaining the commercial value of pineapple.
Transcription factors regulate the ability of cells to express different genes and thereby control development (Riechmann et al., 2000; Levine and Tjian, 2003). Growth-regulating factor (GRF) proteins are widespread transcription factors in plants, and their highly conserved N-terminal QLQ and WRC domains are their most prominent feature (Van der Knaap et al., 2000). These two domains have different functions; the QLQ domain is involved in protein interaction and is rich in aromatic/hydrophobic amino acids (Kim and Kende, 2004), whereas the WRC domain mediates DNA-binding ability (Osnato et al., 2010; Kim et al., 2012; Kuijt et al., 2014).
Since the first GRF protein was discovered in rice (Oryza sativa) (Van der Knaap et al., 2000; Omidbakhshfard et al., 2015), GRFs have been shown to influence almost all plant growth and developmental processes, including leaf growth (Kim and Kende, 2004; Kim and Lee, 2006; Arvidsson et al., 2011; Debernardi et al., 2014; Wu et al., 2014; Zhang et al., 2021), floral organ development (Arvidsson et al., 2011; Pu et al., 2012; Liang et al., 2013; Liu H. et al., 2014; Pajoro et al., 2014; Lee et al., 2017), root development (Bao et al., 2014), seed oil content (Bao et al., 2014), plant lifespan, and stress responses (Kim et al., 2012; Casadevall et al., 2013; Liu J. et al., 2014). Understanding the roles of GRFs is therefore important for plant growth research and genetic improvement.
Recent studies have shown that AtGRFs from the model plant Arabidopsis thaliana can significantly improve the transgenic transformation efficiency of a variety of crops, in addition to performing some of the functions above. For example, overexpression of AtGRF5 and its orthologs increases transformation efficiency, callus cell proliferation, and transgenic bud formation in Beta vulgaris, corn (Zea mays), soybean (Glycine max), and Helianthus annuus (Debernardi et al., 2020; Kong et al., 2020). Expression of a GRF4–GIF1 fusion protein significantly increased the regeneration efficiency, regeneration rate, and somatic embryogenesis of wheat (Triticum aestivum) and rice (Oryza sativa) (Debernardi et al., 2020). Although there are multiple literature reports on the functions of GRF family members, detailed genome-wide phylogenetic and functional studies of GRF genes are not yet available for pineapple or other bromeliads.
To better understand the evolutionary dynamics of GRF genes in pineapple and explore their potential regulatory roles in flower and fruit development and hormone and stress responses, we identified eight AcGRF genes in pineapple and performed a series of analyses, documenting their chromosome locations, motif compositions, evolutionary relationships, genomic collinearity, and selection pressure. We also investigated the potential roles of these AcGRFs using interaction network prediction and gene expression analysis. These results provide insights into the functions of GRF family members in pineapple growth and development.
2 Materials and methods
2.1 Data sources and sequence retrieval
All protein sequences of pineapple were obtained from the Pineapple Genome Project (Ming et al., 2015). Sequences of nine A. thaliana GRF genes and 12 rice GRF genes were obtained from previous studies (Kim et al., 2003; Choi et al., 2004). The corresponding protein sequences were downloaded from the Arabidopsis Information Resource Library (TAIR) (http://www.arabidopsis.org/) and the UniProt Database (https://www.uniprot.org) (Boutet et al., 2016; Cheng et al., 2017). Protein sequences from Phalaenopsis equestris and Nymphaea colorata were obtained from recent studies (Zhang et al., 2017; Zhang and Chen, 2019), and the Amborella trichopoda proteins were downloaded from the PLAZA database (https://bioinformatics.psb.ugent.be/plaza/) (Van Bel et al., 2017). Protein sequences for Vitis vinifera, Sorghum bicolor, Musa acuminata, and other species were obtained from Phytozome (http://www.phytozome.net/).
2.2 Identification and classification of GRFs
We used two strategies to identify GRF genes in pineapple. First, we performed a local BlastP search of pineapple protein sequences using GRF protein sequences from A. thaliana as queries. We then obtained hidden Markov models of the QLQ (PF08880) and WRC (PF08879) domains from the Pfam database (http://pfam.xfam.org/) and used them to query the pineapple protein files using TBtools with an E value of 1e−10 (Chen C. et al., 2020). We identified 21 protein sequences as candidate GRFs in pineapple. We confirmed the presence of GRF core sequences using the Batch CD-Search and SMART programs, further examining all candidate genes that appeared to contain QLQ and WRC domains in the BlastP and HMMER search results. Each candidate gene was then manually checked, and the structurally annotated gene was corrected to ensure that there were conserved heptapeptide sequences at the N terminus of the predicted QLQ and WRC domains. Thirteen protein sequences with no or incomplete QLQ or WRC domains were removed. We identified GRF genes in 23 additional species using the same method, and the complete set of GRF protein sequences was used to study their evolutionary relationships. The ExPASy ProtParam database (https://web.expasy.org/protparam/) was used to analyze the physicochemical properties of the AcGRF genes/proteins, including coding region length, number of amino acids, molecular weight (MW), and theoretical isoelectric point (pI).
The subcellular localizations of the AcGRFs were predicted using Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/#), the transmembrane domains using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), and the signal peptides using SignalP (http://www.cbs.dtu.dk/services/SignalP/).
We next characterized the evolutionary relationships among GRF genes from various species and assigned the putative pineapple GRF genes to specific subfamilies. Multiple-sequence alignments of amino acid sequences were constructed using MAFFT with default parameters, and a species-tree of 26 taxons was constructed using their protein sequences with the maximum likelihood method and 1,000 bootstrap replicates in OrthoFinder with the following parameters: orthofinder -f dataset -M msa -S diamond -t fasttree -t 16 -a 16.
2.3 Chromosome locations, gene structures, and conserved motifs of the AcGRFs
We obtained chromosome locations and genetic structures of each AcGRF gene from the pineapple genome annotation file (Ming et al., 2015). The data were then integrated and plotted using TBtools (Chen C. et al., 2020). We identified conserved motifs shared among the AcGRF proteins (Bailey et al., 2009) using MEME tools with the following parameters: maximum number of cardinal orders, 10; minimum width, 20; maximum width, 50.
2.4 Duplication, collinearity, and evolutionary analysis of the AcGRF gene family
We performed collinearity analysis using the method described in TBtools (Chen et al., 2022). We first prepared the pineapple genome and annotation file, then used it as input for collinearity analysis using the “One Step MCScanX Wrapper” function with the following parameters: CPUs for BlastP, 8; e-value, 1e−3; number of blast hits, 10. Dispersed, proximal, tandem, and segmental/WGD duplicates in the AcGRF family were identified using TBtools. We connected segmentally duplicated gene pairs by red and green arcs in the Circos plot. We also performed collinearity analysis of GRF genes in pineapple and other plant species, including A. thaliana, V. vinifera, A. trichopoda, N. colorata, O. sativa, S. bicolor, M. acuminata, Phalaenopsis equestris, and Spirodela polyrhiza. TBtools was used to visualize a portion of the results.
2.5 Identification of cis-elements in the AcGRF promoters
The 2,000-bp sequence upstream of each AcGRF gene was extracted using TBtools and defined as the promoter region. PlantCARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was then used to predict the cis-elements in the promoter region (Lescot, 2002), and the results were visualized using TBtools.
2.6 Expression profiles of AcGRF genes in different pineapple tissues
Earlier transcriptome studies have generated data on AcGRF gene expression in floral organs at different developmental stages (Wang et al., 2020) and in various fruit tissues during development (Mao et al., 2018). The transcriptome data from both projects were downloaded from NCBI using the project IDs PRJEB38680 and PRJNA483249. The two datasets were analyzed separately using FPKM values. TBtools was used to transform the FPKM values to log2(FPKM+1) values and generate an expression heatmap for the relevant genes.
2.7 Hormone and stress treatments, RNA extraction, and RT-PCR
Well-grown “Shenwan” calli were sampled after exposure to 0.1 mM jasmonic acid (JA), 0.1 mM abscisic acid (ABA), 0.1 mM auxin (IAA), 0.1 mM gibberellin (GA), or 150 mM NaCl in suspension culture medium for 0, 4, 8, 16, 24, 36, and 48 h. All materials were immediately frozen in liquid nitrogen and stored at −80°C for subsequent RNA extraction.
The CWBIO RNApure Plant Kit (DNase I) was used to extract total RNA. RNA quality was checked by agarose gel electrophoresis, and RNA concentration was estimated using a Nanodrop ND-1000 spectrophotometer. First-strand cDNA was synthesized from DNA-free RNA using the HiScript II First-Strand cDNA Synthesis Kit (YEASEN) according to the manufacturer’s protocol. qRT-PCR was performed on a Roche LightCycler 480 instrument with SYBR Green Master Mix (YEASEN). The total reaction volume was 10 μL and contained 5 μL SYBR mix, 0.4 μL upstream and downstream primer mix (2.5 μM), 0.5 μL cDNA template, and 4.1 μL ddH2O. Each reaction was performed in triplicate. The reaction conditions were 95°C for 30 s, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Relative gene expression was calculated by the 2−ΔΔCt method with β-actin as the internal reference gene. Primer sequences used in this study are provided in Table S10.
2.8 Protein–protein interaction network prediction
All AcGRF protein sequences were submitted to the STRING website (http://string-db.org), and their rice orthologs were selected as a reference. After completing the BLAST step, the network was built using the proteins with the highest scores (bitscores). Proteins that were not predicted to interact with any other proteins were removed.
2.9 miRNA target prediction for AcGRFs
Pineapple miRNAs were obtained from previous studies (Yusuf et al., 2015), and AcGRF genes targeted by miRNAs were predicted using psRNATarget (https://www.zhaolab.org/psRNATarget/) with default parameters while selecting target accessibility as described in Dai et al. (2018). We visualized the interactions among the predicted miRNAs and the corresponding target AcGRF genes using TBtools v1.106.
3 Results and analysis
3.1 Physicochemical properties of AcGRF transcription factors
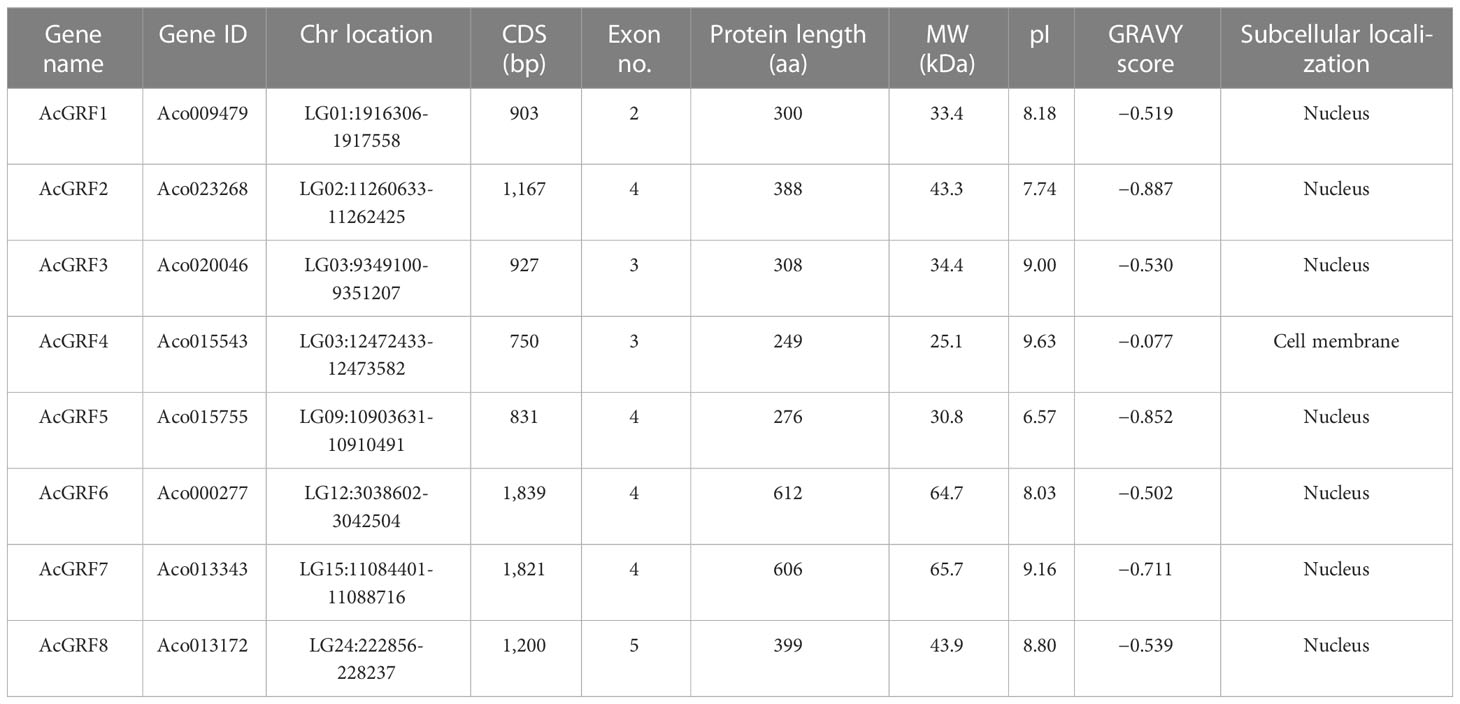
We identified eight GRF gene family members with intact QLQ and WRC domains in pineapple and named them AcGRF1 to AcGRF8 based on their chromosome locations (Table 1). The eight GRFs encoded proteins with 249 (AcGRF4) to 612 (AcGRF6) amino acids, molecular weights (MWs) from 25.1 kDa (AcGRF4) to 65.7 kDa (AcGRF7), and isoelectric points (pIs) from 6.57 (AcGRF5) to 9.63 (AcGRF4). All AcGRF proteins had a negative hydrophilicity score (GRAVY), indicating that they are hydrophilic proteins. Subcellular localization predictions indicated that seven AcGRF proteins were localized to the nucleus and AcGRF4 was localized to the cell membrane.
3.2 Characterization of AcGRF sequences
All the pineapple GRFs contained a highly conserved WRC protein domain with the RTDGKKWRC motif. Seven contained the canonical QLQ (Gln-Leu-Gln) motif; the exception was AcGRF5, in which Leu was replaced by Met (Gln-Met-Gln) (Figure 1A). A similar result was reported in A. thaliana, in which the Leu in AtGRF9 is replaced by Phe (Kim et al., 2003).
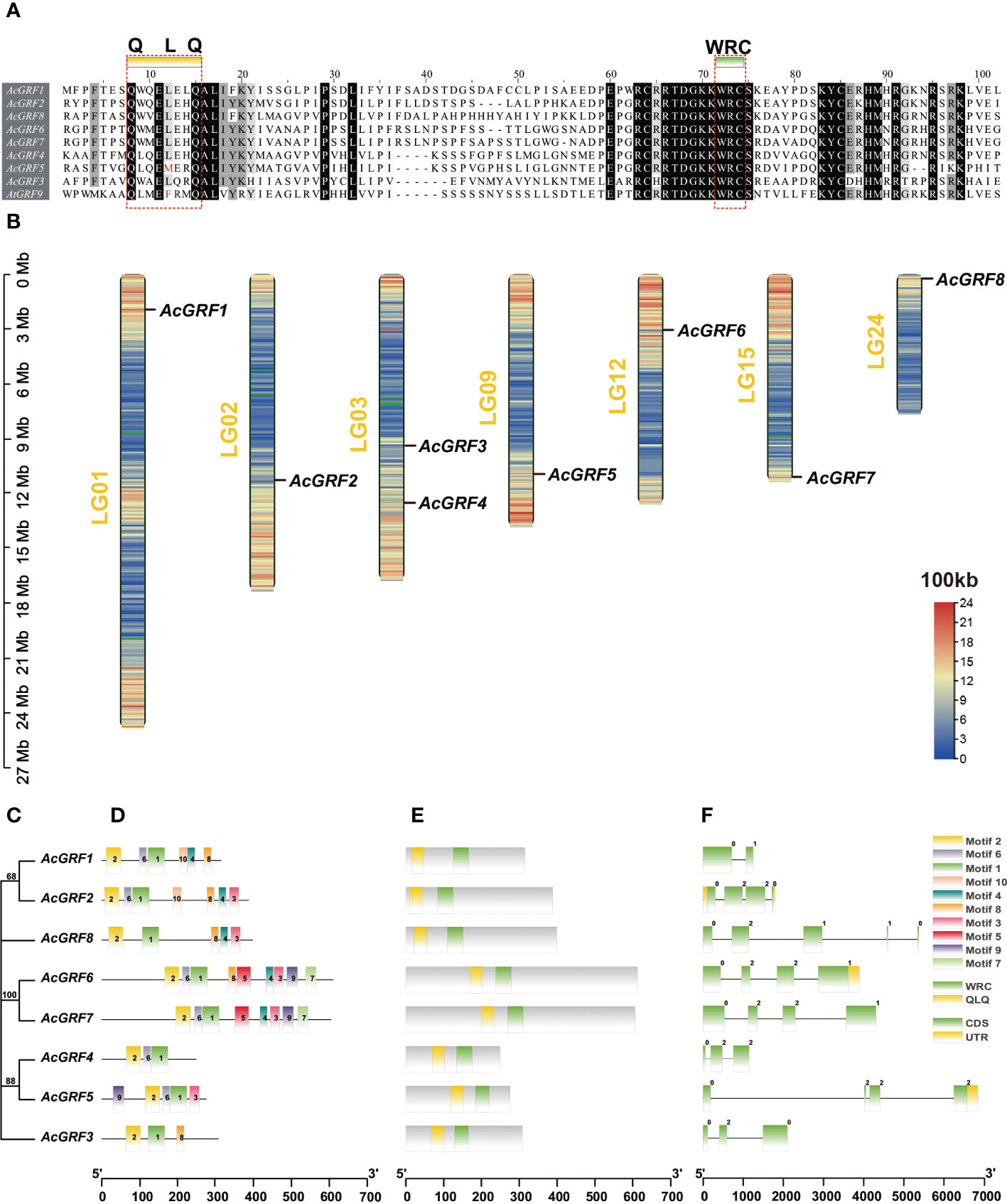
Figure 1 Conserved sequences, chromosomal distribution, gene structures, and motif patterns of AcGRF genes and their encoded proteins. (A) Conserved sequences in AcGRF genes. The QLQ and WRC domains are marked with a red rectangular box, and the amino acids replaced in the QLQ domain are indicated in a red font. (B) Chromosomal distribution of AcGRF genes. The sliding window size was set to 100 kb, and the color from red to blue indicates high to low gene density. Blank areas on chromosomes are genetic regions for which information on gene distribution is lacking.(C) Phylogenetic clustering of AcGRF proteins. (D) Motif patterns of AcGRF proteins. (E) Conserved domains in AcGRF proteins. (F) AcGRF gene structures. Yellow boxes indicate 5′ and 3′ UTR regions; green boxes indicate exons; black lines represent introns; and the number (0–2) indicates the intron phase.
The scattered distribution of AcGRF gene members on different chromosomes is shown in Figure 1B. There are two GRF genes on chromosome 3, but they are physically distant. Most AcGRF genes tend to cluster in regions with higher gene density.
A conserved motif search was performed on the eight AcGRF proteins, and the number of conserved motifs in individual proteins ranged from 3 to 9. Motifs 1 and 2 were components of the WRC and QLQ domains, respectively, and were present in all GRF proteins, whereas other motifs were only present in certain members. AcGRF proteins in the same subgroup had similar motif compositions (Figures 1C–E, S1). Exon numbers ranged from 2 to 5, and most GRF genes did not have 5′ or 3′ UTR annotation information (Figure 1F). Four was the most common number of exons (4 AcGRF genes), followed by three (2 genes).
3.3 Phylogenetic relationships of the AcGRFs
To reveal the evolutionary relationships among GRF genes from various species, we constructed a phylogenetic tree of GRFs from 26 species (Figure 2A). GRF genes were identified from 23 additional plant species and two algal species using the same method applied to pineapples (Figures 2B, C). Most species had around 10 GRF genes; exceptions included Marchantia polymorpha and Physcomitrella patens (1), Musa acuminata (19), and Gossypium raimondii (41) (Figure 2B). We constructed phylogenetic trees of the GRF protein sequences from these species using the maximum likelihood method, dividing them into specific subfamilies (Figures 2C, S2; Table S1). Pineapples contained members of all subfamilies except subfamily II: AcGRF6/7 in subfamily I, AcGRF1/2 in subfamily III, AcGRF4/5 in subfamily V, and AcGRF3 and AcGRF8 in subfamilies IV and VI, respectively (Figure S2). Across all species, subfamily I had the most members, followed by subfamilies III, VI, and V (Figure 2C), which accounted for 21.38%, 18.42%, 17.10%, and 16.78% of all GRF genes in the 26 species, respectively. Interestingly, GRF genes from bryophytes and ferns were not grouped into any of the six subfamilies but were present in a separate outer group. GRFs from basal angiosperms (i.e., N. colorata and A. trichopoda) were found in three of the six subfamilies, and some were in the outer group. Banana (no subfamily IV) and pineapple (no subfamily II) contained members of five GRF subfamilies, and other monocots contained members of only four subfamilies (no subfamilies II or IV). All dicots contained members of all six subfamilies, with the exception of soybean, which lacked members of subfamily V.
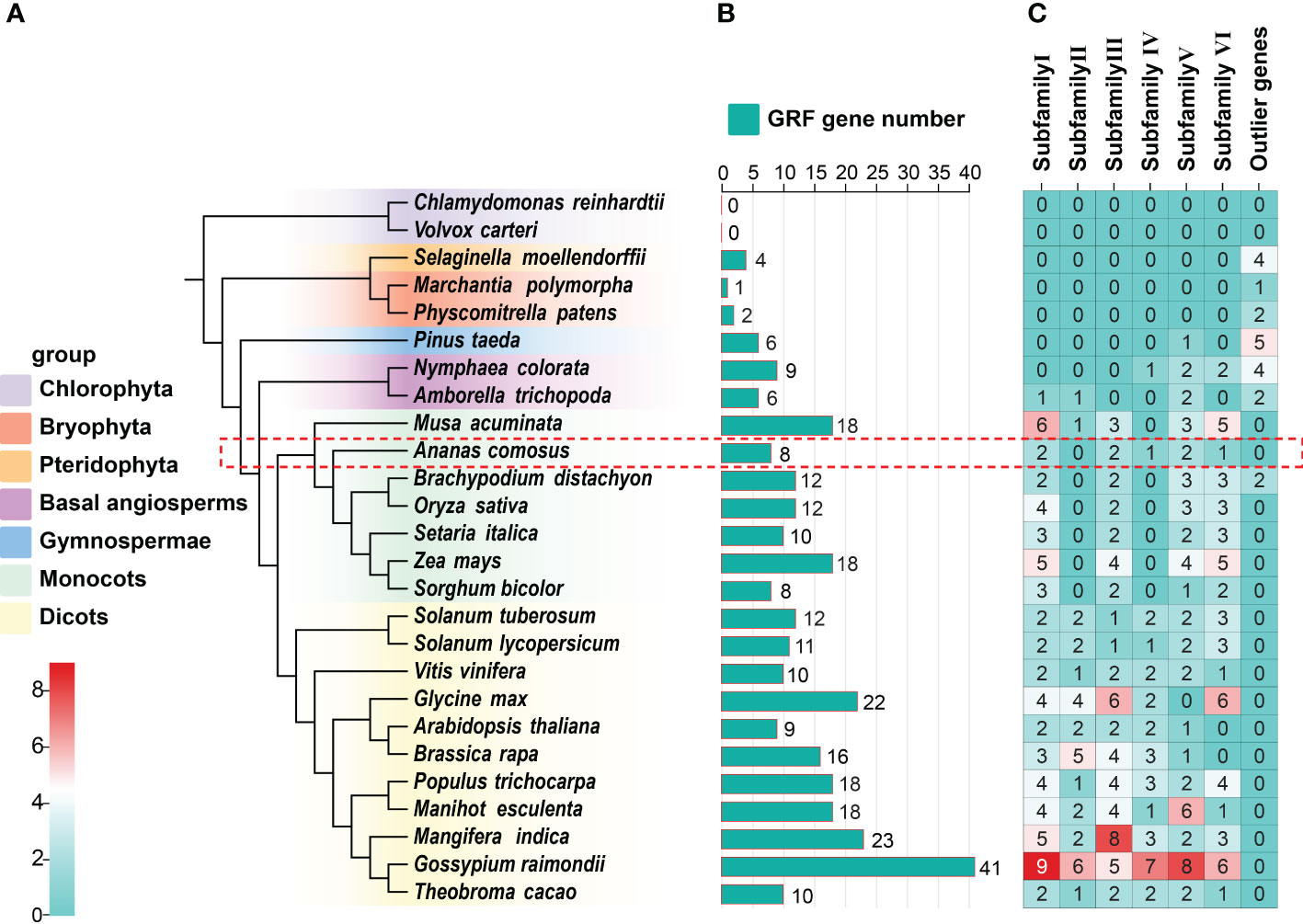
Figure 2 Evolutionary relationships among 26 species and their GRF gene compositions. (A) Phylogenetic relationships among 26 species. (B) Numbers of GRF genes in different species. (C) Distribution of GRF genes in various subfamilies. Phylogenetic analyses of 26 species were performed using OrthoFinder (Emms and Kelly, 2019). The GRF genes in pineapple are marked with a red rectangular box.
3.4 Origins of AcGRF gene members
Gene duplication is thought to be the main driver of species evolution and a direct cause of gene family expansion (Lynch and Conery, 2000; Moore and Purugganan, 2003; Maere et al., 2005). Pineapple experienced two whole-genome duplication (WGD) events in its early evolution, corresponding to two peaks in Figure S3. We searched for tandem and segmental duplicates among the AcGRFs (Tables S2, S3) and visualized them using the Circos plot (Figure 3A). Expansion of the AcGRF gene family appeared to have occurred mainly through segmental duplication, and no tandem duplication events were found. There was one segmentally duplicated gene pair (Figure 3A); the AcGRF1–AcGRF2 gene pair was associated with the WGD events of pineapple.
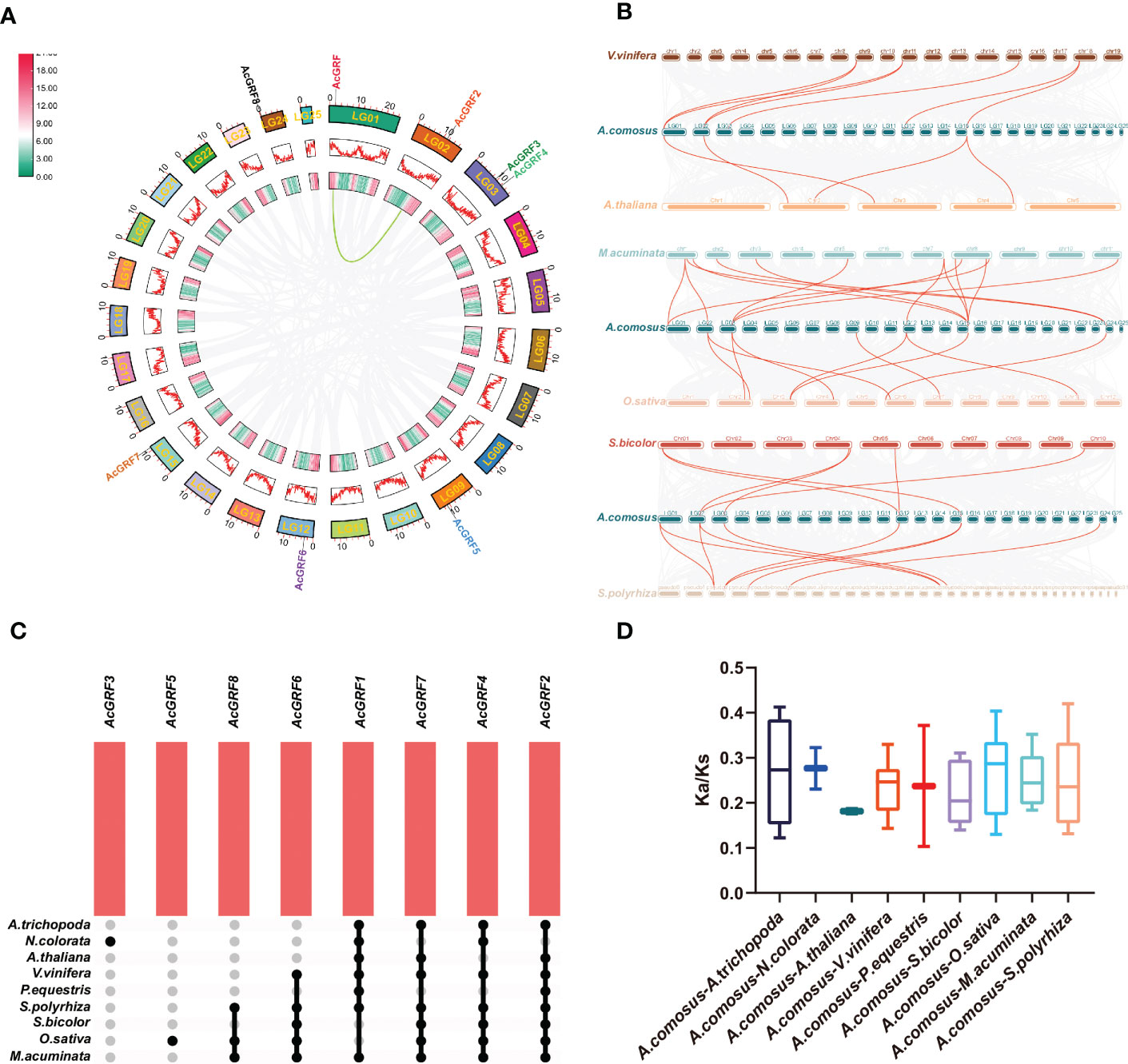
Figure 3 Collinearity and Ka/Ks analyses of pineapple AcGRFs and their homologs in other species. (A) Circos plot of pineapple collinear homologous genes. Genome-wide collinear blocks are set against a gray background, and duplicate AcGRF gene pairs are highlighted by red and green curves. Each pineapple chromosome is accompanied by 100 kb of gene density information, depicted by heat maps and waveform maps. (B) Collinearity of GRF genes in pineapple and six representative species. Same-line blocks are set to a gray background, and collinear GRF genes are highlighted with red curves. (C) Venn diagram of non-redundant collinear GRF genes in pineapple and other species. (D) Box plot of Ka/Ks ratios of GRF orthologs.
To understand the ancestral relationships among AcGRF genes and GRF genes from other species, we examined collinear relationships among genes from pineapple and two core eudicots (A. thaliana, V. vinifera), two basal angiosperms (A. trichopoda, N. colorata), and five core monocots (O. sativa, S. bicolor, P. equestris, M. acuminata, and S. polyrhiza). Four GRF genes from A. thaliana had collinear relationships with AcGRF genes from pineapple; the numbers of collinear genes in other species were 7 in V. vinifera, 14 in M. acuminata, 9 in O. sativa, 6 in S. bicolor, 9 in S. polyrhiza, 2 in P. equestris, and 3 in N. colorata (Table S4; Figures 3B, C). AcGRF2 had collinear relationships with gene(s) in all species except N. colorata; the collinear block in which AcGRF1 is located is present in species other than rice (O. sativa) and S. bicolor, and the collinear block in which AcGRF4 is located is present in species except A. thaliana and P. equestris. Interestingly, AcGRF5-related collinear blocks were found only in O. sativa, and AcGRF3-related blocks were found only in water lilies (N. colorata). We next calculated the Ka/Ks (non-synonymous/synonymous substitution ratio) values of AcGRF genes and their orthologs in nine other species (Tables S4, S5; Figure 3D). All AcGRF orthologous gene pairs had Ka/Ks values less than 1, indicating that the AcGRF gene family has experienced strong purifying selection during evolution.
3.5 Promoter analysis and expression of AcGRF genes
Gene promoters interact with DNA or regulatory proteins to control gene expression (Biłas et al., 2016). We extracted promoter sequences from the pineapple GRF genes (2,000 bp upstream of the start codon) and identified their cis-acting elements. A total of 156 cis-elements were detected in the promoter regions of the AcGRF genes and were divided into three classes (Figures 4A, B and Table S6). Hormone response elements included those responsive to IAA (4; 2.56%), GA (4; 2.56%), ABA (14; 8.97%), JA (10; 6.41%), and salicylic acid (5; 3.21%). With the exception of subfamily III members (AcGRF1/2), the promoter regions of all other subfamily members contained JA and ABA response elements. Growth- and development-related elements included those related to meristem expression (3; 1.92%; present in AcGRF1 and AcGRF8), endosperm expression (4; 2.56%; present in AcGRF1/2/7), light response (72; 46.15%; present in all AcGRF genes), and circadian rhythm control (1; 0.64%; present in AcGRF4). Stress response elements included those related to drought induction (5; 3.21%), low temperature response (2; 1.28%), and anaerobic induction (25; 16.03%). Anaerobic induction elements were present in all genes except AcGRF5, and other response elements were unevenly distributed among the AcGRFs.
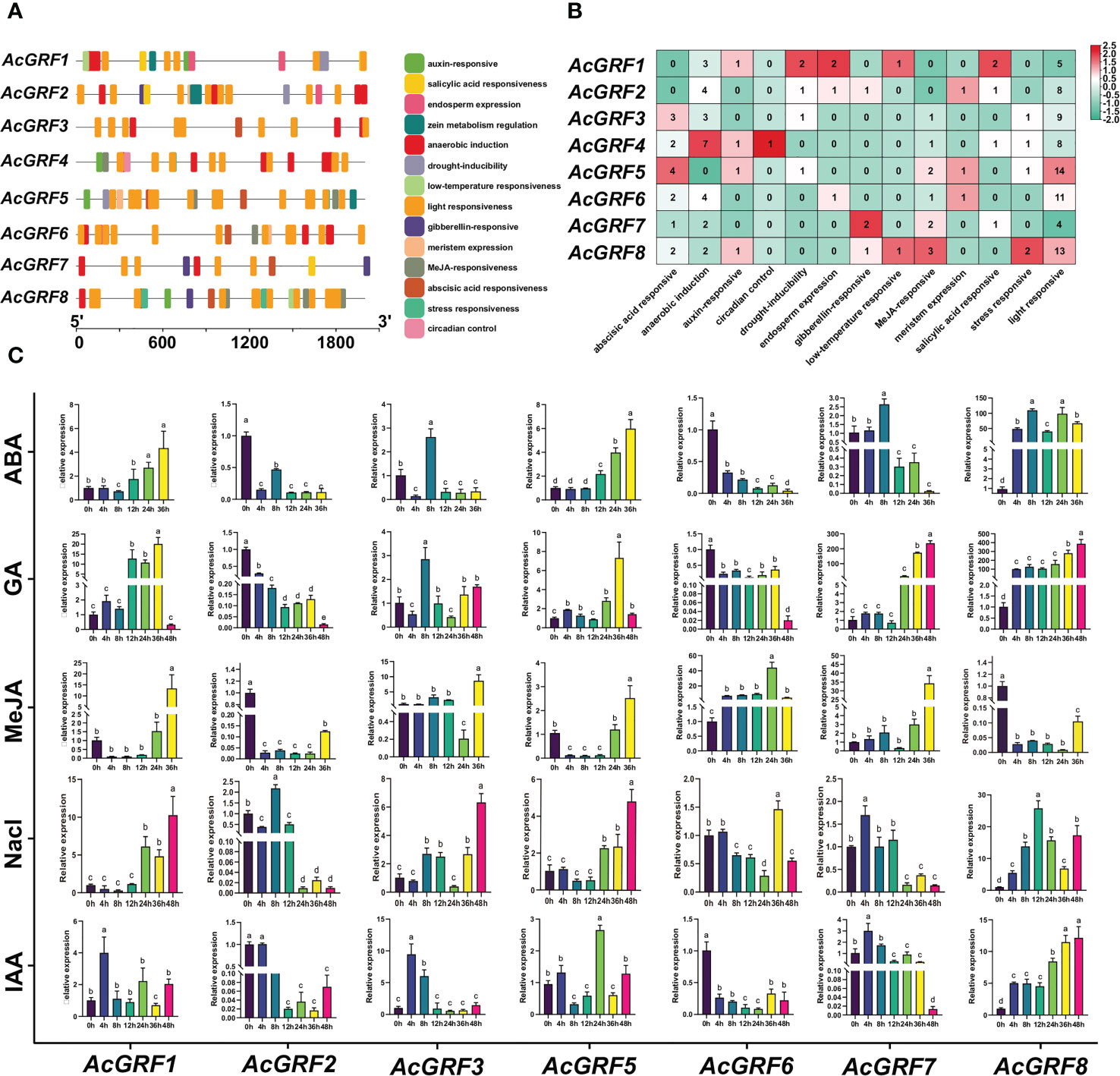
Figure 4 cis-Elements in AcGRF gene promoters and AcGRF expression profiles under hormone and salt stress treatments. (A) cis-Acting elements in the 2,000-bp region upstream of the AcGRF genes. (B) Distribution of cis-Acting elements of the AcGRF genes. (C) AcGRF gene expression profiles under different hormone and salt stress treatments. Data were normalized to the expression of β-actin and expressed as mean ± standard deviation. Different lowercase letters indicate significant differences between treatment times (P < 0.01, F test).
Studies have reported that GRF genes have a positive regulatory effect on callus proliferation (Kong et al., 2020), and GRF genes regulate plant growth and development by regulating GA, IAA, and ABA (Kim et al., 2012; Chen Y. et al., 2020; Huang et al., 2022). To investigate whether the expression of AcGRF genes is affected by abiotic stress and hormone treatments, we measured the expression of seven AcGRF family members by qRT-PCR after exposure to different hormones or 150 mM NaCl (Figure 4C). AcGRF1 expression was induced by all tested treatments. The expression of AcGRF1 and AcGRF5 gradually increased with ABA exposure time, whereas that of AcGRF6 showed the opposite trend. GA significantly induced AcGRF1, AcGRF7, and AcGRF8 expression and inhibited that of AcGRF2 and AcGRF6. JA significantly induced the expression of AcGRF6. IAA significantly induced the expression of AcGRF8 but inhibited that of AcGRF6. Most AcGRF genes increased in expression with increasing duration of NaCl exposure. AcGRF8 had the highest expression levels during ABA and GA treatment, with relative expression levels of 110 and 387, respectively, and AcGRF7 had a relative expression level of 237 after GA treatment. These findings suggest potential roles for AcGRF genes under different hormone treatments and stress conditions.
We next analyzed the expression profiles of GRF genes at different developmental stages of pineapple tissues using our previously reported transcriptome data (Mao et al., 2018) (Figure 5 and Table S7). The analysis included roots, stems, leaves, petals, stamens, pistils, discs, peduncles, ovules, ovaries, fruit hearts, bracts, sepals, and placenta (Figure 5A). In general, GRF genes from the same clade tended to exhibit similar expression patterns. Subfamily I members AcGRF6 and AcGRF7 showed a similar expression in nearly all tissues tested. Interestingly, almost all AcGRF genes were relatively highly expressed in the early stages of floral organ development (Figure 5A), with the exception of AcGRF5, which was relatively highly expressed in the late developmental stages of most tissues.
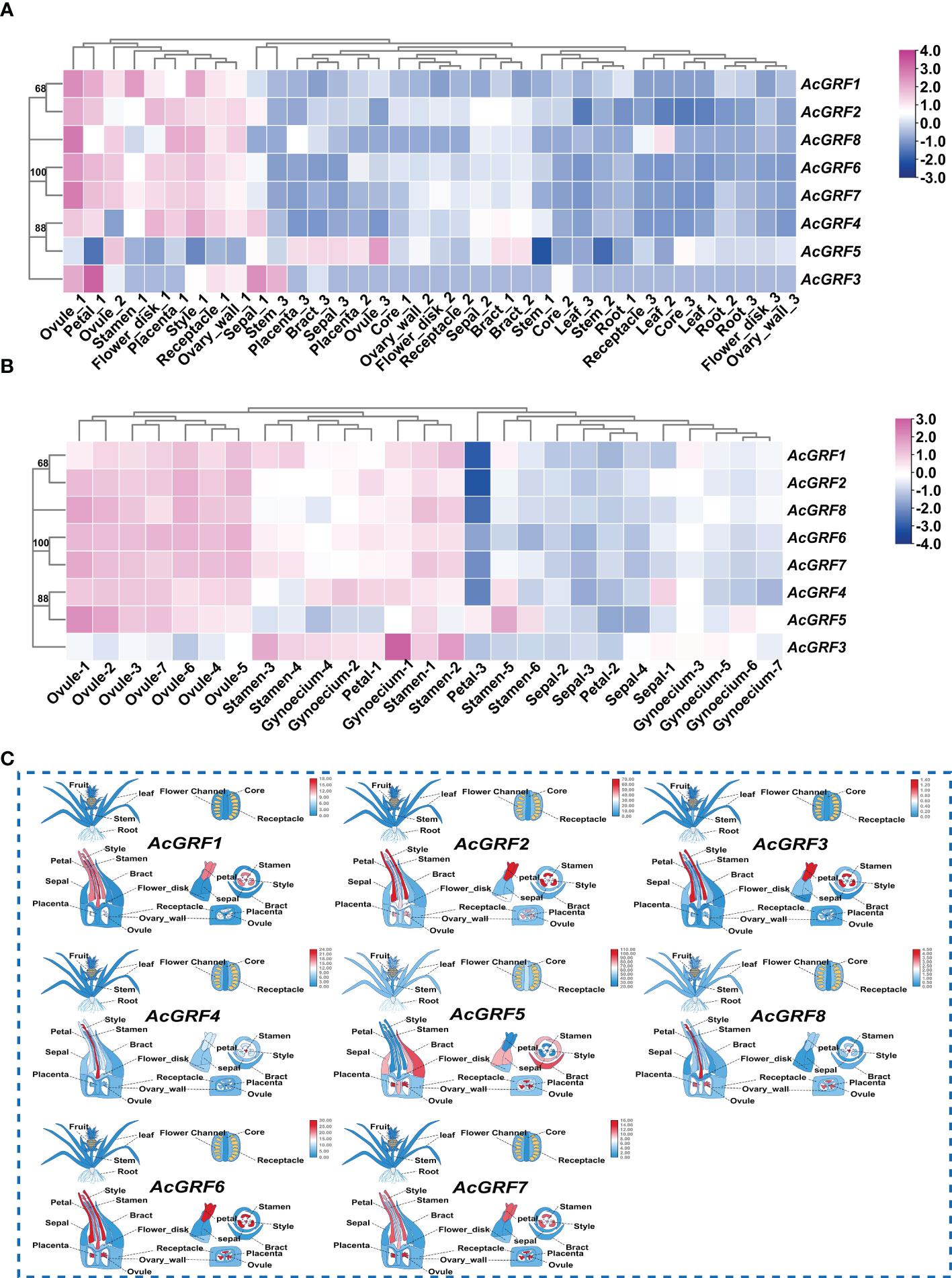
Figure 5 Expression of AcGRF genes in different tissues of pineapple. (A) Clustered heat map showing the expression patterns of AcGRF genes in various tissues at different developmental stages. (B) Expression heat map of AcGRF genes in five floral organs (stamens, pistils, ovules, sepals, and petals) of pineapple at different developmental stages: four stages of sepals (S1–S4), three of petals (S1–S3), five of stamens (S1–S5), and seven of pistils (S1–S7) and ovules (S1–S7). Relative gene expression is quantified as log2(FPKM+1). Blue, white, and red indicate low, medium, and high expression levels. (C) Cartoon heatmap showing the expression patterns of AcGRF genes from different clades in different tissues.
To determine whether individual AcGRF genes are associated with specific developmental stages of pineapple floral organs, we analyzed transcriptome data from five floral organs (sepals, petals, stamens, pistils, and ovules) at different developmental stages (Wang et al., 2020). Almost all AcGRF genes showed the highest expression in ovules at all stages of development, followed by stamens; the only exception was AcGRF3, which was specifically expressed in pistils (Figure 5B and Table S8).
To better visualize the different expression patterns of AcGRF genes in various tissues, we created cartoon heatmaps of relevant gene expression from all pineapple GRFs (Figure 5C). Subfamily III member AcGRF1 was highly expressed in petals, stamens, and pistils. AcGRF2 was highly expressed in petals and ovules, and subfamily VI member AcGRF8 was expressed at higher levels in pistils and ovules. Subfamily I members AcGRF6 and AcGRF7 were highly expressed in petals, pistils, and ovules. AcGRF4 from subfamily V was highly expressed in pistils, and AcGRF5 was highly expressed in bracts, sepals, and ovules. Subfamily IV member AcGRF3 showed relatively high expression in petals. The expression patterns of AcGRF genes thus differ among pineapple tissues, suggesting their potential regulatory roles in pineapple development.
3.6 Predicted protein interaction network of AcGRFs
A predicted protein interaction network indicated that AcGRFs had multiple interaction partners (Figure 6A), including growth regulator interaction factor 1 (GIF1), the TCP (teosinte branched1/cincinnata/proliferating cell factor) transcription factor PCF5, and the YABBY transcription factor YAB2. With the exception of AcGRF1/3/4, all other members interacted with one or more proteins. AcGRF2/5/6/7/8 were predicted to interact with 6, 2, 5, 4, and 2 proteins, respectively. There were also predicted interactions between the AcGRF proteins themselves.
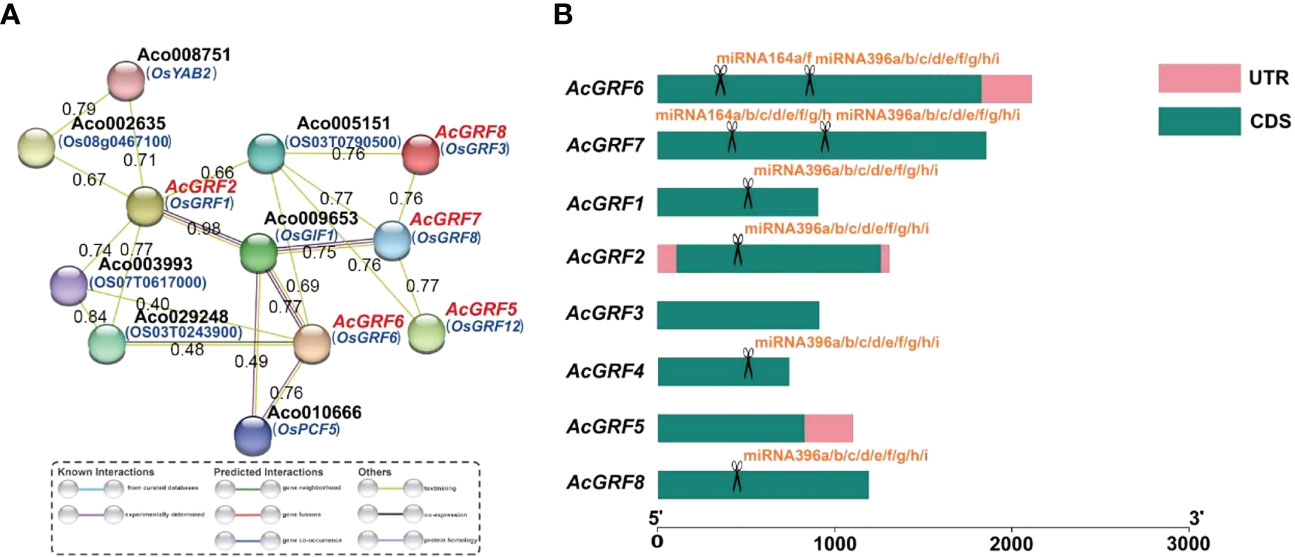
Figure 6 Predicted protein interaction network of AcGRF proteins and miRNA target sites in AcGRF genes. (A) Protein interaction network predicted using AcGRF orthologs from rice. (B) Predicted miRNA targets in the AcGRF genes. The scissors represent the miRNA and its targeted AcGRF gene, with penalty score ≤5 and lower expectations indicating higher prediction accuracy.
We next performed miRNA target site prediction for the AcGRF genes. As shown in Figure 6B, six AcGRF genes were predicted to be targeted by two miRNAs (miRNA164/miRNA396). AcGRF6/7 contained targets for both miRNAs, whereas AcGRF1/2/4/8 contained targets for miRNA396 only (Table S9). Interestingly, all miRNA target sites were located in the CDS regions.
4 Discussion
GRF proteins regulate important processes in plant biology, including leaf growth, floral organ development, root development, seed oil content, plant lifespan, and stress response (Omidbakhshfard et al., 2015). Pineapple is the third most important tropical fruit crop in the world after bananas and citrus, and various biotic and abiotic stresses have a significant impact on its yield and quality. Although GRF genes have been studied in other plant species, they have not yet been characterized in the bromeliad family. We therefore identified the GRF genes in pineapple, analyzed their evolutionary relationships, compared their sequence features, and analyzed their expression patterns in response to hormone and salt stress treatments.
4.1 Identification, classification, and characteristics of the AcGRF genes
We identified eight AcGRF proteins from the pineapple genome and assigned them to five subfamilies on the basis of phylogenetic analysis (Figures 2A, C). Pineapple has fewer GRF genes than the monocots banana (18) and maize (18) but a similar number to Setaria italica and S. bicolor. This suggests that GRF family expansion in monocots may be associated mainly with lineage-specific WGD events. Pineapple contained members of five of the six major GRF subfamilies, similar to rice (four major subfamilies), banana (5), B. distachyon (4), A. thaliana (5), soybean (5), and B. rapa (5) and more than non-angiosperm species such as M. polymorpha (0), S. moellendorffii (0), and Pinus taeda (1). Notably, genes that fell outside the main subfamilies were found only in algae, ferns, gymnosperms, basal angiosperms, and Brachypodium distachyon; they may have been preserved from more ancient duplication events or acquired only in specific clades. Among monocots, there were five GRF subfamilies in banana (missing subfamily IV) and pineapple (missing subfamily II) and four subfamilies in other species (missing both subfamily II and subfamily IV). By contrast, all dicots except soybean (missing subfamily V) contained members of all six subfamilies. These results suggest that the preservation of specific subfamilies may vary among species with different evolutionary histories. Several genomic evolutionary models have been proposed in model species on the basis of comparative genomic analysis (Wolfe and Shields, 1997; Moore and Purugganan, 2003; Hurley et al., 2005). If duplicated genes are retained, they tend to diverge in their regulatory and coding regions, and differences in coding regions, especially those that alter gene function, may cause amino acid changes or substitutions or changes in exon/intron structure (Xu et al., 2012). In general, proteins from the same family have relatively conserved intron/exon structures, such as β-expansins in A. thaliana and rice (Lee and Kende, 2001). However, the number and length of introns are not conserved in GRF genes from pineapple, A. thaliana, and rice (Figure 1C), even within the same subfamily, and GRF genes are randomly distributed in the genome (Table 1 and Figure 1B). This suggests that no recent duplication events have occurred in the GRF gene family and that expansions in this gene family reflect early duplication events.
Gain and loss of exons/introns and differences in exon/intron length can result from chromosome rearrangement and fusion (Zan et al., 2020). In pineapple, AcGRF2 contains three introns, whereas its paralog AcGRF1 contains only one intron. Likewise, intron lengths were much greater in AcGRF5 than in its paraphyletic homologous gene AcGRF4. We therefore speculate that differences in intron/exon length or number may have led to different biological functions for AcGRF4 and AcGRF5. Previous studies have reported that the N-terminal QLQ domain is involved in protein–protein interactions (Kim et al., 2003; Kim and Kende, 2004). Here, we found that the QLQ motif of AcGRF5 is actually Gln-Met-Gln, with a Met substituted for Leu (Figure 1A), similar to that of A. thaliana AtGRF9 in which Phe is substituted for Leu (Kim et al., 2003). The mutated QLQ domain of AcGRF5 may lead to alterations in its protein interaction activity.
4.2 Evolution of the AcGRF genes
Gene duplication, including tandem, segmental, and whole-genome duplication, is one of the main drivers of genome evolution (Moore and Purugganan, 2003), and most seed plants have experienced one or more WGD events in their evolutionary history (Rensing et al., 2007). Pineapple has undergone at least two WGD events (Ming et al., 2015; Ming et al., 2016) (Figure S3), and its gene family expansion reflects the effects of WGDs, as well as tandem and segmental duplications. In this study, we analyzed the presence of GRF genes in 26 species, including algae, mosses, angiosperms, and gymnosperms (Figures 2B, C). GRF genes were present in all species except algae, suggesting that GRF genes appeared in land plants and may have supported their terrestrial adaptation. Two of the eight pineapple GRF genes were associated with segmental duplication events (Figure 3A), consistent with findings in wheat that 26 of 30 GRF genes were associated with such events (Zan et al., 2020). This suggests that segmental duplication may have played an important role in the early expansion of the GRF gene family. The number of AcGRF genes was significantly lower in pineapple than in banana (18) compared with other plants used for collinearity analysis, and it was similar to the number of GRF genes in other plant species (Osnato et al., 2010; Kim et al., 2012; Debernardi et al., 2014; Kuijt et al., 2014; Omidbakhshfard et al., 2015). The large number of banana GRFs may thus reflect banana-specific duplication events. Notably, there was no significant correlation between the number of GRF genes and genome size. For example, A. thaliana has nine GRFs and pineapple has eight, despite the pineapple genome being 3.2 times the size (375 Mb) (Ming et al., 2015) of the A. thaliana genome (115 Mb) (Hou et al., 2022). After identifying non-redundant GRF genes that showed collinear relationships between pineapple and nine other species, we found that an AcGRF5 ortholog was present only in pineapple and rice and an AcGRF3 ortholog was present only in pineapple and water lily (Figure 3C). This suggests that the retention of gene family members can vary among species with different evolutionary histories. All AcGRF orthologous genes had Ka/Ks values less than 1 (Figure 3D), indicating that GRFs experienced strong purifying selection during evolution.
4.3 Inference of biological functions of AcGRF genes
Because of the importance of pineapple as a tropical fruit crop and ornamental plant, the mechanisms that regulate its flower and fruit development are of significant interest. The biological functions of pineapple AcGRF genes remain to be clarified, but identification of their presumed orthologs in different species can provide insight into their functions (Nehrt et al., 2011). GRF genes have previously been studied in other species to characterize their regulatory network and understand their functions in a wide range of biological processes (Omidbakhshfard et al., 2015). Here, we used GRFs with known functions in other species to infer potential functions of their pineapple orthologs (Figure S4 and Figure 6). Clade 2 includes AcGRF1/2, AtGRF6, and OsGRF1. Previous studies have shown that AtGRF6 and OsGRF1 have a role in controlling leaf size (Kim et al., 2003; Luo et al., 2005) and OsGRF1 also influences rice stem elongation, juvenile growth, and panicle extension (Van der Knaap et al., 2000; Luo et al., 2005). AcGRF1/2 in the same clade were highly expressed in early developmental stages of petals and ovules, suggesting that they may have similar functions in pineapple. ZaGRF6, OsGRF3, and AcGRF8 were included in clade 3. Heterologous overexpression of ZaGRF6 increased branching and chlorophyll synthesis and delayed aging in transgenic tobacco, and ectopic overexpression of OsGRF3 in rice reduced tiller numbers and induced the formation of ectopic roots and shoots on the nodes (Kuijt et al., 2014; Zhang et al., 2021). Promoter analysis revealed the presence of two ABA response elements and one gibberellin response element in the AcGRF8 gene promoter, and qRT-PCR results showed that AcGRF8 expression responded significantly to ABA and GA treatment. These results suggest that AcGRF8 may regulate pineapple aging and root and shoot formation through the abscisic acid and gibberellin pathways. Clade 5 included AcGRF4/5, AtGRF9, OsGRF10, and ZmGRF10. In previous studies, overexpression of ZmGRF10 in maize led to reductions in leaf size and plant height, and knockout and overexpression of AtGRF9 made petals and other organs of A. thaliana larger and smaller, respectively (Wu et al., 2014; Omidbakhshfard et al., 2018). Here, AcGRF4 was preferentially expressed in pistils and AcGRF5 in bracts, sepals, and ovules. AcGRF4 and AcGRF5 have IAA response elements, and their expression responded to increasing durations of IAA treatment. We therefore suggest that AcGR4/5 may participate in the regulation of floral organ and leaf size, plant height, and other traits through the auxin pathway. Clade 6 included AcGRF3, AtGRF7, and AtGRF8. Leaves of an AtGRF7 single-allele mutant were reported to be smaller than those of the wild type (Kim et al., 2012), but this phenomenon was not observed by Lee et al. (2022). AtGRF8 has been reported to positively regulate cell proliferation (Tsukaya, 2021). We found that AcGRF3 was preferentially expressed in petals and contained GA-responsive elements in its promoter. We therefore speculate that AcGRF3 may regulate pineapple cell proliferation, leaf size, and petal growth through the GA pathway. Clade 8 included AcGRF6/7, PagGRF15, and OsGRF7/8. PagGRF15 regulates leaf size through its effects on cell expansion during poplar leaf development, and OsGRF7/8 influence plant structure and leaf growth by regulating gibberellin and indole-3-acetic acid metabolism. Here, AcGRF6 and AcGRF7 were preferentially expressed in petals, pistils, and ovules. qRT-PCR experiments showed that GA significantly induced the expression of AcGRF7 but inhibited that of AcGRF6, and IAA significantly inhibited AcGRF6. AcGRF6/7 may therefore regulate pineapple flower and leaf development and plant structure through GA and IAA pathways.
5 Conclusion
We identified eight GRF gene family members on seven chromosomes of the pineapple genome and classified them into five of six known subfamilies on the basis of phylogenetic analysis. In contrast to other gene families, the GRFs showed little conservation of gene structure and motif composition. Collinearity analysis showed that early segmental duplications promoted expansion of the pineapple GRF gene family, and purifying selection was the main force acting on the GRF genes. The paralogs AcGRF1 and AcGRF2 had different expression profiles in different floral organs, perhaps related to their differences in structure. Transcriptome data suggested that all AcGRF genes were involved in regulation of floral organ development, and AcGRF1/2/3/6/7 may have functionally redundant roles in petal development. The AcGRF promoters contained cis-acting elements involved in hormone response, developmental regulation, and stress response, and changes in expression of AcGRFs under ABA, GA, IAA, JA, and NaCl treatments may reflect their different regulatory effects. Protein interactions between AcGRF proteins or with YABBY, TCP, and AcGRF-miRNA may contribute to the formation of more complex regulatory networks. These results support further research on the functions and regulatory mechanisms of AcGRF transcription factors during pineapple growth and development.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YH, CC conceived the project. WY and JW performed the experiments. WY, AL, JW, CL and WZ performed data analysis. WY and AL wrote the original manuscript draft. All authors participated in discussion and revised the manuscript. All authors reviewed and approved the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China(32272677, 31901355), Special Project of Rural Revitalization Strategy in Guangdong Province(2022-NPY-00-031), the National Key R&D Program of China (2018YFD1000500).
Acknowledgments
We thank Guiyang Watch Biotechnology for their advice on gene family data analyses. TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript. The authors thank lab members for their assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1159223/full#supplementary-material
References
Arvidsson, S., Pérez-Rodríguez, P., Mueller-Roeber, B. (2011). A growth phenotyping pipeline for arabidopsis thaliana integrating image analysis and rosette area modeling for robust quantification of genotype effects. New Phytol. 191, 895–907. doi: 10.1111/j.1469-8137.2011.03756.x
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bao, M., Bian, H., Zha, Y., Li, F., Sun, Y., Bai, B., et al. (2014). miR396a-mediated basic helix–Loop–Helix transcription factor bHLH74 repression acts as a regulator for root growth in arabidopsis seedlings. Plant Cell Physiol. 55, 1343–1353. doi: 10.1093/pcp/pcu058
Baruwa, O. I. (2013). Profitability and constraints of pineapple production in osun state, Nigeria. J. Hortic. Res. 21, 59–64. doi: 10.2478/johr-2013-0022
Biłas, R., Szafran, K., Hnatuszko-Konka, K., Kononowicz, A. K. (2016). Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Culture 127, 269–287. doi: 10.1007/s11240-016-1057-7
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-prot, the manually annotated section of the UniProt KnowledgeBase: How to use the entry view. Plant Bioinform.: Methods, 1374. 23–54. doi: 10.1007/978-1-4939-3167-5_2
Casadevall, R., Rodriguez, R. E., Debernardi, J. M., Palatnik, J. F., Casati, P. (2013). Repression of growth regulating factors by the MicroRNA396 inhibits cell proliferation by UV-b radiation in arabidopsis leaves. Plant Cell 25, 3570–3583. doi: 10.1105/tpc.113.117473
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y., Dan, Z., Gao, F., Chen, P., Fan, F., Li, S. (2020). Rice GROWTH-REGULATING FACTOR7 modulates plant architecture through regulating GA and indole-3-Acetic acid metabolism. Plant Physiol. 184, 393–406. doi: 10.1104/pp.20.00302
Chen, C., Wu, Y., Xia, R. (2022). A painless way to customize circos plot: From data preparation to visualization using TBtools. iMeta 1 (3), e35. doi: 10.1002/imt2.35
Cheng, C.-Y., Krishnakumar, V., Chan, A. P., Thibaud-Nissen, F., Schobel, S., Town, C. D. (2017). Araport11: A complete reannotation of the arabidopsis thaliana reference genome. Plant J. 89, 789–804. doi: 10.1111/tpj.13415
Choi, D., Kim, J. H., Kende, H. (2004). Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa l.). Plant Cell Physiol. 45, 897–904. doi: 10.1093/pcp/pch098
Dai, X., Zhuang, Z., Zhao, P. X. (2018). psRNATarget: A plant small RNA target analysis server, (2017 release). Nucleic Acids Res. 46, W49–W54. doi: 10.1093/nar/gky316
Debernardi, J. M., Mecchia, M. A., Vercruyssen, L., Smaczniak, C., Kaufmann, K., Inze, D., et al. (2014). Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 79, 413–426. doi: 10.1111/tpj.12567
Debernardi, J. M., Tricoli, D. M., Ercoli, M. F., Hayta, S., Ronald, P., Palatnik, J. F., et al. (2020). A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279. doi: 10.1038/s41587-020-0703-0
Emms, D. M., Kelly, S. (2019). OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14. doi: 10.1186/s13059-019-1832-y
Hou, X., Wang, D., Cheng, Z., Wang, Y., Jiao, Y. (2022). A near-complete assembly of an arabidopsis thaliana genome. Mol. Plant 15, 1247–1250. doi: 10.1016/j.molp.2022.05.014
Huang, Y., Chen, J., Li, J., Li, Y., Zeng, X. (2022). Genome-wide identification and analysis of the growth-regulating factor family in zanthoxylum armatum DC and functional analysis of ZaGRF6 in leaf size and longevity regulation. Int. J. Mol. Sci. 23, 9043. doi: 10.3390/ijms23169043
Hurley, I., Hale, M. E., Prince, V. E. (2005). Duplication events and the evolution of segmental identity. Evol. Dev. 7, 556–567. doi: 10.1111/j.1525-142x.2005.05059.x
Kim, J. H., Choi, D., Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313x.2003.01862.x
Kim, J. H., Kende, H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in arabidopsis. Proc. Natl. Acad. Sci. U. States America 101, 13374–13379. doi: 10.1073/pnas.0405450101
Kim, J. H., Lee, B. H. (2006). GROWTH-REGULATING FACTOR4 of arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 49, 463–468. doi: 10.1007/bf03031127
Kim, J.-S., Mizoi, J., Kidokoro, S., Maruyama, K., Nakajima, J., Nakashima, K., et al. (2012). Arabidopsis GROWTH-REGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid– and osmotic stress–responsive genes, including DREB2A. Plant Cell 24, 3393–3405. doi: 10.1105/tpc.112.100933
Kong, J., Martin-Ortigosa, S., Finer, J., Orchard, N., Gunadi, A., Batts, L. A., et al. (2020). Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocot species. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.572319
Kuijt, S. J. H., Greco, R., Agalou, A., Shao, J., ‘t Hoen, C. C. J., Övernäs, E., et al. (2014). Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 164, 1952–1966. doi: 10.1104/pp.113.222836
Lee, Y., Kende, H. (2001). Expression of β-expansins is correlated with internodal elongation in deepwater rice. Plant Physiol. 127, 645–654. doi: 10.1104/pp.010345
Lee, G.-H., Lee, B. H., Jung, J.-H., Lee, S.-J., Mai, T.-T., Kim, J. H. (2022). Systematic assessment of the positive role of arabidopsis thaliana GROWTH-REGULATING FACTORs in regulation of cell proliferation during leaf growth. J. Plant Biol. 65, 413–422. doi: 10.1007/s12374-022-09366-1
Lee, S.-J., Lee, B. H., Jung, J.-H., Park, S. K., Song, J. T., Kim, J. H. (2017). Growth-regulating factor and grf-interacting factor specify meristematic cells of gynoecia and anthers. Plant Physiol. 176, 717–729. doi: 10.1104/pp.17.00960
Lescot, M. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Levine, M., Tjian, R. (2003). Transcription regulation and animal diversity. Nature 424, 147–151. doi: 10.1038/nature01763
Liang, G., He, H., Li, Y., Wang, F., Yu, D. (2013). Molecular mechanism of microRNA396 mediating pistil development in arabidopsis. Plant Physiol. 164, 249–258. doi: 10.1104/pp.113.225144
Liu, H., Guo, S., Xu, Y., Li, C., Zhang, Z., Zhang, D., et al. (2014). OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 165, 160–174. doi: 10.1104/pp.114.235564
Liu, J., Rice, J. H., Chen, N., Baum, T. J., Hewezi, T. (2014). Synchronization of developmental processes and defense signaling by growth regulating transcription factors. PloS One 9, e98477. doi: 10.1371/journal.pone.0098477
Lobo, M., Paull, R. (2017). Handbook of pineapple technology: production, postharvest science, processing and nutrition (John Wiley & Sons).
Luo, A.-D., Liu, L., Tang, Z.-S., Bai, X.-Q., Cao, S.-Y., Chu, C.-C. (2005). Down-regulation of OsGRF1 gene in rice rhd1 mutant results in reduced heading date. J. Integr. Plant Biol. 47, 745–752. doi: 10.1111/j.1744-7909.2005.00071.x
Lynch, M., Conery, J. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U. States America 102, 5454–5459. doi: 10.1073/pnas.0501102102
Mao, Q., Chen, C., Xie, T., Luan, A., Liu, C., He, Y. (2018). Comprehensive tissue-specific transcriptome profiling of pineapple (Ananas comosus) and building an eFP-browser for further study. PeerJ 6, e6028. doi: 10.7717/peerj.6028
Ming, R., VanBuren, R., Wai, C. M., Tang, H., Schatz, M. C., Bowers, J. E., et al. (2015). The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 47, 1435–1442. doi: 10.1038/ng.3435
Ming, R., Wai, C. M., Guyot, R. (2016). Pineapple genome: A reference for monocots and CAM photosynthesis. Trends Genet. 32, 690–696. doi: 10.1016/j.tig.2016.08.008
Moore, R. C., Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. U. States America 100, 15682–15687. doi: 10.1073/pnas.2535513100
Nehrt, N. L., Clark, W. T., Radivojac, P., Hahn, M. W. (2011). Testing the ortholog conjecture with comparative functional genomic data from mammals. PloS Comput. Biol. 7, e1002073. doi: 10.1371/journal.pcbi.1002073
Omidbakhshfard, M. A., Fujikura, U., Olas, J. J., Xue, G.-P., Balazadeh, S., Mueller-Roeber, B. (2018). GROWTH-REGULATING FACTOR 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PloS Genet. 14, e1007484. doi: 10.1371/journal.pgen.1007484
Omidbakhshfard, M. A., Proost, S., Fujikura, U., Mueller-Roeber, B. (2015). Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 8, 998–1010. doi: 10.1016/j.molp.2015.01.013
Osnato, M., Stile, M. R., Wang, Y., Meynard, D., Curiale, S., Guiderdoni, E., et al. (2010). Cross talk between the KNOX and ethylene pathways is mediated by intron-binding transcription factors in barley. Plant Physiol. 154, 1616–1632. doi: 10.1104/pp.110.161984
Pajoro, A., Madrigal, P., Muiño, J. M., Matus, J., Jin, J., Mecchia, M. A., et al. (2014). Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 15, R41. doi: 10.1186/gb-2014-15-3-r41
Pu, C.-X., Ma, Y., Wang, J., Zhang, Y.-C., Jiao, X.-W., Hu, Y.-H., et al. (2012). Crinkly4 receptor-like kinase is required to maintain the interlocking of the palea and lemma, and fertility in rice, by promoting epidermal cell differentiation. Plant J. 70, 940–953. doi: 10.1111/j.1365-313x.2012.04925.x
Rensing, S. A., Ick, J., Fawcett, J. A., Lang, D., Zimmer, A., Van de Peer, Y., et al. (2007). An ancient genome duplication contributed to the abundance of metabolic genes in the moss physcomitrella patens. BMC Evol. Biol. 7, 130. doi: 10.1186/1471-2148-7-130
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C.-Z., Keddie, J., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Sanewski, G., Bartholomew, D., Paull, R. (2018). The pineapple: botany, production and uses (CAB International).
Tsukaya, H. (2021). The leaf meristem enigma: The relationship between the plate meristem and the marginal meristem. Plant Cell 33, 3194–3206. doi: 10.1093/plcell/koab190
Van Bel, M., Diels, T., Vancaester, E., Kreft, L., Botzki, A., Van de Peer, Y., et al. (2017). PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46, D1190–D1196. doi: 10.1093/nar/gkx1002
Van der Knaap, E., Kim, J. H., Kende, H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122, 695–704. doi: 10.1104/pp.122.3.695
Wang, L., Li, Y., Jin, X., Liu, L., Dai, X., Liu, Y., et al. (2020). Floral transcriptomes reveal gene networks in pineapple floral growth and fruit development. Commun. 3, 500. doi: 10.1038/s42003-020-01235-2
Wolfe, K., Shields, D. (1997). Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713. doi: 10.1038/42711
Wu, L., Zhang, D., Xue, M., Qian, J., He, Y., Wang, S. (2014). Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 56, 1053–1063. doi: 10.1111/jipb.12220
Xu, G., Guo, C., Shan, H., Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U. States America 109, 1187–1192. doi: 10.1073/pnas.1109047109
Yusuf, N. H. M., Ong, W. D., Redwan, R. M., Latip, M. A., Kumar, S. V. (2015). Discovery of precursor and mature microRNAs and their putative gene targets using high-throughput sequencing in pineapple (Ananas comosus var. comosus). Gene 571, 71–80. doi: 10.1016/j.gene.2015.06.050
Zan, T., Zhang, L., Xie, T., Li, L. (2020). Genome-wide identification and analysis of the growth-regulating factor (GRF) gene family and GRF-interacting factor family in triticum aestivum l. Biochem. Genet. 58, 705–724. doi: 10.1007/s10528-020-09969-8
Zhang, L., Chen, F. (2019). The water lily genome and the early evolution of flowering plants. Nature 577, 79–84. doi: 10.1038/s41586-019-1852-5
Zhang, G.-Q., Liu, K.-W., Li, Z., Lohaus, R., Hsiao, Y.-Y., Niu, S.-C., et al. (2017). The apostasia genome and the evolution of orchids. Nature 549, 379–383. doi: 10.1038/nature23897
Keywords: pineapple, GRF gene family, phylogeny analysis, flower organ, expression analysis
Citation: Yi W, Luan A, Liu C, Wu J, Zhang W, Zhong Z, Wang Z, Yang M, Chen C and He Y (2023) Genome-wide identification, phylogeny, and expression analysis of GRF transcription factors in pineapple (Ananas comosus). Front. Plant Sci. 14:1159223. doi: 10.3389/fpls.2023.1159223
Received: 05 February 2023; Accepted: 17 March 2023;
Published: 14 April 2023.
Edited by:
Eleni Tani, Agricultural University of Athens, GreeceReviewed by:
Dhananjay K. Pandey, Amity University, Jharkhand, IndiaErpei Lin, Zhejiang Agriculture and Forestry University, China
Copyright © 2023 Yi, Luan, Liu, Wu, Zhang, Zhong, Wang, Yang, Chen and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yehua He, heyehua@hotmail.com; Chengjie Chen, ccj0410@gmail.com
†These authors have contributed equally to this work
 Wen Yi
Wen Yi Aiping Luan2†
Aiping Luan2† Chaoyang Liu
Chaoyang Liu Chengjie Chen
Chengjie Chen Yehua He
Yehua He