- Institute of Plant Protection, Hebei Academy of Agricultural and Forestry Sciences, Integrated Pest Management Center of Hebei Province, Key Laboratory of IPM on Crops in Northern Region of North China, Ministry of Agriculture, Baoding, China
Fusarium pseudograminearum is a soil-borne pathogen that is capable of causing a highly destructive crown disease in wheat. Secondary metabolites (SMs), especially deoxynivalenol (DON), are the primary virulence factors during infection. Here, we characterised the global regulator FpLaeB, an orthologue of LaeB protein function, to regulate the SM in Aspergillus nidulans. Through the utility of the gene targeting approach, we found that the vegetative growth of the FpLaeB deletion mutant was drastically reduced compared to that of the wild type. FpLaeB was also important for conidiation because the FpLaeB deletion mutant formed fewer conidia in induced medium. In addition, the sensitivity of the FpLaeB deletion mutant to the cell wall integrity inhibitor was decreased, while its growth was more severely inhibited by the cell membrane inhibitor sodium dodecyl sulfate (SDS) than that of the wild type. More importantly, the virulence was decreased when the FpLaeB deletion mutant was inoculated onto the wheat stem base or head. Through genome-wide gene expression profiling, FpLaeB was found to regulate several processes related to the above phenotypes such as the carbohydrate metabolic process, which is an integral and intrinsic component of membranes, especially SMs. Furthermore, the generation of DON was impaired in the FpLaeB deletion mutant via ultraperformance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) assay. These results showed that FpLaeB plays an important role in the growth, development, and maintenance of the cell wall, and in membrane integrity. More importantly, FpLaeB is required for SMs and full virulence in F. pseudograminearum.
Introduction
The soil-borne pathogen Fusarium pseudograminearum is capable of causing Fusarium crown rot, a highly destructive worldwide disease resulting in yield losses of up to 10%–35% in a normal year in Australia and the Northwestern United States (Smiley et al., 2005; Murray and Brennan, 2009). In particular, this chronic disease is an increasing concern in the Huanghuai region of China including Henan, Hebei, and Shandong provinces (Li et al., 2012; Ji et al., 2016). The colonisation of F. pseudograminearum seems to occur at the coleoptile. Then, the infectious growth spreads to leaf sheaths and subcrown internodes with extensive browning. Severely diseased plants may result in white heads containing either no or shrivelled grains (Kazan and Gardiner, 2018). Similar to other Fusaria, some secondary metabolites (SMs) like the trichothecene toxin deoxynivalenol (DON) can contribute to the virulence in F. pseudograminearum (Monds et al., 2005; Tunali et al., 2012; Powell et al., 2017). Thus, the analyses of genes or regulation related to SM could reveal potential roles for the development or pathogenic life cycles of F. pseudograminearum. Fungal SM biosynthesis has been regulated in a complex process. The different regulations include signal transduction pathways, epigenetic modifications, and pathway-specific and global regulators (Brakhage, 2013). Global regulators including response to ambient light, carbon and nitrogen sources, and pH have been identified in several fungi (Chen et al., 2019). LaeA is a global regulator for sterigmatocystin and penicillin biosynthesis found in Aspergillus nidulans (Bok and Keller, 2004; Bayram et al., 2008). In addition, the regulation of secondary metabolism by LaeA has been characterised in other fungi, such as gliotoxin biosynthesis in A. fumigatus and lovastatin biosynthesis in A. terreus (Brakhage, 2013). In Fusarium graminearum, the expression of seven TRI genes was reduced in the FgLaeA deletion mutant. The accumulation of 15A-DON was abolished as well. The deletion of FgLaeA also leads to a 30-fold reduction of Zearalenone (Hee-Kyoung et al., 2013). Recently, a new global regulator, LaeB, involved in regulating sterigmatocystin production similar to LaeA, was identified using a forward genetic screening in A. nidulans. The LaeB protein contains a transcription initiation factor IIA (TFIIA) domain and a G-protein pathway suppressor domain. The two domains have low homology (Pfannenstiel et al., 2017). The LaeB deletion mutant exhibited a clear colour change compared to the wild type in A. nidulans. The majority of metabolites decreased or disappeared in the LaeB deletion mutant comprising the recipient strain. Meanwhile, some newly produced compounds were detectable (Lin et al., 2018). All these results suggested that most SM gene clusters should be regulated by LaeB in A. nidulans.
In light of the regulation effects of LaeB on SMs in A. nidulans, the biological functions of the plant pathogenic fungus F. pseudograminearum need to be determined to understand the intricate roles of SMs—important virulence factors that are regulated by its homologue—and in which manner this regulation may occur. Functional analysis of LaeB might provide a novel insight to understand the development and pathogenicity of F. pseudograminearum.
In this study, the effect of the LaeB orthologous gene FpLaeB in vegetative growth, conidiation, virulence, sensibility of abiotic stresses, and expression of SM genes was investigated in F. pseudograminearum. In addition to attenuated growth, FpLaeB has made a difference in conidiation and maintenance of cell wall and cell membrane integrity. Moreover, the FpLaeB gene disruption mutant drastically impaired virulence. FpLaeB was found to regulate the expression of genes related to the above phenotype such as the carbohydrate metabolic process and the integral and intrinsic component of membranes, especially SMs. The generation of DON was impaired in the FpLaeB deletion mutant as well. These results indicate that FpLaeB is involved in the growth, development, virulence, and SMs of F. pseudograminearum.
Materials and methods
Strains and growth conditions
The wild-type strain of F. pseudograminearum named 2035 was preserved by the Laboratory of Fungi Diseases in the Institute of Plant Protection, Hebei Academy of Agricultural and Forestry Sciences, PRC. The wild-type and mutant strains were activated and cultured on potato dextrose agar (PDA, 20% potato extract, 2% dextrose, and 1.5% agar) medium in this study.
The growth rates of different strains were expressed as colony radius per day on PDA medium at 25°C. For the conidiation assay, different strains were grown on carboxymethylcellulose sodium (CMC) medium for 4 days. A hemocytometer was used to determine the concentration of conidia (Chen et al., 2021). To assay stress responses, mutants and wild-type strains were grown on synthetic medium (STM) [0.05% yeast extract, 0.5% (NH4)2SO4, salts (0.15% KH2PO4, 0.06% CaCl2, and 0.06% MgSO4), and trace amounts of metals (0.0005% FeSO4 7H2O, 0.00016% MnSO4 H2O, 0.00037% CoCl2, and 0.00014% ZnSO4 7H2O)] containing NaCl (0.7 M), H2O2 (3 mM), Congo red (CR; 200 mg/L), or SDS (0.01%). Colony diameter was measured after incubation for 4 days. During gene deletion or complementarity, TB3 (0.3% yeast extract, 0.3% casamino acids, 20% sucrose, and 1.5% agar) medium mended with hygromycin B (250 μg/ml, Calbiochem, La Jolla, CA) or geneticin (250 μg/ml, Sigma, St. Louis, MO) has been used to select resistant transformants.
FpLaeB gene deletion and complementarity
The LaeB homology protein FpLaeB was identified via querying the F. pseudograminearum genomic sequence (GenBank accession NC_031951.1). The conserved domains of FpLaeB were predicted via the Conserved Domain Search Service (CD Search) in the National Center for Biotechnology Information (NCBI). The phylogenetic tree of FpLaeB and its homology proteins were constructed via the neighbour-joining method with the MEGA version 7.02 software package (Xia et al., 2021).
Open reading frame (ORF) was replaced by the hygromycin phosphotransferase gene to construct the deletion mutant of FpLaeB. Two primer pairs FpLaeB-1F/2R and FpLaeB-3F/4R were used to amplify the upstream and downstream flanking fragment of the FpLaeB gene. The hygromycin phosphotransferase (hph) gene was amplified via the primer pair HYG-F/R. The replacement fragment was constructed by joining the three fragments via double-joint polymerase chain reaction (PCR) (Yu et al., 2004). The FpLaeB replacement fragment was transformed into protoplasts of wild-type 2035 by the polyethylene glycol (PEG) approach (Liu and Friesen, 2012). Following screening by hygromycin, the transformants were screened and confirmed using PCR and Southern blot analyses, respectively (Tang et al., 2018). For complementation assays, XhoI-digested pFL2 and the FpLaeB fragments with promoters cotransformed into yeast strain XK1-25. FpLaeB–pFL2 plasmid was constructed by the yeast gap repair method (Zhang et al., 2017). Then, FpLaeB–pFL2 was transformed into the protoplasts of the FpLaeB deletion mutant by the PEG approach as well. After geneticin screening, the primer pair FpLaeB-5F/6R was used to confirm the complementation strain from geneticin-resistant transformants. Primers used for deletion, complementarity, and gene expression are listed in Supplementary Table S1.
Plant infection assays
For virulence on the wheat stem base, conidia were collected from CMC according to the method used in strains and culture conditions and then diluted to a concentration of 105 conidia/ml. The inoculation procedure was as described by Li et al. (2009) with the following modifications: Seeds of susceptible cultivar Shixin 828 were germinated on wet filter paper saturated in Petri dishes. Germinated seeds were immersed in the spore suspension for 1 min. Then, treated seeds were sown in a pot with a diameter of 15 cm containing sterile soil mix. There were three replicates, with each pot containing 20 seedlings. The severity of Fusarium crown rot was assessed at 35 days post-inoculation (dpi) using a 0–5 scale (Li et al., 2009).
For virulence on wheat heads, conidia of different strains were diluted to a concentration of 105 conidia/ml in 0.01% (vol/vol) Tween 20. A 20-μl aliquot of conidial suspension was injected into a floret of a wheat head of susceptible cultivar Shixin 828 at early anthesis. There were 30 replicates for each strain. The severity of head blight used a scale of 0–4 (Wang et al., 2015).
The severity of plants was determined using the disease index. The disease index (DI) was calculated as follows: DI = [∑ (number of diseased plants in this scale × value of this scale)/(total number of plants investigated × highest value of scale)] × 100.
RNA-seq and bioinformatics analysis
Both wild-type and mutant strains were transferred to PDA and incubated at 25°C for 3 days. Mycelia samples of three biological repetitions were collected from the surface of the colony. Total RNA of the wild-type and FpLeaB deletion mutant were extracted using an RNA extraction kit. The process followed the manufacturer’s instructions (Qiagen, Hilden, Germany). Novogene Co., Ltd. (Tianjin, China) conducted the library preparation and sequencing procedure. Clean reads for each sample were mapped on the reference genome of F. pseudograminearum CS3096 (Gardiner et al., 2012) using the TopHat 2.0.8 software with default parameters. The mapped read counts were used to determine the number of reads per kilobase per million reads (RPKM). The HTSeq v0.9.1 software was used to identify the different expressions of genes between mutant and wild-type strains (Trapnell et al., 2010). The DESeq2 software was used to isolate the differentially expressed genes (DEGs) with false discovery rate adjusted p < 0.05 (Love et al., 2014). The RPKM value of the same gene was used to calculate the fold change (FC) in log2(FC) greater than 1.0 between mutant and wild type. Gene ontology (GO) annotation was implemented by the GOseq package software (Young et al., 2010). The clusterProfiler v3.8.1 software was used to analyse the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment with p < 0.05.
Quantitative real-time PCR (qRT-PCR) was used to determine the transcript levels of SM genes (Wang et al., 2020). Total RNA was isolated from the mycelium using the TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. First-strand complementary deoxyribo nucleic acid (cDNA) was derived from total RNA using the Fermentas 1st cDNA synthesis kit (Hanover, MD) according to the manufacturer’s instructions. All values were calculated and normalised using the 2−ΔΔCT method and FpActin (FPSE_04141) gene, respectively (Livak and Schmittgen, 2001; Xia et al., 2021). Mean and standard deviation of data were collected from three biological replicates. Fisher’s least significant difference (LSD) in the Statistical Package for the Social Sciences (SPSS) was used for statistical analysis (p < 0.05). The primers are listed in Supplementary Table S1.
Determination of DON production
Three 6-mm-diameter agar plugs taken from the edge of the colony were inoculated into a 150-ml Erlenmeyer flask containing 30 ml of trichothecene biosynthesis induction (TBI) medium (Gardiner et al., 2009). After cultivating at 180 rpm in a shaker at 28°C for 3 days, the mycelium was collected for expression analysis of TRI5. The fermentation broth was filtered with a 0.22-μm aqueous filter at 14 dpi. DON was detected via ultraperformance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) (Soleimany et al., 2012).
Results
Deletion and complementarity of FpLaeB
The FpLaeB protein (accession number XP_009259110.1) contains 738 amino acids (aa) and was identified to be 47.99% homologous to A. nidulans LaeB (AN4699). A polyadenylate binding protein domain (PABP-1234) and Sec24-related protein domain (PTZ00395) were predicted at 141–251 and 68–188 aa via CD Search in NCBI, respectively. The coding gene sequence of FpLaeB was interrupted by three introns at 289–378, 1,487–2,200, and 2,875–2,924 bp (Figure 1A). Phylogenetic analysis showed that FpLaeB is a fungal LaeB homologue with a very close genetic relationship to that of F. graminearum (Figure 1B).
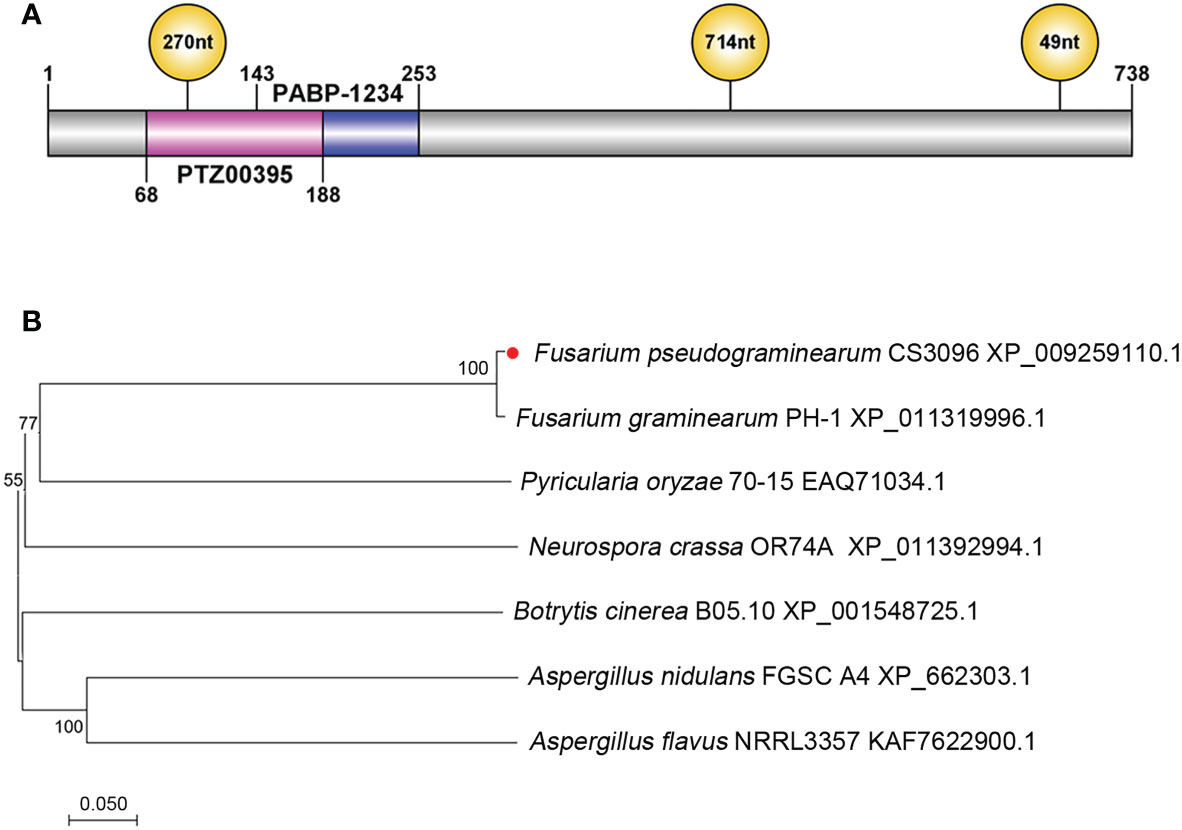
Figure 1 Structures and phylogenetic analysis of LaeB/LaeB of Fusarium pseudograminearum. (A) Location of conserved domains and intron in the FpLaeB protein and gene, respectively. Blue and pink bars represent polyadenylate binding protein domain (PABP-1234) and Sec24-related protein domain (PTZ00395), respectively. Three introns interrupted the FpLaeB gene at different positions. (B) Phylogenetic analysis of LaeB in F. pseudograminearum and other fungi. The neighbour-joining method used to analyse the amino acid sequences by the MEGA version 7.02 software package. The numbers at branches represent the supporting percentage of 1,000 bootstrap replicates.
In order to generate the FpLaeB deletion mutant (LDM), we used the hygromycin B phosphotransferase (hph) gene to replace the entire ORF of FpLaeB (Figure 2A). The transformants were confirmed by PCR amplification after preliminary screening by hygromycin. There is no PCR product when the primer pairs of ORF (FpLaeB-5F/6R) were used to amplify FpLaeB deletion mutants (Figure 2B). The genomic DNA of wild-type and mutant strains were further hybridised using the hph probe (Probe h). We found that only one 6.3-kb fragment band presented in the FpLaeB deletion mutant (Figure 2C). Hence, a homologous recombination event occurred in a single locus in the FpLaeB deletion mutant. The complementarity strain of FpLaeB deletion mutants showed an expected band (LDM-C, Figure 2D).
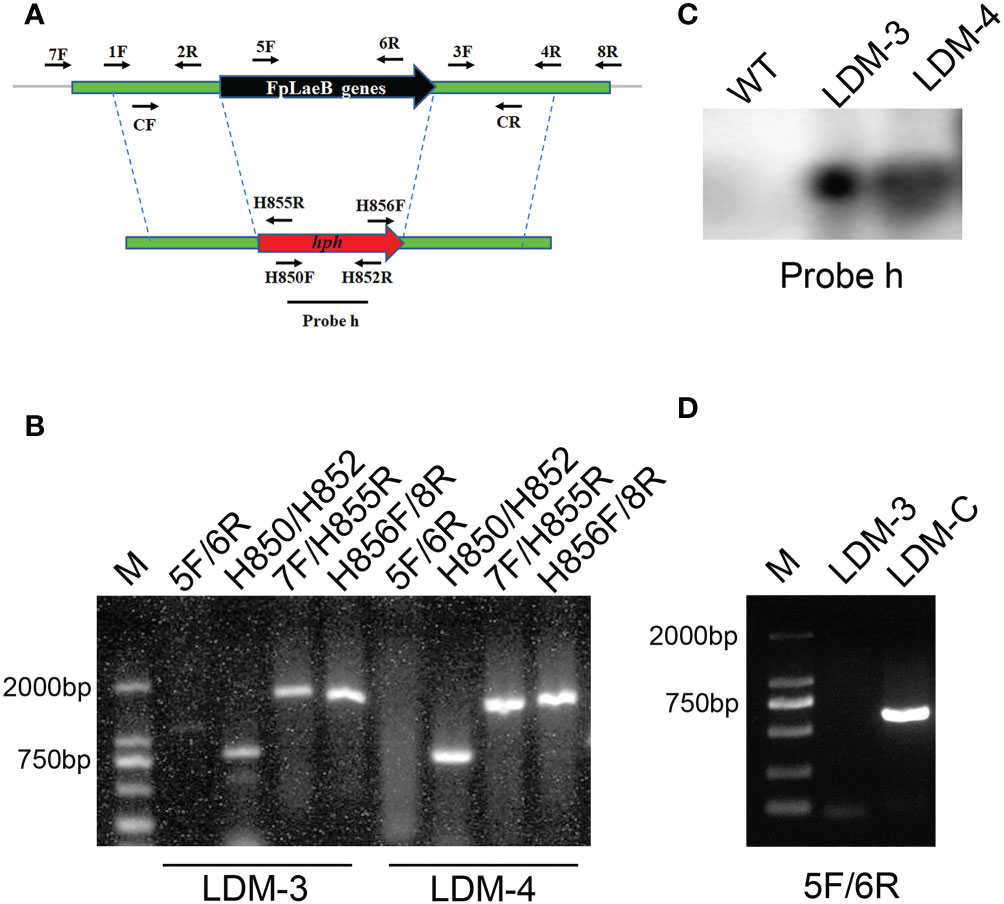
Figure 2 Deletion and complementation of the FpLaeB gene in Fusarium pseudograminearum. (A) The double-joint method was used to generate the FpLaeB gene replacement fragment. The arrows mark the positions and directions of primer pairs used for amplifying fragment and detection by PCR. (B) The genomic DNA of two deletion mutants was detected using four primer pairs, namely, FpLaeB-5F/6R, H850/H852, FpLaeB-7F/H855R, and H856F/FpLaeB-8R. Four lanes present the target gene, hph, and the recombination of upstream and downstream, respectively. (C) Genomic DNA of wild-type and two deletion mutants was hybridised using probe h (hph) in Southern blots. (D) PCR confirmation of complementation using primer FpLaeB-5F/6R.
FpLaeB is important for vegetative growth and conidiation
To evaluate whether FpLaeB was involved in the morphology formation of F. pseudograminearum, the deletion mutant of FpLaeB and wild-type strains were cultivated on PDA medium. The colonial morphology of FpLaeB deletion mutant strains was dramatically affected compared to wild type. There is scarcely any aerial hyphal growth in deletion mutants in contrast to the wild type. The colonies displayed a more compact appearance and a shorter peripheral edge in deletion mutants. Also, the hyphae are tenuous and curved under the microscope (Figure 3A). The hyphal growth rate of the FpLaeB deletion mutant was quantified to be 23% that of the wild type (Figure 3B). When the FpLaeB genes were reintroduced into the deletion mutant strain, the morphology formation of the complemented mutant was restored to that of the wild type (Figures 3A, B). Thus, the FpLaeB showed an important role on the growth phenotype of F. pseudograminearum.
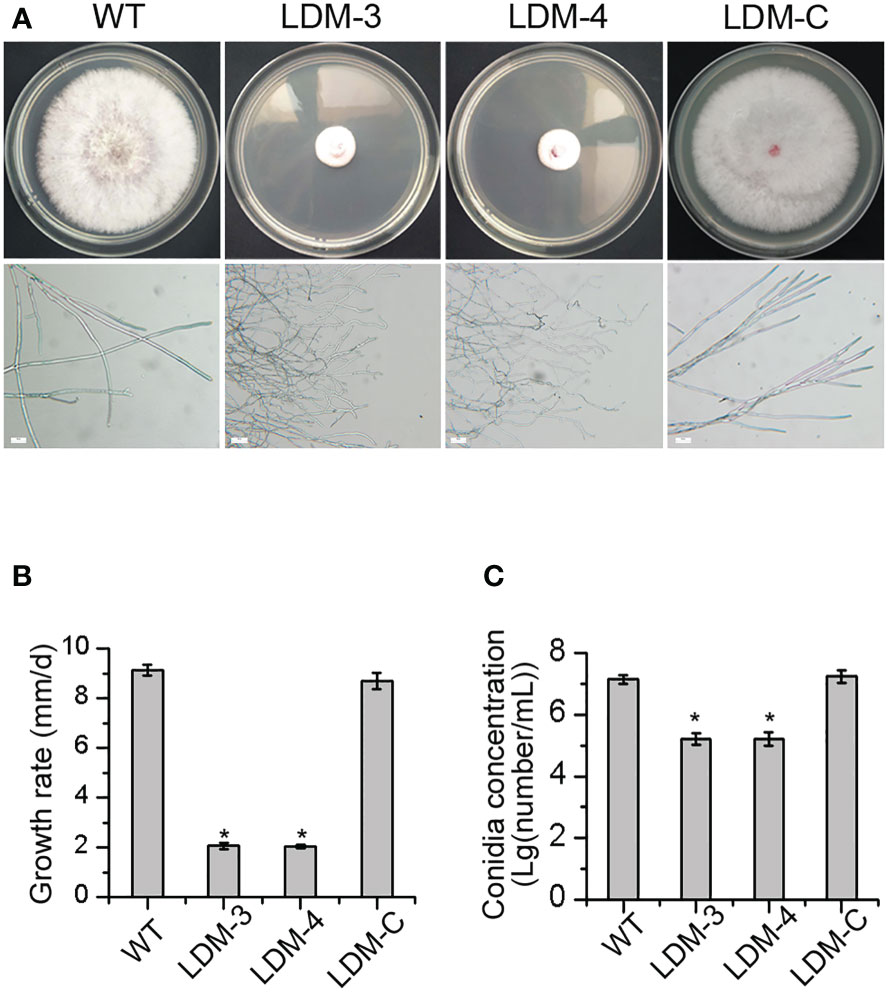
Figure 3 Effects of FpLaeB deletion on the morphology and conidiation of F pseudograminearum. (A) Colonies of wild-type and FpLaeB deletion mutant strains on potato dextrose agar (PDA) at 3 days post-inoculation. The photos of hyphal morphology were taken under a 200 times microscope. Bar = 20 μm. (B) Growth rate was presented by growth radius per day, which calculated radial growth between 2 and 3 dpi. (C) The logarithm of number of conidia per millilitre after 4 dpi in induced medium. Mean and standard deviation of data were calculated from three biological replicates. Statistical analysis was performed using Fisher’s least significant difference (LSD) in the Statistical Package for the Social Sciences (SPSS). Asterisks represent a significant difference between mutants and wild type (p < 0.05).
To test whether FpLaeB affected conidiation, we assayed the conidia concentration of different strains in induced media. After 4 dpi, the number of conidia was 107/ml in the wild-type strain. However, this amount was 105/ml in the deletion mutant at the same time. When reintroducing the FpLaeB genes into deletion mutant strains, the phenomenon could be reversed. This result showed the regulated effect of FpLaeB on conidiation in F. pseudograminearum (Figure 3C).
FpLaeB affects sensibility to cell membrane and the cell wall integrity inhibitor
To characterise whether FpLaeB affects sensibility to abiotic stress, the deletion mutants and wild-type strains were inoculated on STM mended with 3 mM H2O2 (oxidative stress), 0.7 M NaCl (Na+, osmotic pressure), 0.01% SDS (cell membrane damaging agent), or 200 mg/L CR (cell wall inhibitor, Figure 4A). We assayed the inhibition rates of FpLaeB deletion mutants, which were higher than those of wild type on STM mended with 0.01% SDS. The inhibition rates of FpLaeB deletion mutants were lower than those of the wild-type strain on STM mended with 200 mg/L CR. There are no significant differences in sensibility between deletion mutants and the wild-type strain when cultured on NaCl and H2O2 media (Figure 4B). The phenomenon was restored when the FpLaeB genes were reintroduced into the deletion mutant strain. These results indicated that FpLaeB is important to maintain cell wall and membrane integrity in F. pseudograminearum.
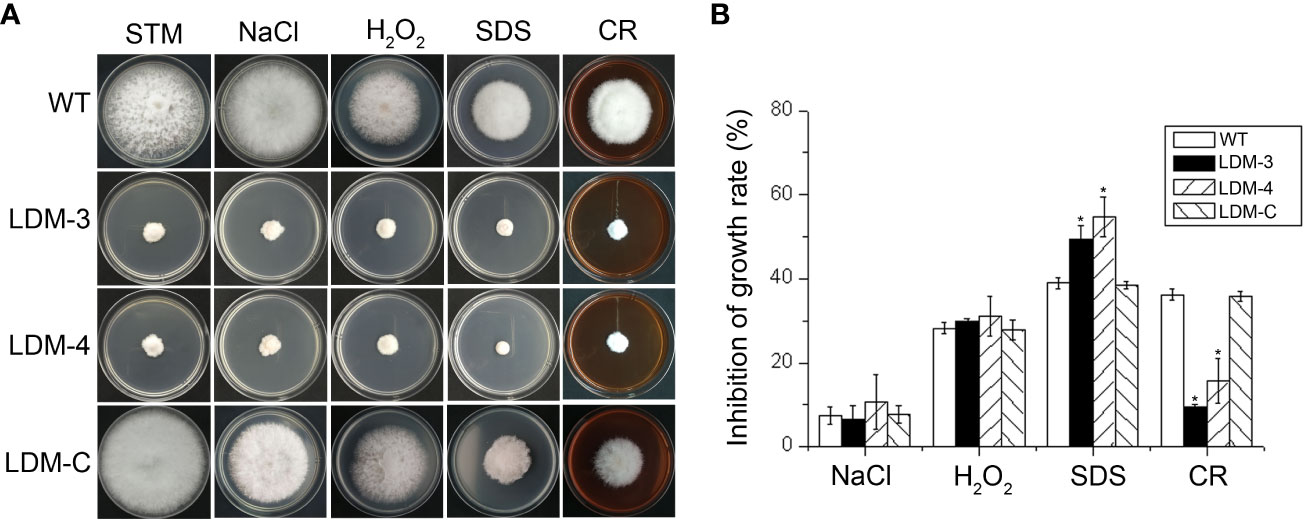
Figure 4 Effects of FpLaeB deletion on sensibility to abiotic stress in F pseudograminearum. (A) The wild-type, FpLaeB deletion mutant, and complemented mutant strains grew on synthetic medium (STM) mended with NaCl, H2O2, SDS, or Congo red (CR). Pictures were taken after 3 dpi on stress media. (B) Inhibition rates were calculated by the growth rate of stress media compared with that of the STM without inhibitor. Mean and standard deviation of data were calculated from three biological replicates. Statistical analysis was performed using Fisher’s LSD in SPSS. Asterisks on the bars represent statistically significant difference compared to wild type (p < 0.05).
FpLaeB is required for plant infection
We assay pathogenicity tests with stem base and flowering wheat heads to characterise the role of FpLaeB in disease development. The wild-type strain developed typical crown rot and head blight symptoms at the stem base and heads at 21 dpi, respectively. Under the same condition, limited discolouration appeared only at the inoculation site of the FpLaeB deletion mutant (Figure 5A). The disease index of the FpLaeB deletion mutant was reduced by approximately 95% in the crown and head compared to that of wild type. To confirm these findings, we performed a complementation assay with the FpLaeB gene. The complementation led to a normal phenotype in both cases (Figure 5B, 5C). This result confirmed that FpLaeB is involved in virulence in F. pseudograminearum.

Figure 5 Effects of FpLaeB deletion in plant infection. (A) Images were taken when wild-type, FpLaeB deletion mutant, and complemented mutant strains were inoculated to the stem base during the seedling stage at 21 days. (B) Flowering wheat heads inoculated with the different strains were photographed at 21 dpi. (C) Virulence is represented by the disease index. The disease index is calculated from plants with different disease grades. The experiments were repeated three times. Statistical analysis was performed by using Fisher’s LSD in SPSS. Asterisks on the bars represent statistically significant difference compared to wild type (p < 0.05).
FpLaeB regulates gene expression including membrane and secondary metabolism
We analysed transcriptomes (RNA-seq) from wild type and the FpLaeB deletion mutant [raw sequence data for RNA-seq data are available in the NCBI Sequence Read Archive (SRA), accession number: PRJNA914495] to reveal the regulatory role of FpLaeB at a genome-wide scale. The absence of FpLaeB caused a significant change in expression levels in more than 3,200 genes in the F. pseudograminearum genome. The number of differentially downregulated genes was 1,748, while 1,456 genes showed an increase in their expression [all the expression analysis at p < 0.05, log2(FC) >1 or <−1] (Supplementary Table S2). Three classes, namely, “molecular function”, “cellular components”, and “biological process”, of the gene product were used to define the GO. In the class cellular components, the two most populated categories were “integral component of membrane” and “intrinsic component of membrane”. This result showed that the majority of both upregulated and downregulated DEGs were significantly associated with “membrane” (Figure 6A; Supplementary Table S3). We observed a broad range of transcripts encoding the biosynthesis of SMs that were enriched during Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Figure 6B; Supplementary Table S4). There were 62 gene-encoded SMs with significantly downregulated expression while 53 genes were upregulated in FpLaeB (Figure 6C). To validate the expression profiles of these SMs, the expression levels of nine genes encoding SMs were used to verify the accuracy of transcriptomes. The results show that the expression levels (five downregulation and four upregulation) were basically the same between transcriptomes and qRT-PCR (Figure 6D).
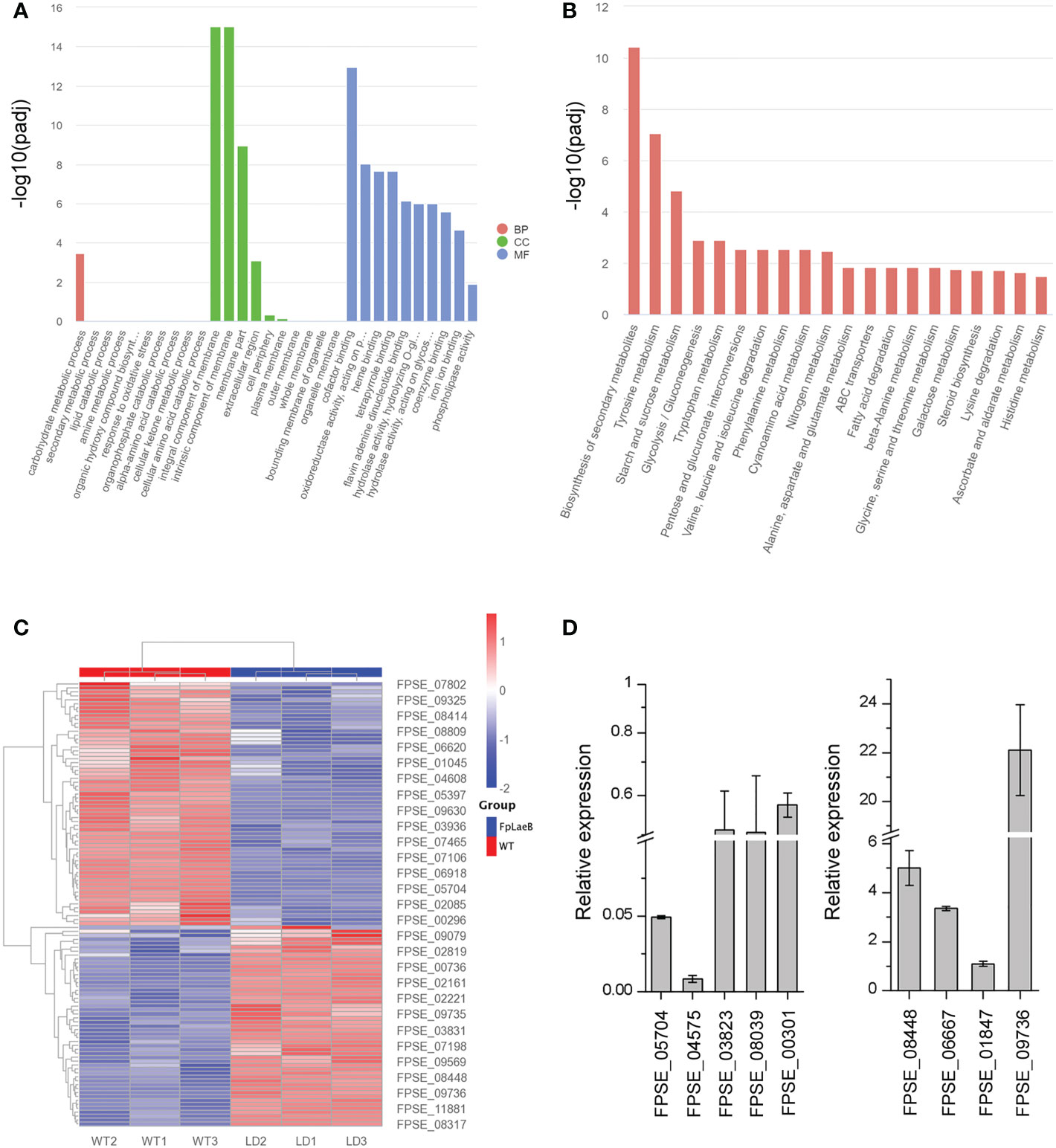
Figure 6 Transcriptome analysis of differentially expressed genes (DEGs) between the FpLaeB deletion mutant and wild type. (A) Bar plot of Gene ontology (GO) annotation of DEGs. The DEGs were categorised into three main categories: biological process (BP), cellular component (CC), and molecular function (MF). The DEGs were log2(FC) greater than 1.0 with a threshold at p and corrected p < 0.05. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs. (C) Heatmap for differentially regulated genes encoding secondary metabolites (SMs). The different colour scale presents the counts of expression normalised by Z-score. (D) Real-time qPCR for nine DEGs encoding SM including five upregulated genes and four downregulated genes between wild-type and FpLaeb mutants. Cycle threshold of FpActin gene was used to normalise the two samples. Expression levels of these genes in wild type were arbitrarily given a value of 1. Mean and standard deviation of data were calculated from three biological replicates. Statistical analysis was performed using Fisher’s LSD in SPSS.
FpLaeB is important for DON production
Furthermore, we monitored the transcription level of the TRI5 gene and the production of DON in the FpLaeB deletion mutant and the wild-type strain in inducing medium. In comparison with the wild type, the expression levels of the TRI5 gene were downregulated 20 times in the FpLaeB deletion mutant (Figure 7A). After induced culture, the DON concentration of the culture solution in the deletion mutant strain was 60.34 µg/L. This value was significantly lower than that of the wild-type strain, which was 1,143.83 µg/L (Figures 7B, C). Therefore, FpLaeB appears to play an important role in regulating the TRI gene expression and DON production of F. pseudograminearum.
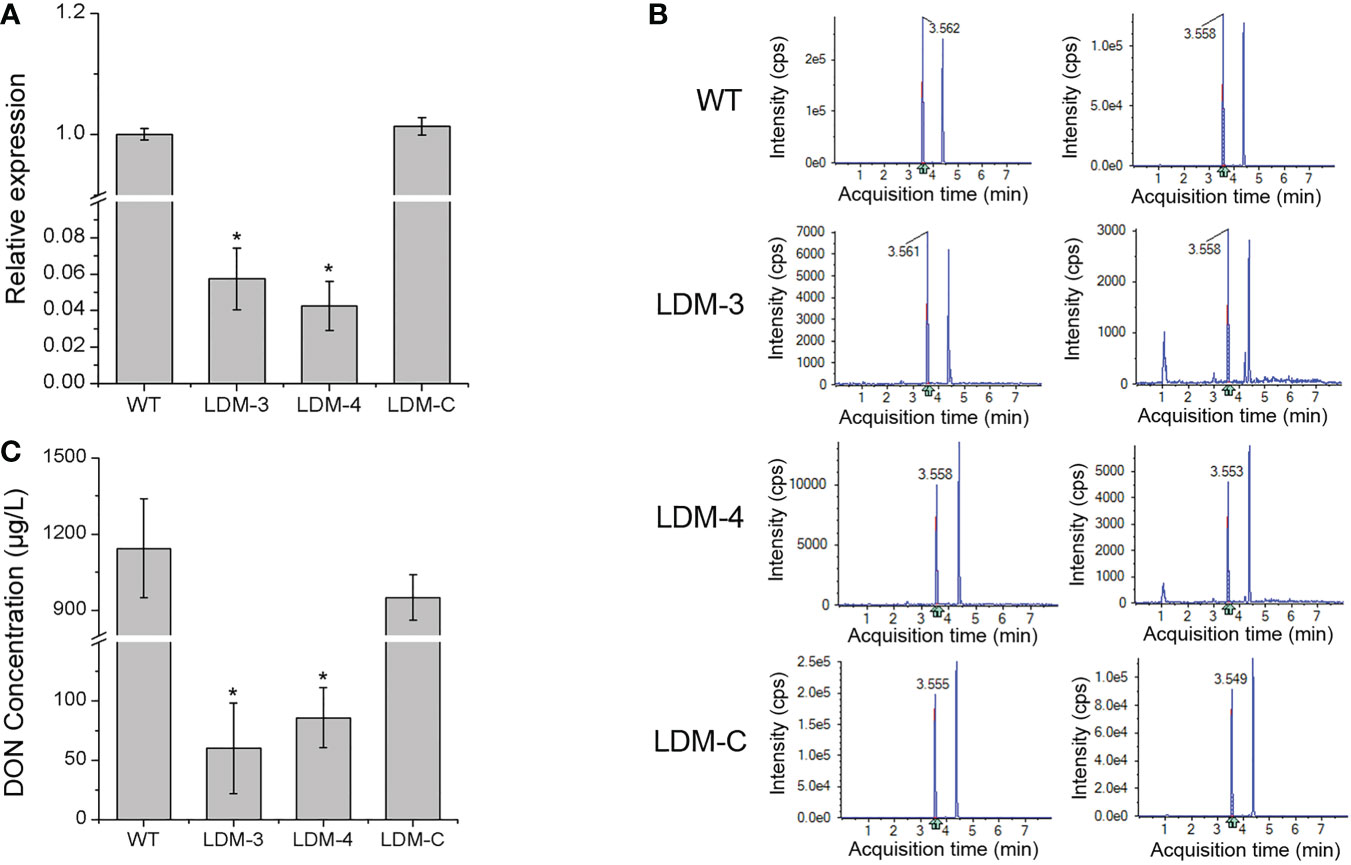
Figure 7 Transcription level of the TRI5 gene and DON concentration in inducing medium. (A) Relative transcript abundances of the TRI5 gene in mycelium in inducing medium were compared between the wild-type and FpLaeB mutant strain at 7 dpi. Cycle threshold of the FpActin gene was used to normalise different samples. Expression levels of wild type were arbitrarily given a value of 1. (B) UPLC-MS/MS chromatogram of DON at inducing media of different strains. (C) DON concentration of different strains cultured in inducing medium for 14 days. Mean and standard deviation of data were calculated from three biological replicates. Statistical analysis was performed using Fisher’s LSD in SPSS.
Discussion
The LaeB protein is identified as a novel transcriptional regulator of the sterigmatocystin in A. nidulans. It is required for the transcription of aflR, the transcriptional regulator of the sterigmatocystin (Woloshuk et al., 1994; Pfannenstiel et al., 2017). In order to explore the regulating function of LaeB in the secondary metabolism (SM) of F. pseudograminearum, we set out to assess the importance of a putative LaeB homologue, FpLaeB, based on its homology to A. nidulans. This study found that the deletion of FpLaeB has drastically impacted the growth rate of mycelia in F. pseudograminearum. This result differs from that of a previous study in which the deletion of LaeB did not impact the growth rate of A. nidulans (Lin et al., 2018). Another homologous protein of LaeB named RsdA has been identified through the genome-wide deletion of regulators in the endophytic fungus Pestalotiopsis fici. Deletion of RsdA resulted in moderate growth reduction (Zhou et al., 2019). This finding suggests that the function of LaeB could be species-specific in fungus. The roles of LaeB could be divergent in different fungi as well.
In addition to affecting growth rate, FpLaeB also affected the conidiation, sensitivity to SDS and CR stress, and virulence in F. pseudograminearum. The regulation of virulence by LaeB is of great significance in the study of pathogenic fungi. In the present study, we demonstrated that FpLaeB plays crucial roles in virulence. Because the deletion mutant impaired growth and development, the reduction in virulence was partially due to the growth defect of the deletion mutant. However, the reduction of the disease index in the stem base and head (approximately 95%) is not proportional to the 77% reduction in growth rate on PDA. Therefore, other reasons should contribute to the reduction of virulence. As a global regulator, the regulatory role of LaeB is extensive in fungi. Other global regulators such as LaeA and velvet complex proteins played crucial roles in the regulation of morphology, development, SM, and virulence in several fungi (Bok et al., 2005; Sarikaya-Bayram et al., 2015; Wang et al., 2019; Maor et al., 2021). They are involved in the regulation of different metabolic pathways, the most significant of which was the effect on secondary metabolism. For example, the expression of at least 9.5% of genes of the A. fumigatus genome was regulated by LaeA, wherein the positive control SM biosynthesis genes such as polyketide synthases, P450, nonribosomal peptide synthetases (NRPSs), and monooxygenases amounted to 20% to 40% (Perrin et al., 2007). Also, transcriptomic and proteomic analyses indicated that VmLaeA performs both SM transport and biosynthesis in Valsa mali (Feng et al., 2020). In addition, FgVeA is involved in various cellular processes including soluble N-ethylmaleimide – sensitive factor attachment protein receptors (SNARE) interactions in vesicular transport and peroxisome biogenesis pathway in F. graminearum (Jiang et al., 2011). In the present study, the deletion of FpLaeB can affect a quarter of the genes in the F. pseudograminearum genome. This result coincided with the severe impact of FpLaeB in multiple developmental processes. The highlighted DEGs of the biological process involved the carbohydrate metabolic process according to the GO enrichment statistics (Supplementary Table S3). In Penicillium expansum, the disturbing carbohydrate metabolic process could lead to growth defect (Lai et al., 2021). Hence, the regulation of the carbohydrate metabolic process by FpLaeB is perhaps associated with growth rate in F. pseudograminearum. The most severe DEGs of the FpLaeB mutant were “membrane” genes. The most DEGs regulated by FpLaeB were membrane protein-coding genes, including “intrinsic component of membrane”, “integral component of membrane”, and “membrane part”. This finding seemed to be consistent with the increased sensitivity to SDS of mutants.
LaeB is indispensable for the biosynthesis of aflatoxin in A. flavus and sterigmatocystin in A. nidulans (Pfannenstiel et al., 2017). On the other hand, some novel polyketides have been discovered by the deletion of LaeB in A. nidulans (Lin et al., 2018). Although this regulator is conserved in Aspergilli, the function is not sterigmatocystin specific. In P. fici, the deletion of rsdA significantly reduces SMs such as asperpentyn, ficiolide A, and chloroisosulochrin. In contrast, in the rsdA deletion mutant, six known compounds were isolated, including a new non-ribosomal peptide that was isolated from P. fici for the first time. In addition, melanin was significantly accumulated in the mycelium. All these results showed that the deletion of rsdA results in significant differences in SM (Zhou et al., 2019). The regulation of SM by FpLaeB was proven again in this study. Not only did the highest expression levels and the most differential genes involve biosynthesis of the SM pathway, but also the decrement of pathogenicity-related DON was further clarified in the FpLaeB deletion mutant. DON was positively regulated by FpLaeB, and this regulation seemed to be involved in virulence in F. pseudograminearum. The model of RsdA regulation of fungal SM was proposed based on the genome-wide expression profile and with reference to the SM regulatory network in P. fici. It is proposed as an upstream regulator of velvet complex proteins for the regulation of SM, although velvet complex proteins were confirmed to be involved in the regulation of morphology, development, SM, and virulence in both F. graminearum and F. pseudograminearum (Jiang et al., 2011; Merhej et al., 2012; Gardiner et al., 2021). More research is needed to confirm this model in F. pseudograminearum.
In conclusion, we have identified that FpLaeB is important for the vegetative growth, conidiation, sensibility of cell wall and membrane inhibitors, and virulence in F. pseudograminearum. Also, a lot of downstream genes have been detected by genome-wide associations of gene expression analysis. In particular, the positive regulation of SM including DON could be related to virulence. Future research should focus on the regulation model of FpLaeB for SM.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be at NCBI BioProject accession number: PRJNA914495.
Author contributions
YXW and LK designed the experiments and managed the projects. YXW, YJW, SH, and QL performed the experiments. YXW, YJW, SH performed the data analysis. YXW wrote the first draft of the manuscript, and LK edited and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Hebei (No. C2021301042), Basic Research Funds of Hebei Academy of Agriculture and Forestry Sciences (2021120201), and the HAAFS Agriculture Science and Technology Innovation Project (2022KJCXZX-ZBS-7).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1132507/full#supplementary-material
Supplementary Table 1 | Primers used in this study.
Supplementary Table 2 | All differentially expressed genes between FpLeaB deletion mutant and wild type.
Supplementary Table 3 | The main categories of differentially expressed genes on the GO annotations.
Supplementary Table 4 | The enriched pathway of differentially expressed genes on the KEGG annotations.
Supplementary Figure 1 | Standard curve used to calculate DON content. Regression equation: y = 475.79926x + 158.33036 (r = 0.99925, r² = 0.99850).
References
Bayram, O., Krappmann, S., Ni, M., Bok, J. W., Helmstaedt, K., Valerius, O., et al. (2008). VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320 (5882), 1504–1506. doi: 10.1126/science.1155888
Bok, J. W., Balajee, S. A., Marr, K. A., Andes, D., Nielsen, K. F., Frisvad, J. C., et al. (2005). LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryotic Cell 4 (9), 1574–1582. doi: 10.1128/EC.4.9.1574-1582.2005
Bok, J. W., Keller, N. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryotic Cell 3 (2), 527–535. doi: 10.1128/EC.3.2.527-535.2004
Brakhage, A. A. (2013). Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11 (1), 21–32. doi: 10.1038/nrmicro2916
Chen, Y., Kistler, H. C., Ma, Z. (2019). Fusarium graminearum trichothecene mycotoxins: Biosynthesis, regulation, and management. Annu. Rev. Phytopathol. 57, 1.1–1.25. doi: 10.1146/annurev-phyto-082718-100318
Chen, L., Ma, Y., Peng, M., Chen, W., Xia, H., Zhao, J., et al. (2021). Analysis of apoptosis-related genes reveals that apoptosis functions in conidiation and pathogenesis of Fusarium pseudograminearum. mSphere 6 (1), e01140–e01120. doi: 10.1128/mSphere.01140-20
Feng, Y., Yin, Z., Wu, Y., Xu, L., Du, H., Wang, N., et al. (2020). LaeA controls virulence and secondary metabolism in apple canker pathogen Valsa mali. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.581203
Gardiner, D. M., Kazan, K., Manners, J. M. (2009). Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum. Fungal Genet. Biol. 46 (8), 604–613. doi: 10.1016/j.fgb.2009.04.004
Gardiner, D. M., McDonald, M. C., Covarelli, L., Solomon, P. S., Rusu, A. G., Marshall, M., et al. (2012). Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PloS Pathog. 8 (9), e1002952. doi: 10.1371/journal.ppat.1002952
Gardiner, D. M., Rusu, A., Benfield, A. H., Kazan, K. (2021). Map-based cloning identifies velvet a as a critical component of virulence in Fusarium pseudograminearum during infection of wheat heads. Fungal Biol. 125 (3), 191–200. doi: 10.1016/j.funbio.2020.10.012
Hee-Kyoung, K., Seunghoon, L., Seong-Mi, J., Mccormick, S. P., Butchko, R., Proctor, R. H., et al. (2013). Functional roles of FgLaeA in controlling secondary metabolism, sexual development, and virulence in Fusarium graminearum. PloS One 8 (7), e68441. doi: 10.1371/journal.pone.0068441
Ji, L., Kong, L., Li, Q., Wang, L., Chen, D., Ma, P. (2016). First report of Fusarium pseudograminearum causing fusarium head blight of wheat in hebei province, China. Plant Dis. 100 (1), 220–220. doi: 10.1094/PDIS-06-15-0643-PDN
Jiang, J., Liu, X., Yin, Y., Ma, Z. (2011). Involvement of a velvet protein FgVeA in the regulation of asexual development, lipid and secondary metabolisms and virulence in Fusarium graminearum. PloS One 6 (11), e28291. doi: 10.1371/journal.pone.0028291
Kazan, K., Gardiner, D. M. (2018). Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: recent progress and future prospects. Mol. Plant Pathol. 19 (7), 1547–1562. doi: 10.1111/mpp.12639
Lai, T., Sun, Y., Liu, Y., Li, R., Chen, Y., Zhou, T. (2021). Cinnamon oil inhibits Penicillium expansum growth by disturbing the carbohydrate metabolic process. J. Fungi (Basel) 7 (2), 123. doi: 10.3390/jof7020123
Li, H., Yuan, H., Fu, B., Xing, X., Sun, B., Tang, W. (2012). First report of Fusarium pseudograminearum causing crown rot of wheat in henan, China. Plant Dis. 96 (7), 1065–1065. doi: 10.1094/PDIS-01-12-0007-PDN
Li, H. B., Zhou, M. X., Liu, C. J. (2009). A major QTL conferring crown rot resistance in barley and its association with plant height. Tagtheoretical Appl. Genet. 118 (5), 903–910. doi: 10.1007/s00122-008-0948-3
Lin, H., Lyu, H., Zhou, S., Yu, J., Keller, N. P., Chen, L., et al. (2018). Deletion of a global regulator LaeB leads to the discovery of novel polyketides in Aspergillus nidulans. Organic Biomolecular Chem. 16 (27), 4973–4976. doi: 10.1039/c8ob01326h
Liu, Z., Friesen, T. L. (2012). “Polyethylene glycol (PEG)-mediated transformation in filamentous fungal pathogens,” in Plant fungal pathogens: Methods and protocols. Eds. Bolton, M. D., Thomma, B. P. H. J. (Totowa, NJ: Humana Press), 365–375.
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Maor, U., Barda, O., Sadhasivam, S., Bi, Y., Levin, E., Zakin, V., et al. (2021). Functional roles of LaeA, polyketide synthase, and glucose oxidase in the regulation of ochratoxin a biosynthesis and virulence in aspergillus carbonarius. Mol. Plant Pathol. 22 (1), 117–129. doi: 10.1111/mpp.13013
Merhej, J., Urban, M., Dufresne, M., Hammond-Kosack, K. E., Richard-Forget, F., Barreau, C. (2012). The velvet gene, FgVe1, affects fungal development and positively regulates trichothecene biosynthesis and pathogenicity in Fusarium graminearum. Mol. Plant Pathol. 13 (4), 363–374. doi: 10.1111/j.1364-3703.2011.00755.x
Monds, R. D., Cromey, M. G., Lauren, D. R., Di, M. M., Marshall, J. (2005). Fusarium graminearum, F. cortaderiae and F. pseudograminearum in new Zealand: molecular phylogenetic analysis, mycotoxin chemotypes and co-existence of species. Mycological Res. 109, 410–420. doi: 10.1017/S0953756204002217
Murray, G. M., Brennan, J. P. (2009). Estimating disease losses to the Australian wheat industry. Australas. Plant Pathol. 38 (6), 558–570. doi: 10.1071/AP09053
Perrin, R. M., Fedorova, N. D., Bok, J. W., Cramer, R. A., Jr., Wortman, J. R., Kim, H. S., et al. (2007). Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PloS Pathog. 3 (4), e50. doi: 10.1371/journal.ppat.0030050
Pfannenstiel, B. T., Zhao, X., Wortman, J., Wiemann, P., Throckmorton, K., Spraker, J. E., et al. (2017). Revitalization of a forward genetic screen identifies three new regulators of fungal secondary metabolism in the genus Aspergillus. mBio 8 (5), e01246–e01217. doi: 10.1128/mBio.01246-17
Powell, J. J., Carere, J., Fitzgerald, T. L., Stiller, J., Covarelli, L., Xu, Q., et al. (2017). The Fusarium crown rot pathogen Fusarium pseudograminearum triggers a suite of transcriptional and metabolic changes in bread wheat (Triticum aestivum l.). Ann. Bot. 119 (5), 853–867. doi: 10.1093/aob/mcw207
Sarikaya-Bayram, O., Palmer, J. M., Keller, N., Braus, G. H., Bayram, O. (2015). One Juliet and four romeos: VeA and its methyltransferases. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00001
Smiley, R. W., Gourlie, J. A., Easley, S. A., Patterson, L.-M., Whittaker, R. G. (2005). Crop damage estimates for crown rot of wheat and barley in the pacific Northwest. Plant Dis. 89 (6), 595–604. doi: 10.1094/PD-89-0595
Soleimany, F., Jinap, S., Faridah, A., Khatib, A. (2012). A UPLC–MS/MS for simultaneous determination of aflatoxins, ochratoxin a, zearalenone, DON, fumonisins, T-2 toxin and HT-2 toxin, in cereals. Food control 25 (2), 647–653. doi: 10.1016/j.foodcont.2011.11.012
Tang, G., Zhang, C., Ju, Z., Zheng, S., Wen, Z., Xu, S., et al. (2018). The mitochondrial membrane protein FgLetm1 regulates mitochondrial integrity, production of endogenous reactive oxygen species and mycotoxin biosynthesis in Fusarium graminearum. Mol. Plant Pathol. 19 (7), 1595–1611. doi: 10.1111/mpp.12633
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28 (5), 511–515. doi: 10.1038/nbt.1621
Tunali, B., Obanor, F., Erginbas, G., Westecott, R. A., Nicol, J., Chakraborty, S. (2012). Fitness of three fusarium pathogens of wheat. FEMS Microbiol. Ecol. 81 (3), 596–609. doi: 10.1111/j.1574-6941.2012.01388.x
Wang, L., Xie, Y., Cui, Y., Xu, J., He, W., Chen, H., et al. (2015). Conjunctively screening of biocontrol agents (BCAs) against fusarium root rot and fusarium head blight caused by fusarium graminearum. Microbiol. Res. 177, 34–42. doi: 10.1016/j.micres.2015.05.005
Wang, L., Xie, S., Zhang, Y., Kang, R., Zhang, M., Wang, M., et al. (2020). The FpPPR1 gene encodes a pentatricopeptide repeat protein that is essential for asexual development, sporulation, and pathogenesis in Fusarium pseudograminearum. Front. Genet. 11. doi: 10.3389/fgene.2020.535622
Wang, G., Zhang, H., Wang, Y., Liu, F., Li, E., Ma, J., et al. (2019). Requirement of LaeA, VeA, and VelB on asexual development, ochratoxin a biosynthesis, and fungal virulence in Aspergillus ochraceus. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02759
Woloshuk, C. P., Foutz, K. R., Brewer, J. F., Bhatnagar, D., Cleveland, T. E., Payne, G. A. (1994). Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60 (7), 2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994
Xia, H., Chen, L., Fan, Z., Peng, M., Zhao, J., Chen, W., et al. (2021). Heat stress tolerance gene FpHsp104 affects conidiation and pathogenicity of Fusarium pseudograminearum. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.695535
Young, M., Wakefield, M., Smyth, G., Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Yu, J.-H., Hamari, Z., Han, K.-H., Seo, J.-A., Reyes-Domínguez, Y., Scazzocchio, C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41 (11), 973–981. doi: 10.1016/j.fgb.2004.08.001
Zhang, Y., Gao, X., Sun, M., Liu, H., Xu, J. (2017). The FgSRP1 SR-protein gene is important for plant infection and pre-mRNA processing in Fusarium graminearum. Environ. Microbiol. 19 (10), 4065–4079. doi: 10.1111/1462-2920.13844
Keywords: Triticum aestivum, deletion mutant, conidiation, secondary metabolite, deoxynivalenol
Citation: Wu Y, Wang Y, Han S, Li Q and Kong L (2023) The global regulator FpLaeB is required for the regulation of growth, development, and virulence in Fusarium pseudograminearum. Front. Plant Sci. 14:1132507. doi: 10.3389/fpls.2023.1132507
Received: 27 December 2022; Accepted: 07 February 2023;
Published: 22 February 2023.
Edited by:
Yanan Wang, Hebei Agricultural University, ChinaReviewed by:
Wenjun Zhu, Wuhan Polytechnic University, ChinaHai-Lei Wei, Institute of Agricultural Resources and Regional Planning (CAAS), China
Copyright © 2023 Wu, Wang, Han, Li and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingxiao Kong, konglingxiao163@163.com
 Yuxing Wu
Yuxing Wu Yajiao Wang
Yajiao Wang Sen Han
Sen Han Qiusheng Li
Qiusheng Li