- 1Institute of Forestry and Pomology, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 2Beijing Engineering Research Center for Strawberry, Beijing, China
- 3Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (North China), Ministry of Agriculture, Beijing, China
Strawberry (Fragaria × ananassa Duch) are sensitive to salt stress, and breeding salt-tolerant strawberry cultivars is the primary method to develop resistance to increased soil salinization. However, the underlying molecular mechanisms mediating the response of strawberry to salinity stress remain largely unknown. This study evaluated the salinity tolerance of 24 strawberry varieties, and transcriptomic and metabolomic analysis were performed of ‘Sweet Charlie’ (salt-tolerant) and ‘Benihoppe’ (salt-sensitive) to explore salt tolerance mechanisms in strawberry. Compared with the control, we identified 3412 differentially expressed genes (DEGs) and 209 differentially accumulated metabolites (DAMs) in ‘Benihoppe,’ and 5102 DEGs and 230 DAMs in ‘Sweet Charlie.’ DEGs Gene Ontology (GO) enrichment analyses indicated that the DEGs in ‘Benihoppe’ were enriched for ion homeostasis related terms, while in ‘Sweet Charlie,’ terms related to cell wall remodeling were over-represented. DEGs related to ion homeostasis and cell wall remodeling exhibited differential expression patterns in ‘Benihoppe’ and ‘Sweet Charlie.’ In ‘Benihoppe,’ 21 ion homeostasis-related DEGs and 32 cell wall remodeling-related DEGs were upregulated, while 23 ion homeostasis-related DEGs and 138 cell wall remodeling-related DEGs were downregulated. In ‘Sweet Charlie,’ 72 ion homeostasis-related DEGs and 275 cell wall remodeling-related DEGs were upregulated, while 11 ion homeostasis-related DEGs and 20 cell wall remodeling-related DEGs were downregulated. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses showed only four KEGG enriched pathways were shared between ‘Benihoppe’ and ‘Sweet Charlie,’ including flavonoid biosynthesis, phenylalanine metabolism, phenylpropanoid biosynthesis and ubiquinone, and other terpenoid-quinone biosynthesis. Integrating the results of transcriptomic and metabolomics analyses showed that adenosine triphosphate-binding cassette (ABC) transporters and flavonoid pathway genes might play important roles in the salt stress response in strawberry, and DAMs and DEGs related to ABC transporter and flavonoid pathways were differentially expressed or accumulated. The results of this study reveal that cell wall remodeling and ABC transporters contribute to the response to salt stress in strawberry, and that related genes showed differential expression patterns in varieties with different salt tolerances. These findings provide new insights into the underlying molecular mechanism of strawberry response to salt stress and suggest potential targets for the breeding of salt-tolerant strawberry varieties.
Introduction
Soil salinization is a significant environmental factor threatening food production and food security. Currently, soil salinization affects 800 million hectares of farmland worldwide, and every year about 1–2% of available acreage for farming is compromised due to soil salinity (Etesami and Beattie, 2018; Bomle et al., 2021). Furthermore, with temperature and precipitation changes related to climate change, irrigation, increasing sea level, and the use of wastewater all increase agricultural soil salinization worldwide (Koohbor et al., 2019; Shah et al., 2022). Saline soils lead to osmotic, ionic, and oxidative stress in plants, affecting overall plant growth and yield (Arora et al., 2020; Bomle et al., 2021). Strawberry is highly sensitive to salt stress, and previous studies have shown that under increased salt stress, there is a decrease in plant biomass and chlorophyll content, and negative impacts on organic acids, soluble solids, and the physical appearance of fruits (Kaya et al., 2019). Generally, the yield loss and commodity fruit rate reduction caused by salt stress significantly reduce economic benefits (Keutgen and Pawelzik, 2007; Suarez and Grieve, 2013; Ferreira et al., 2019). However, mild salt stress has a positive impact on certain desirable metabolites such as anthocyanins and phenolic compounds, compounds that are involved in plant stress responses (Vanessa et al., 2016). The additional cost of a water purification system will significantly raise the cost of production, so the effective breeding of salt tolerant varieties is an important way to counteract soil salinization for the strawberry industry. The use of salt tolerant varieties such as ‘Albion,’ ‘San Andreas,’ and ‘Camarosa’ in a breeding program can improve the performance of offspring under saline conditions (Sun et al., 2015). In our breeding program, F1 offspring, ‘Jingtaoxiang’ (‘Darselect’ × ‘Akihime’) (Wang et al., 2018b) showed significantly higher salt tolerance compared to ‘Akihime,’ the salt sensitive parent. Thus, further study on the molecular mechanisms involved in the response of different strawberry varieties to salt stress is needed to provide a theoretical basis for breeding.
Flavonoids are major non-enzymatic scavengers of reactive oxygen species (ROS), with antioxidant capacities. These compounds play a major role when plants suffer from salt stress. The flavonoid biosynthetic genes CHS, CHI, and F3H function as positive regulators to modulate plant salt stress tolerance by increasing the accumulation of flavonoids (Mahajan and Yadav, 2014; Chen et al., 2015; Wang et al., 2018a; Jayaraman et al., 2021). AtROS1-mediated demethylation improves the expression levels of flavonoid biosynthesis and antioxidant-related genes to increase salt stress tolerance (Bharti et al., 2015). In rice, SQD2.1 acts in the glycosylation of flavonoids and improves the scavenging of ROS to improve salt stress tolerance (Zhan et al., 2019). CrUGT87A1 from Carex rigescens is a positive regulator of plant salt tolerance, and can enhance plant antioxidation capability through increased flavonoid accumulation to improve salt tolerance in plants (Zhang et al., 2021b). The R2R3-MYB transcription factors are core regulators of flavonoid biosynthesis and can modulate plant salt stress tolerance by controlling flavonoid biosynthesis and accumulation (Li et al., 2019a; Wang et al., 2020, 2021).
At the cellular level, the cell wall is the first barrier for plants to respond to salt stresses. Decreased turgor pressure induced by salt stress restricts cell expansion and division and the cell wall responds to salt stress by resisting cell turgor changes (Zhao et al., 2021b). Components of the cell wall such as cellulose, lignin, and xyloglucan have been implicated in the salt tolerance response. OsCSLD4, a major regulator of cell wall polysaccharide synthesis, is involved in the response to salt stress in rice by affecting abscisic acid biosynthesis to regulate osmotic stress tolerance (Zhao et al., 2021a). Companion of Cellulose Synthase 1 (CC1) promotes plant growth under salt stress via control microtubule bundling and dynamics (Kesten et al., 2019). Xyloglucan endotransglucosylase-hydrolase 30 (XTH30) can alter cellulose synthesis and negatively affect salt tolerance (Yan et al., 2019). MdSND1 and AgNAC1 participate in the response to salt stress by regulating lignin biosynthesis or accumulation (Chen et al., 2020; Duan et al., 2020). Lignin biosynthesis and accumulation make a critical contribution to the adaptation of plants to high-salt stress (Chun et al., 2019), and the MdMYB46 transcription factor enhanced salt stress tolerance by activating lignin biosynthesis-related genes (Chen et al., 2019).
Significant progress has been made, but further studies to identify regulatory factors are needed to understand the underlying mechanisms of the strawberry salt stress response and to facilitate salt tolerant breeding efforts. In this study, the salt tolerances of 24 strawberry varieties were investigated with varying NaCl concentrations. Based on the salt damage indices, tested varieties were clustered as salt tolerant, salt sensitive, or salt hypersensitive. To explore the molecular mechanism of strawberry response to salt stress, two strawberry varieties with different salt sensitivity were used to perform transcriptomic and metabolomic analysis. We hypothesized the presence of common and distinct pathways in the response to salt stress in different varieties. To ensure we evaluated varieties with an intact salt response network, we selected a salt-sensitive variety instead of a salt-hypersensitive variety to unravel the mechanism of the strawberry response to salt stress. ‘Sweet Charlie’ (salt-tolerant) and ‘Benihoppe’ (salt-sensitive), the top two varieties with the largest planting area and the highest genetic contribution value in strawberry breeding in China (Chang et al., 2018), were selected in this study. The results further our understanding of the mechanism of the response of strawberry to salt stress and provide a theoretical basis for further study into regulation of salt stress tolerance in strawberry.
Materials and methods
Plant growth and salt stress treatment
In this study, 24 strawberry varieties from the National Strawberry Germplasm Repository (Beijing, China) were used to investigate salt tolerance under different concentrations of NaCl solution, which included four day-neutral varieties: ‘Albion,’ ‘San Andreas,’ ‘Portola,’ and ‘Monterey,’ and twenty short-day varieties: ‘Benihoppe,’ ‘Akihime,’ ‘Ssanta,’ ‘Tokun,’ ‘Kinuama,’ ‘Tochiotome,’ ‘Sweet Charlie,’ ‘Tianxiang,’ ‘Yanxiang,’ ‘Shuxiang,’ ‘Hongxiutianxiang,’ ‘Jingyixiang,’ ‘Jingchengxiang,’ ‘Jingquanxiang,’ ‘Jingzhangxiang,’ ‘Jingliuxiang,’ ‘Jingtaoxiang,’ ‘Pink Princess,’ ‘Snow White,’ and ‘Yanli,’ Strawberry seedlings were cultivated in 2.5-L containers with peat moss and perlite (2:1 v/v ratio) in a greenhouse with a 16-h photoperiod provided by supplemental lighting, 25/16°C (day/night) temperature cycle, 60% relative humidity, and 200 μmol m–2 s–1 light intensity. Salt stress treatments were conducted when the plants were at the 6–7 leaf stages. Plants were irrigated with four different concentrations of salt solutions: 50, 100, 150, or 200 mM NaCl for 50 days. To do this, 100 ml of the salt solution was applied once per week, and the same volume of fresh water was applied in the control treatment. Plants salt damage index (SDI) was measured as described by Zhong et al. (2021), and the SDI grades were scaled into six levels from 0 to 5, according to different damage symptoms observed in the leaves after salt treatment. The classification standard of plant salt damage was as follows: Grade 0: no symptoms of salt damage; Grade1: about 1/3 of the leaves showed wilting phenotype at leaf tips and leaf margins; Grade 2: about 1/2 of the leaves showed wilting and burn phenotype at leaf tips and leaf margins; Grade 3: about 2/3 of the leaves showed burn phenotype and the burn area was about 1/3; Grade 4: all leaves showed burn phenotype and the burn area was more than 1/2; Grade 5: all leaves were scorched. The SDI was counted as = Σ (salt damage series × number of plants with the corresponding salt damage level)/total number of plants tested. Biomass accumulation was measured as the dry weight of the whole plant. The experimental design included three randomized replicate blocks, with ten plants in each replicate block. The experiments were carried out in January, 2017 and January, 2018. To explore the molecular mechanism of strawberry response to salt stress, ‘Sweet Charlie’ and ‘Benihoppe’ were selected as strawberry varieties with different salt sensitivities and were subjected to transcriptomic and metabolomic analysis. ‘Sweet Charlie’ and ‘Benihoppe’ plants with 6–7 leaves were treated with 0 mM (control) or 100 mM NaCl solution, respectively. After 12 h of treatment, the young leaves were collected for ribonucleic acid (RNA) and metabolite extraction. After 10 days of treatment, the young leaves were collected for physiological indices determination.
Measurement of physiological indices
To evaluate the effects of salt stress on physiological performance of ‘Sweet Charlie’ and ‘Benihoppe,’ biochemical indices of malondialdehyde (MDA), superoxide anion (H2O2 and O2•–) content, and superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) activities were measured. Each experiment was performed with three biological replicates.
To evaluate the degree of membrane lipid peroxidation, the content of MDA was measured as described by Feng et al. (2022). Briefly, 0.5-gram samples of leaves were ground in buffer solution, and then supernatants were mixed with equivalent volumes of thiobarbituric acid (TBA) and incubated at 100°C for 10 min. Finally, the supernatant absorbance was read at 450, 532, and 600 nm.
The content of H2O2 was detected using a hydrogen peroxide assay kit (Solarbio, China) (Wang et al., 2019). Briefly, 0.1-gram leaves were homogenized in l ml cold acetone, and then supernatants were mixed with 5% titanium sulfate. The resulting precipitates were dissolved in 2 M sulfuric acid and then the absorbance was read at 415 nm.
The concentration of O2•– (superoxide anion) was determined using a superoxide anion assay kit (Solarbio, China) (Fang et al., 2021). Briefly, 0.1-gram leaves were ground in phosphate buffer solution, and then the supernatant was mixed with potassium phosphate buffer and hydroxylamine hydrochloride before reaction at 25°C for 20 min. Next p-aminobenzene sulfonic acid and 1-naphthylamine were added, the mixture was well-mixed and incubated at 30°C for 30 min, and then the absorbance was read at 530 nm.
To determine the activities of SOD, POD, and CAT, approximately 0.2-grams of leaves were ground and homogenized in sodium phosphate buffer and the resulting supernatants were assayed (Yu et al., 2021). SOD activity was determined according to the ability to inhibit the reduction of nitro blue tetrazolium (NBT) under light. POD activity was detected by measuring the oxidation of guaiacol, and CAT activity was measured by monitoring the consumption of hydrogen peroxide at 240 nm.
Transcriptome sequencing and data analysis
Total RNA was extracted from leaves using TRIzol® Reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions. The quality and concentration of RNA were determined by an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, United States) and a DS-11 Spectrophotometer (DeNovix, Wilmington, DE, United States), respectively. RNA-seq transcriptome library was constructed using TruSeqTM RNA sample preparation Kit (Illumina, San Diego, CA, United States) following the instructions of the manufacturer. An RNA-seq sequencing library was sequenced on the Illumina HiSeq X Ten sequencing platform by using paired-end technology using three biological replicates for each treatment.
After removing and filtering low-quality sequences, the clean reads were separately aligned to the reference genome (Fragaria x ananassa Camarosa Genome v1.0) using TopHat software.1 The determination of fragments per kilobase of exon per million mapped reads (FRKM) was employed to calculate the expression level of each transcript. RSEM2 was used to quantify gene abundances. R statistical package software Empirical analysis of Digital Gene Expression in R (EdgeR)3 was utilized for differential expression analysis. P-value < 0.05 and |log2FC| > 1 were set as the threshold for significantly differential expression.
Functional-enrichment GO and KEGG analyses were performed to identify DEGs significantly enriched in GO terms and metabolic pathways at a Bonferroni-corrected P-value of ≤0.05 compared with the whole-transcriptome background. GO functional enrichment and KEGG pathway analyses were carried out using Goatools4 and KOBAS.5
Metabolite profiling analysis
For the extraction of metabolites, fifty milligrams of strawberry leaves were mixed with 0.4 ml of an 80% aqueous methanol solution containing 0.02 mg/mL L-2-chlorophenylalanin as an internal standard. Chromatographic separation of the metabolites was performed on an UHPLC system (Thermo Fisher, Carlsbad, CA, United States) equipped with an ACQUITY UPLC HSS T3 (Waters, Milford, United States). The mass spectrometric data was collected using a Thermo UHPLC-Q Exactive HF-X Mass Spectrometer equipped with an ESI (electrospray ionization) source operating in either positive or negative ion mode. The metabolite profiling experiments were performed with six biological replicates for each treatment.
After UPLC-MS analyses, the raw data were imported into the Progenesis QI 2.3 (Non-linear Dynamics, Waters, United States) for peak detection and alignment. The preprocessing results generated a data matrix that was utilized for subsequent analyses. Human metabolome database6 (HMDB) and Metlin database7 were used for metabolite identification. Principal component analysis (PCA) and orthogonal partial least squares discriminate analysis (OPLS-DA) were performed on Majorbio Cloud Platform.8 Variable importance in the projection (VIP) were calculated in OPLS-DA model, and p values were estimated with paired Student’s t-test in single dimensional statistical analysis. Differentially accumulated metabolites (DAMs) between groups were selected with VIP ≥ 1, FC (fold change) ≥ 1 or FC ≤ 1 and p value ≤ 0.05. Differential metabolites between two groups were summarized and mapped into their biochemical pathways through metabolic enrichment and pathway analysis based on database search (KEGG9).
Validation of quantitative real-time polymerase chain reaction differentially expressed genes
Quantitative real-time polymerase chain reaction (qPCR) was performed to validate the accuracy of the RNA-seq results. To do this, qPCR was performed on a 7500 Real-time PCR System (Applied Biosystems, Carlsbad, CA, United States), with the following cycling profile: 95°C for 20 s followed by 40 cycles at 95°C for 5 s, 60°C for 10 s, and 72°C for 20 s; followed by a melting curve. The relative gene expression levels were quantified by the 2–ΔΔCT method. FaRPS1 (Merlaen et al., 2020) was selected as the internal control gene. The primer pairs are listed in Supplementary Table 1.
Statistical analysis
Statistical analysis was conducted using SPSS 20.0 software, and statistical comparisons were performed using t-test in SPSS. Data are presented as the mean ± standard deviation (SD) values with at least three biological replicates.
Results
Evaluation of strawberry salt tolerance
With increasing salt concentration, all 24 strawberry varieties showed a salt injury phenotype with the salt injury index positively correlated with salt concentration. Based on the salt damage indices, tested varieties could be clustered into three groups: salt tolerant, salt sensitive, and salt hypersensitive (Figure 1). The five varieties with the highest salt tolerance were ‘Portola,’ ‘Tokun,’ ‘Sweet Charlie,’ ‘Jingliuxiang,’ and ‘Pink Princess.’ Three varieties were extremely salt tolerant, ‘Portola,’ ‘Tokun,’ and ‘Sweet Charlie,’ with the plants neither dying nor withering at the highest test concentration of 200 mM NaCl. The varieties ‘Akihime,’ ‘Jingzhangxiang,’ ‘Monterey,’ ‘Jingchengxiang,’ and ‘Jingquanxiang’ were salt hypersensitive, with these varieties showing moderate to a severe salt damage phenotype under the 100 mM NaCl treatment. The other 16 varieties were salt sensitive varieties, with biomass accumulation and growth and development obviously inhibited under salt stress. In this study, the total biomass of 24 strawberry varieties decreased with the increase of NaCl concentration, but there were differences among different varieties that fell into three classes (Figure 2). Compared with the control treatment, the biomass was slightly increased and then decreased with increasing NaCl concentration for ‘Tokun,’ ‘Sweet Charlie,’ ‘Pink Princess,’ ‘Jingliuxiang,’ ‘Jingyixiang,’ ‘Hongxiutianxiang,’ ‘Tochiotome,’ ‘Jingtaoxiang,’ ‘Jingzangxiang,’ and ‘Akihime’; the biomass of ‘Yanli,’ ‘Benihoppe,’ ‘Ssanta,’ ‘Kinuama,’ ‘San Andreas,’ ‘Shuxiang,’ ‘Tianxiang,’ ‘Snow White,’ ‘Albion,’ ‘Yanxiang,’ ‘Jingchengxiang,’ and ‘Monterey’ steadily decreased with increasing NaCl concentration, and there was little difference in the biomass of ‘Portola’ under different concentrations of NaCl.
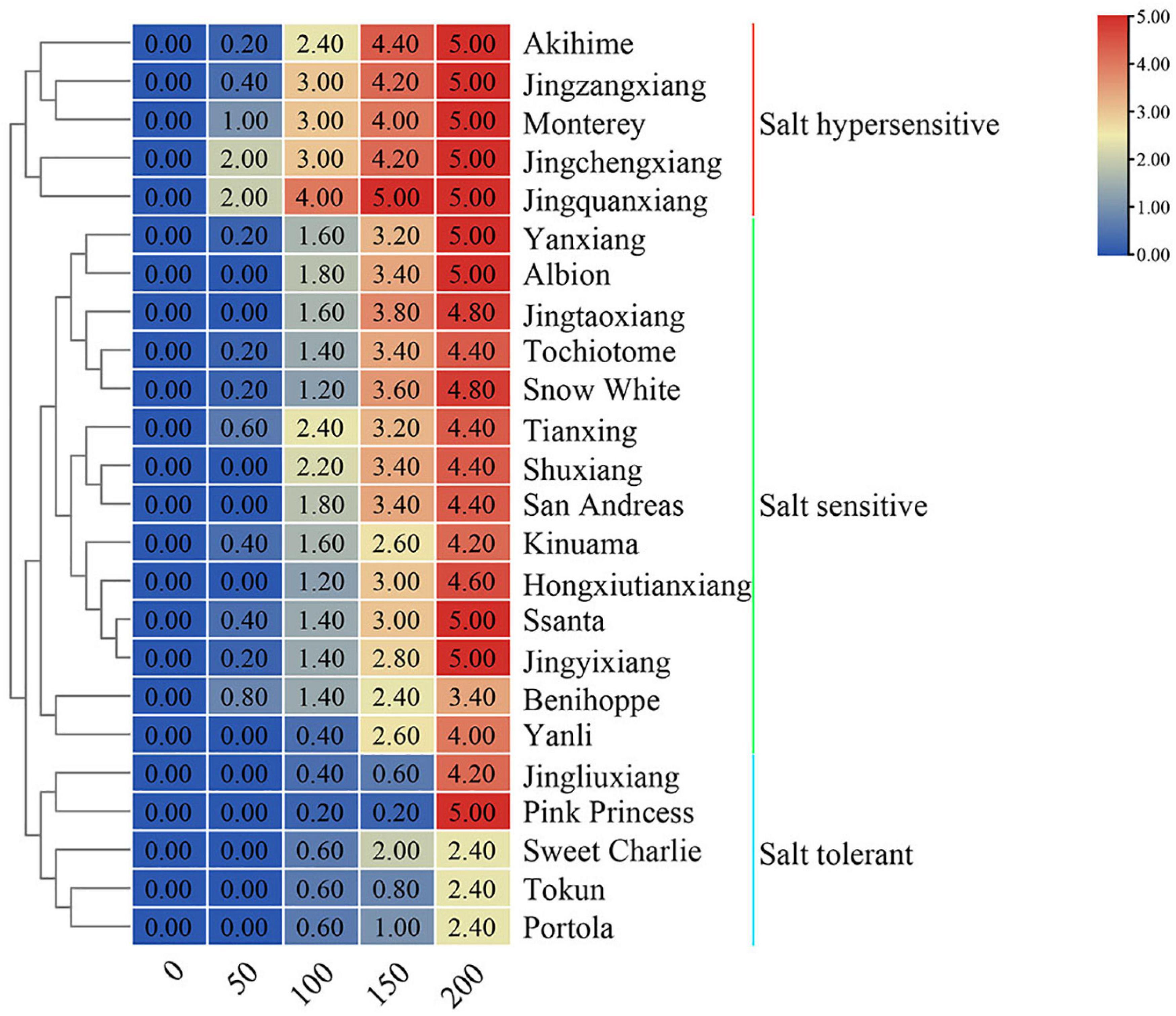
Figure 1. Salt damage index of 24 strawberry varieties under different concentration of NaCl. Strawberry plants with 6–7 leaves were irrigated with five different concentrations of salt solutions: 0, 50, 100, 150, or 200 mM NaCl for 50 days. The salt damage index grades were scaled into six levels from 0 to 5, according to different damage symptoms in the leaves after salt treatment. Based on the salt damage indices, tested varieties were clustered into three groups: salt tolerant, salt sensitive, and salt hypersensitive.
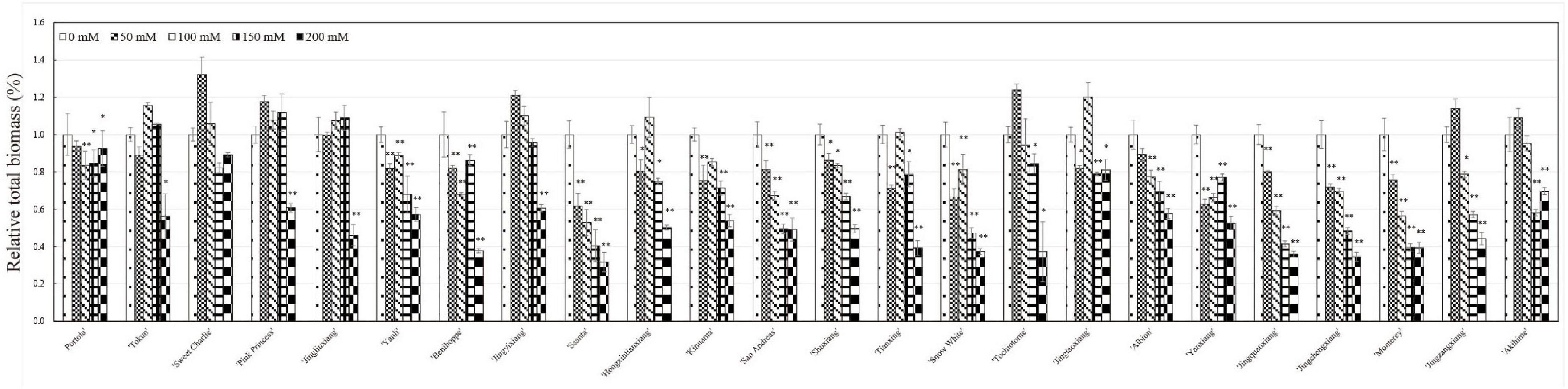
Figure 2. Relative total biomass of 24 strawberry varieties under different concentrations of NaCl. Strawberry plants with 6–7 leaves were irrigated with five different concentrations of salt solutions: 0, 50, 100, 150, or 200 mM NaCl for 50 days. Relative biomass accumulation was counted as the dry weight of the whole plant treated with different concentrations of salt solutions (50, 100, 150, or 200 mM) divided by the dry weight of the whole plant treated with fresh water. Data are means (±SD) of three independent experiments, *p < 0.05 or **p < 0.01.
‘Benihoppe’ is more sensitive to salt stress than ‘Sweet Charlie’
To investigate the physiological changes in the two representative varieties of ‘Sweet Charlie’ and ‘Benihoppe’ under salt stress, strawberry plants at the 6–7 leaf stage, were treated with 100 mM NaCl for 10 days. Next, several physiological parameters were measured. Under normal conditions, the content of MDA in leaves did not differ between ‘Sweet Charlie’ and ‘Benihoppe.’ However, after salt stress, the content of MDA in ‘Benihoppe’ was higher than that of ‘Sweet Charlie’ (Figure 3A). Excessive accumulation of ROS is a consequence of salt stress and leads to membrane lipid peroxidation. MDA is a marker of membrane lipid peroxidation. In addition, the accumulation of hydrogen peroxide and superoxide anion were increased in both ‘Benihoppe’ and ‘Sweet Charlie’ under salt stress condition, with higher values in ‘Benihoppe’ compared to the levels in ‘Sweet Charlie’ (Figures 3B,C). The opposite trend was observed for SOD, POD, and CAT activities, with all three parameters significantly higher in ‘Sweet Charlie’ than in ‘Benihoppe’ under salt stress condition (Figures 3D–F). Thus, the results indicate greater salt sensitivity of ‘Benihoppe’ than ‘Sweet Charlie.’
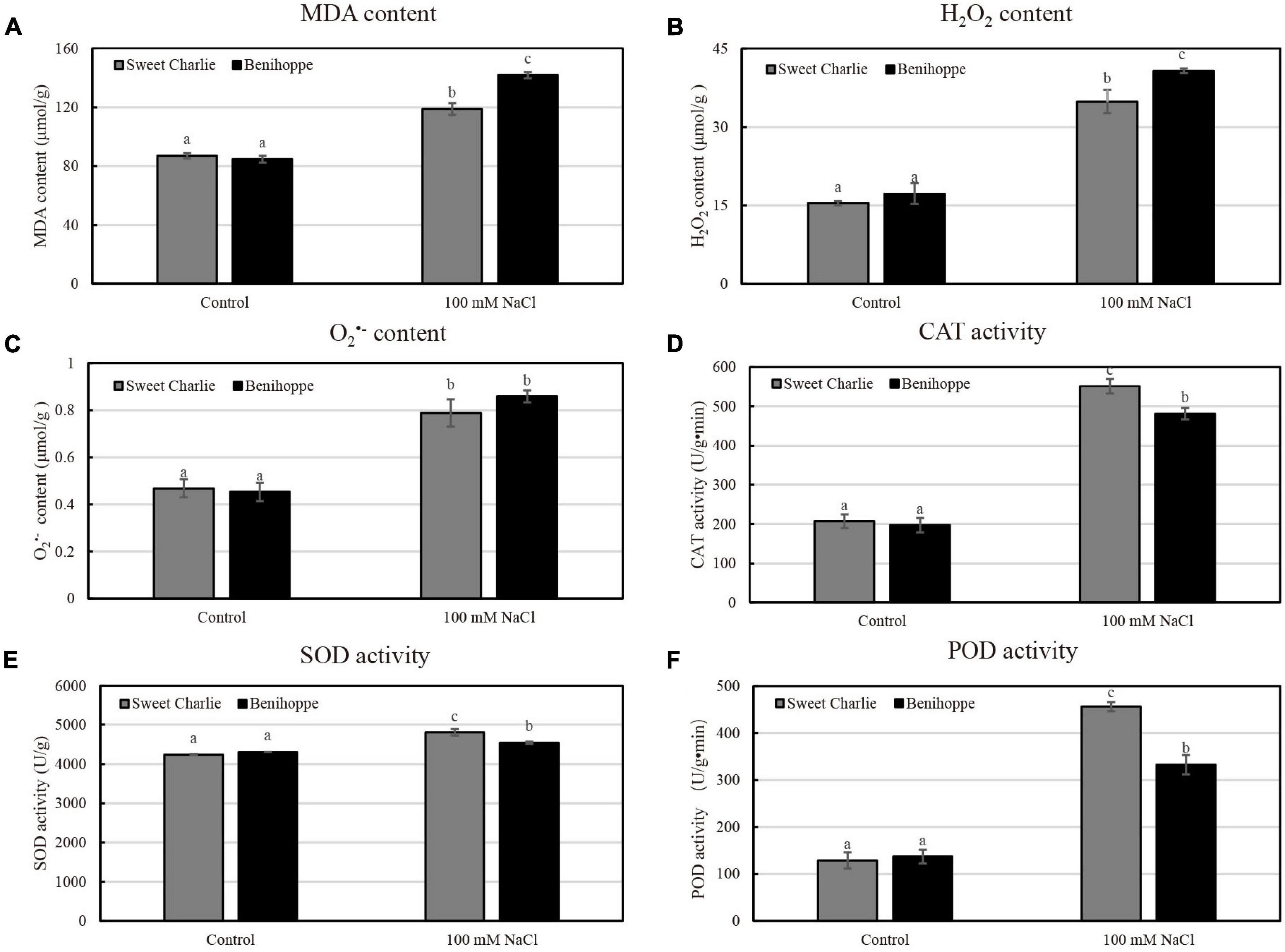
Figure 3. Physiological indices analysis of strawberry under salt stress. ‘Sweet Charlie’ and ‘Benihoppe’ plants with 6–7 leaves were treated with 0 mM (control) or 100 mM NaCl solution, respectively. After 10 days of treatment, the young leaves were collected for physiological indices determination. The content of MDA (A), H2O2 (B), and O2•– (C) under control and salt stress treatment in strawberry leaves. The activities of CAT (D), SOD (E), and POD (F) under control and salt stress treatment in strawberry leaves. Data are means (±SD) of three independent experiments. Different letters indicate significant differences (P < 0.05).
Transcriptomic profiles in ‘Benihoppe’ and ‘Sweet Charlie’ under salt stress
The genetic and biochemical responses of ‘Benihoppe’ and ‘Sweet Charlie’ to salt stress were further explored using RNA-seq analysis. A summary of the sequencing data is presented in Supplementary Table 2. Principal component analysis (PCA) showed clear separations between each group (Supplementary Figure 1). In ‘Benihoppe,’ compared with normal condition, 3412 genes were differentially expressed under 100 mM NaCl treatment, with 1869 were upregulated and 1543 downregulated (Supplementary Figure 2 and Supplementary Table 3). In ‘Sweet Charlie,’ compared with normal condition, 5102 genes were differentially expressed under 100 mM NaCl treatment, with 4396 upregulated and 706 downregulated (Supplementary Figure 2 and Supplementary Table 4). GO and GO enrichment analysis were performed to explore the functional significance of DEGs. In ‘Benihoppe,’ 522 GO terms were annotated and 297 significant GO terms were obtained, with several ion homeostasis related terms ranked in the top 20 enriched terms (Figure 4A and Supplementary Table 5). In ‘Sweet Charlie,’ 572 GO terms were annotated and 339 significant GO terms were obtained, with a suite of cell wall-related terms ranked in the top 20 enriched terms (Figure 4B and Supplementary Table 5). KEGG pathway classification was performed to reveal the active biological pathways in strawberry in response to salt stress. In ‘Benihoppe,’ the DEGs mapped to 116 KEGG pathways, with 21 pathways were significantly enriched (Figure 4C and Supplementary Table 6). In ‘Sweet Charlie,’ the DEGs were mapped to 120 KEGG pathways, and 24 pathways were significantly enriched (Figure 4D and Supplementary Table 6). Only four KEGG enriched pathways were shared between ‘Benihoppe’ and ‘Sweet Charlie,’ including flavonoid biosynthesis, phenylpropanoid biosynthesis, phenylalanine metabolism and ubiquinone biosynthesis, and other terpenoid-quinone biosynthesis. This lack of substantial overlap indicates that ‘Sweet Charlie’ and ‘Benihoppe’ respond to salt stress using different pathways.
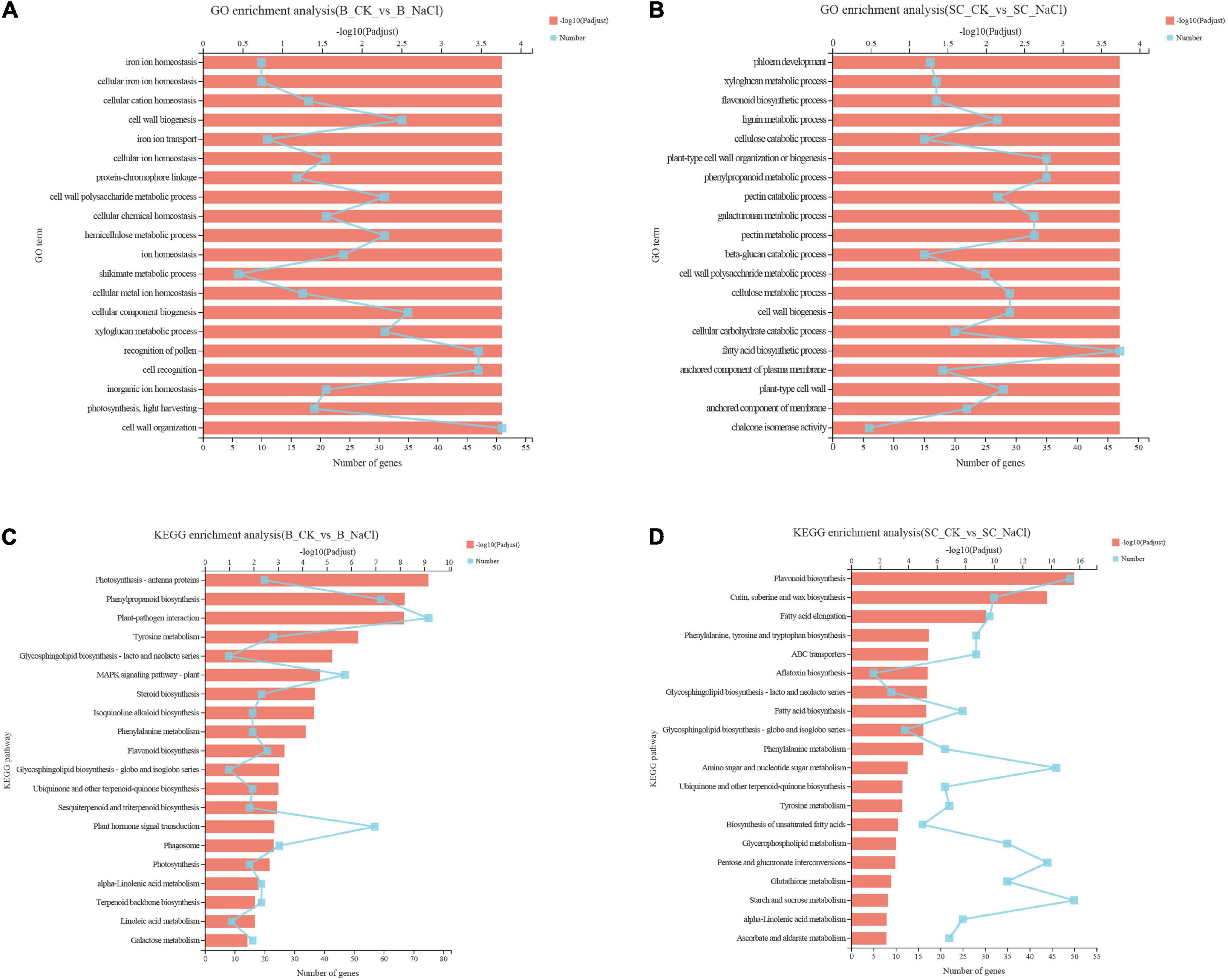
Figure 4. Gene Ontology enrichment analysis and KEGG pathway analysis of differentially expressed genes in strawberry leaves under salt stress. ‘Sweet Charlie’ and ‘Benihoppe’ plants with 6–7 leaves were treated with 0 mM (control) or 100 mM NaCl solution, respectively. After 12 h of treatment, the young leaves were collected for RNA-seq analysis. GO enrichment analysis and KEGG pathway analysis were performed to explore the functional significance of DEGs. Top 20 GO enrichment terms of differentially expressed genes in ‘Benihoppe’ (A) and ‘Sweet Charlie’ (B) under salt stress. Top 20 KEGG pathways of differentially expressed genes in ‘Benihoppe’ (C) and ‘Sweet Charlie’ (D) under salt stress.
Validation of ribonucleic acid-seq results by quantitative real-time polymerase chain reaction
Twelve genes were randomly selected to validate the RNA-seq results via qRT-PCR, testing levels of six downregulated genes and six upregulated genes. The qRT-PCR analysis results showed similar gene expression trends as the observed changes in the RNA-seq data (Figure 5). These results confirmed the reliability of RNA-seq in this study.
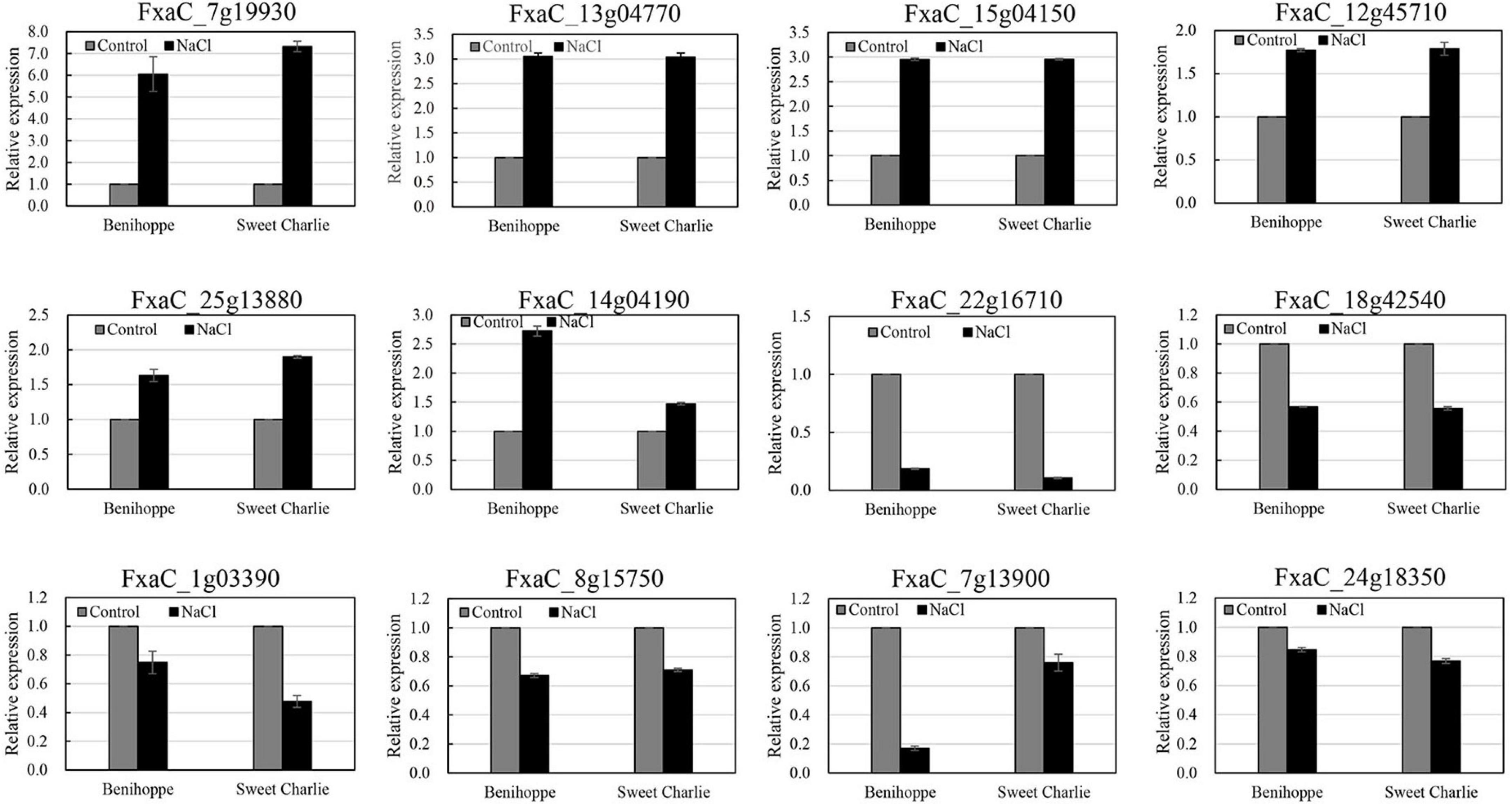
Figure 5. Verification of differentially expressed genes by qRT-PCR. ‘Sweet Charlie’ and ‘Benihoppe’ plants with 6–7 leaves were treated with 0 mM (control) or 100 mM NaCl solution, respectively. After 12 h of treatment, the young leaves were collected for RNA extraction. Twelve genes were randomly selected to validate the RNA-seq results via qRT-PCR. Data are means (±SD) of three independent experiments.
Metabolomic profiles in ‘Benihoppe’ and ‘Sweet Charlie’ under salt stress
Non-targeted metabolomic analysis was performed to expose the metabolomic changes of ‘Benihoppe’ and ‘Sweet Charlie’ in response to salt stress. In ‘Benihoppe,’ 3105 metabolites and 209 DAMs were identified (80 upregulated and 129 downregulated, Figure 6A and Supplementary Table 7). The DAMs were mapped to 25 KEGG pathways, and seven pathways were significantly enriched (Figure 6C and Supplementary Table 8). In ‘Sweet Charlie,’ 3381 metabolites and 230 DAMs were identified (85 upregulated and 145 downregulated, Figure 6B and Supplementary Table 7), and the DAMs were mapped to 42 KEGG pathways with 11 pathways significantly enriched under salt stress (Figure 6D and Supplementary Table 8). Among the enriched KEGG pathways, five pathways were shared between ‘Benihoppe’ and ‘Sweet Charlie,’ including flavonoid biosynthesis, aminoacyl-tRNA biosynthesis, cyanoamino acid metabolism, adenosine triphosphate-binding cassette (ABC) transporters, and tyrosine metabolism.
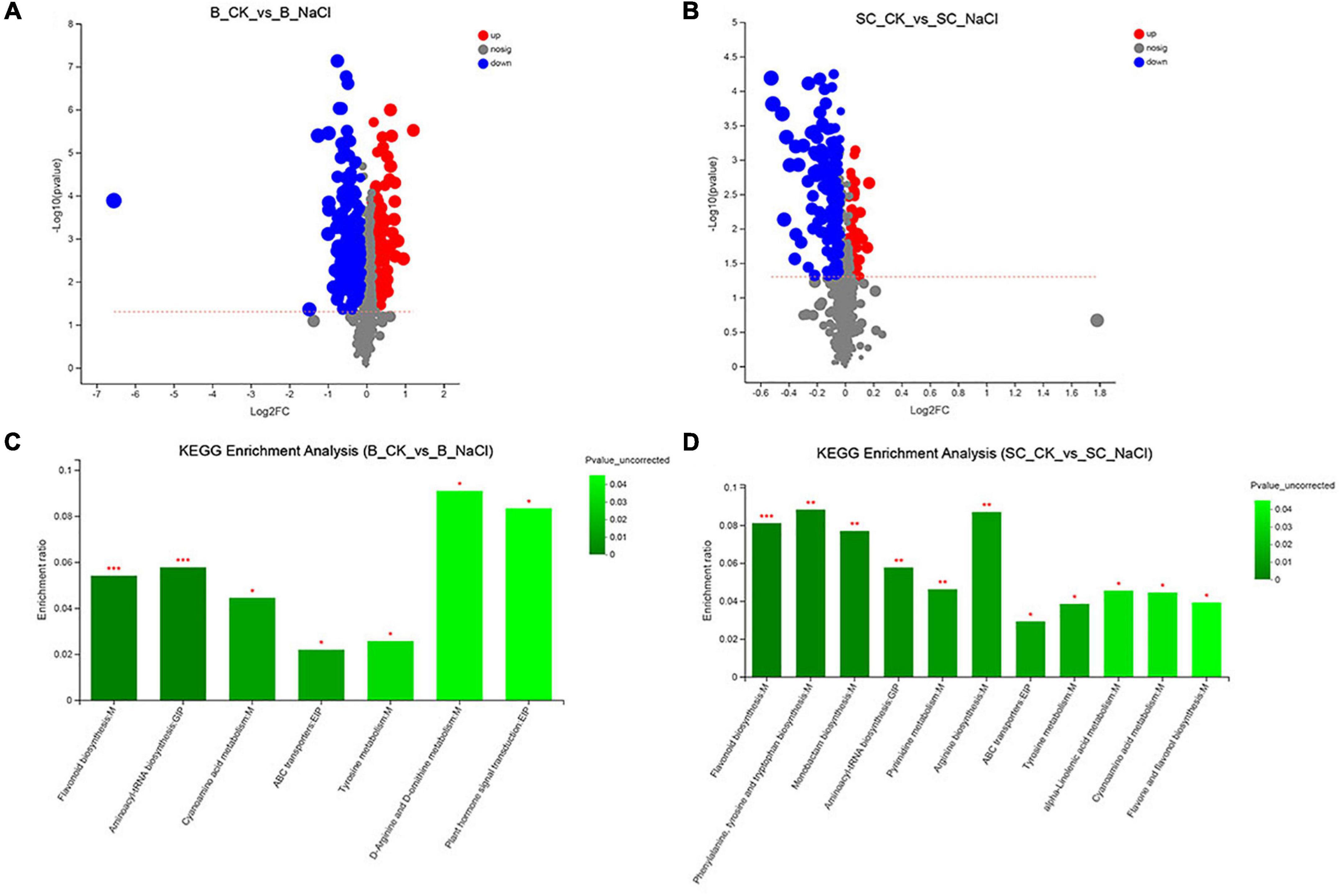
Figure 6. The statistics of differentially accumulated metabolites and KEGG enrichment under salt stress in strawberry. ‘Sweet Charlie’ and ‘Benihoppe’ plants with 6–7 leaves were treated with 0 mM (control) or 100 mM NaCl solution, respectively. After 12 h of treatment, the young leaves were collected for metabolite extraction. Non-targeted metabolomic analysis was performed to expose the metabolomic changes of ‘Benihoppe’ and ‘Sweet Charlie’ in response to salt stress. The volcano map of differentially accumulated metabolites in ‘Benihoppe’ (A) and ‘Sweet Charlie’ (B) leaves under salt stress. KEGG pathways of differentially accumulated metabolites in ‘Benihoppe’ (C) and ‘Sweet Charlie’ (D) under salt stress.
Ion homeostasis regulation under salt stress
Ion channels and ion transporters play vital roles in maintaining ion homeostasis under salt stress. In this study, ion channels and ion transporters related genes showed differential response patterns to salt stress in ‘Benihoppe’ and ‘Sweet Charlie.’ In ‘Benihoppe,’ 19 cyclic nucleotide-gated ion channel (CGNCs), two S-type anion channel (SLAH2), one potassium channel encode gene, two mechanosensitive ion channel protein encoding genes, five potassium transporter encoding genes, three vacuolar cation/proton exchanger (CHXs), and one cation/H+ antiporter (CHA) gene were upregulated, while one CNGC gene, one mechanosensitive ion channel protein encoding gene, one aluminum-activated malate transporter (ALMT) gene, and one CHA gene were downregulated under salt stress. In ‘Sweet Charlie,’ 10 CGNCs, three mechanosensitive ion channel protein encoding genes, three potassium transporter encoding genes, one ALMT gene, six CHXs, and one CHA were upregulated, while six CNGCs, two potassium transporter encoding genes, three ALMTs, and one CHX gene were downregulated under salt stress (Figure 7 and Supplementary Table 9). Aquaporins play important roles in responsive to salt tolerance. In ‘Benihoppe,’ one plasma membrane intrinsic protein (PIP) gene was upregulated and one PIP and eight tonoplast intrinsic protein (TIP) genes were downregulated under salt stress. In ‘Sweet Charlie,’ seven TIPs, two PIPs, two nodulin-like intrinsic protein (NIPs), and one small and basic intrinsic proteins (SIP) gene were upregulated, while one NIP gene was downregulated (Figure 7 and Supplementary Table 9). NPF (NRT1/PTR FAMILY) belong to the large PTR (peptide transporter) family, and plant NPFs can identify a wide variety of substrates, including NO3–, NO2, Cl–, abscisic acid (ABA), auxin (IAA), and gibberellins (GAs) (Corratge-Faillie and Lacombe, 2017). Under salt stress, nine NPFs were upregulated, and eight NPFs were downregulated in ‘Benihoppe,’ with nine NPFs upregulated and three NPFs downregulated in ‘Sweet Charlie’ (Figure 7 and Supplementary Table 9). ABC transporters utilize the energy released from ATP hydrolysis to transport substrates and participate in the response to salt stress. Under salt stress, seven ABCs were upregulated and four ABCs were downregulated in ‘Benihoppe,’ while 38 ABCs were upregulated and one ABC was downregulated in ‘Sweet Charlie.’ In ‘Benihoppe,’ the differentially expressed ABCs belong to B, C, G, and F subfamilies, while in ‘Sweet Charlie,’ the differentially expressed ABCs belong to A, B, C, and G subfamilies. In ‘Benihoppe,’ three DAMs related to the ABC transporter pathway were identified, including L-Arginine, Cytidine, and L-Phenylalanine with L-Phenylalanine downregulated, and L-Arginine and Cytidine upregulated under salt stress. In ‘Sweet Charlie,’ four DAMs related to ABC transporter pathway were identified, including L-Arginine, Uridine, Xanthosine, and L-Aspartic Acid, with L-Arginine upregulated and Uridine, Xanthosine, and L-Aspartic Acid downregulated under salt stress (Figure 7 and Supplementary Table 9).
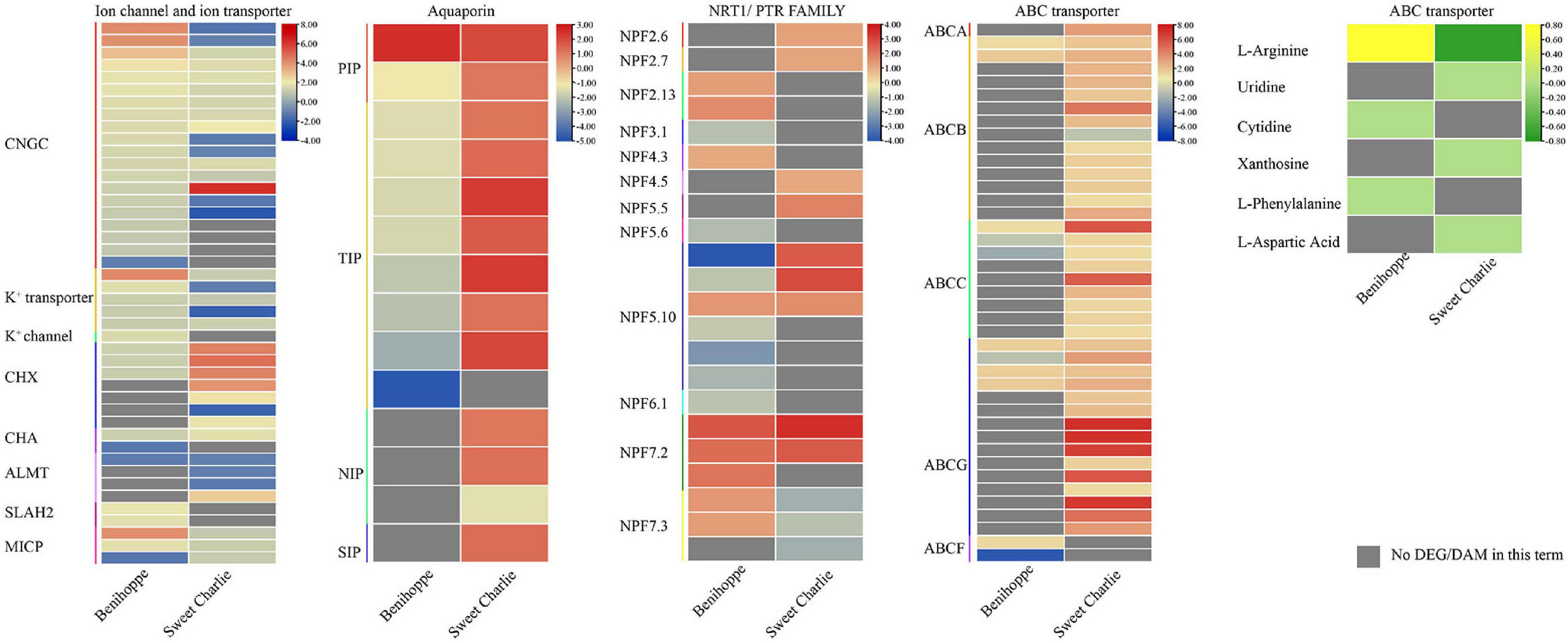
Figure 7. Differentially expressed genes related to ion channel and ion transporter, aquaporin, NRT1/PTR FAMILY, and ABC transporters, and differentially accumulated metabolites related to ABC transporters in ‘Benihoppe’ (left) and ‘Sweet Charlie’ (right) under salt stress. The relative expression levels as described by log2FC are represented by a color gradient from low (blue) to high (red). Under salt stress, the DEGs with log2FC > 0 represent upregulated expression, log2FC < 0 represents downregulated expression, and log2FC = 0 represents unchanged expression. Accumulation of metabolites as described by log2FC is represented by a color gradient from low (green) to high (yellow). Under salt stress, the DAMs with log2FC > 0 represent upregulated accumulation, log2FC < 0 represents downregulated accumulation, and log2FC = 0 represents unchanged accumulation. Gray squares indicate no differentially expressed genes in this term.
Cell wall remodeling genes under salt stress
In this study we identified multiple cell wall-related genes that exhibited differential expression in strawberry in response to salt stress. Xyloglucan endotransglucosylase/hydrolase (XHT) (Sharples et al., 2017), expansin (EXP) (Hepler et al., 2020), alpha-galactosidase (α-Gal A) (Chrost et al., 2007), and L-ascorbate oxidase (AO) (Green and Fry, 2005) participate in cell wall loosening. In‘Benihoppe,’ two XHTs, four EXPs, and one AO encoding genes were upregulated, while 29 XHTs, 10 EXPs, and six AO encoding genes were downregulated under salt stress. In ‘Sweet Charlie,’ 13 XHTs, 16 EXPs, and seven AO encoding genes were upregulated, while two EXPs and two α-Gal A encoding genes were downregulated under salt stress. Cellulase (Cel), endoglucanase, endo-beta-1,4-glucanase, endo-1,4-beta-glucanase, beta xylosidase (BXL), and beta-D-xylosidase (XYL) are well-known cell wall-degrading enzymes. In ‘Benihoppe,’ one endoglucanase encoding gene and three XYLs were downregulated under salt stress. In ‘Sweet Charlie,’ two Cels, 12 endoglucanase encoding genes, four endo-beta-1,4-glucanase encoding genes, three endo-1,4-beta-glucanase encoding genes, three BXLs, and three XYLs were upregulated under salt stress (Figure 8 and Supplementary Table 10). Pectate lyase (PL), Pectinesterase (PE), and Polygalacturonase (PG) are pectin-degrading enzymes (Chen et al., 2018c,2021; Zhang et al., 2021a), and galacturonosyltransferase (GAUTs) are involved in pectin synthesis (Mohnen et al., 2012). In ‘Benihoppe,’ four PEs and two GAUTs were upregulated, while 11 PLs and five PEs were downregulated under salt stress. In ‘Sweet Charlie,’ 17 PLs, 20 PEs, 19 PGs, and four GAUTs were upregulated, and four PEs were downregulated under salt stress (Figure 8 and Supplementary Table 10). Cellulose synthase A (CesA), cellulose synthase-like (Csl), and COBRA-like (COBL) are involved in cellulose synthase (Li et al., 2019b; Daras et al., 2021). In ‘Benihoppe,’ two Csls were downregulated under salt stress. In ‘Sweet Charlie,’ eight CesAs, five Csls, and five COBLs were upregulated (Figure 8 and Supplementary Table 10). Laccase (LAC) and peroxidase (PER) are well-characterized lignin-related enzymes and omega-hydroxypalmitate O-feruloyl transferase (HHT1) participate in lignin monomer synthesis. In ‘Benihoppe,’ three LACs, 10 peroxidase encoding genes, and one omega-hydroxypalmitate O-feruloyl transferase encoding gene were upregulated, while 16 LACs, 14 peroxidase encoding genes, and one HHT1 encoding gene were downregulated under salt stress. In ‘Sweet Charlie,’ 35 LACs, nine peroxidase encoding genes, and three HHT1 were upregulated, while one LAC, seven peroxidase encoding genes, and three HHT1 encoding genes were downregulated under stress (Figure 8 and Supplementary Table 10). Glycosyltransferases, xylan alpha-glucuronosyltransferase (GUX), and DUF579 domain containing proteins IRX15-L are linked with xylan biosynthesis, and IRX7/9/10 are members of the glycosyltransferase family (Oikawa et al., 2010; Jensen et al., 2011, 2014). Glucuronoxylan 4-O-methyltransferase (GXM) involves xylan methylation, ALTERED XYLOGLUCAN 4 (AXY4) involves xylan acetylation, and GDSL esterase/lipase (GSDL) affects xylan deacetylation. In ‘Benihoppe,’ one GDSL was upregulated and one AXY4 and 23 GDSLs were downregulated under salt stress. In ‘Sweet Charlie,’ four GUXs encoding genes, three IRX15-L, two IRX7, three IRX9, two IRX10, three GXM encoding genes, three AXY4s, and 52 GDSLs were upregulated under salt stress (Figure 8 and Supplementary Table 10). Inositol oxygenase is linked to the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides (Kanter et al., 2005). In ‘Sweet Charlie,’ seven inositol oxygenase encoding genes were up regulated under salt stress (Figure 8 and Supplementary Table 10). The members of the PHI-1/EXO/EXL protein family participate in regulating secondary cell wall thickening and composition, and lignification (Sousa et al., 2020). Under salt stress, 11 EXL3s were downregulated in ‘Benihoppe,’ and five EXL3s were upregulated in ‘Sweet Charlie’ (Figure 8 and Supplementary Table 10). The galactoside 2-alpha-L-fucosyltransferase FUT1 adds a fucose residue to the 2-O position of terminal galactosyl residues on XyG side chains (Rocha et al., 2016). Under salt stress, one galactoside 2-alpha-L-fucosyltransferase encode gene was upregulated in ‘Benihoppe,’ and three galactoside 2-alpha-L-fucosyltransferase encoding genes were upregulated in ‘Sweet Charlie’ (Figure 8 and Supplementary Table 10).
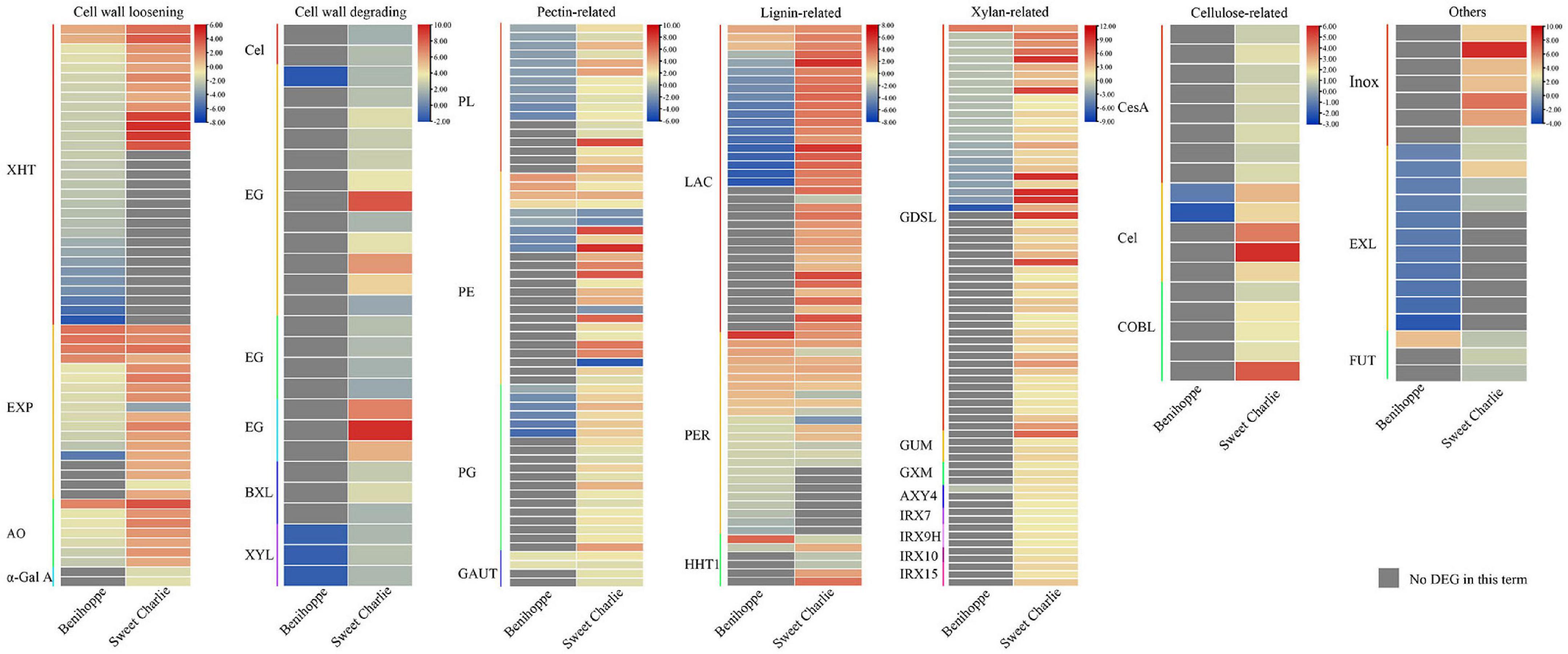
Figure 8. Differentially expressed genes related to cell wall remodeling in ‘Benihoppe’ (left) and ‘Sweet Charlie’ (right) under salt stress. The relative expression levels as described by log2FC are represented by a color gradient from low (blue) to high (red). Under salt stress, DEGs with log2FC > 0 represent upregulated expression, log2FC < 0 represents downregulated expression, and log2FC = 0 represents unchanged expression. Gray squares indicate no differentially expressed gene or differentially accumulated metabolite in this term.
Flavonoid pathway under salt stress
Integrative analysis of transcriptomic and metabolomic data showed that the flavonoid pathway is involved in the salt stress response in strawberry. In ‘Benihoppe,’ the expression level of FLSs was reduced by salt stress, the expression of F3’H, DFR, ANS, LAR, and PGT1 was induced by salt stress, and the content of epigallocatechin, taxifolin, and phloridzin decreased under salt stress. In ‘Sweet Charlie,’ salt stress induced expression of CHS, CHI, F3H, FLS, F3’H, DFR, ANS, ANR, LAR, and PGT1; while the levels of dihydrokaempferol, epicatechin, phloridzin, and pinocembrin were reduced by salt stress; and an increase in the content of pelargonidin was induced by salt stress (Figure 9 and Supplementary Table 11).
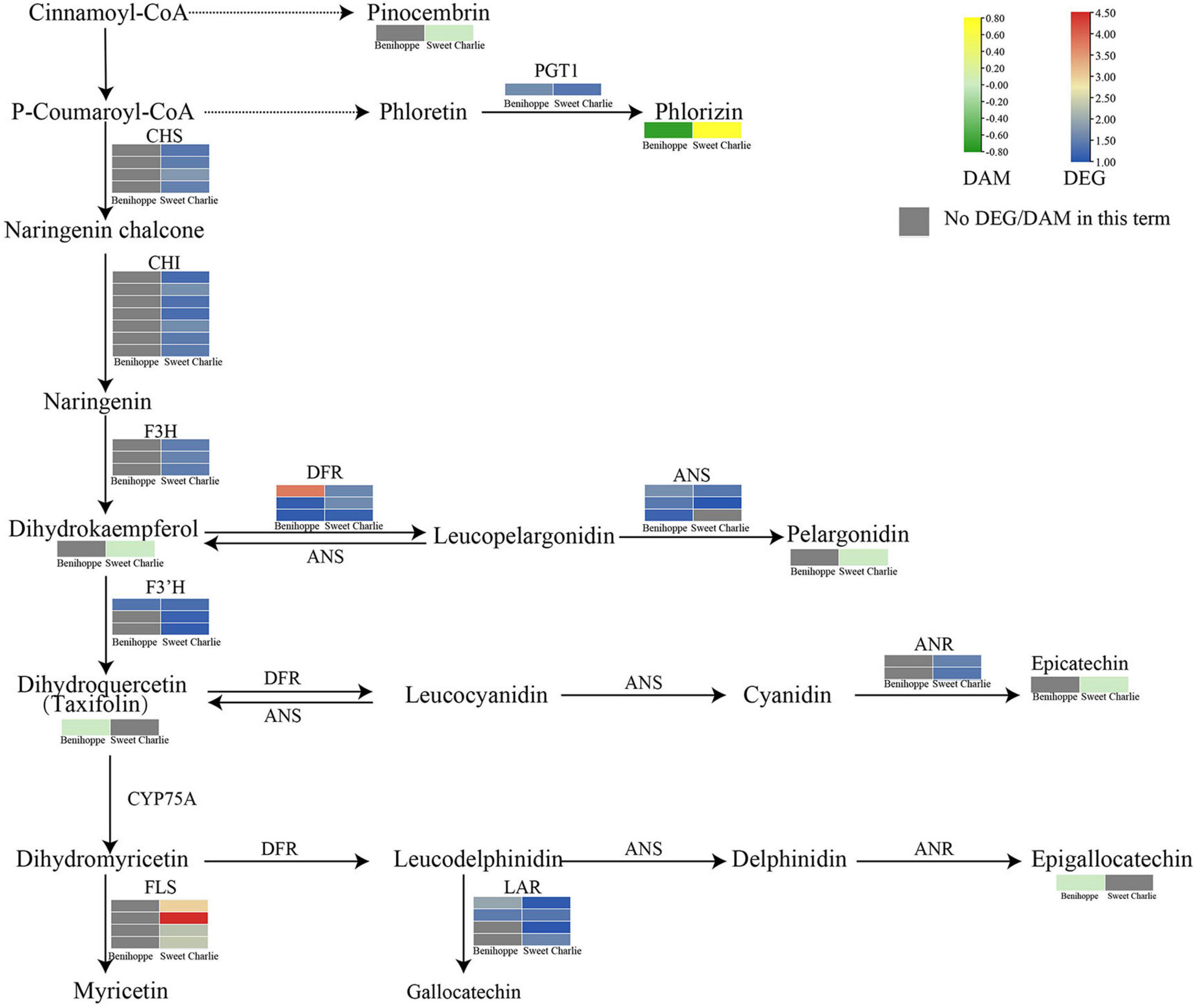
Figure 9. Differentially expressed genes and differentially accumulated metabolites related to flavonoid pathways in ‘Benihoppe’ (left) and ‘Sweet Charlie’ (right) under salt stress. The relative expression levels as described by log2FC are represented by a color gradient from low (blue) to high (red). Under salt stress, the DEGs with log2FC > 0 represent upregulated expression, log2FC < 0 represents downregulated expression, and log2FC = 0 represents unchanged expression. The accumulated of metabolites as described by log2FC are represented by a color gradient from low (green) to high (yellow). Under salt stress, the DAMs with log2FC > 0 represent upregulated accumulation, log2FC < 0 represents downregulated accumulation, and log2FC = 0 represents unchanged accumulation. Gray squares indicate no differentially expressed gene or differentially accumulated metabolite in this term.
Discussion
Salt stress disturbs cellular ionic homeostasis through the excessive accumulation of Na+ and Cl–. Plants have evolved efficient ion transport networks that enable them to maintain ion homeostasis, and maintaining a high K+/Na+ ratio in the cytosol is a key determinant of salinity tolerance (Almeida et al., 2017). CGNC, CHX, CHA, potassium channel, and potassium transporters play core roles in this process. The Arabidopsis genes AtCNGC19 and AtCNGC20 positively regulate plant salt tolerance, and AtCNGC10 participates in K+ and Na+ uptake and long-distance transport (Guo et al., 2008). The CHX protein plays a critical role in maintaining K+ and Na+ homeostasis. In soybean, GmCHX1 and GmCHX20a presented different expression patterns and opposite effects to salt tolerance (Jia et al., 2021). Chloride ions are a major factor contributing to ion toxicity and decreased production in strawberry (Suarez and Grieve, 2013; Devinder et al., 2019; Ferreira et al., 2019). To date, the most commonly reported gene families responsible for Cl– transport are Cl– channels (CLC), cation chloride channels (CCC), SLAH, ALMT, and NPF (Wu and Li, 2019). In this study, changes in the expression of different ion channel encoding genes and ion transporters were detected in ‘Benihoppe’ and ‘Sweet Charlie,’ with significant differences in Cl– transport related genes. These results suggest different regulatory mechanisms of ion homeostasis between ‘Benihoppe’ and ‘Sweet Charlie’ (Figure 7 and Supplementary Table 9).
Aquaporins are a large family of transmembrane channel proteins that participate in the transport of water and nutrients. The aquaporins include five subfamilies of PIP, TIP, SIP, NIP, and X intrinsic proteins (XIP). Aquaporins help plants maintain water and ionic homeostasis and respond to salt stress. In wheat, TaTIP4;1 serves as a positive regulator of salt tolerance by modulating water relations and the accumulation of Na+ (Wang et al., 2022). ZxPIP1;3 conferred transgenic plants with salt tolerance by regulating water status and reducing ion toxicity (Li et al., 2021). MaPIP1;1 enhanced banana salt tolerance via affecting the contents of Na+ and K+ (Xu et al., 2021). In this study, we found that PIP and TIPs participate in response to salt stress in ‘Benihoppe,’ while TIP, PIPs, NIPs, and SIP participate in response to salt stress in ‘Sweet Charlie.’ This suggests that different aquaporins are required for response to salt stress in different varieties, but TIP and PIP are the main aquaporins involved in response to salt stress in strawberry (Figure 7 and Supplementary Table 9).
The ABC transporters are one of the largest families of transporter proteins and are classified into eight subfamilies of ABCA-ABCI (ABCH transporters have not been identified in plants). ABC transporters are responsible for the transport of hormones, xenobiotics, amino acids, sugar, and ions, and are involved in plant growth and development regulation, nutrient uptake, response to biotic and abiotic stresses, and plant interactions with the environment. Recent findings have revealed roles of ABC transporters in response to salt stress. TsABCG11 improved salt tolerance in transgenic Arabidopsis seedlings (Chen et al., 2018b). In Arabidopsis, AtMRP5, a member of the ABC transporter family, can regulate Arabidopsis salt tolerance by altering K+ homeostasis (Lee et al., 2004), and AtABCG36/AtPDR8 improved salt resistance by reducing sodium content (Kim et al., 2010). In rice, OsABCG5 acts in the accumulation of essential and non-essential minerals and modulated rice salinity tolerance by affecting Na+/K+ homeostasis (Leng et al., 2014). ABCG and ABCB subfamilies tightly correlate with abiotic stress in plants (Linjun et al., 2019; Zhang et al., 2020). The ABCG subfamily is the largest group of the ABC transporter family and possesses the most complex function in response to abiotic stresses. In this study, ABCGs were more active in responses to salt stress, with one ABCA, 14 ABCB, nine ABCC, and 15 ABCG participating in salt stress responses in ‘Sweet Charlie,’ and two ABCB, three ABCC, four ABCG, and two ABCF participating in salt stress responses in ‘Benihoppe.’ Integrative analysis of transcriptomic and metabolomic data revealed that ABC transport plays an important role in response to salt stress in strawberry (Figure 7 and Supplementary Table 9).
Cell wall remodeling is a key aspect of plant acclimation to salt stress. Plant cell wall components are dynamically regulated in response to various environmental stresses. Salt stress can modify the deposition of cellulose, matrix, polysaccharides, and lignin. Under salt stress, the levels of cellulose and matrix polysaccharide were reduced but the content of lignin was increased in maize (Oliveira et al., 2020). In soybean, a salt-tolerant cultivar possessed a higher level of pectin than the sensitive line. Under salt stress, sodium ions affected pectin cross-links and disrupted microtubule stability to change cellulose deposition (An et al., 2014). Pectin usually presents in a highly methylesterified form. In Populus tremula, high methylated pectin within the cell wall was increased by salt stress (Muszyńska et al., 2014). Changes caused by salt stress results in cell wall remodeling to maintain cell wall integrity. Cell wall-loosening proteins such as EXPs and XTH play essential roles in response to salt stress. Overexpression of EXPs confers enhanced tolerance to salt stress in plants (Chen et al., 2017, 2018a; Jadamba et al., 2020). XTH positively regulated plant salt stress tolerance by regulating plant architecture (Ishida and Yokoyama, 2022). COBL9 and COBL7, two COBRA-like family genes, are required for salinity tolerance and COBL9 positively regulated root hair elongation and salinity tolerance in Arabidopsis; the rice counterparts of these genes, OsBC1L1 and OsBC1L8, function redundantly in response to salinity stress (Li et al., 2022). Pectin plays an important role in adaptation to salt stress by regulating cell adhesion and tissue cohesion. In Arabidopsis, Pectin methylesterase 31 (PME31) positively modulated salt stress tolerance (Yan et al., 2018). Enhanced cell wall lignification is one of the main salinity tolerance strategies in the roots of halophytes. LAC4 plays an important role in response to early stages of salt stress via affecting specialized protoxylem lignification in undifferentiated root tips (Barzegargolchini et al., 2017). In this study, more cell wall related genes were identified in ‘Sweet Charlie’ than in ‘Benihoppe,’ and those genes presented opposite expression patterns under salt stress. These results suggested that cell wall remodeling is a key driver of plant salt tolerance (Figure 8 and Supplementary Table 10).
In this study, we evaluated the salinity tolerance of 24 strawberry varieties and then clustered the varieties into three groups according to the salt damage indices. Physiological index analysis showed that ‘Benihoppe’ was more sensitive to salt stress than ‘Sweet Charlie,’ and this was consistent with the analysis of salt damage indices. Combined transcriptomic and metabolomic analysis showed that different pathways respond to salt stress in different varieties, and ABC transporters and cell wall remodeling play crucial roles in response to salt stress. Our results provide a foundation for the better understanding of the regulation mechanisms of the response of strawberry to salt stress.
Data availability statement
The data presented in this study are deposited in the NCBI (National Center for Biotechnology Information) SRA repository, accession number PRJNA837123.
Author contributions
SL, GW, YZ, and JS conceived this project and designed the research. SL, LC, and RS performed most of the experiments. JD, CZ, YG, HZ, LW, and YW participated in this work. SL and JS analyzed the data and wrote the manuscript. All authors discussed the manuscript, read, and approved the final manuscript.
Funding
This work was supported by grants from National Key R&D Program of China (2020YFD1000105), China Agriculture Research System of MOF and MARA (CARS-24-A-14), Beijing Academy of Agriculture and Forestry Special Fund for the Construction of Scientific and Technological Innovation Capability (KJCX20200114), Beijing Postdoctoral Research Foundation (2021-ZZ-132), and Beijing Academy of Agriculture and Forestry Special Fund for the Construction of Scientific and Technological Innovation Capability.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.996765/full#supplementary-material
Supplementary Figure 1. Principal component analysis (PCA) plots between individual samples for control and NaCl treatments.
Supplementary Figure 2. Volcano map of differentially expressed genes in ‘Benihoppe’ (A) and ‘Sweet Charlie’ (B) leaves under salt stress.
Footnotes
- ^ http://tophat.cbcb.umd.edu
- ^ http://deweylab.biostat.wisc.edu/rsem
- ^ http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html
- ^ https://github.com/tanghaibao/Goatools
- ^ http://kobas.cbi.pku.edu.cn/index.php
- ^ http://www.hmdb.ca/
- ^ https://metlin.scripps.edu/
- ^ https://cloud.majorbio.com
- ^ http://www.genome.jp/kegg/
References
Almeida, D. M., Oliveira, M. M., and Saibo, N. J. M. (2017). Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 40, 326–345. doi: 10.1590/1678-4685-GMB-2016-0106
An, P., Li, X., Zheng, Y., Matsuura, A., Abe, J., Eneji, A. E., et al. (2014). Effects of NaCl on root growth and cell wall composition of two soya bean cultivars with contrasting salt tolerance. J. Agron. Crop Sci. 200, 212–218. doi: 10.1111/jac.12060
Arora, N. K., Fatima, T., Mishra, J., Mishra, I., Verma, S., Verma, R., et al. (2020). Halo-tolerant plant growth promoting rhizobacteria for improving productivity and remediation of saline soils. J. Adv. Res. 26, 69–82. doi: 10.1016/j.jare.2020.07.003
Barzegargolchini, B., Movafeghi, A., Dehestani, A., and Mehrabanjoubani, P. (2017). Increased cell wall thickness of endodermis and protoxylem in Aeluropus littoralis roots under salinity: The role of LAC4 and PER64 genes. J. Plant Physiol. 218, 127–134. doi: 10.1016/j.jplph.2017.08.002
Bharti, P., Mahajan, M., Vishwakarma, A. K., Bhardwaj, J., and Yadav, S. K. (2015). AtROS1 overexpression provides evidence for epigenetic regulation of genes encoding enzymes of flavonoid biosynthesis and antioxidant pathways during salt stress in transgenic tobacco. J. Exp. Bot. 66, 5959–5969. doi: 10.1093/jxb/erv304
Bomle, D. V., Kiran, A., Kumar, J. K., Nagaraj, L. S., Pradeep, C. K., Ansari, M. A., et al. (2021). Plants saline environment in perception with rhizosphere bacteria containing 1-Aminocyclopropane-1-Carboxylate deaminase. Int. J. Mol. Sci. 22:11461. doi: 10.3390/ijms222111461
Chang, L., Dong, J., Zhong, C. F., Sun, J., Sun, R., Shi, K., et al. (2018). Pedigree analysis of strawberry cultivars released in China. J. Fruit Sci. 35, 158–167. doi: 10.13925/j.cnki.gsxb.20170279
Chen, K., Guo, Y., Song, M., Liu, L., Xue, H., Dai, H. Y., et al. (2020). Dual role of MdSND1 in the biosynthesis of lignin and in signal transduction in response to salt and osmotic stress in apple. Hortic. Res. 7:204. doi: 10.1038/s41438-020-00433-7
Chen, K., Song, M., Guo, Y., Liu, L., Xue, H., Dai, H. Y., et al. (2019). MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol. J. 17, 2341–2355. doi: 10.1111/pbi.13151
Chen, L., Guo, H., Lin, Y., and Cheng, H. (2015). Chalcone synthase EaCHS1 from Eupatorium adenophorum functions in salt stress tolerance in tobacco. Plant Cell Rep. 34, 885–894. doi: 10.1007/s00299-015-1751-7
Chen, Y., Zhang, S., Lin, H., Sun, J., Lin, Y., Wang, H., et al. (2018c). Phomopsis longanae chi-induced changes in activities of cell wall-degrading enzymes and contents of cell wall components in pericarp of harvested longan fruit and its relation to disease development. Front. Microbiol. 9:1051. doi: 10.3389/fmicb.2018.01051
Chen, N., Song, B., Tang, S., He, J., Zhou, Y., Feng, J. C., et al. (2018b). Overexpression of the ABC transporter gene TsABCG11 increases cuticle lipids and abiotic stress tolerance in Arabidopsis. Plant Biotechnol. Rep. 12, 303–313. doi: 10.1007/s11816-018-0495-6
Chen, L., Zou, W., Fei, C., Wu, G., Li, X., Lin, H. H., et al. (2018a). alpha-Expansin EXPA4 positively regulates abiotic stress tolerance but negatively regulates pathogen resistance in Nicotiana tabacum. Plant Cell Physiol. 59, 2317–2330. doi: 10.1093/pcp/pcy155
Chen, Y., Han, Y., Kong, X., Kang, H., Ren, Y., and Wang, W. (2017). Ectopic expression of wheat expansin gene TaEXPA2 improved the salt tolerance of transgenic tobacco by regulating Na+/K+ and antioxidant competence. Physiol. Plant. 159, 161–177. doi: 10.1111/ppl.12492
Chen, Y., Li, W., Turner, J. A., and Anderson, C. T. (2021). PECTATE LYASE LIKE12 patterns the guard cell wall to coordinate turgor pressure and wall mechanics for proper stomatal function in Arabidopsis. Plant Cell 33, 3134–3150. doi: 10.1093/plcell/koab161
Chrost, B., Kolukisaoglu, U., Schulz, B., and Krupinska, K. (2007). An alpha-galactosidase with an essential function during leaf development. Planta 225, 311–320. doi: 10.1007/s00425-006-0350-9
Chun, H. J., Baek, D., Cho, H. M., Lee, S. H., Jin, B. J., Yun, D. J., et al. (2019). Lignin biosynthesis genes play critical roles in the adaptation of Arabidopsis plants to high-salt stress. Plant Signal. Behav. 14:8. doi: 10.1080/15592324.2019.1625697
Corratge-Faillie, C., and Lacombe, B. (2017). Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 68, 3107–3113. doi: 10.1093/jxb/erw499
Daras, G., Templalexis, D., Avgeri, F., Tsitsekian, D., Karamanou, K., and Rigas, S. (2021). Updating insights into the catalytic domain properties of plant cellulose synthase (CesA) and cellulose synthase-like (Csl) proteins. Molecules 26:4335. doi: 10.3390/molecules26144335
Devinder, S., Manju, V. P., Jorge, F. S. F., Xuan, L., Andrew, P., Grover, K. K., et al. (2019). Variable salinity responses and comparative gene expression in woodland strawberry genotypes. Sci. Hortic. 254, 61–69. doi: 10.1016/j.scienta.2019.04.071
Duan, A., Tao, J., Jia, L., Tan, G., Liu, J., Li, T., et al. (2020). AgNAC1, a celery transcription factor, related to regulation on lignin biosynthesis and salt tolerance. Genomics 112, 5254–5264. doi: 10.1016/j.ygeno.2020.09.049
Etesami, H., and Beattie, G. A. (2018). Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Fronti. Microbiol. 9:148. doi: 10.3389/fmicb.2018.00148
Fang, H., Zhou, Q., Cheng, S., Zhou, X., Wei, B., Zhao, Y., et al. (2021). 24-Epibrassinolide alleviates postharvest yellowing of broccoli via improving its antioxidant capacity. Food Chem. 365:130529. doi: 10.1016/j.foodchem.2021.130529
Feng, Y., Wang, A., Fu, W., and Song, D. (2022). Growth performance, antioxidant response, biodegradation and transcriptome analysis of Chlorella pyrenoidosa after nonylphenol exposure. Sci. Total Environ. 806:150507. doi: 10.1016/j.scitotenv.2021.150507
Ferreira, J. F. S., Liu, X., and Suarez, D. L. (2019). Fruit yield and survival of five commercial strawberry cultivars under field cultivation and salinity stress. Sci. Hortic. 243, 401–410. doi: 10.1016/j.scienta.2018.07.016
Green, M. A., and Fry, S. C. (2005). Apoplastic degradation of ascorbate: Novel enzymes and metabolites permeating the plant cell wall. Plant Biosyst. 139, 2–7. doi: 10.1080/11263500500056849
Guo, K., Babourina, O., Christopher, D. A., Borsics, T., and Rengel, Z. (2008). The cyclic nucleotide-gated channel, AtCNGC10, influences salt tolerance in Arabidopsis. Physiol. Plant. 134, 499–507. doi: 10.1111/j.1399-3054.2008.01157.x
Hepler, N. K., Bowman, A., Carey, R. E., and Cosgrove, D. J. (2020). Expansin gene loss is a common occurrence during adaptation to an aquatic environment. Plant J. 101, 666–680. doi: 10.1111/tpj.14572
Ishida, K., and Yokoyama, R. (2022). Reconsidering the function of the xyloglucan endotransglucosylase/hydrolase family. J. Plant Res. 135, 145–156. doi: 10.1007/s10265-021-01361-w
Jadamba, C., Kang, K., Paek, N., Lee, S. I., and Yoo, S. (2020). Overexpression of rice Expansin7 (Osexpa7) confers enhanced tolerance to salt stress in rice. Int. J. Mol. Sci. 21:454. doi: 10.3390/ijms21020454
Jayaraman, K., Raman, V. K., Sevanthi, A. M., Sivakumar, S. R., Gayatri, Viswanathan, C., et al. (2021). Stress-inducible expression of chalcone isomerase2 gene improves accumulation of flavonoids and imparts enhanced abiotic stress tolerance to rice. Environ. Exp. Bot. 190:104582. doi: 10.1016/j.envexpbot.2021.104582
Jensen, J. K., Johnson, N. R., and Wilkerson, C. G. (2014). Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J. 80, 207–215. doi: 10.1111/tpj.12641
Jensen, J. K., Kim, H., Cocuron, J., Orler, R., Ralph, J., and Wilkerson, C. G. (2011). The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J. 66, 387–400. doi: 10.1111/j.1365-313X.2010.04475.x
Jia, Q., Li, M. W., Zheng, C., Xu, Y., Sun, S., Li, Z., et al. (2021). The soybean plasma membrane-localized cation/H+ exchanger GmCHX20a plays a negative role under salt stress. Physiol. Plant. 171, 714–727. doi: 10.1111/ppl.13250
Kanter, U., Usadel, B., Guerineau, F., Li, Y., Pauly, M., and Tenhaken, R. (2005). The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides. Planta 221, 243–254. doi: 10.1007/s00425-004-1441-0
Kaya, C., Akram, N. A., and Ashraf, M. (2019). Influence of exogenously applied nitric oxide on strawberry (Fragaria x ananassa) plants grown under iron deficiency and/or saline stress. Physiol. Plant. 165, 247–263. doi: 10.1111/ppl.12818
Kesten, C., Wallmann, A., Schneider, R., McFarlane, H. E., Diehl, A., Khan, G. A., et al. (2019). The companion of cellulose synthase 1 confers salt tolerance through a Tau-like mechanism in plants. Nat. Commun. 10:857. doi: 10.1038/s41467-019-08780-3
Keutgen, A., and Pawelzik, E. (2007). Modifications of taste-relevant compounds in strawberry fruit under NaCl salinity. Food Chem. 105, 1487–1494. doi: 10.1016/j.foodchem.2007.05.033
Kim, D., Jin, J., Alejandro, S., Martinoia, E., and Lee, Y. (2010). Overexpression of AtABCG36 improves drought and salt stress resistance in Arabidopsis. Physiol. Plant. 139, 170–180. doi: 10.1111/j.1399-3054.2010.01353.x
Koohbor, B., Fahs, M., Ataie-Ashtiani, B., Belfort, B., Simmons, C. T., and Younes, A. (2019). Uncertainty analysis for seawater intrusion in fractured coastal aquifers: Effects of fracture location, aperture, density and hydrodynamic parameters. J. Hydrol. 571, 159–177. doi: 10.1016/j.jhydrol.2019.01.052
Lee, E. K., Kwon, M., Ko, J. H., Yi, H. C., Hwang, M. G., Chang, S. C., et al. (2004). Binding of sulfonylurea by AtMRP5, an Arabidopsis multidrug resistance-related protein that functions in salt tolerance. Plant Physiol. 134, 528–538. doi: 10.1104/pp.103.027045
Leng, P., Yuan, B., and Guo, Y. (2014). The role of abscisic acid in fruit ripening and responses to abiotic stress. J. Exp. Bot. 65, 4577–4588. doi: 10.1093/jxb/eru204
Li, B., Fan, R., Guo, S., Wang, P., Zhu, X., Fan, Y. T., et al. (2019a). The Arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis. Environ. Exp. Botany 166:103807. doi: 10.1016/j.envexpbot.2019.103807
Li, M., Li, M., Li, D., Wang, S. M., and Yin, H. (2021). Overexpression of the Zygophyllum xanthoxylum aquaporin, ZxPIP1;3, Promotes plant growth and stress tolerance. Int. J. Mol. Sci. 22:2112. doi: 10.3390/ijms22042112
Li, P., Liu, Y., Tan, W., Chen, J., Zhu, M., Lv, Y., et al. (2019b). Brittle culm 1 encodes a COBRA-Like protein involved in secondary cell wall cellulose biosynthesis in sorghum. Plant Cell Physiol. 60, 788–801. doi: 10.1093/pcp/pcy246
Li, Z., Zhou, T., Sun, P., Chen, X., Gong, L., Sun, P., et al. (2022). COBL9 and COBL7 synergistically regulate root hair tip growth via controlling apical cellulose deposition. Biochem. Biophys. Res. Commun. 596, 6–13. doi: 10.1016/j.bbrc.2022.01.096
Linjun, A., Qing, M., Jinxia, D., Miao, Y., Fangrui, L., Jiayu, L., et al. (2019). Preliminary classification of the ABC transporter family in Betula halophila and expression patterns in response to exogenous phytohormones and abiotic stresses. Forests 10:722. doi: 10.3390/f10090722
Mahajan, M., and Yadav, S. K. (2014). Overexpression of a tea flavanone 3-hydroxylase gene confers tolerance to salt stress and Alternaria solani in transgenic tobacco. Plant Mol. Biol. 85, 551–573. doi: 10.1007/s11103-014-0203-z
Merlaen, B., De Keyser, E., and Van Labeke, M. (2020). The jasmonic acid pathway, rather than abscisic acid, may partly explain contrasting stomatal responses in two strawberry cultivars under osmotic stress. Plant Physiol. Biochem. 151, 21–33. doi: 10.1016/j.plaphy.2020.02.041
Mohnen, D., Atmodjo, M., Tan, L., Amos, R., Zhua, X., Atwood, J. A., et al. (2012). Synthesis of the plant cell wall’s most complex glycan: Pectin-surprises in glycosyltransferase processing and anchoring in the Golgi. FASEB J. 26:349.3.
Muszyńska, A., Jarocka, K., and Kurczynska, E. U. (2014). Plasma membrane and cell wall properties of an aspen hybrid (Populus tremula×tremuloides) parenchyma cells under the influence of salt stress. Acta Physiol. Plant. 36, 1155–1165. doi: 10.1007/s11738-014-1490-3
Oikawa, A., Joshi, H. J., Rennie, E. A., Ebert, B., Manisseri, C., Heazlewood, J. L., et al. (2010). An integrative approach to the identification of arabidopsis and rice genes involved in xylan and secondary wall development. PLoS One 5:e15481. doi: 10.1371/journal.pone.0015481
Oliveira, D. M., Mota, T. R., Salatta, F. V., Sinzker, R. C., Koncitikova, R., Kopecny, D., et al. (2020). Cell wall remodeling under salt stress: Insights into changes in polysaccharides, feruloylation, lignification, and phenolic metabolism in maize. Plant Cell Environ. 43, 2172–2191. doi: 10.1111/pce.13805
Rocha, J., Cicéron, F., Lerouxel, O., Breton, C., and de Sanctis, D. (2016). The galactoside 2-α-l-fucosyltransferase FUT1 from Arabidopsis thaliana: Crystallization and experimental MAD phasing. Acta Crystallogr. F Struct. Biol. Commun. 72(Pt 7) 564–568. doi: 10.1107/S2053230X16009584
Shah, S. H. H., Wang, J., Hao, X., and Thomas, B. W. (2022). Modelling soil salinity effects on salt water uptake and crop growth using a modified denitrification-decomposition model: A phytoremediation approach. J. Environ. Manage. 301:113820. doi: 10.1016/j.jenvman.2021.113820
Sharples, S. C., Nguyen-Phan, T. C., and Fry, S. C. (2017). Xyloglucan endotransglucosylase/hydrolases (XTHs) are inactivated by binding to glass and cellulosic surfaces, and released in active form by a heat-stable polymer from cauliflower florets. J. Plant Physiol. 218, 135–143. doi: 10.1016/j.jplph.2017.07.022
Sousa, A. O., Camillo, L. R., Assis, E., Lima, N. S., Silva, G. O., Kirch, R. P., et al. (2020). EgPHI-1, a phosphate-induced-1 gene from Eucalyptus globulus, is involved in shoot growth, xylem fiber length and secondary cell wall properties. Planta 252:45. doi: 10.1007/s00425-020-03450-x
Suarez, D. L., and Grieve, C. M. (2013). Growth, yield, and ion relations of strawberry in response to irrigation with chloride-dominated waters. J. Plant Nutr. 36, 1963–1981. doi: 10.1080/01904167.2013.766210
Sun, Y., Niu, G., Wallace, R., Masabni, J., and Gu, M. (2015). Relative salt tolerance of seven strawberry cultivars. Horticulturae 1, 27–43. doi: 10.3390/horticulturae1010027
Vanessa, G., Rafael, D. S. M., Ellen, C. P., Joyce, M. B., Adilson, L. B., Rombaldi, C. V., et al. (2016). Mild salt stress improves strawberry fruit quality. LWT Food Sci. Technol. 73, 693–696. doi: 10.1016/j.lwt.2016.07.001
Wang, G., Sun, R., Dong, J., Zhong, C., Sun, J., Zhang, H., et al. (2018b). A new strawberry cultivar‘Jingtaoxiang’. Acta Hortic. Sin. 45, 1215–1216. doi: 10.16420/j.issn.0513-353x.20170870
Wang, F., Ren, G., Li, F., Qi, S., Xu, Y., Wang, B., et al. (2018a). A chalcone synthase gene AeCHS from Abelmoschus esculentus regulates flavonoid accumulation and abiotic stress tolerance in transgenic Arabidopsis. Acta Physiol. Plant. 40:97. doi: 10.1007/s11738-018-2680-1
Wang, J., Wang, F., Jin, C., Tong, Y., and Wang, T. (2020). A R2R3-MYB transcription factor VvMYBF1 from grapevine (Vitis vinifera L.) regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis. J. Hortic. Sci. Biotechnol. 95, 147–161. doi: 10.1080/14620316.2019.1665480
Wang, Y., Liang, C., Meng, Z., Li, Y., Abid, M. A., Askari, M., et al. (2019). Leveraging Atriplex hortensis choline monooxygenase to improve chilling tolerance in cotton. Environ. Exp. Bot. 162, 364–373. doi: 10.1016/j.envexpbot.2019.03.012
Wang, Y., Zhang, Y., An, Y., Wu, J., He, S., Sun, L. R., et al. (2022). Wheat TaTIP4;1 confers enhanced tolerance to drought, salt and osmotic stress in Arabidopsis and rice. Int. J. Mol. Sci. 23:2085. doi: 10.3390/ijms23042085
Wang, Z., Li, Y., Hu, L., Ye, Y., Chen, J., Li, J. A., et al. (2021). The cotton GhMYB4 gene enhances salt and drought tolerance in transgenic Arabidopsis. Agron. J. 113, 4762–4776. doi: 10.1002/agj2.20906
Wu, H., and Li, Z. (2019). The importance of Cl- exclusion and vacuolar Cl- sequestration: Revisiting the role of Cl- transport in plant salt tolerance. Front. Plant Sci. 10:1418. doi: 10.3389/fpls.2019.01418
Xu, Y., Liu, J., Jia, C., Hu, W., Song, S., Xu, B., et al. (2021). Overexpression of a banana aquaporin gene MaPIP1;1 enhances tolerance to multiple abiotic stresses in transgenic banana and analysis of its interacting transcription factors. Front. Plant Sci. 12:780544. doi: 10.3389/fpls.2021.780544
Yan, J., He, H., Fang, L., and Zhang, A. (2018). Pectin methylesterase31 positively regulates salt stress tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 496, 497–501. doi: 10.1016/j.bbrc.2018.01.025
Yan, J., Huang, Y., He, H., Han, T., Di, P., Sechet, J., et al. (2019). Xyloglucan endotransglucosylase-hydrolase30 negatively affects salt tolerance in Arabidopsis. J. Exp. Bot. 70, 5495–5506. doi: 10.1093/jxb/erz311
Yu, Y., Zhang, H., Xing, H., Cui, N., Liu, X., Meng, X., et al. (2021). Regulation of growth and salt resistance in cucumber seedlings by Hydrogen-Rich water. J. Plant Growth Regul. doi: 10.1007/s00344-021-10536-7
Zhan, X., Shen, Q., Chen, J., Yang, P., Wang, X., and Hong, Y. (2019). Rice sulfoquinovosyltransferase SQD2.1 mediates flavonoid glycosylation and enhances tolerance to osmotic stress. Plant Cell Environ. 42, 2215–2230. doi: 10.1111/pce.13554
Zhang, K., Sun, Y., Li, M., and Long, R. (2021b). CrUGT87A1, a UDP-sugar glycosyltransferases (UGTs) gene from Carex rigescens, increases salt tolerance by accumulating flavonoids for antioxidation in Arabidopsis thaliana. Plant Physiol. Biochem. 159, 28–36. doi: 10.1016/j.plaphy.2020.12.006
Zhang, G., Hou, X., Wang, L., Xu, J., Chen, J., Fu, X., et al. (2021a). Photo-sensitive leaf rolling 1encodes a polygalacturonase that modifies cell wall structure and drought tolerance in rice. New Phytol. 229, 890–901. doi: 10.1111/nph.16899
Zhang, Z., Tong, T., Fang, Y., Zheng, J., Zhang, X., Niu, C. Y., et al. (2020). Genome-wide identification of barley ABC genes and their expression in response to abiotic stress treatment. Plants (Basel, Switzerland) 9:1281. doi: 10.3390/plants9101281
Zhao, S., Zhang, Q., Liu, M., Zhou, H., Ma, C., Wang, P., et al. (2021b). Regulation of plant responses to salt stress. Int. J. Mol. Sci. 22:4609. doi: 10.3390/ijms22094609
Zhao, H., Li, Z., Wang, Y., Wang, J., Xiao, M., Liu, H., et al. (2021a). Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance. Plant Biotechnol. J. 20, 468–484. doi: 10.1111/pbi.13729
Keywords: ABC transporter, cell wall remodeling, salt stress, salinity tolerance, strawberry
Citation: Li S, Chang L, Sun R, Dong J, Zhong C, Gao Y, Zhang H, Wei L, Wei Y, Zhang Y, Wang G and Sun J (2022) Combined transcriptomic and metabolomic analysis reveals a role for adenosine triphosphate-binding cassette transporters and cell wall remodeling in response to salt stress in strawberry. Front. Plant Sci. 13:996765. doi: 10.3389/fpls.2022.996765
Received: 18 July 2022; Accepted: 28 July 2022;
Published: 06 September 2022.
Edited by:
Iftikhar Ali, State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology (CAS), ChinaReviewed by:
Xiaodong Zheng, Qingdao Agricultural University, ChinaKe Duan, Shanghai Academy of Agricultural Sciences, China
Copyright © 2022 Li, Chang, Sun, Dong, Zhong, Gao, Zhang, Wei, Wei, Zhang, Wang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuntao Zhang, zhytao1963@126.com; Guixia Wang, wgxia1972@163.com; Jian Sun, sjroad@126.com
†These authors have contributed equally to this work
 Shuangtao Li1,2,3†
Shuangtao Li1,2,3† Linlin Chang
Linlin Chang Rui Sun
Rui Sun Jing Dong
Jing Dong Chuanfei Zhong
Chuanfei Zhong Yongshun Gao
Yongshun Gao Lingzhi Wei
Lingzhi Wei Yuntao Zhang
Yuntao Zhang Guixia Wang
Guixia Wang Jian Sun
Jian Sun