- 1Department of Plant Pathology, Nanjing Agricultural University, Key Laboratory of Biological Interaction and Crop Health, Key Laboratory of Integrated Management of Crop Diseases and Pests (Ministry of Education), Nanjing, China
- 2College of Plant Protection, Anhui Agricultural University, Hefei, China
- 3Department of Plant Pathology, Washington State University, Pullman, WA, United States
- 4Shandong Linyi Tobacco Co., Ltd., Linyi, China
- 5Tobacco Research Institute of Chinese Academy of Agricultural Sciences, Qingdao, China
Microbial necrosis and ethylene-inducing peptide 1 (Nep1)-like proteins (NLPs) act as cytolytic toxins and immunogenic patterns in plants. Our previous work shows that cytolytic NLPs (i.e., PyolNLP5 and PyolNLP7) from the biocontrol agent Pythium oligandrum enhance plant resistance against Phytophthora pathogens by inducing the expression of plant defensins. However, the relevance between PyolNLP-induced necrosis and plant resistance activation is still unclear. Here, we find that the necrosis-inducing activity of PyolNLP5 requires amino acid residues D127 and E129 within the conserved “GHRHDLE” motif. However, PyolNLP5-mediated plant disease resistance is irrelevant to its necrosis-inducing activity and the accumulation of reactive oxygen species (ROS). Furthermore, we reveal the positive role of non-cytotoxic PyolNLPs in enhancing plant resistance against Phytophthora pathogens and the fugal pathogen Sclerotinia sclerotiorum. Similarly, non-cytotoxic PyolNLPs also activate plant defense in a cell death-independent manner and induce defensin expression. The functions of non-cytotoxic PyolNLP13/14 rely on their conserved nlp24-like peptide pattern. Synthetic Pyolnlp24s derived from both cytotoxic and non-cytotoxic PyolNLPs can induce plant defensin expression. Unlike classic nlp24, Pyolnlp24s lack the ability of inducing ROS burst in plants with the presence of Arabidopsis nlp24 receptor RLP23. Taken together, our work demonstrates that PyolNLPs enhance plant resistance in an RLP23-independent manner, which requires the conserved nlp24-like peptide pattern but is uncoupled with ROS burst and cell death.
Introduction
Millions of years of coevolution of plants and microbial pathogens have shaped the antagonistic ability of both parties. Their interactions upgrade both pathogen invasion approaches and plant defense mechanisms (Jones and Dangl, 2006; Ottmann et al., 2009). Early-stage plant-pathogen interactions take place in the apoplast (Lo Presti et al., 2015; Ma et al., 2017), where microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs) released from bacteria, fungi, oomycetes, or nematodes are recognized by pattern recognition receptors (PRRs) at the plasma membrane (Yu et al., 2021).
Hitherto, only a limited number of MAMP/PAMP-recognizing PRRs have been documented (Tang et al., 2017; Wang et al., 2018). PRRs are often leucine-rich repeat receptor-like kinases (LRR-RLKs). The well-known Arabidopsis thaliana LRR-RLK FLAGELLIN-SENSITIVE 2 (FLS2) binds flg22, an 22-amino-acid epitope at the N-terminal of bacterial flagellin (Chinchilla et al., 2006). The bacterial PAMP elongation factor thermo unstable (EF-Tu) is recognized by Arabidopsis LRR-RLK EFR via its conserved N-terminal N-acetylated epitope elf18 (Zipfel et al., 2006). The tomato (Solanum lycopersicum) LRR-RLK CORE is a high-affinity receptor for the bacterial cold shock protein (CSP) epitope csp22 (Wang et al., 2016). PRRs may also be LRR receptor-like proteins (RLPs) which lack the kinase domain. For example, ReMAX, RLP30, RLP42/RBPG1, ELR, RXEG1 and NbEIX2 are PRRs recognizing Xanthomonas eMAX, Sclerotinia sclerotiorum SCFE1, fungal endopolygalacturonases (endoPG), Phytophthora elicitin INF1, Phytophthora sojae XEG1 and Verticillium dahlia VdEIX3, respectively (Jehle et al., 2013; Zhang et al., 2013, 2014; Du et al., 2015; Tang et al., 2017; Wang et al., 2018; Wan et al., 2019; Yin et al., 2021; Yu et al., 2021). The subsequent immune activation after PRR-RLK/RLP recognition is referred to as MAMP- or PAMP-triggered immunity (MTI or PTI), which leads to the rise of cytosolic Ca2+ level, production of extracellular reactive oxygen species (ROS) and activation of mitogen-activated protein kinase (MAPK) cascades (Couto and Zipfel, 2016). As a major early signaling product, ROS has been proposed to act as defense molecules that kill pathogens as well as signaling molecules that activate additional immune responses (Qi et al., 2017; Yuan et al., 2021). MTI/PTI, ROS accumulation and the downstream signaling cascades trigger various defense mechanisms to defend pathogen invasion (Poland et al., 2009; Yang and Fernando, 2021).
Microbial necrosis and ethylene-inducing peptide 1 (Nep1)-like proteins (NLPs) act as both MAMPs and toxin-like virulence factors in plant-microbe interactions (Qutob et al., 2006). NLPs are produced by bacteria, fungi or oomycetes to induce necrosis and ethylene production in eudicot plants (Gijzen and Nurnberger, 2006; Oome and Van den Ackerveken, 2014; Azmi et al., 2018). Phylogenetic analysis of their amino acid sequences distinguishes Type I, Type II, and Type III NLPs which have one, two and three pairs of conserved cysteines, respectively. All three types of NLPs can be found in bacteria and fungi whereas oomycetes only produce Type I or Type II NLPs (Gijzen and Nurnberger, 2006; Oome and Van den Ackerveken, 2014; Seidl and Van den Ackerveken, 2019). Most plant pathogenic oomycetes, including P. sojae, Pythium ultinum and Pythium aphanidermatum, encode only type I NLPs. Both cytolytic and non-cytolytic Type II NLPs are found in non-pathogenic oomycetes such as Pythium oligandrum and Pythium periplocum (PyolNLPs/PypeNLPs). Oomycete NLPs carry a pattern of 20 or 24 amino acid residues (nlp20 or nlp24), which are precepted by Arabidopsis PRR RLP23 to trigger plant immune responses such as MAPK cascade activation and ROS burst (Bohm et al., 2014; Oome et al., 2014; Albert et al., 2015).
Necrosis and ethylene-inducing peptide 1-like proteins of pathogenic oomycetes Pythium aphanidermatum and Phytophthora parasitica are structurally conserved with cytolytic and pore-forming actinoporins of marine organisms (Ottmann et al., 2009; Azmi et al., 2018). The bindings of cytotoxic oomycete NLPs to glycosylinositol phosphorylceramide (GIPC) sphingolipids induce necrosis in eudicots but not in monocots (Lenarcic et al., 2017; Seidl and Van den Ackerveken, 2019). NLPs of the oomycete pathogen Hyaloperonospora arabidopsidis (HaNLPs) lack the ability to cause necrosis in dicot plants (Cabral et al., 2012), but can induce defense responses such as PATHOGENESIS-RELATED GENE 1 (PR1) expression in Arabidopsis (Oome et al., 2014). Recent study discloses that the functional difference between cytolytic PyaNLP and non-cytolytic HaNLP3 protein is in GIPC headgroup recognition. In contrast to PyaNLP, the HaNLP3 protein does not bind to GIPCs alone, consistent with its inability to cause necrosis of tobacco leaves (Lenarcic et al., 2019).
Cytotoxic NLPs in certain hemibiotrophic plant pathogens such as Phytophthora capsici and Verticillium dahliae are essential for their full virulence and the transition to necrotrophic stages during infection (Dong et al., 2012; Zhou et al., 2012). Hemibiotrophic fungus Colletotrichum orbiculare expressing a mutated NLP1 lacking cytotoxic activity loses its ability to infect cucumber (Azmi et al., 2018). Conlp24, a synthetic peptide derived from C. orbiculare NLP1, elicits ROS generation in Arabidopsis. This ability can be abolished by mutating its first four amino acids (AIMY) to alanine (Conlp24Mut) (Azmi et al., 2018). Furthermore, NLPs typically share a conserved NPP1 domain that contains a heptapeptide “GHRHDWE” motif (Fellbrich et al., 2002; Santhanam et al., 2013; Seidl and Van den Ackerveken, 2019). Mutation of D104 or E106 residue in the motif abolishes the cytolytic activity of NLPPcc from the pathogenic bacterium Pectobacterium carotovorum (Ottmann et al., 2009). The results above suggest that both “AIMY” and “GHRHDWE” motifs may be important for NLP function.
We previously reported that PyolNLPs/PypeNLPs from non-pathogenic P. oligandrum and P. periplocum contain a unique “G/AHxF” motif found in the N-terminal of the nlp24 pattern. In contrast, the “AIMY” motif is typically found in Type I and Type II pathogenic NLPs (Yang et al., 2021). Mutation of the “G/AHxF” or “GHRHDLE” motif impairs PyolNLP5/7-mediated resistance against P. capsici in solanaceous plants, suggesting the crucial role of nlp24 in the function of PyolNLPs. In addition, cytotoxic PyolNLP5 enhances resistance by inducing plant defensin in a non-ROS-injury manner (Yang et al., 2021). However, the possible linkage between PyolNLP-induced necrosis and defense remains enigmatic.
Here, we use mutation analysis to determine Asparticacid (D) and Glutamicacid (E) in the “GHRHDLE” motif of Group 1 PyolNLPs as the two key residues for their necrosis-inducing activity. Using PyolNLP5 as an example, we showed that its resistance enhancing function is independent of necrosis induction and ROS burst. Furthermore, we explore the positive role of non-cytotoxic PyolNLPs in enhancing plant resistance against Phytophthora pathogens and the fugal pathogen S. sclerotiorum. Non-cytotoxic PyolNLPs also activate plant defense in a cell death-independent manner and induce defensin expression. The functions of non-cytotoxic PyolNLP13/14 rely on their conserved nlp24-like peptide pattern. Synthetic Pyolnlp24s derived from both cytotoxic and non-cytotoxic PyolNLPs can induce plant defensin expression. Unlike classic nlp24, Pyolnlp24s lack the ability of inducing ROS burst in plants with the presence of Arabidopsis nlp24 receptor RLP23. Taken together, our work demonstrates that both cytotoxic and non-cytotoxic PyolNLPs enhance plant resistance in an RLP23-independent manner, which requires the conserved nlp24-like peptide pattern but is uncoupled with ROS burst and cell death.
Materials and Methods
Microbial Strains, Plants, and Culture Conditions
Phytophthora nicotianae isolate 025 and Phytophthora capsica isolate LT263 used in this study were routinely cultured at 25°C in the dark on 10% (v/v) V8 juice medium (Zhou et al., 2021). S. sclerotiorum strain WMA1 used in this study was routinely cultured at 25°C in the dark on PDA medium (Wei et al., 2020). Nicotiana benthamiana plants was grown at 25°C with a 16-h light and 8-h dark photoperiod in an environmentally controlled growth room. Arabidopsis plants were grown at 23°C with a 10-h light/14-h dark photoperiod. N. benthamiana seedling of 4–8 weeks old and Arabidopsis seedling aged at 4–6 weeks were used for experiments (Li et al., 2019).
DNA Cloning, Plasmid Construction and Peptide Synthesis
Full-length cDNAs of all PyolNLPs were amplified from P. oligandrum strain CBS 530.74 by polymerase chain reaction (PCR). Fragments used to generate PyolNLP-M24 mutants were synthesized by Sangon Biotech (Shanghai, China). Gene mutated at key locus was cloned using the overlap method. Amplified fragments were cloned into pBINHA, a plasmid vector containing a C-terminal Hybrid Access (HA) tag under the control of the CaMV 35S promoter, using In-Fusion® HD Cloning Kit (Clontech, Mountain View, CA, United States) (Yang et al., 2019). Peptides were ordered from Sangon Biotech and prepared as 2 mM stock solutions in Ultra-pure water before use. Primers used in this work were listed in Supplementary Table 1.
Agrobacterium-Mediated Transient Gene Expression in Nicotiana benthamiana
Constructs were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. Successful transformants were confirmed by PCR amplification using indicated primers (Supplementary Table 1). Transformed Agrobacterium strains were cultured, washed, and re-suspended in infiltration buffer (10 mM MgCl2, 500 mM MES, 100 mM acetosyringone) to make an appropriate optical density (OD) of 0.3 at 600 nm. Four-week-old N. benthamiana leaves were infiltrated with a 1:1 mixture of resuspended Agrobacterium containing the respective constructs and RNA silencing suppressor P19 (Circelli et al., 2010; Green et al., 2012; Lu et al., 2017). Agro-infiltrated leaf samples were collected at given time intervals and immediately frozen with liquid nitrogen before being stored for gene expression analysis.
Western Blot
Proteins from the sample lysate were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) and then electrotransferred to an Immobilon-PSQ polyvinylidene difluoride membrane using transfer buffer (20 mM Tris, 150 mM glycine). The membrane was blocked for 30 min at room temperature by shaking at 50 rpm (Revolutions Per Minute) with phosphate-buffered saline (PBS; pH 7.4) containing 3% non-fat dry milk. After washed with PBST (PBS with 0.1% Tween 20), the membrane was incubated for 90 min with PBSTM (PBS with 0.1% Tween 20 and 3% non-fat dry milk) containing anti-HA (1:2000, Abmart) antibody. After three rounds of washes (5 min each) with PBST, the membrane was then incubated with goat anti-mouse IRDye 800CW antibody (Odyssey) at a ratio of 1:10,000 in PBSTM for 30 min. The membrane was finally washed with PBST and visualized with excitations at 700 and 800 nm (Ai et al., 2021).
Pathogenicity Assay
Detached leaves from 6-week-old N. benthamiana plants were inoculated with mycelia plugs of P. capsici isolate LT263 or P. nicotianae isolate 025, and then incubated at 25°C in the dark. Inoculated leaves were photographed under bright or UV light at 36 and 48 hpi (hours post inoculation). Lesion diameters were measured with the ImageJ software (Ai et al., 2020). S. sclerotiorum infection was examined at 24 and 36 hpi under white light. Three biological replicates were performed for each assay with at least 12 leaves per replicate.
Diaminobenzidine Staining and Reactive Oxygen Species Burst Measurement
For 3,3′-Diaminobenzidine (DAB) staining, N. benthamiana leaves were stained with 1 mg/mL DAB solution for 8 h in the dark at 12 hpi and then decolored with ethanol for light microscopy examination. DAB staining was quantified as intensity per unit area using the ImageJ software (Song et al., 2015). For ROS burst, 0.125 cm2 leaf disks were collected using a cork-borer set (Sigma) and floated in a 96-well plate (1 disk per well) containing 200 μL double distilled water (ddH2O) overnight. Just before measurement with a luminometer (Tecan F200), ddH2O was replaced with a substrate solution containing 20 μM L-012 (Waco), 20 μg/ml horseradish peroxidase (Sigma) and 1 μM purified protein. Light emission was measured at 1 min intervals (Yin et al., 2021).
Electrolyte Leakage Assay
Cell death was determined by measuring ion leakage from leaf disks. For each measurement, five leaf disks (9-mm diameter) were floated with abaxial side up on 5 ml of distilled water for 3 h at room temperature (RT). After incubation, conductivity of the bathing solution, referred to as value A, was measured with a Consort conductivity meter (Con 700; Consort, Turnhout, Belgium). The leaf disks were then incubated with the original bathing solution in sealed tubes at 95°C for 25 min. After being cooled down to room temperature, bathing solution was measured for conductivity again and the result was referred to as value B. For each measurement, ion leakage was expressed as percentage of (value A / value B) × 100. All assays were repeated three times (Yu et al., 2012; Nie et al., 2019).
Defensins Gene Expression and qRT-PCR Analysis
For defense gene expression, leaf samples infiltrated with 1 μM nlp24-like synthetic peptides were collected at 12 hpi. Total RNA samples were extracted from N. benthamiana leaves by using the RNA-simple Total RNA Kit (Tiangen) according to manufacturer’s instructions. cDNA was synthesized using the HiScript 1st Strand cDNA Synthesis Kit (Vazyme). Real-Time PCR was performed by using the ChamQ SYBR qPCR Master Mix Kit (Vazyme) and the ABI Prism 7500 Fast Real-Time PCR system following manufacturer’s instructions (Dong et al., 2020). Gene-specific primers used for qRT-PCR and their purposes are listed in Supplementary Table 1.
Statistical Analysis
The SPSS 22 software was used for statistical analysis of all data. After using a median-edition Levene’s test to determine the homogeneity of variances across groups, the results were then analyzed by one-way ANOVA with a post hoc Tukey’s range test for groups with equal variances, or Kruskal—Wallis test for groups with unequal variance (*p < 0.05; **p < 0.01; ns, no significant differences). Results are expressed as means ± SD of replicates (Yang et al., 2021).
Results
Conserved D and E in the “GHRHDLE” Motif Are Essential for the Necrosis-Inducing Activity of Group 1 PyolNLPs
We previously identified and cloned 25 Type II NLP genes in P. oligandrum and P. periplocum (Yang et al., 2021). However, the key residues that determine the necrosis-inducing activity of PyolNLPs remain unknown. Asparticacid (D) and Glutamicacid (E) in the central heptapeptide motif “GHRHDWE” are two key amino acid residues required for necrosis induction (Ottmann et al., 2009). Our previous study found that five PyolNLPs can induce strong necrosis. Multiple sequence alignment analysis found that these five PyolNLPs were very conserved (Supplementary Figure 1), and evolutionary analysis found that they were all located in Group 1 (Supplementary Figure 2). Meanwhile, we also found two key amino acid residues (Aspartic acid and Glutamic acid) are also conserved among PyolNLP3∼7 (Figure 1A and Supplementary Figure 1). With the mutation of their D or E residue in the conserved “GHRHDLE” motif to alanine (A), Group 1e (PyolNLP3/5/6) and Group 1a (PyolNLP4/7) showed abolished and significantly reduced necrosis-inducing activity in agroinfiltrated N. benthamiana leaves, respectively (Figure 1A and Supplementary Figure 2). In this assay, GFP was expressed as a negative control. Wild-type (WT) PyolNLPs without mutation were used as positive controls, which all induced necrosis in the assay (Figure 1B and Supplementary Figure 2).
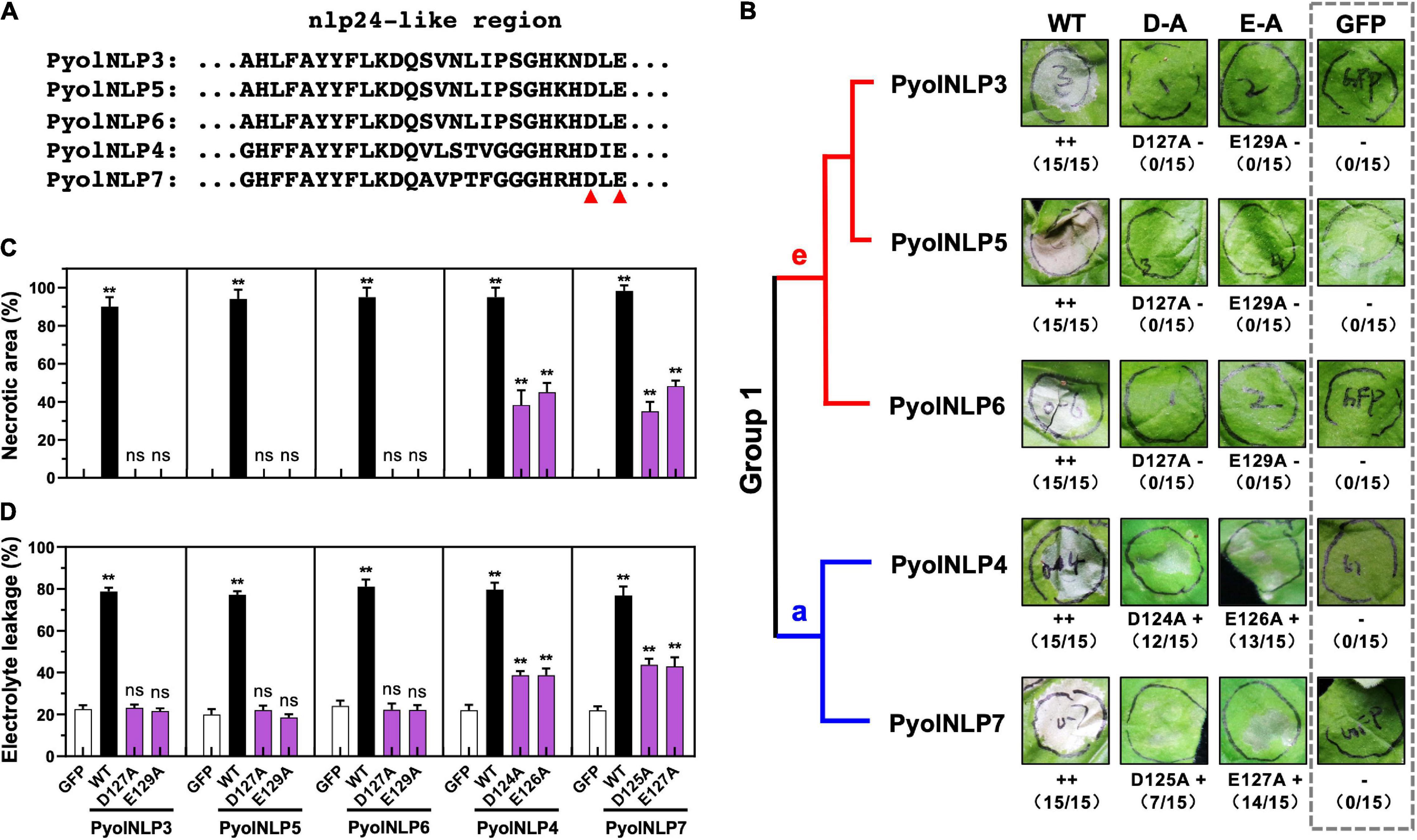
Figure 1. Conserved D and E in the “GHRHDLE” motif are essential for the necrosis-inducing activity of Group 1 PyolNLPs. (A) Schematic representation of nlp24-like regions from PyolNLP3-7. Aspartic acid (D) or glutamic acid (E) residues marked by the red triangle is replaced by alanine (A). (B) The necrosis-inducing activity of wild-type and mutated Group 1 PyolNLPs in agroinfiltrated Nicotiana benthamiana leaves. Necrosis grades are marked (–: no necrosis; + weak necrosis; + + : strong necrosis). The phylogenetic tree of selected Group 1 PyolNLPs are shown on the left. Amino acid mutations, necrosis grades and experiment replicate numbers (necrotic sites versus total infiltration sites) are labeled. The gray dotted rectangle green fluorescent protein (GFP) is shown as a negative control. Photos were taken at 5 days post infiltration (dpi). Quantification of necrosis by measurements of relative (C) necrotic area and (D) electrolyte leakage. Error bar represents mean ± SD (n = 3). Data were analyzed by Median-edition Levene’s test to determine the homogeneity of variance across groups, and then analyzed by one-way ANOVA with post-hoc Tukey’s range test for groups with equal variance (ns, no significant difference; **P < 0.01). All experiments were repeated three times with similar results obtained.
Quantitative measurements showed that all five WT PyolNLPs caused necrosis on more than 90% of the leaf areas (Figure 1C). In contrast, no necrosis was induced by either GFP or mutated PyolNLP3/5/6 of Group 1e (Figure 1C). Mutations on PyolNLP4/7 of Group 1a significantly reduced necrotic leaf areas to 40–60% (Figure 1C). Since ion leakage is positively correlated with cell death (Yu et al., 2012; Nie et al., 2019), this parameter was also measured for infiltrated leaves. Consistent with the necrotic area measurement results, leaves transiently expressing the five WT PyolNLPs exhibited the highest electrolyte leakages of around 80% (Figure 1D). The lowest ion leakages of about 20% were observed in leaves expressing GFP or mutated PyolNLP3/5/6 (Figure 1D). Mutated PyolNLP4/7 led to moderate ion leakages of around 40% in leaves (Figure 1D). Taken together, our results demonstrate that the central heptapeptide motif “GHRHDLE” is required for PyolNLP-triggered necrosis with D and E being two key residues.
PyolNLP5-Mediated Plant Disease Resistance Is Independent of Its Necrosis-Inducing Activity
We previously found that the full-function nlp24-like region is essential for PyolNLP5 to suppress Phytophthora nicotianae and P. capsici infection in N. benthamiana (Yang et al., 2021). To test whether mutations in “GHRHDLE” also impair PyolNLP5-mediated plant disease resistance, PyolNLP5 D127A and E129A mutants, in pair with GFP controls, were transiently expressed in the same N. benthamiana leaves. PyolNLP5-M24, we mutated the conserved sites (the first four amino acids AIMY and the GHRHDWE motif) of the nlp24-like peptide pattern in PyolNLP5 (Yang et al., 2021). Western blots confirmed that all recombinant proteins were properly expressed at the expected sizes in planta (Supplementary Figure 3A). The infiltrated regions were then equally inoculated with fresh mycelia of P. nicotianae isolate 025 or P. capsici isolate LT263. Evaluation of disease development following inoculation clearly showed that both PyolNLP5-D127A and PyolNLP5-E129A retained their suppression capacity toward P. nicotianae or P. capsici infection (Figures 2A,B). In contrast, neither GFP nor the nlp24-loss-of-function mutant PyolNLP5-M24 exhibited disease suppression activity (Figures 2A,B). To evaluate infection precisely, relative Phytophthora biomass in infected N. benthamiana tissues was determined by using qPCR to measure pathogen/plant DNA ratios. Consistent with lesion measurement results, both PyolNLP5-D127A and PyolNLP5-E129A significantly reduced Phytophthora biomass accumulation as compared to GFP and PyolNLP5-M24 (Figure 2C). These results suggest that PyolNLP5-mediated plant resistance against Phytophthora relies on the nlp24-like region, but independent of its necrosis-inducing activity.
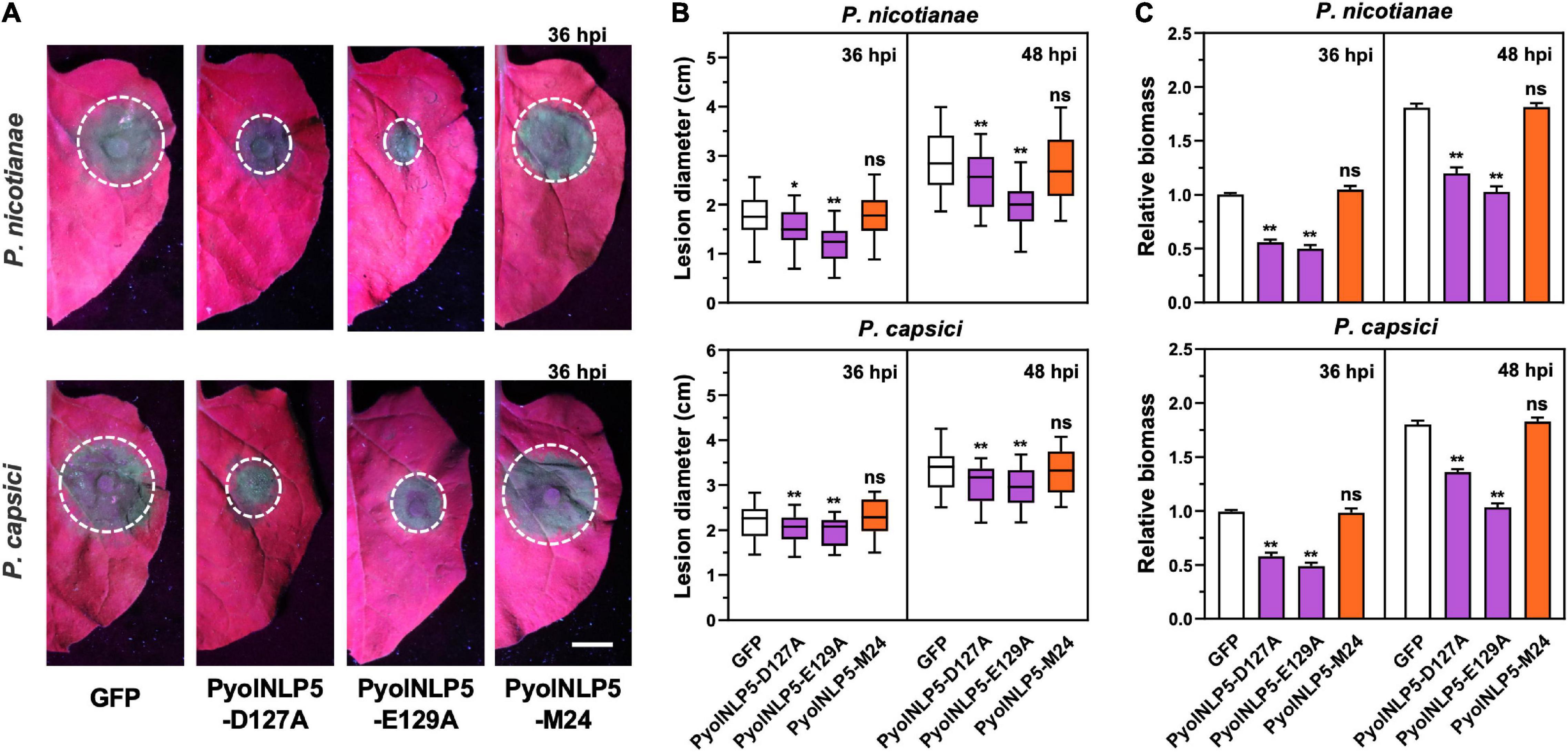
Figure 2. PyolNLP5-mediated plant disease resistance is independent of its necrosis-inducing activity. (A) Infection phenotypes of agroinfiltrated N. benthamiana leaves expressing GFP, PyolNLP5-D127A, PyolNLP5-E129A or PyolNLP5-M24, and followed by the inoculation of P. nicotianae isolate 025 or P. capsici isolate LT263. Photos were taken at 36 hpi (Scale bar, 1 cm). GFP was used as a negative control. (B) Lesion diameters were measured using ImageJ. Data were analyzed from 40 biological replicates. Error bar represents mean ± SD. (C) Relative biomass was determined by quantitative polymerase chain reaction (qPCR). Expression levels of P. capsici and P. nicotianae Actin were determined by qPCR using N. benthamiana EF1α as reference. Error bar represents mean ± SD (n = 3). Data were analyzed by Median-edition Levene’s test to determine the homogeneity of variance across groups, and then analyzed by one-way ANOVA with post-hoc Tukey’s range test for groups with equal variance, or Kruskal-Wallis test analysis for groups with unequal variance (ns, no significant difference; *P < 0.05; **P < 0.01). All experiments were repeated at least three times.
PyolNLP5-Mediated Plant Disease Resistance Is Irrelevant to Reactive Oxygen Species Accumulation
Reactive oxygen species accumulation is an important signal of early plant immune response as well as regulator of plant defense-related gene expression (Li et al., 2019; Wen et al., 2021). Here, the relationship between PyolNLP5-mediated plant resistance and ROS accumulation was explored by DAB staining. Since P. oligandrum oligandrins (Oli-D1 and Oli-D2) are ROS-inducting PAMPs (Ouyang et al., 2015), Oli-D2 was used as a positive control. As shown in Figure 3B, all three PyolNLP5 mutants (PyolNLP5-D127A, PyolNLP5- E129A, and PyolNLP5-M24) lost the ability of stimulating H2O2 accumulation in N. benthamiana with or without the inoculation of P. capsici (Figures 3A,B). Consistent results were obtained from the measurements of relative ROS intensities in the presence of P. capsici (Figure 3C). Taken together, these results show that PyolNLP5-mediated plant resistance is irrelevant to ROS accumulation.
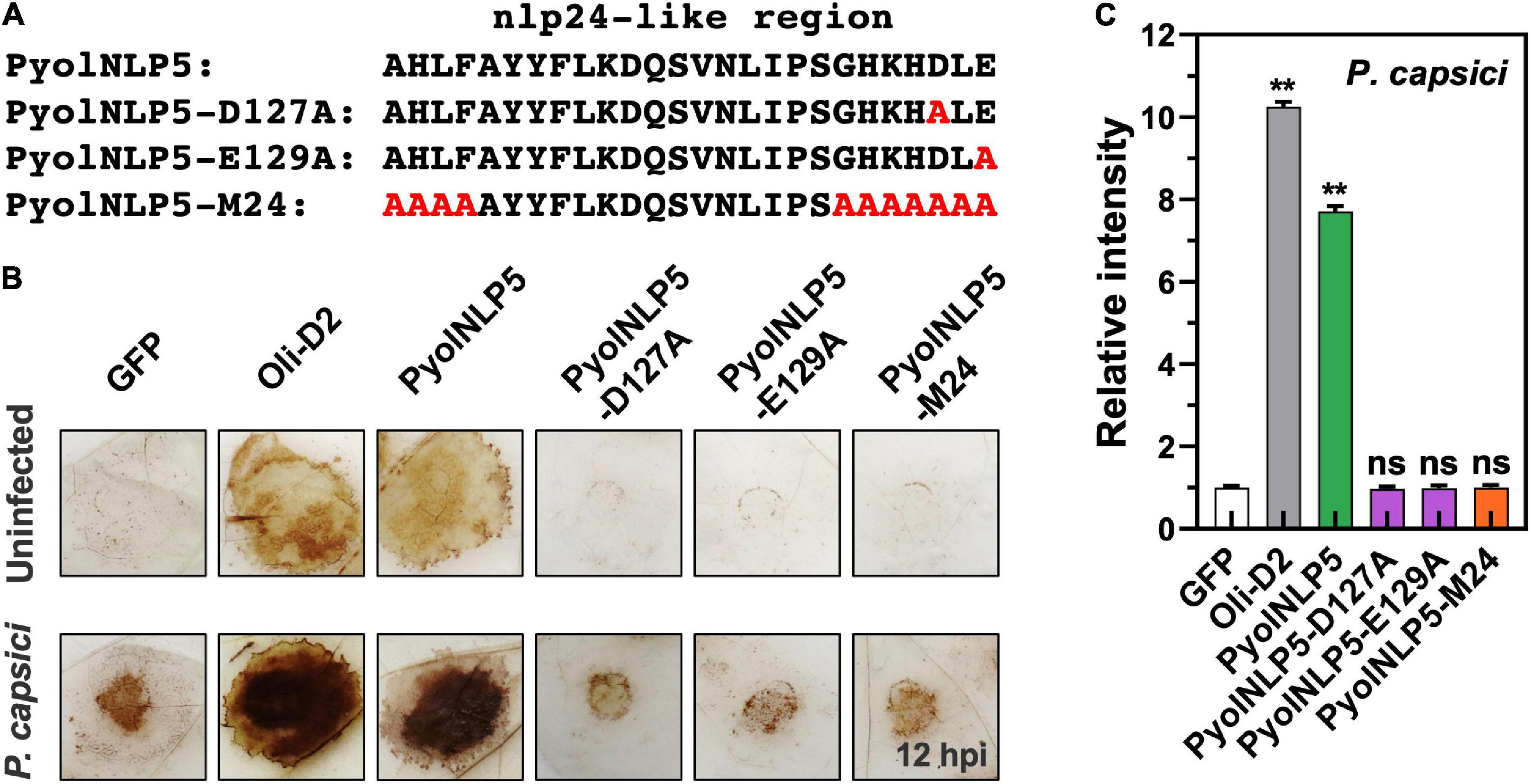
Figure 3. PyolNLP5-mediated plant disease resistance is irrelevant to ROS accumulation. (A) Schematic representation of nlp24-like regions in PyolNLP5, PyolNLP5-D127A, PyolNLP5-E129A and PyolNLP5-M24 with introduced alanine substitutions. (B) H2O2 accumulation on agroinfiltrated N. benthamiana leaves expressing GFP, Oli-D2, PyolNLP5, PyolNLP5-D127A, PyolNLP5-E129A or PyolNLP5-M24. Oli-D2 was used as a positive control. DAB staining was performed at 12 hpi after P. capsici inoculation. (C) Relative ROS intensities in infiltrated N. benthamiana leaves expressing wild-type or mutated PyolNLP5 were measured at 12 hpi using ImageJ. Error bar represents mean ± SD (n = 3). Data were analyzed by Median-edition Levene’s test to determine the homogeneity of variance across groups, and then analyzed by one-way ANOVA with post-hoc Tukey’s range test for groups with equal variance (ns, no significant difference; **P < 0.01). Experiments were repeated three times with similar results obtained.
Non-cytotoxic PyolNLP-Mediated Suppression of Phytophthora Infection Is Irrelevant to Necrosis Induction or Reactive Oxygen Species Accumulation
Cytotoxic PyolNLPs were previously shown to enhance plant resistance independent of their necrosis-inducing activity. However, the roles of non-cytotoxic PyolNLPs in modulating plant resistance are still elusive. We found that non-cytotoxic PyolNLPs are distributed across Groups 1b, 1d, 3a, 3b, 4a, and 4b. One PyolNLP was selected from each of these six subgroups (PyolNLP8/10/11/12/13/14) for pathogenicity assays. Their GFP-fusion constructs and the GFP-only control were carried by Agrobacterium for infiltrations of N. benthamiana leaves, followed by the inoculation of P. nicotianae or P. capsici. Western blots confirmed the proper in planta expression of all recombinant proteins (Supplementary Figure 3B). Lesion and biomass quantification results showed that ectopic expression of PyolNLP8/10/11/13/14 significantly reduced P. nicotianae colonization, with PyolNLP13/14 also delivering resistance to P. capsici (Figures 4A–C). The observation that non-cytotoxic PyolNLPs may enhance plant resistance to certain pathogens further demonstrates the irrelevance between PyolNLP-mediated plant defense and necrosis induction. Furthermore, DAB staining and relative ROS intensity measurement results demonstrated that none of the six non-cytotoxic PyolNLPs are involved in ROS accumulation, which is not affected by P. capsici inoculation (Figures 5A,B).
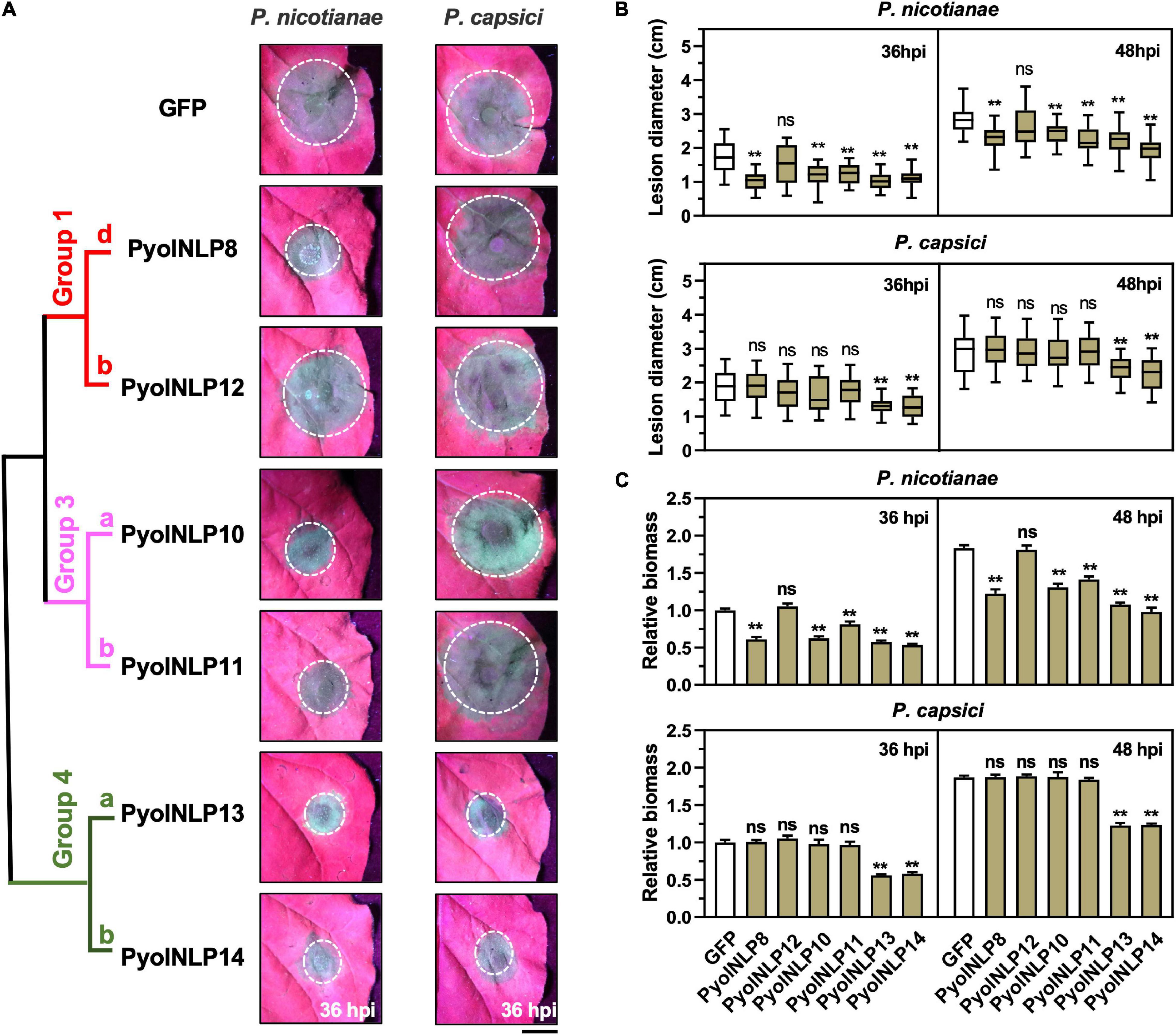
Figure 4. Non-cytotoxic PyolNLPs are able to suppress Phytophthora infection in N. benthamiana. (A) Infection phenotypes of agroinfiltrated N. benthamiana leaves expressing GFP, PyolNLP8, PyolNLP10, PyolNLP11, PyolNLP12, PyolNLP13 or PyolNLP14, and followed by the inoculation of P. nicotianae or P. capsici. Photos were taken at 36 hpi (Scale bar, 1 cm). GFP was used as a negative control. (B) Lesion diameters were measured using ImageJ. Data were analyzed from at least 40 biological replicates. Error bar represents mean ± SD. (C) Relative biomass was determined by qPCR. Expression levels of P. nicotianae and P. capsici Actin were determined by qPCR using N. benthamiana EF1α as reference. Error bar represents mean ± SD (n = 3). Data were analyzed by Median-edition Levene’s test to determine the homogeneity of variance across groups, and then analyzed by one-way ANOVA with post-hoc Tukey’s range test for groups with equal variance, or Kruskal-Wallis test analysis for groups with unequal variance (ns, no significant difference; **P < 0.01). All experiments were repeated at least three times.
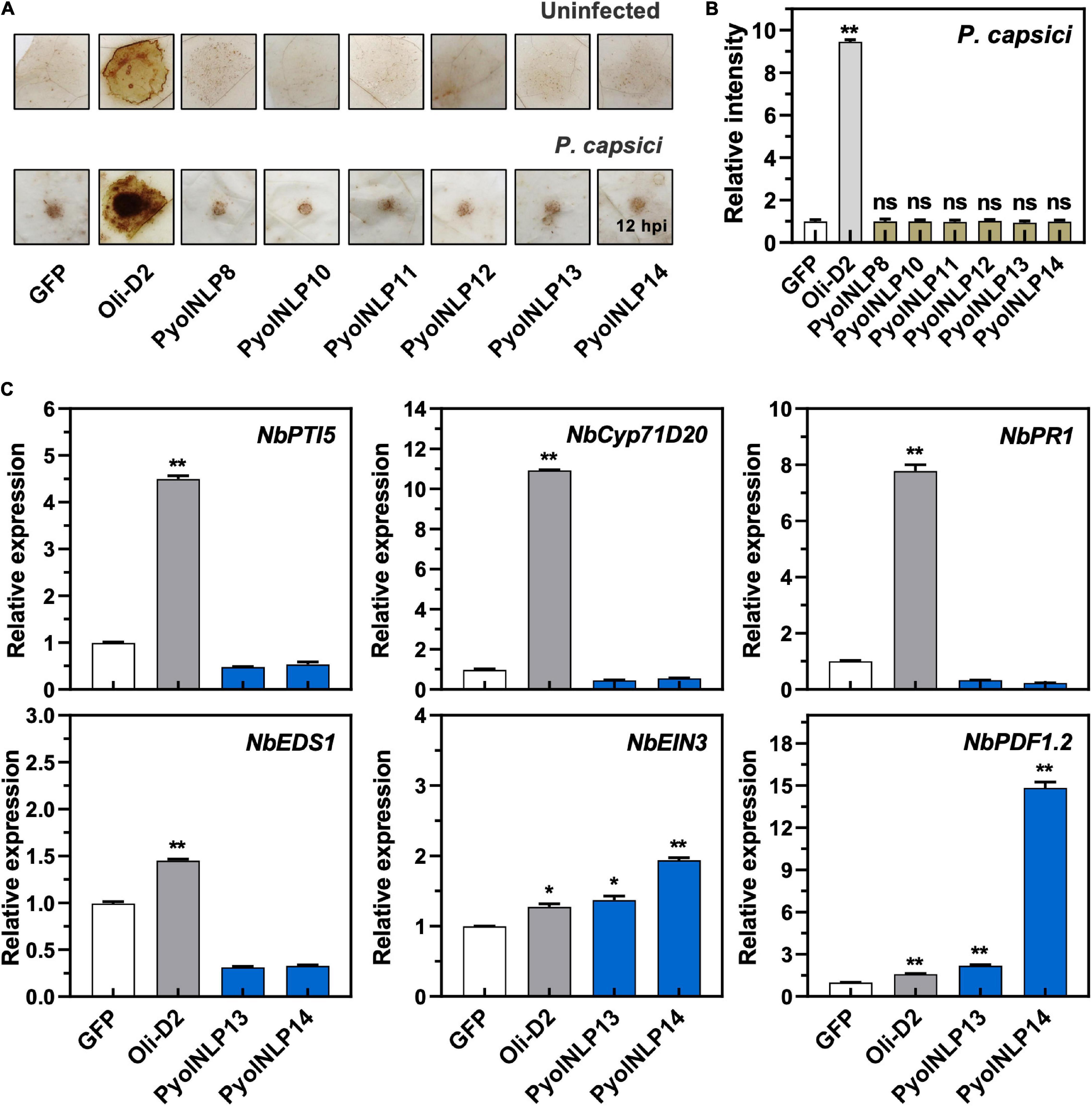
Figure 5. Non-cytotoxic PyolNLPs cannot trigger ROS accumulation and instead induce PDF1.2 expression in N. benthamiana. (A) H2O2 accumulation on infiltrated N. benthamiana leaves expressing GFP, Oli-D2, PyolNLP8, PyolNLP10, PyolNLP11, PyolNLP12, PyolNLP13 or PyolNLP14. was at 0 or 12 hpi. Oli-D2 acts as a positive control. DAB staining was performed at 12 hpi after P. capsici inoculation. (B) Relative ROS intensities in N. benthamiana leaves infiltrated with non-cytotoxic PyolNLPs were measured at 12 hpi using ImageJ. Error bar represents mean ± SD. (n = 3). (C) Relative expression levels of PTI5, Cyp71D20, PR1, EDS1, EIN3 and PDF1.2 in agroinfiltrated N. benthamiana leaves expressing GFP, Oli-D2, PyolNLP13 or PyolNLP14. GFP and Oli-D2 were used as negative and positive controls, respectively. Error bar represents mean ± SD (n = 3). Data were analyzed by Median-edition Levene’s test to determine the homogeneity of variance across groups, and then analyzed by one-way ANOVA with post-hoc Tukey’s range test for groups with equal variance (*P < 0.05;**P < 0.01). All experiments were repeated at least three times.
Non-cytotoxic PyolNLP13/14 Induce PDF1.2 and EIN3 Expression in Nicotiana benthamiana
We further examined whether the non-cytotoxic PyolNLP13/14 could activate plant immunity responses by testing their effect on the expression of six defense-related N. benthamiana genes, including NbPTI5 and NbCyp71D20 involved in PTI, salicylic acid (SA)-dependent ENHANCED DISEASE SUSCEPTIBILITY 1 (NbEDS1) and NbPR1, and ETHYLENE INSENSITIVE 3 (NbEIN3) and PLANT DEFENSIN 1.2 (NbPDF1.2) involved in jasmonic acid and ethylene signaling pathways. Unlike Oli-D2 which induced the expression of all six genes, PyolNLP13/14 could only activate the expression of PDF1.2 and EIN3 (Figure 5C), which is consistent with previous reports that NLPs induce the upregulation of EIN3 and PDF1.2 (Zhou et al., 2012; Yang et al., 2021).
Non-cytotoxic PyolNLPs Also Suppress Pathogen Infection in a nlp24-Dependent Manner
We previously reported that cytotoxic PyolNLP5-mediated suppression of Phytophthora infection requires full function of its nlp24-like region (Yang et al., 2021). To test whether this is also the case for non-cytotoxic PyolNLPs, we mutated conserved sites in the nlp24-like peptide pattern of PyolNLP13/14 to create PyolNLP13/14-M24 (Figure 6A). N. benthamiana leaves were infiltrated with GFP fusion construct of PyolNLP13/14 or PyolNLP13/14-M24 as well as the GFP-only control, followed by the inoculation of P. nicotianae, P. capsici or S. sclerotiorum. Western blot analysis indicated that all recombinant proteins were properly expressed in planta at the expected sizes (Supplementary Figures 3B,C). Lesion and relative pathogen biomass quantification results consistently showed that both PyolNLP13-M24 and PyolNLP14-M24 mutants lost suppression ability on all three oomycete and fungal pathogens as compared to their wild-type counterparts (Figures 6B–D), which suggests the requirement of full-function nlp24-like region for disease resistance mediated by non-cytotoxic PyolNLPs.
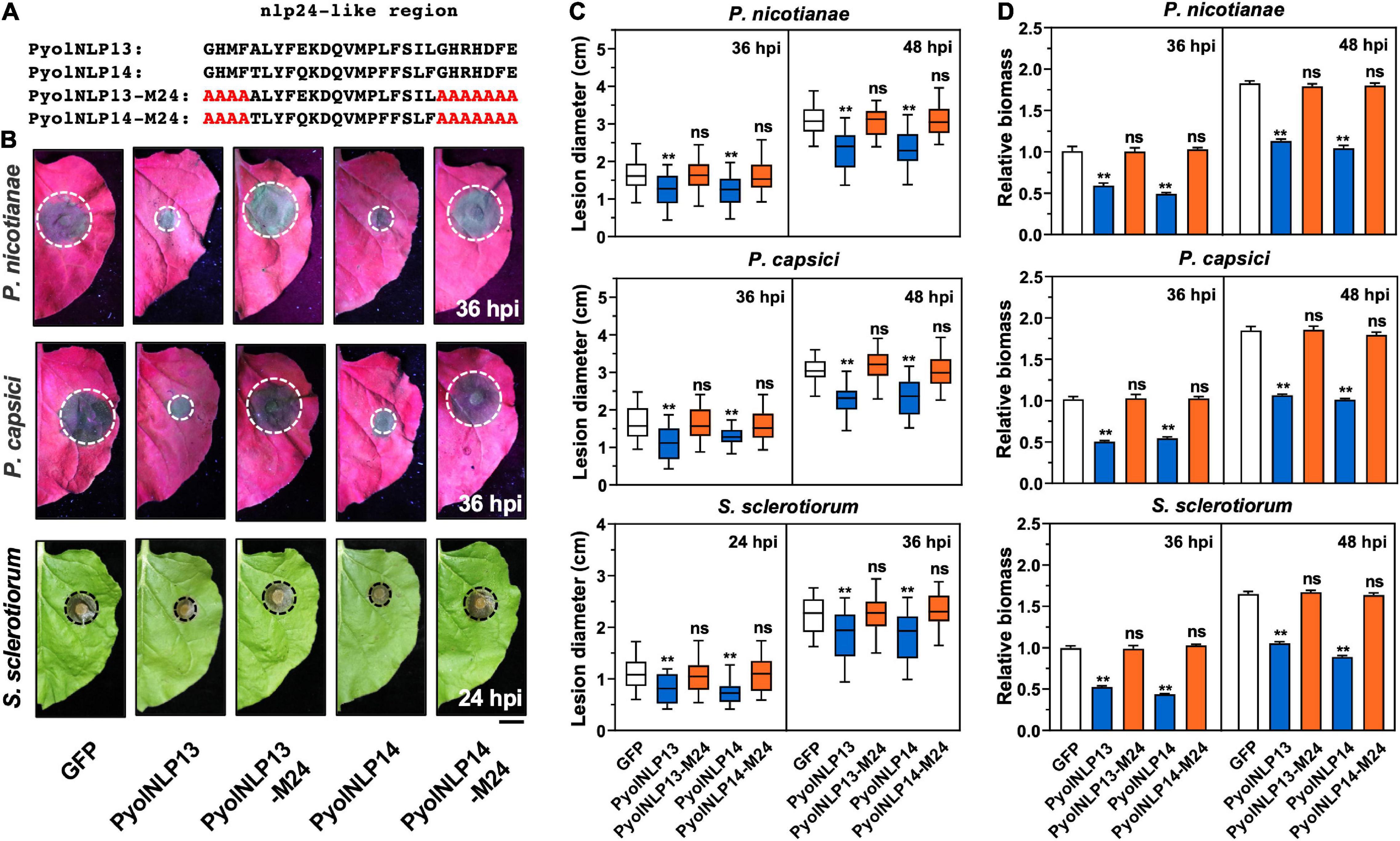
Figure 6. Non-cytotoxic PyolNLPs suppress pathogens infection in N. benthamiana in a nlp24-dependent manner. (A) Schematic representation of nlp24-like regions in PyolNLP13, PyolNLP13-M24, PyolNLP14 and PyolNLP14-M24 with introduced alanine substitutions. (B) Infection phenotypes of infiltrated N. benthamiana leaves expressing GFP, PyolNLP13, PyolNLP13-M24, PyolNLP14 or PyolNLP14-M24, and followed by the inoculation of P. capsici, P. nicotianae or S. sclerotiorum. Photos were taken at 24 and 36 hpi (Scale bar, 1 cm). GFP was used as a negative control. (C) Lesion diameters were measured using ImageJ. Data were analyzed from at least 40 biological replicates. Error bar represents mean ± SD. (D) Relative biomass was determined by qPCR. Expression levels of P. nicotianae, P. capsici and S. sclerotiorum Actin were determined by qPCR using N. benthamiana EF1α as reference. Error bar represents mean ± SD (n = 3). Data were analyzed by Median-edition Levene’s test to determine the homogeneity of variance across groups, and then analyzed by one-way ANOVA with post-hoc Tukey’s range test for groups with equal variance, or Kruskal-Wallis test analysis for groups with unequal variance (ns, no significant difference; **P < 0.01). All experiments were repeated at least three times.
Plant Defensin Expression Induced by Pyolnlp24-Like Pattern Is Irrelevant to the Classic nlp24 Receptor RLP23
We previously found that the nlp24-like pattern is required for PyolNLP5/7-induced expression of plant defensin genes (Yang et al., 2021). Here, we further showed that synthetic peptides of Pyolnlp24-like patterns of PyolNLP5/13/14, flg22 and nlp24 of HaNLP3 are all sufficient to induce the expression of four N. benthamiana defensin genes, including NbPDF1.2, NbDef1.5, NbDef2.1 and NbDef2.2 (Figures 7A,B). However, unlike flg22, none of the nlp24 peptides tested can trigger ROS production in N. benthamiana (Figure 7C). With heterologous expression of Arabidopsis RLP23 (AtRLP23) in N. benthamiana, nlp24 (HaNLP3) but not Pyolnlp24 (PyolNLP5/13/14) can trigger ROS production (Figures 7C,D). Consistently, Pyolnlp24 (PyolNLP5) failed to trigger ROS production in Arabidopsis as compared to flg22 or nlp24 (HaNLP3) (Figure 7E). These data indicate that unlike typical nlp24 patterns such as HaNLP3, Pyolnlp24 peptides can stimulate plant defensin expression but are irrelevant to RLP 23 and ROS burst.
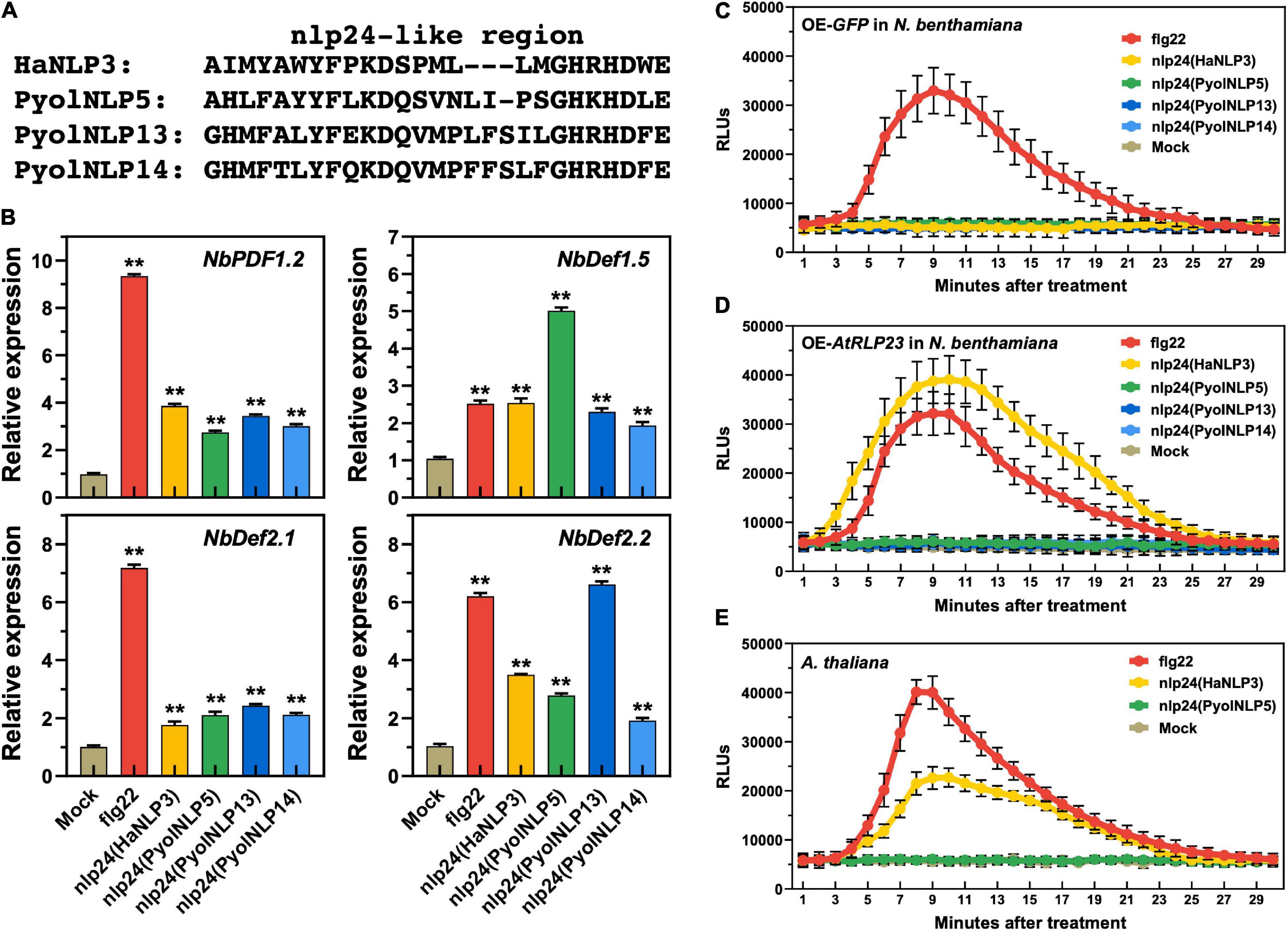
Figure 7. Plant defensin expression induced by Pyolnlp24-Like Pattern is irrelevant to the classic nlp24 receptor RLP23. (A) Schematic representation of nlp24 in HapNLP3 and nlp24-like regions in PyolNLP5, PyolNLP13 and PyolNLP14. (B) Relative expression levels of plant defensin genes in N. benthamiana leaves infiltrated with classic nlp24 (HaNLP3 from Hyaloperonospora parasitica) or Pyolnlp24-like pattern from PyolNLP5, PyolNLP13 or PyolNLP14. ddH2O and flg22 were used as negative and positive controls, respectively. Transcription levels of plant defensin genes NbPDF1.2, NbDef1.5, NbDef2.1 and NbDef2.2 were determined by qRT-PCR using EF1α as reference. Error bar represents mean ± SD (n = 3). Data were analyzed by Median-edition Levene’s test to determine the homogeneity of variance across groups, and then analyzed by one-way ANOVA with post-hoc Tukey’s range test for groups with equal variance (**P < 0.01). (C,D) Dynamics of ROS burst triggered by nlp24 of HaNLP3 in N. benthamiana expressing AtRLP23. (E) Dynamics of ROS burst triggered by nlp24 of HaNLP3 in Arabidopsis thaliana. Leaf disks were treated with water, 500 nM flg22 or nlp24 for 30 min before the detection of relative luminescence units (RLUs) (mean ± SEM, n = 6). All experiments were repeated at least three times.
Discussion
Necrosis and ethylene-inducing peptide 1-like proteins have been proposed to have dual functions in plant-pathogen interactions, acting as both toxin-like virulence factors and triggers of immune responses (Qutob et al., 2006). However, it is unclear whether cytotoxic NLPs directly trigger immune responses or these responses are indirectly induced by cell death. Constitutive expression of a mutant NLP1 lacking cytotoxic activity in the hemibiotrophic pathogen Colletotrichum orbiculare can still block its infection in cucumber (Azmi et al., 2018). In our work, PyolNLP5 mutants with completely abolished necrosis-inducing activity (Figure 1) retain the ability of suppressing Phytophthora infection in N. benthamiana (Figure 2). These consistent results suggest that the cytotoxin and immunity induction activity of NLPs are largely independent.
On the other hand, little is known about the functions of non-cytotoxic NLPs. PiNPP1.2 and PiNPP1.3 from Phytophthora infestans are the first reported non-cytotoxic NLPs (Kanneganti et al., 2006). 11 out of 18 P. sojae NLPs tested cannot induce necrosis (Dong et al., 2012). Among multiple NLPs produced by H. arabidopsidis, none of the tested HaNLPs is cytotoxic (Cabral et al., 2012). In addition to oomycetes, fungi also produce non-cytotoxic NLPs. Such examples have been reported in Colletotrichum higginsianum (Kleemann et al., 2012), V. dahliae (Santhanam et al., 2013) and Magnaporthe oryzae (Fang et al., 2017; Seidl and Van den Ackerveken, 2019). In this work, we find that non-cytotoxic PyolNLP13/14 in Group 4 induce broad resistance to oomycete (P. nicotianae and P. capsici) and fungal (S. sclerotiorum) pathogens in plants (Figures 4, 6, and Supplementary Figure 1). Similar as non-cytotoxic NLP in V. dahliae (VdNLP) (Zhou et al., 2012) and cytotoxic PyolNLP5/7 (Yang et al., 2021), PyolNLP13/14 induce the expression of defensin-encoding gene PDF1.2 (Figure 5C). Our results uncover that both non-cytotoxic and cytotoxic PyolNLPs may promote plant resistance to a wide range of pathogens.
Our work also clarifies that the resistance enhancing activity of both non-cytotoxic and cytotoxic is irrelevant to the accumulation of ROS, which have dual functions of causing cell injury and inducing defense responses in plants (Mittler, 2017). To our knowledge, this is the first report that the resistance- and necrosis-inducing functions of cytotoxic NLPs are largely separate. Non-cytotoxic PyolNLPs can be good genetic engineering targets for enhancing crop disease resistance with no injuries caused by ROS or cell death. We reveal that both non-cytotoxic and cytotoxic PyolNLPs induce the expression of multiple plant defensin genes, which may be the primary downstream pathway responsible for PyolNLP-triggered plant immunity.
The relatively conserved peptide sequence npl24 inside NLPs is recognized as a MAMP by plants (Bohm et al., 2014; Oome et al., 2014), with the heptapeptide “GHRHDWE” motif being a central region (Fellbrich et al., 2002; Santhanam et al., 2013; Seidl and Van den Ackerveken, 2019). Both “AIMY” and “GHRHDWE” motifs are necessary for non-cytotoxic PyolNLP13/14 to suppress pathogen infection (Figure 6). However, how PyolNLPs are perceived by plants remain to be determined. RLP23 is a classic NLP receptor in Arabidopsis (Albert et al., 2015). Genetic complementation tests in Arabidopsis and non-responsive species including tobacco, tomato and potato confirm the requirement of RLP23 for nlp20 pattern recognition (Seidl and Van den Ackerveken, 2019). However, nlp20 can still trigger immunity to the downy mildew pathogen Bremia lactucae in lettuce (Lactuca sativa), which does not have RLP23 (Seidl and Van den Ackerveken, 2019). The synthetic Conlp24 peptide from C. orbiculare triggers ROS burst in Arabidopsis. Its mutant version (Conlp24Mut) loses ROS-inducing ability in Arabidopsis but is still functional in cucumber (Azmi et al., 2018; Seidl and Van den Ackerveken, 2019). In this research, we find that plant defensin expression induced by Pyolnlp24-like pattern is irrelevant to RLP23. These observations suggest the existence of multiple plant NLP receptors, with PyolNLPs being perceived by receptor(s) other than RLP23. Different plant species may harbor distinct sets of receptors to recognize NLPs in a pathogen and NLP-type specific manner.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MJ, KY, and SF designed and wrote the manuscript. KY, CC, SF, YW, JL, and GA, conducted most of the experiments and performed data analysis. YC, HZ, DS, ZY, QS, BW, and WL performed the experiments. DD, YZ, and HP made a proposal and modified the manuscript. All co-authors read and approved it. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31801715, 31721004), Jiangsu Agricultural Science and Technology Innovation Fund [CX(20)3127].
Conflict of Interest
QS, BW, and WL were employed by the company Shandong Linyi Tobacco Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Laura Grenville-Briggs Didymus (Swedish University of Agricultural Sciences) for kindly providing the P. oligandrum strain CBS 530.74 and P. periplocum strain CBS 532.74, Yuanchao Wang, Guoliang Qian, and Yaxin Du (Nanjing Agricultural University) for kind support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.830636/full#supplementary-material
Supplementary Figure 1 | Alignment of the amino acid sequences of PyolNLP3∼7.
Supplementary Figure 2 | The phylogenetic tree of PyolNLPs from P. oligandrium and P. periplocum.
Supplementary Figure 3 | Confirmation of PyolNLP and AtRLP23 expression in N. benthamiana leaves by Western blotting.
Supplementary Table 1 | Primers used in this study.
References
Ai, G., Xia, Q., Song, T., Li, T., Zhu, H., Peng, H., et al. (2021). A Phytophthora sojae CRN effector mediates phosphorylation and degradation of plant aquaporin proteins to suppress host immune signaling. PLoS Pathog 17:e1009388. doi: 10.1371/journal.ppat.1009388
Ai, G., Yang, K., Ye, W. W., Tian, Y. E., Du, Y. X., Zhu, H., et al. (2020). Prediction and characterization of RXLR effectors in pythium species. Mol. Plant Microbe Interact. 33, 1046–1058. doi: 10.1094/Mpmi-01-20-0010-R
Albert, I., Bohm, H., Albert, M., Feiler, C. E., Imkampe, J., Wallmeroth, N., et al. (2015). An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1:15140. doi: 10.1038/nplants.2015.140
Azmi, N. S. A., Singkaravanit-Ogawa, S., Ikeda, K., Kitakura, S., Inoue, Y., Narusaka, Y., et al. (2018). Inappropriate expression of an NLP effector in colletotrichum orbiculare impairs infection on cucurbitaceae cultivars via plant recognition of the C-Terminal region. Mol. Plant Microbe Interact. 31, 101–111. doi: 10.1094/MPMI-04-17-0085-FI
Bohm, H., Albert, I., Oome, S., Raaymakers, T. M., Van den Ackerveken, G., and Nurnberger, T. (2014). A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog 10:e1004491. doi: 10.1371/journal.ppat.1004491
Cabral, A., Oome, S., Sander, N., Kufner, I., Nurnberger, T., and Van den Ackerveken, G. (2012). Nontoxic Nep1-Like proteins of the downy mildew pathogen hyaloperonospora arabidopsidis: repression of necrosis-inducing activity by a surface-exposed region. Mol. Plant Microbe Interact. 25, 697–708. doi: 10.1094/Mpmi-10-11-0269
Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., and Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476. doi: 10.1105/tpc.105.036574
Circelli, P., Donini, M., Villani, M. E., Benvenuto, E., and Marusic, C. (2010). Efficient agrobacterium-based transient expression system for the production of biopharmaceuticals in plants. Bioeng Bugs 1, 221–224. doi: 10.4161/bbug.1.3.11722
Couto, D., and Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Dong, S. M., Kong, G. H., Qutob, D., Yu, X. L., Tang, J. L., Kang, J. X., et al. (2012). The NLP toxin family in phytophthora sojae includes rapidly evolving groups that lack necrosis-inducing activity. Mol. Plant Microbe Interact. 25, 896–909. doi: 10.1094/Mpmi-01-12-0023-R
Dong, Y. M., Jing, M. F., Shen, D. Y., Wang, C. Y., Zhang, M. Q., Liang, D., et al. (2020). The mirid bug Apolygus lucorum deploys a glutathione peroxidase as a candidate effector to enhance plant susceptibility. J. Exp. Bot. 71, 2701–2712. doi: 10.1093/jxb/eraa015
Du, J., Verzaux, E., Chaparro-Garcia, A., Bijsterbosch, G., Keizer, L. C., Zhou, J., et al. (2015). Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 1:15034. doi: 10.1038/nplants.2015.34
Fang, Y. L., Peng, Y. L., and Fan, J. (2017). The Nep1-like protein family of Magnaporthe oryzae is dispensable for the infection of rice plants. Sci. Rep. 7:4372. doi: 10.1038/s41598-017-04430-4430
Fellbrich, G., Romanski, A., Varet, A., Blume, B., Brunner, F., Engelhardt, S., et al. (2002). NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J. 32, 375–390. doi: 10.1046/j.1365-313X.2002.01454.x
Gijzen, M., and Nurnberger, T. (2006). Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67, 1800–1807. doi: 10.1016/j.phytochem.2005.12.008
Green, S. A., Chen, X., and Matich, A. J. (2012). In planta transient expression analysis of monoterpene synthases. Methods Enzymol. 515, 43–61. doi: 10.1016/B978-0-12-394290-6.00003-3
Jehle, A. K., Lipschis, M., Albert, M., Fallahzadeh-Mamaghani, V., Furst, U., Mueller, K., et al. (2013). The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell 25, 2330–2340. doi: 10.1105/tpc.113.110833
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kanneganti, T. D., Huitema, E., Cakir, C., and Kamoun, S. (2006). Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nep1-like protein PiNPP1.1 and INF1 elicitin. Mol. Plant-Microbe Interact. 19, 854–863. doi: 10.1094/Mpmi-19-0854
Kleemann, J., Rincon-Rivera, L. J., Takahara, H., Neumann, U., Ver Loren, van Themaat, E., et al. (2012). Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog 8:e1002643. doi: 10.1371/journal.ppat.1002643
Lenarcic, T., Albert, I., Bohm, H., Hodnik, V., Pirc, K., Zavec, A. B., et al. (2017). Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 358, 1431–1434. doi: 10.1126/science.aan6874
Lenarcic, T., Pirc, K., Hodnik, V., Albert, I., Borisek, J., Magistrato, A., et al. (2019). Molecular basis for functional diversity among microbial Nep1-like proteins. PLoS Pathog 15:e1007951. doi: 10.1371/journal.ppat.1007951
Li, Q., Ai, G., Shen, D. Y., Zou, F., Wang, J., Bai, T., et al. (2019). A Phytophthora capsici effector targets ACD11 binding partners that regulate ROS-Mediated defense response in Arabidopsis. Mol. Plant 12, 565–581. doi: 10.1016/j.molp.2019.01.018
Lo Presti, L., Lanver, D., Schweizer, G., Tanaka, S., Liang, L., Tollot, M., et al. (2015). Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545. doi: 10.1146/annurev-arplant-043014-114623
Lu, J., Bai, M., Ren, H., Liu, J., and Wang, C. (2017). An efficient transient expression system for gene function analysis in rose. Plant Methods 13:116. doi: 10.1186/s13007-017-0268-261
Ma, Z., Zhu, L., Song, T., Wang, Y., Zhang, Q., Xia, Y., et al. (2017). A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 355, 710–714. doi: 10.1126/science.aai7919
Nie, J., Yin, Z., Li, Z., Wu, Y., and Huang, L. (2019). A small cysteine-rich protein from two kingdoms of microbes is recognized as a novel pathogen-associated molecular pattern. New Phytol. 222, 995–1011. doi: 10.1111/nph.15631
Oome, S., and Van den Ackerveken, G. (2014). Comparative and functional analysis of the widely occurring family of Nep1-like proteins. Mol. Plant Microbe Interact. 27, 1081–1094. doi: 10.1094/MPMI-04-14-0118-R
Oome, S., Raaymakers, T. M., Cabral, A., Samwel, S., Bohm, H., Albert, I., et al. (2014). Nep1-like proteins from three kingdoms of life act as a microbe-associated molecular pattern in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 111, 16955–16960. doi: 10.1073/pnas.1410031111
Ottmann, C., Luberacki, B., Kufner, I., Koch, W., Brunner, F., Weyand, M., et al. (2009). A common toxin fold mediates microbial attack and plant defense. Proc. Natl. Acad. Sci. U S A. 106, 10359–10364. doi: 10.1073/pnas.0902362106
Ouyang, Z., Li, X., Huang, L., Hong, Y., Zhang, Y., Zhang, H., et al. (2015). Elicitin-like proteins Oli-D1 and Oli-D2 fromPythium oligandrumtrigger hypersensitive response inNicotiana benthamianaand induce resistance againstBotrytis cinereain tomato. Mol. Plant Pathol. 16, 238–250. doi: 10.1111/mpp.12176
Poland, J. A., Balint-Kurti, P. J., Wisser, R. J., Pratt, R. C., and Nelson, R. J. (2009). Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14, 21–29. doi: 10.1016/j.tplants.2008.10.006
Qi, J., Wang, J., Gong, Z., and Zhou, J. M. (2017). Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 38, 92–100. doi: 10.1016/j.pbi.2017.04.022
Qutob, D., Kemmerling, B., Brunner, F., Kufner, I., Engelhardt, S., Gust, A. A., et al. (2006). Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 18, 3721–3744. doi: 10.1105/tpc.106.044180
Santhanam, P., van Esse, H. P., Albert, I., Faino, L., Nurnberger, T., and Thomma, B. P. H. J. (2013). Evidence for functional diversification within a fungal NEP1-Like protein family. Mol. Plant Microbe Interact. 26, 278–286. doi: 10.1094/Mpmi-09-12-0222-R
Seidl, M. F., and Van den Ackerveken, G. (2019). Activity and phylogenetics of the broadly occurring family of microbial Nep1-Like proteins. Annu. Rev. Phytopathol. 57, 367–386. doi: 10.1146/annurev-phyto-082718-100054
Song, T., Ma, Z., Shen, D., Li, Q., Li, W., Su, L., et al. (2015). An oomycete CRN effector reprograms expression of plant HSP genes by targeting their promoters. PLoS Pathog 11:e1005348. doi: 10.1371/journal.ppat.1005348
Tang, D., Wang, G., and Zhou, J. M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29, 618–637. doi: 10.1105/tpc.16.00891
Wan, W. L., Frohlich, K., Pruitt, R. N., Nurnberger, T., and Zhang, L. (2019). Plant cell surface immune receptor complex signaling. Curr. Opin. Plant Biol. 50, 18–28. doi: 10.1016/j.pbi.2019.02.001
Wang, L., Albert, M., Einig, E., Furst, U., Krust, D., and Felix, G. (2016). The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat. Plants 2:16185. doi: 10.1038/nplants.2016.185
Wang, Y., Xu, Y., Sun, Y., Wang, H., Qi, J., Wan, B., et al. (2018). Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat. Commun. 9:594. doi: 10.1038/s41467-018-03010-3018
Wei, W., Pierre-Pierre, N., Peng, H., Ellur, V., Vandemark, G. J., and Chen, W. (2020). The D-galacturonic acid catabolic pathway genes differentially regulate virulence and salinity response in Sclerotinia sclerotiorum. Fungal Genet. Biol. 145:103482. doi: 10.1016/j.fgb.2020.103482
Wen, Q., Sun, M., Kong, X., Yang, Y., Zhang, Q., Huang, G., et al. (2021). The novel peptide NbPPI1 identified from Nicotiana benthamiana triggers immune responses and enhances resistance against Phytophthora pathogens. J Integr Plant Biol. 63, 961–976. doi: 10.1111/jipb.13033
Yang, B., Wang, Y., Guo, B., Jing, M., Zhou, H., Li, Y., et al. (2019). The Phytophthora sojae RXLR effector Avh238 destabilizes soybean Type2 GmACSs to suppress ethylene biosynthesis and promote infection. New Phytol. 222, 425–437. doi: 10.1111/nph.15581
Yang, C. C., and Fernando, W. G. D. (2021). Analysis of the oxidative burst and its relevant signaling pathways in leptosphaeria maculans-brassica napus pathosystem. Int. J. Mol. Sci. 22:4812. doi: 10.3390/ijms22094812
Yang, K., Dong, X., Li, J., Wang, Y., Cheng, Y., Zhai, Y., et al. (2021). Type 2 Nep1-Like proteins from the biocontrol oomycete Pythium oligandrum suppress Phytophthora capsici infection in solanaceous plants. J. Fungi (Basel) 7:496. doi: 10.3390/jof7070496
Yin, Z., Wang, N., Pi, L., Li, L., Duan, W., Wang, X., et al. (2021). Nicotiana benthamiana LRR-RLP NbEIX2 mediates the perception of an EIX-like protein from Verticillium dahliae. J. Integr. Plant Biol. 63, 949–960. doi: 10.1111/jipb.13031
Yu, T. Y., Sun, M. K., and Liang, L. K. (2021). Receptors in the induction of the plant innate immunity. Mol. Plant Microbe Interact. 34, 587–601. doi: 10.1094/MPMI-07-20-0173-CR
Yu, X., Tang, J., Wang, Q., Ye, W., Tao, K., Duan, S., et al. (2012). The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol. 196, 247–260. doi: 10.1111/j.1469-8137.2012.04241.x
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-3316
Zhang, L., Kars, I., Essenstam, B., Liebrand, T. W., Wagemakers, L., Elberse, J., et al. (2014). Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164, 352–364. doi: 10.1104/pp.113.230698
Zhang, W., Fraiture, M., Kolb, D., Loffelhardt, B., Desaki, Y., Boutrot, F. F., et al. (2013). Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227–4241. doi: 10.1105/tpc.113.117010
Zhou, B. J., Jia, P. S., Gao, F., and Guo, H. S. (2012). Molecular characterization and functional analysis of a necrosis- and ethylene-inducing, protein-encoding gene family from Verticillium dahliae. Mol. Plant Microbe Interact. 25, 964–975. doi: 10.1094/Mpmi-12-11-0319
Zhou, Y., Yang, K., Yan, Q., Wang, X., Cheng, M., Si, J., et al. (2021). Targeting of anti-microbial proteins to the hyphal surface amplifies protection of crop plants against Phytophthora pathogens. Mol. Plant 14, 1391–1403. doi: 10.1016/j.molp.2021.05.007
Keywords: Nep1-like proteins, Pythium oligandrum, Phytophthora, necrosis-inducing activity, ROS burst, RLP23
Citation: Yang K, Chen C, Wang Y, Li J, Dong X, Cheng Y, Zhang H, Zhai Y, Ai G, Song Q, Wang B, Liu W, Yin Z, Peng H, Shen D, Fang S, Dou D and Jing M (2022) Nep1-Like Proteins From the Biocontrol Agent Pythium oligandrum Enhance Plant Disease Resistance Independent of Cell Death and Reactive Oxygen Species. Front. Plant Sci. 13:830636. doi: 10.3389/fpls.2022.830636
Received: 07 December 2021; Accepted: 12 January 2022;
Published: 04 March 2022.
Edited by:
Xiao-Ren Chen, Yangzhou University, ChinaReviewed by:
Gregor Anderluh, National Institute of Chemistry (Slovenia), SloveniaBiao Gu, Northwest A&F University, China
Copyright © 2022 Yang, Chen, Wang, Li, Dong, Cheng, Zhang, Zhai, Ai, Song, Wang, Liu, Yin, Peng, Shen, Fang, Dou and Jing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maofeng Jing, jingmf@njau.edu.cn; orcid.org/0000-0003-3086-3379; Song Fang, fangsong@caas.cn
†These authors have contributed equally to this work
 Kun Yang1†
Kun Yang1† Ying Zhai
Ying Zhai Hao Peng
Hao Peng Danyu Shen
Danyu Shen Daolong Dou
Daolong Dou Maofeng Jing
Maofeng Jing