- 1Technology Center, China Tobacco Hunan Industrial Co., Ltd., Changsha, China
- 2Key Laboratory for Tobacco Gene Resources, Tobacco Research Institute, Chinese Academy of Agricultural Sciences, Qingdao, China
- 3Shandong Peanut Research Institute, Qingdao, China
- 4Graduate School of Chinese Academy of Agricultural Sciences, Beijing, China
The NAC (NAM, ATAF1/2, and CUC2) family acts as one of the largest families of the transcription factor in the plant kingdom and was revealed to function as the important regulators in various environmental stresses. However, a few studies were reported about the biofunctions of the NAC transcription factor in tobacco. In the current study, we characterized a novel NAC transcription factor encoding the gene NtNAC053 in tobacco, which was significantly up-regulated when exposed to salt and drought treatments. The results of cis-acting elements analysis suggested that the promoter region of NtNAC053 gene possesses a number of stress-responsive elements, and this gene could be induced by exogenous abscisic acid (ABA) treatment. Moreover, the NtNAC053–GFP fusion protein was localized in the cell nucleus and possessed a transactivation domain in its C-terminal, implying that NtNAC053 may undertake as a transcriptional activator in tobacco. Notably, the overexpression of NtNAC053 in tobacco resulted in hypersensitivity to ABA treatment. Furthermore, these overexpression lines showed significantly enhanced tolerances to drought and salt stresses. Under salt and drought stresses, these overexpression lines possessed higher superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) activities. Interestingly, the expressions of putative stress-related genes, including NtCOR15A, NtRAB18, NtDREB1A, NtERF5, NtKAT2, and NtERD11, were up-regulated in these overexpression lines when subjected to salt and drought stresses. The clues provided in our study suggested that the NtNAC053 gene encodes a novel NAC transcription factor and could confer the drought and salt stress tolerances by inspiring the downstream stress-responsive genes and antioxidant system in tobacco.
Introduction
Transcription factors play a pivotal role in regulating the expression of target genes involved in plant growth, development, and response to environmental stresses (Qu and Zhu, 2006). The NAC protein family has been reported to be one of the largest plant-specific transcription factor families. Its name is based on three transcription factors: no apical meristem (NAM), Arabidopsis transcription activation factor (ATAF1,2), and cup-shaped cotyledon (CUC) (Souer et al., 1996; Aida et al., 1997). NAC transcription factors harbor a highly conserved DNA-binding domain at the N-terminus and a variable transcription regulation region (TRR) at the C-terminus (Ernst et al., 2004; Olsen et al., 2005). Generally, the N-terminal DNA-binding domain of NAC protein possesses approximately 150 amino acid residues divided into five subdomains (A-E), which are related to the formation of dimer, functional diversity, and DNA-binding activity (Olsen et al., 2005). The C-terminal TRR of NAC protein functions in conferring regulation diversity of transcriptional activation activity (Yamaguchi et al., 2010; Puranik et al., 2011). Furthermore, the transmembrane domain is also found in several NAC proteins, which function in plasma membrane anchoring (Kim et al., 2010).
The NAC gene was first identified from petunias, in which it was involved in the shoot apical meristems development and primordium formation (Souer et al., 1996). In the plant kingdom, the NAC proteins have been reported to play important roles in various biological processes, including lateral root development (Xie et al., 2000; He et al., 2005; Zhang et al., 2018), vascular development (Zhao et al., 2016), development of shoot apical meristem (Takada et al., 2001; Larsson et al., 2012), the formation of the adventitious shoot (Hibara et al., 2003), seed germination (Park et al., 2011), flower formation (Hendelman et al., 2013), secondary wall synthesis (Mitsuda et al., 2007; Zhao et al., 2010), and leaf senescence (Guo and Gan, 2006). So far, the NAC family has been identified in a large number of plants, including Arabidopsis (105) (Ooka et al., 2003), tobacco (Nicotiana tabacum) (154) (Li et al., 2018), potato (Solanum tuberosum) (110) (Singh et al., 2013), white pear (Pyrus bretschneideri) (183) (Gong et al., 2019), maize (Zea mays) (152) (Shiriga et al., 2014), rice (Oryza sativa) (151) (Nuruzzaman et al., 2010), and peanut (Arachis hypogaea) (132) (Li P. et al., 2021).
Furthermore, several NAC members have been reported to participate in response to abiotic stresses in Arabidopsis and other species. Stress-inducible NAC transcription factors ANAC019, ANAC055, and ANAC072/RD26 were found to improve the drought tolerance via regulating the expression of the ERD1 gene (Tran et al., 2004). Furthermore, ANAC072 was identified to be involved in the ABA-dependent signaling pathway (Fujita et al., 2004). Notably, the expression of ANAC002/ATAF1 was induced by drought and ABA treatments. Moreover, the ataf1 knockout mutants displayed a lower survival ratio than wild-type plants under drought conditions, indicating that ATAF1 functions as a regulator in drought stress tolerance (Wu et al., 2009). The ANAC062/NTL6 was reported to play an important role in inducing a drought-resistance response and the overexpression of NTL6 plants exhibit enhanced resistance to water-deficit conditions (Kim et al., 2012). In addition, the loss of ANAC035/LOV1 function results in hypersensitivity to cold temperature, whereas overexpression of this gene could enhance the cold tolerance (Yoo et al., 2007). In rice, the expression of OsNAC5 gene could be significantly induced by various stresses, including drought, salt, cold, ABA, and Me-JA. Whereas the overexpression of OsNAC5 resulted in the improvement of salt stress tolerance (Takasaki et al., 2010). Moreover, the ONAC066 was reported to promote the tolerances against drought and oxidative stresses by activating the transcription of OsDREB2A in transgenic rice (Yuan et al., 2019). In wheat, the overexpression of TaNAC47 in Arabidopsis increases the tolerance to drought stress and sensitivity to ABA (Zhang et al., 2015). Besides, the overexpression of TaNAC2a improved drought tolerance in transgenic tobacco (Tang et al., 2012).
Tobacco is one of the most widely cultivated crops worldwide and serviced as a model plant that possesses various primary and secondary metabolisms. However, tobacco has been at a continuous risk from multiple environmental stresses, affecting both yield and quality. In our previous study, the systematic identification of the NAC family was performed in tobacco (Li et al., 2018). However, a few studies were reported about the biofunctions of the NAC transcription factor in tobacco. Notably, in our previous study, the expression of StNAC053 was significantly up-regulated after salt and drought treatments. The overexpression of this potato gene could enhance the salt and drought tolerance in transgenic Arabidopsis (Wang et al., 2021). Considering the close genetic distance of potato and tobacco, the homolog of StNAC053 might also confer salt and drought tolerances in tobacco. To verify this hypothesis, we cloned and characterized an NAC transcription factor, NtNAC053, from tobacco, and systematic analysis was performed to explore the function of this gene in salt and drought stress responses.
Materials and Methods
Plant Materials and Treatments
Cultivated tobacco K326 was used to analyze the expression pattern of NtNAC053 in the current study. The root, root tip, stem, stem tip, young leaf, and senescent leaf were collected. For salt, drought, and ABA treatments, tobacco seedlings were transferred to MS liquid medium supplemented with 100 mM NaCl, 300 mM mannitol, and 10 μM ABA for 0, 1, 3, and 6 h, respectively. For cold treatment, tobacco seedlings were treated at 4°C for 0, 1, 3, and 6 h. All of the harvested samples were stored at –80°C before RNA extraction was performed. Three biological replicates were used for each sample.
RNA Extraction and qPCR Analysis
Ultrapure RNA Kit (cwbiotech, Beijing, China) was used to extract total RNA, then the first cDNA strand was synthesized using Evo M-MLV Mix Kit with gDNA Clean for qPCR (Accurate Biotechnology, Changsha, China). Expression of stress-responsive genes (NtDREB1A, NtERF5, NtKAT2, NtCOR15A, NtRAB18, NtERD11, NtNHX1, and NtSOS1) and ABA signaling genes from tobacco was detected. The qRT-PCR reactions were carried out in Roche LightCycler 480 Real-Time PCR instrument with SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology, Changsha, China). The tobacco ribosomal protein gene L25 (GenBank No. L18908) was used as a control (Li Z. et al., 2021). All experimental data were obtained through three technical repetitions, and the relative expression level was calculated by 2–ΔΔCT method (Livak and Schmittgen, 2001). The details of the primers in this study are provided in Supplementary Table 1.
Cloning and Sequence Analysis of NtNAC053
BLASTP search with an E-value cutoff of 0.0001 was carried out in SGN1 using potato StNAC053 protein sequence as a query. The candidate gene (Nitab4.5_0000322g0110.1) was obtained and then named as NtNAC053. The CDS of NtNAC053 was amplified by RT-PCR. The purified PCR products were subcloned into the pEASY-Blunt vector, and the constructed vector was then transformed into Escherichia coli 5α competent cells (Trans Gen, Beijing, China). After sequencing, the recombinant plasmid was named Blunt-NtNAC053. The physicochemical properties of the NtNAC053 protein, including molecular weight (MW) and isoelectric point (pI), were predicated using the ProtParam online tool2. The nuclear localization signal (NLS) of NAC proteins was predicted via cNLS Mapper3. The DNAMAN software (Lynnon Biosoft) was used to perform multiple sequence alignment. A neighbor-joining tree was generated using MEGA 6 with 1,000 replications. The NAC protein sequences were obtained from the TAIR4, SGN (see text footnote 1), and NCBI. The conserved motifs of NAC protein sequences were analyzed via MEME5 with default parameters. The collinearity relationship between NtNAC053 and StNAC053 was analyzed using TBtools (Chen et al., 2020).
To assess the promoter cis-acting elements of the NtNAC053 gene, the sequences 2,000 bp upstream of the NtNAC053 gene were extracted from the genome database. The obtained sequences were subjected to PlantCARE6 platform analysis to further search for the putative cis-elements in their promoter regions.
Subcellular Localization and Transactivation Assay
The CDS (without stop codon) of NtNAC053 gene was amplified from Blunt-NtNAC053 using PCR. Then the purified PCR products was inserted into the PYG57 vector, which started by the CaMV-35S promoter and contained the GFP fragment. The construct was transformed into Agrobacterium competent cell GV3101, and transiently expressed in the leaves of Nicotiana benthamiana. Simultaneously, the empty vector injected leaves as a control. Three days after the injection, the leaves were soaked in a DAPI staining solution to determine the location of the nucleus. The confocal laser microscope (Leica SP8) was used to observe the fluorescence signal of the fusion protein.
The CDS of NtNAC053 was amplified and inserted into the EcoRI site of the pBridge vector, using an In-fusion HD Cloning Kit (Takara, Shiga, Japan) to fuse with a GAL4 DNA binding domain. The constructs and the control vector were introduced into the yeast strain AH109 separately, followed by growing yeasts on SD/-Trp, and SD/-Trp supplemented with X-Gal for 4 days at 30°C. The transcriptional activation activities were evaluated based on the growth status of different transformants.
Generation of NtNAC053 Transgenic Tobacco Plants
The coding sequence of NtNAC053 gene was amplified from the cDNA and inserted into the pCHF3 vector, which was driven by the CaMV-35S promoter, to complete the construction of the overexpression vector. The pCHF3 plasmid containing the NtNAC053 gene was transformed into tobacco leaves by the Agrobacterium-mediated method (Buschmann, 2016). T0 transgenic seeds were screened on MS medium containing 50 mg/L kanamycin to obtain NtNAC053 overexpression plants. T3 generation homozygous seedlings were used for further experiments.
Seedling Growth Assays
For seed germination assays, T3 generation seeds and wild-type (K326) seeds were sterilized and evenly sown on MS medium containing 1 μM ABA, 100 mM NaCl, and 150 mM NaCl, respectively. After cultivation for 14 days in a growth chamber (25°C, continuous light), the germination rates were calculated with three replications.
For root elongation assays, tobacco seedlings were grown in vertical MS medium for 14 days, then transferred to MS medium with NaCl (100 and 150 mM) and ABA (1 μM), respectively. The length of the primary root was measured after 14 days with three replications.
Drought Stress Tolerance Assays of NtNAC053 Transgenic Tobacco Plants
For the drought tolerance assays, T3 transgenic and wild-type (K326) tobacco seeds were sown into the soil mixture evenly and kept in normal conditions. Six weeks later, the tobacco seedlings were treated by drought stress for 10 days. Then the seedlings were re-watered for 3 days, and the survival rates of tobacco seedlings were calculated. The leaves of the seedlings were collected and air-dried for 3 h, then leaf weight was recorded at five time points (0, 45, 90, 135, and 180 min). The rate of water loss was calculated based on the leaves’ weight changes.
Physiological Measurements
The physiological parameters, including contents of MDA and hydrogen peroxide, and the enzymatic activities of superoxide (SOD), peroxidase (POD), and catalase (CAT) were measured as previously described (He et al., 2019a). Three biological replicates were performed.
Yeast One Hybrid Assay
For yeast one-hybrid assay, the full-length CDS sequence of NtNAC053 transcription factor was inserted into pGAD424 to construct a pGAD424-NtNAC053 recombinant vector. Furthermore, the pGAD424-NtNAC053 recombinant vector was co-transformed with different LacZ reporter gene constructs containing three tandem reporter sequences into yeast strain YM4271. The β-galactosidase activity was assessed according to the manufacturer’s instruction (Clontech, United States).
Statistical Analysis
The GraphPad Prism 8 (t-test) was used to analyze the statistical significance. P < 0.05, 0.01, or 0.001 were regarded as significantly different from the control. All data were obtained from three replicates.
Results
Isolation and Characterization of the NtNAC053 Gene
The NAC transcription factors were reported to play multiple roles in response to abiotic stresses. In our previous study, the StNAC053 was found to confer salt and drought tolerance in transgenic Arabidopsis (Wang et al., 2021). To identify the homolog of StNAC053 in tobacco, the BLASTP search was carried out in SGN (see text footnote 1) using the StNAC053 protein sequence as a query. The candidate gene (Nitab4.5_0000322g0110.1) was obtained and then named as NtNAC053. Furthermore, the NtNAC053 gene was cloned from cultivated tobacco K326. The full-length coding sequence of NtNAC053 is 897 bp, which encodes a 298 amino acid protein. The molecular weight (MW) and theoretical isoelectric point (pI) of the protein are 34.247 kDa and 6.56, respectively. Multiple sequence alignments persuaded that NtNAC053 possesses a conserved NAC domain (8-162) in the N-terminal region and a variable domain at the C-terminal region. The NAC domain consists of five subdomains, including subdomain A-E, which have been predicted to function as the DNA-binding domain together. Furthermore, a novel nuclear localization signal (NLS) was identified in the subdomain D (Supplementary Figure 1).
Phylogenetic and Syntenic Analysis of the NtNAC053
To explore the phylogenetic relationship of NtNAC053, a neighbor-joining (NJ) tree containing the selected novel NAC transcription factor, including seven members from Arabidopsis, three from potato, and two from wheat. The results persuaded that NtNAC053 was clustered with StNAC053, and both of these two members fall into the ATAF subfamily (Supplementary Figure 2A). Furthermore, the conserved motifs of NtNAC053 and these selected NAC members were predicted by the MEME tools. As a result, 15 conserved motifs were identified (Supplementary Figure 3). Notably, motif 3, 1, 2, and 4 represented the NAC domain and were shared by all tested members (Supplementary Figure 2B). Besides, it was found that motif 12 may be specific to the dicotyledonous members of ATAF subfamily.
To further study the evolutionary relationship of NtNAC053 gene, syntenic analysis was performed for tobacco and potato (Supplementary Figure 2C). The result showed that NtNAC053 was the ortholog gene of StNAC053.
Expression Patterns and Promoter Analysis of NtNAC053
To explore the potential function of NtNAC053, the cis-acting elements and the expression pattern of NtNAC053 in different tissues were investigated. Interestingly, the cis-acting elements analysis revealed that five elements related to ABA responsiveness (ABRE) were identified in the promoter region of the NtNAC053 gene (Supplementary Figure 4). As shown in Figure 1A, the expression of NtNAC053 had a higher level in tobacco root and young leaves, and had a lower level in the root tip, stem, and stem tip. Interestingly, the expression of NtNAC053 had a high level in senescent leaf. Furthermore, we examined the expression pattern of NtNAC053 under various treatments, including drought, salt, cold, and ABA. For the drought and ABA treatments, the expression levels of NtNAC053 were up-regulated before the first 6 h, respectively (Figures 1B,D). For the salt treatment, the expression levels of NtNAC053 were also up-regulated and reached the peak at 3 h (Figure 1E). For the cold treatment, no significant alteration was detected (Figure 1C).
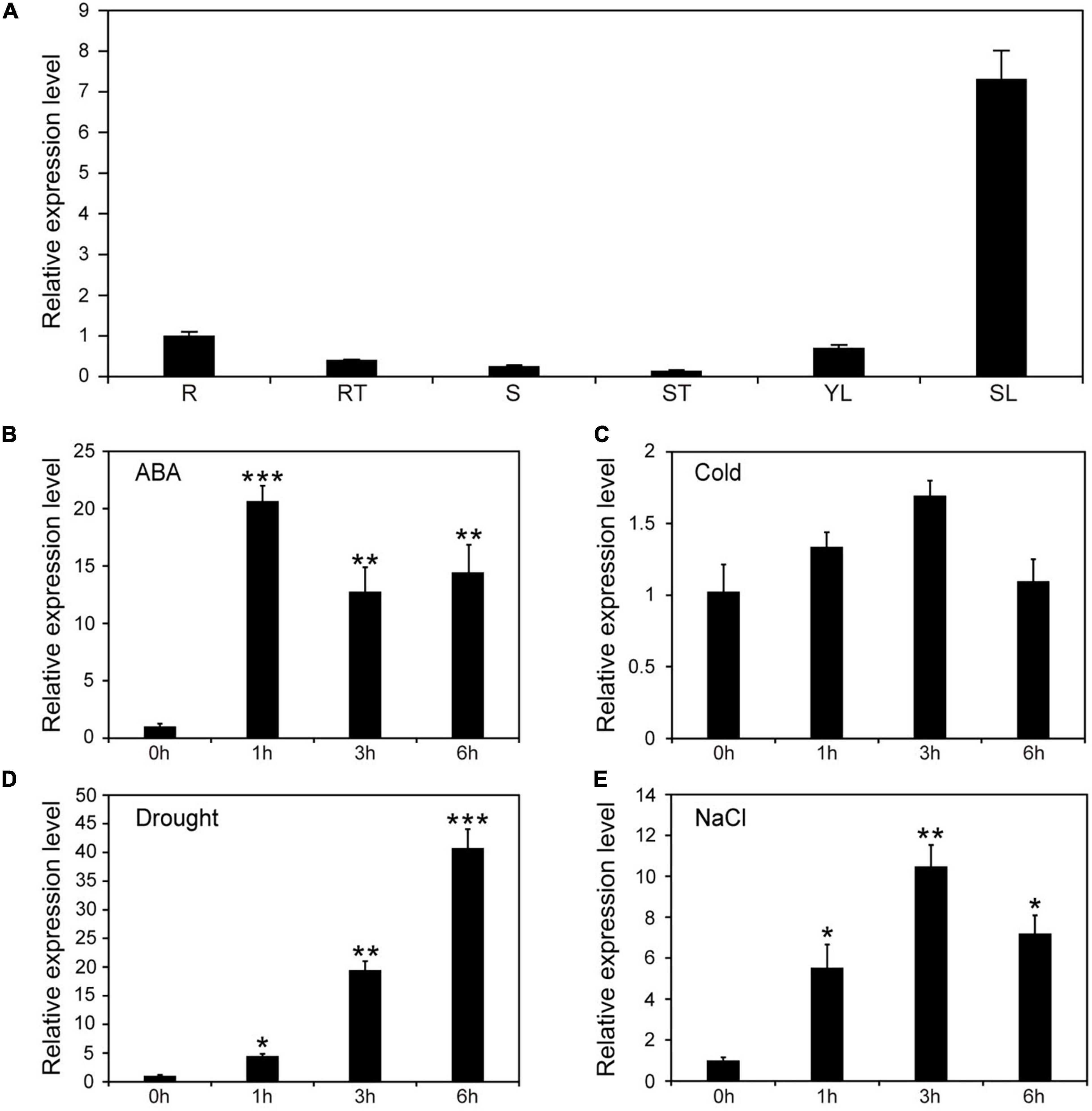
Figure 1. Expression patterns of NtNAC053 in tobacco. (A) The expression pattern of NtNAC053 in tested tissues. R: root; RT: root tip; S: stem; ST: stem tip; YL: young leaf; SL: senescent leaf; Expression pattern of NtNAC053 under ABA (B), cold (C), drought (D), and salt (E) treatments. The data were means ± SD from three independent replications. * p < 0.05, ** p < 0.01, *** p < 0.001 (t-tests).
Subcellular Localization and Transactivation Activity Assay
To further identify the function of NtNAC053, the subcellular localization was investigated. Generally, the coding sequence of NtNAC053 gene was fused with GFP and transiently expressed in the N. benthamiana leaves. As a result, the fluorescence signals of 35S:GFP control displayed as spreading throughout the cells (Figure 2). Meanwhile, the green fluorescent signal of the fused NtNAC053-GFP was only observed in the nucleus. This result illustrated that NtNAC053 protein was a nuclear-localized protein.
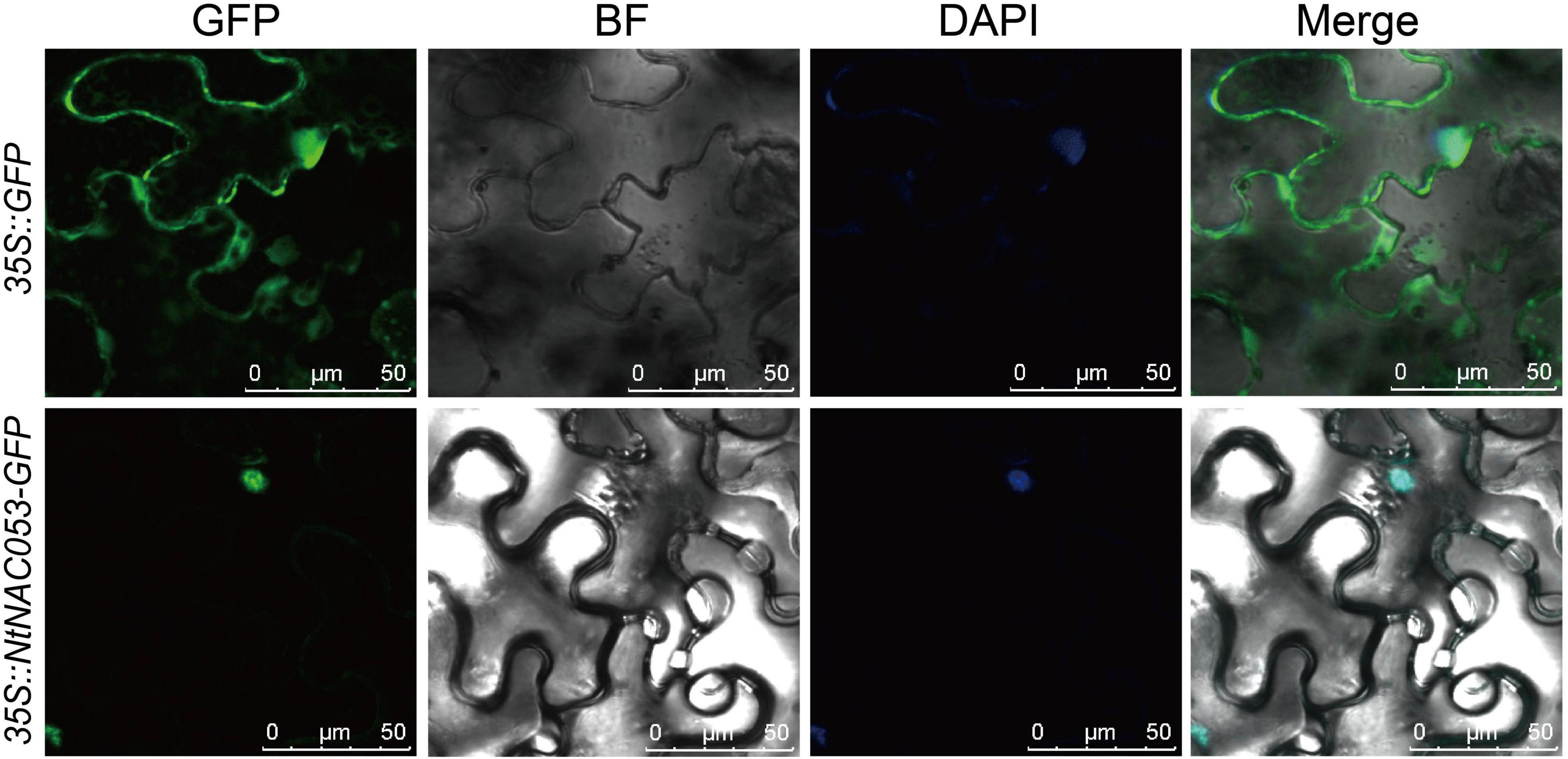
Figure 2. The subcellular localization of the NtNAC053 protein in Nicotiana benthamiana epidermal cells. Transient expression of GFP and NtNAC053-GFP in N. benthamiana epidermal cells. Bar = 50 μM, DAPI, 4,6-diamidino-2-phenylindole for nuclear staining.
To determine the transcriptional activity of NtNAC053, the full length of NtNAC053 and two truncated variants, including N-terminal region and C-terminal region, were inserted into the pBridge vector, respectively (Figure 3A). The pBridge empty vector and these three recombinant constructs were then transformed into yeast competent cells. The transformants containing the full length of NtNAC053 and C-terminal region of NtNAC053 were found to show β-galactosidase activity and turn blue on the SD/-Trp/X-gal medium. Whereas the transformants with pBridge empty vector and N-terminal region of NtNAC053 did not show β-galactosidase activity (Figure 3B). Therefore, NtNAC053 could function as a transcriptional activator, and the C-terminal region is critical for this activity.
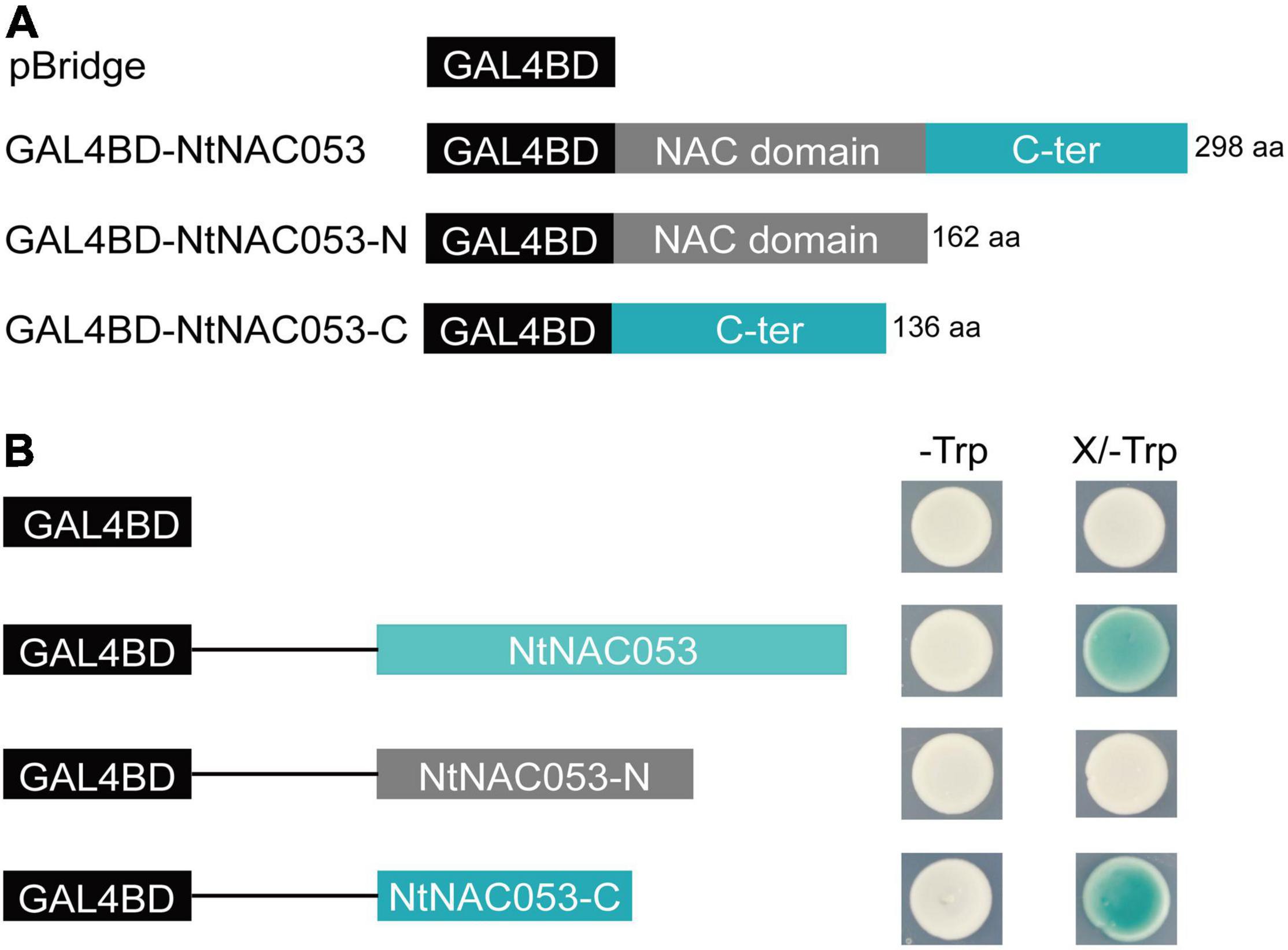
Figure 3. The analysis of transcriptional activation activity of NtNAC053. (A) The fusion proteins with the GAL4 DNA-binding domain. (B) Transcriptional activation activity assay of NtNAC053 in the yeast strain AH109. The β-galactosidase activity was detected on the SD/-Trp/X-gal medium. NtNAC053-N represented the N-terminal region of NtNAC053 (1-162 aa); NtNAC053-C represented the C-terminal region of NtNAC053 (163-298 aa).
The Overexpression of NtNAC053 in Tobacco Confers Abscisic Acid Hypersensitivity
To clarify the potential function of NtNAC053, the transgenic overexpression lines of NtNAC053 gene were generated in tobacco. The qRT-PCR analysis was used to detect the expression levels of NtNAC053 gene in these lines. The results revealed the expression levels of this gene in OE6 and OE8 reached 47.7- and 61.8-fold than that in wild type (WT) (Supplementary Figure 5). These two lines were then selected to perform phenotyping under different treatments. In our previous survey, the NtNAC053 gene could be induced by exogenous ABA. Hence, the germination assay and root elongation assay were carried out to assess the ABA sensitivity in the overexpressed lines. As a result, the germination rates seemed to be identical in the germination assay between the overexpressed lines and WT plants. Interestingly, the overexpressed lines had significantly lower germination rates than WT plants when exposed under ABA treatment (Figures 4A,B). Furthermore, the root length seemed to be equivalent in the root elongation assay between the overexpressed lines and WT plants. Notably, the overexpressed lines seemed to be more sensitive than WT plants when treated by exogenous ABA (Figures 4C,D).
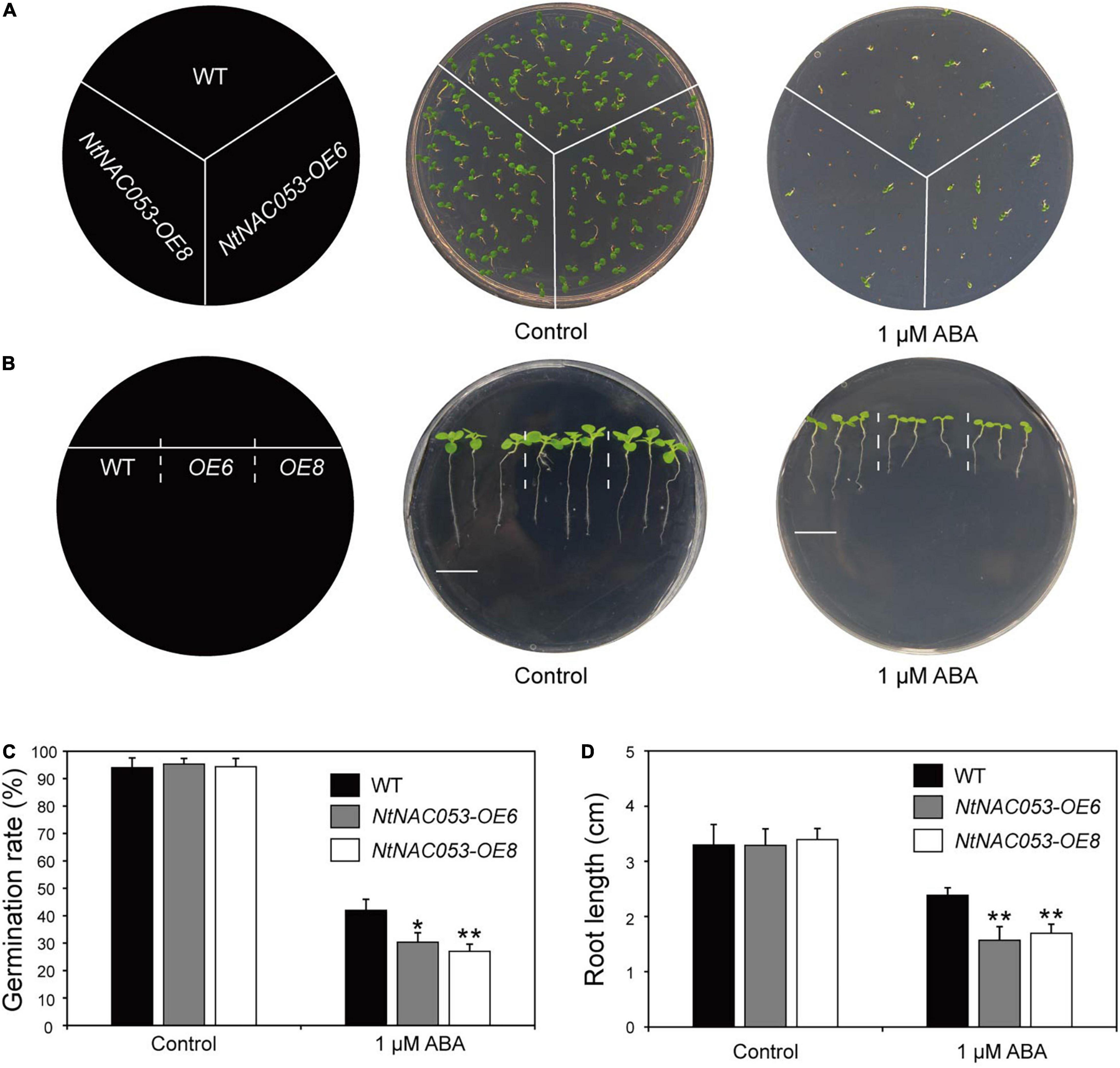
Figure 4. Overexpression of the NtNAC053 gene confers ABA hypersensitivity. (A) Seed germination assay between overexpression lines and wild type under 1 μM ABA treatment. (B) Root elongation assay between overexpression lines and wild type under 1 μM ABA treatment. Bar = 1.5 cm. (C) The statistics of germination rates under normal conditions and 1 μM ABA treatment. (D) The statistics of primary root length under normal conditions and 1 μM ABA treatment. WT, wild type. The data were means ± SD from three independent replications. * p < 0.05, ** p < 0.01 (t-tests).
NtNAC053 Overexpression Enhances Salt Tolerance in Transgenic Tobacco
To explore whether NtNAC053 regulates the tolerance of tobacco plants to salt stress, we performed a seed germination assay and root elongation assay on MS media with salt treatment. As shown in Figure 5, the seed germination rates of both the overexpression lines and WT were decreased under salt treatment, whereas the overexpression lines displayed higher germination rates than WT seeds under salt treatments.
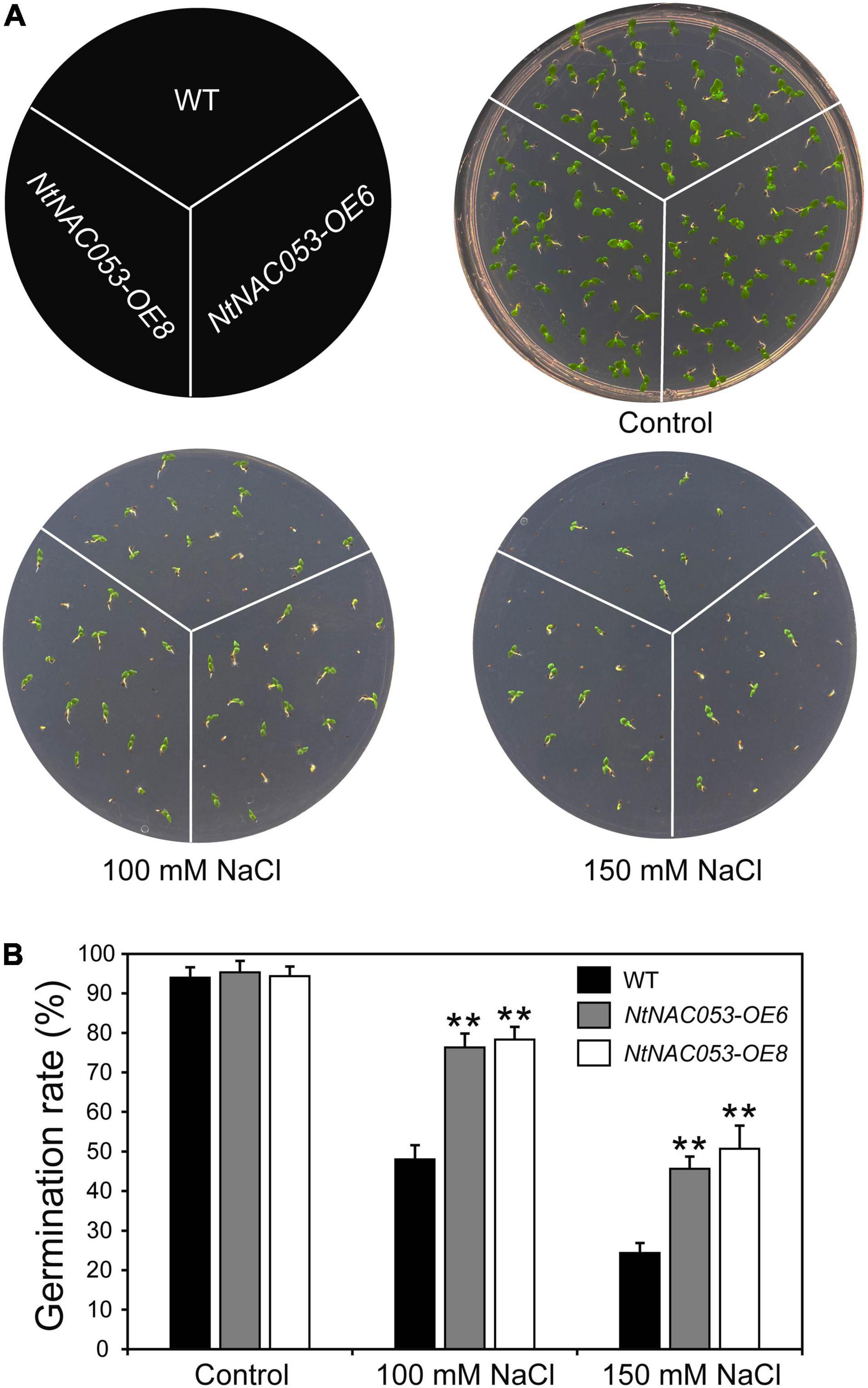
Figure 5. Overexpression of the NtNAC053 gene promotes seed germination under salt stress treatments. (A) Seed germination assay between overexpression lines and wild type under NaCl treatments. (B) The statistics of seed germination under normal conditions and NaCl treatments. WT, wild type. The data were means ± SD from three independent replications. ** p < 0.01 (t-tests).
As shown in Figure 6, no significant difference was observed in root length between overexpression lines and WT seedlings. However, the primary roots of overexpression seedlings were longer than that of WT on the MS media supplemented with tested concentrations of NaCl.
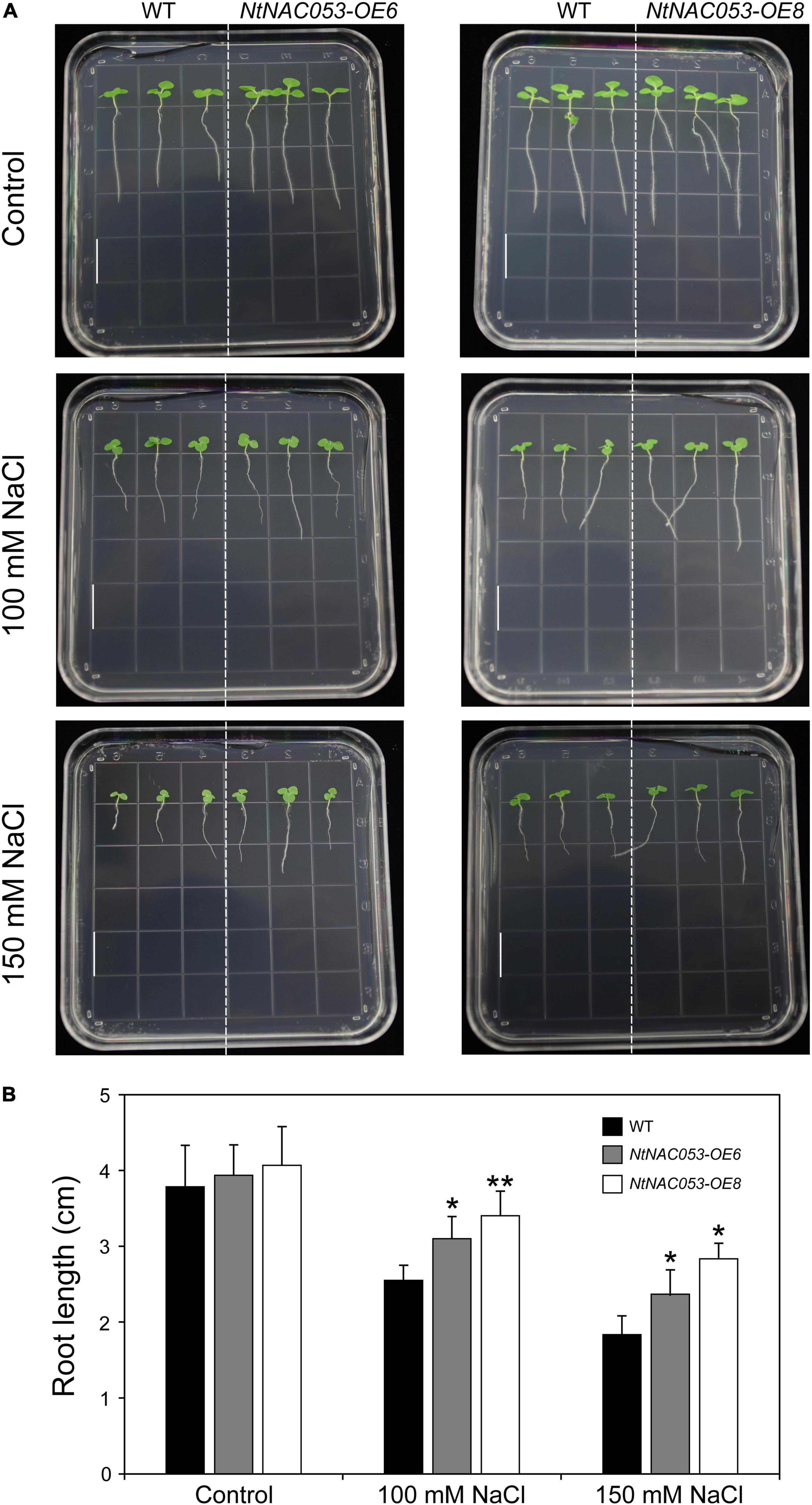
Figure 6. Comparison of root growth between overexpression lines and wild-type tobacco under salt stress treatments. (A) The primary root length of overexpression lines and wild-type tobacco under 100 mM and 150 mM NaCl treatments. The primary root length was recorded 7 days after growth. Bar = 1.5 cm. (B) The statistics of primary root length under normal conditions and NaCl treatments. WT, wild type. The data were means ± SD from three independent replications. * p < 0.05, ** p < 0.01 (t-tests).
NtNAC053 Overexpression Improves Drought Tolerance in Transgenic Tobacco
To further explore the function of NtNAC053 gene, the overexpression lines and WT seedlings were treated by the drought stress with no watering for approximately 10 days. The growth of overexpression lines and WT seedlings was almost the same under normal growth conditions. After the drought treatment, overexpression lines and WT seedlings displayed leaf wilting phenotype, whereas WT seedlings were much more extreme than that in overexpression lines (Figure 7A). After 3 days of re-watering, the survival rates of the OE6 lines (72%) and OE8 lines (85%) were significantly higher than those of the WT seedlings (35%) (Figure 7B). Additionally, the overexpression lines displayed higher water content than WT seedlings during the 3 h dehydration (Figure 7C). Besides, the drought stress-responsive gene NtAQP1 was selected to explore the expression in wild type and overexpression lines. Under drought conditions, the expression of NtAQP1 was significantly higher in overexpression lines (Supplementary Figure 6). These findings suggested that the overexpression of NtNAC053 gene might enhance the drought tolerance of the tobacco plants.
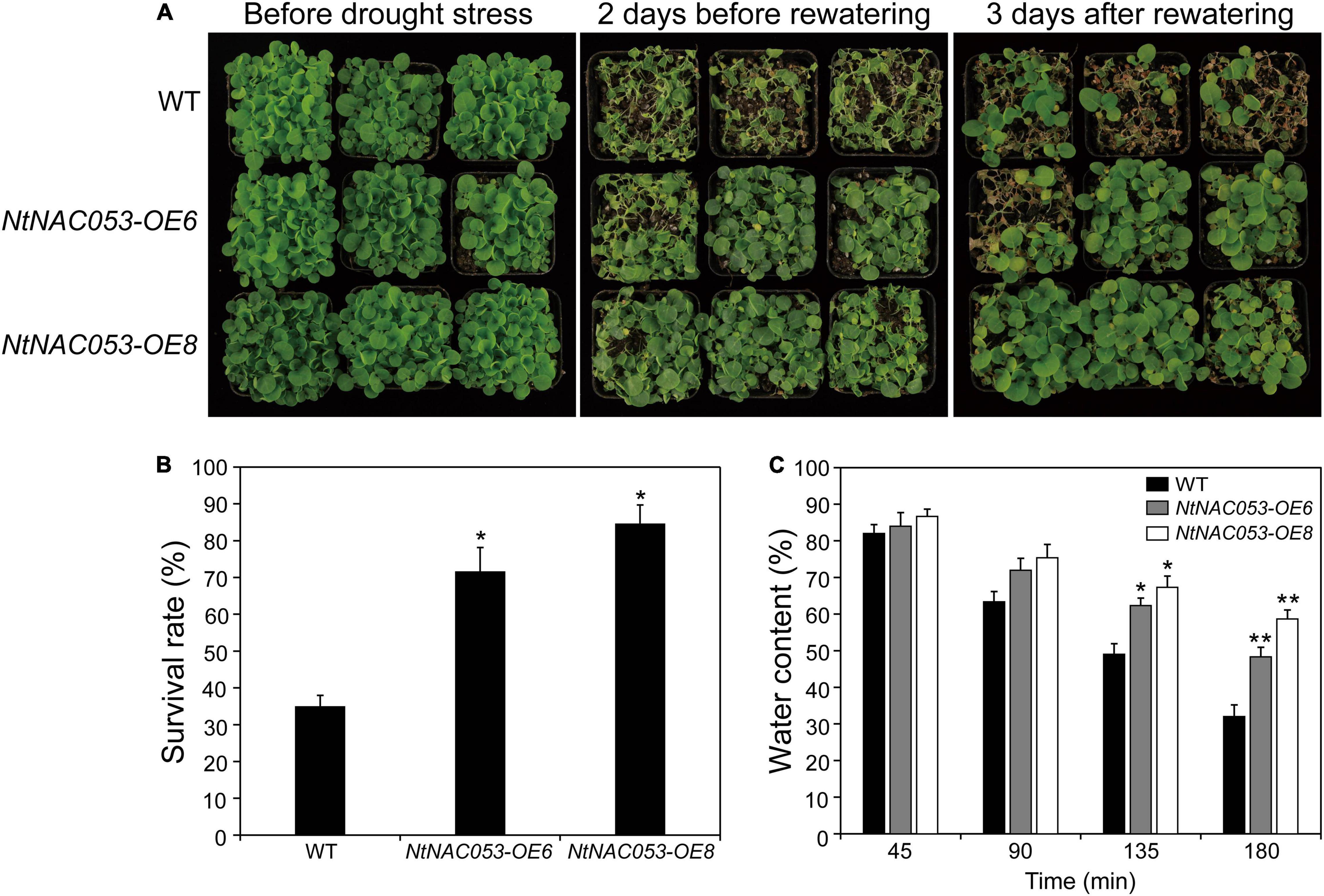
Figure 7. Drought tolerance assay of transgenic NtNAC053 tobacco. (A) Phenotypes of overexpression lines and wild-type tobacco under drought stress. Under normal conditions, the overexpression lines and wild-type tobacco were normally grown for 6 weeks. Overexpression lines displayed higher drought tolerance after 10 days of drought stress and 3 days of recovery. (B) The survival rate of the overexpression lines and wild-type tobacco were determined. (C) The water content of detached leaves of overexpression lines and wild-type tobacco. WT, wild type. The data were means ± SD from three independent replications. * p < 0.05, ** p < 0.01 (t-tests).
NtNAC053 Overexpression Enhances ROS-Scavenging Capability in Transgenic Tobacco
To further investigate the function of NtNAC053, the stress-related physiological parameters, including catalase (CAT), peroxidase (POD), superoxide (SOD), malonic dialdehyde (MDA), and hydrogen peroxide (H2O2), were detected (Figure 8 and Supplementary Figure 7). After salt and drought treatments, the content of MDA and H2O2 in overexpression lines was significantly lower than those in WT plants. However, the activities of CAT, POD, and SOD in overexpression lines were significantly higher than those in WT plants.
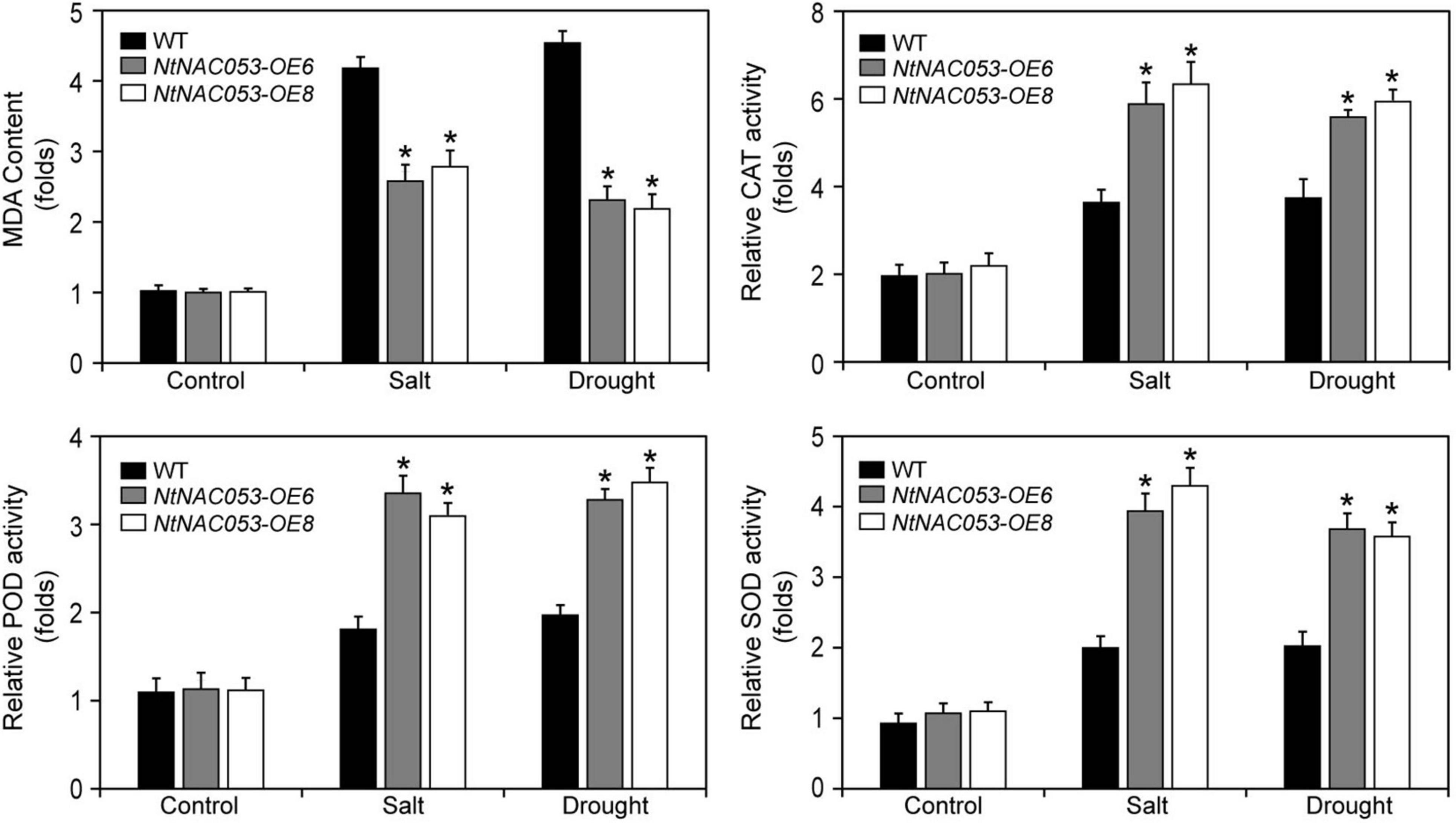
Figure 8. Analysis of the antioxidant enzyme activities and MDA contents in overexpression lines and wild-type tobacco under salt and drought stresses. MDA, malonic dialdehyde; CAT, catalase; POD, peroxidase; SOD; superoxide. WT, wild type. The MDA content and enzyme activity were calculated as folds relative to the level of before treatments. The data were means ± SD from three independent replications. * p < 0.05 (t-tests).
NtNAC053 Regulates the Expression of Stress-Responsive Genes
To further explore the function of NtNAC053 gene, the expression patterns of stress-responsive genes were detected in overexpression lines and WT plants using qRT-PCR. The expression pattern of the selected stress-responsive genes was calculated as folds relative to the expression level of none treatments. Exposed to salt and drought stresses, the expression levels of several genes were significantly induced in overexpression lines compared to WT plants, including NtCOR15A, NtRAB18, NtDREB1A, NtERF5, NtKAT2, and NtERD11 (Figure 9). Besides, we selected two salt stress-responsive genes (NtNHX1 and NtSOS1) and compared their expressions in overexpression lines and WT plants. Under salt conditions, the expressions of NtNHX1 and NtSOS1 were significantly higher in overexpression lines (Supplementary Figure 8). Notably, ABA signaling-related genes (NtPYL6 and NtSnRK2.4) were selected for further study. In overexpression lines, these gene expressions were much higher compared to WT plants under drought and salt conditions (Supplementary Figure 9).
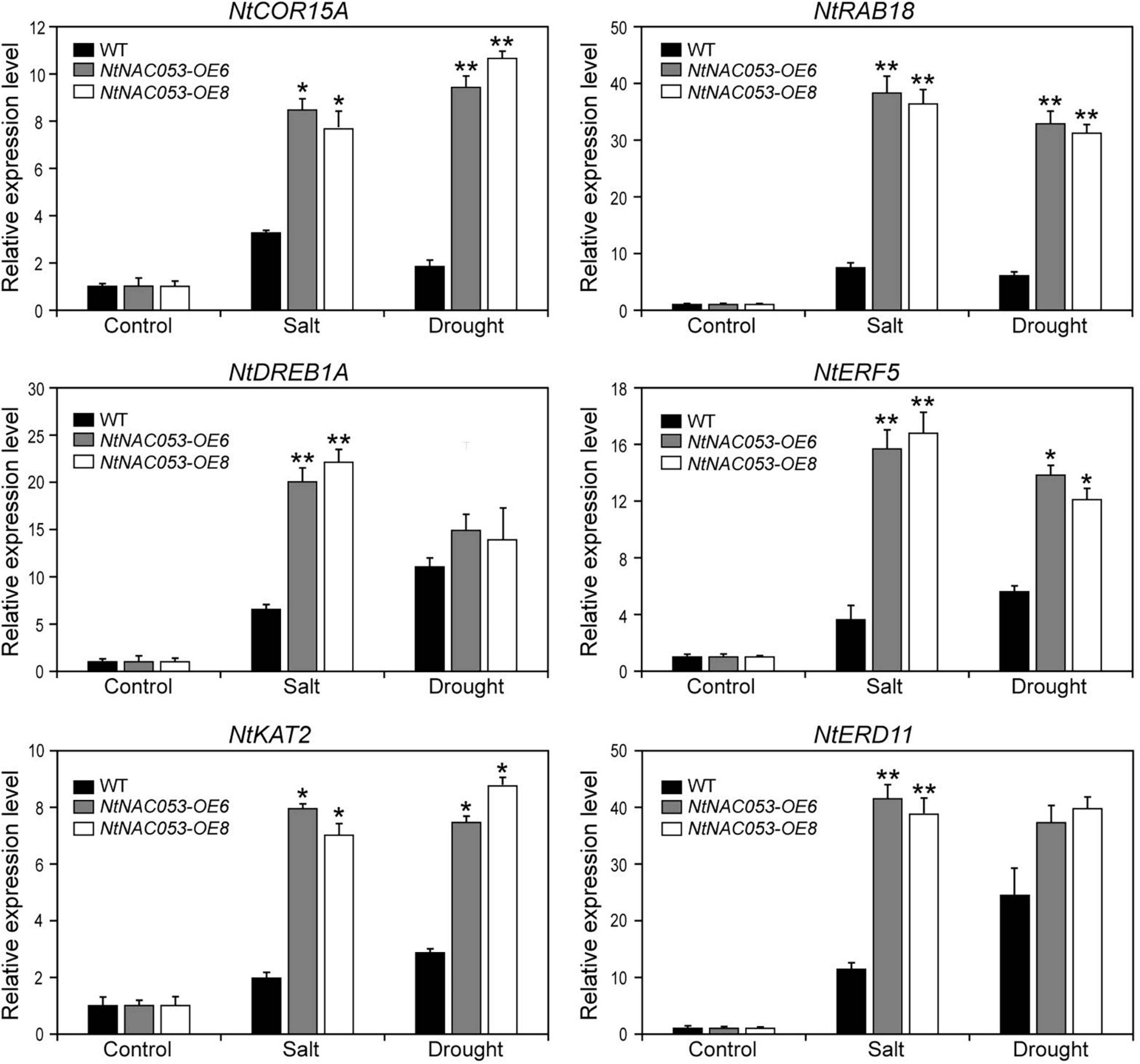
Figure 9. Expression of stress-related genes in overexpression lines and wild-type tobacco under salt and drought stresses. The ratios of gene expression levels were calculated relative to the control, respectively. WT, wild type. The data were means ± SD from three independent replications. * p < 0.05, ** p < 0.01 (t-tests).
NtNAC053 Binds to NACRS Sequence
The NAC transcription factors mainly bind to the NAC recognition sequence (NACRS) with a core sequence of CACG to regulate gene expression, while the reverse complementary sequence is CGT[A/G]. To verify the putative DNA-binding sequences of NtNAC053, the NACRS sequence and the reverse complementary sequence were used as reporters (R1 and R2), while the base-substituted fragment was used as the mutant reporter (RM). The activator vector pGAD-NtNAC053 was co-transformed into the yeast strain with three reporter vectors, respectively. If the NtNAC053 can bind to the tandem sequence of the reporter construct, the GAD will be able to direct LacZ expression, resulting in the blue color accumulation. As shown in Figure 10, these yeast one-hybrid experiments confirmed the ability of NtNAC053 to bind to the NACRS sequence in vivo.
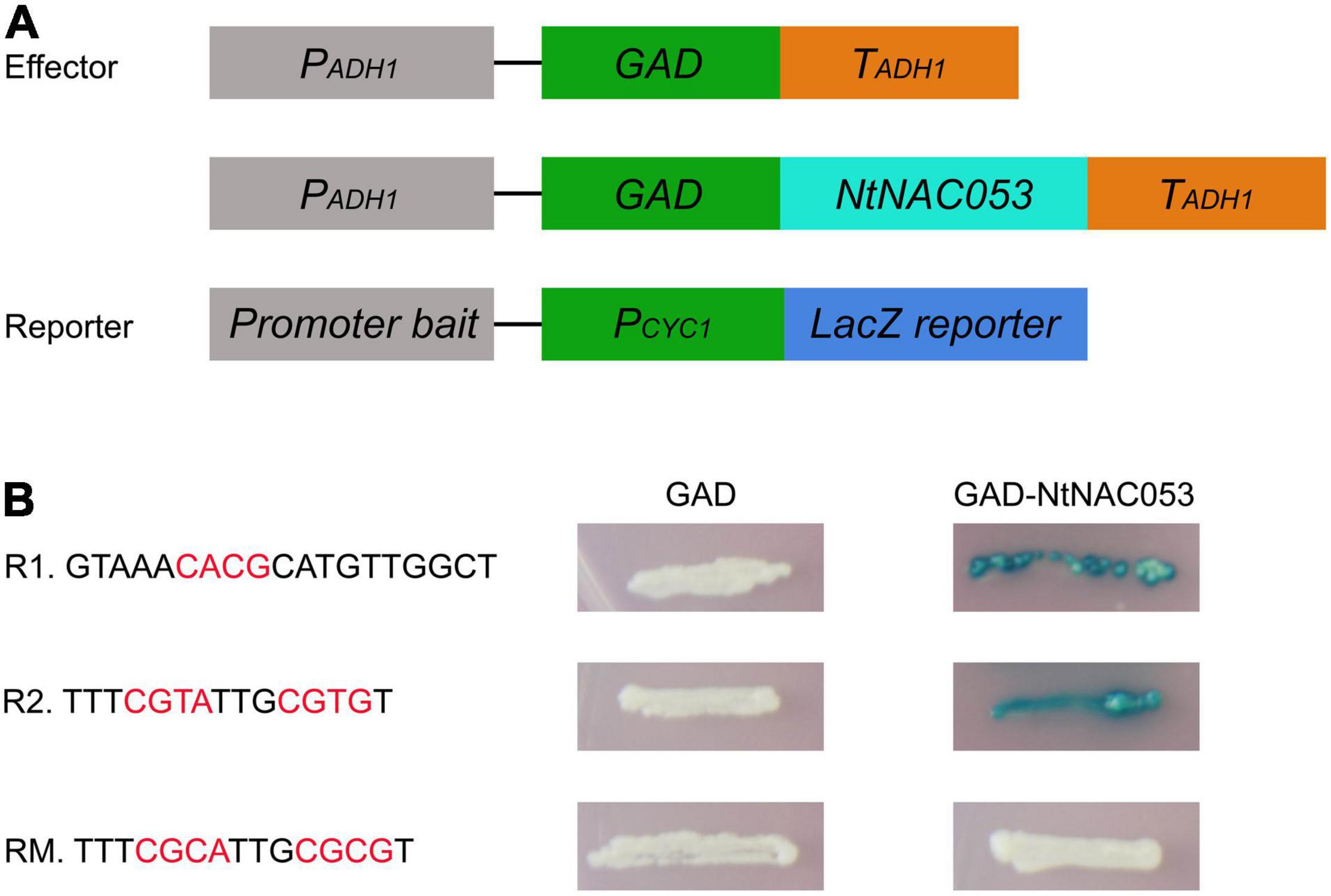
Figure 10. Binding activity of NtNAC053. (A) Diagrams illustrating the structure of reporter and effector constructs. (B) The sequence of the NACRS (R1 and R2) and mutated NACRS (RM). Different combinations of effector and reporter constructs were co-expressed in yeast strain YM4271.
Discussion
Plants are suffering from a combination of abiotic and biotic stresses. The abiotic stresses, including salt and drought, affect plant growth and development. In previous studies, several NAC members have been revealed to participate in responses to abiotic stress through ABA signaling (Nakashima et al., 2012; Shao et al., 2015). In the current study, promoter analysis showed that the NtNAC053 promoter possesses five ABRE motifs, which is an important cis-acting element related to ABA signal transduction, indicating this gene might be involved in the ABA signaling pathway (Supplementary Figure 4). To clarify this inference, the expression pattern analysis and overexpression survey were carried out. Interestingly, it was shown that NtNAC053 was significantly induced by exogenous ABA treatments (Figure 1B). Furthermore, the seed germination rate and root lengths of overexpression lines were significantly lower than those of WT plants, suggesting that overexpression of NtNAC053 conferred transgenic tobacco plants hypersensitive to ABA (Figure 4). Notably, the expressions of ABA signaling genes, including NtPYL6 and NtSnRK2.4, were much higher in overexpression lines compared to WT plants under salt and drought conditions (Supplementary Figure 9).
It has been reported that NAC transcription factors play critical roles in various abiotic stress responses. However, a few studies were reported about the biofunctions of the NAC transcription factor in tobacco. In our previous work, a tobacco NAC gene was found to be dramatically induced by multiple stress treatments (Li et al., 2018), which was identified as the homolog of StNAC053. In our previous study, we found that the overexpression of StNAC053 gene could enhance the salt and drought tolerance of transgenic Arabidopsis (Wang et al., 2021). Considering the close genetic distance of potato and tobacco, the homolog of StNAC053 might confer salt and drought tolerance in tobacco. To verify this hypothesis, a systematic analysis of NtNAC053 was performed. The StNAC053 had been reported to fall into the ATAF subfamily. Hence, several reported ATAF family members from potato, wheat, and Arabidopsis were selected to perform the phylogenetic analysis. Phylogenetic analysis showed that NtNAC053 was clustered with StNAC053 in ATAF subfamily, implying that NtNAC053 served as a homolog of StNAC053. In tobacco, we found that NtNAC053 was significantly up-regulated when subjected to salt and drought stresses, suggesting that NtNAC053 may function as a regulator in response to salt and drought stresses (Figure 1). Furthermore, the seed germination rate and root lengths of WT plants were significantly lower than those of overexpression lines under salt treatment (Figures 5, 6). Meanwhile, overexpression lines showed higher survival rates compared to WT plants when exposed to drought stress (Figures 7A,B). In addition, the leaves of overexpression lines had a lower water loss rate than those of WT plants under drought conditions (Figure 7C). Considering these results, the overexpression of NtNAC053 gene could enhance the tolerance to salt and drought stresses in tobacco.
Additionally, massive amounts of ROS were produced when plants were exposed to multiple stresses. Generally, excessive ROS could destroy antioxidant systems and the integrity of cell membrane, resulting in the accumulation of MDA (Moore and Roberts, 1998; Gill and Tuteja, 2010). Plants have developed intricate defense mechanisms to cope with excess ROS, which include an antioxidant enzyme system. Notably, several enzymes, including CAT, POD, and SOD, played significant roles in the antioxidant enzyme system (Gill and Tuteja, 2010). Relative to wild type, the contents of MDA and hydrogen peroxide in overexpression lines were significantly decreased when exposed to salt and drought stresses, suggesting that there is lower oxidative damage in overexpression lines (Figure 8 and Supplementary Figure 7). Furthermore, the enzyme activities of CAT, POD, and SOD in overexpression lines were significantly higher than those of wild type under salt and drought stresses, suggesting that overexpression lines increased the capacity of the ROS-scavenging system (Figure 8). It was reported that NAC transcription factors usually participate in response to abiotic stresses through regulating the expression of downstream stress-responsive genes (He et al., 2019b). AtNAP functions to negatively regulate salt stress tolerance through repressing ARBE1 (Seok et al., 2017). In this study, we found that NtNAC053 encoded a nuclear localization protein and possessed transcriptional activation ability (Figures 2, 3). Many aquaporins genes were reported to be responsive to abiotic stress during different developing stages. In tobacco, over-expression of the NtAQP1 gene improves water use efficiency, hydraulic conductivity, and yield production under abiotic stress (Sade et al., 2010). Notably, under drought stress treatments, the drought stress-responsive gene NtAQP1 was significantly higher in the NtNAC053 overexpression lines (Supplementary Figure 6). Furthermore, NtNAC053 could significantly activate several stress-responsive genes, including NtCOR15A, NtRAB18, NtDREB1A, NtERF5, NtKAT2, and NtERD11 (Figure 9). Besides, several ion channels encoding genes, including NHX1 and SOS1, have been reported to be involved in the ion homeostasis and salt stress responses. These genes were significantly higher in overexpression lines (Supplementary Figure 8). In addition, promoter analysis revealed that all these stress-responsive genes promoters possessed NAC-binding sites (Supplementary Table 2). Yeast one-hybrid assay implied that NtNAC053 might be combined with NACRS (Figure 10), suggesting NtNAC053 could activate these stress-responsive genes by binding their promoter directly.
Conclusion
In this study, a typical NAC gene NtNAC053 was cloned from tobacco K326 and functionally verified in overexpressing tobacco. NtNAC053 overexpression lines showed significantly increased resistance to salt and drought stresses, while the overexpression lines seemed to be more sensitive to ABA treatment. Taken together, the transcription factor NtNAC053 might be involved in salt and drought stress responses via ABA-mediated signaling in tobacco.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
XL, QW, and CG conducted the research and participated in drafting the manuscript. JS, ZL, YW, AY, WP, and YG assisted in data collection and analysis. JG and LW conceived this research, designed the experiments, and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Tobacco Genome Project [110202001029 (JY-12), and 110202101003 (JY-03)], the China Tobacco Hunan Industrial Co., Ltd., Science and Technology Project (KY2022YC0010 and KY2021YC0005), and the Key Science and Technology Program of Hunan Provincial Tobacco Corporation (Grant No. 19-23Aa01).
Conflict of Interest
XL, YW, WP, and JG were employed by China Tobacco Hunan Industrial Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.817106/full#supplementary-material
Supplementary Figure 1 | Multiple sequence alignment of NtNAC053 with reported NAC proteins from Arabidopsis and potato, including ANAC002, ANAC081, ANAC029, ANAC072, ANAC055, ANAC019, and StNAC053. The black lines indicated five subdomains (A–E) and nuclear location signal (NLS).
Supplementary Figure 2 | The phylogenetic, syntenic analysis and motif organization of NtNAC053. (A) The phylogenetic tree was generated based on the amino acid sequences of NtNAC053 and other reported NAC proteins. (B) The motif organizations were predicted by MEME. (C) The gray line in the background represents the collinear blocks between tobacco and potato, while the red line exhibits the syntenic NAC gene pairs.
Supplementary Figure 3 | The putative conserved motifs in NAC proteins.
Supplementary Figure 4 | The cis-acting element analysis of the promoter regions of NtNAC053 gene.
Supplementary Figure 5 | The expression level of the NtNAC053 gene in wild type and two overexpression lines, the ratios of gene expression levels were calculated relative to the wild type.
Supplementary Figure 6 | Expression of NtAQP1 in overexpression lines and wild-type tobacco under drought stress. The ratios of gene expression levels were calculated relative to the control, respectively.
Supplementary Figure 7 | Analysis of H2O2 contents in overexpression lines and wild-type tobacco under salt and drought stresses. H2O2, hydrogen peroxide. WT, wild type.
Supplementary Figure 8 | Expression of NtNHX1 and NtSOS1 in overexpression lines and wild-type tobacco under salt stress. The ratios of gene expression levels were calculated relative to the control, respectively. WT, wild type.
Supplementary Figure 9 | The expressions of ABA signaling genes in overexpression lines and wild-type tobacco under salt and drought stresses. The ratios of gene expression levels were calculated relative to the control, respectively. WT, wild type.
Footnotes
- ^ http://solgenomics.net/
- ^ https://web.expasy.org/protparam/
- ^ https://www.novopro.cn/tools/nls-signal-prediction.html
- ^ https://www.arabidopsis.org/
- ^ http://meme-suite.org/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Buschmann, H. (2016). Plant cell division analyzed by transient Agrobacterium-mediated transformation of tobacco bY-2 cells. Methods Mol. Biol. 1370, 17–25. doi: 10.1007/978-1-4939-3142-2_2
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Ernst, H. A., Olsen, A. N., Larsen, S., and Lo Leggio, L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303. doi: 10.1038/sj.embor.7400093
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Gong, X., Zhao, L., Song, X., Lin, Z., Gu, B., Yan, J., et al. (2019). Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri). BMC Plant Biol. 19:161. doi: 10.1186/s12870-019-1760-8
Guo, Y., and Gan, S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. doi: 10.1111/j.1365-313X.2006.02723.x
He, K., Zhao, X., Chi, X., Wang, Y., Jia, C., Zhang, H., et al. (2019a). A novel Miscanthus NAC transcription factor MlNAC10 enhances drought and salinity tolerance in transgenic Arabidopsis. J. Plant Physiol. 233, 84–93. doi: 10.1016/j.jplph.2019.01.001
He, L., Bian, J., Xu, J., and Yang, K. (2019b). Novel maize NAC transcriptional repressor ZmNAC071 confers enhanced sensitivity to ABA and osmotic stress by downregulating stress-responsive genes in transgenic Arabidopsis. J. Agric. Food Chem. 67, 8905–8918. doi: 10.1021/acs.jafc.9b02331
He, X. J., Mu, R. L., Cao, W. H., Zhang, Z. G., Zhang, J. S., and Chen, S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. doi: 10.1111/j.1365-313X.2005.02575.x
Hendelman, A., Stav, R., Zemach, H., and Arazi, T. (2013). The tomato NAC transcription factor SlNAM2 is involved in flower-boundary morphogenesis. J. Exp. Bot. 64, 5497–5507. doi: 10.1093/jxb/ert324
Hibara, K., Takada, S., and Tasaka, M. (2003). CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 36, 687–696. doi: 10.1046/j.1365-313x.2003.01911.x
Kim, M. J., Park, M. J., Seo, P. J., Song, J. S., Kim, H. J., and Park, C. M. (2012). Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 448, 353–363. doi: 10.1042/BJ20120244
Kim, S. G., Lee, S., Seo, P. J., Kim, S. K., Kim, J. K., and Park, C. M. (2010). Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95, 56–65. doi: 10.1016/j.ygeno.2009.09.003
Larsson, E., Sundstrom, J. F., Sitbon, F., and von Arnold, S. (2012). Expression of PaNAC01, a picea abies CUP-SHAPED COTYLEDON orthologue, is regulated by polar auxin transport and associated with differentiation of the shoot apical meristem and formation of separated cotyledons. Ann. Bot. 110, 923–934. doi: 10.1093/aob/mcs151
Li, P., Peng, Z., Xu, P., Tang, G., Ma, C., Zhu, J., et al. (2021). Genome-wide identification of NAC transcription factors and their functional prediction of abiotic stress response in peanut. Front. Genet. 12:630292. doi: 10.3389/fgene.2021.630292
Li, W., Li, X., Chao, J., Zhang, Z., Wang, W., and Guo, Y. (2018). NAC family transcription factors in tobacco and their potential role in regulating leaf senescence. Front. Plant Sci. 9:1900. doi: 10.3389/fpls.2018.01900
Li, Z., Chao, J., Li, X., Li, G., Song, D., Guo, Y., et al. (2021). Systematic analysis of the bZIP family in tobacco and functional characterization of NtbZIP62 involvement in salt stress. Agronomy 11:148. doi: 10.3390/agronomy11010148
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., et al. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270–280. doi: 10.1105/tpc.106.047043
Moore, K., and Roberts, L. J. (1998). Measurement of lipid peroxidation. Free Radic. Res. 28, 659–671. doi: 10.3109/10715769809065821
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nuruzzaman, M., Manimekalai, R., Sharoni, A. M., Satoh, K., Kondoh, H., Ooka, H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44. doi: 10.1016/j.gene.2010.06.008
Olsen, A. N., Ernst, H. A., Leggio, L. L., and Skriver, K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. doi: 10.1093/dnares/10.6.239
Park, J., Kim, Y. S., Kim, S. G., Jung, J. H., Woo, J. C., and Park, C. M. (2011). Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 156, 537–549. doi: 10.1104/pp.111.177071
Puranik, S., Bahadur, R. P., Srivastava, P. S., and Prasad, M. (2011). Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet [Setaria italica (L.) P. Beauv]. Mol. Biotechnol. 49, 138–150. doi: 10.1007/s12033-011-9385-7
Qu, L. J., and Zhu, Y. X. (2006). Transcription factor families in Arabidopsis: major progress and outstanding issues for future research. Curr. Opin. Plant Biol. 9, 544–549. doi: 10.1016/j.pbi.2006.07.005
Sade, N., Gebretsadik, M., Seligmann, R., Schwartz, A., Wallach, R., and Moshelion, M. (2010). The role of tobacco Aquaporin1 in improving water use efficiency, hydraulic conductivity, and yield production under salt stress. Plant Physiol. 152, 245–254. doi: 10.1104/pp.109.145854
Seok, H. Y., Woo, D. H., Nguyen, L. V., Tran, H. T., Tarte, V. N., Mehdi, S. M., et al. (2017). Arabidopsis AtNAP functions as a negative regulator via repression of AREB1 in salt stress response. Planta 245, 329–341. doi: 10.1007/s00425-016-2609-0
Shao, H., Wang, H., and Tang, X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6:902. doi: 10.3389/fpls.2015.00902
Shiriga, K., Sharma, R., Kumar, K., Yadav, S. K., Hossain, F., and Thirunavukkarasu, N. (2014). Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta. Gene 2, 407–417. doi: 10.1016/j.mgene.2014.05.001
Singh, A. K., Sharma, V., Pal, A. K., Acharya, V., and Ahuja, P. S. (2013). Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res. 20, 403–423. doi: 10.1093/dnares/dst019
Souer, E., van Houwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. doi: 10.1016/s0092-8674(00)81093-4
Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. doi: 10.1007/s004290100164
Takasaki, H., Maruyama, K., Kidokoro, S., Ito, Y., Fujita, Y., Shinozaki, K., et al. (2010). The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 284, 173–183. doi: 10.1007/s00438-010-0557-0
Tang, Y., Liu, M., Gao, S., Zhang, Z., Zhao, X., Zhao, C., et al. (2012). Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol. Plant 144, 210–224. doi: 10.1111/j.1399-3054.2011.01539.x
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Wang, Q., Guo, C., Li, Z., Sun, J., Deng, Z., Wen, L., et al. (2021). Potato NAC transcription factor StNAC053 enhances salt and drought tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 22:2568. doi: 10.3390/ijms22052568
Wu, Y., Deng, Z., Lai, J., Zhang, Y., Yang, C., Yin, B., et al. (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19, 1279–1290. doi: 10.1038/cr.2009.108
Xie, Q., Frugis, G., Colgan, D., and Chua, N. H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. doi: 10.1101/gad.852200
Yamaguchi, M., Ohtani, M., Mitsuda, N., Kubo, M., Ohme-Takagi, M., Fukuda, H., et al. (2010). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22, 1249–1263. doi: 10.1105/tpc.108.064048
Yoo, S. Y., Kim, Y., Kim, S. Y., Lee, J. S., and Ahn, J. H. (2007). Control of flowering time and cold response by a NAC-domain protein in Arabidopsis. PLoS One 2:e642. doi: 10.1371/journal.pone.0000642
Yuan, X., Wang, H., Cai, J., Bi, Y., Li, D., and Song, F. (2019). Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 19:278. doi: 10.1186/s12870-019-1883-y
Zhang, L., Yao, L., Zhang, N., Yang, J., Zhu, X., Tang, X., et al. (2018). Lateral root development in potato is mediated by stu-mi164 regulation of NAC transcription factor. Front. Plant Sci. 9:383. doi: 10.3389/fpls.2018.00383
Zhang, L., Zhang, L., Xia, C., Zhao, G., Jia, J., and Kong, X. (2015). The novel wheat transcription factor TaNAC47 enhances multiple abiotic stress tolerances in transgenic plants. Front. Plant Sci. 6:1174. doi: 10.3389/fpls.2015.01174
Zhao, J., Liu, J. S., Meng, F. N., Zhang, Z. Z., Long, H., Lin, W. H., et al. (2016). ANAC005 is a membrane-associated transcription factor and regulates vascular development in Arabidopsis. J. Integr. Plant Biol. 58, 442–451. doi: 10.1111/jipb.12379
Keywords: tobacco, NtNAC053, ABA, abiotic stress, antioxidant system
Citation: Li X, Wang Q, Guo C, Sun J, Li Z, Wang Y, Yang A, Pu W, Guo Y, Gao J and Wen L (2022) NtNAC053, A Novel NAC Transcription Factor, Confers Drought and Salt Tolerances in Tobacco. Front. Plant Sci. 13:817106. doi: 10.3389/fpls.2022.817106
Received: 17 November 2021; Accepted: 21 March 2022;
Published: 04 May 2022.
Edited by:
Mukesh Jain, Jawaharlal Nehru University, IndiaReviewed by:
X. Deng, Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences, ChinaZhan Qi Wang, Huzhou University, China
Copyright © 2022 Li, Wang, Guo, Sun, Li, Wang, Yang, Pu, Guo, Gao and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junping Gao, gaojp0104@hngytobacco.com; Liuying Wen, wenliuying@caas.cn
†These authors have contributed equally to this work
 Xiaoxu Li
Xiaoxu Li Qi Wang
Qi Wang Cun Guo
Cun Guo Jinhao Sun
Jinhao Sun Zhiyuan Li
Zhiyuan Li Yaofu Wang1
Yaofu Wang1 Aiguo Yang
Aiguo Yang Yongfeng Guo
Yongfeng Guo Liuying Wen
Liuying Wen