- 1College of Life Science, Hunan Normal University, Changsha, China
- 2Hunan Provincial Key Laboratory for Microbial Molecular Biology, Changsha, China
- 3The National & Local Joint Engineering Laboratory of Animal Peptide Drug Development, Changsha, China
- 4Hunan Academy of Agricultural Sciences, Changsha, China
MicroRNAs (miRNAs), a group of small noncoding RNAs (approximately 20-24 nucleotides), act as essential regulators affecting endogenous gene expression in plants. MiR482/2118 is a unique miRNA superfamily in plants and represses NUCLEOTIDE BINDING SITE-LEUCINE-RICH REPEAT (NBS-LRR) genes to function in plant resistance to pathogens. In addition, over the past several years, it has been found that miR482/2118 not only targets NBS-LRRs but also acts on other molecular mechanisms to affect plant resistance. miR482/2118-5ps, phased small interfering RNAs (phasiRNAs) and long noncoding RNAs (lncRNAs) play important roles in plant disease resistance. This review summarizes the current knowledge of the interactions and links between miR482/2118 and its new interacting molecules, miR482/2118-5p, phasiRNAs and lncRNAs, in plant disease resistance. Here, we aim to provide a comprehensive view describing the new molecular mechanism associated with miR482/2118 in the plant immune system.
Introduction
MicroRNAs (miRNAs), approximately 22 nucleotides (nt) of endogenous small noncoding RNA, have been discovered to act as master regulators affecting endogenous gene expression in plants (Reinhart et al., 2002; Rhoades et al., 2002). In plant miRNAs, miR482/2118 is a unique miRNA superfamily consisting of two mature miRNA isoforms, miR482 and miR2118 (both 22 nt in length) (Shen et al., 2020). For miR482/2118 biogenesis, first, MIR482/2118 genes are transcribed by RNA polymerase II (Pol II) into long primary miR482/2118 (pri-miR482/2118). pri-miR482/2118 are polyadenylated and stranded RNA molecules that fold into hairpin-like structures and produce precursors of miR482/2118 (pre-miR482/2118) by the RNase III family enzyme DICER-LIKE1 (DCL1). By DCL1, HYPONASTIC LEAVES 1 (HYL1) and Serrate (SE), pre-miR482/2118 is then processed into a miR482/2118 duplex consisting of miR482/2118 and complementary miR482/2118*. MiR482/2118 strand is called the guide strand, and miR482/2118* strand is called the passenger strand. The liberated strands have also been defined as miR482/2118-3ps and miR482/2118-5ps, according to the 5’ and 3’arms of the hairpin precursor, after renaming by the miRBase registry. Once liberated from the duplex, mature miR482/2118-3p or miR482/2118-5p is commonly incorporated into specific ARGONAUTE (AGO)-associated RNA-induced silencing complexes (RISCs) and guides RISCs to their targets to mediate gene silencing (Xiao and Luan, 2014; Liu et al., 2017; Zhang et al., 2022) (Figure 1A, B).
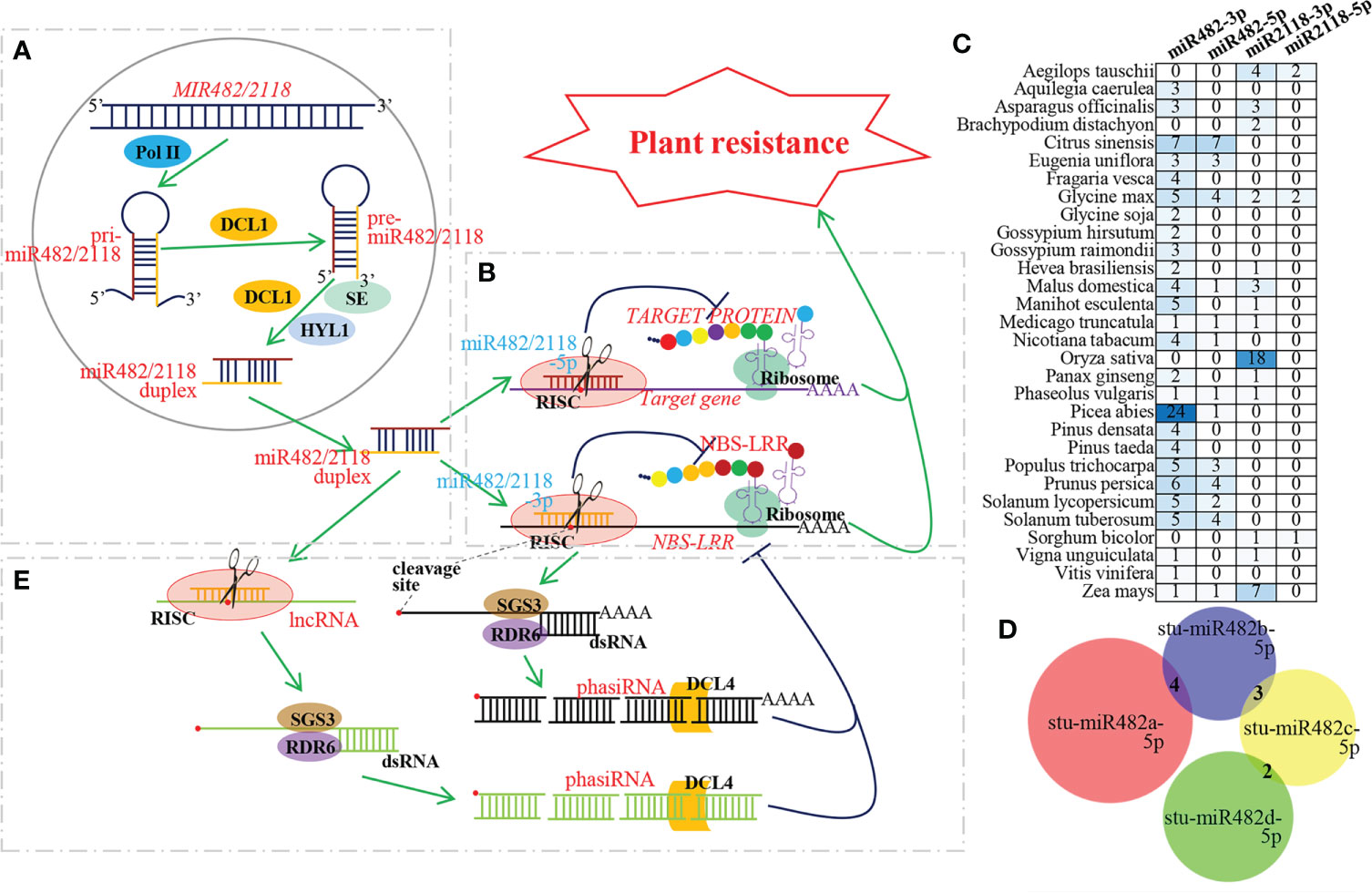
Figure 1 Biogenesis and mechanism of miR481/2118 superfamily during plant resistance to pathogen. (A) Biogenesis pathway of miR482/2118. MIR482/2118 genes are transcribed by Pol II into pri-miR482/2118. Then, pri-miR482/2118 of hairpin-like structure produces pre-miR482/2118 by DCL1. By DCL1, HYL1 and SE, pre-miR482/2118 is then processed into a miR482/2118-3p/miR482/2118-5p duplex. The mature miR482/2118-3p or miR482/2118-5p liberated from the duplex is commonly incorporated into RISCs to their targets to mediate gene silencing. (B) MiR482/2118-3p and -5p co-regulating plant resistance. MiR482/2118-3p and -5p cleave their target genes to inhibit translation of resistant protein, thus, regulating plant resistance. (C) Quantity variation of miR482/2118-3p and -5p in different plant species. MiRNA resources are downloaded from miRBase database (https://www.mirbase.org/). The numbers in the heat map represent the number of miRNAs. (D) Number of the identical target genes among Solanum tuberosum miR482s (stu-miR482). The target genes are predicted by psRNAtarget (https://www.zhaolab.org/psRNATarget/) with Expectation ≤ 4. Venn diagram is performed by DeepVenn (http://www.deepvenn.com/). (E) PhasiRNA biogenesis and function during plant resistance to pathogen. 3’ fragment of cleaved target gene or lncRNA are converted by RDR6 and SGS3 into double-stranded RNAs, and then they are processed into phasiRNAs through continuous DCL4 chopping.
The plant immune system, a ‘zig-zag-zig’ model, is composed of two layers of defense responses that provide protection against pathogens, including pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones and Dangl, 2006; Fei et al., 2016). PTI is the first layer of defense. Membrane-localized pattern recognition receptors (PRRs) function in the recognition of PAMPs to cause plant immune responses, such as stomatal closure and a burst of reactive oxygen species. ETI acts more strongly in its amplitude of defense. ETI is activated by nucleotide-binding domain leucine-rich repeat containing receptors (NLRs) recognizing effectors secreted by pathogens, leading to hypersensitive response (HR). PTI and ETI do not function independently but interdependent and mutually reinforcing. The production of reactive oxygen species by the NADPH oxidase RBOHD is a critical early signalling event connecting PTI and ETI, and the potentiation of PTI is an indispensable component of ETI during pathogen infection (Yuan et al., 2021). moreover, in Oryza sativa, the deubiquitinase PICI1 is identified as an immunity hub for PTI and ETI. PICI1 is targeted for degradation by blast fungal effectors to dampen PTI. NLRs protect PICI1 from effector-mediated degradation to reboot the methionine-ethylene cascade (Zhai et al., 2022). Nucleotide-binding site leucine-rich repeat (NBS-LRR) resistance proteins are important members of NLR family. MiR482/2118 members target conserved sequences encoding the P-loop of NBS-LRR genes, thus inhibiting the expression of NBS-LRR genes (Jiang et al., 2018b). This suggests that the miR482/2118-NBS-LRR module is involved in the ETI.
Many previous studies have shown that the miR482/2118-NBS-LRR module acts in the resistance of various plants to pathogens. For example, the increased susceptibility of stu-miR482e-overexpressing potato plants to Verticillium dahliae infection can be explained by the enhancement of stu-miR482e-mediated silencing of NBS-LRR disease-resistance genes (Yang et al., 2015). During infection with Phytophthora infestans and Botrytis cinerea, sly-miR482b inhibits the expression of its target genes, NBS-LRRs, and transgenic tomato and Arabidopsis overexpressing sly-miR482b show decreased resistance (Jiang et al., 2018a; Wu et al., 2021). In addition, over the past several years, miRNA-5p, phased small interfering RNAs (phasiRNAs) and long noncoding RNAs (lncRNAs) have been involved in plant resistance to pathogens (Xia et al., 2015; Cui et al., 2017; Liu et al., 2017). pre-miR482/2118 can produce miR482/2118-5p; miR482/2118s target the transcripts to trigger phasiRNA production; lncRNAs inhibit miR482/2188s expression, and miR482/2188s target lncRNAs (Canto-pastor et al., 2019; Jiang et al., 2019; Jiang et al., 2020; Liu et al., 2022a). Here, we aim to provide a comprehensive view describing the possible integration of miR482/2118-5p, phasiRNAs and lncRNAs into the plant immune system associated with miR482/2118.
MiR482/2118-3p and -5p co-regulateplant resistance
MiRNA-5p was originally thought to be a nonfunctional and degradable byproduct formed during miRNA biogenesis, since the accumulation of miRNA-5p is much lower than that of miRNA-3p in plants (Liu et al., 2017). However, an increasing number of studies have demonstrated that miRNA-5p, as a regulatory factor, plays important roles in a variety of other biological processes, including plant resistance to pathogens (Zhang et al., 2011; Nie et al., 2019).
The miRBase database (https://www.mirbase.org/) contains 191 members of the miR482/2118 superfamily from 30 plant species, including 153 miR482/2118-3ps and 38 miR482/2118-5ps (Figure 1C). Previous studies have shown that the target genes of miR482/2118-3ps are mainly members of the NBS-LRR family (Jiang et al., 2018b). In contrast, the target genes of miR482/2118-5p superfamily members vary due to their short conserved sequence. There are few identical target genes among members of the same genus. For example, Solanum tuberosum contains four members of the stu-miR482/2118-5p superfamily, stu-miR482a/b/c/d-5p. There were four identical target genes between stu-miR482a-5p and stu-miR482b-5p, three between stu-miR482b-5p and stu-miR482c-5p, and two between stu-miR482c-5p and stu-miR482d-5p, but there were no identical target genes among all four members due to the diversification of stu-miR482 family and the diversification of stu-miR482 family and the nucleotide diversity of the P-loop motif of NBS-LRRs, especially the wobble position of the codons in the target site of miR482/2118 (Zhang et al., 2022) (Figure 1D).
Tomato miR482d-3p/-5p and miR482e-3p/-5p can respond to pathogen infection. Sly-miR482e-5p is significantly downregulated in both Moneymaker tomato (susceptible cultivar to Fusarium oxysporum) and Motelle tomato (resistant cultivar to F. oxysporum) upon F. oxysporum infection. Levels of sly-miR482e-3p and sly-miR482d-3p are suppressed in Motelle but increased in Moneymaker after F. oxysporum treatment. However, sly-miR482d-5p presents the opposite pattern, showing decreased levels in Moneymaker and increased amounts in Motelle after infection with F. oxysporum (Ji et al., 2018). After infection with P. infestans, both sly-miR482e-3p and -5p are downregulated in Zaofen No. 2 tomato, but the expression level of sly-miR482e-5p is even higher than that of sly-miR482e-3p. Transgenic plants that overexpressed sly-miR482e-3p or -5p show susceptibility to the pathogen, lower expression levels of PR1 and PR5 genes, more number of necrotic cells and the reactive oxygen species (ROS) accumulation, while these results are reversed after miR482e-3p or miR482e-5p silencing. These results suggest that sly-miR482e-3p and -5p induces PR gene expression and reduces the ROS accumulation to protect against cell membrane injury, leading to enhanced resistance to P. infestans. (Liu et al., 2022a; Liu et al., 2022b).
These results suggest that miR482/2118-3p and -5p coregulate plant resistance.
MiR482/2118 mediates phasiRNA generation
In addition to targeting genes, it has been demonstrated that some miRNA-mediated cleavages of transcripts can trigger the production of phasiRNA (Gai et al., 2018; Bélanger et al., 2020; López-Márquez et al., 2021; Shi et al., 2022). PhasiRNAs, another major class of small RNAs in plants, are involved in the control of plant biological processes (Guo et al., 2018). The biogenesis of phasiRNAs occurs after cleavage of the targets by miRNA. After cleavage, the 3’fragment is converted to dsRNA via the activity of RNA-DEPENDENT RNA POLYMERASE6 (RDR6), assisting by SUPPRESSOR OF GENE SILENCING3 (SGS3). The resulting dsRNA is iteratively cleaved by a member of Dicer protein family, such as DCL4, from the 5’ end of strand containing the cleavage site, yielding duplexes of phasiRNAs (Liu et al., 2020; Zhang et al., 2022). These phasiRNAs can, in turn, cis-cleave their precursor or trans-regulate other target genes simultaneously (Liu et al., 2020) (Figure 1E). There are two pathways for the biogenesis of phasiRNAs, named “one-hit” and “two-hit” pathways. The one-hit pathway is typified by a single target site for a 22-nucleotide miRNA that results in downstream processing of the target transcript into 21-nucleotide phasiRNAs, while the two-hit pathway is typified by two target sites of a 21-nucleotide miRNA that results in processing upstream of the 3’ site. (Fei et al., 2013; Liu et al., 2020).
In eudicots, miR482/2118 superfamily members can target a number of NBS-LRRs and trigger phasiRNA production, and the resulting phasiRNAs can play important roles in enhancing the silencing effects of miR482/2118 on NBS-LRRs (Canto-Pastor et al., 2019; Shivaprasad et al., 2012; Zhai et al., 2011). As observed in several eudicots, NBS-LRR genes comprise the largest class of genes producing phasiRNAs (PHAS genes) in spruce, and miR482/miR2118, encoded in spruce by at least 24 precursor loci, targets NBS-LRR genes to trigger phasiRNA production (Xia et al., 2015) (Figure 1E).
In addition, miR482/2118 superfamily members can cleave lncRNAs to trigger the generation of phasiRNAs. Spruce miR482/2118 targets noncoding PHAS loci to trigger phasiRNA production, with the latter enriched in reproductive tissues (Xia et al., 2015). Similarly, in litchi, miR482 directs phasiRNA generation from long noncoding genes via alternative splicing ions from long noncoding genes via alternative splicing (AS) and alternative polyadenylation (APA) (Ma et al., 2018). A lncRNA formed by a rearrangement of several CNLs and TNLs, named TAS5, was identified in tomato. TAS5 is targeted by miR2118b, triggering the generation of phasiRNAs that act in trans to regulate multiple NBS-LRRs (Canto-Pastor et al., 2019; Zhang et al., 2022) (Figure 1E).
These results suggest that phasiRNAs triggered by members of the miR482/2118 superfamily may act in regulating plant resistance by affecting NBS-LRR levels.
CeRNAs inhibits MIR482/2118 expression
Competitive endogenous RNAs (ceRNAs) were first proposed by Salmena (Salmena et al., 2011). Many research papers suggest that lncRNAs can act as ceRNAs through competitive binding of miRNAs, releasing or attenuating repression by sequestering miRNAs away from target mRNAs. The binding sites of miRNAs among these lncRNAs are called endogenous target mimics (eTMs). Previous studies have shown that lncRNAs act in the ‘zig-zag-zig’ model of the plant immune system by decoying miR482/2118s, enhancing target gene NBS-LRR levels (an essential component of ETI) (Jiang et al., 2019; Liu et al., 2022b).
Jiang and her colleagues provide the predicted rules of ceRNAs and identified many lncRNAs as ceRNAs that play important roles in tomato resistance to P. infestans (Jiang et al., 2019; Cui et al., 2020). Three tomato lncRNAs, lncRNA23468, lncRNA01308 and lncRNA13262, contain conserved eTM sites for sly-miR482b. When lncRNA23468 is overexpressed in tomato, sly-miR482b expression is significantly decreased, and the expression of the NBS-LRR target genes is significantly increased, resulting in enhanced resistance to P. infestans. Silencing lncRNA23468 in tomato leads to the increased accumulation of sly-miR482b and decreased accumulation of NBS-LRRs, as well as reduced resistance to P. infestans (Jiang et al., 2019) (Figure 2A). Meanwhile, tomato lncRNA08489 acts as a ceRNA of sly-miR482e-3p to suppress sly-miR482e-3p expression and increase the expression level of its target gene NBS-LRR (Liu et al., 2022b) (Figure 2). Four lncRNAs (MSTRG.2115, MSTRG.30601, MSTRG.30599 and MSTRG.31962) respond to root−knot nematodes by acting as a decoy of a member of the miR482/2118 superfamily, gma-MIR482c-p5_2ss12GA19CT, in peanut (Xu et al., 2022) (Figure 2B).
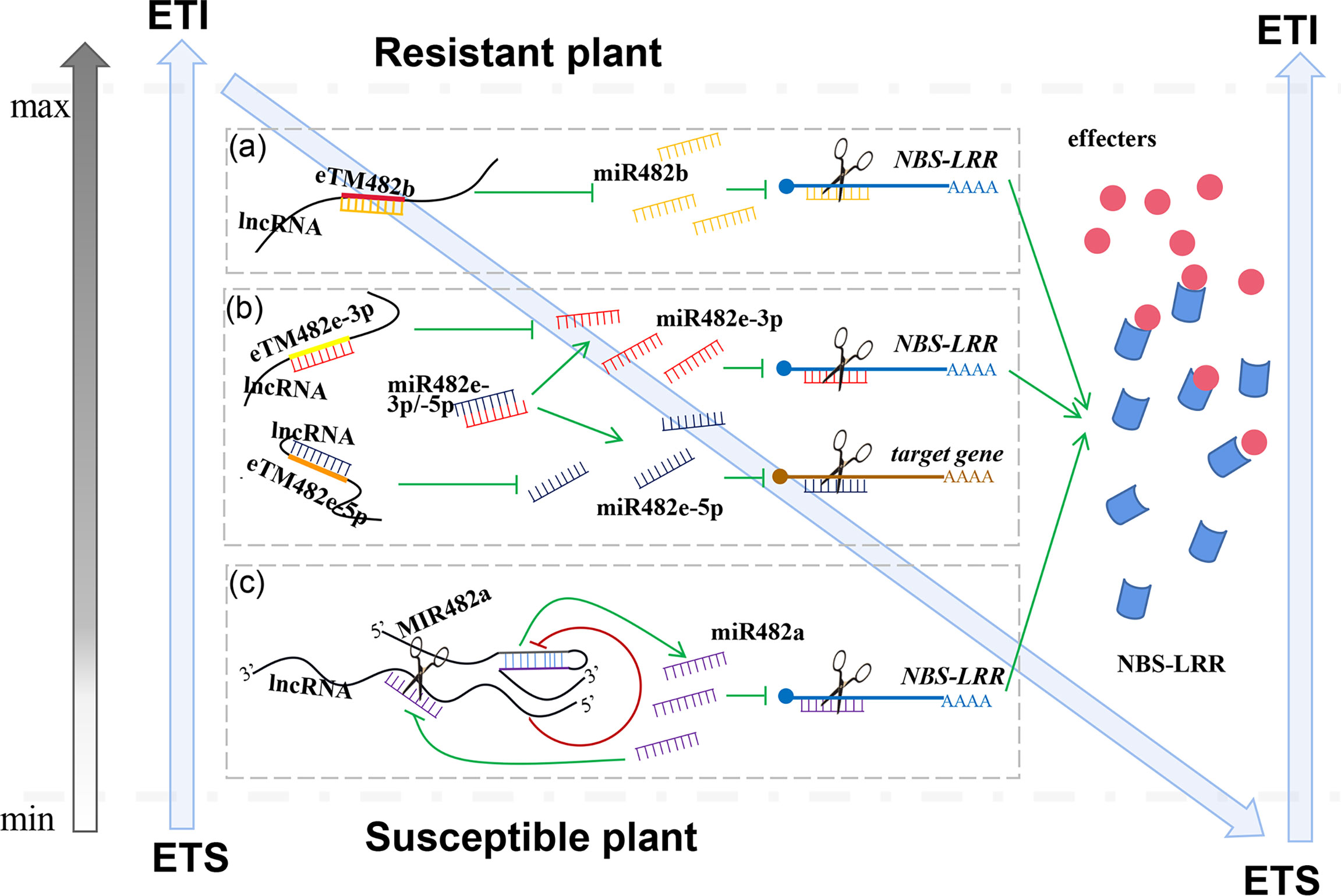
Figure 2 miR482/2118 and lncRNAs are important components in the effector-triggered immunity (ETI) for the plant immune response. In this extended model, lncRNAs function as ceRNAs to modulate resistant genes by decoying miR482b-3p (A), miR482e-3p and -5p (B) in ETI for the plant innate immune response. Other lncRNAs as NATs suppress the levels of pre-miR482a to affect the expression of mature miR482a, which leads to accumulation of resistant genes, NBS-LRRs is increased (C). When excessive amounts of resistant genes accumulate, mature miR482a cleaves NATs to relieve the suppression of pre-miR482a, leading to an increase in accumulation of mature miR482a and decreased accumulation of resistant genes, thus maintaining resistant genes homeostasis.
In addition to silencing miR482-3p as a “sponge”, lncRNAs also repress miR482-5p expression. Due to the eTM site for sly-miR482e-5p in lncRNA39298, lncRNA39298 decoys sly-miR482e-5p to inhibit its expression, resulting in increased plant resistance to P. infestans (Liu et al., 2022a) (Figure 2B).
Taken together, these findings suggest that lncRNAs act in the plant immune system by decoying miR482/2118, regulating plant resistance.
MIR482/2118 gene expression affected by lncRNA
Another interaction mechanism between lncRNAs and miR482/2118 was also found. Tomato MIR482a was determined to be located on chromosome 3 in the tomato genome. After further analysis of this sequence, it was found that lncRNA15492 is located on the antisense sequence of MIR482a as a natural antisense transcript (NAT). Gain- and loss-of-function experiments revealed that lncRNA15492 suppressed the expression of MIR482a to regulate mature miR482a levels. Thus, once the expression of mature sly-miR482a was suppressed, the accumulation of NBS-LRR was increased, and tomato resistance was also enhanced. Interestingly, mature sly-miR482a can also target lncRNA15492. When excessive amounts of NBS-LRR accumulate, mature miR482a cleaves lncRNA15492 to relieve the suppression of MIR482a, leading to an increase in the accumulation of mature sly-miR482a and a decrease in the accumulation of NBS-LRR, thus maintaining NBS-LRR homeostasis (Jiang et al., 2020) (Figure 2C). This finding of maintaining NBS balance in plants during infection with pathogens is very significant. However, at present, this regulatory mechanism is only found in tomato MIR482a, not in all other MIR482/2118 superfamily members. The same mechanism was found in a study of Arabidopsis ath-miR398 function. NATs, NAT398b and NAT398c are located on the antisense sequences of MIR398b and MIR398c, respectively. Knock down of NAT398b and NAT398c upregulates MIR398b and MIR398c; overexpression of NAT398b and NAT398c represses the processing of ath-miR398 (Li et al., 2020).
Conclusions and future perspectives
This review expands our knowledge about the intertwined regulatory role of miR482/2118 in plant resistance to pathogens. New molecules interacting with miR482/2118-3p, including miR482/2118-5p, phasiRNAs, and lncRNAs, and their regulatory mechanisms participating in plant resistance to pathogens were summarized and described in this review. Some research has shown that miR482/2118-3p and -5p coregulate plant resistance, and phasiRNAs triggered by miR482/2118 superfamily members act in regulating plant resistance by affecting NBS-LRR levels. LncRNAs not only act as ceRNAs to silence miR482/2118 but also affect MIR482a located on its antisense sequence during plant resistance to pathogens. However, many important issues remain to be answered, such as whether all mechanisms work together during plant resistance to pathogens and how they work; whether these disease resistance mechanisms respond to the infection of all pathogens or a certain pathogen and whether there are specific proteins or other chemical tags involved in these plant resistance mechanisms associated with miR482/2118. Overall, further research is needed to address these issues and to better understand the resistance pathway associated with miR482/2118. We believe that future studies on resistance mechanisms will provide additional insight into plant immunity and offer effective approaches for the improvement of plant disease resistance.
Author contributions
LL and BX wrote the manuscript. LL, PG and JC collected data. JC and NJ contributed in revising manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from the Natural Science Foundation of Hunan Province (No.2021JJ30441).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bélanger, S., Pokhrel, S., Czymmek, K., Meyers, B. C. (2020). Premeiotic, 24-nucleotide reproductive PhasiRNAs are abundant in anthers of wheat and barley but not rice and maize. Plant Physiol. 184, 1407–1423. doi: 10.1104/pp.20.00816
Canto-Pastor, A., Santos, B. A. M. C., Valli, A. A., Summers, W., Schornack, S., Baulcombe, D. C. (2019). Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proc. Natl. Acad. Sci. U S A. 116, 2755–2760. doi: 10.1073/pnas.1814380116
Cui, J., Jiang, N., Hou, X., Wu, S., Zhang, Q., Meng, J., et al. (2020). Genome-wide identification of lncRNAs and analysis of ceRNA networks during tomato resistance to Phytophthora infestans. Phytopathology 110, 456–464. doi: 10.1094/PHYTO-04-19-0137-R
Cui, J., Luan, Y., Jiang, N., Bao, H., Meng, J. (2017). Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to phytophthora infestans by co-expressing glutaredoxin. Plant J. 89, 577–589. doi: 10.1111/tpj.13408
Fei, Q., Xia, R., Meyers, B. C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25, 2400–2415. doi: 10.1105/tpc.113.114652
Fei, Q., Zhang, Y., Xia, R., Meyers, B. C. (2016). Small RNAs add zing to the zig-Zag-Zig model of plant defenses. Mol. Plant Microbe Interact. 29, 165–169. doi: 10.1094/MPMI-09-15-0212-FI
Gai, Y. P., Yuan, S. S., Zhao, Y. N., Zhao, H. N., Zhang, H. L., Ji, X. L. (2018). A novel LncRNA, MuLnc1, associated with environmental stress in mulberry (Morus multicaulis). Front. Plant Sci. 9, 669. doi: 10.3389/fpls.2018.00669
Guo, G., Liu, X., Sun, F., Cao, J., Huo, N., Wuda, B., et al. (2018). Wheat miR9678 affects seed germination by generating phased siRNAs and modulating abscisic Acid/Gibberellin signaling. Plant Cell 30, 796–814. doi: 10.1105/tpc.17.00842
Jiang, N., Cui, J., Hou, X., Yang, G., Xiao, Y., Han, L., et al. (2020). Sl-lncRNA15492 interacts with sl-miR482a and affects Solanum lycopersicum immunity against Phytophthora infestans. Plant J. 103, 1561–1574. doi: 10.1111/tpj.14847
Jiang, N., Cui, J., Meng, J., Luan, Y. (2018b). A tomato nucleotide binding sites-Leucine-Rich repeat gene is positively involved in plant resistance to Phytophthora infestans. Phytopathology 108, 980–987. doi: 10.1094/PHYTO-12-17-0389-R
Jiang, N., Cui, J., Shi, Y., Yang, G., Zhou, X., Hou, X., et al. (2019). Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction. Hortic. Res. 6, 28. doi: 10.1038/s41438-018-0096-0
Jiang, N., Meng, J., Cui, J., Sun, G., Luan, Y. (2018a). Function identification of miR482b, a negative regulator during tomato resistance to Phytophthora infestans. hortic. Res. 5, 9. doi: 10.1038/s41438-018-0017-2
Ji, H. M., Zhao, M., Gao, Y., Cao, X. X., Mao, H. Y., Zhou, Y., et al. (2018). FRG3, a target of slmiR482e-3p, provides resistance against the fungal pathogen Fusarium oxysporum in tomato. Front. Plant Sci. 9, 26. doi: 10.3389/fpls.2018.00026
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Li, Y., Li, X., Yang, J., He, Y. (2020). Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat. Commun. 11, 5351. doi: 10.1038/s41467-020-19186-x
Liu, W., Cui, J., Luan, Y. (2022a). SlmiR482e-5p regulates tomato resistance to Phytophthora infestans infection along with slmiR482e-3p via sllncRNA39298-mediated inhibition. Physiol. Mol. Plant Pathol. 121, 101875. doi: 10.1016/j.pmpp.2022.101875
Liu, W., Cui, J., Luan, Y. (2022b). Overexpression of lncRNA08489 enhances tomato immunity against Phytophthora infestans by decoying miR482e-3p. Biochem. Biophys. Res. Commun. 587, 36–41. doi: 10.1016/j.bbrc.2021.11.079
Liu, W., Meng, J., Cui, J., Luan, Y. (2017). Characterization and function of MicroRNA∗s in plants. Front. Plant Sci. 8, 2200. doi: 10.3389/fpls.2017.02200
Liu, Y., Teng, C., Xia, R., Meyers, B. C. (2020). PhasiRNAs in plants: Their biogenesis, genic sources, and roles in stress responses, development, and reproduction. Plant Cell 32, 3059–3080. doi: 10.1105/tpc.20.00335
López-Márquez, D., Del-Espino, Á., López-Pagán, N., Rodríguez-Negrete, E. A., Rubio-Somoza, I., Ruiz-Albert, J., et al. (2021). miR825-5p targets the TIR-NBS-LRR gene MIST1 and down-regulates basal immunity against pseudomonas syringae in Arabidopsis. J. Exp. Bot. 72, 7316–7334. doi: 10.1093/jxb/erab354
Ma, W., Chen, C., Liu, Y., Zeng, M., Meyers, B. C., Li, J., et al. (2018). Coupling of microRNA-directed phased small interfering RNA generation from long noncoding genes with alternative splicing and alternative polyadenylation in small RNA-mediated gene silencing. New Phytol. 217, 1535–1550. doi: 10.1111/nph.14934
Nie, P., Chen, C., Yin, Q., Jiang, C., Guo, J., Zhao, H., et al. (2019). Function of miR825 and miR825* as negative regulators in Bacillus cereus AR156-elicited systemic resistance to botrytis cinerea in arabidopsis thaliana. Int. J. Mol. Sci. 20, 5032. doi: 10.3390/ijms20205032
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B., Bartel, D. P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. doi: 10.1101/gad.1004402
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Salmena, L., Poliseno, L., Tay, Y., Kats, L., Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 146, 353–358. doi: 10.1016/j.cell.2011.07.014
Shen, E., Chen, T., Zhu, X., Fan, L., Sun, J., Llewellyn, D. J., et al. (2020). Expansion of MIR482/2118 by a class-II transposable element in cotton. Plant J. 103, 2084–2099. doi: 10.1111/tpj.14885
Shivaprasad, P. V., Chen, H. M., Patel, K., Bond, D. M., Santos, B. A., Baulcombe, D. C. (2012). A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell 24, 859–874. doi: 10.1105/tpc.111.095380
Shi, C., Zhang, J., Wu, B., Jouni, R., Yu, C., Meyers, B. C., et al. (2022). Temperature-sensitive male sterility in rice determined by the roles of AGO1d in reproductive phasiRNA biogenesis and function. New Phytol. 236, 1529–1544. doi: 10.1111/nph.18446
Wu, F., Xu, J., Gao, T., Huang, D., Jin, W. (2021). Molecular mechanism of modulating miR482b level in tomato with botrytis cinerea infection. BMC Plant Biol. 21, 496. doi: 10.1186/s12870-021-03203-2
Xiao, Y., Luan, Y. (2014). MiR482 mediates the biotic stress response of plants. Plant Physilogy J. 50, 741–748. doi: 10.13592/j.cnki.ppj.2014.0141
Xia, R., Xu, J., Arikit, S., Meyers, B. C. (2015). Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol. Biol. Evol. 32, 2905–2918. doi: 10.1093/molbev/msv164
Xu, P., Li, H., Wang, X., Zhao, G., Lu, X., Dai, S., et al. (2022). Integrated analysis of the lncRNA/circRNA-miRNA-mRNA expression profiles reveals novel insights into potential mechanisms in response to root-knot nematodes in peanut. BMC Genomics 23, 239. doi: 10.1186/s12864-022-08470-3
Yang, L., Mu, X., Liu, C., Cai, J., Shi, K., Zhu, W., et al. (2015). Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 57, 1078–1088. doi: 10.1111/jipb.12348
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Zhai, J., Jeong, D. H., Paoli, E. D., Park, S., Rosen, B. D., Li, Y., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553. doi: 10.1101/gad.177527.111
Zhai, K., Liang, D., Li, H., Jiao, F., Yan, B., Liu, J., et al. (2022). NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature 601, 245–251. doi: 10.1038/s41586-021-04219-2
Zhang, Y., Waseem, M., Zeng, Z., Xu, J., Chen, C., Liu, Y., et al. (2022). MicroRNA482/2118, a miRNA superfamily essential for both disease resistance and plant development. New Phytol. 233, 2047–2057. doi: 10.1111/nph.17853
Keywords: MIR482/2118, miR482/2118-3p/5p, phasiRNA, lncRNA, resistance
Citation: Liao L, Xie B, Guan P, Jiang N and Cui J (2022) New insight into the molecular mechanism of miR482/2118 during plant resistance to pathogens. Front. Plant Sci. 13:1026762. doi: 10.3389/fpls.2022.1026762
Received: 24 August 2022; Accepted: 17 October 2022;
Published: 28 October 2022.
Edited by:
Cheng-Guo Duan, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Chen Zhu, Anhui Normal University, ChinaJian-Hua Zhao, Chinese Academy of Sciences (CAS), China
Copyright © 2022 Liao, Xie, Guan, Jiang and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ning Jiang, jiangning0806@outlook.com; Jun Cui, cuijun@hunnu.edu.cn
†These authors have contributed equally to this work
 Lijuan Liao
Lijuan Liao Biao Xie
Biao Xie Peipei Guan
Peipei Guan Ning Jiang
Ning Jiang Jun Cui
Jun Cui