- 1Department of Applied Biosciences, Kyungpook National University, Daegu, South Korea
- 2Mountain Research for Field Crops Khudwani, Sher-e Kashmir University of Agricultural Sciences and Technology of Kashmir, Srinagar, Jamu and Kashmir, India
Bacterial adhesion potential constitutes the transition of bacteria from the planktonic to the static phase by promoting biofilm formation, which plays a significant role in plant-microbial interaction in the agriculture industry. In present study, the adhesion potential of five soil-borne bacterial strains belonging to different genera was studied. All bacterial strains were capable of forming colonies and biofilms of different levels of firmness on polystyrene. Significant variation was observed in hydrophobicity and motility assays. Among the five bacterial strains (SH-6, SH-8, SH-9, SH-10, and SH-19), SH-19 had a strong hydrophobic force, while SH-10 showed the most hydrophilic property. SH-6 showed great variability in motility; SH-8 had a swimming diffusion diameter of 70 mm, which was three times higher than that of SH-19. In the motility assay, SH-9 and SH-10 showed diffusion diameters of approximately 22 mm and 55 mm, respectively. Furthermore, among the five strains, four are predominately electron donors and one is electron acceptors. Overall, positive correlation was observed among Lewis acid base properties, hydrophobicity, and biofilm forming ability. However, no correlation of motility with bacterial adhesion could be found in present experimental work. Scanning electron microscopy images confirmed the adhesion potential and biofilm ability within extra polymeric substances. Research on the role of adhesion in biofilm formation of bacteria isolated from plants is potentially conducive for developing strategies such as plant–microbial interaction to mitigate the abiotic stress.
Introduction
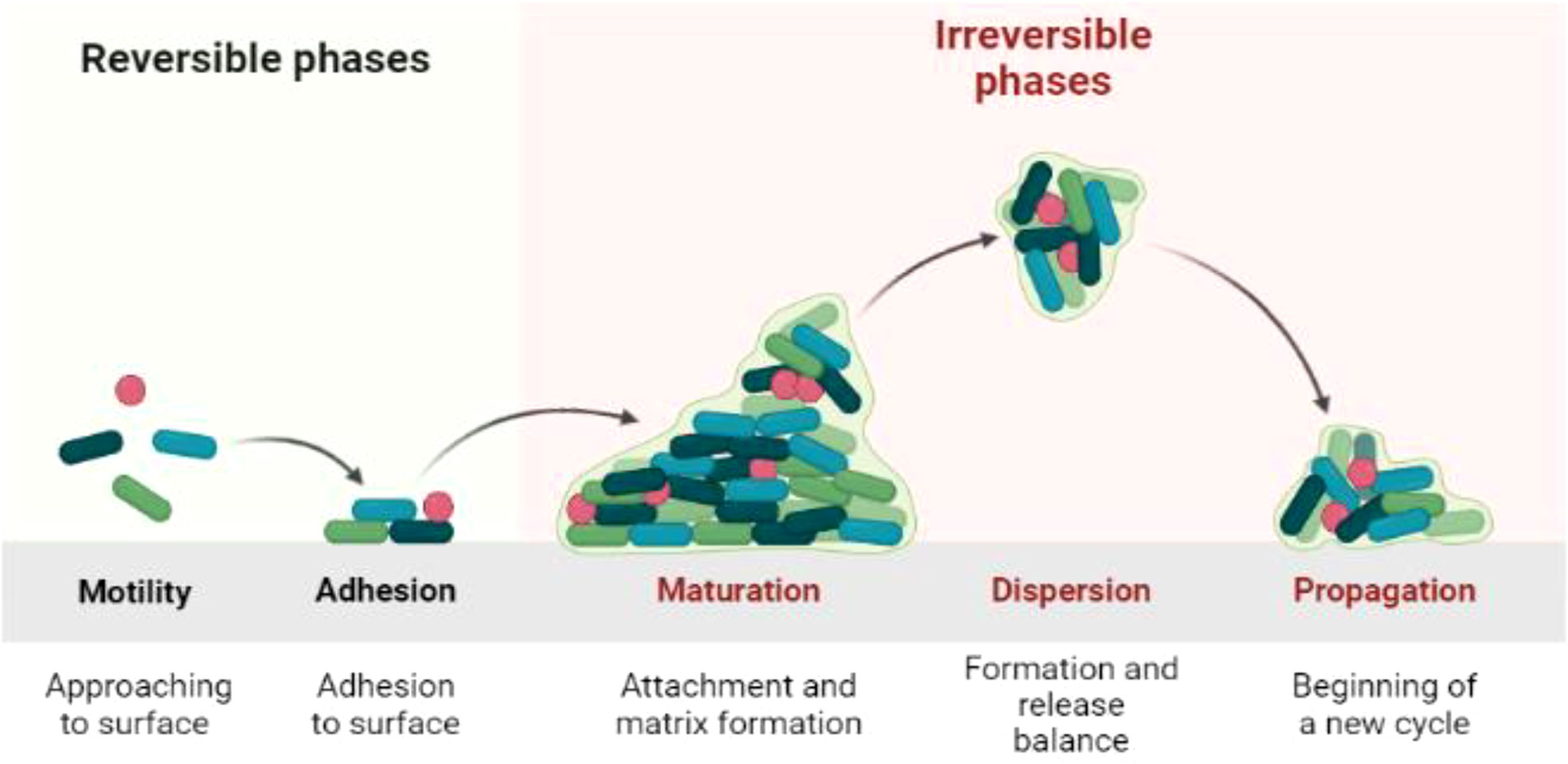
Biofilm is the product of microbial developmental procedure. It is a syntrophic relationship of the microorganism with any biotic or abiotic surface (Watnick and Kolter, 2000; Flemming and Wingender, 2010). The microbes become adherent to an extracellular polymeric substance. During interaction with the surface, microbes produce long chain exopolysaccharides and form a 3D coordinated functional community (O’toole et al., 2000; Lewandowski and Beyenal, 2019). The biofilm lifecycle of the microbes consists of a series of steps and events. The first step is attachment. The microbes are in the planktonic cell mode of biofilm formation with phenotype alteration by quorum sensing (QS) signaling pathway, and 90% of the cell biomass is produced in this phase (Rittmann and Mccarty, 1980; Tolker-Nielsen, 2015). Microbes attach to a surface using weak van der Waals forces. The QS signaling pathway is responsible for the formation of colonies and sub-colonies depending upon the temperature, p, and nutrient availability status. The colonization may extend from unicellular to multicellular cells, where microbes share traits related to nutrition, shelter etc. (Van Loosdrecht et al., 2002; Allison, 2003; Flemming et al., 2011). The bacterial life cycle can be categorized into four phases: log, lag, stationary, and decline phase. The lag phase is explained as the state where there is no proliferation of living bacteria as shown in the Figure 1. In the log phase, the bacteria start to grow rapidly, whereas, in the stationary phase, the death rate and production of bacteria are at a constant rate (Finkel, 2006; Saikia et al., 2022). Decline is the last phase where there are more dead bacterial cells than living (Andrade et al., 2006).The adhesion potential of bacteria can be observed from the log phase to the stationary phase (Swinnen et al., 2004; Jaishankar and Srivastava, 2017; Chevallereau et al., 2022).
After forming colonies, bacteria produce long chain polysaccharides that enclose the bacterial biofilm (BBF) as the cells mature. The life cycle of bacteria is completed in the biofilm, which then becomes ready for dispersion (Flemming et al., 2007; Renner and Weibel, 2011). The BBF disperses with the help of the enzymes dispersin B and deoxyribonuclease. The biofilms are unique owing to the differential expression of genes within these biofilms (Stoodley et al., 1999; Donlan, 2001; Teughels et al., 2006).
Biofilm forming microbes play a noteworthy role in biofertilization, mineralization, development, and maintenance of plant and soil fertility. Biofilms provide a biologically active metabolite exchange platform for plants and soil (Turhan et al., 2019; Kour et al., 2020; Jiang et al., 2021). They may form symbiotic, parasitic, or mutualistic relationships with the ecosystem. Microbes form colonies and biofilm on the leaves, roots, stem, rhizosphere, endosphere, rhizoplane, and phyllosphere of plants (Seneviratne et al., 2010; Seneviratne et al., 2011; Nayak et al., 2020). BBF affects growth attributes directly or indirectly via induced systemic resistance (ISR). Plants secret exudates that form the best surface for microbial attachment owing to high nutrient availability (Velmourougane et al., 2017; Kumar and Singh, 2020; Rheinheimer Dos Santos et al., 2020). Plant cell surface is the favorable site of attachment for bacterial establishment in tropical ecosystems (Bhat K and Bhat Panemangalore, 2022).
BBF is important in abiotic stress tolerance for providing an attachment site for bacteria and enhancing their potential to evolve during plant–microbial interaction (Zhang et al., 2020; Shaffique et al., 2022b). Microbes form dynamic interactions with plants and improve plant productivity, and physiological and molecular processes through a cascade of events such as production of osmolytes, organic acids, volatile compounds, siderophores, exopolysaccharides, and phytohormones and promotion of gene expression. Various recent studies have supported plant–microbial interaction in enhancing growth, development, and stress tolerance (Bhagat et al., 2021; Ciofu et al., 2022; Sachdev and Ansari, 2022). Scanning electron microscopy (SEM) is used to determine biofilm investigation. It provides information on the size, structure, location within the biofilm, and bacterial contact on extra polymeric substances (EPS) (Priester et al., 2007; El Abed et al., 2012).
Agriculture laboratories are commonly known for developing expedient technological properties in the production of beneficial microbes and their interaction with plants, but these microbes could also be involved in plant pathogenesis. It is well known that plant–microbial interactions occur through biofilms (Bednarek and Osbourn, 2009; Berg, 2009). However, only fewer studies have been accompanied on the adhesion potential of bacteria and their association with plants. For this reason, five protagonist bacterial isolates, namely, SH-6, SH-8, SH-9, SH-10, and SH-19, from plant-related sources were considered in the present experimental study.
The current study aim’s is to estimate the 1) adhesion potential of five bacterial isolates from plant-related sources, 2) characterize their cell properties including motility and surface hydrophobicity, 3) determine the correlation between adhesion potential and cell surface hydrophobicity, and 4) create SEM images to visualize the biofilm. The present study aims to enrich the speculative background of microbial biofilm formation and provide evidence on the adhesion potential for further research progress in plant–microbial interaction.
Materials and methods
Bacterial isolation
Seventy-three microbes originating from the rhizospheric soil of Artemisia princeps Pamp. were collected and isolated for further screening assays. The rhizospheric soil was collected from Pohang beach in South Korea, at an elevation of 9.9 m. The sample was preserved in a polyethylene bag in an icebox for transportation to the crop physiology lab, College of Applied Biosciences, Agriculture department, Kyungpook National University, South Korea. One gram of soil removed from the roots of the plants was mixed with 1 M saline solution, and the sample was retained in a shaker at 25°C, for 6 h. The sample was considered as a stock solution. From this stock solution, a serial dilution 10-1-10-9 was made for further analysis. Lysogeny broths were prepared, and aliquots of the serial dilutions were added. The plates were sealed with Parafilm and incubated for 24-72 h at 25°C ± 3°C. Immediately after the emergence of colonies, they were re-plated on LB plates until to obtain pure colonies. These colonies were identified by their size, shape, color, chain length, and growth pattern. The samples were then sent to Solgent Company for identification. The procedure followed is described as in published studies (Chung et al., 2005; Rashid et al., 2012).
Molecular identification and phylogenetic analysis
Inocula preparation
The bacterial strains were individually cultured at 37°C for 18 h in 5 mL of LB. The cells were then harvested by centrifugation at 5000 rpm for 5 min at 4°C. The supernatant was removed, and the cells were washed thrice with saline phosphate buffer solution (pH 7.2) and resuspended in saline phosphate buffer solution to maintain a constant pH. The final inoculum of 103 CFU/mL was prepared as described previously (Ramirez-Chavarin et al., 2013; Li et al., 2020).
Biofilm assay
For each inoculum of 150 µL, 20 µL from freshly prepared sample and 130 µL from pre-prepared inoculum were combined in a 96-well polystyrene microplate. The samples were incubated at 25-30°C for 5 days. Freshly prepared autoclaved media devoid of bacteria were used as the negative control. After 5 days, the microplate was rinsed three times with 0.1% saline phosphate buffer solution and air dried for 30 min. Then, 100 µL crystal violet stain was added and washed three times with phosphate buffer solution. At the end of the procedure, 0.2 mL of 95% ethanol was added to the plates, and the absorbance was measured at 570 nm. The procedure was followed as described previously (Coffey and Anderson, 2014; Shukla and Rao, 2017).
Hydrophobicity assay
Equal amounts of solvents (chloroform, ethyl acetate, and xylene) were mixed with the bacterial suspension to formulate a two-phase system. The solutions were vortexed for 4 minutes and placed at room temperature for 30 min to allow the hydrocarbon phase to increase. The optical density (OD) was calculated at 600 nm before and after the adhesion. The hydrophobicity was calculated using the following equation:
where A0 is considered as the initial optical density, and A1 is the final optical density of the aqueous phase, as described previously (Rahman et al., 2008; Chao et al., 2014).
Motility assay
The motility assay (swimming and swarming) was performed on soft agar plates. The plates for the swimming assay were prepared by mixing 2.5 g/L glucose, 5 g/L sodium chloride, 10 g/L tryptone, and 0.3% agar, while those for the swarming assay contained, 0.5 g/L glucose, 25 g/L Luria-Bertani and 0.5% agar. Two microliter aliquots of each isolates were mottled onto the soft-agar plates surface and then incubated at 25-30°C for 15 min to ensure better absorption. The diameter (mm) of each strain was measured in order to estimate the motility (Morales-Soto et al., 2015; Sun et al., 2018).
SEM microscopy
Bacterial isolates were allowed to grow in the LB plates for 48 h after which pure isolates together with the media were cut (W × D × H, 5 × 2 × 5 mm) and placed into Eppendorf tubes. Then, 0.1 M solution of phosphate buffer (pH 7.3) was added to the tubes and washed three times. The samples were precleared with 2.5% glutaraldehyde solution for 24 h at 4°C. The fixation sample was placed into 0.1 M sodium phosphate buffer solution (pH 7.3) and washed again with 0.1 M sodium phosphate buffer solution. Then, 1% osmium tetroxide in 0.1 M cacodylate buffer was added to the sample for 90 min and washed with sodium phosphate buffer for 40 min. After washing, the samples were dehydrated with ethanol. Excess water was removed using a critical point device, carbon tapes were attached, and the samples were observed under a scanning electron microscope (Hitachi-s-3500N), as previously described (Chen et al., 2022; Kuzmina et al., 2022).
Statistical analysis
All the experiments were repeated five times. The GraphPad Prism software (version 5.8) was used to perform statistical analysis. The mean values P ≤ 0.05 were considered significant using Duncan’s multiple range test (DMRT) in SAS (version 9.1) to find out the statistical analysis.
Results
Biofilm assay
Xylene is considered as polar. All isolates showing strong affinity toward xylene were labeled as hydrophobic. SH-10 had the lowest affinity toward xylene, at less than 20%, which shows the strongest electron acceptor ability. Isolates showing moderate affinity toward chloroform were electron donors (Figure 2).
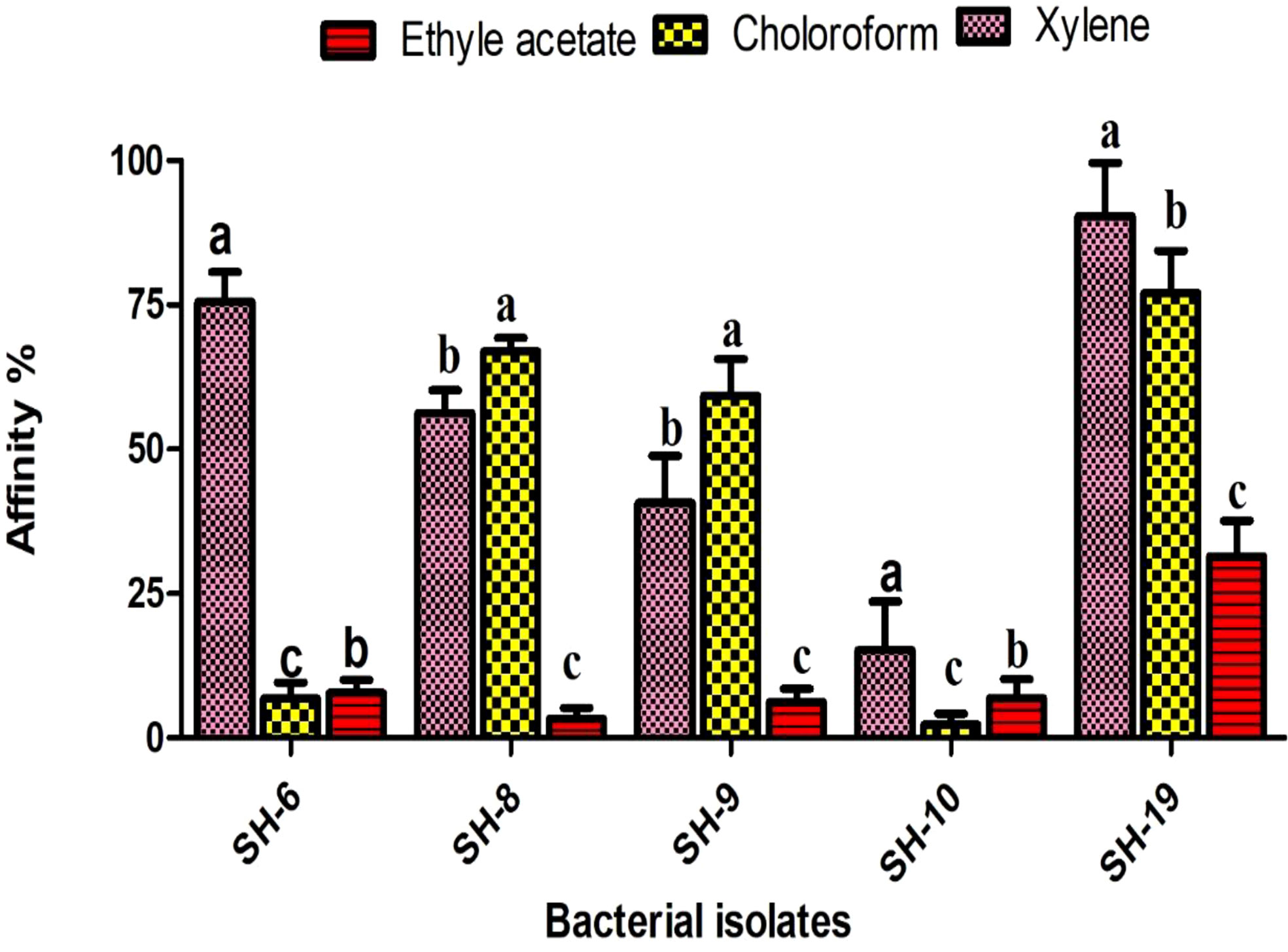
Figure 2 Information of the five selected isolates and their affinity for different solvents. The letter on each bar is the significant different between treatments at P ≤ 0.05. The error bar represents the standard error among the replicates.
Cell-surface characteristic assay
All bacterial isolates possessed adhesion potential for the 96-well plate with different levels of firmness. However, SH-19 showed excellent biofilm formation compared to that by the other four bacterial isolates. Thus, SH-6, SH-8, and SH-9 are moderate biofilm-producing bacterial isolates, while SH-10 had the lowest biofilm-forming ability (Figure 3).
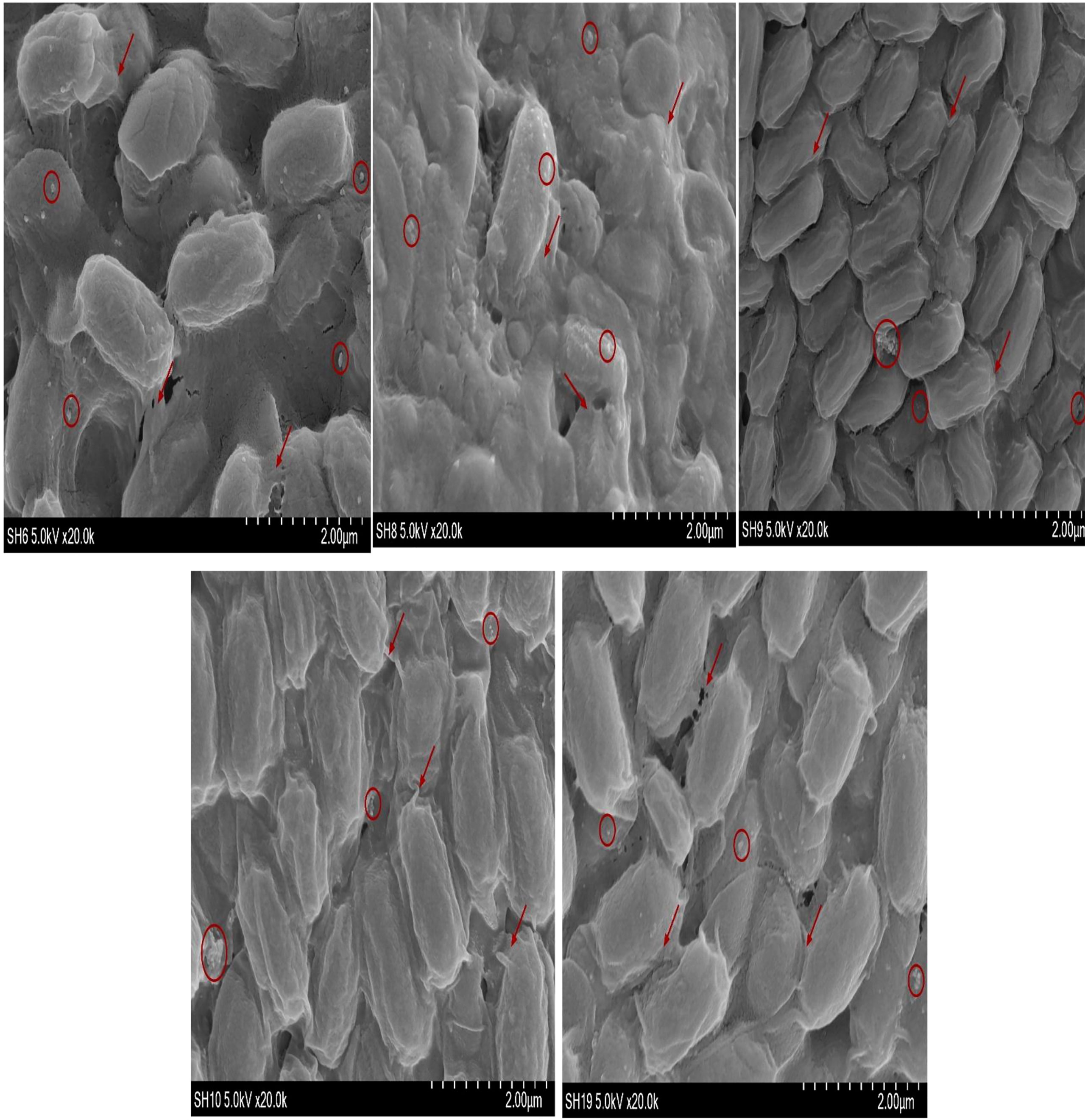
Observation of bacterial biofilm by SEM
The SEM shown in Figure 4 confirms the presence of biofilm whereas the ↗○ represents the location and position of exopolysacchrides
Discussion
BBF formation plays an significant role in plant–microbial interaction in sustainable agriculture industry (Ramey et al., 2004; Bogino et al., 2013). This study involved 5 protagonist soil-borne bacterial isolates for biofilm assay. The optimal temperature of each bacterial isolate varied with an average of 25°C and which was considered as the optimal temperature.
The short and long-term biofilms were evaluated in a time-dependent manner by regulating the incubation time by 24-72 h. The results exhibited that all bacterial isolates were capable to attach on the polystyrene surface with some variation which exposed the isolates belongs to different species. SH-19 exhibited the best adhesion ability regardless of time and temperature, while a moderate amount of biofilm formation was displayed by SH-10, SH-8, SH-6, and SH-9. All strains formed considerable amounts of biofilm at 25-30°C after 24 h. However, for 72 h incubation, a higher temperature was most appropriate for bacterial cell adhesion. The entire different behaviors shown by SH-9 in comparison with SH-19 confirmed that all isolates from a single source do not necessarily have the same characteristics.
SH-19 is an extremophile that can exist at extreme temperatures and produce and maintain biofilms at 50°C. However, the surface of the 96-well polystyrene plate that was used for the present in vitro experimental study is significantly different from that of the plant epidermis, and thus, the activities of related signaling pathways could be limited. Biofilm formation was temperature-dependent as shown in Figure 3. At 24 h, four of the five bacterial isolates displayed a higher attachment at above 30°C than 40°C; after 72 h, only SH-19 showed significant biofilm formation. A further increase in temperature to 50°C resulted in the formation of considerable amount of biofilm by SH-19. This is a well-known phenomenon and occurs probably owing to the diversity of optimal temperatures of the isolates (Rozanov et al., 2021).
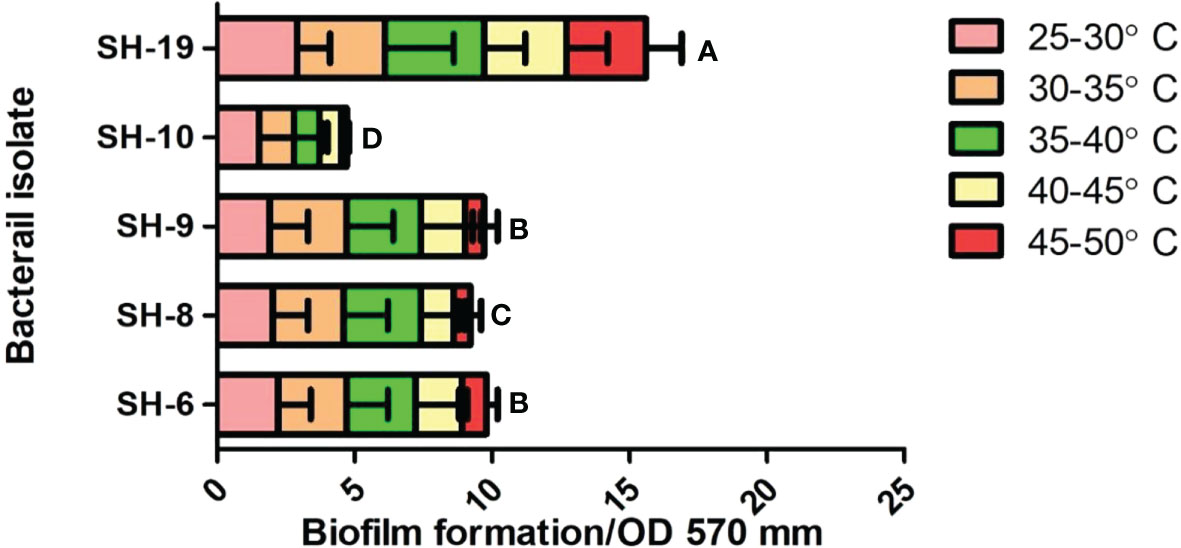
Figure 3 Biofilm information by the five selected isolates on 96-well polystyrene microplates under 22–50°C for 24 h. The letter on each bar is the significant different between treatments at P ≤ 0.05. The error bar represents the standard error among the replicates.
Massive cell density in culture media possibly results in providing an assistance for communication of bacteria, thus, improving the bacterial attachment. Additionally, less biofilm formation by SH-9, SH-6, SH-8, and SH-10 at 50°C after 24 h is probably owing to biofilm dispersal at the end of the incubation periods. The outcomes of study also indicated that temperature and time influenced biofilm formation.
In one complete biofilm development cycle, dispersion is the final step, in which cells detach from the established biofilm at high temperatures (Rumbaugh and Sauer, 2020). After 72 h, the restricted biofilm may be owing to limited nutrition perfusion. The period during which bacteria show impaired adhesion may be owing to the dispersion phase. At 25-30°C, mostly the isolates grow fast over time and form mature biofilms. At 35°C, after 24 h, there is a decreased amount of biofilm by 72 h. This partially proved that higher temperature was preferable for most bacteria. Attention must be given to the existence of particularity in biofilm formation.
Cell adhesion entails contact with the interacting surface of the bacterial cell envelope. Motile properties are important at the docking phase when two objects meet (Ottemann and Miller, 1997). Swimming and swarming motility was investigated in the present study. Swimming is related to the motile potential of individual bacterium, whereas swarming is related to colony formation (Kearns, 2010; Servant et al., 2015; Be’er and Ariel, 2019). SH-6 and SH-8 showed considerable motility and developed large diffusion circles with diameters more than 70 mm, three times larger than those of SH-19 and SH-10.
SH-9 exhibited moderate swimming capacity with a diameter of 22 cm. A lower disparity level was found among the isolates in the swarming assay. The five bacterial isolates showed reserved swarming ability compared to their swimming potential. These results suggest that fluidity of the medium may influence the diffusion of bacterial isolates.
Flowing states might favor plant–microbe interaction and subsequent development of biofilm. Bacterial motility is flagella dependent. Swimming and swarming potential of microbes play important role in their adhesion and biofilm formation (Daniels et al., 2004; Venieraki et al., 2016).
Because different strains of different species possess a diverse range of flagellar numbers, great variations are noted in motility. SH-10 showed a diffusion circle of 55 mm with remarkable motility but were not strong in biofilm formation. Furthermore, no correlation was observed between biofilm formation and motility of the isolates as shown in Table 1.
The function of flagella cannot be ignored. Studies using video microscopy have shown that flagella help bacteria reach the proximate surface, but in the present study, a partial positive correlation was observed.
The surface hydrophobicity is determined by xylene, a polar solvent. High affinity to xylene was noticed in SH-19 and SH-8, representing hydrophobicity, while SH-10 showed hydrophilicity, under 20%, toward xylene.
SH-6 and SH-10 showed competitor of hydrophobicity. Various recent studies have suggested that hydrophobic characteristics enhance biofilm formation potential. This is in line with the results of the present study in which we show a positive correlation and that hydrophobicity enhances biofilm formation.
Lewis acid base properties were also found by using two other solvents, chloroform and ethyl acetate. SH-10 showed strong affinity toward chloroform and ethyl acetate implying electron acceptor capacity. SH-6 showed almost equal affinity toward ethyl acetate and chloroform, which is predominately an electron donor and weak electron acceptor. The other bacterial isolates showed better affinity to chloroform than ethyl acetate representing their greater ability to donate electrons.
The bacterial isolates with electron donor capacity attached easily to the surface of the 96-well polystyrene plate. A positive correlation was noted between hydrophobicity and biofilm formation. Thus, Lewis acid base interaction shows an energetic role in biofilm formation.
SEM utilizes the high-power energy of electron beams to visualize the properties of bacterial cells such as their size, shape, homogeneity, and topological information. Additionally, it provides useful information about biofilm life cycle, from attachment to dispersion (Baldotto and Olivares, 2008; Nongkhlaw and Joshi, 2014). SEM also displayed the observation of bacterial adhesion properties to a surface and their ability to form biofilms. Bacterial adhesion to biofilm formation is the condition in which bacteria attach and adhere firmly to a surface and completes one life cycle (El Abed et al., 2012; Relucenti et al., 2021).
It is important to estimate the adhesion potential of the bacterial isolates because adhesion potential of bacteria provides the transition from planktonic phase to static phase (Singh et al., 2011; Xu and Siedlecki, 2022). The transition of microbes from one phase to another phase is responsible for the development of biofilm. Adhesion is the first step of biofilms which is liable for the docking and locking phase of the biofilm (Chu et al., 2022; Prabu et al., 2022). Adhesion potential has been quantified using a number of automatic tools such as by measuring the production of bacteria in Congo red assay or under a conventional scanning microscope (Taj et al., 2012). SEM is considered as the gold standard to identify biofilm under different resolutions. Bacterial strains are also visualized by SEM. This is an advanced microbiology technique that can measure bacterial cell attachment and provide clear images of bacterial flagellum (Moreira et al., 2012). High-magnification SEM of SH-6 showed small spherical structures and numerous tubular projections on the surface of the growing vegetative cells. The bacterial architecture of SH-8 showed the presence of spherical structure trapped in the exopolymeric substances (Figures 4, 5), while that of SH-9 and SH-10 showed a mat-like structure. SH-19 were spherical bacteria. All the bacteria showed the presence of exopolysaccharides in which the bacteria were trapped. Bacteria interact with plant cells in diverse ways (Shaffique et al., 2022c). The main feature of this interaction is to make colonies, in which the microbes adhere to plant cells as individual cells or in the form of clusters (Morris and Monier, 2003; Tshikantwa et al., 2018). The adherent bacterial populations, defined as biofilms, display structural alignments of several magnitudes. Each plant part has distinguishing levels of saturation such as nutrient availability and cellular chemistry, which affects the formation of biofilm. It is important to evaluate the adhesion potential to measure plant–microbial interaction, which is important in the mitigation of abiotic stress. The adhesive biofilm ability of the bacteria provides the platform to colonies on plant cells with aid of self-producing matrix (EPS) to provide the protective environment against various abiotic stress such as drought, salinity and heat stress etc. (Sharma et al., 2021; Shaffique et al., 2022a).
Beneficial microbes facilitates the mutualistic interaction which is Important in providing the plant microbial interaction. The plant microbial interaction augments the production of the metabolites which improve the plant productivity (Ben Zineb et al., 2022; Mafa‐Attoye et al., 2022). Several studies suggested that biofilm producing bacteria’s are helpful in not only mitigation of abiotic stress but also prevent phytopathogen (Chandwani and Amaresan, 2022; Singh and Chauhan, 2022). They produce the exopolysacchrides which gives the adhesion potential and facilitates the plant microbial interaction (Kant, 2022; Kapadia et al., 2022). They play imperative role in neutralizing the abiotic stress and improving crop yield and quality (Admassie et al., 2022; Siddique et al., 2022) as shown in Figure 5.
Conclusion and future prospective
We concluded that the five soil-borne bacterial isolates were capable of colonization on the 96-well polystyrene plate surface. Cell surface hydrophobicity was positively correlated with biofilm formation, whereas swimming and swarming were negatively correlated with biofilm formation. However, further research on larger scale is needed to clarify this. The biofilm architecture of bacterial isolates was observed by SEM. The results revealed that bacteria were trapped in the exopolymeric substances. The biofilm-forming bacterial isolates strongly influence plant–microbial interaction to mitigate the abiotic stress. Ongoing studies on newly bacterial isolates will provide the new experimental frame work to mitigate the stress and helps in providing the sustainable agronomy.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.
Author contributions
SS write the original draft paper did the experiment and formal analysis as a part of her dissertation. MI and MAK did the software analysis. S-MK and AA did the graphical designing. I-JL did the supervision, provide funding and validate the results. SW did the critical review editing. All authors contributed to the article and approved the submitted version.
Funding
National Research Foundation(NRF) grant funded by the korea goverment(MIST)(No.2022R1A2C1008993).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
SEM, Scanning electron microscope; EPS, Exopolysacchrides; BBF, Bacterial biofilm; OD, Optical density; QS, Quorum sensing; CFU, Colony forming unit.
References
Admassie, M., Woldehawariat, Y., Alemu, T., Gonzalez, E., Jimenez, J. F. (2022). The role of plant growth-promoting bacteria in alleviating drought stress on pepper plants. Agric. Water Manage. 272, 107831. doi: 10.1016/j.agwat.2022.107831
Andrade, J. M., Cairrão, F., Arraiano, C. M. (2006). RNase r affects gene expression in stationary phase: regulation of ompA. Mol. Microbiol. 60, 219–228. doi: 10.1111/j.1365-2958.2006.05092.x
Baldotto, L. E. B., Olivares, F. L. (2008). Phylloepiphytic interaction between bacteria and different plant species in a tropical agricultural system. Can. J. Microbiol. 54, 918–931. doi: 10.1139/W08-087
Bednarek, P., Osbourn, A. (2009). Plant-microbe interactions: chemical diversity in plant defense. Science 324, 746–748. doi: 10.1126/science.1171661
Be’er, A., Ariel, G. (2019). A statistical physics view of swarming bacteria. Movement Ecol. 7, 1–17. doi: 10.1186/s40462-019-0153-9
Ben Zineb, A., Gargouri, M., López-Ráez, J. A., Trabelsi, D., Aroca, R., Mhamdi, R. (2022). Interaction between p fertilizers and microbial inoculants at the vegetative and flowering stage of medicago truncatula. Plant Growth Regul. 1–14. doi: 10.1007/s10725-022-00886-x
Berg, G. (2009). Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. doi: 10.1007/s00253-009-2092-7
Bhagat, N., Raghav, M., Dubey, S., Bedi, N. (2021).bacterial exopolysaccharides: Insight into their role in plant abiotic stress tolerance. 31(8):1045–59. doi: doi: 10.4014/jmb.2105.05009
Bhat K, U., Bhat Panemangalore, D. (2022). “Microbes–surfaces interactions,” in Application of microbes in environmental and microbial biotechnology (Springer, Singapore: Springer), 473–499.
Bogino, P. C., Oliva, M. D. L. M., Sorroche, F. G., Giordano, W. (2013). The role of bacterial biofilms and surface components in plant-bacterial associations. Int. J. Mol. Sci. 14, 15838–15859. doi: 10.3390/ijms140815838
Chandwani, S., Amaresan, N. (2022). Role of ACC deaminase producing bacteria for abiotic stress management and sustainable agriculture production. Environ. Sci. pollut. Res., 20, 1–17. doi: 10.1007/s11356-022-18745-7
Chao, Y., Guo, F., Fang, H. H., Zhang, T. (2014). Hydrophobicity of diverse bacterial populations in activated sludge and biofilm revealed by microbial adhesion to hydrocarbons assay and high-throughput sequencing. Colloids Surf. B: Biointerfaces 114, 379–385. doi: 10.1016/j.colsurfb.2013.10.028
Chen, L., Cheng, Q., Zhang, X., Zhu, M., Hartley, W., Zhu, F. (2022). Novel plant growth-promoting bacteria isolated from bauxite residue: The application for revegetation. Bull. Environ. Contam. Toxicol., 24, 1–10. doi: 10.1007/s00128-021-03433-y
Chevallereau, A., Pons, B. J., Van Houte, S., Westra, E. R. (2022). Interactions between bacterial and phage communities in natural environments. Nat. Rev. Microbiol. 20, 49–62. doi: 10.1038/s41579-021-00602-y
Chung, H., Park, M., Madhaiyan, M., Seshadri, S., Song, J., Cho, H., et al (2005). Isolation and characterization of phosphate solubilizing bacteria from the rhizosphere of crop plants of Korea. Soil Biol. Biochem. 37, 1970–1974. doi: 10.1016/j.soilbio.2005.02.025
Chu, X., Yang, F., Tang, H. (2022). Recent advance in polymer coatings combating bacterial adhesion and biofilm formation. Chin. J. Chem. 40 doi: 10.1002/cjoc.202200434
Ciofu, O., Moser, C., Jensen, P., Høiby, N. (2022). Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol., 3, 1–15. doi: 10.1038/s41579-022-00682-4
Coffey, B. M., Anderson, G. G. (2014). “Biofilm formation in the 96-well microtiter plate,” in Pseudomonas methods and protocols (New York, NY: Springer), 631–641.
Daniels, R., Vanderleyden, J., Michiels, J. (2004). Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 28, 261–289. doi: 10.1016/j.femsre.2003.09.004
Donlan, R. M. (2001). Biofilm formation: a clinically relevant microbiological process. Clin. Infect. Dis. 33, 1387–1392. doi: 10.1086/322972
El Abed, S., Ibnsouda, S. K., Latrache, H., Hamadi, F. (2012). “Scanning electron microscopy (SEM) and environmental SEM: suitable tools for study of adhesion stage and biofilm formation,” in Scanning electron microscopy (Intechopen, London).
Finkel, S. E. (2006). Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 4, 113–120. doi: 10.1038/nrmicro1340
Flemming, H.-C., Neu, T. R., Wozniak, D. J. (2007). The EPS matrix: the “house of biofilm cells”. J. bacteriol. 189, 7945–7947. doi: 10.1128/JB.00858-07
Flemming, H.-C., Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Flemming, H.-C., Wingender, J., Szewzyk, U. (2011). Biofilm highlights (Springer Science & Business Media).
Jaishankar, J., Srivastava, P. (2017). Molecular basis of stationary phase survival and applications. Front. Microbiol. 82000. doi: 10.3389/fmicb.2017.02000
Jiang, Y., Liu, Y., Zhang, X., Gao, H., Mou, L., Wu, M., et al. (2021). Biofilm application in the microbial biochemicals production process. Biotechnol. Adv. 107724. doi: 10.1016/j.biotechadv.2021.107724
Kant, S. (2022). Mechanisms of stress adaptation by bacterial communities. 247–58. Academic Press. doi: 10.1016/B978-0-323-99900-7.00013-4
Kapadia, C., Patel, N., Rana, A., Vaidya, H., Alfarraj, S., Ansari, M. J., et al. (2022). Evaluation of plant growth-promoting and salinity ameliorating potential of halophilic bacteria isolated from saline soil. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.946217
Kearns, D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644. doi: 10.1038/nrmicro2405
Kour, D., Rana, K. L., Kaur, T., Yadav, N., Yadav, A. N., Rastegari, A. A., et al. (2020). “Microbial biofilms: functional annotation and potential applications in agriculture and allied sectors,” in New and future developments in microbial biotechnology and bioengineering: Microbial biofilms (Elsevier), 283–301.
Kumar, A., Singh, J. (2020). Biofilms forming microbes: Diversity and potential application. Plant Microbiome. Sustain. Agric. 25, 173. doi: 10.1007/978-3-030-38453-1_6
Kuzmina, L. Y., Gilvanova, E., Galimzyanova, N., Arkhipova, T., Ryabova, A., Aktuganov, G., et al. (2022). Characterization of the novel plant growth-stimulating strain advenella kashmirensis IB-K1 and evaluation of its efficiency in saline soil. Microbiology 91, 173–183. doi: 10.1134/S0026261722020072
Lewandowski, Z., Beyenal, H. (2019). Fundamentals of biofilm research (London, New York: CRC press).
Li, M., Wang, Y., Cui, H., Li, Y., Sun, Y., Qiu, H.-J. (2020). Characterization of lactic acid bacteria isolated from the gastrointestinal tract of a wild boar as potential probiotics. Front. vet. Sci. 7, 49. doi: 10.3389/fvets.2020.00049
Mafa-Attoye, T. G., Borden, K. A., Alvarez, D. O., Thevathasan, N., Isaac, M. E., Dunfield, K. E. (2022). Roots alter soil microbial diversity and interkingdom interactions in diversified agricultural landscapes. Oikos. doi: 10.1111/oik.08717
Morales-Soto, N., Anyan, M. E., Mattingly, A. E., Madukoma, C. S., Harvey, C. W., Alber, M., et al. (2015). Preparation, imaging, and quantification of bacterial surface motility assays. JoVE (Journal Visualized Experiments), 98, e52338. doi: 10.3791/52338
Moreira, C. A., Oliveira, L. C. D., Mendes, M. S., Santiago, T. D. M., Barros, E. B., Carvalho, C. B. M. D. (2012). Biofilm production by clinical staphylococci strains from canine otitis. Braz. J. Microbiol. 43, 371–374. doi: 10.1590/S1517-83822012000100044
Morris, C. E., Monier, J.-M. (2003). The ecological significance of biofilm formation by plant-associated bacteria. Annu. Rev. Phytopathol. 41, 429. doi: 10.1146/annurev.phyto.41.022103.134521
Nayak, S. K., Nayak, S., Patra, J. K. (2020). Rhizobacteria and its biofilm for sustainable agriculture: a concise review. New Future Dev. Microbial. Biotechnol. Bioeng.: Microbial. Biofilms, 1, 165–175. doi: 10.1016/B978-0-444-64279-0.00013-X
Nongkhlaw, F. M. W., Joshi, S. (2014). Distribution pattern analysis of epiphytic bacteria on ethnomedicinal plant surfaces: A micrographical and molecular approach. J. Micros. Ultrastruct. 2, 34–40. doi: 10.1016/j.jmau.2014.02.003
O'toole, G., Kaplan, H. B., Kolter, R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. doi: 10.1146/annurev.micro.54.1.49
Ottemann, K. M., Miller, J. F. (1997). Roles for motility in bacterial–host interactions. Mol. Microbiol. 24, 1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x
Prabu, R., Mohanty, A., Balakrishnan, S. S., Jayalakshmi, G., Sundar, K. (2022). Molecular docking and simulation of IcaC protein as O-succinyltransferase function in staphylococcus epidermidis biofilm formation. Curr. Res. Struct. Biol. 4, 78–86. doi: 10.1016/j.crstbi.2022.03.002
Priester, J. H., Horst, A. M., Van De Werfhorst, L. C., Saleta, J. L., Mertes, L. A., Holden, P. A. (2007). Enhanced visualization of microbial biofilms by staining and environmental scanning electron microscopy. J. Microbiol. Methods 68, 577–587. doi: 10.1016/j.mimet.2006.10.018
Rahman, M., Kim, W.-S., Kumura, H., Shimazaki, K.-I. (2008). Autoaggregation and surface hydrophobicity of bifidobacteria. World J. Microbiol. Biotechnol. 24, 1593–1598. doi: 10.1007/s11274-007-9650-x
Ramey, B. E., Koutsoudis, M., Von Bodman, S. B., Fuqua, C. (2004). Biofilm formation in plant–microbe associations. Curr. Opin. Microbiol. 7, 602–609. doi: 10.1016/j.mib.2004.10.014
Ramirez-Chavarin, M., Wacher, C., Eslava-Campos, C., Perez-Chabela, M. (2013). Probiotic potential of thermotolerant lactic acid bacteria strains isolated from cooked meat products. Int. Food Res. J. 20:991–1000.
Rashid, S., Charles, T. C., Glick, B. R. (2012). Isolation and characterization of new plant growth-promoting bacterial endophytes. Appl. Soil Ecol. 61, 217–224. doi: 10.1016/j.apsoil.2011.09.011
Relucenti, M., Familiari, G., Donfrancesco, O., Taurino, M., Li, X., Chen, R., et al. (2021). Microscopy methods for biofilm imaging: focus on SEM and VP-SEM pros and cons. Biology 10, 51. doi: 10.3390/biology10010051
Renner, L. D., Weibel, D. B. (2011). Physicochemical regulation of biofilm formation. MRS Bull. 36, 347–355. doi: 10.1557/mrs.2011.65
Rheinheimer Dos Santos, D., Monteiro De Castro Lima, J. A., Paranhos Rosa De Vargas, J., Camotti Bastos, M., Santanna Dos Santos, M. A., Mondamert, L., et al. (2020). Pesticide bioaccumulation in epilithic biofilms as a biomarker of agricultural activities in a representative watershed. Environ. Monit. Assess. 192, 1–14.
Rittmann, B. E., Mccarty, P. L. (1980). Model of steady-state-biofilm kinetics. Biotechnol. bioeng. 22, 2343–2357. doi: 10.1002/bit.260221110
Rozanov, A., Shekhovtsov, S., Bogacheva, N., Pershina, E., Ryapolova, A., Bytyak, D., et al. (2021). Production of subtilisin proteases in bacteria and yeast. Vavilov J. Genet. Breed. 25, 125. doi: 10.18699/VJ21.015
Rumbaugh, K. P., Sauer, K. (2020). Biofilm dispersion. Nat. Rev. Microbiol. 18, 571–586. doi: 10.1038/s41579-020-0385-0
Sachdev, S., Ansari, M. I. (2022). “Role of plant microbiome under stress environment to enhance crop productivity,” in Augmenting crop productivity in stress environment (Springer, Singapore), 205–221.
Saikia, D., Jadhav, P., Hole, A. R., Krishna, C. M., Singh, S. P. (2022). Growth kinetics monitoring of gram-negative pathogenic microbes using raman spectroscopy. Appl. Spectrosc., 00037028221109624.
Seneviratne, G., Jayasekara, A., De Silva, M., Abeysekera, U. (2011). Developed microbial biofilms can restore deteriorated conventional agricultural soils. Soil Biol. Biochem. 43, 1059–1062.
Seneviratne, G., Weerasekara, M., Seneviratne, K., Zavahir, J., Kecskés, M., Kennedy, I. (2010). “Importance of biofilm formation in plant growth promoting rhizobacterial action,” in Plant growth and health promoting bacteria (Springer), 81–95.
Servant, A., Qiu, F., Mazza, M., Kostarelos, K., Nelson, B. J. (2015). Controlled in vivo swimming of a swarm of bacteria-like microrobotic flagella. Adv. Mater. 27, 2981–2988.
Shaffique, S., Khan, M. A., Imran, M., Kang, S.-M., Park, Y.-S., Wani, S. H., et al. (2022a). Research progress in the field of microbial mitigation of drought stress in plants. Front. Plant Sci. 13, 870626–870626.
Shaffique, S., Khan, M. A., Wani, S. H., Imran, M., Kang, S.-M., Pande, A., et al. (2022b). Biopriming of maize seeds with a novel bacterial strain SH-6 to enhance drought tolerance in south Korea. Plants 11, 1674.
Shaffique, S., Khan, M. A., Wani, S. H., Pande, A., Imran, M., Kang, S.-M., et al. (2022c). A review on the role of endophytes and plant growth promoting rhizobacteria in mitigating heat stress in plants. Microorganisms 10:1286.
Sharma, S., Chandra, D., Sharma, A. K. (2021). “Rhizosphere plant–microbe interactions under abiotic stress,” in Rhizosphere biology: Interactions between microbes and plants (Singapore: Springer), 195–216.
Shukla, S. K., Rao, T. S. (2017). An improved crystal violet assay for biofilm quantification in 96-well microtitre plate. Biorxiv 100214.
Siddique, I., Shah, T., Ali, A., Ahmad, I., D'amato, R., Munsif, F. (2022). Arbuscular mycorrhizal fungi and biofilm forming bacteria act synergistically to modulate proline metabolism, antioxidant defense system and aquaporin genes expression under drought stress.
Singh, A., Chauhan, P. S. (2022). N-acyl homoserine lactone mediated quorum sensing exhibiting plant growth-promoting and abiotic stress tolerant bacteria demonstrates drought stress amelioration. J. Pure Appl. Microbiol., 1, 669–684.
Singh, A. V., Vyas, V., Patil, R., Sharma, V., Scopelliti, P. E., Bongiorno, G., et al. (2011). Quantitative characterization of the influence of the nanoscale morphology of nanostructured surfaces on bacterial adhesion and biofilm formation. PloS One 6, e25029.
Stoodley, P., Boyle, J. D., Debeer, D., Lappin-Scott, H. M. (1999). Evolving perspectives of biofilm structure. Biofouling 14, 75–90. doi: 10.1080/08927019909378398
Sun, E., Liu, S., Hancock, R. E. (2018). Surfing motility: a conserved yet diverse adaptation among motile bacteria. J. Bacteriol. 200, e00394–e00318. doi: 10.1128/JB.00394-18
Swinnen, I., Bernaerts, K., Dens, E. J., Geeraerd, A. H., Van Impe, J. (2004). Predictive modelling of the microbial lag phase: a review. Int. J. Food Microbiol. 94, 137–159. doi: 10.1016/j.ijfoodmicro.2004.01.006
Taj, Y., Essa, F., Aziz, F., Kazmi, S. U. (2012). Study on biofilm-forming properties of clinical isolates of staphylococcus aureus. J. Infect. Dev. Countries 6, 403–409. doi: 10.3855/jidc.1743
Teughels, W., Van Assche, N., Sliepen, I., Quirynen, M. (2006). Effect of material characteristics and/or surface topography on biofilm development. Clin. Oral. implants Res. 17, 68–81. doi: 10.1111/j.1600-0501.2006.01353.x
Tolker-Nielsen, T. (2015). Biofilm development. Microbiol. Spectr. 3, 2. doi: 10.1128/microbiolspec.MB-0001-2014
Tshikantwa, T. S., Ullah, M. W., He, F., Yang, G. (2018). Current trends and potential applications of microbial interactions for human welfare. Front. Microbiol. 91156. doi: 10.3389/fmicb.2018.01156
Turhan, E., Erginkaya, Z., Korukluoğlu, M., Konuray, G. (2019). “Beneficial biofilm applications in food and agricultural industry,” in Health and safety aspects of food processing technologies (Cham: Springer), 445–469.
Van Loosdrecht, M., Heijnen, J., Eberl, H., Kreft, J., Picioreanu, C. (2002). Mathematical modelling of biofilm structures. Antonie van Leeuwenhoek 81, 245–256. doi: 10.1023/A:1020527020464
Velmourougane, K., Prasanna, R., Saxena, A. K. (2017). Agriculturally important microbial biofilms: present status and future prospects. J. basic Microbiol. 57, 548–573. doi: 10.1002/jobm.201700046
Venieraki, A., Tsalgatidou, P. C., Georgakopoulos, D., Dimou, M., Katinakis, P. (2016). Swarming motility in plant-associated bacteria. Hellenic Plant Prot. J. 9, 16–27. doi: 10.1515/hppj-2016-0002
Watnick, P., Kolter, R. (2000). Biofilm, city of microbes. J. bacteriol. 182, 2675–2679. doi: 10.1128/JB.182.10.2675-2679.2000
Xu, L. C., Siedlecki, C. A. (2022). Submicron topography design for controlling staphylococcal bacterial adhesion and biofilm formation. J. Biomed. Mater. Res. Part A 110, 1238–1250. doi: 10.1002/jbm.a.37369
Keywords: biofilm, motility, sem, hydrophobicity, bbf
Citation: Shaffique S, Imran M, Wani SH, Khan MA, Kang S-M, Adhikari A and Lee I-J (2022) Evaluating the adhesive potential of the newly isolated bacterial strains in research exploitation of plant microbial interaction. Front. Plant Sci. 13:1004331. doi: 10.3389/fpls.2022.1004331
Received: 27 July 2022; Accepted: 06 October 2022;
Published: 21 October 2022.
Edited by:
Iftikhar Ali, State Key Laboratory of Molecular Developmental Biology (CAS), ChinaReviewed by:
Hafiz Muhammad Ahmad, Government College University, Faisalabad, PakistanMuhammad Azhar Nadeem, Sivas University of Science and Technology, Turkey
Copyright © 2022 Shaffique, Imran, Wani, Khan, Kang, Adhikari and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: In-Jung Lee, ijlee@knu.ac.kr
 Shifa Shaffique
Shifa Shaffique Muhammad Imran
Muhammad Imran Shabir Hussain Wani
Shabir Hussain Wani Muhamad Aqil Khan
Muhamad Aqil Khan Sang-Mo Kang1
Sang-Mo Kang1 Arjun Adhikari
Arjun Adhikari In-Jung Lee
In-Jung Lee