- 1Department of Botany, University of British Columbia, Vancouver, BC, Canada
- 2Michael Smith Laboratories, University of British Columbia, Vancouver, BC, Canada
The outer epidermal cell walls of plant shoots are covered with a cuticle, a continuous lipid structure that provides protection from desiccation, UV light, pathogens, and insects. The cuticle is mostly composed of cutin and cuticular wax. Cuticular wax synthesis is synchronized with surface area expansion during plant development and is associated with plant responses to biotic and abiotic stresses. Cuticular wax deposition is tightly regulated by well-established transcriptional and post-transcriptional regulatory mechanisms, as well as post-translationally via the ubiquitin-26S proteasome system (UPS). The UPS is highly conserved in eukaryotes and involves the covalent attachment of polyubiquitin chains to the target protein by an E3 ligase, followed by the degradation of the modified protein by the 26S proteasome. A large number of E3 ligases are encoded in the Arabidopsis genome, but only a few have been implicated in the regulation of cuticular wax deposition. In this study, we have conducted an E3 ligase reverse genetic screen and identified a novel RING-type E3 ubiquitin ligase, AtARRE, which negatively regulates wax biosynthesis in Arabidopsis. Arabidopsis plants overexpressing AtARRE exhibit glossy stems and siliques, reduced fertility and fusion between aerial organs. Wax load and wax compositional analyses of AtARRE overexpressors showed that the alkane-forming branch of the wax biosynthetic pathway is affected. Co-expression of AtARRE and candidate target proteins involved in alkane formation in both Nicotiana benthamiana and stable Arabidopsis transgenic lines demonstrated that AtARRE controls the levels of wax biosynthetic enzymes ECERIFERUM1 (CER1) and ECERIFERUM3 (CER3). CER1 has also been confirmed to be a ubiquitination substrate of the AtARRE E3 ligase by an in vivo ubiquitination assay using a reconstituted Escherichia coli system. The AtARRE gene is expressed throughout the plant, with the highest expression detected in fully expanded rosette leaves and oldest stem internodes. AtARRE gene expression can also be induced by exposure to pathogens. These findings reveal that wax biosynthesis in mature plant tissues and in response to pathogen infection is controlled post-translationally.
Introduction
The primary aerial surfaces of land plants are covered with a cuticle, a continuous lipidic layer that restricts transpirational water loss, reflects harmful UV light, and prevents organ fusions during development (Reicosky and Hanover, 1978; Sieber et al., 2000; Riederer and Schreiber, 2001; Riederer, 2006). The cuticle also serves as a protective barrier against pathogens and insects (Müller, 2018; Ziv et al., 2018) and is involved in drought-stress signaling (Wang et al., 2011).
The cuticle is mostly composed of cutin and cuticular wax (Samuels et al., 2008). Cutin is a polymer of oxidized 16- and 18-carbon (C16 and C18) fatty acids and glycerol (Beisson et al., 2012), which forms the structural scaffold of the cuticle. Cuticular wax embeds and overlays this cutin matrix and is composed of very long-chain fatty acids (VLCFAs; C20-C38) and their derivatives, including alkanes, aldehydes, primary and secondary alcohols, ketones, and esters. Small amounts of triterpenoids, flavonoids, or sterols may also be present (Jetter et al., 2006; Buschhaus and Jetter, 2011). Wax composition varies among plant species, as well as between different organs, tissues, and developmental stages of the same plant species. These variations in wax composition affect the biochemical and physical properties of the plant surface, which helps the plant adapt to different environments.
Cuticular wax is synthesized by epidermal cells. C16 and C18 fatty acids are made in the plastid and activated to acyl-CoA thioesters, which are translocated to the endoplasmic reticulum (ER) for further elongation to VLC acyl-CoA wax precursors by a fatty acid elongase (FAE) complex (Haslam and Kunst, 2013a). In addition, the ECERIFERUM2-LIKE (CER2-LIKE) family of proteins is required for the formation of C30 to C34 VLC acyl-CoAs (Haslam et al., 2017). Following elongation, VLC acyl-CoAs are modified by one of two pathways, either the acyl reduction pathway, which generates primary alcohols and wax esters, or the alkane-forming pathway, which produces aldehydes, alkanes, secondary alcohols, and ketones (Samuels et al., 2008). In Arabidopsis (Arabidopsis thaliana) leaves and stems, cuticular wax is predominantly derived from the alkane-forming pathway. As the major wax component, alkanes represent over 70 and 50% of the total wax load in leaves and stems, respectively (Bourdenx et al., 2011). It has been proposed that the formation of alkanes is catalyzed by a multiprotein complex comprising CER1, CER3, and a cytochrome B5 protein (CYTB5) that converts VLC acyl-CoAs to alkanes with aldehydes as intermediates (Rowland et al., 2007; Bourdenx et al., 2011; Bernard et al., 2012). CYTB5 isoforms interact with CER1 and provide the electron(s) required for this redox-dependent reaction. The CER1 and CER3 proteins are integral membrane proteins with 35% amino acid identity that contain eight conserved His clusters in their N-terminal domain and an uncharacterized WAX2 domain at their C-terminus (Aarts et al., 1995; Chen et al., 2003; Bernard et al., 2012). In Arabidopsis stems, alkanes can be further oxidized to secondary alcohols and ketones by a cytochrome P450 enzyme, the MID-CHAIN ALKANE HYDROXYLASE1 (MAH1; Greer et al., 2007).
Wax biosynthesis is tightly controlled throughout plant development and in response to biotic and abiotic stresses. Forward and reverse genetic studies in Arabidopsis, barley (Hordeum vulgare), maize (Zea mays), rice (Oryza sativa), and tomato (Solanum lycopersicum) have significantly improved our understanding of cuticular wax deposition and regulatory pathways controlling this process (Samuels et al., 2008; Yeats and Rose, 2013). Production of cuticular waxes is primarily under transcriptional regulation. Several independent studies have demonstrated that the WAX INDUCER1/SHINE1 (WIN1/SHN1) transcription factor, known to predominantly regulate cutin production, also indirectly affects wax synthesis (Aharoni et al., 2004; Broun et al., 2004; Kannangara et al., 2007). Other transcription factors, including MYB16, MYB30, MYB94, MYB96, MYB106, and WRINKLED4, have been reported to positively regulate wax synthesis in Arabidopsis stems and leaves (Raffaele et al., 2008; Seo et al., 2011; Oshima et al., 2013; Lee et al., 2014; Lee and Suh, 2015b; Park et al., 2016). Conversely, the DEWAX and DEWAX2 transcription factors act as repressors of wax production in Arabidopsis (Go et al., 2014; Kim et al., 2018).
In addition to the transcriptional regulation described above, characterization of the Arabidopsis CER7 gene and suppressors of the cer7 mutant resulted in the discovery of a post-transcriptional regulatory mechanism that affects stem wax deposition during inflorescence development. It involves CER7-mediated CER3 gene silencing by trans-acting small interfering RNAs (tasiRNAs; Hooker et al., 2007; Lam et al., 2012, 2015). Recently, another type of small RNAs, microRNAs (miRNAs), were also shown to participate in the regulation of wax synthesis. Specifically, miR156 targets the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 (SPL9) transcription factor that positively regulates the expression of the alkane-forming enzyme CER1 through direct binding to the CER1 promoter. Furthermore, SPL9 was shown to be involved in the optimization of diurnal wax production in Arabidopsis stems and leaves by direct protein–protein interaction with a negative regulator of wax synthesis DEWAX (Li et al., 2019).
Work by several groups demonstrated that wax biosynthesis in Arabidopsis is also post-translationally controlled by the ubiquitin-proteasome system (UPS). The UPS involves two distinct steps: the covalent attachment of a polyubiquitin chain consisting of at least four ubiquitin residues to the protein target, followed by the degradation of the modified protein by the 26S proteasome. Ubiquitination is catalyzed by three enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). Among these proteins, E3 ligases play key roles in determining substrate specificity (Hershko and Ciechanover, 1998; Vierstra, 2009). Several Arabidopsis E3 ligases have been shown to be involved in regulating cuticular wax deposition. Characterization of the wax-deficient cer9 mutant and isolation of the CER9 gene revealed that it encodes a putative E3 ligase, although its enzyme activity and ubiquitination substrate have not been determined (Lü et al., 2012). More recently, MYB30-INTERACTING E3 LIGASE 1 (MIEL1) has been shown to negatively regulate cuticular wax biosynthesis in Arabidopsis stems by targeting MYB30 and MYB96 transcription factors for degradation (Lee and Seo, 2016; Gil et al., 2017). Additionally, F-box protein SAGL1 targets wax biosynthetic enzyme CER3 for degradation thereby negatively regulating cuticular wax production in response to changes in ambient humidity (Kim et al., 2019). In rice, the DROUGHT HYPERSENSITIVE E3 ligase negatively regulates wax production by targeting the RICE OUTERMOST CELL-SPECIFIC GENE4 (ROC4) transcription factor involved in drought-stress response for degradation by the UPS (Wang et al., 2018b).
Based on the presence of over 1,400 putative E3 ligases encoded in the Arabidopsis genome (Kraft et al., 2005) and their importance in the regulation of plant responses to environmental stress, we reasoned that additional E3 ligases may be involved in the control of cuticular wax deposition. Here, we report the identification of a RING-type E3 ubiquitin ligase named ABA-related RING-type E3 ligase (AtARRE) that negatively regulates wax production by promoting the degradation of wax biosynthetic enzymes CER1 and CER3. This E3 ligase was previously reported to be involved in abscisic acid (ABA) signaling, but its ubiquitination target has not been identified (Wang et al., 2018a). Our results demonstrate that Arabidopsis plants overexpressing AtARRE display glossy stems and siliques, markedly reduced wax loads, and often aerial organ fusions and reduced fertility. Co-expression of AtARRE and candidate substrates in both Nicotiana benthamiana and stable Arabidopsis transgenic lines indicates that CER1 and CER3 wax biosynthetic enzymes are targeted by the AtARRE for degradation via the 26S proteasome. The AtARRE gene is highly expressed in tissues that exhibit no or low wax production, such as roots and cotyledons in older developing seedlings, as well as fully expanded rosette leaves and older internodes at the bottom of the stem in mature plants. AtARRE expression can also be induced by pathogen infection. Taken together, our results suggest that AtARRE acts as a quick and efficient switch for turning off wax biosynthesis in tissues where it is no longer needed and upon exposure to pathogens.
Materials and Methods
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) wild type was used in this study. Arabidopsis T-DNA insertion lines atarre-1 (SALK_094303), atarre-2 (SALK_034426C; Alonso et al., 2003), and the cer1-4 and cer4-4 mutants were obtained from the Arabidopsis Biological Resource Center (ABRC).1 GABI-KAT T-DNA line atarre-3 (GABI_383G01) was obtained from gabi-kat.de (Kleinboelting et al., 2012). cer3-6 was a gift from Dr. Takuji Wada (RIKEN, Japan). AtARRE overexpression lines in Col-0 background were identified from the snc1-influencing plant E3 ligase reverse (SNIPER) genetic screen (Tong et al., 2017).
Arabidopsis seeds were germinated on Arabidopsis thaliana (AT) medium (Haughn and Somerville, 1986) supplemented with 1% (w/v) agar and appropriate antibiotics for transgene selection. Seven-day-old seedlings were transplanted to soil (Sunshine Mix 4 or 5, SunGro, Canada) supplemented with liquid AT medium and grown in a plant growth chamber at 20°C under continuous light [100μmolm−2 s−1 of photosynthetically active radiation (PAR)]. Arabidopsis seeds grown for Agrobacterium-mediated transformation were directly spread on the soil supplemented with liquid AT medium at a density of 100 seeds/6″ pot and grown as described above.
Nicotiana benthamiana seeds were sown directly on soil (Sunshine Mix 4 or 5, SunGro, Canada) supplemented with liquid AT medium at a density of 1 seed/3.5″ square pot. Plants were grown under a 14-h light (25°C with 100μmolm−2 s−1 PAR) and 10-h dark (20°C) cycle. For the transient expression assay, 4- to 5-week-old plants were taken out of the growth chamber and left at room temperature for 3 to 4h before infiltration.
RNA Isolation, RT-PCR, and qPCR
Plant tissues were collected and immediately frozen in liquid nitrogen. RNA was extracted from Arabidopsis leaves, stems, flowers, seedlings, and roots using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s protocol. RNA was isolated from Arabidopsis siliques using a phenol:chloroform:isoamyl extraction and precipitated by lithium chloride and sodium acetate (Wilkins and Smart, 1996). RNA integrity was examined on a 1% standard agarose gel, and RNA was quantified using a NanoDrop 8000 (Thermo Scientific). Genomic DNA was removed by DNase I treatment (New England Biolabs) following the manufacturer’s protocol, and single-stranded cDNA was synthesized from equal amounts of purified RNA using iScript RT Supermix (Bio-Rad). ACTIN1 was used as an internal control. The iQ SYBR Green Supermix (Bio-Rad) was used in 20μl reactions to perform qPCR in an iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad) as specified by the manufacturer. Four technical replicates were performed for each sample, and gene expression levels were analyzed using the Pfaffl method (Pfaffl, 2001).
Cloning of Genes in Plant Expression Vectors
Standard methods were used for cloning, and all primer sequences are given in Supplementary Table 1. All constructs were confirmed by sequencing.
The pGreenST/35S:HA-AtARRE construct, which was prepared for the SNIPER screen (Tong et al., 2017), was used as the site-directed mutagenesis template. Of the five splice variants known for the AtARRE gene (AT5G66070), AT5G66070.1 was used for the work described here. The 35S:HA-AtARRE(H197Y,H200Y) site-directed mutagenesis construct was generated using primers H197200Y_F and H197200Y_R designed using the one-step site-directed mutagenesis method (Zheng et al., 2004). The PCR amplification was carried out using Phusion High Fidelity Polymerase (Thermo Fisher Scientific). The PCR products were separated by gel electrophoresis, purified using a PCR Purification Kit (BioBasic), and were further treated with restriction enzyme DpnI (New England Biolabs). The mutations in the 35S:HA-AtARRE(H197Y,H200Y) construct were confirmed by sequencing.
The 35S:GFP-CER3 construct was prepared using Gateway cloning (Thermo Fisher) and destination vectors from Nakagawa et al. (2007). The coding sequence of the CER3 gene was amplified from WT cDNA using CER3cDNA_attbF and CER3cDNA_attbR_WSTOP primers and recombined into the entry vector pDONR221. The insert was then transferred into the destination vector pGWB6 to generate pGWB6/35S:GFP-CER3 and into pGWB15 to generate pGWB15/35S:HA-CER3. The CER3 coding sequence without stop codon was also amplified using CER3cDNA_attbF and CER3cDNA_attbR_NoSTOP primers, recombined to pDONR221, and then transferred to the destination vector pGWB5 to generate pGWB5/35S:CER3-GFP.
The 35S:CER1-GFP construct was made and provided by Dr. Hugo Zheng (McGill University, Canada). The coding region of the CER1 gene was subcloned into the vector pVKH18/35S:GFPC (Dean et al., 2007) to produce the C-terminal CER1-GFP fusion under the control of the enhanced 35S promoter. The 35S promoter fragment was then removed from the vector pVKH18/35S:CER1-GFP and replaced with the CER6 promoter using HindIII and XbaI to generate pVKH18/CER6p:CER1-GFP. The construct pBIN/35S:HDEL-mCherry was provided by Dr. Mathias Schuetz (Nelson et al., 2007), and the construct pGreenST/35S:HA-SNIPER2 was described previously (Wu et al., 2020).
To generate the construct AtARREp:GUS, a fragment of 808bp immediately upstream of the putative AtARRE start codon, which includes the 5′ UTR of AtARRE, as well as 3′ UTR and the last intron of the previous gene, was amplified from WT genomic DNA using LP_attb1_AtARRE and RP1_attb2_AtARRE and recombined into pDONR221 before being introduced into pGWB3 (Nakagawa et al., 2007; Vincent et al., 2018) using GATEWAY cloning (Thermo Fisher).
Cloning of Genes in Bacterial Expression Vectors
Standard methods were used for cloning, and all primer sequences are given in Supplementary Table 1. All constructs were confirmed by sequencing.
To generate the construct pET28b/AtARRE-HIS for the in vitro ubiquitination assay, a 326bp fragment of coding sequence downstream of the transmembrane domains and upstream of the stop codon of AtARRE was amplified from WT cDNA using AtARRETMdel_F_EcoRI_28b and AtARRETMdel_R_SalI. This PCR product was ligated into the pET28b vector using EcoRI and SalI restriction sites to generate pET28b/AtARRE-HIS.
To reconstitute the plant ubiquitination cascade in Escherichia coli, Duet expression vectors (kindly provided by Dr. Dongping Lu, Chinese Academy of Science, China) pCDFDuet/MBP-ABI3-HA-AtUBA1-S, pCDFDuet/AtUBA1-S, pACYCDue/AIP2-Myc-UBC8-S, and pET28a/FLAG-UBQ were used to generate target co-expression constructs (Han et al., 2017). A 326bp fragment of coding sequence downstream of the transmembrane domains and upstream of the stop codon of AtARRE was amplified from WT cDNA and ligated into the BamHI and StuI-digested pACYCDue/AIP2-Myc-UBC8-S vector to generate pACYCDue/AtARRE-Myc-UBC8-S. An 837bp fragment of coding sequence downstream of the transmembrane domains and upstream of the stop codon of CER1 was amplified from WT cDNA was ligated into the EcoRI and StuI-digested pCDFDuet/MBP-ABI3-HA-AtUBA1-S vector to generate pCDFDuet/MBP-CER1-HA-AtUBA1-S.
Agrobacterium-Mediated Plant Transformation
To produce transgenic lines for E3 ubiquitin ligase activity test, degradation assay, and GUS assay, 35S:HA-AtARRE(H197Y,H200Y), CER6p:CER1-GFP, AtARREp:GUS, and AtARREp:AtARRE-GUS were introduced into Agrobacterium tumefaciens GV3101 cells carrying the pMP90 Ti plasmid. The pGreenST plasmid 35S:HA-AtARRE(H197Y,H200Y) was co-transformed with the helper plasmid pSOUP (Hellens et al., 2000). Transformation of WT or cer1-4 plants was carried out using the floral spray method (Chung et al., 2000). T1 transgenic seeds were harvested and screened on AT medium supplemented with 1% (w/v) agar and appropriate antibiotics.
Transient Expression in Nicotiana benthamiana
Transient expression in N. benthamiana was carried out using 4- to 5-week-old plants. Agrobacterium cultures were grown overnight in 3ml of LB medium under antibiotic selection and diluted 1/20 in LB medium with antibiotics and 50μm acetosyringone and incubated for a further 3–5h. During this time, plants were taken out of the growth chamber and left at room temperature before infiltration. Cultures were centrifuged and resuspended in resuspension medium (4.43g/L MS, 10mm MES, and 150μm acetosyringone) at an optical density of 0.6 at A600. For co-expression of multiple constructs, suspensions were mixed in equal ratios. Agrobacterium suspension mixtures were infiltrated using a 1-ml syringe into the abaxial side of the N. benthamiana leaves. A permanent marker was used to mark the infiltrated area on the leaf. Infiltrated plants were incubated at room temperature for 48h, and then, leaf samples were collected for microscopic imaging and/or protein extraction.
Cuticular Wax Extraction and Analysis by GC-FID
Cuticular wax extraction was performed using the method described by Haslam and Kunst (2013b). Briefly, the top 10cm of 4- to 6-week-old inflorescence stems were cut and photographed to allow stem surface area to be calculated by measuring the number of pixels of the two-dimensional area in Photoshop (Adobe), converting the values to cm2, and multiplying by π. After imaging, stems were submerged for 30s in chloroform containing 10μg tetracosane as an internal standard. After wax extraction, chloroform was evaporated under a stream of nitrogen gas and wax components were silylated in 10μlN, O-Bis (trimethylsilyl) trifluoroacetamide (BSTFA; Sigma), and 10μl pyridine for 1h at 80°C. After derivatization, the solvent was evaporated under nitrogen gas and waxes were re-dissolved in 30μl of chloroform for GC analysis. Samples were analyzed on an Agilent 7890A gas chromatograph equipped with a flame ionization detector (GC-FID) using an HP1 column (Agilent) in a 2.7:1 split mode with H2 as the carrier gas at a flow rate of 30ml/min. The gas chromatography program used was as follows: oven temperature was set at 50°C for 2min, raised by 40°C/min to 200°C and held for 1min, and then raised by 3°C/min to 320°C and held for 15min. Wax components were identified by comparing their retention times with those of the internal standards. Four biological replicates were processed for each line.
Microscopy
Fluorescence signals of transiently expressed constructs in N. benthamiana were detected using a Perkin Elmer Ultraview VoX Spinning Disk Confocal Microscope. N. benthamiana leaf discs were mounted in distilled water and immediately imaged using a glycerol immersion lens. GFP was excited using a 488nm laser with an 515/30nm emission filter, and mCherry was excited using a 561nm laser with an 595/50nm emission filter. Confocal images were processed using the Volocity software (Perkin Elmer). GFP signal in infiltrated N. benthamiana leaf was also observed using a Nikon Eclipse 80i Scanning Laser Confocal Microscope excited with a 488nm laser with an 515/30nm emission filter.
For scanning electron microscopy (SEM), segments from the apical 1cm of dry stems were mounted onto stubs and sputter-coated with gold particles for 10min at 40mA in an SEM Prep 2 sputter coater (Nanotech). The coated samples were viewed using an S4700 field emission SEM (Hitachi) with an accelerating voltage of 5kV and a working distance of 12mm.
GUS Histochemical Assay
Tissues at different developmental stages from transgenic lines expressing AtARREp:GUS constructs were immersed in GUS staining buffer {100mM Na-phosphate, 10mM EDTA, pH 7.0, 0.5mM K3[Fe(CN)6], 0.5mM K4[Fe(CN)6], 0.1% (v/v) Triton X-100, and 1mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-gluc)} and incubated for 1 to 3h or overnight. The reaction was stopped by removing the GUS buffer and adding the 70% (v/v) ethanol. Chlorophyll was removed by incubating samples in 70–90% (v/v) ethanol before samples were examined under a Nikon SMZ18 Digital Microscope (Nikon, Japan).
Protein Extraction and Immunoblotting
Plant tissues were ground in liquid nitrogen, and total proteins were extracted in buffer containing 50mM Tris-HCl, pH 7.5, 150mM NaCl, 1mM EDTA, 10% (v/v) glycerol, 1% Triton X-100, 1mM PMSF, and 1X Halt™ protease inhibitor cocktail (Thermo Fisher Scientific). After centrifugation at 18,000g for 20min at 4°C, the supernatant was transferred to a new tube and the concentration of protein extract was determined using the Bradford reagent (Bio-Rad).
For SDS-polyacrylamide gel electrophoresis (SDS-PAGE), 4X SDS loading buffer (200mM Tris-HCl, pH 6.8, 8% (w/v) SDS, 0.4% (w/v) bromophenol blue, 40% glycerol, and 400mM DTT) was added to solubilized protein samples, and 10–35μl of each protein sample was separated on a 10% acrylamide gel with 1% SDS at 200V constant voltage for 50–60min before being transferred to nitrocellulose membrane using a semi-dry blotting system (Bio-Rad) with Bjerrum Schafer-Nielsen buffer (Bio-Rad). Transfer was carried out at a constant voltage of 15V for 50min before the membrane was stained with Ponceau S, imaged, washed, and then blocked with 5% skim milk powder in Tris-buffered saline with 0.1% tween 20 (TBS-T). For immunoblotting, membranes were incubated with primary antibody for 1h at room temperature. Primary antibodies used were anti-GFP (dilution 1:5,000; mouse IgG; Roche), anti-HA (dilution 1:2,500; rat IgG; Roche), anti-HIS (dilution 1:1,000; mouse IgG; Santa Cruz Biotechnology), anti-FLAG (dilution 1:5,000; mouse IgG; Sigma), anti-GST (dilution 1:1,000; rabbit IgG; Sigma), anti-Myc (dilution 1:1,000; Invitrogen), and anti-Ub (dilution 1:1,000; mouse IgG; Sigma). Membranes were then washed three times for 10min each wash with TBS-T and then incubated with appropriate secondary antibodies, including anti-rabbit (dilution 1:10,000; Santa Cruz Biotechnology), anti-mouse (dilution 1:25,000; Santa Cruz Biotechnology), and anti-rat (dilution 1:10,000; Santa Cruz Biotechnology), for 1h at room temperature. The membrane was washed three times as above with TBS-T before horseradish peroxidase was detected with the ECL Prime western blotting detection kit (GE).
Cell-Free Degradation Assay
Plant-derived protein degradation assays were performed as described in Wang et al. (2009), with modifications as follows. Total proteins were extracted from 8-day-old CER6pro:CER1-GFP/cer1-4 transgenic seedlings and quantified. 80μl protein extracts were then incubated with or without 40μM MG132 (Sigma) at 30°C. Samples were taken at select time points, and the reaction was stopped by adding 5μl 4X SDS loading buffer. CER1-GFP protein abundance in each sample was determined by immunoblotting using anti-GFP antibody.
In vitro Ubiquitination Assay
In vitro ubiquitination assays were performed as described in Zhao et al. (2013), with modifications as follows. The plasmid pET28b/AtARRE-HIS was transformed into E. coli strain Rosetta™2 (DE3) for protein production. A 500ml culture was grown in Terrific Broth (TB) medium [1.2% (w/v) tryptone, 2.4% (w/v) yeast extract, 0.4% (v/v) glycerol, 100mM K-PO4] until the exponential phase (OD600 =0.6–1.0) before protein production was induced by adding 0.5mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). After growth overnight at 16–18°C, cells were collected by centrifugation at 6,000g for 5min and frozen in liquid nitrogen and stored at −80°C. Lysis buffer [50mM NaPO4, pH 7.5, 200mM NaCl, 0.1% (v/v) Triton X, 5% glycerol, 1mM PMSF, 1X Halt™ protease inhibitor cocktail, and 1mg/ml lysozyme] was added to the frozen sample pellets, thawed at 37°C for 1min, and resuspended. Lysate was cleared by centrifugation at 14,000g for 15min at 4°C followed by filtration through a 0.45 μm filter. The AtARRE-HIS recombinant proteins were purified using HisPur Ni-NTA Resin (Thermo Fisher) according to the manufacturer’s protocol. Purified recombinant proteins AtUBA2-His and GST-AtUBC8 were kindly provided by Dr. Oliver Xiao'ou Dong (Dong et al. 2018).
In vivo Ubiquitination Assay in Bacteria
In vivo ubiquitination assays in bacteria were carried out using the system described by Han et al. (2017). E. coli strain BL21 (DE3) containing different combinations of the expression vectors were grown in 2ml of TB liquid medium with appropriate antibiotics at 37°C. When the culture A600 nm reached 0.4–0.6, 0.5mM IPTG was added to induce the recombinant protein expression. After induction, bacteria were further grown at 28°C for 10–12h, stored at 4°C overnight, and then harvested from 300μl of culture by centrifugation at 12,000g for 5min. The pellets were resuspended in 100μl 1x SDS loading buffer and boiled at 95°C for 5min followed by immunoblotting.
Results
Overexpression of AtARRE Results in Reduced Wax Accumulation on Arabidopsis Stems and Leaves
To identify novel E3 ligases involved in plant immunity, a SNIPER genetic screen has been carried out (Tong et al., 2017). In this screen, E3-ligase encoding genes induced during plant defense were overexpressed in the wild-type background. Unexpectedly, a number of independent transgenic plants with glossy bright green stems were uncovered among the T1 progeny, suggestive of altered cuticular wax accumulation (Figure 1A). In these plants, the AtARRE/At5g66070 gene encoding a RING-type E3 ubiquitin ligase was expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Supplementary Figures S1A,B). Wax analysis of three representative AtARRE overexpression (AtARREOX) lines by gas chromatography demonstrated that they accumulated only 10–50% of the WT inflorescence stem wax and only ~65% of the WT leaf wax (Figures 1B,C). The stem wax phenotype was further evaluated by scanning electron microscopy (SEM); wild-type stem surface was densely and uniformly covered with column-, vertical plate-, and rod-shaped wax crystals, whereas AtARREOX lines displayed considerably lower density of all types of wax structures (Figure 1D).
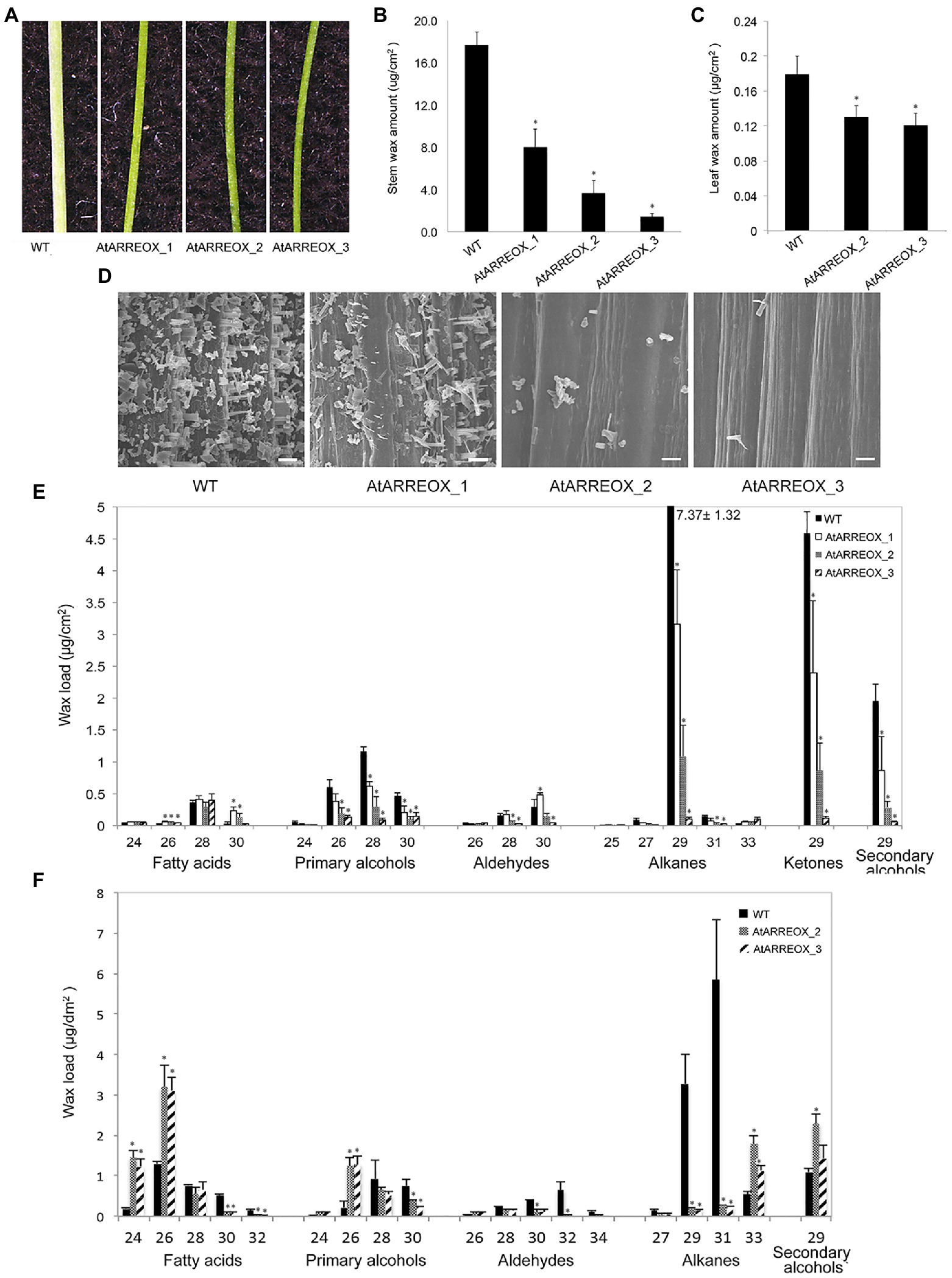
Figure 1. Characterization of the wax-deficient phenotypes of AtARREOX lines. (A) Stems of 6-week-old wild type (WT) and three representative AtARREOX lines. Total stem (B) and leaf (C) wax load of WT and AtARREOX lines as determined by GC-FID. Error bars represent means±SD (n=4). (D) SEM images of WT and AtARREOX inflorescence stem surfaces. Scale bar=20μm. The stem (E) and rosette leaf (F) wax composition of WT and AtARREOX lines as measured by GC-FID. Error bars represent means±SD (n=4). Statistically significant differences between the WT and each AtARREOX line were determined by Student’s t test and are indicated by asterisks (p<0.05).
The wax deficiency uncovered in the AtARREOX lines prompted us to examine the wax phenotypes of atarre mutants. We obtained three T-DNA insertion lines of AtARRE in the Col-0 ecotype and determined AtARRE gene expression in mutant alleles by qPCR. Even though we detected reduced levels of AtARRE transcript in all three T-DNA lines (Supplementary Figures S1A,B), we found no major differences in the total stem wax load or composition with respect to the wild type (Supplementary Figures S1C,D).
AtARREOX Lines Exhibit Altered Wax Composition and Abnormal Organ Morphogenesis
To further investigate the role of AtARRE in cuticular wax biosynthesis, we carried out a detailed analysis of wax composition of AtARREOX lines. We found that amounts of all stem wax components were altered in AtARREOX plants in comparison with the wild type, and detected considerable changes in their relative proportions. In particular, there was a prominent decrease in absolute amounts of alkane pathway-derived compounds that could be attributed primarily to C29 alkanes (68–98.7% decrease), C29 ketones (50–97.6% decrease), and C29 secondary alcohols (56–97% decrease). Conversely, the amounts of fatty acids on AtARREOX stems were higher than in the wild type (115–167% increase) and so was the relative proportion of fatty acids, aldehydes, and primary alcohols (Figure 1E, Supplementary Figure S1E). We also examined the wax composition of AtARREOX rosette leaves. As observed with stem wax, leaf wax also contained significantly reduced amounts of C29 and C31 alkanes relative to the wild type, but also increased amounts of C33 alkanes (Figure 1F). In addition, we detected a major increase in C24 and C26 fatty acids, as well as C26 primary alcohols (Figure 1F). Collectively, these data suggest that wax production by the alkane-forming pathway is impaired in AtARRE overexpressors.
AtARREOX lines with the most severe wax deficiency displayed additional phenotypes including abnormal organ morphogenesis, dwarfism, organ fusions, and reduced fertility over multiple generations (Figure 2). For example, the inflorescence stems of AtARRE overexpressors were considerably shorter than the wild type. Additionally, these plants also exhibited organ fusions between flower buds, flowers, and siliques, as well as between flowers and leaves (Figures 2C,D). Reduced fertility was also often detected. In most cases, fertility could be restored by growing plants under high humidity, except in individuals with severe floral organ fusions (Supplementary Figure S2). Similar phenotypes have previously been reported for several Arabidopsis wax-deficient eceriferum mutants (Koornneef et al., 1989).

Figure 2. Abnormal organ morphogenesis in the AtARREOX lines. (A) Dry stems of WT (left) and a T3 AtARREOX_2 individual (right). (B) 6-week-old WT plant (left) and a T3 AtARREOX_3 plant (right). (C) Flowers of WT (Col-0). (D) Organ fusions between flower buds of a T3 AtARREOX_3 individual. Scale bars=2cm.
AtARRE E3 Ubiquitin Ligase Activity Is Required for Its Function in Wax Deposition
AtARRE (AT5G66070) encodes a predicted polypeptide of 221 amino acids with a molecular mass of 27kDa containing three transmembrane domains located at the N-terminus and a RING domain located at the C-terminus (Figure 3A). RING domain proteins act as E3 ligases by binding to an E2-ubiquitin thioester and catalyzing ubiquitin transfer (Deshaies and Joazeiro, 2009). Whether the RING domain-related E3 ubiquitin ligase activity of the AtARRE protein is required for its function in cuticular wax metabolism is not known. RING domain E3 ligases are known to undergo self-ubiquitination in the absence of their native substrate (Lorick et al., 1999). We used this feature of RING E3 ligases to determine whether AtARRE has E3 ligase activity. Unfortunately, the insolubility of the full-length AtARRE protein upon expression in E. coli prohibited purification of enough protein for the self-ubiquitination assay. Therefore, an AtARRE protein fragment without the N-terminal transmembrane domains was used to produce the recombinant AtARRE-HIS protein. Incubation of the purified AtARRE-HIS recombinant protein with E1, E2, ubiquitin, and ATP resulted in a laddering pattern characteristic of ubiquitination on a protein blot when anti-HIS antibodies were used for AtARRE-HIS detection. This laddering is indicative of a range of molecular weights for AtARRE-HIS as it carries ubiquitin chains of different lengths (Figure 3B). Such a laddering pattern was also detected when anti-Flag antibodies that were used for FLAG-Ub detection of Ub chains bound to AtARRE-HIS. Thus, AtARRE exhibits E3 ligase activity and undergoes self-ubiquitination in vitro. The self-ubiquitination of AtARRE was not observed when E1, E2, or ubiquitin were omitted from the assays.
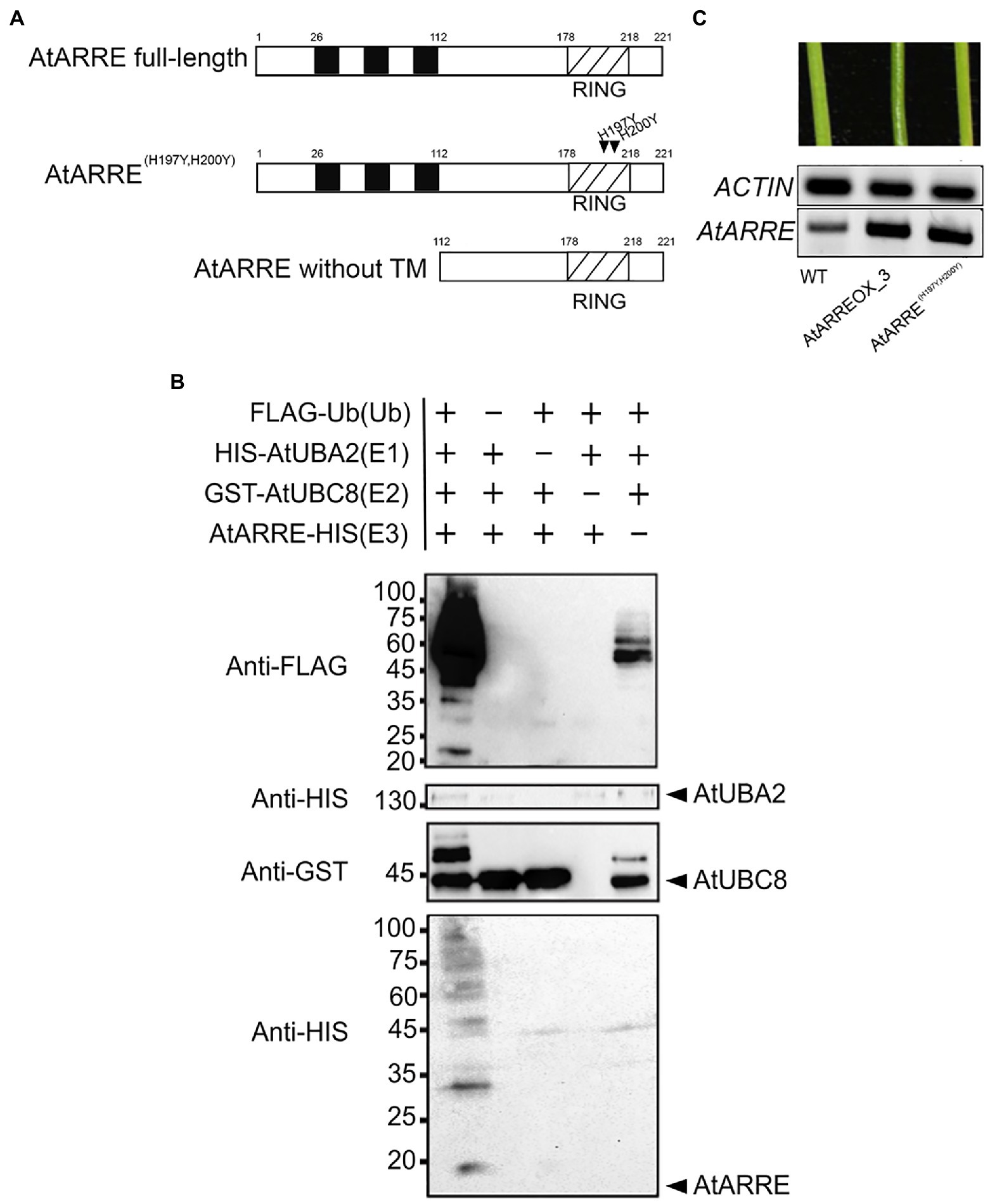
Figure 3. AtARRE exhibits E3 ubiquitin ligase activity. (A) Schematic representations of the full-length AtARRE protein (top), AtARRE protein with mutated RING domain (middle) and truncated AtARRE protein without transmembrane domains used for self-ubiquitination assay (bottom). Arrows show the point mutations in the RING domain. (B) In vitro ubiquitination assays were performed in the presence (+) or absence (−) of AtUBA2 (E1; 140kDa), AtUBC8 (E2; 44kDa), AtARRE (E3; 14.4kDa), and ubiquitin (Ub; 9.5kDa). Ubiquitination of AtARRE was detected by immunoblotting using an anti-FLAG antibody or anti-HIS antibody. AtUBC8 and AtUBA2 were detected by immunoblotting using anti-GST and anti-HIS antibody, respectively. Molecular mass markers are indicated on the left. (C) Stems of 6-week-old WT, AtARREOX_3 (T3 generation), and representative AtARRE(H197YH200Y). AtARRE transcript accumulation in each sample was measured by RT-PCR. ACTIN was used as an internal control.
Conserved Cys and His residues in the RING domain are critical for the E3 ligase activity (Deshaies and Joazeiro, 2009). Their substitution disrupts the RING domain and results in a dominant-negative form of E3 ligase predicted to confer the same phenotype as the loss of E3 ligase function. To further verify whether AtARRE E3 ligase activity is required for its function, we replaced the conserved AtARRE RING domain His-197 and His-200 with Tyr residues by site-directed mutagenesis and expressed the modified protein in wild-type plants. In contrast to AtARRE overexpression which caused wax deficiency, overexpression of the double mutant AtARRE(H197YH200Y) protein had no effect on the stem wax load (Figure 3C). Thus, the E3 ligase activity of AtARRE is required for its function in stem wax deposition.
AtARRE Overexpression Phenotypes Mimic cer1 and cer3 Wax-Deficient Mutants
E3 ligase-mediated ubiquitination of proteins in most cases results in their degradation by the 26S proteasome. Because the most conspicuous result of AtARRE overexpression was reduced cuticular wax accumulation on Arabidopsis inflorescence stems, we hypothesized that AtARRE may act as a negative regulator of wax deposition by ubiquitinating, and thus targeting for degradation, a key player involved in wax biosynthesis. If this is the case, identifying the ubiquitination substrate of AtARRE is critical for determining its biological function. As a first step in uncovering potential candidate ubiquitination substrates, we compared stem wax compositional changes of AtARRE overexpressors with those of ecerifierum (cer) Arabidopsis mutants caused by loss-of-function mutations in wax biosynthetic genes (Figure 4, Supplementary Figure S3). AtARRE overexpressors displayed dramatically reduced alkane, secondary alcohols, and ketone levels on their stem surfaces, similar to null mutants disrupted in CER1 and CER3 genes required for the production of waxes by the alkane-forming branch of wax biosynthesis, but not the mutants with lesions in the CER4 gene required for the production of waxes by the acyl reduction pathway. The distinguishing feature between cer1 and cer3 is that cer1 has slightly increased amounts of aldehydes and reduced primary alcohol levels, whereas cer3 exhibits a major reduction in aldehydes and similar amounts of primary alcohols to the wild type. Thus, the wax profile of AtARREOX lines is most similar to that of the cer1 mutant. Besides cuticular wax changes, some AtARREOX lines additionally display reduced plant height and reduced fertility previously described for the cer1-1 and cer3-1 mutant alleles (Koornneef et al., 1989), and organ fusion phenotypes characteristic of cer3, but not cer1 mutants (Aarts et al., 1995; Chen et al., 2003; Bourdenx et al., 2011).
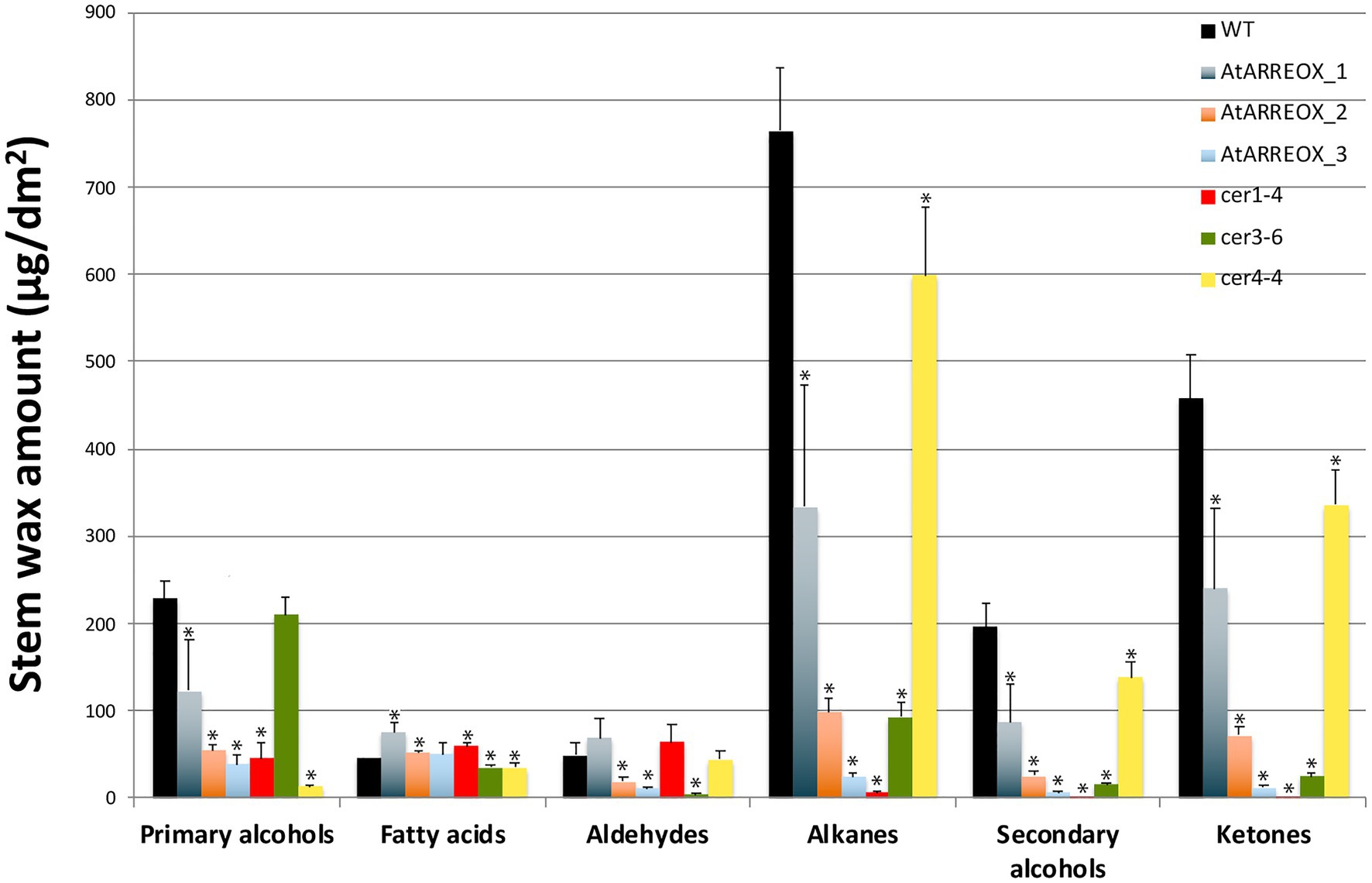
Figure 4. A comparison of stem wax load and composition of AtARRE overexpression lines, and cer1-4, cer3-6, and cer4-4 mutants. Stem wax load and composition of 6-week-old WT, AtARREOX lines, cer1-4, cer3-6, and cer4-4 were determined by GC-FID. Values are means of four biological replicates, and error bars represent SD. Statistically significant differences of wax component amounts between the WT and different genotypes (p<0.05) were determined by Student’s t test and are indicated by asterisks.
AtARRE Promotes CER1 Degradation by the 26S Proteasome
To determine whether CER1 is subjected to 26S proteasome-dependent degradation, we performed a modified cell-free degradation assay. For this purpose, we made CER6pro:CER1-GFP/cer1-4 transgenic lines in which the CER6pro:CER1-GFP transgene complemented the cer1-4 wax deficiency (Supplementary Figure S4A). Total proteins extracted from 8-day-old CER6pro:CER1-GFP/cer1-4 seedlings were incubated with or without the 26S proteasome inhibitor MG132 for 90min and protein levels determined by immunoblotting. The CER1-GFP amounts decreased rapidly in the absence of MG132, but in the presence of MG132, the levels of CER1-GFP remained notably higher over time, suggesting that the 26S proteasome is involved in CER1 proteolysis (Figure 5A).
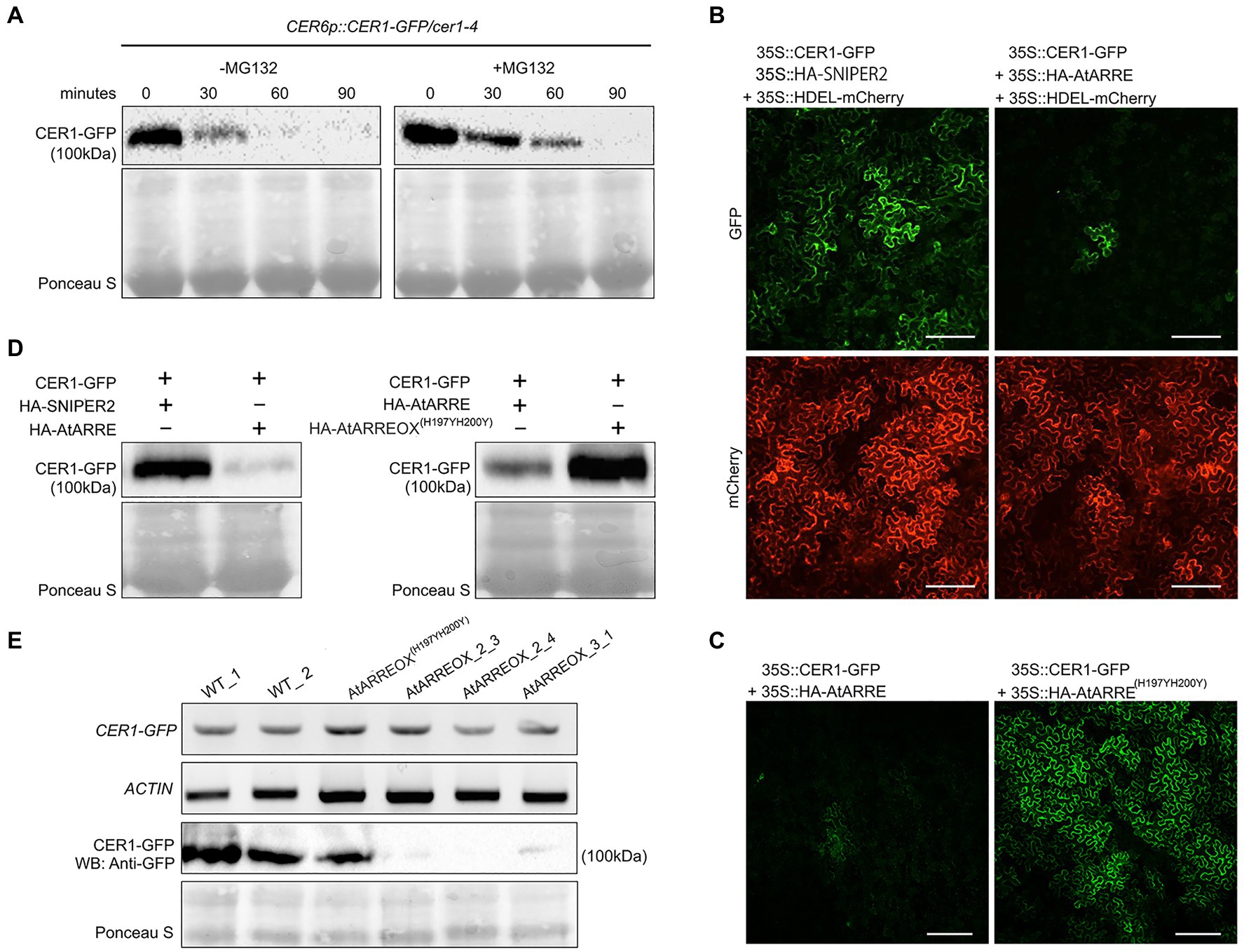
Figure 5. AtARRE promotes CER1 degradation in the Nicotiana benthamiana transient expression system and in stable transgenic lines of Arabidopsis. (A) CER1 is turned over by the 26S proteasome in planta. Total proteins were extracted from 8-day-old CER6pro:CER1-GFP/cer1-4 transgenic plants and incubated with (+) or without (−) 40μM MG132. CER1-GFP protein levels were detected over time by immunoblotting using anti-GFP antibody. (B,C) Transient expression in N. benthamiana leaf epidermal cells. CER1-GFP and an internal control HDEL-mCherry were co-expressed with HA-SNIPER2 E3 ligase or HA-AtARRE E3 ligase. CER1-GFP was co-expressed with HA-AtARRE or HA-AtARRE(H197YH200Y) with mutated RING domain. GFP fluorescence and mCherry fluorescence were examined by confocal microscopy. Scale bars=100μm. 4 technical replicates and 16 biological replicates have been performed. (D) Amounts of CER1-GFP protein were determined by immunoblotting using anti-GFP in the same leaves assayed for fluorescence in (B,C). (E) The AtARRE-dependent CER1 degradation in stable transgenic lines of Arabidopsis. RNA was extracted from 4-week-old Arabidopsis leaves. The CER1-GFP steady-state transcript levels were determined by RT-PCR, and ACTIN was used as an internal control (top two rows). Total protein was extracted from the 4-week-old plant leaves. The CER1-GFP protein level was determined by immunoblotting using anti-GFP antibody; Ponceau S staining shows equal protein loading (bottom two rows).
We next carried out an Agrobacterium-mediated transient expression assay in N. benthamiana leaves. Agrobacterium cell cultures expressing 35Spro:CER1-GFP and 35Spro:HA-AtARRE were co-infiltrated on one half of a N. benthamiana leaf. Cell cultures expressing 35Spro:CER1-GFP and 35Spro:HA empty vector or 35Spro:CER1-GFP and the 35pro:HA-SNIPER2 vector containing the SNIPER2 E3 ligase gene not involved in wax deposition were co-infiltrated symmetrically on the other half of the same leaf as negative controls (Supplementary Figure S4B). 35Spro:HDEL-mCherry was also included in each sample as a marker for ER visualization. In N. benthamiana cells transformed with CER1-GFP and 35Spro:HA empty vector, the GFP signal could be easily detected after 48h, persisted past 72h and was undetectable 96h after infiltration (Supplementary Figure S4C). Bright GFP fluorescence was also detected in cells co-expressing CER1-GFP and the SNIPER2 E3 ligase control 3days post-infiltration, but not in those cells co-expressing CER1-GFP and AtARRE (Figure 5B). In contrast, similar intensity of mCherry fluorescence from the ER-localized HDEL-mCherry marker was detected in all infiltrated regions on both sides of the leaf (Figure 5B). Immunoblot analysis confirmed that CER1-GFP protein level was much lower in the presence of AtARRE than in the negative control sample expressing SNIPER2 E3 ligase (Figure 5D). Unlike the native AtARRE protein, co-expression of CER1-GFP and the AtARRE(H197YH200Y) protein with mutated RING domain did not affect CER1 protein levels, indicating that catalytic activity of AtARRE is required for CER1 degradation (Figures 5C,D). When the substrate CER1 was replaced with CER2, a component of VLCFAs elongation machinery (Haslam et al., 2012), GFP tagged CER2 fluorescence signal intensity was found to be indistinguishable in the presence and absence of AtARRE. Immunoblot results were consistent with microscopy data (Supplementary Figures S4D,E) and demonstrate that AtARRE specifically targets CER1 for degradation by the 26S proteasome.
The AtARRE-dependent degradation of CER1 was further verified in stable transgenic lines of Arabidopsis. Plants harboring the CER1-GFP transgene were crossed with the wild type, AtARREOX, and AtARREOX(H197YH200Y) lines, and the abundance of the CER1-GFP in F1 progeny was examined by immunoblotting. Whereas CER1-GFP transcript accumulation was similar in the F1 progeny from all the crosses, the CER1-GFP protein level in the AtARREOX lines was much lower than observed in the wild type and AtARREOX(H197YH200Y) lines (Figure 5E). Collectively, these results confirm that AtARRE promotes the degradation of CER1, suggesting that CER1 is the ubiquitination substrate of AtARRE.
CER1 Is Ubiquitinated by AtARRE in a Reconstituted E. coli System
To directly test whether CER1 is a ubiquitination substrate of the AtARRE E3 ligase, we performed an in vivo ubiquitination assay in a heterologous E. coli system that expresses the Arabidopsis ubiquitination cascade (Han et al., 2017). In this experiment, recombinant ubiquitination components E1 (AtUBA1-S), E2 (AtUBC8-S), E3 (AtARRE-Myc), ubiquitin (His-Flag-AtUBQ10), and the presumed ubiquitination substrate (MBP-CER1-HA) were co-expressed in E. coli (Figure 6A), and bacterial lysates were analyzed by immunoblotting. Our results show that in the presence of all ubiquitination components, a smear indicative of CER1 ubiquitination can be detected by an anti-HA antibody (Figure 6B). Using an anti-Myc antibody, AtARRE-Myc recombinant protein also shows a laddering pattern indicative of AtARRE self-ubiquitination. These data support the in vitro ubiquitination assay results that AtARRE is an active E3 ligase and demonstrate that it can ubiquitinate CER1 in vivo.
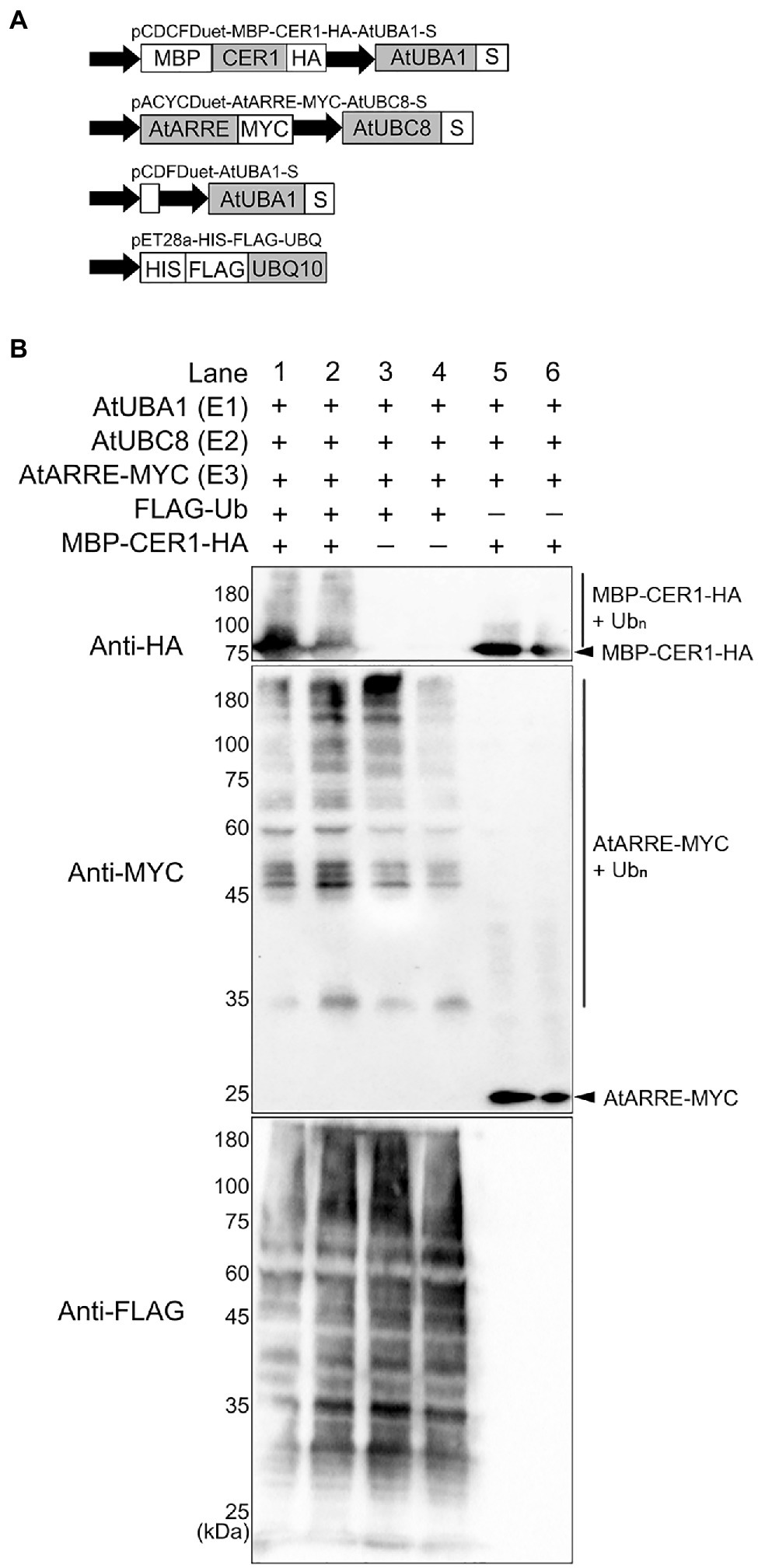
Figure 6. Ubiquitination of CER1 by AtARRE in a heterologous Escherichia coli system. (A) Schematic representation of the plasmids used in the assay. MBP, maltose-binding protein; HA, hemagglutinin. These constructs were transformed into the E. coli Rosetta (DE3) strain to reconstitute the ubiquitination cascade. (B) Bacterial lysates from E. coli strains expressing (+) or missing (−) combinations of AtUBA1-S (E1), AtUBC8-S (E2), AtARRE-MYC (E3), His-FLAG-UBQ10 (Ub), and MBP-CER1-HA (substrate) (+), and strains missing Ub or CER1 (−) were analyzed by immunoblotting. Anti-HA and anti-MYC antibodies were used to detect ubiquitinated CER1 and self-ubiquitinated AtARRE, respectively. Anti-FLAG antibody was used to detect all Ub conjugates. Two replicates for each combination of constructs are shown.
AtARRE Promotes CER3-GFP Degradation
Even though stem wax profiles of AtARRE overexpressors are more similar to the cer1 than the cer3 mutant, the overexpression lines also exhibit organ fusions previously detected in cer3, but not in cer1 mutants. Because CER1 and CER3 proteins are highly related and share 35% sequence identity (Bernard et al., 2012), it is tempting to speculate that in addition to CER1, CER3 may also be a target of the AtARRE-mediated ubiquitination and UPS proteolysis resulting in the dual cer1- and cer3-like phenotypic features of AtARRE overexpression lines. To investigate whether this is the case, we tested whether AtARRE affects the protein levels of CER3-GFP in N. benthamiana leaf epidermal cells. Agrobacterium cells expressing 35Spro:CER3-GFP, the 35Spro:HA-AtARRE, and the 35Spro:HDEL-mCherry transgenes were co-infiltrated on one half of a N. benthamiana leaf, while Agrobacterium cells harboring 35Spro:CER3-GFP, 35Spro:HA-SNIPER2, and 35Spro:HDEL-mCherry were co-infiltrated on the other side of the same leaf as a negative control. Similar to CER1-GFP, AtARRE promoted CER3-GFP degradation as indicated by considerably reduced CER3-GFP fluorescence in the presence of transiently co-expressed AtARRE in comparison with the control co-expressing CER3-GFP with HA-SNIPER2 E3 ligase. By contrast, the signal of the internal control HDEL-mCherry was prominent and indistinguishable on both halves of the leaf (Figure 7). These results demonstrate that CER3 is likely also the AtARRE ubiquitination target. We tried to confirm that AtARRE controls the levels of CER3 in transgenic lines of Arabidopsis, but consistently failed to detect CER3-GFP protein on immunoblots.
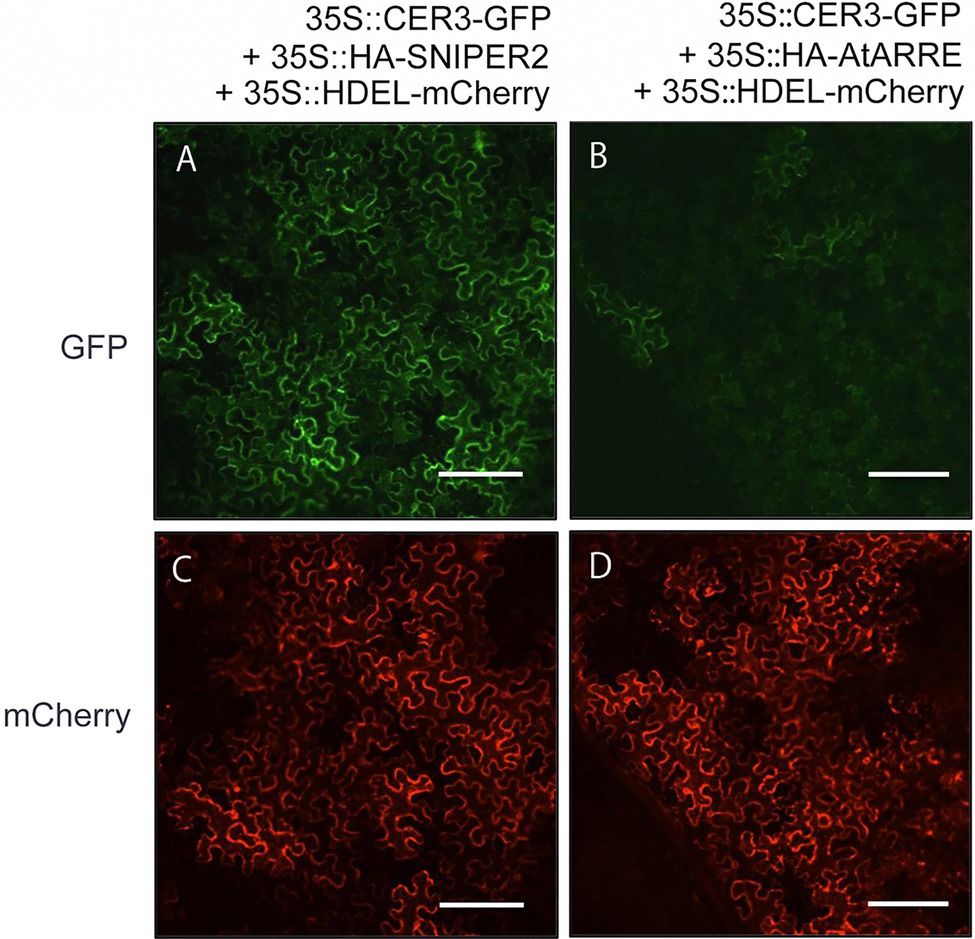
Figure 7. AtARRE promotes CER3 degradation in the N. benthamiana transient expression system. (A,C) 35Spro:CER3-GFP was co-expressed with 35Spro:HA-SNIPER2 or (B,D) 35Spro:HA-AtARRE. 35Spro:HDEL-mCherry was included as an internal control and ER marker. GFP fluorescence (A,B) and mCherry fluorescence (C,D) signals of leaf epidermal cells were examined by confocal microscopy 72h post-infiltration. 12 biological replicates were performed. Scale bars=100μm.
AtARRE Gene Is Expressed in Tissues That Exhibit Low Wax Production and Upon Exposure to Pathogens
To obtain clues regarding the functional significance of the AtARRE-mediated protein degradation of wax biosynthetic enzymes, we investigated the transcription profile of AtARRE gene in different plant tissues using qRT-PCR (Figure 8A). The AtARRE gene was expressed in all tissues examined, with higher expression in rosette leaves and lower expression in flowers and siliques. To further determine the AtARRE expression pattern in different cell types and at different Arabidopsis developmental stages, an 808bp fragment of genomic sequence immediately upstream of the AtARRE coding region was fused to the β-glucuronidase (GUS) reporter gene, and this AtARREpro:GUS reporter was transformed into wild-type Arabidopsis. GUS activity was examined in tissue samples from mature plants and in developing seedlings of ten independent transgenic lines.
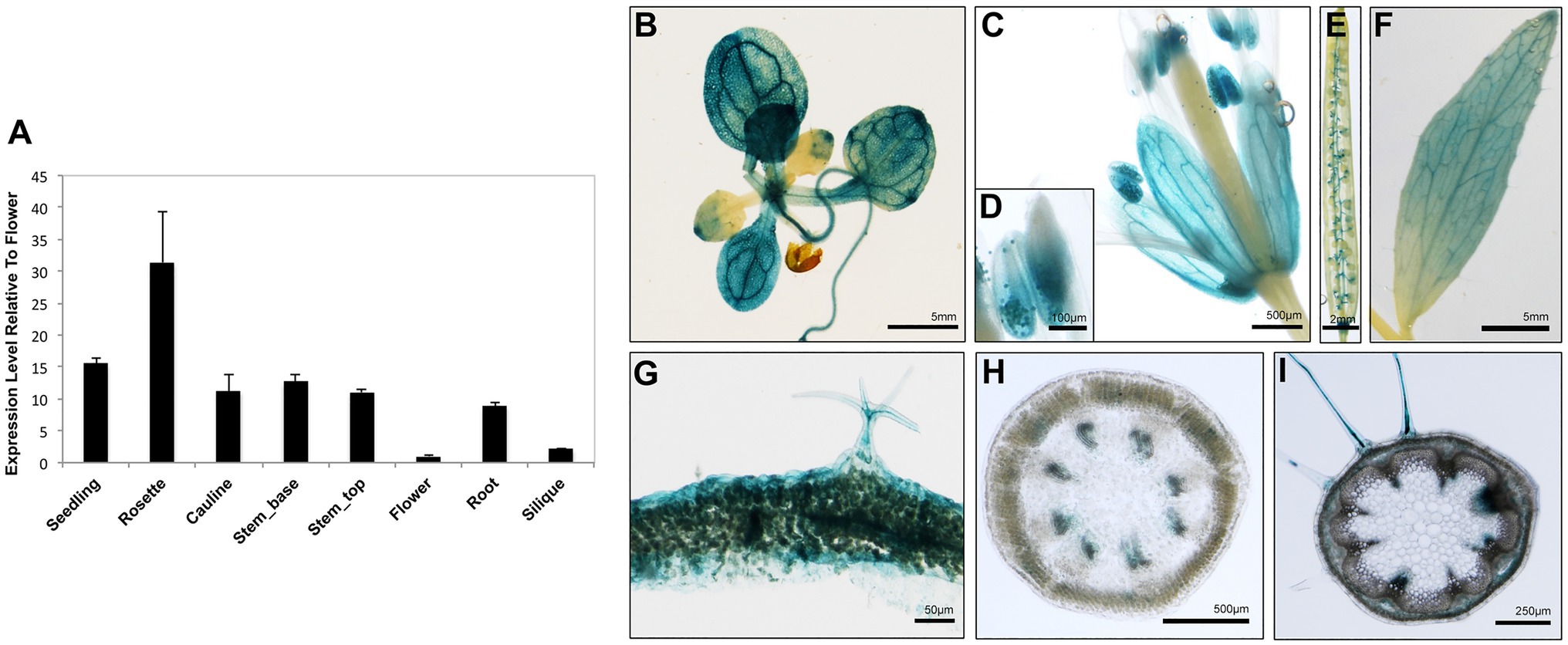
Figure 8. Expression pattern of AtARRE gene in different plant tissues and cell types. (A) qRT-PCR analysis of AtARRE gene expression in the WT relative to flowers and normalized to ACTIN. Seedling and root tissue samples were derived from 10-day-old WT seedlings, and all other aerial tissues samples were derived from 6-week-old WT. Error bars represent means±SD (n=4). (B–I) Putative AtARRE promoter activity determined by GUS assay in transgenic lines expressing the AtARREpro:GUS reporter. Promoter activity was examined after GUS staining in 14-day-old seedlings (B), flowers (C), anthers containing pollen grains (D), siliques (E), cauline leaves (F), cross section of rosette leaf from a 6-week-old plant (G), cross section of the top 3cm (H) and the bottom 3cm (I) of the inflorescence stem of a 6-week-old plant.
In mature plants, high GUS activity was detected in the fully expanded rosette leaves, sepals, pollen grains, and cauline leaves (Figures 8B–F). These results are consistent with the published RNA-seq data showing high AtARRE expression in mature leaves and sepals (Klepikova et al., 2016). Strong GUS signal was also detected in, but not specific to, the epidermal cells of rosette leaves, as well as trichomes, which are specialized epidermal cells (Figure 8G). Surprisingly, no expression of AtARRE was detected in the epidermal cell layer of the stem (Figures 8H,I). This may be due to the fact that the 5′ promoter fragment used in the AtARREpro:GUS construct does not contain the regulatory element required for the AtARRE expression in stem epidermal cell layer. In fact, according to published RNA-seq and microarray data, AtARRE shows the highest expression level in the senescent first internode of the stem, and higher expression was detected in the epidermal cell layer at the bottom of the inflorescence stem compared to the top of the stem (Suh et al., 2005; Klepikova et al., 2016). During seedling development, GUS activity was first detected in roots in 3-day-old seedlings and cotyledons at 4days after imbibition (Supplementary Figure S5), but the GUS staining was much more pronounced in both organs in 5-day-old seedlings.
The observed expression profile fits the role of AtARRE as a negative regulator of wax biosynthesis in tissues that exhibit no or low wax production, such as mature cotyledons in older developing seedlings, as well as fully expanded rosette leaves and older internodes at the bottom of the stem in mature plants. Unexpectedly, AtARRE was also found to be expressed in roots, even though CER1 and CER3 genes encoding AtARRE ubiquitination substrates are not expressed in roots or are expressed at a very low level, respectively (Bourdenx et al., 2011).
Because AtARRE was discovered in a reverse genetic screen conducted to identify novel plant immunity-related E3 ligases, we were interested in determining whether AtARRE is induced upon exposure to pathogens. An earlier study has shown that AtARRE is upregulated after treatment with chitin, a potent elicitor of plant defense responses (Libault et al., 2007). Our qRT-PCR analysis revealed that elicitation with flg22, a peptide derived from bacterial flagellin, also results in a major increase in AtARRE expression (Figure 9A). In addition, infiltration of plants with the type III secretion deficient bacterial strain Pseudomonas syringae pv. tomato (P.s.t.) DC3000 hrcC− and the virulent bacterial strain P. syringae pv. maculicola (P.s.m.) ES4326 (Figures 9B,C) strongly upregulated AtARRE expression. Collectively, these data suggest that AtARRE may be involved in regulating cuticular wax biosynthesis in response to pathogen attack.
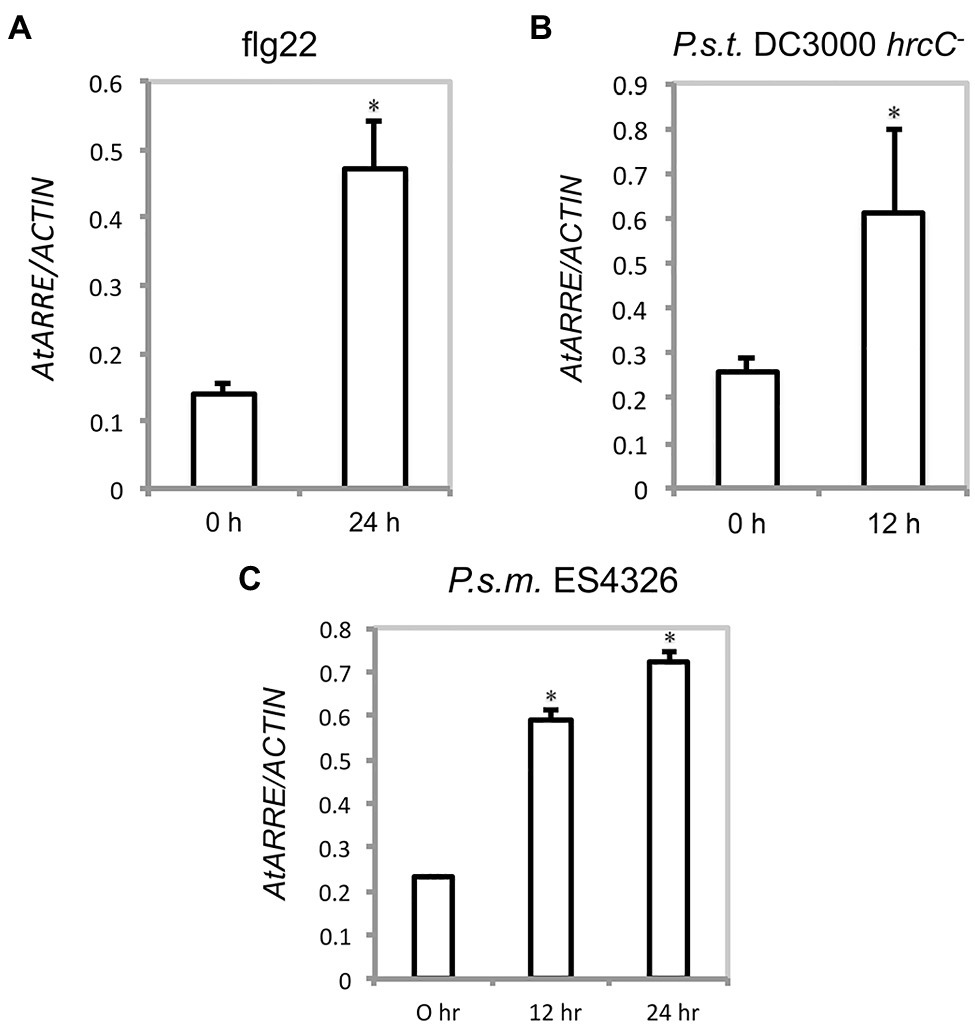
Figure 9. AtARRE gene expression is induced by flg22 and pathogen treatment. (A) Four-week-old WT plants were infiltrated with 100nM flg22, (B) P.s.t. DC3000 hrcC− at a dose of OD600=0.002, or (C) P.s.m. ES4326 at a dose of OD600=0.0002. Samples were collected 0, 12, or 24h after treatment. Expression level of AtARRE was measured by qRT-PCR, and values were normalized to the level of ACTIN. Error bars represent means±SD (n=4). Significant difference compared with WT was determined by Student’s t test and indicated by an asterisk (p<0.01).
Discussion
As an integral part of the cuticle, wax protects plants against diverse biotic and abiotic stress factors present in their environment. To fulfill this protective role, wax composition and wax load need to be continuously adjusted in response to changing environmental conditions (Shepherd and Wynne Griffiths, 2006; Bernard and Joubès, 2013). This is accomplished by extensive transcriptional, post-transcriptional, and post-translational regulation of cuticular wax metabolism (Lee and Suh, 2015a).
VLC alkanes are the major cuticular wax component in many plant species including Arabidopsis, where they comprise more than 70% of the total wax amount in leaves and 50% in stems (Bourdenx et al., 2011). VLC alkane production is catalyzed by CER1 and CER3 enzymes, which together with the cytochrome B5 form a multiprotein ER-membrane-associated complex (Bourdenx et al., 2011; Bernard et al., 2012) Not surprisingly, both enzymes act as key control points for wax biosynthesis. Several transcription factors including MYB30, MYB94, MYB96, DEWAX, and DEWAX2 regulate the expression of CER1 and/or CER3 in specific organs of Arabidopsis or in response to environmental stress (Raffaele et al., 2008; Seo et al., 2011; Go et al., 2014; Lee and Suh, 2015b). Components involved in chromatin remodeling are also required for the transcriptional regulation of CER1 or CER3. HISTONE MONOUBIQUITINATION 1 and 2 (HUB1 and HUB2) are two E3 ligases that are involved in histone monoubiquitination and active chromatin formation, which leads to the transcriptional activation of wax biosynthetic genes, such as CER1 (Ménard et al., 2014). GENERAL CONTROL NON-REPRESSED PROTEIN5 (GCN5) is a histone acetyltransferase that positively modulates CER3 expression via histone acetylation (Wang et al., 2018c). In addition to transcriptional regulation, two classes of small RNAs, tasiRNA and miRNA, post-transcriptionally control CER3 and CER1 transcript levels, respectively (Lam et al., 2012, 2015; Li et al., 2019). Finally, studies on the E3 ligase MIEL1 have shown that it negatively regulates wax accumulation in aerial plant organs (Gil et al., 2017). MIEL1 controls the stability of wax-associated transcription factors MYB96 and MYB30 and thereby indirectly affects the expression of their downstream targets CER1 and CER3 (Marino et al., 2013; Lee and Seo, 2016; Gil et al., 2017). Even though the regulatory framework governing wax accumulation has been established, the intricacies of this process remain poorly understood.
Here, we demonstrate that CER1 and CER3 protein levels, and thus alkane formation, are also controlled directly by the AtARRE RING-type E3 ubiquitin ligase that we identified in our SNIPER genetic screen (Tong et al., 2017). We found that AtARRE overexpression in wild-type Arabidopsis results in phenotypes characteristic of cuticular wax deficiency, including glossy stems and siliques, reduced fertility and fusions between aerial organs, suggesting that AtARRE is a negative regulator of wax biosynthesis (Figures 1A, 2). The wax analysis of AtARRE overexpression lines confirmed that they have reduced stem and leaf wax loads (Figures 1B–F) and revealed that their wax profile most closely resembled that of the cer1 mutant (Figure 4). Functional characterization of AtARRE showed that this protein has E3 ubiquitin ligase activity and that this activity depends on the integrity of the key amino acid residues in its RING domain. When these conserved residues in the RING domain were mutated, the AtARRE protein no longer had any effect on the stem wax accumulation when it was overexpressed in Arabidopsis (Figure 3). These results demonstrate that E3 ligase activity of AtARRE is essential for its function in wax biosynthesis.
Because wax composition of the AtARRE overexpressors was most similar to the cer1 mutant, we tested whether CER1 was the AtARRE ubiquitination substrate. An in vivo ubiquitination assay in a heterologous E. coli system expressing the Arabidopsis ubiquitination cascade confirmed AtARRE-mediated CER1 ubiquitination (Figure 6). Furthermore, transient co-expression of AtARRE and CER1 in both N. benthamiana leaves and in stable transgenic lines of Arabidopsis resulted in AtARRE-dependent degradation of CER1. Thus, AtARRE E3 ligase negatively regulates cuticular wax accumulation by ubiquitinating VLC alkane biosynthetic enzyme CER1 and targeting it for degradation by the 26S proteasome (Figure 5).
It is well-established that the UPS is redundant and that individual proteins may be targeted by multiple E3 ligases. Conversely, a single E3 ligase may have the ability to target multiple substrates for degradation (Iconomou and Saunders, 2016). CER1 and CER3, two key VLC alkane biosynthetic enzymes, share 35% amino acid identity, and both are integral membrane proteins with eight conserved His clusters at their N-terminus and an uncharacterized WAX2 domain at their C-terminus (Bernard et al., 2012). Due to their sequence similarity, it was possible that both of these proteins were substrates of the AtARRE E3 ligase. This fact, together with the observation that AtARRE overexpressors exhibit organ fusions similar to those detected in cer3, but not in cer1 mutants, prompted us to investigate whether AtARRE additionally controls CER3 levels. As previously demonstrated for CER1, AtARRE co-expression with CER3 in the N. benthamiana leaf indeed stimulated CER3 degradation (Figure 7). Thus, CER3 is also likely an AtARRE ubiquitination substrate.
The expression analysis of AtARRE gene revealed that it is preferentially expressed in tissues that exhibit no or low wax production such as roots and cotyledons in the later stages of seedling development, as well as older rosette leaves and inflorescence stem sections (Figure 8). These data suggest that the primary role of AtARRE may be to target CER1 and CER3 proteins for degradation in order to terminate wax production via the alkane pathway in tissues where it is no longer needed. Additionally, AtARRE gene transcription is upregulated upon exposure to bacterial pathogen P. syringae (Figure 9). Previous analysis of the cer1 mutant showed that reduced VLC alkane levels in cuticular wax are associated with increased cuticle permeability, but have no major effect on plant immunity. In contrast, CER1 overexpression resulted in alkane overproduction and decreased cuticle permeability, but surprisingly also in greater susceptibility to P. syringae (Bourdenx et al., 2011). Similarly, cucumber (Cucumis sativus) lines overexpressing CER3 exhibited enhanced susceptibility to the fungal pathogen Botrytis cinerea (Wang et al., 2015). Thus, induction of AtARRE upon exposure to pathogens and the resulting degradation of CER1 and CER3 may serve as a regulatory mechanism aimed at decreasing VLC alkane levels and optimizing cuticular wax composition to enhance plant resistance to bacterial pathogens. However, further work is needed to clearly define the role of AtARRE in plant immunity.
Recently, AtARRE was also reported to be induced by sodium chloride and ABA treatments (Wang et al., 2018a). Analyses of seed germination, stomatal closure, root elongation, and expression of ABA-responsive genes in atarre mutants showed that all these processes were hypersensitive to ABA, whereas AtARRE overexpressors exhibited reduced ABA sensitivity, leading the authors to propose that AtARRE is a negative regulator of ABA-dependent abiotic stress responses in plants (Wang et al., 2018a). This study did not identify the AtARRE ubiquitination target or propose the molecular mechanism governing these ABA stress responses, but it is unlikely that the AtARRE-mediated CER1 degradation by the proteasome that controls attenuation of cuticular wax biosynthesis in the shoot described here is involved. On the contrary, water deficit, salt, and ABA treatment have been shown to cause a large increase in wax amount in the leaf cuticle, predominantly due to an increase in VLC alkane content (Kosma et al., 2009). A huge induction of the CER1 alkane biosynthetic gene consistent with the elevated alkane amounts was also observed, presumably resulting in greater CER1 protein levels. Thus, even though the ABA signaling would not be expected to rely on the AtARRE-CER1 module, it is conceivable that AtARRE function in ABA-dependent signal transduction requires destruction of a different and as yet undiscovered protein target. A good example of an E3 ligase with multiple physiological roles that depend on proteasomal degradation of different protein substrates is MIEL1. MIEL1 controls seed germination and cuticular wax accumulation in Arabidopsis stems by primarily targeting MYB96 transcription factor for degradation, but in addition attenuates pathogen defense responses by promoting degradation of MYB30 (Marino et al., 2013; Lee and Seo, 2016).
Conclusion
Our findings indicate that AtARRE E3 ubiquitin ligase negatively regulates cuticular wax accumulation in Arabidopsis shoots by promoting degradation of CER1 and CER3 VLC alkane biosynthetic enzymes. Based on its expression in mature and senescing tissues and its induction upon plant exposure to pathogens, we propose that AtARRE serves as an efficient regulatory switch that terminates wax biosynthesis via the alkane-forming pathway when it is no longer required, or to optimize wax composition in response to pathogen infection.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
LK and XL planned and designed research. SL, MT, and LZ performed the experiments. SL, LZ, XL, and LK analyzed and discussed the data. SL and LK wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Natural Sciences and Engineering Research Council of Canada Discovery Grants to XL and LK, and scholarships from the China Scholarship Council to SL and MT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing cer 1-4 and cer4-4 mutants and T-DNA insertion alleles, Dr. Takuji Wada (RIKEN, Japan) for the cer3-6 mutant, and the BioImaging Facility at the University of British Columbia for assistance with microscopy. We are grateful to Dr. Oliver Xiao’ou Dong (UBC) for advice on ubiquitination assays, Dr. Yuli Ding (UBC) for help with pathogen treatment, and Drs. Tegan Haslam and Gillian Dean (UBC) for thoughtful comments on our manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.752309/full#supplementary-material
Footnotes
References
Aarts, M. G., Keijzer, C. J., Stiekema, W. J., and Pereira, A. (1995). Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7, 2115–2127. doi: 10.1105/tpc.7.12.2115
Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16, 2463–2480. doi: 10.1105/tpc.104.022897
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. doi: 10.1126/science.1086391
Beisson, F., Li-Beisson, Y., and Pollard, M. (2012). Solving the puzzles of cutin and suberin polymer biosynthesis. Curr. Opin. Plant Biol. 15, 329–337. doi: 10.1016/j.pbi.2012.03.003
Bernard, A., Domergue, F., Pascal, S., Jetter, R., Renne, C., Faure, J.-D., et al. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24, 3106–3118. doi: 10.1105/tpc.112.099796
Bernard, A., and Joubès, J. (2013). Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog. Lipid Res. 52, 110–129. doi: 10.1016/j.plipres.2012.10.002
Bourdenx, B., Bernard, A., Domergue, F., Pascal, S., Leger, A., Roby, D., et al. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156, 29–45. doi: 10.1104/pp.111.172320
Broun, P., Poindexter, P., Osborne, E., Jiang, C. Z., and Reichmann, J. L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 101, 4706–4711. doi: 10.1073/pnas.0305574101
Buschhaus, C., and Jetter, R. (2011). Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J. Exp. Bot. 62, 841–853. doi: 10.1093/jxb/erq366
Chen, X., Goodwin, S. M., Boroff, V. L., Liu, X., and Jenks, M. A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15, 1170–1185. doi: 10.1105/tpc.010926
Chung, M. H., Chen, M. K., and Pan, S. M. (2000). Floral spray transformation can efficiently generate Arabidopsis. Transgenic Res. 9, 471–486. doi: 10.1023/A:1026522104478
Dean, G. H., Zheng, H., Tewari, J., Huang, J., Young, D. S., Hwang, Y. T., et al. (2007). The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19, 4007–4021. doi: 10.1105/tpc.107.050609
Deshaies, R. J., and Joazeiro, C. A. P. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434. doi: 10.1146/annurev.biochem.78.101807.093809
Dong, O. X., Ao, K., Xu, F., Johnson, K. C. M., Wu, Y., Li, L., et al. (2018). Individual components of paired typical NLR immune receptors are regulated by distinct E3 ligases. Nat. Plants 4, 699–710. doi: 10.1038/s41477-018-0216-8
Gil, H. L., Kim, J., Suh, M. C., and Seo, P. J. (2017). The MIEL1 E3 ubiquitin ligase negatively regulates cuticular wax biosynthesis in Arabidopsis stems. Plant Cell Physiol. 58, 1249–1259. doi: 10.1093/pcp/pcx065
Go, Y. S., Kim, H., Kim, H. J., and Suh, M. C. (2014). Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 26, 1666–1680. doi: 10.1105/tpc.114.123307
Greer, S., Wen, M., Bird, D., Wu, X., Samuels, L., Kunst, L., et al. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145, 653–667. doi: 10.1104/pp.107.107300
Han, Y., Sun, J., Yang, J., Tan, Z., Luo, J., and Lu, D. (2017). Reconstitution of the plant ubiquitination cascade in bacteria using a synthetic biology approach. Plant J. 91, 766–776. doi: 10.1111/tpj.13603
Haslam, T. M., Gerelle, W. K., Graham, S. W., and Kunst, L. (2017). The unique role of the ECERIFERUM2-LIKE clade of the BAHD acyltransferase superfamily in cuticular wax metabolism. Plan. Theory 6:23. doi: 10.3390/plants6020023
Haslam, T. M., and Kunst, L. (2013a). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210, 93–107. doi: 10.1016/j.plantsci.2013.05.008
Haslam, T. M., and Kunst, L. (2013b). Wax analysis of stem and rosette leaves in Arabidopsis thaliana. Bioprotocols 3:e782. doi: 10.21769/BioProtoc.782
Haslam, T. M., Mañas-Fernández, A., Zhao, L., and Kunst, L. (2012). Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol. 160, 1164–1174. doi: 10.1104/pp.112.201640
Haughn, G. W., and Somerville, C. R. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204, 430–434. doi: 10.1007/BF00331020
Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S., and Mullineaux, P. M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. doi: 10.1023/A:1006496308160
Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. doi: 10.1146/annurev.biochem.67.1.425
Hooker, T. S., Lam, P., Zheng, H., and Kunst, L. (2007). A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19, 904–913. doi: 10.1105/tpc.106.049304
Iconomou, M., and Saunders, D. N. (2016). Systematic approaches to identify E3 ligase substrates. Biochem. J. 473, 4083–4101. doi: 10.1042/BCJ20160719
Jetter, R., Kunst, L., and Samuels, A. L. (2006). “Composition of plant cuticular waxes,” in Annual Plant Reviews. eds. M. Riederer and C. Müller (Oxford, UK: Blackwell Publishing Ltd), 145–181.
Kannangara, R., Branigan, C., Liu, Y., Penfield, T., Rao, V., Mouille, G., et al. (2007). The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19, 1278–1294. doi: 10.1105/tpc.106.047076
Kim, H., Go, Y. S., and Suh, M. C. (2018). DEWAX2 transcription factor negatively regulates cuticular wax biosynthesis in Arabidopsis leaves. Plant Cell Physiol. 59, 966–977. doi: 10.1093/pcp/pcy033
Kim, H., Yu, S., Jung, S. H., Lee, B., and Suh, M. C. (2019). The F-box protein SAGL1 and ECERIFERUM3 regulate cuticular wax biosynthesis in response to changes in humidity in Arabidopsis. Plant Cell 31, 2223–2240. doi: 10.1105/tpc.19.00152
Kleinboelting, N., Huep, G., Kloetgen, A., Viehoever, P., and Weisshaar, B. (2012). GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 40, D1211–D1215. doi: 10.1093/nar/gkr1047
Klepikova, A. V., Kasianov, A. S., Gerasimov, E. S., Logacheva, M. D., and Penin, A. A. (2016). A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88, 1058–1070. doi: 10.1111/tpj.13312
Koornneef, M., Hanhart, C. J., and Thiel, F. (1989). A genetic and phenotypic description of Eceriferum (cer) mutants in Arabidopsis thaliana. J. Hered. 80, 118–122. doi: 10.1093/oxfordjournals.jhered.a110808
Kosma, D. K., Bourdenx, B., Bernard, A., Parsons, E. P., Lü, S., Joubès, J., et al. (2009). The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 151, 1918–1929. doi: 10.1104/pp.109.141911
Kraft, E., Stone, S. L., Ma, L., Su, N., Gao, Y., Lau, O. S., et al. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139, 1597–1611. doi: 10.1104/pp.105.067983
Lam, P., Zhao, L., Eveleigh, N., Yu, Y., Chen, X., and Kunst, L. (2015). The exosome and trans-acting small interfering RNAs regulate cuticular wax biosynthesis during Arabidopsis inflorescence stem development. Plant Physiol. 167, 323–336. doi: 10.1104/pp.114.252825
Lam, P., Zhao, L., McFarlane, H. E., Aiga, M., Lam, V., Hooker, T. S., et al. (2012). RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiol. 159, 1385–1395. doi: 10.1104/pp.112.199646
Lee, S. B., Kim, H., Kim, R. J., and Suh, M. C. (2014). Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Rep. 33, 1535–1546. doi: 10.1007/s00299-014-1636-1
Lee, H. G., and Seo, P. J. (2016). The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96. Nat. Commun. 7:12525. doi: 10.1038/ncomms12525
Lee, S. B., and Suh, M. C. (2015a). Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 34, 557–572. doi: 10.1007/s00299-015-1772-2
Lee, S. B., and Suh, M. C. (2015b). Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 56, 48–60. doi: 10.1093/pcp/pcu142
Li, R.-J., Li, L.-M., Liu, X.-L., Kim, J.-C., Jenks, M. A., and Lü, S. (2019). Diurnal regulation of plant epidermal wax synthesis through antagonistic roles of the transcription factors SPL9 and DEWAX. Plant Cell 31, 2711–2733. doi: 10.1105/tpc.19.00233
Libault, M., Wan, J., Czechowski, T., Udvardi, M., and Stacey, G. (2007). Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant-Microbe Interact. 20, 900–911. doi: 10.1094/MPMI-20-8-0900
Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S., and Weissman, A. M. (1999). RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 96, 11364–11369. doi: 10.1073/pnas.96.20.11364
Lü, S., Zhao, H., Des Marais, D. L., Parsons, E. P., Wen, X., Xu, X., et al. (2012). Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water satus. Plant Physiol. 159, 930–944. doi: 10.1104/pp.112.198697
Marino, D., Froidure, S., Canonne, J., Ben Khaled, S., Khafif, M., Pouzet, C., et al. (2013). Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat. Commun. 4:1476. doi: 10.1038/ncomms2479
Ménard, R., Verdier, G., Ors, M., Erhardt, M., Beisson, F., and Shen, W. H. (2014). Histone H2B monoubiquitination is involved in the regulation of cutin and wax composition in Arabidopsis thaliana. Plant Cell Physiol. 55, 455–466. doi: 10.1093/pcp/pct182
Müller, C. (2018). Plant–insect interactions on cuticular surfaces. Annu. Plant. Rev. 23, 398–422. doi: 10.1002/9781119312994.apr0241
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. doi: 10.1263/jbb.104.34
Nelson, B. K., Cai, X., and Nebenführ, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Oshima, Y., Shikata, M., Koyama, T., Ohtsubo, N., Mitsuda, N., and Ohme-Takagi, M. (2013). MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25, 1609–1624. doi: 10.1105/tpc.113.110783
Park, C. S., Go, Y. S., and Suh, M. C. (2016). Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 88, 257–270. doi: 10.1111/tpj.13248
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Raffaele, S., Vailleau, F., Leger, A., Joubès, J., Miersch, O., Huard, C., et al. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20, 752–767. doi: 10.1105/tpc.107.054858
Reicosky, D. A., and Hanover, J. W. (1978). Physiological effects of surface waxes. Plant Physiol. 62, 101–104. doi: 10.1104/pp.62.1.101
Riederer, M. (2006). “Introduction: biology of the plant cuticle” in Biology of the Pant Cuticle. eds. M. Riederer and C. Müller (Oxford, UK: Blackwell), 4–5.
Riederer, M., and Schreiber, L. (2001). Protecting against water loss: analysis of the barrier properties of plant cuticles. J. Exp. Bot. 52, 2023–2032. doi: 10.1093/jexbot/52.363.2023
Rowland, O., Lee, R., Franke, R., Schreiber, L., and Kunst, L. (2007). The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett. 581, 3538–3544. doi: 10.1016/j.febslet.2007.06.065
Samuels, L., Kunst, L., and Jetter, R. (2008). Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59, 683–707. doi: 10.1146/annurev.arplant.59.103006.093219
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., and Park, C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23, 1138–1152. doi: 10.1105/tpc.111.083485
Shepherd, T., and Wynne Griffiths, D. (2006). The effects of stress on plant cuticular waxes. New Phytol. 171, 469–499. doi: 10.1111/j.1469-8137.2006.01826.x
Sieber, P., Schorderet, M., Ryser, U., Buchala, A., Kolattukudy, P., Métraux, J.-P., et al. (2000). Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12, 721–737. doi: 10.1105/tpc.12.5.721
Suh, M. C., Samuels, A. L., Jetter, R., Kunst, L., Pollard, M., Ohlrogge, J., et al. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139, 1649–1665. doi: 10.1104/pp.105.070805
Tong, M., Kotur, T., Liang, W., Vogelmann, K., Kleine, T., Leister, D., et al. (2017). E3 ligase SAUL1 serves as a positive regulator of PAMP-triggered immunity and its homeostasis is monitored by immune receptor SOC3. New Phytol. 215, 1516–1532. doi: 10.1111/nph.14678
Vierstra, R. D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. 10, 385–397. doi: 10.1038/nrm2688
Vincent, N. G., Charette, J. M., and Baserga, S. J. (2018). The SSU processome interactome in Saccharomyces cerevisiae reveals novel protein subcomplexes. RNA 24, 77–89. doi: 10.1261/rna.062927.117
Wang, B., Li, C., Kong, X., Li, Y., Liu, Z., Wang, J., et al. (2018a). AtARRE, an E3 ubiquitin ligase, negatively regulates ABA signaling in Arabidopsis thaliana. Plant Cell Rep. 37, 1269–1278. doi: 10.1007/s00299-018-2311-8
Wang, W., Liu, X., Gai, X., Ren, J., Liu, X., Cai, Y., et al. (2015). Cucumis sativus L. WAX2 plays a pivotal role in wax biosynthesis, influencing pollen fertility and plant biotic and abiotic stress responses. Plant Cell Physiol. 56, 1339–1354. doi: 10.1093/pcp/pcv052
Wang, Z., Tian, X., Zhao, Q., Liu, Z., Li, X., Ren, Y., et al. (2018b). The E3 ligase DROUGHT HYPERSENSITIVE negatively regulates cuticular wax biosynthesis by promoting the degradation of transcription factor ROC4 in rice. Plant Cell 30, 228–244. doi: 10.1105/tpc.17.00823
Wang, T., Xing, J., Liu, X., Yao, Y., Hu, Z., Peng, H., et al. (2018c). GCN5 contributes to stem cuticular wax biosynthesis by histone acetylation of CER3 in Arabidopsis. J. Exp. Bot. 69, 2911–2922. doi: 10.1093/jxb/ery077
Wang, Z.-Y., Xiong, L., Li, W., Zhu, J.-K., and Zhu, J. (2011). The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell 23, 1971–1984. doi: 10.1105/tpc.110.081943
Wang, F., Zhu, D., Huang, X., Li, S., Gong, Y., Yao, Q., et al. (2009). Biochemicali insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21, 2378–2390. doi: 10.1105/tpc.108.065433
Wilkins, T. A., and Smart, L. B. (1996). “Isolation of RNA from plant tissue,” in A Laboratory Guide to RNA. ed. P. A. Krieg (New York: Wiley-Liss), 21–41.
Wu, Z., Tong, M., Tian, L., Zhu, C., Liu, X., Zhang, Y., et al. (2020). Plant E3 ligases SNIPER1 and SNIPER2 broadly regulate the homeostasis of sensor NLR immune receptors. EMBO J. 39:e104915. doi: 10.15252/embj.2020104915
Yeats, T. H., and Rose, J. K. C. (2013). The formation and function of plant cuticles. Plant Physiol. 163, 5–20. doi: 10.1104/pp.113.222737
Zhao, Q., Tian, M., Li, Q., Cui, F., Liu, L., Yin, B., et al. (2013). A plant-specific in vitro ubiquitination analysis system. Plant J. 74, 524–533. doi: 10.1111/tpj.12127
Zheng, L., Baumann, U., and Reymond, J.-L. (2004). An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32:e115. doi: 10.1093/nar/gnh110
Keywords: Arabidopsis, cuticle, cuticular wax biosynthesis, AtARRE, E3 ligase, CER1, CER3
Citation: Liu S, Tong M, Zhao L, Li X and Kunst L (2021) The ARRE RING-Type E3 Ubiquitin Ligase Negatively Regulates Cuticular Wax Biosynthesis in Arabidopsis thaliana by Controlling ECERIFERUM1 and ECERIFERUM3 Protein Levels. Front. Plant Sci. 12:752309. doi: 10.3389/fpls.2021.752309
Edited by:
Isabel Molina, Algoma University, CanadaReviewed by:
Mi Chung Suh, Sogang University, South KoreaShiu Cheung Lung, The University of Hong Kong, Hong Kong, SAR China
Copyright © 2021 Liu, Tong, Zhao, Li and Kunst. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ljerka Kunst, ljerka.kunst@ubc.ca
 Shuang Liu
Shuang Liu Meixuezi Tong1,2
Meixuezi Tong1,2 Lifang Zhao
Lifang Zhao Ljerka Kunst
Ljerka Kunst