- 1Guangxi Key Laboratory of Sugarcane Biology, State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, College of Agriculture, Guangxi University, Nanning, China
- 2College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
The glycine-rich domain proteins (GRDP) have been functionally implicated in the cell wall structure, biotic, and abiotic stress responses. However, little is known about GRDP genes in female gametophyte development of Arabidopsis. This study shows that GRDP2, a GRDP, plays a crucial role in female gametophyte development. In GRDP2 overexpression lines, grdp2-3, the embryo sacs were arrested at FG1 and no nucleus stages. Furthermore, callose staining shows that cell plate formation during megasporogenesis is disturbed in grdp2-3. In contrast, the pollen development is not affected in grdp2-3. The expression patterns of auxin-specific marker lines in female gametophytes showed that the auxin distribution and transport were significantly changed during megagametogenesis in grdp2-3. In addition, compared with the membrane-localized pattern of PIN1, PIN2, and PIN7 in WT, the signals were detected in the cytoplasm in grdp2-3. Together, our data suggest that GRDP2 plays an essential role in auxin-mediated female gametophyte development.
Introduction
Proteins with particular glycine-rich regions have been reported in seed coat and cell wall of a wide variety of plants; for example, soybean seed coat contains 21% of glycine, the cell wall of milkweed stem has 31%, and coat coleoptile cells have 27% of glycine (de Oliveira et al., 1990). Glycine-rich proteins (GRPs) have also been isolated from pumpkin seed coat (Varner and Cassab, 1986) and strawberry fruit (Reddy and Poovaiah, 1987) with approximately 47% and 49% glycine residues, respectively. GRP proteins contain characteristic repetitive glycine stretches, amino-terminal sequences characterized by a high content of glycine (∼40–70%), and a repetitive sequence of residues arranged in (Gly)n-X motifs (Sachetto-Martins et al., 2000; Mousavi and Hotta, 2005). At present, five groups of GRPs [i.e., (I) (GGX)n, (II) (GGXXXGG)n, (III) (GXGX)n, (IV) glycine-rich domain with additional motifs such as RNA-recognition motif (RRM) or a cold-shock domain (CSD), CCHC zinc-fingers and (V) GGX/GXGX] have been suggested based on the general structure and the arrangement of the repeated glycine signatures as well as the presence of conserved motifs and domains (Bocca et al., 2005; Mangeon et al., 2010; Ortega-Amaro et al., 2014).
Glycine-rich proteins genes exhibit tissue-specific expression patterns and regulate many developmental processes in plants. Previous studies suggest that GRPs are specifically expressed in the xylem, phloem, epidermis, anther tapetum, and roots (Fusaro and Sachetto-Martins, 2007). GRPs are generally induced by physical, chemical, and biological factors. GRPs function in the plant cell wall structure, plant defense response, regulation of plant flowering time, transcriptional regulation, signal transduction, protein–protein interaction, stress response, etc. (Keller et al., 1988; Bocca et al., 2005; Mangeon et al., 2010; Ortega-Amaro et al., 2014). The PvGRP18, a glycine-enriched protein in legumes, was reported as an agglutinant for cell wall synthesis which is involved in protoxylem growth (Keller et al., 1988; Ryser et al., 1997). PtGRP1 is located between the cell wall and cell membrane, and its expression level in stem and leaf decreases with the increase of tissue development age (Condit, 1993). Phytohormones control several aspects of plant growth, development, and stress response, also regulate GRP expression. For example, GRPs accumulate significantly in auxin deficient strawberry fruits (Reddy and Poovaiah, 1987). In pea, GRP1 gets enriched in fruits and seeds; however, ABA induction results in its increase in the pistil and root (Urbez et al., 2006). Overexpression of alfalfa GRP leads to salt and ABA sensitivity in Arabidopsis thaliana (Long et al., 2013).
In Eucalyptus, apart from the canonical GRPs, another type, the glycine-rich domain (GRDPs), was reported (Bocca et al., 2005; Rodríguez-Hernández et al., 2014). GRDPs have a short glycine-rich domain located in the C terminal of the protein along with repeated GCGXXCXGXCG. Besides, a comparison of GRDPs in 16 species indicated two additional conserved domains in this protein: Domain of Unknown Function (DUF1399) at the N-terminal and a putative RNA binding motif (RNP) in the middle of the protein sequence. In Arabidopsis, there are two members of the GRDPs, GRDP1 and GRDP2. The glycine-rich domain (GRD) is located at the C-terminus of the GRDP1 and GRDP2 proteins, with five glycine regions in the consensus regions XSGCGXXCXGXCG (GRDP1) or GCGXXCXGXCGXXCG (GRDP2) (Rodríguez-Hernández et al., 2014). Besides, two highly conserved domains, DUF1399 at the N-terminus and the putative RNP in the middle, which were also detected in GRDPs. Previously, it was reported that GRDP1 plays a regulatory role in ABA signaling and abiotic stress tolerance (Rodríguez-Hernández et al., 2014), whereas GRDP2 regulates development and stress responses possibly through an auxin-dependent mechanism in Arabidopsis, and common bean (Ortega-Amaro et al., 2014, 2016; Rodríguez-Hernández et al., 2014).
Phytohormones such as auxin, gibberellin, cytokinin, ABA, ethylene, and brassinosteroids play vital roles in plant reproduction. They regulate the development of both male and female reproductive organs, including ovules and gynoecia (Nambara and Van Wees, 2021). For example, auxin is involved in regulating the development of male and female reproductive organs of plants, including ovule and pistil (Liu et al., 2020). It plays an essential role in developing seeds and fruits (Dorcey et al., 2009; Figueiredo et al., 2016). Auxin response is mediated by a series of auxin response factors (ARFs) that bind to specific auxin response elements (AUXRE) in the promoters of their target genes (Ulmasov et al., 1997; Roosjen et al., 2018). At low auxin concentrations, responses are blocked by Aux/IAA repressor proteins, which form dimers with activating ARFs, thus preventing them from regulating their target genes. When auxin levels rise, the phytohormone facilitates binding between the DII domain of the Aux/IAA proteins and TIR1/AFB F-box proteins that are part of an E3 ubiquitin ligase complex (Larsson et al., 2017), resulting in the targeting of the Aux/IAAs for degradation by the 26S proteasome, thereby releasing the ARFs from repression (Roosjen et al., 2018). In 2004, a group of genes regulated explicitly by indole-3-acetic acid (IAA) had been identified through gene microarray, including GRDP2 (Goda et al., 2004). Therefore, it is speculated that the auxin signaling could regulate the expression of the GRDP2 gene.
In this study, we performed molecular characterization of GRDP2 (At4g37900), both p35S:GRDP2 and pUBQ10:GRDP2 overexpressing lines and grdp2-3 mutants were analyzed. Expression profiles analysis of the GRDP2 gene in Arabidopsis showed that it is highly expressed in floral organs. We further showed that GRDP2 overexpression results in two types of ovule development defects in Arabidopsis thaliana, the abnormal arrest at one nucleus stage (FG1 stage) and the nucleus degeneration. Auxin distribution analysis during the gametogenesis in grdp2-3 ovules suggests that auxin may play an essential role in grdp2-3 ovule development.
Materials and Methods
Plant Material and Growth Conditions
The mutant and transgenic lines used in this study were generated in the Arabidopsis thaliana ecotype Columbia 0 (Col-0) background. Arabidopsis seeds of each line were surface-sterilized for 10 min with 75% ethanol, 5 min with absolute ethyl alcohol, and rinsed five times in sterile distilled water. Aseptic stratified seeds (2 days at 4°C) were germinated and grown on agar plates containing 1/2 Murashige and Skoog (MS) medium, pH = 5.7, 0.5% (w/v) sucrose, and 0.8% (w/v) agar (Murashige and Skoog, 1962). Plates were incubated in a growth chamber with a photoperiod of 16 h/8 h, light/dark cycle at a temperature of 22 ± 1°C. Afterward, plants were transferred to soil pots in a growth chamber at 22 ± 1°C with a 16 h light/8 h dark photoperiod (Zhao et al., 2014). For the root growth transfer experiment, WT and mutants’ seeds were sterilized and planted on Hoagland media as detailed previously (Aslam et al., 2020). After 5 days, plants were transferred to Hoagland media supplemented with different IAA or NPA concentrations (15 nM and 30 nM). The plates were then marked, and after 48 h of vertical culture, root length was recorded.
Identification of the T-DNA Insertion Lines and RT-qPCR
Three T-DNA mutant lines Sail_387_D04, SALK_412A10, and SALK_112794C for the GRDP2 (At4g37900) gene were acquired from the Arabidopsis Biological Resource Center1. pDR5:GFP, pPIN1:PIN1-GFP, pPIN2:PIN2-GFP, pPIN7:PIN7-GFP, and pYUCCA5-GFP-GUS (from Dr. Xu Chen, Fujian Agriculture and Forestry University, Fuzhou, China). pSPL:GUS, pKNU:KNU-VENUS, pAKV:H2B-YFP, pDD45:GFP, and pDD65:GFP (from Dr. Yuan Qin, Fujian Agriculture and Forestry University, Fuzhou, China). The T-DNA insertion was identified by primers (Supplementary Table 1) obtained from the SIGnAL website2. GRDP2 expression level in T-DNA mutant lines was confirmed by RT-qPCR using the primers listed in Supplementary Table 1.
Total RNA was extracted from inflorescence tissues using the RNA extraction Kit (Omega Bio-Tek, Shanghai, China) following the manufacturer’s protocol. EasyScript® cDNA Synthesis SuperMix (Transgen, Beijing, China) was used for the cDNA preparation. The RT-qPCR was conducted using TransStart® Top Green qPCR SuperMix (Transgen, Beijing, China). HK2 (AT5G35750) gene was used as a reference gene (Supplementary Table 1; Zhao et al., 2018). These assays were conducted for three biological replicates, and the results are shown as the mean ± standard deviations.
Histological Analysis of Female Gametophyte Development
Differential interference contrast (DIC) microscopy was used for the histological analysis of grdp2 mutant ovule development. The whole inflorescences of grdp2 and WT plants were fixed with FAA solution (6 alcohol:3 chloroform:1 acetic acid) for more than 16 h at room temperature. Then siliques were then dissected longitudinally with hypodermic needles (1 ml insulin syringes), and all the ovules were exposed outside the carpels. Finally, cleared ovules were observed on a Zeiss Axioimager M1 microscope (Carl Zeiss Canada, Toronto, ON, Canada) under DIC optics, and images were acquired with a Zeiss AxioCam MR digital camera. All images were processed for publication using Adobe Photoshop (Adobe Systems, San Jose, CA, United States).
Vector Construction and Plant Transformation
Using a high-fidelity polymerase kit (QIAGEN, United States), the open reading frame of GRDP1 and GRDP2 was amplified from an inflorescence cDNA sample using gene-specific primers, and the AtGRDP1 and AtGRDP2 promoter region (2 kb upstream sequence) was amplified from the Arabidopsis genomic DNA (Supplementary Table 1). The PCR fragments were verified by DNA sequencing and then cloned into the pENTR/D-TOPO vector (Invitrogen, Dusseldorf, Germany). Then the promoter fragments were recombined into the destination vector pGWB633, and pGWB604; and the CDS were recombined into the destination vector pGWB602 (p35S:GRDP2-OE), and pGWB501 (pUBQ10:GRDP1-OE; pUBQ10:GRDP2-OE), using LR Clonase II (Invitrogen, Thermo Fisher Scientific, CA, United States). The plasmids were extracted using the E.Z.N.A.® Plasmid Maxi Kit (Omega Bio-Tek, Norcross, GA, United States) following the manufacturer’s procedure.
An Agrobacterium tumefaciens strain (GV3101) carrying the fragments as described before was introduced into Arabidopsis thaliana WT plants by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected in soil by spraying with Basta (Basta:H2O = 1:1000) or on the MS media and selected by hygromycin. Multiple independent T2 transgenic lines were used for phenotype observation.
Confocal Laser Scanning Microscopy
Dissected carpels were mounted with 40% glycerol. Confocal laser scanning microscopy (CLSM) analysis was performed using a Leica TCS SP8X Meta confocal microscope. To detect the GFP signal, a 488 nm wavelength laser was used for excitation, and a BP 505–550 nm filter was applied for GFP emission. For FM 4-64 FX dye application (Invitrogen), an additional BP 575–615 nm filter was used. For ovule morphological analyses, pistils were fixed as described earlier (Braselton et al., 1996). Samples were excited using a 532 nm laser, and emission was detected between 570 and 740 nm.
Alexander Staining of Pollen Grains
Mature flower buds were collected before anthesis, and non-dehiscent anthers (stage 12 flower buds) were fixed in FAA (6 alcohol:3 chloroform:1 acetic acid) solution for a minimum of 2 h (Peterson et al., 2010). Then the buds were placed on a microscope slide and the fixative was removed with absorbent paper. Two to four drops of the stain solution were applied before the sample completely dries, and the buds were dissected to release the anthers. The sample in the stain was slowly heated on an alcohol burner in a fume hood until the stain solution reached near-boiling. The samples were washed three times with 40% glycerol and mounted for analysis using the ZEISS microscope (ZEISS, Germany).
Selection of CRISPR/Cas9 Target Sites and Molecular Analysis and Detection of Mutations
All possible CRISPR target sites within GRDP1 and GRDP2 are identified with the online tool SSC3. Then three candidates of CRISPR target sites were selected based on the genomic location and potential off-target scores, and the sgRNA targets two target genes simultaneously. Each of the targeting portions was synthesized as a pair of reverse complementary oligonucleotides, then assembled the natural fading primers into the pAtU6-M vector by T4 ligase (Thermo Fisher, Waltham, MA, United States). assembled into the pCambia1300-UBQ: Cas9-P2A-GFP-rbcS-E9t vector by GoldenGate reaction assembly (NEB, Hitchin, United Kingdom). These constructs were confirmed by DNA sequencing and transformed into Arabidopsis by Agrobacterium strain GV3101 separately.
The primers of hygromycin were used to identify transgenic plants containing the T-DNA fragment fused into the genome (Supplementary Table 1). The positive plants were deep sequenced to identify any insertions or deletions within the targeted region. The phenotypes of the transgenic plants were photographed and described in section “Results.”
Results
Overexpression of GRDP2 Causes Reduced Plant Fertility
In Arabidopsis, glycine-rich domain protein (GRDP) is encoded by a single copy gene GRDP2, and its genomic organization consists of five exons and four introns. To investigate the function of GRDP2 in Arabidopsis, we obtained three independent T-DNA inserted alleles of GRDP2 from the Arabidopsis Biological Resource Center (ABRC); grdp2-1 (SALK_387D04), grdp2-2 (SALK_412A10), and grdp2-3 (SALK_112794C). The T-DNA insertion site of these mutants is shown in Figure 1A. The GRDP2 expression level of these mutants was quantified through the RT-qPCR. The results showed that grdp2-1 is a knockout line and grdp2-2 is a knockdown line, while the GRDP2 expression is much higher than WT plants in grdp2-3 (Figure 1B). Phenotypic analysis of the grdp2 T-DNA mutant lines showed a distinct low seed set (51%) of the grdp2-3 line with short siliques than WT (Figures 1C,D).
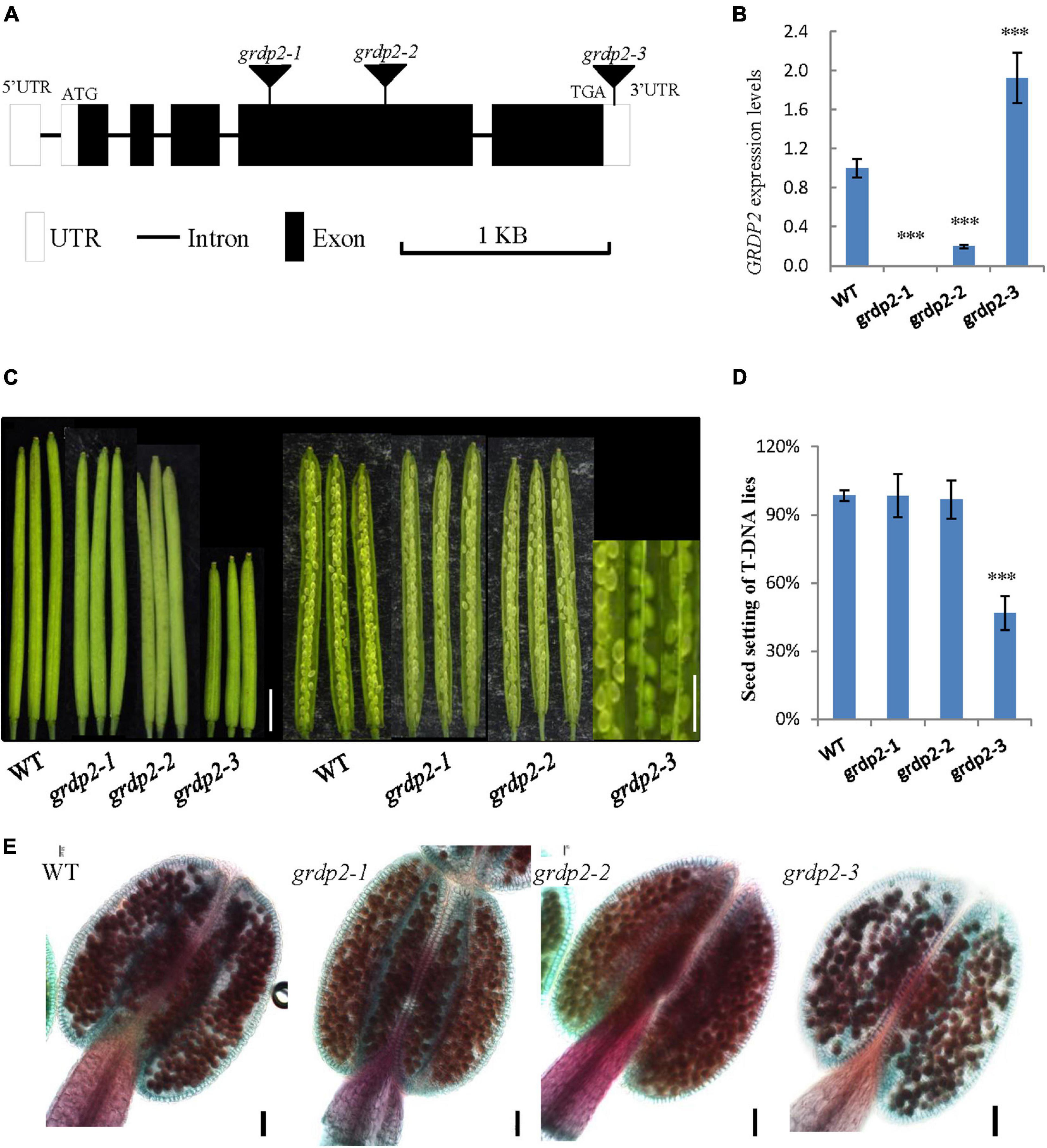
Figure 1. Phenotype analysis of grdp2 T-DNA mutant. (A) Gene structure of GRDP2 and T-DNA insertion sites of the mutant alleles in grdp2. White boxes show UTRs; gray lines show introns; black boxes show exons. (B) Expression level of the three T-DNA insertion lines as determined by real-time RT-PCR. (C) Seed development in wild-type (WT) plants and the grdp2 mutant alleles. Bar = 1 cm. (D) Seed setting statistics of three T-DNA insertion lines. (E) Alexander’s staining of pollen from WT and three grdp2 mutant alleles. The red color indicates viable pollen. Bar = 50 μm. The asterisks show the significance level (∗∗∗p < 0.01) as judged by the Student’s t-test.
We conducted a genetic analysis to determine whether the mutation of GRDP2 caused gametophyte sterility or embryo lethality. A reciprocal cross experiment showed that grdp2-3 has 54.48% sterile or aborted ovules when grdp2-3 was used as the mother plant and pollinated by WT pollen. Conversely, when WT pistils were pollinated with grdp2-3 pollen, only 8.84% of ovules were abnormal (Table 1). This result indicated that the female gametophyte development has a problem, and the pollen development is normal, as shown by Alexander’s staining of pollen grains (Figure 1E). These results indicate that the abnormal development of female gametophytes causes the reduced seed setting rate of grdp2-3.
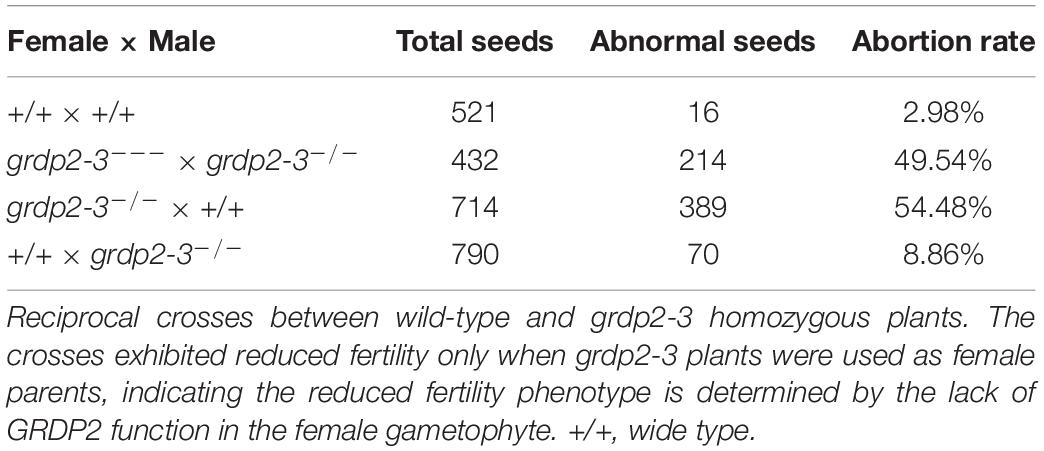
Table 1. Analysis of seed development in siliques from selfing and reciprocal crosses of grdp2-3 plants.
Differential Expression Patterns of GRDP2 in Arabidopsis
To better study the functions of GRDP2, its expression pattern was observed in root, stem, leaf, siliques, and inflorescence tissues using quantitative PCR. The result showed that GRDP2 has a higher expression in inflorescence than root, stem, leaves, and siliques (Figure 2A). We then fused the GRDP2 promoter with GFP/GUS protein to detect the expression patterns of GRDP2 in inflorescence tissue. GUS expression was detected in the whole inflorescence. However, only immature flowers showed the GUS signals, especially in the young pollen grains (Figures 2B–D). GUS signal was also detected in ovules during the female gametophyte development, and no GUS signal was detected at early stages (Figure 2E). A weak GUS signal was detected in the embryo sac and funiculus of ovules at the mature stage (Figure 2F).
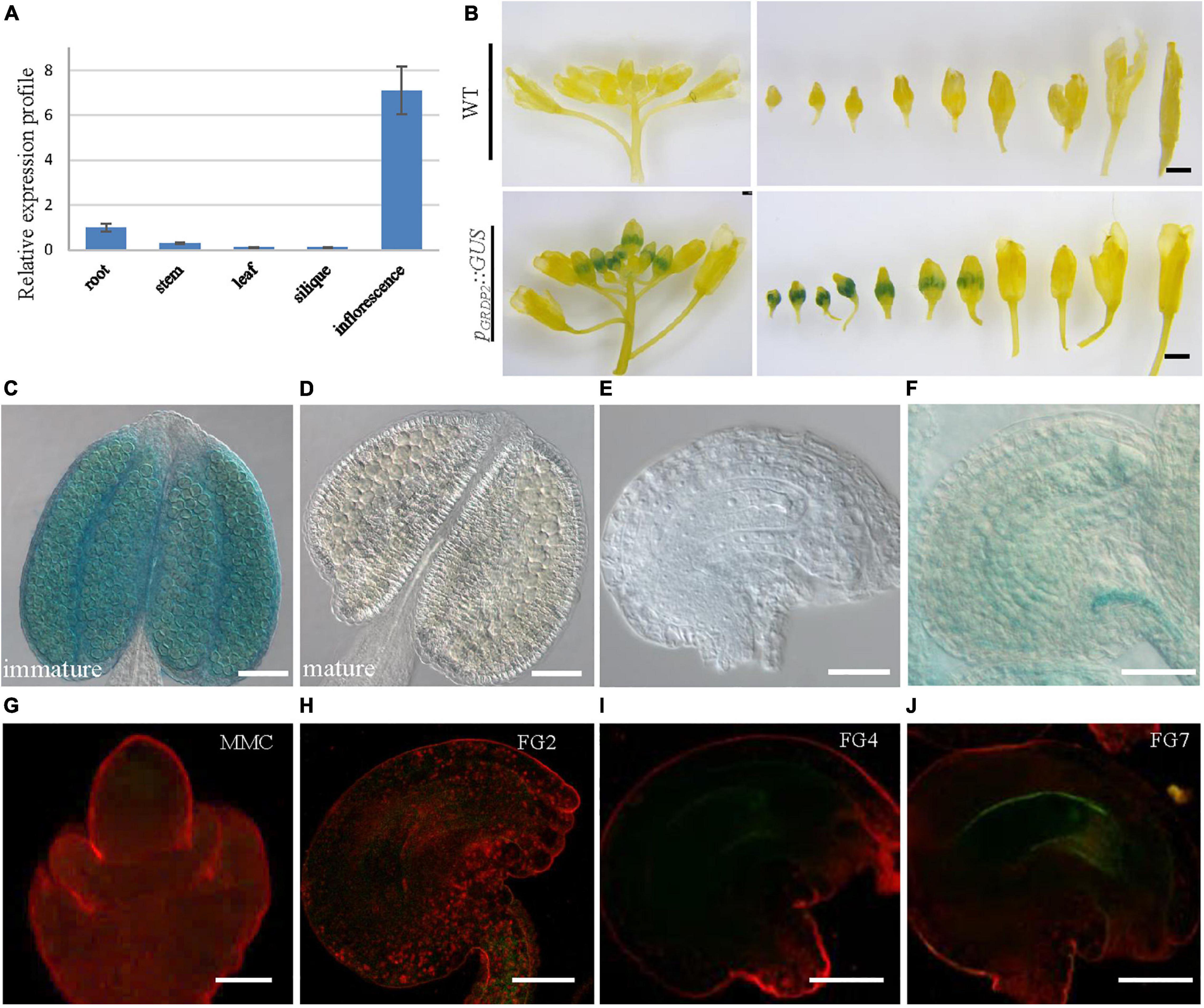
Figure 2. The expression profiles of GRDP2 in Arabidopsis. (A) Real-time PCR to quantify GRDP2 expression levels in different tissue of root, stem, leaf, siliques, and inflorescence. (B) pGRDP2:GUS expression pattern in floral buds. Bars = 1 mm. (C,D) pGRDP2:GUS expression pattern in anthers. Bars = 50 μm. (E,F) pGRDP2:GUS expression pattern in ovules. Bars = 20 μm. (G–J) pGRDP2:GFP expression pattern in different ovule development stages. The developmental stages of the ovules are indicated at the top right corner of each panel. Bars = 20 μm.
Similarly, the GFP reporter gene was also used to visualize the expression pattern of GRDP2. No GFP signals were detected in the ovules during the megasporogenesis stage and early megagametogenesis stage (Figures 2G–I). The mature FG7 ovules expressed a weak GFP signal (Figure 2J), suggesting GRDP2 express only in mature ovules.
Overexpression of GRDP2 Results in Abnormal Ovule Development in Arabidopsis thaliana
To further uncover the underlying mechanisms of female gametophyte development defects, we investigated the female gametophyte structure in WT and grdp2-3 mutants via DIC microscopy. In general, female gametophyte development is divided into two main steps, megasporogenesis and megagametogenesis (Schneitz et al., 1995; Christensen et al., 1997). During megasporogenesis, the megaspore mother cell (MMC) (Figure 3A) undergoes meiosis and produces four haploid megaspores. Three of the megaspores degenerates, and only one persists to become the functional megaspore (FM) (Figure 3B). Subsequently, the FM undergoes three consecutive mitotic divisions (FG1–FG4), nuclear fusion, and cellularization (FG5–FG6) that lead to the formation of the mature embryo sac (FG7) (Figures 3C–F). In the FG7, WT female gametophyte consists of a central cell and an egg cell (Figure 3F).
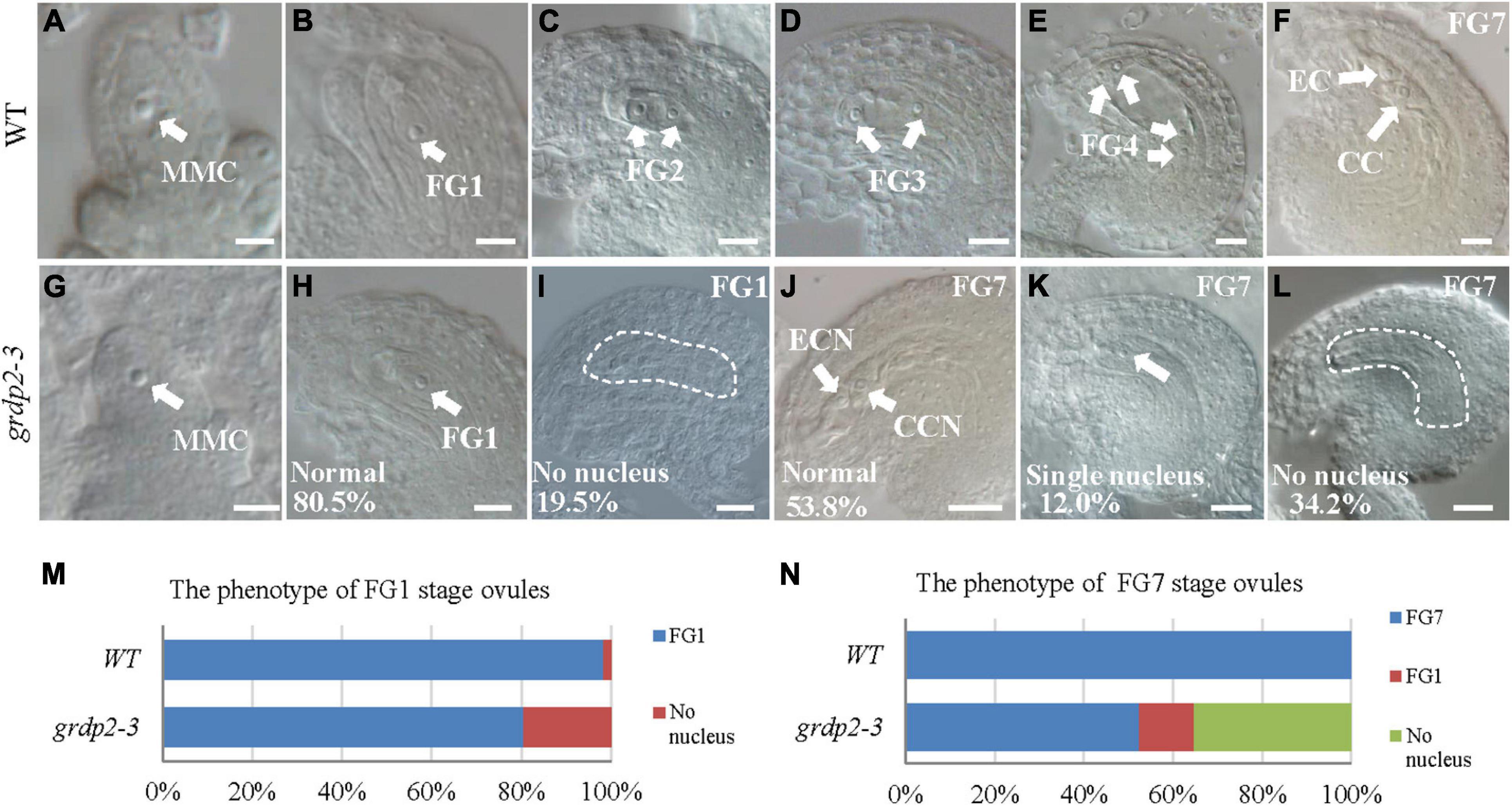
Figure 3. Female gametogenesis in WT and grdp2-3 plants examined by differential interference contrast microscope (DIC). Bar = 50 μm. (A) Megaspore mother cell (MMC) of WT plants. (B) 98.3% of 121 examined FG1 stage from WT normal ovules. (C–F) Female gametogenesis from FG2 to FG7 was normal in WT plants. (G) Megaspore mother cell (MMC) of grdp2-3 plants. (H) 80.5% of 221 examined FG1 stage ovules from grdp2-3 plants were normal. (I) 19.5% of 221 examined FG1 stage ovules from grdp2-3 plants showed no nucleus. (J) 53.8% of 179 examined FG7 stage ovules from grdp2-3 plants were normal. (K) 12.0% 179 examined FG7 stage ovules from grdp2-3 plants showed single nucleus. (L) 34.2% of 179 examined FG7 stage ovules from grdp2-3 plants showed no nucleus. (M) The phenotype analysis of FG1 stage ovules in WT and grdp2-3 plants. (N) The phenotype analysis of FG7 stage ovules in WT and grdp2-3 plants. CCN, central cell nuclei; EC, egg cell nuclei; FG, female gametophyte; white arrows mean nuclei; white dotted lines mean embryo sac.
In contrast, 34.2% of mutant ovules exhibited an absence of the female gametophyte, and 12.0% of mutant ovules have only one abnormal nucleus (Figures 3J–L,N) that may consequently cause an inability of the pollen tube to penetrate the ovule leading to abortion. We examined the female gametophyte structure of the grdp2-3 mutant plants at the early development stages to identify the exact stage of development defects. The result showed normal MMC (Figure 3G), while 19.5% of the functional megaspore produced after meiosis were abnormal and resulted in non-nucleus ovules (Figures 3H,I,M). These results suggest that defects in the mutant manifest during female meiosis before the functional megaspore is formed.
The expression analysis and GRDP2 phenotype in different T-DNA lines (Figure 1B) suggested that overexpression of GRDP2 results in abnormal ovule development. Therefore, we overexpressed GRDP2 and verified our results using p35S:GRDP2-OE and pUBQ10:GRDP2-OE overexpression lines. The positive homozygous plants of the T2 generation were analyzed for their phenotype. Expression of the GRDP2 gene was significantly up-regulated in three randomly selected independent lines of overexpression plants (Figures 4A,H). The seed setting was decreased considerably (about 50%) in the GRDP2 overexpressing lines (Figures 4B,C,I,J). Further phenotypic observation of the ovules of p35S:GRDP2-OE-26#, and pUBQ10:GRDP2-OE-6# plants at the stage of FG7 showed that there were two types of abortions; 30% of the ovules had no embryo sac, and 15% of the ovules were arrested at FG1 stage (Figures 4D,E,K,L). Consistent with the phenotype of grdp2-3 T-DNA insertion plants, Alexander’s staining of pollen showed that the pollen development in these overexpression plants was similar to WT (Figures 4F,G,M,N). Taken together, these results suggest that the accumulation of GRDP2 in Arabidopsis results in abnormal ovule development.
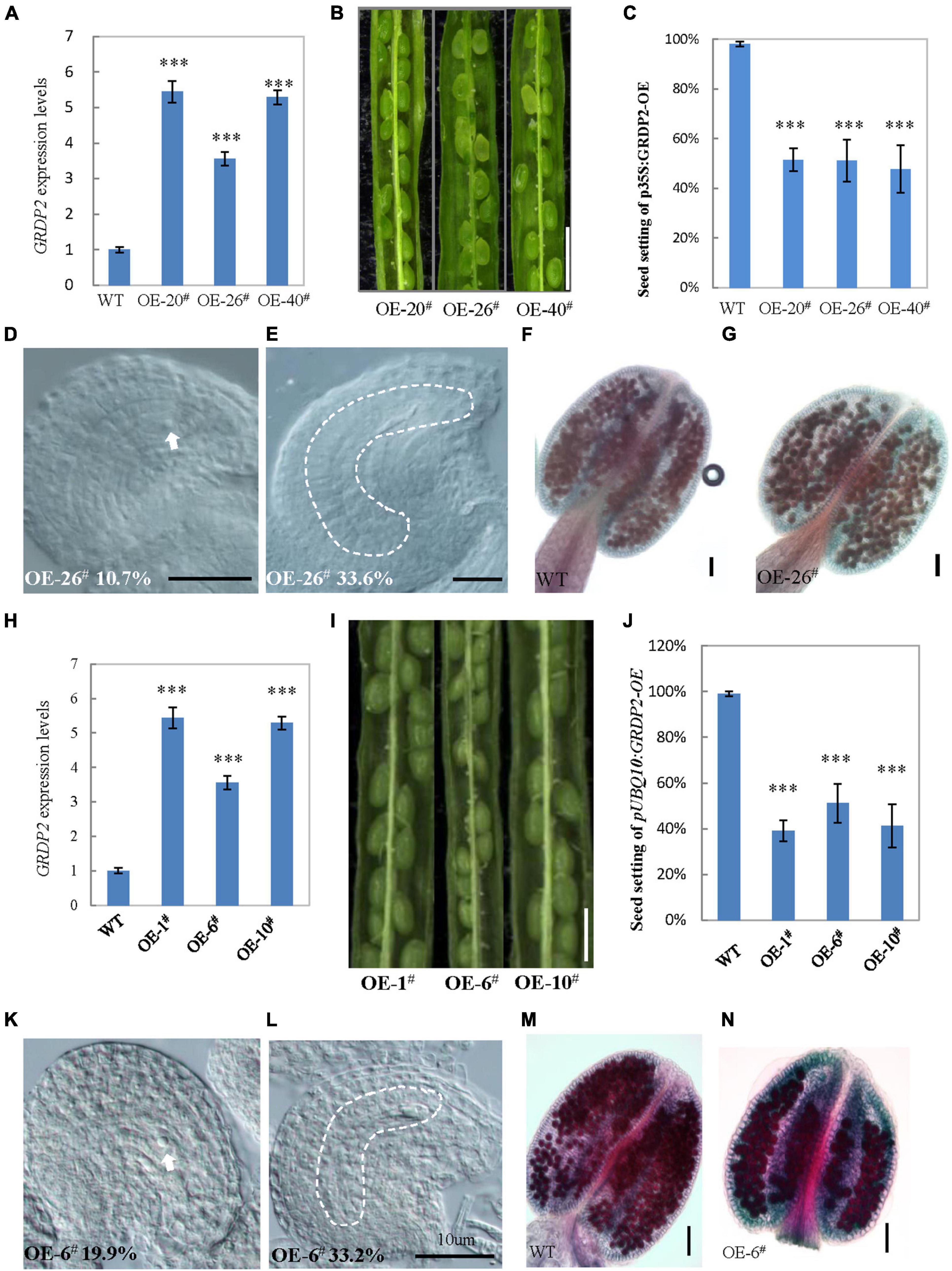
Figure 4. Phenotype analysis of p35S:GRDP2-OE and pUBQ10:GRDP2-OE lines. (A) Expression levels of GRDP2 in WT, p35S:GRDP2-OE-20#, p35S:GRDP2-OE-26#, and p35S:GRDP2-OE-40# as determined by real-time RT-PCR. (B) Seed development in three p35S:GRDP2-OE line plants. Bar = 1 cm. (C) Seed setting statistic of the WT, p35S:GRDP2-OE-20#, p35S:GRDP2-OE-26#, and p35S:GRDP2-OE-40# lines. (D,E) 10.7% of 316 examined FG7 stage ovules from p35S:GRDP2-OE-26# line plants showed single nucleus (D), 33.6% of 316 examined FG7 stage ovules from p35S:GRDP2-OE-26# line plants showed no nucleus (E). (F,G) Alexander’s staining of pollen from WT (F) and p35S:GRDP2-OE-26# (G) line plants. The red color indicates viable pollen. Bar = 50 μm. (H) Seed development in wild-type (WT) and three pUBQ10:GRDP2-OE line plants. Bar = 1 cm. (I) Expression levels of GRDP2 in WT, pUBQ10:GRDP2-OE-1#, pUBQ10:GRDP2-OE-6#, and pUBQ10:GRDP2-OE-10# as determined by real-time RT-PCR. (J) Seed setting statistics of the WT, pUBQ10:GRDP2-OE-1#, pUBQ10:GRDP2-OE-6#, and pUBQ10:GRDP2-OE-10# lines. (K,L) The FG7 stage ovules from pUBQ10:GRDP2-OE-26# line plants. (M,N) Alexander’s staining of pollen from WT (M) and pUBQ10:GRDP2-OE-6# (N) line plants. The red color indicates viable pollen. Bar = 50 μm. Vertical bars represent the mean ± SD as determined by two technical replicates of three biological replicates; asterisks show the significance level judged by Student’s t-test (∗∗∗p < 0.01). White dotted lines mean embryo sac.
Megasporogenesis Is Affected in grdp2-3 Mutants
As stated above that abnormalities were probably appearing in the meiosis of grdp2-3 mutants. Therefore, we investigated whether female meiocytes in the grdp2-3 ovules were competent to enter meiosis in the WT and grdp2-3 ovules at stage 2-IV (Figure 5A). The result revealed that female meiocytes in the grdp2-3 mutant entered meiosis similar to WT. Furthermore, to determine whether subsequent steps in meiosis were also affected in the grdp2-3 mutant, we examined callose deposition during meiosis progression. The result showed that in WT ovule, the callose signal first appears at the cell plate that separates the two daughter cells of female meiocyte in a dyad (Figure 5C, Dyad), followed by accumulation at the cell plates that separate the cells of a triad and a tetrad (Figure 5C, tetrad 1). After meiosis, the callose signal disappeared from the cell plate separating the four megaspores forming tetrad 2 and tetrad 3 (Figure 5C, tetrad 2 and tetrad 3). When a functional megaspore was formed, the callose signal almost disappeared in WT (Figure 5C, FM). However, in the grdp2-3 mutant, callose deposition during megasporogenesis differed from those observed in the WT ovules. grdp2-3 exhibited abnormal callose staining (Figure 5E), suggesting that cell plate formation during megasporogenesis is distorted in the grdp2-3 mutants. In addition to the aberrant callose-staining signal during the meiosis, we observed that callose fluorescence persisted in grdp2-3 ovules, even at later development stages (Figure 5E, FM). An increase at triad 1 stage in the grdp2-3 (31.2%) mutant compared to WT (18.8%) and a decrease in tetrad 2 (3.3%) and tetrad 3 (0.82%) (Figures 5B,D) were observed in the ovules at stage 2-IV, implying that the progression of meiosis is disturbed in the grdp2-3 mutant ovules. Also, 36.9% of ovules exhibited abnormal callose staining, suggesting cell plate formation during megasporogenesis is affected in the grdp2-3 mutants (Figures 5B,E).
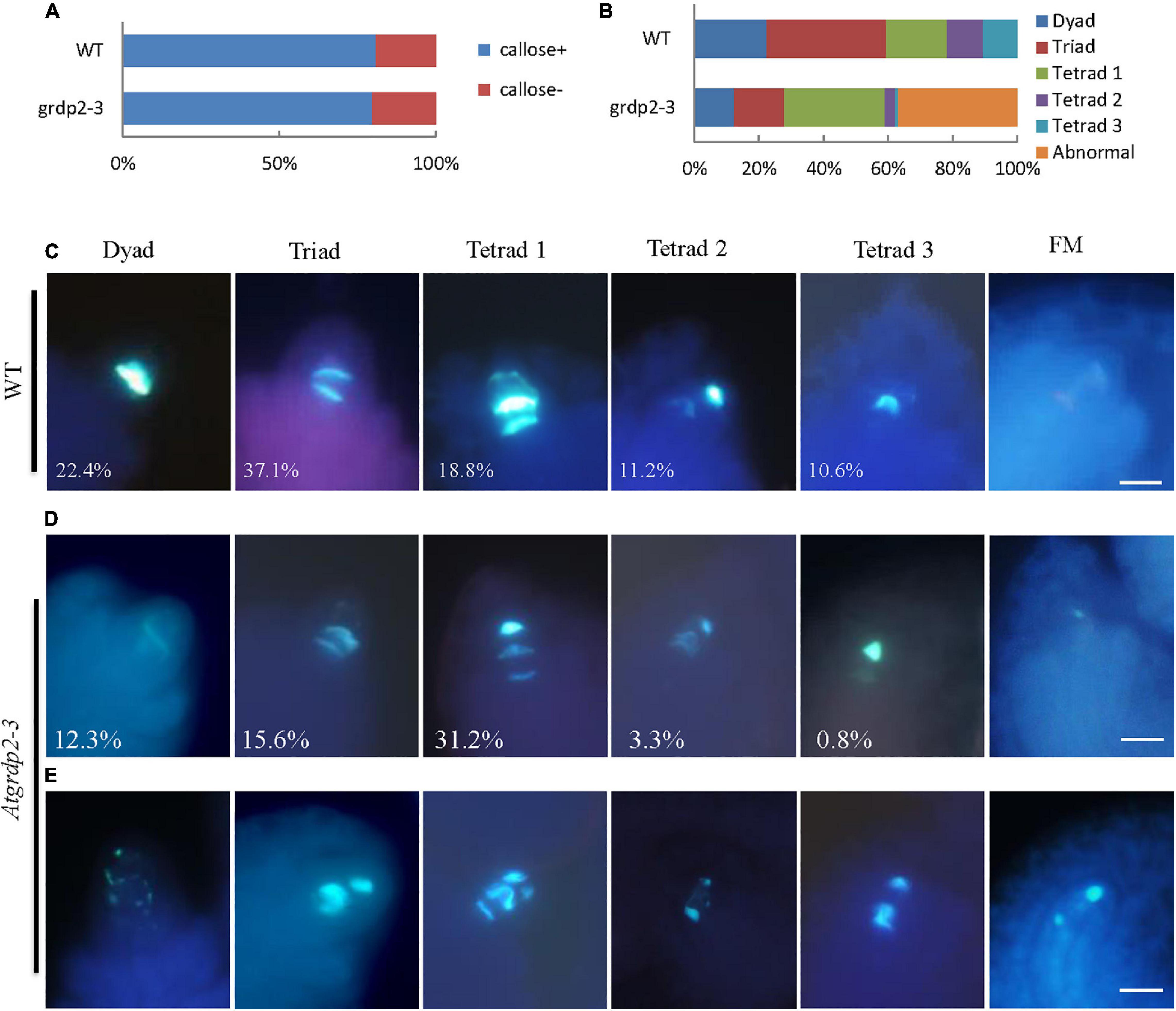
Figure 5. Callose deposition during megasporogenesis are defective in grdp2-3 mutant ovules. (A) Quantification of callose staining-positive (+) and callose staining-negative (–) ovules at stage 2IV in more than 100 ovules for each sample. (B) Quantitative profile of callose deposition at various stages of meiosis in more than 100 ovules for each sample. (C) Callose-stained wall deposition in WT ovules at stage 2IV. (D) Callose-stained wall deposition in grdp2-3 ovules at stage 2IV. (E) Abnormal callose staining is observed in grdp2-3 ovules during meiosis.
Female Gametophyte Specific Maker Genes Reveal Abnormal Ovule Development of grdp2-3
Previous studies have reported various female gametophyte-specific marker genes to detect the developmental process in the female gametophyte, such as the pSPL-GUS, pKNU:KNU-VENUS (Payne et al., 2004), pAKV:H2B-YFP (Schmidt et al., 2011; Cai et al., 2021), pMYB98-GFP (Kasahara et al., 2005; Steffen et al., 2007), pDD45-GFP (Steffen et al., 2007), and pDD65-GFP (Steffen et al., 2007). We introduced the different marker lines in the grdp2-3 background and then analyzed their expression in the wild-type and mutant backgrounds. SPL encodes a novel nuclear protein with limited homology to MADS-box transcription factors and has an early function during male and female sporogenesis. Therefore, we could see the GUS signal in the nucellus at the early gametogenesis stage in the WT background (Figure 6A); the signal in grdp2-3 was the same as WT (Figure 6B). Compared to WT, the expression of MMC-specific marker pKNU:KNU-VENUS was also normal (Figures 6C,D) in grdp2-3. These results suggest that the female gametophyte development at an early stage is not affected in grdp2-3. pAKV:H2B-YFP is a nucleus marker gene during the mitosis process of female gametophyte development. In the WT background, center cell, egg cell, synergid cell, and antipodal cell were present in the embryo sac at the mature FG7 stage (86.4%, n = 189) (Figure 6E). However, in grdp2-3 only 46.8% (n = 173) normal ovules were present (Figure 6F), and a large proportion of no-nucleus (34.7%, n = 173) (Figure 6G) and one nucleus ovules (18.5%, n = 173) (Figure 6H) were seen. Consistently, in WT background at FG7 stage, the synergid cell marker pMYB98-GFP (96.4%, n = 138), egg cell marker pDD45-GFP (96.3%, n = 107) and central cell marker pDD65-GFP (95.0%, n = 199) showed normal GFP signal in synergid cell, egg cell, and central cell, respectively (Figures 6I,L,O), while in grdp2-3 mutant there were only 47.9% (n = 144), 45.5% (n = 110), and 53.9% (n = 204) ovules with GFP signal at FG7 stage (Figures 6J,K,M,N,P,Q). These results indicate that grdp2 is involved in the female gametophyte development in Arabidopsis.
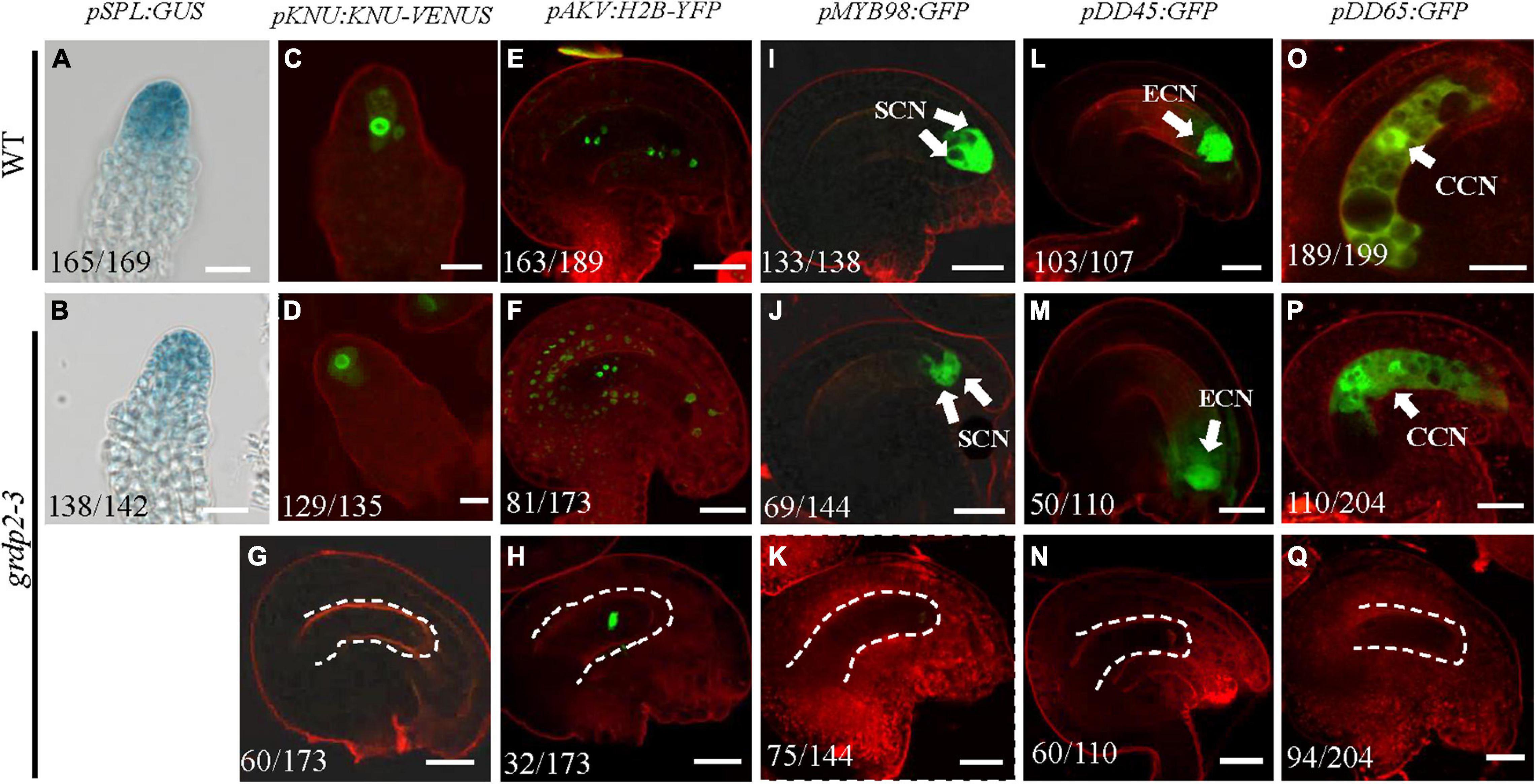
Figure 6. Expression of female gametophyte specific markers. (A,B) pSPL:GUS expression in (A) WT at stage 2-I ovules prior to entering meiosis (165/169) and (B) grdp2-3 stage 2-I ovules (138/142). (C,D) pKNU:KNU-VENUS expression in (C) WT at MMC stage ovules (151/156) and (D) grdp2-3 at MMC stage ovules (129/135). (E) pAKV:H2B-YFP expression in WT at FG6/FG7 stage ovules (163/189), the rest ovules are in the FG5 stage. (F–H) pAKV:H2B-YFP expression in (F) grdp2-3 at FG6/FG7 stage ovules (81/173) ovules are same as the WT; (G) 60/173 ovules have no nucleus; and (H) 32/173 ovule have one nucleus. (I) pMYB98:GFP expression in WT at FG7 stage ovules (133/138). (J,K) pMYB98:GFP expression in grdp2-3 at FG7 stage ovules (J) 69/144 ovules are same as the WT and (K) 75/144 ovules have no GFP signal. (L) pDD45:GFP expression in WT at FG7 stage ovules (103/107). (M,N) pDD45:GFP expression in grdp2-3 at FG7 stage ovules (M) 50/110 ovules are same as the WT and (N) 60/110 ovules have no GFP signal. (O) pDD65:GFP expression in WT at FG7 stage ovules (189/199). (P,Q) pDD65:GFP expression in grdp2-3 at FG7 stage ovules (P) 110/204 ovules are same as the WT and (Q) 96/204 ovules have no GFP signal. Bar = 25 μm. CCN, central cell nuclei; ECN, egg cell nuclei; SCN, synergid cell nuclei; white arrows mean nuclei; white dotted lines mean embryo sac.
Auxin Distribution During Gametogenesis Is Disrupted in grdp2 Ovule
Previous studies have shown that auxin plays an essential role in plant growth and female gametophyte development (Ceccato et al., 2013; Kneuper et al., 2020; Lv et al., 2020). To determine whether auxin distribution is altered in the grdp2-3 mutant, we crossed the DR5:GFP reporter line and examined the spatial distribution of auxin (Benkova et al., 2003) in the grdp2-3 plants. In the WT ovules, the weak DR5:GFP signal was detected in the epidermal L1 cell layer of the ovule primordium starting from stage 2-II (Figure 7A), and it got stronger in stage 2-IV (Figure 7B). From stage 2-V, the auxin response was detected in the epidermal cell and the funiculus pro-vascular cells until stage 3-II (Figures 7C–E). While after stage 3-IV, the DR5:GFP signal could only be found in the funiculus (Figures 7F,G). In contrast, in the grdp2-3 ovules, the DR5:GFP signal pattern was significantly disrupted, and the signal appeared later than that of WT in the epidermal L1 cell layer of the ovule primordium starting from stage 2-IV (Figure 7B’). The DR5 signal continued until the mature stage 3-VI (Figure 7G’), and in the funiculus, DR5:GFP signal appeared later than WT (Figures 7E’–G’).
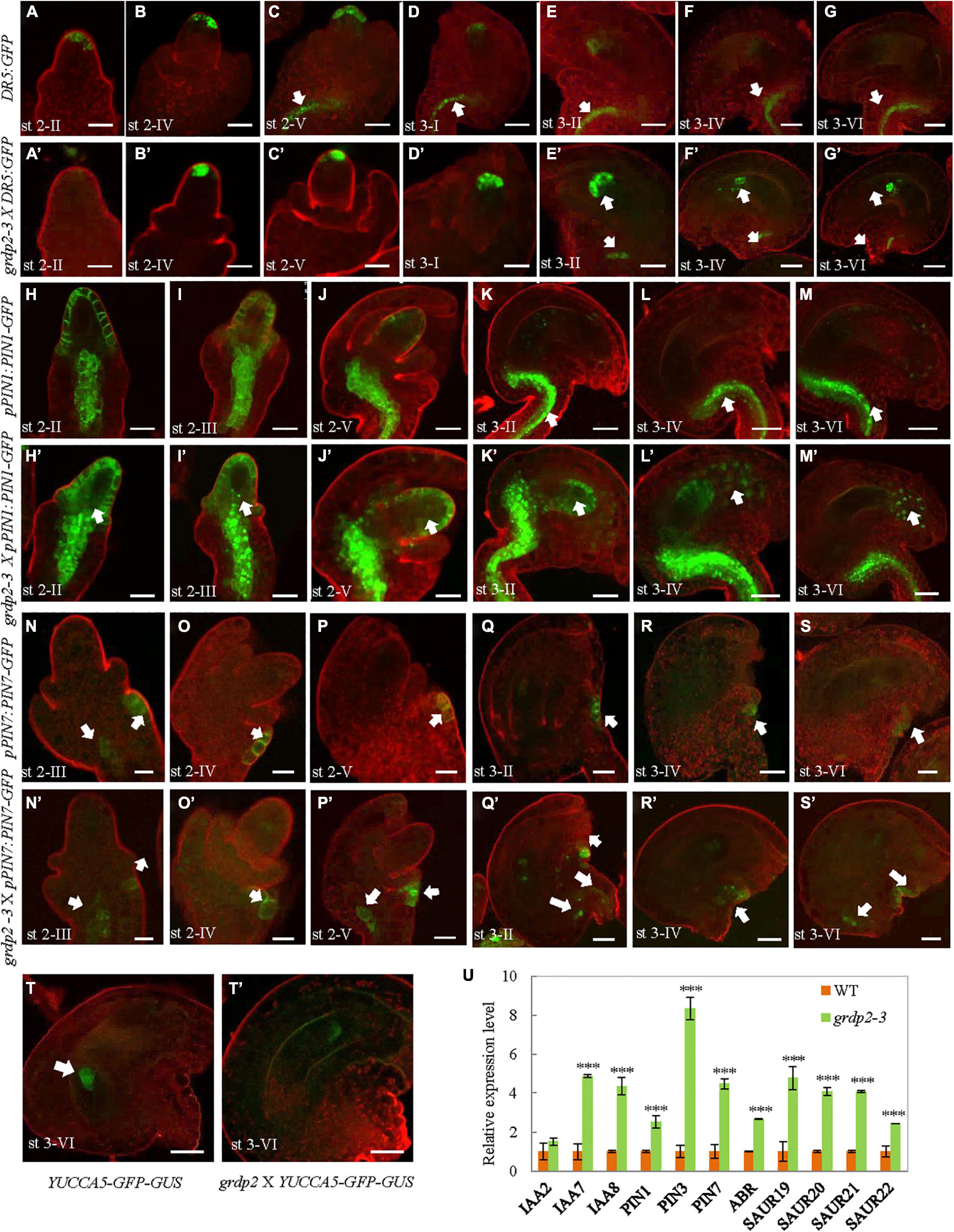
Figure 7. Auxin distribution in developing ovules from WT and grdp2-3 siliques using DR5:GFP, pPIN1:PIN1-GFP, pPIN7:PIN7-GFP, and YUCCA5-GFP-GUS as an auxin reporter. (A–G) WT DR5:GFP ovules were analyzed at different developmental stages. (A) The cytoplasmatic GFP signal is first detected in the hypodermal L1 cell layer of the ovule primordium from stage 2-II, (E) until stage 3-II. (C) DR5:GFP signal is also detected in the pro-vascular cells of the funiculus from stage 2-V to (G) stage 3-VI. (A’–G’) grdp2-3 DR5:GFP ovules were analyzed at different developmental stages. (B’) The cytoplasmatic GFP signal is first detected in the hypodermal L1 cell layer of the ovule primordium from stage 2-III which is later than WT (A, stage 2-II), and (G’) last until the mature stage 3-VI. White arrows in (E’–G’) showed DR5:GFP signal in the pro-vascular cells of the funiculus is also later than WT. (H–M) WT pPIN1:PIN1-GFP ovules were analyzed at different developmental stages. (H) The cytoplasmatic GFP signal is first detected in the hypodermal L1 cell layer of the ovule primordium and the pro-vascular cells of the funiculus from stage 2-II, until (J) stage 2-V. (white arrows in K–M) GFP signal was only detected in the funiculus from stage 3-II to stage 3-IV. (H’–M’) grdp2-3 pPIN1:PIN1-GFP ovules were analyzed at different developmental stages. Except the hypodermal L1 cell layer of the ovule primordium and the pro-vascular cells of the funiculus, the GFP signal also detected in the nucellus at all developmental stages stage 2-II to stage 3-IV. (N–S) WT pPIN7:PIN7-GFP ovules were analyzed at different developmental stages. (N) The cytoplasmatic GFP signal is only detected in the outer integument of the ovule primordium from stage 2-III, (S) until stage 3-VI. (N’–S’) grdp2-3 pPIN7:PIN7-GFP ovules were analyzed at different developmental stages. (N’) The cytoplasmatic GFP signal can be detected not only in the outer integument of the ovule primordium but also the pro-vascular cells of the funiculus from 2-III, until (S’) stage 3-VI. (T) YUC5:GFP-GUS is expressed only in the chalaza at stage 3-VI. (T’) grdp2-3 YUC5:GFP-GUS ovules were analyzed at stage 3-VI, the GFP expression in the embryo sac which is a big difference with WT. In (A–T) and (A’–T’), cell membranes were stained with FMH 4-64 FX. Scale bars = 25 μm. (U) Relative mRNA levels of auxin-related genes by qRT-PCR. Data are means ± SD (n = 3 biological replicates; ***p < 0.01, Student’s t-test).
To analyze if the spatial discrepancies in ovules may be facilitated by polar auxin transport (PAT) away from the sites of synthesis, we assessed the expression of members of the PINFORMED (PIN) family of auxin efflux carriers (Friml et al., 2003). Both pPIN1:PIN1-GFP, and pPIN7:PIN7-GFP constructs were used to detect different female gametophyte development stages in the WT and grdp2-3 backgrounds. The result showed that PIN1-GFP was strongly expressed in the chalaza and the epidermal cell at stage 2-II in WT ovules (Figure 7H). During the subsequent development stages, the PIN1-GFP signals at the epidermal cell were getting weaker and weaker, and no signal was detected at the mature stage. Simultaneously, the PIN1-GFP had a strong expression at all stages in the chalaza and funiculus (Figures 7I–M). However, the expression of PIN1-GFP was more robust in grdp2-3 ovules and appeared in the nucellus except for the epidermal cell and chalaza (Figures 7H’,I’). Furthermore, the micropylar signal existed in the subsequent development stages in the WT (Figures 7J’–M’). The pPIN7:PIN7-GFP expression pattern did not change during the megasporogenesis (Figures 7N,O,N’,O’); however, there was a considerable difference in PIN7 expression during megagametogenesis between WT and grdp2-3. In addition to the outer integument (Figures 7P–S,P’–S’), pPIN7:PIN7-GFP signal was detected in the chalaza (Figures 7P’–S’). In addition, compared with the membrane-localized PIN1 and PIN7 in WT, the signals were detected in the cytoplasm in grdp2-3 (Figures 7H,H’,I,I’,O,P,O’,P’). Consistently, an auxin bio-synthesis gene pYUCCA5-GFP-GUS was used to detect the auxin synthesis in grdp2-3 ovules. The result showed that auxin biosynthesis at stage 3-VI ovules of grdp2-3 was misregulated (Figures 7T,T’). Also, we found both the patterns and intensity of DR5, PIN1, PIN2, and PIN7 were altered in grdp2-3 roots (Supplementary Figure 1). These changes in expression patterns of auxin response and bio-synthesis genes suggest that auxin biosynthesis and transport may be affected in grdp2-3.
We further determined the altered auxin signaling and transport-related defects in grdp2 by analyzing the transcript levels of auxin-related genes in WT and grdp2-3 by qRT-PCR. The auxin efflux carriers PIN1, PIN3, PIN7, and ABR; auxin-responsive genes SAUR19, SAUR20, SAUR21, and SAUR22, and auxin signaling genes IAA2, IAA7, and IAA8 were analyzed. For most of the genes, there was a significant increase in their transcript level in the inflorescence of grdp2-3, and no substantial increase was detected for IAA2 (Figure 7U). We further studied the auxin response in the grdp2-3 mutants by exogenously applying the auxin (IAA) and auxin transport inhibitor (NPA) and observed the growth of the grdp2-3 roots after treatment. The results indicate that auxin accumulation in grdp2-3 results in more rapid root growth (Supplementary Figure 2). These observations suggest that the auxin signaling and transport are disrupted in the grdp2 mutant.
Overexpression of GRDP1 Results in a Similar Phenotype Like GRDP2
GRDP1 and GRDP2 are the two members of GRDPs in Arabidopsis. Their protein sequences share 64% identity (Rodriguez-Hernandez et al., 2017). Therefore, we speculated that there could be similarities in gene function between the two genes. The grdp1 T-DNA (SALK_079708) insertion site and GRDP1 expression level are shown in Supplementary Figure 3A. The development of the seed in grdp1 mutant was normal (Supplementary Figures 3B,C). We fused the GRDP1 promoter with GFP and GUS protein to detect the expression patterns of GRDP1 in different stages of ovule development. The results showed that GFP and GUS signals have the same expression pattern during ovule development. At first, the signals appear in nucellus tissues at the MMC stage (Figures 8A,F). With the development, the signals can be detected in both the embryo sac and nucellus tissue from FG1 to FG2 stage (Figures 8B,C,G,H). During the subsequent developmental stages, the signals appeared at the chalaza end and in integument tissues (Figures 8D,E,I,J). We also detected the expression pattern of GRDP1 in pollen and found that the GRDP1 expressed in mature anther (Figures 8K,L), which is different from the expression pattern of GRDP2. We also use the CRISPR/Cas9 genome-editing system to get the grdp1grdp2 double mutant (Wang and Chen, 2019), the result showed that the seed setting is normal in Crispr-7# (grdp1grdp2 double mutant) line (Supplementary Figures 4A–D).
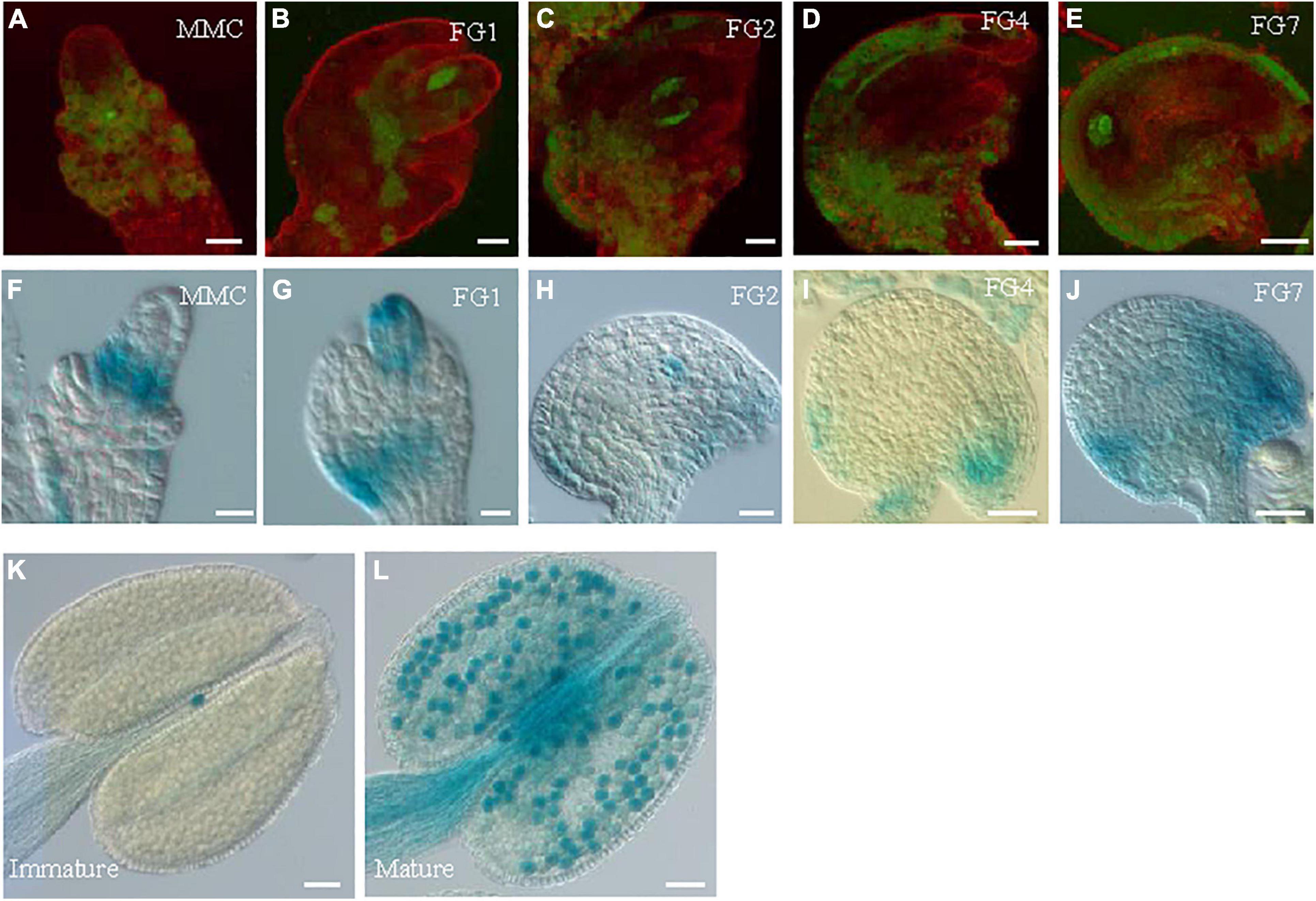
Figure 8. The expression profiles of GRDP1 in Arabidopsis. (A–E) pGRDP1:GFP expression pattern in different ovule development stages. The developmental stages of the ovules are indicated at the top right corner of each panel. Bars = 20 μm. (F–J) pGRDP1:GUS expression pattern in ovules. Bars = 20 μm. (K,L) pGRDP1:GUS expression pattern in pollen. Bars = 50 μm.
The overexpression lines using UBQ10 derived GRDP1 (pUBQ10:GRDP1-OE-2#, pUBQ10:GRDP1-OE-12#, and pUBQ10:GRDP1-OE-23#) showed significant up-regulation in GRDP1 expression (Figure 9A). The seed setting was decreased considerably (50.5%,47.8%, and 36.2%, respectively) in GRDP1 overexpressing lines (Figures 9B,C). Further phenotypic observation of the ovules of pUBQ10:GRDP1-OE-2# plants at FG7 stage showed that there were two types of abortions; 30.6% of the ovules had no embryo sac, and 24.1% of the ovules were arrested at FG1 stage (Figures 9D–G). These results indicate that GRDP1 and GRDP2 have functional redundancy in the regulation of ovule development.
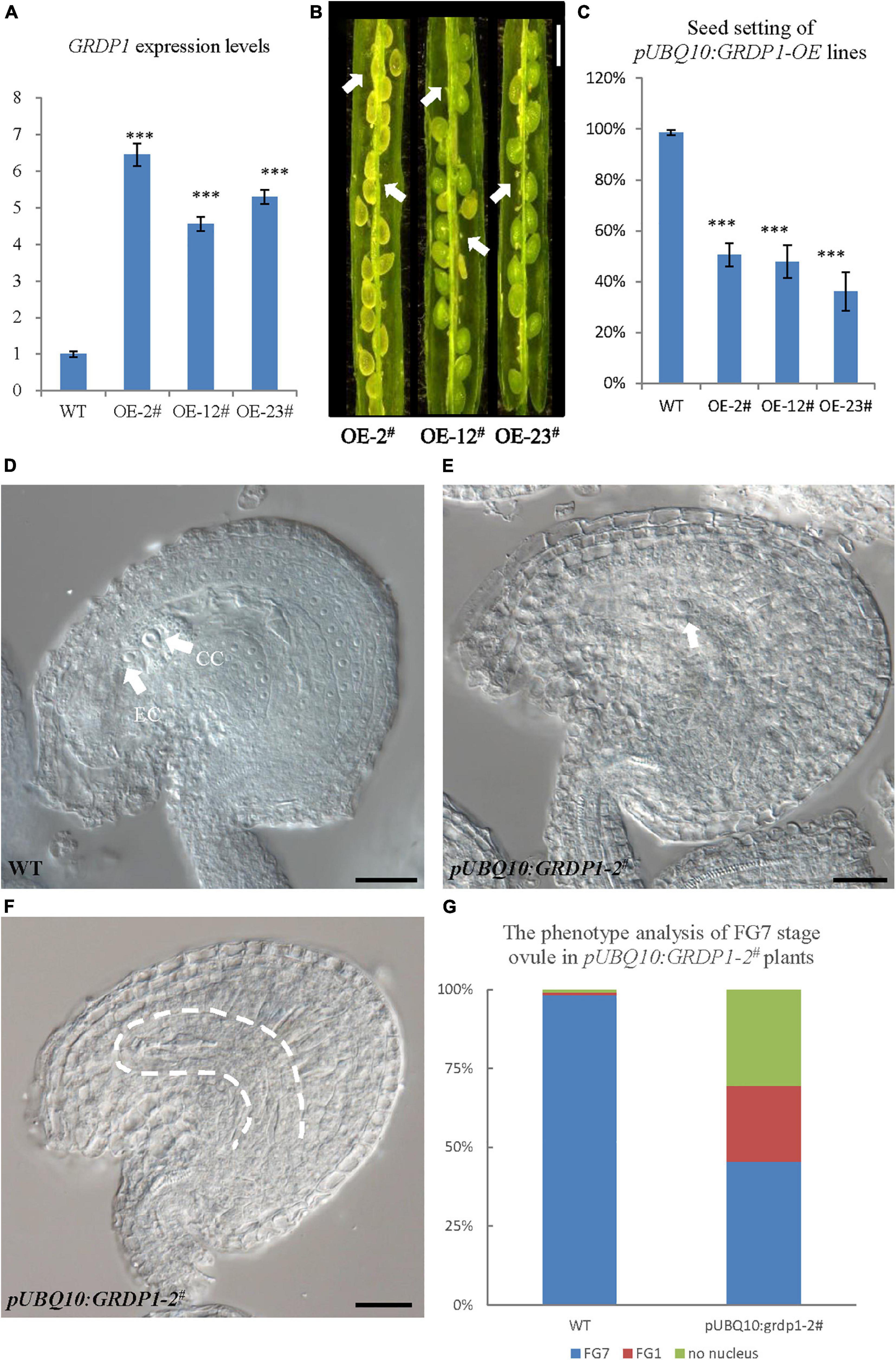
Figure 9. Phenotype analysis of pUBQ10:GRDP1-OE lines. (A) Expression levels of GRDP1 in WT, pUBQ10:GRDP1-OE-2#, pUBQ10:GRDP1-OE-12#, and pUBQ10:GRDP1-OE-23# as determined by real-time RT-PCR. (B) Seed development in three pUBQ10:GRDP1-OE line plants. Bar = 0.5 cm. (C) Seed setting statistics of the WT, pUBQ10:GRDP1-OE-2#, pUBQ10:GRDP1-OE-12#, and pUBQ10:GRDP1-OE-23# lines. (D) The FG7 stage ovules from WT plants. (E,F) The FG7 stage ovules from pUBQ10:GRDP1-OE-2# plants, 24.1% of 196 examined FG7 stage ovules showed single nucleus, 30.6% of 196 examined FG7 stage ovules showed no nucleus. (G) The phenotype analysis of FG7 stage ovules in WT and pUBQ10:GRDP1-OE-2# plants. Vertical bars represent the mean ± SD as determined by two technical replicates of three biological replicates; asterisks show the significance level judged by Student’s t-test (∗∗∗p < 0.01). CC, central cell; EC, egg cell. White arrows mean nuclei; white dotted lines mean embryo sac.
Discussion
The canonical glycine-rich proteins contain a high glycine percentage (from 40% to 70%), with arranged (Gly)n-X repetitions (Sachetto-Martins et al., 2000; Mousavi and Hotta, 2005). Besides the canonical GRPs, there are GRDPs containing a short glycine-rich region. Several transcripts encoding GRDPs have been reported to be induced under abiotic stress (Bocca et al., 2005; Ortega-Amaro et al., 2014; Rodríguez-Hernández et al., 2014). The expression patterns of some canonical plant GRPs have been reported in floral organs. The oleosin-like protein GRP17 was identified as a component of the pollen wall of Arabidopsis thaliana and is needed for rapid initiation of pollination (Suzuki et al., 1999; Mayfield and Preuss, 2000). Another GRP, GRP23, is a novel nuclear PPR domain protein critical for early embryogenesis (Ding et al., 2006). Transcripts of Arabidopsis GRPs (GRP1 and GRP2) express abundantly in flowers, suggesting their essential functions in floral organ development (de Oliveira et al., 1990). These studies indicate that GRPs play a variety of roles in developing flower organs, male and female gametophytes. In this study, we describe the GRDP2 gene function in female gametophyte development. GRDP2 encodes a non-canonical glycine-rich protein of unknown function containing a DUF1399 domain, a putative RNA-binding motif, and a glycine-rich domain. The GRDP2 gene is regulated, with exceptionally high mRNA levels in inflorescence than root, stem, leaf, and immature siliques, implying that GRDP2 participate in plant reproductive development process (Figure 2A). The different expression patterns of GUS or GFP in ovules and pollen indicate the different functions of GRDP2 in male or female gametophytes development.
The phenotype analysis of the three mutant alleles grdp2-1, grdp2-2, and grdp2-3 shared different developmental defects in seed sets, indicating the significant difference between the expression level of GRDP2 in different alleles may account for severe phenotypes in seed sets of grdp2-3. Our results showed that the expression level of GRDP2 in grdp2-1 and grdp2-2 mutants were knockout or knockdown, but the expression level of GRDP2 in grdp2-3 was significantly increased (Figures 1A,B). We further found that grdp2-3 overexpression lines caused reduced plant fertility (50%), but the pollen development was not affected (Figures 1C–E). The female gametogenesis of grdp2-3 mutant was disrupted with abnormal embryo sac degradation and nuclear number at the FG7 stage (Figures 2K,L). Similar to grdp2-3, the p35S:GRDP2-OE and pUBQ10:GRDP2-OE overexpression lines exhibited low seed sets and female gametophyte abortion (Figure 4). These data show that GRDP2 overexpression leads to abnormal female gametogenesis. DIC observation (Figures 3H,I), and callose staining (Figures 5C–E) showed that the megasporogenesis process was also affected by the overexpression of GRDP2. Unlike the typical GRP proteins, the GRDP2 function is mainly in the female gametophyte development process. This may be due to the differences in the structure of the glycine-rich domain.
Hormones, such as auxins, are involved in the complex molecular network that regulates the coordinated development of plant organs (Bennett et al., 1996; Benkova et al., 2003). Genes controlling ovule patterning have been identified and studied in detail (Qin et al., 2014; Zhao et al., 2018; Su et al., 2020); however, the roles of auxin in ovule development are largely unknown. Based on our data on local auxin synthesis, PIN localization, and auxin response patterns, we found that overexpression of GRDP2 leads to defective auxin distribution in the whole ovule development process (Figure 7). Interestingly, it has been reported that the GRDP2 belongs to a group of genes regulated explicitly by indole-3-acetic acid (IAA) (Goda et al., 2004). We analyzed IAA expression levels in WT and grdp2-3 lines, our result of qRT-PCR verified that grdp2-3 lines accumulated the higher auxin levels (Figure 7U). The abnormal auxin responses at the embryo sac might explain defective embryo sac development in grdp2-3 because auxin is critical for embryo sac patterning and gamete specification (Pagnussat et al., 2009). Our results reveal that overexpression of GRDP2 results in abnormal ovule development possibly through an auxin-dependent mechanism.
The different expression patterns of GRDP1 (Figures 8K,L) and GRDP2 (Figures 2C,D) in anther indicate that they may have different roles in pollen development. However, the overexpression lines of pUBQ10:GRDP1 show two types of abnormal ovules at FG1 stage and FG7 stage (Figures 9D–G), which is similar to GRDP2 overexpression plants suggesting that GRDP1 and GRDP2 could have redundant functions during ovule development in Arabidopsis thaliana.
Taken together, our results indicate that the excess accumulation of GRDP2 is harmful to ovule development in Arabidopsis, resulting in disorder of auxin distribution in the ovule and abnormal ovule development.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
LW performed most of the experiments, analyzed the research results, and wrote the manuscript. YL and BJ contributed to data analysis. HC, MA, and YQ made the critical revision. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the Science and Technology Major Project of Guangxi (Gui Ke 2018-266-Z01), the National Natural Science Foundation of China (31970333 to YQ), and the Science and Technology Program of Fujian Province (2019N5008).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.698487/full#supplementary-material
Footnotes
References
Aslam, M., Sugita, K., Qin, Y., and Rahman, A. (2020). Aux/IAA14 regulates microRNA-Mediated cold stress response in arabidopsis roots. Int. J. Mol. Sci. 21:8441. doi: 10.3390/ijms21228441
Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/S0092-8674(03)00924-3
Bennett, M. J., Marchant, A., Green, H. G., May, S. T., Ward, S. P., Millner, P. A., et al. (1996). Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950. doi: 10.1126/science.273.5277.948
Bocca, S. N., Magioli, C., Mangeon, A., Junqueira, R. M., Cardeal, V., Margis, R., et al. (2005). Survey of glycine-rich proteins (GRPs) in the Eucalyptus expressed sequence tag database (ForEST). Genet. Mol. Biol. 28, 608–624. doi: 10.1590/S1415-47572005000400016
Braselton, J. P., Wilkinson, M. J., and Clulow, S. A. (1996). Feulgen staining of intact plant tissues for confocal microscopy. Biotech. Histochem. 71, 84–87. doi: 10.3109/10520299609117139
Cai, H., Liu, L., Zhang, M., Chai, M., Huang, Y., Chen, F., et al. (2021). Spatiotemporal control of miR398 biogenesis via chromatin remodeling and kinase signaling ensures proper ovule development. Plant Cell 33, 1530–1553. doi: 10.1093/plcell/koab056
Ceccato, L., Masiero, S., Roy, D. S., Bencivenga, S., Roig-Villanova, I., Ditengou, F. A., et al. (2013). Maternal control of PIN1 is required for female gametophyte development in arabidopsis. PLoS One 8:e66148. doi: 10.1371/journal.pone.0066148
Christensen, C. A., King, E. J., Jordan, J. R., and Drews, G. N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod. 10, 49–64. doi: 10.1007/s004970050067
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Condit, C. M. (1993). Developmental expression and localization of petunia glycine-rich protein 1. Plant Cell 5, 277–288. doi: 10.1105/tpc.5.3.277
de Oliveira, D. E., Seurinck, J., Inzé, D., Van Montagu, M., and Botterman, J. (1990). Differential expression of five arabidopsis genes encoding glycine-rich proteins. Plant Cell 2, 427–436. doi: 10.1105/tpc.2.5.427
Ding, Y. H., Liu, N. Y., Tang, Z. S., Liu, J., and Yang, W. C. (2006). Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18, 815–830. doi: 10.1105/tpc.105.039495
Dorcey, E., Urbez, C., Blazquez, M. A., Carbonell, J., and Perez-Amador, M. A. (2009). Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J. 58, 318–332. doi: 10.1111/j.1365-313X.2008.03781.x
Figueiredo, D. D., Batista, R. A., Roszakt, P. J., Hennig, L., and Kohler, C. (2016). Auxin production in the endosperm drives seed coat development in Arabidopsis. Elife 5:e20542. doi: 10.7554/eLife.20542.028
Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., et al. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153. doi: 10.1038/nature02085
Fusaro, A. F., and Sachetto-Martins, G. (2007). Blooming time for plant glycine-rich proteins. Plant Signal Behav. 2, 386–387. doi: 10.4161/psb.2.5.4262
Goda, H., Sawa, S., Asami, T., Fujioka, S., Shimada, Y., and Yoshida, S. (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134, 1555–1573. doi: 10.1104/pp.103.034736
Kasahara, R. D., Portereiko, M. F., Sandaklie-Nikolova, L., Rabiger, D. S., and Drews, G. N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17, 2981–2992. doi: 10.1105/tpc.105.034603
Keller, B., Sauer, N., and Lamb, C. J. (1988). Glycine-rich cell wall proteins in bean: gene structure and association of the protein with the vascular system. EMBO J. 7, 3625–3633. doi: 10.1002/j.1460-2075.1988.tb03243.x
Kneuper, I., Teale, W., Dawson, J. E., Tsugeki, R., Katifori, E., Palme, K., et al. (2020). Auxin biosynthesis and cellular efflux act together to regulate leaf vein patterning. J. Exp. Bot. 72, 1151–1165. doi: 10.1093/jxb/eraa501
Larsson, E., Vivian-Smith, A., Offringa, R., and Sundberg, E. (2017). Auxin homeostasis in arabidopsis ovules is anther-dependent at maturation and changes dynamically upon fertilization. Front. Plant Sci. 8:1735. doi: 10.3389/fpls.2017.01735
Liu, L., Zhao, L., Chen, P., Cai, H., Hou, Z., Jin, X., et al. (2020). ATP binding cassette transporters ABCG1 and ABCG16 affect reproductive development via auxin signalling in Arabidopsis. Plant J. 102, 1172–1186. doi: 10.1111/tpj.14690
Long, R., Yang, Q., Kang, J., Zhang, T., Wang, H., Li, M., et al. (2013). Overexpression of a novel salt stress-induced glycine-rich protein gene from alfalfa causes salt and ABA sensitivity in Arabidopsis. Plant Cell Rep. 32, 1289–1298. doi: 10.1007/s00299-013-1443-0
Lv, B., Zhu, J., Kong, X., and Ding, Z. (2020). Light participates in the auxin-dependent regulation of plant growth. J. Integr. Plant Biol. 63, 819–822. doi: 10.1111/jipb.13036
Mangeon, A., Junqueira, R. M., and Sachetto-Martins, G. (2010). Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal Behav. 5, 99–104. doi: 10.4161/psb.5.2.10336
Mayfield, J. A., and Preuss, D. (2000). Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat. Cell Biol. 2, 128–130. doi: 10.1038/35000084
Mousavi, A., and Hotta, Y. (2005). Glycine-rich proteins - A class of novel proteins. Appl. Biochem. Biotechnol. 120, 169–174. doi: 10.1385/ABAB:120:3:169
Murashige, T., and Skoog, F. A. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plantarum 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nambara, E., and Van Wees, S. C. M. (2021). Plant hormone functions and interactions in biological systems. Plant J. 105, 287–289. doi: 10.1111/tpj.15151
Ortega-Amaro, M. A., Rodriguez-Hernandez, A. A., Rodriguez-Kessler, M., Hernandez-Lucero, E., Rosales-Mendoza, S., Ibanez-Salazar, A., et al. (2014). Overexpression of AtGRDP2, a novel glycine-rich domain protein, accelerates plant growth and improves stress tolerance. Front. Plant Sci. 5:782. doi: 10.3389/fpls.2014.00782
Ortega-Amaro, M. A., Rodríguez-Kessler, M., Rodríguez-Hernández, A. A., Becerra-Flora, A., Rosales-Mendoza, S., and Jiménez-Bremont, J. F. (2016). Overexpression of AtGRDP2 gene in common bean hairy roots generates vigorous plants with enhanced salt tolerance. Acta Physiol. Plantarum 38:66. doi: 10.1007/s11738-016-2083-0
Pagnussat, G. C., Alandete-Saez, M., Bowman, J. L., and Sundaresan, V. (2009). Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324, 1684–1689. doi: 10.1126/science.1167324
Payne, T., Johnson, S. D., and Koltunow, A. M. (2004). KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131, 3737–3749. doi: 10.1242/dev.01216
Peterson, R., Slovin, J. P., and Chen, C. (2010). A simplified method for differential staining of aborted and non-aborted pollen grains. Int. J. Plant Biol. 1, 66–69. doi: 10.4081/pb.2010.e13
Qin, Y., Zhao, L. H., Skaggs, M. I., Andreuzza, S., Tsukamoto, T., Panoli, A., et al. (2014). ACTIN-RELATED PROTEIN6 regulates female meiosis by modulating meiotic gene expression in arabidopsis. Plant Cell 26, 1612–1628. doi: 10.1105/tpc.113.120576
Reddy, A. S. N., and Poovaiah, B. W. (1987). Accumulation of a glycine rich protein in auxin-deprived strawberry fruits. Biochem. Biophys. Res. Commun. 147, 885–891. doi: 10.1016/S0006-291X(87)80153-5
Rodriguez-Hernandez, A. A., Muro-Medina, C. V., Ramirez-Alonso, J. I., and Jimenez-Bremont, J. F. (2017). Modification of AtGRDP1 gene expression affects silique and seed development in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 486, 252–256. doi: 10.1016/j.bbrc.2017.03.015
Rodríguez-Hernández, A. A., Ortega-Amaro, M. A., Delgado-Sánchez, P., Salinas, J., and Jiménez-Bremont, J. F. (2014). AtGRDP1 gene encoding a glycine-rich domain protein is involved in germination and responds to ABA signalling. Plant Mol. Biol. Reporter 32, 1187–1202. doi: 10.1007/s11105-014-0714-4
Roosjen, M., Paque, S., and Weijers, D. (2018). Auxin Response Factors: output control in auxin biology. J. Exp. Bot. 69, 179–188. doi: 10.1093/jxb/erx237
Ryser, U., Schorderet, M., Zhao, G. F., Studer, D., Ruel, K., Hauf, G., et al. (1997). Structural cell-wall proteins in protoxylem development: evidence for a repair process mediated by a glycine-rich protein. Plant J. 12, 97–111. doi: 10.1046/j.1365-313X.1997.12010097.x
Sachetto-Martins, G., Franco, L. O., and de Oliveira, D. E. (2000). Plant glycine-rich proteins: a family or just proteins with a common motif? Biochim. Biophys. Acta 1492, 1–14. doi: 10.1016/S0167-4781(00)00064-6
Schmidt, A., Wuest, S. E., Vijverberg, K., Baroux, C., Kleen, D., and Grossniklaus, U. (2011). Transcriptome analysis of the arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biol. 9:e1001155. doi: 10.1371/journal.pbio.1001155
Schneitz, K., Hulskpamp, M., and Puritt, R. E. (1995). Wild-type ovule development in Arabidopsis thaliana a light microscope study of cleared whole mount tissue. Plant J. 7, 731–749. doi: 10.1046/j.1365-313X.1995.07050731.x
Steffen, J. G., Kang, I. H., Macfarlane, J., and Drews, G. N. (2007). Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51, 281–292. doi: 10.1111/j.1365-313X.2007.03137.x
Su, Z., Wang, N., Hou, Z., Li, B., Li, D., Liu, Y., et al. (2020). Regulation of female germline specification via small RNA mobility in arabidopsis. Plant Cell 32, 2842–2854. doi: 10.1105/tpc.20.00126
Suzuki, M., Watanabe, T. K., Fujiwara, T., Nakamura, Y., Takahashi, E., and Tanigami, A. (1999). Molecular cloning, expression, and mapping of a novel human cDNA, GRP17, highly homologous to human gadd45 and murine MyD118. J. Hum. Genet. 44, 300–303. doi: 10.1007/s100380050164
Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. doi: 10.1105/tpc.9.11.1963
Urbez, C., Cercos, M., Perez-Amador, M. A., and Carbonell, J. (2006). Expression of PsGRP1, a novel glycine rich protein gene of Pisum sativum, is induced in developing fruit and seed and by ABA in pistil and root. Planta 223, 1292–1302. doi: 10.1007/s00425-005-0178-8
Varner, J. E., and Cassab, G. I. (1986). A new protein in petunia. Nature 323:110. doi: 10.1038/323110a0
Wang, J., and Chen, H. (2019). A novel CRISPR/Cas9 system for efficiently generating Cas9-free multiplex mutants in Arabidopsis. aBIOTECH 1, 6–14. doi: 10.1007/s42994-019-00011-z
Zhao, L., Cai, H., Su, Z., Wang, L., Huang, X., Zhang, M., et al. (2018). KLU suppresses megasporocyte cell fate through SWR1-mediated activation of WRKY28 expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E526–E535. doi: 10.1073/pnas.1716054115
Keywords: GRDP2, female gametophyte, ovule development, auxin, embryo sac
Citation: Wang L, Liu Y, Aslam M, Jakada BH, Qin Y and Cai H (2021) The Glycine-Rich Domain Protein GRDP2 Regulates Ovule Development via the Auxin Pathway in Arabidopsis. Front. Plant Sci. 12:698487. doi: 10.3389/fpls.2021.698487
Received: 21 April 2021; Accepted: 11 October 2021;
Published: 29 October 2021.
Edited by:
Zhaojun Ding, Shandong University, Qingdao, ChinaReviewed by:
Irma Roig-villanova, Universitat Politecnica de Catalunya, SpainHao Wang, South China Agricultural University, China
Copyright © 2021 Wang, Liu, Aslam, Jakada, Qin and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan Qin, yuanqin@fafu.edu.cn; Hanyang Cai, caihanyang123@163.com
 Lulu Wang1
Lulu Wang1 Mohammad Aslam
Mohammad Aslam Hanyang Cai
Hanyang Cai