- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, China
- 2College of Horticulture, Hebei Agricultural University, Baoding, China
- 3College of Enology, Northwest A&F University, Yangling, China
Voltage-dependent anion channels (VDACs) are the most abundant proteins in the mitochondrial outer membranes of all eukaryotic cells. They participate in mitochondrial energy metabolism, mitochondria-mediated apoptosis, and cell growth and reproduction. Here, the chromosomal localizations, gene structure, conserved domains, and phylogenetic relationships were analyzed. The amino acid sequences of VDACs were found to be highly conserved. The tissue-specific transcript analysis from transcriptome data and qRT-PCR demonstrated that grapevine VDACs might play an important role in plant growth and development. It was also speculated that VDAC3 might be a regulator of modulated leaf and berry development as the expression patterns during these developmental stages are up-regulated. Further, we screened the role of all grape VDACs’ response to pathogen stress and found that VDAC3 from downy mildew Plasmopara viticola-resistant Chinese wild grapevine species Vitis piasezkii “Liuba-8” had a higher expression than the downy mildew susceptible species Vitis vinifera cv. “Thompson Seedless” after inoculation with P. viticola. Overexpression of VpVDAC3 resulted in increased resistance to pathogens, which was found to prevent VpVDAC3 protein accumulation through protein post-transcriptional regulation. Taken together, these data indicate that VpVDAC3 plays a role in P. viticola defense and provides the evidence with which to understand the mechanism of grape response to pathogen stress.
Introduction
Grape is one of the most important fruit crops worldwide. Most grape lines are derived from parental or ancestral Vitis vinifera which are highly susceptible to pathogens that cause serious pre- and post-harvest diseases. These infections have negative effects on grape quality, yield, production, processing, and export. Downy mildew is a common and destructive systemic grape infection caused by the biotrophic oomycete Plasmopara viticola (Wong et al., 2001). It is the foremost threat to grape production and can destroy vineyards in most grape-growing regions of the world (Vercesi et al., 1999).
Hypersensitive response (HR) is a type of programmed cell death (PCD) where in reactive oxygen species (ROS) such as O2– and H2O2 accumulate, which is characterized by DNA fragmentation and chromatin condensation and clustering along nuclear membranes (Levine et al., 1996; Wang et al., 1996). In addition, HR is also used as a defense mechanism triggered by several pathogens, such as bacteria, fungi, insects, and viruses (Wei et al., 1992), which is manifested by rapid curtailing of water and nutrient supply to the affected tissue in the attempt to prevent pathogen proliferation and contact with healthy cells, therefore, it is also a typical feature of immune responses in plants (Dangl and Jones, 2001; Lam et al., 2001). Zamzami (1995) found that mitochondria exhibited reduced transmembrane potential and ROS accumulation before initiating the cell death biochemical reactions, and alteration in mitochondrial permeability is an early event in apoptosis and occurs before nuclear DNA fragmentation, chromatin condensation, and other biochemical processes that culminate in PCD (Kroemer et al., 1997). Genes in mitochondria regulating HRs may also participate in various apoptotic pathways (Johal et al., 1995; Greenberg, 1996).
Voltage-dependent anion channels (VDACs) are some of the most abundant and well-studied proteins in the mitochondrial outer membrane. They were first reported in paramecia and participates in transport across mitochondrial outer membranes (Mayer et al., 1993; Robert et al., 2012). VDACs may also control mitochondrial and cellular energy conversion as well as mitochondrial outer membrane permeability (Colombini, 2004; Young et al., 2007). VDACs may also interact with mitochondrial and cytoplasmic proteins and regulate several mitochondria-related resistance responses (Fiek et al., 1982; Cheng et al., 2003). Hence, VDACs may play complex roles in cell physiology and PCD.
Plant VDACs comprise a small multigene family that might be involved in plant stress responses by regulating cell death (Vianello et al., 2012; Takahashi and Tateda, 2013). A previous study showed that overexpression of rice OsVDAC4 induces apoptosis by a mechanism resembling that of human HVDAC1 (Godbole et al., 2013). Thus, it triggers PCD in mammalian cells (Godbole et al., 2013). PCD was detected in tobacco BY2 cells and leaves overexpressing OsVDAC4 and in rice overexpressing Pennisetum glaucum VDAC (Desai et al., 2006; Godbole et al., 2013). VDAC transcript and protein were upregulated in response to biotic and/or abiotic stress and mediated PCD (Benz, 1994; Lacomme and Santa Cruz, 1999; Desai et al., 2006; Lee et al., 2009; Tateda et al., 2009). Several Arabidopsis VDACs were implicated in biotic stress and upregulated in response to Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) (Robert et al., 2012). However, there is no clear evidence of the biological function of the VDAC family in grape or its response to pathogen stress.
The aim of this study was to identify VDACs in the grapevine genome, and to analyze the biological function of VDAC proteins in response to pathogen stress in grapevine. The results of our study are intended to lay a theoretical foundation for further construction of the regulation network of VDACs in grape and other fruit trees, and provide candidate gene resources for creating a new grape germplasm that is high in quality and disease resistant.
Materials and Methods
Plant Materials
“Thompson Seedless” (V. vinifera) and “Liuba-8” (Vitis piasezkii) were obtained from the grapevine germplasm orchard at Northwest A&F University, Yangling, Shaanxi, China. The plants were cultivated under natural environmental conditions and conventional management. Nicotiana benthamiana plants were grown in a greenhouse at 25°C daytime and 20°C nighttime. Arabidopsis thaliana ecotype Columbia-0 (Col-0; wild-type) was raised in a growth chamber at 22°C, 70% RH, 16/8 h light/dark.
Bioinformatic Analysis of VDACs
The mitochondrial porin signature (MPS) motif regions of VDACs were aligned using the DNAMAN 9.0 software. PyMOL was used to visualize the three-dimensional structures of the VvVDACs protein.
Phylogenetic Analysis and Gene Structures of VDACs in Plants
Phylogenetic characterization of VDACs was performed using MEGA 7.0 with the default options (Kumar et al., 2016). Phylogenetic trees were constructed using the maximum likelihood method. The bootstrap values of 1000 replications were performed, keeping the other parameters as the default. GSDS (Gene structure display server) V2.01 was employed to identify the gene structure of VvVDAC genes.
RNA Isolation, RT-PCR, and qRT-PCR
Total RNA was extracted with an E.Z.N.A.® plant RNA kit (Omega Bio-tek, Norcross, GA, United States). First-strand cDNA was synthesized from 1 μg total RNA in 20 μL reaction mixture using a Prime Script RT reagent kit (Takara Bio Inc., Kusatsu, Shiga, Japan). Real-time PCR (RT-PCR) was conducted in a PCR detection system (Bio-Rad Laboratories, Hercules, CA, United States). Quantitative real-time PCR (qRT-PCR) was performed in the IQ5 real-time PCR system (Bio-Rad Laboratories, Hercules, CA, United States) with SYBR Green qRT-PCR Super Mix (Transgen Biotech, Beijing, China) and using standard protocols. The thermal profile was 95°C for 5 min followed by 45 cycles of 95°C for 10 s, 58°C for 30 s, and 72°C for 10 s. The standard curve was used to calculate relative transcript quantities. Gene expression was calculated relative to Actin as the internal reference. The qRT-PCR primers are listed in Supplementary Table 1.
Expression Profiles of VvVDACs in Different Organs and Tissues
For the expression profile of VvVDAC genes, we utilized the RNA-seq data from National Center for Biotechnology Information (NCBI) gene expression omnibus (GEO) datasets with series entry GSE361282 (Fasoli et al., 2012), which obtained from a V. vinifera cv “Corvina” (clone48) in various organs and tissues at different developmental stages. Heat maps were generated using Heatmap Illustrator, version 1.0.
Pathogen Strains and Inoculations
Plasmopara viticola was maintained on V. vinifera “Thompson Seedless” and prepared as a sporangial suspension (Xu et al., 2014). The inoculation was performed according to the reconstruction method of Liu et al. (2015).
Pseudomonas syringae pv. tomato DC3000 was cultivated in King’s B (KB) liquid medium supplemented with 25 μg mL–1 rifampin at 28°C for 48 h. The bacteria were collected and resuspended in infiltration buffer containing 10 mM MgCl2 and Silwet L-77 at OD600 = 0.002. Either bacterial suspension or 10 mM MgCl2 (mock-inoculation control) was infiltrated with a 1 mL needleless syringe into the laminae of 4-week-old transgenic and wild-type plants. Inoculated plants were then covered with a transparent plastic film for 3 days and photographed (Canon EOS 200D II, Japan).
Transient N. benthamiana Leaf Transformation
Full-length cDNA corresponding to VpVDAC was cloned into the binary vector pCambia 1307 containing 3FLAG tags driven by the 35S promoter. The recombinant plasmids were introduced into Agrobacterium tumefaciens strain GV3101. The latter was then used in transient N. benthamiana transformation by the leaf infiltration method (Goodin et al., 2008). GV3101 cultures bore the construct p35S:VpVDAC or pCambia 1307-3FLAG (empty vector; negative control). The proapoptotic positive control Bax (Lacomme and Santa Cruz, 1999) and the RNA silencing inhibitor P19 were also cultured. All cultures were grown until OD600 ∼1.0. They were then centrifuged at 5000 × g for 10 min and their supernatants were discarded. The pellets were resuspended in acetosyringone (AS) medium [10 mM MES (pH 5.7), 10 mM MgCl2, and 150 μM acetosyringone]. OD600 was then adjusted to 0.8 and working suspensions were prepared by mixing all four strains with equal volumes. The mixed suspensions were incubated for 1 h at 22°C and infiltrated into N. benthamiana leaves with a needleless syringe. The plants were transferred to a greenhouse for 1 day and moved to a dark room at 22°C for 2 days until 3,3′-diaminobenzidine (DAB) analysis (Thordal-Christensen et al., 1997). The stained leaves were transferred to a destaining solution consisting of acetic acid:glycerol:ethanol (1:1:3, v/v/v) in a water bath at 95°C for 10 min, then kept in 70% ethanol until imaging (Canon EOS 200D II, Japan) was performed.
Transient Grapevine Leaf Transformation and Plasmopara viticola Infection
The coding sequence of VpVDAC3 was cloned into the pCambia 2300 binary vector containing YFP driven by the 35S promoter. The recombinant plasmid and pCambia 2300 empty vector were introduced into A. tumefaciens strain GV3101. A transient transformation was performed via vacuum infiltration. Three weeks old plants were filled with bacterial suspensions [10 mM MES (pH 5.7), 10 mM MgCl2, and 150 μM acetosyringone] with OD600 = 0.8 and placed in a vacuum chamber attached to a circulating water vacuum pump (Model#SHZ-D, Shanghai, China) and exposed to four 20 min vacuum/5 min vacuum release periods (0.08 MPa) to aid plant uptake of the suspension. The infiltrated explants were co-cultivated for 12 h at 22°C and a relative humidity of 80% in darkness, then transferred to a regular 16-h-light/8-h-dark cycle. They were then induced after 24 h via spraying with 50 mM β-estradiol and 0.1% Tween. The agro-infiltrated leaves were then inoculated with P. viticola. After 1 day post-inoculation (dpi), 4 dpi, and 6 dpi, leaf discs were punched out of the transformed leaf areas, and H2O2 was assayed using a fluorimetric hydrogen peroxide assay kit (Sigma-Aldrich, MAK165-1KT, United States) following the protocols provided by the manufacturer. The final fluorescence of the experimental group was obtained by deducting the measured fluorescence of the control group from the measured fluorescence of the experimental group. The average sporangia density of P. viticola in two different genotypes at 6 dpi (hours post-inoculation) was calculated.
Arabidopsis Transformation
The A. tumefaciens floral dip transformation method was used for generating VpVDAC3 overexpressing lines (Clough and Bent, 1998). pCambia 2300 binary vector containing p35S:VpVDAC3-YFP and pCambia 2300 empty vector were transformed into the Col-0 background. T3- or T4-generation homozygous plants were used for phenotype analysis.
Protein Extraction and Immunoblot Analysis
The leaves were excised, flash-frozen in liquid nitrogen, and pulverized. Total protein was extracted with denaturing buffer [4 M urea, 65 mM Tris–HCl (pH 7.3), 3% (v/v) sodium dodecyl sulfate (SDS), 10% (v/v) glycerol, 0.05% (w/v) bromophenol blue, and 10 mM dithiothreitol (DTT)]. The protein concentrations in the supernatants were determined with a bicinchoninic acid (BCA) protein assay kit (CW0014; CWBiotech, Beijing, China). The proteins were loaded onto 10% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes which were then incubated with a monoclonal α-GFP antibody (Roche; Cat-No: 11814460001, monoclonal, mouse, dilution 1:2000) and a FLAG antibody (Sigma-Aldrich; Cat-No: F4042, monoclonal, mouse, dilution 1:3000). Anti-mouse horseradish peroxidase (HRP) secondary antibody (Abcam, Cat-No: ab205719, dilution 1:10,000) was used for detection. The PVDF membranes were then incubated in Ponceau S solution [40% (v/v) methanol, 15% (v/v) acetic acid, and 0.25% (w/v) Ponceau S] and destained with deionized water. Images were captured with ChemiDocTM XRS 170-8070 (Bio-Rad Laboratories, Hercules, CA, United States).
Plant Damage and Histochemical Assays
Leaves were stained with DAB to investigate H2O2 accumulation. The leaves were excised, incubated in 1 mg mL–1 DAB (Sigma-Aldrich Corp., St. Louis, MO, United States) in the dark overnight, and destained with 95% (w/v) ethanol until clear. Images were then captured under a microscope (Olympus BX−51, Tokyo, Japan).
Cell death was visualized by trypan blue staining. Inoculated leaves were soaked in 0.125 mg mL–1 trypan blue solution [phenol/lactic acid/glycerol/water (1:1:1:1)], boiled in a water bath for 2 min, left at room temperature overnight, destained with 95% (w/v) ethanol, and observed under a light microscope (Olympus BX−51, Tokyo, Japan).
Electrolyte Leakage
Six round infiltrated leaf disks (1 cm in diameter) were excised from each infiltrated leaf, washed in ddH2O for 30 min, and placed in test tubes each containing 10 mL water. Solution conductivity was measured with a digital conductivity meter (Leici Corporation, China). Electrolyte leakage was measured at 0, 6, 12, 24, and 48 h. Data are representative of three independent experiments.
Availability of Data and Material
Sequence data are cataloged in the NCBI database under the following accession numbers: AtVDAC1 (At3g01280), AtVDAC2 (At5g67500), AtVDAC3 (At5g15090), AtVDAC4 (At5g57490), and AtVDAC5 (At3g49920).
Statistical Analysis
Data are representative of three independent experiments. The data were processed in SPSS v. 17.0 for Windows (IBM Corp., Armonk, NY, United States). One-way analysis of variance was used to determine the significance of the differences among treatment means. Error bars indicate standard deviation (SD) and significant differences are indicated with “∗” (P < 0.05) or “∗∗” (P < 0.01). Graphs were plotted with OriginPro 2015 (OriginLab, Northampton, MA, United States).
Results
Mapping the VDAC Family in V. vinifera
All VvVDAC genes were mapped to island cotton chromosomes based on genome annotations. The annotation revealed that the six grape VDAC (VvVDAC1–VvVDAC6) and one variant were dispersed on five chromosomes (Supplementary Table 2). And VvVDAC1, VvVDAC4, and VvVDAC5 located in chromosome 1, 11, and 14, respectively. VvVDAC2 and VvVDAC3 located in chromosome 7. VvVDAC6.1 and its splice variant VvVDAC6.2 were located on chromosome 17. The members of this alternative splicing pair differed by 18 amino acids.
Phylogenetic analysis of VvVDACs from V. vinifera showed that all VvVDACs were clustered into five distinct categories. VvVDAC1 and VvVDAC5 were clustered together, and the other three members (VvVDAC2, VvVDAC3, and VvVDAC4) also aligned together (Supplementary Figure 1). VvVDAC6 clustered separately from the other five members. To further understand the structural evolution of the VvVDAC genes, these genes were subjected to conduct the exon–intron organization (Supplementary Figure 1). Most VvVDAC family members contained five intron and six exons, while the VvVDAC4 had one more intron.
Cluster analysis of the VvVDAC amino acid sequences showed that full length of the grapevine VvVDAC gene family is highly diverse (Supplementary Table 3). Amino acid sequence alignment disclosed that the VvVDAC family members shared 7.6–91.7% similarity. The VvVDAC2 shared least similarity with other VvVDACs.
3D Structure Prediction and MPS Analysis of the VDAC Family in V. vinifera
The predicted 3D structure of the VDAC family of V. vinifera is shown in Figure 1. VDAC proteins are generally rich in β-sheets (Abrecht et al., 2000; Colombini, 2009). The predicted 3D structures of VvVDAC1, VvVDAC3, VvVDAC4, and VvVDAC5 resemble those of mammalian VDAC1 and zebrafish VDAC2 with 19 transmembrane β-sheets forming β-barrels, and N-terminals forming internal α-helices (Messina et al., 2012; Schredelseker et al., 2014). VvVDAC1 has N-terminals with internal α-helices, but only 16 and 9 transmembrane β-sheets structures, respectively. The 3D structures of VvVDAC6.1 and VvVDAC6.2 lacked the N-terminal α-helices but resembled β-barrels. Among them, VvVDAC2 did not have complete structure, the issues could be due to problems with the available sequence (Figure 1A).
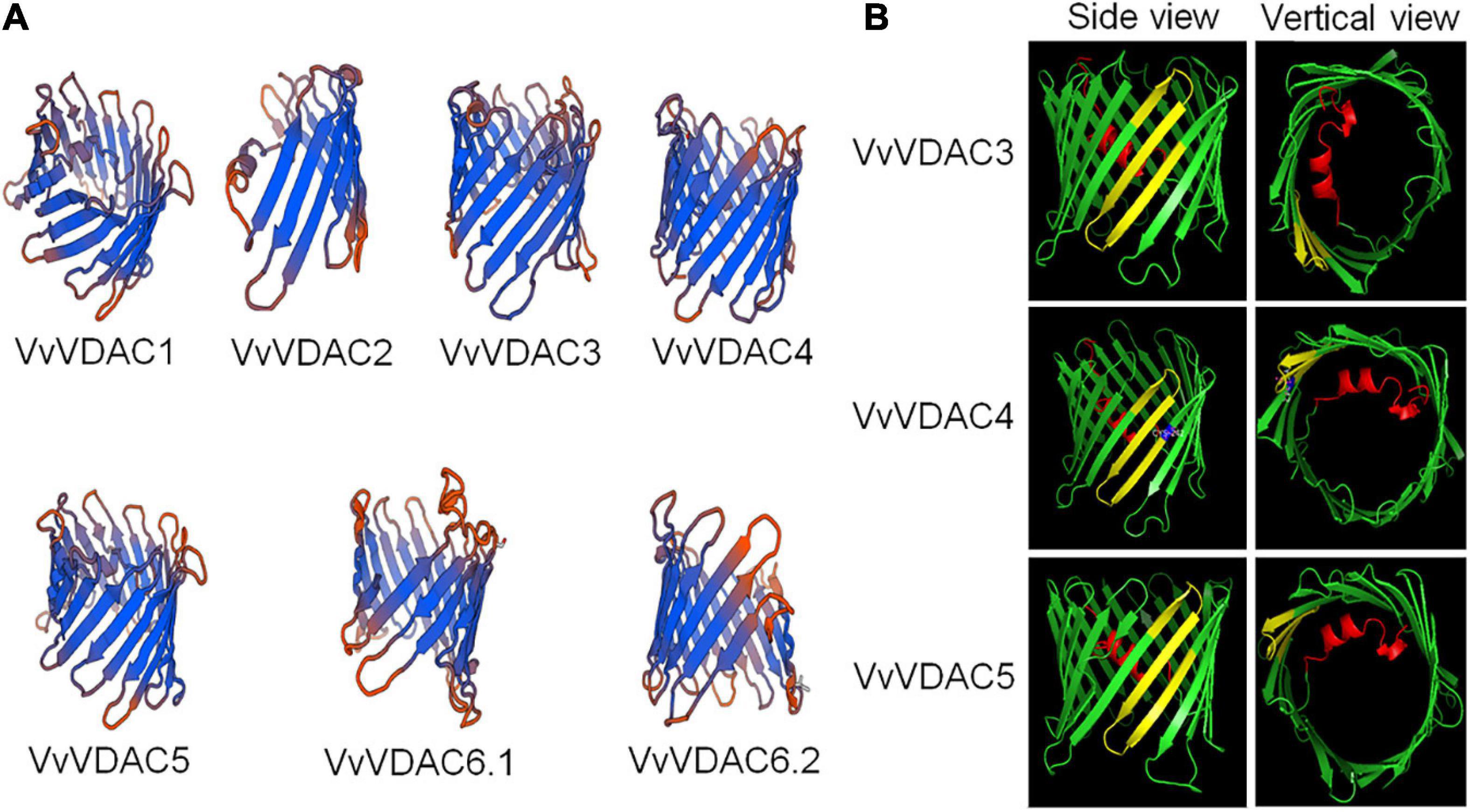
Figure 1. Three-dimensional structure prediction and MPS domain localization of VvVDACS. (A) Three-dimensional structure of VvVDACS is represented by a ribbon diagram. (B) Prediction of MPS motif structures for VvVDAC3, VvVDAC4, and VvVDAC5 (side and vertical views). VvVDAC is a β-barrel (in green) formed by anti-parallel β-strands; the strands are connected by loops (in green). The N-terminal domain (in red) is arranged in an α-helix and is located inside the pore’s lumen. The cysteine residue at amino acid 242 of VvVDAC4 is marked in blue.
Based on the descriptions of structure from VDAC members, eukaryotic organisms contain 23 conservative amino acid MPS in their C-terminal regions: [YH]-X(2)-D-[SPCAD]-X-[STA]-X(3)-[TAG]-[KR]-[LIVMF]-[DNSTA]-[DNS]-X(4)-[GSTAN]-[LIVMA]-X-[LIVMY], X represents any amino acid (Tateda et al., 2011). We analyzed the sequence of MPS domain of VvVDACs from Vitis, several plant species and mouse (Supplementary Figure 2). As previous mentioned, the results here showed that Arabidopsis AtVDAC1 and AtVDAC3 contained the conserved MPS motif in the C-terminal region (Tateda et al., 2009), whereas AtVDAC2 and AtVDAC4 do not. Vitis VvVDAC3 and VvVDAC5 also carried a conserved MPS structure domain. In addition, VvVDAC4 has a similar MPS domain but differs by one amino acid residue at its last position 242. Other VvVDACs, including VvVDAC1 and VvVDAC6, which are similar with AtVDAC2, have divergent MPS sequences. Importantly, VvVDAC2 did not contain MPS domain, which suggests that the sequence of VvVDAC2 might not be the full length of VDAC gene and we excluded it out in the following experiments.
The 3D model (Figure 1B) superimposes very well on the structure of VvVDAC3, VvVDAC4, and VvVDAC5 protein containing conserved MPS domain (side and vertical views). The MPS motifs are shown in yellow, while the N-terminals with α-helix are displayed in red. The cysteine residue at amino acid 242 described in VvVDAC4 is marked in blue.
Phylogenetic Analysis of VDAC in V. vinifera and Other Plants
The phylogenetic relationships between from grapevine and other plants are depicted in Figure 2. The unrooted phylogenetic tree indicates that the VvVDACs clusters into main five distinct groups. In particular, VvVDAC1 is closely related to tobacco NtVDAC3. VvVDAC3 belongs to the same cluster and is closely related to Arabidopsis AtVDAC2 and AtVDAC5. VvVDAC4, VvVDAC5, and VvVDAC6 are grouped into three clusters and are separate from the other VDACs, indicating their complexity and sequence diversity. The two VvVDAC6 splice variants VvVDAC 6.1 and VvVDAC6.2 shared very high similarity with 91.70%.
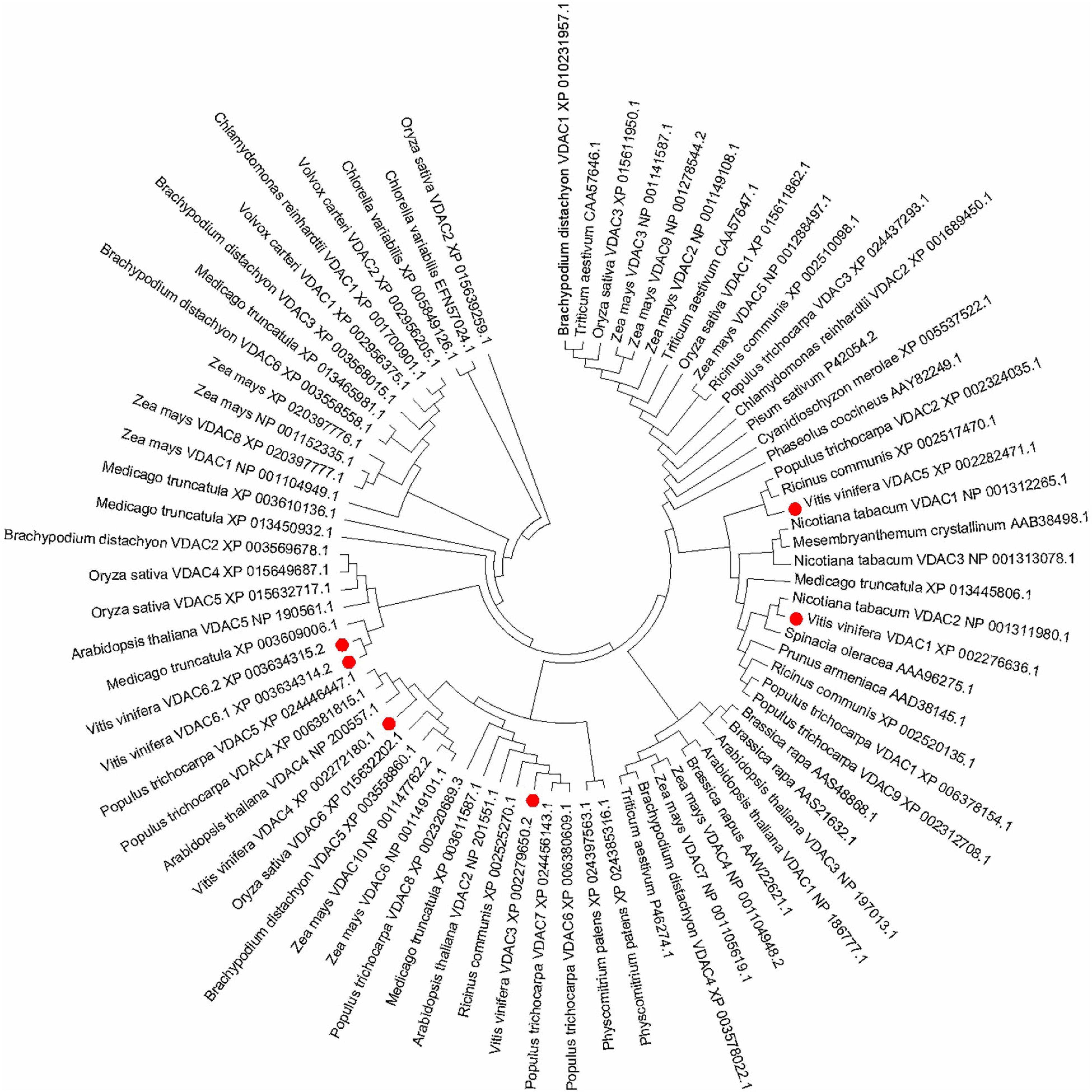
Figure 2. Phylogenetic relationship tree of VDAC genes in plants. The phylogenetic tree was constructed in MEGA 7.0 using the maximum likelihood method (1000 bootstrap).
Expression Patterns of VvVDAC Genes in Specific Organs in Downy Mildew-Susceptible V. vinifera
The expression patterns of all six VvVDAC genes involved in grapevine development were analyzed in the V. vinifera cv. Corvina global gene expression atlas from the GEO Datasets (GSE36128), with 54 organs and tissues at different developmental stages analyzed by RNA-sequencing based on previous reports (Fasoli et al., 2012). As shown in Figure 3, VvVDAC genes were expressed in numerous grapevine tissues and organs during the growth and developmental stages. VvVDAC1 exhibited similar and high expression profiles in nearly all tissues and organs. VvVDAC6 had either moderate or weak expression abundance in all selected tissues and organs, and only showed higher transcript accumulation in the seed fruit set stage, suggesting its limited response in grapevine. VvVDAC4 and VvVDAC5 were expressed across all tissues but displayed a typically lower expression in comparison with VvVDAC1, which had higher expression in buds and pollen than in other tissues, suggesting their participation in the functioning of buds and pollen. Moreover, VvVDAC3 maintained a relative higher abundance in leaves and all berry tissues as the berries developed from fruit set stage to berry pericarp post-harvest withering III (3rd month) stage, which suggests that VvVDAC might have functions in berry and leaves development. This divergent distribution implies the functional divergence of VvVDAC genes for grapevine development.
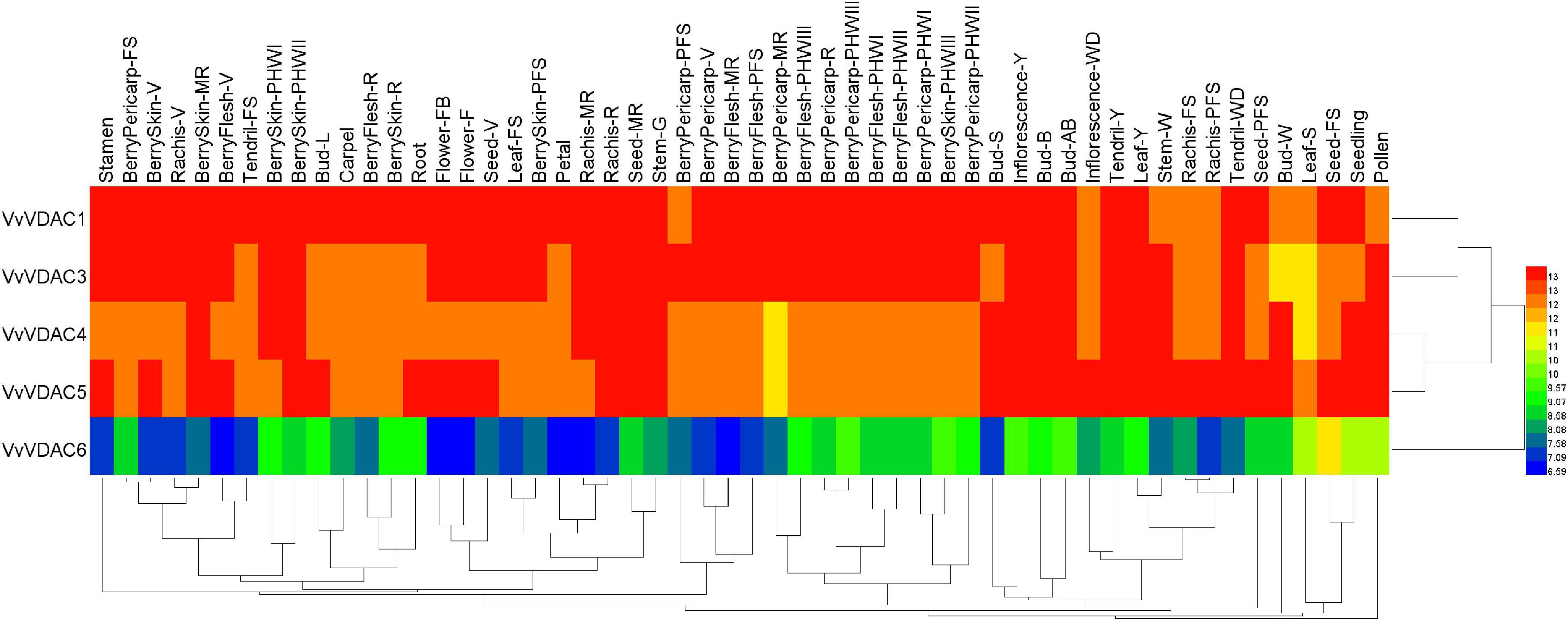
Figure 3. Expression profiles of the VvVDAC genes in the tissues/organs of grapevine during the developmental stages. Data were normalized based on the mean expression value of each gene in all tissues analyzed. Red and green boxes indicate high and low expression levels, respectively. BerryPericarp-FS, berry pericarp fruit set; BerryPericarp-PFS, berry pericarp post-fruit set; BerryPericarp-V, berry pericarp véraison; BerryPericarp-MR, berry pericarp mid-ripening; BerryPericarp-R, berry pericarp ripening; Bud-S, bud swell; Bud-B, bud burst (green tip); Bud-AB, bud after burst (rosette of leaf tips visible); Bud-L, latent bud; Bud-W, winter bud; BerryFlesh-PFS, berry flesh post fruit set; BerryFlesh-V, berry flesh véraison; BerryFlesh-MR, berry flesh mid-ripening; BerryFlesh-R, berry flesh ripening; BerryFlesh-PHWI, berry flesh post-harvest withering I (1st month); BerryFlesh-PHWII, berry flesh post-harvest withering II (2nd month); BerryFlesh-PHWIII, berry flesh post-harvest withering III (3rd month); Inflorescence-Y, young inflorescence; Inflorescence-WD, well-developed inflorescence; Flower-FB, flowering begins; Flower-F, flowering; Leaf-Y, young leaf; Leaf-FS, mature leaf; Leaf-S, senescing leaf; BerryPericarp-PHWI, berry pericarp post-harvest withering I (1st month); BerryPericarp-PHWII, berry pericarp post-harvest withering II (2nd month); BerryPericarp-PHWIII, berry pericarp post-harvest withering III (3rd month); Rachis-FS, rachis fruit set; Rachis-PFS, rachis post-fruit set; Rachis-V, rachis véraison; Rachis-MR, rachis mid-ripening; Rachis-R, rachis ripening; Seed-V, seed véraison; Seed-MR, seed mid-ripening; Seed-FS, seed fruit set; Seed-PFS, seed post-fruit set; BerrySkin-PFS, berry skin post-fruit set; BerrySkin-V, berry skin véraison; BerrySkin-MR, berry skin mid-ripening; BerrySkin-R, berry skin ripening; BerrySkin-PHWI, berry skin post-harvest withering I (1st month); BerrySkin-PHWII, berry skin post-harvest withering II (2nd month); BerrySkin-PHWIII, berry skin post-harvest withering III (3rd month); Stem-G, green stem; Stem-W, woody stem; Tendril-Y, young tendril; Tendril-WD, well-developed tendril; Tendril-FS, mature tendril.
Expression Pattern Analysis of VDAC Family in Downy Mildew-Resistant V. piasezkii
The VpVDAC genes expression levels in V. piasezkii “Liuba-8” leaves, roots, stems, and tendrils are shown in Figure 4. The expression levels of VpVDAC1, VpVDAC3, and VpVDAC4 were higher in the leaves than in the other organs. The expression level of VpVDAC6.1 was highest in the roots and ≥50% greater than those in the other organs. In the stems, however, only VpVDAC6.2 was highly expressed. All other VpVDACs were expressed at very low levels. In the tendrils, VpVDAC3 and VpVDAC6.2 were highly expressed. VpVDAC5 was also upregulated in the tendrils whereas it was relatively downregulated in the other organs. VpVDAC3 showed higher abundance in the leaves than in the roots, stems, and tendrils, suggesting that it might play important role in leaf development.
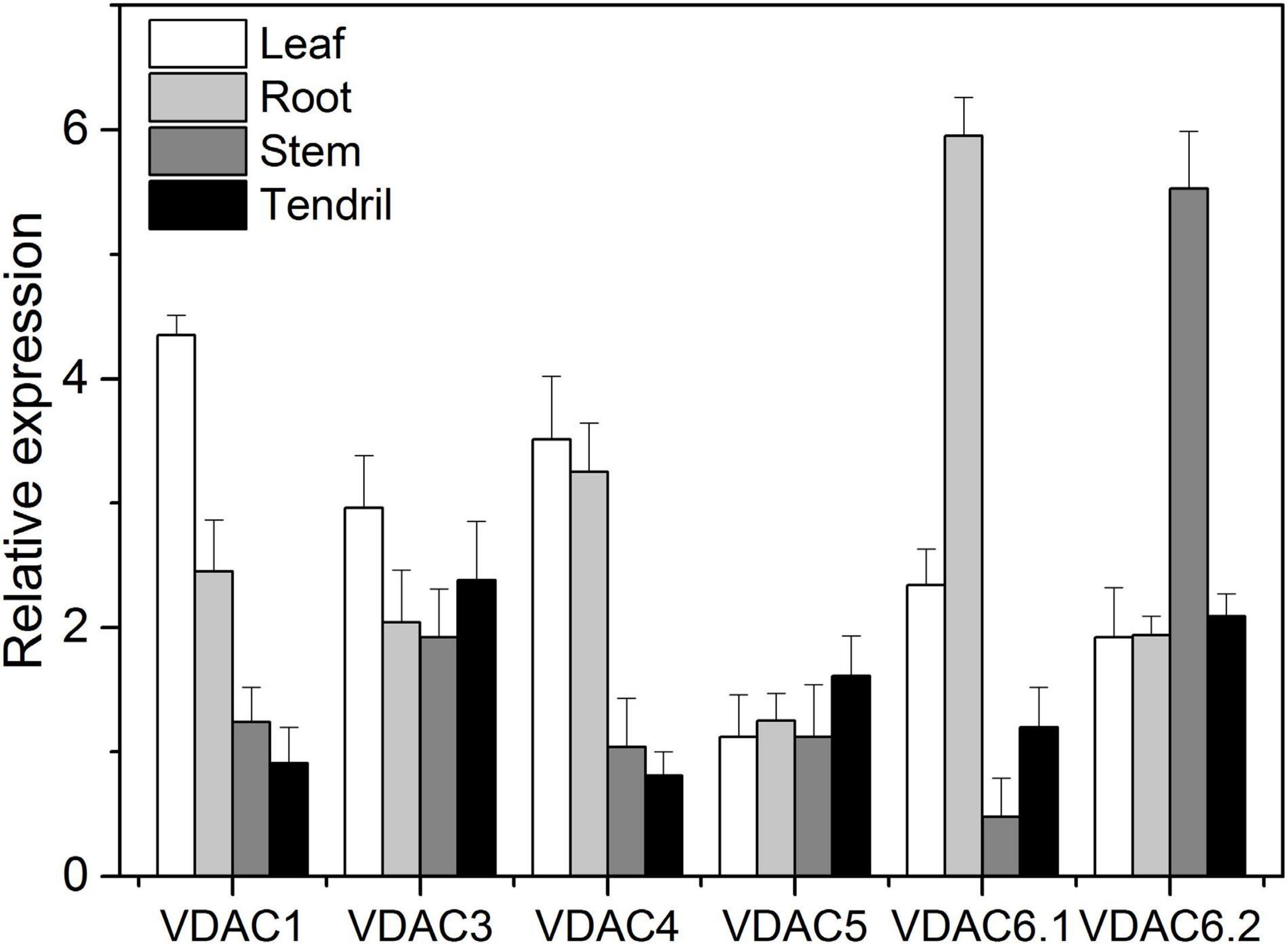
Figure 4. VpVDACs expression in grape tissues. Relative VpVDAC expression in the leaves, roots, stems, and tendrils of “Liuba-8” (V. piasezkii). Error bars indicate the SE (n = 3) from three independent biological replicates.
H2O2 Accumulation in Transient VpVDAC3-Transformed N. benthamiana Leaves
Hydrogen peroxide (H2O2) is an important signaling molecule that initiates transcriptional responses in land plants under biotic and abiotic stress. VpVDAC1 and VpVDAC3 overexpression promoted H2O2 accumulation relative to the empty vector. VpVDAC3 presented a higher H2O2 content than VpVDAC1. Negligible H2O2 accumulation was detected in the other VpVDAC infiltration area (Supplementary Figure 3).
Grapevine VDAC3 Expression in Response to Downy Mildew P. viticola
Figure 5 shows the temporal VDAC3 expression patterns in V. vinifera cv. “Thompson Seedless” susceptible to downy mildew (P. viticola) and Chinese wild grape V. piasezkii “Biuba-8,” which is resistant to this pathogen. We sprayed the leaves either with a mock solution or a P. viticola sporangial suspension. After inoculation, the VDAC3 transcript levels were substantially increased in both “Thompson Seedless” and “Liuba-8” and reached a peak at 48 hpi (Figure 5). Meanwhile, grapevine VDAC3 expression was considerably higher in the resistant “Liuba-8” than it was in the susceptible “Thompson Seedless.” Therefore, we speculated VpVDAC3 might contribute to stronger resistance in Chinese wild grape V. piasezkii “Biuba-8.”
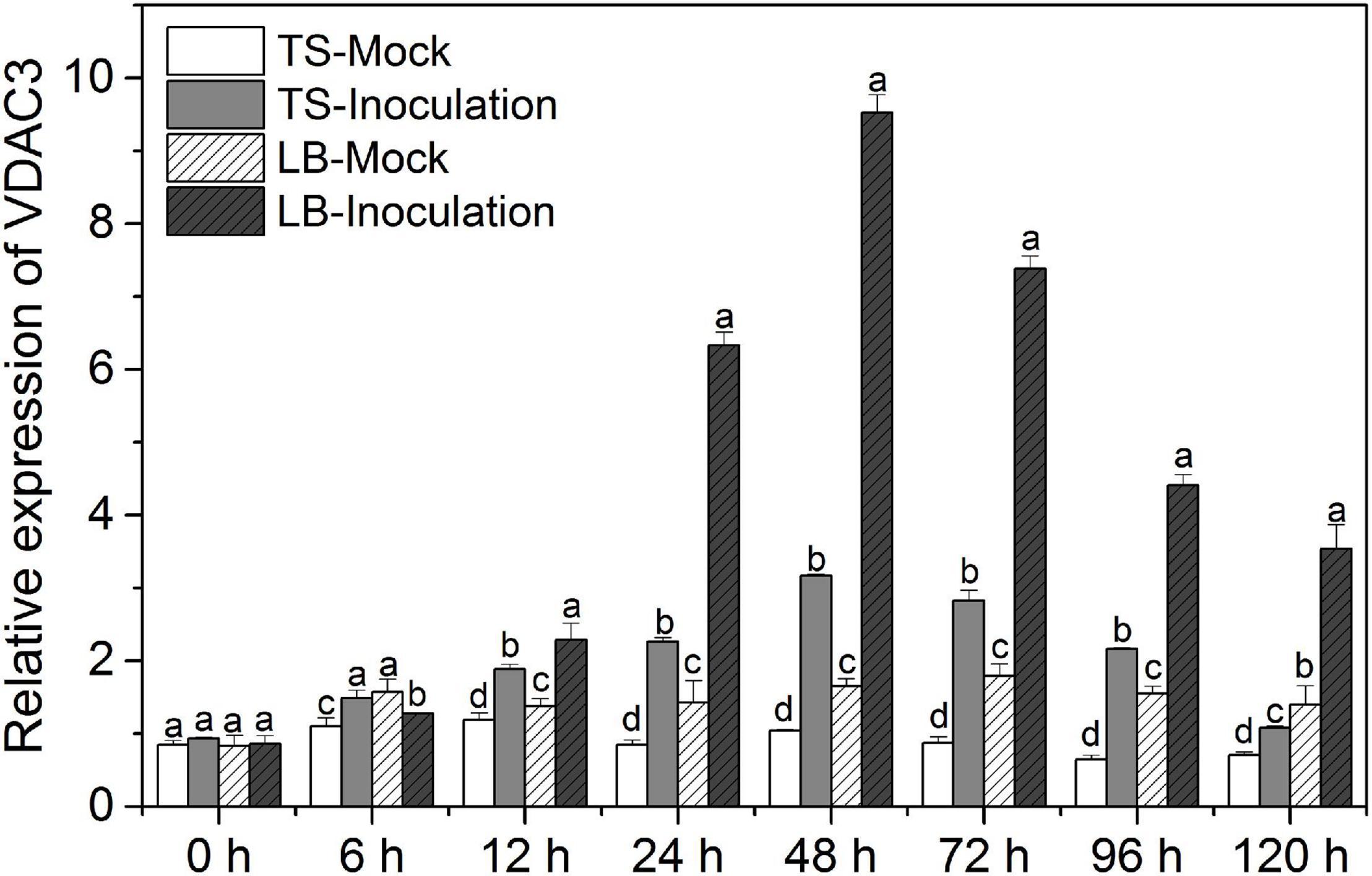
Figure 5. Grapevine VDAC3 responses to P. viticola. Leaves of downy mildew-susceptible “Thompson Seedless” (V. vinifera) and downy mildew-resistant “Liuba-8” (V. piasezkii) were inoculated either with P. viticola or water suspension (mock). TS Mock, “Thompson Seedless” sprayed with water suspension; TS Inoculation, “Thompson Seedless” sprayed with P. viticola; LB Mock, “Liuba-8” sprayed with water suspension; LB Inoculation, “Liuba-8” sprayed with P. viticola. Samples were harvested for qRT-PCR analysis after 0, 6, 12, 24, 48, 72, 96, and 120 h treatment. Error bars indicate the SE (n = 3) from three independent biological replicates. Different letters (a–d) in the same time represent significant difference at P < 0.05 level.
VpVDAC3 Exhibited a Higher Resistance to P. viticola
To evaluate the role of VpVDAC3 in response to downy mildew, we transiently overexpressed VpVDAC3 in the leaves of downy mildew-susceptible “Thompson Seedless” (V. vinifera) and checked P. viticola disease progression using a leaf disc assay. An empty 35S vector was also transformed as a control. Compared with the control, only a few downy mildew symptoms were observed at 4 days after transformation with VpVDAC3; more symptoms were observed with the empty vector (Figure 6A). The functionality of VpVDAC3 was further validated by agro-mediated transient expression in the leaves with P. viticola infection after 6 dpi. VpVDAC3 transient grapevine leaves were confirmed by protein expression (Figure 6B). Meanwhile, the density and sporangiospore number of downy mildew on the leaf discs of VpVDAC3 OX were obviously lower than that of the controls (Figure 6C). We further examined the accumulation of H2O2 between the VpVDAC3-overexpressing leaves and the controls at P. viticola inoculation sites at 1, 4, and 7 dpi. The results showed that the rate of H2O2-level rise in the VpVDAC3-overexpressing leaves was statistically higher compared to control (Figure 6D).
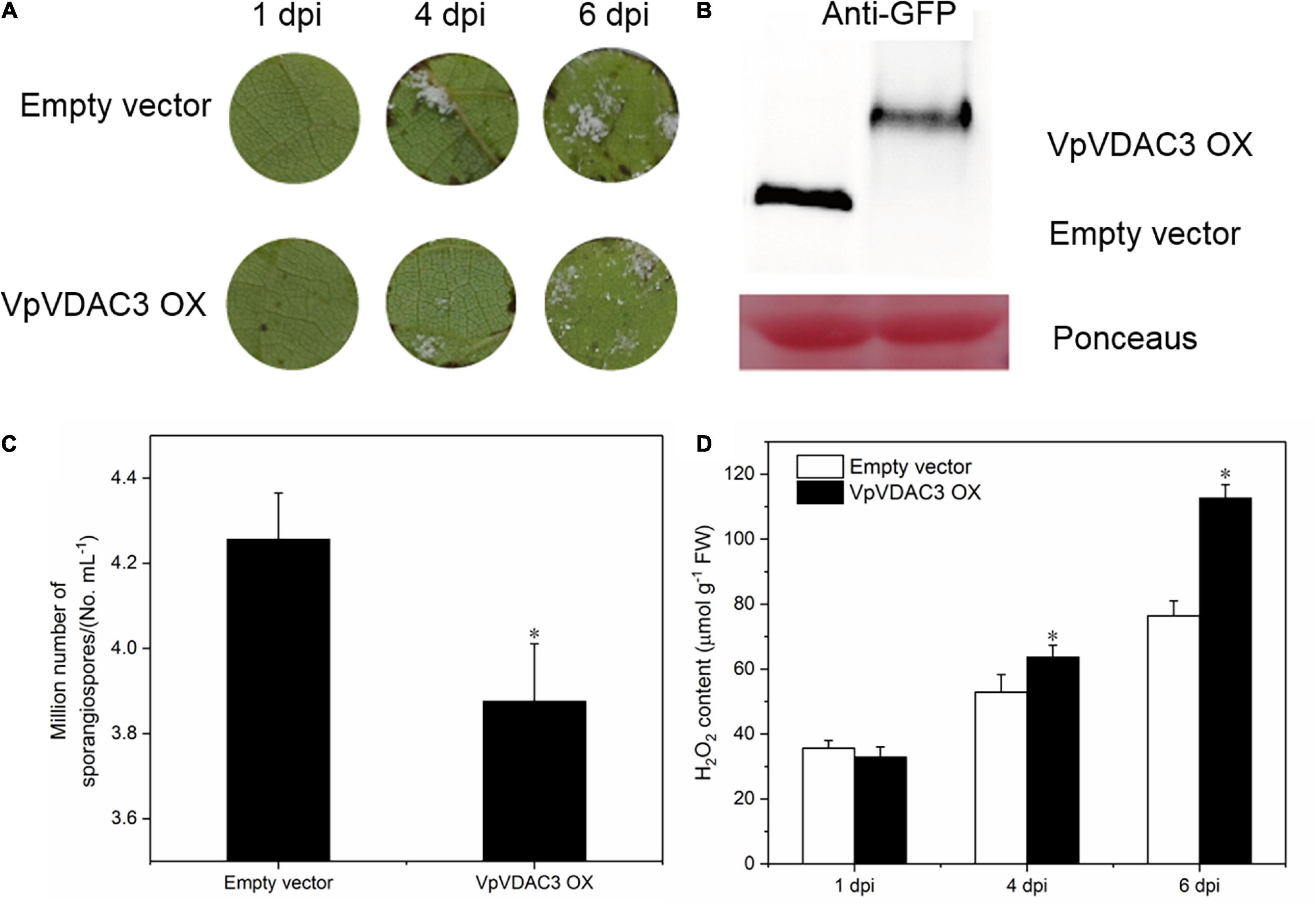
Figure 6. VpVDAC3 enhances H2O2 accumulation during P. viticola infection. (A) p35S: VpVDAC3 and the empty vector transiently transformed in V. vinifera “Thompson Seedless” with P. viticola inoculation. (B) Immunoblot detection of protein expression in transiently transformed grapevine leaves after 6 dpi. (C) Average sporangia density of P. viticola from different genotypes at 6 dpi. (D) H2O2 accumulation. Error bars indicate the SE (n = 3) from three independent biological replicates. Asterisks indicate significant difference from the control (empty vector) (*P < 0.05).
Overexpression of VpVDAC3 Leads to Increased Resistance Against Pst DC3000
Two stable transgenic lines exhibiting elevated VpVDAC3 mRNA and protein expression were subjected to further analysis (Figures 7A,B). The transgenic lines were morphologically identical to wild-type (WT) plants under normal growing conditions (Figure 7C). We evaluated the response of these transgenic lines to the Pst DC3000, which is a kind of bacterial pathogen (Figure 8). Leaves from VpVDAC3-overexpressing and WT (Col-0) Arabidopsis were inoculated with Pst DC3000. Water-soaked spots and severe chlorosis were visible in Col-0 and the leaves exhibited yellowing after 3 days of inoculation (3 dpi). However, the VpVDAC3 overexpressing lines displayed slight chlorosis and little yellowing and were healthier at examined time points, suggesting that VpVDAC3 ectopic expression had greater Pst DC3000 resistance than Col-0 (Figure 8A).
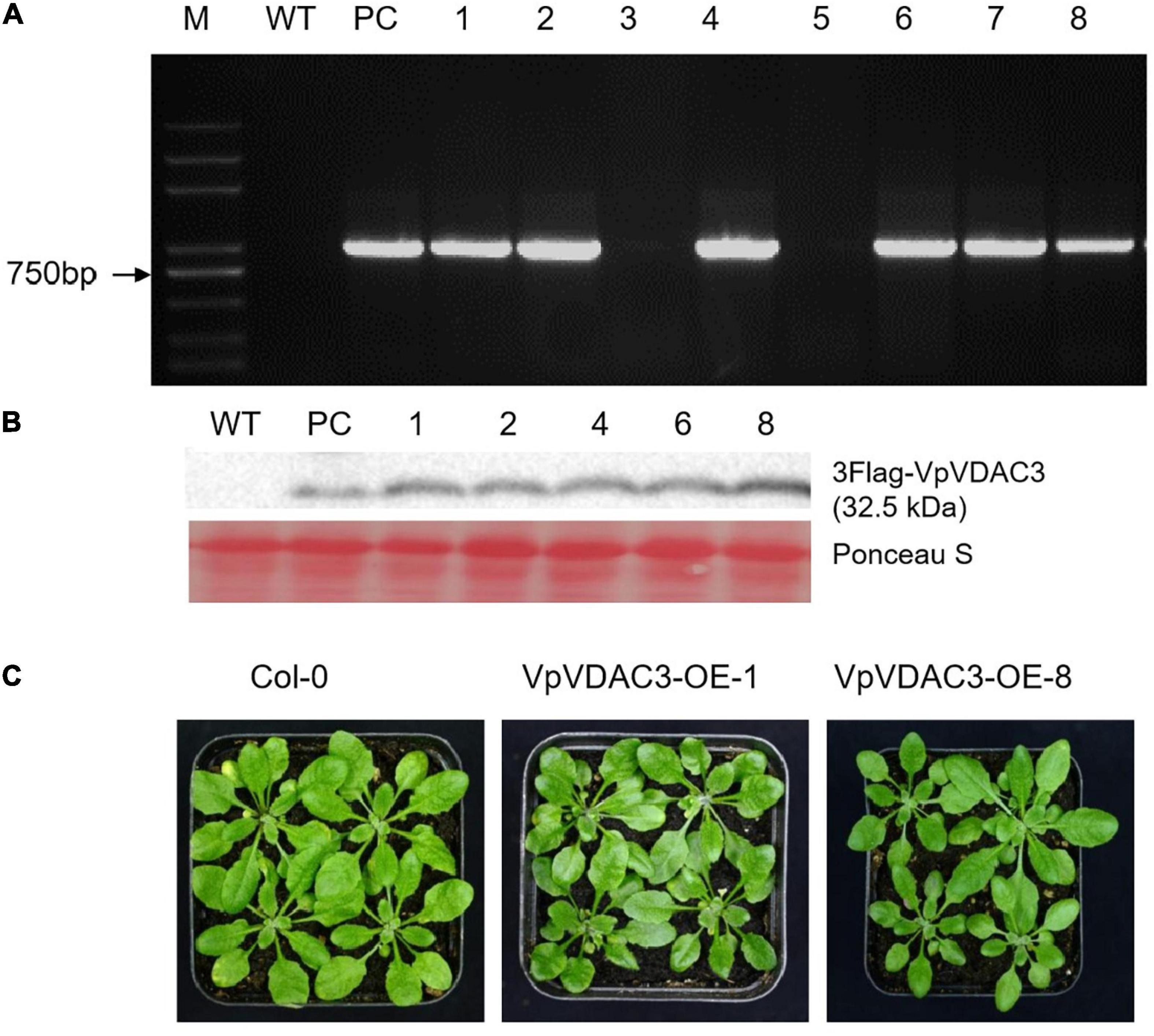
Figure 7. Morphology and VpVDAC3 expression in transgenic Arabidopsis lines. (A) RT-qPCR detection of VpVDAC3 in two independent stable transgenic p35S:VpVDAC3 Arabidopsis lines. (B) Immunoblot detection of VpVDAC3 proteins expressed in stable transgenic lines. (C) Morphology of WT (Col-0) and two independent transgenic Arabidopsis lines grown under normal conditions. M, marker; WT, wild-type Col-0; PC, positive control.
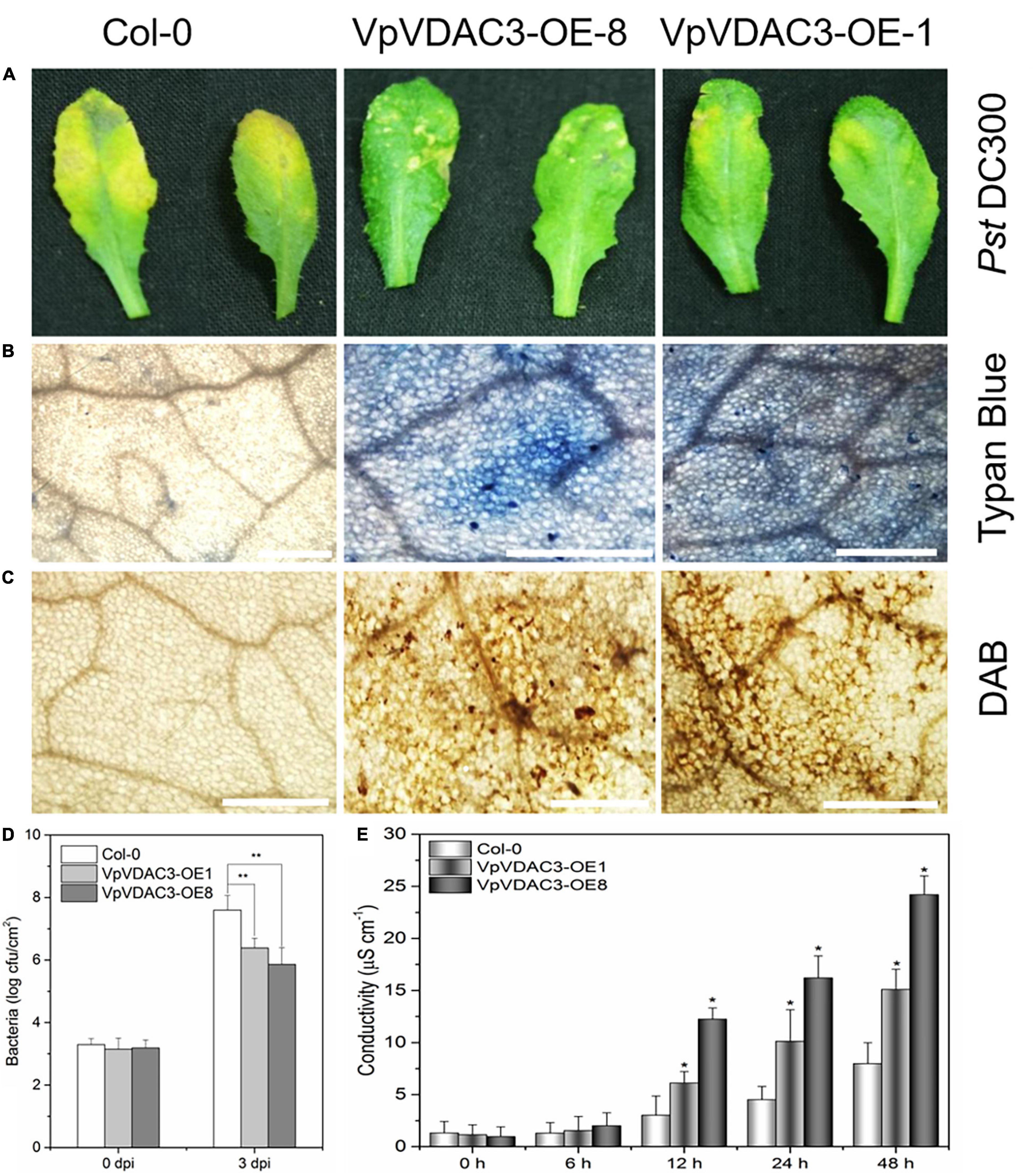
Figure 8. Enhanced Pst DC3000 resistance in VpVDAC3-OE transgenic Arabidopsis. (A) Disease symptoms on leaves 72 h after inoculation. (B) Micrographs of leaves stained with trypan blue 24 h after infiltration. (C) Micrographs of leaves stained with DAB 24 h after inoculation. (D) Bacterial growth in leaves inoculated with Pst DC3000, Bacterial populations were quantified at 0, and 2 dpi. Vertical bars indicate the standard errors for three independent experiments. (E) Ion leakage from leaf tissues inoculated with Pst DC3000. Error bars are the SD of three replications results. Error bars indicate the SE (n = 3) from three independent biological replicates. An asterisk (*) indicates significant differences compared to the Col-0 line as determined by t-tests (*P < 0.05, **P < 0.01) based on three independent replication experiments.
Programmed cell death is a HR that is closely related to plant disease resistance, which was visualized using trypan blue, a vital stain that selectively accumulates in dead cells turning them blue (van Wees, 2008). To investigate the capacity of VpVDAC3 ectopic expression cells to recognize and respond to pathogenic bacteria Pst DC3000, trypan blue staining was used. Figure 8B showed more necrotic cells in the transgenic lines overexpressing VpVDAC3 compared to the controls after Pst DC3000 infection. ROS levels are also closely associated with a pathogen infection. We also used ROS reactive dyes histochemical 3,3′-diaminobenzidine (DAB) to test H2O2 contents in infected rosette leaves. H2O2 is visualized as reddish brown stain formed by the reaction of DAB with the endogenous H2O2. The results showed that VpVDAC3 transgenic leaves stained strongly and accumulated more H2O2 than the WT (Figure 8C), demonstrating that VpVDAC3 could enhance resistance to Pst DC3000. Less bacterial growth were also observed in VpVDAC3 ectopic expression leaves and which was corrected with stronger resistance (Figure 8D). The leaf conductivity represents the extent of membrane injury. Transgenic plants infected with Pst DC3000 demonstrated significantly higher electrolyte release responses and greater leakage than Col-0 (Figure 8E).
Expression of VpVDAC3 protein levels during Pst DC3000 infection are confirmed in Figure 9. The protein levels in mock-treated leaves did not significantly differ at various time points. However, the VpVDAC3 protein level accumulated after Pst DC3000 infection, especially at time points 5 dpi. Thus, VpVDAC3 protein was stabilized during pathogen infection and involved in the increased-resistance in response to pathogen. Together, our results support the idea that VpVDAC3 plays a role in increasing plant resistance against Pst DC3000 through protein post-transcriptional regulation like preventing accumulation after Pst DC3000 pathogen infection.
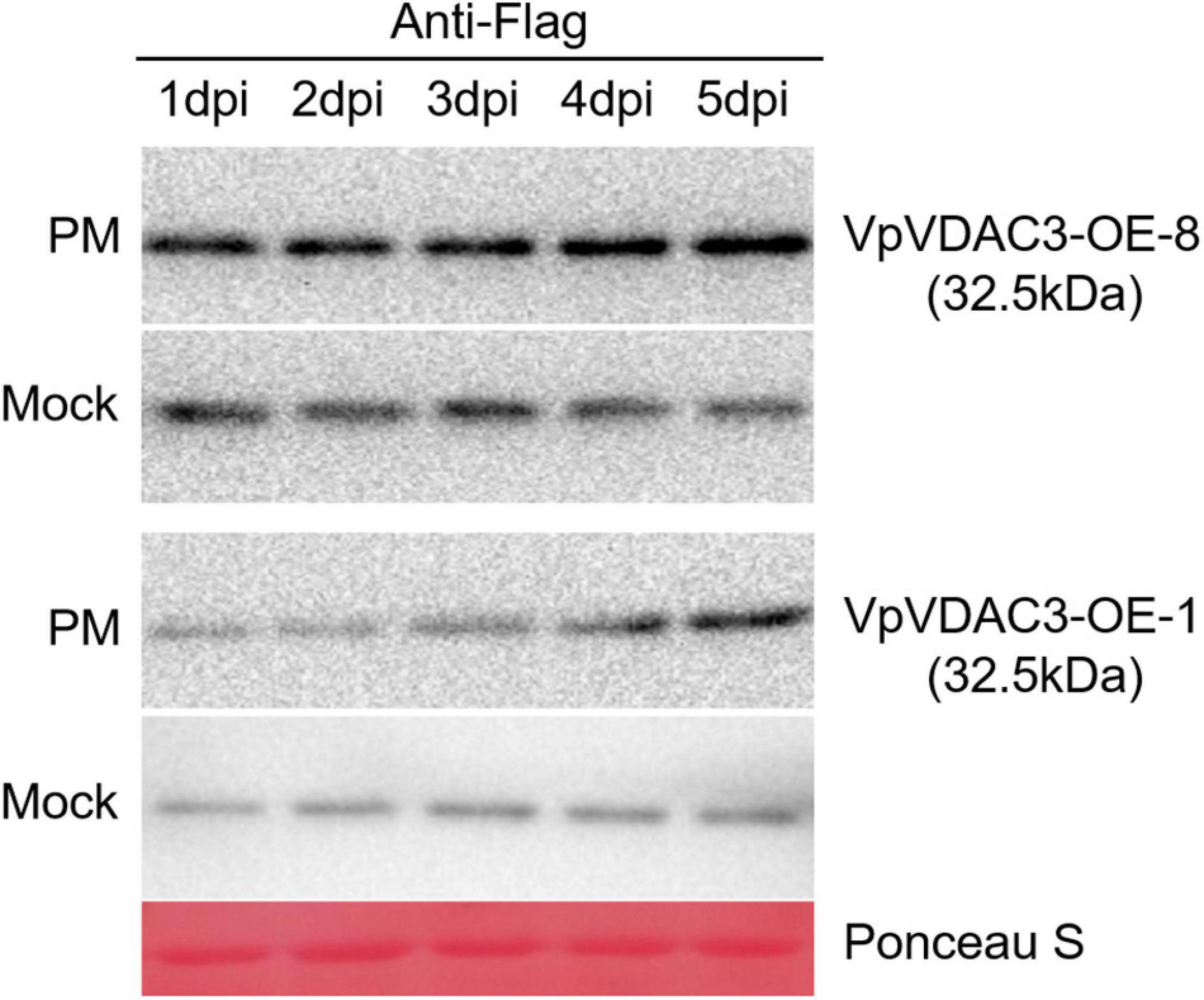
Figure 9. VpVDAC3 protein accumulation during Pst DC3000 infection. Western blot analysis of two stable p35S:VpVDAC3 transgenic Arabidopsis lines (VpVDAC-OE-3 and VpVDAC-OE-8) after Pst DC3000 inoculation. Protein samples were extracted after 1, 2, 3, 4, and 5 dpi. Data are representative of three independent experiments.
Discussion
Voltage-dependent anion channels are the most abundant proteins in the outer mitochondrial membrane. The latter regulates the movement of small mitochondrial and cytoplasmic molecular metabolites and controls other physiological functions (Blachly-Dyson and Forte, 2001; Noskov et al., 2016). Previous studies on VDACs focused mainly on their functions in mammalian cells. The number of VDACs is generally higher in plant cells than mammalian cells (Takahashi and Tateda, 2013). Nevertheless, VDAC functions might be more diverse in the latter. Moreover, there are few studies regarding the importance of VDACs in plant pathogens and stress resistance, and more detailed research in these areas is still required. The present study mainly explored the expression patterns and potential pathogen signaling roles of VDACs in grapevine.
A previous study showed that there are three VDAC protein isoforms in mammals (Messina et al., 2012). Then plant VDACs were also constantly identified. Three VDACs were identified in tobacco (Nicotiana tabacum) (Tateda et al., 2009), five in Lotus japonicus, five in Arabidopsis (Tateda et al., 2011), and eight in rice (Oryza sativa) (Xu et al., 2015). Here, we showed that grape (V. vinifera) has five VDAC isoforms and two variants. Our molecular phylogenetic analysis disclosed that VvVDACs belong to different clusters and these duplications may indicate that they play various functions in grapevine. Evolutionary analysis of VDAC sequences in plants, animals, and fungi revealed that VDAC polygenic families formed by VDAC replication in the genome may occur independently after differentiation (Homblé et al., 2012). In Arabidopsis, VDACs responded to cold, drought, salinity, and pathogens (Lee et al., 2009), and the two of them also have been reported to exhibit the effect of plant growth, the atvdac1 mutant exhibited faster germination kinetics (Li et al., 2013) and the atvdac2 mutant produced an abscisic acid (ABA)-insensitive phenotype (Yan et al., 2009). Therefore, we assumed that the characterized various VDAC isoforms might play roles in several serious grapevine diseases like Erysiphe necator, P. viticola, Botrytis cinerea, or have effects on grape development such as berry size and color. Related research need to do further studies to understand and parse their functions.
Sequence similarity alignments of animals, fungi, and plants demonstrated that VDAC proteins exhibited low sequence identity. However, the sequences still shared a few common features. Each one had a MPS motif near the C-terminal region and a β-signal motif at the C-terminal of the protein (Homblé et al., 2012). We discovered that grapevine VDAC3, VDAC4, and VDAC5 each contained a conserved MPS motif that were also reported for 30 other eukaryotic VDAC sequences. In this study, however, VvVDAC1 and VvVDAC6 carried a divergent MPS motif. Young et al. (2007) stated that the conserved MPS motif is absent in certain VDAC sequences. Takahashi and Tateda (2013) analyzed MPS motif conservation in the VDAC sequences of 75 different dicots and divided them into two groups. One displayed a highly conserved MPS motif while the other presented with a high divergence rate. Grapevine VvVDAC3, VvVDAC4, and VvVDAC5 might belong to the former group whereas VvVDAC1 and VvVDAC6 could be in the latter.
In addition, the research on the MPS of VDACs from other plants species exhibited that VDACs contained a canonical MPS motif are exclusively localized to mitochondria, while the divergent ones are localized not only in mitochondria but also in other unidentified cellular regions. Arabidopsis VDAC1 and VDAC3 with well-conserved MPS motif were detected primarily in mitochondria (Tateda et al., 2011). Further analysis the cellular localization of Pisum sativum and L. japonicus VDAC isoforms carrying a conserved MPS motif showed the similar situation as Arabidopsis (Clausen et al., 2004; Wandrey et al., 2004). However, VDACs bear the divergent MPS motif in the region present additionally in numerous small vesicles at the cell periphery (Wandrey et al., 2004). Further, Tateda et al. (2011) mutated the conserved MPS motif of AtVDAC1 and found that the subcellular localization of AtVDAC1-m2 exhibited the same as that of AtVDAC2 (containing a divergent MPS), indicating that MPS motif is a key domain functioning to restrict VDAC localization to mitochondria.
Voltage-dependent anion channel protein amino acid sequences were not conserved among different species. Nevertheless, the structural framework of VDAC protein may be relatively conservative as the positions and/or lengths of the β-sheets and the α-helices were conserved in all species studied (Young et al., 2007). The four VvVDACs contained 19 β-sheets and α-helices at the N-terminal region in the sequence. Similar basic properties were reported for the predicted structures of mouse and human VDAC1 and plant VDACs (Homblé et al., 2012). Though VDAC proteins have diverged during evolution, their protein structure was conserved. Therefore, the motif has common and important roles among various kingdoms. However, its functional significance remains to be determined. The present work lays the foundation for future investigations into the relationship between the structure and special functions of VDAC protein.
Previous reports have shown that the levels of VDAC genes expression in various plant tissues are different (Elkeles et al., 1995; Al Bitar et al., 2003). Furthermore, gene/protein function may become specialized via changes in the gene expression pattern. We analyzed the expression profile of VDACs in various tissues and organs in downy mildew-susceptible “Thompson Seedless” (V. vinifera) and downy mildew-resistant “Liuba-8” (V. piasezkii). The variant expression patterns observed for each VDAC suggest that genome redundancy partially accounts for the observed duplication and higher VDAC copy number in plants. In addition, VpVDAC3 was expressed in all plant parts whereas it displayed a peak transcript abundance in some tissues, like, for instance, the leaves and berries during the developmental stages, indicating its important role in these tissues and organs’ functions. The different organ-specific expression pattern was also reported for VDACs in developing wheat and rice (Elkeles et al., 1995; Xu et al., 2015). In addition, some VvVDACs only displayed high expression in specific tissues, indicating that it might have a unique function in specific developmental stages. Taken together, most VvVDACs were expressed in most tissues and most developmental stages, which reveals that VvVDAC genes play multiple important roles in various developmental and biological processes.
Reactive oxygen species (ROS) such as hydrogen peroxide (H2O2) are destructive molecules formed in the cell by oxidative metabolism (Gough and Cotter, 2011). Here, we used a transient assay on N. benthamiana leaves to overexpress VpVDACs and examine their roles in the oxidative burst. When VpVDAC3 was upregulated, mitochondrial ROS production and accumulation increased (Supplementary Figure 3). Earlier studies showed that VDAC is vital to mitochondrial physiology and affects ROS production (Veenman et al., 2008). VpVDAC3 overexpressing Arabidopsis plants had a stronger host defense response to Pst DC3000 infection than WT plants. Similar results were obtained for detection in transient grapevine leaves after P. viticola infection correlating with H2O2 accumulation detected in leaves overexpressing VpVDAC3. However, the mechanism by which VDACs regulate H2O2 production remained unknown. The roles of animal VDACs in ROS homeostasis are well documented (Reina et al., 2010). In contrast, little is known about the functions of plant VDACs in ROS regulation. To clarify the mechanism of VpVDAC3 response to Pst DC3000, we evaluated the changes in VpVDAC3 protein level following pathogenesis. The VpVDAC3 protein level were higher after Pst DC3000 infection (Figure 9), which might be caused by the protein post-transcriptional modification when response to pathogen. A previous study showed that Arabidopsis AtVDAC1–AtVDAC4 were upregulated following Pst DC3000 inoculation (Tateda et al., 2009). Another study showed that Pst DC3000 challenge upregulated VDACs (Lee et al., 2009). A subsequent report confirmed that both virulent Pst DC3000 and avirulent Pst DC3000 avrRpt2 upregulated VDACs (Tateda et al., 2011).
Changes in VDAC expression were documented in response to biotic stress such as a HR to pathogen attack (Lee et al., 2009). To establish the role of VpVDAC3 in biotic stress response, we investigated grapevine resistance to the highly destructive downy mildew caused by P. viticola (Kennelly et al., 2007). We found that VpVDAC3 participated in downy mildew resistance and was strongly expressed in the P. viticola-resistant V. piasezkii “Biuba-8.” Transient overexpression of VpVDAC3 in grapevine leaves increased the plant’s resistance to P. viticola infection and led to greater accumulation of H2O2 content. Our previous study indicated that VpVDAC3 plays important roles in the biological functions of grapevine. VDAC3 from V. piasezkii “Biuba-8” interacted with PR10.1 and triggered pathogen resistance (Ma et al., 2018), which is similar with Arabidopsis VDAC1, contributes to disease resistance by regulating the production of H2O2, which is also implicated in plant disease resistance (Tateda et al., 2011). Meanwhile, the expression of grapevine VvVDAC3 in the susceptible “Thompson Seedless” can also be induced slightly by P. viticola. Therefore, we cannot excluded that VvVDAC3 might also contribute to some resistance in V. vinifera. The foregoing studies have helped elucidate the functions of VDACs response to pathogens, providing insight into how regulation pathways may have been adapted to improve the resistance to downy mildew pathogen stress in grapevine.
Conclusions
In conclusion, the findings of this study demonstrate that grapevine VDAC3 could enhance the tolerance of downy mildew pathogen stress in grapevine correlating with H2O2 accumulation. And during the process, the transcript level of VpVDAC3 was induced highly and the protein level was accumulated by post-transcriptional regulation. The results are expected to provide more sustainable production systems for viticulture and new strategies of protection against downy mildew in grapevine through the development of a resistant grapevine germplasm, which in turn could lead to the discovery of new genetic resources and the diversification of the regulatory mechanisms of resistance genes.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YX conceived and designed the experiments. TX, XW, HM, and LS performed the experiments. TX, XW, HM, and WW analyzed the data. TX, JM, and XW wrote the manuscript. All authors participated in this research and approved the manuscript.
Funding
This research was supported by the National Key Research and Development Program of China (Grant No. 2018YFD1000307) and Team Introduction Program of Ningxia (2020RXTDLX08).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.670505/full#supplementary-material
Footnotes
References
Abrecht, H., Goormaghtigh, E., Ruysschaert, J. M., and Homblé, F. (2000). Structure and orientation of two voltage-dependent, anion-selective channel isoforms an attenuated total reflection Fourier-transform infrared spectroscopy study. J. Biol. Chem. 275, 40992–40999. doi: 10.1074/jbc.M006437200
Al Bitar, F., Roosens, N., Smeyers, M., Vauterin, M., Van Boxtel, J., Jacobs, M., et al. (2003). Sequence analysis, transcriptional and posttranscriptional regulation of the rice VDAC family. Biochim. Biophys. Acta 1625, 43–51. doi: 10.1016/S0167-4781(02)00590-0
Benz, R. (1994). Permeation of hydrophilic solutes through mitochondrial ou ter membranes, review on mitochondrial porins. Biochim. Biophys. Acta 1197, 167–196. doi: 10.1016/0304-4157(94)90004-3
Blachly-Dyson, E., and Forte, M. (2001). VDAC channels. IUBMB Life 52, 113–118. doi: 10.1080/15216540152845902
Cheng, E. H. Y., Sheiko, T. V., Fisher, J. K., Craigen, W. J., and Korsmeyer, S. J. (2003). VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science 301, 513–517. doi: 10.1126/science.1083995
Clausen, C., Ilkavets, I., Thompson, R., Philippar, K., Vojta, A., Möhlmann, T., et al. (2004). Intracellular localization of VDAC proteins in plants. Planta 220, 30–37. doi: 10.1007/s00425-004-1325-3
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Colombini, M. (2004). VDAC: the channel at the interface between mitochondria and the cytosol. Mol. Cell. Biochem. 256, 107–115. doi: 10.1023/B:MCBI.0000009862.17396.8d
Colombini, M. (2009). The published 3D structure of the VDAC channel: native or not? Trends Biochem. Sci. 34, 382–389. doi: 10.1016/j.tibs.2009.05.001
Dangl, J. L., and Jones, J. D. (2001). Plant pathogens and integrated defense responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Desai, M. K., Mishra, R. N., Verma, D., Nair, S., Sopory, S. K., and Reddy, M. K. (2006). Structural and functional analysis of a salt stress inducible gene encoding voltage-dependent anion channel (VDAC) from pearl millet (Pennisetum glaucum). Plant Physiol. Biochem. 44, 483–493. doi: 10.1016/j.plaphy.2006.08.008
Elkeles, A., Devos, K. M., Graur, D., Zizi, M., and Breiman, A. (1995). Multiple cDNAs of wheat voltage–dependent anion channels (VDAC): isolation, differential expression, mapping and evolution. Plant Mol. Biol. 29, 109–124. doi: 10.1007/BF00019123
Fasoli, M., Dal Santo, S., Zenoni, S., Tornielli, G. B., Farina, L., Zamboni, A., et al. (2012). The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24, 3489–3505. doi: 10.2307/41692821
Fiek, C., Benz, R., Roos, N., and Brdiczka, D. (1982). Evidence for identity between the hexokinase-binding protein and the mitochondrial porin in the outer membrane of rat liver mitochondria. Biochim. Biophys. Acta 688, 429–440. doi: 10.1016/0005-2736(82)90354-6
Godbole, A., Dubey, A. K., Reddy, P. S., Udayakumar, M., and Mathew, M. K. (2013). Mitochondrial VDAC and hexokinase together modulate plant programmed cell death. Protoplasma 250, 875–884. doi: 10.1007/s00709-012-0470-y
Goodin, M. M., Zaitlin, D., Naidu, R. A., and Lommel, S. A. (2008). Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol. Plant Microbe Interact. 21, 1015–1026. doi: 10.1094/MPMI-21-8-1015
Gough, D. R., and Cotter, T. G. (2011). Hydrogen peroxide: a Jekyll and Hyde signalling molecule. Cell Death Dis. 2:213. doi: 10.1038/cddis.2011.96
Greenberg, J. T. (1996). Programmed cell death: a way of life for plants. Proc. Natl. Acad. Sci. U.S.A. 93, 12094–12097. doi: 10.1073/pnas.93.22.12094
Homblé, F., Krammer, E. M., and Prévost, M. (2012). Plant VDAC: facts and speculations. Biochim. Biophys. Acta 1818, 1486–1501. doi: 10.1016/j.bbamem.2011.11.028
Johal, G. S., Hulbert, S. H., and Briggs, S. P. (1995). Disease lesion mimics of maize: a model for cell death in plants. Bioessays 17, 685–692.
Kennelly, M. M., Gadoury, D. M., Wilcox, W. F., Magarey, P. A., and Seem, R. C. (2007). Primary infection, lesion productivity, and survival of sporangia in the grapevine downy mildew pathogen Plasmopara viticola. Phytopathology 97, 512–522. doi: 10.1094/PHYTO-97-4-0512
Kroemer, G., Zamzami, N., and Susin, S. A. (1997). Mitochondrial control of apoptosis. Immunol. Today 18, 44–51. doi: 10.1016/S0167-5699(97)80014-X
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lacomme, C., and Santa Cruz, S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. U.S.A. 96, 7956–7961. doi: 10.1073/pnas.96.14.7956
Lam, E., Kato, N., and Lawton, M. (2001). Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411, 848–853. doi: 10.1038/35081184
Lee, S. M., Hoang, M. H. T., Han, H. J., Kim, H. S., Lee, K., Kim, K. E., et al. (2009). Pathogen inducible voltage-dependent anion channel (AtVDAC) isoforms are localized to mitochondria membrane in Arabidopsis. Mol. Cells 27, 321–327. doi: 10.1007/s10059-009-0041-z
Levine, A., Pennell, R. I., Alvarez, M. E., Palmer, R., and Lamb, C. (1996). Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr. Biol. 6, 427–437. doi: 10.1016/S0960-9822(02)00510-9
Li, Z. Y., Xu, Z. S., He, G. Y., Yang, G. X., Chen, M., Li, L. C., et al. (2013). The voltage-dependent anion channel 1 (AtVDAC1) negatively regulates plant cold responses during germination and seedling development in Arabidopsis and interacts with calcium sensor CBL1. Int. J. Mol. Sci. 14, 701–713. doi: 10.3390/ijms14010701
Liu, R. Q., Wang, L., Zhu, J., Chen, T. T., Wang, Y. J., and Xu, Y. (2015). Histological responses to downy mildew in resistant and susceptible grapevines. Protoplasma 252, 259–270. doi: 10.1007/s00709-014-0677-1
Ma, H., Xiang, G. Q., Li, Z. Q., Wang, Y. T., Dou, M. R., Su, L., et al. (2018). Grapevine VpPR10.1 functions in resistance to Plasmopara viticola through triggering a cell death-like defense response by interacting with VpVDAC3. Plant Biotechnol. J. 16, 1488–1501. doi: 10.1111/pbi.12891
Mayer, A., Lill, R., and Neupert, W. (1993). Translocation and insertion of precursor proteins into isolated outer membranes of mitochondria. J. Cell Biol. 121, 1233. doi: 10.1083/jcb.121.6.1233
Messina, A., Reina, S., Guarino, F., and Vito De Pinto, D. V. (2012). VDAC isoforms in mammals. Biochim. Biophys. Acta 1818, 1466–1476. doi: 10.1016/j.bbamem.2011.10.005
Noskov, S. Y., Rostovtseva, T. K., Chamberlin, A. C., Teijido, O., Jiang, W., and Bezrukov, S. M. (2016). Current state of theoretical and experimental studies of the voltage-dependent anion channel (VDAC). Biochim. Biophys. Acta 1858, 1778–1790. doi: 10.1016/j.bbamem.2016.02.026
Reina, S., Palermo, V., Guarnera, A., Guarino, F., Messina, A., Mazzoni, C., et al. (2010). Swapping of the N-terminus of VDAC1 with VDAC3 restores full activity of the channel and confers anti-aging features to the cell. FEBS Lett. 584, 2837–2844. doi: 10.1016/j.febslet.2010.04.066
Robert, N., d’Erfurth, I., Marmagne, A., Erhardt, M., Allot, M., and Boivin, K. (2012). Voltage-dependent anion channels (VDACs) in Arabidopsis have a dual localization in the cell but show a distinct role in mitochondria. Plant Mol. Biol. 78, 431–446. doi: 10.1007/s11103-012-9874-5
Schredelseker, J., Paz, A., López, C. J., Altenbach, C., Leung, C. S., Drexler, M. K., et al. (2014). High resolution structure and double electron-electron resonance of the zebrafish voltage-dependent anion channel 2 reveal an oligomeric population. J. Biol. Chem. 289, 12566–12577. doi: 10.1074/jbc.m113.497438
Takahashi, Y., and Tateda, C. (2013). The functions of voltage-dependent anion channels in plants. Apoptosis 18, 917–924. doi: 10.1007/s10495-013-0845-3
Tateda, C., Watanabe, K., Kusano, T., and Takahashi, Y. (2011). Molecular and genetic characterization of the gene family encoding the voltage-dependent anion channel in Arabidopsis. J. Exp. Bot. 62, 4773–4785. doi: 10.1093/jxb/err113
Tateda, C., Yamashita, K., Takahashi, F., Kusano, T., and Takahashi, Y. (2009). Plant voltage-dependent anion channels are involved in host defense against Pseudomonas cichorii and in Bax-induced cell death. Plant Cell Rep. 28, 41–51. doi: 10.1007/s00299-008-0630-x
Thordal-Christensen, H., Zhang, Z. G., Wei, Y. D., Collinge, D. B., Zhang, Z. G., and Wei, Y. D. (1997). Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
van Wees, S. (2008). Phenotypic analysis of Arabidopsis mutants: trypan blue stain for fungi, oomycetes, and dead plant cells. Cold Spring Harb. Protoc. 8:pdb.prot4982. doi: 10.1101/pdb.prot498
Veenman, L., Shandalov, Y., and Gavish, M. (2008). VDAC activation by the 18-kDa translocator protein (TSPO), implications for apoptosis. J. Bioenerg. Biomembr. 40, 199–205. doi: 10.1007/s10863-008-9142-1
Vercesi, A., Tornaghi, R., Sant, S., Burruano, S., and Faoro, F. (1999). A cytological and ultrastructural study on the maturation and germination of oospores of Plasmopara viticola from overwintering vine leaves. Mycol. Res. 103, 193–202. doi: 10.1017/S095375629800700X
Vianello, A., Casolo, V., Petrussa, E., Peresson, C., Patui, S., Bertolini, A., et al. (2012). The mitochondrial permeability transition pore (PTP)-an example of multiple molecular exaptation? Biochim. Biophys. Acta 1817, 2072–2086. doi: 10.1016/j.bbabio.2012.06.620
Wandrey, M., Trevaskis, B., Brewin, N., and Udvardi, M. K. (2004). Molecular and cell biology of a family of voltage-dependent anion channel porins in Lotus japonicus. Plant Physiol. 134, 182–193. doi: 10.1104/pp.103.031484
Wang, H., Li, J., Bostock, R. M., and Gilchrist, D. G. (1996). Apoptosis: a functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 8, 375–391. doi: 10.1105/tpc.8.3.375
Wei, Z. M., Laby, R. J., Zumoff, C. H., Bauer, D. W., He, S. Y., Collmer, A., et al. (1992). Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257, 85–88. doi: 10.1126/science.1621099
Wong, F. P., Burr, H. N., and Wilcox, W. F. (2001). Heterothallism in Plasmopara viticola. Plant Pathol. 50, 427–432. doi: 10.1046/j.1365-3059.2001.00573.x
Xu, T. F., Zhao, X. C., Jiao, Y. T., Wei, J. Y., Wang, L., and Xu, Y. (2014). A pathogenesis related protein, VpPR-10.1, from Vitis pseudoreticulata: an insight of its mode of antifungal activity. PLoS One 9:e95102. doi: 10.1371/journal.pone.0095102
Xu, X., Tan, Y. P., Cheng, G., Liu, X. Q., Xia, C. J., Luo, F. Y., et al. (2015). Genomic survey and gene expression analysis of the VDAC gene family in rice. Genet. Mol. Res. 14, 15683–15696. doi: 10.4238/2015.December.1.20
Yan, J., He, H., Tong, S., Zhang, W., Li, X., and Yang, Y. (2009). Voltage-dependent anion channel 2 of Arabidopsis thaliana (AtVDAC2) is involved in ABA-mediated early seedling development. Int. J. Mol. Sci. 10, 2476–2486. doi: 10.3390/ijms10062476
Young, M. J., Bay, D. C., Hausner, G., and Court, D. A. (2007). The evolutionary history of mitochondrial porins. BMC Evol. Biol. 7:31. doi: 10.1186/1471-2148-7-31
Keywords: Plasmopara viticola, Vitis, reactive oxygen species, voltage-dependent anion channels, VpVDAC3
Citation: Xu T, Wang X, Ma H, Su L, Wang W, Meng J and Xu Y (2021) Functional Characterization of VDACs in Grape and Its Putative Role in Response to Pathogen Stress. Front. Plant Sci. 12:670505. doi: 10.3389/fpls.2021.670505
Received: 21 February 2021; Accepted: 14 May 2021;
Published: 16 June 2021.
Edited by:
Gabor Jakab, University of Pécs, HungaryReviewed by:
Wolfgang Moeder, University of Toronto, CanadaWen-Ming Wang, Sichuan Agricultural University, China
Copyright © 2021 Xu, Wang, Ma, Su, Wang, Meng and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Xu, yan.xu@nwafu.edu.cn
†These authors have contributed equally to this work
 Tengfei Xu
Tengfei Xu Xiaowei Wang
Xiaowei Wang Hui Ma2
Hui Ma2 Jiangfei Meng
Jiangfei Meng