- 1 UPR2357 CNRS, Institut de Biologie Moléculaire des Plantes, Strasbourg, France
- 2 Department of Plant Physiology, Zürich-Basel Plant Science Center, Part of the Swiss Plant Science Web, Botanical Institute of the University of Basel, Basel, Switzerland
Viruses encode viral suppressors of RNA silencing (VSRs) to counteract RNA silencing, a major antiviral defense response in plants. Recent studies indicate a role of virus-derived siRNAs in manipulating the expression of specific host genes and that certain plant viral movement proteins (MPs) can act as viral enhancers of RNA silencing (VERs) by stimulating the spread of silencing between cells. This suggests that viruses have evolved complex responses capable to efficiently hijack the host RNA silencing machinery to their own advantage. We draw here a dynamic model of the interaction of plant viruses with the silencing machinery during invasion of the host. The model proposes that cells at the spreading front of infection, where infection starts from zero and the VSR levels are supposedly low, represent potential sites for viral manipulation of host gene expression by using virus- and host-derived small RNAs. Viral MPs may facilitate the spread of silencing to produce a wave of small RNA-mediated gene expression changes ahead of the infection to increase host susceptibility. When experimentally ascertained, this hypothetical model will call for re-defining viral movement and the function of viral MPs.
Viruses depend on intricate interactions with their hosts to fulfill their life cycle. To infect plants systemically, viruses encode specialized movement proteins (MPs) that allow their movement from cell to cell through plasmodesmata and to distant tissues via the phloem. While virus spread depends on host factors that support replication and movement, the final outcome of infection relies on the ability of the virus to cope with RNA silencing, a major antiviral plant defense mechanism that targets the viral genome for degradation (Bayne et al., 2005; Dunoyer and Voinnet, 2005; Ding and Voinnet, 2007; Ding, 2010). Indeed, the viral RNAs are processed by the RNA silencing machinery into viral small RNAs (vsRNAs) that can potentially be loaded into specific ARGONAUTE effectors (AGOs) that further target viral RNAs for degradation and/or for translational repression. Because small RNAs (sRNAs) are mobile and RNA silencing can propagate from cell to cell (Chitwood and Timmermans, 2010; Dunoyer et al., 2010a,b; Brosnan and Voinnet, 2011; Molnar et al., 2011), antiviral silencing was proposed to involve the movement of vsRNAs that immunize naïve cells ahead of the infection (Ding and Voinnet, 2007; Figure 1A). As part of the ongoing host–virus arms race, viruses have evolved potent suppressors of RNA silencing (VSRs; Figure 1B). The VSRs of different viruses inhibit different RNA silencing components suggesting that they evolved independently (Diaz-Pendon and Ding, 2008; Wu et al., 2010; Burgyan and Havelda, 2011). The antiviral RNA silencing pathway shares components with the endogenous microRNA (miRNA) and trans-acting siRNA (ta-siRNA) pathways (Vazquez et al., 2004; Dunoyer et al., 2005; Gasciolli et al., 2005; Xie et al., 2005) that regulate gene expression and important developmental processes (Pasquinelli and Ruvkun, 2002; Voinnet, 2005). Thus, indirect interference of VSRs with important host sRNA-mediated developmental pathways was identified as a potential molecular explanation for virus-induced “disease symptoms,” i.e., the aberrant plant growth and development observed during viral infection (Kasschau et al., 2003; Chapman et al., 2004; Diaz-Pendon and Ding, 2008; Figure 2). Indeed, based on a recent study developmental disease symptoms triggered by different viruses are presumed to occur via an effect of VSRs on miR167 activity, which results in an increased abundance of the transcription factor AUXIN RESPONSE FACTOR 8 (ARF8; Jay et al., 2011).
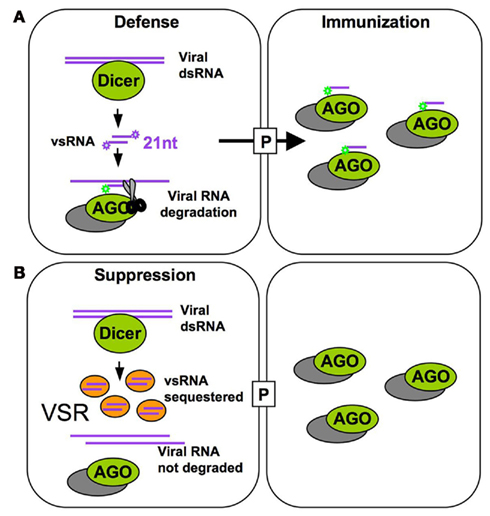
Figure 1. Host antiviral defense and TMV counter defense. (A) The 21 nt vsRNAs generated by Dicer processing (primarily DCL4) of double-stranded viral RNA associate with RISC to further guide vRNA degradation in the absence of the VSR. Intercellular mobility of vsRNAs allows their spreading through plasmodesmata (P) to neighboring cells ahead of the virus front and might serve to immunize naïve cells by programming RISC against the incoming virus. (B) In cells expressing the VSR, vsRNAs are sequestered and allow accumulation of viral RNA.
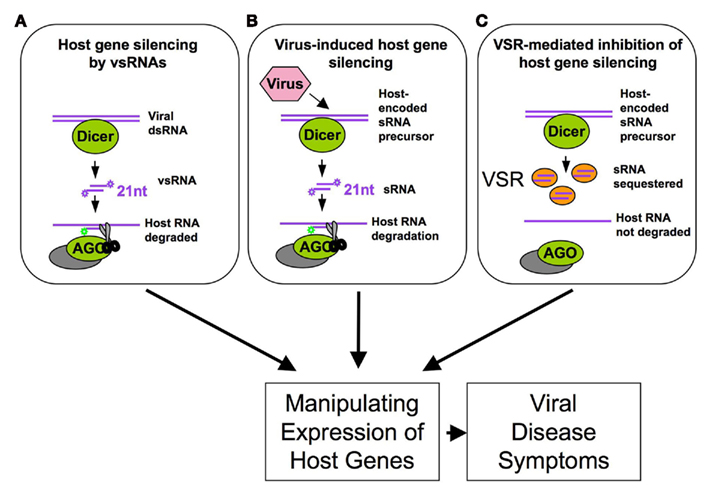
Figure 2. Viral manipulation of host RNA silencing leads to changes in host gene expression and disease symptoms by (A) inducing functional virus-derived vsRNAs complementary to host genes and by (B) inducing or (C) suppressing the production of functional host sRNAs (e.g., here: sequestration of host sRNAs).
Tobacco mosaic virus (TMV) and related tobamoviruses encode a VSR that resides in the small replicase subunit (Kubota et al., 2003; Ding et al., 2004; Csorba et al., 2007; Vogler et al., 2007). This subunit is dispensable for replication but accumulates to high amounts and increases the virus titers (Ishikawa et al., 1986; Lewandowski and Dawson, 2000). Its capacity to suppress silencing was correlated with its ability to bind siRNA and miRNA duplexes in vitro and to interfere with vsRNA and sRNA methylation in vivo (Blevins et al., 2006; Csorba et al., 2007; Kurihara et al., 2007; Vogler et al., 2007). This modus operandi was also reported for several other VSRs like the Hc-Pro of potyviruses and P19 of tombusviruses (Lakatos et al., 2006). Consistently, plants infected with tobamoviruses like TMV, TMV-Cg, cr-TMV, or Oilseed rape mosaic virus (ORMV) generally exhibit increased levels of miRNAs (Blevins et al., 2006; Bazzini et al., 2007; Csorba et al., 2007; Tagami et al., 2007; Vogler et al., 2007). Strikingly, these increased miRNA levels do not lead to the expected decreases in target mRNA levels (Csorba et al., 2007; Tagami et al., 2007; Hu et al., 2011) indicating that sRNA binding by tobamovirus replicases interferes with the use of induced sRNAs in RISC loading or activity. This model is also supported by in vitro observations showing that binding of sRNA duplexes by cr-TMV replicase inhibits RISC assembly (Csorba et al., 2007). Although sRNA accumulation was observed in vivo, it is yet unclear whether this is indeed due to a binding and stabilization of sRNAs by the tobamoviral suppressor or whether this is caused by other yet unidentified virus-induced mechanisms. Moreover, it is reasonable to speculate that virus-encoded or virus-induced sRNAs may not always be inhibited by viral suppressors during the viral life cycle but may have a functional role at some stages of the infection process. Although further studies are necessary, several experimental arguments support this possibility, as discussed below.
Viruses Generate Specific sRNA Profiles
First, it is well documented that several mammalian viruses encode miRNAs to use the silencing machinery of their host to their own advantage. To date, over 200 virus-encoded miRNAs have been identified. Current evidences indicate that mammalian viruses use these miRNAs to manipulate both the cellular and viral gene expression. Moreover, mammalian viral infection can exert a profound impact on the cellular miRNA expression profile and certain mammalian RNA viruses interact directly with cellular miRNAs and use these miRNAs to support their replication (Skalsky and Cullen, 2010). This demonstrates that mammalian viruses, and potentially also plant viruses, have evolved means to subvert the RNA silencing machinery to regulate gene expression of the host.
Moreover, although plant viruses do not encode miRNAs, they generate an abundant and complex pool of vsRNAs by Dicer-like processing, and many of the vsRNAs share complementarity to the host genome and thus could potentially target host mRNAs (Moissiard and Voinnet, 2006; Qi et al., 2009; Llave, 2010; Figure 2A). For example, vsRNAs derived from the translational leader of the Cauliflower mosaic virus 35S RNA, which forms an extensive stem-loop structure, have sequence complementarity to Arabidopsis transcripts. It was shown that at least one of these vsRNAs can act as a bona fide siRNA in infected turnip (Moissiard and Voinnet, 2006). Another important case of vsRNA activity in the regulation of host gene expression was reported for Cucumber mosaic virus (CMV). Its non-coding subviral Y satellite RNA produces specific siRNAs, one of which was shown to specifically affect the expression of the host mRNAs encoding the magnesium protoporphyrin chelatase subunit I, a key enzyme of the chlorophyll biosynthesis pathway, and to be responsible for disease symptoms caused by CMV infection (Shimura et al., 2011). Our own analysis has revealed about 13,000 sequence regions in the Arabidopsis genome that can potentially be targeted by the vsRNAs of ORMV (Kørner and Heinlein, unpublished results). Plant viruses may also utilize host-encoded sRNAs to target host genes (Figure 2B). Plant cells produce various sRNAs, including miRNAs, ta-siRNAs, repeat-associated siRNAs, nat-siRNAs, lsiRNAs, long-miRNAs and chromatin-associated siRNAs to regulate the expression of their genomes and ensure their proper inheritance (Meyers et al., 2008; Vaucheret, 2008; Vazquez et al., 2010). However, viruses may go beyond the normal set of host sRNAs by generating their own host-encoded sRNA environment. For example, tobamoviruses produce a strong and specific enrichment of sRNAs that are processed from miRNA and ta-siRNA precursors (Bazzini et al., 2007; Csorba et al., 2007; Tagami et al., 2007; Vogler et al., 2007; Hu et al., 2011). Deep sequencing of ORMV-infected Arabidopsis plants revealed a specific pattern of host sRNA enrichment that may either be due to replicase binding or to virus-induced activation of host-encoded processing or effector proteins (Hu et al., 2011). Thus, the plethora of virus- and host-derived sRNAs represents a regulatory reservoir that might be used by the virus to promote silencing of host transcripts. Since the virus encodes a silencing suppressor that sequesters sRNAs (Figures 1 and 2C), this activity is highly controlled during infection and likely limited to cells in which the viral silencing suppressor is not yet present or scarce. As summarized in Figure 2, the ability of viruses to influence host gene expression via vsRNAs, host sRNAs, or silencing suppressor activity may provide an important explanation for the development of viral disease symptoms in infected plants.
Viruses may Subvert RNA Silencing at the Leading Front of Infection
It is assumed that VSR proteins must be sufficiently abundant in the considered cells to exert a dominant effect and lead to silencing suppression. This certainly applies to silencing suppressors that act by sRNA sequestration. The tobamoviral VSR shows binding affinity to both siRNAs and miRNAs in vitro (Csorba et al., 2007; Kurihara et al., 2007) and tobamoviral infection causes widespread stabilization of both viral and host sRNAs (Hu et al., 2011). However, as the virus spreads through leaf tissues, infection of a new cell always starts from zero. Thus, the amount of suppressor necessary to efficiently suppress RNA silencing in a given cell can only be reached after a certain delay. As an important consequence of this scenario, efficient RNA silencing suppression can be expected to be limited to cells behind the infection front. Thus, virus-encoded siRNAs and/or virus-induced host sRNAs that remain non-sequestered in leading front cells may be potent to trigger silencing of host genes. It appears likely, therefore, that virus:host interactions change from early to later stages of infection in that the virus first subverts RNA silencing at the leading front of infection and subsequently controls this activity as soon as the silencing suppressor reaches the critical level.
Viruses may Actively Spread RNA Silencing into Cells Ahead the Leading Front of Infection
We and other groups have shown that the MPs of certain viruses act as viral enhancers of RNA silencing (VERs) by promoting the propagation of RNA silencing from cell to cell (Vogler et al., 2008; Zhou et al., 2008; Lacombe et al., 2010). When transiently or transgenically expressed, the MP of TMV causes broadening of the rim of silenced cells around GFP-agroinfiltrated patches in the GFP-transgenic N. benthamiana line 16c (Figure 3A). The MP also enhances the systemic spread of GFP silencing when transgenically expressed. Because these effects of MP were not accompanied by significant changes in the pattern of GFP siRNAs, this suggested that MP promotes silencing signal transmission from incipient to recipient cells (Vogler et al., 2008). It is unclear whether MP enhances silencing spread by its sequence-independent single-stranded nucleic acid binding activity (Citovsky et al., 1990) and/or by increasing the size exclusion limit of plasmodesmata (Wolf et al., 1989), or by affecting the activity of RNA silencing components. Whatever the underlying mechanism, it is likely that this MP activity supports the intercellular spread of sRNAs at the leading front of spreading infection sites. Evidences supporting this conclusion came from studies using a GFP-tagged TMV mutant carrying a single amino acid exchange in the silencing suppressor. This mutant virus caused the formation of infection sites in leaves similar as the wild type virus. However, because the silencing suppressor was impaired, the virus and GFP expression became silenced in the central part of the infection sites (Figure 3B). Similar observations were previously made with Tomato mosaic virus (Kubota et al., 2003). Interestingly, this “central silencing” did no longer occur when the viral MP was deleted (Vogler et al., 2008; Figure 3B). This unraveled the active contribution of MP to the silencing of the virus in the absence of silencing suppressor. This finding is consistent with the previous proposal that antiviral silencing involves the movement of vsRNAs (Ding and Voinnet, 2007). By potentiating vsRNA movement into cells ahead of infection, the MP might provoke an earlier, and consequently stronger, antiviral response in the absence of silencing suppressor. In normal circumstances where the suppressor is present, the virus is not silenced in the center of infection sites. However, at the leading front, where silencing suppressor levels are low, vsRNAs could still spread and immunize naïve cells ahead of the infection. This activity could enhance the quality of the virus:host interaction by preventing the over-accumulation of the virus, which would otherwise be detrimental for the host. Moreover, given that the MP bears a sequence non-specific nucleic acid binding activity and facilitates the spread of transgene silencing, the protein might also spread vsRNAs and host-derived sRNAs complementary to host genes. Permitting local silencing of host genes in cells ahead of the replicating virus front may represent a crucial activity of MP to facilitate the spread of infection.
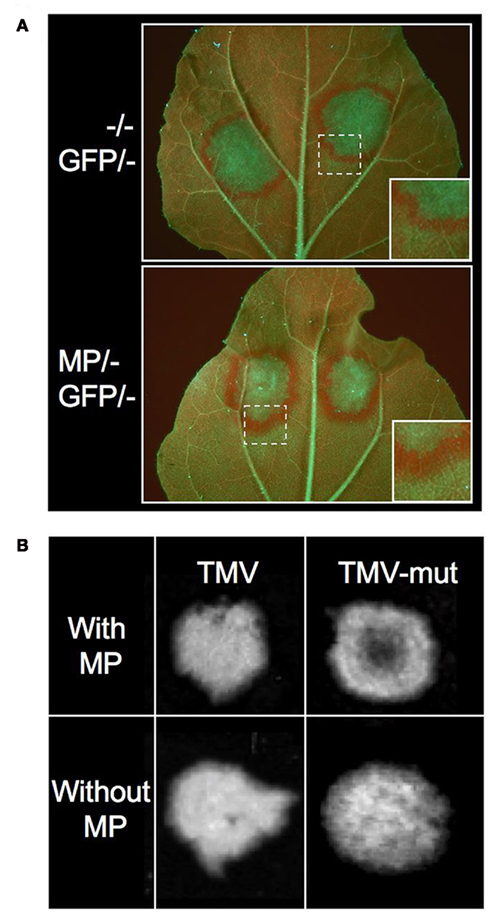
Figure 3. The MP of TMV is a Viral Enhancer of RNA silencing (VER). (A) GFP expression in leaves of GFP-transgenic Nicotiana benthamiana line 16c upon transient expression of 35S::GFP. GFP overexpression induces GFP silencing in the agroinfiltrated patch and triggers the formation of a GFP silencing signal that spreads into surrounding cells. The disappearance of GFP leads to the exposure of strong red chlorophyll autofluorescence. The rim of red fluorescence around the agroinfiltrated patch is broader in tissues expressing the MP (stably or transiently). This indicates that MP enhances the spread of silencing from incipient to recipient cells. (B) GFP-fluorescent TMV infection sites in MP-transgenic plants. Inactivation of the VSR by a missense mutation in the small replicase subunit (TMV-mut) results in silencing of the virus in the center of spreading infection sites in leaves. This central silencing no longer occurs when the MP is deleted. This indicates that viral MP acts by enhancing antiviral silencing. (Figures derived from Vogler et al., 2008, under the Creative Commons Attribution License.)
Another VER is the Rice yellow mottle virus (RYMV) protein P1 (Lacombe et al., 2010). Similar to MP, P1 enhances the spread of GFP silencing in local and systemically infected leaves. Interestingly, P1 can also act as a VSR. This suggests that the virus suppresses or enhances silencing through tight regulation of this single protein. Since P1 also represents the MP of the virus it is conceivable that the spread of RYMV in infected plants critically depends on the coordinated manipulation of RNA silencing by the VSR and VER functions of the protein. VER activity has also been demonstrated for the P8 and P9 proteins of Turnip crinkle virus (TCV; Zhou et al., 2008). Like for TMV, the RNA silencing promoting functions of these proteins were only detected in the absence of the viral silencing suppressor, the p38 coat protein.
A Time-Resolved Model for Manipulation of Host Gene Expression during Virus Cell to Cell Spread
The MP of TMV, which is known to bind RNAs in a sequence-independent manner and to promote the spread of RNA silencing, may facilitate the spread of vsRNAs and host sRNAs in addition to the viral RNA genome. Very low amounts of the protein are sufficient to increase the size exclusion limit of plasmodesmata and to allow the spread of infection between cells (Arce-Johnson et al., 1995). Thus, MP proteins likely act to facilitate the spread of viral RNAs and vsRNAs/host sRNAs into non-infected cells soon after a new cell of the leading front of infection is infected. This activity will likely be restricted to a short time window. Indeed, the protein accumulates only transiently during the early stages of infection (Watanabe et al., 1984; Heinlein et al., 1998) and is subsequently degraded (Reichel and Beachy, 2000). Moreover, its ability to increase plasmodesmatal size exclusion limit is restricted to cells at the leading front of infection (Oparka et al., 1997). In contrast, the silencing suppressor of the virus, which commonly sequesters sRNAs, is continuously expressed (Watanabe et al., 1984) and likely must accumulate to reasonable amounts before efficiently blocking vsRNA/host sRNA function. Thus, our model shown in Figure 4 predicts that MP already acts in low amounts at the leading front of infection, whereas the VSR must first accumulate and, therefore, interferes with RNA silencing with a delay and, thus, only behind the infection front. Together with the above-mentioned observations this leads us to suggest that MP potentiates viral movement not only by facilitating the spread of viral RNA but also by spreading of virus-induced host sRNAs and virus-encoded vsRNAs in order to influence host cell gene expression and thus to enhance the susceptibility of the host cell for the incoming viral RNA. Previous studies with different viral systems have indicated transient changes in host gene expression at or ahead of the spreading virus front (Wang and Maule, 1995; Aranda et al., 1996; Aranda and Maule, 1998; Havelda and Maule, 2000) which is consistent with this hypothesis. The MP supported spreading of vsRNAs could further enhance the probability of a successful virus:host interaction by providing already the recipient cell with the capacity to control the virus to some extent.
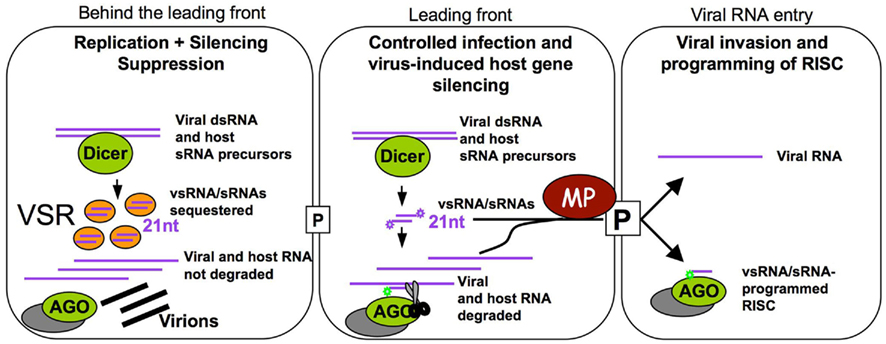
Figure 4. Proposed time-resolved model for TMV infection. Cells at the leading front of infection sites (middle cell) undergo initial stages of infection. The virus induces the biogenesis of vsRNAs and specific host sRNAs. In these cells, VSR levels are still low and vsRNAs and host sRNAs can be loaded into RISC to guide translation repression and cleavage of the virus and of specific host transcripts. In these cells, MP acts at very low concentration to transport vsRNAs, host sRNAs and viral RNA into non-infected cells. The vsRNAs and host sRNAs can program RISC to manipulate the expression of host transcripts and allow recipient cells to control the incoming virus. Our model also suggests that the tight viral control of its own accumulation and the manipulation of host gene expression could represent essential mechanisms sustaining successful infection. Once infection is initiated, the virus replicates and produces a sufficient amount of the VSR. Thus, in cells behind the leading front of infection, vsRNAs and host sRNAs are sequestered and inactivated, allowing the virus to accumulate and produce virions. Meanwhile, the leading front has moved into new cells.
The time-resolved model shown in Figure 4 emphasizes that virus-induced immunization and silencing of host genes is certainly transient as it will subsequently be suppressed by the increasing levels of the VSR. This subsequent silencing suppression allows virus propagation in cells behind the infection front and may also prevent the occurrence of developmental effects that may otherwise be caused by continued virus-induced host gene silencing. This time-resolved model also suggests that virus:host relationships in which VER and VSR functions are tightly controlled and balanced may represent the basis for symptomless infections. Such highly evolved virus:host relationships may be widespread and even mutualistic in nature whereas virus:host relationships causing disease symptoms might rather be an exception limited to agricultural monocultures and specific model pathosystems studied in the laboratory (Roossinck, 2005, 2011; Niehl and Heinlein, 2009). The cell autonomous and cell non-autonomous contribution of the virus-induced sRNA profile to antiviral defense and host genome modification has profound implications for our understanding of viral pathogenicity and host specificity in plants. Understanding the mechanisms by which plants remain healthy during virus infection also has important implications for agriculture and for the agrobiotech industries.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors are supported by grants of the Human Frontier Science Program Organization (grant RGP0022/2006), the Agence Nationale de la Récherche (grant ANR-08-BLAN-0244), and the Swiss National Science Foundation (grant 31003A-124940) to Manfred Heinlein, and a Swiss National Science Foundation Ambizione grant (PZ00P3-126329) to Franck Vazquez.
References
Aranda, M., and Maule, A. (1998). Virus-induced host gene shutoff in animals and plants. Virology 243, 261–267.
Aranda, M. A., Escaler, M., Wang, D., and Maule, A. J. (1996). Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc. Natl. Acad. Sci. U.S.A. 93, 15289–15293.
Arce-Johnson, P., Kahn, T. W., Reimann-Philipp, U., Rivera-Bustamente, R., and Beachy, R. N. (1995). The amount of movement protein produced in transgenic plants influences the establishment, local movement, and systemic spread of infection by movement protein-deficient Tobacco mosaic virus. Mol. Plant Microbe Interact. 3, 415–423.
Bayne, E. H., Rakitina, D. V., Morozov, S. Y., and Baulcombe, D. C. (2005). Cell-to-cell movement of Potato potexvirus X is dependent on suppression of RNA silencing. Plant J. 44, 471–482.
Bazzini, A. A., Hopp, H. E., Beachy, R. N., and Asurmendi, S. (2007). Infection and coaccumulation of Tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc. Natl. Acad. Sci. U.S.A. 104, 12157–12162.
Blevins, T., Rajeswaran, R., Shivaprasad, P. V., Beknazariants, D., Si-Ammour, A., Park, H. S., Vazquez, F., Robertson, D., Meins, F. Jr., Hohn, T., and Pooggin, M. M. (2006). Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34, 6233–6246.
Brosnan, C. A., and Voinnet, O. (2011). Cell-to-cell and long-distance siRNA movement in plants: mechanisms and biological implications. Curr. Opin. Plant Biol. 14, 580–587.
Burgyan, J., and Havelda, Z. (2011). Viral suppressors of RNA silencing. Trends Plant Sci. 16, 265–272.
Chapman, E. J., Prokhnevsky, A. I., Gopinath, K., Dolja, V. V., and Carrington, J. C. (2004). Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18, 1179–1186.
Citovsky, V., Knorr, D., Schuster, G., and Zambryski, P. (1990). The P30 movement protein of Tobacco mosaic virus is a single-stranded nucleic acid binding protein. Cell 60, 637–647.
Csorba, T., Bovi, A., Dalmay, T., and Burgyan, J. (2007). The p122 subunit of Tobacco mosaic virus replicase is a potent silencing suppressor and compromises both small interfering RNA- and microRNA-mediated pathways. J. Virol. 81, 11768–11780.
Diaz-Pendon, J. A., and Ding, S. W. (2008). Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46, 303–326.
Ding, X. S., Liu, J., Cheng, N. H., Folimonov, A., Hou, Y. M., Bao, Y., Katagi, C., Carter, S. A., and Nelson, R. S. (2004). The Tobacco mosaic virus 126-kDa protein associated with virus replication and movement suppresses RNA silencing. Mol. Plant Microbe Interact. 17, 583–592.
Dunoyer, P., Brosnan, C. A., Schott, G., Wang, Y., Jay, F., Alioua, A., Himber, C., and Voinnet, O. (2010a). An endogenous, systemic RNAi pathway in plants. EMBO J. 29, 1699–1712.
Dunoyer, P., Schott, G., Himber, C., Meyer, D., Takeda, A., Carrington, J. C., and Voinnet, O. (2010b). Small RNA duplexes function as mobile silencing signals between plant cells. Science 328, 912–916.
Dunoyer, P., Himber, C., and Voinnet, O. (2005). DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37, 1356–1360.
Dunoyer, P., and Voinnet, O. (2005). The complex interplay between plant viruses and host RNA-silencing pathways. Curr. Opin. Plant Biol. 8, 415–423.
Gasciolli, V., Mallory, A. C., Bartel, D. P., and Vaucheret, H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15, 1494–1500.
Havelda, Z., and Maule, A. J. (2000). Complex spatial responses to Cucumber mosaic virus infection in susceptible Cucurbita pepo cotyledons. Plant Cell 12, 1975–1986.
Heinlein, M., Padgett, H. S., Gens, J. S., Pickard, B. G., Casper, S. J., Epel, B. L., and Beachy, R. N. (1998). Changing patterns of localization of the Tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10, 1107–1120.
Hu, Q., Hollunder, J., Niehl, A., Kørner, C. J., Gereige, D., Windels, D., Arnold, A., Kuiper, M., Vazquez, F., Pooggin, M., and Heinlein, M. (2011). Specific impact of tobamovirus infection on the Arabidopsis small RNA profile. PLoS ONE 6, e19549. doi: 10.1371/journal.pone.0019549
Ishikawa, M., Meshi, T., Motoyoshi, F., Takamatsu, N., and Okada, Y. (1986). In vitro mutagenesis of the putative replicase genes of Tobacco mosaic virus. Nucleic Acids Res. 14, 8291–8305.
Jay, F., Wang, Y., Yu, A., Taconnat, L., Pelletier, S., Colot, V., Renou, J. P., and Voinnet, O. (2011). Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis. PLoS Pathog. 7, e1002035. doi: 10.1371/journal.ppat.1002035
Kasschau, K. D., Xie, Z., Allen, E., Llave, C., Chapman, E. J., Krizan, K. A., and Carrington, J. C. (2003). P1/HC-pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217.
Kubota, K., Tsuda, S., Tamai, A., and Meshi, T. (2003). Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J. Virol. 77, 11016–11026.
Kurihara, Y., Inaba, N., Kutsuna, N., Takeda, A., Tagami, Y., and Watanabe, Y. (2007). Binding of tobamovirus replication protein with small RNA duplexes. J. Gen. Virol. 88, 2347–2352.
Lacombe, S., Bangratz, M., Vignols, F., and Brugidou, C. (2010). The Rice yellow mottle virus P1 protein exhibits dual functions to suppress and activate gene silencing. Plant J. 61, 371–382.
Lakatos, L., Csorba, T., Pantaleo, V., Chapman, E. J., Carrington, J. C., Liu, Y. P., Dolja, V. V., Calvino, L. F., Lopez-Moya, J. J., and Burgyan, J. (2006). Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25, 2768–2780.
Lewandowski, D. J., and Dawson, W. O. (2000). Functions of the 126- and 183-kDa proteins of Tobacco mosaic virus. Virology 271, 90–98.
Llave, C. (2010). Virus-derived small interfering RNAs at the core of plant-virus interactions. Trends Plant Sci. 15, 701–707.
Meyers, B. C., Axtell, M. J., Bartel, B., Bartel, D. P., Baulcombe, D., Bowman, J. L., Cao, X., Carrington, J. C., Chen, X., Green, P. J., Griffiths-Jones, S., Jacobsen, S. E., Mallory, A. C., Martienssen, R. A., Poethig, R. S., Qi, Y., Vaucheret, H., Voinnet, O., Watanabe, Y., Weigel, D., and Zhu, J. K. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20, 3186–3190.
Moissiard, G., and Voinnet, O. (2006). RNA silencing of host transcripts by Cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 19593–19598.
Molnar, A., Melnyk, C., and Baulcombe, D. C. (2011). Silencing signals in plants: a long journey for small RNAs. Genome Biol. 12, 215.
Niehl, A., and Heinlein, M. (2009). Impact of RNA virus infection on plant cell function and evolution. Ann. N. Y. Acad. Sci. 1178, 120–128.
Oparka, K. J., Prior, D. A. M., Santa Cruz, S., Padgett, H. S., and Beachy, R. N. (1997). Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of Tobacco mosaic virus. Plant J. 12, 781–789.
Pasquinelli, A. E., and Ruvkun, G. (2002). Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol. 18, 495–513.
Qi, X., Bao, F. S., and Xie, Z. (2009). Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS ONE 4, e4971. doi: 10.1371/journal.pone.0004971
Reichel, C., and Beachy, R. N. (2000). Degradation of the Tobacco mosaic virus movement protein by the 26S proteasome. J. Virol. 74, 3330–3337.
Roossinck, M. J. (2005). Symbiosis versus competition in plant virus evolution. Nat. Rev. Microbiol. 3, 917–924.
Roossinck, M. J. (2011). The good viruses: viral mutualistic symbioses. Nat. Rev. Microbiol. 9, 99–108.
Shimura, H., Pantaleo, V., Ishihara, T., Myojo, N., Inaba, J., Sueda, K., Burgyan, J., and Masuta, C. (2011). A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 7, e1002021. doi: 10.1371/journal.ppat.1002021
Skalsky, R. L., and Cullen, B. R. (2010). Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64, 123–141.
Tagami, Y., Inaba, N., Kutsuna, N., Kurihara, Y., and Watanabe, Y. (2007). Specific enrichment of miRNAs in Arabidopsis thaliana infected with Tobacco mosaic virus. DNA Res. 14, 227–233.
Vazquez, F., Legrand, S., and Windels, D. (2010). The biosynthetic pathways and biological scopes of plant small RNAs. Trends Plant Sci. 15, 337–345.
Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A. C., Hilbert, J. L., Bartel, D. P., and Crété, P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16, 69–79.
Vogler, H., Akbergenov, R., Shivaprasad, P. V., Dang, V., Fasler, M., Kwon, M. O., Zhanybekova, S., Hohn, T., and Heinlein, M. (2007). Modification of small RNAs associated with suppression of RNA silencing by tobamovirus replicase protein. J. Virol. 81, 10379–10388.
Vogler, H., Kwon, M. O., Dang, V., Sambade, A., Fasler, M., Ashby, J., and Heinlein, M. (2008). Tobacco mosaic virus movement protein enhances the spread of RNA silencing. PLoS Pathog. 4, e1000038. doi: 10.1371/journal.ppat.1000038
Voinnet, O. (2005). Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6, 206–220.
Wang, D., and Maule, A. J. (1995). Inhibition of host gene expression associated with plant virus replication. Science 267, 229–231.
Watanabe, Y., Emori, Y., Ooshika, I., Meshi, T., Ohno, T., and Okada, Y. (1984). Synthesis of TMV-specific RNAs and proteins at the early stage of infection in tobacco protoplasts: transient expression of 30k protein and its mRNA. Virology 133, 18–24.
Wolf, S., Deom, C. M., Beachy, R. N., and Lucas, W. J. (1989). Movement protein of Tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246, 377–379.
Wu, Q., Wang, X., and Ding, S. W. (2010). Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe 8, 12–15.
Xie, Z., Allen, E., Wilken, A., and Carrington, J. C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 102, 12984–12989.
Keywords: RNA silencing, silencing suppressor, silencing enhancer, host:pathogen interactions, viruses, miRNA, siRNA, Tobacco mosaic virus
Citation: Amari K, Vazquez F and Heinlein M (2012) Manipulation of plant host susceptibility: an emerging role for viral movement proteins? Front. Plant Sci. 3:10. doi: 10.3389/fpls.2012.00010
Received: 29 November 2011; Accepted: 11 January 2012;
Published online: 30 January 2012.
Edited by:
Shoko Ueki, State University of New York at Stony Brook, USAReviewed by:
Igor Kovalchuk, University of Lethbridge, CanadaBrion Duffy, Agroscope Changins-Wädenswil, Switzerland
Copyright: © 2012 Amari, Vazquez and Heinlein. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Manfred Heinlein, Institut de Biologie Moléculaire des Plantes, 12, rue du Général Zimmer, 67084 Strasbourg Cedex, France. e-mail: manfred.heinlein@ibmp-cnrs.unistra.fr
†Present address: Khalid Amari, Department of Plant Physiology, Botanical Institute of the University of Basel, Hebelstrasse 1, CH-4056 Basel, Switzerland.
