- 1Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha, China
- 2Institute of Clinical Pharmacology, Hunan Key Laboratory of Pharmacogenetics, Central South University, Changsha, China
- 3Engineering Research Center of Applied Technology of Pharmacogenomics, Ministry of Education, Changsha, China
- 4National Clinical Research Center for Geriatric Disorders, Changsha, China
- 5Department of Anesthesiology, Xiangya Hospital, Central South University, Changsha, China
- 6Department of Pharmaceutical Sciences, School of Pharmacy, University of Maryland at Baltimore, Baltimore, MD, United States
Lenvatinib is the latest and promising agent that has demonstrated a significant improvement of progression-free survival in advanced hepatocellular carcinoma (HCC). However, resistance emerges soon after initial treatment, limiting the clinical benefits of lenvatinib. Therefore, understanding the mechanism of resistance is necessary for improving lenvatinib efficacy. YRDC promotes the proliferation of hepatocarcinoma cells via regulating the activity of the RAS/RAF/MEK/ERK pathway, which was the primary pathway of the anticancer effect of lenvatinib. The purpose of this study is to investigate whether YRDC modulates the sensitivity of lenvatinib in hepatocarcinoma cells. Using the CCK-8 cell viability assay, wound-healing assay and clone formation assay in cell models, and xenograft assay in null mouse, we demonstrated that Huh7 cells with YRDC knockdown showed decreased susceptibility to lenvatinib than their control cells. Furthermore, we found that lenvatinib inhibited the expression of YRDC in a time-dependent manner. This effect may aggravate resistance to lenvatinib in hepatocarcinoma cells and may be an underlying cause of resistance, which emerges soon after lenvatinib initial treatment. To investigate how YRDC modulates the sensitivity of lenvatinib, we assessed the effect of tRNA with different t6A levels on the translation of the KRAS gene by in vitro rabbit reticulocyte translation system and measured the expression levels of the KRAS gene by western blot together with qPCR. We found that YRDC regulates the protein translation of KRAS in cell models, and the tRNA with low t6A modification level reduces the translation of the KRAS in the in vitro translation system. These results suggested that YRDC mediates the resistance of lenvatinib in hepatocarcinoma cells via modulating the translation of the KRAS. In this study, YRDC was confirmed to be a potential novel predictive biomarker of lenvatinib sensitivity in HCC.
Introduction
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer death worldwide and has a higher growth incidence (Bray et al., 2018). The estimated incidence of new cases is approximately 500,000–1,000,000 annually, causing 600,000 mortalities globally per year. More than half of all cases are from China (Zheng et al., 2018). About 30–40% of patients are diagnosed at early stages. Potentially curative treatment of these patients, such as locoregional therapies and surgical therapies, can achieve 5-year survival rates up to 60–70% (European Association for the Study of the Liver, 2018; Llovet et al., 2018; Nault et al., 2018). However, there are few effective therapies for patients with advanced stage or with tumor progression after locoregional, leading to an extremely poor prognosis. Traditional chemotherapy drugs have little effect on survival in patients with advanced HCC (European Association for the Study of the Liver, 2018).
Sorafenib, an oral multikinase inhibitor (MKI), was approved as a unique target drug for advanced HCC in 2007 and has been used as a first-line systemic treatment for patients with unresectable HCC (uHCC) (Llovet et al., 2008). However, after clinical application of sorafenib for over a decade, the long-term survival for patients with uHCC is still suboptimal (Llovet et al., 2008; El-Serag, 2011; Zhu, 2012). In the last few years, several MKIs have been developed and clinically tested for the treatment of uHCC, but none of them has exhibited superior efficacy to sorafenib (European Association for the Study of the Liver, 2018; Heimbach et al., 2018; Vogel et al., 2018; Al-Salama et al., 2019).
Lenvatinib is an emerging MKI that targets the vascular endothelial growth factor receptor (VEGFR) one to three, fibroblast growth factor receptors (FGFR) one to four, platelet-derived growth factor receptor (PDGFR) α, and proto-oncogenes RET and KIT (Tohyama et al., 2014; Yamamoto et al., 2014). Firstly, it was approved for the treatment of advanced renal cell carcinoma (RCC) and differentiated thyroid cancer (DTC) (Schlumberger et al., 2015). In June 2017, the American Society of Clinical Oncology reported that lenvatinib was the first drug to show noninferiority to sorafenib in terms of overall survival in the phase III REFLECT trial in patients with uHCC. In addition, lenvatinib demonstrated a significantly higher objective response rate, longer progression-free survival, and time to progression compared to sorafenib (Al-Salama et al., 2019). Based on all these promising results from the REFLECT trial, lenvatinib is promptly approved for the first-line treatment of patients with uHCC in the USA, the EU, Japan, and China in 2018 total population (Inc, 2018). In fact, despite therapy, most patients with HCC or DTC develop resistance to lenvatinib after a period of time (Capozzi et al., 2019; Bangaru et al., 2020; Fu et al., 2020). For these reasons, it is urgent to identify a biomarker to predict the response or resistance to lenvatinib.
YRDC belongs to the universal family with the sua5-yciO-yrdC domain (PF01300, Swiss-Prot/TrEMBL database) and is highly conserved from E. coli to Homo sapiens (Jia et al., 2002). YrdC/Sua5 family is involved in the N6-threonylcarbamoyladenosine (t6A) biosynthesis of tRNA that recognize ANN codons in E. coli and yeast. The t6A modification is one of the 15 universally conserved modification located at position in tRNA decoding ANN codons (Cantara et al., 2011). Structural studies showed that t6A modification could strengthen the codon–anticodon interaction and promote translational fidelity by the ribosomes (Weissenbach and Grosjean, 1981; Grosjean et al., 1995; Yarian et al., 2000). These findings implicate that YrdC/Sua5 plays a critical role in protein translation at the level of codon recognition.
Our previous study reported that the expression of YRDC was dysregulated in hepatocellular carcinoma tissues. The results of in vitro and in vivo experiments showed that the expression of YRDC was positively associated with the proliferation and metastasis of HCC cells, and the mechanism exploration demonstrated that YRDC probably activates the MEK/ERK signaling pathway (Huang et al., 2019). Lenvatinib inhibits RTK receptors and subsequently blocks its downstream RAS/RAF/MEK/ERK signal transduction pathway, eventually impeding tumor cells proliferation and tumor angiogenesis (Leonetti et al., 2017; Cabanillas and Takahashi, 2018). These information suggested that HCC cells or tumor with high expression of YRDC, containing higher basal activity of the MEK/ERK, might be more sensitive, or responsive, to lenvatinib, and that those with low expression of YRDC might be resistant to lenvatinib. However, whether YRDC expression contributes to lenvatinib resistance has not yet been reported. Therefore, we conducted this study to investigate the association of the efficacy of lenvatinib with the expression of YRDC and explore the mechanism detail behind this association.
Materials and Methods
Cell Culture and Establishment of Stable Cell Lines
Huh7 (TCHu182) and HepG2 (TCHu72) cell lines were purchased from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. The culture medium of these 2 cell lines was Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, United States). The cells were cultured in an incubator at 37°C and 5% CO2. Short hairpin (sh) RNAs were purchased from Shanghai GenePharma Co., Ltd. The sequences of shYRDC and shNC were used as follows: shYRDC: 5′- CATTCGGATTCCTGATCAT -3′; and shNC: 5′- TTCTCCGAACGTGTCACGT -3′. Then, shYRDC and shNC were cloned, respectively, into the BamHI and EcoRI sites of the pGLV3 lentiviral vectors (GenePharm). The YRDC gene was amplified with oligonucleotides to add the NotI and BamHI sites with the primers 5′- AGGGTTCCAAGCTTAAGCGGCCGCGCCACCATGTCTCCGGCGCGTCGGTGCAGGGGGATG -3′ and 5′- ATCAGTAGAGAGTGTCGGATCCTCACAGGTAGGACGCATGTGAGGGGAGCAGTCC -3′. The amplified products were cloned into the pGLV5 lentiviral vector (GenePharm). The empty pGLV5 lentiviral vector was used as a control. Huh7 or HepG2 cells were seeded at 2×104 cells/well in 24-well plates, and 24 h after seeding were treated with a linear range of puromycin concentration. Lentivirus was diluted 10-fold with DMEM (supplemented with 10% FBS) for transduction. Cells were incubated with diluted lentivirus solution (Liu YB. et al., 2018). After 24 h, the culture medium was changed to a complete medium and selected with puromycin for 4 days. Until control, the Huh7 cells or HepG2 cells completely died, the YRDC stably knockout and overexpression cells were established. The optimal screening concentration of puromycin was 1 μg/ml. The lentivirus infected cells were observed under a fluorescence microscope (EVOS M5000, Invitrogen) and then collected for analysis by quantitative real-time PCR (qPCR) and western blot.
Survival Assay
The Cell Counting Kit-8 (Bimake) assay was used to estimate the viability of the cells. Huh7 and HepG2 cells were seeded in 96-well plates (5,000 cells/well). Next, different concentrations of lenvatinib (Selleck) or BGJ (Selleck) were added into each well separately and incubated for 48 h. After that, the culture medium containing lenvatinib or BGJ was sucked out. The cells were washed with phosphate buffer solution (PBS). Then, the mixture of 10 μl CCK-8 solvent and 90 μl of fresh medium was added to each well and incubated for 60 min. Finally, the absorbance at 450 nm was measured using a microplate reader (EON, BioTek Instruments, Inc.). The cell survival rate was calculated. The experiment was repeated four times.
Wound-Healing Assay
The cells were seeded in 6-well plates, and 2 ml of cell suspension with a density of 2×105 cells/ml was added to each well. When the cell growth density was around 90%, a 100 μl pipette tip was used to gently scrape off the cells to create a scratch. PBS was added to wash the floating cells, and subsequently, the medium with lenvatinib was added into each well. Photos were taken with the Leica 3000 microscope, using a magnification of 100 times, at 48 and 72 h. ImageJ 1.8.0 (Bethesda, United States) was used for image processing. The experiment was repeated three times.
Clone Formation Assay
The cells were seeded in 6-well plates at a density of 1,000 cells/per well. After the cells were stabilized, half of the wells were treated with 0.1 μM lenvatinib for 10 days. Lastly, the colonies were fixed with 4% paraformaldehyde (Beyotime, CHN) for 15 min and stained with crystal violet (Beyotime, CHN) overnight. Fifty cells are defined as a colony. The experiment was repeated three times.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated from the cells and tissues using RNAiso Plus (Takara) and reverse-transcribed using a PrimeScript™ RT reagent kit (Takara); the amount of RNA used in this experiment is 1,000 ng. The reverse-transcribed products were used as templates for qPCR with SYBR Green (Takara). Then, qPCR was performed using LightCycler® 480 Instrument (Roche, Basel, Switzerland). Data were analyzed using the 2-∆∆ct method, and the expression of GAPDH was used as normalization control. The primers (synthesized by Sangon Biotech, CHN) used in this study were listed as follows: YRDC, forward: 5′-GCAAGCGGACCCTCAAACAT-3′; reverse:5′-GCTCAACAAGGACCTAAACCCT-3′; GAPDH, forward: 5′-AGATCATCAGCAATGCCTCCTG-3′; reverse: 5′-TTGGCATGGACTGTGGCATG-3′; KRAS, forward: 5′- GGGGAGGGCTTTCTTTGTGTA-3′; reverse: 5′- GTCCTGAGCCTGTTTTGTGTC-3′; FGFR2, forward: 5′-AGTCAAGTGGATGGCTCCAG-3′; and reverse: 5-ACAGTT-CGTTGGTGCAGTTG-3′. The experiment was repeated at least three times.
Animal Experiments
All animal experiments were performed in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Review Board of Central South University (Changsha, China). Forty female 5-week-old BALB/C nude mice (Beijing Vital River Laboratory Animal Technology Co., Ltd.) were fed under specific pathogen-free conditions at the Department of Laboratory Animals, Central South University. The mice were housed in a temperature-controlled environment of 23 ± 1°C with a 12 h light/dark cycle (lights on 08:00 AM) and given a standard mouse diet, with water provided ad libitum. Forty female 5-week-old BALB/C nude mice were randomly divided into four groups of 10 each. YRDC–KD, YRDC–OE, and their control Huh7 cells (NC–KD or NC–OE, 1 × 107) were mixed with the Matrigel in a serum-free medium and then injected subcutaneously into the right lower quadrant of the nude mice. After 14 days, each group of mice was divided into two groups again. Lenvatinib (30 mg/kg) and solvent were intragastrically administered once a day. And, the tumor volume was measured every 2 days. All mice of YRDC–OE and its control were euthanized after 16 days; however, all mice of YRDC–KD and its control were euthanized after 26 days. Tumor nodules were removed and weighed. According to the formula: V = L × W2/2, where V is the volume (mm3), L is the maximum diameter (mm), and W is the shortest diameter (mm).
High PerformanceLiquid Chromatography–Mass Spectrometry Analysis of t6A Modification
The isolated bulk tRNA was enzymatically hydrolyzed to nucleosides as described (Crain, 1990). In brief, we added 0.01 units of snake venom phosphodiesterase (Sigma), 10 units of nuclease P1 (Sigma), and 3 μl alkaline phosphatase (Sigma) in a 100 μl volume to digest approximately 100 μg of bulk tRNA. The digested extracts were directly injected in a HPLC–MS system for the separation and identification of the nucleosides as previously described (Pomerantz and McCloskey, 1990). HPLC–MS was performed with an ExionLC AD instrument (AB SCIEX, USA) coupled to a mass spectrometer (Triple Quad 6,500+, AB SCIEX, USA). A Discovery C18 (250 mm, 4.6 mm, 5 μm) reverse-phase column (Waters) equipped with an Ultrasphere ODS guard column (Beckman) was used for lipid separation. The nucleosides were eluted at a flow rate of 1 ml/min and a column temperature of 30 ℃ with a gradient consisting of 10 mM ammonium acetate (A) and 40% acetonitrile (B) as follows: 0–7 min 99% A, 7–30 min 95% A, 30–40 min 85% A, 40–50 min 95% A, and 50–60 min 99% A. The mass spectrometer records in the cation mode. Using multiple/selected response monitoring modes (MRM/SRM), the collision energy is found to be 10 eV. For adenosine, the mass shifts from m/z 268–m/z 136, and for t6A, the mass shifts from m/z 413.2–m/z 281. The experiment was repeated three times.
In vitro Transcription
The cDNA of human KRAS was synthesized and inserted between EcoRI and SalI sites of pET-28a (+) by GeneChem. The constructed pET-28a (+)-KRAS plasmid was linearized by dissecting with XhoI. The KRAS mRNA was transcribed from 1 μg linearized plasmid DNA in vitro using the MEGAscript T7 kit (Ambion) in the presence of m7G (5′) ppp (5′) G (CAP) as described by the manufacturer. The KRAS mRNA was purified by phenol/chloroform extraction, ethanol precipitation, and quantified by UV spectroscopy.
Chromatographic Depletion of Endogenous tRNAs
The specific depletion of endogenous tRNAs from commercial Flexi rabbit reticulocyte lysate (RRL) system (L4540, Promega) was achieved by ethanolamine–sepharose column chromatography according to the method of Jackson et al. (2001). All experimental steps were performed at 4 °C. 100 μl of 50% ethanolamine–sepharose slurry in buffer A (0.1 mM EDTA, 25 mM KCl, 1.1 mM MgCl2, 10 mM NaCl, 10 mM HEPES-KOH, and pH 7.2) with 50 mM KOAc and 0.25 mM Mg(OAc)2 was incubated with commercial RRL for 45 min at 4°C for the depletion of tRNA in commercial RRL. The desired supernatant (RRL depleted of endogenous tRNA) was collected by brief centrifugation at 1,500 g and was either used immediately for translation reactions or stored at −80 ℃.
In vitro Translation
Cellular tRNAs were prepared from four kinds of Huh7 cells by the procedure of Dittmar et al. (2006). In vitro translation was performed using a RRL system (L4540, Promega). 1 μg of luciferase mRNA (L4561, Promega) or 2 μg of KRAS mRNA was added to RRL containing 70 mM KCl, 2 mM DTT, 10 μM minus leucine, 10 μM minus methionine, and 40 U/μl RNase ribonuclease inhibitor. For RRL that is depleted of endogenous tRNA, 2.5 μg of extra tRNA isolated from the four kinds of Huh7 cells was respectively added to the reaction mixture. A total volume of 50 μL from this reaction mixture was incubated for 60 min at 30 ℃. To avoid aminoacyl tRNAs producing background bands (∼25 kDa), we added 0.2 mg/ml RNase A to the reaction (after completion) and incubate for 5 min at 30 ℃. Luciferase assay was performed according to the protocol by using a luminometer (Sirius, Berthold) to quantify the luciferase produced in the in vitro translation system. The KRAS protein produced in the in vitro translation system was measured by the western blot. The experiment was repeated three times.
Western Blot
The cells were collected from the cell flask into a 1.5 ml EP tube, and it was found that the number of cells is about 1 × 107. Then, 1 ml RIPA lysis solution containing 10 μl protease inhibitor and 10 μL phenylmethanesulfonyl fluoride (P0013B, Beyotime) was added. The mixture was added to the cells and tissues under the conditions at 4 ℃ and fully dissolved for half an hour for protein extraction. The lysate was centrifuged at 8,000 g at 4°C for 15 min. Then, the supernatant was collected and the protein concentration was measured by BCA method. The protein samples in in vitro translation system were prepared by a special method. Once 50 μl translation reaction is completed, the 5 μl aliquot was removed, and it was added to 15 μl SDS sample buffer and heated at 75°C for 15 min to denature the proteins. The protein lysate (30 μg) was subjected to 10% SDS-PAGE and then electrotransferred onto the polyvinylidene difluoride membrane (PVDF, Millipore IPVH00010, Solarbio). And, the membrane was transferred to 5% skimmed milk for blocking for 1 h, followed by a membrane cutting process. Different bands were placed in an incubation box overnight in different primary antibodies. The main antibodies are as follows: β-actin (ab6276, Abcam), YRDC (sc-390477, SANTA CRUZ), and KRAS (12063-1-AP, Proteintech), and the ratios of them to the primary antibody dilution buffer (P0256, Beyotime) are 1:10,000, 1:800, and 1:5,000. The bands were washed the next day, and the secondary antibodies were incubated for 1 hour at room temperature. The ratio of β-actin and YRDC’s secondary antibody (SA00001-1, Proteintech) to the secondary antibody dilution buffer (P0258, Beyotime) is 1:10,000 and 1:5,000, and the ratio of KRAS’s secondary antibody (SA00001-2, Proteintech) to the secondary antibody dilution buffer (P0258, Beyotime) is 1:2,000. The bands were washed and soaked in the ECL kit (36208ES60, yeasen), and then ChemiDoc XRS + image analyzer (Bio-Rad, USA) was used to visualize the protein band, and Image Lab was used for immunoblot densitometric analysis. The experiment was repeated three times.
Statistical Analysis
All data were described as mean ± standard deviation (mean ± SD). The comparison of two groups is carried out by Student’s t-test. All the other data were analyzed with one-way ANOVA followed by LSD when equal variances were assumed. The half maximal inhibitory concentration (IC50) was calculated by GraphPad Prism six software. All statistical analyses were performed by SPSS 22.0 or GraphPad Prism 6 (GraphPad Software, Inc. La Jolla, CA, United States). p < 0.05 was considered statistically significant.
Results
Effects of Lenvatinib on Cell Proliferation, Migration, and Clone Formation In Vitro Are Significantly Correlated With the Expression Level of YRDC
Stable YRDC knockdown (YRDC-KD), YRDC overexpression (YRDC–OE) and their respective control HCC cells (NC–KD or NC–OE), were successfully established in our previous study (Huang et al., 2019). The effects of lenvatinib on cell proliferation were measured by the CCK-8 cell viability assay in these four kinds of Huh7 or HepG2 cell models. Figure 1A showed that the Huh7 cells with YRDC knockdown were significantly less sensitive to lenvatinib-mediated growth inhibition than their control cells (IC50: 1.379 vs 0.821 μM). However, the Huh7 cells with YRDC overexpression were a little higher, but not significantly sensitive to lenvatinib than their control cells (Figure 1B). The HepG2 cells had the same pattern as the Huh7 cells, but the IC50 of lenvatinib to the former is higher than the latter (Figures 1C,D). Wound-healing assays revealed that the migration inhibition of lenvatinib to Huh7 cells was decreased by YRDC knockdown (Figures 2A–C). The clone formation assay further confirmed that Huh7 cells with YRDC knockdown had a less sensitivity to lenvatinib than their control cells (Figures 3A–C). These data suggested that the knockdown of YRDC in Huh7 or HepG2 cells allowed the cells to become more resistant to lenvatinib.
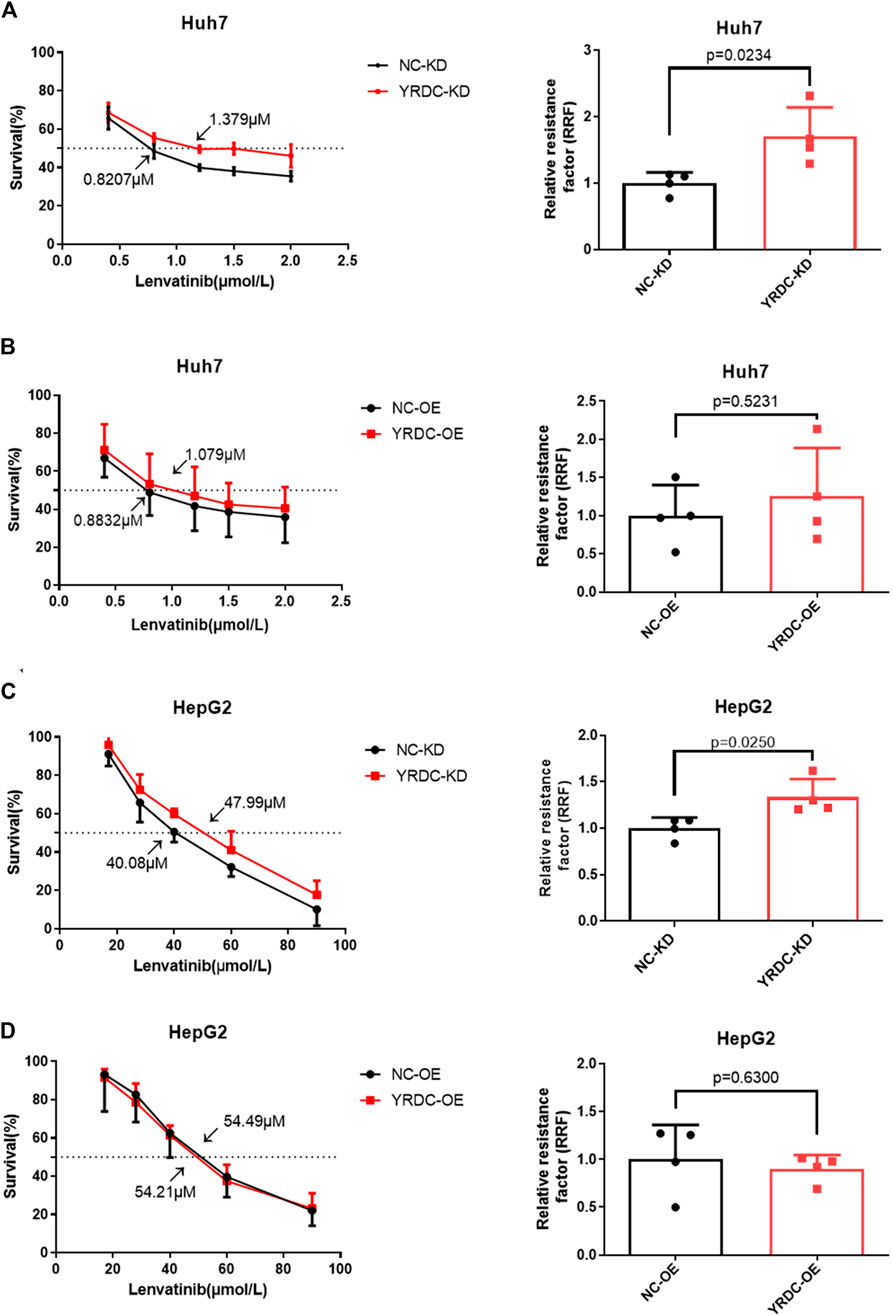
FIGURE 1. The knockdown of YRDC attenuates cellular sensitivity to lenvatinib in in vitro cell viability assay. (A, B) YRDC stable knockdown and overexpression in Huh7 cell were subjected to treatment with lenvatinib at various concentrations. The half maximal inhibitory concentration (IC50) was calculated by GraphPad Prism six software from four independent experiments. The relative resistance factor (RRF) of lenvatinib was calculated by dividing the IC50 of the control cells and by that of the cells with YRDC knockdown or overexpression. (C, D) YRDC stable knockdown and overexpression in HepG2 cell were subjected to treatment with lenvatinib at various concentrations. Dots in the graphs mean biological repetitions, n = 4.
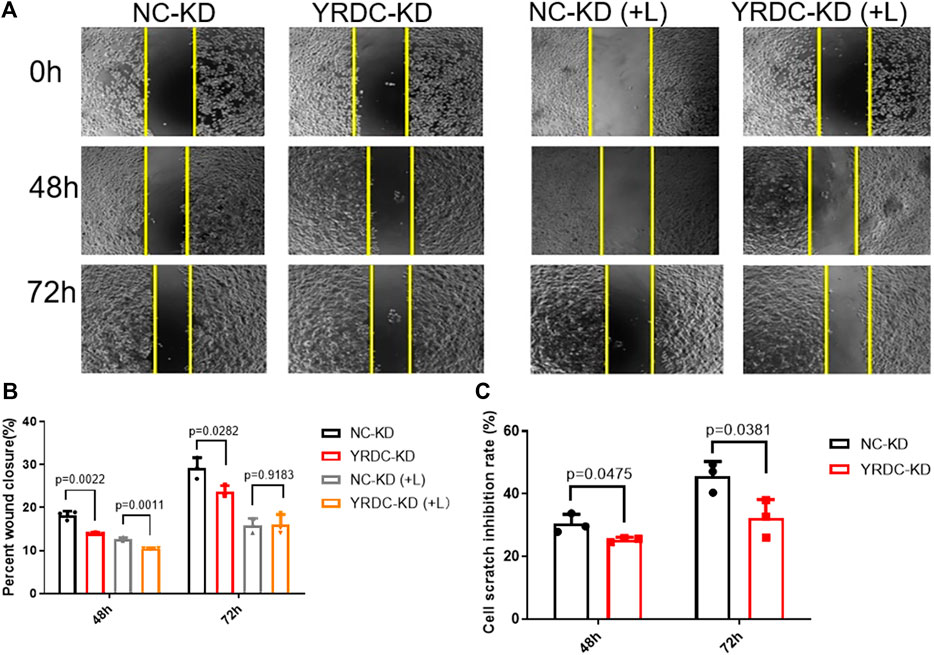
FIGURE 2. The knockdown of YRDC attenuates cellular sensitivity to lenvatinib in a wound-healing assay. (A) The knockdown of YRDC attenuates the antimigration effect of lenvatinib (0.8 μM) through the scratch experiment. (B) The migrating areas were measured at 48 and 72 h after the introduction of wounds in Huh7 cells with lenvatinib. (C) The cell scratch inhibition rate refers to the inhibition rate after lenvatinib addition. Dots in the graphs mean biological repetitions, n = 3.
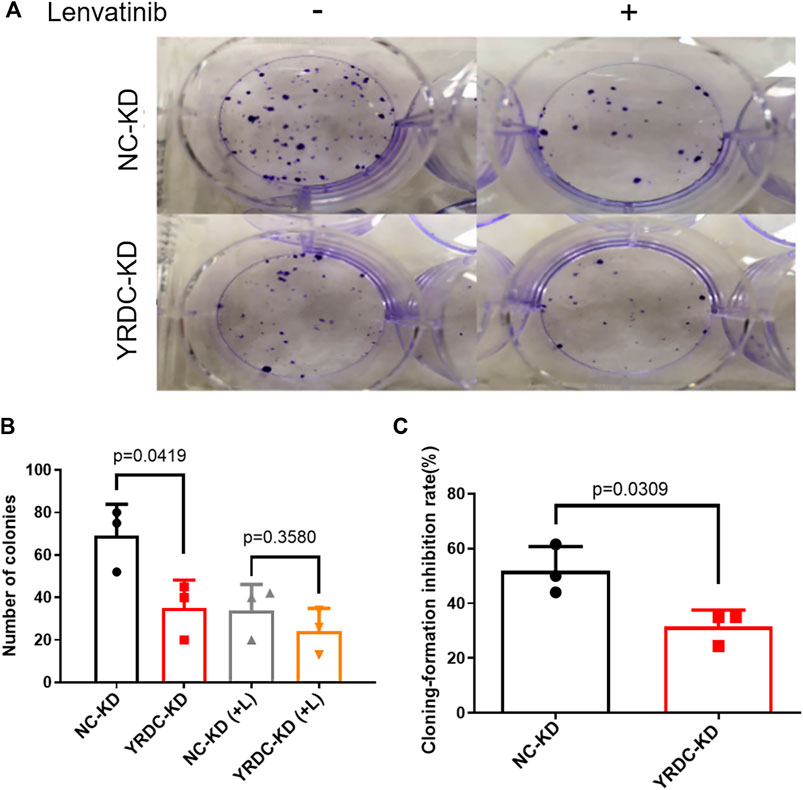
FIGURE 3. The knockdown of YRDC attenuates cellular sensitivity to lenvatinib in the clone formation assay. (A) The knockdown of YRDC attenuates the anti–colony-formation effect of lenvatinib (0.1 μM). (B) The clonal cell number was measured after 15 days in Huh7 cells with lenvatinib. (C) The cloning-formation inhibition rate refers to the inhibition rate after lenvatinib addition. Dots in the graphs mean biological repetitions, n = 3.
Effects of Lenvatinib on Tumor Growth In Vivo Are Significantly Correlated With the Expression Level of YRDC
To test the effect of YRDC on lenvatinib resistance in Huh7 cells in vivo, four kinds of stable Huh7 cells with different expression of YRDC were injected into nude mice for the construction of xenograft models. In vivo growth of xenograft tumors was significantly inhibited by lenvatinib at a dose of 30 mg/kg in all models (Figure 4A). The results indicated that YRDC knockdown or lenvatinib treatment significantly inhibited the tumor growth, exhibited as declined tumor volume (Figure 4B), and decreased tumor weight (Figure 4C). However, the knockdown of YRDC could weaken the lenvatinib-mediated tumor growth inhibition, mainly reflecting in the inhibition of tumor weight (66 vs. 91%, Figure 4C). Meanwhile, YRDC overexpression cells showed more sensitivity to lenvatinib compared with its control cells when evaluated by tumor weight (78 vs. 67%) (Figure 4C), however, not significantly different when evaluated by tumor volume (Figure 4B). These results suggested YRDC expression could affect the lenvatinib sensitivity on tumor growth in vivo, which mainly reflect in tumor weight.
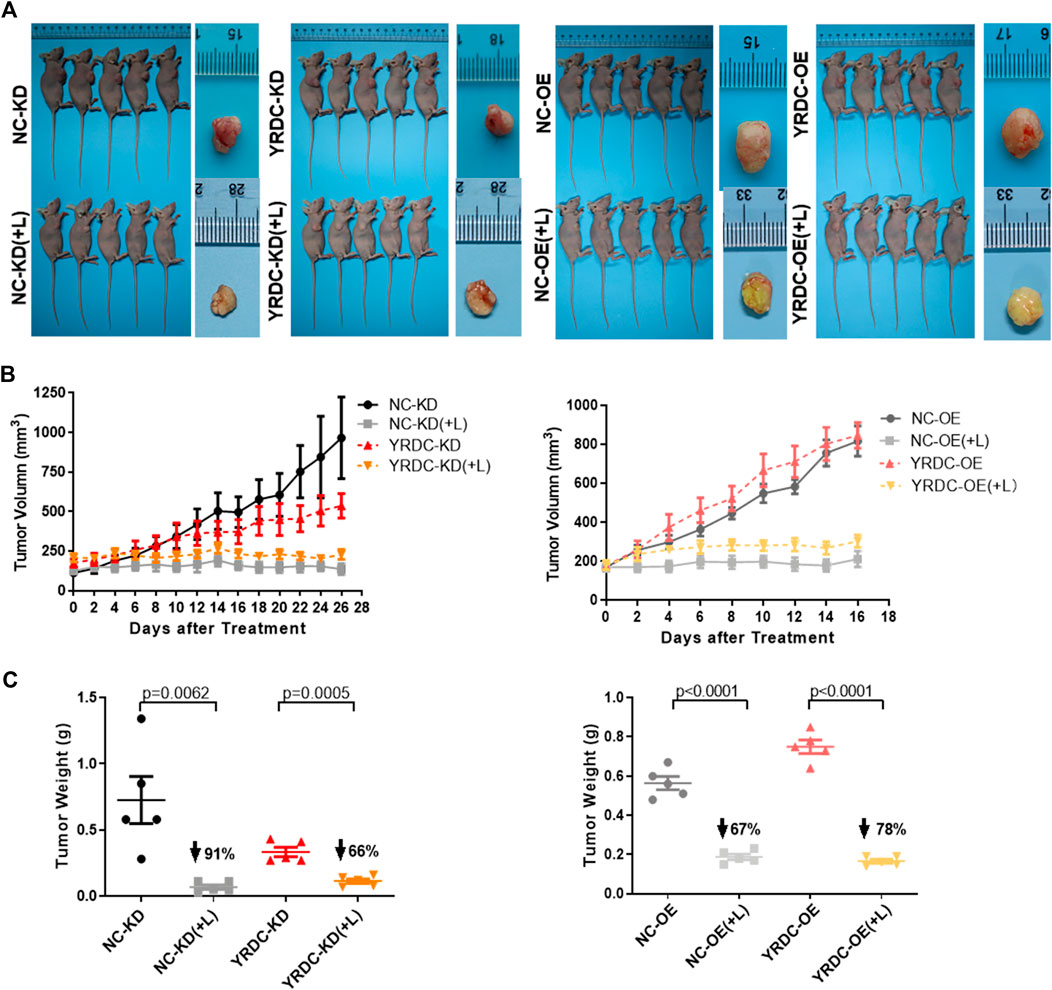
FIGURE 4. The effects of lenvatinib on tumor growth in vivo are significantly correlated with the expression level of YRDC. (A) The in vivo growth of xenograft tumors was significantly inhibited by lenvatinib at a dose of 30 mg/kg in all Huh7 cell models. (B) The tumor growth curves of YRDC knockdown or overexpression and their control cells after lenvatinib. (C) The difference of tumor weight with YRDC knockdown or overexpression cells after lenvatinib was compared with their controls. Dots in the graphs mean biological repetitions, n = 5.
Lenvatinib Inhibits the Expression of YRDC in A Time-Dependent Manner
Since YRDC could be induced under stress, like the ischemia–reperfusion condition (Jiang et al., 2005), more evidence suggested that the resistance emerges soon after the initial treatment with lenvatinib (Fu et al., 2020). We wonder how YRDC changed under the treatment of lenvatinib. Interestingly, the mRNA and protein expression level of YRDC was remarkedly inhibited by lenvatinib in a time-dependent manner in Huh7 cells (Figures 5A,B). Subsequently, we further confirmed the inhibition effect of lenvatinib on YRDC expression in xenograft models (Figures 5C,D). Combined with the results above, we speculated that the inhibition effect of lenvatinib on YRDC could aggravate lenvatinib resistance in HCC cells with a low original expression of YRDC. Additionally, due to the inhibition of lenvatinib on YRDC, the expression of YRDC would be lower after lenvatinib treatment in original sensitive HCC cells (Figures 5A,B), and it may be an underlying cause due to which the resistance emerges soon after lenvatinib initial treatment. However, the detailed mechanisms of YRDC involved in lenvatinib resistance in HCC cells were still unknown.
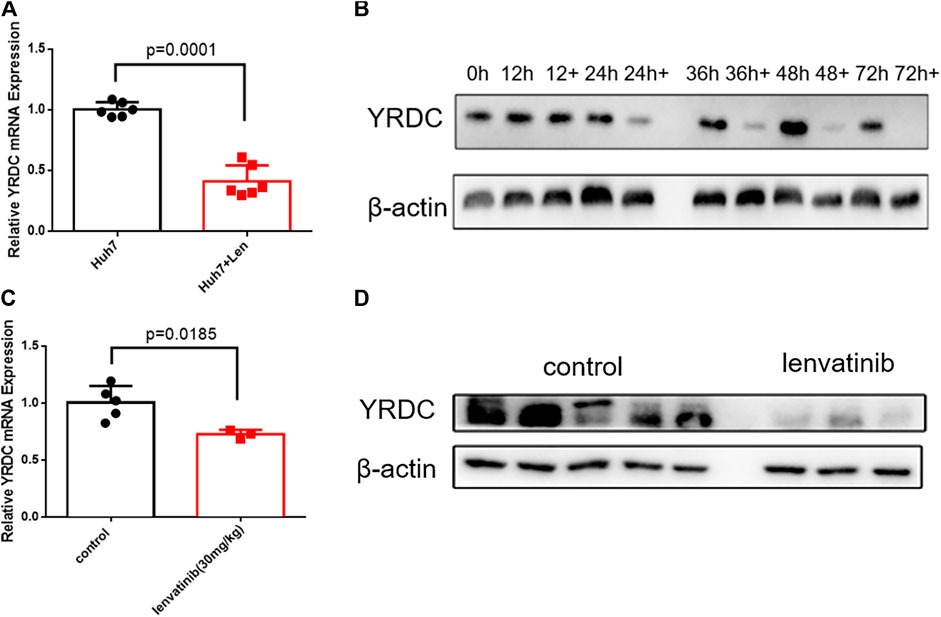
FIGURE 5. YRDC might be the target of anticancer of lenvatinib. (A) qPCR analysis for levels of YRDC in Huh7 cells with lenvatinib (0.8 μM) as well as the corresponding vehicle control, n = 6. (B) Lenvatinib (0.8 μM) inhibits the expression of YRDC in a time-dependent manner in Huh7 cells. (C, D) Lenvatinib inhibits the expression of YRDC in the xenograft of Huh7 cells in null mice, n = 5.
YRDC Regulates the Protein Translation of KRAS With a High Ratio of ANN Codons via Participating in the t6A Modification in NNU-tRNAs
The results mentioned above suggested that YRDC mediated the resistance of lenvatinib in HCC cells. Our previous study reported that the expression level of YRDC was positively correlated with the proliferation, migration, and invasion of Huh7 cells. These suggested that YRDC could be the target and the executor of lenvatinib antitumor. As we know, the antitumor effect of lenvatinib was realized via inhibiting tumor angiogenesis and directly blocking the RAS/RAF/MEK/ERK signaling pathway. Considering our previous finding that YRDC promotes HCC cells proliferation via regulating the activity of the RAS/RAF/MEK/ERK pathway, we speculated that this pathway may involve in YRDC-mediated lenvatinib resistance. Further experiments were conducted to investigate the correlation between them. Trametinib, a selective MEK inhibitor, was used in our following in vitro experiments. It showed that knockdown of YRDC attenuates cellular sensitivity to trametinib (Figure 6A). When combined with lenvatinib treatment, the cell mortality did not decrease any further than trametinib treatment alone (Figure 6A). In other words, there was no difference in the sensitivity of lenvatinib in YRDC knockdown cells and its control cells after trametinib treatment. All these results confirmed that the RAS/RAF/MEK/ERK pathway was involved in the YRDC-mediated lenvatinib resistance.
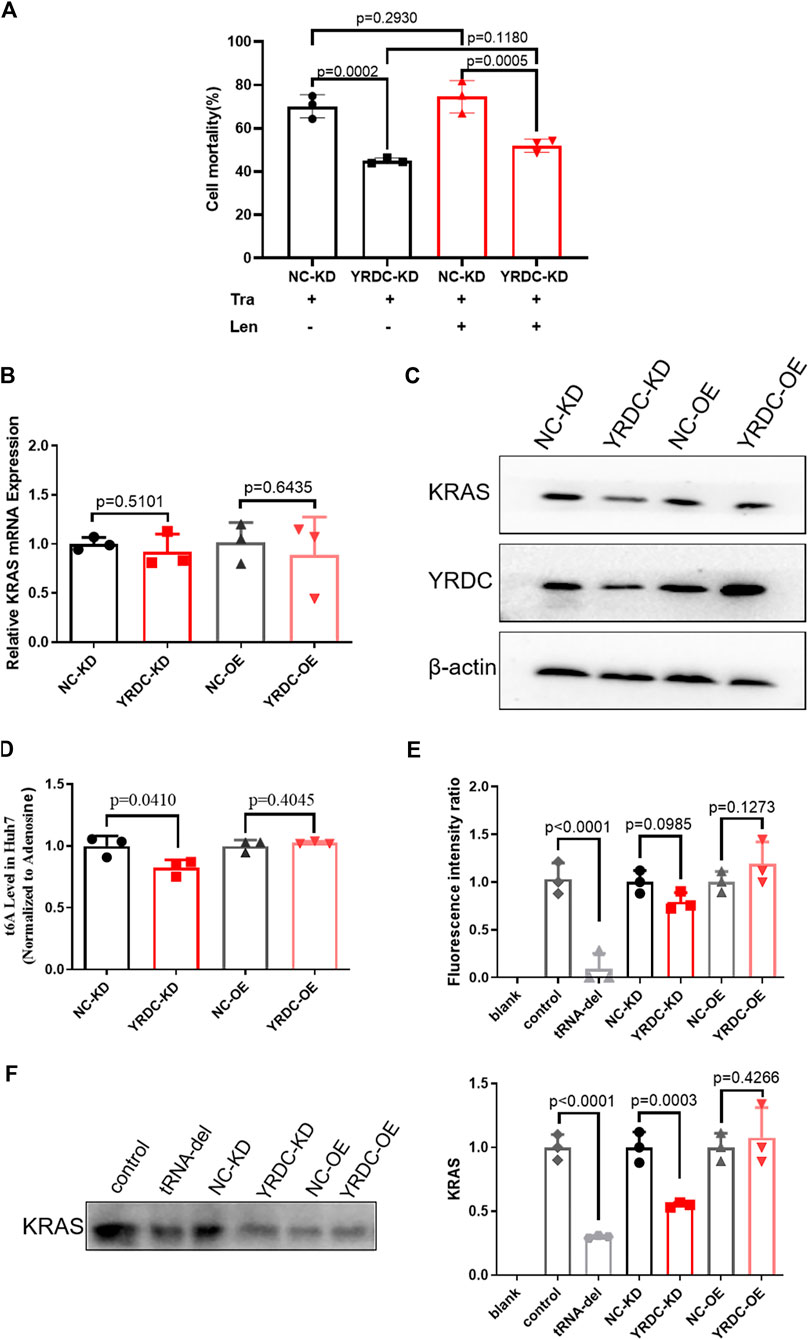
FIGURE 6. The anti-cancer effect of YRDC might affect the KRAS protein levels, but not mRNA levels. (A) The knockdown of YRDC attenuates cellular sensitivity to trametinib. After inhibiting the activity of the MAPK pathway, the expression of YRDC no longer affected the sensitivity of lenvatinib. (B) The mRNA expression levels of KRAS in overexpression, knockdown, and control in Huh7 cell. (C) The protein expression levels of YRDC and KRAS in overexpression, knockdown, and control in the Huh7 cells. (D) t6A levels in four kinds of Huh7 cells with different YRDC expression. (E) The fluorescent activity in addition of tRNAs from four kinds of Huh7 cells with different expressions in vitro reticulocytes translation system. (F) The KRAS level in addition of tRNAs from four kinds of Huh7 cells with different expressions in vitro reticulocytes translation system. Dots in the graphs mean biological repetitions, n = 3.
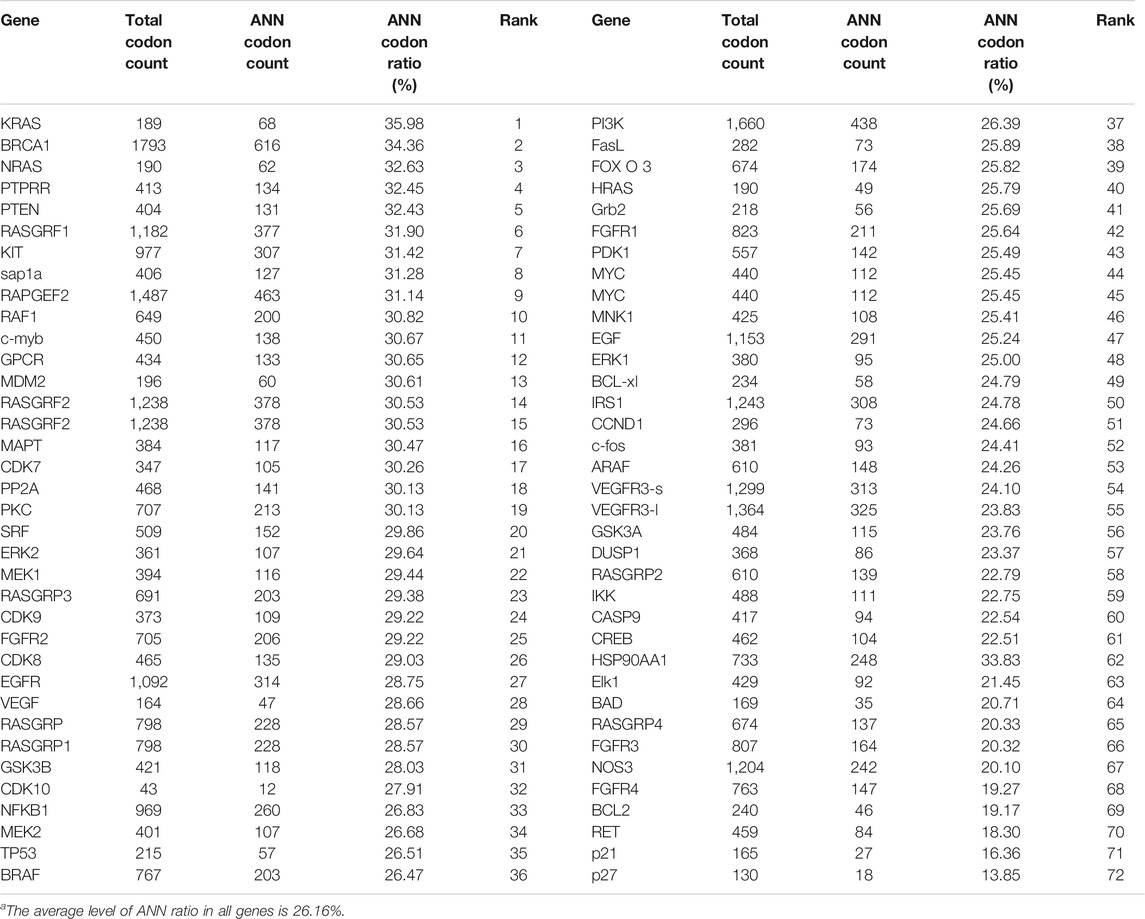
YRDC was an NNU-tRNA t6A modification enzyme. The low t6A modification of NNU-tRNA reduced the translation rate or increased the possiblity of the translation error. We analyzed the codon composition of all genes related in the lenvatinib antitumor pathway and found that the KRAS genes have the highest ANN codon ratio (35.98%, Table 1). In the four kinds of Huh7 cell models, the mRNA expression of the KRAS had no difference (Figure 6B); however, the protein level of the KRAS in YRDC knockdown was significantly lower compared with its control (Figure 6C). The protein level of KRAS in YRDC overexpression cells was slightly higher compared with its control cells. These results suggested that the low expression of YRDC may mainly reduce the level of NNU-tRNA t6A modification and further decrease the protein synthesis of KRAS gene with a high ANN codon ratio.
Then we analyzed the t6A modification level in the four kinds of Huh7 cell models with different YRDC expressions. As expected, the t6A level in YRDC knockdown cells was significantly lower by 18% when compared with its control cells, and the t6A level in YRDC overexpression cells was not significantly changed when compared with its control cells (Figure 6D).
To further confirm this point, we established the in vitro rabbit reticulocyte translation system. Firstly, we eliminated the endogenous tRNA from the rabbit reticulocytes translation system and performed the in vitro translation of the luciferase gene under the addition of tRNA isolated from the four kinds of Huh7 cells. The fluorescent activity in tRNA from YRDC knockdown was slightly lower than that of its control (p = 0.0985). The situation reversed between YRDC overexpression and its control (Figure 6E). The ANN ratio in the luciferase gene was 28%, which is close to the average level 26.1% in all genes expressed in humans. This result indicated that YRDC knockdown could reduce general protein synthesis.
We further performed the in vitro translation of KRAS. The cDNA of KRAS was cloned into the pET-28a (+) plasmid. The KRAS mRNA was obtained by the reverse transcription using pET-28a (+)-KRAS plasmid as a template. In the in vitro rabbit reticulocyte translation system, the level of the KRAS protein was significantly lower when added with the tRNA isolated from YRDC knockdown cells compared with the tRNA from its control cells (0.55 vs. 1.0). However, there is no significance in the KRAS synthesis in addition of tRNA from YRDC overexpression cells compared with tRNA from its corresponding control cells (Figure 6F).
Discussion
In the current study, we firstly found that the YRDC expression was associated with the sensitivity of lenvatinib in hepatocellular carcinoma cells. We also found that lenvatinib inhibited the expression of YRDC in a time-dependent manner. This effect may be an underlying cause due to which the resistance emerges soon after lenvatinib initial treatment. To investigate how YRDC modulates the sensitivity of lenvatinib, we assessed the effect of tRNA with different t6A levels on the translation of KRAS gene by the in vitro rabbit reticulocyte translation system and measured the expression levels of KRAS in vitro and in vivo experiments by western blot together with qPCR. We found that YRDC regulates the protein translation of KRAS in cells model, and the tRNA with low t6A modification level reduces the translation of KRAS in in vitro translation system. These results suggested that YRDC mediates the resistance of lenvatinib in HCC cells via modulating the translation of KRAS.
Based on the impressive finding from the REFLECT clinical trial, lenvatinib is now approved as the first-line therapy for patients with advanced HCC in Japan, the European Union, the United States, and China. However, the HCC patients with response to lenvatinib are still limited. There are almost half of HCC patients who develop resistance to lenvatinib in China (Fu et al., 2020). The understanding of how lenvatinib resistance happens seems to be more urgent in China. Our previous study found that YRDC promotes the proliferation of HCC cells via regulating the activity of the RAS/RAF/MEK/ERK pathway, which was the primary pathway of anticancer effect of lenvatinib. In our current study, we found that low YRDC expression significantly decreased the susceptibility of lenvatinib in Huh7/HepG2 cell models and in xenograft models. In addition, we found that lenvatinib remarkedly inhibited the expression of YRDC in a time-dependent manner. The inhibition effect of lenvatinib on YRDC could aggravate lenvatinib resistance in HCC cells. And, it may be an underlying cause of resistance emerges soon after lenvatinib initial treatment.
Receptors for growth factor, including VEGFR, PDGFR, and FGFR, were continuously activated in HCC and were critical to HCC development, progression, and metastasis. Lenvatinib has direct antitumor effects, as well as antiangiogenic properties, through inhibition of these receptors and their downstream cascade reactions, such as the RAS/RAF/MEK/ERK signaling pathway (Matsui et al., 2008a; Matsui et al., 2008b; Ikuta et al., 2009; Okamoto et al., 2013). Currently, we found that lenvatinib could inhibit the expression of YRDC in a time-dependent manner. Our previous study found that the expression of YRDC was dysregulated in the hepatocellular carcinoma tissues. The overall survival of patients with a high-level YRDC expression was shorter than that with low-level in patients. The low YRDC expression could significantly inhibit the activity of MEK/ERK and inhibit the proliferation of HCC cells in vitro as well as the tumor growth in vivo. All these results suggested that YRDC might be a target and an executor molecule of anticancer activity of lenvatinib. However, the detailed mechanisms of YRDC involved in lenvatinib resistance in HCC cells are still unknown. Exploring the detail mechanisms would provide the strategy for overcoming lenvatinib resistance in HCC and provide the biomarker for the prediction of lenvatinib outcome in HCC patients.
YRDC is involved in t6A biosynthesis of tRNA that recognizes ANN codons in E. coli and yeast. There are 16 kinds of these tRNA, including tRNAIleUAU, tRNAIleGAU, tRNAIleAAU, tRNALysUUU, tRNALysCUU, tRNAAsnGUU, tRNAAsnAUU, tRNAArgUCU, tRNAArgCCU, tRNASerGCU, tRNASerACU, tRNAThrUGU, tRNAThrGGU, tRNAThrCGU, tRNAThrAGU, and tRNAMetCAU (Cantara et al., 2011; Kimura et al., 2014). We analyzed the ratio of these ANN codons corresponding to NNU-tRNA in all genes involved in the pathways of anticancer activity of lenvatinib. The ANN ratio of KRAS was ranked at top 1 (35.98%). The average ANN ratio in all genes was 26.16%. The protein content of KRAS in Huh7 cells with YRDC knockdown and in vitro translation system with tRNA isolated from YRDC knockdown was significantly lower compared with that in control cells. These results suggested that YRDC modulated the translation of key genes (KRAS) in pathways of anticancer activity of lenvatinib, which could be a partial reason that YRDC knockdown caused HCC cells resistance to lenvatinib. Other key genes with an enriched ANN codon, involved in the pathways of anticancer activity of lenvatinib, might be affected by a similar mechanism and should be explored in our future study. In the current study, we firstly provided the direct evidence on YRDC which modulated the translation of KRAS using the in vitro reticulocyte translation system. In our another study that is currently under the review, we also found that the translation of general protein in mouse with the liver-YRDC knockdown decreased, in particular, the proteins with a high ANN ratio decreased much more than those with a low ANN ratio.
Several multikinase inhibitors, including sorafenib, lenvatinib, sunitinib, ramucirumab, regorafenib, and cabozantinib, have recently been introduced into clinical practice for HCC (Abou-Alfa et al., 2018; Bruix et al., 2017; De Luca et al., 2020; Liu D. et al., 2018). In our current study, we give high priority to investigating sorafenib and lenvatinib, which are the first-line systemic therapies for HCC. Compared with sorafenib, lenvatinib has a distinctive feature, i.e., the potent activity against FGFR1-4. More evidence suggests that the activation of FGF signaling pathways in HCC contributes to its malignancy (Futami et al., 2017; Hagel et al., 2015; Miura et al., 2012; Paur et al., 2015; Sawey et al., 2011; Schmidt et al., 2016). The major downstream cascade reaction associated with FGFR in HCC is the RAS/RAF/MEK/ERK pathway (Li et al., 2016). In our study, the Huh7 cells with different expressions of YRDC only demonstrated the varied sensitivity to lenvatinib, but not to sorafenib (Supplementary Figure S1A, B). And, we also found that the Huh7 cells with different YRDC expressions showed the same response manner to pan-FGFR inhibitor BGJ (Supplementary Figure S1C). Furthermore, as mentioned above, the inhibition of lenvatinib on YRDC expression did not exist anymore after the treatment with the MEK inhibitor (Figure 6A). It suggested that the FGFR-related RAS/RAF/MEK/ERK pathway was mainly involved in YRDC-mediated lenvatinib resistance. This result might also be related to a recent finding that the expression level of FGF19 was correlated with lenvatinib susceptibility in HCC cells. In their study, the researchers found that FGF19 was downregulated in lenvatinib-resistant HCC cell lines, and the FGFR pathway plays a critical role in lenvatinib resistance (Myojin et al., 2021). However, whether there is a connection between YRDC and FGF19-mediated lenvatinib resistance is unknown. We could investigate that in the future study.
In addition to the mechanisms mentioned above, there are few other reported mechanisms of lenvatinib resistance in HCC. Lenvatinib-resistant HCC cells, established by culturing with long-term exposure to lenvatinib, were commonly used cell models in these studies. In vitro results showed that the vascular endothelial growth factor (VEGF), VEGFR2, platelet-derived growth factor-AA (PDGF-AA), and angiogenin were increased significantly in lenvatinib-resistant cells (Ao et al., 2021; Zhao et al., 2021). Both activation of the MAPK/MEK/ERK signaling pathway and the upregulation of epithelial mesenchymal transition (EMT) markers were observed in lenvatinib-resistant cells (Ao et al., 2021; Zhao et al., 2021). However, the researchers did not explore the detailed mechanism in these studies. The interferon regulatory factor 2 (IRF2) is a constitutive transcription factor associated with the development of various cancers by regulating the cancer cell growth, apoptosis, and drug resistance. A recent study found that IRF2 promoted proliferation, inhibited apoptosis, and increased lenvatinib resistance of HCC cells by regulating β-catenin expression (Guo et al., 2021). In brief, the mechanisms underlying lenvatinib resistance are still very limited and are worth exploring.
In conclusion, lenvatinib is a promising option in anticancer treatment for advanced HCC. However, lenvatinib resistance limits the clinical use in HCC. YRDC expression was positively correlated with the sensitivity of HCC to lenvatinib in cell models and in vivo xenograft model. The detail of the mechanism is that YRDC mediates the resistance of lenvatinib in hepatocarcinoma cells via modulating the translation of KRAS. The mRNA expression and protein expression level of YRDC in tissue-biopsy would be measured by qPCR and IHC-P in clinical. In future study, we would further investigate whether the expression of YRDC or functional SNP of YRDC might be regarded as a biomarker for the prediction of lenvatinib efficacy in clinical practice.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by the Institutional Review Board of Central South University.
Author Contributions
QL, HZ, WZ, and YS designed and supervised the research. JG and PZ performed the in vitro and in vivo experiments, analysed data, and wrote the manuscript. ZY, MW, HY, and SH were involved in data acquisition. QL revised the manuscript. All authors approved the version to be submitted.
Funding
This research was supported by the National Natural Science Foundation (NNSF) of China (81973401) and the National Development of Key Novel Drugs for Special Projects of China (2019ZX09201-002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.744578/full#supplementary-material
References
Abou-Alfa, G. K., Meyer, T., Cheng, A. L., El-Khoueiry, A. B., Rimassa, L., Ryoo, B. Y., et al. (2018). Cabozantinib in Patients with Advanced and Progressing Hepatocellular Carcinoma. N. Engl. J. Med. 379, 54–63. doi:10.1056/NEJMoa1717002
Al-Salama, Z. T., Syed, Y. Y., and Scott, L. J. (2019). Lenvatinib: A Review in Hepatocellular Carcinoma. Drugs 79, 665–674. doi:10.1007/s40265-019-01116-x
Ao, J., Chiba, T., Shibata, S., Kurosugi, A., Qiang, N., Ma, Y., et al. (2021). Acquisition of Mesenchymal-Like Phenotypes and Overproduction of Angiogenic Factors in Lenvatinib-Resistant Hepatocellular Carcinoma Cells. Biochem. Biophys. Res. Commun. 549, 171–178. doi:10.1016/j.bbrc.2021.02.097
Bangaru, S., Marrero, J. A., and Singal, A. G. (2020). Review Article: New Therapeutic Interventions for Advanced Hepatocellular Carcinoma. Aliment. Pharmacol. Ther. 51, 78–89. doi:10.1111/apt.15573
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 68, 394–424. doi:10.3322/caac.21492
Bruix, J., Qin, S., Merle, P., Granito, A., Huang, Y. H., Bodoky, G., et al. (2017). Regorafenib for Patients with Hepatocellular Carcinoma Who Progressed on Sorafenib Treatment (RESORCE): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 389, 56–66. doi:10.1016/S0140-6736(16)32453-9
Cabanillas, M. E., and Takahashi, S. (2018). Managing the Adverse Events Associated with Lenvatinib Therapy in Radioiodine-Refractory Differentiated Thyroid Cancer. Semin. Oncol. 46 (1), 57–64. doi:10.1053/j.seminoncol.2018
Cantara, W. A., Crain, P. F., Rozenski, J., McCloskey, J. A., Harris, K. A., Zhang, X., et al. (2011). The RNA Modification Database, RNAMDB: 2011 Update. Nucleic Acids Res. 39, D195–D201. doi:10.1093/nar/gkq1028
Capozzi, M., De Divitiis, C., Ottaiano, A., von Arx, C., Scala, S., Tatangelo, F., et al. (2019). Lenvatinib, a Molecule with Versatile Application: From Preclinical Evidence to Future Development in Anti-Cancer Treatment. Cancer Manag. Res. 11, 3847–3860. doi:10.2147/CMAR.S188316
Crain, P. F. (1990). Preparation and Enzymatic Hydrolysis of DNA and RNA for Mass Spectrometry. Methods Enzymol. 193, 782–790. doi:10.1016/0076-6879(90)93450-y
De Luca, E., Marino, D., and Di Maio, M. (2020). Ramucirumab, A Second-Line Option for Patients with Hepatocellular Carcinoma: A Review of the Evidence. Cancer Manag. Res. 12, 3721–3729. doi:10.2147/CMAR.S216220
Dittmar, K. A., Goodenbour, J. M., and Pan, T. (2006). Tissue-Specific Differences in Human Transfer RNA Expression. Plos Genet. 2, e221. doi:10.1371/journal.pgen.0020221
El-Serag, H. B. (2011). Hepatocellular Carcinoma. N. Engl. J. Med. 365, 1118–1127. doi:10.1056/NEJMra1001683
European Association for the Study of the Liver (2018). EASL Clinical Practice Guidelines: Management of Hepatocellular Carcinoma. J. Hepatol. 69, 182–236. doi:10.1016/j.jhep.2018.03.019
Fu, R., Jiang, S., Li, J., Chen, H., and Zhang, X. (2020). Activation of the HGF/c-MET axis Promotes Lenvatinib Resistance in Hepatocellular Carcinoma Cells with High C-MET Expression. Med. Oncol. 37, 24. doi:10.1007/s12032-020-01350-4
Futami, T., Okada, H., Kihara, R., Kawase, T., Nakayama, A., Suzuki, T., et al. (2017). ASP5878, a Novel Inhibitor of FGFR1, 2, 3, and 4, Inhibits the Growth of FGF19-Expressing Hepatocellular Carcinoma. Mol. Cancer Ther. 16, 68–75. doi:10.1158/1535-7163.MCT-16-0188
Grosjean, H., Sprinzl, M., and Steinberg, S. (1995). Posttranscriptionally Modified Nucleosides in Transfer RNA: Their Locations and Frequencies. Biochimie 77, 139–141. doi:10.1016/0300-9084(96)88117-x
Guo, Y., Xu, J., Du, Q., Yan, Y., and Geller, D. A. (2021). IRF2 Regulates Cellular Survival and Lenvatinib-Sensitivity of Hepatocellular Carcinoma (HCC) through Regulating β-Catenin. Transl Oncol. 14, 101059. doi:10.1016/j.tranon.2021.101059
Hagel, M., Miduturu, C., Sheets, M., Rubin, N., Weng, W., Stransky, N., et al. (2015). First Selective Small Molecule Inhibitor of FGFR4 for the Treatment of Hepatocellular Carcinomas with an Activated FGFR4 Signaling Pathway. Cancer Discov. 5, 424–437. doi:10.1158/2159-8290.CD-14-1029
Heimbach, J. K., Kulik, L. M., Finn, R. S., Sirlin, C. B., Abecassis, M. M., Roberts, L. R., et al. (2018). AASLD Guidelines for the Treatment of Hepatocellular Carcinoma. Hepatology 67, 358–380. doi:10.1002/hep.29086
Huang, S., Zhu, P., Sun, B., Guo, J., Zhou, H., Shu, Y., et al. (2019). Modulation of YrdC Promotes Hepatocellular Carcinoma Progression via MEK/ERK Signaling Pathway. Biomed. Pharmacother. 114, 108859. doi:10.1016/j.biopha.2019.108859
Ikuta, K., Yano, S., Trung, V. T., Hanibuchi, M., Goto, H., Li, Q., et al. (2009). E7080, a Multi-Tyrosine Kinase Inhibitor, Suppresses the Progression of Malignant Pleural Mesothelioma with Different Proangiogenic Cytokine Production Profiles. Clin. Cancer Res. 15, 7229–7237. doi:10.1158/1078-0432.CCR-09-1980
Inc, E. (2018). Lenvima® (Lenvatinib Mesylate) Capsules: Chinese Product Specifications. Available at: http://202.96.26.102/index/detai l/id/543 (Accessed Mar 26, 2019).
Jackson, R. J., Napthine, S., and Brierley, I. (2001). Development of a tRNA-Dependent In Vitro Translation System. RNA 7, 765–773. doi:10.1017/s1355838201002539
Jia, J., Lunin, V. V., Sauvé, V., Huang, L. W., Matte, A., and Cygler, M. (2002). Crystal Structure of the YciO Protein from Escherichia coli. Proteins 49, 139–141. doi:10.1002/prot.10178
Jiang, W., Prokopenko, O., Wong, L., Inouye, M., and Mirochnitchenko, O. (2005). IRIP, a New Ischemia/reperfusion-Inducible Protein that Participates in the Regulation of Transporter Activity. Mol. Cel Biol 25, 6496–6508. doi:10.1128/MCB.25.15.6496-6508.2005
Kimura, S., Miyauchi, K., Ikeuchi, Y., Thiaville, P. C., Crécy-Lagard, Vd., and Suzuki, T. (2014). Discovery of the β-Barrel-Type RNA Methyltransferase Responsible for N6-Methylation of N6-Threonylcarbamoyladenosine in tRNAs. Nucleic Acids Res. 42, 9350–9365. doi:10.1093/nar/gku618
Leonetti, A., Leonardi, F., Bersanelli, M., and Buti, S. (2017). Clinical Use of Lenvatinib in Combination with Everolimus for the Treatment of Advanced Renal Cell Carcinoma. Ther. Clin. Risk Manag. 13, 799–806. doi:10.2147/TCRM.S126910
Li, L., Zhao, G. D., Shi, Z., Qi, L. L., Zhou, L. Y., and Fu, Z. X. (2016). The Ras/Raf/MEK/ERK Signaling Pathway and its Role in the Occurrence and Development of HCC. Oncol. Lett. 12, 3045–3050. doi:10.3892/ol.2016.5110
Liu, D., Qi, X., Manjunath, Y., Kimchi, E. T., Ma, L., Kaifi, J. T., et al. (2018a). Sunitinib and Sorafenib Modulating Antitumor Immunity in Hepatocellular Cancer. J. Immunol. Res. Ther. 3, 115–123.
Liu, Y. B., Mei, Y., Tian, Z. W., Long, J., Luo, C. H., and Zhou, H. H. (2018b). Downregulation of RIF1 Enhances Sensitivity to Platinum-Based Chemotherapy in Epithelial Ovarian Cancer (EOC) by Regulating Nucleotide Excision Repair (NER) Pathway. Cell Physiol Biochem 46, 1971–1984. doi:10.1159/000489418
Llovet, J. M., Montal, R., Sia, D., and Finn, R. S. (2018). Molecular Therapies and Precision Medicine for Hepatocellular Carcinoma. Nat. Rev. Clin. Oncol. 15, 599–616. doi:10.1038/s41571-018-0073-4
Llovet, J. M., Ricci, S., Mazzaferro, V., Hilgard, P., Gane, E., Blanc, J. F., et al. (2008). Sorafenib in Advanced Hepatocellular Carcinoma. N. Engl. J. Med. 359, 378–390. doi:10.1056/NEJMoa0708857
Matsui, J., Funahashi, Y., Uenaka, T., Watanabe, T., Tsuruoka, A., and Asada, M. (2008a). Multi-Kinase Inhibitor E7080 Suppresses Lymph Node and Lung Metastases of Human Mammary Breast Tumor MDA-MB-231 via Inhibition of Vascular Endothelial Growth Factor-Receptor (VEGF-R) 2 and VEGF-R3 Kinase. Clin. Cancer Res. 14, 5459–5465. doi:10.1158/1078-0432.CCR-07-5270
Matsui, J., Yamamoto, Y., Funahashi, Y., Tsuruoka, A., Watanabe, T., Wakabayashi, T., et al. (2008b). E7080, a Novel Inhibitor that Targets Multiple Kinases, Has Potent Antitumor Activities against Stem Cell Factor Producing Human Small Cell Lung Cancer H146, Based on Angiogenesis Inhibition. Int. J. Cancer 122, 664–671. doi:10.1002/ijc.23131
Miura, S., Mitsuhashi, N., Shimizu, H., Kimura, F., Yoshidome, H., Otsuka, M., et al. (2012). Fibroblast Growth Factor 19 Expression Correlates with Tumor Progression and Poorer Prognosis of Hepatocellular Carcinoma. BMC cancer 12, 56. doi:10.1186/1471-2407-12-56
Myojin, Y., Kodama, T., Maesaka, K., Motooka, D., Sato, Y., Tanaka, S., et al. (2021). ST6GAL1 Is a Novel Serum Biomarker for Lenvatinib-Susceptible FGF19-Driven Hepatocellular Carcinoma. Clin. Cancer Res. 27, 1150–1161. doi:10.1158/1078-0432.CCR-20-3382
Nault, J. C., Galle, P. R., and Marquardt, J. U. (2018). The Role of Molecular Enrichment on Future Therapies in Hepatocellular Carcinoma. J. Hepatol. 69, 237–247. doi:10.1016/j.jhep.2018.02.016
Okamoto, K., Kodama, K., Takase, K., Sugi, N. H., Yamamoto, Y., Iwata, M., et al. (2013). Antitumor Activities of the Targeted Multi-Tyrosine Kinase Inhibitor Lenvatinib (E7080) against RET Gene Fusion-Driven Tumor Models. Cancer Lett. 340, 97–103. doi:10.1016/j.canlet.2013.07.007
Paur, J., Nika, L., Maier, C., Moscu-Gregor, A., Kostka, J., Huber, D., et al. (2015). Fibroblast Growth Factor Receptor 3 Isoforms: Novel Therapeutic Targets for Hepatocellular Carcinoma? Hepatology 62, 1767–1778. doi:10.1002/hep.28023
Pomerantz, S. C., and McCloskey, J. A. (1990). Analysis of RNA Hydrolyzates by Liquid Chromatography-Mass Spectrometry. Methods Enzymol. 193, 796–824. doi:10.1016/0076-6879(90)93452-q
Sawey, E. T., Chanrion, M., Cai, C., Wu, G., Zhang, J., Zender, L., et al. (2011). Identification of a Therapeutic Strategy Targeting Amplified FGF19 in Liver Cancer by Oncogenomic Screening. Cancer cell 19, 347–358. doi:10.1016/j.ccr.2011.01.040
Schlumberger, M., Tahara, M., Wirth, L. J., Robinson, B., Brose, M. S., Elisei, R., et al. (2015). Lenvatinib versus Placebo in Radioiodine-Refractory Thyroid Cancer. N. Engl. J. Med. 372, 621–630. doi:10.1056/NEJMoa1406470
Schmidt, B., Wei, L., DePeralta, D. K., Hoshida, Y., Tan, P. S., Sun, X., et al. (2016). Molecular Subclasses of Hepatocellular Carcinoma Predict Sensitivity to Fibroblast Growth Factor Receptor Inhibition. Int. J. Cancer 138, 1494–1505. doi:10.1002/ijc.29893
Tohyama, O., Matsui, J., Kodama, K., Hata-Sugi, N., Kimura, T., Okamoto, K., et al. (2014). Antitumor Activity of Lenvatinib (e7080): An Angiogenesis Inhibitor that Targets Multiple Receptor Tyrosine Kinases in Preclinical Human Thyroid Cancer Models. J. Thyroid Res. 2014, 638747. doi:10.1155/2014/638747
Vogel, A., Cervantes, A., Chau, I., Daniele, B., Llovet, J. M., Meyer, T., et al. (2018). Hepatocellular Carcinoma: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 29, iv238–iv255. doi:10.1093/annonc/mdy308
Weissenbach, J., and Grosjean, H. (1981). Effect of Threonylcarbamoyl Modification (t6A) in Yeast tRNA Arg III on Codon-Anticodon and Anticodon-Anticodon Interactions. A Thermodynamic and Kinetic Evaluation. Eur. J. Biochem. 116, 207–213. doi:10.1111/j.1432-1033.1981.tb05320.x
Yamamoto, Y., Matsui, J., Matsushima, T., Obaishi, H., Miyazaki, K., and Nakamura, K. (2014). Lenvatinib, an Angiogenesis Inhibitor Targeting VEGFR/FGFR, Shows Broad Antitumor Activity in Human Tumor Xenograft Models Associated with Microvessel Density and Pericyte Coverage. J. Vasc. Cel 6, 18. doi:10.1186/2045-824X-6-18
Yarian, C., Marszalek, M., Sochacka, E., Malkiewicz, A., Guenther, R., Miskiewicz, A., et al. (2000). Modified Nucleoside Dependent Watson-Crick and Wobble Codon Binding by tRNALysUUU Species. Biochemistry 39, 13390–13395. doi:10.1021/bi001302g
Zhao, Z., Zhang, D., Wu, F., Tu, J., Song, J., Xu, M., et al. (2021). Sophoridine Suppresses Lenvatinib-Resistant Hepatocellular Carcinoma Growth by Inhibiting RAS/MEK/ERK Axis via Decreasing VEGFR2 Expression. J. Cel Mol Med 25, 549–560. doi:10.1111/jcmm.16108
Zheng, R., Qu, C., Zhang, S., Zeng, H., Sun, K., Gu, X., et al. (2018). Liver Cancer Incidence and Mortality in China: Temporal Trends and Projections to 2030. Chin. J. Cancer Res. 30, 571–579. doi:10.21147/j.issn.1000-9604.2018.06.01
Keywords: YRDC, Lenvatinib, HCC, t6A modification, KRAS
Citation: Guo J, Zhu P, Ye Z, Wang M, Yang H, Huang S, Shu Y, Zhang W, Zhou H and Li Q (2021) YRDC Mediates the Resistance of Lenvatinib in Hepatocarcinoma Cells via Modulating the Translation of KRAS. Front. Pharmacol. 12:744578. doi: 10.3389/fphar.2021.744578
Received: 20 July 2021; Accepted: 27 August 2021;
Published: 01 October 2021.
Edited by:
Olivier Feron, Université catholique de Louvain, BelgiumReviewed by:
Yuan Tang, University of Toledo, United StatesRatnakar Tiwari, Northwestern University, United States
Copyright © 2021 Guo, Zhu, Ye, Wang, Yang, Huang, Shu, Zhang, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Li, liqing9251026@csu.edu.cn
†These authors have been contributed equally to this work.
 Jun Guo1,2,3,4†
Jun Guo1,2,3,4† Peng Zhu
Peng Zhu Zhi Ye
Zhi Ye Haijun Yang
Haijun Yang Yan Shu
Yan Shu Wei Zhang
Wei Zhang Honghao Zhou
Honghao Zhou Qing Li
Qing Li