- 1Department of Surgery, Ruijin Hospital, Shanghai Institute of Digestive Surgery, Shanghai Key Laboratory of Gastric Neoplasms, Shanghai, China
- 2Department of Disease Prevention and Control, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
Exploring ERBB2-related pathways will help us finding sensitive molecules and potential combined therapeutic targets of ERBB2-targeted therapy for ERBB2+ gastric cancer (GC). In this study, we performed a cross-databases study focused on ERBB2+ GC. The data of ERBB2+ GC deposited in the cancer genome atlas (TCGA), gene expression omnibus (GEO), InBio MapTM, cancer cell line encyclopedia (CCLE), and cancer therapeutics response portal (CTRP) were analyzed. The correlation of expression levels of candidate and IC50 of candidate genes-targeted drugs were verified on NCI-N87 and MKN-45 GC cell lines. We found that RARA, THRA, CACNB1, and TOP2A are drug sensitive biomarkers of ERBB2-targeted treatment with FDA-approved drugs. All these genes act through Myc signaling pathway. Myc is the downstream hub gene of both ERBB2 and RARA. The expression of RARA, THRA, and CACNB1 were negatively correlated with Myc activation, while ERBB2 and TOP2A positively correlated with Myc activation. SH3BGRL3, SH3BGRL, and NRG2 were identified as potential ligands of ERBB2. The ERBB2+ GC with RARA amplification demonstrated better prognosis than those without RARA amplification, while overexpression of NRG2 and SH3BGRL correlated with poor prognosis in ERBB2+ GC. About 90% of ERBB2+ GC was compatible with chromosome instability (CIN) subtype of TCGA, which overlaps with intestinal-type GC in Lauren classification. In validating experiments, combination of Lapatinib and all-trans retinoic acid (ATRA) synergistically suppresses cell growth, and accompanied by decreased expression of MYC. In conclusions, we identified several predicting biomarkers for ERBB2-targeted therapy and corresponding histological features of ERBB2+ GC. Combination of ERBB2 antagonist or RARA agonist may be effective synergistic regimens for ERBB2+ GC.
Introduction
Gastric cancer (GC) is one of the most common cancers and the third leading cause of mortality worldwide (Ferlay et al., 2015). In China, both of the morbidity and mortality of GC rank the second (Chen et al., 2016). The major therapeutic approaches for GC are surgery, adjuvant chemotherapy and targeted therapy. The targeted drugs approved by FDA are trastuzumab (for ERBB2), cetuximab (for EGFR), and ramucirumab (for VEGF2) (Van Cutsem et al., 2012; Wilke et al., 2014). Lapatinib, a small-molecule inhibitor of ERBB2, plus paclitaxel demonstrated activity in the second-line treatment of ERBB2+ GC (Hecht et al., 2016). In clinics, ERBB2+ GC is defined by scoring 3+ by immunohistochemistry or copy number amplification by FISH (He et al., 2013). In general, ERBB2+ GC accounts for about 13% of all GC cases (Bass et al., 2014). A phase III ToGA study showed that the incidence of ERBB2+ GC is up to 22% (Bang et al., 2010). Because the prognosis of ERBB2+ GC is poor, clarifying the mechanisms of drug sensitivity of ERBB2+ GC will be of clinical significance in ERBB2-targeted therapy (Wang et al., 2017). Currently, trastuzumab plus fluorouracil and platinum can effectively improve overall survival of ERBB2+ GC patients, but the response rate was only 32–68% (Ock et al., 2015). Finding additional therapeutic targets for combined therapy will benefit more ERBB2+ GC patients.
ERBB2 is located in chromosome 17q21.2, where some common oncogenes or tumor suppressor genes, such as TOP2A, TAU, p53, and HIC-1 are located (Zhang and Yu, 2011). Our previous studies confirmed that re-activation of tumor suppressor HIC-1 by small-activating RNAs inhibits cell division, growth and invasion (Zhao et al., 2014, 2015). Retinoic acid receptor alpha (RARA) is another gene located on chromosome 17q21.2. Paroni et al. (2012) reported that combination of RARA agonist and ERBB2-targeted drug demonstrated a synergistic anticancer activity in breast cancer. It suggested that some novel therapeutic targets for ERBB2+ GC may harbor on chromosome 17. In this study, we analyzed ERBB2-related pathways and explored potential drug sensitivity biomarkers that could be used as reference for targeted therapy of ERBB2+ GC.
Materials and Methods
Data Extraction and Data Mining
The data of gene expression, copy number variation, tissue images and clinical information of 413 GC cases was extracted from The Cancer Genome Atlas database (TCGA1) and cBioPortal database2 (Cerami et al., 2012; Gao et al., 2013; Bass et al., 2014). The data of gene expression and copy number variation of GC was also extracted from GSE62717 and GSE57302 in gene expression omnibus (GEO) database3 (Qian et al., 2014; Cristescu et al., 2015). The data of protein-protein interaction was used in InBio MapTM database4 (Li et al., 2017). The information of Lapatinib IC50 of 17 GC cell lines were extracted from database of Cancer Cell Line Encyclopedia (CCLE5) (Barretina et al., 2012). The information of Afatinib IC50 of 16 GC cell lines was extracted from database of Cancer Therapeutics Response Portal (CTRP6) (Basu et al., 2013). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database is used for explaining functions and biology of genes7. The analytic results were confirmed for proteins expression using The Human Protein Atlas8.
Gastric Cancer Cell Lines and Cell Culture
Human GC cell lines NCI-N87 and MKN45 were purchased from the type Culture Collection of Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in RPMI-1640 containing 10% fetal bovine serum (FBS) at 37°C in a humidified incubator with 95% air and 5% CO2.
Cell Viability Assay
Firstly, 5000/well NCI-N87 or MKN45 cells were placed in 96 well plates (100 μl/well). Cells were treated by different concentrations of Lapatinib (Selleck, Houston, TX, United States) and All-Trans Retinoic Acid (ATRA, Selleck, Houston, TX, United States), respectively. CCK-8(10 μl/well) was used and OD value was measured at 450 nm by spectrophotometry (BioTek, VT, United States) at different time points.
Western Blot
The cytoplasmic protein and nuclear protein were extracted, respectively by extraction kit (Cat. P0027, Beyotime, Shanghai, China) according to the protocols. Western blot was performed as previously described (Duan et al., 2016). The antibodies used in this study were as following: HRP-conjugated mouse monoclonal GAPDH (1: 5000, Cat. HRP-60004, Proteintech Group, Inc. Wuhan, China), rabbit monoclonal Histone H3 (1: 5000, Cat. ab1791, Abcam, Cambridge, United Kingdom), and rabbit monoclonal c-MYC (1:1000, Cat. ab32072, Abcam, Cambridge, United Kingdom). Of those, GAPDH and Histone H3 were used as internal controls for cytoplasmic protein and nuclear protein, respectively.
Statistical Analysis
EdgeR package in R language was used to screen differentially expressed genes (Robinson et al., 2010). We plotted OncoPrinter map based on gene copy number data from TCGA and cBioPortal platforms. Pheatmap package in R language was utilized to plot heatmap. Receiver operating characteristic (ROC) was plotted and an optimal cutoff value was selected for biomarkers from clinical samples. The median value was used as cutoff value for cancer cell lines. Survival rate was calculated by Kaplan-Meier method and log-rank test. GSEA software was applied to perform gene set enrichment analysis (Subramanian et al., 2005). Pearson correlation between RNA levels and IC50 was analyzed by Pearson test. CompuSyn software (ComboSyn Inc., Paramus, NJ, United States) was used to calculating combination index (CI) of drugs synergistic effects. CI < 1, =1, and >1 indicate synergistic, additive and antagonistic effects, respectively (Yu et al., 2018). Other statistical analysis was performed by using GraphPad Prism 6.0 (Inc., La Jolla, CA, United States). P-value less 0.05 was considered statistically significant.
Results
ERBB2+ GC Showed Amplification of RARA, THRA, CACNB1, and TOP2A
Among 413 cases of GC from TCGA dataset, ERBB2+ GC accounted for 14.29% (59/413 cases), and the others were ERBB2- GC (354/413 cases). We compared the differential expressed genes and copy number variations between ERBB2+ and ERBB2- GC groups. Forty-seven differential expressed genes were selected based on the likelihood ratio (LR) and P-value (LR > 100, P < 0.0001; Figure 1A). Four out of these 47 genes are the molecular targets of FDA-approved drugs. They are CACNB1 (calcium voltage-gated channel auxiliary subunit beta 1) for amlodipine, TOP2A (DNA topoisomerase II alpha) for etoposide, RARA for ATRA and THRA (thyroid hormone receptor, alpha) for tiratricol. These genes were selected as potential targets of combination therapy with ERBB2-targeted drugs.
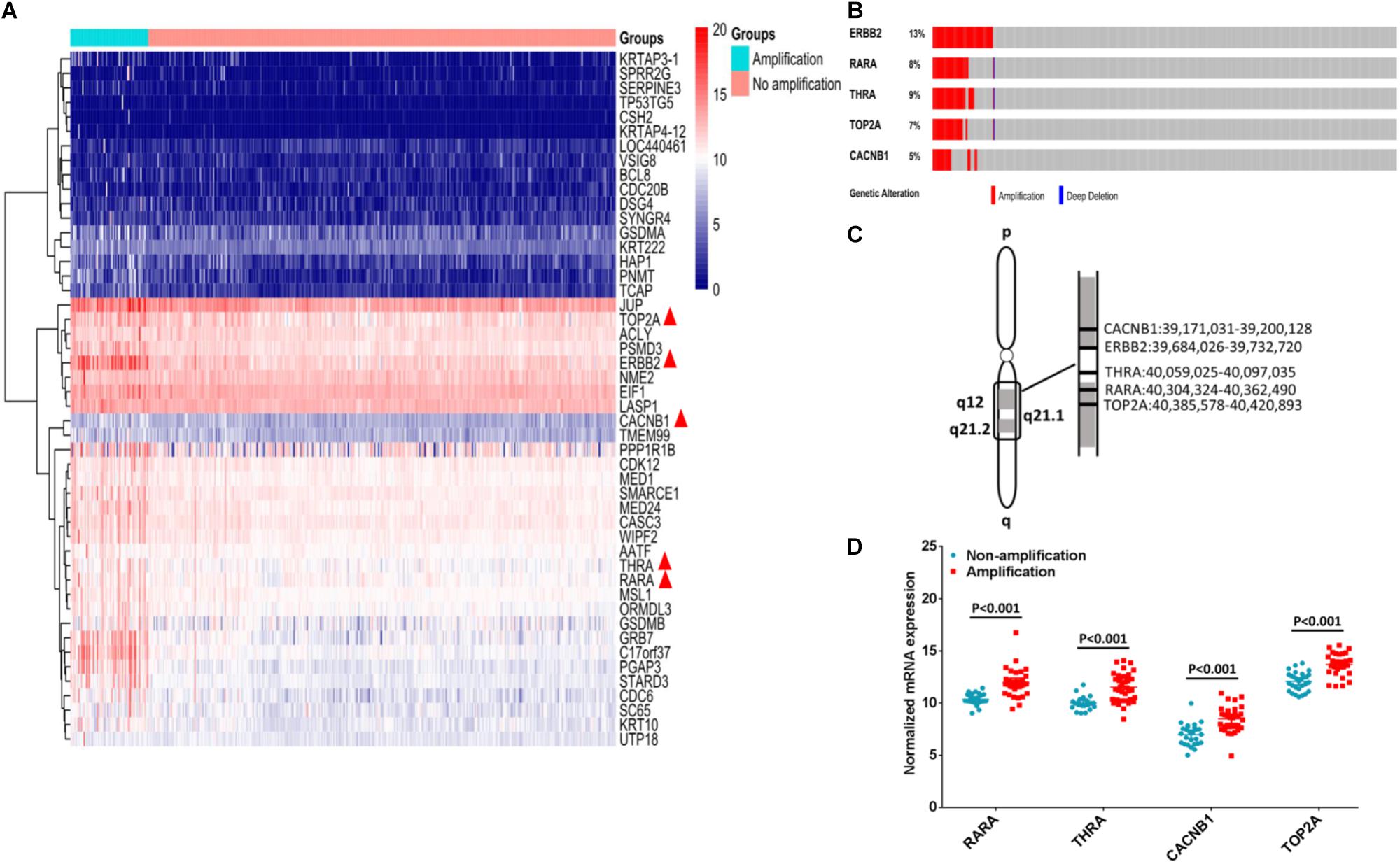
FIGURE 1. Identification of potential targets for combination therapy of ERBB2+ GC. (A) Forty-seven genes were up-regulated in ERBB2+ GC (n = 59 vs. n = 354, P < 0.0001). Among them, five genes are molecular targets of FDA approved drugs (indicated by red triangle). (B) The gene copy number amplification of ERBB2, RARA, THRA, TOP2A, and CACNB1. (C) The five genes ERBB2, RARA, THRA, TOP2A, and CACNB1 located in adjacent region of chromosomal 17q. (D). The mRNA expression of RARA, THRA, CACNB1, and TOP2A in ERBB2+ GC and ERBB2– GC (P < 0.001).
Gene amplification of RARA, THRA, CACNB1, and TOP2A was often accompanied by ERBB2 amplification (Figure 1B). These genes were located in adjacent region of chromosomal 17q (Figure 1C). In ERBB2+ GC, the rate of gene amplification was 57.63% (34 out of 59) for RARA, 62.71% (37 out of 59 (62.71%) for THRA, 50.85% (30 out of 59) for TOP2A, and 54.24% (32 out of 59) for CACNB1. The elevated mRNAs were also identified for cases with gene amplification, compared to cases without gene amplification (Figure 1D, P < 0.001).
Amplified Genes of Chromosome 17q Were Myc Pathway-Related Genes
Gene set enrichment analysis (GSEA) is a visualized method of the gene ontology analysis, which could help researchers finding a group of functional genes in a signaling pathway. Through GSEA, we found that all amplified genes of 17q are Myc-related genes. Of those, RARA, THRA, and CACNB1 were negatively correlated with Myc activation, which revealed minus value of enrichment score (ES). For instance, the ES value of RARA is −1.93 (FDR = 0.017, P = 0.004, Figure 2A), THRA is −1.85 (FDR = 0.035, P = 0.010, Figure 2B), and CACNB1 is −1.89 (FDR = 0.028, P = 0.004, Figure 2C). TOP2A and ERBB2 were positively associated with Myc activation (ES = 2.05, FDR = 0.002, P < 0.001, Figure 2D and (ES = 1.88, FDR = 0.077, P = 0.018, Figure 2E). These results implied that agonists of RARA, THRA, and CACNB1 or inhibitors of TOP2A or ERBB2 might be combined together for the treatment of ERBB2+ GC.
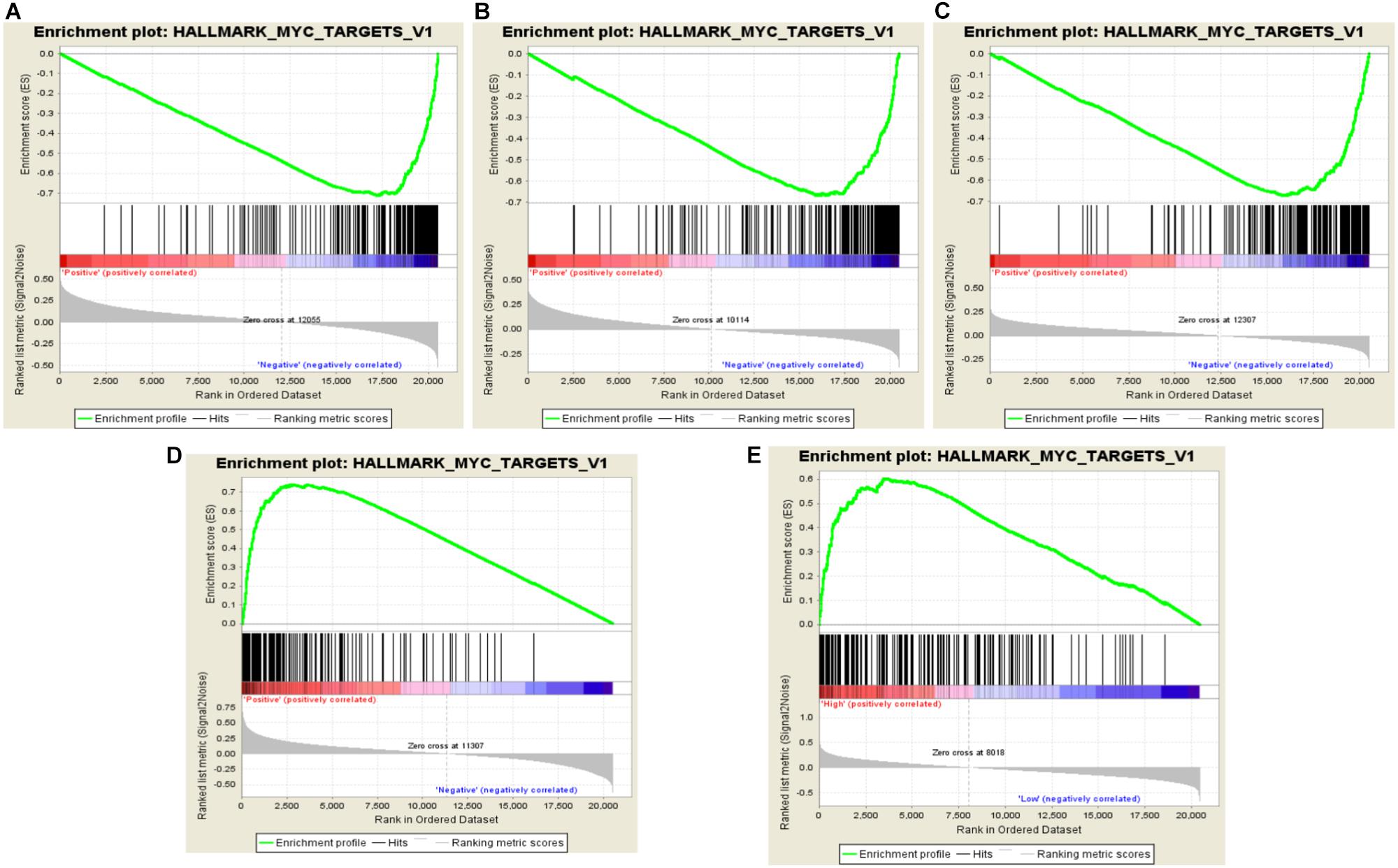
FIGURE 2. Identification of RARA, THRA, CACNB1, and TOP2A related pathways in ERBB2+ GC. (A–C) RARA, THRA, and CACNB1 are negatively related to Myc pathway activation with minus ES value. (D,E) TOP2A and ERBB2 are positively related to Myc pathway activation with plus ES value. The green curve represents enrichment score. The highest point is the enrichment score value in Myc pathway. The ES value indicates the correlation between gene and Myc pathway. The black bar codes represent genes in pathway, which are ordered according to their expression levels. The left end or right end genes are leading edge subset strongly contributed to ES value. The bottom numbers represent order of expression levels from high to low in genome.
The Association Between Amplified Genes and Biological Significance
The association and pathway of amplified genes were analyzed by InBio MapTM. SH3BGRL3, SH3BGRL, and Neuregulin 2 (NRG2) are three candidate ligands of ERBB2. Both RARA and ERBB2 are upstream genes in Myc pathway (Figure 3A). To clarify the clinical significance of amplified genes in ERBB2+ GC, these genes in two datasets from TCGA and GEO were further analyzed. The tendency of better prognosis was noted in RARA amplification group, compared to non-RARA amplification group of ERBB2+ GC in both TCGA cohort (HR = 0.628, 95% CI 0.242–1.539, P = 0.295, Figure 3B), and GEO cohort (HR = 0.312, 95% CI 0.104–0.942, P = 0.04, Figure 3C). In ERBB2+ GC of TCGA cohort, high levels of NRG2 and SH3BGRL were significantly related to poor prognosis (P = 0.009 and P = 0.042, respectively, Figures 3D,E), but not for SH3BGRL3 (P = 0.267, Figure 3F). These results implied the ligand function of NRG2 and SH3BGRL in cancer progression of ERBB2+ GC.
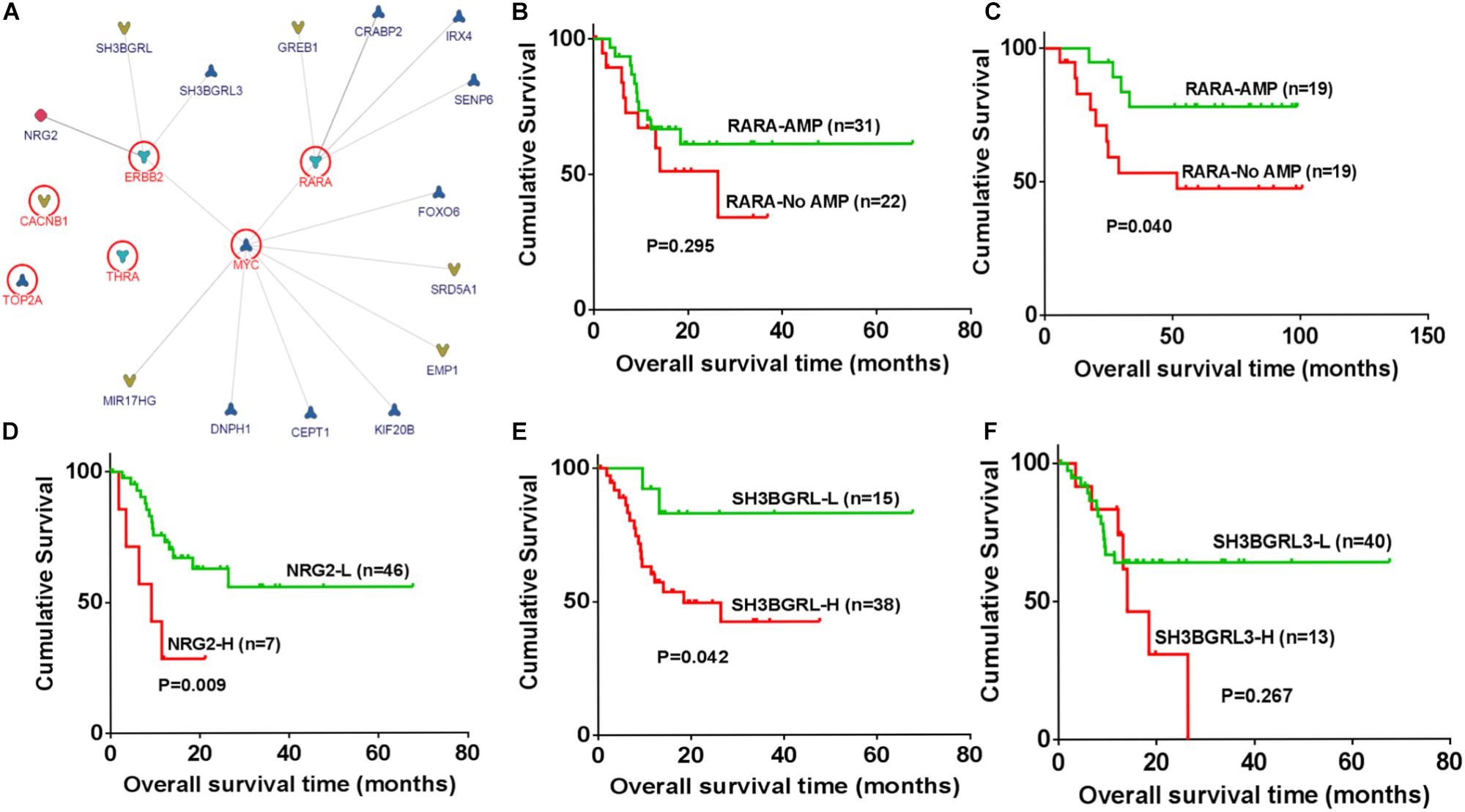
FIGURE 3. Identification of regulatory association for amplified genes and biological significances. (A) SH3BGRL3, SH3BGRL, and NGR2 are identified as potential ligands of ERBB2, while Myc is the downstream hub of ERBB2 and RARA based on InBio MapTM analysis. (B) In ERBB2+ GC, RARA amplification shown better prognosis in TCGA cohort (HR = 0.628, 95% CI 0.242–1.539; P = 0.295). (C) In ERBB2+ GC of GEO datasets, RARA amplification shown better prognosis than that without RARA amplification (HR = 0.312, 95% CI 0.104–0.942; P = 0.040). (D) In ERBB2+ GC of TCGA cohort, overexpression of NRG2 was significantly correlated with poor prognosis (NRG2-L vs. NRG2-H, HR = 0.286, 95% CI 0.265–0.592, P = 0.009). (E) High expression of SH3BGRL was significantly correlated with poor prognosis (SH3BGRL-L vs. SH3BGRL-H, HR = 0.248, 95% CI 0.144–0.960, P = 0.042). (F) No significance was found for SH3BGRL3 (SH3BGRL3-L vs. SH3BGRL3-H, HR = 0.600, 95% CI 0.202–1.55, P = 0.267).
The Relationship Between Amplified Genes and Sensitivity to ERBB2-Targeted Drugs
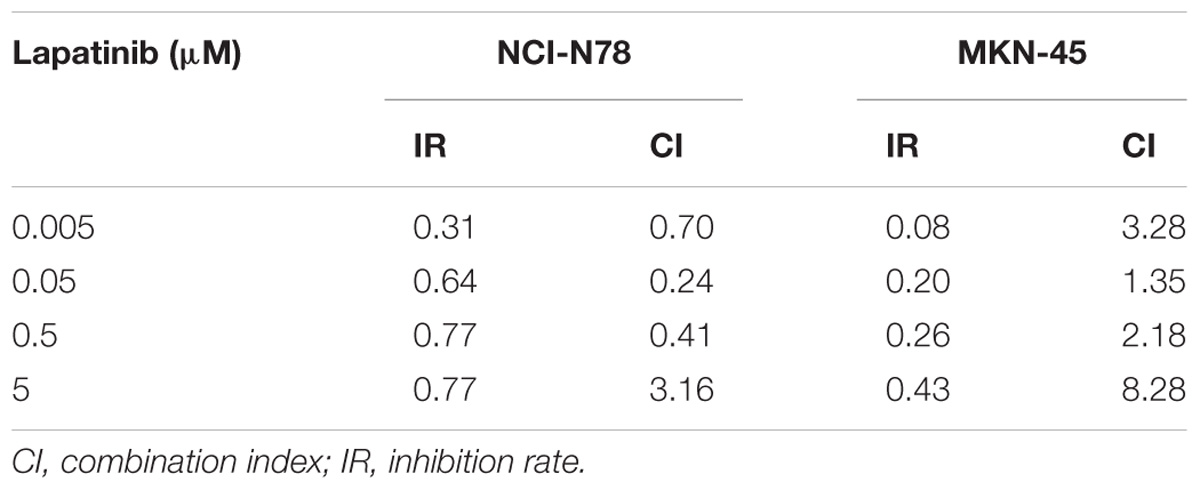
Since the gene copy number, mRNA level and IC50 value to ERBB2-targeted drugs of multiple cancer cell lines were deposited in CCLE database, the drug responsive status of 17 GC cell lines was analyzed. Among those, four cell lines (NCI-N87, KE39, NUGC-4, and MKN-7) were ERBB2-amplified cell lines that showed lower IC50 (μM) to drug Lapatinib than other GC cell lines (P = 0.031, Figure 4A, left). The ERBB2-amplified cell lines also showed lower IC50 (μM) to drug Afatinib than other cell lines (P = 0.027, Figure 4A, right). The CNV of ERBB2 or RARA and their responses to targeted drugs Lapatinib or Afatinib of GC cell lines were listed in Table 1. Cell line NCI-N87 were more sensitive to ERBB2-targeted drugs, which is related to dual amplification of ERBB2 and RARA, while MKN7 was not sensitive to ERBB2-targeted drugs, which is related to non-amplification of RARA.
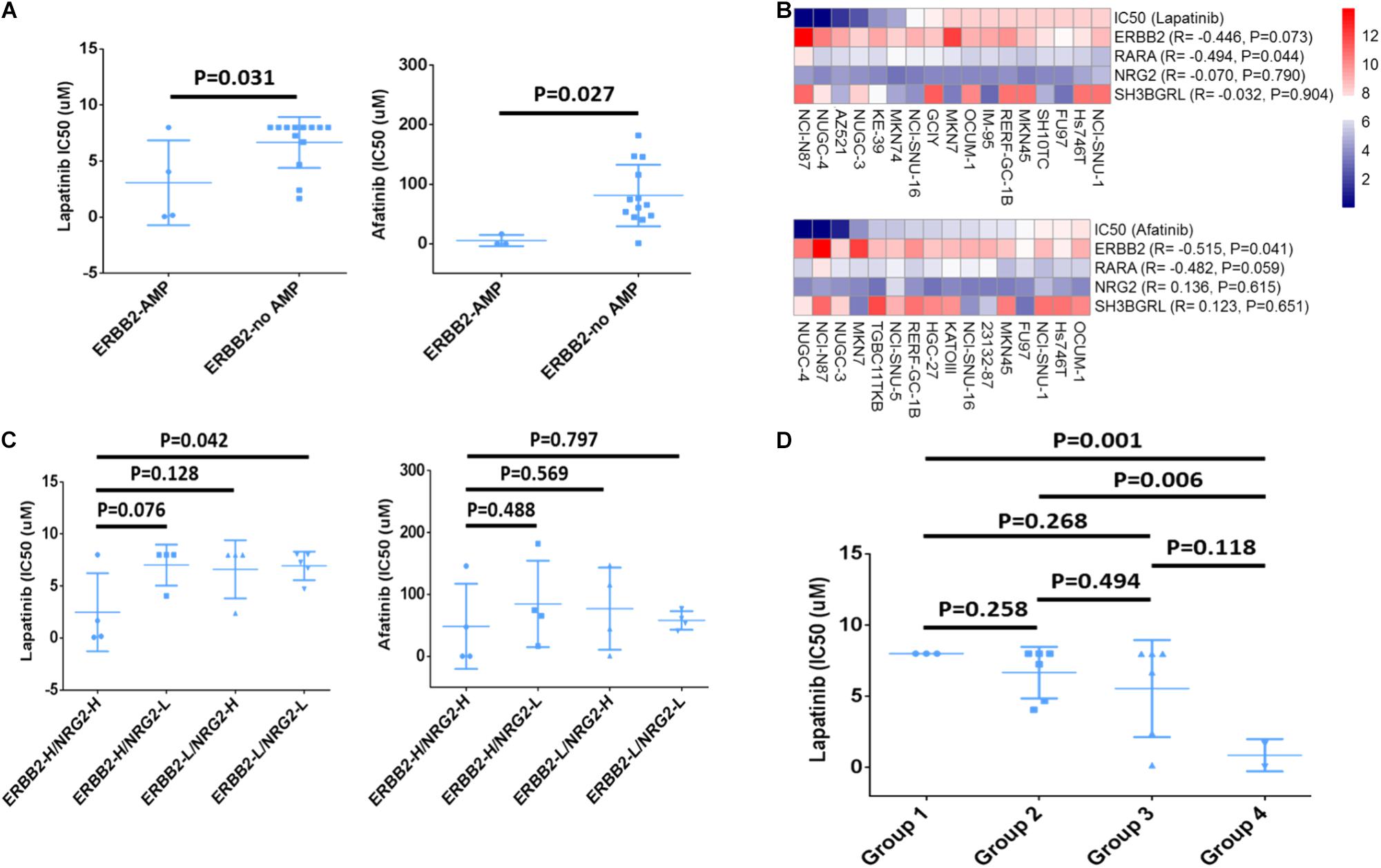
FIGURE 4. Drug sensitivity analysis of ERBB2, RARA, NRG2, and SH3BGRL. (A) GC cells with ERBB2 amplification showed lower IC50 (μM) to Lapatinib (P = 0.031) and Afatinib (P = 0.027) than those without ERBB2 amplification. (B) Relationship between IC50 of ERBB2-targeted drugs Lapatinib and Afatinib and mRNA expression of ERBB2, RARA, NRG2, and SH3BGRL. (C) IC50 of ERBB2-targeted drugs Lapatinib and Afatinib in cells with different mRNA expression, including ERBB2-H/NRG2-H, ERBB2-H/NRG2-L, ERBB2-L/NRG2-H, and ERBB2-L/NRG2-L group. (D) IC50 of Lapatinib on GC cells with different mRNA expression, including Group 1: NRG2-L/ERBB2-L/RARA-L (n = 3); Group 2: higher expression of any one out of NRG2, ERBB2, and RARA (n = 6); Group 3: lower expression of any one out of NRG2, ERBB2, and RARA (n = 6); and Group 4: NRG2-H/ERBB2-H/RARA-H (n = 2).

TABLE 1. The correlation of CNV of ERBB2 or RARA with sensitivity to Lapatinib or Afatinib in ERBB2+ GC cells.
The Relationship of Expression of ERBB2, RARA, NRG2, and SH3BGRL and Sensitivity to ERBB2-Targeted Drugs
ERBB2 mRNA overexpression was often significantly related to lower IC50 to Lapatinib (R = −0.446, P = 0.073) and Afatinib (R = −0.515, P = 0.041). Similarly, overexpression of RARA mRNA was significantly associated with lower IC50 to Lapatinib (R = −0.494, P = 0.044) and Afatinib (R = −0.482, P = 0.059). However, there was no significant correlation for NRG2 and SH3BGRL mRNA expression and drug sensitivity (Figure 4B). Nevertheless, GC cases with overexpression of ERBB2 and NRG2 (ERBB2-H/NRG2-H) were more sensitive to Lapatinib, compared to low gene expression of ERBB2-L/NRG2-L group (P = 0.042, Figure 4C, left). However, there was no significant difference between ERBB2-H/SH3BGRL-H group and ERBB2-L/NRG2-L group for drug Afatinib (Figure 4C, right).
The relationship of simultaneous overexpression of NRG2, ERBB2, and RARA with Lapatinib sensitivity was listed in Table 2. Group 1: NRG2-L/ERBB2-L/RARA-L; Group 2: high expression of any one of NRG2, ERBB2, or RARA; Group 3: lower expression of any one of NRG2, ERBB2, or RARA; and Group 4: NRG2-H/ERBB2-H/RARA-H. The IC50 to Lapatinib of Group 4 was the lowest (P = 0.001). Group 3 is better than that in Group 2 for sensitivity to Lapatinib (Figure 4D).
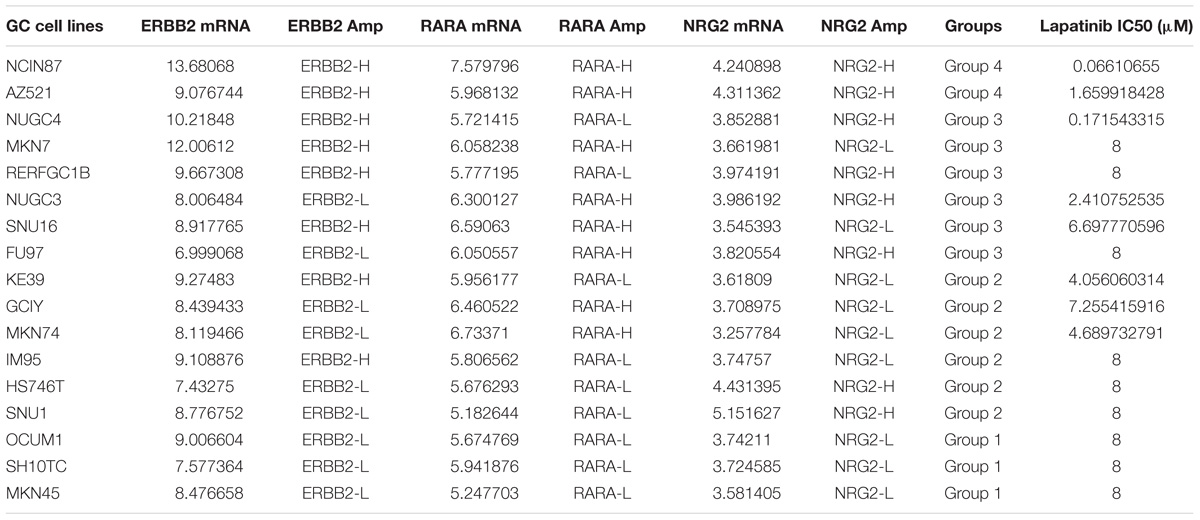
TABLE 2. The association of Lapatinib sensitivity, mRNA expression and gene amplification of ERBB2, RARA, and NRG2.
ERBB2+ GC Strongly Associates With CIN Subtype of TCGA and Intestinal-Type of Lauren Classification
The clinical details and histology of TCGA cohort were recorded in cBioPortal database9, which provided opportunity to analyze correlation between genomic information and clinicopathological characteristics. For 59 cases of ERBB2+ GC, the pathology reports and slide images of 46 cases were uploaded in database (Figure 5A). Fifty-three cases out of 59 ERBB2+ GC (89.83%) belong to chromosome instability (CIN) molecular subtype, and the others include three Epstein-Bar Virus-related (EBV) subtype, two genome stable (GS) subtype, and one microsatellite instability (MSI) subtype (Figure 5B). Based on their pathological reports of ERBB2+ GC, all of those are intestinal-type GC in traditional Lauren classification, such as tubular adenocarcinoma, papillary adenocarcinoma, and a few mucinous adenocarcinomas.
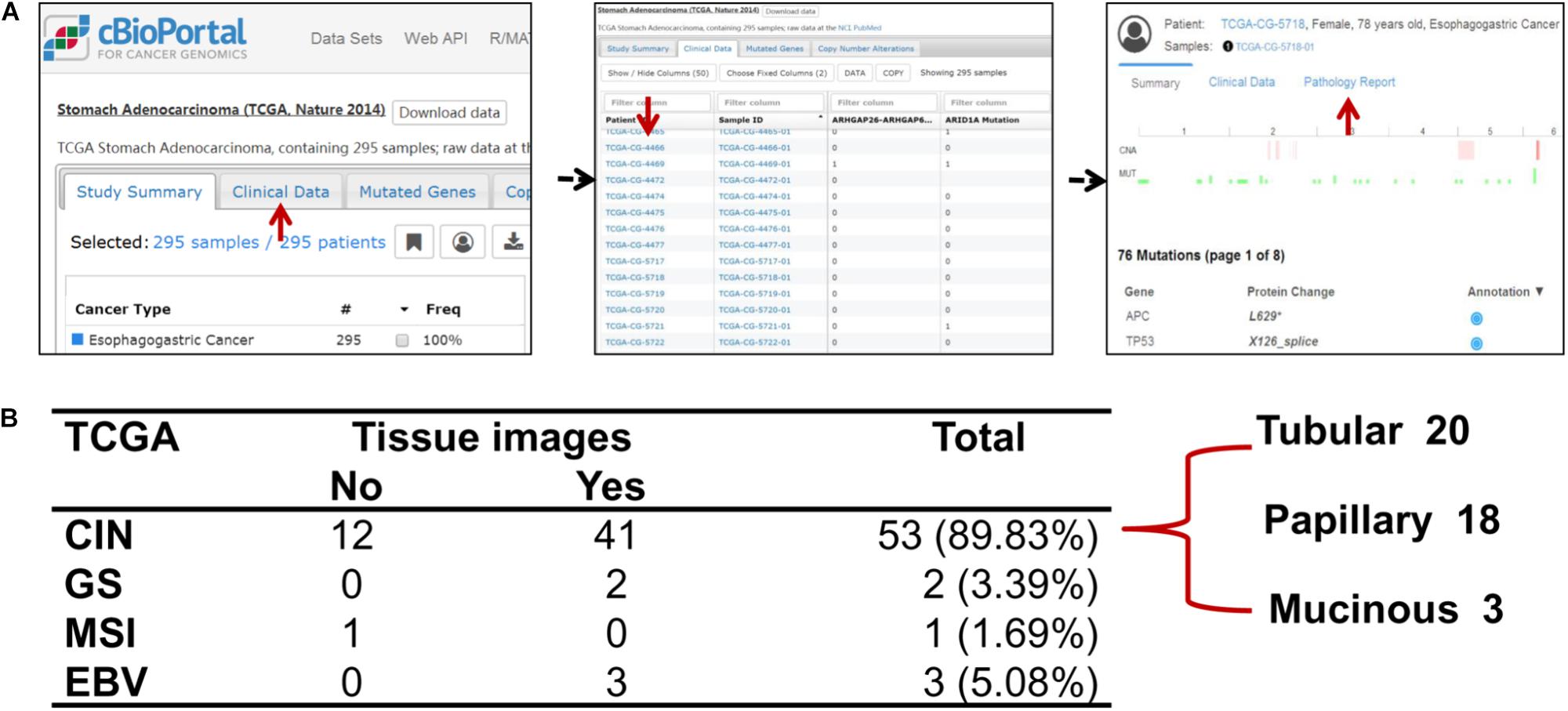
FIGURE 5. The relationship of molecular classification and clinicopathological types of ERBB2+ GC. (A) The flowchart of extracting clinicopathological information of TCGA cohort. (B) The molecular classification features of ERBB2+ GC and corresponding clinicopathological types to Lauren classification in TCGA cohort. The predominant molecular classification is CIN subtype, which overlaps with intestinal-type GC of Lauren classification.
Validating Study of Lapatinib and ATRA Drugs on Different GC Cell Lines
Based on data analysis of multiple GC cell lines (Table 2), cell line NCI-N87 (ERBB2-H/RARA-H) and MKN-45 (ERBB2-L/RARA-L) were selected for further validating experiments. The IC50s of Lapatinib (targeted to ERBB2) and ATRA (targeted to RARA) were assayed by CCK8 method on NCI-N87 and MKN-45 cell lines. As showed in Figure 6A, cell line NCI-N87 was more sensitive than cell line MKN-45 to drug Lapatinib (IC50 0.88μM vs. 15.38 μM, P < 0.001), while there was no significant difference of IC50s of ATRA in both GC cell lines (71.26μM vs. 67.70 μM, P = 0.281). By further CI analysis, synergistic effect of ATRA (25 μM) and Lapatinib (between 0.005 and 0.5 μM) was observed on NCI-N87 cell line, but not in MKN-45 cell line (Table 3).To verify the effect on Myc signaling pathway, the cytoplasmic protein and nuclear protein were extracted separately, and examined expressing levels after incubating Lapatinib, ATRA or both. As results, after 2 h incubation of Lapatinib (0.05 μM), ATRA (25 μM), or both on NCI-N87 and MKN-45 cells, obvious down-regulation of MYC protein levels of cytoplasm and nucleus was found in Lapatinib treated NCI-N87 cells, but bot ATRA did. Importantly, incubation of Lapatinib and ATRA simultaneously further reduced nuclear protein level of MYC in NCI-N87 cells (Figure 6B, right). The similar effects were also observed on NCI-N87 cells after 48 h incubation by Lapatinib and ATRA, or both (Figure 6C, right). However, these effects were not observed in MKN-45 cells (Figures 6B,C, left).
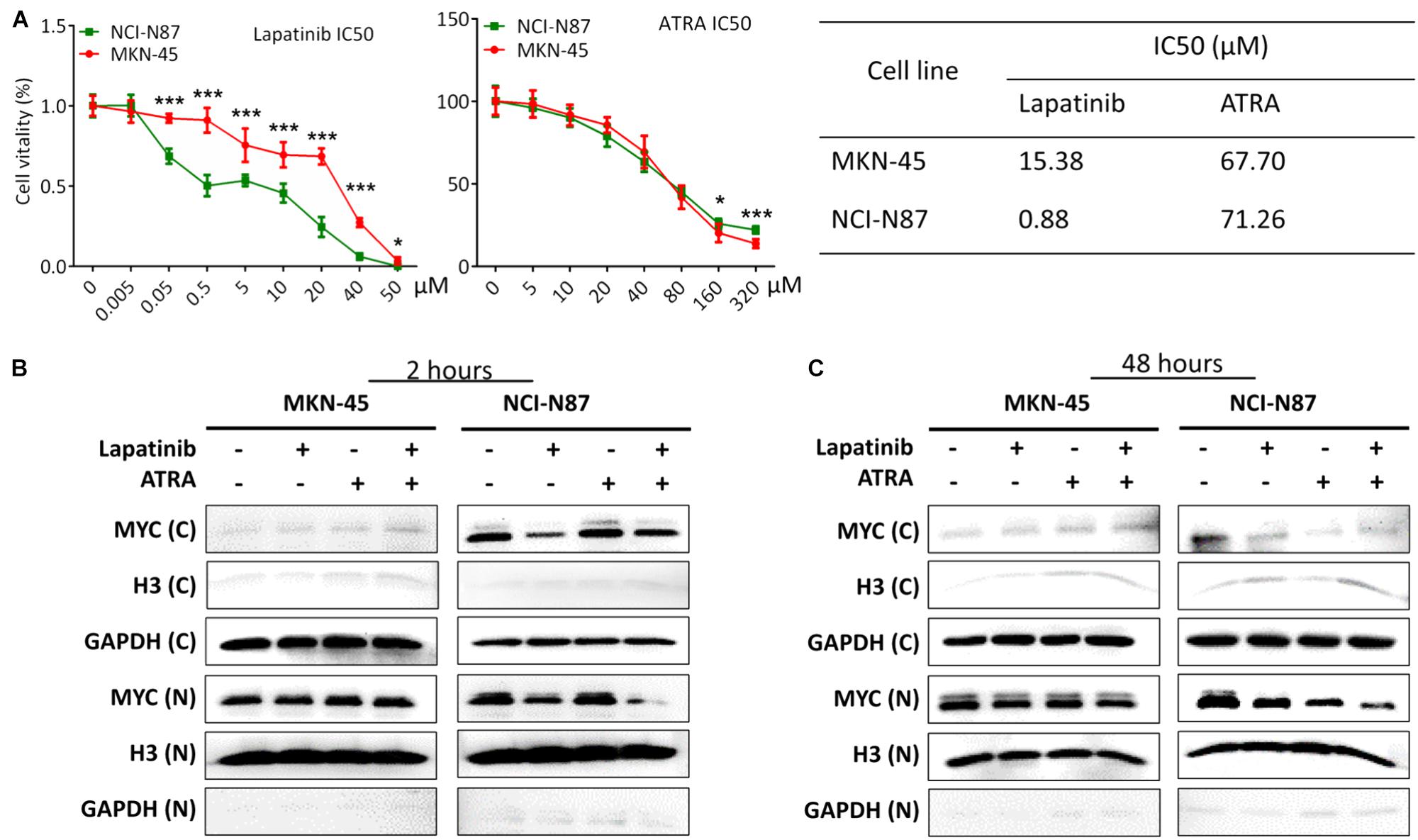
FIGURE 6. The validating study on NCI-N87 and MKN-45 GC cell lines based on drug Lapatinib and ATRA treatment. (A) Analyzing IC50 of Lapatinib and ATRA on MKN-45 and NCI-N87 cell lines. NCI-N87 is a cell line with ERBB2 and RARA amplification, while MKN-45 is a cell line without ERBB2 and RARA amplification. (B) Examination of MYC protein levels on cytoplasm and nucleus after incubation by Lapatinib (0.05 μM) or ATRA (50 μM) or both drugs for 2 h. (C) Examination of MYC protein levels on cytoplasm and nucleus after incubation by Lapatinib (0.05 μM) or ATRA (50 μM) or both drugs for 48 h. “C” represents cytoplasmic protein. “N” represents nuclear proteins. ∗P < 0.05; ∗∗∗P < 0.001.
Discussion
ERBB2 plays a critical role in carcinogenesis and cancer progression and has become a targeted therapeutic molecule in breast cancer and GC. Exploring ERBB2 related pathways and mechanisms of resistance will help oncologist to overcome drug resistance and enhance therapeutic effect of ERBB2-targeted therapy (Shi et al., 2016). Clinically, not all ERBB2+ GC respond well to ERBB2-targeted treatment due to innate resistance or acquired resistance (Kim et al., 2014; Piro et al., 2016). Therefore, exploring resistant mechanism of ERBB2+ GC is desirable. Han et al found that SRC inhibitor combined with trastuzumab can synergistically inhibit proliferation of ERBB2+ GC in vitro (Han et al., 2014).
Two decades passed since trastuzumab was used for treatment of ERBB2+ breast cancer (Goldenberg, 1999; Shak, 1999; Sakamoto and Mitsuyama, 2000). Some ERBB family inhibitors, such as Lapatinib and Afatinib are also developed for treatment of ERBB2+ breast cancer or GC (Geuna et al., 2012; Huang and Rizzo, 2012; Lin et al., 2012; Schuler et al., 2012; Hecht et al., 2016). Large amount of clinical information as well as their corresponding genomic changes has been submitted to open databases. Cross-database analysis is a challenging work, but it helps researchers to get an overview of interested problems. In this study, we performed a cross-database data mining for factors involved in sensitivity or resistance to ERBB2-targeted therapy in ERBB2+ GC. We found that both ERBB2 and RARA are involved in Myc signaling pathway. Myc is the important hub gene of both ERBB2 and RARA. By data mining, a total of 47 genes were outlined as potential targets of treatment for ERBB2+ GC. We noticed that RARA, THRA, CACNB1, and TOP2A are targets of FDA-approved drugs. For instance, CACNB1 is a calcium channel protein and the inhibitor amlodipine is one of the most popular medications for treatment of high blood pressure (Gregg et al., 1993; Nussberger et al., 2008). TOP2A is the target of etoposide, which has been used in cancer treatment (Slater et al., 2001; Bartlett et al., 2015). RARA, which participates in regulation of cell development, differentiation, apoptosis and granulopoiesis. ATRA is a RARA agonist, which revealed clinical efficacy in leukemia treatment (Degos and Wang, 2001). THRA is thyroid hormone receptor alpha, whose ligand tiratricol was used in treatment of hypothyroidism (Yazdanparast et al., 2006; Tylki-Szymanska et al., 2015). GSEA is a powerful tool to find biological pathways for associated genes. By GSEA method, all amplified genes ERBB2, RARA, THRA, CACNB1, and TOP2A are identified to be related to Myc signaling pathway. Myc plays important roles in carcinogenesis and cancer progression (Sun et al., 2011; Stine et al., 2015). In gastric carcinogenesis, pathogen infection of H. pylori and EBV could activate Myc pathway (Calcagno et al., 2008). Our cross-database analysis suggests that inhibitor of TOP2A and agonist of RARA, THRA, or CACNB1 may have synergistic efficacy in ERBB2-targeted therapy for ERBB2+ GC. Li and coworkers found that ERBB2 activated downstream Myc and promoted cells proliferation by phosphorylating AKT1 and Erk1/2 in cervical cancer (Li et al., 2018). RARA could interact with Myc and regulate RARA-dependent genes expression in leukemia cells (Uribesalgo et al., 2012).
In this study, three potential ligands (SH3BGRL3, SH3BGRL, and NGR2) of ERBB2 have been proposed. The latter has been reported as a ligand for ERBB3/ERBB4 (Chang et al., 1997; Schulze et al., 2005; Chiang et al., 2015). Slattery and coworkers found that NRG2 as a growth factor was involved in progression of breast cancer (Slattery et al., 2013). Sun and colleagues found that both NRG1 and NRG2 functioned as ligands of ERBB family and involved in progression of non-small cell lung cancer (Sun et al., 2009). Hitherto, the relationship of NRG2 and ERBB2 in GC is not clear yet. Our analysis disclosed that overexpression of NRG2 is significantly associated with poor prognosis of ERBB2+ GC. Some reports suggested that NRG2 can promote phosphorylation of ERBB2, ERBB3, and ERBB4 directly or indirectly (Sliwkowski et al., 1994; Chang et al., 1997; Mill et al., 2011). Therefore, NRG2 as a potential ligand of ERBB2 is worth further explored. Regarding to SH3BGRL3 and SH3BGRL, there is no any report on the relationship of these genes with ERBB2. In the present study, the simultaneous overexpression of ERBB2, NRG2, and RARA were correlated with increased sensitivity to Lapatinib. We speculate that ERBB2, NRG2, and RARA might act together as a functional network.
It has been reported that there are some histopathological features of ERBB2+ GC (Park et al., 2016). ERBB2 positivity is more often observed in intestinal-type GC (He et al., 2013; Irkkan et al., 2017). Up-to-date, there is no correlation analysis on both levels of genomics and clinical histology for ERBB2+ GC. TCGA database provides a good resource for comparative analysis on both levels because it stores perfect genomics and histological information for GC samples. In 59 cases of ERBB2+ GC, 46 cases were accompanied by whole slide tissue images. ERBB2+ GC revealed characteristic histopathology. All 46 cases were classified into intestinal-type GC using Lauren classification, mainly including tubular adenocarcinoma, papillary adenocarcinoma, and a few mucinous adenocarcinomas. The majority (near 90%) of ERBB2+ GC was classified into CIN subtype on genomic level using TCGA molecular classification (Bass et al., 2014). These findings provide an important clue for clinical selection of ERBB2-targeted therapy.
In our validating study, we confirmed that GC cell line with amplified ERBB2 and RARA (NCI-N87) was sensitive to Lapatinib relative to GC cell without amplified ERBB2 and RARA (MKN-45). The sensitivity to ATRA on both cell lines did not show significant difference, which may attribute to some alternative pathways for ATRA-mediated apoptosis (Patrad et al., 2018). We found that Lapatinib and ATRA could synergistically inhibit cell growth in NCI-N87 cell, but not in MKN-45 cell. These biological effects were involved in interfering Myc signaling pathway. The similar effect was reported for breast cancer (Paroni et al., 2012).
In summary, by cross-database analysis, GC with increased expression of ERBB2, RARA, and NRG2 simultaneously should be taken as sensitive category to Lapatinib therapy. The current study proposed that NRG2-ERBB2-MYC is cross-interfaced with RARA-MYC pathway. Intervention of both routes will regulate hub gene Myc, and then inhibit cell growth for ERBB2+ GC (Figure 7). Moreover, combination of RARA agonist ATRA with ERBB2-targeted drug disclosed synergistic anticancer effect for ERBB2+ GC in vitro.
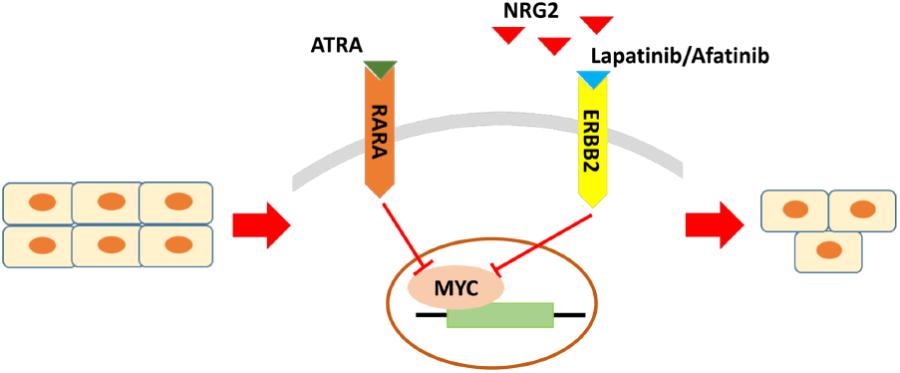
FIGURE 7. The crucial molecules and regulatory pathways of ERBB2+ GC. The main regulatory pathway is NRG2-ERBB2-MYC, which is cross-interfaced with RARA-MYC pathway. For ERBB2+ GC, beyond ERBB2 itself, RARA and NRG2 are potential drug sensitive biomarkers too for predicting response to ERBB2-targeted therapy. Combination of NRG2 antagonist or RARA agonist with ERBB2-targeted drug may be effective regimens for ERBB2+ GC.
Author Contributions
YY, XH, and ZZ were involved in concept and design. ZX, JW, and JZ acquired the data and performed data mining. ZX, JJ, and RY performed the experiments. WC and YY supported the analysis. All authors wrote, reviewed, and revised the manuscript.
Funding
This project was supported by National Key R&D Program of China (2017YFC0908303 and 2016YFC1303202), the National Natural Science Foundation of China (81772505 and 81372644), Shanghai Science and Technology Committee (18411953100), the Cross-Institutes Research Fund of Shanghai Jiao Tong University (YG2017ZD01 and YG2015MS62), Innovation Foundation of Translational Medicine of Shanghai Jiao Tong University School of Medicine (15ZH4001, TM201617, and TM 201702), and Technology Transfer Project of Science & Technology Dept. Shanghai Jiao Tong University School of Medicine.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge multiple open databases of TCGA, GEO (GSE62717 and GSE57302), InBio MapTM, CCLE, KEGG and The Human Protein Atlas.
Footnotes
- ^ https://cancergenome.nih.gov/
- ^ http://www.cbioportal.org/
- ^ https://www.ncbi.nlm.nih.gov/geo/
- ^ https://www.intomics.com/inbio/map/#home
- ^ http://www.broadinstitute.org/ccle
- ^ http://portals.broadinstitute.org/ctrp.v2.1/
- ^ http://www.genome.jp/kegg/pathway.html
- ^ https://www.proteinatlas.org/
- ^ http://www.cbioportal.org/study?id=stad_tcga_pub#clinical
References
Bang, Y. J., Van Cutsem, E., Feyereislova, A., Chung, H. C., Shen, L., Sawaki, A., et al. (2010). Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 376, 687–697. doi: 10.1016/S0140-6736(10)61121-X
Barretina, J., Caponigro, G., Stransky, N., Venkatesan, K., Margolin, A. A., Kim, S., et al. (2012). The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607. doi: 10.1038/nature11003
Bartlett, J. M., McConkey, C. C., Munro, A. F., Desmedt, C., Dunn, J. A., Larsimont, D. P., et al. (2015). Predicting anthracycline Benefit: TOP2A and CEP17-not only but also. J. Clin. Oncol. 33, 1680–1687. doi: 10.1200/JCO.2013.54.7869
Bass, A. J., Thorsson, V., Shmulevich, I., Reynolds, S. M., Miller, M., Bernard, B., et al. (2014). Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209. doi: 10.1038/nature13480
Basu, A., Bodycombe, N. E., Cheah, J. H., Price, E. V., Liu, K., Schaefer, G. I., et al. (2013). An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154, 1151–1161. doi: 10.1016/j.cell.2013.08.003
Calcagno, D. Q., Leal, M. F., Assumpcao, P. P., Smith, M. A., and Burbano, R. R. (2008). MYC and gastric adenocarcinoma carcinogenesis. World J. Gastroenterol. 14, 5962–5968. doi: 10.3748/wjg.14.5962
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. doi: 10.1158/2159-8290.CD-12-0095
Chang, H., Riese, D. J., Gilbert, W., Stern, D. F., and McMahan, U. J. (1997). Ligands for ErbB-family receptors encoded by a neuregulin-like gene. Nature 387, 509–512. doi: 10.1038/387509a0
Chen, W., Zheng, R., Baade, P. D., Zhang, S., Zeng, H., Bray, F., et al. (2016). Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115–132. doi: 10.3322/caac.21338
Chiang, C. Y., Pan, C. C., Chang, H. Y., Lai, M. D., Tzai, T. S., Tsai, Y. S., et al. (2015). SH3BGRL3 protein as a potential prognostic biomarker for urothelial carcinoma: a novel binding partner of epidermal growth factor receptor. Clin. Cancer Res. 21, 5601–5611. doi: 10.1158/1078-0432.CCR-14-3308
Cristescu, R., Lee, J., Nebozhyn, M., Kim, K. M., Ting, J. C., Wong, S. S., et al. (2015). Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 21, 449–456. doi: 10.1038/nm.3850
Degos, L., and Wang, Z. Y. (2001). All trans retinoic acid in acute promyelocytic leukemia. Oncogene 20, 7140–7145. doi: 10.1038/sj.onc.1204763
Duan, Y., Gao, Y., Zhang, J., Chen, Y., Jiang, Y., Ji, J., et al. (2016). Mitochondrial aldehyde dehydrogenase 2 protects gastric mucosa cells against DNA damage caused by oxidative stress. Free Radic Biol. Med. 93, 165–176. doi: 10.1016/j.freeradbiomed.2016.02.001
Ferlay, J., Soerjomataram, I., Dikshit, R., Eser, S., Mathers, C., Rebelo, M., et al. (2015). Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386. doi: 10.1002/ijc.29210
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:1. doi: 10.1126/scisignal.2004088
Geuna, E., Montemurro, F., Aglietta, M., and Valabrega, G. (2012). Potential of afatinib in the treatment of patients with HER2-positive breast cancer. Breast Cancer 4, 131–137. doi: 10.2147/BCTT.S25868
Goldenberg, M. M. (1999). Trastuzumab, a recombinant DNA-derived humanized monoclonal antibody, a novel agent for the treatment of metastatic breast cancer. Clin. Ther. 21, 309–318. doi: 10.1016/S0149-2918(00)88288-0
Gregg, R. G., Powers, P. A., and Hogan, K. (1993). Assignment of the human gene for the beta subunit of the voltage-dependent calcium channel (CACNLB1) to chromosome 17 using somatic cell hybrids and linkage mapping. Genomics 15, 185–187. doi: 10.1006/geno.1993.1029
Han, S., Meng, Y., Tong, Q., Li, G., Zhang, X., Chen, Y., et al. (2014). The ErbB2-targeting antibody trastuzumab and the small-molecule SRC inhibitor saracatinib synergistically inhibit ErbB2-overexpressing gastric cancer. mAbs 6, 403–408. doi: 10.4161/mabs.27443
He, C., Bian, X. Y., Ni, X. Z., Shen, D. P., Shen, Y. Y., Liu, H., et al. (2013). Correlation of human epidermal growth factor receptor 2 expression with clinicopathological characteristics and prognosis in gastric cancer. World J. Gastroenterol. 19, 2171–2178. doi: 10.3748/wjg.v19.i14.2171
Hecht, J. R., Bang, Y. J., Qin, S. K., Chung, H. C., Xu, J. M., Park, J. O., et al. (2016). Lapatinib in combination with capecitabine plus oxaliplatin in human epidermal growth factor receptor 2-positive advanced or metastatic gastric. J. Clin. Oncol. 34, 443–451. doi: 10.1200/JCO.2015.62.6598
Huang, Y., and Rizzo, R. C. (2012). A water-based mechanism of specificity and resistance for lapatinib with ErbB family kinases. Biochemistry 51, 2390–2406. doi: 10.1021/bi2016553
Irkkan, C., Balci, S., Guler Tezel, G., Akinci, B., Yalcin, B., and Guler, G. (2017). Comparison of clinicopathologic parameters and survivals between epstein-barr virus-positive and her2-positive gastric cancers. Appl. Immunohistochem. Mol. Morphol. 25, 609–614. doi: 10.1097/PAI.0000000000000353
Kim, J., Fox, C., Peng, S., Pusung, M., Pectasides, E., Matthee, E., et al. (2014). Preexisting oncogenic events impact trastuzumab sensitivity in ERBB2-amplified gastroesophageal adenocarcinoma. J. Clin. Invest. 124, 5145–5158. doi: 10.1172/JCI75200
Li, S., Ma, Y. M., Zheng, P. S., and Zhang, P. (2018). GDF15 promotes the proliferation of cervical cancer cells by phosphorylating AKT1 and Erk1/2 through the receptor ErbB2. J. Exp. Clin. Cancer Res. 37:80. doi: 10.1186/s13046-018-0744-0
Li, T., Wernersson, R., Hansen, R. B., Horn, H., Mercer, J., Slodkowicz, G., et al. (2017). A scored human protein-protein interaction network to catalyze genomic interpretation. Nat. Methods 14, 61–64. doi: 10.1038/nmeth.4083
Lin, N. U., Winer, E. P., Wheatley, D., Carey, L. A., Houston, S., Mendelson, D., et al. (2012). A phase II study of afatinib (BIBW 2992), an irreversible ErbB family blocker, in patients with HER2-positive metastatic breast cancer progressing after trastuzumab. Breast Cancer Res. Treat. 133, 1057–1065. doi: 10.1007/s10549-012-2003-y
Mill, C. P., Zordan, M. D., Rothenberg, S. M., Settleman, J., Leary, J. F., and Riese, D. J. (2011). ErbB2 Is Necessary for ErbB4 Ligands to Stimulate Oncogenic Activities in Models of Human Breast Cancer. Genes Cancer 2, 792–804. doi: 10.1177/1947601911431080
Nussberger, J., Aubert, J. F., Bouzourene, K., Pellegrin, M., Hayoz, D., and Mazzolai, L. (2008). Renin inhibition by aliskiren prevents atherosclerosis progression: comparison with irbesartan, atenolol, and amlodipine. Hypertension 51, 1306–1311. doi: 10.1161/HYPERTENSIONAHA.108.110932
Ock, C. Y., Lee, K. W., Kim, J. W., Kim, J. S., Kim, T. Y., Lee, K. H., et al. (2015). Optimal patient selection for trastuzumab treatment in her2-positive advanced gastric cancer. Clin. Cancer Res. 21, 2520–2529. doi: 10.1158/1078-0432.CCR-14-2659
Park, C. K., Park, J. S., Kim, H. S., Rha, S. Y., Hyung, W. J., Cheong, J. H., et al. (2016). Receptor tyrosine kinase amplified gastric cancer: clinicopathologic characteristics and proposed screening algorithm. Oncotarget 7, 72099–72112. doi: 10.18632/oncotarget.12291
Paroni, G., Fratelli, M., Gardini, G., Bassano, C., Flora, M., Zanetti, A., et al. (2012). Synergistic antitumor activity of lapatinib and retinoids on a novel subtype of breast cancer with coamplification of ERBB2 and RARA. Oncogene 31, 3431–3443. doi: 10.1038/onc.2011.506
Patrad, E., Niapour, A., Farassati, F., and Amani, M. (2018). Combination treatment of all-trans retinoic acid (ATRA) and gamma-secretase inhibitor (DAPT) cause growth inhibition and apoptosis induction in the human gastric cancer cell line. Cytotechnology 70, 865–877. doi: 10.1007/s10616-018-0199-3
Piro, G., Carbone, C., Cataldo, I., Di Nicolantonio, F., Giacopuzzi, S., Aprile, G., et al. (2016). An FGFR3 autocrine loop sustains acquired resistance to trastuzumab in gastric cancer patients. Clin. Cancer Res. 22, 6164–6175. doi: 10.1158/1078-0432.CCR-16-0178
Qian, Z., Zhu, G., Tang, L., Wang, M., Zhang, L., Fu, J., et al. (2014). Whole genome gene copy number profiling of gastric cancer identifies PAK1 and KRAS gene amplification as therapy targets. Genes Chromosomes Cancer 53, 883–894. doi: 10.1002/gcc.22196
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Sakamoto, G., and Mitsuyama, S. (2000). New molecule-targeting therapy with herceptin (trastuzumab), an anti-HER2 (c-erB-2) monoclonal antibody. Breast Cancer 7, 350–357. doi: 10.1007/BF02966404
Schuler, M., Awada, A., Harter, P., Canon, J. L., Possinger, K., Schmidt, M., et al. (2012). A phase II trial to assess efficacy and safety of afatinib in extensively pretreated patients with HER2-negative metastatic breast cancer. Breast Cancer Res. Treat. 134, 1149–1159. doi: 10.1007/s10549-012-2126-1
Schulze, W. X., Deng, L., and Mann, M. (2005). Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol. Syst. Biol. 1:0008. doi: 10.1038/msb4100012
Shak, S. (1999). Overview of the trastuzumab (Herceptin) anti-HER2 monoclonal antibody clinical program in HER2-overexpressing metastatic breast cancer. Herceptin Multinational Investigator Study Group. Semin. Oncol. 26(4 Suppl. 12), 71–77.
Shi, H., Zhang, W., Zhi, Q., and Jiang, M. (2016). Lapatinib resistance in HER2+ cancers: latest findings and new concepts on molecular mechanisms. Tumour Biol. [Epub ahead of print]. doi: 10.1007/s13277-016-5467-2
Slater, L. M., Stupecky, M., Sweet, P., Osann, K., Eklof, A., and Arquilla, E. R. (2001). Etoposide induction of tumor immunity in Lewis lung cancer. Cancer Chemother. Pharmacol. 48, 327–332. doi: 10.1007/s002800100357
Slattery, M. L., John, E. M., Stern, M. C., Herrick, J., Lundgreen, A., Giuliano, A. R., et al. (2013). Associations with growth factor genes (FGF1, FGF2, PDGFB, FGFR2, NRG2, EGF, ERBB2) with breast cancer risk and survival: the Breast Cancer Health Disparities Study. Breast Cancer Res. Treat. 140, 587–601. doi: 10.1007/s10549-013-2644-5
Sliwkowski, M. X., Schaefer, G., Akita, R. W., Lofgren, J. A., Fitzpatrick, V. D., Nuijens, A., et al. (1994). Coexpression of erbB2 and erbB3 proteins reconstitutes a high affinity receptor for heregulin. J. Biol. Chem. 269,14661–14665.
Stine, Z. E., Walton, Z. E., Altman, B. J., Hsieh, A. L., and Dang, C. V. (2015). MYC, Metabolism, and Cancer. Cancer Discov. 5, 1024–1039. doi: 10.1158/2159-8290.CD-15-0507
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Sun, M., Behrens, C., Feng, L., Ozburn, N., Tang, X., Yin, G., et al. (2009). HER family receptor abnormalities in lung cancer brain metastases and corresponding primary tumors. Clin. Cancer Res. 15, 4829–4837. doi: 10.1158/1078-0432.CCR-08-2921
Sun, T., Wang, C., Xing, J., and Wu, D. (2011). miR-429 modulates the expression of c-myc in human gastric carcinoma cells. Eur. J. Cancer 47, 2552–2559. doi: 10.1016/j.ejca.2011.05.021
Tylki-Szymanska, A., Acuna-Hidalgo, R., Krajewska-Walasek, M., Lecka-Ambroziak, A., Steehouwer, M., Gilissen, C., et al. (2015). Thyroid hormone resistance syndrome due to mutations in the thyroid hormone receptor alpha gene (THRA). J. Med. Genet. 52, 312–316. doi: 10.1136/jmedgenet-2014-102936
Uribesalgo, I., Benitah, S. A., and Di Croce, L. (2012). From oncogene to tumor suppressor: the dual role of Myc in leukemia. Cell Cycle 11, 1757–1764. doi: 10.4161/cc.19883
Van Cutsem, E., de Haas, S., Kang, Y. K., Ohtsu, A., Tebbutt, N. C., Ming, et al. (2012). Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: a biomarker evaluation from the AVAGAST randomized phase III trial. J. Clin. Oncol. 30, 2119–2127. doi: 10.1200/JCO.2011.39.9824
Wang, Y., He, L., and Cheng, Y. (2017). An independent survival prognostic role for human epidermal growth factor receptor 2 in gastric cancer: evidence from a meta-analysis. Clin. Transl. Oncol. 20, 212–220. doi: 10.1007/s12094-017-1711-5
Wilke, H., Muro, K., Van Cutsem, E., Oh, S. C., Bodoky, G., Shimada, Y., et al. (2014). Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol. 15, 1224–1235. doi: 10.1016/S1470-2045(14)70420-6
Yazdanparast, P., Carlsson, B., Oikarinen, A., Risteli, J., Lavin, T., and Faergemann, J. (2006). Action of topical thyroid hormone analogue, triiodothyroacetic acid in reversing glucocorticoid-induced skin atrophy in humans. Thyroid 16, 1157–1162. doi: 10.1089/thy.2006.16.1157
Yu, M., Liu, T., Chen, Y., Li, Y., and Li, W. (2018). Combination therapy with protein kinase inhibitor H89 and Tetrandrine elicits enhanced synergistic antitumor efficacy. J. Exp. Clin. Cancer Res. 37:114. doi: 10.1186/s13046-018-0779-2
Zhang, W., and Yu, Y. (2011). The important molecular markers on chromosome 17 and their clinical impact in breast cancer. Int. J. Mol. Sci. 12, 5672–5683. doi: 10.3390/ijms12095672
Zhao, F., Pan, S., Gu, Y., Guo, S., Dai, Q., Yu, Y., et al. (2014). Small activating RNA restores the activity of the tumor suppressor HIC-1 on breast cancer. PLoS One 9:e86486. doi: 10.1371/journal.pone.0086486
Keywords: gastric cancer, HER2, RARA, lapatinib, ATRA
Citation: Xiang Z, Huang X, Wang J, Zhang J, Ji J, Yan R, Zhu Z, Cai W and Yu Y (2018) Cross-Database Analysis Reveals Sensitive Biomarkers for Combined Therapy for ERBB2+ Gastric Cancer. Front. Pharmacol. 9:861. doi: 10.3389/fphar.2018.00861
Received: 21 May 2018; Accepted: 17 July 2018;
Published: 03 August 2018.
Edited by:
Dong-Hua Yang, St. John’s University, United StatesReviewed by:
Qi Liu, Tongji University, ChinaRu Wen, Stanford University, United States
Zhi Shi, Jinan University, China
Copyright © 2018 Xiang, Huang, Wang, Zhang, Ji, Yan, Zhu, Cai and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Cai, cw11109@rjh.com.cn; Yingyan Yu, yingyan3y@sjtu.edu.cn; ruijinhospitalyyy@163.com
†These authors have contributed equally to this work.
 Zhen Xiang
Zhen Xiang Xia Huang2†
Xia Huang2† Yingyan Yu
Yingyan Yu