Metabolic Phenotype and Microbiome of Infants Fed Formula Containing Lactobacillus paracasei Strain F-19
- 1Department of Food Science and Technology, University of California, Davis, Davis, CA, United States
- 2Department of Pediatrics, Peking University Third Hospital, Beijing, China
- 3Arla Foods amba, Arla Innovation Center, Skejby, Denmark
- 4Department of Child Health Care, Children's Hospital, Fudan University, Shanghai, China
- 5Department of Child Health Care, Children's Hospital of Nanjing Medical University, Nanjing, China
- 6Department of Clinical Sciences, Pediatrics, Umeå University, Umeå, Sweden
- 7Department of Nutrition, University of California, Davis, Davis, CA, United States
Early childhood nutrition drives the development of the gut microbiota. In contrast to breastfeeding, feeding infant formula has been shown to impact both the gut microbiota and the serum metabolome toward a more unfavorable state. It is thought that probiotics may alter the gut microbiota and hence create a more favorable metabolic outcome. To investigate the impact of supplementation with Lactobacillus paracasei spp. paracasei strain F-19 on the intestinal microbiota and the serum metabolome, infants were fed a formula containing L. paracasei F19 (F19) and compared to a cohort of infants fed the same standard formula without the probiotic (SF) and a breast-fed reference group (BF). The microbiome, as well as serum metabolome, were compared amongst groups. Consumption of L. paracasei F19 resulted in lower community diversity of the gut microbiome relative to the SF group that made it more similar to the BF group at the end of the intervention (4 months). It also significantly increased lactobacilli and tended to increase bifidobacteria, also making it more similar to the BF group. The dominant genus in the microbiome of all infants was Bifidobacterium throughout the intervention, which was maintained at 12 months. Although the serum metabolome of the F19 group was more similar to the group receiving the SF than the BF group, increases in serum TCA cycle intermediates and decreases in several amino acids in the metabolome of the F19 group were observed, which resulted in a metabolome that trended toward the BF group. Overall, L. paracasei F19 supplementation did not override the impact of formula-feeding but did impact the microbiome and the serum metabolome in a way that may mitigate some unfavorable metabolic impacts of formula-feeding.
Introduction
It is well established that early childhood nutrition can impact long-term health. Development of the gut microbiome is important for ensuring proper gut function and development of the immune system (1), as well as development of other organs including the brain (2). There are many factors that influence the development of the microbiome, including infant diet. It has been shown that formula-fed (FF) and breast-fed (BF) infants harbor distinct microbiomes (3–8), with BF infants having a microbiome dominated by bifidobacteria and lactobacilli. Formula-feeding has been linked with metabolic stress that includes metabolic and immune alterations such as higher serum insulin coupled with higher serum amino acids, altered cytokines and blood lipids (5, 7, 9–11), as well as higher infection rates during the first year of life (12) compared with BF infants. Furthermore, FF infants are more likely to develop obesity and metabolic dysfunction later in life than BF infants (13, 14).
Probiotics are microorganisms thought to confer a health benefit when consumed through altering the composition of the intestinal microbiota. For premature infants, they are becoming more accepted as prophylaxis against necrotizing enterocolitis (15, 16). Indeed, evidence has shown that provision of probiotics, such as Bifidobacterium animalis lactis, B. bifidum, B. infantis, Lactobacillus acidophilus, L. reuteri, and L. rhamnosus, in preterm and term infants as well as rhesus monkey infants has a significant impact on the microbial community structure (4, 17–19). Probiotic effects are also population-specific due to differences in the basic commensal bacteria and environment (20). Among probiotic strains, L. paracasei subsp. paracasei F19 (F19) is a GRAS-approved (GRN No. 840) strain that exhibits genetic stability throughout production (21). F19 survives gastric transit in infants (22), and actively interacts with the gut epithelium and immune system while exhibiting antioxidative and proteolytic activities in the gut (23). Its main glucose fermentation product is lactic acid, a metabolite that is known to have antimicrobial, immune-modulating, and intestinotrophic effects (24). Clinically, research on F19 has largely focused on gastrointestinal health (25–27) and immune modulation (28), and to a lesser extent on protection against obesity (23).
We recently conducted a trial to investigate the impact of feeding an infant formula containing a probiotic strain, Lactobacillus paracasei spp. paracasei strain F-19 (F19) in infants from 21 days (± 7 days) until 4 months of age (29). Overall, the F-19 supplemented formula was well-tolerated, with few adverse effects (29). To understand the impact of supplementation with F-19 more fully, we report on the serum metabolome and fecal microbiome in the same cohort of infants.
Results
Study Participants
Demographic data on study participants are shown in Table 1. Two sets of subjects were randomly chosen for serum metabolomics analysis at 4 months and microbiome analysis at three timepoints: baseline, 4 months, and 12 months (with some subjects overlapping in the two sets). Approximately equal numbers of male and female subjects comprised the breast-fed reference cohort (BF) and the cohort provided the Lactobacillus paracasei spp. paracasei strain F-19 probiotic (F19); however, there were more female than male subjects in the standard formula group (SF) in our dataset. Additionally, a greater number of infants were delivered vaginally in the BF group compared to the F19 or SF groups, and the F19 group had a greater number of vaginally born infants compared to the SF group. Mean birthweight was not significantly different amongst the groups. We previously reported that both formula groups experienced similar infectious episodes during the intervention, but only the SF group had significantly more days and episodes of fever than the BF group (29). For these subsets of infants, we did not find any difference in the frequency of antibiotic use or diarrheal episodes among groups. We observed significantly higher serum IFN-γ in the SF group compared to the BF and F19 groups, which is what was published previously for this cohort (30). For the subset of infants used for metabolomics, we also observed higher serum IL-2 (general T cell stimulation) in the F19 group compared to the BF group, but this was not significant for the subset of infants used for microbiota analysis (data not shown).
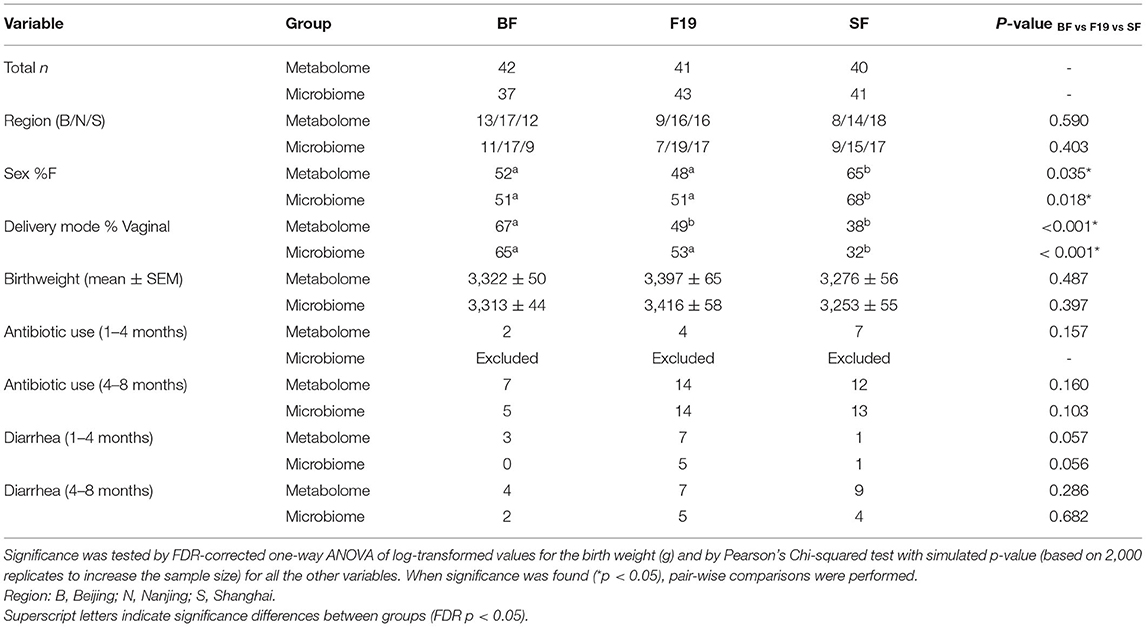
Table 1. Characteristics of infants used for analysis of the serum metabolome and the fecal microbiome.
Impact of F19 on the Fecal Microbiome
Based on the weighted UniFrac distance matrix, principal coordinates analysis (PCoA) revealed that at baseline (3 wk ± 7 d), the infants assigned to the F19 group were significantly different from the BF group (pairwise PERMANOVA q = 0.033; n = 999 permutations) but were not different from the SF group (q = 0.174). At the end of the intervention, when infants were 4 months of age, the two formula groups were not different and together were significantly different from the BF group (pairwise PERMANOVA q = 0.015 (BF-SF), 0.015 (BF-F19); n = 999 permutations). By 12 months of age, fecal microbiota was indistinguishable between all three groups (Figure 1A).
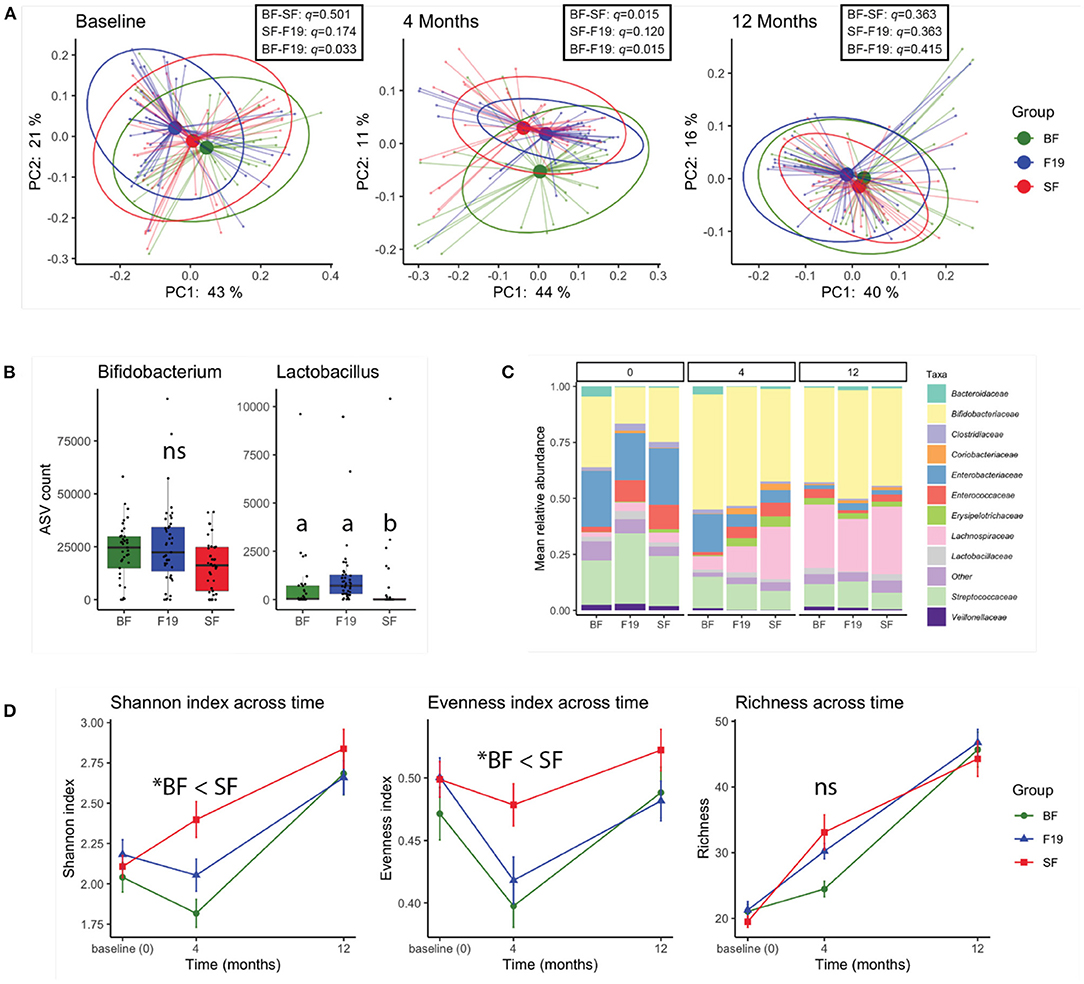
Figure 1. 16S rRNA analysis reveals a moderate effect of L. paracasei F19 supplementation. (A) Principal coordinates analysis (PCoA) of fecal microbiota at baseline (prior to the intervention), 4 months (at the end of the intervention), and 12 months based on the weighted UniFrac distance metric. Results of Pairwise PERMANOVA are provided in the table insert. (B) Amplicon sequence variant (ASV) counts of Bifidobacterium and Lactobacillus genera for each group at 4 months of age. The different letters indicate statistical significance (ANCOM FDR < 0.05). (C) Bar plots of the mean relative abundance at family level of taxonomy in each group at baseline (0), 4, and 12 months of age. Unclassified family or the family with < 2.5% relative abundance were grouped as Other. (D) Fecal microbial alpha-diversity represented as the Shannon, Evenness, and Richness indices over time (*pairwise Kruskal-Wallis q < 0.05). Values are shown as mean ± SE. In all panels, the L. paracasei F19 supplemented group is represented in blue, the standard formula group in red, and the breastfed reference group in green. A total of 121 samples (n = 37 BF, n = 41 SF, n = 43 F19) were analyzed.
Differential abundance analysis revealed that at 4 months, the feces of infants in the F19 group were enriched in members of the Lactobacillus genus compared to the SF group (ANCOM FDR < 0.05, tested on relative abundance), and were not significantly different from the BF group (Figure 1B). Aside from higher relative abundance of Lactobacillus, no differences at the genus level were observed in the F19 group relative to the SF group. Interestingly, the relative abundance of Bifidobacterium was not significantly different between the groups (Figure 1B). Aside from higher Lactobacillaceae in the F19 group compared to the SF group at 4 months, the relative abundances of bacterial families were comparable between the formula groups. Both formula groups were different from the BF reference group with respect to the relative abundance of several bacterial families including Lachnospiraceae, Veillonellaceae, Enterobacteriaceae, Erysipelotrichaceae, and Enterococcaceae (Figure 1C). Notably, group differences were no longer apparent at 12 months, as the relative abundance of bacterial families including Lactobacillaceae were similar in all three groups (ANCOM FDR>0.05). This showed that increased Lactobacillus through F19 supplementation did not persist 8 months after the end of intervention. Throughout the study, Bifidobacterium was a dominant genus in all groups. Analysis of the diversity of the microbiome revealed that BF infants had significantly lower Shannon diversity at all timepoints compared to the SF group, with lower evenness at all three timepoints, and lower growth of richness from baseline to 4 months (Figure 1D). The F19 group tended to have lower Shannon diversity than the SF group at 4 and 12 months, which was reflected as lower evenness at both timepoints, and was not significantly different from the BF group at either time point.
Impact of F19 Supplementation on the Serum Metabolome
We have previously shown that in both the post-prandial and semi-fasting states, there are profound differences in the serum metabolome between BF and FF infants (4–7, 9, 11). In this study, as expected, the SF group differed from the BF reference group (Figures 2A,B), with metabolome differences similar to what was previously reported. Here, significantly higher levels of several amino acids (including valine, isoleucine, leucine, lysine, threonine, phenylalanine, asparagine, tyrosine, alanine, and histidine) and other metabolites [3-hydroxyisobutyrate, 2-hydroxybutyrate, 3-hydroxyisovalerate, creatine, carnitine, dimethyl sulfone (DMSO2), galactonate, and urea], and lower levels of 1,2-propanediol, ketone bodies (3-hydroxybutyrate and acetone), glutamine, methanol, formate, fumarate, citrate, succinate, pyroglutamate, serine, dimethylamine, creatinine, myo-inositol, 2-oxoglutarate, and N,N-dimethylglycine were observed in the serum of SF infants compared to BF infants (p < 0.05). Comparison of F19 infants to BF infants revealed a similar pattern in metabolites but with fewer significantly different metabolites and lower magnitudes of effect sizes than comparison of SF infants to BF infants (Figure 2C). Metabolites higher in F19 infants compared to BF infants included 3-hydroxyisobutyrate, DMSO2, creatine, urea, 2-hydroxybutyrate, and several amino acids (including valine, isoleucine, threonine, lysine, and leucine). Compared to BF infants, F19 infants exhibited lower levels of glutamine, 1,2-propanediol, 3-hydroxybutyrate, myo-inositol, serine, creatinine, formate, dimethylamine, N,N-dimethylglycine, betaine, proline, methanol, ornithine, and taurine.
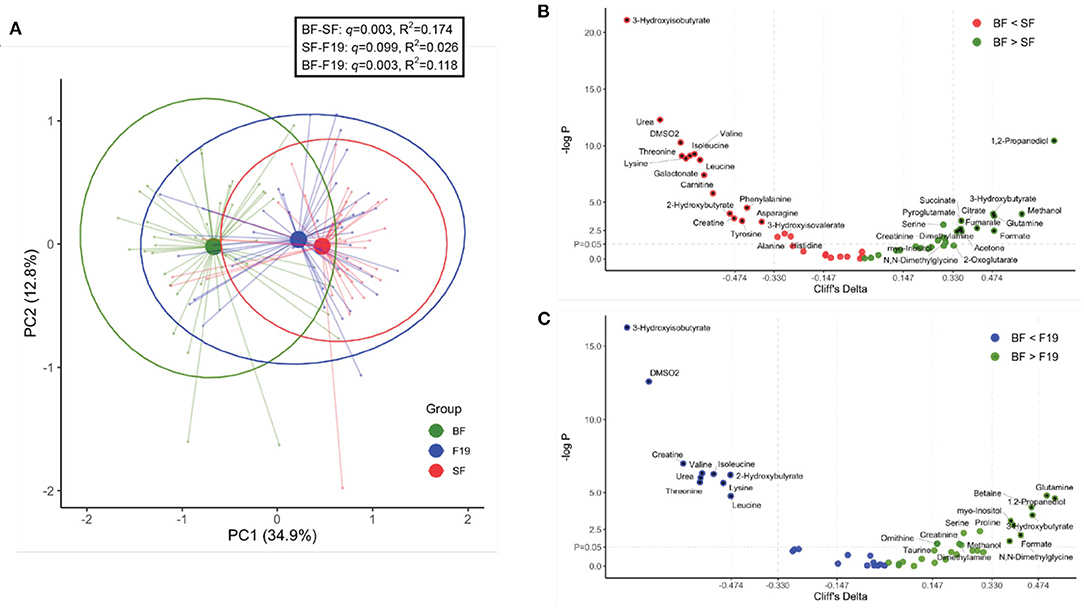
Figure 2. Comparison of the serum metabolome at 4 months of age of infants fed standard formula (SF; n = 40) or standard formula supplemented with L. paracasei F19 (F19; n = 41) and breast-fed infants (BF; n = 42). (A) Principal components analysis (PCA) of the generalized log-transformed serum metabolite concentration obtained from 1H NMR analysis. The L. paracasei F19 supplemented group is represented in blue, the standard formula group in red, and the breast-fed reference group in green. Results of Pairwise PERMANOVA are provided in the table insert. (B) Volcano plot showing Cliff's delta effect sizes vs. the log-transformed p-value comparing the standard formula group (red) and the breast-fed reference group (green). A p-value of 0.05 is indicated by the horizontal line. The vertical lines correspond to the cutoff between small and medium as well as medium and large effect sizes. Asterisks correspond to a p-value < 0.05 and at least a medium effect size after correcting for hospital. The color represents whether the concentration is higher in breast-fed infants (green) or infants fed standard formula (red). (C) Volcano plot showing Cliff's delta effect sizes versus the log-transformed p-value comparing the L. paracasei F19 supplemented group (blue) to the breast-fed reference group (green).
Given that the F19 infants had a serum metabolome more similar to SF infants than to BF infants, we sought to determine the difference between the F19 and SF groups. Using a linear mixed-effect model, with hospital as a random effect, p-values and Cliff delta effect sizes were calculated and are summarized in a volcano plot (Figure 3A). Compared to the SF group, the F19 group had higher levels citric acid cycle intermediates (citrate, succinate, and fumarate) and methanol, and lower levels of several amino acids (tyrosine, lysine, phenylalanine, histidine, proline, leucine, and threonine), 3-hydroxyisovalerate, ornithine, betaine, urea, taurine, and mannose. The metabolites with at least a medium effect size and a p < 0.05 were summarized in Figure 3B. One of the branched chain amino acids, leucine, as well as taurine and urea were lower in F19 infants compared to SF infants with medium effect sizes, but these were not significant (p < 0.1).
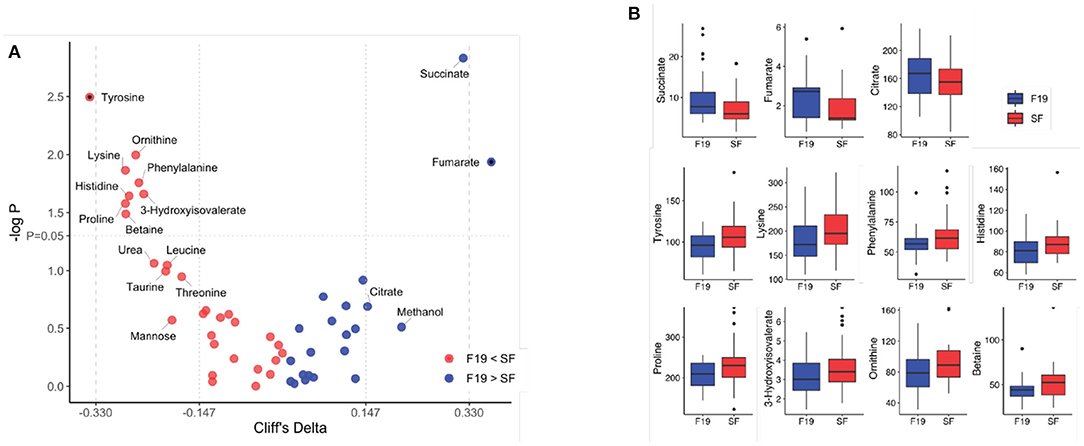
Figure 3. Comparison of the serum metabolome of infants fed standard formula with infants fed standard formula containing L. paracasei F19 at 4 months of age. (A) Volcano plot showing Cliff's delta effect sizes versus the log-transformed p-value comparing the standard formula group (red) and the L. paracasei F19 supplemented group (blue). A p-value of 0.05 is indicated by the horizontal line. The vertical lines correspond to the cutoff between small and medium as well as medium and large effect sizes. Asterisks correspond to a p-value < 0.05 and at least a medium effect size after correcting for hospital. The color represents whether the concentration is higher in infants fed standard formula (red) or infants provided the L. paracasei F19 probiotic (blue). (B) Box plots of metabolite concentrations in μmol/L comparing infants fed standard formula (red) with infants provided the L. paracasei F19 probiotic (blue).
Several SF (n = 6) and F19 (n = 12) infants had serum metabolomes consistent with a BF infant's metabolome, with substantial levels of 1,2-propanediol [a metabolite that is formed by fermentation of milk oligosaccharides by select strains of bifidobacteria that contain the genes to catabolize fucose (31–33)] and ketone bodies, with significantly lower levels of branched chain amino acids. Further, there were a few infants in the BF group that had levels of these metabolites that were consistent with FF infants (4–7, 9, 11). Removal of these samples from analysis resulted in no difference in our reported results suggesting that their presence in the dataset did not inadvertently skew the results.
Discussion
Probiotics, including Lactobacillus and Bifidobacterium, have been studied as additives to infant formulas for many years [reviewed in (34–37)]; however, few studies have directly measured the metabolic impact. We previously studied the role of Bifidobacterium animalis subsp. lactis (B. lactis) supplementation on the rhesus infant metabolome and microbiome (4), and observed significant impacts on the fecal, urine, and serum metabolomes as well as microbiome compared to rhesus infants fed a standard formula. However, despite these changes, B. lactis supplementation did not override the impact of formula-feeding, nor pushed the metabolome or microbiome to a state more similar to BF infant (4).
In the current study, we assessed the impact of supplementation with L. paracasei strain F19 in human infants on the microbiome and the metabolome. As reported previously for this cohort (29, 30), no adverse effects of the F19 probiotic were observed in infants when the probiotic was consumed with a standard infant formula; however, a significant cytokine response at 4 months of age [higher IL-2 in the F19 group relative to BF, and lower IFN-γ in the F19 group relative to the SF group (with no difference to BF)] was observed (30). The reason for this difference could be due to the probiotic itself, the probiotic-modulated gut microbiota, and/or the probiotic and microbiota-associated metabolites that interact with immune cells and epithelial cells. Indeed, it is known that gut microbiota can alter immune responses (38–46).
As expected, at 4 months (the end of the supplementation period), the SF and F19 infants had a similar microbial community structure (besides the higher relative abundance of Lactobacillus in the F19 group), which was significantly different from BF infants. We have observed this in other studies (3, 4, 6, 7), and this shows that there is a distinct microbial composition for BF infants compared to FF infants. As we have also shown in previous studies (3, 6), at 12 months, infants from all three groups were indistinguishable from one another, which suggests that current diet, rather than previous diet, impacts the microbial composition measured through 16S sequencing.
Prior to the start of the study (at baseline), the microbial community structure of infants recruited to the F19 group was significantly different from the BF reference group. One reason could be the higher rate of Caesarian section births (47% in the F19 vs. 35% in the BF group); however, the SF group had more Caesarian section births (68%) than the F19 group, so it is unlikely that this was a contributing factor. A more likely explanation is that infants in the FF groups were already receiving formula and possibly more infants in the F19 group had a greater consumption of formula at this age than those in in the SF group, but the amount of formula consumption at recruitment was not recorded. Nonetheless, at baseline, the Evenness Index and Shannon Index were similar for the F19 and SF group, and higher than the BF reference group (Figure 1D).
Interestingly, after the intervention, microbial community diversity in the F19 group was significantly lower than in the SF group, and comparable to the BF group. Whether community diversity is associated with its stability or instability is controversial (47), but lower microbial diversity over the first few months of life has been consistently associated with breastfeeding (48). Human milk contains selective growth substrates for specific microbes (i.e., human milk oligosaccharides) that have the additional property of inhibiting growth of undesirable bacteria (49). It is not known whether it is the HMOs, the Bifidobacterium, or a combination of the two that are responsible for limiting the type and number of organisms colonizing the infant gut, but it would appear in this case that L. paracasei F19 has the ability to alter the evenness (but not the richness) of commensal bacteria. This makes sense as the overall composition would be dictated by the diet (i.e., formula-fed), but the relative abundance of each species could be modulated by L. paracasei F19. This may occur through the production of antimicrobial compounds including lactic acid and bacteriocins from L. paracasei F19, or could arise due to competition for nutrients (50). It is also interesting that although the relative abundance of Firmicutes was higher in the F19 group compared with the BF group, it was not higher than the SF group despite supplementation with the probiotic. Remarkably, supplementation with F19 tended to increase the average relative abundance of Actinobacteria, specifically the genus Bifidobacterium, in the F19 group to levels similar to the BF group, although it is unlikely that there are similar species / strains of Bifidobacterium in the BF and F19 groups since HMOs were not present in the formula. To understand if F19 has an ecological relationship with species of Bifidobacterium in the gut of formula-fed infants, future analyses of bacterial species and strains or metagenomics should be performed. Nonetheless, this is an important observation as bifidobacterial species in general have been shown to have many health benefits, including immunomodulatory effects (41–43), which could together with L. paracasei F19 drive some of the changes in the levels of IL-2 and/or IFN-γ reported previously in this cohort (30). It is interesting to note that similar observations were not observed in an infant cohort provided L. paracasei F19 as part of weaning foods (28, 51), thus the changes observed here may be specific to early supplementation.
Serum metabolites provide a reflection of recent food intake as well as information regarding metabolic status. Supplementation with L. paracasei F19 resulted in differences in the serum metabolome when compared with SF fed infants and suggests that F19 supplementation may mitigate reported metabolic discrepancies between BF and FF infants (9, 11). Specifically, BF infants compared to FF infants exhibited observed lower levels of serum amino acids and higher levels of citric acid cycle intermediates. Here, F19 supplementation lowered the median concentrations of several essential amino acids (tyrosine, lysine, leucine, phenylalanine, histidine) and 3-hydroxyisovalerate, and increased levels of citrate, succinate, and fumarate in the serum of FF infants, resulting in concentrations comparable to the BF group. The median concentrations of ornithine and betaine were similar in the SF and BF groups, but were lower in the F19 group. Changes in plasma amino acid concentrations with L. paracasei probiotic supplementation have been reported (52). Specifically, consumption of plant protein with an L. paracasei probiotic increased several amino acids in the blood, which was interpreted as changes in digestion of the plant protein (52). In a humanized mouse model, metabolic changes were observed in liver, plasma, fecal and urine when mice were provided an L. paracasei supplement compared with control (53). Supplementation with L. paracasei F19 in infant formula may modulate protein digestion / utilization and/or amino acid metabolism, as observed with reduced levels of some amino acids and nitrogen waste products in serum. This trait is a metabolic characteristic of BF infants that may benefit insulin sensitivity and metabolic function later in life (5, 7, 9–11) and whether this would yield any long-term consequence needs further investigation. Although not analyzed in this study, the fecal metabolome or metagenomics could provide more insights on how bacterial metabolism is interrelated with the host metabolome with L. paracasei F19 supplementation.
A limitation of the current study includes a lack of dietary records of potential other foods consumed by infants (other than breast milk or study formulas), although it was recommended for parents not to provide other foods during the intervention. In conclusion, our study provides insights on how probiotic supplementation with L. paracasei F19 induces changes in host nutrient utilization and energy metabolism, characterized by increased citric acid cycle metabolites and reduced protein degradation products, suggesting a shift in metabolism closer to BF infants. While the L. paracasei F19 supplementation significantly increased the relative abundance of Lactobacillus, it did not induce significant compositional changes in the other bacteria compared to the infants consuming the standard formula. However, there was a noticeable change in the microbial diversity and the tendency of harboring more Bifidobacterium in the gut. To date, the clinical outcomes of this observation are not known, and the long-term effects need to be assessed in future studies.
Materials and Methods
Subjects
This clinical trial was a multicenter double-blind, randomized controlled trial with infants from 21 ± 7 days until the end of the 4th month of age. Infants were followed until the age of 1 year. The design and clinical results of the study were previously published (29). The trial was approved by the Institutional Review Board (IRB) at the University of California Davis and the regional Ethical Review Boards in Nanjing, Shanghai, and Beijing in China (ClinicalTrials.gov Identifier: NCT01755481). Inclusion criteria encompassed healthy infants born full term with a birthweight between 2,500 g and 4,000 g. For infants in the breast-fed reference group (BF), inclusion criteria also included exclusive breastfeeding from birth, with mothers intending to breast-feed for at least 5 months and providing at least 80% of the calories to their infants from breast milk. Additional inclusion criteria for the two formula groups (FF) included infants of mothers who either could not breast-feed, or voluntarily resigned from breast-feeding by 28 days.
Except for the addition of the probiotic Lactobacillus paracasei ssp. paracasei strain F19 to one of the formulas [at a dose of 108 CFU/L; 8.33 × 107 CFU/day based on an average formula consumption of 833 mL/day (29)] (F19), the composition of the study formulas was exactly the same. The formulas were manufactured in Hohot, China using a bovine milk powder provided by Arla Foods amba, Denmark, and the L. paracasei ssp. paracasei strain F19 provided by Chr. Hansen, Denmark. The nutrient composition of the formula was previously published (29).
Prior to the intervention, infants in the FF groups were provided standard formula (SF) if formula feeding had been initiated. Between the 5th and 6th month of life, infants in both FF groups were provided SF. For BF infants, if milk supply was insufficient, SF was used, but was not to exceed 20% of total calorie intake based on a monthly three-day intake record (29). Although feeding of other foods or formulas was not recommended during the intervention, data on consumption of other foods or formulas was not recorded. Parents were recommended to introduce complementary foods to the infants no later than 6.5 months of age. The group code was blinded from study staff and enrolled participants, and was not broken until sample extraction, lab analysis, and generation of concentration/relative abundance data had been completed.
To confirm consumption of L. paracasei ssp. paracasei strain F19 in the F19 group, the amount in the stool was determined using quantitative PCR using the same primers and conditions as described by Sieuwerts and Håkansson (54).
Serum Metabolomics Analysis
A total of 150 subjects (n = 50 per group) were randomly chosen from 179 BF, 167 SF and 167 F19 infants completing the study (29), and serum samples collected from those infants at 4 months of age were used for metabolomics analysis. Out of the potential 150 samples, 6 samples had too small (< 50 μl) or no volume, 15 samples were hemolyzed (identified by a bright red color), 3 samples from the F19 group were from infants with no detectable levels of L. paracasei ssp. paracasei strain F19 in their stool, and 3 samples had noisy NMR spectra or potential contamination. Exclusion of these samples left 123 samples (n = 42 BF, n = 40 SF, and n = 41 F19) for metabolomics analysis.
Serum samples were prepared by filtering through 3,000 MW cutoff Amicon filters (Merck Millipore, MA, USA) followed by the addition of potassium phosphate buffer (pH 6.7) and an internal standard containing 5 mM 3-(trimethylsilyl)-1-propanesulfonic acid–d6 (DSS-d6) and 0.2% NaN3 in 99.8% D2O as previously described (6). NMR spectra were acquired on a Bruker Avance 600 MHz NMR spectrometer at 25°C using the Bruker noesypr1d experiment as previously described (7). NMR spectra were processed using Chenomx NMR Suite v8.2 Processor (Chenomx Inc., Edmonton, Canada) (RRID:SCR_014682), and metabolites were identified and quantified using Chenomx NMR Suite Profiler v8.2 based on the internal standard DSS-d6 as described (55). Quantified metabolite concentrations were corrected for dilution and are expressed as absolute concentrations in μmol/L.
Fecal Microbiome Sequencing
One hundred and fifty-one subjects (n = 51 BF, n = 50 SF and n = 50 F19) from the original cohort were randomly chosen for 16s rRNA gene sequencing, and a total of 453 fecal samples representing three time points [baseline (21±7 d), end of the 4th month and 12 months] from 151 infants were used. Fecal samples were collected by a parent or guardian from the infant's diaper, and placed in a provided container, which was placed in a freezer bag and stored at −20°C in a home freezer until the hospital visit (29). At the hospital, samples were placed in a −20°C freezer and stored until transport on dry ice to the Netherlands Organization for Applied Scientific Research (TNO) for analysis.
DNA from fecal samples was isolated and V4 hypervariable region of the bacterial 16S rRNA gene was amplified with universal bacterial 515F−806R primer pair as described (6). Prepared libraries were sequenced on the paired-end 2 × 200 bases Illumina MiSeq platform with a MiSeq Reagent Kit v2 (MS-102-2003, Illumina).
Paired sequence reads from 453 selected samples (BF n = 51, SF n = 50, F19 n = 50) and 48 quality control samples were pre-processed in QIIME2 (version 2019.4; https://qiime2.org/) as described (6). The following subject's samples were excluded from sequencing analyses: (1) one subject in the F19 group identified as quantitative-PCR negative for the F19 strain at 4 months; (2) 13 non-exclusive breastfed infants in the BF group; and (3) infants treated with antibiotics during the intervention (3 BF, 9 SF, and 6 F19), leaving a total of 37 BF, 43 F19, and 41 SF. Raw sequence and processed files have been deposited through QIITA (study ID: 12874) in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB38295 (https://www.ebi.ac.uk/ena/browser/view/PRJEB38295).
Statistical Analysis
To determine associations in the microbiota with diet, principal coordinate analysis (PCoA) was performed. Comparisons for beta diversity metric estimates were made by computing pairwise Permutational Multivariate Analysis of Variance (PERMANOVA) (56) based on 999 permutations. Kruskal-Wallis tests were used for comparing alpha diversity estimates between categorical variables. Longitudinal analyses were performed on changes in alpha diversity estimates over time using paired-difference testing with the q2-longitudinal plugin in QIIME2, followed by visualization using qiime2R (ver. 0.99.1) and ggplot2 (ver. 3.1.1) packages in R. Taxonomic assignments of representative sequences were conducted for 515F/806R primer pair through a naïve-Bayes classifier trained on the Greengenes ver. 13_8 database at 99% OTU similarities. Differential abundance analysis of each taxon was determined using ANCOM (Analysis of composition of microbiomes) (57). Taxa bar plots were generated using vegan (ver. 2.5-5) and ggplot2 packages in R.
For metabolomics, statistical computation was performed in R (ver. 3.5.3.) and visualized using the ggplot2 package unless stated otherwise. Principal component analysis (PCA) (prcomp function; mean-centered, non-scaled, and log-transformed data) was used to visualize the data. Groups were compared using pairwise PERMANOVA on Euclidean distances. Two-way Analysis of Variance (ANOVA, aov function; log-transformed data) were computed to test for significant effects of diet, sex, region, hospital, and mode of delivery as previously described (6). The magnitude of the diet effect on the metabolome was estimated by calculating Cliff's Delta effect size estimates (cliff.delta function).
Data Availability Statement
The original contributions presented in the study are publicly available. The microbiome data can be found at: ENA, PRJEB38295 (https://www.ebi.ac.uk/ena/browser/view/PRJEB38295?show=reads). The metabolome data supporting the conclusions of this article will be made available on request by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board (IRB) at the University of California Davis and the Regional Ethical Review Boards in Nanjing, Shanghai, and Beijing in China (ClinicalTrials.gov Identifier: NCT01755481). Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
ZL, BC, YP, XL, OH, and BL designed and ZL, YP, and XL performed the clinical trial. HL analyzed samples for metabolomics and performed all metabolome, microbiota, and subject data analyses. Manuscript written by HL and CS. OH and BL edited, and all authors approved the final manuscript.
Funding
The authors declare that this study received funding from Arla Foods amba. The funder was involved in the study design, but not in the conduct of the study, collection of samples, analysis or interpretation of the data, nor in the writing of this article or the decision to submit it for publication. This work was also supported by the USDA National Institute of Food and Agriculture Hatch project 1021411, and the Kinsella Endowed Chair in Food, Nutrition, and Health (to CMS). The 600 MHz NMR is supported through NIH grant 1S10RR011973-01.
Conflict of Interest
BC is employed by Arla Foods amba. BL and OH have served as consultants for Arla Foods amba.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge and thank Jos. M.B.M. van der Vossen, Anita M.T. Ouwens, and Hakim Rahaoui from TNO for performing DNA extraction and 16S rRNA gene sequencing for the fecal samples.
References
1. Collado MC, Cernada M, Baüerl C, Vento M, Pérez-Martínez G. Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes. (2012) 3:352–65. doi: 10.4161/gmic.21215
2. Needham BD, Kaddurah-Daouk R, Mazmanian SK. Gut microbial molecules in behavioural and neurodegenerative conditions. Nat Rev Neurosci. (2020) 21:717–31. doi: 10.1038/s41583-020-00381-0
3. He X, Parenti M, Grip T, Lönnerdal B, Timby N, Domellöf M, et al. Fecal microbiome and metabolome of infants fed bovine MFGM supplemented formula or standard formula with breast-fed infants as reference: a randomized controlled trial. Sci Rep. (2019) 9:11589. doi: 10.1038/s41598-019-47953-4
4. He X, Slupsky CM, Dekker JW, Haggarty NW, Lönnerdal B. Integrated role of Bifidobacterium animalis subsp. lactis supplementation in gut microbiota, immunity, and metabolism of infant rhesus monkeys. mSystems. (2016) 1:e00128–16. doi: 10.1128/mSystems.00128-16
5. He X, Sotelo-Orozco J, Rudolph C, Lonnerdal B, Slupsky CM. The role of protein and free amino acids on intake, metabolism, and gut microbiome: a comparison between breast-fed and formula-fed rhesus monkey infants. Front Pediatr. (2019) 7:563. doi: 10.3389/fped.2019.00563
6. Lee H, Slupsky CM, Heckmann AB, Christensen B, Peng Y, Li X, et al. Milk fat globule membrane as a modulator of infant metabolism and gut microbiota: a formula supplement narrowing the metabolic differences between breastfed and formula-fed infants. Mol Nutr Food Res. (2020) 65:e2000603. doi: 10.1002/mnfr.202000603
7. O'Sullivan A, He X, McNiven EMS, Haggarty NW, Lönnerdal B, Slupsky CM. Early diet impacts infant rhesus gut microbiome, immunity, and metabolism. J Proteome Res. (2013) 12:2833–45. doi: 10.1021/pr4001702
8. Roggero P, Liotto N, Pozzi C, Braga D, Troisi J, Menis C, et al. Analysis of immune, microbiota and metabolome maturation in infants in a clinical trial of Lactobacillus paracasei CBA L74-fermented formula. Nat Commun. (2020) 11:2703. doi: 10.1038/s41467-020-16582-1
9. He X, Parenti M, Grip T, Domellöf M, Lönnerdal B, Hernell O, et al. Metabolic phenotype of breast-fed infants, and infants fed standard formula or bovine MFGM supplemented formula: a randomized controlled trial. Sci Rep. (2019) 9:339. doi: 10.1038/s41598-018-36292-5
10. Prentice P, Koulman A, Matthews L, Acerini CL, Ong KK, Dunger DB. Lipidomic analyses, breast- and formula-feeding, and growth in infants. J Pediatr. (2015) 166:276–81. doi: 10.1016/j.jpeds.2014.10.021
11. Slupsky CM, He X, Hernell O, Andersson Y, Rudolph C, Lönnerdal B, et al. Postprandial metabolic response of breast-fed infants and infants fed lactose-free vs regular infant formula: a randomized controlled trial. Sci Rep. (2017) 7:3640. doi: 10.1038/s41598-017-03975-4
12. Timby N, Hernell O, Vaarala O, Melin M, Lönnerdal B, Domellöf M. Infections in infants fed formula supplemented with bovine milk fat globule membranes. J Pediatr Gastroenterol Nutr. (2015) 60:384–9. doi: 10.1097/MPG.0000000000000624
13. Rito AI, Buoncristiano M, Spinelli A, Salanave B, Kunešová M, Hejgaard T, et al. Association between characteristics at birth, breastfeeding and obesity in 22 countries: the WHO European childhood obesity surveillance initiative-COSI 2015/2017. Obes Facts. (2019) 12:226–43. doi: 10.1159/000500425
14. Stuebe A. The risks of not breastfeeding for mothers and infants. Rev Obstet Gynecol. (2009) 2:222–31.
15. Deshmukh M, Patole S. Prophylactic probiotic supplementation for preterm neonates-A systematic review and meta-analysis of nonrandomized studies. Adv Nutr. (2021) 12:1411–23. doi: 10.1093/advances/nmaa164
16. Underwood MA. Impact of probiotics on necrotizing enterocolitis. Sem Perinatol. (2017) 41:41–51. doi: 10.1053/j.semperi.2016.09.017
17. Alcon-Giner C, Dalby MJ, Caim S, Ketskemety J, Shaw A, Sim K, et al. Microbiota supplementation with Bifidobacterium and Lactobacillus modifies the preterm infant gut microbiota and metabolome: an observational study. Cell Rep Med. (2020) 1:100077. doi: 10.1016/j.xcrm.2020.100077
18. Martí M, Spreckels JE, Ranasinghe PD, Wejryd E, Marchini G, Sverremark-Ekström E, et al. Effects of Lactobacillus reuteri supplementation on the gut microbiota in extremely preterm infants in a randomized placebo-controlled trial. Cell Rep Med. (2021) 2:100206. doi: 10.1016/j.xcrm.2021.100206
19. van Best N, Trepels-Kottek S, Savelkoul P, Orlikowsky T, Hornef MW, Penders J. Influence of probiotic supplementation on the developing microbiota in human preterm neonates. Gut Microbes. (2020) 12:16. doi: 10.1080/19490976.2020.1826747
20. Grześkowiak Ł, Grönlund M-M, Beckmann C, Salminen S, Berg Av, Isolauri E. The impact of perinatal probiotic intervention on gut microbiota: Double-blind placebo-controlled trials in Finland and Germany. Anaerobe. (2012) 18:7–13. doi: 10.1016/j.anaerobe.2011.09.006
21. Morelli L, Campominosi E. Genetic stability of Lactobacillus paracasei subsp. paracasei F19. Microb Ecol Health Dis. (2009) 14:14–6. doi: 10.1080/089106002760003297
22. Crittenden R, Saarela M, Mättö J, Ouwehand AC, Salminen S, Pelto L, et al. Lactobacillus paracasei subsp. paracasei F19: Survival, ecology and safety in the human intestinal tract - A survey of feeding studies within the PROBDEMO project. Microb Ecol Health Dis. (2009) 14:22–6. doi: 10.1080/089106002760003314
23. Di Cerbo A, Palmieri B. Lactobacillus paracasei subsp. paracasei F19; a farmacogenomic and clinical update. Nutr Hosp. (2013) 28:1842–50. doi: 10.3305/nh.2013.28.6.6831
24. Garrote GL, Abraham AG, Rumbo M. Is lactate an undervalued functional component of fermented food products? Front Microbio. (2015) 6:629. doi: 10.3389/fmicb.2015.00629
25. Lombardo L, Blanco I. Clinical evaluation of treatment with Lactobacillus paracasei subsp paracasei F19 in IBS patients. Digest Liver Dis. (2008) 40:S187–S. doi: 10.1016/S1590-8658(08)60501-0
26. Lombardo L, Vernetto A, Blanco I. Clinical evaluation of Lactobacillus paracasei subsp. paracasei F19 with gluco-oligosaccharides in the short-term treatment of irritable bowel syndrome. Microb Ecol Health Dis. (2009) 21:28–32. doi: 10.1080/08910600802610815
27. Zampieri N, Pietrobelli, A, Biban, P, Soffiati, M, Dall'agnola, A, Camoglio, FS,. Lactobacillus paracasei subsp. paracasei F19 in Bell's stage 2 of necrotizing enterocolitis. Minerva Pediatr. (2013) 65:353–60. Available online at: https://www.minervamedica.it/en/journals/minerva-pediatrics/article.php?cod=R15Y2013N04A0353#
28. West CE, Gothefors L, Granström M, Käyhty H, Hammarström M-LKC, Hernell O. Effects of feeding probiotics during weaning on infections and antibody responses to diphtheria, tetanus and Hib vaccines. Pediatr Allergy Immunol. (2008) 19:53–60. doi: 10.1111/j.1399-3038.2007.00583.x
29. Li X, Peng Y, Li Z, Christensen B, Heckmann AB, Stenlund H, et al. Feeding infants formula with probiotics or milk fat globule membrane: a double-blind, randomized controlled trial. Front Pediatr. (2019) 7:503S. doi: 10.3389/fped.2019.00347
30. Li X, Peng Y, Li Z, Christensen B, Heckmann AB, Lagerqvist C, et al. Serum cytokine patterns are modulated in infants fed formula with probiotics or milk fat globule membranes: a randomized controlled trial. PLoS ONE. (2021) 16:e025. doi: 10.1371/journal.pone.0251293
31. Ruiz-Moyano S, Totten SM, Garrido DA, Smilowitz JT, German JB, Lebrilla CB, et al. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl Environ Microbiol. (2013) 79:6040–9. doi: 10.1128/AEM.01843-13
32. Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun. (2016) 7:11939. doi: 10.1038/ncomms11939
33. Garrido D, Ruiz-Moyano S, Kirmiz N, Davis JC, Totten SM, Lemay DG, et al. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596 .Sci Rep. (2016) 6:35045. doi: 10.1038/srep35045
34. Almeida CC, Mendonça Pereira BF, Leandro KC, Costa MP, Spisso BF, Conte-Junior CA. Bioactive compounds in infant formula and their effects on infant nutrition and health: a systematic literature review. Int J Food Sci. (2021) 2021:8850080. doi: 10.1155/2021/8850080
35. Sun S, Chang G, Zhang L. The prevention effect of probiotics against eczema in children: an update systematic review and meta-analysis. J Dermatolog Treat. (2021) 1–11. doi: 10.1080/09546634.2021.1925077
36. Tan-Lim CSC, Esteban-Ipac NAR, Recto MST, Castor MAR, Casis-Hao RJ, Nano ALM. Comparative effectiveness of probiotic strains on the prevention of pediatric atopic dermatitis: a systematic review and network meta-analysis. Pediatr Allergy Immunol. (2021) 32:1255–70. doi: 10.1111/pai.13514
37. Tremblay A, Xu X, Colee J, Tompkins TA. Efficacy of a multi-strain probiotic formulation in pediatric populations: a comprehensive review of clinical studies. Nutrients. (2021) 13:1908. doi: 10.3390/nu13061908
38. Alemao CA, Budden KF, Gomez HM, Rehman SF, Marshall JE, Shukla SD, et al. Impact of diet and the bacterial microbiome on the mucous barrier and immune disorders. Allergy. (2021) 76:714–34. doi: 10.1111/all.14548
39. Botía-Sánchez M, Alarcón-Riquelme ME, Galicia G. B cells and microbiota in autoimmunity. IJMS. (2021) 22:4846. doi: 10.3390/ijms22094846
40. Cristofori F, Dargenio VN, Dargenio C, Miniello VL, Barone M, Francavilla R. Anti-inflammatory and immunomodulatory effects of probiotics in gut inflammation: a door to the body. Front Immunol. (2021) 12:578386. doi: 10.3389/fimmu.2021.578386
41. Henrick BM, Chew S, Casaburi G, Brown HK, Frese SA, Zhou Y, et al. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr Res. (2019) 86:749–57. doi: 10.1038/s41390-019-0533-2
42. Henrick BM, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. (2021) 184:3884–98.e11. doi: 10.1016/j.cell.2021.05.030
43. Lim HJ, Shin HS. Antimicrobial and immunomodulatory effects of Bifidobacterium strains: a review. J Microbiol Biotechnol. (2020) 30:1793–800. doi: 10.4014/jmb.2007.07046
44. Rhoads JM, Fatheree NY, Norori J, Liu Y, Lucke JF, Tyson JE, et al. Altered fecal microflora and increased fecal calprotectin in infants with colic. J Pediatr. (2009) 155:823–8.e1. doi: 10.1016/j.jpeds.2009.05.012
45. Tourkochristou E, Triantos C, Mouzaki A. The influence of nutritional factors on immunological outcomes. Front Immunol. (2021) 12:665968. doi: 10.3389/fimmu.2021.665968
46. Zouali M. B lymphocytes, the gastrointestinal tract and autoimmunity. Autoimmun Rev. (2021) 20:102777. doi: 10.1016/j.autrev.2021.102777
47. Shade A. Diversity is the question, not the answer. ISME J. (2017) 11:1–6. doi: 10.1038/ismej.2016.118
48. Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat Commun. (2018) 9:4169. doi: 10.1038/s41467-018-06473-x
49. Wicinski M, Sawicka E, Gebalski J, Kubiak K, Malinowski B. Human milk oligosaccharides: health benefits, potential applications in infant formulas, and pharmacology. Nutrients. (2020) 12:266. doi: 10.3390/nu12010266
50. Vieco-Saiz N, Belguesmia Y, Raspoet R, Auclair E, Gancel F, Kempf I, et al. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front Microbio. (2019) 10:57. doi: 10.3389/fmicb.2019.00057
51. West CE, Hernell O, Andersson Y, Sjöstedt M, Hammarström M-LKC. Probiotic effects on T-cell maturation in infants during weaning. Clin Exp Allergy. (2012) 42:540–9. doi: 10.1111/j.1365-2222.2011.03941.x
52. Jäger R, Zaragoza J, Purpura M, Iametti S, Marengo M, Tinsley GM, et al. Probiotic administration increases amino acid absorption from plant protein: a placebo-controlled, randomized, double-blind, multicenter, crossover study. Probiotics Antimicrob Proteins. (2020) 12:1330–9. doi: 10.1007/s12602-020-09656-5
53. Martin F-PJ, Wang Y, Sprenger N, Yap IKS, Lundstedt T, Lek P, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. (2008) 4:157. doi: 10.1038/msb4100190
54. Sieuwerts S, Håkansson J. Development of a standardized method for the quantification of Lactobacillus Paracasei F19 In stool samples of various ages. EC Nutrition. (2016) 3:633–42. Available online at: https://www.ecronicon.com/ecnu/nutrition-ECNU-03-000084.php
55. Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. (2006) 78:4430–42. doi: 10.1021/ac060209g
56. Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. (2001) 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Keywords: gut microbiota, metabolome, infant formula, Lactobacillus, probiotics
Citation: Lee H, Li Z, Christensen B, Peng Y, Li X, Hernell O, Lönnerdal B and Slupsky CM (2022) Metabolic Phenotype and Microbiome of Infants Fed Formula Containing Lactobacillus paracasei Strain F-19. Front. Pediatr. 10:856951. doi: 10.3389/fped.2022.856951
Received: 17 January 2022; Accepted: 16 March 2022;
Published: 26 April 2022.
Edited by:
Ana Isabel Lopes, University Hospital Center Lisbon Norte, PortugalReviewed by:
Yuying Liu, University of Texas Health Science Center at Houston, United StatesBethany M. Henrick, Evolve Biosystems, INC., United States
Copyright © 2022 Lee, Li, Christensen, Peng, Li, Hernell, Lönnerdal and Slupsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carolyn M. Slupsky, cslupsky@ucdavis.edu
 Hanna Lee
Hanna Lee Zailing Li
Zailing Li Britt Christensen3
Britt Christensen3  Xiaonan Li
Xiaonan Li Olle Hernell
Olle Hernell Bo Lönnerdal
Bo Lönnerdal Carolyn M. Slupsky
Carolyn M. Slupsky