- 1Key Laboratory of Carcinogenesis, National Health Commission, Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, China
- 2Cancer Research Institute, Central South University, Changsha, China
- 3The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Central South University, Changsha, China
- 4Hunan Key Laboratory of Oncotarget Gene, Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, China
- 5The First Clinical College of Changsha Medical University, Changsha, China
Yin-Yang 1 (YY1) is a member of the GLI-Kruppel family of zinc finger proteins and plays a vital dual biological role in cancer as an oncogene or a tumor suppressor during tumorigenesis and tumor progression. The tumor microenvironment (TME) is identified as the “soil” of tumor that has a critical role in both tumor growth and metastasis. Many studies have found that YY1 is closely related to the remodeling and regulation of the TME. Herein, we reviewed the expression pattern of YY1 in tumors and summarized the function and mechanism of YY1 in regulating tumor angiogenesis, immune and metabolism. In addition, we discussed the potential value of YY1 in tumor diagnosis and treatment and provided a novel molecular strategy for the clinical diagnosis and treatment of tumors.
1 Introduction
Within the tumor microenvironment (TME), cancer cells coexist with noncancerous adjacent cells and components that constitute the TME and impact tumor growth and progression through diverse mechanisms. In addition to cancer cells, the TME comprises immune cells, fibroblasts, bone marrow-derived inflammatory cells, as well as noncellular components such as abnormal tumor blood vessels, signaling molecules, and extracellular matrix (1, 2). As the forces of the tumor cells grow, the microenvironment that originally suppressed tumor growth and protected the survival of normal cells has also been gradually transformed into a place suitable for tumor cells to survive. Furthermore, the microenvironment provides essential nutrients, amino acids, and nucleotides to the tumor, facilitating tumor growth. At the same time, it forms a protective barrier against the pernicious effect of immune cells and drugs during tumor treatment (3). Yin-Yang 1 (YY1) is a ubiquitous transcription factor with multiple roles in tumorigenesis, functioning as both a transcriptional activator and repressor (4, 5). Moreover, its function depends on its interacting partners, promoter environment, and chromatin structure (6, 7), thereby regulating transcriptional activation and repression of approximately 10% of the total human gene set associated with a variety of cellular biological processes (8, 9). Several recent reports suggest that YY1 mostly acts as an oncogene in most types of cancers but also acts as a tumor suppressor in some other types of cancer (10–13). Therefore, this review focuses on the expression patterns of YY1 in different tumors, its influence on tumor angiogenesis, tumor metabolism and the tumor immune microenvironment, and its potential value in tumor diagnosis and treatment. This review expands the understanding of the roles and mechanisms of YY1 in the TME and provides a novel molecular strategy for the clinical diagnosis and treatment of tumors.
2 Expression of YY1 in different tumors
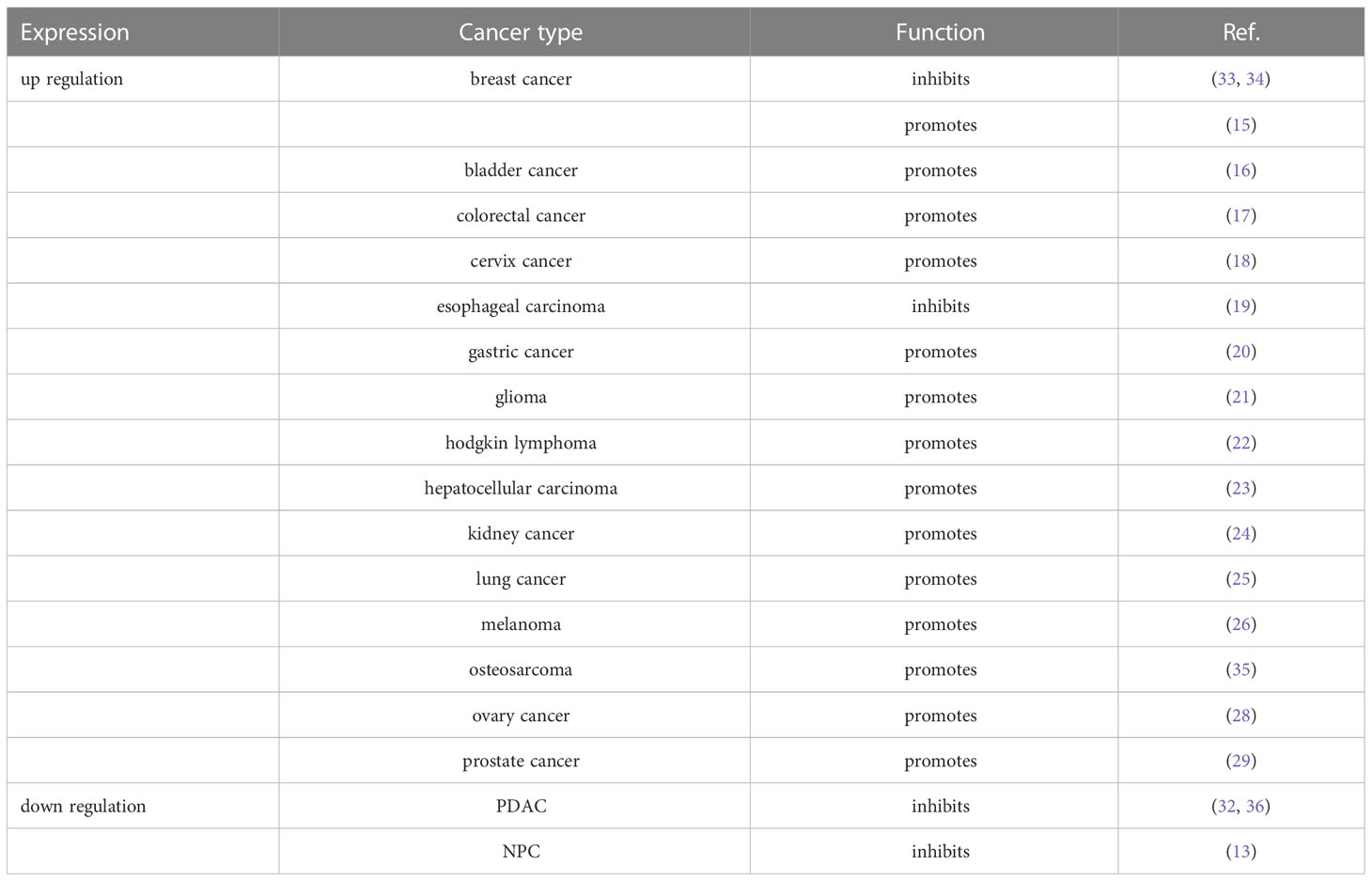
YY1 is widely expressed in various tissues, and its upregulation in different types of tumors shows different clinical implications. To investigate the differential expression of YY1 in different types of tumors, studies have analyzed GEO datasets and found that YY1 has high mRNA expression in most tumor tissues (14). Likewise, several studies have shown that YY1 is highly expressed in breast cancer (15), bladder cancer (16), colorectal cancer (17), cervix cancer (18), esophageal carcinoma (19), gastric cancer (20), glioma (21), hodgkin lymphoma (22), hepatocellular carcinoma (23), kidney cancer (24), lung cancer (25), melanoma (26), osteosarcoma (27), ovary cancer (28), and prostate cancer (29). In addition, in the studies evaluated for all paired primary and metastatic samples, YY1 expression appeared to be increased in metastatic tissues compared with matched primary tumor tissues. These results indicate that YY1 is closely related to tumor invasion and metastasis and is most likely involved in regulating the TME. More intriguingly, In osteosarcoma. YY1 presents lower expression levels in pediatric osteosarcomas compared to normal human osteoblasts (30). In addition, YY1 is also not always highly expressed in tumors, such as melanoma, YY1 expression is significantly lower in metastatic melanoma than in normal melanocytes (31). Furthermore, we have also recently revealed an additional contribution of YY1 in nasopharyngeal carcinoma (NPC). Compared with the expression of YY1 in normal NPE tissues and NPC tissues, most patients exhibited a significant decrease, and its expression negatively correlated with increasing clinical TNM stage and positively correlated with the OS rate with NPC (13). It is same with pancreatic ductal adenocarcinoma (PDAC), several reports indicate that YY1 is downregulated in PDAC tissues compared with adjacent normal pancreatic tissues, and the high expression of YY1 represents a better prognosis in patients with PDAC (32). These results suggest that YY1 has two distinct expression patterns (high or low) in different tumor types, and playing a dual role as an oncogene or tumor suppressor to facilitate tumorigenesis or tumor progression (Table 1).
3 Role of YY1 in the tumor microenvironment
3.1 YY1 in tumor angiogenesis
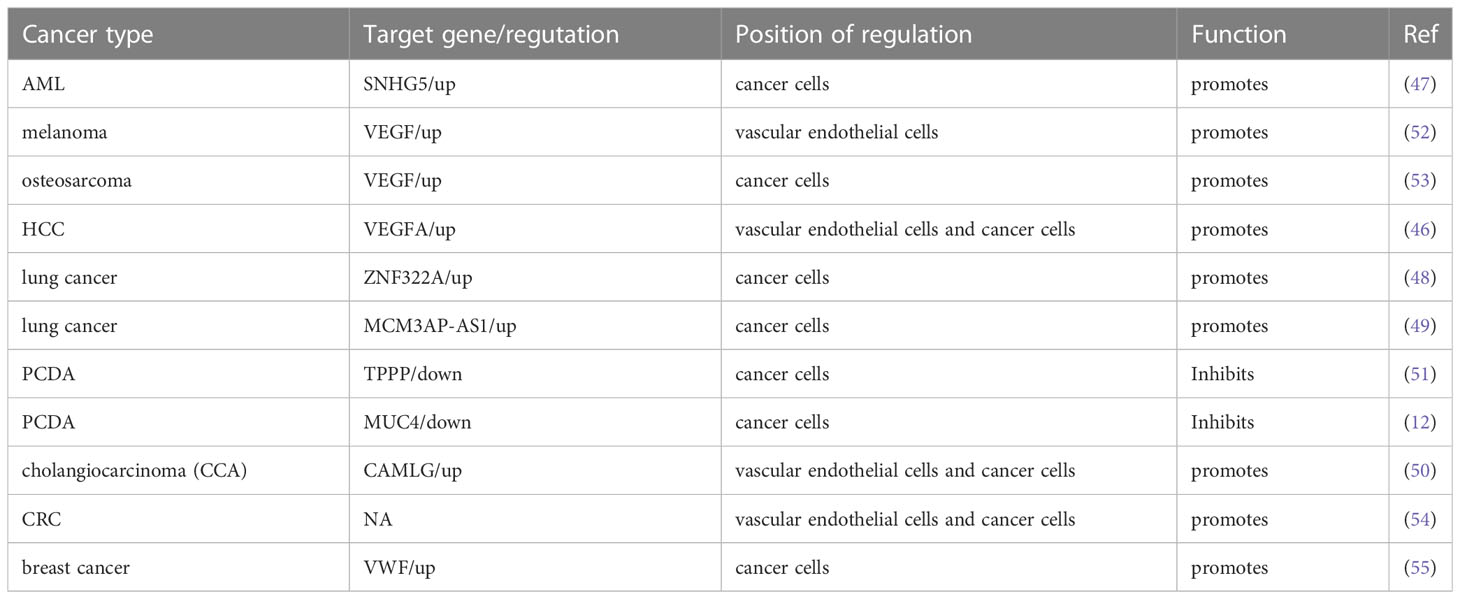
Tumor vasculature is a critical component of the TME, which is the critical channel of oxygen and nutrient delivery for maintaining tumor growth but is also a pathway of tumor cell metastasis (37). Due to insufficiencies in the tumor vasculature, tumor cells are often exposed to a hostile TME with low nutrient and oxygen levels (38). Tumor cells secrete angiogenic factors to induce neovascularization to satisfy tumor cell oxygen and nutrient requirements (39). Vascular endothelial growth factor A (VEGFA) is the most critical and primary component of the VEGF family, which is a multipurpose cytokine active on blood vessel cells (40). Through a paracrine mechanism induces tumor vascularization to meet the increased requirement of oxygen and nutrients and remove metabolic wastes (41, 42). VEGF is usually regulated by HIF-1α or the CXCR4/SDF-1 axis in tumors (43). The molecular mechanism between the YY1 and VEGF was discovered in multiple studies. De et al. found that YY1 was a critical component of the HIF-1α complex and binds its target sequences on the regulatory regions of VEGFA, -B, and -C to upregulates the transcriptional activity and expression of them (44). In addition, studies have shown that the expression of YY1 was upregulated in the malignancy of osteosarcoma, and silencing the expression of YY1 can significantly downregulate the activation of the VEGF/CXCR4 axis, thereby inhibiting the angiogenesis, invasion and metastasis (45). HCC is a well-known typical angiogenesis-dependent solid tumor with rich blood vessels, and YY1 is reported to be involved in regulating tumor malignancy in HCC. Yang et al. found that microvessel density (MVD) was positively correlated with YY1 and poor prognosis in HCC, and overexpression of YY1 promoted VEGFA transcriptional activity by binding to the VEGFA promoter (46). These results highlight a new mechanism by which YY1 plays an essential role though inducing VEGFA transcription in HCC angiogenesis. In addition to direct regulation, the indirect regulation between YY1 and VEGF has gradually been discovered. Li et al. found that YY1 contributes to angiogenesis via SNHG5/miR-26b/CTGF/VEGFA axis in AML (47). Moreover, a VEGF-independent mechanism to promote angiogenesis was discovered. In the presence of KRAS mutations, YY1 promoted neovascularization by targeting the ZNF322A promoter to increase its expression. It is significant to note that ZNF322A increases the sonic hedgehog (Shh) expression, which encodes a secreted factor that turns on pro-angiogenic responses in endothelial cells. According to these results, disruption of the Kras/YY1/ZNF322A/Shh transcriptional axis encourages lung cancer neoangiogenesis and cancer development (48). Not only protein-encoding genes but YY1 can target the transcription of lncRNA MCM3AP-AS1, further targeting the miR-340-5p/KPNA4 axis stimulated lung cancer angiogenesis and progression (49). Similarly, in cholangiocarcinoma, it is also found that the Circ-CCAC1/miR-514a-5p/YY1/CAMLG axis disrupts endothelial barrier integrity and induces angiogenesis (50).
YY1 plays dual biological roles in the initiation and progression of various cancers. Likewise, YY1 exhibits promote or inhibit angiogenesis. Several previous studies have reported that YY1 plays a tumor suppressor role in PCDA. For example, YY1 inhibited angiogenesis by downregulating the TPPP-mediated p38/MAPK and PI3K/AKT pathways opening a new horizon in PCDA research. ChIP sequencing results showed that YY1 directly binds to the promoter region of TPPP and thus inhibits pancreatic cancer cell migration, invasion and angiogenesis (51). Beyond that, latterly the discovery of YY1 suppressed the invasion and metastasis of PCDA cells by downregulating the expression of MMP10 through the MUC4/ErbB2/p38/MEF2C regulatory axis (12, 32). Overall, this is strong evidence that YY1 plays an inhibitory role in neo-angiogenesis.
In general, existing evidence indicates that YY1 acts as a double-edged sword that alters the TME by promoting or inhibiting angiogenesis in different tumor types (Figure 1), where some of the position of YY1 regulating target genes related to angiogenesis occur in cancer cells, which indirectly affected the formation of blood vessels by regulating the expression of angiogenesis related factors. Some regulation positions take place in vascular endothelial cells and directly affects the formation of blood vessels. The effect and mechanism of YY1 on angiogenesis in various tumors are shown in Table 2.
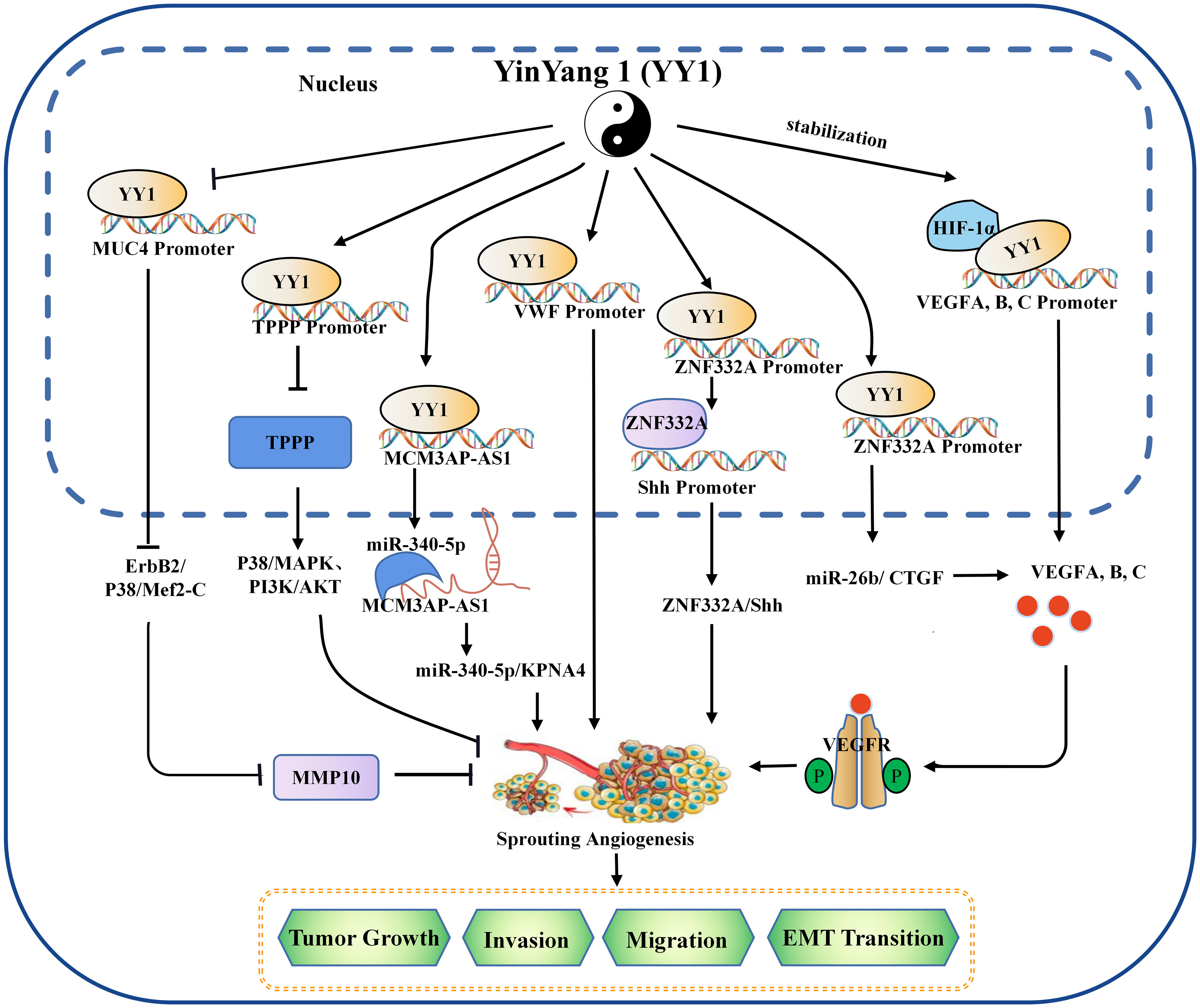
Figure 1 The mechanism of YY1 regulating angiogenesis in the tumor microenvironment. YY1 regulates the expression of crucial proteins associated with tumor neovascularization (such as SNHG5, VWF, MUC4, ZNF332a, TPPP, MCM3AP-AS1, VEGF family, et al.), by interacting with their promoters, thus to regulate the proliferation, migration, invasion, and EMT transition of cancer cells in an oncogene or tumor suppressor role. YY1, Yin Yang 1; HIF-1α, hypoxia-inducible factor 1-α; VEGF, vascular endothelial growth factor; VEGFA, vascular endothelial growth factor A; TPPP, tubulin polymerization promoting protein; MMP 10, matrix metallopeptidase 10; EMT, epithelial-mesenchymal transition.
3.2 YY1 in immunity
Human cells are constantly under surveillance by the immune system. The TME is the main battleground between tumor cells and the host immune system, and tumors can escape immune surveillance by initiating various immunosuppressive cells or establishing an immunosuppressive microenvironment (56, 57). Recent studies have shown that YY1 regulates broad general processes throughout all stages of T-cell and B-cell differentiation (58, 59).
3.2.1 T cells
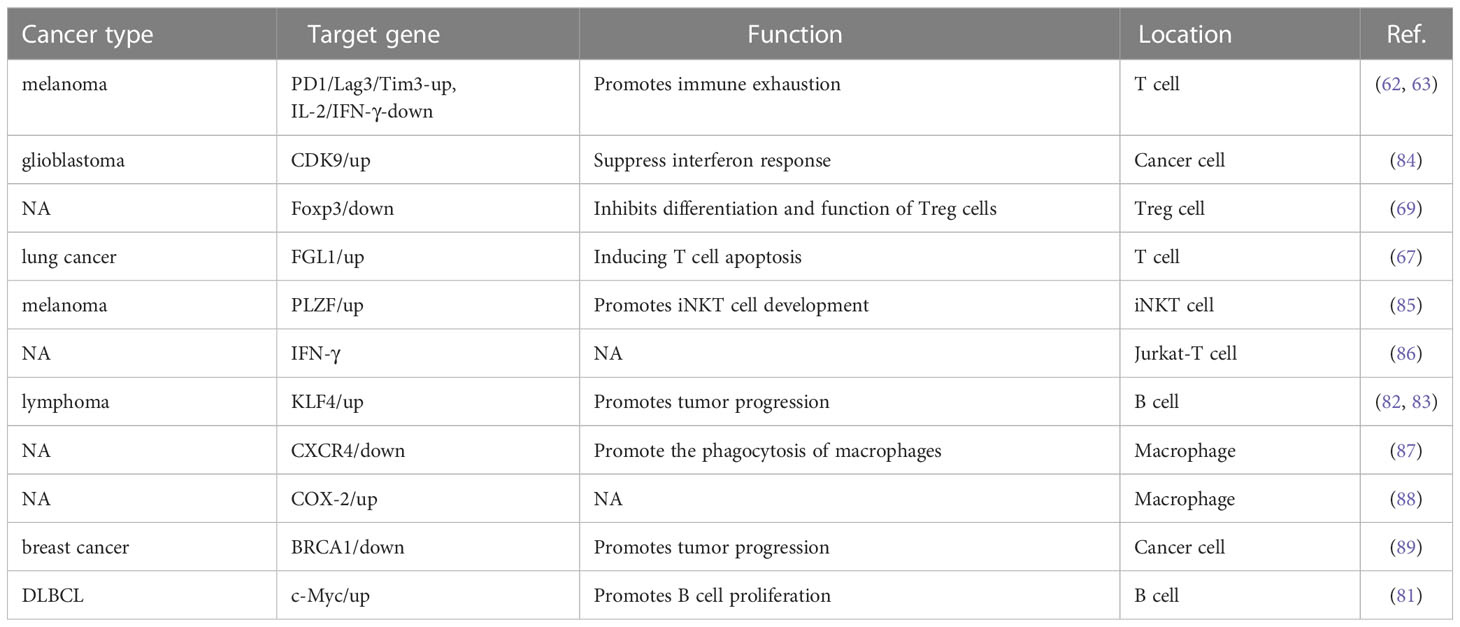
Immune checkpoint activation is a critical mechanism by which tumor cells evade clearance by immune cells. In the TME, where tumor cells are located, the surface antigens of tumor cells cannot be presented to T cells, resulting in tumor cells evading immune killing. Programmed death receptor-1 (PD-1) and cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) are the most common immune checkpoint receptors expressed on the surface of T lymphocytes (60). YY1 has been shown to positively regulate immune checkpoint receptors such as PD1 and CTLA-4 (61), and YY1 was upregulated in PD1-positive T cells infiltrating lymphocytes in melanoma tumors and directly regulated the expression of PD1 and LAG3 by binding to the promoter region (62, 63). PD-1 is a critical receptor expressed on activated T cells that inhibits T cell-mediated immune responses upon binding to its ligand programmed cell death ligand 1 (PD-L1) (64). YY1 can increase PD-L1 expression through various signaling pathways. As a negative regulator of p53, YY1 inhibits the expression of miR-34a downstream of p53, while downregulation of miR-34a expression leads to an increase of PD-L1 expression by binding to the PD-L1 3’UTR (65). YY1 has also been shown to promote PD-L1 expression by downregulating PTEN via p53 and activating the PI3K/Akt/mTOR pathway (66). YY1 has been reported to be associated with T cell apoptosis. FGL1 promotes the secretion of interleukin-2 by T cells and induces their apoptosis. Indeed, YY1 is the upstream molecule of FGL1, which was found to be transcriptionally regulated by YY1 (67). These results suggest that YY1 may hinder T-cell-mediated tumor immunotherapy by interfering PD-1/PD-L1 checkpoint.
As the tumor progresses, the TIME is constantly changing, and the speed of tumor development depends on the proportion and characteristics of T cells within the TIME. As early as ten years ago, evidence in the field of tumor immunity suggested that the TME limits the accumulation of T cells among tumor cells. Regulatory T (Treg) cells are essential for maintaining immune homeostasis (60, 68). Hwang et al. showed that the expression of YY1 in Treg cells was lower than that in Tconv cells, and the overexpression of YY1 led to a significant decrease in the expression of Foxp3, making the suppressive function of Treg cells ineffective (69). As one of the most concerned phenotypes in the CD4 T cell family, Treg cells are an immunosuppressive subtype in nature, but with a dual personality as an angel side, Treg cells can regulate the immune system, inhibit the pathological immune response caused by the overactivation of autoreactive T cells, and control inflammation. It plays a key role in maintaining autoimmune tolerance and immune homeostasis. However, in the tumor microenvironment, it shows a devil side. It inhibits the proliferation, metabolism and killing function of CD8 T cells by secreting inhibitory factors such as IL-10, TGF-β and affecting the maturation of dendritic cells (DC). It induces the immune escape of tumor cells by inhibiting the anti-tumor immune response, thus promoting the growth and proliferation of tumors (70). Foxp3 is the critical transcription factor for Treg cell function and differentiation (71). YY1 physically interacts with Foxp3 to directly switch on Foxp3 target genes and interfere with Foxp3-dependent target gene expression. Thus, YY1 inhibits the Treg cell’s differentiation and function by blocking Foxp3 (69, 72). YY1 is a polycomb group (PcG) protein and an original member of the local group of sequence-specific DNA-binding PcG proteins in mammals (73). PcG proteins silence target genes through histone modification, especially methylation modification (74). By contrast, YY1 promotes early T cell survival through its PcG function. In the thymus, T cell development begins when double negative (DN) progenitor T cells lacking CD4 and CD8 expression become CD4+CD8+ double positive pre-T cells (DP) and then differentiate into mature single CD4+ or CD8+ positive cells, DN cells have four stages (75–77). Ana et al. demonstrated that YY1 promotes DN1 to DN2 T cell transition and early T cell survival independent of PcG/REPO domain function (78). Overall, YY1 is crucial for T-cell differentiation, function, and development, but YY1 has opposite effects on T-cell development and function (Figure 2).
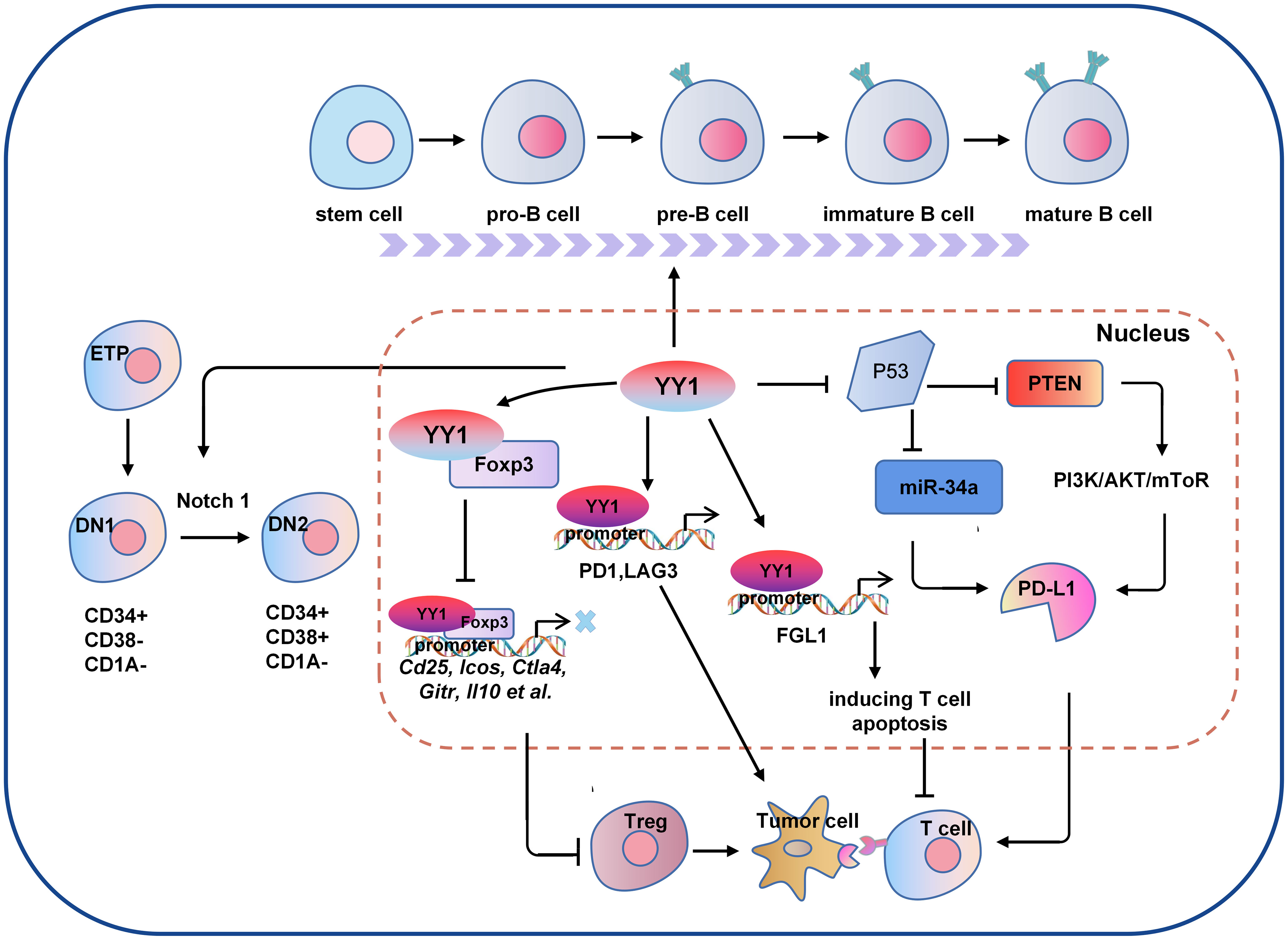
Figure 2 The molecular mechanism of YY1 regulating B cell and T cell-mediated tumor immunity. Several signaling pathways crosstalk exist between the regulations of YY1 and T cell function. For example, YY1 inhibits the function of T cells by blocking Foxp3 mediated transcription of its downstream genes Cd25, Icos, Ctla4, Gitr and Il10 et al., or directly regulating the expression of PD1 and LAG3 by binding to their promoter regions. In addition, YY1 promotes the DN1-to-DN2 T cell transition by activating Notch1 pathway. For B cells, YY1 is essential for all stages of B cell differentiation, thus to be involved in B cells-mediated immune escape. DN1, double negative 1; DN2, double negative 2; PD-L1: programmed death ligand 1; Foxp3, forkhead box p3.
3.2.2 B cells
B lymphocytes are derived from bone marrow hematopoietic stem cells through a series of differentiation stages. Activated B cells produce many cytokines involved in immune regulation, the inflammatory response and hematopoiesis (79). YY1 plays an important role in Ig rearrangement and B-cell development. Studies have shown that conditional knockdown of YY1 prevented the transition of pro-B cells to cells in the pre-B-cells and early developmental stages of B cells (58). Subsequent proof found that YY1 regulates the germinal center B-cells (GC B cells) transcriptional program (59). In specific deletion of YY1 in GC B cells, B-cell numbers in both unimmunized and immunized conditions are severely reduced. In addition, YY1 also affects the immunoglobulin class switch recombination (CSR) process in splenic B cells. These studies strongly suggest that YY1 is essential for B cell differentiation in all stages and is required for the survival and proliferation of B cells (Figure 2), B-NHL is a heterogeneous lymphoma derived from a B-cell mutation (80). Most intravascular lymphomas are large B-cell lymphomas, it is an aggressive lymphoma with a poor prognosis. The molecular characteristics are similar to germinal center or post-germinal center B cells. such as diffuse large B-cell lymphoma (DLBCL), which is the most common subtype. Ramkumar et al. found that the Smurf2-YY1-c-Myc axis has become an important regulatory mechanism in suppression of B-cell proliferation and consequently lymphomagenesis (81). The anti-tumor effects of B cells depend mainly on the secretion of anti-tumor-associated antigen (TAA) antibodies and the provision of co-stimulatory signals to TAA-specific CD4+T cells to activate T cells. Kruppel-like factor 4 (KLF4) is a member of the zinc finger-containing KLF transcription factor family involved in regulating apoptosis, proliferation, and differentiation of B-cells and B-cell malignancies (82). Martinez et al. identified two YY1-binding sites in the KLF4 promoter region, and overexpressing YY1 resulted in significant promotion of KLF4 (83). However, whether the regulatory role of YY1 in lymphoma is suitable for the regulation of tumor by B cells in the tumor microenvironment needs to be further confirmed in future studies. the direct regulation of YY1 on tumor immune microenvironment in various tumors are shown in Table 3.
3.2.3 Other immune-related cells
The TME has appreciable numbers of immune cells, including adaptive immune responses, such as T cells and B cells, and innate immune cells, such as macrophages and neutrophils (90, 91). Tumor-associated macrophages (TAMs) infiltrate tumor tissue and play an essential role in the TME (92). Joo et al. reported that YY1 and STAT1 were upregulated in macrophages stimulated by oxidized LDL and subsequently were translocated into the nucleus to upregulated miR-29a, thereby reducing proinflammatory cytokine secretion and dampening inflammatory responses in macrophages. Cyclooxygenase 2 (COX-2) has been reported to be involved in inflammatory environments and lung cancer pathogenesis, and lipopolysaccharide treatment induced YY1 binding to the homologous site of endogenous COX-2 promoter YY1 and enhances its transcriptional activity in macrophages. Mechanically, YY1 was reported to interact with p300 and HDAC1/2 to affect the acetylation status of YY1. However, lipopolysaccharide treatment disrupted this interaction and binding to the COX-2 promoter. Indicated that it may be a competitive relationship between YY1 and COX-2 promoter, p300 and HDAC1/2 after lipopolysaccharide treatment (88). In addition, Xu et al. indicates that miR-301a/YY1/CXCR4 signaling pathway plays an important role in macrophage migration and phagocytosis (87). Hence, these results strongly suggest that YY1 is critically important for the tumor immune microenvironment.
3.3 Role of YY1 in tumor metabolism
As we all know, one of the critical features of the TME is metabolic abnormalities. Due to the rapid growth of tumors, cancer cells have increased anabolic and energy demands (93, 94). Therefore, extracellular nutrients determine the proliferation and growth rate of tumor cells. However, unlike normal cells, cancer cells have more remarkable metabolic plasticity, which forces them to adapt to poor environmental conditions (95, 96). Tumor metabolism abnormalities occur mainly in tumor cells, and cellular metabolites are a vital part of the TME and play an essential role in the formation of the TME. In addition to tumor cells, the interactions between different cells in the TME shape the unique metabolic characteristics of the microenvironment and maintain tumor growth, such as CAF, immune cells, and stromal cells, but the metabolism of YY1 in these components has rarely been reported. As an essential transcription factor, YY1 was reported to be involved in glucose, glutamine and lipid metabolism by regulating the transcriptional activation and expression of some key molecules related to these metabolic processes.
3.3.1 YY1 in glucose metabolism
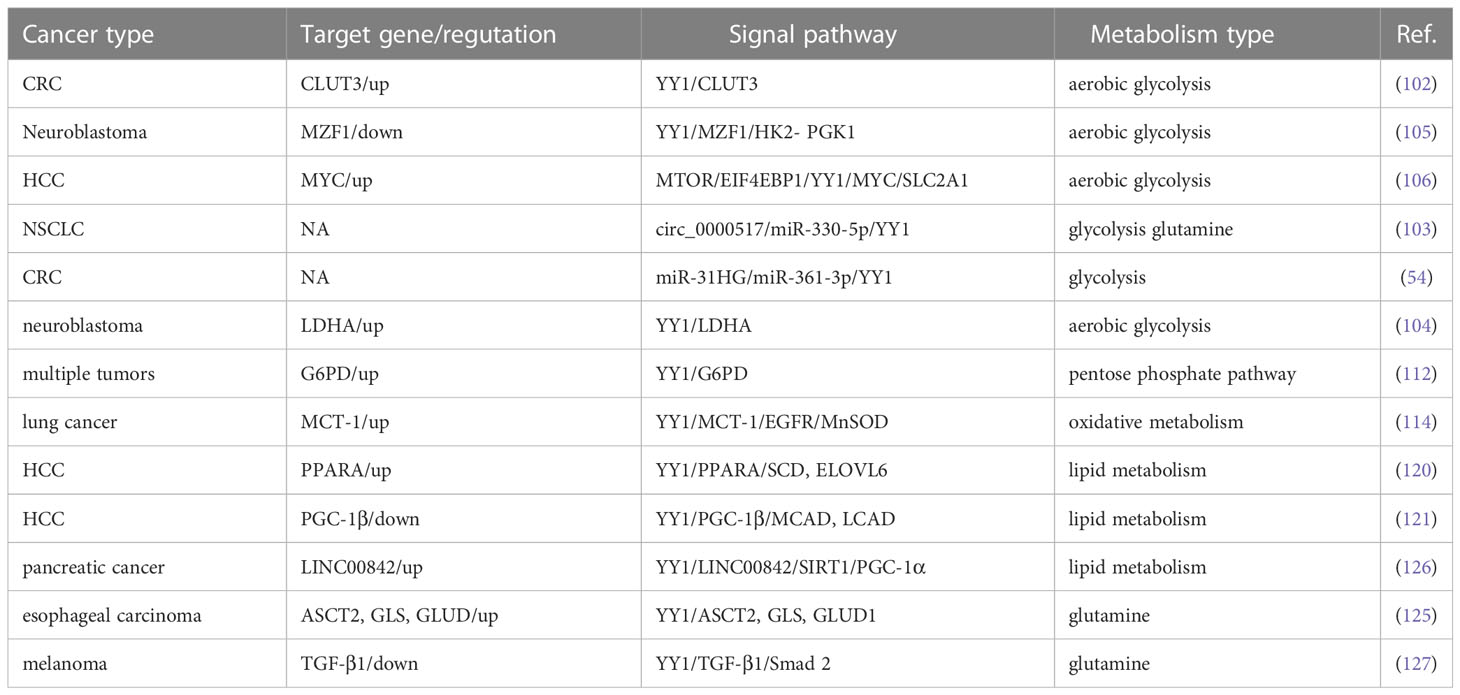
Since glucose is the primary energy source of organisms used in aerobic and anaerobic respiration (97, 98), which is maintains a balance between catabolic glycolysis/oxidative phosphorylation (OXPHOS) and anabolic gluconeogenesis/glycogen production (99). Abnormal glucose metabolism is integral to tumor metabolic reprogramming in the TME (100). YY1 is closely related to the glycolysis process. The glucose transporter mediated by the glucose transporter family (GLUT) is a pacemaker of aerobic glycolysis and, thus, is essential for tumor cell metabolism. YY1 may indirectly participate in reprogramming glucose metabolism in tumor cells by promoting the stability of HIF-1α under hypoxic conditions to enhance GLUT1 and GLUT3 expression (101). Wang et al. also reported that YY1 activates GLUT3 transcription by directly binding to its promoter while enhancing aerobic glycolysis in colon carcinoma cells (102). In NSCLC cells, circ_0000517/miR-330-5p/YY1 axis promotes the proliferation, glycolysis, and glutamine catabolism by regulating the expression of glycolysis and glutamine catabolism-related genes HK2, LDHA, ASCT2 and GLS1 (103). LDHA is a glycolytic enzyme that converts glucose-derived pyruvate into lactic acid. YY1 directly bound to the promoter LDHA, and overexpression of LDHA reverses the inhibitory effect of sh-YY1 on aerobic glycolysis and proliferation of neuroblastoma cells, indicating that YY1 induces aerobic glycolysis and proliferation (104). Glycolysis-related genes are critical to the glycolysis process. In neuroblastoma (NB), researchers searched public neuroblastoma datasets to identify transcriptional regulators and their glycolysis-related genes. They found that the zinc finger domain of YY1 binds a 21 amino acid peptide encoded by an open reading frame upstream of Myeloid zinc transcription factor 1 (MZF1), called MZF1-upep. MZF1 has been found to exert independent prognostic and promotes the aerobic glycolysis and tumor progression. Notably, the combination of these leads to decreased transcriptional activity of YY1 and inhibited transcription of MZF1 and downstream glycolytic genes HK2 and PGK1, therefore inhibiting aerobic glycolysis and NB progression (105). Hepatitis B virus (HBV) pre-S2 mutant induces HCC by inducing endoplasmic reticulum stress and activating the mTOR signaling pathway. Research has found that pre-S2 mutants initiate the glycolytic via activate mTOR signal pathway and activate SLC2A1 in YY1-dependent transcription, contributing to increased aerobic glycolysis, glucose uptake, lactic acid production and release in advanced tumorigenesis of HCC (106). In general, the above studies confirmed that YY1 alters the hypoxic microenvironment of tumor cells by regulating the whole process of glycolysis through different molecular mechanisms in various tumors, thus promoting tumorigenesis.
Since Otto Warburg discovered the characteristic of high-level glycolysis in tumor cell metabolism programming, which not only upregulates the adaptive potential of tumor cells to fluctuating oxygen tension but also produces glycolytic intermediates and increases the pentose phosphate pathway (PPP) (107–109), the primary significance of the PPP can generate nicotinamide adenine dinucleotide phosphate (NADPH) and a variety of monosaccharides. NADPH is the only enzyme specifically used to produce reactive oxygen species (ROS) and is essential for cellular antioxidant defense (110, 111). A recent study by Wu et al. identified that YY1 directly binds to the promoter of G6PD and promotes its transcription activity and expression, therefore, stimulating the PPP and protecting tumor cell from oxidative stress and inducing tumor progression (112). In conclusion, research on the molecular mechanisms of the G6PD-mediated PPP is closely related to YY1-induced tumor cell proliferation and tumorigenesis.
One of the typical features of metabolic changes in cancer cells is high levels of oxidative stress, and higher cellular oxidative stress levels favor the activation of glycolysis and promote tumorigenesis by affecting the signal transduction system, depriving them of normal contact inhibition, and promoting invasion and metastasis (113). The functions of these are complementary to each other. Tseng et al. found that p53 accumulation was reduced by MCT-1 overexpression. However, the levels of manganese-dependent superoxide dismutase (MnSOD) were upregulated through the YY1-EGFR signaling pathway, protecting cells from oxidative damage. On the other hand, limiting ROS production and inhibiting YY1 in lung cancer cells prevented MCT-1-induced cell invasiveness and the EGFR-MnSOD signaling pathway (114). However, ROS and YY1 do not have a definite positive association. According to research, low levels of ROS promoted the production of YY1, while high levels of ROS clearly inhibited it. YY1 was recruited to the antioxidant-responsive element binding site when its expression was stimulated, working in concert with other coregulators and then boosted NRF2-mediated ARE transcription, thereby shielding cells from damage by amplifying the antioxidant response (115). Overall, the available data indicate that YY1 can significantly activate cell glycolysis and ROS metabolism and play a critical role in remodeling the TME (Figure 3).
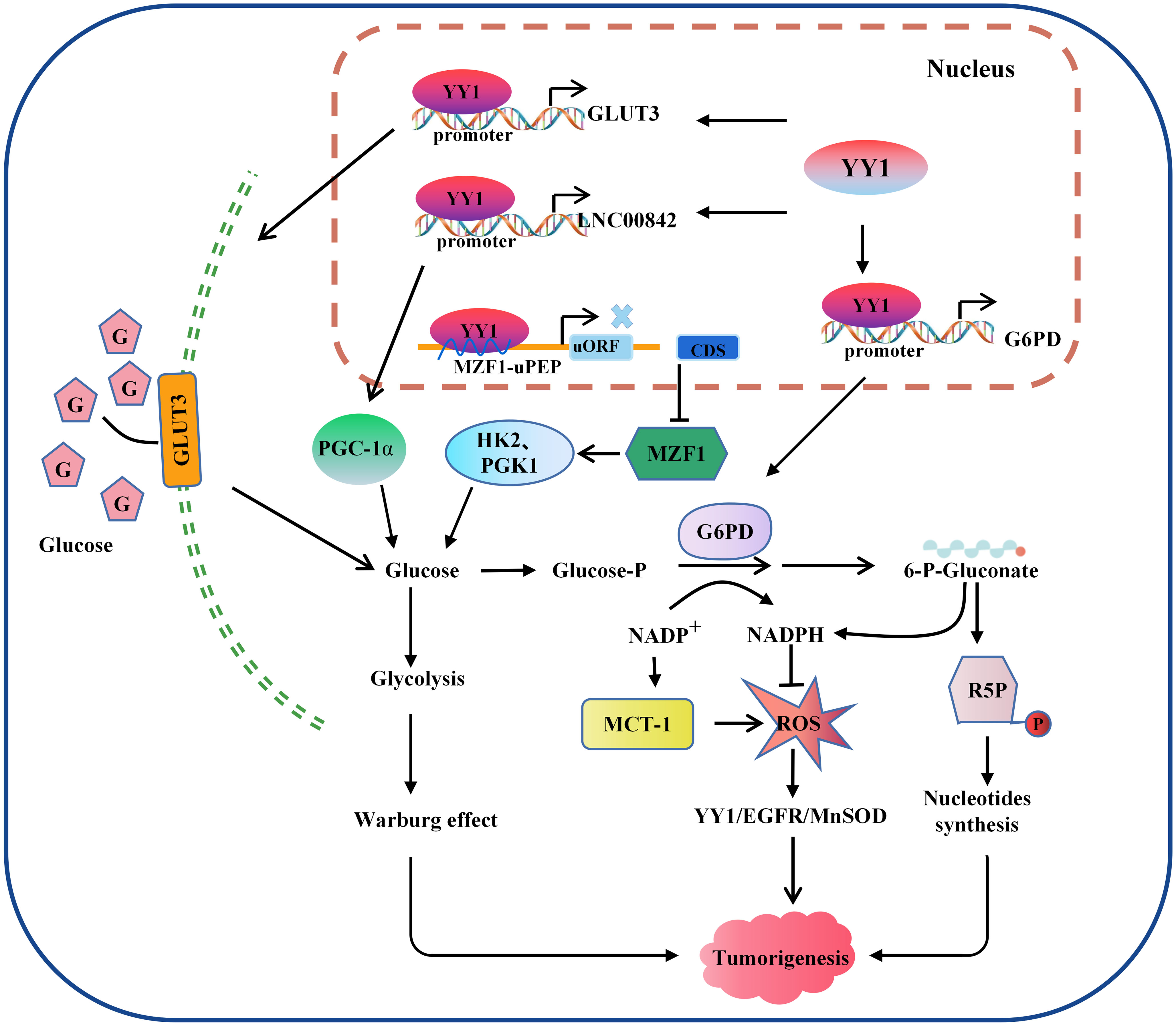
Figure 3 Model of the mechanism through which YY1 regulates glucose metabolism. YY1 is involved in regulating glucose metabolism in tumor cells by affecting the transcription activation of some key molecules. For example, activation of GLUT3 and LNC00842 transcription enhances aerobic glycolysis in tumor cells, while downregulation of YY1 by MZF1 inhibits the transcription of its downstream glycolytic genes HK2 and PGK1, thereby inhibiting aerobic glycolysis. In addition, YY1 activates G6PD transcription and stimulates the pentose phosphate pathway to enhance nucleotide generation and DNA synthesis and reduce intracellular reactive oxygen species levels, thus promoting tumorigenesis. YY1, Yin Yang 1; G6PD, glucose-6-phosphate dehydrogenase; GLUT, glucose transporter; ROS, reactive oxygen species; HK2, hexokinase 2; PGK1, phosphoglycerate kinase 1; R5P, Ribulose 5-phosphate.
3.3.2 YY1 in lipid metabolism
It has been widely observed that lipid metabolic reprogramming during cancer development confers cancer cells the ability to survive through enhanced lipid synthesis, storage, and catabolism, even under nutrient-limiting conditions. Abnormal lipid metabolism has always been observed in various tumors, and chronic liver disease and nonalcoholic fatty liver disease (NAFLD) are closely related to lipid homeostasis disorder. Accumulating evidence suggests that the hallmark feature of NAFLD is an increase in intrahepatic TG content (116). To investigate the crosstalk between lipid metabolism and YY1 in liver cancer cells, the authors found that several key transcription factors of lipid metabolism and their coactivators were significantly reduced in YY1-knockout HCC cells, such as PPARA/RXRA complex member CHREBP, SREBF2 and FXR and HNF family member HNF4A, FOXA1 and FOXA2 (117–119). Moreover, silencing YY1 suppresses the expression of crucial transcription factors markedly reduced their cooperation at various regulatory levels and various regulatory regions, which are involved in hepatic lipid metabolism regulated by these proteins. Especially the expression of SCD and ELOVL6, which encode key enzymes for adipogenesis, is regulated cooperatively by YY1 and PPARA/RXRA complexes on their promotors (120). In normal cells, cellular fatty acid concentration balance is controlled by the normal synthesis and breakdown of fatty acids. But tumor cells modify lipid metabolism by speeding up fatty acid synthesis while blocking fatty acid breakdown, leading to intracellular lipid buildup, in order to meet the demands of their rapid multiplication and growth. Li et al. demonstrated that YY1 was crucial for alterations in lipid metabolism in HCC cells because it suppresses the expression of the PGC-1β, which is a transcriptional activator of MCAD and LCAD that boosts both enzymes’ expression. The MCAD and LCAD are key enzymes required for FAO and crucial for fostering carcinogenesis (121). Therefore, YY1 upregulation prevented fatty acid β-oxidation, which increased triglyceride levels and lipid buildup in HCC cells and demonstrated the promising potential to accelerate the growth of tumors. Wu et al. also demonstrated that miR-122 could promote lipid droplet formation and triacylglycerol (TG) accumulation in vitro by reducing the stability of YY1 mRNA and upregulating FXR-SHP signaling. Suggesting that YY1 plays an essential role in regulating the accumulation of hepatic lipids, especially hepatic triglycerides (TGs) in the TME (122) (Figure 4).
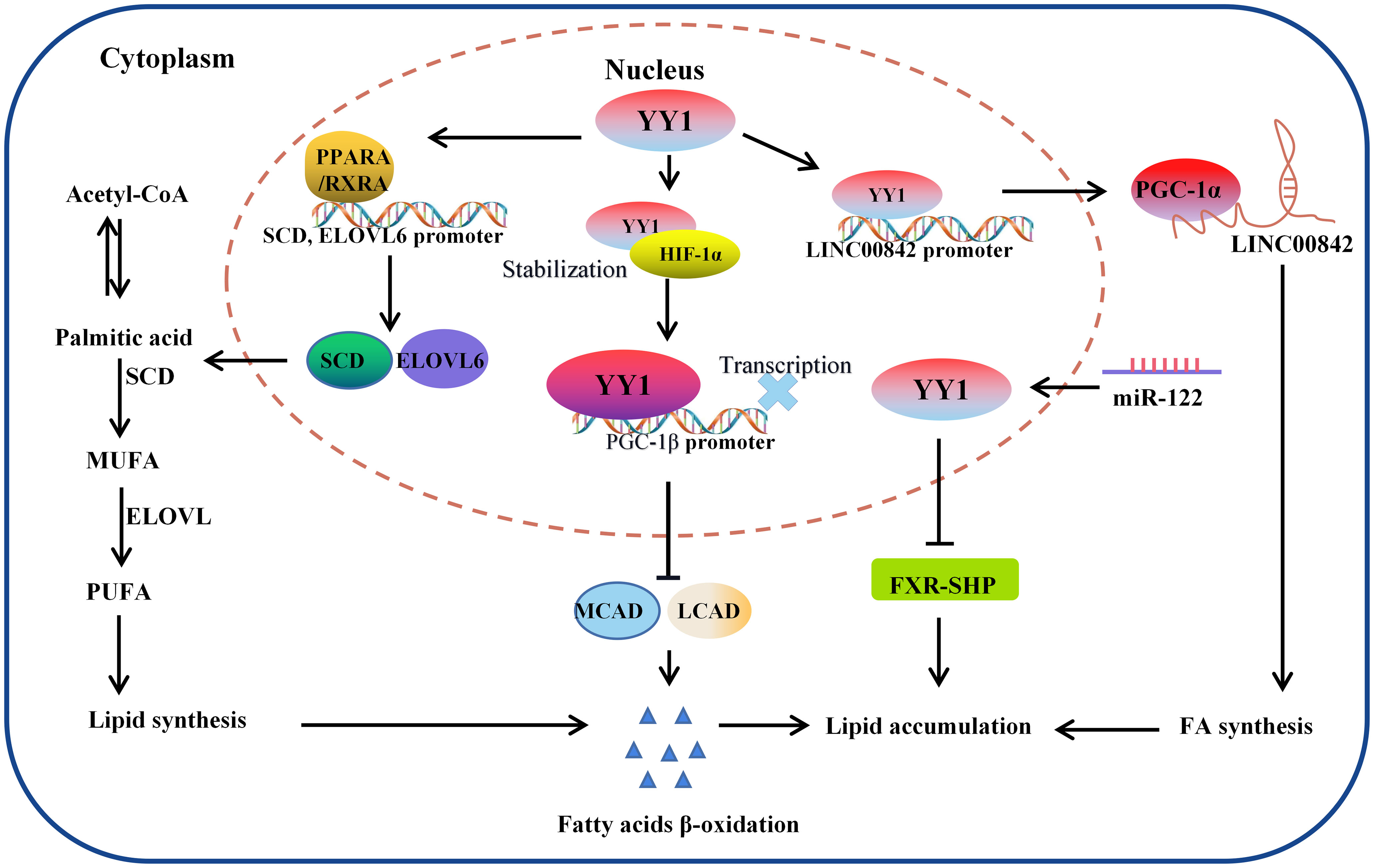
Figure 4 The molecular mechanism of YY1 regulating lipid metabolism. YY1 promotes triglyceride accumulation by regulating lipid metabolism-related molecule transcription. In this process, YY1 suppressed fatty acid β-oxidation, further inhibiting the expression of medium-chain acyl-CoA dehydrogenase (MCAD) and long-chain acyl-CoA dehydrogenase (LCAD) which is the key enzymes necessary for FAO. Additionally, miR-122 could promote lipid droplet formation and triacylglycerol (TG) accumulation in vitro by reducing the stability of YY1 mRNA and upregulating FXR-SHP signaling. Furthermore, YY1 also leads to lipid accumulation through the PPARA, RXRA/SCD, ELOVL6 signaling pathway and regulated the expression of LINC00842. LCAD, long-chain acyl-CoA dehydrogenase; MCAD, medium-chain acyl-CoA dehydrogenase; PGC-1β: peroxisome proliferator-activated receptor gamma coactivator-1β; SCD: PGC-1α: peroxisome proliferator-activated receptor gamma coactivator-1α.
3.3.3 YY1 in glutamine metabolism
Glutamine is an essential amino acid to maintain normal cell biological function, including biosynthesis, cell signaling, and preventing antioxidant damage (100). Of note, glutamine metabolism can provide the energy and biomacromolecules required by tumor cells for rapid growth and proliferation, helping them maintain intracellular redox homeostasis and intracellular signaling (123), and their anabolic/catabolic actions are dysregulated in cancer. Following glucose and fatty acid metabolism, oncogene-mediated abnormal glutamine metabolism in tumor cells has emerged as a new energy source (124). The researchers analyzed the effect of the YY1 on biological processes using the DAVID method, and these results suggested that the promotion of glutamine metabolism by YY1 may promote the progression of ESCC by affecting cancer pathways. Furthermore, the expression of key rate-limiting enzymes ASCT2, GLS and GLUT1 showed an upward trend and led to glutamine uptake and production were significantly increased. Briefly, these data support a model that YY1 promotes glutamine metabolism and regulates the development of ESCC (125). We also noted that YY1 promotes the expression of vital rate-limiting enzymes HK2, LHDA, PCNA, ASCT2 and GLS in A549 cells. Accordingly, YY1 obviously promoted glucose-to-glutamate conversion (103). In summary, these studies illustrated the role of YY1 in glutamine metabolism in the tumor, and the role of YY1 and its molecular mechanism on tumor metabolism is shown in Table 4.
4 Clinical potentials of YY1 and the microenvironment
4.1 The clinical value of YY1 in cancer diagnosis
In recent years, molecular diagnosis has become an important strategy for clinical tumor diagnosis and prognosis evaluation based on the expression specificity and clinical relevance of YY1 in different tumors. On the one hand, YY1 is highly expressed in prostate cancer, gastric cancer, ovarian cancer, breast cancer, multiple myeloma, liver cancer, and lung cancer etc. It especially shows increased expression in metastases compared with primary tumors, suggesting that high expression of YY1 may be a molecular marker for early diagnosis and prognosis evaluation of these tumors. On the other hand, the expression of YY1 was decreased in PDAC, NPC, pediatric osteosarcoma, etc., and negatively correlated with its clinical progression and a poor prognosis, suggesting that the low YY1 expression predict poor prognosis of these tumors. Tumor cells evade the immune response by producing a TME that suppresses the immune response, specifically a TME involving the T cell checkpoint receptor PD-1. It is a mechanism that prevents overstimulation of the immune system. Studies have shown that YY1 expression is positively correlated with PD-L1 expression, so elevated YY1 expression is expected to be a molecular marker for diagnosing T cell-mediated tumor immune escape (66). In addition, the expression level of YY1 in Burkitt lymphoma and DLBCL is higher than that in normal B cells and low-grade lymphoma (128). As mentioned above, YY1 may be a valuable diagnostic and prognostic marker for chemotherapy or immunotherapy regimens.
4.2 YY1 is an important molecular target for tumor therapy
The TME is a facilitator of tumor drug resistance (129), and the TME influences the progression and metastasis of solid tumors. Most current therapies target the tumor cells themselves, ignoring the local microenvironment around them. However, it is becoming clear that the most effective approach for cancer treatment involves therapies targeting both tumor cells and the TME (130, 131). Based on the functions of YY1 in tumor angiogenesis, metabolism and tumor-related microenvironment reconstruction, targeting YY1 are expected to become an important molecular strategy for tumor therapy (132). Previous studies have proven that many clinical drugs targeting YY1 have sound therapeutic effects on YY1-mediated tumor drug resistance or malignant progression. For example, the clinical drug oxaliplatin was reported to suppress colon carcinoma cell proliferation and angiogenesis by inhibiting the YY1/GLUT3 axis (102). Another study proved that YY1 plays an important role in angiogenesis and bevacizumab resistance by inducing VEGFA transcriptional activity and expression, and YY1 may be a potential molecular therapeutic target for anti-angiogenic therapy in HCC (46). In addition, cisplatin has always been one of the drugs of choice for head and neck squamous cell carcinoma (HNSCC), but most tumors that are initially responsive to cisplatin later acquire resistance. Zhao et al. found that targeting YY1 or PP2A enhanced the efficiency of cisplatin chemotherapy in HNSCC (133). Therefore, YY1 plays a crucial role in regulating TME-mediated chemoresistance in tumor cells, which has important implications for diagnosing and treating patients.
Immunotherapy has become a mainstay of cancer treatment in many malignancies, and clinical evaluation confirmed that the expression of the oncogene YY1 was upregulated in PD-1+ T cells infiltrating lymphocytes in tumors, suggesting that YY1 may hinder T-cell-mediated tumor immunotherapy, especially since the effect on PD-1/PD-L1 is not fully blocked. Therefore, exploring the underlying mechanism of regulating PD-L1 expression on tumor cells is a way to make tumor cells effective for PD-1/PD-L1 antibody therapy. Multiple signaling crosstalk pathways have been reported to be involved in the regulatory relationship between YY1 and PD-L1 (66). In addition, remodeling the immunosuppressive microenvironment and enhancing immunotherapy response are effective strategies for the clinical treatment of cancer. Qiu et al. confirmed that YY1-dependent glioblastoma stem cells were sensitive to the CDK9 inhibitors (Alvocidib and Dinaciclib), and the combination of Alvocidib and anti-PD-1 can more significantly reshape the tumor immune microenvironment, enhance the immunotherapy response and inhibit the occurrence of glioma (84). Based on these findings, combining strategies targeting YY1 and the above pathways may enhance cell-mediated antitumor cell responses and reverse the resistance observed with checkpoint inhibitors alone. Therefore, YY1 is a critical target for immunotherapy and chemotherapy of cancers. However, YY1 acts as a tumor suppressor gene in pancreatic cancer and nasopharyngeal cancer, and specific clinical drugs targeting YY1 have not yet been discovered. We look forward to having made better progress in improving tumor treatment efficacy in the future.
5 Conclusions and perspectives
YY1 is a critical transcription regulator and is closely related to the remodeling and regulation of the TME, and it has two distinct expression patterns (high or low) in different tumor types. It is upregulated in most types of tumors, such as prostate cancer, gastrointestinal cancer, ovarian cancer and breast cancer, and downregulated in a few tumors, including nasopharyngeal carcinoma, thus, it plays a dual role as an oncogene or tumor suppressor in tumorigenesis and tumor progression. YY1 plays critical roles in tumor angiogenesis, glucose metabolism, lipid metabolism, and immune regulation, which provides an important perspective for understanding the mechanism YY1 is involved in tumorigenesis and development. Based on the specific expression patterns of YY1 in tumors and its role in the formation and regulation of the TME, targeting YY1 and its related TME would be a potential molecular strategy for the clinical diagnosis and treatment of tumors. Although YY1 as a target has demonstrated a valuable application prospect in the diagnosis and treatment of tumors, and certain drugs or inhibitors could indeed alleviate the clinical malignant phenotype or drug resistance of tumor patients by reversing YY1 expression, there is still no specific and inhibitor or targeted drug to YY1 used for the clinical treatment of tumors, further efforts are needed for researchers to develop specific inhibitors or drugs targeting YY1.
Author contributions
MZ, ML, WX, FT and GL designed and conducted this study. ML and JW drafted the manuscript. CX, XZ, SC, LZ, YD and HD revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (grant no. 82172592), the Free Exploration Program of Central South University (grant nos. 2020zzts225 and 2022zzts0935), the program of Introducing Talents of Discipline to Universities (grant no. 111-2-12).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
YY1, Yin Yang 1; TME, tumor microenvironment; NPC, nasopharyngeal carcinoma; PDAC, pancreatic ductal adenocarcinoma; VEGF, vascular endothelial growth factor; MVD, micro vessel density; HCC, Hepatocellular carcinoma; GLUT, glucose transporter; MZF1, myeloid zinc finger 1; HK2, hexokinase 2; PGK1, phosphoglycerate kinase 1; SLC2A1, solute carrier family 2, member 1; PPP, pentose phosphate pathway; NADPH, nicotinamide adenine dinucleotide phosphate; ROS, reactive oxygen species; G6PD, glucose-6-phosphate dehydrogenase; MnSOD, manganese-dependent superoxide dismutase; TIME, immunosuppression microenvironment; PD-1, Programmed death receptor-1; CTLA-4, Cytotoxic T lymphocyte-associated antigen -4; Treg, Regulatory T; PcG, Polycomb Group; LPS, lipopolysaccharide; COX-2, cyclooxygenase-2; HIF-1α, hypoxia-inducible factor 1-α.
References
2. Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med (2011) 17:320–9. doi: 10.1038/nm.2328
3. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med (2013) 19:1423–37. doi: 10.1038/nm.3394
4. Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription. vitro Nat (1991) 354:241–5. doi: 10.1038/354241a0
5. Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell (1991) 67:377–88. doi: 10.1016/0092-8674(91)90189-6
6. Satyamoorthy K, Park K, Atchison ML, Howe CC. The intracisternal a-particle upstream element interacts with transcription factor YY1 to activate transcription: Pleiotropic effects of YY1 on distinct DNA promoter elements. Mol Cell Biol (1993) 13:6621–8. doi: 10.1128/mcb.13.11.6621-6628.1993
7. Xiao R, Chen JY, Liang Z, Luo D, Chen G, Lu ZJ, et al. Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell (2019) 178:107–21.e118. doi: 10.1016/j.cell.2019.06.001
8. Figiel M, Gorecki A. Physical interaction of human yin yang 1 protein with DNA. Crit Rev Oncog (2017) 22:75–97. doi: 10.1615/CritRevOncog.2017020976
9. Li L, Williams P, Ren W, Wang MY, Gao Z, Miao W, et al. YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat Chem Biol (2021) 17:161–8. doi: 10.1038/s41589-020-00695-1
10. Bonavida B. Therapeutic YY1 inhibitors in cancer: ALL in ONE. Crit Rev Oncog (2017) 22:37–47. doi: 10.1615/CritRevOncog.2017020472
11. Zhang JJ, Zhu Y, Xie KL, Peng YP, Tao JQ, Tang J, et al. Yin yang-1 suppresses invasion and metastasis of pancreatic ductal adenocarcinoma by downregulating MMP10 in a MUC4/ErbB2/p38/MEF2C-dependent mechanism. Mol Cancer (2014) 13:130. doi: 10.1186/1476-4598-13-130
12. Nicholson S, Whitehouse H, Naidoo K, Byers RJ. Yin yang 1 in human cancer. Crit Rev Oncog (2011) 16:245–60. doi: 10.1615/critrevoncog.v16.i3-4.80
13. Li M, Liu Y, Wei Y, Wu C, Meng H, Niu W, et al. Zinc-finger protein YY1 suppresses tumor growth of human nasopharyngeal carcinoma by inactivating c-myc-mediated microRNA-141 transcription. J Biol Chem (2019) 294:6172–87. doi: 10.1074/jbc.RA118.006281
14. Zaravinos A, Spandidos DA. Yin yang 1 expression in human tumors. Cell Cycle (2010) 9:512–22. doi: 10.4161/cc.9.3.10588
15. Yang W, Feng B, Meng Y, Wang J, Geng B, Cui Q, et al. FAM3C-YY1 axis is essential for TGFβ-promoted proliferation and migration of human breast cancer MDA-MB-231 cells via the activation of HSF1. J Cell Mol Med (2019) 23:3464–75. doi: 10.1111/jcmm.14243
16. Agarwal N, Dancik GM, Goodspeed A, Costello JC, Owens C, Duex JE, et al. GON4L drives cancer growth through a YY1-androgen receptor-CD24 axis. Cancer Res (2016) 76:5175–85. doi: 10.1158/0008-5472.can-16-1099
17. Zhang N, Li X, Wu CW, Dong Y, Cai M, Mok MT, et al. microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene (2013) 32:5078–88. doi: 10.1038/onc.2012.526
18. Wang W, Yue Z, Tian Z, Xie Y, Zhang J, She Y, et al. Expression of yin yang 1 in cervical cancer and its correlation with e-cadherin expression and HPV16 E6. PLoS One (2018) 13:e0193340. doi: 10.1371/journal.pone.0193340
19. Luo J, Zhou X, Ge X, Liu P, Cao J, Lu X, et al. Upregulation of ying yang 1 (YY1) suppresses esophageal squamous cell carcinoma development through heme oxygenase-1. Cancer Sci (2013) 104:1544–51. doi: 10.1111/cas.12248
20. Kang W, Tong JH, Chan AW, Zhao J, Dong Y, Wang S, et al. Yin yang 1 contributes to gastric carcinogenesis and its nuclear expression correlates with shorter survival in patients with early stage gastric adenocarcinoma. J Transl Med (2014) 12:80. doi: 10.1186/1479-5876-12-80
21. Baritaki S, Chatzinikola AM, Vakis AF, Soulitzis N, Karabetsos DA, Neonakis I, et al. YY1 over-expression in human brain gliomas and meningiomas correlates with TGF-beta1, IGF-1 and FGF-2 mRNA levels. Cancer Invest (2009) 27:184–92. doi: 10.1080/07357900802210760
22. Dukers DF, van Galen JC, Giroth C, Jansen P, Sewalt RG, Otte AP, et al. Unique polycomb gene expression pattern in hodgkin's lymphoma and hodgkin's lymphoma-derived cell lines. Am J Pathol (2004) 164:873–81. doi: 10.1016/s0002-9440(10)63175-6
23. Kim JS, Son SH, Kim MY, Choi D, Jang IS, Paik SS, et al. Diagnostic and prognostic relevance of CP2c and YY1 expression in hepatocellular carcinoma. Oncotarget (2017) 8:24389–400. doi: 10.18632/oncotarget.15462
24. Zapata-Tarres M, Juarez-Villegas LE, Maldonado-Valenzuela A, Baay-Guzman GJ, Lopez-Perez TV, Cabrera-Muñoz L, et al. Expression of YY1 in wilms tumors with favorable histology is a risk factor for adverse outcomes. Future Oncol (2019) 15:1231–41. doi: 10.2217/fon-2018-0764
25. Huang T, Wang G, Yang L, Peng B, Wen Y, Ding G, et al. Transcription factor YY1 modulates lung cancer progression by activating lncRNA-PVT1. DNA Cell Biol (2017) 36:947–58. doi: 10.1089/dna.2017.3857
26. Zhao G, Li Q, Wang A, Jiao J. YY1 regulates melanoma tumorigenesis through a miR-9 ~ RYBP axis. J Exp Clin Cancer Res (2015) 34:66. doi: 10.1186/s13046-015-0177-y
27. de Nigris F, Botti C, de Chiara A, Rossiello R, Apice G, Fazioli F, et al. Expression of transcription factor yin yang 1 in human osteosarcomas. Eur J Cancer (2006) 42:2420–4. doi: 10.1016/j.ejca.2006.06.008
28. Matsumura N, Huang Z, Baba T, Lee PS, Barnett JC, Mori S, et al. Yin yang 1 modulates taxane response in epithelial ovarian cancer. Mol Cancer Res (2009) 7:210–20. doi: 10.1158/1541-7786.mcr-08-0255
29. Kashyap V, Bonavida B. Role of YY1 in the pathogenesis of prostate cancer and correlation with bioinformatic data sets of gene expression. Genes Cancer (2014) 5:71–83. doi: 10.18632/genesandcancer.12
30. Sadikovic B, Yoshimoto M, Chilton-MacNeill S, Thorner P, Squire JA, Zielenska M. Identification of interactive networks of gene expression associated with osteosarcoma oncogenesis by integrated molecular profiling. Hum Mol Genet (2009) 18:1962–75. doi: 10.1093/hmg/ddp117
31. Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res (2006) 19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x
32. Chen Q, Zhang JJ, Ge WL, Chen L, Yuan H, Meng LD, et al. YY1 inhibits the migration and invasion of pancreatic ductal adenocarcinoma by downregulating the FER/STAT3/MMP2 signaling pathway. Cancer Lett (2019) 463:37–49. doi: 10.1016/j.canlet.2019.07.019
33. Wang J, Zhou L, Li Z, Zhang T, Liu W, Liu Z, et al. YY1 suppresses FEN1 over-expression and drug resistance in breast cancer. BMC Cancer (2015) 15:50. doi: 10.1186/s12885-015-1043-1
34. Lee MH, Lahusen T, Wang RH, Xiao C, Xu X, Hwang YS, et al. Yin yang 1 positively regulates BRCA1 and inhibits mammary cancer formation. Oncogene (2012) 31:116–27. doi: 10.1038/onc.2011.217
35. de Nigris F, Zanella L, Cacciatore F, De Chiara A, Fazioli F, Chiappetta G, et al. YY1 overexpression is associated with poor prognosis and metastasis-free survival in patients suffering osteosarcoma. BMC Cancer (2011) 11:472. doi: 10.1186/1471-2407-11-472
36. Liu D, Zhang J, Wu Y, Shi G, Yuan H, Lu Z, et al. YY1 suppresses proliferation and migration of pancreatic ductal adenocarcinoma by regulating the CDKN3/MdM2/P53/P21 signaling pathway. Int J Cancer (2018) 142:1392–404. doi: 10.1002/ijc.31173
37. Jiang X, Wang J, Deng X, Xiong F, Zhang S, Gong Z, et al. The role of microenvironment in tumor angiogenesis. J Exp Clin Cancer Res (2020) 39:204. doi: 10.1186/s13046-020-01709-5
38. Choi KS, Bae MK, Jeong JW, Moon HE, Kim KW. Hypoxia-induced angiogenesis during carcinogenesis. J Biochem Mol Biol (2003) 36:120–7. doi: 10.5483/bmbrep.2003.36.1.120
39. Barker HE, Paget JT, Khan AA, Harrington KJ. The tumour microenvironment after radiotherapy: Mechanisms of resistance and recurrence. Nat Rev Cancer (2015) 15:409–25. doi: 10.1038/nrc3958
40. Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol (2006) 7:359–71. doi: 10.1038/nrm1911
41. Matsumoto K, Ema M. Roles of VEGF-a signalling in development, regeneration, and tumours. J Biochem (2014) 156:1–10. doi: 10.1093/jb/mvu031
42. Melincovici CS, Boşca AB, Şuşman S, Mărginean M, Mihu C, Istrate M, et al. Vascular endothelial growth factor (VEGF) - key factor in normal and pathological angiogenesis. Rom J Morphol Embryol (2018) 59:455–67. doi: 10.1007/978-3-319-28140-7_58
43. Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: Alternative therapeutic targets. Angiogenesis (2017) 20:409–26. doi: 10.1007/s10456-017-9562-9
44. de Nigris F, Crudele V, Giovane A, Casamassimi A, Giordano A, Garban HJ, et al. CXCR4/YY1 inhibition impairs VEGF network and angiogenesis during malignancy. Proc Natl Acad Sci USA (2010) 107:14484–9. doi: 10.1073/pnas.1008256107
45. de Nigris F, Rossiello R, Schiano C, Arra C, Williams-Ignarro S, Barbieri A, et al. Deletion of yin yang 1 protein in osteosarcoma cells on cell invasion and CXCR4/angiogenesis and metastasis. Cancer Res (2008) 68:1797–808. doi: 10.1158/0008-5472.can-07-5582
46. Wang W, Li D, Sui G. YY1 is an inducer of cancer metastasis. Crit Rev Oncog (2017) 22:1–11. doi: 10.1615/CritRevOncog.2017021314
47. Li ZJ, Cheng J, Song Y, Li HH, Zheng JF. LncRNA SNHG5 upregulation induced by YY1 contributes to angiogenesis via miR-26b/CTGF/VEGFA axis in acute myelogenous leukemia. Lab Invest (2021) 101:341–52. doi: 10.1038/s41374-020-00519-9
48. Lin CC, Kuo IY, Wu LT, Kuan WH, Liao SY, Jen J, et al. Dysregulated Kras/YY1/ZNF322A/Shh transcriptional axis enhances neo-angiogenesis to promote lung cancer progression. Theranostics (2020) 10:10001–15. doi: 10.7150/thno.47491
49. Li X, Yu M, Yang C. YY1-mediated overexpression of long noncoding RNA MCM3AP-AS1 accelerates angiogenesis and progression in lung cancer by targeting miR-340-5p/KPNA4 axis. J Cell Biochem (2020) 121:2258–67. doi: 10.1002/jcb.29448
50. Xu Y, Leng K, Yao Y, Kang P, Liao G, Han Y, et al. Cholangiocarcinoma-associated circular RNA 1, contributes to cholangiocarcinoma progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology (2021) 73:1419–35. doi: 10.1002/hep.31493
51. Chen Q, Yang C, Chen L, Zhang JJ, Ge WL, Yuan H, et al. YY1 targets tubulin polymerisation-promoting protein to inhibit migration, invasion and angiogenesis in pancreatic cancer via p38/MAPK and PI3K/AKT pathways. Br J Cancer (2019) 121:912–21. doi: 10.1038/s41416-019-0604-5
52. Liu H, Qiu Y, Pei X, Chitteti R, Steiner R, Zhang S, et al. Endothelial specific YY1 deletion restricts tumor angiogenesis and tumor growth. Sci Rep (2020) 10:20493. doi: 10.1038/s41598-020-77568-z
53. Infante T, Mancini FP, Lanza A, Soricelli A, de Nigris F, Napoli C. Polycomb YY1 is a critical interface between epigenetic code and miRNA machinery after exposure to hypoxia in malignancy. Biochim Biophys Acta (2015) 1853:975–86. doi: 10.1016/j.bbamcr.2015.01.009
54. Guo T, Liu D, Peng S, Wang M, Li Y. A positive feedback loop of lncRNA MIR31HG-miR-361-3p -YY1 accelerates colorectal cancer progression through modulating proliferation, angiogenesis, and glycolysis. Front Oncol (2021) 11:684984. doi: 10.3389/fonc.2021.684984
55. Tao Q, Lu Y, Qi Y, Yu D, Gu J, Zhu Y, et al. Hypoxia promotes the expression of Von willebrand factor in breast cancer cells by up-regulating the transcription factor YY1 and down-regulating the hsa-miR-424. Eur J Pharmacol (2022) 934:175308. doi: 10.1016/j.ejphar.2022.175308
56. de Miguel M, Calvo E. Clinical challenges of immune checkpoint inhibitors. Cancer Cell (2020) 38:326–33. doi: 10.1016/j.ccell.2020.07.004
57. Hashimoto M, Kamphorst AO, Im SJ, Kissick HT, Pillai RN, Ramalingam SS, et al. CD8 T cell exhaustion in chronic infection and cancer: Opportunities for interventions. Annu Rev Med (2018) 69:301–18. doi: 10.1146/annurev-med-012017-043208
58. Kleiman E, Jia H, Loguercio S, Su AI, Feeney AJ. YY1 plays an essential role at all stages of b-cell differentiation. Proc Natl Acad Sci USA (2016) 113:E3911–3920. doi: 10.1073/pnas.1606297113
59. Banerjee A, Sindhava V, Vuyyuru R, Jha V, Hodewadekar S, Manser T, et al. YY1 is required for germinal center b cell development. PloS One (2016) 11:e0155311. doi: 10.1371/journal.pone.0155311
60. Li C, Jiang P, Wei S, Xu X, Wang J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol Cancer (2020) 19:116. doi: 10.1186/s12943-020-01234-1
61. Schiano C, Soricelli A, De Nigris F, Napoli C. New challenges in integrated diagnosis by imaging and osteo-immunology in bone lesions. Expert Rev Clin Immunol (2019) 15:289–301. doi: 10.1080/1744666x.2019.1561283
62. Balkhi MY, Wittmann G, Xiong F, Junghans RP. YY1 upregulates checkpoint receptors and downregulates type I cytokines in exhausted, chronically stimulated human T cells. iScience (2018) 2:105–22. doi: 10.1016/j.isci.2018.03.009
63. Kwiatkowska D, Mazur E, Reich A. YY1 is a key player in melanoma Immunotherapy/Targeted treatment resistance. Front Oncol (2022) 12:856963. doi: 10.3389/fonc.2022.856963
64. Yan Y, Zhang L, Zuo Y, Qian H, Liu C. Immune checkpoint blockade in cancer immunotherapy: Mechanisms, clinical outcomes, and safety profiles of PD-1/PD-L1 inhibitors. Arch Immunol Ther Exp (Warsz) (2020) 68:36. doi: 10.1007/s00005-020-00601-6
65. Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, et al. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst (2016) 108. doi: 10.1093/jnci/djv303
66. Hays E, Bonavida B. YY1 regulates cancer cell immune resistance by modulating PD-L1 expression. Drug Resist Update (2019) 43:10–28. doi: 10.1016/j.drup.2019.04.001
67. Tang XY, Xiong YL, Zhao YB, Yang J, Shi AP, Zheng KF, et al. Dual immunological and proliferative regulation of immune checkpoint FGL1 in lung adenocarcinoma: The pivotal role of the YY1-FGL1-MYH9 axis. Front Immunol (2022) 13:1014053. doi: 10.3389/fimmu.2022.1014053
68. Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: Mechanisms of differentiation and function. Annu Rev Immunol (2012) 30:531–64. doi: 10.1146/annurev.immunol.25.022106.141623
69. Hwang SS, Jang SW, Kim MK, Kim LK, Kim BS, Kim HS, et al. YY1 inhibits differentiation and function of regulatory T cells by blocking Foxp3 expression and activity. Nat Commun (2016) 7:10789. doi: 10.1038/ncomms10789
70. Su S, Liao J, Liu J, Huang D, He C, Chen F, et al. Blocking the recruitment of naive CD4(+) T cells reverses immunosuppression in breast cancer. Cell Res (2017) 27:461–82. doi: 10.1038/cr.2017.34
71. Wing JB, Tanaka A, Sakaguchi S. Human FOXP3(+) regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity (2019) 50:302–16. doi: 10.1016/j.immuni.2019.01.020
72. Hwang SS, Kim YU, Lee S, Jang SW, Kim MK, Koh BH, et al. Transcription factor YY1 is essential for regulation of the Th2 cytokine locus and for Th2 cell differentiation. Proc Natl Acad Sci USA (2013) 110:276–81. doi: 10.1073/pnas.1214682110
73. Srinivasan L, Pan X, Atchison ML. Transient requirements of YY1 expression for PcG transcriptional repression and phenotypic rescue. J Cell Biochem (2005) 96:689–99. doi: 10.1002/jcb.20562
74. Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell (2004) 14:637–46. doi: 10.1016/j.molcel.2004.05.009
75. Shah DK, Zúñiga-Pflücker JC. An overview of the intrathymic intricacies of T cell development. J Immunol (2014) 192:4017–23. doi: 10.4049/jimmunol.1302259
76. Itahashi K, Irie T, Nishikawa H. Regulatory T-cell development in the tumor microenvironment. Eur J Immunol (2022) 52:1216–27. doi: 10.1002/eji.202149358
77. Ciofani M, Zúñiga-Pflücker JC. The thymus as an inductive site for T lymphopoiesis. Annu Rev Cell Dev Biol (2007) 23:463–93. doi: 10.1146/annurev.cellbio.23.090506.123547
78. Assumpção A, Fu G, Singh DK, Lu Z, Kuehnl AM, Welch R, et al. A lineage-specific requirement for YY1 polycomb group protein function in early T cell development. Development (2021) 148. doi: 10.1242/dev.197319
79. Downs-Canner SM, Meier J, Vincent BG, Serody JS. B cell function in the tumor microenvironment. Annu Rev Immunol (2022) 40:169–93. doi: 10.1146/annurev-immunol-101220-015603
80. Arribas Arranz J, Winter DN, Drexler HG, Eberth S. Suitability of yin yang 1 transcript and protein levels for biomarker studies in b cell non-Hodgkin lymphoma. Biomark Res (2018) 6:11. doi: 10.1186/s40364-018-0126-y
81. Ramkumar C, Cui H, Kong Y, Jones SN, Gerstein RM, Zhang H. Smurf2 suppresses b-cell proliferation and lymphomagenesis by mediating ubiquitination and degradation of YY1. Nat Commun (2013) 4:2598. doi: 10.1038/ncomms3598
82. Montecillo-Aguado M, Morales-Martínez M, Huerta-Yepez S, Vega MI. KLF4 inhibition by kenpaullone induces cytotoxicity and chemo sensitization in b-NHL cell lines via YY1 independent. Leuk Lymphoma (2021) 62:1422–31. doi: 10.1080/10428194.2020.1869960
83. Morales-Martinez M, Valencia-Hipolito A, Vega GG, Neri N, Nambo MJ, Alvarado I, et al. Regulation of krüppel-like factor 4 (KLF4) expression through the transcription factor yin-yang 1 (YY1) in non-Hodgkin b-cell lymphoma. Oncotarget (2019) 10:2173–88. doi: 10.18632/oncotarget.26745
84. Qiu Z, Zhao L, Shen JZ, Liang Z, Wu Q, Yang K, et al. Transcription elongation machinery is a druggable dependency and potentiates immunotherapy in glioblastoma stem cells. Cancer Discov (2022) 12:502–21. doi: 10.1158/2159-8290.cd-20-1848
85. Ou X, Huo J, Huang Y, Li YF, Xu S, Lam KP. Transcription factor YY1 is essential for iNKT cell development. Cell Mol Immunol (2019) 16:547–56. doi: 10.1038/s41423-018-0002-6
86. Ye J, Young HA, Ortaldo JR, Ghosh P. Identification of a DNA binding site for the nuclear factor YY1 in the human GM-CSF core promoter. Nucleic Acids Res (1994) 22:5672–8. doi: 10.1093/nar/22.25.5672
87. Xu J, Fu L, Deng J, Zhang J, Zou Y, Liao L, et al. miR-301a deficiency attenuates the macrophage migration and phagocytosis through YY1/CXCR4 pathway. Cells (2022) 11. doi: 10.3390/cells11243952
88. Joo M, Wright JG, Hu NN, Sadikot RT, Park GY, Blackwell TS, et al. Yin yang 1 enhances cyclooxygenase-2 gene expression in macrophages. Am J Physiol Lung Cell Mol Physiol (2007) 292:L1219–1226. doi: 10.1152/ajplung.00474.2006
89. Lei JH, Lee MH, Miao K, Huang Z, Yao Z, Zhang A, et al. Activation of FGFR2 signaling suppresses BRCA1 and drives triple-negative mammary tumorigenesis that is sensitive to immunotherapy. Adv Sci (Weinh) (2021) 8:e2100974. doi: 10.1002/advs.202100974
90. Kim J, Bae JS. Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediators Inflamm (2016) 2016:6058147. doi: 10.1155/2016/6058147
91. Li MO, Wolf N, Raulet DH, Akkari L, Pittet MJ, Rodriguez PC, et al. Innate immune cells in the tumor microenvironment. Cancer Cell (2021) 39:725–9. doi: 10.1016/j.ccell.2021.05.016
92. Pan Y, Yu Y, Wang X, Zhang T. Tumor-associated macrophages in tumor immunity. Front Immunol (2020) 11:583084. doi: 10.3389/fimmu.2020.583084
93. Reina-Campos M, Moscat J, Diaz-Meco M. Metabolism shapes the tumor microenvironment. Curr Opin Cell Biol (2017) 48:47–53. doi: 10.1016/j.ceb.2017.05.006
94. DeBerardinis RJ. Tumor microenvironment, metabolism, and immunotherapy. N Engl J Med (2020) 382:869–71. doi: 10.1056/NEJMcibr1914890
95. Li Y, Zhao L, Li XF. Hypoxia and the tumor microenvironment. Technol Cancer Res Treat (2021) 20:15330338211036304. doi: 10.1177/15330338211036304
96. Ocaña MC, Martínez-Poveda B, Quesada AR, Medina M. Metabolism within the tumor microenvironment and its implication on cancer progression: An ongoing therapeutic target. Med Res Rev (2019) 39:70–113. doi: 10.1002/med.21511
97. Bose S, Le A. Glucose metabolism in cancer. Adv Exp Med Biol (2018) 1063:3–12. doi: 10.1007/978-3-319-77736-8_1
98. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab (2016) 23:27–47. doi: 10.1016/j.cmet.2015.12.006
99. Lu J, Tan M, Cai Q. The warburg effect in tumor progression: Mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett (2015) 356:156–64. doi: 10.1016/j.canlet.2014.04.001
100. Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci (2016) 73:377–92. doi: 10.1007/s00018-015-2070-4
101. Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol (2005) 202:654–62. doi: 10.1002/jcp.20166
102. Wang Y, Wu S, Huang C, Li Y, Zhao H, Kasim V. Yin yang 1 promotes the warburg effect and tumorigenesis via glucose transporter GLUT3. Cancer Sci (2018) 109:2423–34. doi: 10.1111/cas.13662
103. Bing ZX, Zhang JQ, Wang GG, Wang YQ, Wang TG, Li DQ. Silencing of circ_0000517 suppresses proliferation, glycolysis, and glutamine decomposition of non-small cell lung cancer by modulating miR-330-5p/YY1 signal pathway. Kaohsiung J Med Sci (2021) 37:1027–37. doi: 10.1002/kjm2.12440
104. Wang Q, Fan W, Liang B, Hou B, Jiang Z, Li C. YY1 transcription factor induces proliferation and aerobic glycolysis of neuroblastoma cells via LDHA regulation. Exp Ther Med (2023) 25:37. doi: 10.3892/etm.2022.11736
105. Fang E, Wang X, Wang J, Hu A, Song H, Yang F, et al. Therapeutic targeting of YY1/MZF1 axis by MZF1-uPEP inhibits aerobic glycolysis and neuroblastoma progression. Theranostics (2020) 10:1555–71. doi: 10.7150/thno.37383
106. Teng CF, Hsieh WC, Wu HC, Lin YJ, Tsai HW, Huang W, et al. Hepatitis b virus pre-S2 mutant induces aerobic glycolysis through mammalian target of rapamycin signal cascade. PloS One (2015) 10:e0122373. doi: 10.1371/journal.pone.0122373
107. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab (2008) 7:11–20. doi: 10.1016/j.cmet.2007.10.002
108. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
109. Pouysségur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature (2006) 441:437–43. doi: 10.1038/nature04871
110. Kroemer G, Pouyssegur J. Tumor cell metabolism: Cancer's achilles' heel. Cancer Cell (2008) 13:472–82. doi: 10.1016/j.ccr.2008.05.005
111. Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol (2011) 13:310–6. doi: 10.1038/ncb2172
112. Wu S, Wang H, Li Y, Xie Y, Huang C, Zhao H, et al. Transcription factor YY1 promotes cell proliferation by directly activating the pentose phosphate pathway. Cancer Res (2018) 78:4549–62. doi: 10.1158/0008-5472.can-17-4047
113. Shi DY, Xie FZ, Zhai C, Stern JS, Liu Y, Liu SL. The role of cellular oxidative stress in regulating glycolysis energy metabolism in hepatoma cells. Mol Cancer (2009) 8:32. doi: 10.1186/1476-4598-8-32
114. Tseng HY, Chen YA, Jen J, Shen PC, Chen LM, Lin TD, et al. Oncogenic MCT-1 activation promotes YY1-EGFR-MnSOD signaling and tumor progression. Oncogenesis (2017) 6:e313. doi: 10.1038/oncsis.2017.13
115. Liu W, Guo Q, Zhao H. Oxidative stress-elicited YY1 potentiates antioxidative response via enhancement of NRF2-driven transcriptional activity: A potential neuronal defensive mechanism against ischemia/reperfusion cerebral injury. BioMed Pharmacother (2018) 108:698–706. doi: 10.1016/j.biopha.2018.09.082
116. Kawano Y, Cohen DE. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol (2013) 48:434–41. doi: 10.1007/s00535-013-0758-5
117. Lu Y, Ma Z, Zhang Z, Xiong X, Wang X, Zhang H, et al. Yin yang 1 promotes hepatic steatosis through repression of farnesoid X receptor in obese mice. Gut (2014) 63:170–8. doi: 10.1136/gutjnl-2012-303150
118. Lai CY, Lin CY, Hsu CC, Yeh KY, Her GM. Liver-directed microRNA-7a depletion induces nonalcoholic fatty liver disease by stabilizing YY1-mediated lipogenic pathways in zebrafish. Biochim Biophys Acta Mol Cell Biol Lipids (2018) 1863:844–56. doi: 10.1016/j.bbalip.2018.04.009
119. Yuan X, Chen J, Cheng Q, Zhao Y, Zhang P, Shao X, et al. Hepatic expression of yin yang 1 (YY1) is associated with the non-alcoholic fatty liver disease (NAFLD) progression in patients undergoing bariatric surgery. BMC Gastroenterol (2018) 18:147. doi: 10.1186/s12876-018-0871-2
120. Pan G, Diamanti K, Cavalli M, Lara Gutiérrez A, Komorowski J, Wadelius C. Multifaceted regulation of hepatic lipid metabolism by YY1. Life Sci Alliance (2021) 4. doi: 10.26508/lsa.202000928
121. Li Y, Kasim V, Yan X, Li L, Meliala ITS, Huang C, et al. Yin yang 1 facilitates hepatocellular carcinoma cell lipid metabolism and tumor progression by inhibiting PGC-1beta-induced fatty acid oxidation. Theranostics (2019) 9:7599–615. doi: 10.7150/thno.34931
122. Liu H, He B, Hu W, Liu K, Dai Y, Zhang D, et al. Prenatal dexamethasone exposure induces nonalcoholic fatty liver disease in male rat offspring via the miR-122/YY1/ACE2-MAS1 pathway. Biochem Pharmacol (2021) 185:114420. doi: 10.1016/j.bcp.2021.114420
123. Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J Clin Invest (2013) 123:3678–84. doi: 10.1172/jci69600
124. Li T, Le A. Glutamine metabolism in cancer. Adv Exp Med Biol (2018) 1063:13–32. doi: 10.1007/978-3-319-77736-8_2
125. Luo C, Chen X, Bai Y, Xu L, Wang S, Yao L, et al. Upregulation of yin-Yang-1 associates with proliferation and glutamine metabolism in esophageal carcinoma. Int J Genomics (2022) 2022:9305081. doi: 10.1155/2022/9305081
126. Huang X, Pan L, Zuo Z, Li M, Zeng L, Li R, et al. LINC00842 inactivates transcription co-regulator PGC-1α to promote pancreatic cancer malignancy through metabolic remodelling. Nat Commun (2021) 12:3830. doi: 10.1038/s41467-021-23904-4
127. Guendisch U, Loos B, Cheng PF, Dummer R, Levesque MP, Varum S, et al. Loss of YY1, a regulator of metabolism in melanoma, drives melanoma cell invasiveness and metastasis formation. Front Cell Dev Biol (2022) 10:916033. doi: 10.3389/fcell.2022.916033
128. Castellano G, Torrisi E, Ligresti G, Nicoletti F, Malaponte G, Traval S, et al. Yin yang 1 overexpression in diffuse large b-cell lymphoma is associated with b-cell transformation and tumor progression. Cell Cycle (2010) 9:557–63. doi: 10.4161/cc.9.3.10554
129. Erin N, Grahovac J, Brozovic A, Efferth T. Tumor microenvironment and epithelial mesenchymal transition as targets to overcome tumor multidrug resistance. Drug Resist Update (2020) 53:100715. doi: 10.1016/j.drup.2020.100715
130. Huang T, Song C, Zheng L, Xia L, Li Y, Zhou Y. The roles of extracellular vesicles in gastric cancer development, microenvironment, anti-cancer drug resistance, and therapy. Mol Cancer (2019) 18:62. doi: 10.1186/s12943-019-0967-5
131. Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting tumor microenvironment for cancer therapy. Int J Mol Sci (2019) 20. doi: 10.3390/ijms20040840
132. Shi J, Hao A, Zhang Q, Sui G. The role of YY1 in oncogenesis and its potential as a drug target in cancer therapies. Curr Cancer Drug Targets (2015) 15:145–57. doi: 10.2174/1568009615666150131124200
Keywords: YY1, cancer, tumor microenvironment, tumor progression, tumor immune
Citation: Li M, Wei J, Xue C, Zhou X, Chen S, Zheng L, Duan Y, Deng H, Xiong W, Tang F, Li G and Zhou M (2023) Dissecting the roles and clinical potential of YY1 in the tumor microenvironment. Front. Oncol. 13:1122110. doi: 10.3389/fonc.2023.1122110
Received: 12 December 2022; Accepted: 13 March 2023;
Published: 04 April 2023.
Edited by:
Tinghe Yu, Chongqing Medical University, ChinaReviewed by:
Frankie Chi Fat Ko, The University of Hong Kong, Hong Kong SAR, ChinaJie Li, Chinese Academy of Medical Sciences and Peking Union Medical College, China
Copyright © 2023 Li, Wei, Xue, Zhou, Chen, Zheng, Duan, Deng, Xiong, Tang, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Zhou, zhouming2001@163.com
 MengNa Li1,2,3,4
MengNa Li1,2,3,4 Wei Xiong
Wei Xiong FaQing Tang
FaQing Tang GuiYuan Li
GuiYuan Li Ming Zhou
Ming Zhou