- 1UPMC Hillman Cancer Center, Division of Hematology and Oncology, Department of Medicine, University of Pittsburgh, Pittsburgh, PA, United States
- 2Department of Immunology, University of Pittsburgh, Pittsburgh, PA, United States
Immune checkpoint inhibitors (ICIs) have revolutionized cancer treatment including in head and neck squamous cell carcinomas (HNSCCs); however, only a fraction of HNSCC patients respond to ICI, whereas the majority fail to do so. The mechanisms underlying such variable responses remain incompletely understood. A better understanding of such mechanisms may broaden the spectrum of responding patients and enhance the rate of ICI response. HNSCCs exhibit a high level of genetic heterogeneity, manifested as mutations or amplifications of oncogenes (e.g., PIK3CA) and mutations of tumor suppressor genes (e.g., TP53). The immune tumor microenvironment (TME) of HNSCCs also varies significantly in composition and in relative abundance of distinct immune subsets such as CD8 tumor-infiltrating lymphocytes (TILs) or tumor-associated macrophages (TAMs), which represents a high degree of immunological heterogeneity. Here, we briefly discuss how heterogeneous ICI responses may be attributed to tumor-intrinsic factors, including genetic, transcriptional, and functional variations in tumor cells, and host-intrinsic factors, including cellular composition of the TME (e.g., CD8 TILs and TAMs), and host-intrinsic differences in the T cell receptor (TCR) repertoire of CD8 TILs. We also discuss the potential impact of these factors on designing strategies for personalized immunotherapy of HNSCCs.
Introduction
Head and neck cancer (HNC) is a heterogeneous group of cancers arising from the mucosal surfaces of the upper aerodigestive tract including sinonasal and oral cavities, nasopharynx, oropharynx, hypopharynx, and larynx (1). Collectively, HNC is the sixth most prevalent cancer worldwide with 890,000 new cases and 450,000 deaths in 2018 (1, 2). In the United States (US), HNC accounts for 3-4% new cases of all cancer types (3), with 90% of cases being head and neck squamous cell carcinoma (HNSCC). The risk factors for HNSCC include carcinogens (e.g., tobacco and alcohol) and human papilloma virus (HPV) (1, 4). Hence, HNSCC can be classified as HPV+ or HPV− based on the distinct etiological factors. In general, HPV+ HNSCC patients exhibited better overall survival (OS) than HPV− patients, the latter showing worse prognosis (1, 4). During the past few decades, the incidence of HPV+ oropharyngeal HNSCCs has been increasing rapidly in the US (1, 4).
Extensive genomic and multi-omic studies performed using HNSCC patient samples conclude that HNSCCs displayed a high level of tumor heterogeneity including genetic, epigenetic, transcriptional, and functional variations between tumors or within tumors. A comprehensive multi-omic study revealed that HPV− HNSCCs can be clustered into three major subtypes by integrating copy-number, RNA, miRNA, protein, and phosphor-peptide data. The three subtypes include high chromosome instability (CIN), Basal, and Immune (5). CIN cluster was associated with heavy smoking and exhibited the worst prognosis (5). On the other hand, Immune cluster was enriched with tumors where smoking evidence was weak and associated with higher immune scores (5). Another study also employed multi-omic approaches to compare different types of SCCs in lung, cervix and head and neck, and showed that HNSCCs appeared to scatter broadly instead of localizing to discrete TumorMap islands and distributed into distinct iClusters (6). These findings are consistent with other large genomic studies showing that HNSCCs harbor a high level of genetic and epigenetic alterations (7–9).
HNSCC datasets of the Cancer Genome Atlas (TCGA) identified many commonly occurring genetic alterations in both HPV− and HPV+ HNSCCs. The most commonly mutated gene in HNSCC is tumor suppressor gene TP53 (8), encoding a transcription factor regulating DNA repair, cell cycle, senescence, and apoptosis (10). Over 80% of HPV− HNSCCs harbor TP53 mutations; in contrast, TP53 mutations almost never occur in HPV+ HNSCCs (~3%) (8), due to p53 protein degradation induced by the HPV E6 oncoprotein (11, 12). Another commonly mutated gene in HNSCCs is PIK3CA, encoding a catalytic subunit (p110α) of phosphoinositide 3-kinase (PI3K). PIK3CA genetic alterations, including both point mutations and gene amplification, affected both HPV+ and HPV− HNSCCs (56% and 34%, respectively) (8, 13), making PIK3CA the most frequently mutated gene in HPV+ HNSCCs. We evaluated the TCGA HNSCC dataset and found that the patients with PIK3CA amplification and gain (PIK3CAAmp) had a much greater chance of harboring TP53 mutations (14). Moreover, PIK3CAAmp/TP53Mut group exhibited significantly worse survival compared to PIK3CAWT/TP53WT groups (14).
Prior studies have generated murine models that mimicked the alterations of PIK3CA and/or p53 in HNSCCs (15–17); however, none of the previous studies showed that genetic alterations in these two genes can spontaneously induce HNSCC development. We recently established a genetically engineered mouse model by deleting p53 and constitutively activating PIK3CA in mouse keratin 15-expressing (K15+) stem cells, which leads to the spontaneous development of multi-lineage tumors including SCCs, termed keratin-15-p53-PIK3CA (KPPA) tumors (14). Furthermore, we derived transplantable daughter cell lines from KPPA tumors, which may provide a platform for testing new therapeutic strategies in HNSCCs (18).
HNSCCs also exhibited a high level of immunological heterogeneity, evidenced by a highly variable immune tumor microenvironment (TME) (19–21). Prior studies showed that the infiltration extent of CD8 tumor-infiltrating lymphocytes (TILs) correlated with HNSCC prognosis (22–25), while myeloid cell infiltration may contribute to worse survival and metastasis (19). We uploaded RNA-seq data of TCGA-HNSCC patients onto CIBERSORT and found that HNSCCs with PIK3CAAmp/TP53mut have significantly lower expression of gene signatures for CD8 T cells and activated natural killer (NK) cells but significantly higher expression of macrophage gene signature, compared with HNSCCs lacking both mutations (14). HNSCC is also characterized by defects in DNA repair pathways that can be induced by drug perturbation such as PARP inhibitor (26) or by loss of tumor suppressors such as Smad4 (27). SMAD4 loss has been associated with downregulation of FANC/BRCA genes in HNSCC harboring increased genomic instability (26). Genomic instability in HNSCC may generate cytosolic double-stranded DNA (dsDNA), which can be sensed by STING protein (28). STING activation subsequently induces type I interferon (IFN) and TNFα production, and triggers anti-tumor innate immunity (26). It would be of great interest to further elucidate whether a different level of genomic instability influences the level of CD8 TILs in HNSCC.
Differential responses to immune checkpoint inhibitors (ICI) in human HNSCC patients
So far, two ICIs, namely, nivolumab and pembrolizumab, both of which are anti-PD-1 monoclonal antibodies (mAbs), were approved by FDA for treating recurrent/metastatic (R/M) HNSCCs (29–31). However, only a fraction of HNSCC patients (10-20%) responded to ICI while others failed to do so (29–31). KEYNOTE-048 trial tested ICI for treating R/M HNSCCs in first line therapy (32). The study reported positive results in OS, thus, ICI emerged as the new standard-of-care (SOC) therapy (32). According to the observed efficacy and safety, pembrolizumab (pembro) plus platinum and 5-fluorouracil (5-FU) serves as a proper first-line therapy for R/M HNSCC while pembro monotherapy is an appropriate first-line treatment for PD-L1+ R/M HNSCC (combined positive score (CPS)>1). However, many issues remain to be addressed. While pembro plus chemotherapy increased OS, it did not significantly increase overall response rate (ORR) between Pembro+Platinum+5-FU (35.6%) vs. EXTREME (cetuximab+Platinum+5-FU) (36.3%), moreover, it significantly reduced the duration of response (DOR) between Pembro only (22.6 months) vs. Pembro+Platinum+5-FU (6.7 months) (32). Hence, other strategies are worth exploring to improve ORR and extend DOR. With regard to pembro monotherapy, the ORR still remained low, and the progressive disease rate was 40.5% (32); thus, it is critical to enhance treatment efficacy and better stratify and identify patients who would benefit the most from ICI treatment. Besides chemotherapy, HNSCC patients are often treated by radiation therapy (RT), whose critical role has been investigated and reviewed extensively (33–38).
For treating locally advanced HNSCCs, a randomized, double-blind, and placebo-controlled phase 3 trial compared avelumab (anti-PD-L1) plus chemoradiotherapy (CRT) vs. CRT alone, where anti-PD-L1 was administrated concurrently with CRT; however, the trial reported negative results (39). Therefore, more effective, and novel combinatorial strategies are needed to better treat locally advanced HNSCCs. Nivolumab was employed to treat at-risk, previously untreated, resectable HPV+ and HPV− HNSCC in a neoadjuvant setting (40–43). One of the neoadjuvant trials compared HPV+ vs. HPV− HNSCCs and showed that HPV+ HNSCC patients responded better to neoadjuvant nivolumab, although the response rate was low in both HPV+ and HPV− HNSCCs (40) compared to other cancer types (44, 45). Less responsiveness in HNSCC is consistent with its immunosuppressive TME (19, 46). Recent and ongoing ICI trials in HNSCCs were extensively reviewed (31, 47). However, it remains poorly understood why some patients responded to ICIs while others failed to do so (48–50). A better understanding of the mechanisms underlying such variable responses may broaden the spectrum of responding patients and enhance the rate of ICI response in HNSCCs.
It remains unresolved whether HPV+ HNSCC patients responded to ICI better than HPV− ones because clinical studies reported inconsistent results (Table 1). In the KEYNOTE-012 trial with pembro as first-line or subsequent-line treatment, R/M HPV+ HNSCC patients showed a higher ORR than HPV− patients (25% vs. 14%) (51). The expansion cohort of KEYNOTE-012 reported an even higher ORR (32% vs. 14%), and favorable 6-month progression-free survival rate (37% vs. 20%) and 6-month OS rate (70% vs. 56%) in HPV+ HNSCC patients compared to HPV− ones (52). Similarly, a higher ORR was reported for HPV+ patients in CheckMate 141 trial (53) using nivolumab (HPV+ vs. HPV−: 17.2% vs. 14.3%) and in HAWK study (54) using durvalumab (HPV+ vs. HPV−: 29.4% vs. 10.8%). However, Keynote-055 trial using pembro showed a similar ORR in R/M HNSCC patients regardless of HPV status (HPV+ vs. HPV−: 16% vs. 15%) (55). A similar ORR was also reported for HPV+ and HPV− HNSCC patients (HPV+ vs. HPV−, 15% vs. 17%) in NCT01375842 trial using Atezolizumab (56). Two recent meta-analysis integrated all the clinical data and showed that anti-PD-1/PD-L1 therapy favored a higher response rate in HPV+ than HPV− HNSCC patients (57, 58). While these studies collectively suggest an increased sensitivity of HPV+ HNSCCs to ICI treatment, future clinical trials with a greater number of patients probably are needed to completely resolve this issue.
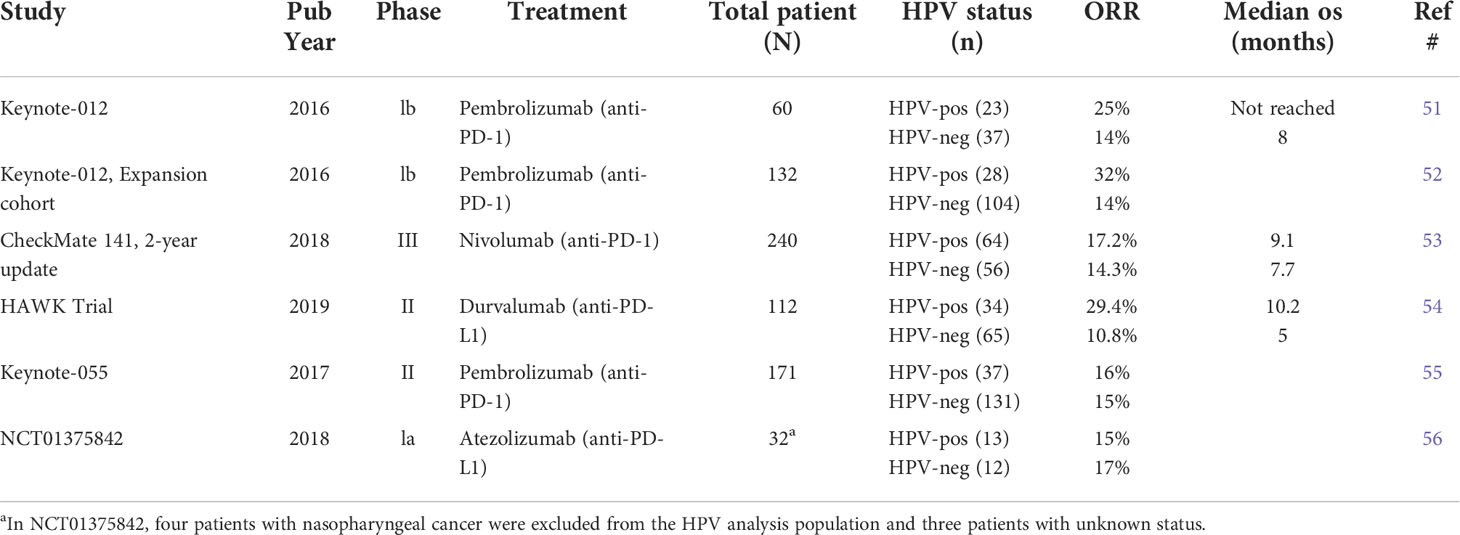
Table 1 Summary of clinical trial studies for differential ICI response in HPV+ vs HPV− HNSCC patients.
Differential responses to ICI in mouse HNSCC models
To better delineate why ICI treatment results into variable responses, we employed another syngeneic mouse model of SCC, namely, the A223 tumor with Smad4 deletion, which has been characterized previously (59–61). While Smad4 mutations do not occur commonly in HNSCCs, Smad4 deletion is frequently observed in a large portion of HNSCC samples (62). Given Smad4 plays a key role in TGFβ signaling and Smad4-deficient SCCs elevated TGFβ level (63), we investigated whether Smad4-/- SCCs responded to combined TGFβ/PD-L1 blockade differentially. We found that distinct immune TME profiles of therapeutic responders emerge in combined TGFβ/PD-L1 blockade-treated SCC (64). Responders contained more CD8 TILs and these CD8 TILs also exhibited more potent effector functions compared to non-responders (64). Additionally, responders harbored more M1 macrophages and less resident monocytes in the TME, compared to non-responders (64). The expression of major histocompatibility complex (MHC) was higher on responder myeloid cells or dendritic cells than non-responder counterparts (64). Nevertheless, it remains unclear why certain tumor recipients emerged as responders while others as non-responders.
To test whether oncogenic driver mutations affect differential ICI responses, we employed the two established KPPA tumor lines (TAb2 vs. TCh3), both of which harbor TP53 deletion and PIK3CA hyperactivation. When transplanted into C57BL/6 recipients, TAb2 and TCh3 tumors responded to anti-PD-L1 differentially, with the former completely lacking response and the latter being relatively sensitive (18). We employed conventional flow cytometry and single-cell RNA-sequencing to identify the difference in TILs. We found that TAb2 and TCh3 KPPA tumors exhibited heterogeneous immune profiles pre-existing treatment that dictated their unresponsiveness or sensitivity to anti-PD-L1 (18). Others have established murine HNSCC cell lines from primary 4NQO-induced tumors in the tongue of C57BL/6 (B6) mice, which were designated 4MOSC, short for 4NQO-induced murine oral squamous cells (65). Some of the 4MOSC cell lines also exhibited variable responses to anti-PD-1 (65). Overall, ICI treatment can result in heterogeneous outcomes in preclinical models. These findings are consistent with clinical observations.
Tumor-intrinsic factors influence differential ICI responses
Extensive prior studies have suggested a critical role of tumor-intrinsic factors in mediating differential responses to ICI, including tumor mutational burden (TMB), PD-L1 expression, or genetic and epigenetic differences in tumor cells themselves (Figure 1). For instance, TMB was shown to correlate to ICI efficacy in melanoma and non-small cell lung cancer (NSCLC) (66–68). However, the role of TMB in HNSCCs remains controversial. While studies showed that TMBhigh HNSCCs responded to ICI treatment better (69–71), conflicting data showed that TMB did not correlate with ICI response (72, 73). The pathogenesis of HPV− HNSCCs are strongly associated with carcinogens (e.g., tobacco); thus, HPV− HNSCCs contain a high level of TMB yet they failed to respond to neoadjuvant nivolumab treatment as well as melanoma or NSCLCs (40). Overall, these data indicate that the TMB level does not fully explain differential ICI responses in HNSCCs. It remains unclear why the role of TMB in HNSCCs differ from melanoma and NSCLC. It is possible that HNSCC is a type of cancers that are inherently heterogeneous (5, 6). It is also possible that HNSCCs may have a lower level of CD8 TIL infiltration before ICI treatment compared to other cancers such as melanoma.
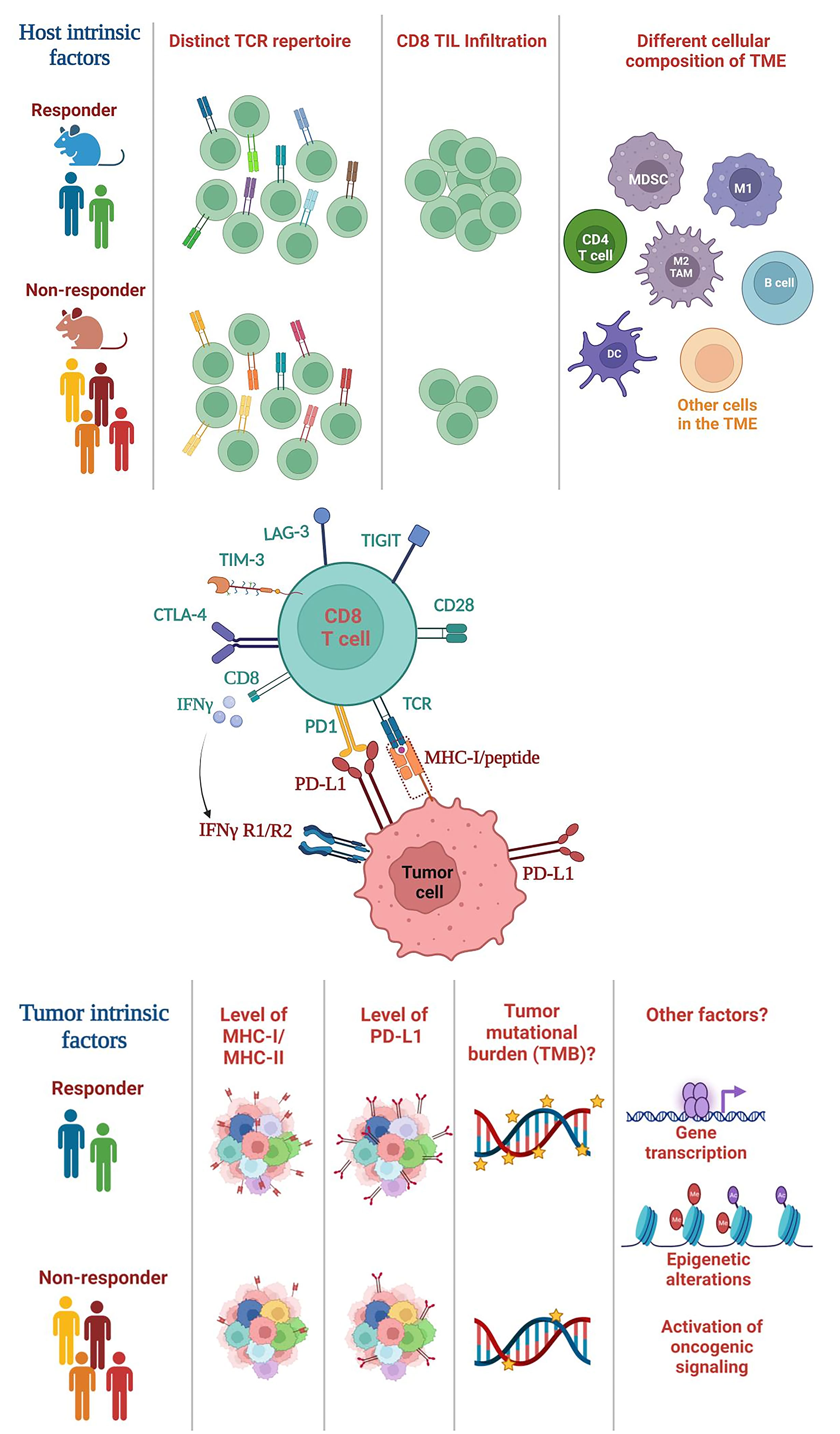
Figure 1 Summary of host-intrinsic and tumor-intrinsic factors that may influence the heterogeneous outcomes of ICI treatment. (Top) Host-intrinsic factors: (1) distinct TCR repertoires in responders vs. non-responders, (2) different level of CD8 TIL infiltration, (3) different cellular composition of the TME. Various subsets of immune cells are shown including CD4 T cells, B cells and myeloid cells, whereas many subsets of other cells in the TME are not shown including NK cells, Tregs, fibroblasts etc. TCR, T cell receptor; TIL, tumor-infiltrating lymphocyte; TME, tumor microenvironment. (Middle) The interaction between CD8 T cells and tumor cells. CD8 T cells recognize the MHC class I/peptide complex present on tumor cell surface. CD8 T cells can express various exhaustion markers including PD-1, TIM-3, LAG-3 etc. CD8 T cells can also secrete cytokines such as IFN-γ, while tumor cells express IFNγ receptor (IFNγR). (Bottom) Tumor-intrinsic factors: (1) differential level of MHC class I or class II expression, (2) PD-L1 expression on tumors (shown) or other cells in the TME (not shown), (3) the level of TMB, and (4) other potential factors such as genetic or epigenetic alterations in tumor cells, differential transcriptomes; activation of different oncogenic signaling pathways. TMB, tumor mutational burden.
PD-L1 expression has been used as a biomarker for correlating ICI responses in HNSCCs. Clinical trial data showed that PD-L1 expression is predictive of the response rate and survival if the CPS was used based on expression of tumor and TME (31). However, the PD-L1 expression based on the tumor proportion score (TPS) does not predict ICI response rate or survival (31), suggesting that tumor-derived PD-L1 expression is less important than combined PD-L1 expression from both tumor and TME. Nevertheless, CPS cannot accurately predict ICI responses and additional accurate biomarkers are needed. Tumors may respond to ICI better if tumor cells are capable of increasing the PD-L1 expression in response to inflammatory stimuli abundant in the immune TME. In line with this idea, studies from melanoma suggest that conserved IFN-γ signaling drives clinical response to ICI treatment (74), although it remains unknown whether this mechanism also operates in HNSCCs. Our studies suggest that the ability of tumor cells to upregulate PD-L1 expression in response to IFN-γ stimulation may serve as a predictive marker for ICI responses (18), consistent with prior findings that IFN-γ and expanded immune gene signatures correlated with better ICI response in HNSCCs (75).
We uncovered tumor-intrinsic differences that may underlie the differential responses to ICI by employing two KPPA tumor lines, TAb2 vs. TCh3 (18). TAb2 tumors failed to respond to anti-PD-L1, whereas TCh3 tumors were relatively sensitive (18). Unresponsive TAb2 tumors were highly enriched with functional tumor-associated macrophages (TAMs), especially M2-TAMs (18). In contrast, sensitive TCh3 tumors contained more CD8 TILs with better effector functions before anti-PD-L1 treatment (18). While anti-PD-L1 did not affect the TME of TAb2 tumors, it significantly increased the number of CD8 TILs in TCh3 tumors (18). These studies suggest that pre-existing immune profiles may dictate the likelihood of a given tumor to respond to anti-PD-L1, consistent with clinical data showing that increased CD8 TILs before ICI treatment correlated with better responses and survival (76).
The obvious question is why these two tumor lines exhibited differential immune profiles before anti-PD-L1 treatment. We performed RNA-seq and whole exome sequencing (WES) and discovered tumor-specific transcriptional, genetic, and epigenetic changes in TAb2 and TCh3 (18). For example, TAb2 tumors expressed a higher level of CSF1, VEGF-C and VEGF-D, and TAb2 tumor cells drastically expanded F4/80+ TAMs from bone marrow precursors in a CSF1 and VEGF dependent manner (18). ICI unresponsive TAb2 tumors upregulated distinct signaling pathways that correlate with aggressive tumor phenotypes such as STAT3 pathway (18). However, it remains unknown what tumor-specific changes account for such differential phenotypes, and further studies are warranted. Our studies also suggest that stratifying cancers according to their genetic alterations alone may not be sufficient and evaluating HNSCC tumor-intrinsic cues together with immune profiles in the TME may help better predict ICI responses.
The effects of host-intrinsic factors on differential ICI responses
Since the adaptive immune system is vastly distinct between different individuals, it is possible that immunological heterogeneity may contribute to the highly variable outcomes of ICI therapy (Figure 1). One of the key features of adaptive immunity is “diversity”, generated via a somatic DNA recombination process, termed V(D)J recombination. V(D)J recombination occurs in a random stochastic manner in the progenitors of T or B cells, thereby creating a vastly diverse T cell receptor (TCR) or B cell receptor (BCR) repertoire in millions of T or B cells, respectively (77, 78). The TCR of most conventional T cells consists of two different protein chains, an alpha (α) chain and a beta (β) chain, encoded by TRA and TRB, respectively, and linked by disulfide bonds. A TCR clonotype consists of a unique TCRα and a TCRβ chain with unique V(D)J usage and complementarity-determining region 3 (CDR3). CDR3 encompasses the highly divergent junction of V(D)J recombination and determines TCR specificity; hence, its unique nucleotide or protein sequences can serve as a barcode for individual TCRs.
Studying the formation and diversity of the human TCR repertoire has been difficult due to limited access to human thymus samples and non-feasibility of manipulating variables in vivo. Therefore, humanized mouse models were generated by implanting immunodeficient mice with human hematopoietic stem cells (HSCs) and human thymus from the same or different donors to study the development of human T cell repertoire (79). Despite receiving identical HSCs and thymi and having same genetic background and environment, human TCR repertoires were formed in a largely stochastic manner and totally divergent in different recipient mice (79). This means that each individual has an almost completely different TCR repertoire, even in the case of identical twins, an observation which may explain why genetically controlled autoimmune diseases exhibit incomplete penetrance in monozygotic twins (80). Similar to the human TCR repertoires, we predict that individual mice will contain an almost completely different thymic TCR repertoire even if they have identical genetic background (e.g., B6). The initially formed TCR repertoire will be continuously shaped by additional factors including immunization of foreign antigens, pathogen infection, or therapeutic interventions (e.g., CRT). Nonetheless, the peripheral TCR repertoire will remain divergent in individual mice. Prior studies showed that many different TCR clonotypes can react to the same MHC/peptide antigens including model or viral antigens (81, 82). These studies collectively implicate a possibility that different mice could mount anti-tumor immune responses against the same tumor antigens utilizing totally distinct TCR clonotypes. Thus, we propose that the intrinsic differences in diverse TCR repertoires may also contribute to heterogeneous anti-tumor immune responses in different hosts (83). If so, this notion may offer a new explanation for why some hosts would harbor T cells that can eradicate tumors, while others would not.
We proposed “a hole in TCR repertoire” hypothesis to explain the differential ICI responses (83). Currently, there is no data to support the existence of “any hole” in the TCR repertoire given it is dynamic and constantly shaped by various factors. However, there are abundant data supporting immunological differences in individual cancer patients or mouse recipients transplanted with tumors. The frequency of CD8 and CD4 TILs differed in patient samples and positively correlated with clinical outcomes in HNSCCs (23). We also found that the percentage of CD8 TILs varied substantially in HNSCCs with a small fraction containing a high level of CD8 TILs while most patient samples were infiltrated with a low to moderate level of CD8 TILs (20), consistent with an immunosuppressive TME of HNSCCs (19, 84). However, it remains unknown whether the TCR repertoires of CD8 TILs differ in tumor-eradicating vs. tumor-progressing hosts. In this regard, our previous studies showed that Smad4−/− SCCs (A223) elicited divergent responses when transplanted into genetically identical WT B6 mice (20). While a small fraction of tumor-bearing recipients spontaneously rejected the A223 tumor (regressor), most of them underwent tumor progression (progressor) (20). Intriguingly, the top TCR clonotypes were almost mutually exclusive between regressors and progressors (20). Furthermore, both regressor and progressor top TCR clonotypes presented in a recipient-specific manner, suggesting a highly individualized anti-tumor immune response (20). Further studies are warranted to investigate whether the TCR repertoires of CD8 TILs differ in ICI responders vs. non-responders, which may be easier to address using syngeneic mouse models since WT B6 mice have a limited number of MHC class I alleles. The differences in TCR repertoires need to be functionally defined and quantified using an antigen-specific system which can test if distinct TCR clonotypes elicit qualitatively or quantitatively different responses against the same tumor-specific antigen.
It is thus conceivable that immunological heterogeneity (e.g., differences in the TIL TCR repertoire) contributes to the highly variable outcomes of ICI treatment. Why has this idea not been discussed before? Likely because the well-established dogma assumes that there would be sufficient TCR clones that can effectively recognize any tumor-antigen in a given individual due to the enormous size of a TCR repertoire: the estimated T cell number is about 3×1011 (85) and the number of TCR clonotypes is about 1010 in a given adult (86). Indeed, our adaptive immune system can recognize millions of different pathogens or foreign antigens. However, the effect of immunological heterogeneity is understudied in the context of anti-tumor immunity, and many fundamental questions remain to be addressed, for instance, the actual factors responsible for the highly variable ICI responses remain elusive. Beside TCR differences, BCR may also differ in responders vs. non-responders. Prior studies showed that characteristics of tumor-infiltrating B cells also varied significantly in HNSCCs and correlated with clinical outcomes (87, 88). Overall, future studies are needed to elucidate whether and how host-specific immunological heterogeneity influences differential responses to ICI.
Discussion
Why are responses to ICI heterogeneous in different cancer patients? What underlying mechanisms lead to such differential responses? These are imperative and fundamental questions for cancer immunology field. Addressing such questions may substantially impact developing new strategies for personalized cancer immunotherapy. We suggest that both tumor-intrinsic and host-intrinsic factors may contribute to differential ICI responses. For instance, by establishing and employing two SCC tumor lines, TAb2 vs. TCh3, both of which harbor TP53 deletion and PIK3CA hyperactivation, we uncovered tumor-intrinsic differences that may underlie the differential responses to ICI (18). However, it still remains to be addressed what tumor-specific genetic or epigenetic changes lead to unresponsiveness in TAb2 or sensitize TCh3 to anti-PD-L1 treatment, and whether such changes are also applicable to the heterogeneous ICI responses in human HNSCCs.
Distinct top TIL TCR clonotypes were found to correlate with tumor eradication vs. tumor progression phenotypes (20). This observation implies that regressor and progressor CD8 TILs might mount drastically different responses by employing distinct TCRs against the same A223 tumor cell line. In line with our observation, prior studies showed that many different TCR clonotypes can react to the same MHC/peptide antigens including model or viral antigens (81, 82). It remains unknown whether the top TCR clonotypes differ in ICI responders vs. non-responders. To address whether the spectrum of TCR clonotypes within regressor or ICI responder provides advantageous recognition of tumor antigens over the spectrum of TCR clonotypes within progressor or ICI non-responder, it would require an antigen-specific model system. Altogether, we propose that stochastic differences in TIL TCR repertoire may be one of several factors that might underlie differential responses to ICI treatment. Of course, this notion does not exclude the contribution of tumor-intrinsic factors, including TMB, tumor immunogenicity, PD-L1 expression or others, to differential ICI responses (21, 50, 89–92); nevertheless, our hypothesis may offer a new perspective to test whether stochastic differences in TCR repertoire contribute to variable ICI responses in different individuals.
Author contributions
Conceptualization: JW; Writing: JW, ZC, and JJ. All authors contributed to the article and approved the submitted version.
Funding
This work is supported by UPMC Hillman Cancer Center startup fund to JW, NIH R01-DE027329, R01-DE028420, and R01-DE031947 to JW. ZC was supported by ACS IRG #16-184-56 from the American Cancer Society. The sponsors or funders have no role in the preparation, review, or approval of the manuscript.
Acknowledgments
We thank Christine Maloney and Dr. Samantha Chen for proofreading the manuscript. We apologize to those whose work was not cited due to length restrictions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers (2020) 6(1):92. doi: 10.1038/s41572-020-00224-3
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
3. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708
4. Cramer JD, Burtness B, Le QT, Ferris RL. The changing therapeutic landscape of head and neck cancer. Nat Rev Clin Oncol (2019) 16(11):669–83. doi: 10.1038/s41571-019-0227-z
5. Huang C, Chen L, Savage SR, Eguez RV, Dou Y, Li Y, et al. Proteogenomic insights into the biology and treatment of HPV-negative head and neck squamous cell carcinoma. Cancer Cell (2021) 39(3):361–79 e16. doi: 10.1016/j.ccell.2020.12.007
6. Campbell JD, Yau C, Bowlby R, Liu Y, Brennan K, Fan H, et al. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep (2018) 23(1):194–212 e6. doi: 10.1016/j.celrep.2018.03.063
7. Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science (2011) 333(6046):1154–7. doi: 10.1126/science.1206923
8. Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature (2015) 517(7536):576–82. doi: 10.1038/nature14129
9. Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science (2011) 333(6046):1157–60. doi: 10.1126/science.1208130
10. Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer (2014) 14(5):359–70. doi: 10.1038/nrc3711
11. Wang Z, Xia RH, Ye DX, Li J. Human papillomavirus 16 infection and TP53 mutation: Two distinct pathogeneses for oropharyngeal squamous cell carcinoma in an Eastern Chinese population. PloS One (2016) 11(10):e0164491. doi: 10.1371/journal.pone.0164491
12. Buitrago-Perez A, Hachimi M, Duenas M, Lloveras B, Santos A, Holguin A, et al. A humanized mouse model of HPV-associated pathology driven by E7 expression. PloS One (2012) 7(7):e41743. doi: 10.1371/journal.pone.0041743
13. Jung K, Kang H, Mehra R. Targeting phosphoinositide 3-kinase (PI3K) in head and neck squamous cell carcinoma (HNSCC). Cancers Head Neck (2018) 3:3. doi: 10.1186/s41199-018-0030-z
14. Chen SMY, Li B, Nicklawsky AG, Krinsky AL, Brunetti T, Woolaver RA, et al. Deletion of p53 and hyper-activation of PIK3CA in keratin-15(+) stem cells lead to the development of spontaneous squamous cell carcinoma. Int J Mol Sci (2020) 21(18):6585. doi: 10.3390/ijms21186585
15. Du L, Chen X, Cao Y, Lu L, Zhang F, Bornstein S, et al. Overexpression of PIK3CA in murine head and neck epithelium drives tumor invasion and metastasis through PDK1 and enhanced TGFβ signaling. Oncogene (2016) 35(35):4641–52. doi: 10.1038/onc.2016.1
16. Savar A, Acin S, Gonzalez CL, El-Sawy T, Mejia O, Li Z, et al. Loss of epithelial p53 and αv integrin cooperate through akt to induce squamous cell carcinoma yet prevent remodeling of the tumor microenvironment. Oncogene (2015) 34(4):516–24. doi: 10.1038/onc.2013.585
17. García-Carracedo D, Cai Y, Qiu W, Saeki K, Friedman RA, Lee A, et al. PIK3CA and p53 mutations promote 4NQO-initated head and neck tumor progression and metastasis in mice. Mol Cancer Res (2020) 18(6):822–34. doi: 10.1158/1541-7786.Mcr-19-0549
18. Chen SMY, Popolizio V, Woolaver RA, Ge H, Krinsky AL, John J, et al. Differential responses to immune checkpoint inhibitor dictated by pre-existing differential immune profiles in squamous cell carcinomas caused by same initial oncogenic drivers. J Exp Clin Cancer Res (2022) 41(1):123. doi: 10.1186/s13046-022-02337-x
19. Chen SMY, Krinsky AL, Woolaver RA, Wang X, Chen Z, Wang JH. Tumor immune microenvironment in head and neck cancers. Mol Carcinog (2020) 59(7):766–74. doi: 10.1002/mc.23162
20. Woolaver RA, Wang X, Krinsky AL, Waschke BC, Chen SMY, Popolizio V, et al. Differences in TCR repertoire and T cell activation underlie the divergent outcomes of antitumor immune responses in tumor-eradicating versus tumor-progressing hosts. J immunother Cancer (2021) 9(1):e001615. doi: 10.1136/jitc-2020-001615
21. Canning M, Guo G, Yu M, Myint C, Groves MW, Byrd JK, et al. Heterogeneity of the head and neck squamous cell carcinoma immune landscape and its impact on immunotherapy. Front Cell Dev Biol (2019) 7:52. doi: 10.3389/fcell.2019.00052
22. Nordfors C, Grun N, Tertipis N, Ahrlund-Richter A, Haeggblom L, Sivars L, et al. CD8+ and CD4+ tumour infiltrating lymphocytes in relation to human papillomavirus status and clinical outcome in tonsillar and base of tongue squamous cell carcinoma. Eur J Cancer (2013) 49(11):2522–30. doi: 10.1016/j.ejca.2013.03.019
23. de Ruiter EJ, Ooft ML, Devriese LA, Willems SM. The prognostic role of tumor infiltrating T-lymphocytes in squamous cell carcinoma of the head and neck: A systematic review and meta-analysis. Oncoimmunology (2017) 6(11):e1356148. doi: 10.1080/2162402X.2017.1356148
24. Ward MJ, Thirdborough SM, Mellows T, Riley C, Harris S, Suchak K, et al. Tumour-infiltrating lymphocytes predict for outcome in HPV-positive oropharyngeal cancer. Br J Cancer (2014) 110(2):489–500. doi: 10.1038/bjc.2013.639
25. Nguyen N, Bellile E, Thomas D, McHugh J, Rozek L, Virani S, et al. Tumor infiltrating lymphocytes and survival in patients with head and neck squamous cell carcinoma. Head Neck (2016) 38(7):1074–84. doi: 10.1002/hed.24406
26. Moutafi M, Economopoulou P, Rimm D, Psyrri A. PARP inhibitors in head and neck cancer: Molecular mechanisms, preclinical and clinical data. Oral Oncol (2021) 117:105292. doi: 10.1016/j.oraloncology.2021.105292
27. Hernandez AL, Young CD, Wang JH, Wang XJ. Lessons learned from SMAD4 loss in squamous cell carcinomas. Mol Carcinog (2019) 58(9):1648–55. doi: 10.1002/mc.23049
28. Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol (2016) 17(10):1142–9. doi: 10.1038/ni.3558
29. Ferris RL, Blumenschein G, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. New Engl J Med (2016) 375(19):1856–67. doi: 10.1056/NEJMoa1602252
30. Hsieh RW, Borson S, Tsagianni A, Zandberg DP. Immunotherapy in Recurrent/Metastatic squamous cell carcinoma of the head and neck. Front Oncol (2021) 11:705614. doi: 10.3389/fonc.2021.705614
31. Borel C, Jung AC, Burgy M. Immunotherapy breakthroughs in the treatment of recurrent or metastatic head and neck squamous cell carcinoma. Cancers (Basel) (2020) 12(9):2691. doi: 10.3390/cancers12092691
32. Burtness B, Harrington KJ, Greil R, Soulieres D, Tahara M, de Castro G Jr., et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet (2019) 394(10212):1915–28. doi: 10.1016/S0140-6736(19)32591-7
33. Karam SD, Raben D. Radioimmunotherapy for the treatment of head and neck cancer. Lancet Oncol (2019) 20(8):e404–e16. doi: 10.1016/S1470-2045(19)30306-7
34. Hernandez AL, Young CD, Bian L, Weigel K, Nolan K, Frederick B, et al. PARP inhibition enhances radiotherapy of SMAD4-deficient human head and neck squamous cell carcinomas in experimental models. Clin Cancer Res (2020) 26(12):3058–70. doi: 10.1158/1078-0432.CCR-19-0514
35. Knitz MW, Bickett TE, Darragh LB, Oweida AJ, Bhatia S, Van Court B, et al. Targeting resistance to radiation-immunotherapy in cold HNSCCs by modulating the treg-dendritic cell axis. J immunother Cancer (2021) 9(4):e001955. doi: 10.1136/jitc-2020-001955
36. Hui C, Chau B, Gan G, Stokes W, Karam SD, Amini A. Overcoming resistance to immunotherapy in head and neck cancer using radiation: A review. Front Oncol (2021) 11:592319. doi: 10.3389/fonc.2021.592319
37. Anderson G, Ebadi M, Vo K, Novak J, Govindarajan A, Amini A. An updated review on head and neck cancer treatment with radiation therapy. Cancers (Basel) (2021) 13(19):4912. doi: 10.3390/cancers13194912
38. Zhou X, Wang X. Radioimmunotherapy in HPV-associated head and neck squamous cell carcinoma. Biomedicines (2022) 10(8):1990. doi: 10.3390/biomedicines10081990
39. Lee NY, Ferris RL, Psyrri A, Haddad RI, Tahara M, Bourhis J, et al. Avelumab plus standard-of-care chemoradiotherapy versus chemoradiotherapy alone in patients with locally advanced squamous cell carcinoma of the head and neck: a randomised, double-blind, placebo-controlled, multicentre, phase 3 trial. Lancet Oncol (2021) 22(4):450–62. doi: 10.1016/S1470-2045(20)30737-3
40. Ferris RL, Spanos WC, Leidner R, Goncalves A, Martens UM, Kyi C, et al. Neoadjuvant nivolumab for patients with resectable HPV-positive and HPV-negative squamous cell carcinomas of the head and neck in the CheckMate 358 trial. J immunother Cancer (2021) 9(6):e002568. doi: 10.1136/jitc-2021-002568
41. Ferrarotto R, Bell D, Rubin ML, Hutcheson KA, Johnson JM, Goepfert RP, et al. Impact of neoadjuvant durvalumab with or without tremelimumab on CD8(+) tumor lymphocyte density, safety, and efficacy in patients with oropharynx cancer: CIAO trial results. Clin Cancer Res (2020) 26(13):3211–9. doi: 10.1158/1078-0432.CCR-19-3977
42. Uppaluri R, Campbell KM, Egloff AM, Zolkind P, Skidmore ZL, Nussenbaum B, et al. Neoadjuvant and adjuvant pembrolizumab in resectable locally advanced, human papillomavirus-unrelated head and neck cancer: A multicenter, phase II trial. Clin Cancer Res (2020) 26(19):5140–52. doi: 10.1158/1078-0432.CCR-20-1695
43. Schoenfeld JD, Hanna GJ, Jo VY, Rawal B, Chen YH, Catalano PS, et al. Neoadjuvant nivolumab or nivolumab plus ipilimumab in untreated oral cavity squamous cell carcinoma: A phase 2 open-label randomized clinical trial. JAMA Oncol (2020) 6(10):1563–70. doi: 10.1001/jamaoncol.2020.2955
44. Amaria RN, Reddy SM, Tawbi HA, Davies MA, Ross MI, Glitza IC, et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat Med (2018) 24(11):1649–54. doi: 10.1038/s41591-018-0197-1
45. Forde PM, Chaft JE, Smith KN, Anagnostou V, Cottrell TR, Hellmann MD, et al. Neoadjuvant PD-1 blockade in resectable lung cancer. New Engl J Med (2018) 378(21):1976–86. doi: 10.1056/NEJMoa1716078
46. Succaria F, Kvistborg P, Stein JE, Engle EL, McMiller TL, Rooper LM, et al. Characterization of the tumor immune microenvironment in human papillomavirus-positive and -negative head and neck squamous cell carcinomas. Cancer immunol immunother: CII (2021) 70(5):1227–37. doi: 10.1007/s00262-020-02747-w
47. Kao HF, Lou PJ. Immune checkpoint inhibitors for head and neck squamous cell carcinoma: Current landscape and future directions. Head Neck (2019) 41 Suppl 1:4–18. doi: 10.1002/hed.25930
48. Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discovery (2018) 8(9):1069–86. doi: 10.1158/2159-8290.CD-18-0367
49. Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med (2018) 50(12):1–11. doi: 10.1038/s12276-018-0191-1
50. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer (2019) 19(3):133–50. doi: 10.1038/s41568-019-0116-x
51. Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol (2016) 17(7):956–65. doi: 10.1016/S1470-2045(16)30066-3
52. Chow LQM, Haddad R, Gupta S, Mahipal A, Mehra R, Tahara M, et al. Antitumor activity of pembrolizumab in biomarker-unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: Results from the phase ib KEYNOTE-012 expansion cohort. J Clin Oncol (2016) 34(32):3838–45. doi: 10.1200/jco.2016.68.1478
53. Ferris RL, Blumenschein G Jr., Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab vs investigator’s choice in recurrent or metastatic squamous cell carcinoma of the head and neck: 2-year long-term survival update of CheckMate 141 with analyses by tumor PD-L1 expression. Oral Oncol (2018) 81:45–51. doi: 10.1016/j.oraloncology.2018.04.008
54. Zandberg DP, Algazi AP, Jimeno A, Good JS, Fayette J, Bouganim N, et al. Durvalumab for recurrent or metastatic head and neck squamous cell carcinoma: Results from a single-arm, phase II study in patients with >/=25% tumour cell PD-L1 expression who have progressed on platinum-based chemotherapy. Eur J Cancer (2019) 107:142–52. doi: 10.1016/j.ejca.2018.11.015
55. Bauml J, Seiwert TY, Pfister DG, Worden F, Liu SV, Gilbert J, et al. Pembrolizumab for platinum- and cetuximab-refractory head and neck cancer: Results from a single-arm, phase II study. J Clin Oncol (2017) 35(14):1542–9. doi: 10.1200/JCO.2016.70.1524
56. Colevas AD, Bahleda R, Braiteh F, Balmanoukian A, Brana I, Chau NG, et al. Safety and clinical activity of atezolizumab in head and neck cancer: results from a phase I trial. Ann Oncol (2018) 29(11):2247–53. doi: 10.1093/annonc/mdy411
57. Xu Y, Zhu G, Maroun CA, Wu IXY, Huang D, Seiwert TY, et al. Programmed death-1/Programmed death-ligand 1-axis blockade in recurrent or metastatic head and neck squamous cell carcinoma stratified by human papillomavirus status: A systematic review and meta-analysis. Front Immunol (2021) 12:645170. doi: 10.3389/fimmu.2021.645170
58. Wang J, Sun H, Zeng Q, Guo XJ, Wang H, Liu HH, et al. HPV-positive status associated with inflamed immune microenvironment and improved response to anti-PD-1 therapy in head and neck squamous cell carcinoma. Sci Rep-Uk (2019) 9(1):13404. doi: 10.1038/s41598-019-49771-0
59. Mishra AK, Kadoishi T, Wang X, Driver E, Chen Z, Wang XJ, et al. Squamous cell carcinomas escape immune surveillance via inducing chronic activation and exhaustion of CD8+ T cells co-expressing PD-1 and LAG-3 inhibitory receptors. Oncotarget (2016) 7(49):81341–56. doi: 10.18632/oncotarget.13228
60. Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest (2009) 119(11):3408–19. doi: 10.1172/JCI38854
61. White RA, Neiman JM, Reddi A, Han G, Birlea S, Mitra D, et al. Epithelial stem cell mutations that promote squamous cell carcinoma metastasis. J Clin Invest (2013) 123(10):4390–404. doi: 10.1172/JCI65856
62. Hernandez AL, Wang Y, Somerset HL, Keysar SB, Aisner DL, Marshall C, et al. Inter- and intra-tumor heterogeneity of SMAD4 loss in head and neck squamous cell carcinomas. Mol Carcinog (2019) 58(5):666–73. doi: 10.1002/mc.22958
63. White RA, Malkoski SP, Wang XJ. TGFbeta signaling in head and neck squamous cell carcinoma. Oncogene (2010) 29(40):5437–46. doi: 10.1038/onc.2010.306
64. Strait AA, Woolaver RA, Hall SC, Young CD, Karam SD, Jimeno A, et al. Distinct immune microenvironment profiles of therapeutic responders emerge in combined TGFbeta/PD-L1 blockade-treated squamous cell carcinoma. Commun Biol (2021) 4(1):1005. doi: 10.1038/s42003-021-02522-2
65. Wang Z, Wu VH, Allevato MM, Gilardi M, He Y, Luis Callejas-Valera J, et al. Syngeneic animal models of tobacco-associated oral cancer reveal the activity of in situ anti-CTLA-4. Nat Commun (2019) 10(1):5546. doi: 10.1038/s41467-019-13471-0
66. Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther (2017) 16(11):2598–608. doi: 10.1158/1535-7163.MCT-17-0386
67. Dousset L, Poizeau F, Robert C, Mansard S, Mortier L, Caumont C, et al. Positive association between location of melanoma, ultraviolet signature, tumor mutational burden, and response to anti-PD-1 therapy. JCO Precis Oncol (2021) 5:PO.21.00084. doi: 10.1200/PO.21.00084
68. Chae YK, Davis AA, Raparia K, Agte S, Pan A, Mohindra N, et al. Association of tumor mutational burden with DNA repair mutations and response to anti-PD-1/PD-L1 therapy in non-Small-Cell lung cancer. Clin Lung Cancer (2019) 20(2):88–96 e6. doi: 10.1016/j.cllc.2018.09.008
69. Haddad RI, Seiwert TY, Chow LQM, Gupta S, Weiss J, Gluck I, et al. Influence of tumor mutational burden, inflammatory gene expression profile, and PD-L1 expression on response to pembrolizumab in head and neck squamous cell carcinoma. J immunother Cancer (2022) 10(2):e003026. doi: 10.1136/jitc-2021-003026
70. Burcher KM, Lantz JW, Gavrila E, Abreu A, Burcher JT, Faucheux AT, et al. Relationship between tumor mutational burden, PD-L1, patient characteristics, and response to immune checkpoint inhibitors in head and neck squamous cell carcinoma. Cancers (Basel) (2021) 13(22):5733. doi: 10.3390/cancers13225733
71. Hanna GJ, Lizotte P, Cavanaugh M, Kuo FC, Shivdasani P, Frieden A, et al. Frameshift events predict anti-PD-1/L1 response in head and neck cancer. JCI Insight (2018) 3(4):e98811. doi: 10.1172/jci.insight.98811
72. McGrail DJ, Pilie PG, Rashid NU, Voorwerk L, Slagter M, Kok M, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol (2021) 32(5):661–72. doi: 10.1016/j.annonc.2021.02.006
73. Xu X, Li R, Zhang L, Zhu G, Ren D, Wu L, et al. Identification of factors related to immunotherapy efficacy and prognosis in patients with advanced head and neck squamous cell carcinoma. Diagn Pathol (2021) 16(1):110. doi: 10.1186/s13000-021-01147-7
74. Grasso CS, Tsoi J, Onyshchenko M, Abril-Rodriguez G, Ross-Macdonald P, Wind-Rotolo M, et al. Conserved interferon-gamma signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell (2020) 38(4):500–15 e3. doi: 10.1016/j.ccell.2020.08.005
75. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest (2017) 127(8):2930–40. doi: 10.1172/JCI91190
76. Li F, Li C, Cai X, Xie Z, Zhou L, Cheng B, et al. The association between CD8+ tumor-infiltrating lymphocytes and the clinical outcome of cancer immunotherapy: A systematic review and meta-analysis. EClinicalMedicine (2021) 41:101134. doi: 10.1016/j.eclinm.2021.101134
77. Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell (2002) 109 Suppl:S45–55. doi: 10.1016/s0092-8674(02)00675-x
78. Jung D, Alt FW. Unraveling V(D)J recombination; insights into gene regulation. Cell (2004) 116(2):299–311. doi: 10.1016/s0092-8674(04)00039-x
79. Khosravi-Maharlooei M, Obradovic A, Misra A, Motwani K, Holzl M, Seay HR, et al. Crossreactive public TCR sequences undergo positive selection in the human thymic repertoire. J Clin Invest (2019) 130:2446–62. doi: 10.1172/JCI124358
80. Redondo MJ, Yu L, Hawa M, Mackenzie T, Pyke DA, Eisenbarth GS, et al. Heterogeneity of type I diabetes: analysis of monozygotic twins in great Britain and the united states. Diabetologia (2001) 44(3):354–62. doi: 10.1007/s001250051626
81. Dash P, Fiore-Gartland AJ, Hertz T, Wang GC, Sharma S, Souquette A, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature (2017) 547(7661):89–93. doi: 10.1038/nature22383
82. Tu AA, Gierahn TM, Monian B, Morgan DM, Mehta NK, Ruiter B, et al. TCR sequencing paired with massively parallel 3’ RNA-seq reveals clonotypic T cell signatures. Nat Immunol (2019) 20(12):1692–9. doi: 10.1038/s41590-019-0544-5
83. Wang JH. Why the outcome of anti-tumor immune responses is heterogeneous: A novel idea in the context of immunological heterogeneity in cancers. Bioessays (2020) 42(10):e2000024. doi: 10.1002/bies.202000024
84. Ferris RL. Immunology and immunotherapy of head and neck cancer. J Clin Oncol (2015) 33(29):3293–304. doi: 10.1200/JCO.2015.61.1509
85. Jenkins MK, Chu HH, McLachlan JB, Moon JJ. On the composition of the preimmune repertoire of T cells specific for peptide-major histocompatibility complex ligands. Annu Rev Immunol (2010) 28:275–94. doi: 10.1146/annurev-immunol-030409-101253
86. Lythe G, Callard RE, Hoare RL, Molina-Paris C. How many TCR clonotypes does a body maintain? J Theor Biol (2016) 389:214–24. doi: 10.1016/j.jtbi.2015.10.016
87. Hladikova K, Koucky V, Boucek J, Laco J, Grega M, Hodek M, et al. Tumor-infiltrating b cells affect the progression of oropharyngeal squamous cell carcinoma via cell-to-cell interactions with CD8(+) T cells. J immunother Cancer (2019) 7(1):261. doi: 10.1186/s40425-019-0726-6
88. Gavrielatou N, Vathiotis I, Economopoulou P, Psyrri A. The role of b cells in head and neck cancer. Cancers (Basel) (2021) 13(21):5383. doi: 10.3390/cancers13215383
89. Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med (2016) 8(328):328rv4. doi: 10.1126/scitranslmed.aad7118
90. Santini FC, Hellmann MD. PD-1/PD-L1 axis in lung cancer. Cancer J (2018) 24(1):15–9. doi: 10.1097/PPO.0000000000000300
91. Jardim DL, Goodman A, de Melo Gagliato D, Kurzrock R. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell (2021) 39(2):154–73. doi: 10.1016/j.ccell.2020.10.001
Keywords: immune checkpoint inhibitors, tumor heterogeneity, immunological heterogeneity, individualized anti-tumor immune responses, TCR repertoire
Citation: Chen Z, John J and Wang JH (2022) Why responses to immune checkpoint inhibitors are heterogeneous in head and neck cancers: Contributions from tumor-intrinsic and host-intrinsic factors. Front. Oncol. 12:995434. doi: 10.3389/fonc.2022.995434
Received: 15 July 2022; Accepted: 03 October 2022;
Published: 18 October 2022.
Edited by:
Anna Pasetto, Oslo University Hospital, NorwayReviewed by:
Haihua Yang, Wenzhou Medical University, ChinaMyrto K. Moutafi, Yale University, United States
Copyright © 2022 Chen, John and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing H. Wang, JHW51@pitt.edu
 Zhangguo Chen1
Zhangguo Chen1 Jing H. Wang
Jing H. Wang