- 1Department of Emergency and Organ Transplantation (D.E.T.O.), Hematology and Stem Cell Transplantation Unit, University of Bari “Aldo Moro”, Bari, Italy
- 2School of Medicine, University of Bari “Aldo Moro”, Bari, Italy
Inflammatory bowel diseases (IBDs) are a group of chronic conditions of the gastrointestinal tract in which nationwide studies have revealed a higher risk of hematological malignancies (HMs). Clonal hematopoiesis (CH) is a premalignant condition defined by the presence of an acquired somatic mutation characterized by a variant allele frequency (VAF) of ≥2%, in a gene frequently associated with HMs. A growing body of evidence suggests a correlation between inflammation and CH; its occurrence in the context of IBD has been previously demonstrated. With the aim to assess CH possible co-occurrence in patients with an IBD associated with HMs, we performed a targeted next-generation sequencing analysis in a cohort of thirteen patients who were referred to our center with IBD associated with HMs. Eleven (85%) patients showed one or more mutations in CH-associated genes; DNMT3A was the most frequently mutated gene, followed by ASXL1 and JAK2. These results may suggest that the mechanisms at the basis of the inflammatory environment could potentially select for the growth of hematopoietic clones harboring specific mutations. In this context, CH emergence may be boosted by the proinflammatory IBD environment, thus acting as a biological link between IBD and the HM onset. If these data are confirmed, IBD patients screened and positive for CH should undergo a hematologic follow-up to assess the risk of developing HM. Future study will clarify the relationship between these conditions.
Introduction
The term inflammatory bowel diseases (IBDs) define a group of chronic conditions of the gastrointestinal tract. Crohn’s disease (CD) and ulcerative colitis (UC) are the most common types. These two conditions differ slightly, both histologically and clinically. IBDs are thought to arise due to host genetics, environmental factors, and immune system function, ultimately resulting in chronic inflammation and consequent clinical symptoms (1). Nationwide studies have revealed a higher risk of hematological malignancies (HMs) in these patients (2–4). In particular, IBDs are associated with the onset of lymphoproliferative disorders. The hypothetical causal link relies on the iatrogenic effect of novel immunosuppressive drugs (such as thiopurines, methotrexate, and biologic agents) adopted to treat IBDs. Notwithstanding, no clear evidence related to specific drugs has emerged (5).
Clonal hematopoiesis (CH) is a premalignant condition defined by the presence of an acquired somatic mutation characterized by a variant allele frequency (VAF) of ≥2%, in a gene frequently associated with HMs, without fulfillment of other diagnostic criteria for a hematologic neoplasm diagnosis (6). CH incidence is age-related, showing a prevalence of about 10% in persons aged > 70 years, while it is less frequent in young and young adults. A growing body of evidence suggests a correlation between inflammation and CH. On the one hand, an inflammatory state may favor the selection and consequent emergence of the mutated hematopoietic clone (7), thus acting as a stress factor on hemopoiesis. On the other hand, the mutated clone itself may increase the expression of inflammatory genes in innate immune cells (8).
IBD patients have an underlying pro-inflammatory state, whose potential effects could intersect the roads of CH, as already demonstrated in atherosclerosis and autoimmune disorders, such that these diseases could boost the emergence of CH (9, 10). Recently, experimental data are unveiling ever more profound interactions between a pro-inflammatory milieu and mutated hematopoietic stem cells (HSCs). These latter show a sort of adaptive capacity, not only conferring resistance to inflammation but ultimately enhancing their ability to outgrow normal HSCs (11).
All this premised, a reflection is needed to investigate the link between IBDs and HMs. Conversely, an iatrogenic trigger (i.e., drugs) could be enough to cause a neoplastic evolution. Moreover, the interplay between the hematopoietic system and inflammation could be more than a supporting cast.
Method
From February 2011 to May 2021, 13 cases (median age 58 years, range 47 – 70) were referred to our center with IBD associated with HMs; their main clinical data are reported in Table 1. In all cases, the IBD diagnosis was confirmed by histological analysis. Before the HM onset, they were all treated with mesalazine except case #5. The study was approved by the local ethics committee “Azienda Ospedaliero Universitaria Policlinico di Bari”. Written, informed consent was obtained from all patients before enrolment in accordance with the Declaration of Helsinki. Their records/information were anonymized and de-identified before analysis.
Next-generation sequencing (NGS) analysis with an AmpliSeq customized panel (Thermo Fisher Scientific), encompassing 26 target genes that are frequently mutated in myeloid malignancies, was performed on genomic DNA extracted from bone marrow (BM) (cases #3 - #13) or peripheral blood (PB) (cases #1, #2) samples, as previously described (12). Quality control reads alignment to the human genome (hg19) and variant calling (using the somatic workflow for single samples and the default parameters) were performed using Torrent Suite Software v5.16 (Thermo Fisher Scientific). Variants were annotated using Ion Reporter Software v5.16 (Thermo Fisher Scientific). Variants located in intronic regions (not in splice sites) or synonymous or present with ≥1% global minor allele frequency (MAF) in the healthy population, according to the Genome aggregation database (gnomAD) v2.1.1, were filtered out. Only variants affecting the CH-associated genes (13), with ≥2% VAF (6), and with a depth of coverage >500x were considered. Selected variants were investigated for a potential pathogenic role using the Catalogue of Somatic Mutations in Cancer (COSMIC v95) and dbSNP databases.
Results
The median latency time between IBD and HM was 71 months (range 4 – 312 months). Overall, 20 variants affecting 9 genes (DNMT3A, ASXL1, JAK2, GATA2, TET2, ETV6, SF3B1, EZH2, KIT) were detected in our cohort (Table and Figure 1); 11/13 (85%) patients showed one or more mutations, with a VAF ranging from 2.6% to 53.0%. Moreover, cases #6 and #7 showed variants of CH-associated genes but in the absence of a BM lymphoma localization. Five variants (25.0%) showed a VAF<10%, whereas, in the 15 others identified (75.0%), the clone was dominant. In accordance with previously published data in the IBD context, DNMT3A was the most frequently mutated gene (4/20, 20.0%) (13, 14), with ASXL1 (4/20, 20.0%), followed by JAK2 (3/20, 15.0%). All mutations are single nucleotide variants (SNV) with different functions: missense variants (13/20, 65.0%), nonsense variants (4/20, 20.0%), and unknown variants affecting a splice site or an untranslated region (3/20, 15.0%). Five cases (45.5%) carried just one variant, whereas the other six (54.5%) showed two or more mutations.
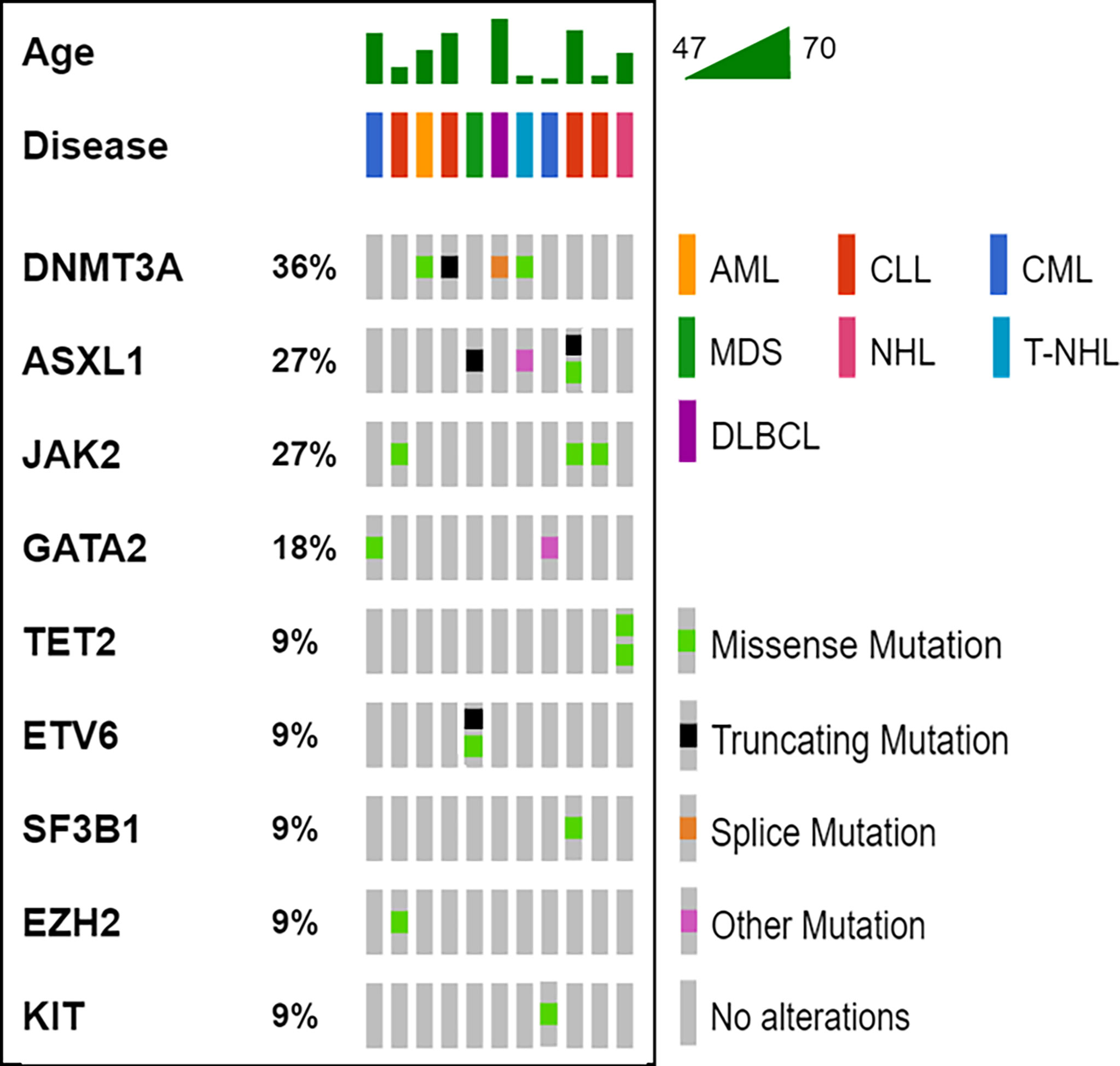
Figure 1 Oncoprinter visualization of all variants identified. For all cases (columns), the age, the hematological disease and the variants identified are reported. The percentage value associated to each gene, indicates its variants occurrence in the cohort analyzed. AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; DLBCL, diffuse large B-cell lymphoma; MDS, myelodysplastic syndrome; NH, non-Hodgkin’s lymphoma; T-NHL, T-cell non-Hodgkin’s lymphoma.
Discussion
Individuals with CH have a higher incidence of HM, coronary heart disease, ischemic stroke, and heart failure (15). Recent works demonstrated that chronic infection, UC and atherosclerosis, can drive the gene loss of function associated with CH (14, 16, 17). In particular, chronic infection and UC may promote the selection of the DNMT3A gene mutation associated with CH by the IFNγ signaling induced in the course of these disorders (14, 16). It is worthy of note that four (36.4%) cases in our series bore DNMT3A gene mutations. According to data previously shown in UC patients (14), this fact may be considered to demonstrate that the mechanisms at the basis of the inflammatory environment potentially select for the growth of CH specific mutations. Thus, CH may act as a possible biological link between IBD and the HMs onset (Figure 2). In fact, CH may be fed by the IBD inflammatory stimuli, and in turn, feed the IBD inflammation. This context can also explain the low median age of our patient series compared to that expected in healthy individuals with CH.
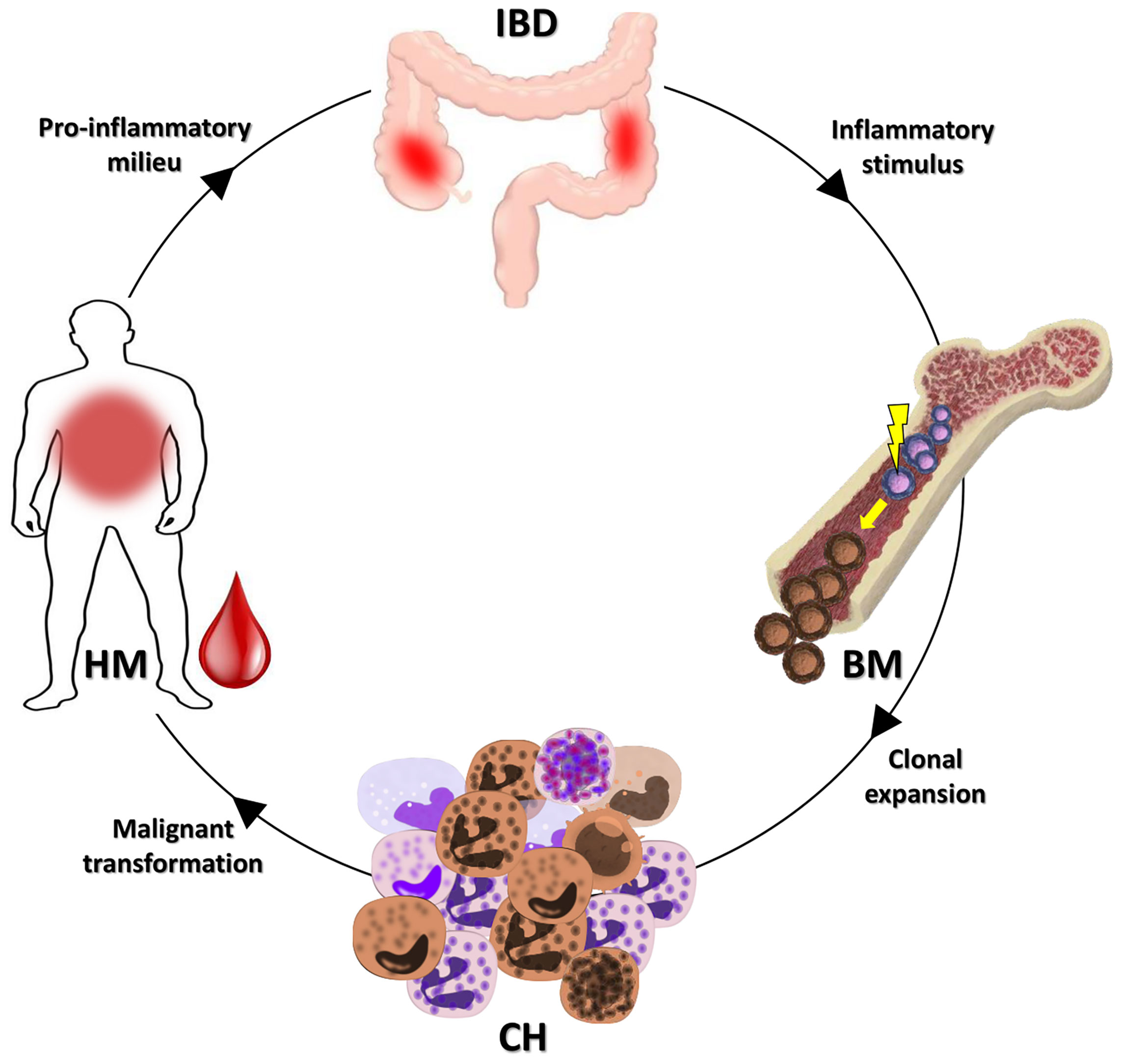
Figure 2 Possible link between IBDs and HMs. As demonstrated, an IBD may promote the CH onset (14). The clone selected could evolve in a subsequent HM or favor the insurgence of an HM. In both cases, the inflammatory milieu of the HM may, in turn, feed the IBD. IBD, inflammatory bowel disease; BM, bone marrow; CH, clonal hematopoiesis; HM, hematological malignancy.
Our main study limitation is that it was not possible to demonstrate that the CH-associated genes variant was present before the onset of the HMs, since no genomic data were available at that time. Notwithstanding, the low median age of our patients, the prevalence of lymphoproliferative disorders (not due to mesalazine administration), and the incidence of typical CH-associated mutations in our cohort seem to suggest a biological link between these conditions.
Interestingly, in our observation 4 patients (#4, #5, #6, and #7) showed a very low VAF of the CH-associated genes. This circumstance could suggest that there may be a distinct role between CH promoted and selected by inflammation and the genomic alterations that drive tumorigenesis and control the HM pathogenesis. Particularly, the low VAF of DNMT3A gene in cases #6 and #7, both patients without evidence of BM lymphoma involvement, is strongly suggestive of CH promoted by IBD. It’s noteworthy that DNMT3A mediated CH should not be seen as an inactive player in the immune system. In fact, it has been shown to alter T cells’ homeostasis and be associated with enhanced T cell activity in recipients of allografts harboring a DNMT3A mutation (18, 19).
Furthermore, cases #1, #2, #8, #10, #11 showed CH-associated gene variants usually rare in chronic myeloid and lymphoid leukemia. It’s noteworthy that we didn’t include two cases of JAK2 V617F myeloproliferative neoplasms in our analysis, preceded by IBD. Both patients were aged <70 and had a multiannual story of IBD at the time of HM onset. In this context, we should consider that, on the one hand, JAK2 is undoubtedly “driving” the disease; on the other hand, it accounts for 3% of CH in the general population (20). Therefore, even in these circumstances, the interplay between IBD, CH and HM cannot be a priori excluded.
In conclusion, our report suggests that CH may be seen as a biological link at the crossroads of IBDs and HMs, whose role is yet to be fully elucidated. Nonetheless, if these data are confirmed, IBD patients screened and positive for CH should undergo hematologic follow-up to assess the risk of developing HM. Further studies are warranted to characterize the biological relationship between IBDs, CH, and HMs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA719027.
Ethics Statement
The studies involving human participants were reviewed and approved by Local ethics committee “Azienda Ospedaliero Universitaria Policlinico di Bari”. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CC, FT, and FA conceived and designed the study and wrote the manuscript. CC and AZ performed the main experiments. LA, CM, NC, GT, LI, EP, MC, and IR performed diagnostic molecular analysis. PC, MD, AG, MCL, TP, and AR provided clinical data. GS, PM, and FA reviewed and edited the manuscript. FA supervised the manuscript preparation. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by “Associazione Italiana contro le Leucemie (AIL)-BARI” and by the association for non-Hodgkin lymphoma research “Il sorriso di Antonio”, Corato – Italy.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Khor B, Gardet A, Xavier RJ. Genetics and Pathogenesis of Inflammatory Bowel Disease. Nature (2011) 474:307–17. doi: 10.1038/nature10209
2. Cheddani H, Dauchet L, Fumery M, Charpentier C, Marie Bouvier A, Dupas JL, et al. Cancer in Elderly Onset Inflammatory Bowel Disease: A Population-Based Study. Am J Gastroenterol (2016) 111:1428–36. doi: 10.1038/ajg.2016.304
3. Jung YS, Han M, Park S, Kim WH, Cheon JH. Cancer Risk in the Early Stages of Inflammatory Bowel Disease in Korean Patients: A Nationwide Population-Based Study. J Crohns Colitis (2017) 11:954–62. doi: 10.1093/ecco-jcc/jjx040
4. Matsushita M, Fukata N, Omiya M, Okazaki K. Nationwide Studies of Hematological Malignancies in Inflammatory Bowel Disease. Am J Gastroenterol (2017) 112:1171–2. doi: 10.1038/ajg.2017.142
5. Lam GY. Lymphoproliferative Disorders in Inflammatory Bowel Disease Patients on Immunosuppression: Lessons From Other Inflammatory Disorders. World J Gastrointest Pathophysiol (2015) 6:181. doi: 10.4291/wjgp.v6.i4.181
6. Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal Hematopoiesis of Indeterminate Potential and its Distinction From Myelodysplastic Syndromes. Blood (2015) 126:9–16. doi: 10.1182/blood-2015-03-631747
7. Cook EK, Luo M, Rauh MJ. Clonal Hematopoiesis and Inflammation: Partners in Leukemogenesis and Comorbidity. Exp Hematol (2020) 83:85–94. doi: 10.1016/j.exphem.2020.01.011
8. Jaiswal S, Ebert BL. Clonal Hematopoiesis in Human Aging and Disease. Science (80- ) (2019) 366:80–99. doi: 10.1126/science.aan4673
9. Lusis AJ. A Vicious Cycle in Atherosclerosis. Cell (2021) 184:1139–41. doi: 10.1016/J.CELL.2021.02.005
10. Savola P, Lundgren S, Keränen MAI, Almusa H, Ellonen P, Leirisalo-Repo M, et al. Clonal Hematopoiesis in Patients With Rheumatoid Arthritis. Blood Cancer J (2018) 8(8):1–5. doi: 10.1038/s41408-018-0107-2
11. Speck NA. A Pernicious Cycle Affecting Premalignant Stem Cells. N Engl J Med (2022) 386:596–8. doi: 10.1056/NEJMCIBR2117528
12. Coccaro N, Zagaria A, Orsini P, Anelli L, Tota G, Casieri P, et al. RARA and RARG Gene Downregulation Associated With EZH2 Mutation in Acute Promyelocytic-Like Morphology Leukemia. Hum Pathol (2018) 80:82–6. doi: 10.1016/j.humpath.2018.02.023
13. Wu H-T, Kalashnikova E, Mehta S, Salari R, Sethi H, Zimmermann B, et al. Characterization of Clonal Hematopoiesis of Indeterminate Potential Mutations From Germline Whole Exome Sequencing Data. J Clin Oncol (2020) 38:1525–5. doi: 10.1200/jco.2020.38.15_suppl.1525
14. Zhang CRC, Nix D, Gregory M, Ciorba MA, Ostrander EL, Newberry RD, et al. Inflammatory Cytokines Promote Clonal Hematopoiesis With Specific Mutations in Ulcerative Colitis Patients. Exp Hematol (2019) 80:36–41.e3. doi: 10.1016/j.exphem.2019.11.008
15. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N Engl J Med (2017) 377:111–21. doi: 10.1056/nejmoa1701719
16. Hormaechea-Agulla D, Matatall KA, Le DT, Kain B, Long X, Kus P, et al. Chronic Infection Drives Dnmt3a-Loss-of-Function Clonal Hematopoiesis via IFN$γ$ Signaling. Cell Stem Cell (2021) 8:1428–42. doi: 10.1016/j.stem.2021.03.002
17. Heyde A, Rohde D, McAlpine CS, Zhang S, Hoyer FF, Gerold JM, et al. Increased Stem Cell Proliferation in Atherosclerosis Accelerates Clonal Hematopoiesis. Cell (2021) 184:1348–61.e22. doi: 10.1016/j.cell.2021.01.049
18. Gibson CJ, Kim HT, Murdock HM, Hambley B, Zhao L, Green L, et al. DNMT3A Clonal Hematopoiesis in Older Donors Is Associated With Improved Survival in Recipients After Allogeneic Hematopoietic Cell Transplant. Blood (2020) 136:26–6. doi: 10.1182/blood-2020-142925
19. Abplanalp WT, Cremer S, John D, Hoffmann J, Schuhmacher B, Merten M, et al. Clonal Hematopoiesis-Driver DNMT3A Mutations Alter Immune Cells in Heart Failure. Circ Res (2021) 2:216–28. doi: 10.1161/CIRCRESAHA.120.317104
Keywords: clonal hematopoiesis, inflammatory bowel diseases, hematological malignancies, DNMT3A, ASXL1, JAK2
Citation: Cumbo C, Tarantini F, Zagaria A, Anelli L, Minervini CF, Coccaro N, Tota G, Impera L, Parciante E, Conserva MR, Redavid I, Carluccio P, Delia M, Giordano A, Longo MC, Perrone T, Rossi AR, Specchia G, Musto P and Albano F (2022) Clonal Hematopoiesis at the Crossroads of Inflammatory Bowel Diseases and Hematological Malignancies: A Biological Link? Front. Oncol. 12:873896. doi: 10.3389/fonc.2022.873896
Received: 11 February 2022; Accepted: 21 March 2022;
Published: 12 April 2022.
Edited by:
Adam Finn Binder, Thomas Jefferson University, United StatesReviewed by:
Xing Fan, National Institutes of Health (NIH), United StatesReuben Kapur, Purdue University Indianapolis, United States
Copyright © 2022 Cumbo, Tarantini, Zagaria, Anelli, Minervini, Coccaro, Tota, Impera, Parciante, Conserva, Redavid, Carluccio, Delia, Giordano, Longo, Perrone, Rossi, Specchia, Musto and Albano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Albano, francesco.albano@uniba.it
†These authors have contributed equally to this work
 Cosimo Cumbo
Cosimo Cumbo Francesco Tarantini
Francesco Tarantini Antonella Zagaria
Antonella Zagaria Luisa Anelli
Luisa Anelli Crescenzio Francesco Minervini
Crescenzio Francesco Minervini Nicoletta Coccaro
Nicoletta Coccaro Giuseppina Tota1
Giuseppina Tota1 Maria Rosa Conserva
Maria Rosa Conserva Immacolata Redavid
Immacolata Redavid Paola Carluccio
Paola Carluccio Mario Delia
Mario Delia Giorgina Specchia
Giorgina Specchia Francesco Albano
Francesco Albano