- 1Department of Pathology, School of Biology & Basic Medical Sciences, Soochow University, Suzhou, China
- 2Section Experimental and Translational Head and Neck Oncology, Department of Otolaryngology, Head and Neck Surgery, University Hospital Heidelberg, Heidelberg, Germany
- 3Department of Pharmacy, The First Affiliated Hospital of University of Science and Technology of China (USTC), Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 4Research Group Molecular Mechanisms of Head and Neck Tumors, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany
Ubiquitin C-terminal hydrolases (UCHs), a subfamily of deubiquitinating enzymes (DUBs), have been found in a variety of tumor entities and play distinct roles in the pathogenesis and development of various cancers including head and neck cancer (HNC). HNC is a heterogeneous disease arising from the mucosal epithelia of the upper aerodigestive tract, including different anatomic sites, distinct histopathologic types, as well as human papillomavirus (HPV)-positive and negative subgroups. Despite advances in multi-disciplinary treatment for HNC, the long-term survival rate of patients with HNC remains low. Emerging evidence has revealed the members of UCHs are associated with the pathogenesis and clinical prognosis of HNC, which highlights the prognostic and therapeutic implications of UCHs for patients with HNC. In this review, we summarize the physiological and pathological functions of the UCHs family, which provides enlightenment of potential mechanisms of UCHs family in HNC pathogenesis and highlights the potential consideration of UCHs as attractive drug targets.
Introduction
Head and neck cancer (HNC) represents the seventh most prevalent human malignancies with an annual incidence of 890,000 new cases worldwide, including 76,000 cases in China and 18,260 cases in Germany (1, 2). Anatomically, HNC occurs at distinct sites including lip, oral cavity, nasal cavity, sinonasal cavity, nasopharynx, oropharynx, hypopharynx, larynx, and salivary glands, and etiologic risk factors, epidemiology, treatment strategies as well as clinical outcome differ among individual subsites (3). Over 90% of cases are diagnosed as head and neck squamous cell carcinoma (HNSCC), which arises from the mucosal epithelia of the upper aerodigestive tract. High incidence areas for oral cavity cancer include Middle and South Asia, Western and Southern Europe as well as South Africa. The incidence of oropharyngeal SCC (OPSCC) is elevated in Europe and North America. Nasopharyngeal cancer (NPC) is most common in East and Southeast Asia, especially in South China (4). Tobacco smoking and heavy alcohol consumption have been identified as the most important risk factors in developed countries (5). In developing countries, risk factors also include EpsteinBarr virus (EBV) infection for NPC, areca nut chewing, consumption of preserved foods, and oral hygiene (6–8). During the past two decades, the overall incidence of HNSCC has gradually decreased in western developed countries. However, a subgroup of HNSCC, particularly OPSCC, has been becoming more prevalent in young adults, which is attributed to high-risk human papillomavirus (HPV) infection, predominantly HPV16 (9). High-risk HPV types comprise two oncogenes, E6 and E7, which inactivate the tumor suppressors p53 and retinoblastoma (RB), respectively. As a result, cell cycle progression and cell death in infected cells are disrupted, as initial steps for HPV-related carcinogenesis (10–13). Besides the viral oncogenes E6/E7, HPV E2, E4, and E5 have been shown to facilitate the synergistic effects of viral oncogenesis, which represents an alternative manner to HPV-induced carcinogenesis (14). It has been well-established that HPV-positive and HPV-negative OPSCC have distinct differences in gene expression profiles, genomic alterations, immune landscape, as well as clinical outcomes (15–18). Due to the more favorable prognosis of HPV-positive OPSCC, clinical trials have been launched to investigate HPV-stratified de-escalation treatment based on currently established protocols. However, final results and definitive conclusions are pending, which might improve the post-treatment quality of HPV-related OPSCC patients (19–22).
Despite advances in multi-disciplinary treatment for HNSCC, including surgical approaches, radiotherapy, chemotherapy, molecular-targeted therapy, and immunotherapy, the overall survival of advanced HNSCC has only improved slightly, and appropriate therapy remains a major challenge. Over the past decade, large-scale genomic profiling and proteomic studies, including The Cancer Genome Atlas (TCGA) projects, have highlighted a comprehensive molecular landscape of changes in DNA copy number, somatic mutations, promoter methylation, and protein and gene expression, indicating the critical components and signal pathway in HNSCC pathogenesis (23–25). A better understanding of these molecular underpinnings may inspire novel drug targets as well as molecular biomarkers for personalized treatment.
The proteome is exceedingly complex and has been regarded as the major driver or actuator of fundamental cellular processes. Protein ubiquitination is a post-translational modification process that plays critical roles in numerous biological processes, including cell growth and differentiation, signal transduction, DNA repair, and oncogenesis (26). The conjugation of ubiquitin (Ub) to target proteins is catalyzed by a cascade of ubiquitinating enzymes, including Ub-activating enzymes (E1s), Ub-conjugating enzymes (E2s), and Ub ligases (E3s). Conjugation of Ub to a substrate lysine, its lysines or its N-terminus, results in the generation of different substrate ubiquitin structures, which can be either a mono- or poly-ubiquitylation process and allows targeted proteins to fulfill a diverse range of functions. However, protein ubiquitination is highly reversible. Deubiquitinases or deubiquitinating enzymes (DUBs) catalyze the removal of ubiquitin from target proteins to generate free monomeric Ub (27). The human genome encodes approximately 100 DUBs categorized into six subfamilies: the ubiquitin C-terminal hydrolases (UCHs), the ubiquitin-specific proteases (USPs), the ovarian tumor proteases (OTUs), the Josephin or Machado-Joseph disease protein domain proteases (MJDs), the Jab1/MPN domain-associated metalloisopeptidase (JAMM), and the monocyte chemotactic protein-induced protein family (MCPIP) (27). Among these families, UBPs are mostly described to date, with 60 proteases in humans, which have been well-reviewed by a range of publications (28–31). Recent studies have revealed the emerging functions of UCHs in the pathogenesis and progression of human malignancies. However, few studies on UCHs in HNC are available. One member of UCHs family, BRCA1-associated protein-1 (BAP1), was identified to be associated with poor outcome following radiation in HPV-negative HNSCC clinical sample by proteomic and transcriptomic analysis (32). Moreover, another member of UCHs family, UCHL1 was demonstrated as a tumor suppressor gene in nasopharyngeal carcinoma (NPC) (33). In this review, we systematically summarize the physiological and pathological functions of the UCHs family in human malignancies, providing enlightenment on potential mechanisms of UCHs family in HNC pathogenesis and the potential consideration of UCHs as novel promising drug targets.
Structures and Functions of UCHs
Among DUBs family, molecular structures of UCHs were the first to be characterized. Four UCHs in humans have been identified: UCHL1/PGP9.5 (protein gene product 9.5), UCHL3, UCHL5/UCH37, and BAP1. All UCHs share a core catalytic domain with 230 amino acids and close homology among family members. They comprise a confined loop that cleaves short ubiquitylated peptides (up to 20–30 amino acids) from the C-terminal glycine residue (Figure 1).
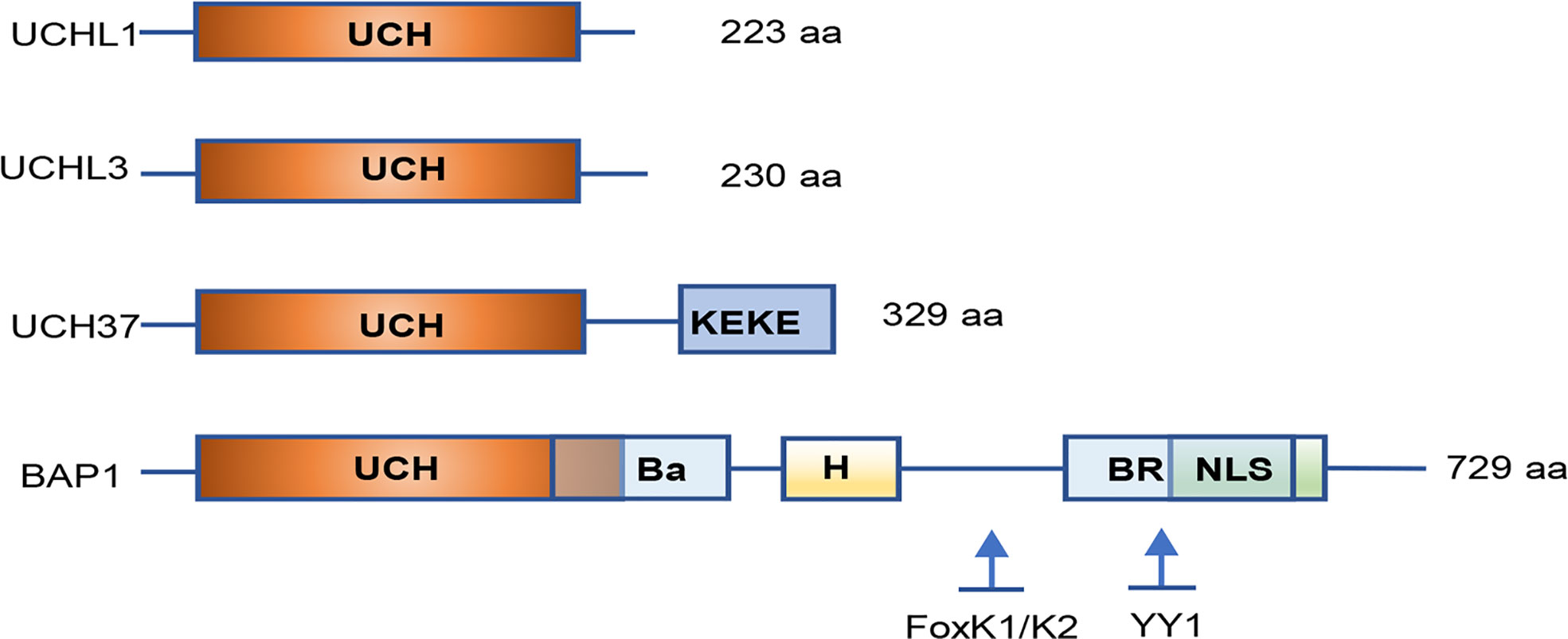
Figure 1 Simplified structure of the UCH family proteins. All UCH members share close homology in their catalytic domains and have a core catalytic domain with 230 amino acids. UCHL3 contains a KEKE motif in the C-terminal tail. BAP1 consists of a long C-terminal extension illustrating numerous functional domains and binding sites for interacting proteins. UCH, ubiquitin C-terminal hydrolase; Ba, BARD1 binding domain; H, HCF-binding motif; BR, BRCA1 binding domain; NLS, nuclear localization signal; YY1, Ying Yang 1 binding region. Inspired by (26).
UCHL1
First identified as a member of UCHs family in the 1980s, UCHL1 is an abundant neuronal protein containing only one UCH domain with very short N- and C- terminal extensions. It possesses one of the most complicated protein knotted structure, which is regarded to protect UCHs from degradation in the proteasome as well as to maintain proper proteasomal function. Besides its DUB function in the recycling of free Ub, UCHL1 is also known to have a ubiquitin ligase activity as a mono-Ub stabilizer by preventing its degradation (34). It was reported that the generation rate of monomeric Ub by UCHL1 in vitro is enhanced by the catalytic residues C90 and H161 (35). Analysis of the crystal structure suggests that UCHL1 preferentially binds monomeric or small adducts of Ub, but does not act on large polymers of Ub (36). Therefore, UCHL1 has the potential for numerous ubiquitination-dependent biological processes.
UCHL1 is predominantly expressed in the brain, where it comprises up to 5% of total neuronal proteins. Although the precise function of UCHL1 is not fully clarified in many pathological processes, better understandings of functional UCHL1 has been largely reported in neuronal dysfunction and neurodegenerative disorders (34, 37–39). For instance, the specific distribution and activity of UCHL1 in human tissues has the potential clinical significance for Parkinson’s disease (PD) and Alzheimer’s disease (AD), might be a major target of reactive oxygen species (ROS) damage (40). Although most cases of PD are sporadic, a small subgroup of PD has been linked to specific genomic mutations (41). Interestingly, The I93M point mutation in UCHL1 has been reported to be associated with PD susceptibility by decreasing hydrolytic activity (42). By contrast, an S18Y variation in UCHL1 shows a protective enzyme with a reduced risk of PD by a reduction of α-synuclein (43). A study has shown that modification of the UCHL1 C152 site decreases injury to gray and white matter, resulting in the recovery of motor function after middle cerebral artery occlusion (44). Another potential feature of UCHL1 is an ATP-independent E3 ligase activity, which promotes Lys63 (K63) polyubiquitination of α-synuclein (34). Moreover, UCHL1 was demonstrated as a novel interactor and substrate of PD linked E3 ubiquitin-protein ligase parkin by the autophagy-lysosome pathway (45).
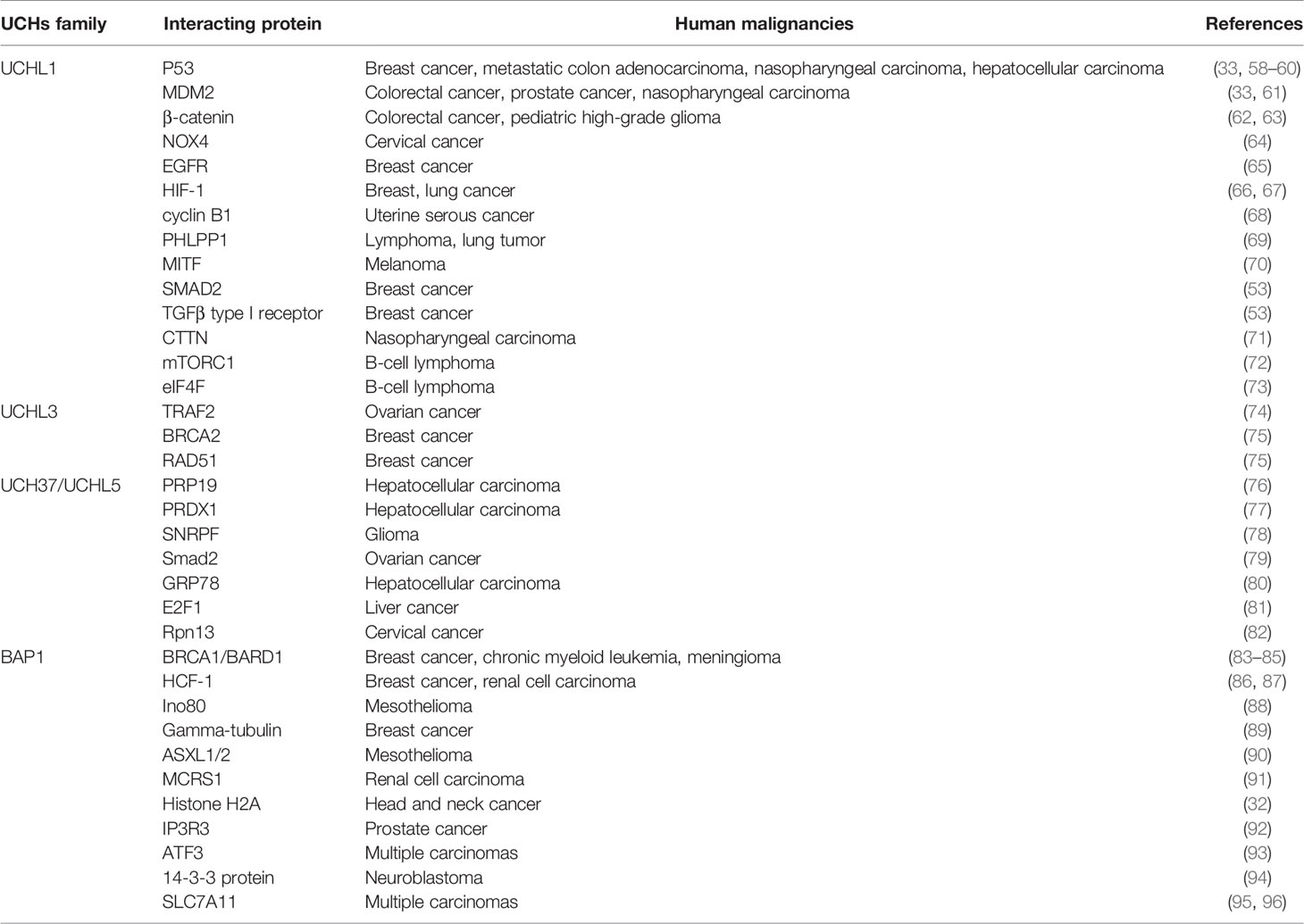
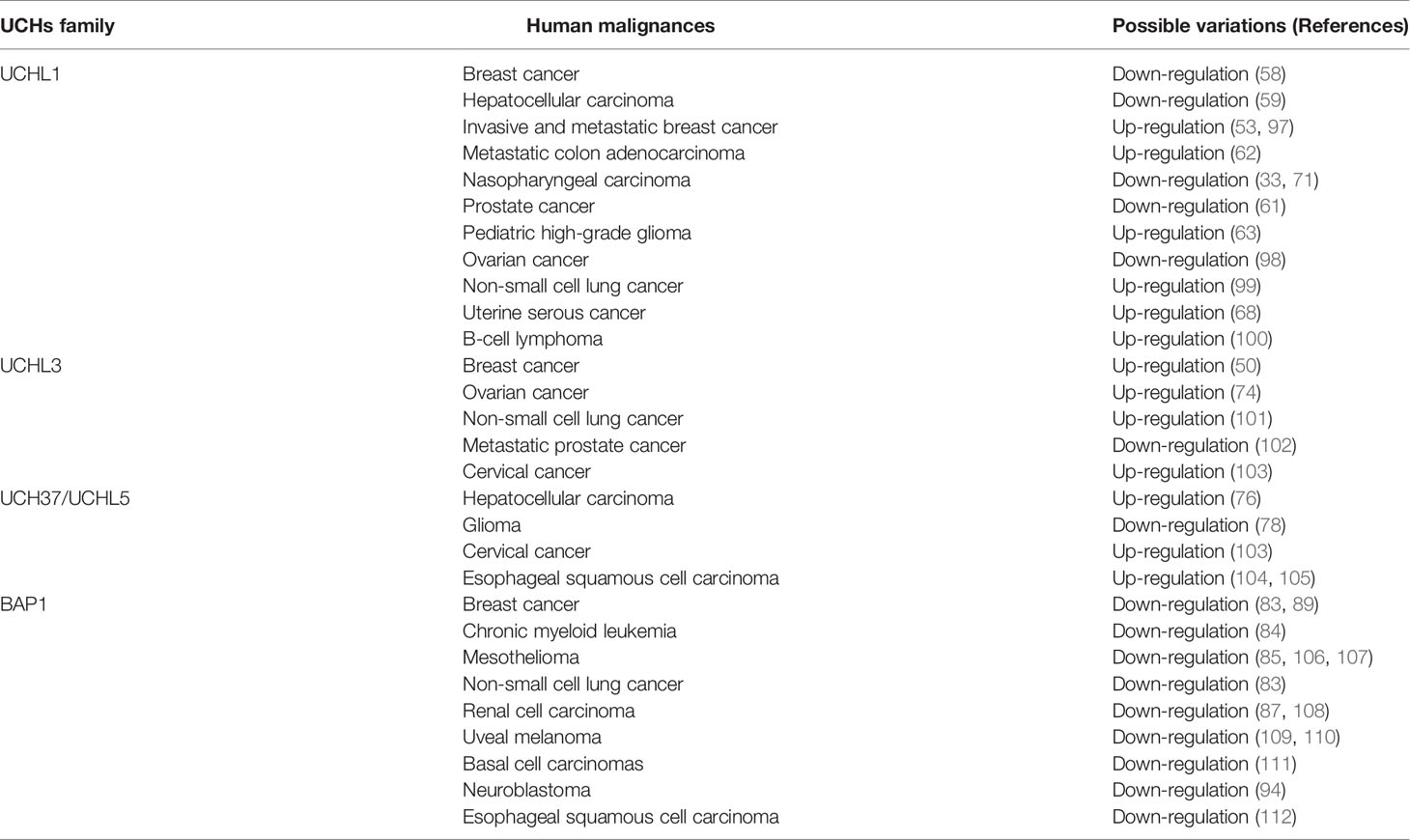
It has also been reported at much lower levels in kidney, breast epithelium, and reproductive tissues (46, 47), and to be expressed context-dependent in individual cells, such as human fibroblasts during wound healing (48). Uniform cytoplasmic staining of UCHL1 was observed in neurons, but UCHL1 can translocate into the nucleus and regulate microtubule dynamics (49). Although it is absent in most other normal tissues, UCHL1 appears to be aberrantly expressed in many non-neuronal tumors, including breast, prostate, colorectal, gastric, head and neck, and pancreatic ductal carcinomas (33, 50–55). The functions and potential mechanisms of UCHL1 in tumorigenesis have been reviewed by several excellent publications (56, 57). The interactive proteins with UCHL1 as well as other UCH family members in human malignancies are summarized in Table 1. In addition, altered expression levels of UCHs in various cancers have also been reviewed in Table 2.
Recent findings have revealed significant functions for UCHL1 in immune response and regulation. UCHL1 was found in mouse kidney, spleen, and bone marrow-derived dendritic cells, and its expression and activity were strongly regulated by the immune stimuli LPS and IFN-γ (113). UCHL1 modulates antigen processing by affecting the colocalization of intracellular MHC I with late endosomal/lysosomal compartments necessary for cross priming of CD8 T cells (113). Interestingly, an induced UCHL1 expression was also demonstrated in multipotent mesenchymal stromal cells (MSCs) upon stimulation with proinflammatory cytokines IFN-γ plus TNF-α, and negatively regulated the immunosuppressive capacity and survival of MSC. This discovery may provide potential MSC-based immunotherapy for inflammatory diseases by modulation of UCHL1 (114).
UCHL3
UCHL3 and UCHL1 have significant structural similarity. However, the biological characteristics of UCHL3 are quite distinct concerning expression patterns and ligase activity. Unlike UCHL1, which is mainly restricted to neuronal and neurosecretory tissues, UCHL3 is more widely expressed throughout mammalian tissues. Interestingly, UCHL3 hardly exhibits ligase activity, while its hydrolytic activity is two-hundred-fold higher than UCHL1 toward a fluorogenic ubiquitin C-terminal amide (34). It was reported that UCHL3 enables to cleave the C-terminus of NEDD8, which is a ubiquitin-like protein that exerts the function of Ub to be conjugated to a lysine residue of the substrate (115). Next, UCHL3 has also been demonstrated to alleviate cryptorchid-induced germ cell apoptosis in gad mice. UCHL3 appears to have dual affinities for ubiquitin and Nedd8, and function as a deNEDDylating enzyme in vivo, suggesting that UCHL3 plays a critical role in germ cell apoptosis (116). Several studies using similar UCHL3 knockout mouse models revealed the significant functions in photoreceptor cell degeneration, neurodegeneration, fertilization and embryogenesis, stress responses in skeletal muscle, diet-induced obesity, and osteoblast differentiation (117–122). It is worth mentioning that level of UCHL3 protein in several neurodegenerative diseases is unchanged, while it hydrolyzes the C-terminal extension of a mutant ubiquitin (UBB+1), contributing to the role in neurodegenerative disorders (123).
An increasing number of studies have demonstrated vital functions of UCHL3 on tumorigenesis, including breast, prostate, ovarian, and non-small cell lung cancer (Table 2) (50, 74, 101, 102). Luo et al. found that UCHL3 deubiquitinates RAD51 and subsequently facilitates RAD51-BRCA2 interaction, which is critical for homologous recombination (HR) and contributes to therapeutic resistance in breast cancer (75). By contrast, UCHL3 is reduced in metastatic prostate cancer cell lines, and knockdown of UCHL3 promotes epithelial-to-mesenchymal transition (EMT), contributing to cancer cell invasion and metastasis (102). In contrast, high UCHL3 expression was reported in ovarian cancer and predicted a worse clinical outcome. The elevated UCHL3 facilitates carcinogenesis and enhances inflammation by deubiquitinating and stabilizing TNF Receptor Associated Factor 2 (TRAF2) (74). Taken together, the UCHL3 function in cancer remains controversial, suggesting the roles of UCHL3 is complicated and context-dependent in individual tumor types.
UCH37
UCH37 (also known as UCHL5) was identified first as a 19S-associated deubiquitinating enzyme in the 1990s, which comprises a C-terminal extension (residues 227-329) in addition to an N-terminal UCH domain (residues 1–226) (124). It is specific for the distal subunit of Lys48-linked poly-Ub chains. Isolated full-length UCH37 displays weak catalytic activity due to autonomic inhibition by the C-terminal extension (125). The proteolytic activity requires a Ub receptor called ADRM1 (named hRpn13 in humans) binding to UCH37 via its C-terminal 46 residues (also called the KEKE motif) (125). In addition, hRpn13 was found to directly enhance the de-ubiquitination activity of UCH37 in vitro (125–127). The hRpn13-UCH37 complex hydrolyzes large Ub conjugates with incorporation into the 19S complex. By contrast, UCH37 is inhibited by the chromatin remodeling complex component INO80G mediated by the N-terminal domain of NFRKB (nuclear factor related to κB, NFRKB) (128). Rpn13 and INO80G share a conserved deubiquitinase adaptor (DEUBAD) domain that interacts with the C-terminal of UCH37, revealing conformational plasticity to regulate deubiquitinating activity on or off, respectively (128). Functionally, UCH37 is reported to perform a crucial role in certain protein-protein interactions involving several physiological and pathological processes, including development, cell proliferation, and apoptosis, hippocampal synaptic plasticity, Alzheimer’s disease, pulmonary fibrosis, as well as human malignancies (26, 129–133).
Wicks and colleagues reported UCH37 interacts with Smad7 to control TGF-β/Smad signaling activity, suggesting that UCH37-mediated deubiquitination might contribute to tumorigenesis (134). The first direct evidence of UCH37 in cancer study was described by a chemistry-based functional proteomics approach in cervical carcinoma. Activity profiling showed UCH37 is induced in the majority of carcinoma tissues and HPV E6/E7 immortalized human keratinocytes, indicating a significant role of UCH37 in tumor transformation (103). Subsequently, an increasing number of studies reported the potential functions in tumor cell proliferation, apoptosis, migration, and invasion, as well as clinical implications (Table 2) (76–78, 135–138).
BAP1
The BAP1 protein consists of 729 amino acids that are encoded by the BAP1 gene located on human chromosome 3p21.1. BAP1 protein was identified as a nuclear-localized DUB. In addition to the N-terminal UCH domain, BAP1 comprises a long C-terminal extension (Figure 1). BAP1 was originally found to interact with the RING finger domain of BRCA1 and to perform the cell growth-suppressive function. BAP1 is also involved in chromatin modification and transcription by deubiquitinating lysine residues in HCF1 and YY1. Both recruit histone-modifying complexes and regulate expression of numerous genes involved in multiple physiological processes (139). Moreover, BAP1 interacts with the transcription factor FOXK1/K2 in a phosphorylation-dependent manner, which represses FOXK2-target genes forming a ternary protein complex in which BAP1 bridges FoxK2 and HCF-1. Loss of BAP1 causes the increase of FoxK2 target genes, which is dependent on the Ring1B-Bmi1 complex (140).
Polycomb group proteins exert critical roles in transcriptional regulation, which contributes to a variety of physiological processes, including embryonic development, differentiation, and self-renewal. Polycomb repressive complexes (PRCs) are responsible for histone ubiquitination and methylation (139). BAP1 interacts with additional sex combs like 1 (ASXL1), forming a polycomb group repressive deubiquitinase complex (PR-DUB). The transcriptional function is regulated through histones modification via ubiquitination by PRCs and deubiquitination by PR-DUB. Thus BAP1 deficiency significantly alters ubiquitination level of histone 2A, leading to the dysregulation of cell cycle and cellular senescence (141). A recent study found cytoplasm BAP1 localizes at the ER, where it regulates type 3 inositol-1,4,5-trisphosphate receptor (IP3R3), modulating calcium (Ca2+) release from the endoplasmic reticulum into the cytosol and mitochondria, promoting apoptosis, which plays a critical role in cellular transformation (92). Another study has identified cystine transporter SLC7A11 as a critical BAP1 target gene in human malignancies, which was repressed by BAP1, causing increasing lipid peroxidation and ferroptosis (95).
BAP1 functions as a tumor suppressor through chromatin modulation, transcriptional regulation, cell cycle control, cellular differentiation, and DNA damage repair (142). Loss or mutation of BAP1 gene is a common event in cancer and serves as a potential pathogenetic mechanism in various human malignancies, including uveal melanoma, mesothelioma, small cell and non-small cell lung carcinomas, renal cell carcinoma (RCC), breast cancer, and hepatocellular carcinoma (Table 2) (107, 143–148). Tumors associated with BAP1 somatic mutations have already been discussed in recent reviews (139, 149). Other alterations in the BAP1 gene have been reported, such as large deletions of exons causing premature protein termination, frameshift mutation, splice site mutations, and base substitutions-induced nonsense and missense mutations (143, 149). BAP1 acts as a tumor suppressor depending on both deubiquitination activity interfered by missense mutations and loss of nuclear localization signal by truncating mutations. Furthermore, several studies showed that BAP1 loss or modification is associated with different tumor phenotypes and clinical outcomes (108, 110, 150–152). For example, BAP1-mutated mesothelioma is significantly correlated with female predominance, younger age at onset, epithelioid differentiation, and better prognosis (153). At the same time, BAP1 mutation is strongly associated with a more aggressive, metastatic phenotype in uveal melanomas (143). BAP1 is frequently mutated in sporadic clear cell RCC with an incidence rate of 6–17%, which is associated with high tumor grade, rhabdoid/sarcomatoid transformation, and poor clinical outcome (154, 155). From a therapeutic standpoint in renal cell carcinoma, inactivation of BAP1 sensitizes tumor cells to irradiation and PARP-inhibitors, which might be due to the impaired ability of double-stranded DNA breaks (87).
UCHs Members in HNC
Although UCHs members have been well investigated in a variety of human malignancies, the exact function of these enzymes in HNSCC pathogenesis and progression remain elusive. Each member of the UCHs family exerts distinct roles depending on the various tumor types. For example, UCHL1 has been controversially considered as a tumor suppressor or tumor promoter in specific tumor types. It was reported that UCHL1 is silenced by promoter CpG hypermethylation in a large panel of primary tumors including HNSCC cell lines and primary tumors, suggesting a tumor-suppressive function (33, 156). The methylation of the CpG locus associated with the UCHL1 gene is dependent on the anatomic site of HNSCC primary tumors, with most hypermethylation of UCHL1 specifically in oral cavity SCC (157). Restored UCHL1 expression significantly suppressed tumor cell proliferation and induced cellular apoptosis through activation of the p14ARF-p53 signaling pathway (33). A more recent study in nasopharyngeal carcinoma revealed a similar conclusion that UCHL1 promoter hypermethylation was validated in nasopharyngeal carcinoma tissues. In addition, restoration of UCHL1 inhibits tumor invasion and metastasis in vitro and in vivo. UCHL1 exerts tumor suppressor function by inducing K48-linked ubiquitination of CTTN (71). Currently, it is widely accepted that high-risk HPV infection is a risk factor for HNSCC, particularly in the oropharynx. High-risk HPV infects the oropharyngeal epithelium causing host immune suppression and evasion (11). UCHL1 does not assist HPV genome replication and viral propagation, but suppresses keratinocyte-mediated production of inflammatory cytokines and chemokines, thereby contributing to immune evasion and HPV persistent infection (158). UCHL1 interacts with tumor necrosis factor receptor-associated factor 3 (TRAF3), which acts as a negative regulator of the alternative NF-κB pathway and antiviral type I IFN activation. TRAF3 has been shown as a tumor suppressor that regulates the malignant phenotype of HPV-positive HNSCC (158).
As a tumor suppressor, BAP1 is critical for promoting DNA repair and cellular recovery from DSB via modulation of H2A ubiquitination (159). BAP1 was found to mediate radioresistance in an in vivo xenograft model and HNSCC cell lines via the deubiquitination of H2A and modulation of HR. Moreover, up-regulation of BAP1 was associated with worse clinical outcome in HNSCC, which indicates BAP1 might serve as a potential therapeutic target in HNSCC (32). In summary, it seems that loss of BAP1 foster genomic instability in tumor pathogenesis, however, the activity of BAP1 promotes tumor cell survival and contributes to therapeutic resistance during irradiation.
Induced activity of UCHL1 and UCHL3 were observed in E6/E7 immortalized primary keratinocytes, indicating the potential function of UCHL1 and UCHL3 in HPV-related HNSCC (103). However, few direct evidences concerning the function of UCHL3 and UCH37 in HNSCC have been reported.
Recently, comprehensive epigenetic and genomic profiling studies have highlighted the most frequently altered genes and signaling pathways in HNSCC. The genomic characterization of 279 HNSCCs including HPV-positive and HPV-negative tumors, has been published (23). Moreover, the molecular profiling data from over 500 HNSCC patients are available at the cBioPortal for Cancer Genomics, which provides interactive exploration and analysis of genetic alterations (160, 161). In addition, the GTEx project provides RNA sequencing data from more than 8,000 normal tissues. Currently, several web-based tools deliver interface-friendly and personalized functions based on TCGA and GTEx data (161, 162). cBioPortal provides visualization for the genomic alteration data. Clinical and genomic analysis of multicohort HNSCC has demonstrated that HPV-positive and HPV-negative tumors present heterogeneity in anatomical regions, mutation profiles, molecular characteristics, immune landscapes, and clinical prognosis. Many evidence revealed the diversity and heterogeneity of HNSCC clinicopathology and therapeutic responses depending on HPV status (163). To better understand the UCHs family mutational landscape in HNSCC, the cBioPortal tool was used to display the types of mutations and their positions in the domain structure of proteins (Figures 2A, B). UCHs member genes are altered in 22 (8%) of queried patients. Of these, 20 cases are HPV-negative, and 2 cases are HPV-positive.
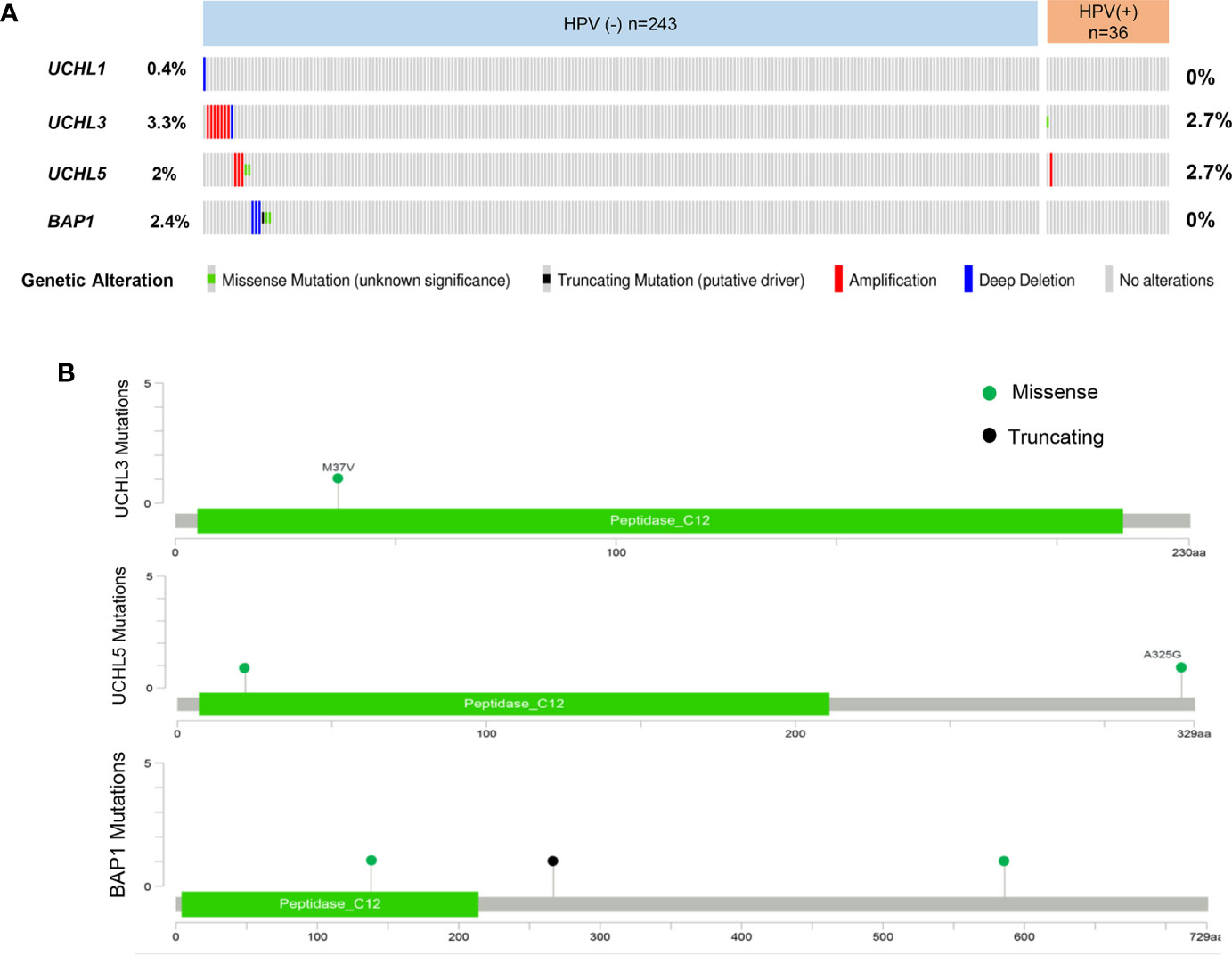
Figure 2 Overview of genetic changes of UCHs family in TCGA HNSCC patients. (A) Oncoprint shows altered UCHs family genes. The colors are associated with one class of variants, and the percentage (%) of patients affected is shown on the graph. (B) cBioPortal predicted mutation maps showing the positions of mutations on the functional domains of UCHL3, UCHL5, and BAP1 proteins.
UCHL1 alterations accounted for 0.4% in HPV-negative subgroup and no genetic alteration in HPV-positive patients, UCHL3 for 3.3% in HPV-negative and 2.7% in HPV-positive, UCHL5 for 2% in HPV-negative and 2.7% in HPV-positive, and BAP1 for 2.4% in HPV-negative and 0 in HPV-positive. Interestingly, there is no samples overlapped. Concerning the mutation type, one missense mutation in UCH-domain of UCHL3, two missense mutations in UCHL5, two missense mutations and one truncating mutation in BAP1. A web-based tool GEPIA (164) analysis revealed UCHL1 gene expression in HNSCC tissues is significantly elevated as compared to normal tissues, which is different from the previous studies in nasopharyngeal carcinoma (71) (Figure 3A). Survival analyses based on gene expression levels was also applied to evaluate the clinical relevance of UCHs family genes (Figures 3B, C). The quartile cut-off method was determined depending on the optimization and visualization of the online web tool. However, numerous problems remain unsolved. We were not able to divide the cohort into two subtypes due to the incompleteness of the HPV status information. More specific subgroups of HNSCC patients for certain phenotypes need to be discovered depending on the protein expression patterns of UCHs family, which may contribute to illuminate the clinical relevance of UCHs family for HNSCC patients. Moreover, the gene networks regulated by UCHs family genes should be identified by analyzing the RNA-sequencing profiling data. Novel signaling pathways and biological processes related to UCHs family in HNSCC are urgent to be clarified. Functional proteomics represents a useful approach to investigate the UCHs family activity-related biological processes in different subtypes of HNSCC. Only BAP1 protein expression data by reverse-phase protein arrays (RPPAs) are available in the TCPA dataset, where BAP1 serves as a strong prognostic predictor for female-related cancer cohorts including samples of invasive breast carcinoma, Ovarian serous cystadenocarcinoma, Uterine Corpus Endometrial Carcinoma (25). More large-scale proteomic profiling data on the other UCHs members are urgent to be produced.
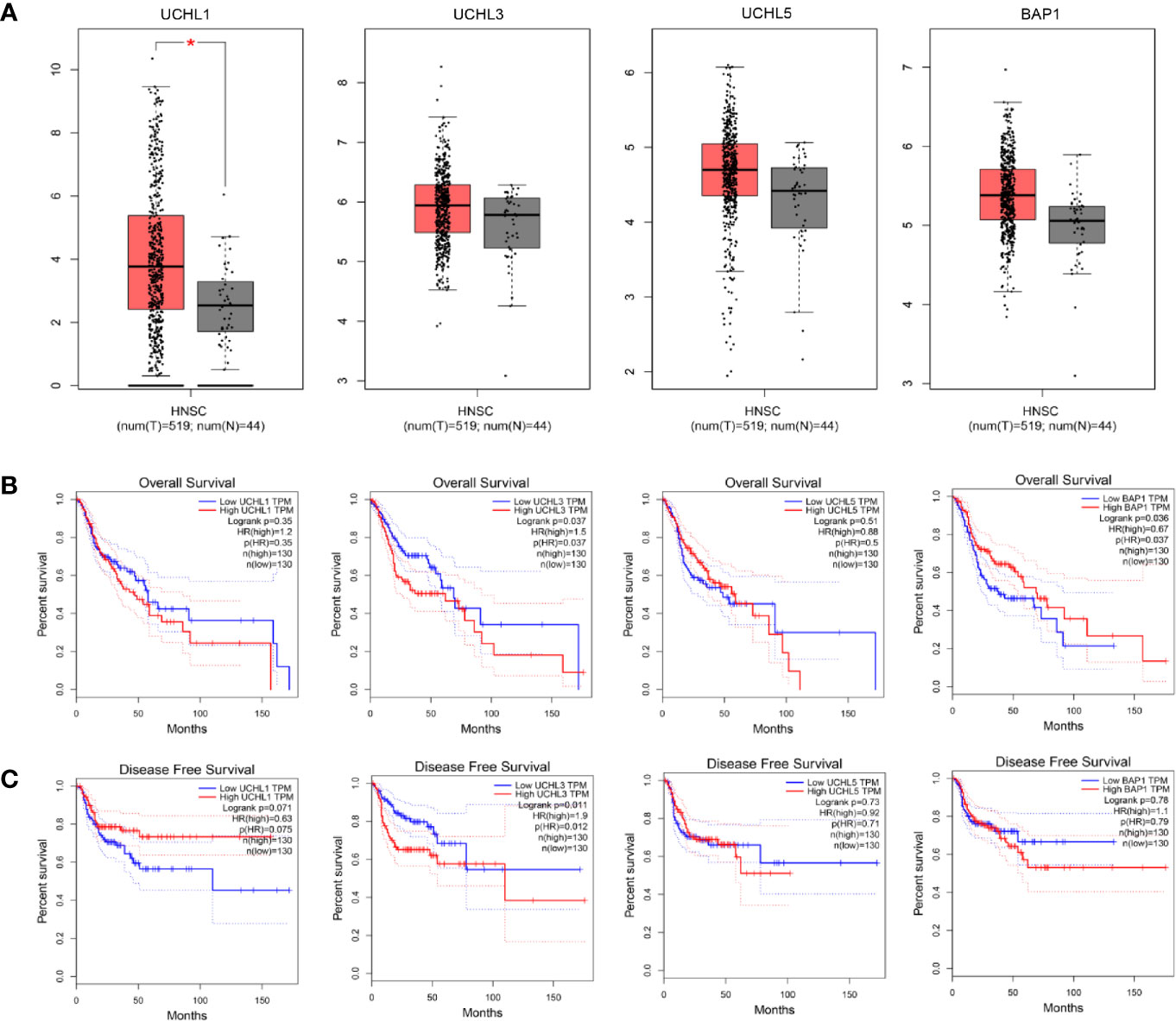
Figure 3 Differential expression analysis of UCH family genes between tumor tissues and normal tissues in the TCGA-HNSC cohort (A). Kaplan-Meier plots analyses show overall (B) and disease-free (C) survival compared by log-rank test. The Cox proportional hazard ratio (HR) is shown in the survival plots.
Therapeutic Implications for HNC Targeting UCHs members
Research on targeting UCHs members in HNSCC therapy is still in the initial period. To our knowledge, there are no studies focusing on UCHs family molecular inhibitors or drugs for clinical trials, which reflects the lack of theoretical and preclinical research. Encouragingly, UCHs family has been shown to predict therapeutic sensitivity and clinical outcomes for various tumors. For example, UCHL1 strengthens tumor cells chemosensitivity in melanoma and colorectal cancer by stabilizing NOXA (165). BAP1 was also reported to modulate cancer cell sensitivity to radiotherapy and the molecular inhibitors including PARP (olaparib) or histone deacetylase inhibitors (panobinostat), which may become potential therapeutic strategies (87, 166). The small molecule b-AP15 as a previously unidentified class of proteasome inhibitor abrogates the activity of two 19S regulatory-particle-associated deubiquitinases, UCH37/UCHL5, and USP14 (167). In vivo b-AP15 prevents tumor progression in four different solid tumor models, including HNSCC, indicating deubiquitinating activity of UCH37/UCHL5 represents a novel therapeutic target for cancer (167).
Over the last decade, the high-risk HPV infection in HNC plays a critical role in staging and prognosis, which promotes personalized therapy and the de-intensification of currently established treatment protocols based on HPV status (168). The underlying mechanisms of UCHs family in HPV-related carcinogenesis remains an enigma. It is worth mentioning that UCHL1 was specifically up-regulated by high-risk HPV in primary keratinocytes to escape innate immunity. Therefore, the precious functions of UCHL1 and other UCHs family members in HPV-related HNSCC need to be disclosed. One of the current therapeutic challenges is to find more suitable biomarkers or surrogate markers for the identity and selection of subpopulation, which would benefit from personalized and therapy. Response rates of HNSCC patients to cetuximab, the only FDA-approved molecularly target-EGFR monoclonal antibody, are only 10% (169). UCHs members have been described to interact with EGFR (170), suggesting the potential of combination therapy with UCHs members for cetuximab treatment in HNSCC.
HNSCC, like other human malignancies, is an immunosuppressive disease. Therefore, immunomodulatory treatment to overcome immune suppressive phenotypes in HNSCC patients has emerged as novel and effective strategies, which include cancer vaccines (e.g., HPV vaccines, tumor peptide antigens), cytokines (e.g., IL2, IFNγ, TNFα), specific monoclonal antibodies (e.g., anti-PD1/PD-L1, CTLA-4 antibodies) (171). Over the past 10 years, the most remarkable therapeutic advances have been achieved in immune checkpoint blockade in HNSCC. FDA approved several target immune checkpoint agents for the treatment of patients with HNSCC. However, the patients revealed different responses to these agents, with only less than 20% of the responder (172, 173). There are many challenges for the immunotherapy of HNC in the future, such as the selection of responding patients, integration into the spectrum of conventional treatment, reduction of immunosuppression in non-responding patients (174). The DUBs are involved in the regulation of innate and adaptive immune response, which sheds light on the immunoregulatory of UCHs family for combination immunotherapy in HNSCC (114, 175). In addition, a variety of patented compounds targeting UCHs members have been developed, which would prepare a path toward the outstanding achievement of genuinely personalized medicine for the treatment of cancers (176–179).
Conclusion and Perspective
In summary, an increasing number of studies suggest that members of UCHs family exert distinct functions in a variety of human malignancies. However, available studies on UCHs in head and neck cancer are limited. It is an exciting time for HNSCC research based on the comprehensive genomic data, as the molecular landscape and altered signaling pathways has been synthetically described. But there are no genetic and proteomic screening tests routinely incorporated into the HNSCC clinically. Emerging evidence has revealed the members of UCHs are associated with the pathogenesis and clinical prognosis of HNSCC, which highlights the prognostic and therapeutic implications of UCHs for patients with HNC. Based on the available data, we have launched a joint project on the expression and function of UCHs in HNSCC, which aims to provide more evidence that UCHs might be the novel prognostic marker and therapeutic target. There are some emerging unresolved issues in HNSCC, such as: what are the precise substrates and regulators of the UCHs family? What are genetic or epigenetic events, and signaling pathways relevant to the UCHs family? Are UCHs family members able to serve as biomarkers for identifying a subset of patients to receive the optimal treatment? Can the agents targeting UCHs family become one of the novel treatment regimens? Optimization of combination regimens of immune checkpoint inhibitors and the agents targeting UCHs family may be a remarkable challenge for immunotherapy of HNSCC. Finally, whether and how the UCHs family members can be translated into the clinical management of HNC remains a formidable mission for the future.
Author Contributions
Conception and design: CR, RZ, and JH. Writing—original draft preparation: CR and RZ. Review of the literature: CR, RZ, and SW. Project supervision: DS and S-LW. Critical revision of the manuscript: JH. Revise and resubmit: CR. All authors contributed to the article and approved the submitted version.
Funding
CR and SW are supported by the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (No. 20KJB310014, No. 18KJB320017), Natural Science Foundation of Jiangsu Province (No. BK20200878). RZ and DS are supported by Wu Jieping Medical Foundation Research Fund Project of Chinese Medical Association (No. 320.6750.19090-19). S-LW is supported by National Key R&D Program of China (Grant No. 2016YFC1303800). This study was also supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge cBioPortal and GEPIA that provide the web-based tools for exploring, visualizing, and analyzing cancer genomic data. We apologize to colleagues whose work was not cited in this review due to space constraints.
Abbreviations
MDM2, murine double minute 2; NOX4, NADPH oxidase 4; EGFR, Epidermal growth factor receptor; HIF-1, Hypoxia-inducible factor 1; PHLPP1, PH Domain and Leucine Rich Repeat Protein Phosphatase 1; MITF, Melanocyte Inducing Transcription Factor; CTTN, Cortactin; mTORC1, mTOR complex 1; eIF4F, Eukaryotic initiation factor 4F; TRAF2, TNF Receptor Associated Factor 2; PRDX1, Peroxiredoxin 1; SNRPF, Small Nuclear Ribonucleoprotein Polypeptide F; Smad2, SMAD Family Member 2; GRP78, 78-kDa glucose-regulated protein; E2F1, E2F Transcription Factor 1; HCF-1, Host cell factor C1; ASXL1/2, ASXL Transcriptional Regulator 1/2; MCRS1, Microspherule Protein 1; IP3R3, type-3 inositol-1,4,5-trisphosphate-receptor; ATF3, Activating Transcription Factor 3; 14-3-3 protein; SLC7A11, Solute Carrier Family 7 Member 11.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer (Oxford Engl 1990) (2018) 103:356–87. doi: 10.1016/j.ejca.2018.07.005
3. Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer (2011) 11(1):9–22. doi: 10.1038/nrc2982
4. Shield KD, Ferlay J, Jemal A, Sankaranarayanan R, Chaturvedi AK, Bray F, et al. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA: Cancer J Clin (2017) 67(1):51–64. doi: 10.3322/caac.21384
5. Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet (2008) 371(9625):1695–709. doi: 10.1016/S0140-6736(08)60728-X
6. Guo X, Johnson RC, Deng H, Liao J, Guan L, Nelson GW, et al. Evaluation of nonviral risk factors for nasopharyngeal carcinoma in a high-risk population of Southern China. Int J Cancer (2009) 124(12):2942–7. doi: 10.1002/ijc.24293
7. Tsao SW, Yip YL, Tsang CM, Pang PS, Lau VMY, Zhang G, et al. Etiological factors of nasopharyngeal carcinoma. Oral Oncol (2014) 50(5):330–8. doi: 10.1016/j.oraloncology.2014.02.006
8. Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet (London England) (2016) 387(10022):1012–24. doi: 10.1016/S0140-6736(15)00055-0
9. Rietbergen MM, van Bokhoven A, Lissenberg-Witte BI, Heideman DAM, Leemans CR, Brakenhoff RH, et al. Epidemiologic associations of HPV-positive oropharyngeal cancer and (pre)cancerous cervical lesions. Int J Cancer J Int Du Cancer (2018) 143(2):283–8. doi: 10.1002/ijc.31315
10. Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Institute (2000) 92(9):709–20. doi: 10.1093/jnci/92.9.709
11. Sabatini ME, Chiocca S. Human papillomavirus as a driver of head and neck cancers. Br J Cancer (2020) 122(3):306–14. doi: 10.1038/s41416-019-0602-7
12. Syrjanen S. Human papillomavirus (HPV) in head and neck cancer. J Clin Virol (2005) 32 Suppl 1:S59–66. doi: 10.1016/j.jcv.2004.11.017
13. Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol (2010) 11(8):781–9. doi: 10.1016/S1470-2045(10)70017-6
14. Ren S, Gaykalova DA, Guo T, Favorov AV, Fertig EJ, Tamayo P, et al. HPV E2, E4, E5 drive alternative carcinogenic pathways in HPV positive cancers. Oncogene (2020) 39(40):6327–39. doi: 10.1038/s41388-020-01431-8
15. Braakhuis BJ, Snijders PJ, Keune WJ, Meijer CJ, Ruijter-Schippers HJ, Leemans CR, et al. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Institute (2004) 96(13):998–1006. doi: 10.1093/jnci/djh183
16. Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, Zhang X, et al. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res An (2006) 12(3 Pt 1):701–9. doi: 10.1158/1078-0432.CCR-05-2017
17. Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. New Engl J Med (2010) 363(1):24–35. doi: 10.1056/NEJMoa0912217
18. Spence T, Bruce J, Yip KW, Liu FF. HPV Associated Head and Neck Cancer. Cancers (Basel) (2016) 8(8). doi: 10.3390/cancers8080075
19. Chera BS, Amdur RJ, Tepper J, Qaqish B, Green R, Aumer SL, et al. Phase 2 Trial of De-intensified Chemoradiation Therapy for Favorable-Risk Human Papillomavirus-Associated Oropharyngeal Squamous Cell Carcinoma. Int J Radiat Oncol Biol Phys (2015) 93(5):976–85. doi: 10.1016/j.ijrobp.2015.08.033
20. Mirghani H, Amen F, Blanchard P, Moreau F, Guigay J, Hartl DM, et al. Treatment de-escalation in HPV-positive oropharyngeal carcinoma: ongoing trials, critical issues and perspectives. Int J Cancer J Int Du Cancer (2015) 136(7):1494–503. doi: 10.1002/ijc.28847
21. Paclitaxel, cisplatin, and cetuximab followed by cetuximab and intensity-modulated radiation therapy in treating patients with HPV-associated stage III or stage IV cancer of the oropharynx that can be removed by surgery; NCT01084083. Available at: http://www.clinicaltrials.gov/.
22. Bigelow EO, Seiwert TY, Fakhry C. Deintensification of treatment for human papillomavirus-related oropharyngeal cancer: Current state and future directions. Oral Oncol (2020) 105:104652. doi: 10.1016/j.oraloncology.2020.104652
23. Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature (2015) 517(7536):576–82. doi: 10.1038/nature14129
24. Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun (2014) 5:3887. doi: 10.1038/ncomms4887
25. Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods (2013) 10(11):1046–7. doi: 10.1038/nmeth.2650
26. Fang Y, Shen X. Ubiquitin carboxyl-terminal hydrolases: involvement in cancer progression and clinical implications. Cancer Metastasis Rev (2017) 36(4):669–82. doi: 10.1007/s10555-017-9702-0
27. Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell (2005) 123(5):773–86. doi: 10.1016/j.cell.2005.11.007
28. Pfoh R, Lacdao IK, Saridakis V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr Relat Cancer (2015) 22(1):T35–54. doi: 10.1530/ERC-14-0516
29. Pal A, Young MA, Donato NJ. Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res (2014) 74(18):4955–66. doi: 10.1158/0008-5472.CAN-14-1211
30. Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem (2009) 78:363–97. doi: 10.1146/annurev.biochem.78.082307.091526
31. Young MJ, Hsu KC, Lin TE, Chang WC, Hung JJ. The role of ubiquitin-specific peptidases in cancer progression. J BioMed Sci (2019) 26(1):42. doi: 10.1186/s12929-019-0522-0
32. Liu X, Kumar M, Yang L, Molkentine DP, Valdecanas D, Yu S, et al. BAP1 Is a Novel Target in HPV-Negative Head and Neck Cancer. Clin Cancer Res (2018) 24(3):600–7. doi: 10.1158/1078-0432.CCR-17-1573
33. Li L, Tao Q, Jin H, van Hasselt A, Poon FF, Wang X, et al. The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clin Cancer Res (2010) 16(11):2949–58. doi: 10.1158/1078-0432.CCR-09-3178
34. Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT Jr. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell (2002) 111(2):209–18. doi: 10.1016/s0092-8674(02)01012-7
35. Larsen CN, Price JS, Wilkinson KD. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry (1996) 35(21):6735–44. doi: 10.1021/bi960099f
36. Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry (1998) 37(10):3358–68. doi: 10.1021/bi972274d
37. Sakurai M, Sekiguchi M, Zushida K, Yamada K, Nagamine S, Kabuta T, et al. Reduction in memory in passive avoidance learning, exploratory behaviour and synaptic plasticity in mice with a spontaneous deletion in the ubiquitin C-terminal hydrolase L1 gene. Eur J Neurosci (2008) 27(3):691–701. doi: 10.1111/j.1460-9568.2008.06047.x
38. Day IN, Thompson RJ. UCHL1 (PGP 9.5): neuronal biomarker and ubiquitin system protein. Prog Neurobiol (2010) 90(3):327–62. doi: 10.1016/j.pneurobio.2009.10.020
39. Setsuie R, Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem Int (2007) 51(2-4):105–11. doi: 10.1016/j.neuint.2007.05.007
40. Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, et al. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem (2004) 279(13):13256–64. doi: 10.1074/jbc.M314124200
41. Chai C, Lim KL. Genetic insights into sporadic Parkinson’s disease pathogenesis. Curr Genomics (2013) 14(8):486–501. doi: 10.2174/1389202914666131210195808
42. Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, et al. The ubiquitin pathway in Parkinson’s disease. Nature (1998) 395(6701):451–2. doi: 10.1038/26652
43. Maraganore DM, Farrer MJ, Hardy JA, Lincoln SJ, McDonnell SK, Rocca WA. Case-control study of the ubiquitin carboxy-terminal hydrolase L1 gene in Parkinson’s disease. Neurology (1999) 53(8):1858–60. doi: 10.1212/wnl.53.8.1858
44. Liu H, Povysheva N, Rose ME, Mi Z, Banton JS, Li W, et al. Role of UCHL1 in axonal injury and functional recovery after cerebral ischemia. Proc Natl Acad Sci U S A (2019) 116(10):4643–50. doi: 10.1073/pnas.1821282116
45. McKeon JE, Sha D, Li L, Chin LS. Parkin-mediated K63-polyubiquitination targets ubiquitin C-terminal hydrolase L1 for degradation by the autophagy-lysosome system. Cell Mol Life Sci (2015) 72(9):1811–24. doi: 10.1007/s00018-014-1781-2
46. Bradbury JM, Thompson RJ. Immunoassay of the neuronal and neuroendocrine marker PGP 9.5 in human tissues. J Neurochem (1985) 44(2):651–3. doi: 10.1111/j.1471-4159.1985.tb05461.x
47. Schumacher U, Mitchell BS, Kaiserling E. The neuronal marker protein gene product 9.5 (PGP 9.5) is phenotypically expressed in human breast epithelium, in milk, and in benign and malignant breast tumors. DNA Cell Biol (1994) 13(8):839–43. doi: 10.1089/dna.1994.13.839
48. Olerud JE, Chiu DS, Usui ML, Gibran NS, Ansel JC. Protein gene product 9.5 is expressed by fibroblasts in human cutaneous wounds. J Invest Dermatol (1998) 111(4):565–72. doi: 10.1046/j.1523-1747.1998.00330.x
49. Bheda A, Gullapalli A, Caplow M, Pagano JS, Shackelford J. Ubiquitin editing enzyme UCH L1 and microtubule dynamics: implication in mitosis. Cell Cycle (2010) 9(5):980–94. doi: 10.4161/cc.9.5.10934
50. Miyoshi Y, Nakayama S, Torikoshi Y, Tanaka S, Ishihara H, Taguchi T, et al. High expression of ubiquitin carboxy-terminal hydrolase-L1 and -L3 mRNA predicts early recurrence in patients with invasive breast cancer. Cancer Sci (2006) 97(6):523–9. doi: 10.1111/j.1349-7006.2006.00202.x
51. Ummanni R, Mundt F, Pospisil H, Venz S, Scharf C, Barett C, et al. Identification of clinically relevant protein targets in prostate cancer with 2D-DIGE coupled mass spectrometry and systems biology network platform. PloS One (2011) 6(2):e16833. doi: 10.1371/journal.pone.0016833
52. Fukutomi S, Seki N, Koda K, Miyazaki M. Identification of methylation-silenced genes in colorectal cancer cell lines: genomic screening using oligonucleotide arrays. Scand J Gastroenterol (2007) 42(12):1486–94. doi: 10.1080/00365520701491173
53. Liu S, Gonzalez-Prieto R, Zhang M, Geurink PP, Kooij R, Iyengar PV, et al. Deubiquitinase Activity Profiling Identifies UCHL1 as a Candidate Oncoprotein That Promotes TGFbeta-Induced Breast Cancer Metastasis. Clin Cancer Res (2019). doi: 10.1158/1078-0432.CCR-19-1373
54. Yamashita K, Park HL, Kim MS, Osada M, Tokumaru Y, Inoue H, et al. PGP9.5 methylation in diffuse-type gastric cancer. Cancer Res (2006) 66(7):3921–7. doi: 10.1158/0008-5472.CAN-05-1511
55. Tezel E, Hibi K, Nagasaka T, Nakao A. PGP9.5 as a Prognostic Factor in Pancreatic Cancer. Clin Cancer Res (2000) 6:4764–7. doi: 10.1038/bjc.2014.417
56. Hurst-Kennedy J, Chin LS, Li L. Ubiquitin C-terminal hydrolase l1 in tumorigenesis. Biochem Res Int (2012) 2012:123706. doi: 10.1155/2012/123706
57. Jara JH, Frank DD, Ozdinler PH. Could dysregulation of UPS be a common underlying mechanism for cancer and neurodegeneration? Lessons from UCHL1. Cell Biochem Biophys (2013) 67(1):45–53. doi: 10.1007/s12013-013-9631-7
58. Xiang T, Li L, Yin X, Yuan C, Tan C, Su X, et al. The ubiquitin peptidase UCHL1 induces G0/G1 cell cycle arrest and apoptosis through stabilizing p53 and is frequently silenced in breast cancer. PloS One (2012) 7(1):e29783. doi: 10.1371/journal.pone.0029783
59. Yu J, Tao Q, Cheung KF, Jin H, Poon FF, Wang X, et al. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology (2008) 48(2):508–18. doi: 10.1002/hep.22343
60. Abdelmaksoud-Dammak R, Saadallah-Kallel A, Miladi-Abdennadher I, Ayedi L, Khabir A, Sallemi-Boudawara T, et al. CpG methylation of ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) and P53 mutation pattern in sporadic colorectal cancer. Tumour Biol (2016) 37(2):1707–14. doi: 10.1007/s13277-015-3902-4
61. Ummanni R, Jost E, Braig M, Lohmann F, Mundt F, Barett C, et al. Ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) is a potential tumour suppressor in prostate cancer and is frequently silenced by promoter methylation. Mol Cancer (2011) 10:129. doi: 10.1186/1476-4598-10-129
62. Zhong J, Zhao M, Ma Y, Luo Q, Liu J, Wang J, et al. UCHL1 acts as a colorectal cancer oncogene via activation of the beta-catenin/TCF pathway through its deubiquitinating activity. Int J Mol Med (2012) 30(2):430–6. doi: 10.3892/ijmm.2012.1012
63. Sanchez-Diaz PC, Chang JC, Moses ES, Dao T, Chen Y, Hung JY. Ubiquitin carboxyl-terminal esterase L1 (UCHL1) is associated with stem-like cancer cell functions in pediatric high-grade glioma. PloS One (2017) 12(5):e0176879. doi: 10.1371/journal.pone.0176879
64. Kim HJ, Magesh V, Lee JJ, Kim S, Knaus UG, Lee KJ. Ubiquitin C-terminal hydrolase-L1 increases cancer cell invasion by modulating hydrogen peroxide generated via NADPH oxidase 4. Oncotarget (2015) 6(18):16287–303. doi: 10.18632/oncotarget.3843
65. Jin Y, Zhang W, Xu J, Wang H, Zhang Z, Chu C, et al. UCH-L1 involved in regulating the degradation of EGFR and promoting malignant properties in drug-resistant breast cancer. Int J Clin Exp Pathol (2015) 8(10):12500–8.
66. Goto Y, Zeng L, Yeom CJ, Zhu Y, Morinibu A, Shinomiya K, et al. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1alpha. Nat Commun (2015) 6:6153. doi: 10.1038/ncomms7153
67. Li X, Hattori A, Takahashi S, Goto Y, Harada H, Kakeya H. Ubiquitin carboxyl-terminal hydrolase L1 promotes hypoxia-inducible factor 1-dependent tumor cell malignancy in spheroid models. Cancer Sci (2019). doi: 10.1111/cas.14236
68. Kwan SY, Au-Yeung CL, Yeung TL, Rynne-Vidal A, Wong KK, Risinger JI, et al. Ubiquitin Carboxyl-Terminal Hydrolase L1 (UCHL1) Promotes Uterine Serous Cancer Cell Proliferation and Cell Cycle Progression. Cancers (Basel) (2020) 12(1). doi: 10.3390/cancers12010118
69. Hussain S, Foreman O, Perkins SL, Witzig TE, Miles RR, van Deursen J, et al. The de-ubiquitinase UCH-L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling. Leukemia (2010) 24(9):1641–55. doi: 10.1038/leu.2010.138
70. Seo EY, Jin SP, Sohn KC, Park CH, Lee DH, Chung JH. UCHL1 Regulates Melanogenesis through Controlling MITF Stability in Human Melanocytes. J Invest Dermatol (2017) 137(8):1757–65. doi: 10.1016/j.jid.2017.03.024
71. Zhao Y, Lei Y, He SW, Li YQ, Wang YQ, Hong XH, et al. Hypermethylation of UCHL1 Promotes Metastasis of Nasopharyngeal Carcinoma by Suppressing Degradation of Cortactin (CTTN). Cells (2020) 9(3). doi: 10.3390/cells9030559
72. Hussain S, Bedekovics T, Liu Q, Hu W, Jeon H, Johnson SH, et al. UCH-L1 bypasses mTOR to promote protein biosynthesis and is required for MYC-driven lymphomagenesis in mice. Blood (2018) 132(24):2564–74. doi: 10.1182/blood-2018-05-848515
73. Hussain S, Bedekovics T, Ali A, Zaid O, May DG, Roux KJ, et al. A cysteine near the C-terminus of UCH-L1 is dispensable for catalytic activity but is required to promote AKT phosphorylation, eIF4F assembly, and malignant B-cell survival. Cell Death Discov (2019) 5:152. doi: 10.1038/s41420-019-0231-1
74. Zhang MH, Zhang HH, Du XH, Gao J, Li C, Shi HR, et al. UCHL3 promotes ovarian cancer progression by stabilizing TRAF2 to activate the NF-kappaB pathway. Oncogene (2020) 39(2):322–33. doi: 10.1038/s41388-019-0987-z
75. Luo K, Li L, Li Y, Wu C, Yin Y, Chen Y, et al. A phosphorylation-deubiquitination cascade regulates the BRCA2-RAD51 axis in homologous recombination. Genes Dev (2016) 30(23):2581–95. doi: 10.1101/gad.289439.116
76. Fang Y, Fu D, Tang W, Cai Y, Ma D, Wang H, et al. Ubiquitin C-terminal Hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim Biophys Acta (2013) 1833(3):559–72. doi: 10.1016/j.bbamcr.2012.11.020
77. Fang Y, He J, Janssen HLA, Wu J, Dong L, Shen XZ. Peroxiredoxin 1, restraining cell migration and invasion, is involved in hepatocellular carcinoma recurrence. J Dig Dis (2018) 19(3):155–69. doi: 10.1111/1751-2980.12580
78. Ge J, Hu W, Zhou H, Yu J, Sun C, Chen W. Ubiquitin carboxyl-terminal hydrolase isozyme L5 inhibits human glioma cell migration and invasion via downregulating SNRPF. Oncotarget (2017) 8(69):113635–49. doi: 10.18632/oncotarget.23071
79. Fukui S, Nagasaka K, Miyagawa Y, Kikuchi-Koike R, Kawata Y, Kanda R, et al. The proteasome deubiquitinase inhibitor bAP15 downregulates TGF-beta/Smad signaling and induces apoptosis via UCHL5 inhibition in ovarian cancer. Oncotarget (2019) 10(57):5932–48. doi: 10.18632/oncotarget.27219
80. Fang Y, Mu J, Ma Y, Ma D, Fu D, Shen X. The interaction between ubiquitin C-terminal hydrolase 37 and glucose-regulated protein 78 in hepatocellular carcinoma. Mol Cell Biochem (2012) 359(1-2):59–66. doi: 10.1007/s11010-011-0999-7
81. Han W, Lee H, Han JK. Ubiquitin C-terminal hydrolase37 regulates Tcf7 DNA binding for the activation of Wnt signalling. Sci Rep (2017) 7:42590. doi: 10.1038/srep42590
82. Randles L, Anchoori RK, Roden RB, Walters KJ. The Proteasome Ubiquitin Receptor hRpn13 and Its Interacting Deubiquitinating Enzyme Uch37 Are Required for Proper Cell Cycle Progression. J Biol Chem (2016) 291(16):8773–83. doi: 10.1074/jbc.M115.694588
83. Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene (1998) 16(9):1097–112. doi: 10.1038/sj.onc.1201861
84. Dkhissi F, Aggoune D, Pontis J, Sorel N, Piccirilli N, LeCorf A, et al. The downregulation of BAP1 expression by BCR-ABL reduces the stability of BRCA1 in chronic myeloid leukemia. Exp Hematol (2015) 43(9):775–80. doi: 10.1016/j.exphem.2015.04.013
85. Shankar GM, Santagata S. BAP1 mutations in high-grade meningioma: implications for patient care. Neuro Oncol (2017) 19(11):1447–56. doi: 10.1093/neuonc/nox094
86. Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem (2009) 284(49):34179–88. doi: 10.1074/jbc.M109.046755
87. Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet (2012) 44(7):751–9. doi: 10.1038/ng.2323
88. Lee HS, Lee SA, Hur SK, Seo JW, Kwon J. Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat Commun (2014) 5:5128. doi: 10.1038/ncomms6128
89. Zarrizi R, Menard JA, Belting M, Massoumi R. Deubiquitination of gamma-tubulin by BAP1 prevents chromosome instability in breast cancer cells. Cancer Res (2014) 74(22):6499–508. doi: 10.1158/0008-5472.CAN-14-0221
90. Daou S, Barbour H, Ahmed O, Masclef L, Baril C, Sen Nkwe N, et al. Monoubiquitination of ASXLs controls the deubiquitinase activity of the tumor suppressor BAP1. Nat Commun (2018) 9(1):4385. doi: 10.1038/s41467-018-06854-2
91. Peng J, Ma J, Li W, Mo R, Zhang P, Gao K, et al. Stabilization of MCRS1 by BAP1 prevents chromosome instability in renal cell carcinoma. Cancer Lett (2015) 369(1):167–74. doi: 10.1016/j.canlet.2015.08.013
92. Bononi A, Giorgi C, Patergnani S, Larson D, Verbruggen K, Tanji M, et al. BAP1 regulates IP3R3-mediated Ca(2+) flux to mitochondria suppressing cell transformation. Nature (7659) 2017) 546:549–53. doi: 10.1038/nature22798
93. Dai F, Lee H, Zhang Y, Zhuang L, Yao H, Xi Y, et al. BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc Natl Acad Sci U S A (2017) 114(12):3192–7. doi: 10.1073/pnas.1619588114
94. Sime W, Niu Q, Abassi Y, Masoumi KC, Zarrizi R, Kohler JB, et al. BAP1 induces cell death via interaction with 14-3-3 in neuroblastoma. Cell Death Dis (2018) 9(5):458. doi: 10.1038/s41419-018-0500-6
95. Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol (2018) 20(10):1181–92. doi: 10.1038/s41556-018-0178-0
96. Zhang Y, Zhuang L, Gan B. BAP1 suppresses tumor development by inducing ferroptosis upon SLC7A11 repression. Mol Cell Oncol (2019) 6(1):1536845. doi: 10.1080/23723556.2018.1536845
97. Luo Y, He J, Yang C, Orange M, Ren X, Blair N, et al. UCH-L1 promotes invasion of breast cancer cells through activating Akt signaling pathway. J Cell Biochem (2018) 119(1):691–700. doi: 10.1002/jcb.26232
98. Okochi-Takada E, Nakazawa K, Wakabayashi M, Mori A, Ichimura S, Yasugi T, et al. Silencing of the UCHL1 gene in human colorectal and ovarian cancers. Int J Cancer J Int Du Cancer (2006) 119(6):1338–44. doi: 10.1002/ijc.22025
99. Ding X, Gu Y, Jin M, Guo X, Xue S, Tan C, et al. The deubiquitinating enzyme UCHL1 promotes resistance to pemetrexed in non-small cell lung cancer by upregulating thymidylate synthase. Theranostics (2020) 10(13):6048–60. doi: 10.7150/thno.42096
100. Bedekovics T, Hussain S, Feldman AL, Galardy PJ. UCH-L1 is induced in germinal center B cells and identifies patients with aggressive germinal center diffuse large B-cell lymphoma. Blood (2016) 127(12):1564–74. doi: 10.1182/blood-2015-07-656678
101. Li G, Jin X, Zheng J, Jiang N, Shi W. UCH-L3 promotes non-small cell lung cancer proliferation via accelerating cell cycle and inhibiting cell apoptosis. Biotechnol Appl Biochem (2020). doi: 10.1002/bab.1909
102. Song HM, Lee JE, Kim JH. Ubiquitin C-terminal hydrolase-L3 regulates EMT process and cancer metastasis in prostate cell lines. Biochem Biophys Res Commun (2014) 452(3):722–7. doi: 10.1016/j.bbrc.2014.08.144
103. Rolen U, Kobzeva V, Gasparjan N, Ovaa H, Winberg G, Kisseljov F, et al. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol Carcinog (2006) 45(4):260–9. doi: 10.1002/mc.20177
104. Chen Y, Fu D, Xi J, Ji Z, Liu T, Ma Y, et al. Expression and clinical significance of UCH37 in human esophageal squamous cell carcinoma. Dig Dis Sci (2012) 57(9):2310–7. doi: 10.1007/s10620-012-2181-9
105. Sha B, Chen X, Wu H, Li M, Shi J, Wang L, et al. Deubiquitylatinase inhibitor b-AP15 induces c-Myc-Noxa-mediated apoptosis in esophageal squamous cell carcinoma. Apoptosis An Int J Programmed Cell Death (2019) 24(9-10):826–36. doi: 10.1007/s10495-019-01561-9
106. Leblay N, Lepretre F, Le Stang N, Gautier-Stein A, Villeneuve L, Isaac S, et al. BAP1 Is Altered by Copy Number Loss, Mutation, and/or Loss of Protein Expression in More Than 70% of Malignant Peritoneal Mesotheliomas. J Thoracic Oncol Off Publ Int Assoc Study Lung Cancer (2017) 12(4):724–33. doi: 10.1016/j.jtho.2016.12.019
107. Yoshikawa Y, Sato A, Tsujimura T, Emi M, Morinaga T, Fukuoka K, et al. Frequent inactivation of the BAP1 gene in epithelioid-type malignant mesothelioma. Cancer Sci (2012) 103(5):868–74. doi: 10.1111/j.1349-7006.2012.02223.x
108. Joseph RW, Kapur P, Serie DJ, Eckel-Passow JE, Parasramka M, Ho T, et al. Loss of BAP1 protein expression is an independent marker of poor prognosis in patients with low-risk clear cell renal cell carcinoma. Cancer (2014) 120(7):1059–67. doi: 10.1002/cncr.28521
109. See TR, Stalhammar G, Phillips S, Grossniklaus HE. BAP1 Immunoreactivity Correlates with Gene Expression Class in Uveal Melanoma. Ocul Oncol Pathol (2020) 6(2):129–37. doi: 10.1159/000502550
110. Kalirai H, Dodson A, Faqir S, Damato BE, Coupland SE. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br J Cancer (2014) 111(7):1373–80. doi: 10.1038/bjc.2014.417
111. Mochel MC, Piris A, Nose V, Hoang MP. Loss of BAP1 Expression in Basal Cell Carcinomas in Patients With Germline BAP1 Mutations. Am J Clin Pathol (2015) 143(6):901–4. doi: 10.1309/AJCPG8LFJC0DHDQT
112. Mori T, Sumii M, Fujishima F, Ueno K, Emi M, Nagasaki M, et al. Somatic alteration and depleted nuclear expression of BAP1 in human esophageal squamous cell carcinoma. Cancer Sci (2015) 106(9):1118–29. doi: 10.1111/cas.12722
113. Reinicke AT, Raczkowski F, Muhlig M, Schmucker P, Lischke T, Reichelt J, et al. Deubiquitinating Enzyme UCH-L1 Promotes Dendritic Cell Antigen Cross-Presentation by Favoring Recycling of MHC Class I Molecules. J Immunol (2019) 203(7):1730–42. doi: 10.4049/jimmunol.1801133
114. Gu Y, Ding X, Huang J, Xue M, Zhang J, Wang Q, et al. The deubiquitinating enzyme UCHL1 negatively regulates the immunosuppressive capacity and survival of multipotent mesenchymal stromal cells. Cell Death Dis (2018) 9(5):459. doi: 10.1038/s41419-018-0532-y
115. Wada H, Kito K, Caskey LS, Yeh ET, Kamitani T. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem Biophys Res Commun (1998) 251(3):688–92. doi: 10.1006/bbrc.1998.9532
116. Kwon J, Wang YL, Setsuie R, Sekiguchi S, Sato Y, Sakurai M, et al. Two closely related ubiquitin C-terminal hydrolase isozymes function as reciprocal modulators of germ cell apoptosis in cryptorchid testis. Am J Pathol (2004) 165(4):1367–74. doi: 10.1016/S0002-9440(10)63394-9
117. Sano Y, Furuta A, Setsuie R, Kikuchi H, Wang YL, Sakurai M, et al. Photoreceptor cell apoptosis in the retinal degeneration of Uchl3-deficient mice. Am J Pathol (2006) 169(1):132–41. doi: 10.2353/ajpath.2006.060085
118. Kurihara LJ, Kikuchi T, Wada K, Tilghman SM. Loss of Uch-L1 and Uch-L3 leads to neurodegeneration, posterior paralysis and dysphagia. Hum Mol Genet (2001) 10(18):1963–70. doi: 10.1093/hmg/10.18.1963
119. Setsuie R, Suzuki M, Tsuchiya Y, Wada K. Skeletal muscles of Uchl3 knockout mice show polyubiquitinated protein accumulation and stress responses. Neurochem Int (2010) 56(8):911–8. doi: 10.1016/j.neuint.2010.03.021
120. Setsuie R, Suzuki M, Kabuta T, Fujita H, Miura S, Ichihara N, et al. Ubiquitin C-terminal hydrolase-L3-knockout mice are resistant to diet-induced obesity and show increased activation of AMP-activated protein kinase in skeletal muscle. FASEB J (2009) 23(12):4148–57. doi: 10.1096/fj.09-132217
121. Suzuki M, Setsuie R, Wada K. Ubiquitin carboxyl-terminal hydrolase l3 promotes insulin signaling and adipogenesis. Endocrinology (2009) 150(12):5230–9. doi: 10.1210/en.2009-0332
122. Kim JY, Lee JM, Cho JY. Ubiquitin C-terminal hydrolase-L3 regulates Smad1 ubiquitination and osteoblast differentiation. FEBS Lett (2011) 585(8):1121–6. doi: 10.1016/j.febslet.2011.03.053
123. Dennissen FJ, Kholod N, Hermes DJ, Kemmerling N, Steinbusch HW, Dantuma NP, et al. Mutant ubiquitin (UBB+1) associated with neurodegenerative disorders is hydrolyzed by ubiquitin C-terminal hydrolase L3 (UCH-L3). FEBS Lett (2011) 585(16):2568–74. doi: 10.1016/j.febslet.2011.06.037
124. Lam YA, DeMartino GN, Pickart CM, Cohen RE. Specificity of the ubiquitin isopeptidase in the PA700 regulatory complex of 26 S proteasomes. J Biol Chem (1997) 272(45):28438–46. doi: 10.1074/jbc.272.45.28438
125. Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol (2006) 8(9):994–1002. doi: 10.1038/ncb1460
126. Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J (2006) 25(19):4524–36. doi: 10.1038/sj.emboj.7601338
127. Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J (2006) 25(24):5742–53. doi: 10.1038/sj.emboj.7601450
128. Sahtoe DD, van Dijk WJ, El Oualid F, Ekkebus R, Ovaa H, Sixma TK. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol Cell (2015) 57(5):887–900. doi: 10.1016/j.molcel.2014.12.039
129. Al-Shami A, Jhaver KG, Vogel P, Wilkins C, Humphries J, Davis JJ, et al. Regulators of the proteasome pathway, Uch37 and Rpn13, play distinct roles in mouse development. PloS One (2010) 5(10):e13654. doi: 10.1371/journal.pone.0013654
130. Yun D, Zhuang Y, Kreutz MR, Behnisch T. The role of 19S proteasome associated deubiquitinases in activity-dependent hippocampal synaptic plasticity. Neuropharmacology (2018) 133:354–65. doi: 10.1016/j.neuropharm.2018.01.043
131. Kikuchi M, Ogishima S, Miyamoto T, Miyashita A, Kuwano R, Nakaya J, et al. Identification of unstable network modules reveals disease modules associated with the progression of Alzheimer’s disease. PloS One (2013) 8(11):e76162. doi: 10.1371/journal.pone.0076162
132. Mahanic CS, Budhavarapu V, Graves JD, Li G, Lin WC. Regulation of E2 promoter binding factor 1 (E2F1) transcriptional activity through a deubiquitinating enzyme, UCH37. J Biol Chem (2015) 290(44):26508–22. doi: 10.1074/jbc.M115.659425
133. Nan L, Jacko AM, Tan J, Wang D, Zhao J, Kass DJ, et al. Ubiquitin carboxyl-terminal hydrolase-L5 promotes TGFbeta-1 signaling by de-ubiquitinating and stabilizing Smad2/Smad3 in pulmonary fibrosis. Sci Rep (2016) 6:33116. doi: 10.1038/srep33116
134. Wicks SJ, Haros K, Maillard M, Song L, Cohen RE, Dijke PT, et al. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene (2005) 24(54):8080–4. doi: 10.1038/sj.onc.1208944
135. Chen Z, Niu X, Li Z, Yu Y, Ye X, Lu S, et al. Effect of ubiquitin carboxy-terminal hydrolase 37 on apoptotic in A549 cells. Cell Biochem Funct (2011) 29(2):142–8. doi: 10.1002/cbf.1734
136. Arpalahti L, Laitinen A, Hagstrom J, Mustonen H, Kokkola A, Bockelman C, et al. Positive cytoplasmic UCHL5 tumor expression in gastric cancer is linked to improved prognosis. PloS One (2018) 13(2):e0193125. doi: 10.1371/journal.pone.0193125
137. Arpalahti L, Hagstrom J, Mustonen H, Lundin M, Haglund C, Holmberg CI. UCHL5 expression associates with improved survival in lymph-node-positive rectal cancer. Tumour Biol (2017) 39(7):1010428317716078. doi: 10.1177/1010428317716078
138. Arpalahti L, Saukkonen K, Hagstrom J, Mustonen H, Seppanen H, Haglund C, et al. Nuclear ubiquitin C-terminal hydrolase L5 expression associates with increased patient survival in pancreatic ductal adenocarcinoma. Tumour Biol (2017) 39(6):1010428317710411. doi: 10.1177/1010428317710411
139. Wang A, Papneja A, Hyrcza M, Al-Habeeb A, Ghazarian D. Gene of the month: BAP1. J Clin Pathol (2016) 69(9):750–3. doi: 10.1136/jclinpath-2016-203866
140. Okino Y, Machida Y, Frankland-Searby S, Machida YJ. BRCA1-associated protein 1 (BAP1) deubiquitinase antagonizes the ubiquitin-mediated activation of FoxK2 target genes. J Biol Chem (2015) 290(3):1580–91. doi: 10.1074/jbc.M114.609834
141. Daou S, Hammond-Martel I, Mashtalir N, Barbour H, Gagnon J, Iannantuono NV, et al. The BAP1/ASXL2 Histone H2A Deubiquitinase Complex Regulates Cell Proliferation and Is Disrupted in Cancer. J Biol Chem (2015) 290(48):28643–63. doi: 10.1074/jbc.M115.661553
142. Di Nunno V, Frega G, Santoni M, Gatto L, Fiorentino M, Montironi R, et al. BAP1 in solid tumors. Future Oncol (2019) 15(18):2151–62. doi: 10.2217/fon-2018-0915
143. Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science (2010) 330(6009):1410–3. doi: 10.1126/science.1194472
144. Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet (2011) 43(10):1022–5. doi: 10.1038/ng.912
145. Fan LH, Tang LN, Yue L, Yang Y, Gao ZL, Shen Z. BAP1 is a good prognostic factor in advanced non-small cell lung cancer. Clin Invest Med (2012) 35(4):E182–9. doi: 10.25011/cim.v35i4.17146
146. Njauw CN, Kim I, Piris A, Gabree M, Taylor M, Lane AM, et al. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PloS One (2012) 7(4):e35295. doi: 10.1371/journal.pone.0035295
147. Farley MN, Schmidt LS, Mester JL, Pena-Llopis S, Pavia-Jimenez A, Christie A, et al. A novel germline mutation in BAP1 predisposes to familial clear-cell renal cell carcinoma. Mol Cancer Res MCR (2013) 11(9):1061–71. doi: 10.1158/1541-7786.MCR-13-0111
148. Hirsch TZ, Negulescu A, Gupta B, Caruso S, Noblet B, Couchy G, et al. BAP1 mutations define a homogeneous subgroup of hepatocellular carcinoma with fibrolamellar-like features and activated PKA. J Hepatol (2019). doi: 10.1016/j.jhep.2019.12.006
149. Murali R, Wiesner T, Scolyer RA. Tumours associated with BAP1 mutations. Pathology (2013) 45(2):116–26. doi: 10.1097/PAT.0b013e32835d0efb
150. Baumann F, Flores E, Napolitano A, Kanodia S, Taioli E, Pass H, et al. Mesothelioma patients with germline BAP1 mutations have 7-fold improved long-term survival. Carcinogenesis (2015) 36(1):76–81. doi: 10.1093/carcin/bgu227
151. Wang K, McDermott JD, Schrock AB, Elvin JA, Gay L, Karam SD, et al. Comprehensive genomic profiling of salivary mucoepidermoid carcinomas reveals frequent BAP1, PIK3CA, and other actionable genomic alterations. Ann Oncol (2017) 28(4):748–53. doi: 10.1093/annonc/mdw689
152. Durante MA, Walter SD, Paez-Escamilla M, Tokarev J, Decatur CL, Dubovy SR, et al. Intraocular Metastasis in Unilateral Multifocal Uveal Melanoma Without Melanocytosis or Germline BAP1 Mutations. JAMA Ophthalmol (2019). doi: 10.1001/jamaophthalmol.2019.3941
153. Farzin M, Toon CW, Clarkson A, Sioson L, Watson N, Andrici J, et al. Loss of expression of BAP1 predicts longer survival in mesothelioma. Pathology (2015) 47(4):302–7. doi: 10.1097/PAT.0000000000000250
154. Wang SS, Gu YF, Wolff N, Stefanius K, Christie A, Dey A, et al. Bap1 is essential for kidney function and cooperates with Vhl in renal tumorigenesis. Proc Natl Acad Sci U S A (2014) 111(46):16538–43. doi: 10.1073/pnas.1414789111
155. Bi M, Zhao S, Said JW, Merino MJ, Adeniran AJ, Xie Z, et al. Genomic characterization of sarcomatoid transformation in clear cell renal cell carcinoma. Proc Natl Acad Sci U S A (2016) 113(8):2170–5. doi: 10.1073/pnas.1525735113
156. Tokumaru Y, Yamashita K, Kim MS, Park HL, Osada M, Mori M, et al. The role of PGP9.5 as a tumor suppressor gene in human cancer. Int J Cancer J Int Du Cancer (2008) 123(4):753–9. doi: 10.1002/ijc.23354
157. Lleras RA, Smith RV, Adrien LR, Schlecht NF, Burk RD, Harris TM, et al. Unique DNA methylation loci distinguish anatomic site and HPV status in head and neck squamous cell carcinoma. Clin Cancer Res (2013) 19(19):5444–55. doi: 10.1158/1078-0432.CCR-12-3280
158. Zhang J, Chen T, Yang X, Cheng H, Spath SS, Clavijo PE, et al. Attenuated TRAF3 Fosters Activation of Alternative NF-kappaB and Reduced Expression of Antiviral Interferon, TP53, and RB to Promote HPV-Positive Head and Neck Cancers. Cancer Res (2018) 78(16):4613–26. doi: 10.1158/0008-5472.CAN-17-0642
159. Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci U S A (2014) 111(1):285–90. doi: 10.1073/pnas.1309085110
160. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal (2013) 6(269):p11. doi: 10.1126/scisignal.2004088
161. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery (2012) 2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095
162. Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet (2013) 45(6):580–5. doi: 10.1038/ng.2653
163. Canning M, Guo G, Yu M, Myint C, Groves MW, Byrd JK, et al. Heterogeneity of the Head and Neck Squamous Cell Carcinoma Immune Landscape and Its Impact on Immunotherapy. Front Cell Dev Biol (2019) 7:52. doi: 10.3389/fcell.2019.00052
164. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45(W1):W98–102. doi: 10.1093/nar/gkx247
165. Brinkmann K, Zigrino P, Witt A, Schell M, Ackermann L, Broxtermann P, et al. Ubiquitin C-terminal hydrolase-L1 potentiates cancer chemosensitivity by stabilizing NOXA. Cell Rep (2013) 3(3):881–91. doi: 10.1016/j.celrep.2013.02.014
166. Piva F, Santoni M, Matrana MR, Satti S, Giulietti M, Occhipinti G, et al. BAP1, PBRM1 and SETD2 in clear-cell renal cell carcinoma: molecular diagnostics and possible targets for personalized therapies. Expert Rev Mol Diagn (2015) 15(9):1201–10. doi: 10.1586/14737159.2015.1068122
167. D’Arcy P, Brnjic S, Olofsson MH, Fryknas M, Lindsten K, De Cesare M, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med (2011) 17(12):1636–40. doi: 10.1038/nm.2536
168. Ventz S, Trippa L, Schoenfeld JD. Lessons Learned from De-escalation trials in favorable risk HPV-associated Squamous Cell Head and Neck Cancer - A Perspective on future trial designs. Clin Cancer Res (2019) 25(24):7281–6. doi: 10.1158/1078-0432.CCR-19-0945
169. Mehanna H, Robinson M, Hartley A, Kong A, Foran B, Fulton-Lieuw T, et al. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): an open-label randomised controlled phase 3 trial. Lancet (2019) 393(10166):51–60. doi: 10.1016/S0140-6736(18)32752-1
170. Bi H-L, Zhang X-L, Zhang Y-L, Xie X, Xia Y-L, Du J, et al. The deubiquitinase UCHL1 regulates cardiac hypertrophy by stabilizing epidermal growth factor receptor. J Sci Adv (2020) 6(16):eaax4826. doi: 10.1126/sciadv.aax4826
171. Azoury SC, Gilmore RC, Shukla V. Molecularly targeted agents and immunotherapy for the treatment of head and neck squamous cell cancer (HNSCC). Discovery Med (2016) 21(118):507–16. doi: 10.1016/S0140-6736(18)32752-1
172. Ferris RL. Immunology and Immunotherapy of Head and Neck Cancer. J Clin Oncol Off J Am Soc Clin Oncol (2015) 33(29):3293–304. doi: 10.1200/JCO.2015.61.1509
173. Whiteside TL. Head and Neck Carcinoma Immunotherapy: Facts and Hopes. Clin Cancer Res An Off J Am Assoc Cancer Res (2017). doi: 10.1158/1078-0432.CCR-17-1261
174. Gotwals P, Cameron S, Cipolletta D, Cremasco V, Crystal A, Hewes B, et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat Rev Cancer (2017) 17(5):286–301. doi: 10.1038/nrc.2017.17
175. Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol (2008) 8(7):501–11. doi: 10.1038/nri2337
176. Farshi P, Deshmukh RR, Nwankwo JO, Arkwright RT, Cvek B, Liu J, et al. Deubiquitinases (DUBs) and DUB inhibitors: a patent review. Expert Opin Ther Pat (2015) 25(10):1191–208. doi: 10.1517/13543776.2015.1056737
177. Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discovery (2018) 17(1):57–78. doi: 10.1038/nrd.2017.152
178. Islam MT, Zhou X, Chen F, Khan MA, Fu J, Chen H. Targeting the signalling pathways regulated by deubiquitinases for prostate cancer therapeutics. Cell Biochem Funct (2019) 37(5):304–19. doi: 10.1002/cbf.3401
Keywords: head and neck cancer, ubiquitin C-terminal hydrolases, deubiquitinating enzymes, genomic alteration, clinical relevance
Citation: Rong C, Zhou R, Wan S, Su D, Wang S-L and Hess J (2021) Ubiquitin Carboxyl-Terminal Hydrolases and Human Malignancies: The Novel Prognostic and Therapeutic Implications for Head and Neck Cancer. Front. Oncol. 10:592501. doi: 10.3389/fonc.2020.592501
Received: 02 September 2020; Accepted: 07 December 2020;
Published: 29 January 2021.
Edited by:
Jessica Lilian Bell, Children’s Cancer Institute Australia, AustraliaReviewed by:
Katerina Strati, University of Cyprus, CyprusTuula Kallunki, Danish Cancer Society Research Center (DCRC), Denmark
Copyright © 2021 Rong, Zhou, Wan, Su, Wang and Hess. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shou-Li Wang, wangshouli@suda.edu.cn
†These authors have contributed equally to this work
 Chao Rong
Chao Rong Ran Zhou
Ran Zhou Shan Wan1
Shan Wan1 Dan Su
Dan Su