- Department of Breast Surgery, Renji Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai, China
Background: We aimed to explore whether the expression of serum miR-222-3p might contribute to early prediction of therapeutic response, clinical outcomes, and adverse events for HER2-positive breast cancer patients receiving neoadjuvant therapy (NAT).
Methods: A total of 65 HER2-positive breast cancer patients receiving NAT were analyzed. The concentration of serum miR-222-3p was detected by quantitative real-time PCR. Logistic regression analysis was used to identify the association of serum miR-222-3p with pathological complete response (pCR). The relationship of serum miR-222-3p with disease-free survival (DFS) and overall survival (OS) was examined via log-rank test and Cox proportional hazards analysis. The ordered logistic regression was applied to evaluate the association between serum miR-222-3p and adverse events.
Results: The miR-222-3p low group was more likely to achieve pCR [odds ratio (OR) = 0.258, P = 0.043]. The interaction between miR-222-3p and presenting Ki67 level was also detected for pCR (OR = 49.230, Pinteraction = 0.025). The miR-222-3p low group was correlated with superior DFS (P = 0.029) and OS (P = 0.0037). The expression of serum miR-222-3p was the independent protective factor for trastuzumab-induced cardiotoxicity (P < 0.05) and anemia (P = 0.013).
Conclusions: Serum miR-222-3p is the potential factor to predict pCR, survival benefit and trastuzumab-induced cardiotoxicity for HER2-positive breast cancer patients receiving NAT.
Introduction
Neoadjuvant chemotherapy (NAC) is increasingly used in primary breast cancer patients, which not only aims to improve the operability or breast-conservability, but also serves as a good platform for in-vivo tests of various drugs (1–3). Trastuzumab, an anti-HER2 monoclonal antibody, is well exemplified in the treatment of HER2-positive breast cancer (4). Manifold data have demonstrated that the addition of trastuzumab to NAC significantly improves the pathological complete response (pCR) rates and thereby results in survival benefit (5–8). However, a majority of HER2-positive breast cancer patients still failed to achieve pCR or even progressed despite trastuzumab-based neoadjuvant therapy (NAT) (9–12). Given the aggressive biological behavior of HER2-positive breast cancer, it hints at a demand to identify potential biomarkers to predict its response to NAT.
On the other hand, adverse events, especially trastuzumab-induced cardiotoxicity, also accompany during or after NAT. The overall incidence of cardiotoxicity was reportedly 3-7% for trastuzumab monotherapy, 13% for trastuzumab with paclitaxel, and as high as 27% for trastuzumab with anthracycline (13, 14). Unfortunately, few reliable biomarkers could help to predict the trastuzumab-induced cardiotoxicity.
Liquid biopsy, as a minimally invasive test, has developed dramatically in recent years. MicroRNAs (miRNAs) belong to a class of noncoding, regulatory, single-stranded RNAs, which have been reported to contribute in early detection of treatment efficacy and adverse reaction (15–23). Our previous research showed that the expression level of miR-222-3p in serum declined after surgery and was an independent prognostic factor for disease-free survival (DFS) in breast cancer (24). Basic studies revealed that miR-222-3p could upregulate HER2 signaling pathway in fulvestrant-resistant breast cancer cells and inhibit the autophagy of cardiac myocytes in mice (25–27). Previous studies have revealed that miR-222-3p was associated with immune invasion and immune resistance in a variety of tumors. Overexpression of miR-222-3p was found to enhance the resistance of tumor cells to tumor infiltrating lymphocytes (TILs) by down-regulating the expression of intercellular cell adhesion molecule-1 (ICAM1) in melanoma, which resulted in ipilimumab (anti-cytotoxic T lymphocyte-associated antigen-4 antibody) resistance in patients with melanoma (28). On the other hand, Ying et al. found that in epithelial ovarian cancer, cancer cell-derived exosomes with high contents of miR-222-3p transferred to the tumor-associated macrophages (TAM) and then induced their polarization to the M2 phenotype via SCOX3/STAT3 pathway (29). The transformation of TAM from M1 to M2 phenotype predicted poor prognosis (30) and contributed to the resistance to anti-HER2/Neu treatment in breast cancer (31). However, it still remains ill-defined whether serum miR-222-3p can serve as a potential biomarker for predicting the response to NAT in HER2-positive breast cancer patients as well as their trastuzumab-induced cardiotoxicity.
On these premises, we hypothesized that the expression of serum miR-222-3p might contribute to early prediction of therapeutic response, clinical outcomes and adverse events for HER2-positive breast cancer patients receiving NAT.
Materials and Methods
Study Procedure
All the enrolled HER2-positive breast cancer patients came from two neoadjuvant clinical trials registered as SHPD001 (NCT02199418) and SHPD002 (NCT02221999) in ClinicalTrials.gov. The SHPD001 and SHPD002 trials were verified and authorized by the Independent Ethical Committee of Renji Hospital, Shanghai Jiaotong University. Each patient signed written informed consent.
The eligibility criteria for these two neoadjuvant trials included women aged ≥18 and ≤70 years old with locally advanced invasive breast cancer (T2-4 or N1-3) confirmed independently by two pathologists based on World Health Organization (WHO) classification. All the patients received paclitaxel 80 mg/m2 on day 1, 8, 15 and 22 and cisplatin 25 mg/m2 on day 1, 8 and 15 every 4 weeks for 4 cycles. For HER2-positive patients, concurrent weekly trastuzumab was given at a loading-dose of 4 mg/kg, followed by maintenance dose of 2 mg/kg, on day 1 for 16 weeks. However, 6 HER2-positive breast cancer patients couldn't afford to receive trastuzumab. In the SHPD002 trial, hormone receptor (HR)-positive patients were randomly assigned to receive preoperative endocrine therapy or not. Endocrine therapy hereinto referred to gonadotropin releasing hormone agonist for premenopausal women and letrozole for postmenopausal counterparts, concurrently with NAC. Tumor assessment was performed every 2 months by physical examination, mammary magnetic resonance imaging (MRI) and ultrasonography. All the patients were required to measure the left ventricular ejection fraction (LVEF) at baseline and every 3 months thereafter. Adverse events were graded according to Common Terminology Criteria for Adverse Events (CTCAE) 4.0. After completion of NAT, the patients underwent surgery. Up until April 2018, 65 HER2-positive breast cancer patients were available for this analysis from these two trials. The results were reported and the analysis was devised according to the Tumor Marker Prognostic Report Recommendation (REMARK) guidelines (32, 33).
Study Outcome
The primary outcome of SHPD001 and SHPD002 was pCR, which was defined as the absence of tumor in the breast tissues and auxiliary lymph nodes removed at the time of surgery (ypT0 ypN0). The secondary outcomes of SHPD001 and SHPD002 included DFS, overall survival (OS) and adverse events. DFS was defined as the time from surgery until first occurrence of locoregional relapse, contralateral breast cancer, distant metastasis, second primaries and death from any cause. OS was defined as the time from surgery until death from any cause.
Pathological Examination
Estrogen receptor (ER), progesterone receptor (PR), HER2 and ki67 were evaluated by immunohistochemistry (IHC) in paraffin-embedded tumor samples from core needle biopsy. ER or PR positive was defined as ≥10% of stained cells. HER2-positive was defined as immunohistochemistry (IHC) 3+ or fluorescence in-situ hybridization (FISH) amplified according to the American Society of Clinical Oncology (ASCO)/College of American Pathologists (CAP) guideline at that time. Clinical or pathological stage for each patient was determined according to the seventh edition of the American Joint Committee on Cancer staging (AJCC-7).
Sample Collection
Before NAT, 5 mL peripheral blood was collected in the coagulation-promoting tubes and let stand at 4°C for at least 1 h. Then it was centrifuged at 1,000 × g for 10 min at 4°C to spin down the blood cells. The supernatants were centrifuged at 12,000 × g for 10 min at 4°C to completely remove cellular components. Subsequently, the serum samples were divided into 300 ul/tube in RNA-free EP tubes and stored at −80°C until use (24).
Extraction of Total RNA
The mirVana PARIS kit (Ambion, Texas, United States) was used to isolate total RNA from 300 μl serum in each patient according to manufacturer's instructions. The extracted RNAs were immediately stored at −80°C until use.
Quantitative Real-Time PCR
cDNA was obtained by reverse transcription of total RNA using a TaqMan Reverse Transcription Kit (Ambion, Texas, United States). For quantitative real-time PCR, the miRNA-specific TaqMan Small RNA Assays (Ambion, Texas, United States) for miR-222-3p and cel-miR-39 (reference microRNA) were used as described by the manufacturer. The TaqMan primers used for hsa-miR-222-3p (RT002276) and cel-miR-39 (RT000200) were obtained from Applied Biosystems. Briefly, 100 ng of total RNA was reverse transcribed using primers specific to each miRNA target followed by real-time PCR on LightCycler® 480 II (Roche, Mannheim, Germany) using TaKaRa probe qPCR kit (RR390A, TaKaRa, Dalian, China) according to the manufacturer's instructions. The expression of miR-222-3p relative to cel-miR-39 was determined using 2 −ΔCT method. ΔCt = mean value Ct (miR-222-3p)-mean value Ct (reference miR-39).
Statistical Analysis
For pCR and survival analysis, the patients were dichotomized into miR-222-3p high (2−ΔCt > 0.03) or low (2−ΔCt ≤ 0.03). Chi-square test or Fisher's test was performed to evaluate the correlation of serum miR-222-3p with clinicopathological characteristics. Logistic regression analysis was used to identify the association of serum miR-222-3p with pCR. Interaction of serum miR-222-3p was also tested with presenting ER or PR status, presenting Ki67 level and the use of neoadjuvant trastuzumab for pCR. The relationship of serum miR-222-3p with DFS and OS was examined via log-rank test and Cox proportional hazards analysis.
For safety analysis, the patients were trichotomized into miR-222-3p low [2−ΔCt ≤ 0.006 (at the 25th percentile)], intermediate (0.006 <2−ΔCt ≤ 0.053) or high [2−ΔCt> 0.053 (at the 75th percentile)]. The ordered logistic regression was applied to evaluate the association between serum miR-222-3p and adverse events. The relative and absolute drop of LVEF from baseline were respectively calculated when the LVEF measured at two time points or more were available. The relative drop of LVEF from baseline (rLVEF) was defined as (LVEFmin-LVEFbaseline)/LVEFbaseline, and the absolute drop of LVEF from baseline (aLVEF) as LVEFmin-LVEFbaseline. Since grade 3/4 LVEF decrease occurred in only a minority of patients, either rLVEF or aLVEF was divided by its quartile.
The statistical analysis was performed by STATA Statistics SE 14 (Stata Corp LP, College Station, TX, USA). All tests were two-tailed and P <0.05 was considered statistically significant.
Results
The Association Between the Expression of Serum miR-222-3p and the Clinicopathological Characteristics
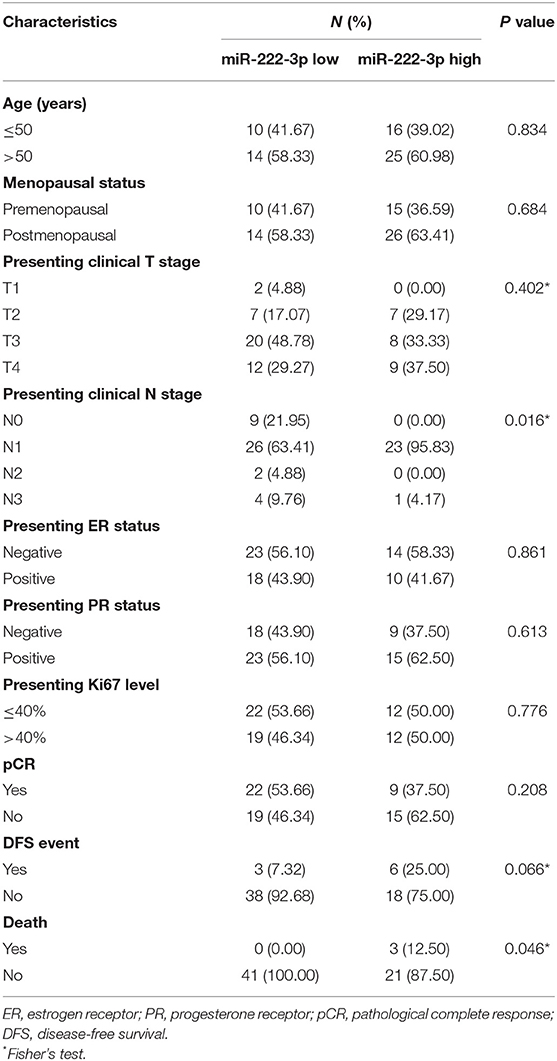
The baseline characteristics of all the patients are listed in Table 1. The expression of serum miR-222-3p was associated with presenting clinical N stage (P = 0.016) and death (P = 0.046). The miR-222-3p low group tended to experience less DFS events (P = 0.066). Significant correlation failed to be discovered between serum miR-222-3p and other clinicopathological characteristics.
The Association Between the Expression of Serum miR-222-3p and the pCR Rate
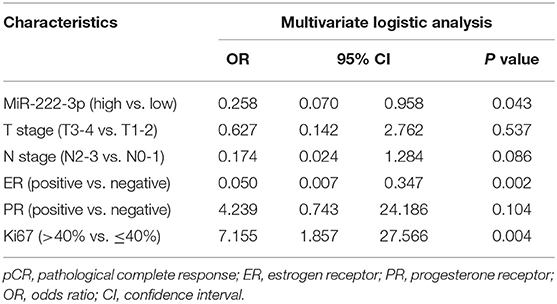
In general, 31 out of 65 (47.69%) patients reached pCR. In the multivariate analysis, the miR-222-3p low group was more likely to achieve pCR [OR = 0.258, 95% confidence interval (CI): 0.070-0.958, P = 0.043]. Meanwhile, patients with lower ER expression (OR = 0.050, 95% CI: 0.007–0.347, P = 0.002) and higher Ki67 (OR = 7.155, 95% CI: 1.857–27.566, P = 0.004) were much easier to achieve pCR (Table 2). The interaction between miR-222-3p and presenting Ki67 level was also detected for pCR (OR = 49.230, 95% CI: 1.624–1492.078, Pinteraction = 0.025; Figure 1). However, no interaction of miR-222-3p was found with presenting ER or PR status and the use of neoadjuvant trastuzumab.
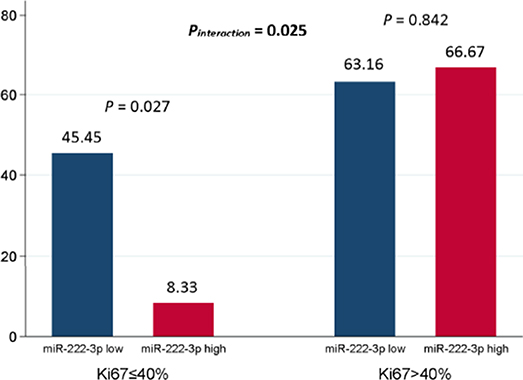
Figure 1. Interaction between serum miR-222-3p and presenting Ki67 level for pCR. The P value for interaction was adjusted by serum miR-222-3p level, presenting clinical T stage, presenting clinical N stage, presenting ER status, presenting PR status and presenting Ki67 level in the multivariate logistic analysis. pCR, pathological complete response; ER, estrogen receptor; PR, progesterone receptor.
Survival Analysis
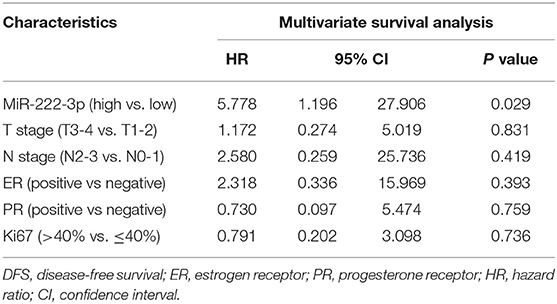
In either univariate (P = 0.0273; Figure 2) or multivariate [hazard ratio (HR) = 5.778, 95% CI: 1.196–27.906, P = 0.029; Table 3] survival analysis, superior DFS was seen in the miR-222-3p low group. Furthermore, the lower expression of serum miR-222-3p was also related with better OS (P = 0.0037; Figure 3).
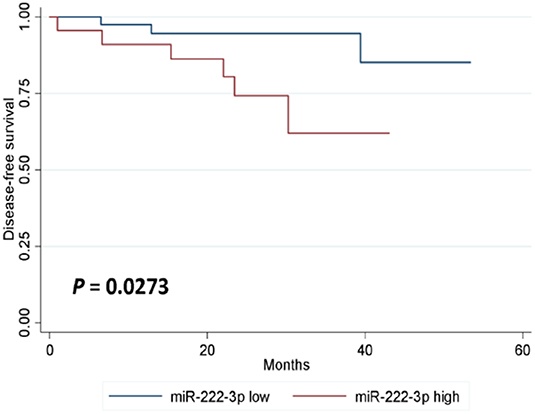
Figure 2. Kaplan-Meier estimates of disease-free survival according to the expression of serum miR-222-3p.
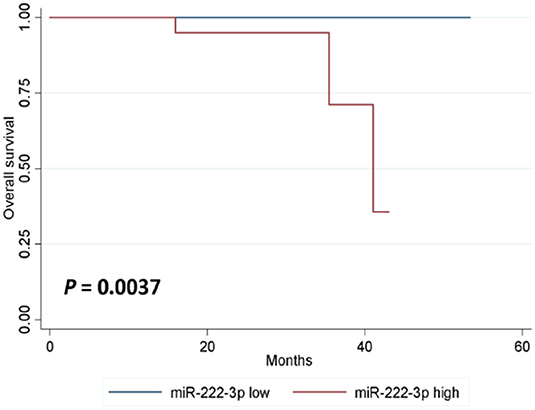
Figure 3. Kaplan-Meier estimates of overall survival according to the expression of serum miR-222-3p.
The Association Between the Expression of Serum miR-222-3p and Adverse Events
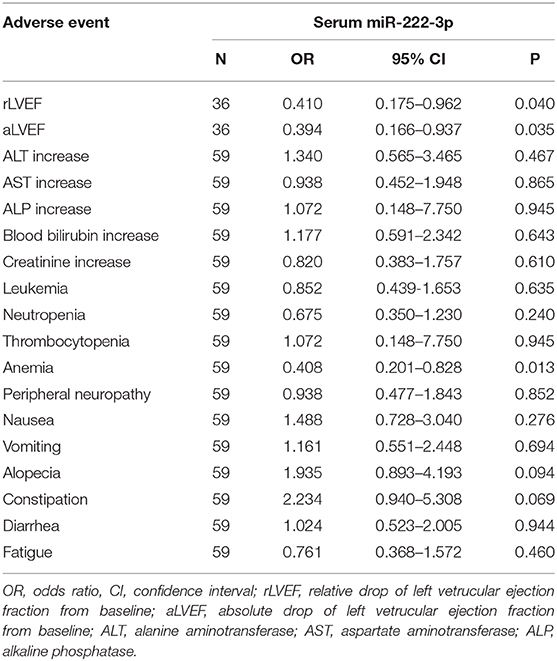
The ordered logistic regression analysis showed that the expression of serum miR-222-3p was the independent protective factor for rLVEF (OR = 0.410, 95% CI: 0.175–0.962, P = 0.040; Table 4), aLVEF (OR = 0.394, 95% CI: 0.166–0.937, P = 0.035; Table 4), and anemia (OR = 0.408, 95% CI: 0.201–0.828, P = 0.013; Table 4).Alopecia (OR = 1.935, 95% CI: 0.893–4.193, P = 0.094) and constipation (OR = 2.234, 95% CI: 0.940–5.308, P = 0.069) were marginally correlated with the expression of serum miR-222-3p (Table 4).
Discussion
This study, to the best of our knowledge, is the first to report the association between the expression of serum miR-222-3p and the response to NAT in the HER2-positive breast cancer patients. This is also the first time to report the relationship between the serum miR-222-3p expression level and adverse events.
Serum miRNAs were identified as biomarkers to diagnose or predict prognosis in various carcinomas (34, 35). Some basic studies demonstrated that tumor can release miRNAs into circulation and serum miRNAs exist in a remarkably stable form protected from endogenous RNase activity (36, 37), while others indicated the presence of miRNA-enriched exosomes secreted by non-cancer cells such as adipose tissue macrophages and mesenchymal stromal cells (38–40). Therefore, certain miRNAs, with cancer-, tissue- or organ-specific functions, might produce endocrine, paracrine or autocrine effect (41–44) on breast cancer patients, directly or indirectly regulating their response to NAT.
Our data reveal that higher level of serum miR-222-3p was associated with inferior pCR rate, which might be attributed to trastuzumab-resistance. So far, two important pathways might contribute to trastuzumab resistance. Firstly, the activation of phosphatase and tension homologs (PTEN) predicts inhibitory effect of trastuzumab, whereas PI3K pathway activation through PTEN loss and PIK3CA mutation confers trastuzumab resistance in breast cancer (45–49). Secondly, PTEN loss leads to SRC hyperactivation, and SRC inhibition appears to overcome trastuzumab resistance (50–53). On the other hand, miR-222 was reported to promote adriamycin or tamoxifen resistance through PTEN/Akt pathway in breast cancer (54, 55). Consequently, miR-222-3p may also induce trastuzumab resistance by modulating the PTEN/PI3K/Akt pathway and the expression of SRC, which prompts further investigation.
We observed the poor prognosis in the patients with higher level of serum miR-222-3p. An increasing number of researches have demonstrated that miR-222 promotes epithelial-to-mesenchymal transition (EMT) (26, 56, 57), G1/S transition of cell cycle (58) and cell proliferation (59, 60) in breast cancer. Therefore, the expression level of miR-222-3p is responsible for the invasion and metastasis of breast cancer. The prognostic value of miR-222 has also been clarified not only in breast cancer (24, 61) but also in many other malignancies including pancreatic cancer, bladder cancer and glioblastoma (62–66). Furthermore, the anti-miR-222 might improve the chemosensitivity and survival, which provides a novel strategy for breast cancer management (67).
Trastuzumab administration is associated with an increased risk of cardiovascular adverse events (68–70). However, the underlying mechanism remains suspended. Trastuzumab-induced cardiotoxicity, regarded as a type II adverse drug reaction, commonly cause reversible dysfunction without cell loss (71). It generally presents as asymptomatic LVEF decline (72–74). Moreover, LVEF decline frequently improves with drug interruption and resuming trastuzumab after recovery is often feasible (71, 74). This study showed that the impairment of LVEF was associated with the expression level of serum miR-222-3p in this study. MiR-222 has been found to participate in many physiological and pathological processes in the cardiovascular system (75). Additionally, miR-222 is necessary for cardiomyocyte growth induced by exercise and is sufficient to protect against adverse cardiac remodeling after ischemic injury (76, 77). Serum miR-222-3p may play an endocrine role similar to that of hormones, reaching every organ, tissue or cell in the body with blood circulation. When miR-222-3p acts on cancer cells, it plays a role in carcinogenesis and drug resistance, and when it acts on cardiac myocytes, it plays a role in protecting the heart. Therefore, the overexpression of miR-222-3p might prevent the heart from trastuzumab-induced injury.
As far as we know, this is the first time that patients with lower expression level of serum miR-222-3p have been reported prone to anemia after receiving NAT. However, the molecular mechanism has not been clarified. The progressively down-regulating of miR-222-3p was observed during the normal erythropoiesis (78). In addition, miR-222-3p inhibited the expansion of erythroblasts and hematopoietic differentiation via down-regulated c-kit expression and miR-222-3p decreased the expression of BLVRA and CRKL to suppress erythroid differentiation (79–81). Consistent with previous basic studies, our data also showed that the patients with high miR-222-3p level have lower baseline hemoglobin level (OR = 0.07, 95% CI 0.005–0.991, P = 0.049). These results suggested that the erythroblasts may be more resistant to chemotherapy in the high miR-222-3p group due to the lower proliferative activity and conversely patients with low level of miR-222-3p might be prone to anemia.
The limitations of this study were as follows. Firstly, the sample size was relatively small. Large-scale studies are needed to confirm the predictive and prognostic value of miR-222-3p. Secondly, some follow-up data are missing on adverse events. Thirdly, as the serum samples were collected before NAT in this retrospective study of prospective trials, it was not possible for us to evaluate the correlation of miR-222-3p changes at different time points during or after NAT with various outcomes including response, survival and safety, which warrants further research.
In conclusion, our study revealed that serum miR-222-3p is the potential factor to predict pCR, survival benefit and trastuzumab-induced cardiotoxicity for HER2-positive breast cancer patients receiving NAT. However, the logic behind it is still open to investigation.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Independent Ethical Committee of Renji Hospital, Shanghai Jiaotong University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SZ, YaoW, JL, and WY contributed conception and design of the study. SZ organized the database. YanW, JP, CY, LZ, SX, YL, YD, FY, JZ, and HD contributed acquisition of the data. SZ, YaoW, and WY performed the statistical analysis. SZ and YaoW wrote the first draft of the manuscript. WY and JL revised the manuscript. All authors read and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China [81172505, 81302302], the Doctoral Programs Foundation of the Ministry of Education of China [20120071120105], the Shanghai Natural Science Foundation [13ZR1452800], the Shanghai Municipal Commission of Health and Family Planning [20144Y0218, 201640006], the Clinical Research Plan of Shanghai Hospital Development Center [16CR3065B, 12016231], the Shanghai Rising Stars of Medical Talent Youth Development Program for Outstanding Youth Medical Talents [2018-16], the Shanghai Rising Stars of Medical Talent Youth Development Program for Youth Medical Talents—Specialist [2018-15], the Shanghai Collaborative Innovation Center for Translational Medicine [TM201908], the Shanghai Natural Science Foundation [19ZR1431100], the Multidisciplinary Cross Research Foundation of Shanghai Jiao Tong University [ZH2018QNA42] and the Nurturing Fund of Renji Hospital [PY2018-IIC-01, PY2018-III-15].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Colomer R, Saura C, Sanchez-Rovira P, Pascual T, Rubio IT, Burgues O, et al. Neoadjuvant management of early breast cancer: a Clinical and investigational position statement. Oncologist. (2019) 24:603–11. doi: 10.1634/theoncologist.2018-0228
2. Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. (2018) 19:27–39. doi: 10.1016/s1470-2045(17)30777-5
3. King TA, Morrow M. Surgical issues in patients with breast cancer receiving neoadjuvant chemotherapy. Nat Rev Clin Oncol. (2015) 12:335–43. doi: 10.1038/nrclinonc.2015.63
4. Prat A, Pascual T, De Angelis C, Gutierrez C, Llombart-Cussac A, Wang T, et al. HER2-enriched subtype and ERBB2 expression in HER2-positive breast cancer treated with dual HER2 blockade. J Natl Cancer Inst. (2019) 112:46–54. doi: 10.1093/jnci/djz042
5. Gianni L, Eiermann W, Semiglazov V, Lluch A, Tjulandin S, Zambetti M, et al. Neoadjuvant and adjuvant trastuzumab in patients with HER2-positive locally advanced breast cancer (NOAH): follow-up of a randomised controlled superiority trial with a parallel HER2-negative cohort. Lancet Oncol. (2014) 15:640–7. doi: 10.1016/s1470-2045(14)70080-4
6. Gianni L, Eiermann W, Semiglazov V, Manikhas A, Lluch A, Tjulandin S, et al. Neoadjuvant chemotherapy with trastuzumab followed by adjuvant trastuzumab versus neoadjuvant chemotherapy alone, in patients with HER2-positive locally advanced breast cancer (the nOAH trial): a randomised controlled superiority trial with a parallel HER2-negative cohort. Lancet. (2010) 375:377–84. doi: 10.1016/s0140-6736(09)61964-4
7. Buzdar AU, Valero V, Ibrahim NK, Francis D, Broglio KR, Theriault RL, et al. Neoadjuvant therapy with paclitaxel followed by 5-fluorouracil, epirubicin, and cyclophosphamide chemotherapy and concurrent trastuzumab in human epidermal growth factor receptor 2-positive operable breast cancer: an update of the initial randomized study population and data of additional patients treated with the same regimen. Clin Cancer Res. (2007) 13:228–33. doi: 10.1158/1078-0432.ccr-06-1345
8. Buzdar AU, Ibrahim NK, Francis D, Booser DJ, Thomas ES, Theriault RL, et al. Significantly higher pathologic complete remission rate after neoadjuvant therapy with trastuzumab, paclitaxel, and epirubicin chemotherapy: results of a randomized trial in human epidermal growth factor receptor 2-positive operable breast cancer. J Clin Oncol. (2005) 23:3676–85. doi: 10.1200/jco.2005.07.032
9. Fumagalli D, Venet D, Ignatiadis M, Azim HA Jr, Maetens M, et al. RNA sequencing to predict response to neoadjuvant anti-HER2 therapy: a Secondary analysis of the neoALTTO randomized clinical trial. JAMA Oncol. (2017) 3:227–34. doi: 10.1001/jamaoncol.2016.3824
10. Tanioka M, Fan C, Parker JS, Hoadley KA, Hu Z, Li Y, et al. Integrated analysis of rNA and dNA from the phase iII trial cALGB 40601 identifies predictors of response to trastuzumab-Based neoadjuvant chemotherapy in HER2-Positive breast cancer. Clin Cancer Res. (2018) 24:5292–304. doi: 10.1158/1078-0432.Ccr-17-3431
11. Salgado R, Denkert C, Campbell C, Savas P, Nuciforo P, Aura C, et al. Tumor-Infiltrating lymphocytes and associations with pathological complete response and event-Free survival in HER2-Positive early-Stage breast cancer treated with lapatinib and trastuzumab: a Secondary analysis of the neoALTTO trial. JAMA Oncol. (2015) 1:448–54. doi: 10.1001/jamaoncol.2015.0830
12. Datta J, Berk E, Xu S, Fitzpatrick E, Rosemblit C, Lowenfeld L, et al. Anti-HER2 cD4(+) t-helper type 1 response is a novel immune correlate to pathologic response following neoadjuvant therapy in HER2-positive breast cancer. Breast Cancer Res. (2015) 17:71. doi: 10.1186/s13058-015-0584-1
13. Seidman A, Hudis C, Pierri MK, Shak S, Paton V, Ashby M, et al. Cardiac dysfunction in the trastuzumab clinical trials experience. J Clin Oncol. (2002) 20:1215–21. doi: 10.1200/jco.2002.20.5.1215
14. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. (2001) 344:783–92. doi: 10.1056/nejm200103153441101
15. Niu J, Xue A, Chi Y, Xue J, Wang W, Zhao Z, et al. Induction of miRNA-181a by genotoxic treatments promotes chemotherapeutic resistance and metastasis in breast cancer. Oncogene. (2016) 35:1302–13. doi: 10.1038/onc.2015.189
16. Fan S, Tian T, Chen W, Lv X, Lei X, Zhang H, et al. Mitochondrial miRNA determines chemoresistance by reprogramming metabolism and regulating mitochondrial transcription. Cancer Res. (2019) 79:1069–84. doi: 10.1158/0008-5472.can-18-2505
17. Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. (2015) 527:472–6. doi: 10.1038/nature15748
18. Qin X, Guo H, Wang X, Zhu X, Yan M, Wang X, et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting cDKN1B and iNG5. Genome Biol. (2019) 20:12. doi: 10.1186/s13059-018-1604-0
19. Leonetti A, Assaraf YG, Veltsista PD, El Hassouni B, Tiseo M, Giovannetti E. MicroRNAs as a drug resistance mechanism to targeted therapies in eGFR-mutated NSCLC: current implications and future directions. Drug Resist Updat. (2019) 42:1–1. doi: 10.1016/j.drup.2018.11.002
20. Cai J, Fang L, Huang Y, Li R, Xu X, Hu Z, et al. Simultaneous overactivation of wnt/beta-catenin and tGFbeta signalling by miR-128-3p confers chemoresistance-associated metastasis in NSCLC. Nat Commun. (2017) 8:15870. doi: 10.1038/ncomms15870
21. Xu Z, Sharma M, Gelman A, Hachem R, Mohanakumar T. Significant role for microRNA-21 affecting toll-like receptor pathway in primary graft dysfunction after human lung transplantation. J Heart Lung Transplant. (2017) 36:331–9. doi: 10.1016/j.healun.2016.08.028
22. Chaudhari U, Nemade H, Gaspar JA, Hescheler J, Hengstler JG, Sachinidis A. MicroRNAs as early toxicity signatures of doxorubicin in human-induced pluripotent stem cell-derived cardiomyocytes. Arch Toxicol. (2016) 90:3087–98. doi: 10.1007/s00204-016-1668-0
23. Amstutz U, Offer SM, Sistonen J, Joerger M, Diasio RB, Largiader CR. Polymorphisms in mIR27A associated with early-Onset toxicity in fluoropyrimidine-based chemotherapy. Clin Cancer Res. (2015) 21:2038–44. doi: 10.1158/1078-0432.ccr-14-2817
24. Wang Y, Yin W, Lin Y, Yin K, Zhou L, Du Y, et al. Downregulated circulating microRNAs after surgery: potential noninvasive biomarkers for diagnosis and prognosis of early breast cancer. Cell Death Discov. (2018) 4:21. doi: 10.1038/s41420-018-0089-7
25. Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. (2011) 30:1082–97. doi: 10.1038/onc.2010.487
26. Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O'Brien C, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. (2011) 4:ra41. doi: 10.1126/scisignal.2001538
27. Su M, Chen Z, Wang C, Song L, Zou Y, Zhang L, et al. Cardiac-Specific overexpression of miR-222 induces heart failure and inhibits autophagy in mice. Cell Physiol Biochem. (2016) 39:1503–11. doi: 10.1159/000447853
28. Galore-Haskel G, Nemlich Y, Greenberg E, Ashkenazi S, Hakim M, Itzhaki O, et al. A novel immune resistance mechanism of melanoma cells controlled by the aDAR1 enzyme. Oncotarget. (2015) 6:28999–9015. doi: 10.18632/oncotarget.4905
29. Ying X, Wu Q, Wu X, Zhu Q, Wang X, Jiang L, et al. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget. (2016) 7:43076–87. doi: 10.18632/oncotarget.9246
30. Honkanen TJ, Tikkanen A, Karihtala P, Mäkinen M, Väyrynen JP, Koivunen JP. Prognostic and predictive role of tumour-associated macrophages in HER2 positive breast cancer. Sci Rep. (2019) 9:10961. doi: 10.1038/s41598-019-47375-2
31. Xu M, Liu M, Du X, Li S, Li H, Li X, et al. Intratumoral delivery of iL-21 overcomes anti-HER2/Neu resistance through shifting tumor-Associated macrophages from m2 to m1 phenotype. J Immunol. (2015) 194:4997–5006. doi: 10.4049/jimmunol.1402603
32. McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. (2005) 97:1180–4. doi: 10.1093/jnci/dji237
33. Sauerbrei W, Taube SE, McShane LM, Cavenagh MM, Altman DG. Reporting recommendations for tumor marker prognostic studies (REMARK): an abridged explanation and elaboration. J Natl Cancer Inst. (2018) 110:803–11. doi: 10.1093/jnci/djy088
34. Gu J, Wang Y, Wu X. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr Pharm Des. (2013) 19:1292–300. doi: 10.2174/138161213804805775
35. Thakral S, Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr Gene Ther. (2015) 15:142–50. doi: 10.2174/1566523214666141224095610
36. Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport rNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. (2008) 10:1470–6. doi: 10.1038/ncb1800
37. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. (2008) 105:10513–8. doi: 10.1073/pnas.0804549105
38. Song H, Li X, Zhao Z, Qian J, Wang Y, Cui J, et al. Reversal of osteoporotic activity by endothelial cell-Secreted bone targeting and biocompatible exosomes. Nano Lett. (2019) 19:3040–8. doi: 10.1021/acs.nanolett.9b00287
39. Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, et al. Adipose tissue macrophage-Derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. (2017) 171:372–84.e12. doi: 10.1016/j.cell.2017.08.035
40. Zhao J, Li X, Hu J, Chen F, Qiao S, Sun X, et al. Mesenchymal stromal cell-Derived exosomes attenuate myocardial ischemia-Reperfusion injury through miR-182-Regulated macrophage polarization. Cardiovasc Res. (2019) 115:1205–16. doi: 10.1093/cvr/cvz040
41. Pakravan K, Babashah S, Sadeghizadeh M, Mowla SJ, Mossahebi-Mohammadi M, Ataei F, et al. MicroRNA-100 shuttled by mesenchymal stem cell-derived exosomes suppresses in vitro angiogenesis through modulating the mTOR/HIF-1alpha/VEGF signaling axis in breast cancer cells. Cell Oncol (Dordr). (2017) 40:457–70. doi: 10.1007/s13402-017-0335-7
42. Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. (2017) 542:450–5. doi: 10.1038/nature21365
43. Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. (2012) 56:1946–57. doi: 10.1002/hep.25873
44. Deng Z, Rong Y, Teng Y, Zhuang X, Samykutty A, Mu J, et al. Exosomes miR-126a released from mDSC induced by dOX treatment promotes lung metastasis. Oncogene. (2017) 36:639–51. doi: 10.1038/onc.2016.229
45. Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, et al. A functional genetic approach identifies the pI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. (2007) 12:395–402. doi: 10.1016/j.ccr.2007.08.030
46. Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of pTEN predicts trastuzumab resistance in patients. Cancer Cell. (2004) 6:117–27. doi: 10.1016/j.ccr.2004.06.022
47. Wang Y, Liu Y, Du Y, Yin W, Lu J. The predictive role of phosphatase and tensin homolog (PTEN) loss, phosphoinositol-3 (PI3) kinase (PIK3CA) mutation, and pI3K pathway activation in sensitivity to trastuzumab in HER2-positive breast cancer: a meta-analysis. Curr Med Res Opin. (2013) 29:633–42. doi: 10.1185/03007995.2013.794775
48. Li Y, Chu J, Feng W, Yang M, Zhang Y, Zhang Y, et al. EPHA5 mediates trastuzumab resistance in HER2-positive breast cancers through regulating cancer stem cell-like properties. Faseb J. (2019) 33:4851–65. doi: 10.1096/fj.201701561RRRR
49. Chandarlapaty S, Sakr RA, Giri D, Patil S, Heguy A, Morrow M, et al. Frequent mutational activation of the PI3K-AKT pathway in trastuzumab-resistant breast cancer. Clin Cancer Res. (2012) 18:6784–91. doi: 10.1158/1078-0432.ccr-12-1785
50. Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, et al. Combating trastuzumab resistance by targeting sRC, a common node downstream of multiple resistance pathways. Nat Med. (2011) 17:461–9. doi: 10.1038/nm.2309
51. Hanker AB, Estrada MV, Bianchini G, Moore PD, Zhao J, Cheng F, et al. Extracellular matrix/Integrin signaling promotes resistance to combined inhibition of HER2 and pI3K in HER2(+) breast cancer. Cancer Res. (2017) 77:3280–92. doi: 10.1158/0008-5472.can-16-2808
52. Peiro G, Ortiz-Martinez F, Gallardo A, Perez-Balaguer A, Sanchez-Paya J, Ponce JJ, et al. Src, a potential target for overcoming trastuzumab resistance in HER2-positive breast carcinoma. Br J Cancer. (2014) 111:689–95. doi: 10.1038/bjc.2014.327
53. Han S, Meng Y, Tong Q, Li G, Zhang X, Chen Y, et al. The ERBB2-targeting antibody trastuzumab and the small-molecule sRC inhibitor saracatinib synergistically inhibit ERBB2-overexpressing gastric cancer. MAbs. (2014) 6:403–8. doi: 10.4161/mabs.27443
54. Shen H, Wang D, Li L, Yang S, Chen X, Zhou S, et al. MiR-222 promotes drug-resistance of breast cancer cells to adriamycin via modulation of pTEN/Akt/FOXO1 pathway. Gene. (2017) 596:110–8. doi: 10.1016/j.gene.2016.10.016
55. Gu J, Wang Y, Wang X, Zhou D, Shao C, Zhou M, et al. Downregulation of lncRNA gAS5 confers tamoxifen resistance by activating miR-222 in breast cancer. Cancer Lett. (2018) 434:1–10. doi: 10.1016/j.canlet.2018.06.039
56. Hwang MS, Yu N, Stinson SY, Yue P, Newman RJ, Allan BB, et al. miR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS ONE. (2013) 8:e66502. doi: 10.1371/journal.pone.0066502
57. Han SH, Kim HJ, Gwak JM, Kim M, Chung YR, Park SY. MicroRNA-222 expression as a predictive marker for tumor progression in hormone receptor-Positive breast cancer. J Breast Cancer. (2017) 20:35–44. doi: 10.4048/jbc.2017.20.1.35
58. Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang Z, et al. miR-221/222 promotes s-phase entry and cellular migration in control of basal-like breast cancer. Molecules. (2014) 19:7122–37. doi: 10.3390/molecules19067122
59. Zhang C, Zhang J, Zhang A, Wang Y, Han L, You Y, et al. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int J Oncol. (2010) 37:1621–6. doi: 10.3892/ijo_00000816
60. Zong Y, Zhang Y, Sun X, Xu T, Cheng X, Qin Y. miR-221/222 promote tumor growth and suppress apoptosis by targeting lncRNA gAS5 in breast cancer. Biosci Rep. (2019) 39:1. doi: 10.1042/bsr20181859
61. Kim C, Go EJ, Kim A. Recurrence prediction using microRNA expression in hormone receptor positive breast cancer during tamoxifen treatment. Biomarkers. (2018) 23:804–11. doi: 10.1080/1354750x.2018.1499131
62. Zhang Z, Pan B, Lv S, Ji Z, Wu Q, Lang R, et al. Integrating microRNA expression profiling studies to systematically evaluate the diagnostic value of microRNAs in pancreatic cancer and validate their prognostic significance with the cancer genome atlas data. Cell Physiol Biochem. (2018) 49:678–95. doi: 10.1159/000493033
63. Tsikrika FD, Avgeris M, Levis PK, Tokas T, Stravodimos K, Scorilas A. miR-221/222 cluster expression improves clinical stratification of non-muscle invasive bladder cancer (TaT1) patients' risk for short-term relapse and progression. Genes Chrom Cancer. (2018) 57:150–61. doi: 10.1002/gcc.22516
64. Zhao H, Shen J, Hodges TR, Song R, Fuller GN, Heimberger AB. Serum microRNA profiling in patients with glioblastoma: a survival analysis. Mol Cancer. (2017) 16:59. doi: 10.1186/s12943-017-0628-5
65. Zhang L, Huang Z, Zhang H, Zhu M, Zhu W, Zhou X, et al. Prognostic value of candidate microRNAs in gastric cancer: a validation study. Cancer Biomark. (2017) 18:221–30. doi: 10.3233/cbm-160091
66. Zheng Z, Li X, Zhu Y, Gu W, Xie X, Jiang J. Prognostic significance of miRNA in patients with diffuse large b-Cell lymphoma: a meta-Analysis. Cell Physiol Biochem. (2016) 39:1891–904. doi: 10.1159/000447887
67. Bliss SA, Sinha G, Sandiford OA, Williams LM, Engelberth DJ, Guiro K, et al. Mesenchymal stem cell-Derived exosomes stimulate cycling quiescence and early breast cancer dormancy in bone marrow. Cancer Res. (2016) 76:5832–44. doi: 10.1158/0008-5472.can-16-1092
68. Bowles EJ, Wellman R, Feigelson HS, Onitilo AA, Freedman AN, Delate T, et al. Risk of heart failure in breast cancer patients after anthracycline and trastuzumab treatment: a retrospective cohort study. J Natl Cancer Inst. (2012) 104:1293–305. doi: 10.1093/jnci/djs317
69. Chen J, Long JB, Hurria A, Owusu C, Steingart RM, Gross CP. Incidence of heart failure or cardiomyopathy after adjuvant trastuzumab therapy for breast cancer. J Am Coll Cardiol. (2012) 60:2504–12. doi: 10.1016/j.jacc.2012.07.068
70. Rochette L, Guenancia C, Gudjoncik A, Hachet O, Zeller M, Cottin Y, et al. Anthracyclines/trastuzumab: new aspects of cardiotoxicity and molecular mechanisms. Trends Pharmacol Sci. (2015) 36:326–48. doi: 10.1016/j.tips.2015.03.005
71. Zagar TM, Cardinale DM, Marks LB. Breast cancer therapy-associated cardiovascular disease. Nat Rev Clin Oncol. (2016) 13:172–84. doi: 10.1038/nrclinonc.2015.171
72. Portera CC, Walshe JM, Rosing DR, Denduluri N, Berman AW, Vatas U, et al. Cardiac toxicity and efficacy of trastuzumab combined with pertuzumab in patients with trastuzumab-insensitive human epidermal growth factor receptor 2-positive metastatic breast cancer. Clin Cancer Res. (2008) 14:2710–6. doi: 10.1158/1078-0432.ccr-07-4636
73. Dang C, Guo H, Najita J, Yardley D, Marcom K, Albain K, et al. Cardiac outcomes of patients receiving adjuvant weekly paclitaxel and trastuzumab for node-Negative, ERBB2-Positive breast cancer. JAMA Oncol. (2016) 2:29–36. doi: 10.1001/jamaoncol.2015.3709
74. Barroso-Sousa R, Santana IA, Testa L, de Melo Gagliato D, Mano MS. Biological therapies in breast cancer: common toxicities and management strategies. Breast. (2013) 22:1009–18. doi: 10.1016/j.breast.2013.09.009
75. Ding S, Huang H, Xu Y, Zhu H, Zhong C. MiR-222 in cardiovascular diseases: physiology and pathology. Biomed Res Int. (2017) 2017:4962426. doi: 10.1155/2017/4962426
76. Vujic A, Lerchenmuller C, Wu TD, Guillermier C, Rabolli CP, Gonzalez E, et al. Exercise induces new cardiomyocyte generation in the adult mammalian heart. Nat Commun. (2018) 9:1659. doi: 10.1038/s41467-018-04083-1
77. Liu X, Xiao J, Zhu H, Wei X, Platt C, Damilano F, et al. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. (2015) 21:584–95. doi: 10.1016/j.cmet.2015.02.014
78. Bruchova H, Yoon D, Agarwal AM, Mendell J, Prchal JT. Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis. Exp Hematol. (2007) 35:1657–67. doi: 10.1016/j.exphem.2007.08.021
79. Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. (2005) 102:18081–6. doi: 10.1073/pnas.0506216102
80. Lee JY, Kim M, Heo HR, Ha KS, Han ET, Park WS, et al. Inhibition of microRNA-221 and 222 enhances hematopoietic differentiation from human pluripotent stem cells via c-KIT upregulation. Mol Cells. (2018) 41:971–8. doi: 10.14348/molcells.2018.0244
Keywords: breast cancer, serum miR-222-3p, neoadjuvant therapy, predictive, prognostic, adverse event
Citation: Zhang S, Wang Y, Wang Y, Peng J, Yuan C, Zhou L, Xu S, Lin Y, Du Y, Yang F, Zhang J, Dai H, Yin W and Lu J (2020) Serum miR-222-3p as a Double-Edged Sword in Predicting Efficacy and Trastuzumab-Induced Cardiotoxicity for HER2-Positive Breast Cancer Patients Receiving Neoadjuvant Target Therapy. Front. Oncol. 10:631. doi: 10.3389/fonc.2020.00631
Received: 04 August 2019; Accepted: 06 April 2020;
Published: 28 April 2020.
Edited by:
Ondrej Slaby, Brno University of Technology, CzechiaReviewed by:
Cindy H. Chau, National Cancer Institute (NCI), United StatesMassimo Fantini, Precision Biologics, Inc., United States
Copyright © 2020 Zhang, Wang, Wang, Peng, Yuan, Zhou, Xu, Lin, Du, Yang, Zhang, Dai, Yin and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjin Yin, followroad@163.com; Jinsong Lu, lujjss@163.com
†These authors have contributed equally to this work
 Shan Zhang†
Shan Zhang† Yaohui Wang
Yaohui Wang Jing Peng
Jing Peng Chenwei Yuan
Chenwei Yuan Wenjin Yin
Wenjin Yin Jinsong Lu
Jinsong Lu