Efficient strategies based on behavioral and electrophysiological methods for epilepsy-related gene screening in the Drosophila model
- 1Department of Neurology, Institute of Neuroscience, Key Laboratory of Neurogenetics and Channelopathies of Guangdong Province and the Ministry of Education of China, The Second Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
- 2The Second Clinical Medicine School of Guangzhou Medical University, Guangzhou, China
- 3The First Clinical Medicine School of Guangzhou Medical University, Guangzhou, China
- 4School of Clinical Medicine, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, Hong Kong SAR, China
Introduction: With the advent of trio-based whole-exome sequencing, the identification of epilepsy candidate genes has become easier, resulting in a large number of potential genes that need to be validated in a whole-organism context. However, conducting animal experiments systematically and efficiently remains a challenge due to their laborious and time-consuming nature. This study aims to develop optimized strategies for validating epilepsy candidate genes using the Drosophila model.
Methods: This study incorporate behavior, morphology, and electrophysiology for genetic manipulation and phenotypic examination. We utilized the Gal4/UAS system in combination with RNAi techniques to generate loss-of-function models. We performed a range of behavioral tests, including two previously unreported seizure phenotypes, to evaluate the seizure behavior of mutant and wild-type flies. We used Gal4/UAS-mGFP flies to observe the morphological alterations in the brain under a confocal microscope. We also implemented patch-clamp recordings, including a novel electrophysiological method for studying synapse function and improved methods for recording action potential currents and spontaneous EPSCs on targeted neurons.
Results: We applied different techniques or methods mentioned above to investigate four epilepsy-associated genes, namely Tango14, Klp3A, Cac, and Sbf, based on their genotype-phenotype correlation. Our findings showcase the feasibility and efficiency of our screening system for confirming epilepsy candidate genes in the Drosophila model.
Discussion: This efficient screening system holds the potential to significantly accelerate and optimize the process of identifying epilepsy candidate genes, particularly in conjunction with trio-based whole-exome sequencing.
1. Introduction
Epilepsy is a prevalent neurological disorder affecting approximately 50 million people worldwide, as reported by the World Health Organization [WHO] (2017). Although up to 30% of epilepsy cases are claimed to have a genetic cause (Weber et al., 2017), the underlying genetic factors remain unknown for a significant number of patients (Weber et al., 2017; Lasko and Luthy, 2021). The trio (two parents and the affected proband)-based study combined with whole-exome sequencing (WES) has emerged as a popular method to identify new epilepsy candidate genes (Dupépé et al., 2019). WES produces a thorough yet precise list of candidate genes at a reasonable price by focusing on the protein-coding regions (Harnish et al., 2019; Qiao et al., 2022a). Although several large-scale genetic diagnostic tests using trio-based WES have yielded significant results (Guan et al., 2021; Zhai et al., 2021), the approach has a high rate of false positive results, like another next-generation sequencing (NGS) techniques (Zhang et al., 2015), highlighting the need for direct experimentation to fully understand the effect of a variant (Lee et al., 2014; XiangWei et al., 2019).
Experimental models, including cultured cells, Drosophila, zebrafish, and rodents such as mice and rats, are widely used to study epilepsy candidate genes. Drosophila is a particularly attractive model due to its short life cycle of only 50 days and its ability to produce a large number of offspring. Due to these factors, spatiotemporal genetic modifications, such as those enabled by the UAS/Gal4 system, can be used to conduct fly research in just a few months (Kepecs et al., 2002; Takai et al., 2020; Guerrini et al., 2021). The discovery of the Drosophila model is also easy to translate to other vertebrate models (Hunt et al., 2019; Hegazi et al., 2022).
Under various seizure-induction assays, adequate seizure-susceptible mutants of Drosophila adults, which correlate with different epilepsy subtypes, exhibit a wide spectrum of seizure-like behaviors. For instance, sudden unexpected death in epilepsy (SUDEP) mutants Shaker and Hyperkinetic display leg-shaking behavior following etherization (Kaplan and Trout, 1969; D’Adamo et al., 2020), while general epilepsy with febrile seizure plus (GEFG+) and Dravet syndrome (DS) knock-in mutations display loss of posture and random wing buzzing after heat application (Sun et al., 2012; Schutte et al., 2014), Na+ channel mutants display spontaneous shuddering (Kaas et al., 2016), whereas bang-sensitive (BS) mutants exhibit seizure-like activity when evoked mechanically. However, only one kind of activity, characterized by seizure and paralysis after a mechanical stimulus and observed in over twenty BS mutants, has been defined as “seizure-like” (Parker et al., 2011a). Therefore, we introduced two novel seizure phenotypes induced by vertex shock in Drosophila to extend the application of mechanical induction assays and further investigate the relationship between epileptic phenotypes and genotypes in both Drosophila and humans with greater precision.
However, a broad applicability screening system is urgently required since enormous epilepsy candidate genes must be verified by functional experiments. Here, we propose a set of simple experiments in Drosophila to evaluate seizure behaviors, brain morphology, and electrophysiology, which can expedite the progress of identifying epilepsy candidate genes. Especially, our approach introduces two novel seizure phenotypes induced by vertex shock and several optimized electrophysiological methods, thereby expanding the range of research methods available for investigating epilepsy in the Drosophila model.
2. Materials and methods
2.1. Drosophila stocks
The flies used in this study were fed standard cornmeal food and maintained in an incubator at 25°C (except for the special statement) and 60–70% humidity in a 12-h light-dark cycle. UAS-Tango14-RNAi (THU1886), UAS-Sbf-RNAi (THU0865), and UAS-Cac-RNAi (THU2586) flies were kindly donated by Tsing Hua Fly Center (https://thfc.zzbd.org/, Tsinghua University, Beijing, China). UAS-Klp3A-RNAi (BDSC: 43230) was purchased from Bloomington Fly Stock Center (Bloomington, IN, USA). Canton-S and tub-Gal4 were kindly donated by Prof. LIU Ji-Yong (Guangzhou Medical University, Guangzhou, China). UAS-EGFP, UAS-mCD8:GFP, and GH146-Gal4 were kindly donated by Prof. KE Ya (the Chinese University of Hong Kong, Hong Kong). Canton-S was used as the wild-type flies in this study.
2.2. Loss-of-function mutation model
The tub-Gal4 line was utilized in the cross-breeding of two RNAi lines, the Klp3A-RNAi and the Cac-RNAi, to generate global knockdown of Klp3A and Cac in Drosophila melanogaster (tub-Gal4 > Klp3A-RNAi and tub-Gal4 > Cac-RNAi). The elav-Gal4 line was utilized in the cross-breeding of two RNAi lines, the Tango14-RNAi and the Sbf-RNAi, to generate neuronal knockdown of Tango14 and Sbf in Drosophila melanogaster (elav-Gal4 > Tango14-RNAi and elav-Gal4 > Sbf-RNAi). The efficiency of knockdown models was detected by qPCR.
2.3. Seizure behaviors
The seizure behavior test was performed on flies 3–5 days after eclosion. Flies were anesthetized with CO2 and transferred to another clean food vial 18–24 h before testing. Approximately two to seven flies were placed in one vial and mechanically stimulated with a vortex mixer (VWR, Radnor, PA, USA) at maximum speed for 20 s. To record the behavior of the flies, a high-resolution camera was utilized. Each genotype was evaluated using at least five trials, with each trial consisting of ten vials of flies. The rates of BS paralysis, which were recorded from the moment of “banging” until flies regained the ability to stand upright (Ganetzky and Wu, 1982), as well as those of late-phase seizure and hyperactive seizure in flies, were documented. The behavior assay was performed using a high-resolution camera (720–1080 P, 30–60 FPS at least), and the records were manually played back on the computer with video software (Baofeng Group Co., Ltd., China) to count the rates of BS.
2.4. Brain morphology
To examine morphological variations in the brain, UAS-mCD8:GFP was used to generate tub-Gal4 > UASmCD8:GFP; UAS-target gene-RNAi knockdown flies and tub-Gal4 > UAS-mCD8:GFP control flies, respectively, that were labeled with membrane GFP. The brain was dissected and fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) with 0.1% Triton X-100 for 1 h at 25°C, then washed three times with 0.3% Triton X-100 PBS. Images were captured using a confocal microscope (SP8; Zeiss, Jena, Germany) and analyzed using the ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.5. Larvae development
The larval stage of flies can be divided into three molting stages, namely, the first, second, and third instar larvae. We utilized the length of fly larvae to evaluate their development. For the experiment, one female and three male flies were placed into a fresh food vial to cross for 24 h. After 24 h, the female flies were transferred to separate fresh food vials and allowed to lay eggs for 12 h (taking note of the exact time of the transfer), while the adult flies were removed. The vials containing the eggs were then incubated under standard conditions for varying durations in order to obtain larvae at the desired developmental stage. Once the larvae had reached the desired stage, 2 ml of a 20% sucrose solution was added to the food vials, and after 5 min, the floating larvae were decanted into a Petri dish. The larvae were then washed by transferring them to a dish containing 1X PBS using a pipette. To devitalize the larvae, they were heated in water bath equipment at 50°C for 10 min. After heating, the larvae were placed on a black card, and photographs of the larvae along with a scale were taken. The length of the larvae was then measured using the ImageJ NIH software.1
2.6. Electrophysiology
2.6.1. Patch-clamp recording
Fly brains were dissected as previously described, transferred to a recording chamber with the fly external solution, and immobilized with a C-sharp holder (Gu and O’Dowd, 2006). The standard external solution contained (mM) 101 NaCl, 1 CaCl2, 4 MgCl2, 3 KCl, 5 glucose, 1.25 NaH2PO4, and 20.7 NaHCO3 (pH 7.2 and 250 mOsm). The internal solution for whole-cell recording contained (mM) 0.085 CaCl2, 1.7 MgCl2, 10 HEPES, 1 EGTA, 103 K-gluconate, 2 Na2-ATP, and 0.4 Na-GTP. The patch pipette for the attached recording was filled with an external solution. Recordings were acquired using the 700B amplifier, Digidata 1440B digital-analog converter, and pClamp 10.5 software (molecular devices).
2.6.2. Evoked EPSP recording
The equipment, brain, and recording solution preparations used were identical to those for whole-cell recording. Evoked EPSPs between the antennal lobe (AL) and the mushroom body (MB) were stimulated by current pulses (0.5 ms; 0.1–0.5 mA) from the stimulation electrode on the antennal lobe (presynaptic structure). Then the evoked EPSPs were recorded by whole-cell recording on the MB neurons (postsynaptic structure) (Qiao et al., 2022b). Different intensities of current pulses were applied for establishing the input-output relationship. The subsequent recording was then performed using the current pulse to induce 50% of the maximum EPSP response. To test the synaptic connection, ten trials were performed with a 1-min interval between each trial. The latency and amplitude of evoked EPSPs were recorded and analyzed using the pClamp 10.5 software.
2.7. Statistical analysis
All quantitative data were presented as mean ± S.D. The Student’s t-test was used to compare two independent or paired samples. Multiple samples were analyzed by one-way ANOVA, and differences between the two groups were evaluated using Tukey’s post hoc test. Statistical analyses were performed with GraphPad Prism 7.00 and SPSS 20. The cutoff value for statistical significance was 0.05. The n number in the behavior data represented the number of trials.
3. Results
3.1. Seizure-like behavior under mechanical stimulation
3.1.1. Hyperactivity seizure behavior
Vortex stimulation is a simple and effective method to induce seizure behavior in Drosophila. Unlike classical BS seizure behavior (Parker et al., 2011a), our investigation of seizure sensitivity in the epilepsy candidate genes Sbf and Tango14 revealed an unreported hyperactivity behavior. We administered the mechanical stimulation to flies with pan-neural knockdown of Tango14 (elav-Gal4 > Tango14-RNAi) and Sbf (elav-Gal4 > Sbf-RNAi), as well as wild-type Canton-S flies. The elav-Gal4 > Sbf-RNAi showed classical BS seizure behavior (**p = 0.0078, n > 5 in each group, one-way ANOVA, Tukey’s multiple comparisons tests, Figure 1C). While the elav-Gal4 > Tango14-RNAi flies exhibited hyperactivity-like behavior that consisted of repeat drop and jump behaviors after seizure (Figure 1A and Supplementary video 1). Notably, this hyperactivity behavior, as defined in our study, differs from the previous definition, which is characterized by intense, uncoordinated motor activity before paralysis (Pavlidis and Tanouye, 1995). Flies typically recover quickly after a classical seizure-like behavior (Parker et al., 2011b; Wang et al., 2021), but in hyperactivity seizures, they repeatedly drop and jump, which may lead to recurring seizures. Such unique behavior was not observed in wild-type or mutant flies without mechanical stimulus. Approximately 66.00% of elav-Gal4 > Tango14-RNAi flies showed this behavior, which cannot be observed in elav-Gal4 > Sbf-RNAi, Canton-S WT files, and Tango14-RNAi flies (****p < 0.0001, n > 5 in each group, one-way ANOVA, Tukey’s multiple comparisons tests, Figure 1B).
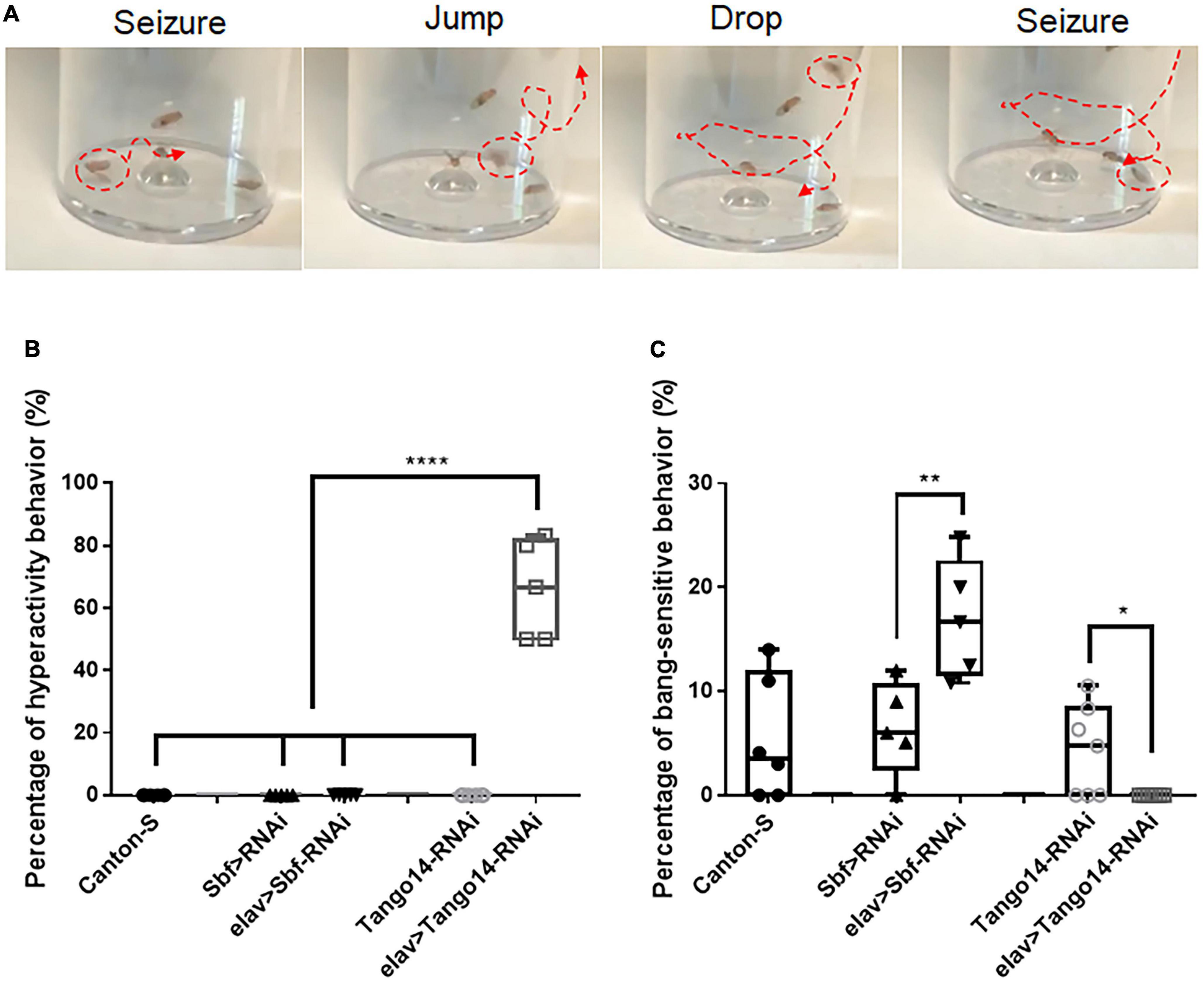
Figure 1. Hyperactivity seizure behavior in Tango14 knockdown flies. (A) Typical hyperactivity seizure behavior is shown. The seizure stages contain the first-phase seizure stage, the jump-and-drop stage, and the second-phase seizure stage. (B) Approximately 70% of Tango14 knockdown files (elav > Tango14-RNAi) showed hyperactivity seizure behavior. Contrarily, the control flies (Canton-S files and Tango-RNAi flies) did not show any hyperactivity seizure behavior. One-way ANOVA and Tukey’s multiple comparison tests. (C) elav > Sbf-RNAi showed a higher BS seizure rate than that of wild-type flies (Sbf > RNAi). While elav > Tango14-RNAi showed no classical BS seizure. **P < 0.01, ****P < 0.001.
3.1.2. Late-phase seizure behavior
In this study, another new type of seizure behavior called late-phase seizure was identified, which is distinct from the classical BS seizure behavior. Specifically, after vortex stimulation, the late-phase seizure behavior displayed a brief freezing or normal phase lasting 1–3 s before the onset of the seizure phase, as shown in Figure 2B and Supplementary video 2. In contrast, the classical BS seizure behavior started immediately after vortex stimulation without a freezing phase, as shown in Figure 2A. The knockdown of the epilepsy candidate gene Klp3A in flies (tub-Gal4 > UAS-Klp3A-RNAi) resulted in a significantly higher occurrence of late-phase seizures (22.27 ± 8.49%, n = 5) compared to the UAS-Klp3A-RNAi flies (0.00 ± 0.00%, n = 5, p = 0.0002, one-way ANOVA, Tukey’s multiple comparisons test). However, there was no significant difference in the occurrence of classical seizures between the two groups (7.50 ± 7.45%, n = 5 vs. 5.67 ± 3.98%, n = 6, p = 0.95). Furthermore, the tub-Gal4 > UAS-Klp3A-RNAi flies exhibited a significant difference in the manifestation of late-phase seizure and classical seizure (22.27 ± 8.49%, n = 5 vs. 7.50 ± 7.45%, n = 5, **p = 0.0036, Figure 2C). These results suggest that the primary phenotype of Klp3A knockdown flies is late-phase seizure rather than classical or hyperactivity seizures.
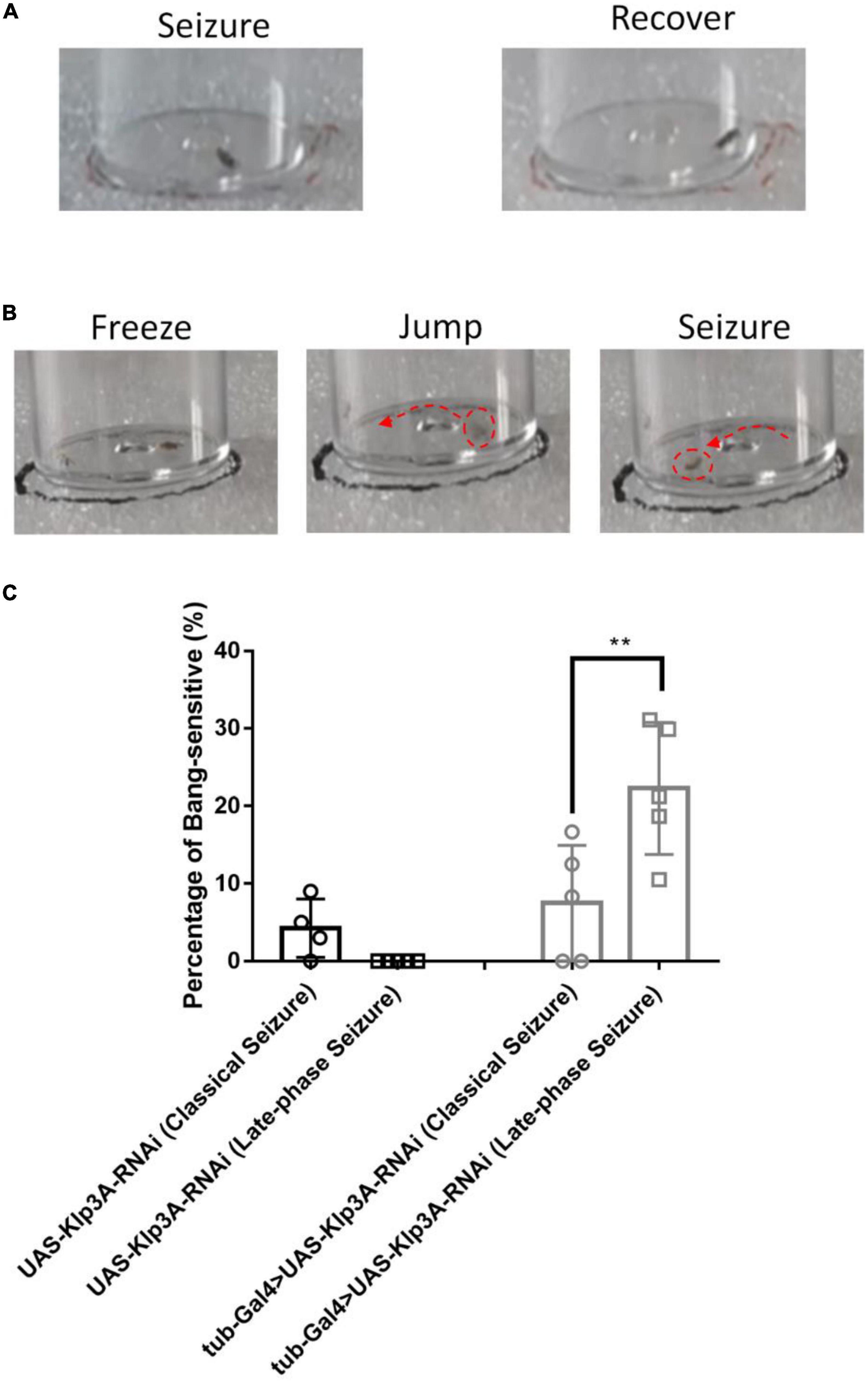
Figure 2. Late-phase seizure behavior is shown in Klp3A knockdown flies. (A) The flies showed seizure behavior immediately in classical seizure behavior. (B) Late-phase seizure behavior contained the freezing stage and jumping stage before seizure behavior. (C) Klp3A showed a higher rate in late-phase seizures than that in classical seizures. One-way ANOVA, Tukey’s multiple comparison tests. **P < 0.01.
3.2. Morphological observation in Drosophila
To visualize the major brain regions that can be affected by epilepsy, we utilized membrane GFP labeling via the tub-Gal4 > UAS-mCD8:GFP construct. These regions, namely, the mushroom body and central complex (Figure 3), play a crucial role in higher-order cognitive functions. Our previous research has found that knockdown/knockout of genes involved in epilepsy or neural disorder, such as UNC13B, LRP1, and YWHAZ, leads to various brain abnormalities, including neuron blurring in mushroom body (Wang et al., 2021), partial destruction in the central complex (Zhang et al., 2022), and gamma lobe mutilation in the mushroom body (Wan et al., 2022). Although not all epilepsy gene defects could induce brain structural abnormalities, the morphological study still can provide valuable insights into the underlying mechanisms of epilepsy caused by specific genes.
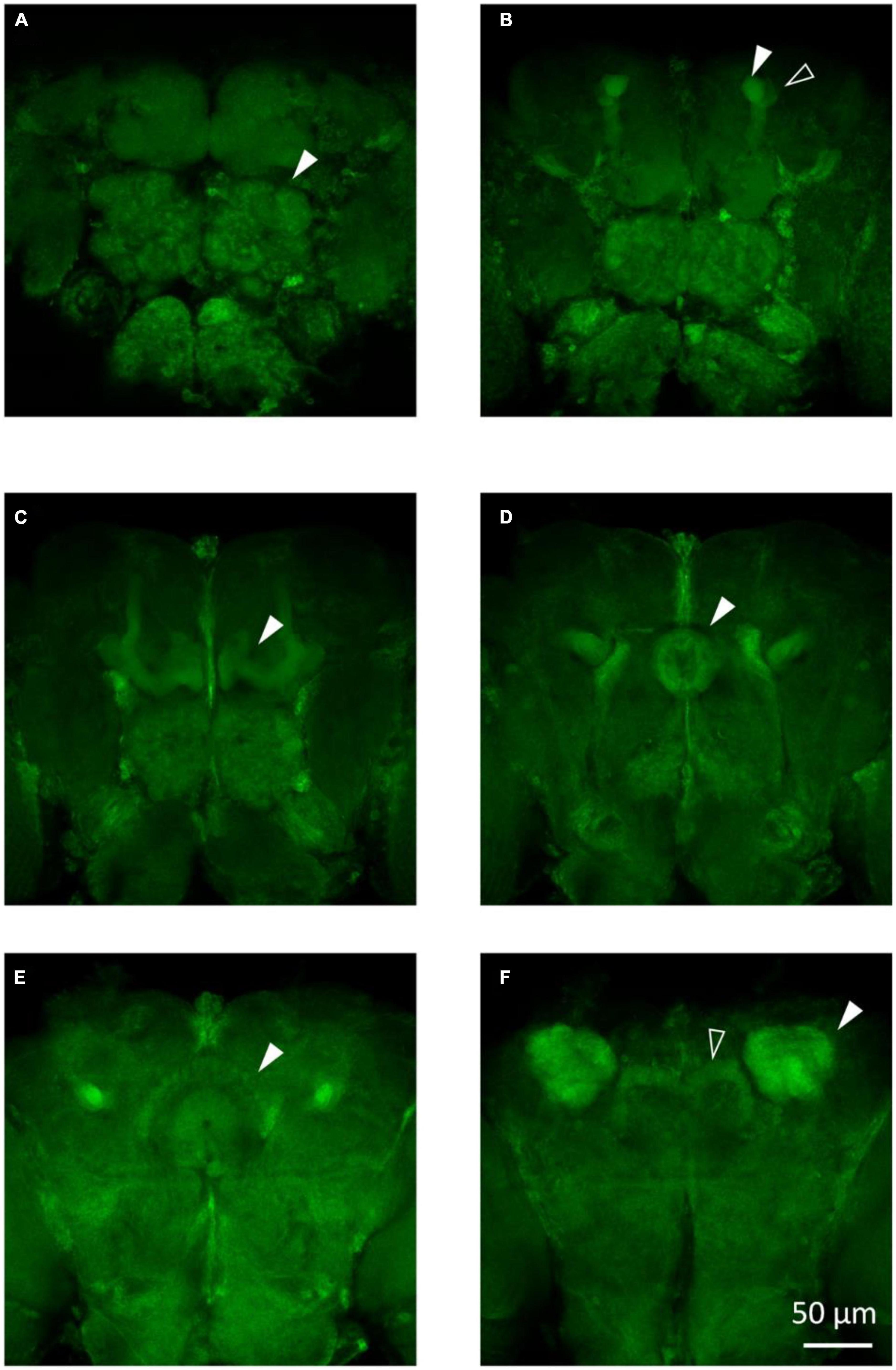
Figure 3. The main structures in the fly brain. (A) Antennal lobes are indicated by the arrow in the anterior view of the tub-Gal4 > UAS-mCD8:GFP fly brain. (B) Alpha lobe (solid arrow) and alpha’ lobe (hollow arrow) of the mushroom body are shown in the anterior view of the fly brain. (C) Gamma lobe of the mushroom body in the tub-Gal4 > UAS-mCD8:GFP fly brain. (D) Ellipsoid body of the central complex in the tub-Gal4 > UAS-mCD8:GFP fly brain. (E) Fan-sharp body of the central complex in the tub-Gal4 > UAS-mCD8:GFP fly brain. (F) Protocerebral bridge (hollow arrow) of the central complex and calyx (solid arrow) of the mushroom body are shown in the posterior view of the fly brain.
3.3. Larvae development in Drosophila
CACNA1A is a well-known epilepsy-associated gene (OMIM*601011). To study the developmental effect of CACNA1A, we used the UAS/Gal4 system to establish calcium channel knockdown (KD) flies (tub-Gal4 > Cac-RNAi). UAS-cac-RNAi flies were used as wild-type (WT) flies The results showed that the global knockdown of Cac did not affect the development of 1st (WT: 0.89 ± 0.21, n = 28 vs. KD: 0.85 ± 0.22, n = 22, p = 0.64, Student’s t-test), 2nd (WT: 1.69 ± 0.35, n = 41 vs. KD: 1.85 ± 0.48, n = 10, p = 0.26), and 3rd (WT: 2.76 ± 0.47, n = 22 vs. KD: 2.81 ± 0.63, n = 8, p = 0.82) larval stage, compared to the wild-type flies (Figure 4).
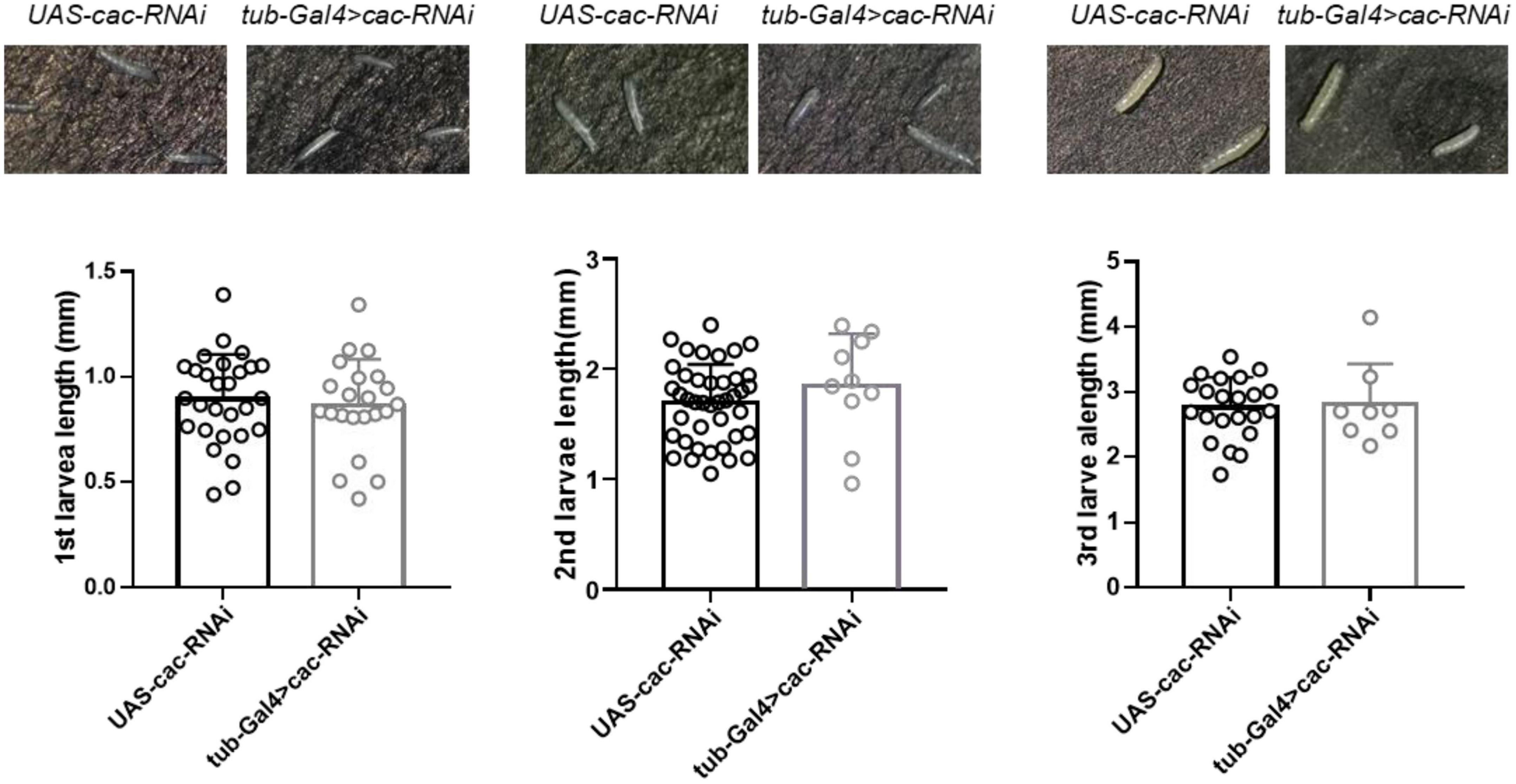
Figure 4. Global knockdown of calcium channel (cac) did not affect the development of Drosophila. The length of the 1st, 2nd, and 3rd larval stages did not show significant differences between wild-type files UAS-Cac-RNAi and calcium channel knockdown flies tub-Gal4 > cac-RNAi.
3.4. EPSCs and action currents in the antennal lobe of Drosophila
The projection neuron located in the antennal lobe (AL) of the fly brain is a significant excitatory neuron group, and these neurons are easily distinguishable using bright field imaging in a patch microscope (Duan et al., 2012), which makes them an ideal model for investigating neuronal excitability in epilepsy-prone flies. Here, we performed whole-cell, voltage-clamp recordings on neurons in AL based on their expression of GFP (Figure 5A) to detect the excitatory postsynaptic currents (EPSCs) of GH146-Gal4 > GFP WT flies. Typical traces of spontaneous EPSCs in WT flies GH146-Gal4 > GFP are presented in Figure 5B. To test the setup in seizures, we established Sbf, another epilepsy candidate gene, knockdown flies GH146-Gal4 > GFP; Sbf-RNAi. The GH146-Gal4 > GFP;Sbf-RNAi flies had a significantly higher frequency of sEPSCs in projection neurons than that of GH146-Gal4 > GFP WT flies (11.71 ± 3.39 Hz [n = 5] vs. 5.71 ± 1.86 Hz [n = 6]; **p = 0.0046) (Figure 5C). There was no significant difference in sEPSC amplitude between GH146-Gal4 > GFP; Sbf-RNAi and GH146-Gal4 > GFP WT flies (6.33 ± 3.15 pA [n = 10] vs. 6.62 ± 2.70 pA [n = 6]; p = 0.85) (Figure 5D).
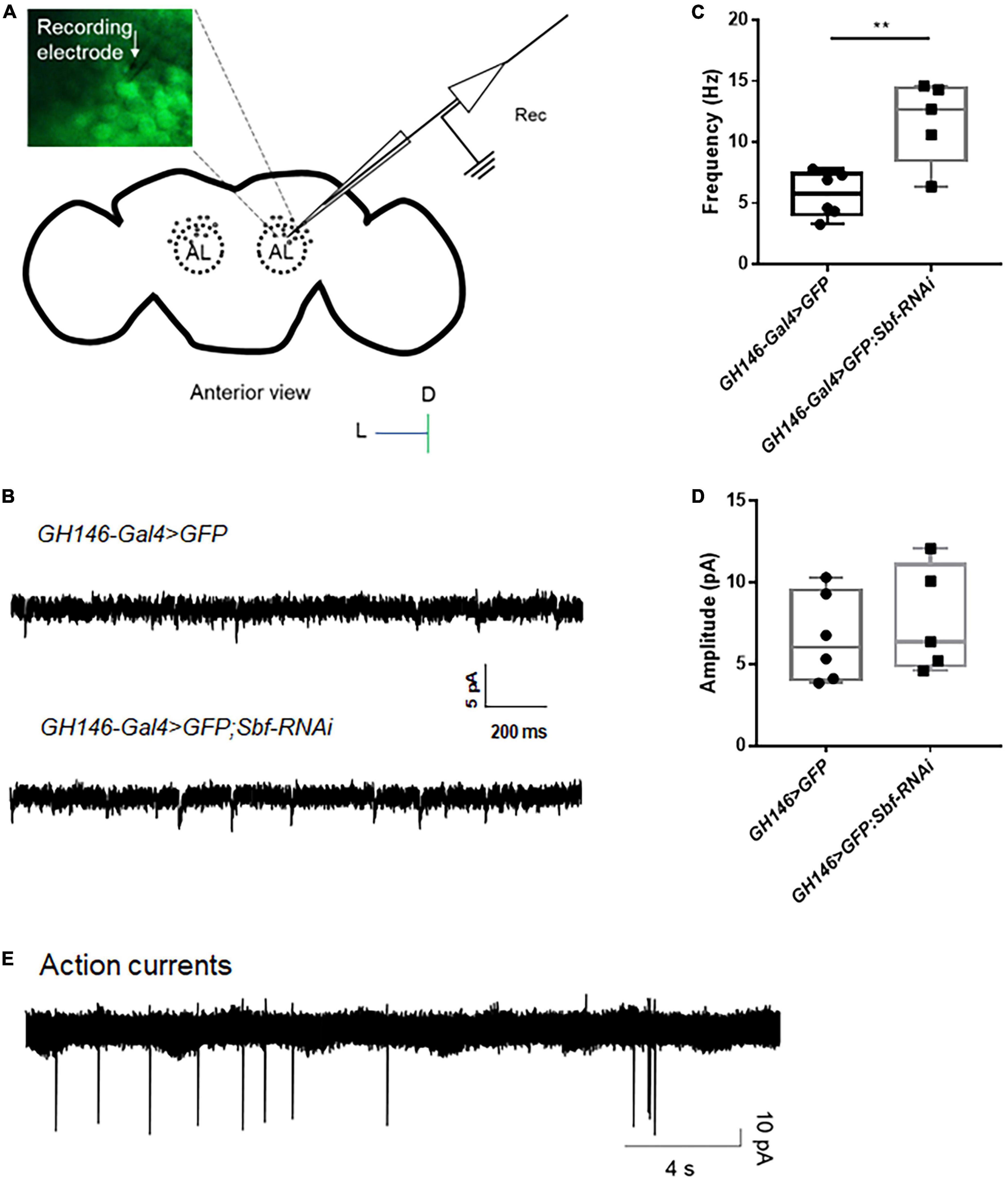
Figure 5. Whole-cell recording and attached recording in the fly brain. (A) Scheme of whole-cell recording or attached recording in the projection neurons of antennal lobes. Antennal lobes were observed in the anterior view of the brain. Neurons were labeled with GFP, and a recording electrode was used to patch the target neurons. (B) Typical trace of EPSCs in projection neurons of the wild-type flies and Sbf knockdown flies. The frequency of EPSCs in GH146-Gal4 > GFP;Sbf-RNAi flies is higher than that in wild-type files (GH146-Gal4 > GFP) (C), but the amplitude of EPSCs showed no difference between knockdown flies and wild-type flies (D). (E) Typical trace of action currents in the projection neurons. **P < 0.01.
The attached recording is an essential electrophysiological technique for investigating neuronal excitability, particularly for analyzing spontaneous action potential currents (Dubin and Harris, 1997; Perkins, 2006). The recording captured and presented in Figure 5E displays the recorded action potential currents. Our previous study has shown that the knockdown of UNC13B induces a higher frequency of action currents in projection neurons (Wang et al., 2021).
3.5. Evoked EPSP from the antennal lobe to the mushroom body of Drosophila
Given the numerous genes related to synaptic structure that has been implicated in epilepsy (Lammertse et al., 2020), it is evident that synaptic connections must be considered when evaluating the mechanisms of epilepsy candidate genes. However, to date, few epilepsy gene studies conducted using the Drosophila model have utilized electrophysiological techniques to directly assess synaptic events. To address this knowledge gap, we established a single-cell-level recording technique to monitor evoked excitatory postsynaptic potentials (EPSPs) from the antennal lobe (AL) to the mushroom body (MB), which is a major synaptic functional connection in the Drosophila central nervous system. The technique involved forming a whole-cell recording on an MB neuron and applying an electrical stimulus to AL neurons to induce evoked EPSPs from AL to MB (Figures 6A, B). The amplitude of the evoked EPSPs increased with increasing stimulus intensity (Figure 6C). The input-output relationship was established to determine the appropriate stimulus intensity, and the current intensity that induced 50% of the maximum EPSP response was used for subsequent recordings. Tubocurarine, a cholinergic receptor inhibitor, was used to confirm the functional connection between AL and MB (Parker et al., 2011b). The latency of the evoked EPSPs varied depending on the different connections between projection neurons and mushroom body neurons. For example, gamma neurons (1471-Gal4 > GFP) exhibited evoked EPSPs with a latency of approximately 1.6 ms (Figure 6D).
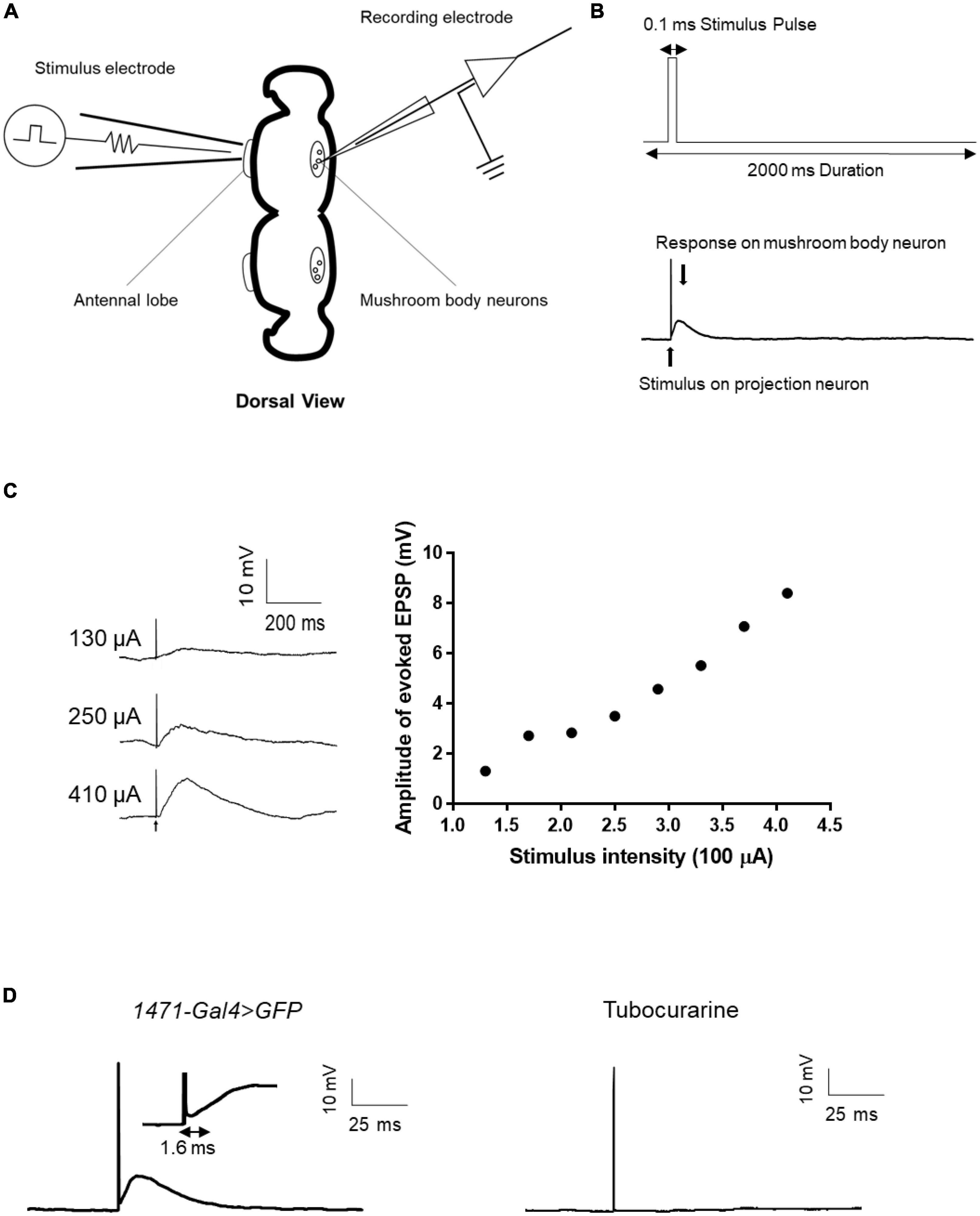
Figure 6. Evoked EPSP recording in the fly brain. (A) Scheme of evoked EPSP recording between the antennal lobe and mushroom body. The stimulation electrode attached to the antennal lobe and recording electrode was used to record evoked EPSP in mushroom body neurons. (B) Stimulus protocol for the stimulation electrode is shown in the upward picture. Stimulus artifact and response signal are shown in the downward picture. (C) Typical trace of evoked EPSP in different stimulus intensities and the input-output curve of evoked EPSP. (D) Latency time of evoked EPSP in gamma neuron. Tubocurarine confirmed that the evoked EPSP is cholinergic.
3.6. Knockdown efficiency detected by RT-qPCR
In terms of knockdown efficiency, tub-Gal4 > Cac-RNAi, tub-Gal4 > Sbf-RNAi, tub-Gal4 > Klp3A-RNAi, and tub-Gal4 > Tango14-RNAi had values of 53.95, 39.53, 49.26, and 38.59%, separately [46.05 ± 2.76% (n = 3) vs. 100 ± 0.00% (n = 3), ****p < 0.0001; 60.47 ± 9.86% (n = 3) vs. 100 ± 0.00% (n = 3), **p = 0.0023; 50.74 ± 11.19% (n = 3) vs. 100 ± 0.00% (n = 3), **p = 0.0016; 61.41 ± 8.99% (n = 6) vs. 100 ± 0.00% (n = 6), ****p < 0.0001, Figure 7]. Cac, Sbf, Klp3A, and Tango14 were knockdown successfully in these Drosophila lines, as shown in Figure 7.
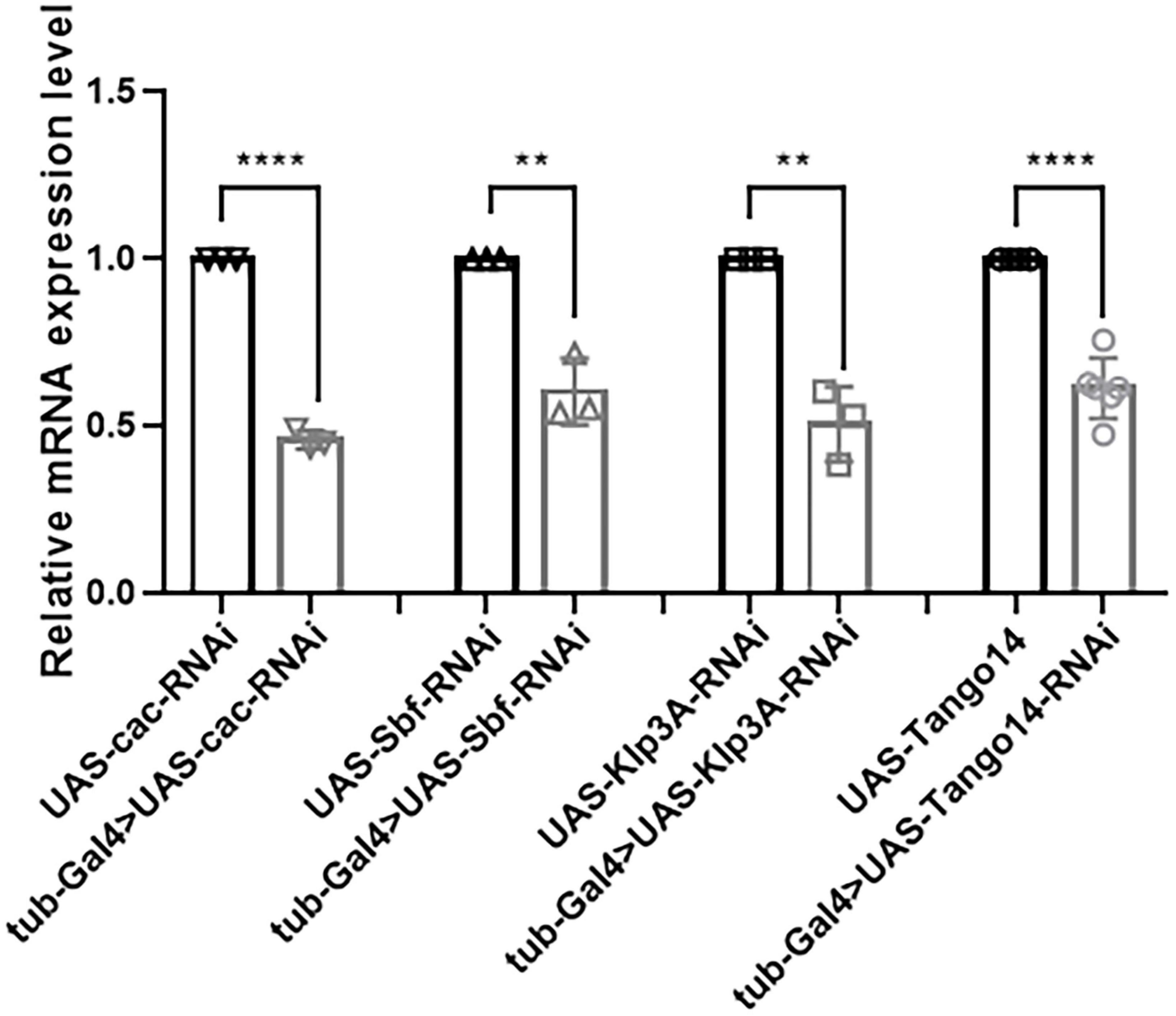
Figure 7. Knockdown efficacy of knockdown fly lines. Relative expression of mRNA in Cac knockdown flies, Sbf knockdown flies, Klp3A knockdown flies, Tango14 knockdown flies, and wild-type flies. **P < 0.01, ****P < 0.001.
4. Discussion
Trio-based WES has been successful in defining the most likely epilepsy-implicated loci and screening out candidate genes, such as UNC13B, CELSR3, and CDK19 in numerous studies (Chung et al., 2020; Wang et al., 2021; Li et al., 2022). Besides, NGS analysis of a large cohort of patients with epilepsy revealed that approximately 14% of sequence changes in 70 common and rare mutations were detected as nonsense or deletion (Lindy et al., 2018), while missense or frameshift mutations can also lead to loss-of-function (LOF) consequences (Chen et al., 2017; Lindy et al., 2018). The Gal4/UAS system allows for precise spatiotemporal manipulations of gene expression for cell labeling or gene function analysis (Del Valle Rodríguez et al., 2011). By crossing Gal4 driver lines to UAS-RNAi lines, the expression of UAS-linked genes can be effectively blocked in the chosen Gal4 pattern. However, some missense mutations lead to gain-of-function (GOF). For example, hundreds of missense mutations, including GOF mutations, in the gene SCN1A can confer a wide spectrum of epileptic phenotypes in Drosophila as they do in humans (Schutte et al., 2014), thus knock-in Drosophila lines mimicking the missense variants should be generated to realize more precise genetic editing (Xue et al., 2014). In our previous study, c.147A > T missense variation of Ywhaz was created using the CRISPR/Cas9 system to confirm its pathogenic effects (Wan et al., 2022). Thus, the knockdown model established by the Gal4/UAS system and the knock-in model established by the CRISPR/Cas9 system can cover most scenarios of gene mutations, including LOF and GOF mutations.
Animal experimentation can be a challenging and time-consuming process for validating the extensive range of candidate genes and variants proposed by WES. However, Drosophila has emerged as an attractive model organism to examine human epilepsy due to its short life span, strong genomic conservation, the abundance of technological resources for genomic manipulations (Shulman, 2015), and the exhibition of seizure behavior similar to humans. Historically, mechanical induction assays in Drosophila have only elicited one seizure behavior. This study expands the range of seizure-like phenotypes in the Drosophila model by discovering two novel seizure behaviors. Knockdown of Tango14 results in hyperactive seizure behavior (Figure 1), while knockdown of Klp3A results in late-phase seizure behavior (Figure 2). Our findings were further validated by the demonstration of hyperactivity seizure behavior in knockdown of the Ywhaz gene (Wan et al., 2022), suggesting that this novel behavior could be applied to verifying other epilepsy candidate genes. These results imply that loss of function in different candidate genes can result in distinct phenotypes. In light of our observation, the hyperactivity seizure, manifesting increased movement and excitement, is comparable to hyperesthesia and highly consistent with the hyperactive-automatism subtype in complex partial epilepsy. The late-phase seizure may correlate with focal epilepsy originating from the sensory cortex. However, further investigation into the genotype-phenotype relationship in Drosophila is required through additional studies.
Electrophysiology has shown to be a valuable tool for the study of epilepsy, with numerous studies reporting its effectiveness in assessing electrical activity in the giant fiber and neuromuscular junction (NMJ) (Kuebler et al., 2001; Marley and Baines, 2011; Parker et al., 2011a,b; Duan et al., 2012; Xue et al., 2014). However, its application in the central nervous system (CNS) studies of epilepsy using the Drosophila model is less common (Guo et al., 2019; Savitsky et al., 2020; Chi et al., 2022). Our study adopted three electrophysiological methods, namely, whole-cell recording, cell-attached recording, and evoked EPSP recording, to directly explore the electrophysiological characteristics in the CNS. Whole-cell/cell-attached recordings allow us to assess the neuronal excitability of mutants compared to wild-type neurons in specific GFP-expressed regions of the Drosophila brain, such as the mushroom body (MB), a major high-level brain structure in flies. We could determine whether the cells are firing action currents and record the firing modes, such as tonic or burst firing. Moreover, the approach of recording evoked EPSP from AL to MB, previously used to reveal input-timing-dependent plasticity in the MB circuit during olfactory learning (Qiao et al., 2022b), has the potential to investigate the synaptic and molecular causes of epilepsy. The latency of evoked EPSP can measure the time interval of monosynaptic transmission in target synapses (Gil and Amitai, 1996). Our preliminary results suggest that the latency of evoked EPSP in PN-gamma neurons is around 1.6 ms (Figure 6D), validating the efficacy of our techniques in accurately detecting the time interval of target synaptic transmission. While we have yet to observe positive results in any epilepsy candidate genes, these methods can provide a strong tool for studying genes involved in synaptic function coding in epilepsy. Combining these electrophysiological methods with the LPF recording technique in the CNS of Drosophila (Iyengar and Wu, 2021) can develop an instrumental platform for investigating the cellular and synaptic mechanisms underlying neural function or dysfunction (Roemmich et al., 2018), with excellent accessibility for studying epilepsy’s pathological and pharmacological aspects (Sheeba et al., 2008).
The prevalence of seizures and epilepsies is alarmingly high among infants and preschoolers, with 10–25% of these young patients suffering from intractable seizures and varying degrees of intellectual/developmental disabilities (ID/DD) (Chung et al., 2020). Understanding the genetic basis of these conditions is crucial for accurate diagnosis, prognosis, genetic counseling, and treatment. Recent advances in NGS have revealed that de novo missense mutations in the CACNA1A gene contribute to ID/DD and therapy-resistant epilepsy (Indelicato and Boesch, 2021). As a result, our development study utilized the knockdown of the Cac gene (the ortholog of CACNA1A) to further investigate the impact of epilepsy candidate genes on development. Although approximately 50% knockdown did not demonstrate a significant effect (Figure 7), the study provided a reliable protocol for examining the impact of epilepsy candidate genes on development. The use of Gal4 drivers in conjunction with Gal80ts, which exhibit minimal activity at 18°C and maximal activity at 29°C (Duffy, 2002), can precisely restrict the gene knockdown to the first, second, and third instar larval or pupal stages through temperature control. By measuring parameters such as larval or pupal weight, pupation rate, eclosion rate, and body length (Zhang et al., 2021), we can conduct a more comprehensive analysis of candidate genes that exclusively express during specific developmental stages.
The potential of the genetic screening system, which models epilepsy using genotypic and phenotypic strategies, could be further demonstrated by applying it to a larger collection of genes implicated in epilepsy, such as a gene set. In addition, it is important to consider the sex of the flies in current experiments, as gender differences in epilepsy exist in both humans and Drosophila (Sharma et al., 2009).
Notably, epilepsy in individuals may be caused by multiple pathogenic variants, both common and rare, with a range of effect sizes (Ferraro and Buono, 2006; Epi25 Collaborative, 2019; Thijs et al., 2019). Drosophila is advantageous in the study of epilepsy due to its ability to map genetic interactions, which is facilitated by powerful resources and techniques such as genome-wide RNAi transgenic stocks (Dietzl et al., 2007; Ni et al., 2011), collections of mapped transposon insertion alleles, Gal4/UAS system, and the recently developed CRISPR/Cas9 system. Notably, integrating the RNAi with Gal4/UAS system could achieve double- or even triple-knockdown in one generation without complicated genetic crosses (Okumura et al., 2015). Drosophila is a premier genetic model system for elucidating mechanisms responsible for many neurological genetic disorders (Jackson et al., 1998; Finelli et al., 2004). By making the most of resources and strategies, Drosophila holds great potential for advancing our understanding of the pathogenesis of epilepsy and other complex polygenic disorders, as well as the development of potential therapies.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
C-QL, J-DQ, and K-CW designed the study and wrote the manuscript. X-CQ, M-FH, D-HL, C-QL, S-MX, X-XZ, Y-ML, and W-JZ performed the behavior experiments. J-DQ performed the electrophysiological experiments. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Guangdong Basic and Applied Basic Research Foundation (grant no. 2022A1515111123 to J-DQ), Guangzhou Medical University (C-QL, funding no. 2022A056 and S-MX, funding no. 2021A021), Plan on enhancing scientific research in GMU (J-DQ), and the Second Affiliated Hospital of Guangzhou Medical University (J-DQ, funding no. 2021004).
Acknowledgments
We thank Tsinghua Fly Center for donating transgenic RNAi lines.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2023.1121877/full#supplementary-material
Footnotes
References
Chen, T., Giri, M., Xia, Z., Subedi, Y. N., and Li, Y. (2017). Genetic and epigenetic mechanisms of epilepsy: A review. Neuropsychiatr. Dis. Treat. 13, 1841–1859.
Chi, W., Iyengar, A., Fu, W., Liu, W., Berg, A., Wu, C., et al. (2022). Drosophila carrying epilepsy-associated variants in the vitamin B6 metabolism gene PNPO display allele- and diet-dependent phenotypes. Proc. Natl. Acad. Sci. U.S.A. 119:e2115524119. doi: 10.1073/pnas.2115524119
Chung, H., Mao, X., Wang, H., Park, Y., Marcogliese, P. C., Rosenfeld, J. A., et al. (2020). De novo variants in CDK19 are associated with a syndrome involving intellectual disability and epileptic encephalopathy. Am. J. Hum. Genet. 106, 717–725. doi: 10.1016/j.ajhg.2020.04.001
D’Adamo, M., Liantonio, A., Rolland, J., Pessia, M., and Imbrici, P. (2020). Kv1.1 channelopathies: Pathophysiological mechanisms and therapeutic approaches. Int. J. Mol. Sci. 21:2935. doi: 10.3390/ijms21082935
Del Valle Rodríguez, A., Didiano, D., and Desplan, C. (2011). Power tools for gene expression and clonal analysis in Drosophila. Nat. Methods 9, 47–55. doi: 10.1038/nmeth.1800
Dietzl, G., Chen, D., Schnorrer, F., Su, K., Barinova, Y., Fellner, M., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. doi: 10.1038/nature05954
Duan, J., Li, W., Yuan, D., Sah, B., Yan, Y., and Gu, H. (2012). Nitric oxide signaling modulates cholinergic synaptic input to projection neurons in Drosophila antennal lobes. Neuroscience 219, 1–9. doi: 10.1016/j.neuroscience.2012.05.068
Dubin, A. E., and Harris, G. L. (1997). Voltage-activated and odor-modulated conductances in olfactory neurons of Drosophila melanogaster. J. Neurobiol. 32, 123–137. doi: 10.1002/(sici)1097-4695(199701)32:1<123::aid-neu11>3.0.co;2-l
Duffy, J. B. (2002). GAL4 system in Drosophila: A fly geneticist’s swiss army knife. Genesis 34, 1–15. doi: 10.1002/gene.10150
Dupépé, E., Kicielinski, K., Gordon, A., and Walters, B. (2019). What is a case-control study? Neurosurgery 84, 819–826.
Epi25 Collaborative (2019). Ultra-rare genetic variation in the epilepsies: A whole-exome sequencing study of 17,606 individuals. Am. J. Hum. Genet. 105, 267–282. doi: 10.1016/j.ajhg.2019.05.020
Finelli, A., Kelkar, A., Song, H., Yang, H., and Konsolaki, M. (2004). A model for studying Alzheimer’s Abeta42-induced toxicity in Drosophila melanogaster. Mol. Cell. Neurosci. 26, 365–375. doi: 10.1016/j.mcn.2004.03.001
Ganetzky, B., and Wu, C. F. (1982). Indirect suppression involving behavioral mutants with altered nerve excitability in Drosophila melanogaster. Genetics 100, 597–614. doi: 10.1093/genetics/100.4.597
Gil, Z., and Amitai, Y. (1996). Properties of convergent thalamocortical and intracortical synaptic potentials in single neurons of neocortex. J. Neurosci. 16, 6567–6578. doi: 10.1523/JNEUROSCI.16-20-06567.1996
Gu, H., and O’Dowd, D. K. (2006). Cholinergic synaptic transmission in adult Drosophila Kenyon cells in situ. J. Neurosci. 26, 265–272. doi: 10.1523/JNEUROSCI.4109-05.2006
Guan, J., Li, J., Chen, G., Shi, T., Lan, L., Wu, X., et al. (2021). Family trio-based sequencing in 404 sporadic bilateral hearing loss patients discovers recessive and de novo genetic variants in multiple ways. Eur. J. Med. Genet. 64:104311. doi: 10.1016/j.ejmg.2021.104311
Guerrini, R., Balestrini, S., Wirrell, E. C., and Walker, M. C. (2021). Monogenic epilepsies: Disease mechanisms, clinical phenotypes, and targeted therapies. Neurology 97, 817–831.
Guo, H., Bettella, E., Marcogliese, P. C., Zhao, R., Andrews, J. C., Nowakowski, T. J., et al. (2019). Disruptive mutations in TANC2 define a neurodevelopmental syndrome associated with psychiatric disorders. Nat. Commun. 10:4679. doi: 10.1038/s41467-019-12435-8
Harnish, J., Deal, S., Chao, H., Wangler, M., and Yamamoto, S. (2019). In vivo functional study of disease-associated rare human variants using Drosophila. J. Vis. Exp. 20:e59658. doi: 10.3791/59658
Hegazi, S., Cheng, A., Krupp, J., Tasaki, T., Liu, J., Szulc, D., et al. (2022). UBR4/POE facilitates secretory trafficking to maintain circadian clock synchrony. Nat. Commun. 13:1594. doi: 10.1038/s41467-022-29244-1
Hunt, L., Stover, J., Haugen, B., Shaw, T., Li, Y., Pagala, V., et al. (2019). A key role for the ubiquitin ligase UBR4 in myofiber hypertrophy in Drosophila and mice. Cell Rep. 28, 1268–1281.e6. doi: 10.1016/j.celrep.2019.06.094
Indelicato, E., and Boesch, S. (2021). From genotype to phenotype: Expanding the clinical spectrum of CACNA1A variants in the era of next generation sequencing. Front. Neurol. 12:639994. doi: 10.3389/fneur.2021.639994
Iyengar, A., and Wu, C. F. (2021). Fly seizure EEG: Field potential activity in the Drosophila brain. J. Neurogenet. 35, 295–305. doi: 10.1080/01677063.2021.1950714
Jackson, G. R., Salecker, I., Dong, X., Yao, X., Arnheim, N., Faber, P. W., et al. (1998). Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron 21, 633–642. doi: 10.1016/s0896-6273(00)80573-5
Kaas, G. A., Kasuya, J., Lansdon, P., Ueda, A., Iyengar, A. S., Wu, C., et al. (2016). Lithium-responsive seizure-like hyperexcitability is caused by a mutation in the Drosophila voltage-gated sodium channel gene paralytic. eNeuro 3:ENEURO.0221-16.2016. doi: 10.1523/ENEURO.0221-16.2016
Kaplan, W. D., and Trout, W. E. III (1969). The behavior of four neurological mutants of Drosophila. Genetics 61, 399–409. doi: 10.1093/genetics/61.2.399
Kepecs, A., van Rossum, M., Song, S., and Tegner, J. (2002). Spike-timing-dependent plasticity: Common themes and divergent vistas. Biol. Cybern. 87, 446–458. doi: 10.1007/s00422-002-0358-6
Kuebler, D., Zhang, H., Ren, X., and Tanouye, M. A. (2001). Genetic suppression of seizure susceptibility in Drosophila. J. Neurophysiol. 86, 1211–1225. doi: 10.1152/jn.2001.86.3.1211
Lammertse, H. C., van Berkel, A., Iacomino, M., Toonen, R. F., Striano, P., Gambardella, A., et al. (2020). Homozygous STXBP1 variant causes encephalopathy and gain-of-function in synaptic transmission. Brain 143, 441–451. doi: 10.1093/brain/awz391
Lasko, P., and Luthy, K. (2021). Investigating rare and ultrarare epilepsy syndromes with Drosophila models. Fac. Rev. 10:10. doi: 10.12703/r/10-10
Lee, H., Deignan, J. L., Dorrani, N., Strom, S. P., Kantarci, S., Quintero-Rivera, F., et al. (2014). Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 312, 1880–1887.
Li, J., Lin, S., Qiao, J., Liu, X., Wang, J., Jiang, M., et al. (2022). CELSR3 variants are associated with febrile seizures and epilepsy with antecedent febrile seizures. CNS Neurosci. Ther. 28, 382–389. doi: 10.1111/cns.13781
Lindy, A. S., Stosser, M. B., Butler, E., Downtain-Pickersgill, C., Shanmugham, A., Retterer, K., et al. (2018). Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia 59, 1062–1071. doi: 10.1111/epi.14074
Marley, R., and Baines, R. A. (2011). Increased persistent Na+ current contributes to seizure in the slamdance bang-sensitive Drosophila mutant. J. Neurophysiol. 106, 18–29. doi: 10.1152/jn.00808.2010
Ni, J., Zhou, R., Czech, B., Liu, L., Holderbaum, L., Yang-Zhou, D., et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8, 405–407. doi: 10.1038/nmeth.1592
Okumura, T., Sasamura, T., Inatomi, M., Hozumi, S., Nakamura, M., Hatori, R., et al. (2015). Class I myosins have overlapping and specialized functions in left-right asymmetric development in Drosophila. Genetics 199, 1183–1199. doi: 10.1534/genetics.115.174698
Parker, L., Howlett, I. C., Rusan, Z. M., and Tanouye, M. A. (2011a). Seizure and epilepsy: Studies of seizure disorders in Drosophila. Int. Rev. Neurobiol. 99, 1–21. doi: 10.1016/B978-0-12-387003-2.00001-X
Parker, L., Padilla, M., Du, Y., Dong, K., and Tanouye, M. A. (2011b). Drosophila as a model for epilepsy: Bss is a gain-of-function mutation in the para sodium channel gene that leads to seizures. Genetics 187, 523–534. doi: 10.1534/genetics.110.123299
Pavlidis, P., and Tanouye, M. A. (1995). Seizures and failures in the giant fiber pathway of Drosophila bang-sensitive paralytic mutants. J. Neurosci. 15, 5810–5819. doi: 10.1523/JNEUROSCI.15-08-05810.1995
Perkins, K. L. (2006). Cell-attached voltage-clamp and current-clamp recording and stimulation techniques in brain slices. J. Neurosci. Methods 154, 1–18. doi: 10.1016/j.jneumeth.2006.02.010
Qiao, J., Li, X., Li, J., Guo, Q., Tang, X., Chen, L., et al. (2022a). Reply: UNC13B and focal epilepsy. Brain 145, e13–e16. doi: 10.1093/brain/awab486
Qiao, J., Yang, S., Geng, H., Yung, W., and Ke, Y. (2022b). Input-timing-dependent plasticity at incoming synapses of the mushroom body facilitates olfactory learning in Drosophila. Curr. Biol. 32, 4869–4880.e4. doi: 10.1016/j.cub.2022.09.054
Roemmich, A. J., Schutte, S. S., and O’Dowd, D. K. (2018). Ex vivo whole-cell recordings in adult Drosophila brain. Bio Protoc. 8:e2467.
Savitsky, M., Solis, G., Kryuchkov, M., and Katanaev, V. (2020). Humanization of Drosophila Gαo to model GNAO1 paediatric encephalopathies. Biomedicines 8:395.
Schutte, R. J., Schutte, S. S., Algara, J., Barragan, E. V., Gilligan, J., Staber, C., et al. (2014). Knock-in model of dravet syndrome reveals a constitutive and conditional reduction in sodium current. J. Neurophysiol. 112, 903–912. doi: 10.1152/jn.00135.2014
Sharma, A., Mohammad, F., and Singh, P. (2009). Gender differences in a Drosophila transcriptomic model of chronic pentylenetetrazole induced behavioral deficit. PLoS One 4:e8136. doi: 10.1371/journal.pone.0008136
Sheeba, V., Gu, H., Sharma, V. K., O’Dowd, D. K., and Holmes, T. C. (2008). Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J. Neurophysiol. 99, 976–988. doi: 10.1152/jn.00930.2007
Shulman, J. M. (2015). Drosophila and experimental neurology in the post-genomic era. Exp. Neurol. 274, 4–13. doi: 10.1016/j.expneurol.2015.03.016
Sun, L., Gilligan, J., Staber, C., Schutte, R. J., Nguyen, V., O’Dowd, D. K., et al. (2012). A knock-in model of human epilepsy in Drosophila reveals a novel cellular mechanism associated with heat-induced seizure. J. Neurosci. 32, 14145–14155. doi: 10.1523/JNEUROSCI.2932-12.2012
Takai, A., Yamaguchi, M., Yoshida, H., and Chiyonobu, T. (2020). Investigating developmental and epileptic encephalopathy using Drosophila melanogaster. Int. J. Mol. Sci. 21:6442. doi: 10.3390/ijms21176442
Wan, R., Liu, Z., Huang, X., Kwan, P., Li, Y., Qu, X., et al. (2022). YWHAZ variation causes intellectual disability and global developmental delay with brain malformation. Hum. Mol. Genet. 32, 462–472. doi: 10.1093/hmg/ddac210
Wang, J., Qiao, J., Liu, X., Liu, D., Chen, Y., Wu, Y., et al. (2021). UNC13B variants associated with partial epilepsy with favourable outcome. Brain 144, 3050–3060.
Weber, Y. G., Biskup, S., Helbig, K. L., Spiczak, S. V., and Lerche, H. (2017). The role of genetic testing in epilepsy diagnosis and management. Expert Rev. Mol. Diagn. 17, 739–750.
World Health Organization [WHO] (2017). Epilepsy. Available online at: https://www.who.int/news-room/fact-sheets/detail/epilepsy (accessed April 11, 2023).
XiangWei, W., Kannan, V., Xu, Y., Kosobucki, G. J., Schulien, A. J., Kusumoto, H., et al. (2019). Heterogeneous clinical and functional features of GRIN2D-related developmental and epileptic encephalopathy. Brain 142, 3009–3027. doi: 10.1093/brain/awz232
Xue, Z., Wu, M., Wen, K., Ren, M., Long, L., Zhang, X., et al. (2014). CRISPR/Cas9 mediates efficient conditional mutagenesis in Drosophila. G3 (Bethesda) 4, 2167–2173. doi: 10.1534/g3.114.014159
Zhai, Y., Zhang, Z., Shi, P., Martin, D. M., and Kong, X. (2021). Incorporation of exome-based CNV analysis makes trio-WES a more powerful tool for clinical diagnosis in neurodevelopmental disorders: A retrospective study. Hum. Mutat. 42, 990–1004. doi: 10.1002/humu.24222
Zhang, J., Chen, M., He, J., Li, Y., Li, Z., Ye, Z., et al. (2022). Ketone body rescued seizure behavior of LRP1 deficiency in Drosophila by modulating glutamate transport. J. Mol. Neurosci. 72, 1706–1714. doi: 10.1007/s12031-022-02026-6
Zhang, J., Wu, J., Wu, M., and Tong, H. (2021). Palmitic acid inhibits the growth, development and glucose and lipid metabolism of Drosophila melanogaster. Mod. Food Sci. Technol. 37, 114–119+213. doi: 10.13982/j.mfst.1673-9078.2021.11.0234
Keywords: epilepsy, trio-based whole-exome sequencing, Drosophila, Gal4/UAS system, electrophysiology
Citation: Liu C-Q, Qu X-C, He M-F, Liang D-H, Xie S-M, Zhang X-X, Lin Y-M, Zhang W-J, Wu K-C and Qiao J-D (2023) Efficient strategies based on behavioral and electrophysiological methods for epilepsy-related gene screening in the Drosophila model. Front. Mol. Neurosci. 16:1121877. doi: 10.3389/fnmol.2023.1121877
Received: 12 December 2022; Accepted: 27 March 2023;
Published: 20 April 2023.
Edited by:
Yasir Ahmed Syed, Cardiff University, United KingdomReviewed by:
Mohammad Farhan, Hamad Bin Khalifa University, QatarMouhamed Alsaqati, Newcastle University, United Kingdom
Copyright © 2023 Liu, Qu, He, Liang, Xie, Zhang, Lin, Zhang, Wu and Qiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing-Da Qiao, Joaquinqjd@163.com; orcid.org/0000-0002-4693-8390
 Chu-Qiao Liu
Chu-Qiao Liu Xiao-Chong Qu
Xiao-Chong Qu Ming-Feng He
Ming-Feng He De-Hai Liang1
De-Hai Liang1  Yong-Miao Lin
Yong-Miao Lin Jing-Da Qiao
Jing-Da Qiao