- 1Department of Biochemistry, Human Metabolomics, Faculty of Natural and Agricultural Sciences, North-West University, Potchefstroom, South Africa
- 2Department of Pediatric Infectious Diseases and Immunology, Pediatric Infectious Diseases and Immunology, Amsterdam University Medical Center, Emma Children's Hospital, Amsterdam, Netherlands
Mycobacterium tuberculosis infection, which claims hundreds of thousands of lives each year, is typically characterized by the formation of tuberculous granulomas — the histopathological hallmark of tuberculosis (TB). Our knowledge of granulomas, which comprise a biologically diverse body of pro- and anti-inflammatory cells from the host immune responses, is based mainly upon examination of lungs, in both human and animal studies, but little on their counterparts from other organs of the TB patient such as the brain. The biological heterogeneity of TB granulomas has led to their diverse, relatively uncoordinated, categorization, which is summarized here. However, there is a pressing need to elucidate more fully the phenotype of the granulomas from infected patients. Newly emerging studies at the protein (proteomics) and metabolite (metabolomics) levels have the potential to achieve this. In this review we summarize the diverse nature of TB granulomas based upon the literature, and amplify these accounts by reporting on the relatively few, emerging proteomics and metabolomics studies on TB granulomas. Metabolites (for example, trimethylamine-oxide) and proteins (such as the peptide PKAp) associated with TB granulomas, and knowledge of their localizations, help us to understand the resultant phenotype. Nevertheless, more multidisciplinary ‘omics studies, especially in human subjects, are required to contribute toward ushering in a new era of understanding of TB granulomas – both at the site of infection, and on a systemic level.
Introduction
Tuberculosis (TB) is a devastating infectious disease of pandemic proportions, caused by the bacterium Mycobacterium tuberculosis (Mtb). For many years, it has been one of the top ten causes of death from a single infectious agent (1). Although there has been a decline in the number of TB cases in some geographical regions, the rise in drug-resistant TB poses new challenges (2). TB is mostly a pulmonary disease, although it can affect other organs of the body, in which case it is known as extra-pulmonary TB (EPTB). The most severe form of EPTB occurs in the central nervous system (CNS-TB), with a prevalence of approximately 1% of the total TB cases reported (3), leaving about half of its patients either dead or permanently neurologically disabled (4).
Granulomas are the body's response to chronic antigenic stimulants. They represent the body's first line of response to Mtb infection and thus serve as a histopathological hallmark of TB. They comprise a combination of specialized immune cells influenced by an orchestrated pro- and anti-inflammatory response (5, 6). Granuloma formation can be affected by the number and location of these cells or other underlying diseases in the host. For example, the presence of a large number of B lymphocytes contribute to tuberculous granuloma formation through the release of chemokines and cytokines (7, 8). Granulomas can develop in any organ of the body where the causative infectious agent resides. They can also be formed in response to other persistent diseases such as; sarcodiosis schistosomiasis, syphilis, Crohn's disease, leprosy and in infections caused by bacteria, fungi and protozoa (9–18), and are either capable of containing the infection (latent TB), or not (active TB) (19–21). The rupture of a mature TB granuloma (the cause of which is not fully understood) in an individual with latent TB typically results in active TB (19, 21, 22). The most widely studied organs with TB granulomas are the lungs (6, 7, 19, 20, 23–28), whereas the characterization of TB granulomas in the brain, a site where infection with Mtb can have devastating consequences, is poorly described and understood (3). An improved understanding of TB granulomas in general could shed new light on improved approaches to TB management (20). There are, however, limited agreement on the conclusions to draw from studies of TB granulomas, such as lack of understanding of how the human granuloma functions. There is also no workable model that depicts the diseased human organ in vitro, as the structure and composition of granulomas vary among different organisms (6, 7, 29, 30). These are handicaps to our fuller understanding of TB. There is also no standard classification for the different types of granulomas identified to date. They are either grouped according to the composition of the immune cells, as primary or secondary granulomas (23, 29), or based on appearance and features such as: solid, caseous and cavitary, necrotizing, and non-necrotizing (5–7, 21, 22, 31). Consequently, the study of the TB granuloma needs to be expanded upon, using research methodologies, such as the ‘omics approaches, including, but not limited to, proteomics and metabolomics. Both proteomics and metabolomics have the potential of being instructive by elaborating upon the phenotype of TB granulomas at the protein and metabolite level, respectively.
This review provides a brief overview of TB; discusses the TB granuloma via function and importance, development, and categorization; and provides insights into the pro- and anti-inflammatory immune responses to Mtb by examining the limited proteomics studies (animal and human) conducted on TB granulomas and identifying specific metabolic indicators of TB progression and/or severity. The primary aim of this review is to indicate how proteomics and metabolomics have contributed, and can be expected to contribute, toward a better understanding of the TB granuloma extracted mainly from animal and human lung tissue. The secondary objective is to identify gaps in the literature that limit our understanding of the disease, in particular that there are few in vitro studies on TB granulomas in the brain. Lastly, inferences from current, as well as directions for future, ‘omics-related research on granulomas, are offered for tuberculous meningitis (TBM).
Brief Overview of TB
Pulmonary TB begins with the transmission of the Mtb bacilli, via the transfer of Mtb-containing aerosols expelled from an infected person and inhaled by a non-infected person. TB affects the lungs more than any other organ of the body, since the lungs are usually the first point of entry into the host (32, 33). When these infected air droplets enter the new host, there is a subsequent activation of the host's innate agents conferring immunity, namely, neutrophils, dendritic cells and alveolar macrophages, which phagocytose the Mtb in the terminal alveoli (6, 34). The infected immune cells from the alveoli migrate to the lymphoid tissue, activating type 1 T-helper cells, producing pro-inflammatory cytokines such as interleukins (IL) and tumor necrosis factor alpha (TNF-α). These initial immune reactions lead to inflammatory changes in the lungs (30, 34–36). The Ghon focus, the primary site of infection in the lungs, enlarges as the disease progresses or the foci heal, leading to dense scars that may calcify (37). In an attempt to contain Mtb, macrophages engulf the bacilli and destroy them. However, if the process of elimination is unsuccessful, there will be continuous multiplication of the engulfed bacilli within the macrophage. Consequently, immune cells continue to work tirelessly in an attempt to contain the spread of the Mtb. In such instances, monocytes are subsequently recruited from the bloodstream to the site of infection, and the cycle of engulfment continues (32, 38). In a further attempt to neutralize the infection, the adaptive response/delayed type hypersensitivity is activated within 2–10 weeks after exposure to the infection (38, 39). This combined immune response to the invading bacilli eventually leads to the formation of a mature granuloma. Depending on the patient's immunity, the bacilli can be contained and the disease kept dormant (latent), not causing any clinical symptoms or disease or, typically in the case of individuals with a compromised immunity, active TB in which the infection spreads in the lungs. TB can progress from a latent form to an active state through the release of Mtb from the host's immune cells that have failed to contain/kill the bacilli (21). The progression from latent to active TB can be a result of several factors, such as immune deficiency, large or recurrent exposure to the disease, insufficient immune response to the pathogen, or exposure to a more virulent Mtb strain (6, 30, 40). Globally, about 23% of the human population have latent TB. The chances of individuals developing active TB from a latent infection are between 2–23%, and are higher (7–33%) in people living with human immunodeficiency viruses and acquired immunodeficiency syndrome (30, 41, 42). Latent TB can be detected only by carrying out tests such as the tuberculin skin test, interferon-gamma (IFN-γ) release assays, or the more recently developed QuantiFERON (Qiagen, Germany) and T.SPOT (Oxford Immunotec, UK) tests. Since these tests measure the resulting immune response in vivo or in vitro only, they cannot differentiate latent TB from resolved infection or active TB (43–45).
Of the 6.4 million TB cases reported by the World Health Organization (WHO) in 2017, 14% represent EPTB cases (1). EPTB occurs as a result of the lympho-hematogenous spread of bacilli to other organs of the body (46, 47). Once Mtb enters the host's immune system, the bacilli can replicate within the immune cells, thereby inducing the rapid production of chemokines and/or cytokines, which can be detected 2 h after infection (6, 7), and can be transported as far as the immune system carrying it travels (47). Mtb escapes the infected immune cells through the region of difference (RD1) locus (this locus is mostly found in virulent strains of Mtb), where the bacilli continue to replicate in the cytosol (38). The lymphatic endothelial cells, activated by IFN-γ, are said to be capable of restricting the RD1 locus of the bacilli from replicating (48), although this does not always occur sufficiently.
Tuberculous meningitis (TBM), the most lethal form of EPTB, is a form of CNS-TB where Mtb invades the meninges of the brain (49). Despite the protective properties of the blood–brain barrier (BBB) and the cerebrospinal fluid (CSF), Mtb are still able to enter the brain. The etiology of TBM is not fully known, but based upon research using animal models, it is proposed that Mtb enters the brain through infected immune cells, such as neutrophils and macrophages (the “Trojan horse” mechanism), and/or bind to endothelial cells of the brain (50–52). Once the bacilli penetrate the CNS barriers, they cause the resident immune cells (microglia) in the brain to be activated. Activation of microglia causes the release of numerous cytokines, which are important in the host's defense against Mtb infection, and which can also mediate inflammation. Microglia, rather than astrocytes, are preferentially infected by Mtb. The resulting neuroinflammatory processes lead to the disruption of the BBB, trigger the formation of vasogenic edema and recruit immune mediators that contribute toward BBB disintegration and the influx of the innate and adaptive immune cells from the periphery into the CNS (53, 54). The subsequent accumulation of these immune cells around the Mtb leads to the brain lesions typically seen in these patients. Rich and McCordock proposed that TBM is caused by the rupture of one of these lesions, the Rich focus; they are found in both the meninges and the brain parenchyma, and usually follow a vascular pattern (55–57). Activation of the Rich foci and the release of Mtb into the subarachnoid space, leads to additional cascades of devastating neuroinflammatory events (58).
During active Mtb infection, regardless of where it occurs in the body, there is a strong immune response involving clusters of differentiation of CD4+ and CD8+ lymphocytes and cytokines, including IFN-γ and TNF-α, produced by the dominant T-helper cell subset associated with the successful control of Mtb (59, 60). The inflammatory response subsequently causes IFN-γ to act in conjunction with TNF-α in order to activate macrophages and dendritic cells. IFN-γ and TNF-α are thought to play a crucial part in the complete eradication of Mtb because of their role in enabling the macrophages to produce reactive nitrogen intermediates and phagolysosome acidification (24, 61) to contain the bacilli. They do this by enhancing, stimulating, activating and mobilizing host defenses against the Mtb. These activated macrophages are essential for the formation of a granuloma (31). The pathogen is, however, merely contained (latent), maintaining a level of supposed inactivity, while the immune system constantly tries to maintain its defense with no clinical manifestations in the host (62–64). Despite this understanding of the initial mechanisms of Mtb infection, the perplexity of TB granulomas, in general, and especially so in the brain, still needs to be elucidated.
The function, importance and development of TB granulomas are comprehensively summarized in the next two sections, in order to provide a backdrop on our existing knowledge and a basis for future work.
TB Granuloma Function and Importance
Granulomas are characterized by aggregates of lymphocytes, monocytes, plasma cells, mono-nuclear phagocytes, epithelioid cells, and giant cells – multi-nucleated fusion of immune cells, formed during chronic inflammation (29, 65, 66), formed in response to an infectious microorganism. The TB granuloma characterizes the host's immune response to an Mtb infection (67), and comprises an array of innate and adaptive immune cells, formed in response to signals given off by the infected macrophages, that trigger the migration of uninfected macrophages to the site of infection, in order to contain the Mtb invasion. However, in the case of failure to contain the bacilli, the TB granulomas can become shells in which the bacilli actually proliferate (6, 29, 65).
This poses the question: who benefits the most from the development of the granuloma – the host or the pathogen? The current precept backs the idea that a granuloma is a mark of an adequate and restrictive host immune response (68). However, studies (69–71) have shown that Mtb has the capacity to use the TB granuloma to its benefit. Upon initial infection, the bacilli recruit additional macrophages to contribute to the further spreading of the disease within the host. Macrophage infection triggers a localized pro-inflammatory response, resulting in the recruitment of activated innate immune components, including the neutrophils and dendritic cells. This then leads to the secretion of antimicrobial peptides (e.g., cathelicidin), cytokines (including IL-1α, IL-1β, TNF-α, IL-6 and IL-12), chemokines and additional macrophages that convene into a TB granuloma – a multicellular structure that cloisters the infecting Mtb from the surrounding tissue (36, 50, 72, 73). The formation of the TB granuloma is controlled by chemokines and cytokines, produced by local tissue cells and infiltrating leukocytes (74, 75). The organized, compact structure of the TB granuloma acts to constrain the Mtb infection and coordinates the activation of the immune cells, although the mechanism(s) surrounding this remains unclear (74). TB granulomas function via an interplay between the innate and cellular adaptive immunity (76), and orchestrate the interaction between immune cells, leading to an effective response to the infection: (1) the inhibition and killing of the Mtb; (2) containment of the infection and preventing the spread of the organism; or (3) localizing the inflammatory response and tissue damage (76, 77). TB granulomas can also serve as a niche for the invading Mtb (the bacilli can survive inside these structures for a relatively long time in a dormant state), while simultaneously protecting the host from active disease (78). Thus, TB granulomas have a duplicitous role. Owing to the dual role of the granuloma, the metabolism of Mtb and its survival within the host has been the subject of intense research. Because the bacilli depend completely on the host for survival, access to nutrients becomes a critical battle for survival (72). The Mtb genome subsequently evinces its metabolic flexibility and autonomy, as well as its capacity to resist nutrient stress (79). Mtb survives by producing sulfatides (which inhibit phagolysosomal fusion), cord factors (which inhibit neutrophil migration and damage mitochondria) and wax D (which controls intramural acidity, thereby making the DNA replication possible), which are all part of the Mtb cell wall (80–82). Mtb can persist and adapt to an in vivo environment by modifying gene expression; its metabolic capabilities enable the pathogen to cope in different stressful conditions within the host – (re)initiating the infectious cycle when environmental conditions are favorable (83, 84).
Changes in the number and the location of macrophages have an effect on the balance of TB granulomas and determine if the TB granuloma will effectively control the spread of the bacilli, or not (6). The structure of TB granulomas also changes as the disease progresses (85). These features are not yet completely understood and have given rise to several research studies using animal models; these, however, have not given a clear understanding of these phenomena in human granulomas (85–89). Furthermore, the TB granuloma studies performed in most animal species use other strains of mycobacteria, for instance, Mycobacterium bovis in sheep (91) and Mycobacterium marinum in zebrafish (90).
While there is much that we know about how the immune system responds to Mtb, much remains unknown. Within the tried-and-tested reductionist scientific approach, we can only answer the questions that we ask. What of the questions that we do not yet know how to ask? A new approach (‘omics) is needed to explore the dark side of the moon.
Development of TB Granulomas
During the early infection phase, the interaction between macrophages and Mtb has a negative effect on the host – dampening the immune response and causing the increased survival of the Mtb bacilli (91). It has been proposed that the most abundant antigens of Mtb could benefit the pathogen rather than the host (92). The initial aggregation of macrophages occurs in response to a persistent stimulus triggered by a granulomatous reaction at its core (93, 94). Macrophages are distinct at the core of the granuloma, undergoing a series of distinguishable morphological changes exhibiting their immunometabolic characteristics, most importantly, epithelioid cell differentiation (94). Furthermore, macrophage polarization into M1 and M2 types is finely regulated by the host for the purpose of managing chronic infection(s), thereby regulating the promotion and formation of the granuloma (65, 94–97). A pro-inflammatory response, promoted by the M1 macrophages, bridges the innate and the adaptive immune response to infection. This functionality is considered the cornerstone of an effective host defense. At the same time, the M2 macrophages promote an anti-inflammatory response, which is crucial to immune regulation, also preventing an aggravated chronic inflammatory state, and simultaneously promoting the maintenance of tissue homeostasis (94, 95, 98). Lipid-laden macrophages, also known as foamy cells, are linked to TB granuloma necrosis, since they promote the formation of caseum – the build-up of necrotic debris at the core of the granuloma. When infected, foamy cells are packed with host lipids and are consumed by Mtb through aerobic glycolysis, thus promoting inflammation as well as transmission of live bacilli and the subsequent development of disease. Studies have shown localization and migration of Mtb toward the host lipid reservoirs (67, 99, 100), and the presence of foamy macrophages in TB granulomas (80, 81, 91, 92). Foamy macrophages seem to sustain intracellular Mtb in a physiological state, which explains the persistence and nutritional advantages to the Mtb bacilli (67, 101).
Natural killer (NK) cells, as well as B and T cells, dendritic cells, and neutrophils, are also recruited and contribute to TB granuloma function (76, 95). NK cells do not require antigen-specific recognition to kill target cells. IFN-γ, produced by CD4+, CD8+ and NK cells, and T-lymphocytes, are important mediators of macrophage activation, effector function and the immune response targeted against Mtb (76, 102). The dendritic cells and macrophages are mainly responsible for proteolytic processing and antigen presentation, and their functions include: regulating the overall immune response, initiating antigen-presenting cell activation, directing T-lymphocytes, activating NK cells, augmenting IFN-γ production, and directing antigen-induced cytotoxicity, (76, 103). The dendritic cells can also serve as a replication niche after ingesting Mtb, and are essential for antigen presentation to the T cells in the draining lymph nodes. However, pathogenic Mtb bacilli have developed mechanisms to prevent both dendritic cell migration and antigen presentation – an evolved adaptation of the pathogen (104, 105). In response to inflammatory stimuli, neutrophils are of the first cells to migrate to the infection site in response to the Mtb, in order to kill the pathogen, aided by the antimicrobial molecules (defensins, lactoferrin, cathelicidin, and lysozymes) contained in their granules (106–108). However, these immune reactions which target the pathogen may also lead to destruction of the surrounding host tissue, due to high levels of neutrophil influx, contributing to the development of a pathology serving as a “Trojan horse” for the Mtb. Although much is known about the immune cells that make up TB granuloma formation, much more remains to be learned about their functions in response to promoting or preventing disease (109–111).
The hallmark of the Mtb bacillus is a thick and waxy cell wall, whose unique design makes the bacilli difficult for the host to kill, which renders it rigid and impermeable, protecting the pathogen from dehydration and also rendering it resistant to conventional antibiotics (80, 84). Of the protective antigens of Mtb, mycolyltransferase and lipoarabinomannan are considered to be the most important. Modulating interactions with the immune system plays a key role in the infection cycle of Mtb, by blocking phagosome maturation and thus preventing lysis and antigen presentation, while also creating a favorable habitable environment for Mtb to replicate (112–115). Furthermore, as the immune system becomes activated, macrophages get stimulated with IFN-γ to increase proficiency against Mtb (83).
The Mtb serine/threonine protein kinase, called PknG, regulates bacterial metabolism as well as the pathogen's ability to survive inside the host by inhibiting phagosome maturation (116, 117). Studies show that PknG expedites Mtb growth and adaptation under in vitro stress conditions, such as nutrient deprivation, acid stress, and hypoxia (118–120). Under aerobic conditions, the bacilli are predicted to access glucose and triacylglycerides as sources of carbon throughout early replication; however, glucose-deficient macrophages trigger a metabolic shift in Mtb to utilize lipids (72, 121). PknG further disrupts host macrophage metabolic homeostasis to promote the accumulation of lipid bodies, complementing the aforementioned lipid use (122).
Hitherto, this review has summarized the basis of TB and its characteristic granuloma. From here on we examine several studies that illustrate the diverse categorization of TB granulomas, followed by the contributions offered by proteomics and metabolomics. The prime point of pretension proffered is that the categorization of TB granulomas is diverse.
Categorization of TB Granulomas in Human or Animal Models
A compact ordered aggregation of macrophages and their derivatives characterize granulomatous inflammation morphologically (40). It is thought that an understanding of these derivatives involved in inflammation (initiation and maintenance) could be important in assessing the development of the lesion, which in turn could help in the design of more effective and selective therapies (123). Subsequently, this has resulted in the categorization of the various TB granulomas, based on the varying derivatives associated with inflammation and/or Mtb interaction (Table 1). One key feature that distinguishes human TB granuloma from animal granuloma types is how they are structurally organized, although, differences are observed within granuloma types, rather than between the types. This could be as a result of the tissue location, cytokines, growth factors and associated cells, while the animal granuloma types may not be as organized as a result of the type of causative agent injected into the animal in order to induce immune responses similar to that observed in human TB granuloma (6, 20, 22). Figures 1, 2 illustrate our depictions of TB granulomas, based upon the information given on the categorizations below.
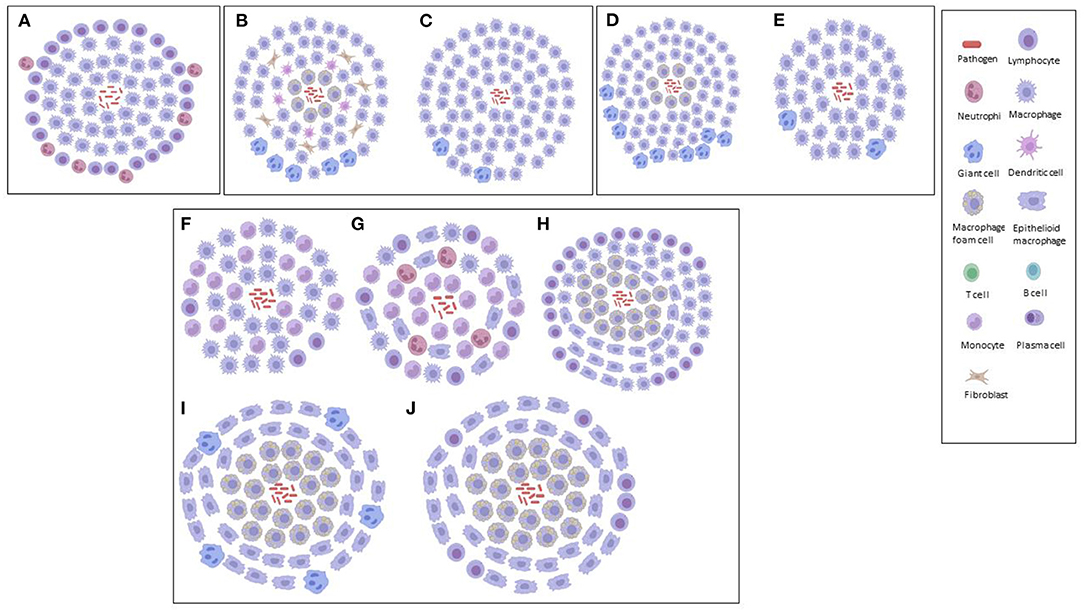
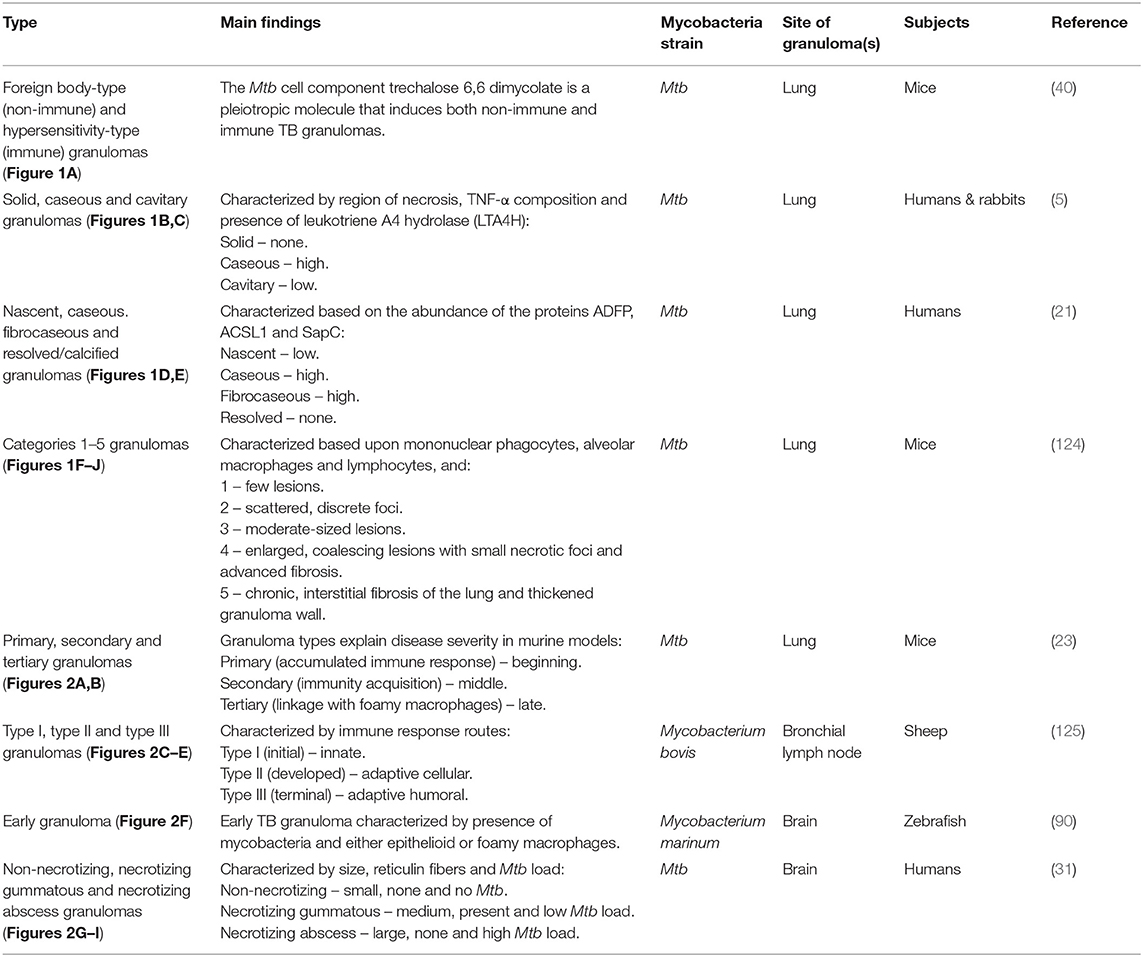
Figure 1. Graphical illustrations of TB granulomas based upon key cellular elements in their categorizations, based upon literature. (A) Foreign body-type and hypersensitivity granuloma. (B) Caseous and cavitary granuloma. (C) Solid granuloma. (D) Caseous and fibrocaseous granuloma. (E) Nasent granuloma. (F) Category 1 granuloma. (G) Category 2 granuloma. (H) Category 3 granuloma. (I) Category 4 granuloma. (J) Category 5 granuloma.
Figure 2. Graphical illustrations of TB granulomas based upon key cellular elements in their categorizations, based upon literature. (A) Primary granuloma. (B) Secondary granuloma. (C) Type 1 granuloma. (D) Type II granuloma. (E) Type III granuloma. (F) Early granuloma. (G) Non-necrotizing granuloma. (H) Necrotizing gummatous granuloma. (I) Necrotizing abscess granuloma.
Foreign Body- and Hypersensitivity-Type Granulomas
The pre-existing classification of lesions was according to their causative agents: infectious granulomas, foreign body granulomas and granulomas of unknown etiology (126, 127). Boros (128) revised this classification into the foreign body-type (non-immune) and hypersensitivity-type (immune) granuloma, in which classification was based on the involvement of antigen-specific lymphocytes in lesion development. The foreign body type was T-cell independent, whereas the hypersensitivity type was T-cell dependent. The hypersensitivity types were formed in response to foreign agents, which would require a delayed type of immunological response and would usually involve a high number of cellular responses. These agents could be bacteria, worms, fungi or viruses. The foreign body type were not formed in response to specific antigens and may be mostly inactive in the absence of specific foreign agents, such as a streptococcal cell wall, cord factor and silica (40, 123). Using a surface glycolipid derived from the cell walls of virulent strains of Mtb – trechalose 6,6 dimycolate (TDM) – Yamagami et al. (40) showed that TDM can induce both foreign body- and hypersensitivity-type granulomas in mouse lung tissue. Hence, both non-immune and immune mechanisms participate in granulomatous inflammation induced by Mtb infection.
Solid, Caseous, and Cavitary Granulomas
Classification of granulomas by Marakalala et al. (5) was conducted on the basis of the histological characteristics of the Mtb-infected lung tissue collected from rabbits and humans. Solid granulomas lacked necrosis and had only one histologically distinct region. The caseous and cavitary granulomas had fused with an airway and both displayed two histologically distinct regions. The enzyme leukotriene A4 hydrolase (LTA4H) was present in abundance in caseous granulomas, less so in cavitary granulomas, and none was present in solid granulomas. LTA4H synthesizes leukotriene B4, a pro-inflammatory eicosanoid associated with the production of TNF-α, a key component of early host control of Mtb growth. From the protein and lipid snapshots of the human and rabbit lesions analyzed, Marakalala et al. (5) hypothesized that the pathological response to TB is shaped by the precise anatomical localization of these inflammatory pathways during the development of the granuloma.
Nascent, Caseous, Fibrocaseous, and Resolved/Calcified Granulomas
Kim et al. (21) performed immunohistological analysis on lung tissues excised from TB patients. Tissues from 32 independent Mtb-infected samples (each sample had multiple granulomas) were categorized into four stages: nascent, caseous, fibrocaseous, and resolved. Their categorization of granulomas was based upon the proteins adipophilin (ADFP), acyl-CoA synthase long-chain family member 1 (ACSL1), and saposin C (SapC), as three key representatives of the lipid-modulating pathways that are impacted by the pro-inflammatory cytokine TNF-α. ADFP is a lipid droplet-associated protein present on the surface and in the core of intracellular lipid droplets formed in foam cells. ACSL1 mediates the formation of fatty acyl-CoA esters and a key component to stepin lipid biosynthesis and fatty acid degradation. SapC is involved in glycosphingolipid metabolism, and is also known to transfer mycobacterial lipid antigens, thereby activating the antigen-specific T cells. Nascent TB granulomas showed little-to-no presence of these three proteins, resolved/calcified TB granulomas showed none, and caseous and fibrocaseous TB granulomas demonstrated abundant ADFP, ACSL1 and SapC. The actual caseum of several TB granulomas revealed an abundance of ADFP, suggesting that the lipids of the caseum were likely to have been derived from the lipid droplets sequestered within foam cells upon the subsequent death of those cells. Kim et al. (21) further hypothesized that the TB granuloma undergoes a shift in lipid metabolism and accumulates host-derived lipids, supported further by the analysis of the lipid constituents of the caseous material isolated from human pulmonary TB granulomas. Hence, Kim et al. (21) proposed that granuloma formation was dependent on host lipid metabolism, mediated by the pathogen.
Categories 1–5 Granulomas
Using mice aerogenically infected with Mtb, Rhoades et al. (124) categorized granulomas into five distinct immunopathological stages based upon the lesions formed in the lungs and their level of progress and development. Category 1 – small, isolated lesions composed of a few adjacent alveoli with thickened septae, scattered throughout infected lungs, consisting primarily of mono-nuclear phagocytes, alveolar macrophages and an occasional lymphocyte. Category 2 – scattered, discrete foci of alveolitis filled with mono-nuclear phagocytes and a few epithelioid macrophages, with perivascular and peribronchiolar lymphocytes. Category 3 – moderately sized granulomatous lesions characterized by sheets of epithelioid and foamy macrophages that fill the alveoli, with mild interstitial fibrosis and tight associations of lymphocytes. Category 4 – enlarged, coalescing granulomatous lesions consisting mainly of macrophages, including epithelioid cells, large foamy macrophages, and an occasional multinucleated giant cell, with small necrotic foci, advanced fibrosis of alveolar septae and a few remaining lymphocytes. Category 5 – late spectrum granuloma degeneration associated with chronic, interstitial fibrosis of the lung and characterized by thickened alveolar septae demarcated areas filled with dead/dying epithelioid and foamy macrophages. A major limitation observed by Rhoades et al. (124) was that the Mtb-infected mice did not develop caseating granulomas as is observed in humans. Rhoades et al. (124) further postulated that in animals the cytokine milieu around the lesions is different, perhaps leading to degeneration of the centers of large granulomas and necrotic caseation, and that the migratory capacity of the mouse lymphocytes is limited.
Primary, Secondary, and Tertiary Granulomas
Also using the aerogenic infection model of mice with Mtb, Cardona et al. (23) simplified the histopathological categorization of TB granulomas in lungs as follows: primary, secondary and tertiary, based upon the immune reactions. A large accumulation of infected macrophages at the beginning of the infection defined primary granulomas, characterized by their large centers, and being surrounded by a lymphocytic mantle over time. After the acquisition of immunity against Mtb, secondary granulomas develop, in which scanty infected macrophages are surrounded by a thick lymphocytic mantle. Neighboring primary or secondary granulomas eventually become linked with foamy macrophages to form tertiary granulomas. Rhoades et al. (124) hypothesized a different histological evolution considering the migration of packed wedges of lymphocytes into granulomas, made by sheets of epithelioid and foamy macrophages, rather than foamy macrophages spreading out to the periphery of secondary granulomas (23).
Type I, Type II, and Type III Granulomas
Using sheep infected with Mycobacterium bovis, Vallejo et al. (125) analyzed the bronchial lymph nodes for the immunohistochemical characterization of TB granulomas, based upon the number of acid-fast bacilli (AFB), T cells, B cells, macrophages and plasma cells each contained. Type I TB granulomas had <10 AFBs, no plasma cells, 1–10 T-lymphocytes, 11–50 B-lymphocytes and predominantly (51–100) macrophages. Type II TB granulomas had 10–20 AFBs, 11–50 plasma cells, 1–10 T-lymphocytes, 51–100 B-lymphocytes and predominantly (>100) macrophages. Type III TB granulomas had 10–20 AFBs, 51–100 plasma cells, no T-lymphocytes, >100 B-lymphocytes, and 51–100 macrophages that appeared peripherally. Thus, in the early/latent stages, an innate immune response would be more prominent. If the bacilli are not contained, there would be an increase in the number of macrophages that would subsequently attract more lymphocytes to the site of infection (adaptive cellular). Finally, a shift to a new phase in the immune response (adaptive humoral) would occur where B-lymphocytes and plasma cells would contain the infection with a humoral component (shift toward type 2 T-helper response) (125). Furthermore, Vallejo et al. (125) demonstrated how sheep are less than ideal models for studies using mycobacteria, since these animals were highly resistant, mostly attributed to their highly effective innate immune response that prevents the spread of the infection in the initial stages.
Early Granuloma
Using an adapted zebrafish model of Mycobacterium marinum infection, van Leeuwen et al. (90) showed that the zebrafish–M. marinum model is particularly suitable for characterizing the early steps in the formation of brain granulomas, their immunological composition and the effect of bacterial virulence factors in the context of TBM, allowing for the detailed analysis of both bacterial and host factors involved in the disease. The brain is a notoriously difficult organ to analyse in terms of TB granulomas; hence, knowledge is limited. Van Leeuwen et al. (90) characterized the initial phases of zebrafish infection at the early developmental stages (innate immunity), with formation of bacterial clusters that were identified as early TB granulomas. All clusters contained both mycobacteria and a population of either epithelioid or foamy macrophages; however, their development was not influenced by changes to the BBB. The results of van Leeuwen et al. (90) suggest that traverse over the BBB occurs even at very low levels of extracellular bacterial loads, which led to their hypothesis that mycobacteria possibly make use of host cells – that is, macrophages – to migrate out of the bloodstream (which supports the concept of a “Trojan horse”).
Non-necrotizing, Necrotizing Gummatous, and Necrotizing Abscess Granulomas
In one of the only studies to categorize TB granulomas in human brain tissue, Zaharie et al. (31) described three different types of granulomas – non-necrotizing, necrotizing gummatous and necrotizing abscess-type granulomas. The non-necrotizing granulomas are the smallest (0.1–0.5 mm) of the three, containing activated macrophages, lymphocytes and plasma cells, with absence of Mtb bacilli. The necrotizing gummatous granulomas (>5 mm) contain reticulin fibers in the center of necrosis and a relatively low Mtb bacilli load. The necrotizing abscess-type granulomas (up to 10 mm) are the largest, containing a large number of neutrophils, and completely absent of reticulin fibers in the center, with a high load of Mtb bacilli. Zaharie et al. (31) postulated that these different types of granuloma are part of the developmental stages of the pathological process. These authors (31) also challenge the classical theory of the Rich focus in TBM (58), by demonstrating that chronic granulomatous inflammation is initiated almost exclusively in the leptomeninges and not in the brain parenchyma, as opposed to the Rich hypothesis, in which the bacilli spread through the subarachnoid space, causing leptomeningitis after an active pulmonary infection.
All of the granuloma types reviewed here are categorized on the basis of the research interests of the researchers and writers. Perhaps if the granulomas are categorized according to disease severity, the organ from which the granulomas are extracted and characteristics of the cells that consist the granulomas, there could be a better understanding of how granulomas function and how such functions can be channeled to better understand the TB disease. The use of the ‘omics approach (discussed next) is a step in the right direction, which will give better insight, understanding and possibly help in the diagnosis of TB in the future. Also, more human post-mortem samples (not only from brain but from other organs where granulomas form) need to be studied in order to gain better insight into how the TB granuloma forms and functions as seen in the study conducted by Zaharie et al. (31).
Challenges to TB Granuloma Categorization
Challenges to studying TB granulomas, specifically in the brain, include the lack of appropriate models and inaccessibility to human brain tissues, which has subsequently lead to the limited literature on the topic. Further limitations are due to the fact that direct inferences from the current animal models to humans cannot be made as conclusive outcomes of the disease since animals may have a different response to the disease and the mode of infection varies with that of humans; hence, the disease itself most likely presents differently. Therefore, there is still need to use human post-mortem tissue samples for the study of the human TB granulomas, as seen in the study of TBM in zebrafish in comparison with the study carried out on actual human post mortem samples, where notable differences were seen to exist between the granuloma developed in the zebrafish and that of human post-mortem samples (31, 90).
Using Proteomics and Metabolomics to Understand TB Granulomas
The increasing demand and search for faster, more accurate methodologies that best describe the biochemical/pathophysiological status of granulomas have channeled efforts in the area of biomedical research recently to achieve better diagnostic and treatment methods (129, 130). From the need to augment work in biomedical research, the ‘omics era was born (131). The two such sub-disciplines that apply in the case of TB granulomas are proteomics and metabolomics.
Proteomics
A proteome is loosely defined as all the proteins that can be expressed in a cell, tissue or in an organism, including the post-translational modified forms of proteins, and their isoforms (132). Proteomics – the study of the proteome – serves as a tool to advance biomedical research, such as vaccinology, drug discovery, and biomarker identification (133). The inherent focus of proteomics is to provide the most detailed insights into various cellular processes, by analyzing proteins, which cannot be captured by genomics or transcriptomics (134–136). Proteomics has come a long way over the last two decades and is currently in transition from its use in basic research to applications in medicine (134). During this period proteomics has contributed significantly to improving our understanding of the human pathogen Mtb, including application of improved intervention and prevention measures for TB, and providing exciting new insights into the adaptive life cycle of Mtb under different conditions, ranging from active metabolism and replication to dormancy (134, 135). Using proteomics, antigens expressed by Mtb under defined conditions with relevance for vaccine development have also been identified (137). Proteomics also enables superior characterization of proteins located in the Mtb cell wall, which previously were extremely difficult to identify (134, 138, 139). The application of proteomics has also contributed toward the understanding of TB granulomas (Table 2).
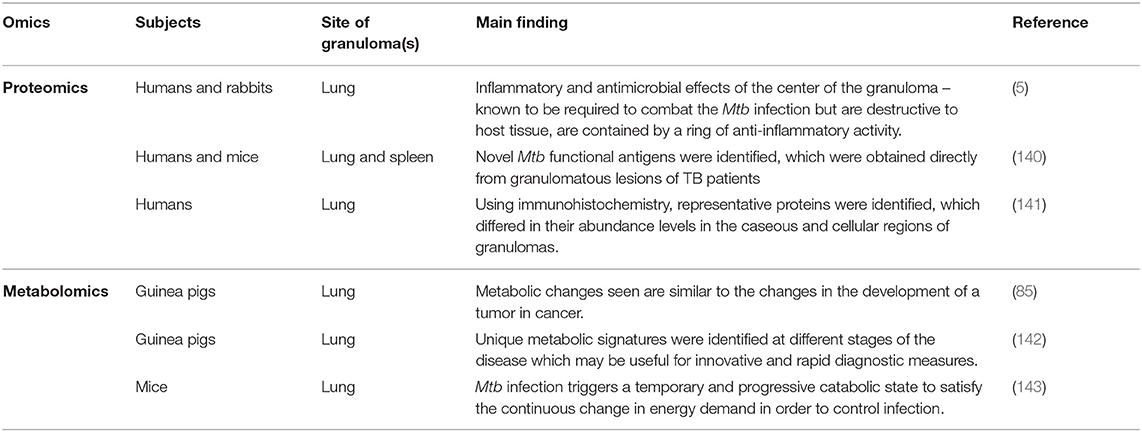
Table 2. Main findings from studies describing the role of proteomes and metabolomes in granulomas in both animal and human subjects infected with Mtb.
In a 2015 proteomics study, Marakalala et al. (5) combined laser-capture microdissection, confocal microscopy, and a Q-Exactive mass spectrometer coupled on-line to a nanoflow ultra-high-pressure liquid chromatograph (LC–MS–MS) to analyze over 3,000 proteins and a limited number of lipids during the different stages of TB granuloma formation in the lungs of humans and rabbits. Their main finding was that the center of the TB granuloma has a pro-inflammatory environment characterized by anti-microbial peptides, reactive oxygen species and pro-inflammatory eicosanoids, and that the tissue surrounding the caseum has an anti-inflammatory signature. Marakalala et al. (5) hypothesized that the pathological response to Mtb is shaped by the precise localization of these inflammatory pathways during the development of the granuloma. Hence, the organization of the TB granulomas promotes antimicrobial activity while limiting host tissue destruction. These results were consistent across both human and rabbit lung tissue samples.
Also in 2015, Yu et al. (140) used free-solution isoelectric focusing to obtain chaperone-rich cell lysates from the granulomatous lung lesions of active TB patients, and combined the technique with high-resolution orbitrap mass spectrometry to identify six Mtb-associated peptides not noted in the control samples. They further identified a peptide (PKAp) derived from Mtb protein kinase, which not only contributed to significant antigen-specific IFN secretion, but also to cytotoxic lymphocyte function and T-cell proliferation. Yu et al. further validated these results in vivo with mice immunized by the PKAp peptide, showing increased cellular IFN-γ in both the lungs and spleen, without causing immunopathogenesis/disease. Mice immunized with the PKAp peptide alone showed increased IFN- γ secretion only in lung CD4+ T cells but not in spleen cells (140).
A recent proteomics study (2020) by Seto et al. (141), applied mass spectrometry-based proteomics combined with laser microdissection, in order to investigate the unique protein markers in human mycobacterial granulomatous lesions. These researchers compared protein abundance (2,812 proteins) in the caseous regions between TB and Mycobacterium avium complex lung disease (MAC-LD) granulomas and demonstrated the expression of mycobacterial proteins by proteomic profiling, and showed a significant change in the abundance of several proteins in MAC-LD caseum relative to those of the TB caseum. Enrichment analysis further revealed that neutrophil proteins were accumulating in the caseum region. Proteins in the proteasome were abundant in the TB cell region, suggesting that antigen processing and presentation by dendritic cells and macrophages actively occur in this region (i.e., active innate immune response).
Considering the above, proteomics has shown that TB granulomas observed in human lungs have specific protein markers unique to each granuloma type; TB granulomas are different in the composition and abundance of cells that are part of the immune response; features depend on the causative agent of the disease (5, 78, 140, 141).
Metabolomics
Metabolomics is capable of quantifying all low molecular weight compounds (metabolites; collectively known as the metabolome) in biofluids, cells, tissues, and whole organisms, and is designed to quantitatively analyze and describe molecular phenotypes (144). Metabolomics is an omics technique that has been adapted for the study of infectious diseases – broadening our knowledge with respect to its use for prognostic and diagnostic purposes, specifically the screening of disease-specific biomarkers (145). Metabolomics has been, and is still being, used to study the biological mechanisms of TB both in vitro and in vivo, in animal models and human patients (146–148). The technique has revealed the carbon sources available to pathogenic Mtb in vivo (149, 150), and has led to the identification of metabolic profiles associated with TB in animal models. It has also been used to characterize human latent and active TB infection by means of NMR-based analysis of serum, as well as MS-based analysis of sputum (151–154). These studies have been reviewed by Du Preez and Luies (146) and Du Preez and Loots (153). Furthermore, metabolomics has also served as an effective research tool for the identification of specific novel TB metabolic markers (155, 156) from patients' sputum, plasma, serum and urine (151, 157–159). However, all of these metabolic markers identified to date are only associated with biofluids, with very few instances where the actual infected tissue (in particular lungs or brain) were analyzed. Hence, there is still a need to isolate the infected tissue (TB granulomas) and study the altered host and Mtb metabolism at the site(s) of infection.
Considering the latter, an ex vivo study using untargeted high-resolution magic angle spinning nuclear magnetic resonance (NMR) spectroscopy was carried out on lung tissue of guinea pigs infected with Mtb (85). The study showed significantly elevated lactate, alanine, acetate and glutamate, aspartate, creatine, phosphocholine, glycerophosphocholine, betaine, trimethylamine N-oxide (TMAO), myo-inositol, scyllo-inositol, and dihydroxyacetone, as well as reduced phosphatidylcholine, and oxidized and reduced forms of glutathione. These altered metabolites indicate utilization of alternate energy sources by the infiltrating cells that generate much of the metabolites in the increasingly necrotic and hypoxic developing granuloma, via the glycolytic, pentose phosphate, and tricarboxylic acid pathways. Furthermore, the metabolic changes seen in their study were very similar to what was previously reported in cancer during tumor development. Somashekar et al. (85) concluded that their non-destructive metabolomics approach is particularly relevant in studying the metabolic fate of Mtb during human infection and that metabolic fingerprints reflect the cycle of active replication and persistence of Mtb during granuloma formation in all stages of infection. In 2012, Somashekar et al. (143), again using NMR metabolomics, analyzed lung tissue collected from guinea pigs infected with the W-Beijing Mtb strain, and indicated a characteristic metabolic difference (in respect of acetate, alanine, glutamate, aspartate, glutathione, phosphocholine, creatine, glutathione, glycerophosphocholine, betaine, myo-inositol, TMAO, dihydroxyacetone, and scyllo-inositol) compared to the control group, which also changed as the disease state progressed. This metabolic profile closely matched that observed in their previous study (85). However, in this investigation they additionally observed an increase in glutathione in response to elevated oxygen radicals produced in the lesions. These researchers also indicated that use of NMR in this study can be used to identify changes to important pathways that occurred as a result of disease progression, and that these metabolic signatures could possibly be used in the development of improved vaccination or therapeutic strategies.
In a study on mice, using high-resolution mass spectrometry coupled with the powerful imaging technology of MALDI, Prideaux et al. (160) indicated that Mtb uses TMAO as an electron acceptor under anaerobic conditions. They additionally reported that high abundances of TMAO are produced by the host, in response to the elevated concentrations of trimethylamine, a substrate supplied by the Mtb after infection. They concluded that the lung tissue metabolome can be greatly altered as TB disease progresses, and untargeted metabolomics is an extremely valuable approach to understand better those metabolic changes associated with site-specific Mtb infection (142).
Considering the above, metabolomics can be expected to be useful in advancing our understanding of granuloma formation in TB, and in determining the unique metabolic signatures of different granuloma and tissue types (such as the brain). Of particular interest, in all three of the metabolomics studies on TB granulomas described above, TMAO was a common metabolite and so worthy of further investigation.
Mind the Gap
Table 1 shows that TB granulomas manifest great diversity, depending on the source material and whether it is associated with inflammation caused by Mtb infection. The table also reveals that our knowledge of these granulomas is predominantly based upon examination of lungs, in both human and animal studies. Only two investigations have reported EPTB granulomas – in the bronchial lymph nodes in sheep (125) and the brain in zebrafish (90). Just one study, by Zaharie et al. in 2020, has examined TB granulomas in the human brain (31). This work, which was based upon immunohistochemistry analyses and three-dimensional computer modeling, challenges the classical theory of the Rich focus (58) in TBM. Zaharie et al. demonstrated that most of the chronic granulomatous inflammation was located in the subarachnoid space (leptomeninges) and to a much lesser extent in the superficial brain parenchyma (adjacent to the pia mater). They hypothesized that the superficial intraparenchymal component is just a transpial or perivascular extension (Virchow-Robin spaces) of a leptomeningeal granuloma. Furthermore, all three granuloma types studied by Zaharie et al. showed no observable differences between adult and children, highlighting that the immune response in a developing CNS that has been impacted by an infection functions in a similar manner to that of a developed CNS; however, hydrocephalus – a physiological complication of infection, occurs in more frequency in children (31). Thus, it is very clear that we need more research on the TB granulomas in the brain, especially in children, in order to understand the immunopathogenesis of TBM – one of the major knowledge gaps identified by the US National Institutes of Health (161) at a 2018 workshop.
The advantage offered by untargeted proteomics and metabolomics is that the exploratory nature of their analyses examines the research subject with an open mind and in a holistic fashion, and permits hypotheses to be generated based upon discoveries. This non-biased research approach is ideal for providing new biological insight(s) that are otherwise difficult to achieve via the traditional reductionist scientific method. Discoveries that have been elucidated are that the metabolite TMAO plays an important role in the lung granuloma of animal models, and that the localization of specific proteins in granulomatous lesions in the lungs of human TB patients are distributed specifically to counter Mtb infection, while containing the contents of the granuloma in order to limit tissue damage. The sustained to-and-fro battle between host and Mtb is immensely complicated, which is reflected in the complexity of the TB granuloma. This complexity is compounded by the biological variability among individuals; hence, each host responds differently to the assault by the pathogen.
Although our data on the role of TB granulomas in the human brain are extremely limited, we predict that applying proteomics and metabolomics to the study of TB granulomas in the brains of human TBM cases should provide new insights into the pathogenesis of the disease. This newfound understanding could then be translated into practical application toward reducing the severity of this devastating, often fatal, neurology.
Concluding Remarks
The WHO aims to eradicate TB completely by the year 2030 (162) – an ambitious task. In order to achieve this, it is necessary to identify those living with latent TB, and to neutralize the threat before they become infectious (45). Identifying specific markers peculiar to patients with latent TB would be beneficial to tackling this challenge. The use of bioinformatics, and the other ‘omics' disciplines, have proved able to identify several potential targets for the survival of Mtb in the latently infected host (163–165).
Proteomics and metabolomics studies, using human and the increasing sophistication of animal modeling have generated new ideas about the roles of TB granulomas; nonetheless, many questions still remain unanswered regarding the host's response to TB. For example, from work on the immunology and immunometabolic events in TB granulomas (166), two major points for future research have been identified, namely: (1) What is the nature and function of the differential spatial organization of immune cells within the TB granuloma and how does the expression of the different anti-TB enzyme systems change?; and (2) as T cells are essential to constrain the spread of infection, what are the metabolic cues during TB-specific T-cell trafficking between TB granulomas and draining lymph nodes? Nevertheless, more multidisciplinary ‘omics studies, such as gene expression studies (167), especially in human subjects, are required to contribute toward ushering in a new era of understanding of TB granulomas – both at the site of infection, and on a systemic level.
Author Contributions
SM conceived the review. AS, AW, DL, and SM planned the outline and structure. AS wrote the manuscript. AW, AT, DL, MK, and SM read various drafts of the manuscript and contributed and incorporated by AS. All authors read and approved of the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. World Health Organization. Global tuberculosis report. Geneva: World Health Organization (2019). (accessed January 2021).
2. Hollo V, Beauté J, Ködmön C, van der Werf M. Tuberculosis notification rate decreases faster in residents of native origin than in residents of foreign origin in the EU/EEA, 2010 to 2015. Eurosurveillance. (2017) 22:30486. doi: 10.2807/1560-7917.ES.2017.22.12.30486
3. Thwaites G, Fisher M, Hemingway C, Scott G, Solomon T, Innes J. British Infection Society guidelines for the diagnosis and treatment of tuberculosis of the central nervous system in adults and children. J Infect. (2009) 59:167–87. doi: 10.1016/j.jinf.2009.06.011
4. Wilkinson RJ, Rohlwink U, Misra UK, Van Crevel R, Mai NTH, Dooley KE, et al. Tuberculous meningitis. Nat Rev Neurol. (2017) 13:581–98. doi: 10.1038/nrneurol.2017.120
5. Marakalala MJ, Raju RM, Sharma K, Zhang YJ, Eugenin EA, Prideaux B, et al. Inflammatory signaling in human tuberculosis granulomas is spatially organized. Nat Med. (2016) 22:531–8. doi: 10.1038/nm.4073
6. Flynn JL, Chan J, Lin P. Macrophages and control of granulomatous inflammation in tuberculosis. Mucosal Immunol. (2011) 4:271–8. doi: 10.1038/mi.2011.14
7. Algood HMS, Chan J, Flynn JL. Chemokines and tuberculosis. Cytokine Growth Factor Rev. (2003) 14:467–77. doi: 10.1016/S1359-6101(03)00054-6
8. Daniel BD, Grace GA, Natrajan M. Tuberculous meningitis in children: Clinical management & outcome. Indian J Med Res. (2019) 150:117–30. doi: 10.4103/ijmr.IJMR_786_17
9. Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, Williams D. The continuing challenges of leprosy. Clin Microbiol Rev. (2006) 19: 338–81. doi: 10.1128/CMR.19.2.338-381.2006
10. Adams LB, Pena MT, Sharma R, Hagge DA, Schurr E, Truman RW. Insights from animal models on the immunogenetics of leprosy: a review. Memorias do Instituto Oswaldo Cruz. (2012) 107:197–208. doi: 10.1590/S0074-02762012000900028
11. Kajdacsy-Balla AA, Howeedy O, Bagasra. Experimental model of congenital syphilis. Infect Immun. (1993) 61: 3559–61. doi: 10.1128/iai.61.8.3559-3561.1993
12. Law DCG, Bernstein KT, Serre ML, Schumacher CM, Leone PA, Zenilman J, et al. Modeling a syphilis outbreak through space and time using the Bayesian maximum entropy approach. Ann Epidemiol. (2006) 16:797–804. doi: 10.1016/j.annepidem.2006.05.003
13. Samokhin AO, Gauthier JY, Percival MD, Brömme D. Lack of cathepsin activities alter or prevent the development of lung granulomas in a mouse model of sarcoidosis. Respir Res. (2011). 12:1–10. doi: 10.1186/1465-9921-12-13
14. Yeager H, Gopalan S, Mathew P, Lawless O, Bellanti J A. Sarcoidosis: can a murine model help define a role for silica? Med Hypotheses. (2012) 78:36–8. doi: 10.1016/j.mehy.2011.09.036
15. Dillman JR, Stidham RW, Higgins PD., Moon DS., Johnson LA., Rubin JM. US elastography–derived shear wave velocity helps distinguish acutely inflamed from fibrotic bowel in a Crohn disease animal model. Radiology. (2013) 267:757–66. doi: 10.1148/radiol.13121775
16. Tlaxca JL, Rychak JJ, Ernst PB, Konkalmatt PR., Shevchenko TI., Pizzaro TT, et al. Ultrasound-based molecular imaging and specific gene delivery to mesenteric vasculature by endothelial adhesion molecule targeted microbubbles in a mouse model of Crohn's disease. J Control Release. (2013) 165, 216–25. doi: 10.1016/j.jconrel.2012.10.021
17. Stavitsky AB. Regulation of granulomatous inflammation in experimental models of schistosomiasis. Infect Immun. (2004) 72:1–12. doi: 10.1128/IAI.72.1.1-12.2004
18. James DG. A clinicopathological classification of granulomatous disorders. Postgrad Med J, 76(898), 457–65. doi: 10.1136/pmj.76.898.457
19. Cyktor JC, Carruthers B, Kominsky RA, Beamer GL, Stromberg P. Turner J IL-10 inhibits mature fibrotic granuloma formation during Mycobacterium tuberculosis infection. J Immunol (2013). (2000) 190:2778–90. doi: 10.4049/jimmunol.1202722
20. Guirado E, Schlesinger L. Modeling the Mycobacterium tuberculosis granuloma–the critical battlefield in host immunity and disease. Front Immunol. (2013) 4:1–7. doi: 10.3389/fimmu.2013.00098
21. Kim MJ, Wainwright HC, Locketz M, Bekker LG, Walther GB, Dittrich C, et al. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol Med. (2010) 2:258–74. doi: 10.1002/emmm.201000079
22. Ndlovu H, Marakalala MJ. Granulomas and inflammation: host-directed therapies for tuberculosis. Front Immunol. (2016) 7:1–11. doi: 10.3389/fimmu.2016.00434
23. Cardona P, Llatjos R, Gordillo S, Diaz J, Ojanguren I, Ariza A, et al. Evolution of granulomas in lungs of mice infected aerogenically with Mycobacterium tuberculosis. Scand J Immunol. (2000) 52:156–63. doi: 10.1046/j.1365-3083.2000.00763.x
24. Algood HMS, Flynn JL. CCR5-deficient mice control Mycobacterium tuberculosis infection despite increased pulmonary lymphocytic infiltration. J Immunol. (2004) 173:3287–96. doi: 10.4049/jimmunol.173.5.3287
25. Gautam US, Foreman TW, Bucsan AN, Veatch AV, Alvarez X, Adekambi T, et al. In vivo inhibition of tryptophan catabolism reorganizes the tuberculoma and augments immune-mediated control of Mycobacterium tuberculosis. PNAS. (2018) 115:E62–71. doi: 10.1073/pnas.1711373114
26. Carow B, Hauling T, Qian X, Kramnik I, Nilsson M, Rottenberg ME. Spatial and temporal localization of immune transcripts defines hallmarks and diversity in the tuberculosis granuloma. Nat Commun. (2019) 10:1–15. doi: 10.1038/s41467-019-09816-4
27. Kathamuthu GR, Kumar NP, Moideen K, Nair D, Banurekha VV, Sridhar R, et al. Matrix metalloproteinases and tissue inhibitors of metalloproteinases are potential biomarkers of pulmonary and extra-pulmonary tuberculosis. Front Immunol. (2020) 11:1–13. doi: 10.3389/fimmu.2020.00419
28. Zar H, Tannenbaum E, Apolles P, Roux P, Hanslo D, Hussey G. Sputum induction for the diagnosis of pulmonary tuberculosis in infants and young children in an urban setting in South Africa. Arch Dis Child. (2000) 82:305–8. doi: 10.1136/adc.82.4.305
29. Ramakrishnan L. Revisiting the role of the granuloma in tuberculosis. Nat Rev Immunol. (2012) 12:352–66. doi: 10.1038/nri3211
30. Saunders BM, Cooper AMJI. Restraining mycobacteria: role of granulomas in mycobacterial infections. Immunol Cell Biol. (2000) 78:334–41. doi: 10.1046/j.1440-1711.2000.00933.x
31. Zaharie S-D, Franken DJ, van der Kuip M, van Elsland S, de Bakker BS, Hagoort J, et al. The immunological architecture of granulomatous inflammation in central nervous system tuberculosis. Tuberculosis. (2020) 125:102016. doi: 10.1016/j.tube.2020.102016
32. Lyon SM, Rossman MD. Pulmonary tuberculosis. Microbiol Spectr. (2017) 5:1–13. doi: 10.1128/microbiolspec.TNMI7-0032-2016
33. Perlman DC, El-Sadr WM, Nelson ET, Matts JP, Telzak EE, Salomon N, et al. Variation of chest radiographic patterns in pulmonary tuberculosis by degree of human immunodeficiency virus-related immunosuppression. Clin Infect Dis. (1997) 25:242–6. doi: 10.1086/514546
34. O'Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP. The immune response in tuberculosis. Annu Rev Immunol. (2013) 31:475–527. doi: 10.1146/annurev-immunol-032712-095939
35. Seddon JA, Tugume L, Solomons R, Prasad K, Bahr NC, Consortium TMIR. The current global situation for tuberculous meningitis: epidemiology, diagnostics, treatment and outcomes. Wellcome Open Res. (2019) 4:167. doi: 10.12688/wellcomeopenres.15535.1
36. Manyelo CM, Solomons RS, Walzl G, Chegou NN. Tuberculous meningitis: pathogenesis, immune responses, diagnostic challenges, and the potential of biomarker-based approaches. J Clin Microbiol. (2021) 59:e01771–20. doi: 10.1128/JCM.01771-20
37. Jeong YJ, Lee KS. Pulmonary tuberculosis: up-to-date imaging and management. Am J Roentgenol. (2008) 191:834–44. doi: 10.2214/AJR.07.3896
38. Leung AN. Pulmonary tuberculosis: the essentials. Radiology. (1999) 210:307–22. doi: 10.1148/radiology.210.2.r99ja34307
39. Dannenberg AM Jr. Delayed-type hypersensitivity and cell-mediated immunity in the pathogenesis of tuberculosis. Immunol Today. (1991) 12:228–33. doi: 10.1016/0167-5699(91)90035-R
40. Yamagami H, Matsumoto T, Fujiwara N, Arakawa T, Kaneda K, Yano I, et al. Trehalose 6, 6′-dimycolate (cord factor) of mycobacterium tuberculosis induces foreign-body-and hypersensitivity-type granulomas in mice. Infect Immun. (2001) 69:810–5. doi: 10.1128/IAI.69.2.810-815.2001
41. Houben RM, Dodd PJ. The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med. (2016) 13:e1002152. doi: 10.1371/journal.pmed.1002152
42. Rajamanickam A, Munisankar S, Dolla CK, Babu S. Undernutrition is associated with perturbations in T cell-, B cell-, monocyte-and dendritic cell-subsets in latent Mycobacterium tuberculosis infection. PLoS ONE. (2019) 14:e0225611. doi: 10.1371/journal.pone.0225611
43. Pai M, Dheda K, Cunningham J, Scano F, O'Brien R. T-cell assays for the diagnosis of latent tuberculosis infection: moving the research agenda forward. Lancet Infect Dis. (2007) 7:428–38. doi: 10.1016/S1473-3099(07)70086-5
44. Haas MK, Belknap RW. Diagnostic tests for latent tuberculosis infection. Clin Chest Med. (2019) 40:829–37. doi: 10.1016/j.ccm.2019.07.007
45. Mwaba P, Chakaya JM, Petersen E, Wejse C, Zumla A, Kapata N. Advancing new diagnostic tests for latent tuberculosis infection due to multidrug-resistant strains of Mycobacterium tuberculosis—End of the road? Int J Infect Dis. (2020) 92:S69–71. doi: 10.1016/j.ijid.2020.02.011
46. Krishnan N, Robertson BD, Thwaites G. The mechanisms and consequences of the extra-pulmonary dissemination of Mycobacterium tuberculosis. Tuberculosis. (2010) 90:361–6. doi: 10.1016/j.tube.2010.08.005
47. Kumar R, Kolloli A, Singh P, Vinnard C, Kaplan G, Subbian S. Thalidomide and phosphodiesterase 4 inhibitors as host directed therapeutics for tuberculous meningitis: insights from the rabbit model. Front cell. (2020) 9:1–9. doi: 10.3389/fcimb.2019.00450
48. Lerner TR, de Souza Carvalho-Wodarz C, Repnik U, Russell MR, Borel S, Diedrich CR, et al. Lymphatic endothelial cells are a replicative niche for Mycobacterium tuberculosis. J Clin Investig. (2017) 126:1093–108. doi: 10.1172/JCI83379
49. Colas RA, Nhat LTH, Thuong NTT, Gómez EA, Ly L, Thanh HH, et al. Proresolving mediator profiles in cerebrospinal fluid are linked with disease severity and outcome in adults with tuberculous meningitis. FASEB J. (2019) 33:13028–39. doi: 10.1096/fj.201901590R
50. Davis AG, Rohlwink UK, Proust A, Figaji AA, Wilkinson RJ. The pathogenesis of tuberculous meningitis. J Leukoc Biol. (2019) 105:267–80. doi: 10.1002/JLB.MR0318-102R
51. Be NA, Kim KS, Bishai WR, Jain SK. Pathogenesis of central nervous system tuberculosis. Curr Mol Med. (2009) 9:94–9. doi: 10.2174/156652409787581655
52. Jain SK, Paul-Satyaseela M, Lamichhane G, Kim KS, Bishai WR. Mycobacterium tuberculosis invasion and traversal across an in vitro human blood-brain barrier as a pathogenic mechanism for central nervous system tuberculosis. J Infect Dis. (2006) 193:1287–95. doi: 10.1086/502631
53. Engelhardt B, Sorokin L editors. The Blood–Brain and the Blood–Cerebrospinal Fluid Barriers: Function and Dysfunction. Semin Immunopathol (2009) Berlin: Springer. doi: 10.1007/s00281-009-0177-0
54. van der Flier M, Hoppenreijs S, van Rensburg AJ, Ruyken M, Kolk AH, Springer P, et al. Vascular endothelial growth factor and blood-brain barrier disruption in tuberculous meningitis. J Pediatr Infect Dis. (2004) 23:608–13. doi: 10.1097/01.inf.0000131634.57368.45
55. Dastur DK, Manghani DK, Udani PM. Pathology and pathogenetic mechanisms in neurotuberculosis. Radiol Clin North Am. (1995) 33:733–52.
56. Donald P, Schaaf H, Schoeman JF. Tuberculous meningitis and miliary tuberculosis: the rich focus revisited. J Infect. (2005) 50:193–5. doi: 10.1016/j.jinf.2004.02.010
57. Rock RB, Olin M, Baker CA, Molitor TW, Peterson PK. Central nervous system tuberculosis: pathogenesis and clinical aspects. Clin Microbiol Rev. (2008) 21:243–61. doi: 10.1128/CMR.00042-07
58. Rich AR, McCordock HA. The pathogenesis of tuberculous meningitis. Bull John Hopkins Hosp. (1933) 52:5–37.
59. Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. (2001) 19:93–129. doi: 10.1146/annurev.immunol.19.1.93
60. Gideon HP, Flynn JL. Latent tuberculosis: what the host “sees”? J Immunol Res. (2011) 50:202–12. doi: 10.1007/s12026-011-8229-7
61. Chan J, Xing Y, Magliozzo RS, Bloom BR. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. (1992) 175:1111–22. doi: 10.1084/jem.175.4.1111
62. Barry CE, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. (2009) 7:845–55. doi: 10.1038/nrmicro2236
63. Young DB, Gideon HP, Wilkinson RJ. Eliminating latent tuberculosis. Trends Microbiol. (2009) 17:183–8. doi: 10.1016/j.tim.2009.02.005
64. World Health Organization. Latent tuberculosis infection: updated and consolidated guidelines for programmatic management. Geneva: World Health Organization (2019). (accessed January 2021).
66. Williams GT, Williams WJ. Granulomatous inflammation–a review. J Clin Pathol. (1983) 36:723–33. doi: 10.1136/jcp.36.7.723
67. Russell DG, Cardona P-J, Kim M-J, Allain S, Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol. (2009) 10:943–8. doi: 10.1038/ni.1781
68. Martinot AJ. Microbial offense vs. host defense: who controls the TB granuloma? Vet Pathol. (2017) 55:14–26. doi: 10.1177/0300985817705177
69. Davis JM, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. (2009) 136:37–49. doi: 10.1016/j.cell.2008.11.014
70. Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. (2002) 17:693–702. doi: 10.1016/S1074-7613(02)00475-2
71. Clay H, Davis JM, Beery D, Huttenlocher A, Lyons SE, Ramakrishnan L. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe. (2007) 2:29–39. doi: 10.1016/j.chom.2007.06.004
72. Warner DF. Mycobacterium tuberculosis metabolism. Cold Spring Harb Perspect Med. (2015) 5:a021121. doi: 10.1101/cshperspect.a021121
73. Yamshchikov AV, Kurbatova EV, Kumari M, Blumberg HM, Ziegler TR, Ray SM, et al. Vitamin D status and antimicrobial peptide cathelicidin (LL-37) concentrations in patients with active pulmonary tuberculosis. Am J Clin Nutr. (2010) 92:603–11. doi: 10.3945/ajcn.2010.29411
74. Ulrichs T, Kaufmann SH. New insights into the function of granulomas in human tuberculosis. J Pathol. (2006) 208:261–9. doi: 10.1002/path.1906
75. Silva Miranda M, Breiman A, Allain S, Deknuydt F, Altare F. The tuberculous granuloma: an unsuccessful host defence mechanism providing a safety shelter for the bacteria? Clin Dev Immunol. (2012) 2012:139127. doi: 10.1155/2012/139127
76. Eley BS, Beatty DW. The Basic Immunology of Tuberculosis. Tuberculosis. Edinburgh: Elsevier (2009). 75–86. p. doi: 10.1016/B978-1-4160-3988-4.00008-1
77. Sia JK, Rengarajan J. Immunology of Mycobacterium tuberculosis infections. Microbiol Spectr. (2019) 7:1–37. doi: 10.1128/microbiolspec.GPP3-0022-2018
78. Kruh NA, Troudt J, Izzo A, Prenni J, Dobos KM. Portrait of a pathogen: the mycobacterium tuberculosis proteome in vivo. PLoS ONE. (2010) 5:e13938. doi: 10.1371/journal.pone.0013938
79. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. (1998) 393:537–44. doi: 10.1038/31159
80. Moon MS, Kim SS, Moon HL, Kim DH. Mycobacterium tuberculosis in spinal tuberculosis. Asian Spine J. (2017) 11:138–49. doi: 10.4184/asj.2017.11.1.138
81. Slonczewski JL, Foster JW. Microbiology: An Evolving Science. Third International Student edition: New York, NY: WW Norton & Company (2013).
82. Saita N, Fujiwara N, Yano I, Soejima K, Kobayashi K. Trehalose 6, 6′-dimycolate (cord factor) of Mycobacterium tuberculosis induces corneal angiogenesis in rats. Infect Immun. (2000) 68:5991–7. doi: 10.1128/IAI.68.10.5991-5997.2000
83. Saviola B. Mycobacterium Tuberculosis Adaptation to Survival in a Human Host: ISBN. London (2013). doi: 10.5772/54956
84. Lima A, Leyva A, Rivera B, Portela MM, Gil M, Cascioferro A, et al. Proteome remodeling in the mycobacterium tuberculosis PknG knockout: molecular evidence for the role of this kinase in cell envelope biogenesis and hypoxia response. J Proteomics. (2021) 2021:104276. doi: 10.1016/j.jprot.2021.104276
85. Somashekar BS, Amin AG, Rithner CD, Troudt J, Basaraba R, Izzo A, et al. Metabolic profiling of lung granuloma in Mycobacterium tuberculosis infected guinea pigs: ex vivo 1H magic angle spinning NMR studies. J Proteome Res. (2011) 10:4186–95. doi: 10.1021/pr2003352
86. Hunter RL. Tuberculosis as a three-act play: a new paradigm for the pathogenesis of pulmonary tuberculosis. Tuberculosis. (2016) 97:8–17. doi: 10.1016/j.tube.2015.11.010
87. Flynn JL. Lessons from experimental Mycobacterium tuberculosis infections. Microbes Infect. (2006) 8:1179–88. doi: 10.1016/j.micinf.2005.10.033
88. Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and non-human primates. Infect Immun. (2008) 76:2333–40. doi: 10.1128/IAI.01515-07
89. Dutta NK, Illei PB, Jain SK, Karakousis PC. Characterization of a novel necrotic granuloma model of latent tuberculosis infection and reactivation in mice. Am J Pathol. (2014) 184:2045–55. doi: 10.1016/j.ajpath.2014.03.008
90. Van Leeuwen LM, Van Der Kuip M, Youssef SA, De Bruin A, Bitter W, van Furth AM, et al. Modeling tuberculous meningitis in zebrafish using Mycobacterium marinum. Dis Models Mech. (2014) 7:1111–22. doi: 10.1242/dmm.015453
91. Kumar R, Singh P, Kolloli A, Shi L, Bushkin Y, Tyagi S, et al. Immunometabolism of phagocytes during Mycobacterium tuberculosis infection. Front Mol Biosci. (2019) 6:1–20. doi: 10.3389/fmolb.2019.00105
92. Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. (2010) 42:498–503. doi: 10.1038/ng.590
93. Izbicki G, Chavko R, Banauch GI, Weiden MD, Berger KI, Aldrich TK, et al. World trade center “sarcoid-like” granulomatous pulmonary disease in New York city fire department rescue workers. Chest. (2007) 131:1414–23. doi: 10.1378/chest.06-2114
94. Wilson JL, Mayr HK, Weichhart T. Metabolic programming of macrophages: implications in the pathogenesis of granulomatous disease. Front Immunol. (2019) 10:1–22. doi: 10.3389/fimmu.2019.02265
95. Jagatia H, Tsolaki AG. The role of complement system and the immune response to tuberculosis infection. Medicina. (2021) 57:84. doi: 10.3390/medicina57020084
96. O'Neill LA, Kishton RJ, Rathmell J, A guide to immunometabolism for immunologists. Nat Rev Immunol. (2016) 16:553–65. doi: 10.1038/nri.2016.70
97. Kaye P. Granulomatous diseases. Int J Exp Pathol. (2000) 81:289–90. doi: 10.1046/j.1365-2613.2000.00171.x
98. Liu X, Liu J, Zhao S, Zhang H, Cai W, Cai M, et al. Interleukin-4 is essential for microglia/macrophage M2 polarization and long-term recovery after cerebral ischemia. Stroke. (2016) 47:498–504. doi: 10.1161/STROKEAHA.115.012079
99. Ordway D, Henao-Tamayo M, Orme IM, Gonzalez-Juarrero M. Foamy macrophages within lung granulomas of mice infected with Mycobacterium tuberculosis express molecules characteristic of dendritic cells and antiapoptotic markers of the TNF receptor-associated factor family. J Immunol. (2005) 175:3873–81. doi: 10.4049/jimmunol.175.6.3873
100. Greenwood DJ, Dos Santos MS, Huang S, Russell MRG, Collinson LM, MacRae JI, et al. Subcellular antibiotic visualization reveals a dynamic drug reservoir in infected macrophages. Science. (2019) 364:1279–82. doi: 10.1126/science.aat9689
101. Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, et al. Foamy macrophages from tuberculous patients' granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. (2008) 4:e1000204. doi: 10.1371/journal.ppat.1000204
102. Ottenhoff THM, Verreck FAW, Lichtenauer-Kaligis EGR, Hoeve MA, Sanal O, van Dissel JT. Genetics, cytokines and human infectious disease: lessons from weakly pathogenic mycobacteria and salmonellae. Nat Genet. (2002) 32:97–105. doi: 10.1038/ng0902-97
103. Van Crevel R, Ottenhoff THM, Van Der Meer JWM. Innate immunity to Mycobacterium tuberculosis. Clin Microbiol Rev. (2002) 15:294–309. doi: 10.1128/CMR.15.2.294-309.2002
104. Dheda K, Schwander SK, Zhu B, van Zyl-Smit RN, Zhang Y. The immunology of tuberculosis: from bench to bedside. Respirology. (2010) 15:433–50. doi: 10.1111/j.1440-1843.2010.01739.x
105. Wolf AJ, Linas B, Trevejo-Nuñez GJ, Kincaid E, Tamura T, Takatsu K, et al. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol. (2007) 179:2509–19. doi: 10.4049/jimmunol.179.4.2509
106. Korbel DS, Schneider BE, Schaible UE. Innate immunity in tuberculosis: myths and truth. Microb Infect. (2008) 10:995–1004. doi: 10.1016/j.micinf.2008.07.039
107. Faurschou M, Borregaard N. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. (2003) 5:1317–27. doi: 10.1016/j.micinf.2003.09.008
108. Martineau AR, Newton SM, Wilkinson KA, Kampmann B, Hall BM, Nawroly N, et al. Neutrophil-mediated innate immune resistance to mycobacteria. J Clin Investig. (2007) 117:1988–94. doi: 10.1172/JCI31097
109. Eruslanov EB, Lyadova IV, Kondratieva TK, Majorov KB, Scheglov IV, Orlova MO, et al. Neutrophil responses to Mycobacterium tuberculosis infection in genetically susceptible and resistant mice. Infect Immun. (2005) 73:1744–53. doi: 10.1128/IAI.73.3.1744-1753.2005
110. Abadie V, Badell E, Douillard P, Ensergueix D, Leenen PJ, Tanguy M, et al. Neutrophils rapidly migrate via lymphatics after Mycobacterium bovis BCG intradermal vaccination and shuttle live bacilli to the draining lymph nodes. Blood. (2005) 106:1843–50. doi: 10.1182/blood-2005-03-1281
111. Nandi B, Behar SM. Regulation of neutrophils by interferon-γ limits lung inflammation during tuberculosis infection. J Exp Med. (2011) 208:2251–62. doi: 10.1084/jem.20110919
112. Schlesinger LS, Hull SR, Kaufman TM. Binding of the terminal mannosyl units of lipoarabinomannan from a virulent strain of Mycobacterium tuberculosis to human macrophages. J Immunol. (1994) 152:4070–9.
113. Brennan PJ. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis. (2003) 83:91–7. doi: 10.1016/S1472-9792(02)00089-6
114. Fratti RA, Chua J, Vergne I, Deretic V. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. PNAS. (2003) 100:5437–42. doi: 10.1073/pnas.0737613100
115. Maeda N, Nigou J, Herrmann J-L, Jackson M, Amara A, Lagrange PH, et al. The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. J Biol Chem. (2003) 278:5513–6. doi: 10.1074/jbc.C200586200
116. Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, et al. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. (2004) 304:1800–4. doi: 10.1126/science.1099384
117. Gil M, Lima A, Rivera B, Rossello J, Urdániz E, Cascioferro A, et al. New substrates and interactors of the mycobacterial Serine/Threonine protein kinase PknG identified by a tailored interactomic approach. J Proteomics. (2019) 192:321–33. doi: 10.1016/j.jprot.2018.09.013
118. Cowley S, Ko M, Pick N, Chow R, Downing KJ, Gordhan BG, et al. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol. (2004) 52:1691–702. doi: 10.1111/j.1365-2958.2004.04085.x
119. Khan MZ, Bhaskar A, Upadhyay S, Kumari P, Rajmani RS, Jain P, et al. Protein kinase G confers survival advantage to Mycobacterium tuberculosis during latency-like conditions. J Biol Chem. (2017) 292:16093–108. doi: 10.1074/jbc.M117.797563
120. Paroha R, Chourasia R, Mondal R, Chaurasiya SK. PknG supports mycobacterial adaptation in acidic environment. Mol Cell Biochem. (2018) 443:69–80. doi: 10.1007/s11010-017-3211-x
121. Boshoff HI, Barry CE III. Tuberculosis - metabolism and respiration in the absence of growth. Nat Rev Microbiol. (2005) 3:70–80. doi: 10.1038/nrmicro1065
122. Singh V, Jamwal S, Jain R, Verma P, Gokhale R, Rao KV. Mycobacterium tuberculosis-driven targeted recalibration of macrophage lipid homeostasis promotes the foamy phenotype. Cell Host Microbe. (2012) 12:669–81. doi: 10.1016/j.chom.2012.09.012
123. Kunkel SL, Chensue SW, Strieter RM, Lynch JP, Remick DG. Cellular and molecular aspects of granulomatous inflammation. Am J Respir Cell Mol Biol. (1989) 1:439–47. doi: 10.1165/ajrcmb/1.6.439
124. Rhoades E, Frank A, Orme IM. Progression of chronic pulmonary tuberculosis in mice aerogenically infected with virulent Mycobacterium tuberculosis. Tuberc Lung Dis. (1997) 78:57–66. doi: 10.1016/S0962-8479(97)90016-2
125. Vallejo R, Marín JFG, Juste RA, Muñoz-Mendoza M, Salguero FJ, Balseiro A. Immunohistochemical characterization of tuberculous lesions in sheep naturally infected with Mycobacterium bovis. BMC Vet Res. (2018) 14:1–7. doi: 10.1186/s12917-018-1476-2
126. Present D, Lindner A, Janowitz H. Granulomatous diseases of the gastrointestinal tract. Ann Rev Med. (1966) 17:243–56. doi: 10.1146/annurev.me.17.020166.001331
127. Warren KS. A functional classification of granulomatous inflammation. Ann NY Acad Sci. (1976) 278:7–18. doi: 10.1111/j.1749-6632.1976.tb47011.x
128. Boros DL. Granulomatous inflammations. Progress Allergy. (1978) 24:183–267. doi: 10.1159/000401230
129. Lima EO, Esteves CZ, Oliveira DN, Guerreiro TM, Catharino RR. Mass spectrometry and metabolomics—New approaches for helminth biochemical studies. Human Helminthiasis: IntechOpen. (2017). doi: 10.5772/65040
130. Hare DJ, New EJ. On the outside looking in: redefining the role of analytical chemistry in the biosciences. ChemComm. (2016) 52:8918–34. doi: 10.1039/C6CC00128A
131. Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. (2017) 18:1–15. doi: 10.1186/s13059-017-1215-1
132. Tyers M, Mann M. From genomics to proteomics. Nature. (2003) 422:193–7. doi: 10.1038/nature01510
133. da Costa JP, Carvalhais V, Ferreira R, Amado F, Vilanova M, Cerca N, et al. Proteome signatures—how are they obtained and what do they teach us? Appl Microbiol Biotechnol. (2015) 99:7417–31. doi: 10.1007/s00253-015-6795-7
134. Gengenbacher M, Mouritsen J, Schubert O, Aebersold R, Kaufmann S. Mycobacterium tuberculosis in the proteomics era. Microbiol Spectr. (2014) 239–60. doi: 10.1128/microbiolspec.MGM2-0020-2013
135. Bisht D, Sharma D, Sharma D, Singh R, Gupta VK. Recent insights into Mycobacterium tuberculosis through proteomics and implications for the clinic. Expert Rev Proteomics. (2019) 16:443–56. doi: 10.1080/14789450.2019.1608185
136. Southan C. Has the yo-yo stopped? An assessment of human protein-coding gene number. Proteomics. (2004) 4:1712–26. doi: 10.1002/pmic.200300700
137. Galassie AC, Link AJ. Proteomic contributions to our understanding of vaccine and immune responses. Proteomics Clin Appl. (2015) 9:972–89. doi: 10.1002/prca.201500054
138. Kelkar DS, Kumar D, Kumar P, Balakrishnan L, Muthusamy B, Yadav AK, et al. Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol Cell Proteomics. (2011) 10:M111.011445. doi: 10.1074/mcp.M111.011627
139. Schubert OT, Mouritsen J, Ludwig C, Röst HL, Rosenberger G, Arthur PK, et al. The Mtb proteome library: a resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe. (2013) 13:602–12. doi: 10.1016/j.chom.2013.04.008
140. Yu Y, Jin D, Hu S, Zhang Y, Zheng X, Zheng J, et al. A novel tuberculosis antigen identified from human tuberculosis granulomas. Mol Cell Proteomics. (2015) 14:1093–103. doi: 10.1074/mcp.M114.045237
141. Seto S, Morimoto K, Yoshida T, Hiramatsu M, Hijikata M, Nagata T, et al. Proteomic profiling reveals the architecture of granulomatous lesions caused by tuberculosis and Mycobacterium avium complex lung disease. Front Microbiol. (2020) 10:1–15. doi: 10.3389/fmicb.2019.03081
142. Fernández-García M, Rey-Stolle F, Boccard J, Reddy VP, García A, Cumming BM, et al. Comprehensive examination of the mouse lung metabolome following Mycobacterium tuberculosis infection using a multiplatform mass spectrometry approach. J Proteome Res. (2020) 19:2053–70. doi: 10.1021/acs.jproteome.9b00868
143. Somashekar BS, Amin AG, Tripathi P, MacKinnon N, Rithner CD, Shanley CA, et al. Metabolomic signatures in guinea pigs infected with epidemic-associated W-Beijing strains of Mycobacterium tuberculosis. J Proteome Res. (2012) 11:4873–84. doi: 10.1021/pr300345x
144. Weckwerth W. Metabolomics: an integral technique in systems biology. Bioanalysis. (2010) 2:829–36. doi: 10.4155/bio.09.192
145. Tounta V, Liu Y, Cheyne A, Larrouy-Maumus G. Metabolomics in infectious diseases and drug discovery. Mol Omics. (2021) 17:376–93. doi: 10.1039/D1MO00017A
146. Du Preez I, Luies L. The application of metabolomics toward pulmonary tuberculosis research. Tuberculosis. (2019) 115:126–39. doi: 10.1016/j.tube.2019.03.003
147. Fernández-García M, Rojo D, Rey-Stolle F, García A, Barbas C. Metabolomic-based methods in diagnosis and monitoring infection progression. Metab Interact Infect. (2018) 109:283–315. doi: 10.1007/978-3-319-74932-7_7
148. Mirsaeidi M, Banoei MM, Winston BW, Schraufnagel DE. Metabolomics: applications and promise in mycobacterial disease. Ann Am Thorac Soc. (2015) 12:1278–87. doi: 10.1513/AnnalsATS.201505-279PS
149. Rhee KY, de Carvalho LPS, Bryk R, Ehrt S, Marrero J, Park SW, et al. Central carbon metabolism in Mycobacterium tuberculosis: an unexpected frontier. Trends Microbiol. (2011) 19:307–14. doi: 10.1016/j.tim.2011.03.008
150. Beste DJV, Nöh K, Niedenführ S, Mendum TA, Hawkins ND, Ward JL, et al. 13C-flux spectral analysis of host-pathogen metabolism reveals a mixed diet for intracellular Mycobacterium tuberculosis. Chem Biol. (2013) 20:1012–21. doi: 10.1016/j.chembiol.2013.06.012
151. Zhou A, Ni J, Xu Z, Wang Y, Lu S, Sha W, et al. Application of 1H NMR spectroscopy-based metabolomics to sera of tuberculosis patients. J Proteome Res. (2013) 12:4642–9. doi: 10.1021/pr4007359
152. Shin JH, Yang JY, Jeon BY, Yoon YJ, Cho SN, Kang YH, et al. 1H NMR-based metabolomic profiling in mice infected with Mycobacterium tuberculosis. J Proteome Res. (2011) 10:2238–47. doi: 10.1021/pr101054m
153. Du Preez I, Loots DT. New sputum metabolite markers implicating adaptations of the host to Mycobacterium tuberculosis, and vice versa. Tuberculosis. (2013) 93:330–7. doi: 10.1016/j.tube.2013.02.008
154. Che N, Cheng J, Li H, Zhang Z, Zhang X, Ding Z, et al. Decreased serum 5-oxoproline in TB patients is associated with pathological damage of the lung. Clin Chim Acta. (2013) 423:5–9. doi: 10.1016/j.cca.2013.04.010
155. Schmidt CW. Metabolomics: what's happening downstream of DNA. Environ Health Perspect. (2004) 112:A410–A5. doi: 10.1289/ehp.112-a410
156. Illig T, Gieger C, Zhai G, Römisch-Margl W, Wang-Sattler R, Prehn C, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. (2010) 42:137–41. doi: 10.1038/ng.507
157. Sun L., Li J-q, Ren N, Qi H, Dong F, Xiao J, et al. Utility of novel plasma metabolic markers in the diagnosis of pediatric tuberculosis: a classification and regression tree analysis approach. J Proteome Res. (2016) 15:3118–25. doi: 10.1021/acs.jproteome.6b00228
158. Luies L, Mienie J, Motshwane C, Ronacher K, Walzl G, Loots DT. Urinary metabolite markers characterizing tuberculosis treatment failure. Metabolomics. (2017) 13:1–10. doi: 10.1007/s11306-017-1261-4
159. Deng J, Liu L, Yang Q, Wei C, Zhang H, Xin H, et al. Urinary metabolomic analysis to identify potential markers for the diagnosis of tuberculosis and latent tuberculosis. Arch Biochem Biophys. (2021) 704:108876. doi: 10.1016/j.abb.2021.108876
160. Prideaux B, Dartois V, Staab D, Weiner DM, Goh A, Via LE, et al. High-Sensitivity MALDI-MRM-MS imaging of moxifloxacin distribution in tuberculosis-infected rabbit lungs and granulomatous lesions. Anal Chem. (2011) 83:2112–8. doi: 10.1021/ac1029049
161. Jain SK, Tobin DM, Tucker EW, Venketaraman V, Ordonez AA, Jayashankar L, et al. Tuberculous meningitis: a roadmap for advancing basic and translational research. Nat Immunol. (2018) 19:521–5. doi: 10.1038/s41590-018-0119-x
162. World Health Organization. The end TB strategy. Geneva: World Health Organization (2015). (accessed January 2021).
163. Pan L, Wei N, Jia H, Gao M, Chen X, Wei R, et al. Genome-wide transcriptional profiling identifies potential signatures in discriminating active tuberculosis from latent infection. Oncotarget. (2017) 8:112907. doi: 10.18632/oncotarget.22889
164. Seidi K, Jahanban-Esfahlan R. A novel approach to eradicate latent TB: based on resuscitation promoting factors. J Medical Hypotheses Ideas. (2013) 7:69–74. doi: 10.1016/j.jmhi.2013.04.002
165. Campaniço A, Harjivan SG, Warner DF, Moreira R, Lopes F. Addressing latent tuberculosis: new advances in mimicking the disease, discovering key targets, and designing hit compounds. Int J Mol Sci. (2020) 21:8854. doi: 10.3390/ijms21228854
166. Qualls JE, Murray PJ editors. Immunometabolism Within the Tuberculosis Granuloma: Amino Acids, Hypoxia, and Cellular Respiration. Semin Immunopathology (2016) Berlin: Springer. doi: 10.1007/s00281-015-0534-0
Keywords: tuberculosis (TB), tuberculous meningitis, innate immunity, adaptive immunity, tuberculous granuloma, metabolomics, proteomics, biomarkers
Citation: Sholeye AR, Williams AA, Loots DT, Tutu van Furth AM, van der Kuip M and Mason S (2022) Tuberculous Granuloma: Emerging Insights From Proteomics and Metabolomics. Front. Neurol. 13:804838. doi: 10.3389/fneur.2022.804838
Received: 29 October 2021; Accepted: 24 February 2022;
Published: 21 March 2022.
Edited by:
Elizabeth W. Tucker, Johns Hopkins Medicine, United StatesReviewed by:
Cornelia A. Deeg, Ludwig Maximilian University of Munich, GermanyVenkata Ramanarao Parasa, Linköping University, Sweden
Copyright © 2022 Sholeye, Williams, Loots, Tutu van Furth, van der Kuip and Mason. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shayne Mason, nmr.nwu@gmail.com
 Abisola Regina Sholeye
Abisola Regina Sholeye Aurelia A. Williams
Aurelia A. Williams Du Toit Loots
Du Toit Loots A. Marceline Tutu van Furth
A. Marceline Tutu van Furth Martijn van der Kuip
Martijn van der Kuip Shayne Mason
Shayne Mason