Challenges in characterization of transcriptomes of extracellular vesicles and non-vesicular extracellular RNA carriers
- 1Faculty of Biology and Biotechnology, HSE University, Moscow, Russia
- 2Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry RAS, Moscow, Russia
- 3Hertsen Moscow Oncology Research Center, Moscow, Russia
- 4Art Photonics GmbH, Berlin, Germany
Since its original discovery over a decade ago, extracellular RNA (exRNA) has been found in all biological fluids. Furthermore, extracellular microRNA has been shown to be involved in communication between various cell types. Importantly, the exRNA is protected from RNases degradation by certain carriers including membrane vesicles and non-vesicular protein nanoparticles. Each type of carrier has its unique exRNA profile, which may vary depending on cell type and physiological conditions. To clarify putative mechanisms of intercellular communication mediated by exRNA, the RNA profile of each carrier has to be characterized. While current methods of biofluids fractionation are continuously improving, they fail to completely separate exRNA carriers. Likewise, most popular library preparation approaches for RNA sequencing do not allow obtaining exhaustive and unbiased data on exRNA transcriptome. In this mini review we discuss ongoing progress in the field of exRNA, with the focus on exRNA carriers, analyze the key methodological challenges and provide recommendations on how the latter could be overcome.
1 Introduction
Since its initial discovery in 2008, exRNA has been found in all human biological fluids including blood, saliva, milk, urine, bile, sweat, sputum, lacrimal, seminal, amniotic, cerebrospinal, synovial fluids, and ascites (Mateescu et al., 2022). In the following years, it became apparent that exRNA represents a complex mixture of RNase-stable RNA biotypes and their parts, including miRNA, tRNA, YRNA, snRNA, sno/scaRNA, vault RNA, piRNA, mRNA, lncRNA, rRNA, and circRNA (Dellar et al., 2022). To remain stable in RNase-rich cell-free milieu, exRNA must be protected by association with a certain carrier (Arroyo et al., 2011; Turchinovich et al., 2011).
To date, experimentally confirmed exRNA carriers include extracellular vesicles (EVs), RNA binding proteins (RBPs), and large protein aggregates designated as “exomeres” and “supermeres” (Figure 1). EVs of various sizes remain the most studied exRNA carriers (Mateescu et al., 2022). Primarily, mammalian cells secrete microvesicles (MVs, also known as shedding vesicles), which are 50–1,000 nm in diameter and are formed by outward budding of the plasma membrane. The second type of EVs, the exosomes, have a diameter of 30–150 nm. They initially arise as intraluminal vesicles budding off the interior of the multivesicular bodies (MVBs) and penetrate into extracellular space upon fusion of MVBs with the plasma membrane (Figure 1) (Van Niel et al., 2022; Costa et al., 2023; Dixson et al., 2023). Cells typically secrete between 10 and 2,000 EVs of different types per cell per day (Barman et al., 2022; Garcia-Martin et al., 2022). Integrins and selectins remain well-confirmed MV markers along with membrane-associated proteins from parental cells. On the contrary, exosomes can be differentiated by the presence of tetraspanins, including CD9, CD63, and CD81 (Escola et al., 1998; Abache et al., 2007). Apoptotic bodies–the third type of membrane vesicles in the extracellular milieu–are typically between 1,000 and 5,000 nm in size and may contain cell organelles as well as nuclear fractions. Finally, the “oncosomes,” membrane vesicles exported by cancer cells, have been referred as a separate type of exRNA carriers by some authors (Di Vizio et al., 2012; Morello et al., 2013; Dixson et al., 2023). Apart from various proteins, the EV cargo typically includes complex RNA transcriptome. Thus, several online databases, including Vesiclepedia (Kalra et al., 2012), EVpedia (Kim et al., 2015), ExoCarta (Keerthikumar et al., 2016), and EV-ADD (Tsering et al., 2022) have been recently created to systematize the content of various EV types in human liquid biopsy samples.
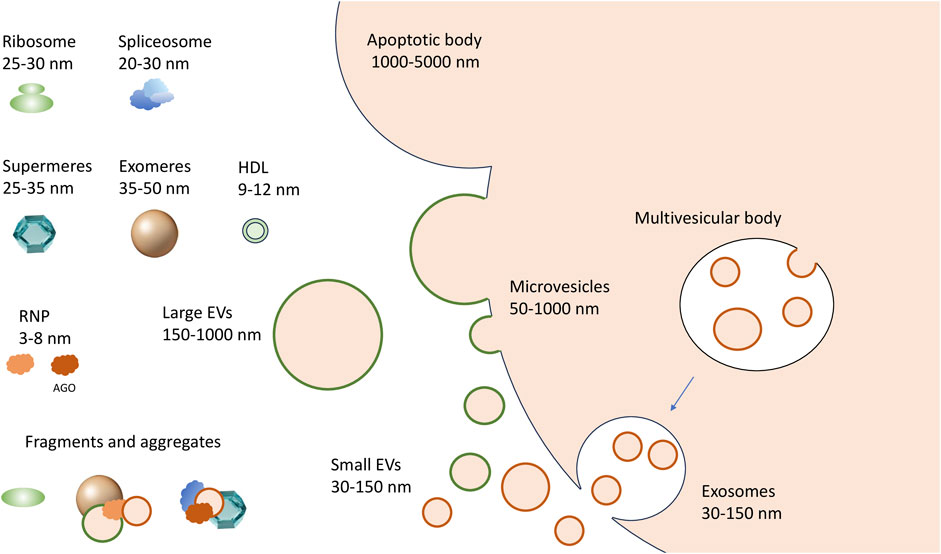
FIGURE 1. A complex spectrum of extracellular RNA carriers. Microvesicles are generated by outward budding (shedding out) from the cell membrane; the exosomes are formed by the fusion of multivesicular bodies with the cell membrane, while larger apoptotic bodies are formed during programmed cell death. The non-vesicular carriers include exomeres and supermeres. The RNP complexes that can be released from dying or damaged cells include ribosomes, spliceosomes and their fragments, and other RNA-protein complexes of different sizes, up to single proteins in complex with RNA, like AGO-miRNA complexes.
Recently, two novel types of non-vesicular nanoparticles–exomeres and supermeres–have been isolated from the conditioned media of different cultured cells. Specifically, 35–50 nm exomeres were purified by asymmetric field-flow fractionation (AF4) (Zhang et al., 2018) or ultracentrifugation at 167,000 g (Zhang et al., 2019). Upon precipitation of exomeres, the supernatant was further subjected to high-speed ultracentrifugation at 367,000 g, resulting in sedimentation of smaller 25–35 nm particles named “supermeres” (Zhang et al., 2021). Both types of particles contained metabolic enzymes, resident cytoplasmic proteins, and significant amounts of short RNA. However, it remains unanswered if exomeres and supermeres are to any extent different from the aggregates of various RNPs which are also abundant in the extracellular environment (Tosar et al., 2022). Finally, certain miRNAs have been found in isolates of lipoprotein particles (HDL, VLDL and LDL) (Vickers et al., 2011; Florijn et al., 2019; Clément et al., 2021), but their association with VLDL and LDL remains controversial (Srinivasan et al., 2019; Rossi-Herring et al., 2023).
To understand the exact role of an exRNA carrier in cell-cell communication and to explore its potential as a disease biomarker, it is crucial to characterize the associated transcriptomes. However, the latter task has proven to be unexpectedly challenging. Primarily, the size of different EV types can be identical, while protein markers specific for each EV type have not been yet established, and apparently may not exist at all (Van Niel et al., 2022; Hendrix et al., 2023). Because only a mixed population of EVs can be obtained in any experimental setting described so far, the terms “large EV” (lEV) and “small EV” (sEV) are now increasingly used instead of “microvesicle” and “exosome,” respectively (Figure 1) (Mathieu et al., 2019). Secondly, certain non-vesicular particles and their aggregates have sizes and densities identical to EVs (Mathieu et al., 2019). Therefore, none of the existing bulk isolation methods secures complete separation of extracellular RNA carriers. In addition, a lack of standardization in applying particular approaches can result in significant discrepancies between the results obtained by different research groups. Combinations of several methods usually give significantly better separation but can be too labor-intensive and require large sample volumes. Thus, the development of more accessible and efficient techniques for separating exRNA carriers, as well as their standardization accepted by the EV community, remains of paramount importance (Théry et al., 2018; Hendrix et al., 2023).
In the following parts of the manuscript, we discuss the reported transcriptomes of various exRNA carriers and key points for their characterization, highlighting current trends, advances and challenges in the field.
2 Transcriptome of extracellular vesicles
To date, hundreds of original research reports have addressed transcriptomes of various biological fluids and individual exRNA carriers. As a result, several databases accumulating information on exRNA profiles in whole biological fluids and individual types of carriers have been created. For instance, the exRNA Atlas, established by the Extracellular RNA Communication Consortium, is the most extensive database predominantly featuring small RNA-seq and RT-qPCR data, encompassing both whole biofluids and EV-specific datasets (Murillo et al., 2019). The EVAtlas includes small RNA-seq data on EV fractions only (Liu et al., 2022), while exoRBase is focused on long RNA content in EV (Lai et al., 2022). Overall, exRNA Atlas data contain miRNAs (19.1%), mRNA fragments (13.8%), tRNAs (10.1%), piRNAs (0.8%) and other genome sequences (45.9%) (rRNAs are excluded on preprocessing stage). According to EVAtlas, EVs from different biological fluids and conditioned media include rRNA fragments (38.12%), miRNAs (26.94%), tRNAs (19.19%), Y RNAs (12.59%), piRNAs (1.51%), snRNAs (1.05%), and snoRNAs (0.6%).
However, the results obtained by different laboratories are highly heterogeneous. Thus, analysis of 2,756 small RNA-seq datasets from 83 studies of EV transcriptomes showed that the distribution of different RNA biotypes in EVs was poorly coherent between reports even for the same biofluid (Wang et al., 2023a). On average, the major RNA fractions in EVs isolated from blood plasma were miRNAs (39.6%), Y RNAs (15.9%), rRNA fragments (10.5%) and tRNAs (3.2%), while snRNA, sno/scaRNA, tRNA, and other RNAs accounted for less than 1%. As expected, the distribution of different exRNA biotypes strongly depended on the EVs isolation method. Notably, according to most reports, the ratio of miRNAs found in EVs was significantly lower than in cells, while tRNA and rRNA proportions were higher (Wang et al., 2023a).
Apart from short RNAs, EVs isolates contain long RNA molecules detected by 3rd-generation sequencing technologies (Padilla et al., 2023). Specifically, EVs have been shown to contain considerable amounts of intact RNA molecules (Rodosthenous et al., 2020; O’Grady et al., 2022), with full-length mRNAs enriched in lEVs (Padilla et al., 2023). mRNAs are represented mainly by the 3′UTRs (Wei et al., 2017). A total of 19,643 mRNAs and 15,646 lncRNAs, as well as pseudogenes and circRNAs were detected by long RNA sequencing in EVs according to the current release of exoRBase (Lai et al., 2022). Full-length tRNAs and YRNAs were also identified in EVs (Shurtleff et al., 2017).
Unlike sEVs, the lEVs transcriptome composition resembles that of parent cells and contains many full-length RNA molecules, including intact rRNA (Jeppesen et al., 2019). However, the reported sEVs and lEVs transcriptomes are highly heterogeneous and poorly reproducible.
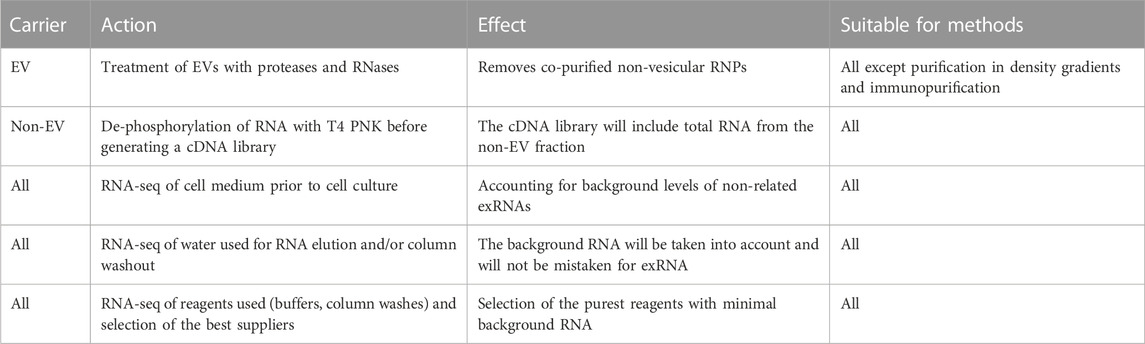
Considerable variations in exRNA profiles obtained by different studies, along with limitations of current methodological approaches, facilitated the development of the so-called computational deconvolution technique, which allows to identify the ratio of different types of exRNA carriers in a given sample based on RNA-seq data from whole biological fluids and “training samples” corresponding to individual kinds of exRNA carriers (Murillo et al., 2019; Srinivasan et al., 2019). However, the precision of the method mentioned above highly depends on the quality of the original training samples, which ideally should be obtained from the cleanest possible media with well-documented identities. In their recent report, LaPlante et al. (2023) proposed an integrative analysis of eCLIP data, which implied the identification of 150 RBPs binding sites in more than 6,000 human EV samples. Specifically, 34 RBPs were indeed detected in blood plasma and conditioned media. Remarkably, the deconvolution method described above enabled attributing RBPs to certain exRNA carriers (LaPlante et al., 2023).
The overall amount of RNA associated with a given exRNA carrier is poorly understood and highly debated. Thus, several authors reported a very low proportion of EV RNA relative to the entire exRNA pool. Furthermore, blood plasma EVs isolates pretreated with protease and RNase (along with SEC-isolated EVs) had very marginal RNA content compared to that in the original plasma (Galvanin et al., 2019). Surprisingly, two research groups reported that, on average, less than one miRNA molecule is associated with sEV (Chevillet et al., 2014; Wei et al., 2017). Recently, RNA derived from gut microbiota and viruses was detected in plasma EV (Galvanin et al., 2019; Wang et al., 2023b). Although EVs with bacterial LPS were indeed found in blood plasma (Tulkens et al., 2020), great care is needed in exogenous EV RNA studies because when RNA concentrations in sequencing preparations are very low, the contribution of contaminating RNAs, which are found in both water, reagents, and columns for RNA isolation, becomes significant (Heintz-Buschart et al., 2018). Therefore, to confirm the conclusions of works such as the ones described above, it is necessary to perform control sequencing of at least water and, preferably, of washes from RNA extraction columns, if they are used, as well as the main reagents.
Nevertheless, cell-cell communication through EV-associated RNA has been hypothesized to occur via subpopulations of EVs with high RNA content, including miRNAs, and these EVs were recently found (Lee et al., 2019a; Barman et al., 2022), suggesting them as promising candidates for intercellular signaling.
3 Transcriptome of non-vesicular exRNA carriers
3.1 Non-vesicular exRNA carriers in sEV fraction
Until recently, differential ultracentrifugation at 100,000 g followed by precipitation remained a gold standard for sEV isolation (Théry et al., 2006). However, careful analysis of sEV pellets in iodixanol gradients revealed the presence of a distinct non-vesicular fraction (NVF) enriched with metabolic enzymes (GAPDH, PKM, ENO1), cytoplasmic proteins (HSP90, tubulins, ribosome proteins and translation factors), histones, and intact vault particles. In addition, all cellular RNA classes have been detected in NVF by small RNA sequencing. Interestingly, NVF RNA profiles differed significantly from both intracellular and sEV ones. Specifically, NVF was highly enriched in vault RNA, while most overrepresented miRNAs were associated with NVF and not sEVs. Furthermore, NVF fraction contained almost all AGO proteins. Long RNA sequencing revealed no significant differences between NVF and sEV exRNA profiles, with more than 40% of all reads attributed to mRNAs (Jeppesen et al., 2019).
3.2 Transcriptome of exomeres and supermeres
While both exomeres and supermeres contain RNA, the latter’s distribution was very uneven. Specifically, only 10% RNA was found in the exomeres, whereas the supermeres fraction contained ∼65%, and the remaining ∼25% RNA was associated with sEV-NVF (precipitate after ultracentrifugation at 167,000 g) (Zhang et al., 2021). However, the RNA content of lEV and supermeres supernatants was not reported, and thus the full spectrum of exRNA remained unaddressed. According to small RNA sequencing data, all major small RNA biotypes have been found in exomeres and supermeres, with miRNA reads significantly prevailed (more than 60% of all reads in the supermeres and 79% in the exomeres). Furthermore, both types of particles were enriched in AGO proteins. Interestingly, expression patterns of miRNAs in exomeres and supermeres were similar but differed significantly from sEV-NVF and cells (Zhang et al., 2018; Jeppesen et al., 2019; Zhang et al., 2019; Zhang et al., 2021).
The mechanisms of biogenesis and the exact biological role of exomeres and supermeres remain to be addressed. Their protein composition resembles that of NVF-fraction, so the question arises whether and to what extent aggregates of exomeres and supermeres form NVFs. Whether exomeres and supermeres represent discrete particles with unique protein sets and distinct biogenesis remains unknown. It is feasible, however, that both exomeres and supermeres are, in fact, continuous series of cellular RNPs of different sizes. The point is that the extracellular environment contains ribosomes and their fragments, as well as fragments of spliceosomes, and both ribosome and spliceosome proteins were detected in supermeres and exomeres (Tosar et al., 2020; Zhang et al., 2021; Tosar et al., 2022). The size of ribosomes and spliceosomes corresponds to the size of supermeres (Figure 1). Therefore, it is feasible that both exomeres and supermeres represent extracellular RNP fractions consisting of a set of intracellular RNP and protein complexes of different sizes and their fragments (Tosar et al., 2022). This, however, does not exclude the possibility of the existence of secreted particles of a certain composition in these fractions (i.e., bona fide exomeres and supermeres) (Jeppesen et al., 2022). Another question is whether the fractions of these particles are free of vesicles. Thus, ultracentrifugation at 200,000 g shows evidence of the presence of very small vesicles in the precipitate corresponding to the exomeres (Lee et al., 2019b). However, whatever non-vesicular exRNA carriers consist of, they are likely to serve as a new page in the story of cell-cell communication: it has already been shown that exomeres can deliver functionally active protein cargo to recipient cells (Zhang et al., 2019). And since it is the non-vesicular exRNA carriers where the bulk of exRNAs, including miRNAs as well as AGO proteins, are concentrated, they may prove to be an important actor in this story. In this regard, it is interesting to note the recent demonstration that non-vesicular miRNA-AGO complexes in the extracellular fluids are indeed functional and capable of efficient silencing (Geekiyanage et al., 2020).
4 Challenges in the exRNA profiling
The heterogeneity of the results obtained by different research groups could stem from methods used for RNA conversion into cDNA. Specifically, widely used RNA-seq library preparation approaches produce different biases for both long (Rodosthenous et al., 2020) and small RNA sequencing (Srinivasan et al., 2019). For instance, a comparison of six different cDNA library preparation methods for long RNA-seq demonstrated drastic variation in mapping efficacy and proportions of different RNA classes detected (Rodosthenous et al., 2020). It should be noted that the vast majority of previous reports on small RNA sequencing of exRNA used standard cDNA library preparation protocols based on the ligation of adapters to both termini of small RNA. By default, such ligation reaction requires the presence of phosphate at the 5′-end of the RNA and hydroxyl at the 3′-end. The ligation-based methods were originally tailored for capturing intracellular miRNAs that are 5′-phosphorylated, and until recently, the biases associated with standard ligation-based commercial kits have not been considered. However, extracellular RNAs are exposed to RNases and as a result are converted to 5′-hydroxyl and 3′-(cyclo) phosphate entities (Sorrentino, 2010; Giraldez et al., 2019; Nechooshtan et al., 2020). Because ssRNA adapters cannot be ligated to such RNAs, the latter will not be included into the final library and sequenced without prior end-repair. Therefore, the distortions of most currently reported exRNA profiles can be very significant. Such distortions can be avoided by prior treatment of samples with T4 polynucleotide kinase (T4 PNK), which phosphorylates the 5′-ends of RNA and removes phosphates/cyclophosphates from the 3′-termini. Thus, deep sequencing of T4 PNK-treated extracellular small RNAs demonstrated that in lEVs, sEVs, NVF and in whole plasma libraries ∼80–90% were rRNA fragments (Wei et al., 2017; Akat et al., 2019; Giraldez et al., 2019; Solaguren-Beascoa et al., 2023). In rRNA-depleted libraries, the percentage of reads corresponding to mRNA and lncRNA fragments was increased up to 10-fold (Giraldez et al., 2019), while lEVs and sEVs contained ∼25% repetitive sequences, and NVF contained ∼45% tRNAs. Thus, most exRNAs are degradation products of various long RNAs and tRNAs, with a very small proportion of miRNAs and other small RNA types.
Importantly, exRNA profiling has several previously underestimated crucial methodological issues that should ideally be addressed in every experimental setting. To facilitate this process, we offer a guide containing the key experiments required for the correct profiling of exRNA carriers (Table 1). Further progress in characterizing the exRNA population is expected upon application of recently emerged RNA-seq library preparation methods such as D-Plex small RNA-seq kit (Diagenode), SMARTer smRNA-seq kit (Takara) and BioLiqX small RNA-seq kit (Heidelberg Biolabs), which enable unbiased incorporation of short RNA fragments with modified nucleotides. Wide application of novel RNA sequencing methods can confirm exRNA transcriptomes of exomeres, supermeres and whole biological fluids.
5 Conclusion and future directions
The experimentally confirmed exRNA carriers list includes lEVs, several types of sEVs, single RBPs, exomeres and supermeres. Biofluids apparently contain a set of membrane vesicles of continuous size range–up to the minimum physically possible, as well as a set of proteins and RNP aggregates ranging up to individual proteins. Furthermore, the concentration of a given exRNA carrier could be labile and prone to change under different physiologic and pathologic conditions. Apart from that, methodological obstacles and biological diversity explain the marked variation in the transcriptomic content of exRNA carriers obtained by different groups. Thus, the introduction of unified experimental standards for exRNA purification and detection is of utmost importance. Further systematic profiling of individual subfractions, such as immunopurified EVs and RNPs, may enable the compilation of a comprehensive extracellular transcriptome map and the creation of relevant training samples for computational deconvolution of the exRNA transcriptome in biofluids.
Author contributions
JM: Conceptualization, Writing–original draft. DM: Writing–review and editing. AT: Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was performed within the framework of the Basic Research Program at the National Research University Higher School of Economics (HSE University).
Conflict of interest
Author AT was employed by company art photonics GmbH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abache, T., Le Naour, F., Planchon, S., Harper, F., Boucheix, C., and Rubinstein, E. (2007). The transferrin receptor and the tetraspanin web molecules CD9, CD81, and CD9P-1 are differentially sorted into exosomes after TPA treatment of K562 cells. J. Cell. Biochem. 102, 650–664. doi:10.1002/jcb.21318
Akat, K. M., Lee, Y. A., Hurley, A., Morozov, P., Max, K. E. A., Brown, M., et al. (2019). Detection of circulating extracellular mRNAs by modified small-RNA-sequencing analysis. JCI Insight 4, e127317. doi:10.1172/jci.insight.127317
Arroyo, J. D., Chevillet, J. R., Kroh, E. M., Ruf, I. K., Pritchard, C. C., Gibson, D. F., et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108, 5003–5008. doi:10.1073/pnas.1019055108
Barman, B., Sung, B. H., Krystofiak, E., Ping, J., Ramirez, M., Millis, B., et al. (2022). VAP-A and its binding partner CERT drive biogenesis of RNA-containing extracellular vesicles at ER membrane contact sites. Dev. Cell 57, 974–994.e8. doi:10.1016/j.devcel.2022.03.012
Chevillet, J. R., Kang, Q., Ruf, I. K., Briggs, H. A., Vojtech, L. N., Hughes, S. M., et al. (2014). Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. U. S. A. 111, 14888–14893. doi:10.1073/pnas.1408301111
Clément, A.-A., Desgagné, V., Légaré, C., Guay, S.-P., Boyer, M., Hutchins, E., et al. (2021). HDL-enriched miR-30a-5p is associated with HDL-cholesterol levels and glucose metabolism in healthy men and women. Epigenomics 13, 985–994. doi:10.2217/epi-2020-0456
Costa, B., Li Calzi, M., Castellano, M., Blanco, V., Cuevasanta, E., Litvan, I., et al. (2023). Nicked tRNAs are stable reservoirs of tRNA halves in cells and biofluids. Proc. Natl. Acad. Sci. U.S.A. 120, e2216330120. doi:10.1073/pnas.2216330120
Dellar, E. R., Hill, C., Melling, G. E., Carter, D. R. F., and Baena-Lopez, L. A. (2022). Unpacking extracellular vesicles: RNA cargo loading and function. J Extracell. Bio 1, e40. doi:10.1002/jex2.40
Di Vizio, D., Morello, M., Dudley, A. C., Schow, P. W., Adam, R. M., Morley, S., et al. (2012). Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am. J. Pathol. 181, 1573–1584. doi:10.1016/j.ajpath.2012.07.030
Dixson, A. C., Dawson, T. R., Di Vizio, D., and Weaver, A. M. (2023). Context-specific regulation of extracellular vesicle biogenesis and cargo selection. Nat. Rev. Mol. Cell Biol. 24, 454–476. doi:10.1038/s41580-023-00576-0
Escola, J.-M., Kleijmeer, M. J., Stoorvogel, W., Griffith, J. M., Yoshie, O., and Geuze, H. J. (1998). Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273, 20121–20127. doi:10.1074/jbc.273.32.20121
Florijn, B. W., Duijs, J. M. G. J., Levels, J. H., Dallinga-Thie, G. M., Wang, Y., Boing, A. N., et al. (2019). Diabetic nephropathy alters the distribution of circulating angiogenic MicroRNAs among extracellular vesicles, HDL, and ago-2. Diabetes 68, 2287–2300. doi:10.2337/db18-1360
Galvanin, A., Dostert, G., Ayadi, L., Marchand, V., Velot, É., and Motorin, Y. (2019). Diversity and heterogeneity of extracellular RNA in human plasma. Biochimie 164, 22–36. doi:10.1016/j.biochi.2019.05.011
Garcia-Martin, R., Wang, G., Brandão, B. B., Zanotto, T. M., Shah, S., Kumar Patel, S., et al. (2022). MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 601, 446–451. doi:10.1038/s41586-021-04234-3
Geekiyanage, H., Rayatpisheh, S., Wohlschlegel, J. A., Brown, R., and Ambros, V. (2020). Extracellular microRNAs in human circulation are associated with miRISC complexes that are accessible to anti-AGO2 antibody and can bind target mimic oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 117, 24213–24223. doi:10.1073/pnas.2008323117
Giraldez, M. D., Spengler, R. M., Etheridge, A., Goicochea, A. J., Tuck, M., Choi, S. W., et al. (2019). Phospho-RNA-seq: a modified small RNA-seq method that reveals circulating mRNA and lncRNA fragments as potential biomarkers in human plasma. EMBO J. 38, e101695. doi:10.15252/embj.2019101695
Heintz-Buschart, A., Yusuf, D., Kaysen, A., Etheridge, A., Fritz, J. V., May, P., et al. (2018). Small RNA profiling of low biomass samples: identification and removal of contaminants. BMC Biol. 16, 52. doi:10.1186/s12915-018-0522-7
Hendrix, A., Lippens, L., Pinheiro, C., Théry, C., Martin-Jaular, L., Lötvall, J., et al. (2023). Extracellular vesicle analysis. Nat. Rev. Methods Prim. 3, 56. doi:10.1038/s43586-023-00240-z
Jeppesen, D. K., Fenix, A. M., Franklin, J. L., Higginbotham, J. N., Zhang, Q., Zimmerman, L. J., et al. (2019). Reassessment of exosome composition. Cell 177, 428–445. doi:10.1016/j.cell.2019.02.029
Jeppesen, D. K., Zhang, Q., Franklin, J. L., and Coffey, R. J. (2022). Are supermeres a distinct nanoparticle? J Extracell. Bio 1, e44. doi:10.1002/jex2.44
Kalra, H., Simpson, R. J., Ji, H., Aikawa, E., Altevogt, P., Askenase, P., et al. (2012). Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 10, e1001450. doi:10.1371/journal.pbio.1001450
Keerthikumar, S., Chisanga, D., Ariyaratne, D., Al Saffar, H., Anand, S., Zhao, K., et al. (2016). ExoCarta: a web-based compendium of exosomal cargo. J. Mol. Biol. 428, 688–692. doi:10.1016/j.jmb.2015.09.019
Kim, D.-K., Lee, J., Simpson, R. J., Lötvall, J., and Gho, Y. S. (2015). EVpedia: a community web resource for prokaryotic and eukaryotic extracellular vesicles research. Seminars Cell and Dev. Biol. 40, 4–7. doi:10.1016/j.semcdb.2015.02.005
Lai, H., Li, Y., Zhang, H., Hu, J., Liao, J., Su, Y., et al. (2022). exoRBase 2.0: an atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res. 50, D118–D128. doi:10.1093/nar/gkab1085
LaPlante, E. L., Stürchler, A., Fullem, R., Chen, D., Starner, A. C., Esquivel, E., et al. (2023). exRNA-eCLIP intersection analysis reveals a map of extracellular RNA binding proteins and associated RNAs across major human biofluids and carriers. Cell Genomics 3, 100303. doi:10.1016/j.xgen.2023.100303
Lee, S.-S., Groot, M., Pinilla-Vera, M., Fredenburgh, L. E., and Jin, Y. (2019a). Identification of miRNA-rich vesicles in bronchoalveolar lavage fluid: insights into the function and heterogeneity of extracellular vesicles. J. Control. Release 294, 43–52. doi:10.1016/j.jconrel.2018.12.008
Lee, S.-S., Won, J.-H., Lim, G. J., Han, J., Lee, J. Y., Cho, K.-O., et al. (2019b). A novel population of extracellular vesicles smaller than exosomes promotes cell proliferation. Cell Commun. Signal 17, 95. doi:10.1186/s12964-019-0401-z
Liu, C.-J., Xie, G.-Y., Miao, Y.-R., Xia, M., Wang, Y., Lei, Q., et al. (2022). EVAtlas: a comprehensive database for ncRNA expression in human extracellular vesicles. Nucleic Acids Res. 50, D111–D117. doi:10.1093/nar/gkab668
Mateescu, B., Jones, J. C., Alexander, R. P., Alsop, E., An, J. Y., Asghari, M., et al. (2022). Phase 2 of extracellular RNA communication consortium charts next-generation approaches for extracellular RNA research. iScience 25, 104653. doi:10.1016/j.isci.2022.104653
Mathieu, M., Martin-Jaular, L., Lavieu, G., and Théry, C. (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 21, 9–17. doi:10.1038/s41556-018-0250-9
Morello, M., Minciacchi, V., De Candia, P., Yang, J., Posadas, E., Kim, H., et al. (2013). Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle 12, 3526–3536. doi:10.4161/cc.26539
Murillo, O. D., Thistlethwaite, W., Rozowsky, J., Subramanian, S. L., Lucero, R., Shah, N., et al. (2019). exRNA atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell 177, 463–477. doi:10.1016/j.cell.2019.02.018
Nechooshtan, G., Yunusov, D., Chang, K., and Gingeras, T. R. (2020). Processing by RNase 1 forms tRNA halves and distinct Y RNA fragments in the extracellular environment. Nucleic Acids Res. 48, 8035–8049. doi:10.1093/nar/gkaa526
O’Grady, T., Njock, M.-S., Lion, M., Bruyr, J., Mariavelle, E., Galvan, B., et al. (2022). Sorting and packaging of RNA into extracellular vesicles shape intracellular transcript levels. BMC Biol. 20, 72. doi:10.1186/s12915-022-01277-4
Padilla, J.-C. A., Barutcu, S., Malet, L., Deschamps-Francoeur, G., Calderon, V., Kwon, E., et al. (2023). Profiling the polyadenylated transcriptome of extracellular vesicles with long-read nanopore sequencing. BMC Genomics 24, 564. doi:10.1186/s12864-023-09552-6
Rodosthenous, R. S., Hutchins, E., Reiman, R., Yeri, A. S., Srinivasan, S., Whitsett, T. G., et al. (2020). Profiling extracellular long RNA transcriptome in human plasma and extracellular vesicles for biomarker discovery. iScience 23, 101182. doi:10.1016/j.isci.2020.101182
Rossi-Herring, G., Belmonte, T., Rivas-Urbina, A., Benítez, S., Rotllan, N., Crespo, J., et al. (2023). Circulating lipoprotein-carried miRNome analysis reveals novel VLDL-enriched microRNAs that strongly correlate with the HDL-microRNA profile. Biomed. Pharmacother. 162, 114623. doi:10.1016/j.biopha.2023.114623
Shurtleff, M. J., Yao, J., Qin, Y., Nottingham, R. M., Temoche-Diaz, M. M., Schekman, R., et al. (2017). Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. U.S.A. 114, E8987-E8995. doi:10.1073/pnas.1712108114
Solaguren-Beascoa, M., Gámez-Valero, A., Escaramís, G., Herrero-Lorenzo, M., Ortiz, A. M., Minguet, C., et al. (2023). Phospho-RNA-seq highlights specific small RNA profiles in plasma extracellular vesicles. IJMS 24, 11653. doi:10.3390/ijms241411653
Sorrentino, S. (2010). The eight human “canonical” ribonucleases: Molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 584, 2194–2200. doi:10.1016/j.febslet.2010.04.018
Srinivasan, S., Yeri, A., Cheah, P. S., Chung, A., Danielson, K., De Hoff, P., et al. (2019). Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell 177, 446–462. doi:10.1016/j.cell.2019.03.024
Théry, C., Amigorena, S., Raposo, G., and Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. CP Cell Biol. 30, Unit 3.22. doi:10.1002/0471143030.cb0322s30
Théry, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell. Vesicle 7, 1535750. doi:10.1080/20013078.2018.1535750
Tosar, J. P., Cayota, A., and Witwer, K. (2022). Exomeres and supermeres: monolithic or diverse? J Extracell. Bio 1, e45. doi:10.1002/jex2.45
Tosar, J. P., Segovia, M., Castellano, M., Gámbaro, F., Akiyama, Y., Fagúndez, P., et al. (2020). Fragmentation of extracellular ribosomes and tRNAs shapes the extracellular RNAome. Nucleic Acids Res. 48, 12874–12888. doi:10.1093/nar/gkaa674
Tsering, T., Li, M., Chen, Y., Nadeau, A., Laskaris, A., Abdouh, M., et al. (2022). EV-ADD, a database for EV-associated DNA in human liquid biopsy samples. J Extracell. Vesicle 11, e12270. doi:10.1002/jev2.12270
Tulkens, J., Vergauwen, G., Van Deun, J., Geeurickx, E., Dhondt, B., Lippens, L., et al. (2020). Increased levels of systemic LPS-positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut 69, 191–193. doi:10.1136/gutjnl-2018-317726
Turchinovich, A., Weiz, L., Langheinz, A., and Burwinkel, B. (2011). Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233. doi:10.1093/nar/gkr254
Van Niel, G., Carter, D. R. F., Clayton, A., Lambert, D. W., Raposo, G., and Vader, P. (2022). Challenges and directions in studying cell–cell communication by extracellular vesicles. Nat. Rev. Mol. Cell Biol. 23, 369–382. doi:10.1038/s41580-022-00460-3
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D., and Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–433. doi:10.1038/ncb2210
Wang, J., Chen, H.-C., Sheng, Q., Dawson, T. R., Coffey, R. J., Patton, J. G., et al. (2023a). Systematic assessment of small RNA profiling in human extracellular vesicles. Cancers 15, 3446. doi:10.3390/cancers15133446
Wang, J., Xie, G.-Y., Zhang, Q., Zhang, X., and Guo, A.-Y. (2023b). Characterization of exogenous sequence fragments in extracellular vesicles from human. Small Struct. 4, 2200406. doi:10.1002/sstr.202200406
Wei, Z., Batagov, A. O., Schinelli, S., Wang, J., Wang, Y., El Fatimy, R., et al. (2017). Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 8, 1145. doi:10.1038/s41467-017-01196-x
Zhang, H., Freitas, D., Kim, H. S., Fabijanic, K., Li, Z., Chen, H., et al. (2018). Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 20, 332–343. doi:10.1038/s41556-018-0040-4
Zhang, Q., Higginbotham, J. N., Jeppesen, D. K., Yang, Y.-P., Li, W., McKinley, E. T., et al. (2019). Transfer of functional cargo in exomeres. Cell Rep. 27, 940–954. doi:10.1016/j.celrep.2019.01.009
Keywords: extracellular vesicles, exosomes, miRNA, extracellular RNA, exomeres, supermeres
Citation: Makarova J, Maltseva D and Tonevitsky A (2023) Challenges in characterization of transcriptomes of extracellular vesicles and non-vesicular extracellular RNA carriers. Front. Mol. Biosci. 10:1327985. doi: 10.3389/fmolb.2023.1327985
Received: 25 October 2023; Accepted: 20 November 2023;
Published: 05 December 2023.
Edited by:
Andrey Turchinovich, German Cancer Research Center (DKFZ), GermanyReviewed by:
Sergei Koshkin, Thomas Jefferson University, United StatesJulia Veryaskina, Institute of Molecular and Cellular Biology (RAS), Russia
Copyright © 2023 Makarova, Maltseva and Tonevitsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia Makarova, jmakarova@hse.ru
 Julia Makarova
Julia Makarova Diana Maltseva
Diana Maltseva Alexander Tonevitsky1,2,4
Alexander Tonevitsky1,2,4