NMR-based metabolomics in Alzheimer’s disease research: a review
- 1Department of Chemistry “Ugo Schiff”, University of Florence, Sesto Fiorentino, Italy
- 2Magnetic Resonance Center (CERM), University of Florence, Sesto Fiorentino, Italy
- 3Consorzio Interuniversitario Risonanze Magnetiche MetalloProteine (CIRMMP), Sesto Fiorentino, Italy
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder and represents the most common cause of dementia in the elderly population worldwide. Currently, there is no cure for AD, and the continuous increase in the number of susceptible individuals poses one of the most significant emerging threats to public health. However, the molecular pathways involved in the onset and progression of AD are not fully understood. This information is crucial for developing less invasive diagnostic instruments and discovering novel potential therapeutic targets. Metabolomics studies the complete ensemble of endogenous and exogenous metabolites present in biological specimens and may provide an interesting approach to identify alterations in multiple biochemical processes associated with AD onset and evolution. In this mini review, we summarize the results from metabolomic studies conducted using nuclear magnetic resonance (NMR) spectroscopy on human biological samples (blood derivatives, cerebrospinal fluid, urine, saliva, and tissues) from AD patients. We describe the metabolic alterations identified in AD patients compared to controls and to patients diagnosed with mild cognitive impairment (MCI). Moreover, we discuss the challenges and issues associated with the application of NMR-based metabolomics in the context of AD research.
1 Introduction
Alzheimer’s disease (AD) represents an irreversible neurodegenerative condition characterized by a gradual deterioration of memory, cognitive abilities, and eventually the capacity to perform basic daily tasks. It stands as the predominant neurodegenerative ailment in the elderly population, impacting approximately 5%–7% of individuals aged 60 and above (Foley et al., 2017). In the clinical practice, the identification of AD-affected individuals is facilitated by measuring the levels of cerebrospinal fluid (CSF) core AD biomarkers, namely total tau (t-tau), threonine-181-phosphorylated-tau (p-tau) proteins, and amyloid beta 1–42 peptide (Aβ42) (Blennow et al., 2010). The variation in concentration of these molecules reflects the key aspects of disease pathogenesis (i.e., neuronal degeneration, tangles formation, and aggregation and deposition of amyloid plaques). During the asymptomatic phase of AD, CSF analysis in affected individuals commonly reveals diminished concentrations of Aβ42, and elevated levels of t-tau and p-tau proteins (Mattsson et al., 2009). Remarkably, these characteristic alterations are evident prior to the onset of clinical symptoms. Thus, the identification of AD-affected individuals is facilitated by the detection of abnormal levels of these CSF core biomarkers, even in the prodromal phase (Dubois et al., 2014).
Despite the clinical utility of these biomarkers, the intricate molecular pathways contributing to the onset and progression of AD remain incompletely elucidated. The imperative to uncover novel molecular targets for AD, with applications in early diagnosis, prognosis, disease trajectory prediction, and therapeutic interventions, underscores the critical need for comprehensive insights into the underlying molecular biochemistry of AD (Paglia et al., 2016).
Metabolomics, a discipline dedicated to the identification, quantification, and characterization of the entire spectrum of endogenous and exogenous metabolites in a biological specimen, emerges as a promising avenue of exploration (Ashrafian et al., 2021). Metabolites, representing the downstream products of the genome, transcriptome, and proteome, as well as the upstream inputs from diverse external factors such as environment, lifestyle, diet, and drug exposure, encapsulate a holistic view of the biochemical landscape (Nicholson and Lindon, 2008).
Given these considerations, metabolomics presents itself as a compelling approach for investigating alterations in multiple biochemical networks throughout the course of AD. This approach holds potential not only for enhancing our understanding of the disease mechanisms but also for paving the way towards the identification of new, effective, and minimally invasive targets for early detection, prognosis, and therapeutic intervention in Alzheimer’s disease.
Mass spectrometry and Nuclear Magnetic Resonance (NMR) spectroscopy are the two main analytical platforms available to perform metabolomic analysis. MS overshadows NMR in terms of sensitivity, having a detection limit in the rage of nano-to picomolar concentrations, which translates in being able to quantify a number of compounds of the order of 103. In contrast, NMR struggles to detect metabolites at concentrations below the micromolar level. On the other hand, NMR is intrinsically quantitative, high-throughput and highly reproducible on a wide dynamic range (Vignoli et al., 2019). To be exhaustive, NMR, performed at an intermediate field such as a 600 MHz (the metabolomics gold standard) and in complex samples with crowded spectra such as biofluids, is limited also by spectral resolution. Considering all the abovementioned aspects, NMR and MS can be considered complementary since the weaknesses of one platform can be compensated by the strengths of the other, and both can contribute to AD research (González-Domínguez et al., 2017). Our review is focused on NMR which in the last years has demonstrated to be a powerful tool for searching novel biomarkers (Emwas et al., 2019) for disease diagnosis, prognosis, monitoring patients during therapeutic treatments and finding novel potential therapeutic targets (Wishart, 2016; Holmes et al., 2018; McCartney et al., 2019; Vignoli et al., 2020b; Jobard et al., 2021; Buergel et al., 2022; Vignoli et al., 2022).
In this review we decided to collect the main finding obtained from metabolomic studies performed using NMR spectroscopy on human biological samples from AD patients. Providing a comprehensive and in-depth methodological description of the use of NMR technique for metabolomic analyses is beyond the scope of this work. However, we refer interested readers to a recently published review by our group that specifically addresses these aspects for both liquid and semi-solid samples (Ghini et al., 2023).
2 Article selection
2.1 Study inclusion and exclusion criteria
The detailed study selection criteria are presented according to the Population, Exposure, Comparison, Outcome and Study design (PECOS) criteria as outlined below:
Inclusion criteria
⁃ P (Participants): Adult patients (>18 years of age) from any geographic location, any age or gender.
⁃ E (Exposure): Patients with confirmed diagnosis of AD.
⁃ C (Comparison): Difference in concentration of metabolites and lipoproteins between AD and other control/pathological groups.
⁃ O (Outcome): Dysregulation of metabolite/lipoproteins concentrations between the study groups.
⁃ S (Study Design): Human-based observational studies (case-control, cohort, or cross-sectional) that performed metabolomics via NMR to quantify the concentrations of metabolites and lipoproteins.
Exclusion criteria
⁃ Targeted metabolomic experiments that are used to validate and translate already identified metabolites from hypothesis generating studies.
⁃ Animal or cell-based studies.
⁃ Non-Observational study designs such as case reports, conference proceedings, letters to editor, reviews, and meta-analysis.
⁃ Metabolites quantified using analytical platforms other than NMR (e.g., mass spectrometry).
⁃ Studies published after October 2023.
2.2 Search and selection strategy
The search was conducted on the Web of Science electronic database in the title (TI) and in the topic (TS) fields, using a combination of keywords paired with the Boolean operator “AND”. The final query was (TI=(alzheimer) AND TS = ((metabolomic* OR metabolite*) AND *NMR*)). The search was conducted the 16th of November 2023.
Applying the abovementioned inclusion and exclusion criteria, study selection was carried out screening titles and abstracts of all publications. Studies that matched the inclusion criteria but had insufficient details in abstracts were further examined by inspecting the complete texts.
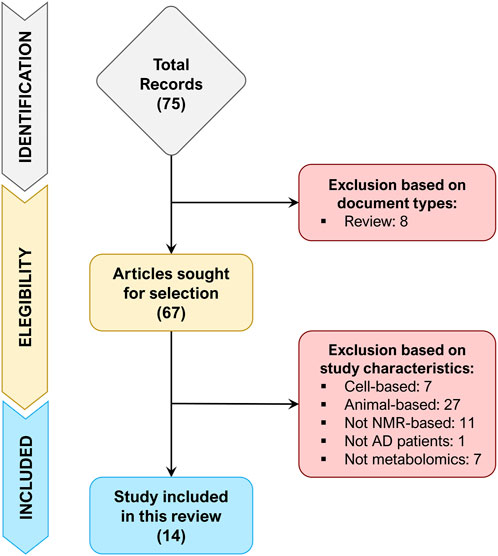
A total of 75 records were retrieved after the identification phase, out of which 8 records were excluded because of document type (review). The remaining 67 records were screened for their title and abstract (or entire text when required), of which 53 records were excluded since did not match the inclusion/exclusion criteria. Figure 1 depicts the flowchart of study selection, summarizing the process of study identification, eligibility, and inclusion.
3 Study characteristics
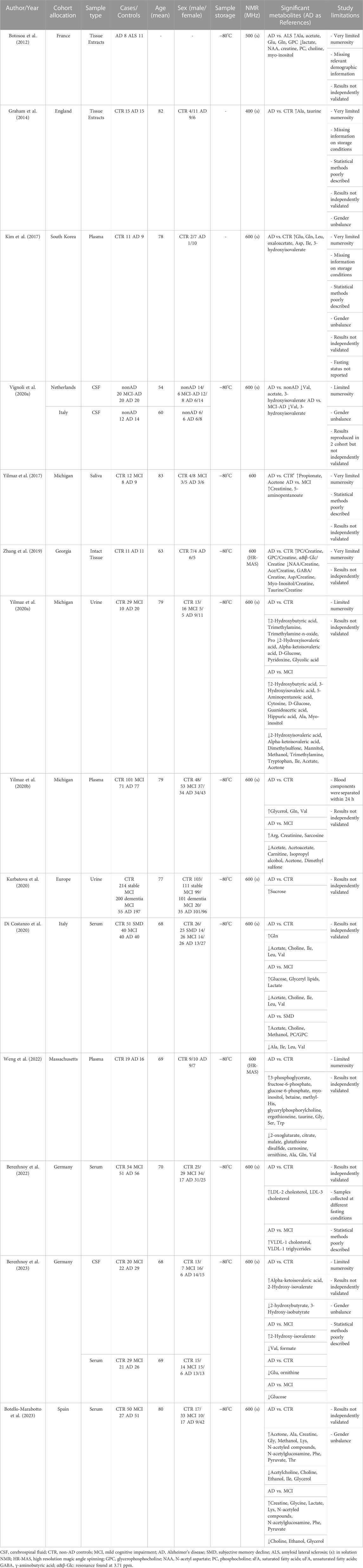
The characteristics of the 14 included studies are presented in Table 1. The selected studies were published in the last 11 years (2012–2023) and were mainly conducted in Europe and United States (86%). Of the studies that reported age and sex, the mean age of the enrolled population ranges from 54 to 82 years, with 47% of them being male. Most of the research compared metabolomics data between AD and healthy controls and MCI. The majority of the studies (7 out of 14) analyzed blood derivatives (plasma or serum), followed by cerebrospinal fluid (CSF), urine, tissues and saliva. Of the studies that reported the information, all samples were stored at −80°C pending NMR analysis as per the best practice for metabolomics, and, except for two studies, all samples were acquired using a spectrometer operating at 600 MHz.
4 Metabolomic/lipoproteomic differences between AD and controls
Although abnormal levels of the CSF core AD biomarkers enable the accurate identification of patients affected by AD, a full comprehension of the underlying molecular mechanisms involved in the onset and progression of this pathology is still distant. For this reason, most of the published studies aim to detect and quantify, in different biospecimens, AD-relevant metabolites and/or lipoproteins in AD patients as compared to cognitively normal individuals (CTR).
The first metabolomic studies via NMR date back to 2012–2014, and both examined post-mortem brain tissue extracts (Botosoa et al., 2012; Graham et al., 2014). Botosoa et al. analyzed frontal cortex extracts of samples collected post-mortem from AD patients and from a control group constituted by amyotrophic lateral sclerosis (ALS) patients. The AD metabolic signature was characterized by high levels of alanine, acetate, glutamate, glutamine and glycerophosphocholine (GPC), and low levels of lactate, creatine, N-AcetyAspartate (NAA), phosphocholine (PC), choline and myo-inositol (Table 1). In particular, NAA is a biomarker of neuronal integrity in the brain and its reduction reflects deficiency and neuronal dysfunction. Therefore, this metabolite could play a pivotal role in AD pathogenesis. Graham et al. analyzed extracts of post-mortem brain Brodmann 7 region samples. Multivariate statistical analysis shows a clear distinction between AD and CTR samples, and the most relevant metabolites in the discrimination were alanine and taurine (Table 1). In 2019 the first NMR-based metabolomic study conducted on intact post-mortem tissues obtained from frontal cortex was published (Zhang et al., 2019). The authors reported that high levels of PC, GPC and low levels of NAA, acetate, GABA, aspartate, myo-inositol and taurine are characteristic features of samples of AD patients as compared to CTR (Table 1).
Going from tissues to biofluids, NMR-based metabolomics of blood derivatives (plasma or serum) has shown the potential to distinguish patients with AD from CTR with optimal results (Kim et al., 2017; Yilmaz et al., 2020b; Di Costanzo et al., 2020; Berezhnoy et al., 2022, Berezhnoy et al., 2023; Weng et al., 2022; Botello-Marabotto et al., 2023). Among metabolites (which include amino acids, carbohydrates, lipids, choline-derived metabolites, keto acids, and fatty acids) and lipoproteins (fraction and subfractions) identified and quantified in the studies, several metabolites and 2 lipoprotein subfraction of LDL cholesterol (Table 1) were described as differentially abundant between AD and CTR. However, there is not a clear consensus on the directions (up or down) of dysregulation of these metabolic alterations and on their significance (Table 1). The high heterogeneity emerged could be ascribed to several factors: the small sample size of most of the studies, the lack of independent validation cohorts in the majority of the studies, relevant differences in sample collection, processing, and analytical method employed (e.g. not all blood samples were collected pre-prandially), and not adequately addressing of potential confounding risk factors (e.g. not all studies enrolled patients and controls age and sex matched). These factors could significantly impact metabolite concentrations and may be leading factors for inconsistencies of reported results.
Two studies (Yilmaz et al., 2020a; Kurbatova et al., 2020) investigated the metabolic phenotype of AD in urine (Table 1). Both studies proposed a combined approach using both 1H NMR and mass spectrometry and showed that the urine phenotype can discriminate AD and CTR with high discrimination accuracy. Yilmaz et al. reported that 2-hydroxybutyric acid, trimethylamine, trimethylamine-n-oxide, proline have higher concentrations in AD patients, whereas 2-hydroxyisovaleric acid, alpha-ketoisovaleric acid, D-glucose, pyridoxine and glycolic acid have lower concentrations. In this case, using available information, it was not possible to distinguish between the metabolites quantified through NMR and those quantified through mass spectrometry; whereas Kurbatova et al. clearly differentiated the quantification assay and the only significant difference emerged by NMR is the increasing of sucrose. One study searched for diagnostic biomarkers of AD in saliva samples (Yilmaz et al., 2017) identifying differences in the concentrations of 22 metabolites in AD and MCI as compared to CTR. Moreover, the authors built two distinct logistic regression models: one, based on creatinine and 5-aminopentanoate, able to discriminate AD from MCI with 0.900 sensitivity and 0.944 specificity, and another, based on propionate and acetone, which discriminates AD from CTR with 0.909 sensitivity and 0.842 specificity (Table 1).
Since brain directly transfers its metabolites into CSF, the latter most likely reflects the brain biochemistry, and thus CSF is obviously the biofluid of choice when it comes to studying neurological disorders. However, the invasiveness of the sample collection procedure and associated ethical issues (especially for control/healthy individuals) have resulted in only one available NMR-based metabolomic study examining AD and CTR (Table 1). In this study (Berezhnoy et al., 2023) it is showed that alpha-ketoisovaleric acid and 2-hydroxy-isovalerate are upregulated in the CSF of AD patients respect to CTR, whereas 2-hydroxybutyrate and 3-hydroxy-isobutyrate are downregulated. Moreover, they identified sex-specific metabolite alterations that underling once more how sex is a relevant confounding factor when one wants to perform metabolomics analyses (Vignoli et al., 2018; Bell et al., 2021; Costanzo et al., 2022).
5 Metabolomic/lipoproteomic differences between AD and MCI
The AD “continuum” starting from cognitively normal subjects, begins with subjective memory decline, progresses to Mild Cognitive Impairment (MCI) and eventually reaches AD (Jessen et al., 2014). However, MCI subjects may not evolve into dementia, indeed only 20%–40% of patients progresses to AD (Tahami Monfared et al., 2022). Understanding the mechanisms underlying this progression could contribute to addressing the still unsolved question of AD pathogenesis and evolution.
Two studies (Vignoli et al., 2020a; Berezhnoy et al., 2023) analyzed cerebrospinal fluid of AD and MCI patients by NMR, revealing a clear distinction between the two groups. Both studies consistently found a reduction in valine levels in the CSF of AD patients (Table 1). Furthermore, Vignoli et al. showed that valine is reduced even in comparison to MCI-AD patients, and that it correlates with patient cognitive decline. In addition to valine, Vignoli et al. also identified in AD patients a reduction in the levels of acetate and 3-hydroxyisovalerate. Conversely, Berezhnoy et al. reported higher CSF levels of 2-hydroxy-isovalerate and lower levels of formate in AD patients.
Focusing the attention from a compartmentalized biofluid such CSF to systemic biofluids, we were able to find five studies conducted on blood derivatives and two studies on urine (Table 1). Among the five studies on plasma/serum, four were focused on metabolite analysis (Yilmaz et al., 2020b; Di Costanzo et al., 2020; Berezhnoy et al., 2023; Botello-Marabotto et al., 2023) and reported 25 differentially abundant metabolites between AD and MCI. As observed in the comparison between AD and CTR, there is some inconsistency among the results (e.g. glucose, a metabolite which is very sensitive to sample collection and pre-analytical procedures, is reported reduced in AD in one study and increased in another study); however, two out of three studies described a significant reduction acetate in AD patients and it has been hypothesized that it could play a role in the compromission of the neurotransmission activity of acetylcholine (Di Costanzo et al., 2020).
Berezhnoy et al. focused their analysis on lipoproteins, using a commercially available quantitative lipoprotein assay based on 600 MHz NMR spectroscopy. They were able to correlate a set of 112 lipoprotein variables with clinical metadata and AD core biomarkers in CSF. They obtained a deeper insight into the pathophysiology of dementia and reported an increase of VLDL-1 cholesterol and VLDL-1 triglycerides in AD patients as compared to MCI (Berezhnoy et al., 2022). It is known that VLDL-1 cholesterol levels are correlated with ApoE4, that in turn is a factor that affect the Aβ levels: when a patient shows altered ApoE4 function, AD-related risks increase multiple-fold (Kloske and Wilcock, 2020). Further, the increase in VLDL-1 triglycerides in the AD group may be correlated with a prediabetic condition. Indeed, individuals with insulin resistance have a higher production of VLDL particles, which are responsible for triglyceride transport, reinforcing the idea of a close interconnection of blood lipoprotein biomarkers of type 2 diabetes and AD: some authors refer to AD as a type 3 diabetes (Accardi et al., 2012, 3). Putting things together, the authors speculated a strong link between VLDL parameters and amyloid plaque formation (Berezhnoy et al., 2022). This study provides a proof of concept that NMR-based lipoprotein analysis, in conjunction with metabolites analysis, is potentially able to provide an in-depth investigations of AD. This approach can be easily extended to further neurological diseases to provide interesting picture of the complex interplay among metabolites, lipoproteins, and clinical outcomes.
Among the two studies conducted on urine sample, only one reported the results of the comparison between AD and MCI (Yilmaz et al., 2020a), whereas in the other study MCI patients were enrolled to compare stable and dementia MCI (Kurbatova et al., 2020). Yilmaz et al. reported 19 urine metabolites significantly different between AD and MCI (Table 1). They used the concentrations of a panel of 10 metabolites (glucose, guanidinoacetate, urocanate, hippuric acid, cytosine, 2- and 3-hydroxyisovalerate, 2-ketoisovalerate, tryptophan, and malonate) to build a model able to discriminate the two groups with 78% sensitivity and 80% specificity. Based on these results, the authors suggests that urine metabolomics may be useful for developing a non-invasive test capable of diagnosing and distinguishing AD from MCI patients (Yilmaz et al., 2020a).
6 Conclusion
The NMR-based metabolomic studies presented in this review, despite their limitations, have demonstrated the existence of metabolic alterations in AD, albeit not consistent. Differences can be detected in brain tissue, blood serum/plasma, CSF, urine, and saliva. Moreover, there is evidence that some metabolic changes could predict the progression of AD. Therefore, NMR-based metabolomics could play a role in AD diagnosis and prognosis, serving as a valuable addition to classical clinical approaches.
NMR, with its ability to provide quantitative and extremely reproducible results, offers a valuable approach to understand the multifaceted and intricate metabolic landscape of AD. In particular, NMR-based metabolomics could play a role in: 1) long-term monitoring of AD evolution; 2) improving diagnosis and prognosis of AD on the base of their metabolic changes; 3) accelerate the discovery of metabolic biomarkers associated with AD; 4) characterize the biochemistry underlying AD with the final aim of identifying potential novel pharmaceutical targets. While this review is entirely focused on the NMR technique and its potential in AD research, we want to make it clear that we are neither implying that this is the sole analytical approach possible nor suggesting that this approach should replace others. On the contrary, the integration of NMR and MS in a multi-omics approach represents a powerful strategy, leveraging the strengths of both techniques. The synergy between these techniques in AD research extends beyond conventional boundaries, and holds promise for dissecting the complexities of the molecular mechanisms underlying neurodegeneration in AD.
Based on our review we foresee the need for improvement in enrolling larger and independent cohorts of patients and for a higher degree of standardization in the recommendations for sample collection, handling, preparation, acquisition, and data processing. These improvements would enable future researchers to obtain more robust, coherent, and interpretable results and facilitate the development of clinical applications for metabolomics.
Author contributions
AV: Conceptualization, Funding acquisition, Investigation, Writing–original draft, Writing–review and editing. LT: Conceptualization, Funding acquisition, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors acknowledge the funding by Associazione Italiana Ricerca Alzheimer Onlus–Airalzh (AGYR2022). The authors acknowledge co-funding from Next-Generation EU, in the context of the National Recovery and Resilience Plan, Investment PE8–Project Age-It: “Ageing Well in an Ageing Society”. This resource was co-financed by the Next-Generation EU (DM 1557 11.10.2022).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The views and opinions expressed are only those of the authors and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them.
References
Accardi, G., Caruso, C., Colonna-Romano, G., Camarda, C., Monastero, R., and Candore, G. (2012). Can alzheimer disease Be a form of type 3 diabetes? Rejuvenation Res. 15, 217–221. doi:10.1089/rej.2011.1289
Ashrafian, H., Sounderajah, V., Glen, R., Ebbels, T., Blaise, B. J., Kalra, D., et al. (2021). Metabolomics: the stethoscope for the twenty-first century. Med. Princ. Pract. 30, 301–310. doi:10.1159/000513545
Bell, J. A., Santos Ferreira, D. L., Fraser, A., Soares, A. L. G., Howe, L. D., Lawlor, D. A., et al. (2021). Sex differences in systemic metabolites at four life stages: cohort study with repeated metabolomics. BMC Med. 19, 58. doi:10.1186/s12916-021-01929-2
Berezhnoy, G., Laske, C., and Trautwein, C. (2022). Quantitative NMR-based lipoprotein analysis identifies elevated HDL-4 and triglycerides in the serum of Alzheimer’s disease patients. Int. J. Mol. Sci. 23, 12472. doi:10.3390/ijms232012472
Berezhnoy, G., Laske, C., and Trautwein, C. (2023). Metabolomic profiling of CSF and blood serum elucidates general and sex-specific patterns for mild cognitive impairment and Alzheimer’s disease patients. Front. Aging Neurosci. 15. doi:10.3389/fnagi.2023.1219718
Blennow, K., Hampel, H., Weiner, M., and Zetterberg, H. (2010). Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat. Rev. Neurol. 6, 131–144. doi:10.1038/nrneurol.2010.4
Botello-Marabotto, M., Martínez-Bisbal, M. C., Calero, M., Bernardos, A., Pastor, A. B., Medina, M., et al. (2023). Non-invasive biomarkers for mild cognitive impairment and Alzheimer’s disease. Neurobiol. Dis. 187, 106312. doi:10.1016/j.nbd.2023.106312
Botosoa, E. P., Zhu, M., Marbeuf-Gueye, C., Triba, M. N., Dutheil, F., Duyckäerts, C., et al. (2012). NMR metabolomic of frontal cortex extracts: first study comparing two neurodegenerative diseases, Alzheimer disease and amyotrophic lateral sclerosis. IRBM 33, 281–286. doi:10.1016/j.irbm.2012.08.002
Buergel, T., Steinfeldt, J., Ruyoga, G., Pietzner, M., Bizzarri, D., Vojinovic, D., et al. (2022). Metabolomic profiles predict individual multidisease outcomes. Nat. Med. 28, 2309–2320. doi:10.1038/s41591-022-01980-3
Costanzo, M., Caterino, M., Sotgiu, G., Ruoppolo, M., Franconi, F., and Campesi, I. (2022). Sex differences in the human metabolome. Biol. Sex. Differ. 13, 30. doi:10.1186/s13293-022-00440-4
Di Costanzo, A., Paris, D., Melck, D., Angiolillo, A., Corso, G., Maniscalco, M., et al. (2020). Blood biomarkers indicate that the preclinical stages of Alzheimer’s disease present overlapping molecular features. Sci. Rep. 10, 15612. doi:10.1038/s41598-020-71832-y
Dubois, B., Feldman, H. H., Jacova, C., Hampel, H., Molinuevo, J. L., Blennow, K., et al. (2014). Advancing research diagnostic criteria for Alzheimer’s disease: the IWG-2 criteria. Lancet Neurol. 13, 614–629. doi:10.1016/S1474-4422(14)70090-0
Emwas, A.-H., Roy, R., McKay, R. T., Tenori, L., Saccenti, E., Nagana Gowda, G. A., et al. (2019). NMR spectroscopy for metabolomics research. Metabolites 9, 123. doi:10.3390/metabo9070123
Foley, S. F., Tansey, K. E., Caseras, X., Lancaster, T., Bracht, T., Parker, G., et al. (2017). Multimodal brain imaging reveals structural differences in Alzheimer’s disease polygenic risk carriers: a study in healthy young adults. Biol. Psychiatry 81, 154–161. doi:10.1016/j.biopsych.2016.02.033
Ghini, V., Meoni, G., Vignoli, A., Di Cesare, F., Tenori, L., Turano, P., et al. (2023). Fingerprinting and profiling in metabolomics of biosamples. Prog. Nucl. Magnetic Reson. Spectrosc. 138 (139), 105–135. doi:10.1016/j.pnmrs.2023.10.002
González-Domínguez, R., Sayago, A., and Fernández-Recamales, Á. (2017). Metabolomics in Alzheimer’s disease: the need of complementary analytical platforms for the identification of biomarkers to unravel the underlying pathology. J. Chromatogr. B 1071, 75–92. doi:10.1016/j.jchromb.2017.02.008
Graham, S. F., Holscher, C., and Green, B. D. (2014). Metabolic signatures of human Alzheimer’s disease (AD): 1H NMR analysis of the polar metabolome of post-mortem brain tissue. Metabolomics 10, 744–753. doi:10.1007/s11306-013-0610-1
Holmes, M. V., Millwood, I. Y., Kartsonaki, C., Hill, M. R., Bennett, D. A., Boxall, R., et al. (2018). Lipids, lipoproteins, and metabolites and risk of myocardial infarction and stroke. J. Am. Coll. Cardiol. 71, 620–632. doi:10.1016/j.jacc.2017.12.006
Jessen, F., Amariglio, R. E., van Boxtel, M., Breteler, M., Ceccaldi, M., Chételat, G., et al. (2014). A conceptual framework for research on subjective cognitive decline in preclinical Alzheimer’s disease. Alzheimer’s Dementia 10, 844–852. doi:10.1016/j.jalz.2014.01.001
Jobard, E., Dossus, L., Baglietto, L., Fornili, M., Lécuyer, L., Mancini, F. R., et al. (2021). Investigation of circulating metabolites associated with breast cancer risk by untargeted metabolomics: a case–control study nested within the French E3N cohort. Br. J. Cancer 124, 1734–1743. doi:10.1038/s41416-021-01304-1
Kim, D. H., Gim, J.-A., Yoon, D., Kim, S., and Kim, H.-S. (2017). Metabolomics and mitochondrial dysfunction in Alzheimer’s disease. Genes Genom 39, 295–300. doi:10.1007/s13258-016-0494-3
Kloske, C. M., and Wilcock, D. M. (2020). The important interface between apolipoprotein E and neuroinflammation in Alzheimer’s disease. Front. Immunol. 11. doi:10.3389/fimmu.2020.00754
Kurbatova, N., Garg, M., Whiley, L., Chekmeneva, E., Jiménez, B., Gómez-Romero, M., et al. (2020). Urinary metabolic phenotyping for Alzheimer’s disease. Sci. Rep. 10, 21745. doi:10.1038/s41598-020-78031-9
Mattsson, N., Zetterberg, H., Hansson, O., Andreasen, N., Parnetti, L., Jonsson, M., et al. (2009). CSF biomarkers and incipient Alzheimer disease in patients with mild cognitive impairment. JAMA 302, 385–393. doi:10.1001/jama.2009.1064
McCartney, A., Vignoli, A., Tenori, L., Fornier, M., Rossi, L., Risi, E., et al. (2019). Metabolomic analysis of serum may refine 21-gene expression assay risk recurrence stratification. NPJ Breast Cancer 5, 26. doi:10.1038/s41523-019-0123-9
Nicholson, J. K., and Lindon, J. C. (2008). Systems biology: metabonomics. Nature 455, 1054–1056. doi:10.1038/4551054a
Paglia, G., Stocchero, M., Cacciatore, S., Lai, S., Angel, P., Alam, M. T., et al. (2016). Unbiased metabolomic investigation of Alzheimer’s disease brain points to dysregulation of mitochondrial aspartate metabolism. J. Proteome Res. 15, 608–618. doi:10.1021/acs.jproteome.5b01020
Tahami Monfared, A. A., Byrnes, M. J., White, L. A., and Zhang, Q. (2022). Alzheimer’s disease: epidemiology and clinical progression. Neurol. Ther. 11, 553–569. doi:10.1007/s40120-022-00338-8
Vignoli, A., Ghini, V., Meoni, G., Licari, C., Takis, P. G., Tenori, L., et al. (2019). High-throughput metabolomics by 1D NMR. Angew. Chem. Int. Ed. 58, 968–994. doi:10.1002/anie.201804736
Vignoli, A., Meoni, G., Ghini, V., Di Cesare, F., Tenori, L., Luchinat, C., et al. (2022). NMR-based metabolomics to evaluate individual response to treatments. Handb. Exp. Pharmacol. doi:10.1007/164_2022_618
Vignoli, A., Paciotti, S., Tenori, L., Eusebi, P., Biscetti, L., Chiasserini, D., et al. (2020a). Fingerprinting Alzheimer’s disease by 1H nuclear magnetic resonance spectroscopy of cerebrospinal fluid. J. Proteome Res. 19, 1696–1705. doi:10.1021/acs.jproteome.9b00850
Vignoli, A., Santini, G., Tenori, L., Macis, G., Mores, N., Macagno, F., et al. (2020b). NMR-based metabolomics for the assessment of inhaled pharmacotherapy in chronic obstructive pulmonary disease patients. J. Proteome Res. 19, 64–74. doi:10.1021/acs.jproteome.9b00345
Vignoli, A., Tenori, L., Luchinat, C., and Saccenti, E. (2018). Age and sex effects on plasma metabolite association networks in healthy subjects. J. Proteome Res. 17, 97–107. doi:10.1021/acs.jproteome.7b00404
Weng, J., Muti, I. H., Zhong, A. B., Kivisäkk, P., Hyman, B. T., Arnold, S. E., et al. (2022). A nuclear magnetic resonance spectroscopy method in characterization of blood metabolomics for Alzheimer’s disease. Metabolites 12, 181. doi:10.3390/metabo12020181
Wishart, D. S. (2016). Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 15, 473–484. doi:10.1038/nrd.2016.32
Yilmaz, A., Geddes, T., Han, B., Bahado-Singh, R. O., Wilson, G. D., Imam, K., et al. (2017). Diagnostic biomarkers of Alzheimer’s disease as identified in saliva using 1H NMR-based metabolomics. J. Alzheimers Dis. 58, 355–359. doi:10.3233/JAD-161226
Yilmaz, A., Ugur, Z., Bisgin, H., Akyol, S., Bahado-Singh, R., Wilson, G., et al. (2020a). Targeted metabolic profiling of urine highlights a potential biomarker panel for the diagnosis of Alzheimer’s disease and mild cognitive impairment: a pilot study. Metabolites 10, 357. doi:10.3390/metabo10090357
Yilmaz, A., Ustun, I., Ugur, Z., Akyol, S., Hu, W. T., Fiandaca, M. S., et al. (2020b). A community-based study identifying metabolic biomarkers of mild cognitive impairment and Alzheimer’s disease using artificial intelligence and machine learning. J. Alzheimer’s Dis. 78, 1381–1392. doi:10.3233/JAD-200305
Keywords: NMR, metabolites, lipoproteins, Alzheimer’s disease, mild cognitive impairment
Citation: Vignoli A and Tenori L (2023) NMR-based metabolomics in Alzheimer’s disease research: a review. Front. Mol. Biosci. 10:1308500. doi: 10.3389/fmolb.2023.1308500
Received: 06 October 2023; Accepted: 21 November 2023;
Published: 30 November 2023.
Edited by:
Fabio Moda, IRCCS Carlo Besta Neurological Institute Foundation, ItalyReviewed by:
Martina Banchelli, National Research Council (CNR), ItalyLuigi Russo, University of Campania Luigi Vanvitelli, Italy
Copyright © 2023 Vignoli and Tenori. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alessia Vignoli, alessia.vignoli@unifi.it; Leonardo Tenori, leonardo.tenori@unifi.it
 Alessia Vignoli
Alessia Vignoli Leonardo Tenori1,2,3*
Leonardo Tenori1,2,3*