CK2β Is a Gatekeeper of Focal Adhesions Regulating Cell Spreading
- 1Univ. Grenoble Alpes, INSERM, CEA, UMR Biosanté, U1292, Grenoble, France
- 2Univ. Grenoble Alpes, INSERM, CEA, UMR Biosanté U1292, CNRS FR 2048, Grenoble, France
- 3Univ. Grenoble Alpes, INSERM U1209, CNRS 5309, Institute for Advanced Biosciences (IAB), Grenoble, France
CK2 is a hetero-tetrameric serine/threonine protein kinase made up of two CK2α/αʹ catalytic subunits and two CK2β regulatory subunits. The free CK2α subunit and the tetrameric holoenzyme have distinct substrate specificity profiles, suggesting that the spatiotemporal organization of the individual CK2 subunits observed in living cells is crucial in the control of the many cellular processes that are governed by this pleiotropic kinase. Indeed, previous studies reported that the unbalanced expression of CK2 subunits is sufficient to drive epithelial to mesenchymal transition (EMT), a process involved in cancer invasion and metastasis. Moreover, sub-stoichiometric expression of CK2β compared to CK2α in a subset of breast cancer tumors was correlated with the induction of EMT markers and increased epithelial cell plasticity in breast carcinoma progression. Phenotypic changes of epithelial cells are often associated with the activation of phosphotyrosine signaling. Herein, using phosphotyrosine enrichment coupled with affinity capture and proteomic analysis, we show that decreased expression of CK2β in MCF10A mammary epithelial cells triggers the phosphorylation of a number of proteins on tyrosine residues and promotes the striking activation of the FAK1-Src-PAX1 signaling pathway. Moreover, morphometric analyses also reveal that CK2β loss increases the number and the spatial distribution of focal adhesion signaling complexes that coordinate the adhesive and migratory processes. Together, our findings allow positioning CK2β as a gatekeeper for cell spreading by restraining focal adhesion formation and invasion of mammary epithelial cells.
Introduction
The serine/threonine protein kinase CK2 has been implicated in the phosphorylation of hundreds of cellular proteins, and more than 10% of the phosphoproteome matches the consensus sequence for CK2 phosphorylation (Meggio and Pinna, 2003; Salvi et al., 2009). CK2 exists as a tetrameric complex made up of two CK2α or CK2αʹ catalytic subunits and two CK2β regulatory subunits (Litchfield, 2003). Extensive studies have established that CK2 regulates the crosstalk among multiple signaling pathways critical for cell differentiation, proliferation, and survival (Ahmed et al., 2002; Litchfield, 2003; Hong and Benveniste, 2021; Roffey and Litchfield, 2021), thereby justifying its potential as a therapeutic target in diverse human diseases (Borgo et al., 2021; Strum et al., 2021). A variety of experimental evidence supports the view that deregulated CK2 is functionally linked to cancer (Chua et al., 2017; Nunez de Villavicencio-Diaz et al., 2017). For instance, CK2α, whose expression is abnormally high in a wide range of tumors, operates as a cancer driver by creating the cellular environment favorable to neoplasia (Guerra and Issinger, 2008; Kreutzer et al., 2010; Schmitt et al., 2021) and the CK2α gene (CSNK2A1) is one of 186 genes making up an “invasiveness signature” (Rozanov et al., 2008). CK2α overexpression in human breast cancers is predictive of metastatic risk and poor outcome (Giusiano et al., 2011). In contrast to other multi-subunit protein kinases, the free CK2α catalytic subunits possess a constitutive activity, while the homodimer of CK2β regulatory subunits operates as a regulatory component, modifying the accessibility of binding substrates to the catalytic site of the holoenzyme (Filhol et al., 2003; Filhol and Cochet, 2009; Deshiere et al., 2011). The spatiotemporal organization of the CK2 individual subunits in living cells coupled with the observation that the free CK2α subunit and the tetrameric holoenzyme have distinct, though overlapping, substrate specificity profiles has led to postulate that such a balance is crucial in the control of the many cellular processes that are governed by this pleiotropic kinase (Filhol et al., 2003; Filhol et al., 2004). Interestingly, a sub-stoichiometric expression of CK2β compared to CK2α in a subset of breast cancer tumors is correlated with the induction of hypoxia and epithelial to mesenchymal transition (EMT) markers, providing evidence that unbalanced expression of CK2 subunits may influence key cellular processes associated with epithelial cell plasticity in breast carcinoma progression (Deshiere et al., 2013; Golden and Cantley, 2014). We further demonstrate that CK2β depletion in non-transformed mammary epithelial cells induced dissociation of cell–cell contacts, led to the acquisition of a mesenchymal cell shape (properties described for EMT-induced cells), and drove breast cell stemness (Deshiere et al., 2008; Deshiere et al., 2011; Deshiere et al., 2013; Golden and Cantley, 2014; Filhol et al., 2015; Duchemin-Pelletier et al., 2017). These disparate effects may be dependent on the levels of free CK2α, which is markedly elevated in metastatic tumors compared to non-transformed cells (Kim et al., 2007; Laramas et al., 2007; Giusiano et al., 2011; Lin et al., 2011; Roelants et al., 2015; Choy et al., 2017; Schmitt et al., 2021). Phenotypic changes in cancer cells are controlled by a network of positive and negative regulators (Qian et al., 2015). In particular, phosphorylation-mediated signaling networks drive tumor progression and regulate inherent and acquired therapeutic resistance (Burridge, 2017). While tyrosine phosphorylation (pTyr) accounts for only 0.1–1% of the total phosphoproteome, 30% of the known oncoproteins are protein tyrosine kinases (PTKs) that regulate a vast array of cellular signaling networks necessary for processes such as survival, growth, migration, and invasion (Hunter, 1995; Blume-Jensen and Hunter, 2001; Olsen et al., 2006; Hochgrafe et al., 2010). Phenotypic changes in epithelial cells such as focal adhesion formation, cell spreading, and cytoskeletal reorganization are associated with pTyr signaling pathways. PTK activity can promote invadopodia formation, invasion, and diverse cellular processes implicated in EMT induction and subsequent steps in the metastatic cascade (Kim et al., 2014; Okayama et al., 2015). Since a CK2β level establishes a critical cell fate threshold in the control of epithelial cell plasticity, the purpose of this study was to evaluate the impact of an unbalanced expression of CK2 subunits on the potential activation of pTyr signaling pathways. We demonstrate that CK2β depletion in MCF10A mammary cells (ΔCK2β) promotes the phosphorylation on tyrosine residues of a number of proteins involved in cell migration and focal adhesion formation, including Src and focal adhesion kinase 1 (FAK1) tyrosine kinases. Our findings reveal that the unbalanced expression of the CK2 subunits, which was observed in a subset of breast tumors (Deshiere et al., 2013), may affect pTyr signaling cascades through activation of the FAK-Src-paxillin axis promoting spreading and migration of mammary epithelial cells.
Material and Methods
Cell Culture and Retroviral Infection
MCF10A cells from ATCC-LGS (Molsheim, France) (CRL-10317) are mammary epithelial cells derived from fibrocystic breast tissue from women with no family history of breast cancer and no evidence of disease. MCF7 cells are invasive breast cancer cells. Stable CK2β silencing was accomplished in MCF10A or MCF7 cells by transduction with lentiviruses pLKO1 (Sigma-Aldrich, St. Louis, MO, United States) followed by puromycin selection as previously described (Deshiere et al., 2013). Stable transfected cells were maintained in puromycin-containing medium for 5–6 days before use and the silencing efficiency was evaluated at the protein level by Western blot analysis. Mock-cells are MCF10A or MCF7 cells transduced with an empty pLKO1 vector and are referred in the article as WT MCF10A cells. They were both cultured as described (Debnath et al., 2003). Transient CK2β-siRNA transfection was performed as previously described (Deshiere et al., 2013).
Western Blot Analysis
Western blot membranes were immunostained with primary antibodies as follows: phosphotyrosine 4G10 clone, #05-321 (Merck); P-FAK1 Y576/577, #3281, P-PAX1 Y118, #2541, P-p130CAS Y410, #4011, and P-EGFR Y1173, #4407 (Cell Signaling, Danvers, MA, United States); P-Src Y418, #11091 (SAB Signaling Antibody, Maryland, United States); Src, #sc-19, and EGFR, #sc-03 (Santa Cruz Biotechnology Inc., Santa Cruz, CA, United States); PAX1, #610052, FAK1, #610088, and p130CAS, #610271 (BD Biosciences, Pont de Claix, France); P-PLCγ Y771, #ab131455, and PLCγ, #ab52200 (Abcam, Paris, France); and GAPDH, #AM4300 (Invitrogen, Thermo Fisher, France). Secondary antibodies were peroxidase-conjugated affinity pure goat anti-rabbit IgG (#111035003) and peroxidase-conjugated affinity pure goat anti-mouse IgG (#115035003) from Jackson Immuno Research. Cells were scrapped in lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.5% NP40, 1 M EDTA, 1 mM EGTA, 10 mM NaF, 2.5 mM Na4P2O7, 1 mM β-glycerophosphate, 1 mM Na2VO4, and protease-inhibitor cocktail, #P8340 (Sigma-Aldrich). The lysate concentrations were normalized after determination of protein concentration using the BCA protein assay kit (Thermo Scientific, Sacley, France). SDS-PAGE was performed using pre-cast 4–12% gradient gel (Bio-Rad, Hercules, CA, United States). Separated proteins at 20–25 μg/lane were transferred to PVDF membranes (100 V for 60 min). Blotted membranes were blocked during 1 h at room temperature with saturation buffer (1% BSA in 10 mM Tris-buffered saline, 0.1% Tween (TBST)) and then incubated with primary antibody diluted in saturation buffer, for 2 h or overnight. After three washes with TBST, appropriate secondary antibodies were added for 1 h at room temperature. Luminata Forte Western HRP substrate (Millipore, Billerica, MA, United States) was added and membranes were read with Fusion Fx7 (PerkinElmer, Waltham, MA, United States). Quantification was performed using ImageJ software.
Immunoaffinity Purification of Tyr-Phosphorylated Cellular Proteins
Subconfluent WT and ΔCK2β MCF10A cells in two 10-cm culture dishes were washed with PBS and then lysed directly in lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.5% NP40, 1 mM EDTA, 1 mM EGTA, 10 mM NaF, 2.5 mM Na4P2O7, 1 mM β-glycerophosphate, 1 mM Na2VO4, and protease-inhibitor cocktail) for 20 min on ice. After centrifugation at 14,000 g for 15 min, cell lysates were normalized for protein concentration and incubated overnight with 50 μl of anti-phosphotyrosine monoclonal antibody covalently coupled to agarose beads (4G10 resin). The beads were then washed three times with lysis buffer and twice with PBS. The adsorbed proteins were eluted in four successive washes of 100 μl containing100 mM phenyl phosphate in 10 mM Tris-HCl pH 7.5, 50 mM NaCl, and 2% glycerol.
Phosphate Metal Affinity Chromatography of Phosphorylated Cellular Proteins
Phosphoprotein enrichment of cell extracts was performed according to the manufacturer’s protocol (TALON PMAC Phosphoprotein Enrichment Kit, Clontech Laboratories, United States). Subconfluent WT and ΔCK2β MCF10A cells in five 10-cm culture dishes were washed with PBS and then lysed directly in extraction/loading buffer A containing 10 mM NaF, 2.5 mM Na4P2O7, 1 mM β-glycerophosphate, 1 mM Na2VO4, and protease-inhibitor cocktail for 20 min on ice. After centrifugation at 14,000 g for 15 min, Na2VO4 which interferes with phosphoprotein binding to PMAC resin was removed by running the cell lysates through PD10 desalting columns. Samples of desalted cell lysates containing 3.5 mg protein were loaded on 1 ml of phosphoprotein affinity column (PMAC resin) and the column was agitated with the sample at 4°C for 2 h on a platform shaker. The column was then washed with 20 ml buffer A. The adsorbed proteins were eluted in three successive washes of 1 ml of buffer B (20 mM sodium phosphate and 0.5 M sodium chloride).
Immunofluorescence
A total of 10,000 cells were seeded on glass coverslips coated with 2.5 μg vitronectin and incubated at 37°C 5% CO2 for 24 h. The cells were fixed with 4% PFA for 15 min at room temperature, washed three times with PBS, and permeabilized for 10 min with PBS containing 0.3% Triton X-100. After blocking with 3% BSA and 1% goat serum in PBS for 1 h, the cells were incubated with the primary antibody diluted in blocking solution for 1 h. The samples were then washed, incubated with a secondary Alexa Fluor-conjugated antibody (Thermo Fisher Scientific) for 1 h in blocking solution and Alexa-Fluor 647-phalloidin (A22287), when required, and finally washed and mounted using Mowiol-containing DAPI (Santa Cruz). Primary antibodies used for immunostaining are the following: P-tyrosine 4G10 (Millipore #05321), P-paxillin (Invitrogen #44-722G), P-FAK (Invitrogen #44-624G), and P-Src (Invitrogen #44-660G).
Images were acquired on an iMIC Andromeda (FEI, Gräfelfing, Germany) microscope at 40× magnification in spinning disk mode. We analyzed at least 50 cells per experiment in 3 independent experiments. Segmentation of focal adhesions was performed using Ilastik (interactive machine learning for (bio)image analysis) (Berg et al., 2019) and subsequently analyzed with Fiji software (Schindelin et al., 2012). Fiji is an open-source platform for biological image analysis. Statistics analyses were performed with R. Significant differences were evaluated by Student test.
Protein Kinase CK2 Assay
Radiometric CK2 assay was performed in a final volume of 18 μl containing 3 μl of cell extract, and a mixture containing 100 μM of peptide substrate (RRREDEESDDEE), 10 mM MgCl2, and 100 μM [γ-32P]-ATP (6,000 Ci/mmol). Assays were performed at room temperature for 5 min before termination by the addition of 60 μl of 4% trichloroacetic acid. Incorporation of 32P in the peptide substrate was determined as previously described (Filhol et al., 1991). All kinase assays were performed in triplicates.
Multiplex Kinase Activity Profiling
PamGene PTK multiplex activity assays were used to investigate TK activity of 196 PTKs. This platform utilizes a high-throughput peptide microarray system by measuring the phosphorylation of peptide representations of targets/substrates (referred hereafter as phosphosites) that are immobilized on the PamChip® microarrays. Generic fluorescent-labeled antibodies that recognize phosphorylated residues are used to visualize the phosphorylation. Whole-cell lysates of WT and ΔCK2β MCF10A cells were prepared according to the manufacturer’s instructions (PamGene International, ’s-Hertogenbosch, the Netherlands). Cell samples were lysed for 30 min on ice and centrifuged for 15 min at 16,000 g at 4°C in a precooled centrifuge. After protein quantification (BCA protein determination, Pierce Scientific), aliquots of the samples were stored immediately in a −80°C freezer. Per array, 4 μg of protein and 100 μM ATP were loaded onto the appropriate PTK PamChip in kinase buffer. Phosphorylation of peptides was monitored by PamStation 12 following the manufacturer’s protocol as described previously (Arni et al., 2017). Technical triplicates were analyzed, and sequence-specific peptide tyrosine phosphorylation was detected by the fluorescein-labeled antibody PY20 (Exalpha, Maynard, MA). Capture of peptide phosphorylation signal is via a computer-controlled CCD camera. Kinomic profiling was performed using the Evolve software (PamGene International). Data were analyzed using the BioNavigator software version 6.3 (PamGene International) for raw data transformation into kinetic and steady-state values. Data were expressed as the average signal intensity (±SD) of the 196 peptide spots based on end levels of the phosphorylation curve. Prior to statistical analysis, fluorescent signal intensities were log2-transformed to satisfy the normality assumptions. Significant differences between two conditions were determined using two-sided Student’s t-test. The differential activity of kinases was predicted using an in-house bioinformatic approach, upstream kinase analysis (UKA) (Chirumamilla et al., 2019). Briefly, UKA identifies kinase activity based on a permutation analysis of the peptide substrate phosphorylation, by using known associations of phosphoproteins and kinases from literature and in silico databases. This permutation analysis gave a specificity score (mapping of peptides to kinases) and a significance score (kinase statistic that indicates the difference between treatment groups). Prior to UKA, the raw signal intensities were transformed using variance stabilizing normalization (VSN) (Huber et al., 2002).
Mass Spectrometry-Based Proteomic Analyses
After phosphoprotein enrichment (both 4G10 and PMAC), the eluates were solubilized in Laemmli buffer before stacking of proteins in the top layer of a 4–12% NuPAGE gel (Invitrogen) for separation followed by R-250 Coomassie blue staining. The gel bands were manually excised and digested using modified trypsin (sequencing grade, Promega) as previously described (Salvetti et al., 2016). After peptide extraction, the samples were split into two parts before drying: one-third for proteome analysis and two-thirds for phosphopeptide enrichment. The phosphopeptides were enriched with TiO2 beads (GL Science) in batch mode with a modified protocol from Jensen and Larsen (2007). Briefly, the samples were mixed for 1 h with 0.6 mg beads in loading buffer (1 M glycolic acid in 80% acetonitrile (v/v) and 5% TFA (v/v)). The beads were washed three times with loading buffer, 80% acetonitrile with 1% TFA (v/v), and finally 10% acetonitrile (v/v) with 0.1% TFA (v/v). The phosphopeptides were eluted with 10% ammonia solution (v/v) for 10 min. After acidification with formic acid, the peptides were desalted using C18 ultra-micro spin columns (Harvard Apparatus) and dried under vacuum. The dried extracted peptides were resuspended in 5% acetonitrile and 0.1% trifluoroacetic acid and analyzed by online nano-liquid chromatography coupled to tandem mass spectrometry (LC–MS/MS) (Ultimate 3000 RSLCnano and the Q-Exactive HF, Thermo Fisher Scientific). The peptides were sampled on a 300-μm internal diameter, 5-mm length PepMap C18 precolumn (Thermo Fisher Scientific) and separated on a 75-μm internal diameter, 250-mm length C18 column (Reprosil-Pur 120 C18-AQ, 1.9 μm particles, Dr. Maisch HPLC GmbH). The column flow rate was 300 nL/min. The mobile phases consisted of solution A (water with 0.1% (v/v) formic acid) and solution B (acetonitrile with 0.1% (v/v) formic acid). For 4G10 samples, the peptides were eluted with a gradient consisting of an increase in solvent B from 5 to 13% in 1.5 min, then from 13 to 31% over 29 min, and from 31 to 41% over 4.5 min. For data-dependent acquisition (DDA), the spray voltage was set at 2 kV and the heated capillary was adjusted to 270°C. Survey full-scan MS spectra (m/z = 400–2,000) were acquired with a resolution of 60,000 after the accumulation of 3 × 106 ions (maximum filling time 250 ms). The 12 most intense ions were fragmented by higher-energy collisional dissociation (HCD) after the accumulation of 1 × 106 ions (maximum filling time 250 ms). For PMAC samples, the nano-LC method consisted of a 120-min multilinear gradient at a flow rate of 300 nl/min (same gradient slopes as for 4G10 samples). Survey full-scan MS spectra (m/z = 400–2,000) were acquired with a resolution of 60,000 after the accumulation of 1 × 106 ions (maximum filling time 200 ms). The 20 most intense ions were fragmented by HCD after the accumulation of 1 × 105 ions (maximum filling time 50 ms). MS and MS/MS data were acquired using the software Xcalibur (Thermo Scientific).
Mass Spectrometry-Based Proteomic Data Processing
The data were processed automatically using Mascot Distiller software (version 2.7.1.0, Matrix Science). The peptides and proteins were identified using Mascot (version 2.8) through concomitant searches against Uniprot (Homo sapiens taxonomy, September 2021 version) and a database of 250 classical contaminants (homemade) and their corresponding reversed databases. Trypsin/P was chosen as the enzyme and two missed cleavages were allowed. Precursor and fragment mass error tolerances were set, respectively, to 10 and 20 ppm. Peptide modifications allowed during the search were cysteine carbamidomethylation (fixed), acetyl (protein N-terminal, variable), methionine oxidation (variable), and serine, threonine, tyrosine phosphorylation (variable). Proline software (version 2.1) (Bouyssie et al., 2020) was used to merge DDA results from proteome analysis and phosphopeptide enrichment. After combination, the results were filtered: conservation of rank 1 peptide-spectrum matches (PSMs) with a minimal length of 6 amino acids and a minimal score of 25. PSM score filtering was then optimized to reach a false discovery rate (FDR) of PSM identification below 1% by employing the target decoy approach. A minimum of one specific peptide per identified protein group was required. Proline was then used to perform MS1-based label-free quantification of the peptides and protein groups from the different samples with cross-assignment activated. Protein abundances were computed as a sum of specific peptide abundances, without using phosphopeptides and their counterparts. The data from 4G10 and PMAC enrichment were processed separately to obtain two different datasets.
Statistical Analysis of Mass Spectrometry-Based Proteomic Data
Statistical analysis on the proteins from 4G10 dataset was performed using ProStaR (Wieczorek et al., 2017) to determine differentially abundant proteins between WT and ΔCK2β MCF10A cells. Protein sets were filtered out if they were not identified and quantified in at least two biological replicates of one condition. Reverse protein sets and contaminants were also filtered out. These filters downsized the dataset to 249 proteins. After log2 transformation, the data were normalized with VSN method. POV missing values were imputed with slsa method and MEC ones with 1-percentile value of each sample. Statistical testing was conducted using limma test. Differentially expressed proteins were sorted out using a log2 (fold change) cut-off of 1 and a p-value inferior to 0.05. Proteins with more than three imputed values in the upregulated condition were invalidated. For each replicate, pTyr-phosphopeptides were declared upregulated in ΔCK2β cells (or, respectively, WT cells) if they were detected only in this condition or if their log2 (fold change) was higher than 1 (respectively, lower than –1). The peptides upregulated in at least two replicates were further considered. For the PMAC protein dataset, after log2 transformation, missing values were imputed with 1-percentile value. Log2 (fold change) was calculated and normalized.
Pathway Enrichment
Biological pathway enrichment analyses were carried out by Gene Set Enrichment Analysis (GSEA, FDR <0.05) from the 249 proteins quantified by proteomic analysis and kept during statistical analysis (Supplementary Table S1). The enrichGO and the cnet (category net plot used for visualization) functions were executed by ClusterProfiler v4.2.2 (Wu et al., 2015) (Bioconductor 3.14, https://www.bioconductor.org/, accessed on XX February 2022, R v4.1.2 (2021-11-01)). The enrichment analyses were performed using the biological pathway annotations from the Gene Ontology (GO, GO. db_v3.14.0 (2021-09-01), Bioconductor v3.14 Rv4.1.2 (2021-11-01), and KEGG databases (release 101, 2022/01) (Kanehisa and Goto, 2000). Gene sets consisting of at least 10 genes and less than 500 genes were retained for analysis. Thousand permutations were performed to compute p-values corrected by the Benjamini–Hochberg method.
Results
Changes in Tyrosine Kinase Activity in Extracts of WT and ΔCK2β MCF10A Cells
Given the interplay between serine/threonine and tyrosine kinases, we evaluated the potential impact of an imbalanced expression of CK2 subunits on global tyrosine kinase activity in WT non-transformed mammary MCF10A or shRNA-mediated CK2β-silenced MCF10A cells (ΔCK2β) (Deshiere et al., 2013; Deshiere et al., 2011; Deshiere et al., 2008; Filhol et al., 2015; Duchemin-Pelletier et al., 2017; Golden and Cantley, 2014). For this purpose, we used the PamChip PTK multiplex activity profiling (PamGene International BV ’s-Hertogenbosch, Netherlands). This methodology has been developed as a screening tool that allows for the robust analysis of kinase activity from cells and tissues (Anderson et al., 2015; Arsenault et al., 2011). A 73% efficiency in CK2β silencing was evaluated at the protein level in ΔCK2β MCF10A cells (Supplementary Figure S1). Equal amounts of protein extracts of WT or ΔCK2β MCF10A cells were then applied to peptide array chips. In a two-group comparison between these two cell lines, the deregulation of phosphorylation for each peptide represents the changes in kinase activity. There was a clear difference in the mean pattern of peptide substrate phosphorylation on comparing WT and ΔCK2β MCF10A cells as illustrated in the volcano plot of Figure 1A, indicating that upon CK2β silencing, the activity of certain signaling pathways driven by distinct PTK was altered (Figure 1B). Based on those phosphorylation patterns, a computational upstream kinase analysis predicted increased activity of PTKs that are known to contribute to invasive progression and metastasis of breast tumor, namely, FLT4, JAK1b, TRKB, and FLT1 (Zhang et al., 2010; Qian et al., 2015; Choy et al., 2017; Wehde et al., 2018). In contrast, the activity of other TKs (Lyn and Fgr) was decreased in ΔCK2β MCF10A cells (Figure 1B). Thus, a significant change in the pattern of Tyr phosphorylation was noted when comparing WT and ΔCK2β MCF10A cells. A ranked list of possible canonical pathways (and networks) responsible for the differences in the peptide phosphorylation was established. CK2β loss induced changes in transmembrane PTK in correlation with biological events such as cell adhesion and migration (Figure 1C).
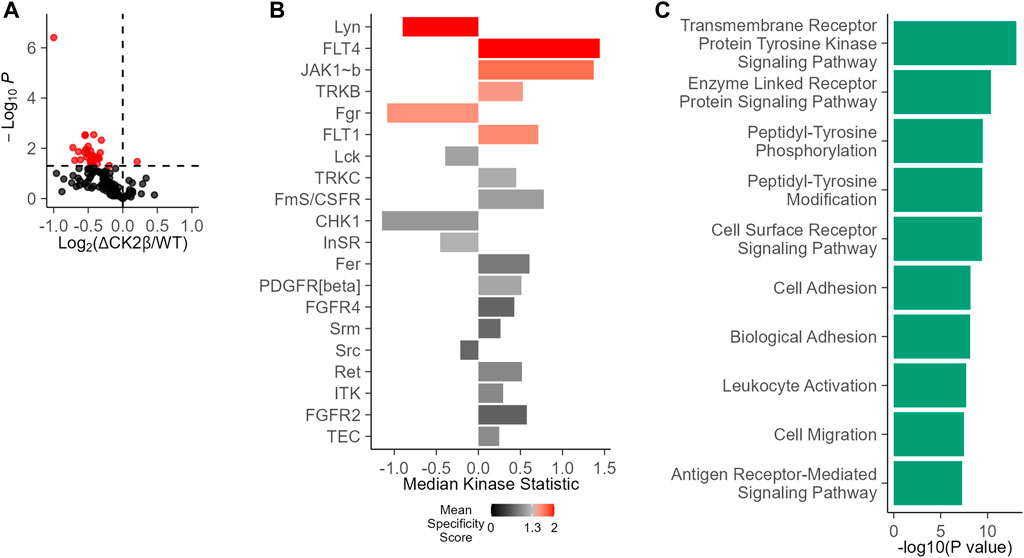
FIGURE 1. Changes in PTK activity in ΔCK2β MCF10A cells compared with WT cells. (A) Functional kinome profiling using the PamChips, which contain multiple copies of individual peptides. Sequence-specific tyrosine phosphorylation is detected by fluorochrome-coupled phospho-specific antibodies and CCD camera. The volcano plot visualizes the result of the tests by plotting, for each test, the effect size (x-axis, LFC or delta) versus significance (y-axis, –log10 (p-value)) of the test. Red spots are peptides that show significant differences compared to control (p < 0.05). (B) PTK score plot highlighting kinases that were modulated in ΔCK2β MCF10A cells compared with WT cells. The plot shows results from the upstream kinase analysis (UKA) tool, a functional scoring tool which rank-orders the top kinases that are differentially active between the two groups. The ranking factor is the final (median) kinase score. This score is based on a combined sensitivity score (difference between “treatment” and “control” groups) and specificity score (for a set of peptides to kinase relationship derived from current databases). The kinase statistics indicates the group differences, with effect size (values) and direction (+ or >0 = activation; – or 0 = inhibition). (C) Signaling pathways and biological process analysis in ΔCK2β MCF10A cells compared with WT cells (GProfiler, Gene Ontology Biological Processes).
Abundant pTyr-Containing Proteins Are Identified in ΔCK2β MCF10A Cells
To obtain an exhaustive landscape of proteins differentially modified on Tyr residues in WT versus ΔCK2β MCF10A cells, we performed a proteomic analysis of pTyr-containing proteins. Since pTyr-containing proteins of low abundance might be difficult to detect within cell extracts, phosphoprotein enrichment was performed using pTyr immunoaffinity chromatography on anti-phosphotyrosine monoclonal antibody covalently coupled to agarose beads (4G10 resin). After extensive washing, the retained proteins were eluted with 100 mM phenyl phosphate, a phosphotyrosine hapten analog (Figure 2A). After trypsin digestion of the enriched proteins, a fraction of peptides was further loaded on a TiO2 resin, to allow the selection of phosphorylated peptides. We identified 542 unique proteins (contaminants filtered) from the total tryptic digest of the proteins retained on the 4G10 resin (Figure 2B, Supplementary Table S1). In addition, by analyzing the tryptic peptides further enriched on the TiO2 resin, 204 phosphosites were identified. Among them, 40 sites from 25 unique proteins were localized on tyrosine with a confidence higher than 75% (Supplementary Table S2). A large part of the pTyr-phosphopeptides was more abundant or specifically detected in ΔCK2β MCF10A cells (Figure 2C, Supplementary Table S2). This was the case not only for the EPH tyrosine receptor A2 (EPHA2) but also for a number of non-receptor tyrosine kinases including FAK1, the signaling pseudokinase pragmin (PRAG1), activated CDC42 kinase 1 (ACK1), glycogen synthase kinase 3α (GSK3A), and mitogen-activated protein kinase 1 (MK01). pTyr-phosphopeptides from signaling proteins, such as p130CAS/BCAR1, paxillin (PAX1), partitioning defective 3 (PARD3), lipoma-preferred partner (LPP), and ARF GTPase-activating protein (GIT1), that are activated by tyrosine phosphorylation, were also enriched or specifically detected in ΔCK2β MCF10A cells (Figure 2C). Interestingly, several of these well-characterized proteins form supra-molecular complexes belonging to adhesive structures at cell membrane known under the name of focal adhesions (FAs), playing important functional roles in integrin signaling (Wozniak et al., 2004; Burridge, 2017) and coordinating the adhesive and migratory processes (Geiger and Bershadsky, 2001). Among them, FAK1 as well as GIT1, ACK1, PAX1, p130CAS, PARD3, and LPP are known substrates of Src kinase (Wang et al., 2006; Sachdev et al., 2009; Meenderink et al., 2010; Totaro et al., 2014; Wu et al., 2015; Ngan et al., 2017; Atashrazm and Ellis, 2021) (Table 1). Of note, even if we did not identify upregulated pTyr-phosphopeptides from their sequences, several proteins such as vimentin (VIME), fibronectin 1 (FINC), sortin nexin-18 (SNX18), BCAR3 adaptor protein (BCAR3), tensin 2 tyrosine protein phosphatase (TNS2), inactive-tyrosine protein kinase PEAK (PEAK1), and growth factor receptor-bound protein 2 (GRB2) were also substantially enriched in ΔCK2β MCF10A cell extracts (Supplementary Table S1). In contrast, pTyr-phosphopeptides from the epidermal growth factor receptor (EGFR) and the SH2 domain-containing adapter protein B (SHB) were downregulated in ΔCK2β MCF10A cells suggesting a de-enrichment of the proliferation signature (Figure 2C, Supplementary Table S2).
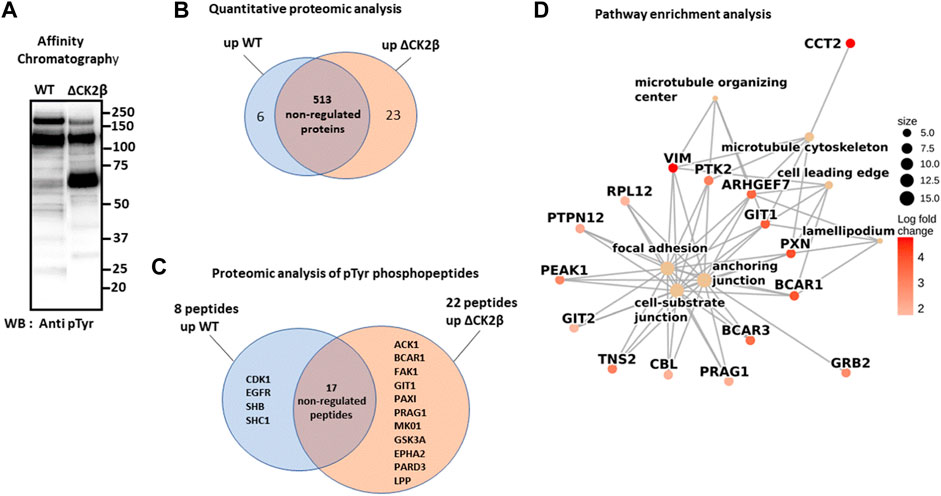
FIGURE 2. LC-MS/MS and GSEA analysis of differentially elevated levels of Tyr-phosphorylated proteins in WT and ΔCK2β MCF10A cells. (A) Proteins immunopurified from WT and ΔCK2β MCF10A cell lysates were analyzed by Western blotting for pTyr proteins using the 4G10 antibody. (B) Venn diagram showing differentially abundant proteins between WT and CK2β-depleted cells in samples obtained by immunoaffinity purification of Tyr-phosphorylated proteins (Supplementary Table S1). (C) Venn diagram showing the proteins in which differentially abundant pTyr-phosphopeptides between WT and CK2β-depleted cells were identified (Supplementary Table S2). These pTyr-phosphopeptides were identified and quantified from the LC-MS/MS analyses of peptide samples coming from a double enrichment starting from the two cell types: immunoaffinity purification of Tyr-phosphorylated proteins followed by the enrichment of their tryptic peptides on a TiO2 resin. (D) Significant gene ontology cellular components enriched from proteomics quantification of phosphoproteins (Supplementary Table S3) according to corrected p-values by the Benjamini–Hochberg method <0.05. Cellular component names are displayed as yellow nodes and bound to core phosphoproteins that led to a significant enrichment by GSEA. The core phosphoproteins are the nodes colored in red according to their log2 fold change value.
A pathway enrichment analysis by GSEA was carried out to identify deregulated molecular processes from proteomics quantification data (Supplementary Table S1). The enrichment analysis was performed from the log2 (fold change) values computed according to protein abundances between both phenotypes (ΔCK2β/WT). Figure 2D shows the seven significant cellular components enriched using the pathway annotation from the GO database (Supplementary Table S3). Significance was defined as each cellular component with a p-value less than 0.05 after the Benjamini–Hochberg correction. We observed that among them, the focal adhesion, anchoring junction, cell-substrate junction, microtubule cytoskeleton, and microtubule organizing center were significantly enriched in upregulated proteins. These enrichment insights support the idea that the overabundance of phosphoproteins involved in these biological processes following CK2β depletion leads to a revamping of the cell adhesion mechanisms. Moreover, the pathway enrichment by GSEA based on the KEGG database (Supplementary Table S4) reveals the upregulation of actin cytoskeleton as the only significant pathway. These results suggest that changes in phosphoprotein abundances following CK2β downregulation may also impair the actin or microtubule cytoskeleton regulation.
Focal Adhesion Proteins Are Hyperphosphorylated in Extracts of ΔCK2β MCF10A Cells
In line with the phosphoproteomic approach, immunoblotting of cell extracts of subconfluent WT and ΔCK2β MCF10A cells with the 4G10 antibody confirmed the upregulation in CK2β-depleted MCF10A cells of diverse tyrosine-phosphorylated proteins (Figure 3A). Notably, Western blot analysis of pTyr proteins released from immobilized phosphotyrosine antibody (4G10 resin) highlighted the enrichment of two major upregulated phosphoproteins (65 and 115 kDa), most highly phosphorylated on Tyr residues in ΔCK2β cell extracts (Figure 3B). Similar patterns of enhanced tyrosine-phosphorylated proteins could be observed in either MCF10A cells which have been transiently depleted for CK2β or in shRNA-mediated CK2β-silenced MCF7 cells (Supplementary Figure 2 and Supplementary Figure S3). To go further, relevant antibodies also revealed the upregulation of additional pTyr-containing proteins in ΔCK2β MCF10A cells. Among them, non-receptor tyrosine kinases were identified, including FAK1 and the Src kinase. Important adhesion complex proteins such as PAX1 and p130CAS were also highly tyrosine phosphorylated (Figure 3C). Immunoblotting as well as immunoprecipitation analysis in these immunoaffinity-purified proteins suggested that the major 65-kDa tyrosine-phosphorylated protein may represent the scaffolding protein PAX1 (Supplementary Figure S4). PAX1 phosphorylation on tyrosine residue (Y118) is a prominent event during EMT and cell migration, through its recruitment into adhesion sites called FAs (Nakamura et al., 2000). PAX1 is also a well-known substrate for the FAK1/Src adhesion protein complex, which phosphorylates its residues Tyr31 and Tyr118 in dynamic adhesions, thus regulating both the assembly and the turnover of FAs (Schindelin et al., 2012). Plasma membranes act as transient trapping sites for signaling molecules playing a pivotal role in biological functions, including signal transduction and cell–ECM communication. Interestingly, it has been reported that the localization of a fraction of CK2 to the plasma membrane is controlled by cell–matrix interactions (Filhol et al., 2015). Moreover, protein–protein interaction between the scaffold protein CKIP-1 and CK2 provided evidence for CK2 targeting to the plasma membrane (Olsten et al., 2004). Therefore, selected differentially tyrosine-phosphorylated proteins known to be localized to the plasma membrane were validated using immunoblotting. This was the case for the activation of EGFR, a key driver in growth factor-dependent signal transduction, and its downstream effector PLCγ in WT and ΔCK2β MCF10A cells. The phosphorylation of EGFR and PLCγ on Tyr1173 and Tyr771, respectively, was decreased in ΔCK2β MCF10A cells compared to WT cells (Figure 3D). Thus, in agreement with the proteomic analysis (Figure 2D), CK2β downregulation significantly inhibited the activated form of EGFR and reduced the activation of its downstream signaling molecule PLCγ. Collectively, these data were consistent with the proteomic analyses, highlighting the phosphorylation-dependent activation of focal adhesion markers of the FAK1-Src-PAX1 axis upon CK2β depletion in MCF10A cells.

FIGURE 3. Upregulation of pTyr proteins in lysates of ΔCK2β MCF10A cells. (A) Lysates from WT and ΔCK2β MCF10A cells were analyzed for pTyr-containing proteins using the 4G10 antibody. GAPDH was used as a loading control. (B) Cell extracts (500 μg of protein) of WT and ΔCK2β MCF10A cells were loaded on 4G10 resin. After extensive washing, the bound proteins were eluted with 100 mM phenyl phosphate. The collected fractions were analyzed by Western blotting with the 4G10 antibody. (C) Proteins immunopurified from WT and ΔCK2β MCF10A cell lysates were analyzed by Western blotting for p-FAK (Y576/577), p-Src (Y418), p-PAX1(Y118), p-p130CAS (Y410), FAK, Src, PAX1, and p130CAS and (D) for p-EGFR (Y1173), EGFR, p-PLCγ (Y771), and PLCγ.
CK2β Depletion Induces Cell Spreading and Increase of Focal Adhesion Number
Early evidence indicated that tyrosine phosphorylation plays an important role in the overall organization of adhesion complexes and their dynamic regulation (Craig and Johnson, 1996; Burridge et al., 1988). In addition, tyrosine phosphorylation events were acknowledged to be concentrated at focal adhesions formed by the cells with the substratum (Maher et al., 1985). It has also been reported that PAX1 and FAK1 enter nascent adhesions as discrete entities and form dynamic molecular complexes within adhesion sites. Moreover, phosphorylation of PAX1 at Y31-Y118 regulates the formation and size of the complexes (Choi et al., 2011). As these focal adhesion markers were identified in ΔCK2β MCF10A cells after pTyr immunoaffinity chromatography, we analyzed the impact of CK2β depletion in the distribution of focal adhesions of isolated MCF10A cells spread onto vitronectin. Vitronectin was chosen as the adhesive substrate since WT or ΔCK2β MCF10A cells plated on vitronectin-coated coverslips clearly display a distinguishable morphology (Deshiere et al., 2013). In particular, ΔCK2β MCF10A cells plated on vitronectin-coated coverslips display higher cell spreading as evidenced by the projected area of actin cytoskeleton (Figure 4A and Figure 4B). This cell spreading is associated with an increase in the number (Figure 4C) and the size (Figure 4D) of focal adhesions giving rise to an increase in the total adhesive surface when compared to the cell surface (Figure 4E). FAK1 phosphorylation is one of the key steps following integrin engagement, either directly or through its role as a scaffolding protein for Src (Tapial Martinez et al., 2020). Based on the increase in cell spreading upon CK2β loss, we investigated whether this increase might be related to a change in focal adhesion organization by immunostaining FAK1, Src, and PAX1 (Supplementary Figure S5A,F,K). Immunofluorescence analyses demonstrated that CK2β depletion increased the Src labeling and changed the spatial distribution of FAK1 and PAX1 in focal adhesion sites. Morphometric analyses showed that CK2β-depleted cells were much larger than WT cells. Moreover, descriptors of focal adhesion morphology such as number and mean size of the focal adhesions were strikingly upregulated in these cells (Supplementary Figure S5). It has been reported that the mean size of focal adhesions uniquely predicts cell speed independently of focal adhesion surface density and molecular composition (Kim and Wirtz, 2013). The regulation of FAs by CK2β is consistent with our previous observations, showing that CK2β-depleted cells displayed a mesenchymal phenotype and a loss of polarity driving a 2D-to-3D morphologic transformation (Deshiere et al., 2008; Deshiere et al., 2013).
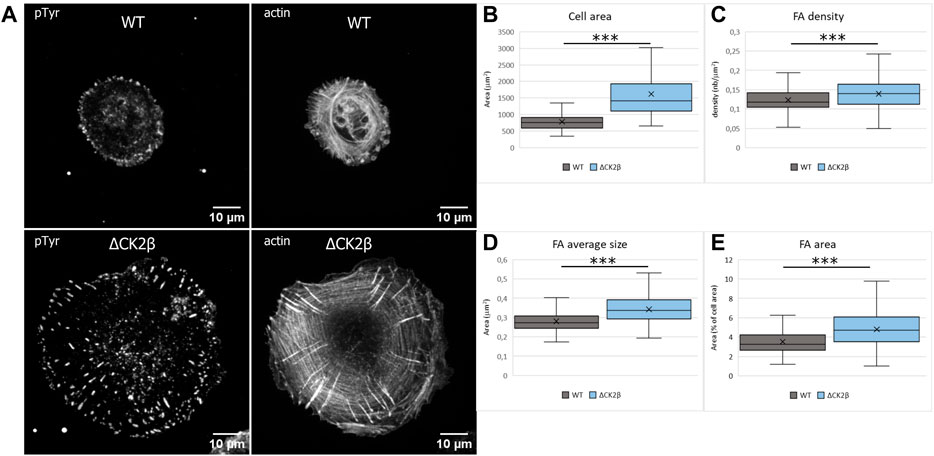
FIGURE 4. Cell spreading and size of focal adhesions are dependent on CK2β. (A) Staining of pTyr and actin was carried out on cells spread on vitronectin-coated cover glass for 24 h. Scale bar represents 10 µm. (B) Quantification of the cell area. (C) Quantification of the number of FAs normalized to the cell area. (D) Quantification of the FA average area. (E) Quantification of the FA area, normalized to the cell area. Error bars represent standard deviation. ***p-value ≤0.0005.
The Catalytic Activity of CK2 Is Not Involved in Enhanced Tyr Phosphorylation in ΔCK2β MCF10A Cells
CK2 is mostly known as a Ser/Thr kinase, yet the enzyme also exhibits tyrosine kinase activity in mammalian cells (Basnet et al., 2014; Borgo et al., 2021). To clarify the potential contribution of CK2 activity in the activation of these Tyr phosphorylations, we analyzed pTyr proteins in the extracts of WT or ΔCK2β MCF10A cells that had been incubated for 5 h with 10 μM of the small chemical CK2 inhibitor CX-4945. CK2 activity measured in whole-cell lysates from WT or ΔCK2β MCF10A cells treated with CX-4945 was inhibited by more than 90% (Figure 5A). Immunoblotting analysis of protein extracts from cells treated under these conditions revealed that the inhibition of CK2 did not abrogate the Tyr phosphorylation of the 65- and 115-kDa endogenous proteins (Figure 5B). In addition, the Tyr phosphorylation of adhesion complex proteins was either not affected (FAK1, p130CAS) or enhanced (Src, PAX1) upon CX-4945 treatment (Figure 5C). The increased Tyr phosphorylation of Src and PAX1 observed in ΔCK2β MCF10A cells after CX-4945 treatment suggests that the activation of these two components of the adhesion complexes could be similarly antagonized by CK2β and CK2α. This would reinforce the notion that any variation in the expression/activity of either CK2 subunits could have significant impacts on focal adhesion signaling pathways.
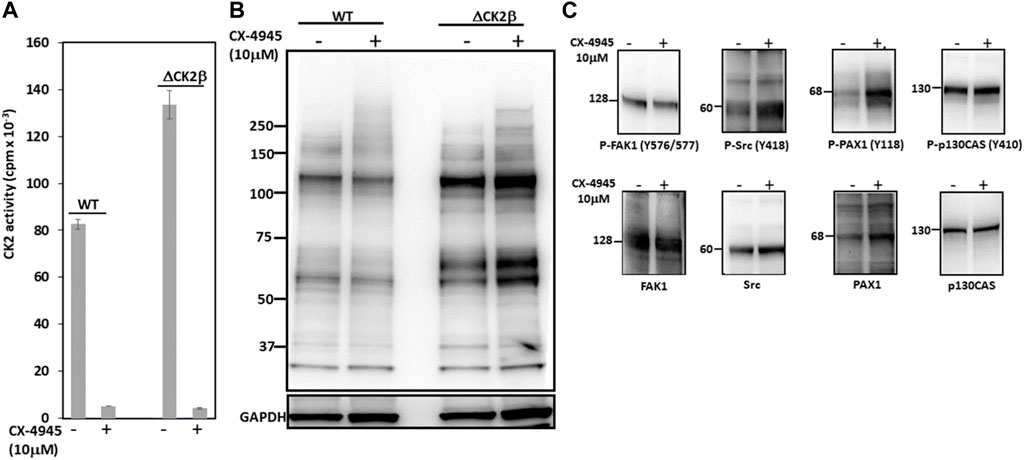
FIGURE 5. CK2 catalytic activity does not contribute to enhanced Tyr phosphorylation in ΔCK2β MCF10A cells. (A) CK2 kinase activity in cell lysates from WT or ΔCK2β MCF10A cells treated for 5 h in the absence or presence of 10 μM CX-4945. (B) Cell lysates as in (A) were Western blotted with 4G10 antibody for the presence of pTyr proteins. GAPDH was used as a loading control. (C) ΔCK2β MCF10A cell lysates were also analyzed by Western blotting for p-FAK (Y576/577), p-Src (Y418), p-PAX1 (Y118), p-p130CAS (Y410), FAK, Src, PAX1, and p130CAS.
Discussion
In the present study, we demonstrate that CK2β downregulation in MCF10A mammary cells triggers the phosphorylation of an array of tyrosine-phosphorylated proteins involved in cell migration and focal adhesion including Src and FAK1 tyrosine kinases. Our findings highlight a crosstalk between CK2, a serine/threonine kinase, and pTyr signaling pathways involved in phenotypic changes of epithelial cells. Such a potential link has been previously evoked. For instance, the catalytic subunits of CK2 are readily phosphorylated in vitro by the Src family protein tyrosine kinases Lyn and c-Fgr with a concomitant threefold increase in catalytic activity of CK2 (Donella-Deana et al., 2003). Interestingly, the presence of CK2β decreased this Lyn/Fgr-mediated phosphorylation, suggesting that at least in vitro, the CK2 holoenzyme is less prone to tyrosine phosphorylation. One could speculate that, within cells, the pool of catalytic subunits that is not incorporated into the holoenzyme would acquire an increased activity toward a subset of specific targets. Indeed, it has been reported that in advanced cancers, the TGFβ receptor (TGFBR) 1 kinase phosphorylated CK2β targeting it for degradation. The resulting CK2α/CK2β subunit imbalance promoted the activation of CK2 and EMT induction (Kim et al., 2018). Thus, a dysregulation in CK2 subunit levels, originating either from shRNA-mediated CK2β silencing or from TGFβ-dependent decrease of CK2β, invariably promotes EMT activation in epithelial cells (Deshiere et al., 2013; Filhol et al., 2015). Consequently, it is conceivable that signaling-mediated drop of CK2β expression could increase intracellular CK2 activity impacting downstream signaling events such as pTyr-relayed signaling cascades. Indeed, our results reveal that spreading-coupled pTyr signaling pathways including FAK1, PAX1, ACK1, EPHA2, PARD3, and LPP are impacted by CK2β depletion (Supplementary Table S2). Of note, the focal adhesion network (www.adhesome.org) which is involved in the regulation of cell shape and focal adhesion complexes is acting at the plasma membrane level. This is consistent with previous observations showing that in epithelial cells, the association of a fraction of CK2 to the plasma membrane is controlled by cell–matrix interactions (Deshiere et al., 2008). Phosphoproteomic analysis revealed that FAK1 is activated through membrane recruitment by growth factors, extracellular matrix, and integrin signaling followed by subsequent autophosphorylation at Tyr 397 exposing an SH2-binding domain, which in turn recruits Src and promotes phosphorylation of FAK1 at Tyr 576/577 (Robertson et al., 2015). The fully active FAK1/Src complex can then recruit, phosphorylate, and activate numerous targets including p130CAS and PAX1, which play critical roles in regulating cell spreading, cell motility, and cytoskeletal modifications (Zhang et al., 2010). In this line, it has been shown that maximal tyrosine phosphorylation of PAX1 by Src and FAK1 is required for the induction of anchorage-independent signal transduction, a characteristic of metastatic cells (Vilk et al., 2008; Anderson et al., 2015). As a signal transducer for ECM–tumor cell interactions, FAK1 expression and/or activation has been found altered in most human epithelial cancers, resulting in enhanced invasive potential and poor overall patient survival (McLean et al., 2005; Luo and Guan, 2010). Similarly, CK2α overexpression in a significant fraction of breast cancers is predictive of metastatic risk (Giusiano et al., 2011). Moreover, suppression of CK2β expression in a subset of breast cancers modulates the expression of EMT-related markers (Deshiere et al., 2013). Our findings now demonstrate that a loss of CK2β leads to changes in the size of the focal adhesions, through the activation of the well-characterized FAK1-Src-PAX1 signaling pathway.
While CK2β is traditionally considered as a regulatory subunit of CK2, some studies also suggest that this subunit might function independently of CK2α (Filhol et al., 2015; Borgo et al., 2019; Lettieri et al., 2019). Indeed, blocking the CK2 catalytic activity in CK2β-depleted cells with CX-4945 did not affect the Tyr phosphorylation of FAK1 and p130CAS while triggering enhanced phosphorylation of Src and PAX1, supporting the hypothesis that CK2β may play additional roles outside of the CK2 tetrameric holoenzyme (Bibby and Litchfield, 2005; Borgo et al., 2019). Our data show that by limiting the focal adhesion formation, CK2β appears as a major molecular break for cell spreading. The impact of CK2β on FAs is supported by the role of CK2 in cytoskeletal alteration (Canton and Litchfield, 2006; Lettieri et al., 2019) and in the regulation of actin and tubulin polymerization (reviewed in D'Amore et al., 2019). For instance, it has been previously reported that epithelial cell polarity and morphology might be controlled by CK2 through the phosphorylation of the Pleckstrin homology domain-containing protein (CKIP-1) and coronins, key proteins playing roles in the functional organization of actin-dependent cellular processes like protrusion formation, migration, and invasion (Canton et al., 2005; Deshiere et al., 2008; Deshiere et al., 2011; Xavier et al., 2012; Filhol et al., 2015).
We have generated proteomics data on samples enriched by PMAC (Supplementary Table S5), which give a global overview of the phosphoproteome, and on the 4G10 affinity resin which enriches for pTyr-modified proteins (Supplementary Table S2). Both approaches provided a complementary view on the phosphoproteins deregulated upon deletion of the CK2β subunit and are also incomplete, as they missed many proteins involved in the Src pathway, for example. To follow specifically the phosphorylation state of proteins of interest, a complementary approach such as targeted proteomics on the proteolytic peptides containing the modified Ser/Thr/Tyr residues of interest or Western blot analysis using specific antibodies would be better suited. Yet targeted proteomics requires time for method development and Western blot analysis is only possible if antibodies of validated specificity are available.
In summary, the data collected here support the notion that CK2β works by more than altering the CK2 activity: it also plays a key role in the negative regulation of focal adhesion maturation, blocking dominant pTyr downstream signaling pathways involved in cell scattering and invasion. Further functional characterization of the molecular mechanism by which CK2β could restrain these pathways in normal human mammary cells is the subject of ongoing work. More broadly, this analysis will cast additional insights into the specific contribution of individual CK2 subunits during breast carcinoma progression.
Data Availability Statement
Publicly available datasets were analyzed in this study. The LC-MS/MS data have been submitted to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository under dataset identifier PXD030991.
Author Contributions
Conceptualization: CC, OF, and DP; investigation: A-MH, OF, CC, and A-PB; formal analysis: CC, OF, DP, FJ, CB, A-MH, CA-R, and A-PB; writing original draft: CC, OF, and DP; funding acquisition: OF, CC, CA-R, and DP; supervision: OF, CC, DP, and CA-R.
Funding
FJ is supported by the KATY EU program Horizon 2020/H2020-SCI-FA-DTS-2020-1 (contract number 101017453). The proteomic experiments were partially supported by Agence Nationale de la Recherche under projects ProFI (Proteomics French Infrastructure, ANR-10-INBS-08) and GRAL, a program from the Chemistry Biology Health (CBH) Graduate School of University Grenoble Alpes (ANR-17-EURE-0003). This research was supported in part by Institut National de la Santé et de la Recherche Médicale (Inserm), Commissariat à l’Energie Atomique et aux Energies Alternatives (CEA), the Ligue Nationale contre le Cancer, the Ligue Comité de la Loire, and Université Grenoble-Alpes (UGA).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Peter Linders for his contribution to PamGene data analysis and Mélodie Malige and Justine Cristante for their technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.900947/full#supplementary-material
References
Abu-Thuraia, A., Goyette, M.-A., Boulais, J., Delliaux, C., Apcher, C., Schott, C., et al. (2020). AXL Confers Cell Migration and Invasion by Hijacking a PEAK1-Regulated Focal Adhesion Protein Network. Nat. Commun. 11, 3586. doi:10.1038/s41467-020-17415-x
Ahmed, K., Gerber, D. A., and Cochet, C. (2002). Joining the Cell Survival Squad: An Emerging Role for Protein Kinase CK2. Trends Cell Biol. 12, 226–230. doi:10.1016/s0962-8924(02)02279-1
Anderson, J. C., Willey, C. D., Mehta, A., Welaya, K., Chen, D., Duarte, C. W., et al. (2015). High Throughput Kinomic Profiling of Human Clear Cell Renal Cell Carcinoma Identifies Kinase Activity Dependent Molecular Subtypes. PLoS One 10, e0139267. doi:10.1371/journal.pone.0139267
Arni, S., LE, T. H. N., DE Wijn, R., Garcia-Villegas, R., Dankers, M., Weder, W., et al. (2017). Ex Vivo Multiplex Profiling of Protein Tyrosine Kinase Activities in Early Stages of Human Lung Adenocarcinoma. Oncotarget 8, 68599–68613. doi:10.18632/oncotarget.19803
Arsenault, R., Griebel, P., and Napper, S. (2011). Peptide Arrays for Kinome Analysis: New Opportunities and Remaining Challenges. Proteomics 11, 4595–4609. doi:10.1002/pmic.201100296
Atashrazm, F., and Ellis, S. (2021). The Polarity Protein PARD3 and Cancer. Oncogene 40, 4245–4262. doi:10.1038/s41388-021-01813-6
Basnet, H., Su, X. B., Tan, Y., Meisenhelder, J., Merkurjev, D., Ohgi, K. A., et al. (2014). Tyrosine Phosphorylation of Histone H2A by CK2 Regulates Transcriptional Elongation. Nature 516, 267–271. doi:10.1038/nature13736
Berg, S., Kutra, D., Kroeger, T., Straehle, C. N., Kausler, B. X., Haubold, C., et al. (2019). Ilastik: Interactive Machine Learning for (Bio)Image Analysis. Nat. Methods 16, 1226–1232. doi:10.1038/s41592-019-0582-9
Bibby, A. C., and Litchfield, D. W. (2005). The Multiple Personalities of the Regulatory Subunit of Protein Kinase CK2: CK2 Dependent and CK2 Independent Roles Reveal a Secret Identity for CK2β. Int. J. Biol. Sci. 1, 67–79. doi:10.7150/ijbs.1.67
Blume-Jensen, P., and Hunter, T. (2001). Oncogenic Kinase Signalling. Nature 411, 355–365. doi:10.1038/35077225
Borgo, C., D’Amore, C., Sarno, S., Salvi, M., and Ruzzene, M. (2021). Protein Kinase CK2: A Potential Therapeutic Target for Diverse Human Diseases. Sig. Transduct. Target Ther. 6, 183. doi:10.1038/s41392-021-00567-7
Borgo, C., Franchin, C., Cesaro, L., Zaramella, S., Arrigoni, G., Salvi, M., et al. (2019). A Proteomics Analysis of CK2β (−/−) C2C12 Cells Provides Novel Insights into the Biological Functions of the Non‐Catalytic β Subunit. FEBS J. 286, 1561–1575. doi:10.1111/febs.14799
Bouyssié, D., Hesse, A.-M., Mouton-Barbosa, E., Rompais, M., Macron, C., Carapito, C., et al. (2020). Proline: An Efficient and User-Friendly Software Suite for Large-Scale Proteomics. Bioinformatics 36, 3148–3155. doi:10.1093/bioinformatics/btaa118
Bridgewater, R. E., Norman, J. C., and Caswell, P. T. (2012). Integrin Trafficking at a Glance. J. Cell Sci. 125, 3695–3701. doi:10.1242/jcs.095810
Brown, M. C., Perrotta, J. A., and Turner, C. E. (1996). Identification of LIM3 as the Principal Determinant of Paxillin Focal Adhesion Localization and Characterization of a Novel Motif on Paxillin Directing Vinculin and Focal Adhesion Kinase Binding. J. Cell Biol. 135, 1109–1123. doi:10.1083/jcb.135.4.1109
Burridge, K., Fath, K., Kelly, T., Nuckolls, G., and Turner, C. (1988). Focal Adhesions: Transmembrane Junctions Between the Extracellular Matrix and the Cytoskeleton. Annu. Rev. Cell. Biol. 4, 487–525. doi:10.1146/annurev.cb.04.110188.002415
Burridge, K. (2017). Focal Adhesions: A Personal Perspective on a Half Century of Progress. FEBS J. 284, 3355–3361. doi:10.1111/febs.14195
Canton, D. A., and Litchfield, D. W. (2006). The Shape of Things to Come: An Emerging Role for Protein Kinase CK2 in the Regulation of Cell Morphology and the Cytoskeleton. Cell. Signal. 18, 267–275. doi:10.1016/j.cellsig.2005.07.008
Canton, D. A., Olsten, M. E. K., Kim, K., Doherty-Kirby, A., Lajoie, G., Cooper, J. A., et al. (2005). The Pleckstrin Homology Domain-Containing Protein CKIP-1 Is Involved in Regulation of Cell Morphology and the Actin Cytoskeleton and Interaction with Actin Capping Protein. Mol. Cell Biol. 25, 3519–3534. doi:10.1128/mcb.25.9.3519-3534.2005
Chen, Z., Oh, D., Biswas, K. H., Yu, C. H., Zaidel-Bar, R., and Groves, J. T. (2018). Spatially Modulated ephrinA1:EphA2 Signaling Increases Local Contractility and Global Focal Adhesion Dynamics to Promote Cell Motility. Proc. Natl. Acad. Sci. U. S. A. 115, E5696–E5705. doi:10.1073/pnas.1719961115
Chirumamilla, C. S., Fazil, M. H. U. T., Perez-Novo, C., Rangarajan, S., DE Wijn, R., Ramireddy, P., et al. (2019). Profiling Activity of Cellular Kinases in Migrating T-Cells. Methods Mol. Biol. 1930, 99–113. doi:10.1007/978-1-4939-9036-8_13
Choi, C. K., Zareno, J., Digman, M. A., Gratton, E., and Horwitz, A. R. (2011). Cross-Correlated Fluctuation Analysis Reveals Phosphorylation-Regulated Paxillin-FAK Complexes in Nascent Adhesions. Biophysical J. 100, 583–592. doi:10.1016/j.bpj.2010.12.3719
Choy, L., Hagenbeek, T. J., Solon, M., French, D., Finkle, D., Shelton, A., et al. (2017). Constitutive NOTCH3 Signaling Promotes the Growth of Basal Breast Cancers. Cancer Res. 77, 1439–1452. doi:10.1158/0008-5472.can-16-1022
Chua, M. M., Ortega, C. E., Sheikh, A., Lee, M., Abdul-Rassoul, H., Hartshorn, K. L., et al. (2017). CK2 in Cancer: Cellular and Biochemical Mechanisms and Potential Therapeutic Target. Pharm. (Basel) 10, 18. doi:10.3390/ph10010018
Craig, S. W., and Johnson, R. P. (1996). Assembly of Focal Adhesions: Progress, Paradigms, and Portents. Curr. Opin. Cell Biol. 8, 74–85. doi:10.1016/s0955-0674(96)80051-2
D'Amore, C., Salizzato, V., Borgo, C., Cesaro, L., Pinna, L. A., and Salvi, M. (2019). A Journey Through the Cytoskeleton with Protein Kinase CK2. Cpps 20, 547–562. doi:10.2174/1389203720666190119124846
Debnath, J., Muthuswamy, S. K., and Brugge, J. S. (2003). Morphogenesis and Oncogenesis of MCF-10A Mammary Epithelial Acini Grown in Three-Dimensional Basement Membrane Cultures. Methods 30, 256–268. doi:10.1016/s1046-2023(03)00032-x
Deshiere, A., Duchemin-Pelletier, E., Spreux, E., Ciais, D., Combes, F., Vandenbrouck, Y., et al. (2013). Unbalanced Expression of CK2 Kinase Subunits Is Sufficient to Drive Epithelial-To-Mesenchymal Transition by Snail1 Induction. Oncogene 32, 1373–1383. doi:10.1038/onc.2012.165
Deshiere, A., Duchemin-Pelletier, E., Spreux, E., Ciais, D., Forcet, C., Cochet, C., et al. (2011). Regulation of Epithelial to Mesenchymal Transition: CK2β on Stage. Mol. Cell Biochem. 356, 11–20. doi:10.1007/s11010-011-0942-y
Deshière, A., Theis-Febvre, N., Martel, V., Cochet, C., and Filhol, O. (2008). Protein Kinase CK2 and Cell Polarity. Mol. Cell Biochem. 316, 107–113. doi:10.1007/s11010-008-9845-y
Donato, D. M., Ryzhova, L. M., Meenderink, L. M., Kaverina, I., and Hanks, S. K. (2010). Dynamics and Mechanism of p130Cas Localization to Focal Adhesions. J. Biol. Chem. 285, 20769–20779. doi:10.1074/jbc.m109.091207
Donella-Deana, A., Cesaro, L., Sarno, S., Ruzzene, M., Brunati, A. M., Marin, O., et al. (2003). Tyrosine Phosphorylation of Protein Kinase CK2 by Src-Related Tyrosine Kinases Correlates with Increased Catalytic Activity. Biochem. J. 372, 841–849. doi:10.1042/bj20021905
Duchemin-Pelletier, E., Baulard, M., Spreux, E., Prioux, M., Burute, M., Mograbi, B., et al. (2017). Stem Cell-Like Properties of CK2β-Down Regulated Mammary Cells. Cancers (Basel) 9, 114. doi:10.3390/cancers9090114
Filhol, O., Cochet, C., Delagoutte, T., and Chambaz, E. M. (1991). Polyamine Binding Activity of Casein Kinase II. Biochem. Biophysical Res. Commun. 180, 945–952. doi:10.1016/s0006-291x(05)81157-x
Filhol, O., and Cochet, C. (2009). Protein Kinase CK2 in Health and Disease. Cell. Mol. Life Sci. 66, 1830–1839. doi:10.1007/s00018-009-9151-1
Filhol, O., Giacosa, S., Wallez, Y., and Cochet, C. (2015). Protein Kinase CK2 in Breast Cancer: The CK2β Regulatory Subunit Takes Center Stage in Epithelial Plasticity. Cell. Mol. Life Sci. 72, 3305–3322. doi:10.1007/s00018-015-1929-8
Filhol, O., Martiel, J. L., and Cochet, C. (2004). Protein Kinase CK2: A New View of an Old Molecular Complex. EMBO Rep. 5, 351–355. doi:10.1038/sj.embor.7400115
Filhol, O., Nueda, A., Martel, V., Gerber-Scokaert, D., Benitez, M. J., Souchier, C., et al. (2003). Live-Cell Fluorescence Imaging Reveals the Dynamics of Protein Kinase CK2 Individual Subunits. Mol. Cell Biol. 23, 975–987. doi:10.1128/mcb.23.3.975-987.2003
Finney, A. C., Scott, M. L., Reeves, K. A., Wang, D., Alfaidi, M., Schwartz, J. C., et al. (2021). EphA2 Signaling within Integrin Adhesions Regulates Fibrillar Adhesion Elongation and Fibronectin Deposition. Matrix Biol. 103-104, 1–21. doi:10.1016/j.matbio.2021.09.001
Galvagni, F., Pennacchini, S., Salameh, A., Rocchigiani, M., Neri, F., Orlandini, M., et al. (2010). Endothelial Cell Adhesion to the Extracellular Matrix Induces C-Src-Dependent VEGFR-3 Phosphorylation Without the Activation of the Receptor Intrinsic Kinase Activity. Circulation Res. 106, 1839–1848. doi:10.1161/circresaha.109.206326
Geiger, B., and Bershadsky, A. (2001). Assembly and Mechanosensory Function of Focal Contacts. Curr. Opin. Cell Biol. 13, 584–592. doi:10.1016/s0955-0674(00)00255-6
Giusiano, S., Cochet, C., Filhol, O., Duchemin-Pelletier, E., Secq, V., Bonnier, P., et al. (2011). Protein Kinase CK2α Subunit Over-Expression Correlates with Metastatic Risk in Breast Carcinomas: Quantitative Immunohistochemistry in Tissue Microarrays. Eur. J. Cancer 47, 792–801. doi:10.1016/j.ejca.2010.11.028
Golden, D., and Cantley, L. G. (2014). Casein Kinase 2 Prevents Mesenchymal Transformation by Maintaining Foxc2 in the Cytoplasm. Oncogene 34, 4702–4712. doi:10.1038/onc.2014.395
Guerra, B., and Issinger, O.-G. (2008). Protein Kinase CK2 in Human Diseases. Cmc 15, 1870–1886. doi:10.2174/092986708785132933
Harte, M. T., Hildebrand, J. D., Burnham, M. R., Bouton, A. H., and Parsons, J. T. (1996). p130Cas, a Substrate Associated with V-Src and V-Crk, Localizes to Focal Adhesions and Binds to Focal Adhesion Kinase. J. Biol. Chem. 271, 13649–13655. doi:10.1074/jbc.271.23.13649
Hochgräfe, F., Zhang, L., O'Toole, S. A., Browne, B. C., Pinese, M., Porta Cubas, A., et al. (2010). Tyrosine Phosphorylation Profiling Reveals the Signaling Network Characteristics of Basal Breast Cancer Cells. Cancer Res. 70, 9391–9401. doi:10.1158/0008-5472.can-10-0911
Hong, H., and Benveniste, E. N. (2021). The Immune Regulatory Role of Protein Kinase CK2 and Its Implications for Treatment of Cancer. Biomedicines 9, 1932. doi:10.3390/biomedicines9121932
Hu, Y.-L., Lu, S., Szeto, K. W., Sun, J., Wang, Y., Lasheras, J. C., et al. (2014). FAK and Paxillin Dynamics at Focal Adhesions in the Protrusions of Migrating Cells. Sci. Rep. 4, 6024. doi:10.1038/srep06024
Huber, W., VON Heydebreck, A., Sültmann, H., Poustka, A., and Vingron, M. (2002). Variance Stabilization Applied to Microarray Data Calibration and to the Quantification of Differential Expression. Bioinformatics 18 (Suppl. 1), S96–S104. doi:10.1093/bioinformatics/18.suppl_1.s96
Hunter, T. (1995). Protein Kinases and Phosphatases: The Yin and Yang of Protein Phosphorylation and Signaling. Cell 80, 225–236. doi:10.1016/0092-8674(95)90405-0
Jensen, S. S., and Larsen, M. R. (2007). Evaluation of the Impact of Some Experimental Procedures on Different Phosphopeptide Enrichment Techniques. Rapid Commun. Mass Spectrom. 21, 3635–3645. doi:10.1002/rcm.3254
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30. doi:10.1093/nar/28.1.27
Kim, D. H., and Wirtz, D. (2013). Focal Adhesion Size Uniquely Predicts Cell Migration. FASEB J. 27, 1351–1361. doi:10.1096/fj.12-220160
Kim, H., Choi, K., Kang, H., Lee, S.-Y., Chi, S.-W., Lee, M.-S., et al. (2014). Identification of a Novel Function of CX-4945 as a Splicing Regulator. PLoS One 9, e94978. doi:10.1371/journal.pone.0094978
Kim, J. S., Eom, J. I., Cheong, J.-W., Choi, A. J., Lee, J. K., Yang, W. I., et al. (2007). Protein Kinase CK2α as an Unfavorable Prognostic Marker and Novel Therapeutic Target in Acute Myeloid Leukemia. Clin. Cancer Res. 13, 1019–1028. doi:10.1158/1078-0432.ccr-06-1602
Kim, S., Ham, S., Yang, K., and Kim, K. (2018). Protein Kinase CK2 Activation Is Required for Transforming Growth Factor β‐Induced Epithelial-Mesenchymal Transition. Mol. Oncol. 12, 1811–1826. doi:10.1002/1878-0261.12378
Kobayashi, T., Hino, S.-i., Oue, N., Asahara, T., Zollo, M., Yasui, W., et al. (2006). Glycogen Synthase Kinase 3 and H-Prune Regulate Cell Migration by Modulating Focal Adhesions. Mol. Cell Biol. 26, 898–911. doi:10.1128/mcb.26.3.898-911.2006
Kreutzer, J. N., Ruzzene, M., and Guerra, B. (2010). Enhancing Chemosensitivity to Gemcitabine via RNA Interference Targeting the Catalytic Subunits of Protein Kinase CK2 in Human Pancreatic Cancer Cells. BMC Cancer 10, 440. doi:10.1186/1471-2407-10-440
Laramas, M., Pasquier, D., Filhol, O., Ringeisen, F., Descotes, J.-L., and Cochet, C. (2007). Nuclear Localization of Protein Kinase CK2 Catalytic Subunit (CK2α) Is Associated with Poor Prognostic Factors in Human Prostate Cancer. Eur. J. Cancer 43, 928–934. doi:10.1016/j.ejca.2006.11.021
Lettieri, A., Borgo, C., Zanieri, L., D'Amore, C., Oleari, R., Paganoni, A., et al. (2019). Protein Kinase CK2 Subunits Differentially Perturb the Adhesion and Migration of GN11 Cells: A Model of Immature Migrating Neurons. Int. J. Mol. Sci. 20, 5951. doi:10.3390/ijms20235951
Lin, K.-Y., Tai, C., Hsu, J.-C., Li, C.-F., Fang, C.-L., Lai, H.-C., et al. (2011). Overexpression of Nuclear Protein Kinase CK2 α Catalytic Subunit (CK2α) as a Poor Prognosticator in Human Colorectal Cancer. PLoS One 6, e17193. doi:10.1371/journal.pone.0017193
Litchfield, D. W. (2003). Protein Kinase CK2: Structure, Regulation and Role in Cellular Decisions of Life and Death. Biochem. J. 369, 1–15. doi:10.1042/bj20021469
Luo, M., and Guan, J.-L. (2010). Focal Adhesion Kinase: A Prominent Determinant in Breast Cancer Initiation, Progression and Metastasis. Cancer Lett. 289, 127–139. doi:10.1016/j.canlet.2009.07.005
Machiyama, H., Hirata, H., Loh, X. K., Kanchi, M. M., Fujita, H., Tan, S. H., et al. (2014). Displacement of p130Cas from Focal Adhesions Links Actomyosin Contraction to Cell Migration. J. Cell Sci. 127, 3440–3450. doi:10.1242/jcs.143438
Maher, P. A., Pasquale, E. B., Wang, J. Y., and Singer, S. J. (1985). Phosphotyrosine-Containing Proteins Are Concentrated in Focal Adhesions and Intercellular Junctions in Normal Cells. Proc. Natl. Acad. Sci. U.S.A. 82, 6576–6580. doi:10.1073/pnas.82.19.6576
Maru, Y., Hanks, S. K., and Shibuya, M. (2001). The Tubulogenic Activity Associated with an Activated Form of Flt-1 Kinase Is Dependent on Focal Adhesion Kinase. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1540, 147–153. doi:10.1016/s0167-4889(01)00127-6
Mclean, G. W., Carragher, N. O., Avizienyte, E., Evans, J., Brunton, V. G., and Frame, M. C. (2005). The Role of Focal-Adhesion Kinase in Cancer - A New Therapeutic Opportunity. Nat. Rev. Cancer 5, 505–515. doi:10.1038/nrc1647
Meenderink, L. M., Ryzhova, L. M., Donato, D. M., Gochberg, D. F., Kaverina, I., and Hanks, S. K. (2010). P130Cas Src-Binding and Substrate Domains Have Distinct Roles in Sustaining Focal Adhesion Disassembly and Promoting Cell Migration. PLoS One 5, e13412. doi:10.1371/journal.pone.0013412
Meggio, F., and Pinna, L. A. (2003). One‐Thousand‐and‐One Substrates of Protein Kinase CK2? FASEB J. 17, 349–368. doi:10.1096/fj.02-0473rev
Miao, H., Burnett, E., Kinch, M., Simon, E., and Wang, B. (2000). Activation of EphA2 Kinase Suppresses Integrin Function and Causes Focal-Adhesion-Kinase Dephosphorylation. Nat. Cell Biol. 2, 62–69. doi:10.1038/35000008
Modzelewska, K., Newman, L. P., Desai, R., and Keely, P. J. (2006). Ack1 Mediates Cdc42-Dependent Cell Migration and Signaling to p130Cas. J. Biol. Chem. 281, 37527–37535. doi:10.1074/jbc.m604342200
Nakamura, K., Yano, H., Uchida, H., Hashimoto, S., Schaefer, E., and Sabe, H. (2000). Tyrosine Phosphorylation of Paxillin α Is Involved in Temporospatial Regulation of Paxillin-Containing Focal Adhesion Formation and F-Actin Organization in Motile Cells. J. Biol. Chem. 275, 27155–27164. doi:10.1016/s0021-9258(19)61492-4
Ngan, E., Stoletov, K., Smith, H. W., Common, J., Muller, W. J., Lewis, J. D., et al. (2017). LPP Is a Src Substrate Required for Invadopodia Formation and Efficient Breast Cancer Lung Metastasis. Nat. Commun. 8, 15059. doi:10.1038/ncomms15059
Nunez DE Villavicencio-Diaz, T., Rabalski, A. J., and Litchfield, D. W. (2017). Protein Kinase CK2: Intricate Relationships within Regulatory Cellular Networks. Pharm. (Basel) 10, 27. doi:10.3390/ph10010027
Okayama, A., Miyagi, Y., Oshita, F., Ito, H., Nakayama, H., Nishi, M., et al. (2015). Identification of Tyrosine-Phosphorylated Proteins Upregulated During Epithelial-Mesenchymal Transition Induced with TGF-β. J. Proteome Res. 14, 4127–4136. doi:10.1021/acs.jproteome.5b00082
Olsen, B. B., Boldyreff, B., Niefind, K., and Issinger, O.-G. (2006). Purification and Characterization of the CK2α′-Based Holoenzyme, an Isozyme of CK2α: A Comparative Analysis. Protein Expr. Purif. 47, 651–661. doi:10.1016/j.pep.2005.12.001
Olsten, M. E. K., Canton, D. A., Zhang, C., Walton, P. A., and Litchfield, D. W. (2004). The Pleckstrin Homology Domain of CK2 Interacting Protein-1 Is Required for Interactions and Recruitment of Protein Kinase CK2 to the Plasma Membrane. J. Biol. Chem. 279, 42114–42127. doi:10.1074/jbc.m407628200
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 47, D442–D450. doi:10.1093/nar/gky1106
Petit, M. M., Meulemans, S. M., Alen, P., Ayoubi, T. A., Jansen, E., and VAN DE Ven, W. J. (2005). The Tumor Suppressor Scrib Interacts with the Zyxin-Related Protein LPP, Which Shuttles Between Cell Adhesion Sites and the Nucleus. BMC Cell Biol. 6, 1. doi:10.1186/1471-2121-6-1
Petit, M. M. R., Meulemans, S. M. P., and VAN DE Ven, W. J. M. (2003). The Focal Adhesion and Nuclear Targeting Capacity of the LIM-Containing Lipoma-Preferred Partner (LPP) Protein. J. Biol. Chem. 278, 2157–2168. doi:10.1074/jbc.m206106200
Petropoulos, C., Oddou, C., Emadali, A., Hiriart-Bryant, E., Boyault, C., Faurobert, E., et al. (2016). Roles of Paxillin Family Members in Adhesion and ECM Degradation Coupling at Invadosomes. J. Cell Biol. 213, 585–599. doi:10.1083/jcb.201510036
Qian, B.-Z., Zhang, H., Li, J., He, T., Yeo, E.-J., Soong, D. Y. H., et al. (2015). FLT1 Signaling in Metastasis-Associated Macrophages Activates an Inflammatory Signature that Promotes Breast Cancer Metastasis. J. Exp. Med. 212, 1433–1448. doi:10.1084/jem.20141555
Robertson, J., Jacquemet, G., Byron, A., Jones, M. C., Warwood, S., Selley, J. N., et al. (2015). Defining the Phospho-Adhesome Through the Phosphoproteomic Analysis of Integrin Signalling. Nat. Commun. 6, 6265. doi:10.1038/ncomms7265
Roelants, C., Giacosa, S., Duchemin-Pelletier, E., Mcleer-Florin, A., Tisseyre, C., Aubert, C., et al. (2015). Dysregulated Expression of Protein Kinase CK2 in Renal Cancer. German: Springer.
Roffey, S. E., and Litchfield, D. W. (2021). CK2 Regulation: Perspectives in 2021. Biomedicines 9, 1361. doi:10.3390/biomedicines9101361
Rozanov, D. V., Savinov, A. Y., Williams, R., Liu, K., Golubkov, V. S., Krajewski, S., et al. (2008). Molecular Signature of MT1-MMP: Transactivation of the Downstream Universal Gene Network in Cancer. Cancer Res. 68, 4086–4096. doi:10.1158/0008-5472.can-07-6458
Sachdev, S., Bu, Y., and Gelman, I. H. (2009). Paxillin-Y118 Phosphorylation Contributes to the Control of Src-Induced Anchorage-Independent Growth by FAK and Adhesion. BMC Cancer 9, 12. doi:10.1186/1471-2407-9-12
Salgia, R., Li, J.-L., Lo, S. H., Brunkhorst, B., Kansas, G. S., Sobhany, E. S., et al. (1995). Molecular Cloning of Human Paxillin, a Focal Adhesion Protein Phosphorylated by P210BCR/ABL. J. Biol. Chem. 270, 5039–5047. doi:10.1074/jbc.270.10.5039
Salvetti, A., Couté, Y., Epstein, A., Arata, L., Kraut, A., Navratil, V., et al. (2016). Nuclear Functions of Nucleolin Through Global Proteomics and Interactomic Approaches. J. Proteome Res. 15, 1659–1669. doi:10.1021/acs.jproteome.6b00126
Salvi, M., Sarno, S., Cesaro, L., Nakamura, H., and Pinna, L. A. (2009). Extraordinary Pleiotropy of Protein Kinase CK2 Revealed by Weblogo Phosphoproteome Analysis. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1793, 847–859. doi:10.1016/j.bbamcr.2009.01.013
Schaller, M. D., Hildebrand, J. D., and Parsons, J. T. (1999). Complex Formation with Focal Adhesion Kinase: A Mechanism to Regulate Activity and Subcellular Localization of Src Kinases. MBoC 10, 3489–3505. doi:10.1091/mbc.10.10.3489
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 9, 676–682. doi:10.1038/nmeth.2019
Schmalzigaug, R., Garron, M., Roseman, J., Xing, Y., Davidson, C., Arold, S., et al. (2007). GIT1 Utilizes a Focal Adhesion Targeting-Homology Domain to Bind Paxillin. Cell. Signal. 19, 1733–1744. doi:10.1016/j.cellsig.2007.03.010
Schmitt, B. M., Boewe, A. S., Gotz, C., Philipp, S. E., Urbschat, S., Oertel, J., et al. (2021). CK2 Activity Mediates the Aggressive Molecular Signature of Glioblastoma Multiforme by Inducing Nerve/Glial Antigen (NG)2 Expression. Cancers (Basel) 13, 1678. doi:10.3390/cancers13071678
Senda, Y., Murata‐Kamiya, N., and Hatakeyama, M. (2016). C‐Terminal Src Kinase‐Mediated EPIYA Phosphorylation of Pragmin Creates a Feed‐Forward C‐Terminal Src Kinase Activation Loop that Promotes Cell Motility. Cancer Sci. 107, 972–980. doi:10.1111/cas.12962
Shen, Y., and Schaller, M. D. (1999). Focal Adhesion Targeting: The Critical Determinant of FAK Regulation and Substrate Phosphorylation. MBoC 10, 2507–2518. doi:10.1091/mbc.10.8.2507
Strum, S. W., Gyenis, L., and Litchfield, D. W. (2021). CSNK2 in Cancer: Pathophysiology and Translational Applications. Br. J. Cancer 126, 994–1003. doi:10.1038/s41416-021-01616-2
Sun, T., Kim, B., and Kim, L. W. (2013). Glycogen Synthase Kinase 3 Influences Cell Motility and Chemotaxis by Regulating PI3K Membrane Localization in Dictyostelium. Dev. Growth Differ. 55, 723–734. doi:10.1111/dgd.12078
Tapial Martínez, P., López Navajas, P., and Lietha, D. (2020). FAK Structure and Regulation by Membrane Interactions and Force in Focal Adhesions. Biomolecules 10. doi:10.3390/biom10020179
Totaro, A., Astro, V., Tonoli, D., and DE Curtis, I. (2014). Identification of Two Tyrosine Residues Required for the Intramolecular Mechanism Implicated in GIT1 Activation. PLoS One 9, e93199. doi:10.1371/journal.pone.0093199
Valdivia, A., Cárdenas, A., Brenet, M., Maldonado, H., Kong, M., Díaz, J., et al. (2020). Syndecan-4/PAR-3 Signaling Regulates Focal Adhesion Dynamics in Mesenchymal Cells. Cell Commun. Signal 18, 129. doi:10.1186/s12964-020-00629-3
Vilk, G., Weber, J. E., Turowec, J. P., Duncan, J. S., Wu, C., Derksen, D. R., et al. (2008). Protein Kinase CK2 Catalyzes Tyrosine Phosphorylation in Mammalian Cells. Cell. Signal. 20, 1942–1951. doi:10.1016/j.cellsig.2008.07.002
Volberg, T., Romer, L., Zamir, E., and Geiger, B. (2001). pp60c-src and Related Tyrosine Kinases: A Role in the Assembly and Reorganization of Matrix Adhesions. J. Cell Sci. 114, 2279–2289. doi:10.1242/jcs.114.12.2279
Wang, Y., DU, D., Fang, L., Yang, G., Zhang, C., Zeng, R., et al. (2006). Tyrosine Phosphorylated Par3 Regulates Epithelial Tight Junction Assembly Promoted by EGFR Signaling. EMBO J. 25, 5058–5070. doi:10.1038/sj.emboj.7601384
Wehde, B. L., Rädler, P. D., Shrestha, H., Johnson, S. J., Triplett, A. A., and Wagner, K.-U. (2018). Janus Kinase 1 Plays a Critical Role in Mammary Cancer Progression. Cell Rep. 25, 2192–2207. e5. doi:10.1016/j.celrep.2018.10.063
Wieczorek, S., Combes, F., Lazar, C., Giai Gianetto, Q., Gatto, L., Dorffer, A., et al. (2017). DAPAR & ProStaR: Software to Perform Statistical Analyses in Quantitative Discovery Proteomics. Bioinformatics 33, 135–136. doi:10.1093/bioinformatics/btw580
Wozniak, M. A., Modzelewska, K., Kwong, L., and Keely, P. J. (2004). Focal Adhesion Regulation of Cell Behavior. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1692, 103–119. doi:10.1016/j.bbamcr.2004.04.007
Wu, J.-C., Chen, Y.-C., Kuo, C.-T., Wenshin Yu, H., Chen, Y.-Q., Chiou, A., et al. (2015). Focal Adhesion Kinase-Dependent Focal Adhesion Recruitment of SH2 Domains Directs SRC into Focal Adhesions to Regulate Cell Adhesion and Migration. Sci. Rep. 5, 18476. doi:10.1038/srep18476
Xavier, C.-P., Rastetter, R. H., Blömacher, M., Stumpf, M., Himmel, M., Morgan, R. O., et al. (2012). Phosphorylation of CRN2 by CK2 Regulates F-Actin and Arp2/3 Interaction and Inhibits Cell Migration. Sci. Rep. 2, 241. doi:10.1038/srep00241
Yin, G., Zheng, Q., Yan, C., and Berk, B. C. (2005). GIT1 Is a Scaffold for ERK1/2 Activation in Focal Adhesions. J. Biol. Chem. 280, 27705–27712. doi:10.1074/jbc.m502271200
Keywords: CK2β depletion, EMT, tyrosine-phosphorylated proteins, FAK1-Src-PAX1 signaling pathway, focal adhesions, epithelial cell spreading
Citation: Filhol O, Hesse A-M, Bouin A-P, Albigès-Rizo C, Jeanneret F, Battail C, Pflieger D and Cochet C (2022) CK2β Is a Gatekeeper of Focal Adhesions Regulating Cell Spreading. Front. Mol. Biosci. 9:900947. doi: 10.3389/fmolb.2022.900947
Received: 21 March 2022; Accepted: 02 May 2022;
Published: 29 June 2022.
Edited by:
Lorenzo Alberto Pinna, National Research Council, ItalyReviewed by:
Chungho Kim, Korea University, South KoreaJose R Bayascas, Universidad Autónoma de Barcelona, Spain
Copyright © 2022 Filhol, Hesse, Bouin, Albigès-Rizo, Jeanneret, Battail, Pflieger and Cochet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claude Cochet, claude.cochet@cea.fr; Delphine Pflieger, delphine.pflieger@cea.fr
†These authors share senior authorship
 Odile Filhol
Odile Filhol Anne-Marie Hesse
Anne-Marie Hesse Anne-Pascale Bouin3
Anne-Pascale Bouin3  Florian Jeanneret
Florian Jeanneret Delphine Pflieger
Delphine Pflieger