Metal Ion Binding in Wild-Type and Mutated Frataxin: A Stability Study
- 1Dipartimento di Fisica, Universitá di Roma Tor Vergata, Rome, Italy
- 2INFN, Sezione di Roma Tor Vergata, Rome, Italy
- 3Dipartimento di Scienze Biochimiche “A. Rossi Fanelli”, Sapienza Universitá di Roma, Rome, Italy
- 4CNR—Istituto di Chimica dei Composti Organometallici, Firenze, Italy
- 5ENEA CR Frascati, Diagnostics and Metrology Laboratory FSN-TECFIS-DIM, Frascati, Italy
- 6Chair of Pharmacology, Section of Medicine, University of Fribourg, Fribourg, Switzerland
- 7Observatoire des Sciences de L’Univers de Grenoble, UAR 832 CNRS, Université Grenoble Alpes, Grenoble, France
- 8Museo Storico della Fisica e Centro Studi e Ricerche E. Fermi, Roma, Italy
This work studies the stability of wild-type frataxin and some of its variants found in cancer tissues upon Co2+ binding. Although the physiologically involved metal ion in the frataxin enzymatic activity is Fe2+, as it is customarily done, Co2+ is most often used in experiments because Fe2+ is extremely unstable owing to the fast oxidation reaction Fe2+ → Fe3+. Protein stability is monitored following the conformational changes induced by Co2+ binding as measured by circular dichroism, fluorescence spectroscopy, and melting temperature measurements. The stability ranking among the wild-type frataxin and its variants obtained in this way is confirmed by a detailed comparative analysis of the XAS spectra of the metal-protein complex at the Co K-edge. In particular, a fit to the EXAFS region of the spectrum allows positively identifying the frataxin acidic ridge as the most likely location of the metal-binding sites. Furthermore, we can explain the surprising feature emerging from a detailed analysis of the XANES region of the spectrum, showing that the longer 81-210 frataxin fragment has a smaller propensity for Co2+ binding than the shorter 90-210 one. This fact is explained by the peculiar role of the N-terminal disordered tail in modulating the protein ability to interact with the metal.
1 Introduction
The study of the role played by metal ions in biology dates back several decades. Among the metals normally present in living systems, transition metals, such as Fe, Co, Cu, and Zn, are the most noteworthy. They usually exist as electropositive chemical elements that can replace the proton of an acid and form complexes with hydroxyl anions. Metals are highly reactive and are involved in many important biological processes (Shi and Chance, 2008; Bertini et al., 2007; Crichton, 2019). An unbalanced concentration of metals can result in severe threats to living systems.
Many mechanisms make metal ions easily available when needed but harmless when stored. In many instances, whether metals are useful or harmful crucially depends on their local concentration. Thus, storing, metabolism, and trafficking of metals must be accurately regulated. The failure of the tuning mechanism of metal delivery is possibly also involved in the development of severe neurodegenerative pathologies such as Alzheimer’s disease, Transmissible Spongiform Encephalopathies, and Parkinson’s disease (De Santis et al., 2015).
One of the proteins required for the fine and correct tuning of cellular iron homeostasis is frataxin (FXN). FXN is highly conserved in eukaryotes and prokaryotes and plays a role in different cellular pathways. FXN specific function(s) is (are) still a matter of debate, while there is a general agreement that binding of iron to FXN is required for the cellular control of iron homeostasis. Nevertheless, the FXN role in helping cells regulate iron chemistry and availability has not been fully elucidated. Among many others, functions proposed for FXN are that it may act as an iron chaperone during heme and iron-sulfur (Fe-S) cluster assembly, serve as an iron-storage reservoir during iron overloading, and/or serve as a factor in lowering the concentration of reactive oxygen species (ROS), thus controlling the cellular oxidative stress (Bencze et al., 2006).
It is unlikely that only a single protein can control, in different cells, so many different pathways. In contrast, it is possible that different functions are not specific for FXN. Nevertheless, a breakdown in FXN production may lead to a complete failure in controlling the cellular iron availability. In particular, the deletion of the FXN gene has been seen to be followed by overloading of mitochondrial iron deposits and a general default of the Fe-S cluster assembly (Babcock et al., 1997; Foury and Cazzalini, 1997; Foury, 1999; Rötig et al., 1991). On these premises, FXN has been proposed as a candidate for the control of the mitochondrial iron efflux (Radisky et al., 1999).
Reduced expression levels of FXN in humans are linked to the development, progression, and severity of Friedreich’s ataxia (FA) (Pastore and Puccio, 2013), a neurodegenerative disease that is supposed to be linked to disproportion in iron concentration (Gentry et al., 2013).
The 210 amino acids long human FXN precursor is imported into the mitochondrion where it undergoes a two-step proteolytic maturation: first into a 19 kDa (42–210) intermediate and then into the final 14 kDa (81–210) form1, in both healthy individuals and FA patients (Pastore and Puccio, 2013).
FXN is known to possess non-structured regions (of which there is no 3D information from X-ray crystallography) that are supposed to play quite an important (direct/indirect) role in iron binding.
To date, FXN is not considered a cancer driver gene (Davoli et al., 2013; Martínez-Jiménez et al., 2020). However, it has been reported by Pastore and Puccio (2013) that, in cancer tissues, several FXN variants, whose stability and functional activity are reduced with respect to the wild-type (wt), are present. The importance of mutations in the onset and progression of neoplastic diseases is not yet clarified. However, mitochondrial functions and the complex process of tumorigenesis are connected at multiple levels. Impaired FXN functions may lead to defective mitochondria, which leads to a reduced assembly of Fe-S clusters, thus enhancing carcinogenesis (Rychtarcikova et al., 2017).
The Fe-FXN coordination mode is not fully elucidated. However, of the three putative ion binding sites, one is found to bring into play the His86 residue located in the disordered N-terminal region, while the other two involve aspartate and glutamate residues in a region of the protein called the “acidic ridge” (Gentry et al., 2013).
In this complex biological framework, the main goal of our research is to help understand the key structural aspects of the interaction of FXN and the three somatic missense variants of D104G, Y123S, and S161I (described in Section 2.2) with Fe2+ mimicked by its akin Co2+ (Maiti et al., 2017; Söderberg et al., 2011; Söderberg et al., 2013).
Following the same strategy already successfully used in by Gentry et al. (2013), Noguera et al. (2015), and Pastore et al. (2007), in the present study, we have replaced Fe2+ with the chemically very similar Co2+ ion. In fact, although the FXN natural ligand ion is Fe2+, the latter is known to be very unstable, spontaneously switching to the oxidized Fe3+ species in aerobic conditions. Instead, Co2+ is not sensitive to aerobic conditions and has been proved to be a reliable surrogate to probe Fe2+ sites in proteins (Maiti et al., 2017) and particularly in frataxin (Söderberg et al., 2011; Söderberg et al., 2013). The use of Co2+ in place of Fe2+ to study the structural properties is also supported by the observation that Co2+ has an ionic radius very similar to that of Fe2+ (Shannon, 1976).
The experimental technique of election for the study of protein-metal interactions is X-ray absorption spectroscopy (XAS) because, even in the case of very diluted metal-protein complexes, it allows providing accurate structural information about the environment of the metal-binding site within a radius of up to about 5 Å (Strange et al., 1993; Ortega et al., 2012)2.
In particular, XAS has shown its valuable power in elucidating the mechanisms by which metal ions interacting with intrinsically disordered proteins can push the latter to either the correct folding or a potentially dangerous misfolding, according to specific physicochemical circumstances (Morante et al., 2004; Morante and Rossi, 2014).
This work compares structural XAS information with data obtained from other complementary experimental techniques: circular dichroism, fluorescence spectroscopy and melting temperature measurements. A set of fairly good consistent results for the structure of the protein-metal complex and the stability ranking among wt-FXN and the three variants of D104G, Y123S, and S161I upon metal binding is obtained in this way.
2 Materials and Methods
2.1 Circular Dichroism and Fluorescence Spectroscopy
Circular dichroism (CD) and fluorescence spectroscopy (FS) techniques (Ősz et al., 2002) are used to monitor conformational changes induced by Co2+ binding in wt-FXN and in its variants D104G, Y123S, and S161I.
CD measurements were carried out with a JASCO-815 spectropolarimeter (Jasco, Easton, MD, United States), and the results are expressed as the mean residue ellipticity (Θ), assuming a mean residue molecular mass of 110 per amino acid residue.
Far-UV (190–250 nm) CD spectra were monitored at 20°C at an FXN concentration ranging from 100 to 140 μg/ml, using a 0.1 cm path length quartz cuvette. Near-UV (350–420 nm) CD spectra were recorded in a 1.0 cm path length quartz cuvette at a protein concentration ranging between 1.00 and 1.30 mg/ml. In both the far- and near-UV CD spectra measurements, the protein was dissolved in 20 mM Hepes, pH 8.0, 20 mM Na2SO4, and 20% glycerol.
Wild-type FXN and variants (with protein concentrations ranging from 100 to 140 μg/ml) were heated from 20 to 95°C and then cooled from 95 to 20°C in a 0.1 cm quartz cuvette with a heating rate of 1.0°/min controlled by a Jasco programmable Peltier element. The dichroic activity at 222 nm and the PMTV were continuously monitored in parallel every 0.5°C (Benjwal et al., 2006). All thermal scans were corrected for the solvent contribution at different temperatures. Melting temperature (Tm) values were calculated by taking the first derivative of the ellipticity at 222 nm with respect to temperature. All denaturation experiments were performed in triplicate.
Intrinsic fluorescence emission spectra were monitored at 50 μg/ml under the identical excitation and emission conditions for all the samples. Right angle light scattering was measured at 20°C with both excitation and emission wavelengths set at 480 nm. Measurements were performed using an LS50B spectrofluorometer (PerkinElmer) with a 1.0 cm path length quartz cuvette. Spectra were recorded in a range from 300 to 450 nm, with a 1 nm sampling interval, and the excitation wavelength was set at 295 nm.
2.2 X-Ray Absorption Spectroscopy
XAS measurements at the Co K-edge were performed at the BM30 beamline of the European Synchrotron Radiation Facility (ESRF; Grenoble, France) Proux et al. (2005).
The beam energy was selected using a Si(220) double-crystal monochromator with a resolution of 0.5 eV. The beam spot on the sample was approximately 240, ×, 150 μm2 (H × V, FWHM). Spectra were recorded in fluorescence mode using a 13-element solid-state Ge detector. To avoid photo-degradation and undesired spectra evolution during data taking, all the samples, held at 77 K since their preparation, were cooled down and kept at 13 K in a helium cryostat during the whole XAS measurements. For the same reason, in the process of data acquisition, the hitting X-ray beam was systematically moved to a different position on the sample after each scan.
As usual, XAS spectra are analyzed by separating the low photons energy XANES (X-ray absorption near edge spectroscopy) region from the high photons energy EXAFS (extended X-ray absorption fine structure) one. The XANES spectra are normalized using the standard software Athena (Ravel and Newville, 2005). EXAFS data are extracted with the help of cubic splines interpolation as implemented in the AUTOBKG algorithm (Newville et al., 1993) of Athena. EXAFS data analysis was performed using the EXCURV98 code (Binsted et al., 1998).
As specified in Table 1, besides the wt-FXN in two different lengths, we considered three somatic missense variants (Petrosino et al., 2019): D104G, Y123S, and S161I. These mutations, whose location in the folded protein is highlighted in Figure 1, have different effects on the Fe-S cluster formation, as shown by absorbance measurements by Petrosino et al. (2019). The reasons for focusing on these somatic missense variants are summarized as follows:
• D104G: this variant is present in liver carcinoma cells. As shown in Figure 1, residue D104 is located in the α1 helix region3 of the protein. In the case of this variant, Fe-S clusters formation is totally inhibited, although the thermodynamic stability and the secondary and tertiary structure arrangements are similar to those of the wt-FXN (Petrosino et al., 2019).
• Y123S: this variant is present in the cells of the digestive tract carcinoma. Residue Y123 is located in the coil region between the β-sheet region and the α1 helix (Figure 1). In this case, activity measurements show that Fe-S clusters formation is only partially inhibited. This FXN variant shows a Tm of about 14°C lower than that of the wt-FXN, suggesting an enhanced unfolding propensity and a significant decrease in thermal stability (Petrosino et al., 2019).
• S161I: this variant has been found in the cells of the endometrium carcinoma. Residue S161 is located in the coil region between the β-sheet region and the α2 helix. Again the thermal stability is decreased, as indicated by a Tm lower, by about 11°C, compared to that of the wt-FXN. Interestingly, with this missense mutation, the Fe-S cluster formation is totally inhibited (Petrosino et al., 2019).
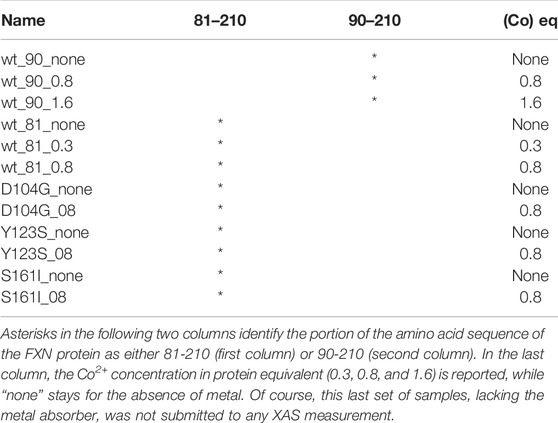
TABLE 1. In the first column, we report the names by which the analyzed samples are denoted in the study.
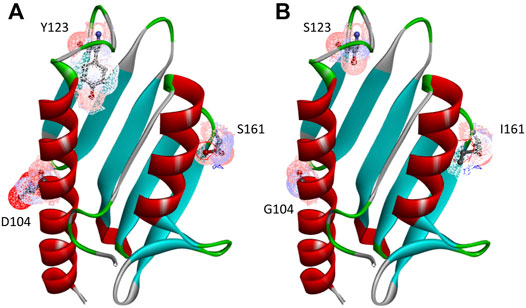
FIGURE 1. Human FXN folded structure. The three point-mutations, D104G, Y123S, and S161I, which are the object of this investigation, are highlighted. (A) Wild-type, (B) variants.
For the purpose of XAS measurements, wt-FXN and variants have been expressed as N-terminal His-tagged proteins using a pET28a vector in E. coli Rosetta cells transformed with the selected plasmid, grown in LB medium, and purified as described by Petrosino et al. (2019).
The samples listed in Table 1 were prepared for the XAS measurements according to the following protocol. For the wt protein and its variants, stocks of 45 μL of 0.9 mM protein solutions were obtained by dissolving the appropriate amount of protein in 20 mM Hepes, pH 8.0, containing 20 mM Na2SO4 and 20% glycerol. A Co2+ 16 mM stock solution was prepared by dissolving CoSO4 in a 20 mM Hepes, pH 8.0, containing 20 mM Na2SO4 with 20% glycerol. In order to get the desired Co2+ final concentrations (see Table 1), to each 45 μL sample holder, the appropriate amount of a Co2+ concentrated stock solution was finally added. The solutions in each sample holder were mixed by pipetting and then immediately frozen in liquid N2, shipped overnight from Rome to Grenoble in a dry shipping Dewar cooled with liquid N2, and immediately transferred to the ESRF beamline upon arrival.
3 Results
3.1 Circular Dichroism, Fluorescence Spectroscopy, and Thermal Shift Analysis
Both near-UV CD and FS suggest that Co2+ interacts preferentially with the short FXN isoform 90-210 rather than with the long isoform 81-210.
In particular, panel A of Figure 2 shows that, by adding Co2+ to FXN 90-210, important changes occur in the regions of 252–256, 265–270, and 288–295 nm. At the same time, fluorescence intensity reduction and redshift are observed (see panel B of Figure 2), indicating consistent conformational changes that may be related to the Co2+ presence with protein-metal complex formation. Right angle light scattering data indicate that aggregation never occurred in any measured samples.
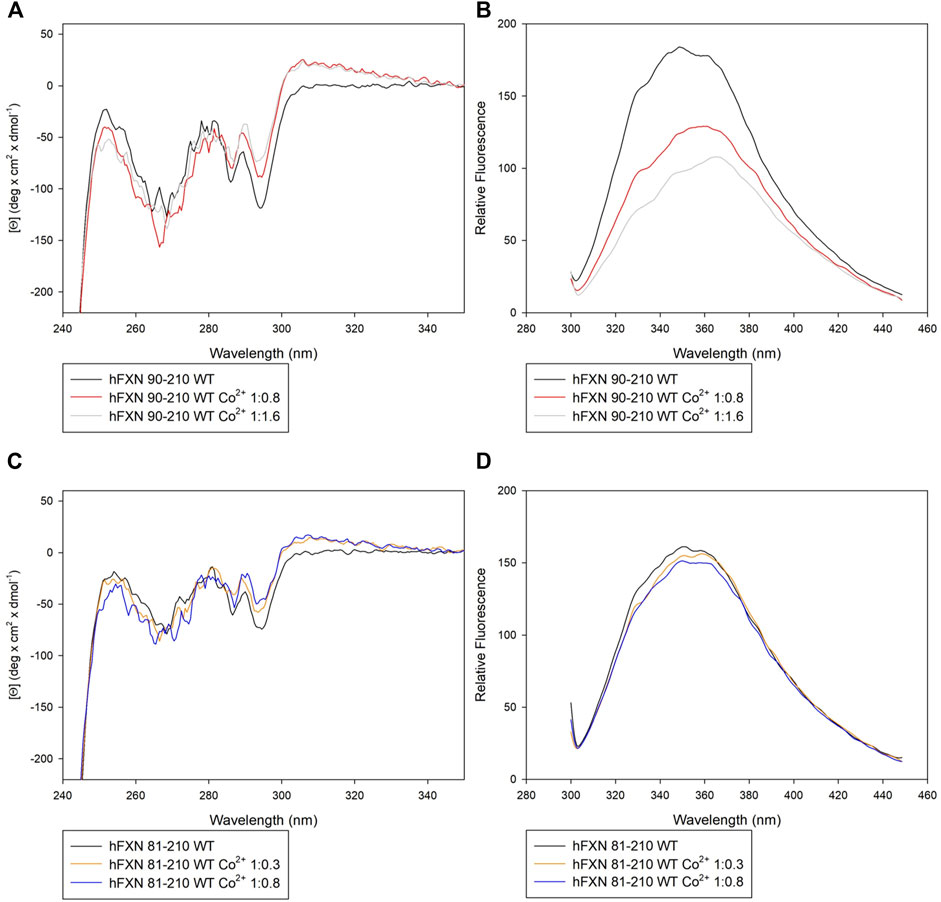
FIGURE 2. Spectroscopic characterization of FXN isoforms with and without Co2+. (A) Near-UV CD spectra of FXN 90-210 in the absence of Co2+ (black) and in the presence of 0.8 (red line) and 1.6 (grey line) Co2+ equiv. (B) Intrinsic fluorescence emission spectra of FXN 90-210 in the absence of Co2+ and in the presence of 0.8 and 1.6 Co2+ equiv (code color is as in panel A). (C) Near-UV CD spectra of FXN 81-210 in the absence of Co2+ (dark purple) and in the presence of 0.3 (orange) and 0.8 (blue) Co2+ equiv. (D) Intrinsic fluorescence emission spectra of FXN 81-210 in the absence of Co2+ and in the presence of 0.3 and 0.8 Co2+ equiv (code color is as in panel C). Fluorescence spectra were monitored at 50 μg/ml under identical excitation and emission conditions.
Furthermore, the longer, FXN 81-210, isoform shows changes in the near-UV CD spectrum in the regions 252–256 and 288–295 nm (see panel C of Figure 2). However, neither fluorescence intensity modification nor redshift is visible (panel D), thus hinting at a lack of protein-metal complex formation.
These indications are confirmed by thermal stability analysis performed on the protein in the presence of Co2+ at the concentrations specified in Table 2 (see Figure 3). Thermal stability measurements are considered a powerful and reliable way to study metal-binding interactions in proteins (Layton and Hellinga, 2011).
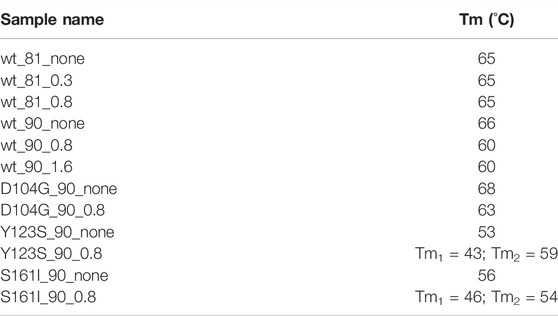
TABLE 2. Melting temperature of wt-FXN and D104G, Y123S, and S161I variants, in the absence of Co2+ and in the presence of 0.8 and 1.6 Co2+ equiv. Sample names are as shown in Table 1.
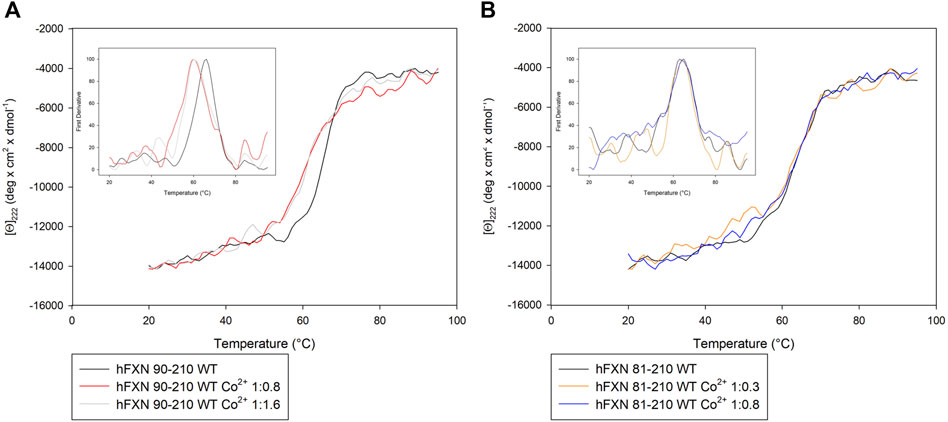
FIGURE 3. Thermal unfolding transition of FXN isoforms with and without Co2+. (A) Thermal unfolding curves of FXN 90-210 in the absence of Co2+ (black) and in the presence of 0.8 (red) or 1.6 (grey) Co2+ equiv. (B) Thermal unfolding curves of FXN 81-210 in the absence of Co2+ (dark purple) and in the presence of 0.3 (orange) or 0.8 (blue) Co2+ equiv. The curves are obtained by monitoring the molar ellipticity at 222 nm [Θ]222, in the range between 20 and 95°C. Insets show the first derivative of the experimental curve.
In order to measure thermal stability, samples with the wt-FXN and samples with the D104G, Y123S, and S161I variants at concentrations ranging from 100 to 140 μg/ml, in the absence of Co2+ and the presence of different concentrations of Co2+, were heated from 20°C to 95°C in a quartz cuvette with a heating rate of 1°C/min, controlled by a Jasco programmable Peltier element as described by Petrosino et al. (2019). We stress that thermal transitions appeared reversible in all the conditions we considered, with no hysteresis during the cooling phase (see Supplementary Material).
Looking at the behavior of the molar ellipticity changes at 222 nm between 20°C and 95°C, we observe a decrease in the Tm values of the wt sample by 6° upon adding 0.8 equiv or 1.6 Co2+ equiv to FXN 90-210 (Table 2; Figure 3A). Instead, Tm does not change when the metal ion is added to FXN 81-210 (see Table 2; Figure 3B). The three variants, D104G, Y123S, and S161I, show conformational changes and a decrease in Tm in the presence of Co2+ (see Figures 4, 5).
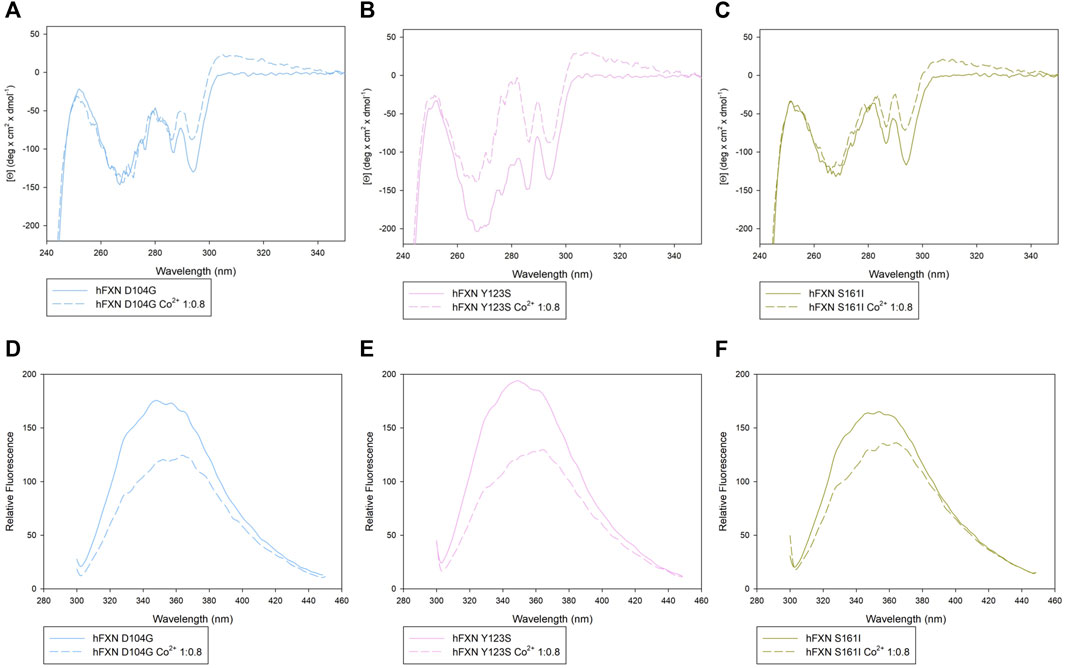
FIGURE 4. Spectroscopic characterization of the D104G, Y123S, and S161I variants with and without Co2+. Upper panels: near-UV CD spectra of D104G (light blue, A), Y123S (light pink, B), and S161I (dark yellow, C) in the absence of Co2+ (continuous line) and in the presence of 0.8 Co2+ equiv (dotted line). Lower panels: intrinsic fluorescence emission spectra of D104G (light blue, D), Y123S (light pink, E), and S161I (dark yellow, F) in the absence of Co2+ (continuous line) and in the presence of 0.8 Co2+ equiv (dotted line). Fluorescence spectra were monitored at 50 μg/ml under identical excitation and emission conditions.
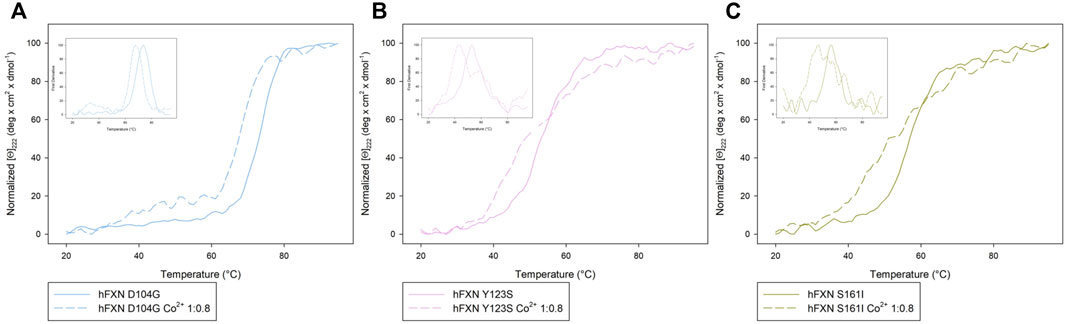
FIGURE 5. Comparison of the Near-UV CD data of [Θ]222 as a function of the temperature of the D104G (light blue), Y123S (light pink), and S161I (dark yellow) variants (A–C) in the absence of Co2+ (continuous curves) and in the presence of 0.8 Co2+ equiv (dotted lines). The insets show the first derivative of the experimental curve with respect to temperature.
In the case of the Y123S and S161I variants, two melting temperatures are reported in Table 2. They correspond to the two local maxima displayed by the dotted curves in the insets of Figures 5B,C. The presence of two slopes in the Θ222 curve is interpreted as an indication of the existence of a two-step thermal denaturation process.
The most significant structural modification induced by Co2+ was observed in the case of the Y123S variant, which involves the tyrosine residue in position 123, in a region usually considered a putative FXN iron-binding site (Nair et al., 2004; Gentry et al., 2013).
In the case of the Y123S variant, the presence of Co2+ induces a dramatic modification in both the near-UV CD and the fluorescence spectra (Figures 4B,E). Indeed, in the presence of 0.8 Co2+ equiv, a significant loss of cooperativity occurs in the thermal unfolding process, as shown by the far-UV CD data of [Θ]222 as a function of the temperature reported in Figures 5B,C and Table 2.
3.2 X-Ray Absorption Spectroscopy
Figure 1 displays the structured region of wt-FXN and the short non-structured region at the C-terminal of the amino acid sequence. A second disordered region is located between residues 81 and 904. For the latter, we propose a peculiar role in modulating metal binding (see Section 4) based on the analysis of XAS data (see below) and the melting temperature results reported in Table 2.
The first step toward a correct interpretation of the physicochemical implications of the FXN-metal interaction is to clarify the structure of the accessible metal-binding sites located along the protein amino acid sequence. For this task, XAS is the technique of election as it allows accurately “taking a picture” of the metal ion atomic environment within a region of up to about 5 Å around the metal absorber.
As customarily done, we shall separately discuss the features of the low energy (XANES) and the high energy (EXAFS) region of the XAS spectrum.
The XANES region is dominated by multiple scattering processes, where the photoelectron undergoes more than one scattering event against the atomic scatterers located within a sphere of a radius of about 5 Å from the absorber. Although extracting accurate geometrical information from the XANES region of the spectrum is computationally extremely difficult, fingerprints of specific geometries can still be identified (see Section 3.2.1). In the EXAFS region, where the extracted photoelectron owns a quite high kinetic energy, single scattering events prevail, thus making, in principle, the extraction of geometrical information comparatively much easier (see Section 3.2.2).
3.2.1 XANES
In this section, we discuss the information one can get from comparing the XANES spectrum of the wt-FXN with the three variants we are focusing our attention on.
In the left panel of Figure 6, the XANES spectra of the wt_81_0.8 and wt_90_0.8 samples are compared with the buffer spectrum. The two peptides differ because of the presence (wt_90_0.8) or absence (wt_81_0.8) of the disordered 81-90 tail.
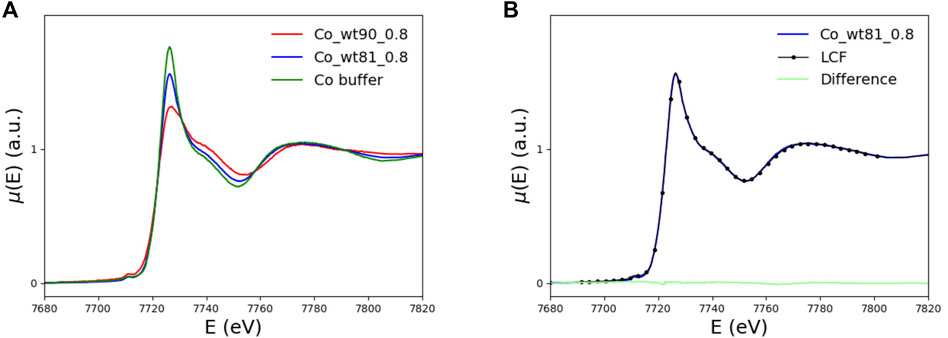
FIGURE 6. (A) XANES spectra of the FXN samples including (wt_81_0.8, blue curve) and not including (wt_90_0.8, red curve) the N-terminal 81-90 disordered region, compared with the buffer spectrum (green curve). (B) XANES spectrum of the long, wt_81_0.8, fragment (blue curve) compared to the spectrum obtained as a linear combination of 43% of wt_90_0.8 and 57% of buffer spectra (black dotted). In light green, we plot the difference spectrum.
We recall that the His86 amino acid, which is supposed to be a binding site for Co2+ (as well as Fe2+) (Gentry et al., 2013), is missing in the shorter fragment wt_90_0.8.
By comparing, in the left panel of Figure 6, the XANES data for the wt_81_0.8 sample with those of the wt_90_0.8 sample and the buffer, one sees that the spectrum of the longer fragment, wt_81_0.8 sample, looks much more similar to the buffer than the spectrum of the wt_90_0.8 sample. This feature suggests that, in the case of wt_81_0.8, a non-negligible amount of Co2+ remains free in the solution.
In order to make this observation more quantitative, we perform a minimization of the difference between the wt_81_0.8 XANES spectrum and the spectrum obtained as a linear combination, point by point, of the wt_90_0.8 and buffer spectra. By fitting the mixing coefficient, we find that the wt_81_0.8 XANES spectrum is well reproduced by a linear combination of 43% of the wt_90_0.8 spectrum plus 57% of the buffer.
We conclude that, in the case of the wt_81_0.8 sample, more than half (57%) of the available Co2+ does not bind to FXN but remains free in solution. This observation is the reason why we did not explore higher metal concentrations. In fact, going at 1.6 eq (or higher) would only increase the uninteresting contribution of Co2+ in the solution.
This finding seems somewhat surprising because it says that, in the presence of an extra putative metal-binding site, a smaller amount of Co2+ is bound to the protein.
We will show in Section 4 that the reason for this, at first sight, strange feature has to do with the peculiar arrangement of the disordered 81-90 tail belonging to the longer wt_81_0.8 fragment. The point is that this tail can “obscure” some of the protein metal-binding sites otherwise available for binding in the case of the shorter wt_90_0.8 peptide (see Figure 13). This interesting interpretation of the puzzling XANES data in Figure 6 is nicely confirmed by the results of melting temperature measurements displayed in Table 2, which show that, in the presence of the metal, the longer 81-210 fragment is more stable than the shorter one.
In Figure 7, the XANES spectra of the D104G (blue), Y123S (pink), and S161I (gold) variants are compared with those of the wt_90_0.8 sample (red) and the buffer (green). We see that the buffer spectrum appears to be significantly different from the spectra of all the variants, thus concluding that all the variants can bind Co2+.
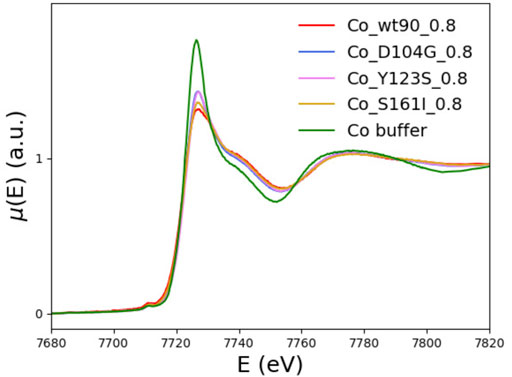
FIGURE 7. The XANES spectra of the D104G (blue), Y123S (violet), and S161I (gold) variants are plotted together with the wt-FXN (red) and the buffer (green) spectra.
The tendency of XANES spectra of D104G and Y123S variants toward that of the buffer indicates a slightly larger contribution to spectra of dissociated forms in these variants than in the wild-type sequence.
Qualitatively, we see that the most significant difference with the wt-FXN spectrum occurs in the case of variants, in which the destabilizing effect due to metal binding (measured by the difference of the melting temperature with and without metal, see Table 2) is large. While destabilization upon metal binding is somewhat surprising in the case of the D104G mutation, as the latter just occurs well inside the alleged metal-binding acidic ridge, the other two mutations, Y123S and S161I, are located far away from it, and the reasons at the basis of the metal-induced destabilization are more difficult to identify. They would require a much more refined investigation, which is outside the scope of the present work.
3.2.2 EXAFS
The first simple observation is that EXAFS spectra show the same ranking of similarity observed for the XANES spectra. In particular, one notices that although the EXAFS spectra of the wt_90_210 and wt_81_210 samples are both definitely different from that of the buffer (see Figure 8), they are also quite different between themselves, with the spectrum of the wt_81_210 sample visibly more similar to that of the buffer than to that of the wt_90_210 sample.
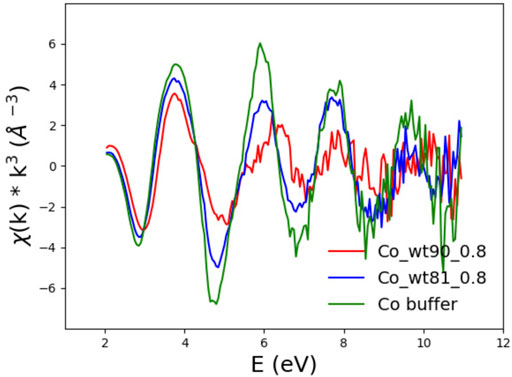
FIGURE 8. EXAFS spectra of FXN wt_90_0.8 (red) and wt_81_0.8 (blue) in the presence of 0.8 Co2+ equiv compared to the buffer spectrum (green).
Structural information about the atomic arrangement around the absorbing metal can be extracted from a fit to the EXAFS region of the spectrum, provided that some “reasonable” assumption about the atomic structure around the Co2+ absorber is available. Unfortunately, not much is known about the precise location of the possible Fe/Co-FXN binding sites (Gentry et al., 2013). In order to overcome this lack of information, we decided to take advantage of the metal ion-binding (MIB) site prediction and docking server (Lin et al., 2016) that, given the protein 3D structure, provides the most probable binding sites for a specific metal ion.
The amino acid patterns to which MIB has assigned the highest scores for Co2+, Fe2+, and Fe3+ binding in FXN 90-210 are reported in Table 3. As expected, the so-called acid ridge is signaled as the most probable region for Co2+, Fe2+, and Fe3+ binding.

TABLE 3. Pairs of residues to which MIB assigns the highest score as Fe2+, Fe3+, and Co2+ metal-binding sites in wt-FXN 90-210.
We limited the MIB search to FXN 90-210 because the FXN 81-210 longer fragment differs from the former for the presence of the disordered 81-90 N-terminal tail, which is not supposed to be able to host a metal. Despite this fact, as discussed in Section 4, this unstructured protein region will be argued to play an important role in the FXN 81-210 ability to bind the metal.
Building on the structural information provided by the MIB search, we are in a position to construct a physically sensible Co2+ binding model configuration from which one can start the theoretical calculation of the EXAFS spectrum. Fitting model parameters against the measured spectrum will allow for accurately determining the atomic structure of the metal-binding site.
As a starting configuration, we took the one with the highest score among those listed in the last column of Table 3. It corresponds to a situation in which Co2+ is coordinated to the residues Glu100 and Asp104.
The structure provided by MIB (see the upper circled inset in the left part of Figure 9) is relaxed and equilibrated at 300 K with classical MD simulations.
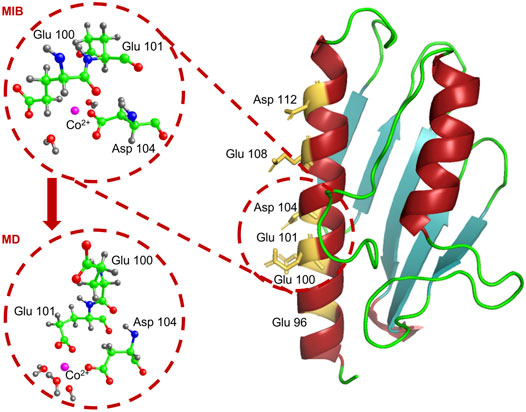
FIGURE 9. We show the atomic structure of the Glu100-Asp104 site around the Co2+ ion, as proposed by MIB (upper circle) and at the end of the MD simulation (lower circle). Co2+ is in pink, carbon in green, oxygen in red, nitrogen in blue, and hydrogen in grey.
MD simulations were performed using the code NAMD (Phillips et al., 2020) with the Co2+ ion forced in its preferred octahedral geometry by employing the dummy atoms method (Pang, 2001; Furlan et al., 2010; La Penna et al., 2013; Duarte et al., 2014; La Penna et al., 2015). Indeed, the octahedral Co2+ geometry, which is at the basis of our MD simulations, is confirmed by the pre-edge feature visible in the XANES spectra of Figure 7, around 7,710 eV (signaling a 1 s → 3 d transition), typical of an octahedral (or pseudo octahedral) site geometry (Bresson et al., 2006).
The dummy atoms method amounts to perform MD simulations where the positive density of the divalent cation is distributed in a blocked octahedral geometry to prevent the negatively charged groups from collapsing over the charged point-like particle of the opposite sign (La Penna and Chelli, 2018).
The metal-protein complex was placed in a 64 × 62 × 74 Å3 box and solvated by adding 9259 TIP3P water molecules to have a layer of water of at least 15 Å around the protein. Na+ and Cl− ions were inserted in the box to get an overall neutral system with the same ion concentration as in the in vitro experiments. At the end of a 4 ns MD simulation, Co2+ is found to be stably located at a binding distance from the Glu101 and Asp104 residues (see lower circled inset in the left panel of Figure 9), thus slightly displaced from its initial position.
A sphere of a 5 Å radius, centered on the Co2+ ion, is excised from the simulation box and used as the starting configuration for the fit of the EXAFS data of the wt_90_08 sample.
The fit to the EXAFS data is performed by employing the program EXCURV98 Binsted et al. (1998). The code implements the so-called constrained refinement strategy, which consists of treating molecules as rigid bodies. In this way, the number of fitting parameters can be significantly reduced (Binsted et al., 1992). In the present case, we considered the amino acids bound to the Co2+ ion rigid bodies, namely, Glu101 and Asp104. Starting from the configuration depicted in the lower circled inset in Figure 9, we refined the Fermi energy shift and the distances of the six oxygen atoms located in the first coordination sphere. These are the two oxygen atoms belonging to the carboxylic group of Glu101, the oxygen belonging to Asp104, and three oxygen atoms belonging to the three nearby water molecules (see Table 4).

TABLE 4. Co2+-ligand distances and Fermi energy shift at the end of the EXAFS fit. Note that the carboxylic group of Glu101 contributes to the octahedral coordination with two oxygen atoms.
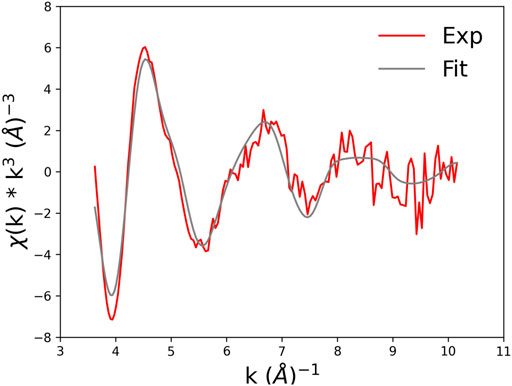
The best fit is displayed in Figure 10, where we show the experimental EXAFS data (in red) superimposed on the fitted curve (in grey). Table 4 gives the O-Co2+ best fit distances. Figure 11 compares the metal-binding site configuration at the end of the MD simulation and after the EXAFS fitting step. We remark that the MD simulation was able to give a pretty good picture of the atomic environment around the metal, as one finds that both before and after the EXAFS fit the six metal ligands (two oxygen atoms from Glu101, one atom from Asp104, and three atoms from water molecules) all fall in a shell between 1.8 and 2.2 Å.
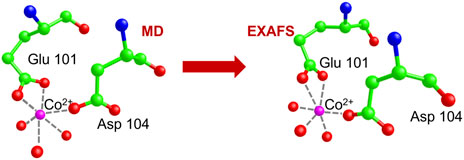
FIGURE 11. Comparison of the Co. binding site configuration at the end of the MD simulation and after the EXAFS fitting step. The color code is as shown in Figure 9. Hydrogen atoms are not shown as they are standardly considered not to contribute to the XAS signal. Broken lines represent the distances of Co2+ from its six oxygen ligands.
We end this section with an observation concerning the second (putative) metal-binding site (Glu108-Asp112) identified by the MIB prediction and docking server (see Table 3). We note that an analysis completely analog to the one performed in the case of the Glu100-Asp104 site shows that the atomic environments of the two sites around the metal are practically identical (data not shown). The main reason is that, in the two sites, the same two ligands, namely, Glu and Asp, via the carboxylate group of the acid chain, are involved in metal binding. Thus, we expect that although the two ligands at the end of the MD simulation can be differently positioned in space, the two binding sites will equally contribute to the EXAFS experimental signal.
4 The Role of the Disordered 81-90 N-Terminal Region
It is very instructive to look at the XANES spectral regions (see Figure 6) in Section 3.2.1 in conjunction with the pattern of the measured melting temperature of wt-FXN and D104G, Y123S, and S161I variants (see Table 2), as quite an interesting feature emerges.
To start the discussion, we recall that the XANES spectrum of the 81-210 fragment is very well reproduced by the sum, point by point, of 43% of the wt_90_0.8 spectrum plus 57% of the buffer spectrum (see Section 3.2.1). This suggests that more than half of Co2+ remains in the solution; it is not bound to the protein.
We also note that, on the one hand, as Figure 12 shows, the XANES spectra of the 90-210 wt-FXN samples with 0.8 and 1.6 Co2+ equiv are well superimposable, proving that, even when the Co2+ is added to the solution at two times the protein concentration, still there is no Co2+ in the solution. This hints at the conclusion that at least two (more or less equivalent) Co2+ binding sites are available in the wt-FXN protein. On the other hand, it is necessary to assume that about half of the amount of Co2+ in the sample has remained free in solution (i.e., not bound to the protein) to get a good fitting of the XANES and EXAFS spectra of the 81-210 FXN.
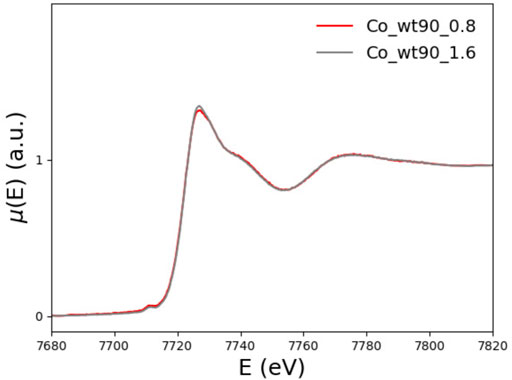
FIGURE 12. XANES spectra of wt_90_0.8 (red) and wt_90_1.6 (grey) in the presence of 0.8 and 1.6 Co2+ equiv, respectively.
A suggestive explanation for this rather puzzling behavior could be that the 81-90 tail present in 81-210 FXN somehow prevents Co2+ from binding to one of the two available coordination sites present in the acidic ridge.
4.1 Experimental Evidence
The disordered 81-90 N-terminal tail in one of its relaxed configurations is geometrically long enough to be able to somehow “obscure” part of the acidic ridge in the α1 helix. In Figure 13, we show one of these typical configurations. They are obtained at the end of a long and careful MD simulation started from an initial all-trans arrangement.
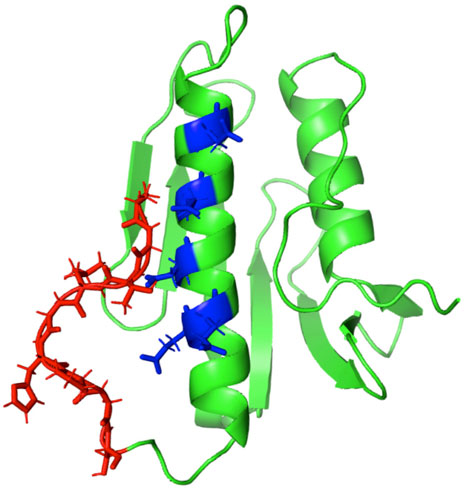
FIGURE 13. We display the configuration of the 81-90 disordered region in red as it results after an elaborated MD relaxation step that was started from an all-trans geometry. The potential metal-binding site locations are highlighted in blue.
This elongated configuration can result from the “sealing” of the Co2+ ion acting between the acidic ridge and the carboxylic end, which helps stabilize the structure. Such a model would explain why, in the case of the long 81-210 wt-FXN, half of Co2+ is found free in solution when the latter is added to the solution at a concentration of twice that of the protein.
The scenario we propose is confirmed by the pattern of the Tm values shown in Table 2. In fact, while the presence of Co2+ does not affect the Tm of the long, 81-210, wt-FXN fragment, it significantly destabilizes the short one, 90-2105. This observation reinforces our hypothesis, according to which, in the presence of the 81-90 disordered tail, metal ion binding sites in the acidic ridge are not fully available. As a result, the destabilizing effect due to metal binding is reduced or not occurring anymore.
5 Conclusion
Based on information extracted from some experimental techniques (CD, FS, XAS) and numerical simulations, we confirm the ability of the wt-FXN and its variants to stably bind metal ions (in our investigation Co2+), strengthening in this way the conjecture according to which FXN is involved in Fe2+ storage and transport.
In particular, XAS data consistently confirm the ranking of thermal stability established by measuring the melting temperatures when wt-FXN is compared to D104G, Y123S, and S161I variants.
Starting from the crystallographic structure of the putative Co2+ binding sites identified by the MIB site prediction and docking server (Lin et al., 2016), we have performed long (up to 4 ns) MD simulations of FXN in the presence of Co2+ to have a reliable atomic geometry around the metal that could be used as an initial configuration in the analysis of our newly collected XAS data. A fit to the EXAFS region of the spectrum allows us to positively identify the FXN acidic ridge as the location of the most likely metal-binding sites.
Furthermore, we can explain the surprising feature emerging from a detailed analysis of the XANES region of the spectrum, according to which the longer 81-210 FXN fragment has a smaller propensity for Co2+ binding than the shorter 90-210 one, despite the presence in the former of the His86 residue, which is supposed to be a specific Co2+ binding site (Gentry et al., 2013).
Indeed, the difference between the XAS spectrum of the long 81-210 FXN fragment compared to that of the shorter 90-210 one that we understand as due to a large fraction (almost a half) of Co2+ remaining in solution in the case of 81-210 FXN, is explained as due to a peculiar behavior of the “disordered” N-terminal region of 81-90 FXN. Our conjecture, which to our knowledge was never considered in the literature, is that the disordered N-terminal tail in the 81-210 FXN sample gets locked in an extended configuration by binding a Co2+ ion, which acts as a bridge between the 81-90 tail and the α1-helix. In this configuration, after a first metal ion is bound, the other metal-binding site allegedly present in the acid ridge is “obscured” by the extended N-terminal tail and becomes unattainable by a second metal ion. The Co2+ “sealing” ability we are invoking here is in agreement with the large body of experimental work that assigns a role to metal ions as structural stabilizers in disordered peptides and protein folding processes (see La Penna and Morante (2021) for a recent review on this issue).
In the future, we intend to develop our investigations along two lines. The first is a simulation study of the relative stability of wt-FXN compared to that of a more extended set of FXN single-point variants (Botticelli et al., 2022) than the ones considered here. The computation is carried out using the MD methods where altruistic metadynamics is employed to generate maximally unbiased sets of protein configurations (Barducci et al., 2006; Laio and Gervasio, 2008). The constrained maximal entropy principle is then exploited to appropriately reweigh the collected configurations in thermal average computations (La Penna et al., 2004).
The second line of investigation is of a more experimental nature and aims to extend to the FXN variants considered by Botticelli et al. (2022), the CD, FS, and XAS measurements described in the present study.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was partially supported by the BIOPHYS initiative (INFN, Italy). We also acknowledge partial financial support from the MIUR (Italy) PRIN-201744NR8S project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Numerical calculations have been made possible owing to a CINECA-INFN agreement, providing us access to resources on MARCONI at CINECA. The ESRF program committee is acknowledged for beamtime allocation (proposal LS-2968).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.878017/full#supplementary-material
Footnotes
1For the readers’ convenience, we report here the amino acid sequence of the mature, 81‐210, FXN. SGTLGHPGSL DETTYERLAE ETLDSLAEFF EDLADKPYTF EDYDVSFGSG VLTVKLGGDL GTYVINKQTP NKQIWLSSPS SGPKRYDWTG. KNWVYSHDGV SLHELLAAEL TKALKTKLDL SSLAYSGKDA. The α1-helix, from Glu92 to Leu113, is in red. The α2-helix, from Leu182 to Ala193, is in green. The β-sheet core, from Asp124 to Trp168 is in blue.
2We may remark here that Co2+ has another property that is extremely useful for our XAS measurements: unlike Fe2+, it is a very resilient species under radiation damage.
3This nomenclature is defined in the footnote in the introductory section.
4The amino acid sequence of the mature, 81-210, wt-FXN is given in the footnote in the introductory section.
5In this analysis, when two Tm are reported (see Table 2), the smallest is considered as already at this temperature unfolding is starting to take place.
References
Babcock, M., de Silva, D., Oaks, R., Davis-Kaplan, S., Jiralerspong, S., Montermini, L., et al. (1997). Regulation of Mitochondrial Iron Accumulation by Yfh1p, a Putative Homolog of Frataxin. Science 276, 1709–1712. doi:10.1126/science.276.5319.1709
Barducci, A., Chelli, R., Procacci, P., Schettino, V., Gervasio, F. L., and Parrinello, M. (2006). Metadynamics Simulation of Prion Protein: β-Structure Stability and the Early Stages of Misfolding. J. Am. Chem. Soc. 128, 2705–2710. doi:10.1021/ja057076l
Bencze, K. Z., Kondapalli, K. C., Cook, J. D., McMahon, S., Millán-Pacheco, C., Pastor, N., et al. (2006). The Structure and Function of Frataxin. Crit. Rev. Biochem. Mol. Biol. 41, 269–291. doi:10.1080/10409230600846058
Benjwal, S., Verma, S., Röhm, K.-H., and Gursky, O. (2006). Monitoring Protein Aggregation during thermal Unfolding in Circular Dichroism Experiments. Protein Sci. 15, 635–639. doi:10.1110/ps.051917406
Binsted, N., Gurman, S., and Campbell, J. (1998). Daresbury Laboratory EXCURV98 Program. Warrington: CLRC Daresbury Laboratory.
Binsted, N., Strange, R. W., and Hasnain, S. S. (1992). Constrained and Restrained Refinement in EXAFS Data Analysis with Curved Wave Theory. Biochemistry 31, 12117–12125. doi:10.1021/bi00163a021
Botticelli, S., La Penna, G., Nobili, G., Rossi, G., Stellato, F., and Morante, S. (2022). Modelling Protein Plasticity: The Example of Frataxin and its Variants. Molecules 27, 1955. doi:10.3390/molecules27061955
Bresson, C., Esnouf, S., Lamouroux, C., Solari, P. L., and Den Auwer, C. (2006). Xas Investigation of Biorelevant Cobalt Complexes in Aqueous media. New J. Chem. 30, 416–424. doi:10.1039/b514454j
Davoli, T., Xu, A. W., Mengwasser, K. E., Sack, L. M., Yoon, J. C., Park, P. J., et al. (2013). Cumulative Haploinsufficiency and Triplosensitivity Drive Aneuploidy Patterns and Shape the Cancer Genome. Cell 155, 948–962. doi:10.1016/j.cell.2013.10.011
De Santis, E., Minicozzi, V., Proux, O., Rossi, G., Silva, K. I., Lawless, M. J., et al. (2015). Cu(II)-Zn(II) Cross-Modulation in Amyloid-Beta Peptide Binding: An X-ray Absorption Spectroscopy Study. J. Phys. Chem. B 119, 15813–15820. doi:10.1021/acs.jpcb.5b10264
Duarte, F., Bauer, P., Barrozo, A., Amrein, B. A., Purg, M., Åqvist, J., et al. (2014). Force Field Independent Metal Parameters Using a Nonbonded Dummy Model. J. Phys. Chem. B 118, 4351–4362. doi:10.1021/jp501737x
Foury, F., and Cazzalini, O. (1997). Deletion of the Yeast Homologue of the Human Gene Associated with Friedreich's Ataxia Elicits Iron Accumulation in Mitochondria. FEBS Lett. 411, 373–377. doi:10.1016/s0014-5793(97)00734-5
Foury, F. (1999). Low Iron Concentration and Aconitase Deficiency in a Yeast Frataxin Homologue Deficient Strain. FEBS Lett. 456, 281–284. doi:10.1016/s0014-5793(99)00961-8
Furlan, S., Hureau, C., Faller, P., and La Penna, G. (2010). Modeling the Cu+ Binding in the 1−16 Region of the Amyloid-β Peptide Involved in Alzheimer's Disease. J. Phys. Chem. B 114, 15119–15133. doi:10.1021/jp102928h
Gentry, L. E., Thacker, M. A., Doughty, R., Timkovich, R., and Busenlehner, L. S. (2013). His86 from the N-Terminus of Frataxin Coordinates Iron and Is Required for Fe-S Cluster Synthesis. Biochemistry 52, 6085–6096. doi:10.1021/bi400443n
I. Bertini, H. B. Gray, E. I. Stiefel, and J. S. Valentine (Editors) (2007). Biological Inorganic Chemistry: Structure and Reactivity (Sausalito, California: University Science Books).
La Penna, G., and Chelli, R. (2018). Structural Insights into the Osteopontin-Aptamer Complex by Molecular Dynamics Simulations. Front. Chem. 6, 2. doi:10.3389/fchem.2018.00002
La Penna, G., Hureau, C., Andreussi, O., and Faller, P. (2013). Identifying, by First-Principles Simulations, Cu[Amyloid-β] Species Making Fenton-Type Reactions in Alzheimer's Disease. J. Phys. Chem. B 117, 16455–16467. doi:10.1021/jp410046w
La Penna, G., Minicozzi, V., Morante, S., Rossi, G. C., and Stellato, F. (2015). A First-Principle Calculation of the XANES Spectrum of Cu2+ in Water. J. Chem. Phys. 143, 124508. doi:10.1063/1.4931808
La Penna, G., and Morante, S. (2021). Aggregates Sealed by Ions. Methods Mol. Biol. 2340, 309–341. doi:10.1007/978-1-0716-1546-1_14
La Penna, G., Morante, S., Perico, A., and Rossi, G. C. (2004). Designing Generalized Statistical Ensembles for Numerical Simulations of Biopolymers. J. Chem. Phys. 121, 10725–10741. doi:10.1063/1.1795694
Laio, A., and Gervasio, F. L. (2008). Metadynamics: A Method to Simulate Rare Events and Reconstruct the Free Energy in Biophysics, Chemistry and Material Science. Rep. Prog. Phys. 71. doi:10.1088/0034-4885/71/12/126601
Layton, C. J., and Hellinga, H. W. (2011). Quantitation of Protein-Protein Interactions by thermal Stability Shift Analysis. Protein Sci. 20, 1439–1450. doi:10.1002/pro.674
Lin, Y.-F., Cheng, C.-W., Shih, C.-S., Hwang, J.-K., Yu, C.-S., and Lu, C.-H. (2016). MIB: Metal Ion-Binding Site Prediction and Docking Server. J. Chem. Inf. Model. 56, 2287–2291. doi:10.1021/acs.jcim.6b00407
Maiti, B. K., Almeida, R. M., Moura, I., and Moura, J. J. G. (2017). Rubredoxins Derivatives: Simple sulphur-rich Coordination Metal Sites and its Relevance for Biology and Chemistry. Coord. Chem. Rev. 352, 379–397. doi:10.1016/j.ccr.2017.10.001
Martínez-Jiménez, F., Muiños, F., Sentís, I., Deu-Pons, J., Reyes-Salazar, I., Arnedo-Pac, C., et al. (2020). A Compendium of Mutational Cancer Driver Genes. Nat. Rev. Cancer 20, 555–572. doi:10.1038/s41568-020-0290-x
Morante, S., González-Iglesias, R., Potrich, C., Meneghini, C., Meyer-Klaucke, W., Menestrina, G., et al. (2004). Inter- and Intra-octarepeat Cu(II) Site Geometries in the Prion Protein. J. Biol. Chem. 279, 11753–11759. doi:10.1074/jbc.m312860200
Morante, S., and Rossi, G. (2014). Metals in Alzheimer’s Disease: A Combined Experimental and Numerical Approach. Advanc. Alz. Res. 2, 100–147.
Nair, M., Adinolfi, S., Pastore, C., Kelly, G., Temussi, P., and Pastore, A. (2004). Solution Structure of the Bacterial Frataxin Ortholog, CyaY. Structure 12, 2037–2048. doi:10.1016/j.str.2004.08.012
Newville, M., Līviņš, P., Yacoby, Y., Rehr, J. J., and Stern, E. A. (1993). Near-edge X-ray-absorption fine Structure of Pb: A Comparison of Theory and experiment. Phys. Rev. B 47, 14126–14131. doi:10.1103/physrevb.47.14126
Noguera, M. E., Roman, E. A., Rigal, J. B., Cousido-Siah, A., Mitschler, A., Podjarny, A., et al. (2015). Structural Characterization of Metal Binding to a Cold-Adapted Frataxin. J. Biol. Inorg. Chem. 20, 653–664. doi:10.1007/s00775-015-1251-9
Ortega, R., Carmona, A., Llorens, I., and Solari, P. L. (2012). X-ray Absorption Spectroscopy of Biological Samples. A Tutorial. J. Anal. Spectrom. 27, 2054–2065. doi:10.1039/c2ja30224a
Ősz, K., Bóka, B., Várnagy, K., Sóvágó, I., Kurtán, T., and Antus, S. (2002). The Application of Circular Dichroism Spectroscopy for the Determination of Metal Ion Speciation and Coordination Modes of Peptide Complexes. Polyhedron 21, 2149–2159.
Pang, Y.-P. (2001). Successful Molecular Dynamics Simulation of Two Zinc Complexes Bridged by a Hydroxide in Phosphotriesterase Using the Cationic Dummy Atom Method. Proteins 45, 183–189. doi:10.1002/prot.1138
Pastore, A., and Puccio, H. (2013). Frataxin: a Protein in Search for a Function. J. Neurochem. 126, 43–52. doi:10.1111/jnc.12220
Pastore, C., Franzese, M., Sica, F., Temussi, P., and Pastore, A. (2007). Understanding the Binding Properties of an Unusual Metal-Binding Protein − a Study of Bacterial Frataxin. FEBS J. 274, 4199–4210. doi:10.1111/j.1742-4658.2007.05946.x
Petrosino, M., Pasquo, A., Novak, L., Toto, A., Gianni, S., Mantuano, E., et al. (2019). Characterization of Human Frataxin Missense Variants in Cancer Tissues. Hum. Mutat. 40, 1400–1413. doi:10.1002/humu.23789
Phillips, J. C., Hardy, D. J., Maia, J. D. C., Stone, J. E., Ribeiro, J. V., Bernardi, R. C., et al. (2020). Scalable Molecular Dynamics on Cpu and Gpu Architectures with NAMD. J. Chem. Phys. 153, 044130. doi:10.1063/5.0014475
Proux, O., Biquard, X., Lahera, E., Menthonnex, J. J., Prat, A., Ulrich, O., et al. (2005). FAME A New Beamline for XRay Absorption Investigations of VeryDiluted Systems of Environmental, Material and Biological Interests. Physica Scripta, 970. doi:10.1238/physica.topical.115a00970
Radisky, D. C., Babcock, M. C., and Kaplan, J. (1999). The Yeast Frataxin Homologue Mediates Mitochondrial Iron Efflux. J. Biol. Chem. 274, 4497–4499. doi:10.1074/jbc.274.8.4497
Ravel, B., and Newville, M. (2005). ATHENA,ARTEMIS,HEPHAESTUS: Data Analysis for X-ray Absorption Spectroscopy usingIFEFFIT. J. Synchrotron Radiat. 12, 537–541. doi:10.1107/s0909049505012719
R. Crichton (Editor) (2019). Biological Inorganic Chemistry: A New Introduction to Molecular Structure and Function. third edition edn (Academic Press). doi:10.1016/B978-0-12-811741-5.00030-8
Rötig, A., de Lonlay, P., Chretien, D., Foury, F., Koenig, M., Sidi, D., et al. (1991). Aconitase and Mitochondrial Iron-sulphur Protein Deficiency in Friedreich Ataxia. Nat. Genet. 17, 215–217. doi:10.1038/ng1097-215
Rychtarcikova, Z., Lettlova, S., Tomkova, V., Korenkova, V., Langerova, L., Simonova, E., et al. (2017). Tumor-initiating Cells of Breast and Prostate Origin Show Alterations in the Expression of Genes Related to Iron Metabolism. Oncotarget 8, 6376–6398. doi:10.18632/oncotarget.14093
Shannon, R. D. (1976). Revised Effective Ionic Radii and Systematic Studies of Interatomic Distances in Halides and Chalcogenides. Acta Cryst. Sect A. 32, 751–767. doi:10.1107/s0567739476001551
Shi, W., and Chance, M. R. (2008). Metallomics and Metalloproteomics. Cell. Mol. Life Sci. 65, 3040–3048. doi:10.1007/s00018-008-8189-9
Söderberg, C. A. G., Rajan, S., Shkumatov, A. V., Gakh, O., Schaefer, S., Ahlgren, E.-C., et al. (2013). The Molecular Basis of Iron-Induced Oligomerization of Frataxin and the Role of the Ferroxidation Reaction in Oligomerization. J. Biol. Chem. 288, 8156–8167. doi:10.1074/jbc.M112.442285
Söderberg, C. A. G., Shkumatov, A. V., Rajan, S., Gakh, O., Svergun, D. I., Isaya, G., et al. (2011). Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering. J. Mol. Biol. 414, 783–797. doi:10.1016/j.jmb.2011.10.034
Strange, R., Morante, S., Stefanini, S., Chiancone, E., and Desideri, A. (1993). Nucleation of the Iron Core Occurs at the Three-fold Channels of Horse Spleen Apoferritin: an EXAFS Study on the Native and Chemically-Modified Protein. Biochim. Biophys. Acta (Bba) - Protein Struct. Mol. Enzymol. 1164, 331–334. doi:10.1016/0167-4838(93)90267-u
Keywords: frataxin, metal ions (Co2+), frataxin mutants, XAS (XAFS, XANES), termal stability
Citation: Morante S, Botticelli S, Chiaraluce R, Consalvi V, La Penna G, Novak L, Pasquo A, Petrosino M, Proux O, Rossi G, Salina G and Stellato F (2022) Metal Ion Binding in Wild-Type and Mutated Frataxin: A Stability Study. Front. Mol. Biosci. 9:878017. doi: 10.3389/fmolb.2022.878017
Received: 17 February 2022; Accepted: 12 April 2022;
Published: 31 May 2022.
Edited by:
Bikash Ranjan Sahoo, Howard Hughes Medical Institute (HHMI), United StatesReviewed by:
Jagadeeswara Rao Bommi, Case Western Reserve University, United StatesErnesto Andrès Roman, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina
Copyright © 2022 Morante, Botticelli, Chiaraluce, Consalvi, La Penna, Novak, Pasquo, Petrosino, Proux, Rossi, Salina and Stellato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: S. Morante , silvia.morante@roma2.infn.it
 S. Morante
S. Morante S. Botticelli
S. Botticelli R. Chiaraluce
R. Chiaraluce V. Consalvi
V. Consalvi G. La Penna
G. La Penna L. Novak3
L. Novak3  A. Pasquo
A. Pasquo M. Petrosino
M. Petrosino O. Proux
O. Proux G. Rossi
G. Rossi F. Stellato
F. Stellato