- 1Department of Microbiology, Abbottabad University of Science and Technology, Abbottabad, Pakistan
- 2College of Animal Science and Technology, Anhui Agricultural University, Hefei, China
- 3Faculty of Veterinary and Animal Sciences, Gomal University, Dera Ismail Khan, Pakistan
- 4Department of Physics, University of Azad Jammu and Kashmir, Muzaffarabad, Pakistan
- 5Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 6Plant Production Department, College of Food & Agriculture Sciences, King Saud University, Riyadh, Saudi Arabia
Staphylococcus aureus is part of normal human flora and is widely associated with hospital-acquired bacteremia. S. aureus has shown a diverse array of resistance to environmental stresses and antibiotics. Methicillin-resistant S. aureus (MRSA) is on the high priority list of new antibiotics discovery and glycopeptides are considered the last drug of choice against MRSA. S. aureus has developed resistance against glycopeptides and the emergence of vancomycin-intermediate-resistant, vancomycin-resistant, and teicoplanin-resistant strains is globally reported. Teicoplanin-associated genes tcaR-tcaA-tcaB (tcaRAB) is known as the S. aureus glycopeptide resistance operon that is associated with glycopeptide resistance. Here, for the first time, the role of tcaRAB in S. aureus persister cells formation, and ΔtcaA dependent persisters’ ability to resuscitate the bacterial population was explored. We recovered a clinical strain of MRSA from a COVID-19 patient which showed a high level of resistance to teicoplanin, vancomycin, and methicillin. Whole genome RNA sequencing revealed that the tcaRAB operon expression was altered followed by high expression of glyS and sgtB. The RNA-seq data revealed a significant decrease in tcaA (p = 0.008) and tcaB (p = 0.04) expression while tcaR was not significantly altered. We knocked down tcaA, tcaB, and tcaR using CRISPR-dCas9 and the results showed that when tcaA was suppressed by dCas9, a significant increase was witnessed in persister cells while tcaB suppression did not induce persistence. The results were further evaluated by creating a tcaA mutant that showed ΔtcaA formed a significant increase in persisters in comparison to the wild type. Based on our findings, we concluded that tcaA is the gene that increases persister cells and glycopeptide resistance and could be a potential therapeutic target in S. aureus.
1. Introduction
Staphylococcus aureus has two major types of resistant strains, namely, methicillin-resistant S. aureus (MRSA) and vancomycin-resistant S. aureus (VRSA). MRSA is considered the major cause of hospital and community-acquired infections (Aqib and Rodriguez-Morales, 2021) and is largely treated with glycopeptides. However, the emergence of teicoplanin resistance, vancomycin-intermediate S. aureus (VISA), and VRSA has made the clinical treatment unsuccessful and increased death rates across the world (Howden et al., 2010). S. aureus resistant to glycopeptides are a serious threat to public health and teicoplanin-resistant (Elsaghier et al., 2002; Szymanek-Majchrzak et al., 2018), VISA, and VRSA strains are globally reported (Appelbaum, 2006; Shariati et al., 2020). A meta-analysis of 155 articles from 2010 to 2019 reported a global prevalence of VRSA of 2.4 and 4.3% for VISA (Shariati et al., 2020), whereas a similar study reported a 7% frequency of VRSA from 2015 to 2020, with a prevalence of 16% in Africa, 5% in Asia, and 4% in America (Wu et al., 2021). Different genes were reported for glycopeptide resistance in S. aureus such as the vanA, yycF, yycG, tcaA, and ccpA (Renzoni et al., 2009). Other include mutations in accessory gene regulator, vraSR, and graSR two-component regulatory systems (Howden et al., 2008; Hu et al., 2016). Likewise, mutations in sigB and trfAB genes also contributed to glycopeptide resistance (Renzoni et al., 2009; Schulthess et al., 2009). According to the Clinical and Laboratory Standards Institute (CLSI) guidelines, S. aureus strains with a minimum inhibitory concentration (MIC) of ≤2 μg/mL would be considered susceptible (Clinical and Laboratory Standards Institute, 2018) and MRSA isolates with a glycopeptide MIC of greater than 2 μg/mL reflect poor clinical outcomes (Chen et al., 2013; Song et al., 2017). Several studies documented that teicoplanin resistance developed earlier than vancomycin resistance (Brunet et al., 1990; Hiramatsu, 2001) and was related to teicoplanin resistance operon tcaRAB (Brandenberger et al., 2000; Maki et al., 2004). A study revealed that teicoplanin resistance resulted in a slight increase in vancomycin resistance (Hiramatsu, 2001). Furthermore, penicillin binding protein-2 (pbp2) ectopic expression was associated with an increase in vancomycin MIC from 1 to 2 μg/mL and teicoplanin MIC from 2 to 8 μg/mL (Sieradzki et al., 1998). Interestingly, both Gram-negative and Gram-positive bacteria use different strategies to evade antibiotic actions. One of the mechanisms bacteria have adopted is the formation of persister cells. Persisters are nongrowing or metabolically less active cells that can survive high antibiotic concentrations without being resistant (Lewis, 2010; Personnic et al., 2023). Upon favorable conditions, persisters become metabolically active, regain their virulence potential, and resuscitate the whole population that remains fully susceptible to antibiotics (Eisenreich et al., 2021). Persistence should not be confused with tolerance and resistance because tolerance is the ability of a bacterial population to survive a transient exposure to antibiotics usually higher than the MIC (Handwerger and Tomasz, 1985), whereas resistance is an inherited trait that develops due to genetic changes in bacteria (Blair et al., 2015). Resistance to antibiotics is quantified by MIC testing and substantially higher than the MIC for a susceptible strain of bacteria, whereas the MIC for a tolerant strain is similar to the susceptible strain. Similarly, a persistent strain has a similar MIC and minimum duration for killing (MDK99) to a susceptible strain, whereas the MDK99 for a tolerant strain is substantially higher than the MDK99 for a susceptible strain. However, the MDK for 99.99% (MDK99.99) of bacterial cells in the population is substantially higher for a persistent strain than the MDK99.99 for a susceptible strain (Brauner et al., 2016). Several factors have been reported in different bacteria that contributed to persister cell formation, including nutrient starvation, acidic pH, accumulation of insoluble proteins, ATP depletion, and antibiotics exposure (Levin et al., 2017; Mohiuddin et al., 2021). Particularly, ATP depletion has induced persister cell formation in S. aureus, E. coli, and P. aeruginosa (Cameron et al., 2018). S. aureus purB and purM mutants showed defective persistence in low pH, heat stress, and through rifampicin treatment (Lin et al., 2020). The work of Wang et al. (2015) revealed the results of the transposon mutant library of a clinical MRSA strain where gene mutations in the tricarboxylic acid cycle, oxidative phosphorylation, and ABC transporters showed a lower number of persisters. For instance, the mutant of succinate dehydrogenase was defective in persister cells formation against levofloxacin (Wang et al., 2015). Shang and coworkers inactivated phoU which decreased vancomycin and levofloxacin persisters (Shang et al., 2020). Tricarboxylic acid cycle genes increased persister cell formation due to reduced ATP level and membrane potential (Wang et al., 2018). Likewise, S. aureus grown in polymicrobial cultures displayed increase antibiotic tolerance, accompanied by low intracellular ATP and membrane potential (Jia et al., 2013). Of note, persister cells can facilitate the evolution of drug resistance because tolerance precedes resistance and can boost the chances for resistance mutations in bacterial populations (Levin-Reisman et al., 2017). Due to the COVID-19 pandemic, numerous bacterial resistant strains were reported worldwide and were associated with antibiotic resistance and treatment failure (Ghanizadeh et al., 2021; Habib et al., 2022, 2023; Tariq et al., 2023). In this study, we assessed teicoplanin- and vancomycin-intermediate-resistant S. aureus recovered from a COVID-19 patient and explored the involvement of tcaABR operon in glycopeptide resistance and persistence. The role of tcaA in glycopeptide resistance has been widely studied but tcaA involvement in persister cell formation has not been discovered. Using CRISPR-dCas-9, the genetic basis was explored that revealed the teicoplanin resistance gene tcaA was associated with an increase in persister cell formation. The inactivation of tcaA has shown persistence and resistance to glycopeptides whereas tcaB and tcaR have no significant effects.
2. Materials and methodology
2.1. Bacterial strains, media, vectors, and growth parameters
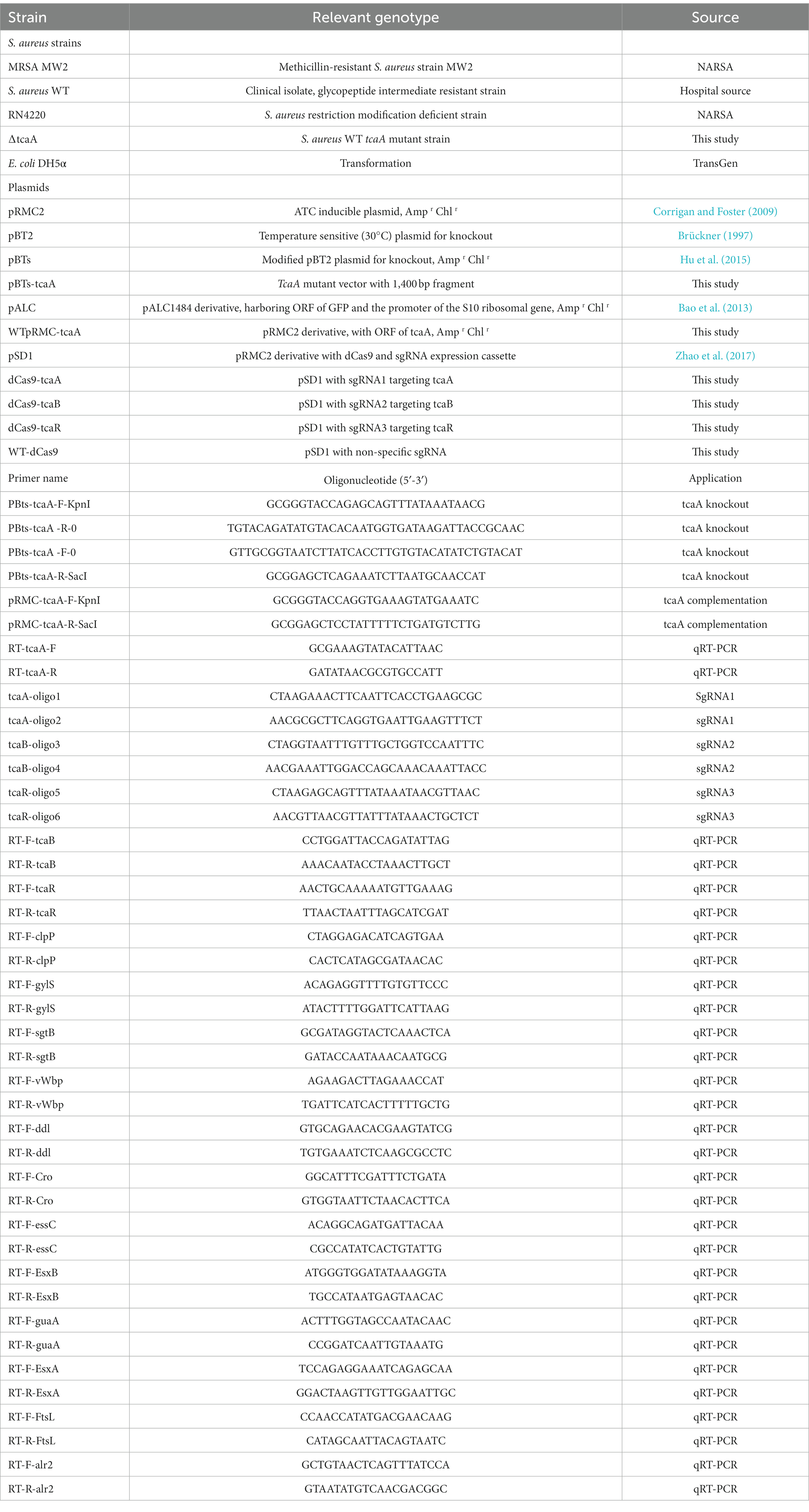
The S. aureus teicoplanin- and vancomycin-intermediate-resistant clinical strain, hereafter referred to as wild-type (WT), was obtained from Lady Reading Hospital Peshawar. Tryptic soy broth (TSB), Luria Bertani broth (LB), and cation-adjusted Mueller Hinton broth (MHB) were used for S. aureus, E. coli, and MIC testing, respectively. Bacterial cultures were refreshed from −80°C and were grown in TSB (S. aureus) and LB (E. coli) at 37°C with shaking at 220 rpm. Bacterial strains were grown in round bottom tubes (14 mL tube with 3 mL culture media) and persister assays were performed in a conical flask (100 mL flask with 30 mL culture media). Cells were washed with ddH2O. Temperature-sensitive (at 30°C) vector pBTs were used for mutant construction in S. aureus, pRMC-2 as an inducible vector for gene expression, and pALC with green fluorescent protein (GFP) gene was used for fluorescence assay. For plasmid maintenance, media were supplemented with appropriate antibiotic concentration, i.e., ampicillin 150 μg/mL and chloramphenicol 20 μg/mL. The pSD1 is a pRMC2 derivative vector that contains dCas9 and sgRNA expression cassettes. S. aureus RN4220 and E. coli DH5α were used for transformation. The primers, plasmids, recombinant vectors, and bacterial strains used in this study are listed in Table 1.
2.2. Whole genome RNA sequencing procedure
We performed RNA sequencing to study the transcriptome of the resistant isolate. S. aureus 24 h culture was 100 times diluted and was grown for 2 h (OD600 = 2) in TSB medium, cells were harvested, washed, and dissolved in RNAiso Plus (Takara Tokyo, Japan) and preserved at −80°C. RNA sequencing was performed in two biological replicates by the core sequence facility of the Institute of Microbiology Chinese Academy of Sciences. A NanoPhotometer® (Implen CA, USA), the Qubit® RNA Assay Kit and Fluorometer (Life Technologies CA, USA), and the RNA Nano 6,000 Assay Kit from the Agilent Bioanalyzer 2,100 system were used to measure RNA purity, concentration, and integrity, respectively. RNA degradation or contamination was checked by agarose gel (1%).
2.2.1. Library construction for transcriptome sequencing
The NEBNext®UltraTM RNA Library Prep Kit from Illumina®(USA) was used for library preparation, and index codes were added to attribute sequences to each sample. Briefly, poly-T oligo-attached magnetic beads were used to purify mRNA from total RNA, then fragmentation was performed using divalent cations under high temperature in NEBNext First-Strand Synthesis Reaction Buffer (5X). Next, the first-strand cDNA strand was synthesized by M-MuLV Reverse Transcriptase and random hexamer primer, whereas the second-strand cDNA strand was synthesized by DNA Polymerase I and RNase H, and the ends of the remaining overhang were converted into blunt ends by the polymerase activity. Further, adenylation of 3′ ends of DNA fragments was performed, and an adaptor (NEBNext) with a hairpin loop structure was ligated, and cDNA fragments of 200-250 bp were preferentially purified by the AMPure XP system (Beckman Coulter, Beverly, USA). The size-selected, adaptor-ligated cDNA was treated with 3 μL USER Enzyme (NEB, USA) at 37°C for 15–20 min and followed by 5 min at 95°C. Next, PCR was performed with Phusion High-Fidelity DNA polymerase, Index (X) Primer, and Universal PCR primers. The PCR product was purified by the AMPure XP system. The Agilent Bioanalyzer 2,100 system was used to assess the quality of the constructed library (Wang Q. et al., 2017).
2.2.2. Clustering and sequencing
The index-coded sample clustering was done on the Illumina cBot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS protocols. After cluster generation, the Illumina Hiseq 2,500 platform was used for library sequencing, and paired-end reads were generated.
2.2.3. Quality assessment and comparative analysis
Raw reads (raw data) of fastq format were processed through in-house Perl scripts and the clean data was obtained by removing reads containing adapter and ploy-N. In addition, the low-quality reads from raw data were also removed, and the clean data Q20, Q30, GC-content, and sequence duplication levels were calculated. Clean data with high quality were used for all the downstream analyses. During data processing, the adaptor sequences and low-quality reads were removed, and clean reads were generated that were mapped to the reference genome sequence. The reads with a perfect match were annotated based on the reference genome. Hisat2 software was used to annotate the reads with the reference genome.
2.2.4. Gene functional and differential expression analysis
Gene functions were annotated with the help of the following databases: Nt (NCBI non-redundant nucleotide sequences); Nr (NCBI non-redundant protein sequences); Pfam (Protein family); KOG/COG (Clusters of Orthologous Groups of proteins); Swiss-Prot (protein sequence database); KO (KEGG Orthologue database); GO (Gene Ontology). Differential expression analysis was performed using the DESeq R package (1.10.1). DESeq is used for determining differential expression in digital gene expression data using the model based on the negative binomial distribution (Anders and Huber, 2010). Benjamini and Hochberg’s approach was used to adjust the p-values to control for the false discovery rate. Genes with an adjusted p < 0.05 found by DESeq were assigned as differentially expressed.
2.2.5. Gene Ontology (GO) enrichment analysis
For GO enrichment analysis of the differentially expressed genes (DEGs), the GOstats and topGO packages based on Wallenius non-central hypergeometric distribution were applied (Young et al., 2010).
2.2.6. KEGG enrichment analysis
The KEGG database is a set of processes to map genes and proteins, etc. to molecular interaction and relation networks, and is used for large-scale molecular datasets analysis generated by genome sequencing1 (Kanehisa et al., 2007). We used KOBAS software to test the statistical enrichment of differential expression genes in KEGG pathways (Mao et al., 2005).
2.3. RNA isolation and RT-qPCR
Staphylococcus aureus cells were collected and dissolved in 1 mL of RNAiso Plus. The cell lysis was performed with the help of 0.1-mm silica beads in the FastPrep-24 automated system, and then the lysate was treated with DNase I to remove the remaining DNA. For reverse transcription and qPCR, the PrimeScript cDNA synthesis kit and SYBR Premix Ex Taq reagent kit (Takara Tokyo, Japan) were used, respectively. The StepOne real-time PCR system was used for RT-qPCR analysis. The hu gene cDNA abundance was used for normalization (Valihrach and Demnerova, 2012).
2.4. Gene knockdown vector pSD1
For gene knockdown, the anhydrotetracycline (ATC) inducible vector pSD1 was used which expresses dcas9 and custom-designed sgRNA. The pSD1 vector was derived from Streptococcus pyogenes cas9 and was used as a CRISPR interference system (CRISPRi) (Zhao et al., 2017). From the vector design, the dcas9 and sgRNA are under the control of the PtetO and PpflB inducible and constitutive promoters, respectively. For tcaA/tcaB/tcaR knockdown, gene specific complementary oligonucleotide sequences were synthesized and cloned into the SapI-digested pSD1 site. The sgRNA 5′ variable region binds to the target gene while the 3′ constant region binds to dCas9. The dCas9 functions as a DNA-binding protein guided by sgRNA which is complementary to each target gene. The pSD1 plasmid without the target gene (WT-dCas9) was used as a negative control that can also express the sgRNA which has no specific target site in S. aureus.
2.5. Construction of the tcaA mutant strain
To create the tcaA mutant, the upstream and downstream regions were amplified by primer pairs PBts-tcaA-F-KpnI and PBts-tcaA-R-0, and PBts-tcaA-F-0 and PBts-tcaA-R-SacI from S. aureus WT genome, respectively, (Table 1). The PBts-tcaA-R-0 and PBts-tcaA-F-0 are the overlap primers that contain the overlap segments, and a 1,400-bp fragment was generated by PBts-tcaA-F-KpnI and PBts-tcaA-R-SacI in Prime Star PCR conditions: 95°C for 5 min, 95°C for 30 s, 55°C for 1.5 min, 72°C for 1 min, 35 cycles, and 72°C for 10 min. A 1400-bp joint fragment was constructed, sequenced, and purified to clone into pBTs, a modified vector of pBT2 (Brückner, 1997; Bae and Schneewind, 2006) via restriction enzymes. The 1,400 bp fragment and pBTs vector were digested with KpnI and SacI for 1 h at 37°C, the mixture was purified by a DNA purification kit separately and ligated into pBTs by DNA ligase. The constructed recombinant vector was initially transferred to DH5α, then transferred to RN4220 for genomic modification, and lastly to S. aureus WT. S. aureus colonies carrying the recombinant vector were selected by tryptic soy agar plus chloramphenicol (20 μg/mL) at 37°C. S. aureus carrying the recombinant vector pBTs-tcaA was grown at 30°C with shaking at 220 rpm for 24–72 h. Daily screening was performed on ATC inducible tryptic soy agar plates at 37°C. PCR was performed to screen the mutant colonies. The tcaA markerless mutant was confirmed by DNA sequencing.
2.6. Antibiotic susceptibility and MIC testing
We followed our previous method of antibiotic susceptibility testing in S. aureus (Habib et al., 2020). Initially, the tcaA complementary strain (C-tcaA) was constructed by amplifying the tcaA gene through primer pairs pRMC-tcaA-F-KpnI and pRMC-tcaA-R-SacI, and the vector WTpRMC-tcaA was constructed and transferred to S. aureus WT. The 24 h fresh culture of S. aureus, ∆tcaA, and C-tcaA (complementary strain) was grown for 2 h in TSB (OD600 = 2.0) and 2 × 109 CFU/mL were challenged with 100 fold MIC of teicoplanin, vancomycin, and azithromycin at 37°C. The inhibition zones were mapped after 24–36 h. MIC was determined in MHB medium using the two-fold broth microdilution method (Clinical and Laboratory Standards Institute, 2018). Briefly, the double dilutions of antibiotics were distributed into microtiter plates wells, and freshly prepared bacterial suspension (5 × 105 CFU/mL) was added to each well, and plates were incubated at 35°C. S. aureus colonies suspension with a density of 5 × 105 CFU/mL required a transfer of 100 μL of the 0.5 McFarland equivalent suspension to 10 mL of broth. Of note, CLSI does not recommend performing vancomycin susceptibility testing using the disk diffusion method.
2.7. Western blot assay
The protein expression levels of tcaA, tcaB, and tcaR were detected by specific antisera for tcaA, tcaB, and tcaR through Western blotting. The dCas9-tcaA, dCas9-tcaB, and dCas9-tcaR were induced at OD600 = 0.4 with ATC 100 ng/mL for 1 h. The cells were collected, and media were removed and washed twice with ddH2O. An equal number of cells were taken and lysed by lysostaphin for 1 h at 37°C. The mixture was washed with normal saline and total proteins were collected in lysis buffer. A 12% SDS-PAGE was used for whole-cell protein separation and proteins were electrotransferred to a polyvinylidene difluoride membrane. The tcaA, tcaB, and tcaR proteins were detected with rabbit anti-tcaA, anti-tcaB, and anti-tcaR antibodies (1:1000) followed by horseradish-peroxidase conjugated antibody (1:10,000 dilution). The membrane was analyzed using a Thermo Fisher chemiluminescent detection kit and spots were detected using an ImageQuant LAS 4000.
2.8. Persister assay
A persistence assay was adopted from our previous protocol (Habib et al., 2020). The cells were grown to OD600 = 0.40 in the TSB medium. The pSD1 containing dCas9-tcaA and dCas9-tcaB were induced with ATC 100 ng/mL for 1 h, the medium was removed, and fresh TSB and antibiotic were added whereas ΔtcaA cells were challenged with antibiotic at OD600 = 0.40. After 12 h, a 200 μL sample was taken and CFU counting was performed. Each antibiotic concentration was 10 fold of MIC, and S. aureus WT and S. aureus WT-dCas9 were used as controls.
2.9. Fluorescence microscopy
Fluorescence microscopy was used to observe the persister cell resuscitation ability of the whole population. S. aureus WT and ΔtcaA were transferred with pALC fluorescence shuttle plasmid with GFP and challenged with 20-fold MIC for 48 h. The cells were harvested, washed with ddH2O, and resuspended in TSB media. The cells were grown for 2 h at 37°C and the S. aureus WT and ΔtcaA were observed under the fluorescence microscope.
2.10. Growth curve analysis
The overnight culture was 100 times diluted in TSB medium and a 200 μL was inoculated into a 96-well microtiter plate. The plates were incubated at 37°C with shaking at 200 rpm. At different time intervals (1–18 and 24 h), OD600 was measured using a microplate reader (Thermo Fisher, Waltham, MA, USA).
2.11. Statistical analyses
Experiments were performed in biological triplicates unless otherwise stated. The data values were analyzed by Student’s t-test for two groups (unpaired, two-tailed) and a one-way analysis of variance for more than two groups. The gene sequences were analyzed by vector NTI advance and data were analyzed by GraphPad prism 8. * p < 0.05; ** p < 0.01; *** p < 0.005.
3. Results
3.1. Characterization of glycopeptide intermediate resistant Staphylococcus aureus (WT)
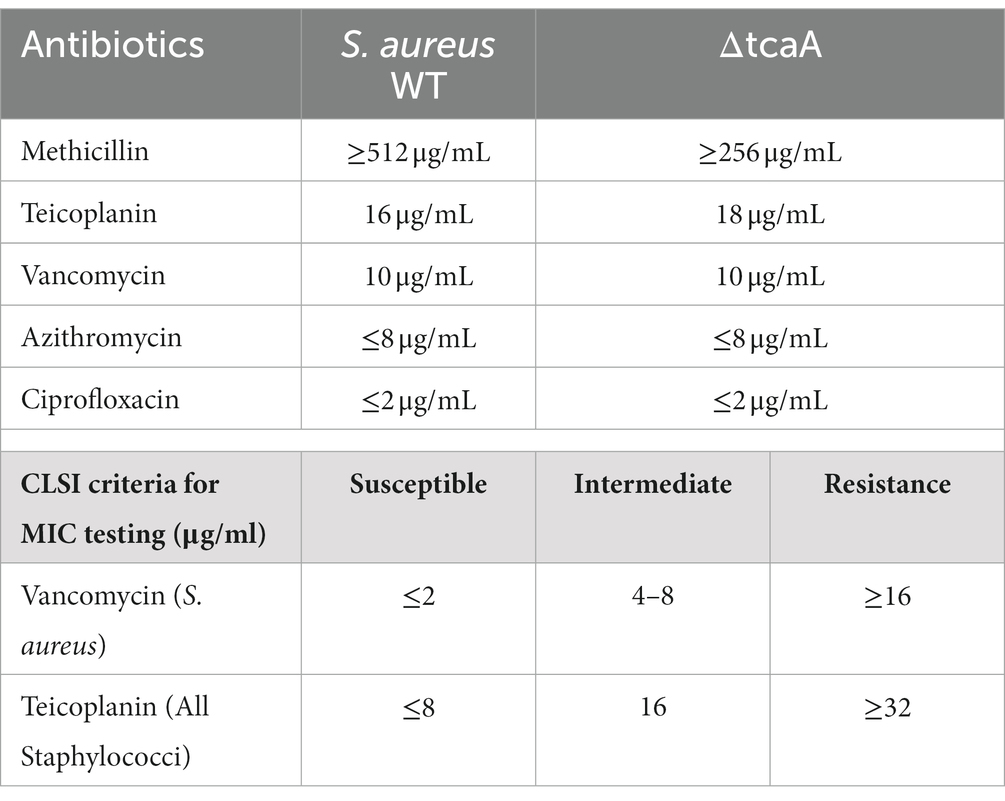
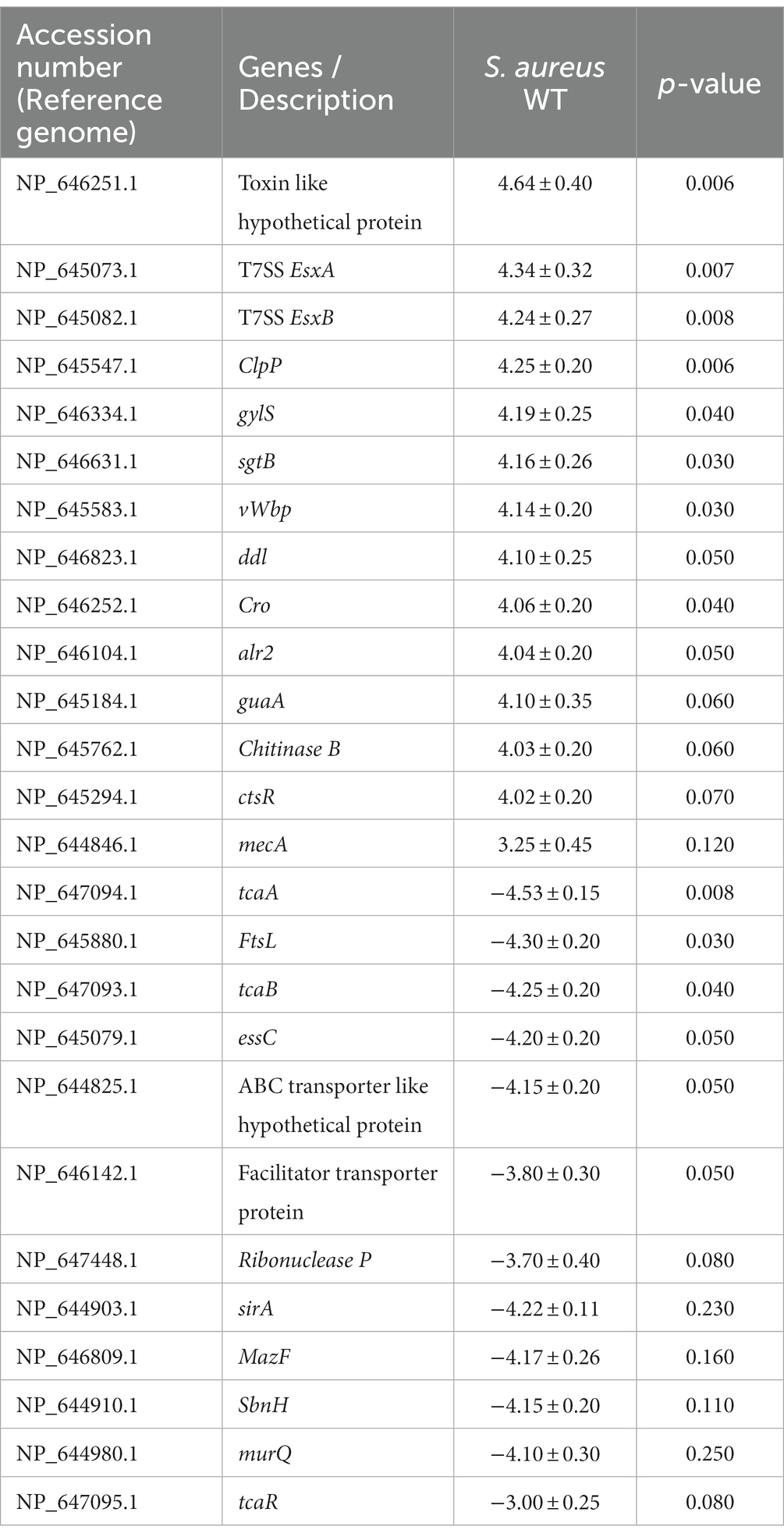
Staphylococcus aureus isolated from a COVID-19 patient was resistant to methicillin, teicoplanin, and vancomycin, while sensitive to azithromycin and ciprofloxacin. The MIC of methicillin was >512 μg/mL, teicoplanin was 16 μg/mL, and vancomycin was 10 μg/mL (Table 2). RNA sequencing results of WT revealed that multiple genes and pathways have been affected, comprising 62 differentially expressed genes. The differential expression of highly affected genes is shown by a heat map (Figure 1A). Among the differential expressed genes, Clusters of Orthologous Genes (COG) involved in cell motility (N), extracellular structures (W), prophages and transposons (V), cell division, and chromosome partitioning (D) were marginally expressed whereas amino acid transport and metabolism (E), inorganic ion transport and metabolism (P), and general function prediction (R) were highly expressed (Figure 1B). The COG group associated with nucleotide transport and metabolism (F), translation (J), transcription (K), secondary metabolites biosynthesis (Q), intracellular trafficking (U), defense mechanism (V), cell wall/membrane/envelope biogenesis (M) were slightly expressed (Figure 1B). Differentially expressed genes such as ClpP, EsxB, EsxA, toxin-like hypothetical protein, glyS, Cro, sgtB, ddl, alr2, and vWbp genes expression was high whereas tcaA, essC, tcaB, FtsL, and ABC transporter permease gene expression was low (Table 3). We screened out the genes responsible for glycopeptide resistance in S. aureus which is reported to be the tcaRAB operon. The tcaRAB is stimulated when S. aureus is exposed to a minimum inhibitory concentration of glycopeptides. In the tcaRAB operon, the tcaA is a 454-residues zinc ribbon domain-containing protein, the tcaB is a 402-residues multidrug efflux MFS transporter, and tcaR is a 151-residues MarR family transcriptional regulator. The WT strain RNA sequencing data revealed a significant decrease in tcaA (p = 0.008) and tcaB (p = 0.04) expression while tcaR was not significantly altered (p = 0.08) (Figure 1A and Table 3). We validated the RNA sequence data by RT-qPCR which confirmed that tcaAB and essC genes were significantly suppressed whereas EsxAB, ClpP, glyS, and sgtB were significantly upregulated (Figure 2 and Table 3). The EsxAB are the secretory protein of type seven secretion system (T7SS), ClpP is involved in proteostasis, glyS is glycine tRNA synthetase, sgtB is mono-functional peptidoglycan glycosyltransferase, ddl codes for D-alanine-D-alanine ligase/synthetase, and alr2 is alanine racemase. From this data, we proposed that EsxAB, ClpP, glyS, and sgtB expression was upregulated when tcaA was suppressed which might be linked with cell wall protection against wall-damaging agents such as glycopeptides and viruses. Besides, the T7SS ATP synthesis machinery gene essC was significantly downregulated (Figure 2) which is associated with S. aureus survival during host infection. Collectively, these results corroborated the RNA sequencing data and confirmed that tcaAB suppression is associated with glycopeptides resistance either to protect the cell wall against wall-piercing agents or to increase survival during host infection.
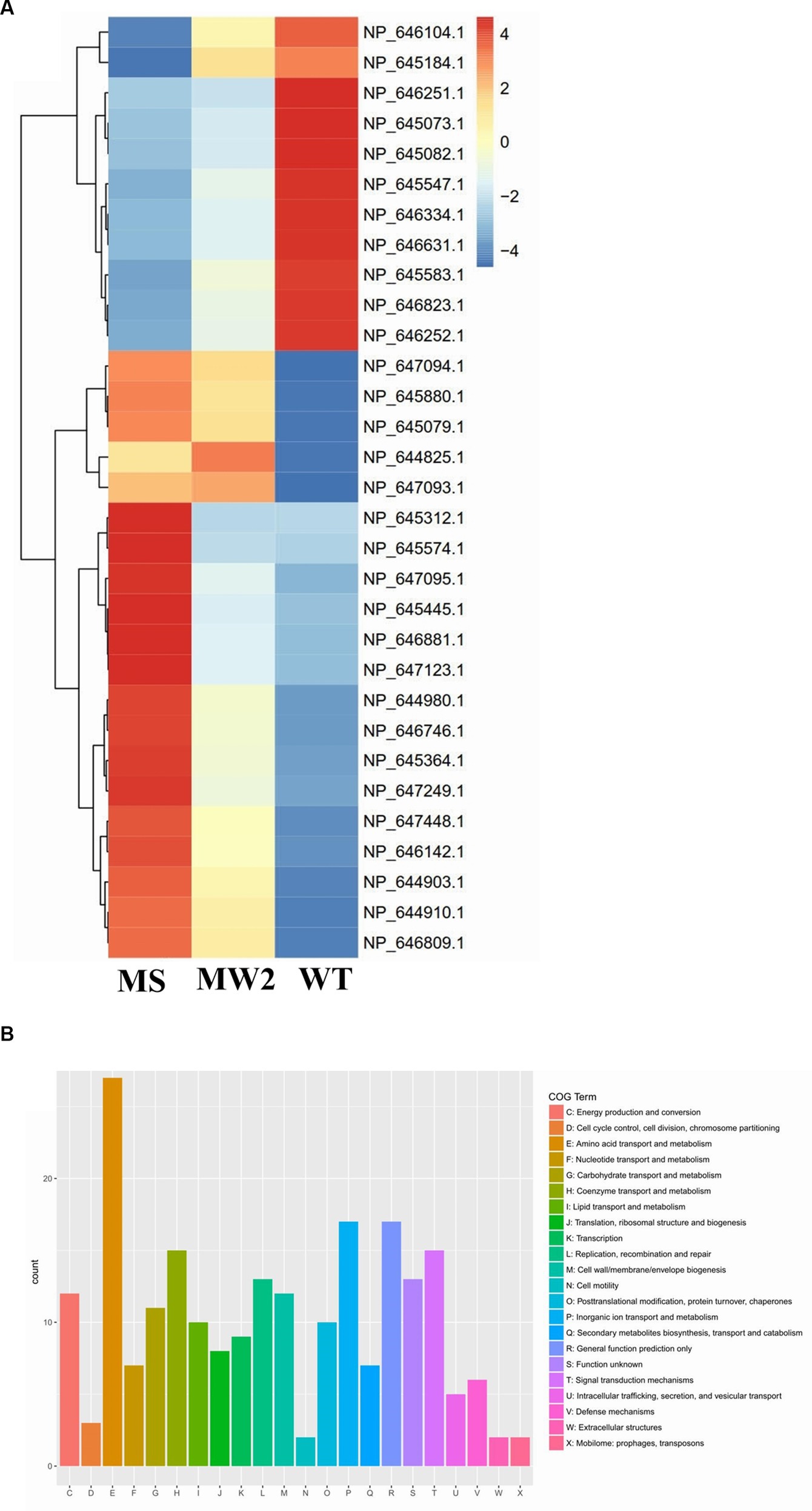
Figure 1. RNA-seq analysis. (A) The heat map of differentially expressed genes. Red represents upregulated genes and blue indicates downregulated genes. The tcaRAB operon genes tcaA (NP_647094.1) and tcaB (NP_647093.1) were significantly downregulated. The differentially expressed genes of S. aureus WT were compared with MS (methicillin-sensitive S. aureus) and MRSA MW2 as reference strains. (B) The COG function classification of differentially expressed genes was performed in S. aureus WT vs. MRSA MW2.
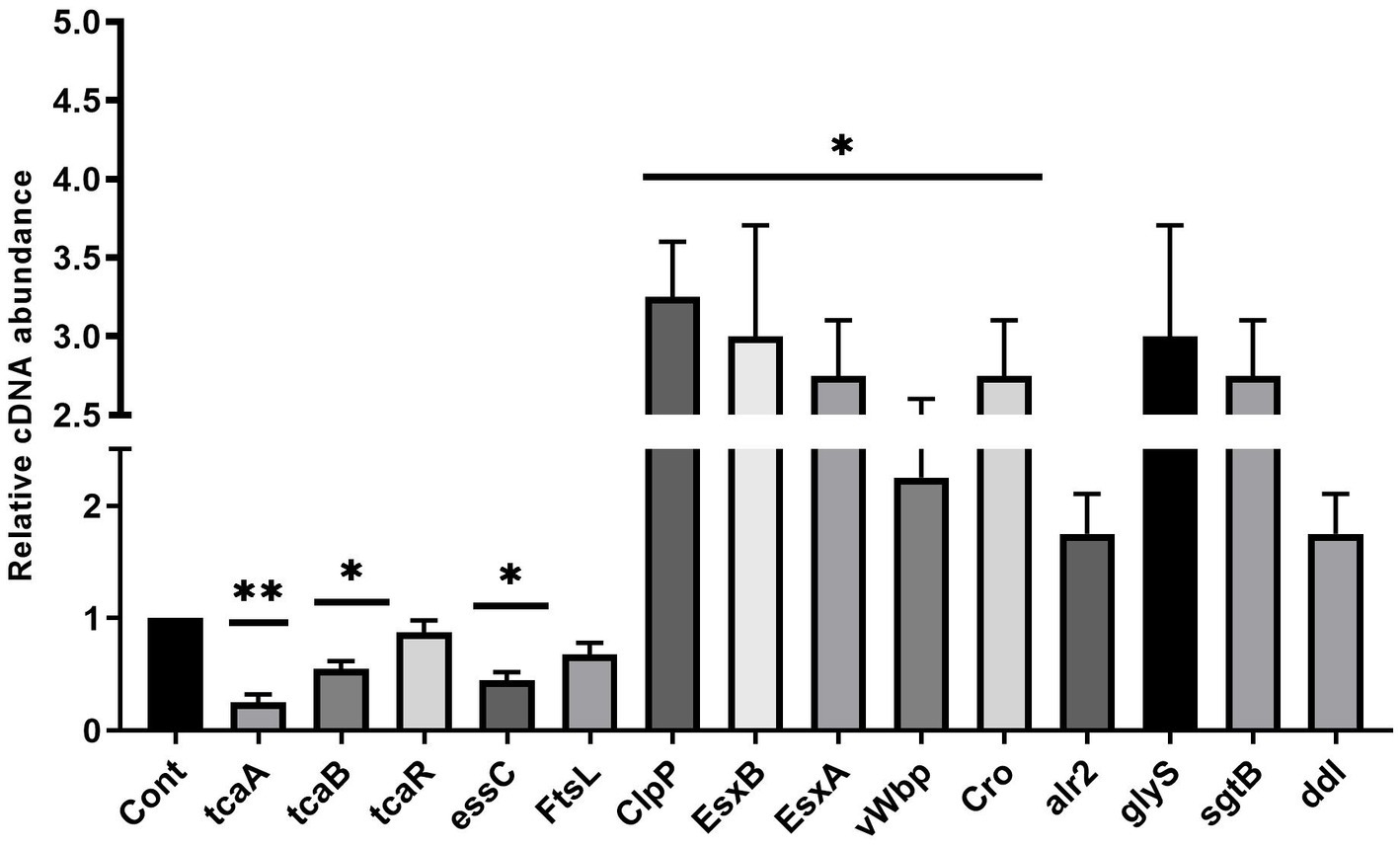
Figure 2. The up and downregulated genes. The RT-qPCR analysis revealed the transcript level of tcaRAB, essC, FtsL, ClpP, EsxB, EsxA, glyS, alr2, sgtB, ddl, Cro, and vWbp in S. aureus WT strain. * p < 0.05; ** p < 0.01.
3.2. Knockdown of tcaA, tcaB, and tcaR By CRISPR-dCas9
We performed CRISPRi-mediated tcaA, tcaB, and tcaR knockdown by dCas9 in S. aureus. Gene-specific sgRNAs were designed that can bind to the target site of tcaA, tcaB, and tcaR (Table 1). Media were supplied with 100 ng/mL of ATC to induce the CRISPR-dCas9 to suppress gene expression. Initially, the dCas9 repression efficiency was determined in both the knockdown and WT strain by RT-qPCR. The data revealed that the strains expressing sgRNA1, sgRNA2, and sgRNA3 exhibited a 22-fold, 20-fold, and 26-fold decrease in the tcaA, tcaB, and tcaR mRNA levels, respectively, (Figure 3A). Further, we analyzed the dCas-tcaA, dCas-tcaB, and dCas-tcaR protein expression level by rabbit anti-tcaA, anti-tcaB, and anti-tcaR antibodies through Western blotting. The immunoblots showed brighter tcaA, tcaB, and tcaR bands in S. aureus WT whereas suppression was seen in dCas-tcaA, dCas-tcaB, and dCas-tcaR carrying S. aureus (Figures 3B–D). The dCas9 was used as a negative control to check the inhibitory effects of the dCas-9 vector on tcaRAB proteins. From Figures 3B–D, it is confirmed that CRISPRi-mediated tcaA, tcaB, and tcaR knockdown was successful and can be used for persister assay.
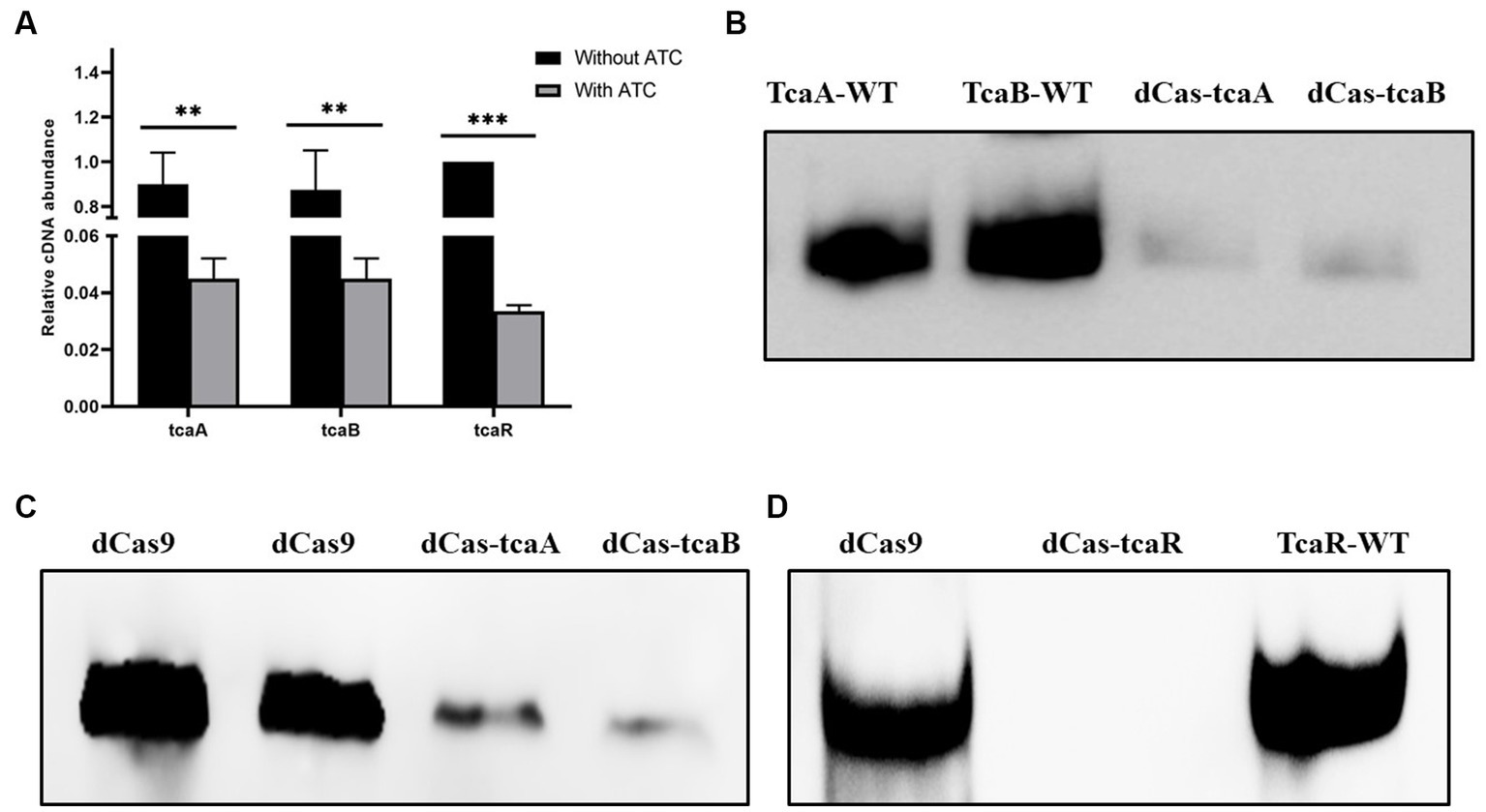
Figure 3. CRISPR-dCas9 mediated suppression. (A) The transcript level of tcaRAB operon before and after ATC induction (100 ng/mL). * p < 0.05, ** p < 0.01, *** p < 0.005. (B) The protein expression level was detected by Western blot before and after ATC induction (100 ng/mL). TcaA-WT and TcaB-WT bands are brighter, whereas dCas-TcaA and dCas-TcaB bands are dim, indicating tcaA and tcaB suppression. (C) The dCas9 was used as negative control (blank vector) and brighter bands were detected, while the dCas-TcaA and dCas-TcaB showed tcaA and tcaB protein suppression. (D) The suppression of tcaR protein was confirmed by dCas-tcaR where no protein band was detected whereas TcaR-WT and dCas9 have clear tcaR protein bands.
3.3. The ΔtcaA increased persistence to glycopeptides antibiotics
We performed a persistence assay with the WT, WT-dCas9, dCas9-tcaA, ΔtcaA, and dCas9-tcaB strains. The WT-dCas9 was the S. aureus WT strain expressing target unspecific sgRNA (control), dCas9-tcaA was the WT strain expressing sgRNA binding to tcaA (sgRNA1), dCas9-tcaB was WT expressing sgRNA binding to tcaB (sgRNA2), and ΔtcaA was tcaA mutant in S. aureus WT. All the strains were challenged with 10-fold MIC of teicoplanin, vancomycin, ciprofloxacin, and azithromycin which revealed that the dCas9-tcaA and ΔtcaA had a higher number of persisters compared to WT-dCas9 and WT strain. After 12 h of teicoplanin treatment, the surviving fraction of cells of the dCas9-tcaA and ΔtcaA strains were 17 and 18 times more than control strains WT-dCas9 and WT, respectively, (Figure 4A). Similarly, upon vancomycin treatment, the dCas9-tcaA and ΔtcaA showed a 15- and 16-fold increase relative to controls, respectively, (Figure 4B). After 48 h, the dCas9-tcaA and ΔtcaA showed a 13- and 14-fold increase in persister cells in response to teicoplanin and a 10- and 11-fold increase toward vancomycin relative to controls, respectively, (Figures 4A,B). The dCas9-tcaB did not induce persister cell formation and was similar to WT-dCas9 and WT strain (Figures 4A,B). When the cells were challenged with azithromycin and ciprofloxacin, the dCas9-tcaA, ΔtcaA, and dCas9-tcaB showed similar results with controls (Figures 4C,D). Overall, neither of the strains showed persister cell formation toward azithromycin and ciprofloxacin that confirmed the emergence of glycopeptides persisters due to tcaA suppression and deletion. Further, we transferred the pALC shuttle vector (with GFP) to S. aureus WT and ΔtcaA competent cells and challenged them with a 20-fold MIC of teicoplanin and vancomycin. After 48 h, the cells were washed and resuspended in a fresh TSB medium without antibiotics, and fluorescence microscopy was performed. The microscopy revealed a high number of cells expressing GFP in ΔtcaA cells compared to WT (Figure 5). This confirmed that persister cells tolerated a high concentration of teicoplanin and vancomycin (20 fold MIC) for 48 h and resuscitated the whole population when the antibiotic was removed. Collectively, the tcaA gene inactivation developed persistence to glycopeptide antibiotics while the tcaB did not influence the persistence phenotype. This data disclosed that S. aureus ΔtcaA formed persister cells and confirmed the RNA-seq and RT-qPCR results where suppression of tcaA was associated with the development of glycopeptides resistance. To date, Brandenberger et al. (2000) and Maki et al. (2004) reported ΔtcaA involvement in glycopeptides resistance, and the present data corroborated their results and reproduced the findings that the ΔtcaA showed resistance to teicoplanin and vancomycin and C-ΔtcaA strain restored the phenotype by expressing the tcaA gene via WT-pRMC-tcaA (Supplementary Figure S1).
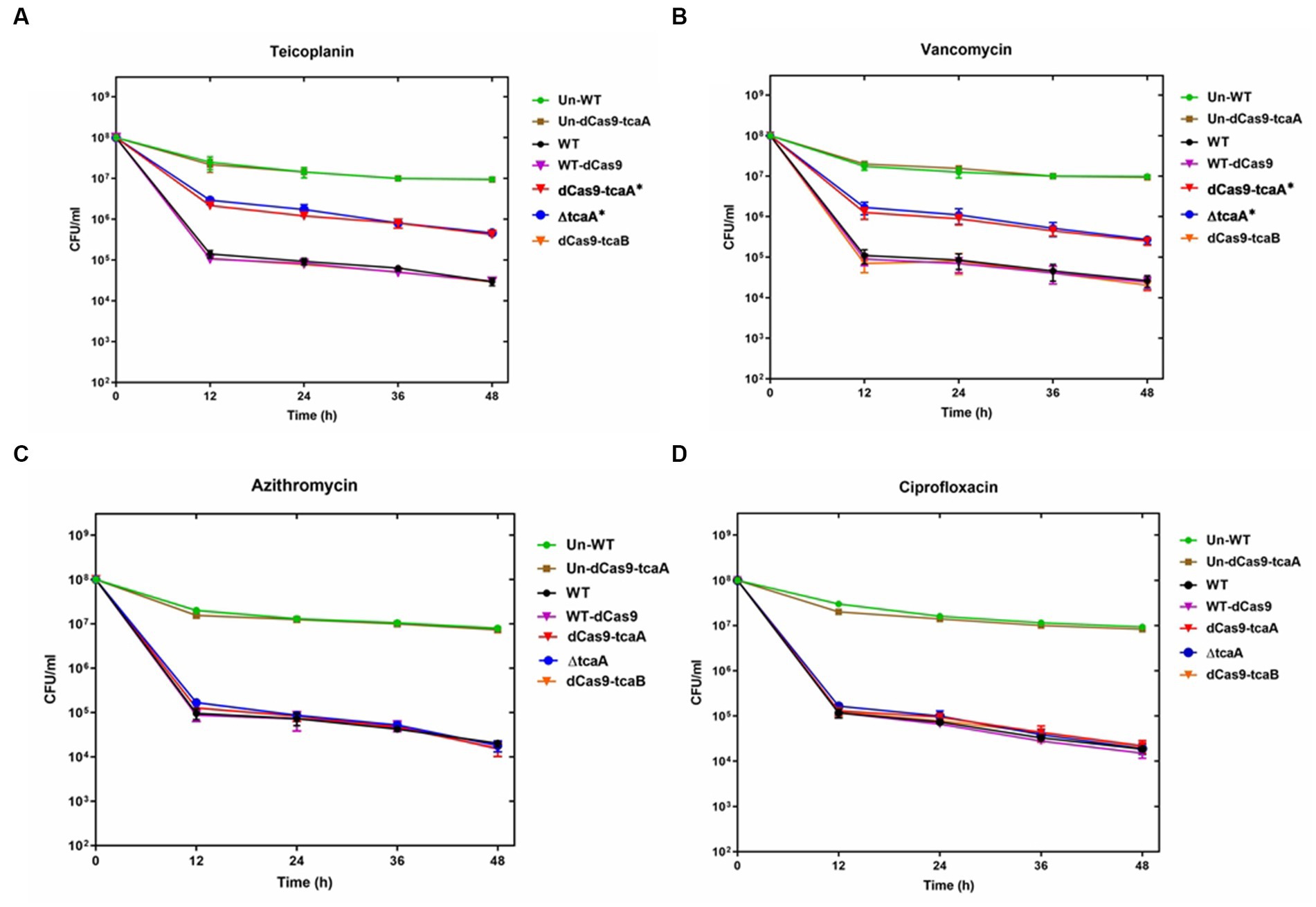
Figure 4. Persister assay. (A–D) The WT, WT-dCas9, dCas9-tcaA, dCas9-tcaB, and ΔtcaA were challenged with 10-fold MIC of teicoplanin for 48 h. The sample was taken after 12 h and CFU counting was performed. The dCas9-tcaA and ΔtcaA significantly increased the number of persister cells in panel (A,B). No significant changes were observed in azithromycin and ciprofloxacin persister assays (C,D). Un-WT and the Un-dCas9-tcaA were used as untreated antibiotic control strains. S. aureus WT and WT-dCas9 were used as controls. Experiments were performed in triplicates and error bars denote standard deviation. Statistical significance was determined using Student’s t-test (control versus treatment). * p < 0.05.
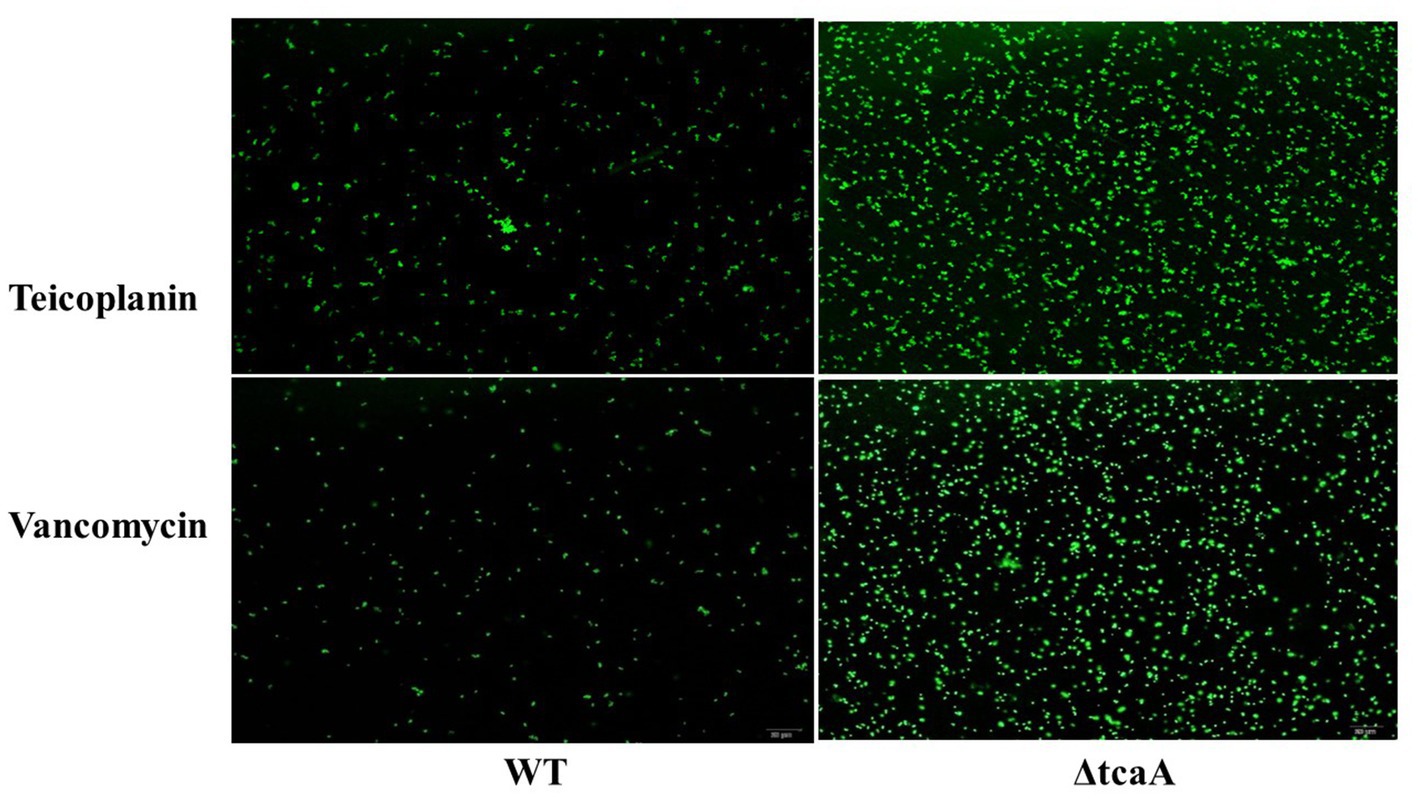
Figure 5. The fluorescence microscopy. S. aureus WT showed very dim GFP expression that revealed a low number of persisters. The ΔtcaA cells have brighter GFP expression that showed a higher number of persister and resuscitation of the bacterial population.
3.4. The ΔtcaA growth analysis
We tested cell growth in the TSB medium with 1–2 mg/L of teicoplanin and vancomycin. The results showed that the MRSA MW2 reference strain displayed slow growth rates at 1 mg/L, whereas the WT, ΔtcaA, and C-ΔtcaA strains displayed fast and steady growth (Figures 6A,B). At 2 mg/L, MRSA MW2 did not show any growth whereas WT, ΔtcaA, and ΔtcaA complementary strain displayed slow growth (Figures 6C,D). These results confirmed a decrease in susceptibility of the WT, C-ΔtcaA, and ΔtcaA to teicoplanin and vancomycin compared to MW2. We conclude that S. aureus WT, ΔtcaA, and C-ΔtcaA strains grow at 1–2 mg/L concentration of glycopeptides which supports the fact that tcaA is involved in the emergence of glycopeptides resistance.
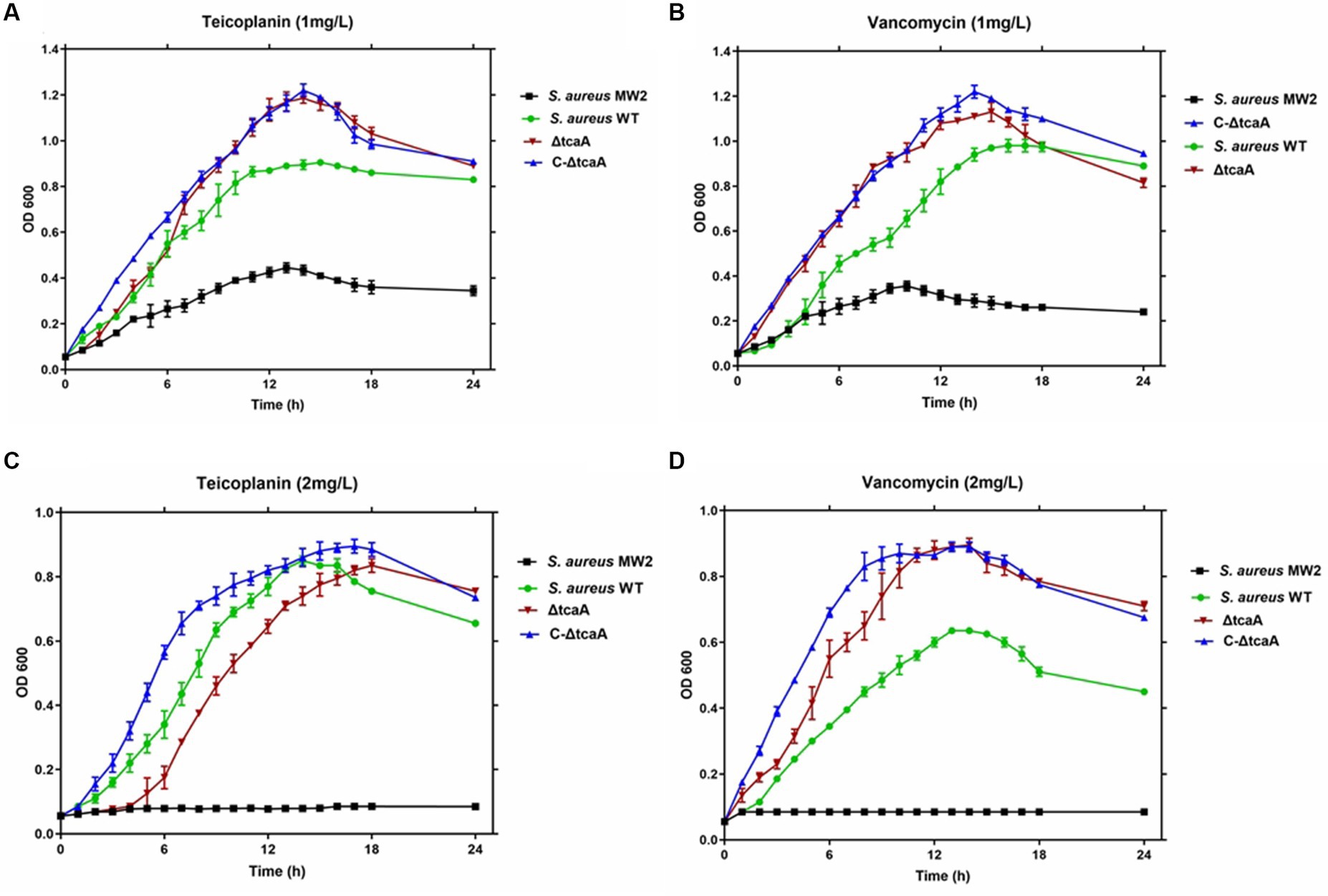
Figure 6. Growth curves. The S. aureus MW2, S. aureus WT, ΔtcaA, and C-ΔtcaA were grown in the TSB medium containing 1 mg/L and 2 mg/L of teicoplanin and vancomycin. (A,B) The growth curves at 1 mg/L of teicoplanin and vancomycin showed slow growth by MW2 and fast by S. aureus WT, ΔtcaA, and C-ΔtcaA. (C,D) At 2 mg/L of teicoplanin and vancomycin, S. aureus MW2 showed no growth, whereas S. aureus WT, ΔtcaA, and C-ΔtcaA displayed a slow growth.
3.5. Inactivation of the tcaA influences the expression of cell wall biosynthesis genes
The present data indicated that tcaA is involved in cell wall-associated glycopeptide resistance and persistence. From RNA-seq data, the tcaA was significantly suppressed and tcaA deletion increased persister cell formation. During tcaA suppression, the glyS, sgtB, ddl, and alr2 transcript level was high. We hypothesized that evaluating these gene expressions in ΔtcaA would validate the RNA-seq data and disclose the correlation. From RT-qPCR analysis, the glyS and sgtB expression was significant in the tcaA mutant while ddl and alr2 transcript level was not significant (Figure 7). The high expression of glyS and sgtB might help in cell wall biogenesis because glyS is glycine tRNA synthetase which is involved in the supply of glycine for incorporation into nascent polypeptides during bacterial cell wall synthesis (Schneider et al., 2004; Giannouli et al., 2009) whereas sgtB is a mono-functional peptidoglycan glycosyltransferase which is involved in peptidoglycan synthesis and also supports the growth of S. aureus in the absence of the main glycosyltransferase pbp2 (Wang et al., 2001; Reed et al., 2015). Collectively, this data revealed that the tcaA inactivation altered the expression of cell wall-associated genes, probably allowing the cell wall to better withstand external pressures, and it would be interesting to explore glyS and sgtB involvement in cell wall biogenesis during glycopeptide treatment or in the persister cell formation.
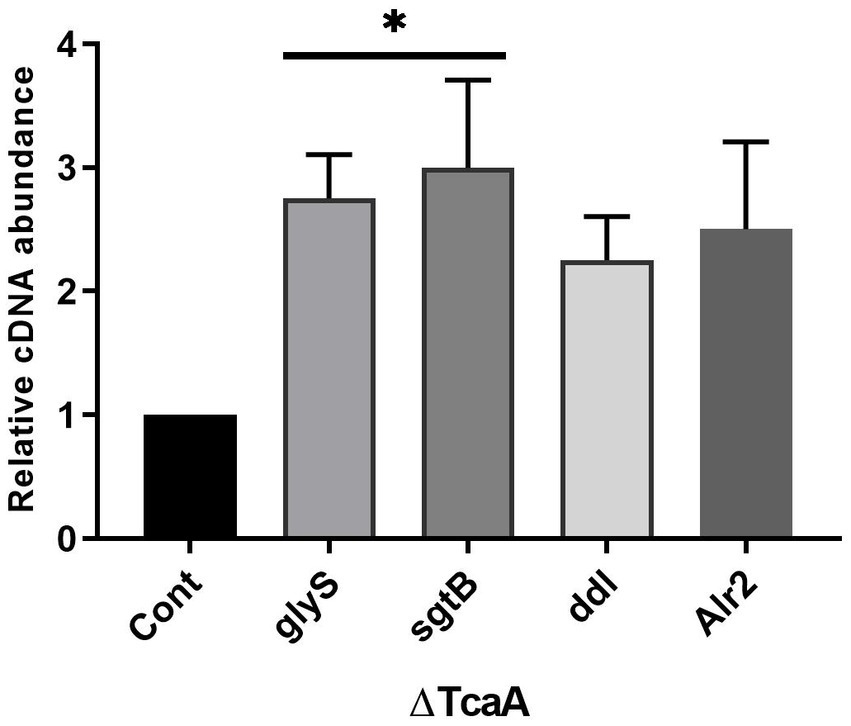
Figure 7. The cell wall-associated genes analysis in ΔtcaA. The glyS and sgtB showed significant expression in the mutant strain relative to S. aureus WT, whereas ddl and Alr2 were not significant. * p < 0.05.
4. Discussion
Glycopeptide resistance in MRSA is a global problem and vancomycin- and teicoplanin-resistant strains are of major clinical relevance. Several studies have reported the emergence of teicoplanin and vancomycin resistance in S. aureus (Wang Y. et al., 2017; Szymanek-Majchrzak et al., 2018; Wu et al., 2021). Typically, teicoplanin and vancomycin bind to D-alanine-D-alanine subunits of the murein monomer and cross resistance could develop between teicoplanin and vancomycin (Bakthavatchalam et al., 2019). The cell wall thickness also contributed to the development of vancomycin and teicoplanin resistance in S. aureus (Cui et al., 2006; Bakthavatchalam et al., 2019). Two-component systems, such as walKR, vraSR, graSR, and tcaRAB operon, are linked to the development of teicoplanin and vancomycin resistance in S. aureus (Howden et al., 2010). Also, vanA and tcaA are considered to give resistance to vancomycin and teicoplanin in S. aureus (Chang et al., 2003; Maki et al., 2004), while a report also claimed that the effects of the tcaA deletion on resistance were strain specific because the tcaA mutant in the S. aureus Col strain had higher MIC than the tcaA mutant in the BB1372 strain (Brandenberger et al., 2000). tcaA has been reported to be upregulated by vancomycin (Kuroda et al., 2003), oxacillin, and teicoplanin treatment (Utaida et al., 2003).The tcaA gives strong resistance when inactivated while its overexpression from an inducible promoter was effective in lowering teicoplanin resistance (Maki et al., 2004). A study reported that pbp2 and pbp4 lead to S. aureus cell wall thickening that reduced vancomycin susceptibility (Sieradzki and Tomasz, 2003). The pbp2 upregulation promoted cell wall synthesis, while pbp4 downregulation resulted in a decrease in murein cross-linking that increased D-alanine-D-alanine production (Gardete and Tomasz, 2014). Importantly, mutations in different genes such as yvqF (Elsaghier et al., 2002), vraSR (Yoo et al., 2013), and rpoB were also associated with teicoplanin and vancomycin resistance (Watanabe et al., 2011). In the present investigation, we detected higher MICs of 16 μg/mL and 10 μg/mL for teicoplanin and vancomycin, respectively, which is in accordance with previous reports where teicoplanin and vancomycin MICs of 16 μg/mL and 2 μg/mL were reported for resistant isolates (Wang Y. et al., 2017). Previously, it was shown that the overexpression of tcaA in clinical strains decreased glycopeptide MICs while its inactivation resulted in glycopeptide resistance (Maki et al., 2004). Here, the data revealed that S. aureus suppressed the tcaA expression under the influence of COVID-19 infection and developed glycopeptide resistance. The results were derived from RNA-seq, RT-qPCR, and Western blot analysis that disclosed tcaA suppression and inactivation. The results were corroborated by creating a tcaA mutant which revealed that tcaA is solely responsible for the emergence of glycopeptide resistance and persistence. The tcaA persistence assay revealed a 10–11 fold and 13–14 fold increase in persister cells toward vancomycin and teicoplanin treatment, respectively. The tcaA-dependent persisters were not detected in azithromycin and ciprofloxacin challenge assays that indicated persisters might be dependent on specific gene function or dysfunction in a specific environment. We also confirmed that tcaA deletion did not affect azithromycin resistance whereas vancomycin and teicoplanin susceptibility was decreased as previously reported (Brandenberger et al., 2000; Maki et al., 2004). The study of Vogwill et al. (2016) investigated the coevolution of resistance and persistence to ciprofloxacin and rifampicin across the genus Pseudomonas and concluded that persistence correlates positively to antibiotic resistance across Pseudomonas strains (Vogwill et al., 2016). From theoretical and experimental analysis, Windels et al. (2019) proposed that persisters facilitated genetic resistance and increased survival and mutation rates that might affect the evolution of clinical resistance in E. coli (Windels et al., 2019). To date, S. aureus persisters increased with the decrease in intracellular ATP (Conlon et al., 2016), while decreasing upon deletion of the msaABCR operon (Pandey et al., 2021). Moreover, phenol-soluble modulin toxins expression reduced persisters in S. aureus (Baldry et al., 2020), and the expression of T7SS facilitated S. aureus survival in persistent infection. S. aureus T7SS is crucial for its virulence and the essC is thought to be a central membrane transporter (Burts et al., 2005; Jäger et al., 2018). Studies have shown that deletion of the entire T7SS or its components (EsxA, EssC, EsxB, EsaD, etc.) decreased S. aureus virulence (Burts et al., 2005, 2008; Anderson et al., 2017). EsxA and EsxB are the secretory proteins of T7SS and are required for establishing S. aureus infection in the host and the EsxAB mutants caused a decrease in abscess formation in mice (Burts et al., 2005). Currently, a high expression of EsxAB and a low expression of essC were detected that might facilitate S. aureus survival during host infection, however, the mechanism behind this phenomenon remained elusive. Even though the EsxAB genes are involved in S. aureus persistence and essC is required for establishing S. aureus infection during lung infection (Ishii et al., 2014), details of their altered chemistry in host infection need further investigation. From current data, we conclude that tcaA suppression and deletion induced persistence in S. aureus, and glyS and sgtB showed increased expression in the tcaA mutant strain that might indicate a cumulative approach toward cell wall protection. Although glyS and sgtB roles in cell wall synthesis in S. aureus are well defined (Wang et al., 2001; Schneider et al., 2004; Giannouli et al., 2009; Reed et al., 2015), their involvement in cell wall protection during host infection and persister cell formation remained obscure. Altogether, this is the first study to reveal persister cell formation due to tcaA inactivation, and conclude that the emergence of resistance might reflect the adaptation mechanism of persister cell genotype in future.
5. Conclusion
It is well known that S. aureus can change its fitness during infection to increase its survival in a hostile environment. In the present study, S. aureus, showing an intermediate level of glycopeptide resistance, was recovered from a COVID-19 patient. Genome analysis revealed a high expression of genes involved in cell wall biosynthesis and a low expression of virulence regulatory genes. The data summarize that tcaA inactivation gave rise to persisters that tolerated a high concentration of glycopeptides and resuscitated the bacterial population. This showed the involvement of genetic determinants in the development of persisters. Here, we conclude that if bacterial genes remain the main culprit of persister cell formation, then there would be concern regarding the potential spread of the persistence-associated genes. Although more remains to be explored concerning the genetic basis of persisters, our data will increase the understanding of the mechanism of persister cell formation during host infection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
GH and HG: conceptualization and writing-original draft. GH, HG, MR, and AH: data curation and formal analysis. GH, HG, HE, PA, and IM: methodology and software. GH, MR, PA, HE, and IM: resources and funding acquisition. HG, MR and AH: supervision and project administration: AH, MR, HG, PA, HE, and IM: review and editing. All authors proofread the article and approved the current version.
Funding
This work was funded by Researchers Supporting Project number (RSPD2023R741), King Saud University.
Acknowledgments
The authors would like to thank the Researchers Supporting Project number (RSPD2023R741), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1241995/full#supplementary-material
Footnotes
References
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Nat. Prec. :1. doi: 10.1038/npre.2010.4282.2
Anderson, M., Ohr, R. J., Aly, K. A., Nocadello, S., Kim, H. K., and Schneewind, C. E. (2017). EssE promotes Staphylococcus aureus ESS-dependent protein secretion to modify host immune responses during infection. J. Bacteriol. 199:e00527-16. doi: 10.1128/JB.00527-16
Appelbaum, P. (2006). The emergence of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 12, 16–23. doi: 10.1111/j.1469-0691.2006.01344.x
Aqib, A., and Rodriguez-Morales, A.J. (2021). Insights into drug resistance in Staphylococcus aureus. London. IntechOpen.
Bae, T., and Schneewind, O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63. doi: 10.1016/j.plasmid.2005.05.005
Bakthavatchalam, Y. D., Babu, P., Munusamy, E., Dwarakanathan, H. T., Rupali, P., Zervos, M., et al. (2019). Genomic insights on heterogeneous resistance to vancomycin and teicoplanin in methicillin-resistant Staphylococcus aureus: a first report from South India. PLoS One 14:e0227009. doi: 10.1371/journal.pone.0227009
Baldry, M., Bojer, M. S., Najarzadeh, Z., Vestergaard, M., Meyer, R. L., Otzen, D. E., et al. (2020). Phenol-soluble modulins modulate persister cell formation in Staphylococcus aureus. Front. Microbiol. 11:573253. doi: 10.3389/fmicb.2020.573253
Bao, Y., Li, Y., Jiang, Q., Zhao, L., Xue, T., Hu, B., et al. (2013). Methylthioadenosine/S-adenosylhomocysteine nucleosidase (Pfs) of Staphylococcus aureus is essential for the virulence independent of LuxS/AI-2 system. Int. J. Med. Microbiol. 303, 190–200. doi: 10.1016/j.ijmm.2013.03.004
Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Brandenberger, M., Tschierske, M., Giachino, P., Wada, A., and Berger-Bächi, B. (2000). Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophys. Acta 1523, 135–139. doi: 10.1016/S0304-4165(00)00133-1
Brauner, A., Fridman, O., Gefen, O., and Balaban, N. Q. (2016). Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330. doi: 10.1038/nrmicro.2016.34
Brückner, R. (1997). Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151, 1–8. doi: 10.1016/S0378-1097(97)00116-x
Brunet, F., Vedel, G., Dreyfus, F., Vaxelaire, J., Giraud, T., and Schremmer, B. (1990). Failure of teicoplanin therapy in two neutropenic patients with staphylococcal septicemia who recovered after administration of vancomycin. Eur. J. Clin. Microbiol. Infect. Dis. 9, 145–147. doi: 10.1007/BF01963643
Burts, M. L., DeDent, A. C., and Missiakas, D. M. (2008). EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol. Microbiol. 69, 736–746. doi: 10.1111/j.1365-2958.2008.06324.x
Burts, M. L., Williams, W. A., DeBord, K., and Missiakas, D. M. (2005). EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. 102, 1169–1174. doi: 10.1073/pnas.0405620102
Cameron, D. R., Shan, Y., Zalis, E. A., Isabella, V., and Lewis, K. (2018). A genetic determinant of persister cell formation in bacterial pathogens. J. Bacteriol. 200:e00303-18. doi: 10.1128/JB.00303-18
Chang, S., Sievert, D. M., Hageman, J. C., Boulton, M. L., Tenover, F. C., Downes, F. P., et al. (2003). Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348, 1342–1347. doi: 10.1056/NEJMoa025025
Chen, K.-Y., Chang, H.-J., Hsu, P.-C., Yang, C.-C., Chia, J.-H., Wu, T.-L., et al. (2013). Relationship of teicoplanin MICs to treatment failure in teicoplanin-treated patients with methicillin-resistant Staphylococcus aureus pneumonia. J. Microbiol. Immunol. Infect. 46, 210–216. doi: 10.1016/j.jmii.2012.06.010
Clinical and Laboratory Standards Institute (2018). “Performance standards for antimicrobial susceptibility testing” in CLSI supplement M100. 28th edn. (Wayne, PA, USA: Clinical and Laboratory Standards Institute). Available at: https://clsi.org/media/1930/m100ed28_sample.pdf
Conlon, B. P., Rowe, S. E., Gandt, A. B., Nuxoll, A. S., Donegan, N. P., Zalis, E. A., et al. (2016). Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 1, 1–7. doi: 10.1038/nmicrobiol.2016.51
Corrigan, R. M., and Foster, T. J. (2009). An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61, 126–129. doi: 10.1016/j.plasmid.2008.10.001
Cui, L., Iwamoto, A., Lian, J.-Q., Neoh, H. M., Maruyama, T., and Horikawa, Y. (2006). Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob. agents chemother. 50, 428–438. doi: 10.1128/AAC.50.2.428-438.2006
Eisenreich, W., Rudel, T., Heesemann, J., and Goebel, W. (2021). Persistence of intracellular bacterial pathogens—with a focus on the metabolic perspective. Front. Cell. Infect. Microbiol. 10:615450. doi: 10.3389/fcimb.2020.615450
Elsaghier, A. A., Aucken, H. M., Hamilton-Miller, J. M., Shaw, S., and Kibbler, C. C. (2002). Resistance to teicoplanin developing during treatment of methicillin-resistant Staphylococcus aureus infection. J. Antimicrob. Chemother. 49, 423–424. doi: 10.1093/jac/49.2.423
Gardete, S., and Tomasz, A. (2014). Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Invest. 124, 2836–2840. doi: 10.1172/JCI68834
Ghanizadeh, A., Najafizade, M., Rashki, S., Marzhoseyni, Z., and Motallebi, M. (2021). Genetic diversity, antimicrobial resistance pattern, and biofilm formation in Klebsiella pneumoniae isolated from patients with coronavirus disease 2019 (COVID-19) and ventilator-associated pneumonia. Biomed. Res. Int. 2021, 1–11. doi: 10.1155/2021/2347872
Giannouli, S., Kyritsis, A., Malissovas, N., Becker, H. D., and Stathopoulos, C. (2009). On the role of an unusual tRNAGly isoacceptor in Staphylococcus aureus. Biochimie 91, 344–351. doi: 10.1016/j.biochi.2008.10.009
Habib, G., Mahmood, K., Ahmad, L., Gul, H., Hayat, A., and Rehman, M. U. (2023). Clinical manifestations of active tuberculosis patients coinfected with severe acute respiratory syndrome coronavirus-2. J. Clin. Tuberc. Other Mycobact. Dis. 31:100359. doi: 10.1016/j.jctube.2023.100359
Habib, G., Mahmood, K., Gul, H., Tariq, M., Ain, Q. U., Hayat, A., et al. (2022). Pathophysiology of methicillin-resistant Staphylococcus aureus superinfection in COVID-19 patients. Pathophysiology 29, 405–413. doi: 10.3390/pathophysiology29030032
Habib, G., Zhu, J., and Sun, B. (2020). A novel type I toxin-antitoxin system modulates persister cell formation in Staphylococcus aureus. Int. J. Med. Microbiol. 310:151400. doi: 10.1016/j.ijmm.2020.151400
Handwerger, S., and Tomasz, A. (1985). Antibiotic tolerance among clinical isolates of bacteria. Annu. Rev. Pharmacol. Toxicol. 25, 349–380. doi: 10.1146/annurev.pa.25.040185.002025
Hiramatsu, K. (2001). Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1, 147–155. doi: 10.1016/S1473-3099(01)00091-3
Howden, B. P., Davies, J. K., Johnson, P. D., Stinear, T. P., and Grayson, M. L. (2010). Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23, 99–139. doi: 10.1128/CMR.00042-09
Howden, B. P., Stinear, T. P., Allen, D. L., Johnson, P. D., Ward, P. B., and Davies, J. K. (2008). Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob. Agents Chemother. 52, 3755–3762. doi: 10.1128/AAC.01613-07
Hu, J., Zhang, X., Liu, X., Chen, C., and Sun, B. (2015). Mechanism of reduced vancomycin susceptibility conferred by walK mutation in community-acquired methicillin-resistant Staphylococcus aureus strain MW2. Antimicrob. Agents Chemother. 59, 1352–1355. doi: 10.1128/AAC.04290-14
Hu, Q., Peng, H., and Rao, X. (2016). Molecular events for promotion of vancomycin resistance in vancomycin intermediate Staphylococcus aureus. Front. Microbiol. 7:1601. doi: 10.3389/fmicb.2016.01601
Ishii, K., Adachi, T., Yasukawa, J., Suzuki, Y., Hamamoto, H., and Sekimizu, K. (2014). Induction of virulence gene expression in Staphylococcus aureus by pulmonary surfactant. Infect. immun. 82, 1500–1510. doi: 10.1128/IAI.01635-13
Jäger, F., Kneuper, H., and Palmer, T. (2018). EssC is a specificity determinant for Staphylococcus aureus type VII secretion. Microbiology 164, 816–820. doi: 10.1099/mic.0.000650
Jia, G., Rao, Z., Zhang, J., Li, Z., and Chen, F. (2013). Tetraether biomarker records from a loess-paleosol sequence in the western Chinese loess plateau. Front. Microbiol. 4:199. doi: 10.3389/fmicb.2013.00199
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2007). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kuroda, M., Kuroda, H., Oshima, T., Takeuchi, F., Mori, H., and Hiramatsu, K. (2003). Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49, 807–821. doi: 10.1046/j.1365-2958.2003.03599.x
Levin-Reisman, I., Ronin, I., Gefen, O., Braniss, I., Shoresh, N., and Balaban, N. Q. (2017). Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830. doi: 10.1126/science.aaj2191
Levin, B. R., McCall, I. C., Perrot, V., Weiss, H., Ovesepian, A., and Baquero, F. (2017). A numbers game: ribosome densities, bacterial growth, and antibiotic-mediated stasis and death. MBio 8:e02253-16. doi: 10.1128/mBio.02253-16
Lewis, K. (2010). Persister cells. Annu. Rev. Microbiol. 64, 357–372. doi: 10.1146/annurev.micro.112408.134306
Lin, L., Wang, X., Wang, W., Zhou, X., and Hargreaves, J. R. (2020). Cleaning up China’s medical cabinet—an antibiotic take-back programme to reduce household antibiotic storage for unsupervised use in rural China: a mixed-methods feasibility study. Antibiotics 9:212. doi: 10.3390/antibiotics9050212
Maki, H., McCallum, N., Bischoff, M., Wada, A., and Berger-Bächi, B. (2004). tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. agents chemother. 48, 1953–1959. doi: 10.1128/AAC.48.6.1953-1959.2004
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Mohiuddin, S. G., Ghosh, S., Ngo, H. G., Sensenbach, S., Karki, P., Dewangan, N. K., et al. (2021). Cellular self-digestion and persistence in Bacteria. Microorganisms 9:2269. doi: 10.3390/microorganisms9112269
Pandey, S., Sahukhal, G. S., and Elasri, M. O. (2021). The msaABCR operon regulates persister formation by modulating energy metabolism in Staphylococcus aureus. Front. Microbiol. 12:657753. doi: 10.3389/fmicb.2021.657753
Personnic, N., Doublet, P., and Jarraud, S. (2023). Intracellular persister: a stealth agent recalcitrant to antibiotics. Front. Cell. Infect. Microbiol. 13:1141868. doi: 10.3389/fcimb.2023.1141868
Reed, P., Atilano, M. L., Alves, R., Hoiczyk, E., Sher, X., Reichmann, N. T., et al. (2015). Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. PLoS Pathog. 11:e1004891. doi: 10.1371/journal.ppat.1004891
Renzoni, A., Kelley, W. L., Barras, C., Monod, A., Huggler, E., and François, P. (2009). Identification by genomic and genetic analysis of two new genes playing a key role in intermediate glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 53, 903–911. doi: 10.1128/AAC.01287-08
Schneider, T., Senn, M. M., Berger-Bächi, B., Tossi, A., Sahl, H. G., and Wiedemann, I. (2004). In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (lipid II-Gly5) of Staphylococcus aureus. Mol. Microbiol. 53, 675–685. doi: 10.1111/j.1365-2958.2004.04149.x
Schulthess, B., Meier, S., Homerova, D., Goerke, C., Wolz, C., Kormanec, J., et al. (2009). Functional characterization of the σB-dependent yabJ-spoVG operon in Staphylococcus aureus: role in methicillin and glycopeptide resistance. Antimicrob. Agents Chemother. 53, 1832–1839. doi: 10.1128/AAC.01255-08
Shang, Y., Wang, X., Chen, Z., Lyu, Z., Lin, Z., Zheng, J., et al. (2020). Staphylococcus aureus PhoU homologs regulate persister formation and virulence. Front. Microbiol. 11:865. doi: 10.3389/fmicb.2020.00865
Shariati, A., Dadashi, M., Moghadam, M. T., van Belkum, A., Yaslianifard, S., and Darban-Sarokhalil, D. (2020). Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci. Rep. 10:12689. doi: 10.1038/s41598-020-69058-z
Sieradzki, K., and Tomasz, A. (2003). Alterations of Cell Wall structure and MetabolismAccompany reduced susceptibility to vancomycin in an IsogenicSeries of clinical isolates of Staphylococcusaureus. J. Bacteriol. 185, 7103–7110. doi: 10.1128/JB.185.24.7103-7110.2003
Sieradzki, K., Villari, P., and Tomasz, A. (1998). Decreased susceptibilities to teicoplanin and vancomycin among coagulase-negative methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 42, 100–107. doi: 10.1128/AAC.42.1.100
Song, K.-H., Kim, M., Kim, C. J., Cho, J. E., Choi, Y. J., Park, J. S., et al. (2017). Impact of vancomycin MIC on treatment outcomes in invasive Staphylococcus aureus infections. Antimicrob. Agents Chemother. 61, e01845–e01816. doi: 10.1128/AAC.01845-16
Szymanek-Majchrzak, K., Mlynarczyk, A., and Mlynarczyk, G. (2018). Characteristics of glycopeptide-resistant Staphylococcus aureus strains isolated from inpatients of three teaching hospitals in Warsaw, Poland. Antimicrob. Resist. Infect. Control 7, 1–6. doi: 10.1186/s13756-018-0397-y
Tariq, F. N., Shafiq, M., Khawar, N., Habib, G., Gul, H., Hayat, A., et al. (2023). The functional repertoire of AmpR in the AmpC β-lactamase high expression and decreasing β-lactam and aminoglycosides resistance in ESBL Citrobacter freundii. Heliyon 9:e19486. doi: 10.1016/j.heliyon.2023.e19486
Utaida, S., Dunman, P., Macapagal, D., Murphy, E., Projan, S., and Singh, V. (2003). Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149, 2719–2732. doi: 10.1099/mic.0.26426-0
Valihrach, L., and Demnerova, K. (2012). Impact of normalization method on experimental outcome using RT-qPCR in Staphylococcus aureus. J. Microbiol. Methods 90, 214–216. doi: 10.1016/j.mimet.2012.05.008
Vogwill, T., Comfort, A., Furió, V., and MacLean, R. (2016). Persistence and resistance as complementary bacterial adaptations to antibiotics. J. Evol. Biol. 29, 1223–1233. doi: 10.1111/jeb.12864
Wang, Q., Lu, F., and Lan, R. (2017). RNA-sequencing dissects the transcriptome of polyploid cancer cells that are resistant to combined treatments of cisplatin with paclitaxel and docetaxel. Mol. BioSyst. 13, 2125–2134. doi: 10.1039/C7MB00334J
Wang, Q. M., Peery, R. B., Johnson, R. B., Alborn, W. E., Yeh, W.-K., and Skatrud, P. L. (2001). Identification and characterization of a monofunctional glycosyltransferase from Staphylococcus aureus. J. Bacteriol. 183, 4779–4785. doi: 10.1128/JB.183.16.4779-4785.2001
Wang, W., Chen, J., Chen, G., Du, X., Cui, P., and Wu, J. (2015). Transposon mutagenesis identifies novel genes associated with Staphylococcus aureus persister formation. Front. Microbiol. 6:1437. doi: 10.3389/fmicb.2015.01437
Wang, Y., Bojer, M. S., George, S. E., Wang, Z., Jensen, P. R., Wolz, C., et al. (2018). Inactivation of TCA cycle enhances Staphylococcus aureus persister cell formation in stationary phase. Sci. Rep. 8:10849. doi: 10.1038/s41598-018-29123-0
Wang, Y., Li, X., Jiang, L., Han, W., Xie, X., Jin, Y., et al. (2017). Novel mutation sites in the development of vancomycin-intermediate resistance in Staphylococcus aureus. Front. Microbiol. 7:2163. doi: 10.3389/fmicb.2016.02163
Watanabe, Y., Cui, L., Katayama, Y., Kozue, K., and Hiramatsu, K. (2011). Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J. Clin. Microbiol. 49, 2680–2684. doi: 10.1128/JCM.02144-10
Windels, E. M., Michiels, J. E., Fauvart, M., Wenseleers, T., Van den Bergh, B., and Michiels, J. (2019). Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 13, 1239–1251. doi: 10.1038/s41396-019-0344-9
Wu, Q., Sabokroo, N., Wang, Y., Hashemian, M., Karamollahi, S., and Kouhsari, E. (2021). Systematic review and meta-analysis of the epidemiology of vancomycin-resistance Staphylococcus aureus isolates. Antimicrob. Resist. Infect. Control 10, 1–13. doi: 10.1186/s13756-021-00967-y
Yoo, J. I., Kim, J. W., Kang, G. S., Kim, H. S., Yoo, J. S., and Lee, Y. S. (2013). Prevalence of amino acid changes in the yvqF, vraSR, graSR, and tcaRAB genes from vancomycin intermediate resistant Staphylococcus aureus. J. Microbiol. 51, 160–165. doi: 10.1007/s12275-013-3088-7
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14–R12. doi: 10.1186/gb-2010-11-2-r14
Keywords: MRSA, tcaA , tcaB , persister cell, glycopeptides resistance, glyS
Citation: Habib G, Gul H, Ahmad P, Hayat A, Rehman MU, Mohamed Moussa I and Elansary HO (2023) Teicoplanin associated gene tcaA inactivation increases persister cell formation in Staphylococcus aureus. Front. Microbiol. 14:1241995. doi: 10.3389/fmicb.2023.1241995
Edited by:
Lucinda Janete Bessa, Egas Moniz Center of Interdisciplinary Research - Egas Moniz School of Health & Science, PortugalReviewed by:
Norma Velazquez-Guadarrama, Federico Gómez Children's Hospital, MexicoGang Zhang, Chinese Academy of Sciences (CAS), China
Wei Wang, China National Center for Food Safety Risk Assessment, China
Copyright © 2023 Habib, Gul, Ahmad, Hayat, Rehman, Mohamed Moussa and Elansary. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gul Habib, gulhabib101@gmail.com; Hosam O. Elansary, helansary@ksu.edu.sa; Ihab Mohamed Moussa, imoussa1@ksu.edu.sa
 Gul Habib
Gul Habib Haji Gul2,3
Haji Gul2,3 Hosam O. Elansary
Hosam O. Elansary