- 1Faculdade de Ciências Biológicas, Universidade Federal do Pará, Altamira, Pará, Brazil
- 2Laboratório de Genética Molecular de Microrganismos, Departamento de Microbiologia Agrícola, Universidade Federal de Viçosa, Viçosa, Minas Gerais, Brazil
- 3Instituto Federal do Sudeste de Minas Gerais—Campus Juiz de Fora, Juiz de Fora, Minas Gerais, Brazil
- 4Departamento de Fitopatologia, Universidade Federal de Viçosa, Viçosa, Minas Gerais, Brazil
Fungi belonging to the genus Trichoderma have been widely recognized as efficient controllers of plant diseases. Although the majority of isolates currently deployed, thus far, have been isolated from soil, endophytic Trichoderma spp. is considered to be a promising option for application in biocontrol. In this study, 30 endophytic Trichoderma isolates—obtained from the leaves, stems, and roots of wild Hevea spp. in the Brazilian Amazon—were analyzed using specific DNA barcodes: sequences of internal transcribed spacers 1 and 2 of rDNA (ITS region), genes encoding translation elongation factor 1-α (TEF1-α), and the second largest subunit of RNA polymerase II (RPB2). The genealogical concordance phylogenetic species recognition (GCPSR) concept was used for species delimitation. A phylogenetic analysis showed the occurrence of Trichoderma species, such as T. erinaceum, T. ovalisporum, T. koningiopsis, T. sparsum, T. lentiforme, T. virens, and T. spirale. Molecular and morphological features resulted in the discovery of four new species, such as T. acreanum sp. nov., T. ararianum sp. nov., T. heveae sp. nov., and T. brasiliensis sp. nov. The BI and ML analyses shared a similar topology, providing high support to the final trees. The phylograms show three distinct subclades, namely, T. acreanum and T. ararianum being paraphyletic with T. koningiopsis; T. heveae with T. subviride; and T. brasiliensis with T. brevicompactum. This study adds to our knowledge of the diversity of endophytic Trichoderma species in Neotropical forests and reveals new potential biocontrol agents for the management of plant diseases.
Introduction
The Amazon biome is recognized worldwide for its high biodiversity of flora and fauna. Thus, the intensification of research into the characterization of the microbial biodiversity found in this biome can provide useful information for a range of biotechnological strategies, including the production of enzymes (Pereira et al., 2015) and antimicrobial compounds (Nuankeaw et al., 2020).
The rubber trees Hevea brasiliensis (Willd. ex A.Juss.) Müll.Arg. and Hevea guianensis Aubl. are Euphorbiaceae species native to the Amazon rainforest. Hevea brasiliensis is the main source of raw materials for the production of natural rubber (Zhu et al., 2015; Meenakumari et al., 2018). Worldwide production, mainly concentrated in Southeast Asia, generates over 10 million tons per year, of which more than 80% is extracted by small producers in a chain that benefits approximately 20 million people (Rivano et al., 2013). However, fungal pathogens, such as Pseudocercospora ulei (Henn.) Hora & Mizubuti (formerly known as Microcyclus ulei), the causal agent of South American leaf blight of rubber (Hora Júnior et al., 2014), have been the major constraints to production in its Neotropical center of origin since the establishment of commercial rubber plantations.
The genus Trichoderma (Hypocreales) is cosmopolitan and ubiquitous in the environment, commonly being found in the microflora of natural and agricultural soils, although some taxa are also able to endophytically colonize plant tissues. Trichoderma species have now been isolated as endophytes from a range of woody plants, including Herrania sp. (Crozier et al., 2006), Aegle marmelos (L.) Corrêa (Gond et al., 2007), Theobroma spp. (Evans et al., 2003; Bae et al., 2009), Hevea spp. (Chaverri et al., 2010; Gazis and Chaverri, 2010), and Coffea spp. (Rodriguez et al., 2021). Moreover, this genus is widely recognized as a parasite of other fungi (Harman et al., 2004; Mukherjee et al., 2013; Rodriguez et al., 2021).
Numerous studies have been conducted on Trichoderma spp., with particular emphasis on mycoparasitic activities and their potential as biocontrol agents of plant diseases, including Rhizoctonia solani Kühn (Almeida et al., 2007), Moniliophthora perniciosa (Stahel) Aime & Phillips-Mora (De Souza et al., 2008), Pyricularia oryzae Cavara (Dey et al., 2013), Phytophthora colocasiae Racib. (Nath et al., 2014), Phytophthora megakarya Brasier & Griffin (Mbarga et al., 2014), Phymatotrichopsis omnivora (Shear) Hennebert (Guigón-López et al., 2015), Sclerotium cepivorum Berk. (Rivera-Méndez et al., 2020), and Hemileia vastatrix Berk. & Broome (Rodriguez et al., 2021). Trichoderma spp. are also known to produce enzymes and secondary metabolites applicable to industrial processes (Saravanakumar et al., 2016; Baldoni et al., 2020), to induce systemic resistance in plants (Vitti et al., 2016) and promote plant growth (Kuswinanti et al., 2015).
Defining species within the genus Trichoderma has proven to be challenging because of their complex and cryptic characteristics that lack distinguishable phenotypic traits (Jaklitsch et al., 2008; Jaklitsch, 2009). Moreover, various morphospecies such as Trichoderma harzianum Rifai represent species complexes, and the taxonomy of this complex was updated by Chaverri et al. (2015), who identified numerous new species. This study also indicated a tendency for habitat specialization; thus, species commonly isolated from soil tend not to be endophytes, and species isolated as endophytes tend not to be isolated from soils.
In several studies, morphological characteristics (stromata) and ecological factors, such as specific fungus/host relationships, have been included to improve the morphological concept and species delimitation (Taylor et al., 2000). Without the use of multilocus molecular phylogenetics, all of these species would have been identified as T. harzianum. New species, such as Trichoderma amazonicum (Chaverri & Gazis), have gradually been recognized within the complex based on subtle phenotypic or biological characteristics (Chaverri et al., 2010). The taxonomy of Trichoderma has substantially expanded through the use of molecular phylogeny (Druzhinina et al., 2006; Singh et al., 2020). Trichoderma spp. are now commonly described using a combination of morphological characteristics (asexual and sexual), cultural features, ecology, phylogenetic analysis, and the genealogical concordance phylogenetic species recognition (GCPSR) concept (Taylor et al., 2000; Cai and Druzhinina, 2021), including the species isolated in this study.
Some endophytic isolates belonging to the genus Trichoderma have recently been recognized as new species (Chaverri et al., 2010; Druzhinina, 2011; Rodriguez et al., 2021; Zheng et al., 2021). This may be the result of rapid speciation of the genus following adaptation to new ecological niches (Chaverri et al., 2010, 2015), or a likely trend is that these researchers were most intensely looking for endophytes.
Since Hevea spp. are the main sources of natural rubber (Zhu et al., 2015), endophytic Trichoderma spp. from these hosts may have potential applications for the biocontrol of the main rubber pathogens, especially Pseudocercospora ulei (Gazis and Chaverri, 2010; Hora Júnior et al., 2014), although few studies have investigated endophytic fungi in rubber trees (Gazis and Chaverri, 2010; Araújo et al., 2018, 2020). Thus, the aim of the present study was to increase our knowledge of Trichoderma endophytes in wild rubber, using morphological and molecular characterization to assess their biodiversity and potentially identify new species, as well as to generate information that could be used in future biotechnological strategies.
Materials and methods
Sample collection, processing, and growth conditions
Endophytic Trichoderma spp. were isolated from leaves, stems, and roots of Hevea spp. trees from natural populations in the Brazilian Amazon (Acre and Amazonas states). The collection has been detailed and described by Araújo et al. (2018, 2020), and fungal isolation was performed following the recommendations suggested by Wirsel et al. (2001), Evans et al. (2003), and Leite et al. (2013) and reported by Araújo et al. (2018, 2020). Currently, these isolates are kept in the collection of fungi at the Laboratory of Molecular Genetics of Microorganisms, BIOAGRO, Universidade Federal de Viçosa, MG, Brazil. A representative sample of each new species was deposited in the Herbarium, the Universidade Federal de Viçosa, (Herbarium VIC), and the isolates were deposited in the culture collection Coleção Octávio de Almeida Drumond (COAD), the Universidade Federal de Viçosa. The holotype is a dried culture of the endophytic fungus growing on potato dextrose agar (PDA).
Morphological observations
The characterization of new species was determined by observations on different media, namely, PDA (500 mL of potato infusion prepared from 200 g of potatoes, 20 g of glucose, and 17 g of agar, and made up with 1 L with distilled water), cornmeal dextrose agar (CMD; Sigma—C1176), and, synthetic nutrient-poor agar (SNA; Nirenberg, 1976). To monitor colony growth rate, CMD, SNA, and PDA plates were incubated at 25, 30, and 35°C, with a 12 h light/dark cycle for 72 h. Photomicrographs and biometric descriptions of the reproductive structures of the isolates were obtained from microculture on SNA, incubated for 24–48 h at 25°C with a 12 h light/dark cycle. Slides were mounted in lacto-glycerol, the fungal structures were examined, and the images were produced using a light microscope (Olympus BX50 and BX53), fitted with a Q-Color 3 digital camera (Olympus PM-C35DX), together with Q-Capture Pro 6 software. Conidiophores and conidia were also measured using ImageJ software (Abramoff et al., 2004). To monitor colony growth rate assays, 5-mm mycelial disks were placed in 9 cm-diameter Petri plates containing 20 mL of PDA, CMD, or SNA media. The plates were cultured for up to 7 days for all assessments, and the mycelial diameter was measured using a caliper. Each culture medium was tested in triplicate.
DNA extraction
Isolates were grown on PDA for 5 days under the conditions described earlier. Approximately 50 mg of mycelium was scraped from the colonies formed on the plates and placed in sterile tubes. A Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, United States) was used according to the manufacturer’s instructions.
PCR, sequencing, and phylogenetic analysis
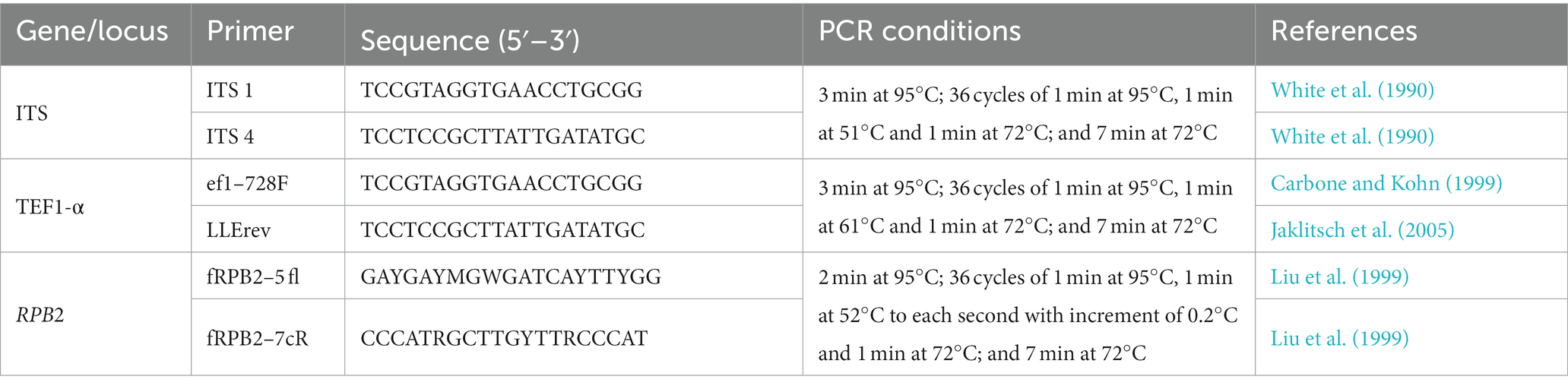
For analysis by PCR and sequencing, the region of nuclear rDNA containing the internal transcribed spacer regions 1 and 2 and the 5.8S rDNA gene region was amplified using the ITS1 and ITS4 primers (White et al., 1990). Translation elongation factor 1-α (TEF1-α) was amplified using ef1–728F and LLErev primers (Carbone and Kohn, 1999; Jaklitsch et al., 2005), and RNA polymerase II subunit B (RPB2) was amplified using fRPB2–5 fl and fRPB2–7cR primers (Liu et al., 1999; Table 1). PCR conditions are presented in Table 1. Amplification products were subjected to 1.2% agarose gel electrophoresis and analyzed. The resulting products were purified and sequenced by Macrogen, South Korea.1 The sequences obtained were edited and corrected manually using the DNA BASER sequence assembly software system2 and compared by a sequence similarity search with the GenBank database using the algorithm for local alignment of nucleotide sequences (Blastn; Altschul et al., 1990) and TrichoBLAST (Kopchinskiy et al., 2005), which allowed the identification of isolates as belonging to species of Trichoderma or just to the genus. Sequence alignments were performed using MUSCLE implemented in MEGA6 (Tamura et al., 2013). The alignments of concatenated TEF1-α and RPB2 sequences were manually adjusted, contained 128 sequences of species of the genus Trichoderma, 30 of which were isolated in the course of this study, and 98 isolates retrieved from GenBank, including the two sequences of the outgroup (Table 2). In total, the dataset comprised 119 partial TEF1-α sequences and 112 partial RPB2 sequences. The NEXUS file was interleaved using Paup 4b10 software (Swofford et al., 2001) to verify whether it was possible to concatenate the amplified TEF1-α and RPB2 sequences. Individual phylogenetic analysis of TEF1-α and RPB2 and concatenated multilocus analysis of all analyzed regions were carried out by Bayesian inference (BI) using MrBayes 3.2.1 (Huelsenbeck and Ronquist, 2001). The best evolutionary models were selected according to the Akaike Information Criterion (AIC) using MrModeltest v2.3 program (Nylander, 2004). Nucleotide substitution models in the single and multilocus trees were GTR + I + G for TEF1-α and SYM + I + G for RPB2. For all trees, the BI was estimated in the CIPRES Science Gateway Platform using Mr. Bayes 3.2.6. Two independent runs with four Markovian chain Monte Carlo (MCMC) procedures were conducted for 10 million generations, and the trees were sampled and retained every 1,000th generation. The first million tree samples were discarded in the burning phase, and the trees were summarized to generate a majority-rule consensus tree.
For maximum likelihood (ML) analyses, the concatenated tree with the two genes was generated in Sequence matrix v1.8125 (Vaidya et al., 2011). The ML analyses for single-loci trees and multilocus were estimated in the CIPRES Science Gateway Platform using RaxML-HPC v.8 (Stamatakis, 2006). For the concatenated dataset, all free modal parameters were estimated by RAxML with an ML estimate of 25 per site rate category. The concatenated dataset was partitioned by loci in the RAxML platform. The RAxML software accommodated the GTR model of nucleotide substitution with the additional options of modeling rate heterogeneity (G) and proportion invariable sites (I).
Phylogenetic trees (BI and ML) were visualized using FigTree.3 In BI, tree clades with posterior probability values below 95% were excluded (Harada et al., 1995; Rannala and Yang, 1996), and bootstrap values (≥70%) are given near nodes. The topologies obtained using the two methods (BI and ML) were then compared, and the phylogram layout was edited with CorelDRAW Graphics Suite 2020.
Genealogical concordance phylogenetic species recognition
The genealogical concordance phylogenetic species recognition (GCPSR) concept was applied to identify independent evolutionary lineages using the tools developed in Perl language by Brankovics et al. (2017) available at GitHub.4 The phylogenetic trees were built side-by-side using the ggtree package in R 4.0 (R Core Team 2021), to highlight the patterns of genealogical concordance and non-concordance.
Results
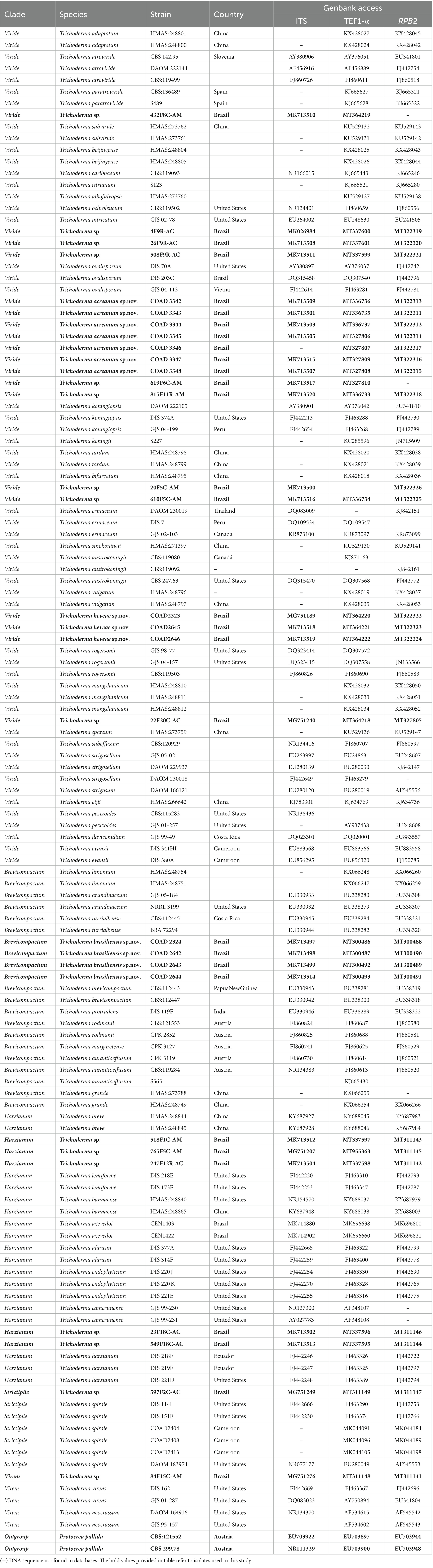
A total of 30 Trichoderma isolates were obtained from leaf, stem, and root samples of Hevea spp. in the Brazilian Amazon Forest. In quantitative terms, 13 from the state of Acre and 17 from the state of Amazonas were isolated (Araújo et al., 2018, 2020). The preliminary sequence similarity search with the ITS region and TEF1-α and RPB2 gene sequences showed that the 30 isolates belong to the Brevicompactum, Harzianum, Virens, Viride, and Strictipile clades. Therefore, as a next step, a phylogenetic analysis was conducted using a single locus of ITS, TEF1-α, and RPB2, and a multilocus dataset of concatenated TEF1-α and RPB2, which revealed the presence of seven different Trichoderma species. Isolates 20F5C–AM and 610F5C–AM were phylogenetically close to Trichoderma erinaceum (Bissett, Kubicek, and Szakacs), the isolates 619F6C–AM and 815F11R–AM were phylogenetically close to Trichoderma koningiopsis (Samuels, Suárez, and Evans), and 4F9R–AC, 26F9R–AC, and 508F9R–AC were phylogenetically close to Trichoderma ovalisporum (Samuels and Schroers). In addition, in the Viride clade, isolate 22F20C–AC was close to T. sparsum, and 432F8C–AM was close to Trichoderma atroviride Karst. In the Harzianum clade, isolates 23F18C–AC and 549F18C–AC were close to Trichoderma azevedoi (Valadares-Inglis & Inglis), 518F1C–AM and 765F5C–AM were close to Trichoderma breve (Chen & Zhuang), and 247F12R–AC was close to Trichoderma lentiforme (Rehm; Chaverri, Samuels & Rocha). The isolate 597F2C–AC was phylogenetically close to T. spirale Bissett (Strictipile clade) and 84F15C–AM was close to Trichoderma virens (Miller, Giddens & Foster) Arx. (Virens clade). These Trichoderma spp. were retained for future research to confirm species-level classification. In addition to the molecular data, the four new species were characterized morphologically, such as Trichoderma acreanum sp. nov., Trichoderma ararianum sp. nov., Trichoderma heveae sp. nov., and Trichoderma brasiliensis sp. nov. Phylogenetic trees and DNA sequence alignment data were deposited in TreeBase (Study 29026).
The clade classification was strongly supported by bootstrap and posterior probability values (Figure 1). The three single loci are shown in Supplementary Figures 1–3. Viride was the clade with the highest number and percentage of isolates (19), accounting for 63.3% of the total. The Harzianum clade included five and the Brevicompactum clade comprised four isolates, accounting for 16.7 and 13.3%, respectively. Strictipile and Virens clades were represented by one isolate each (3.3%).
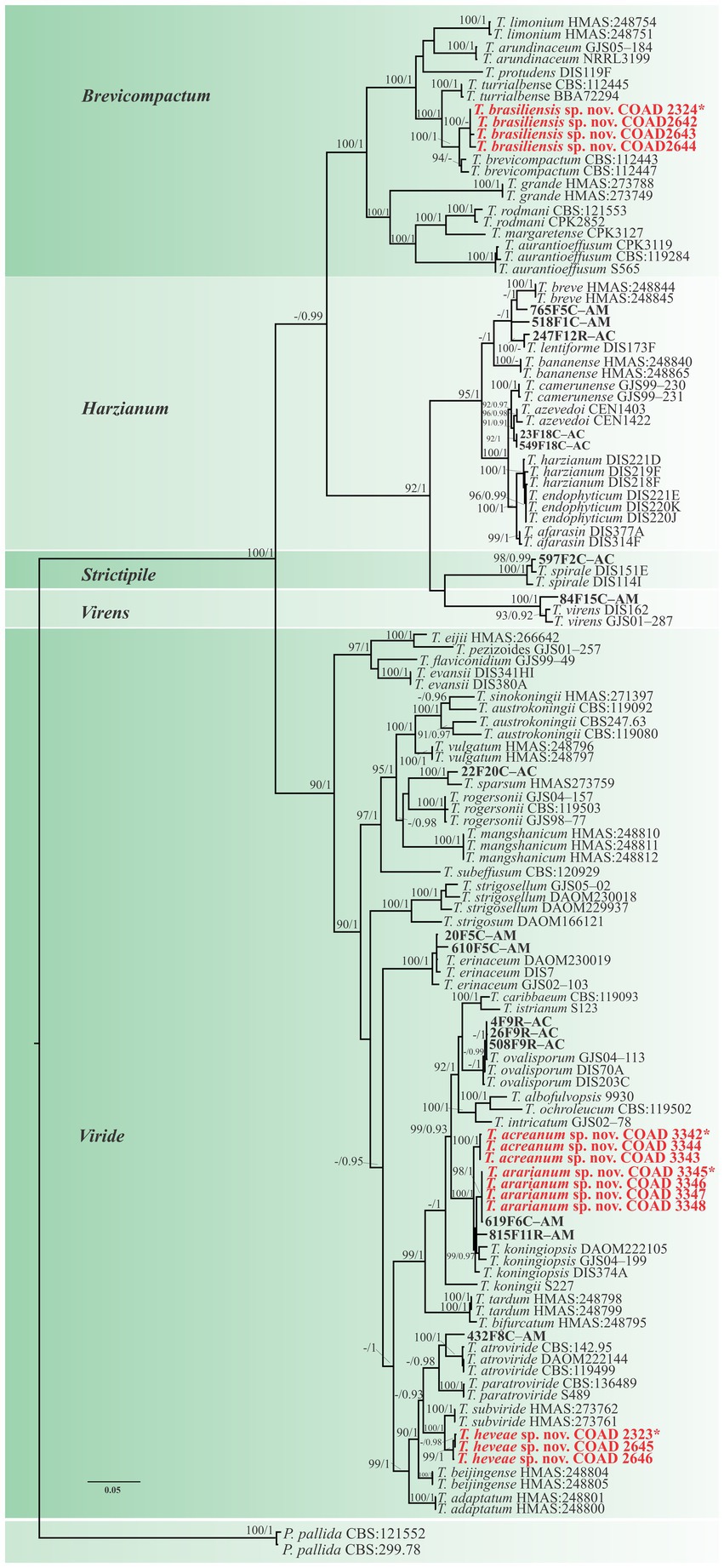
Figure 1. Phylogenetic tree of Trichoderma spp. based on combined RPB2 and TEF1-α sequence datasets. Bootstrap values (≥70%) of the ML analyses, as well as posterior probability scores (≥0.95) from a BI of the same dataset, are indicated at well-supported nodes together with thickened branches. The scale bar represents the number of expected substitutions per site (0.05). The isolates belonging to known species obtained in this study are in bold red. Isolates of new species described in this study are in bold red. Holotype labeled with an asterisk. The tree is rooted in two isolates of Protocrea pallida (CBS:121552 and CBS:299.78).”–” indicates a lack of support.
Isolates were recognized in their respective clades based on the previously accepted GCPSR concept (Taylor et al., 2000; Dettman et al., 2003). Thus, by applying genealogical concordance, the clade was present in the majority of the single-locus topologies (TEF1-α and RPB2), as revealed by the multilocus tree. In contrast, genealogical non-discordance (or discordance) was not observed, since a clade must be well supported, as judged by BI and ML, by at least one single-locus genealogy and not contradicted in any other single-locus topology (Figure 2).
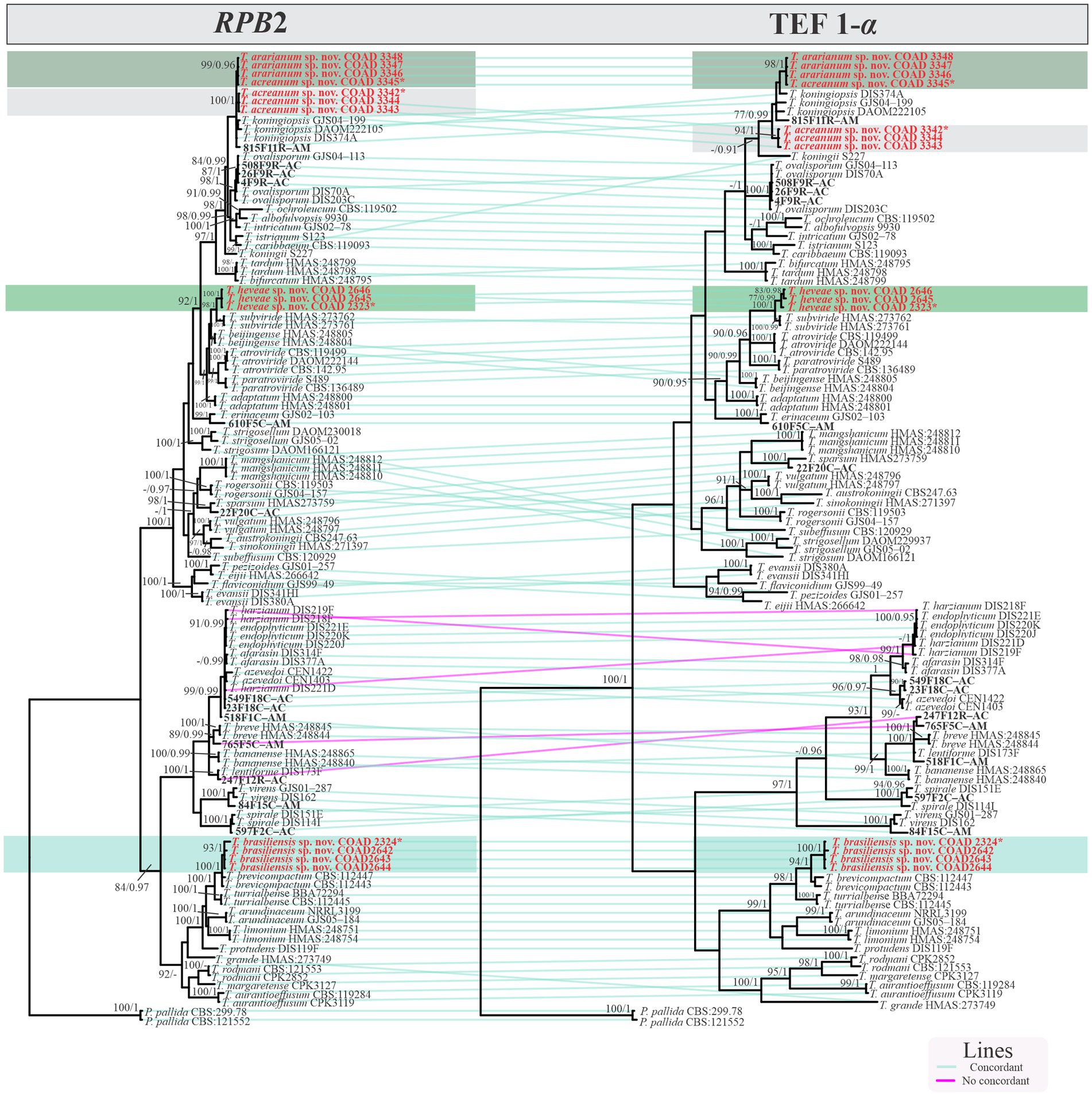
Figure 2. Phylogram GCPSR based on TEF1-α and RPB2 sequence datasets. Simultaneous analysis of two gene genealogies to show the transition from concordance among branches used to diagnose species.
A total of three new species were identified in the Viride clade, such as T. acreanum sp. nov. (COAD3345), T. ararianum sp. nov. (COAD3342), and T. heveae sp. nov. (COAD2323). The other six isolates were identified as Trichoderma sp. and formed a monophyletic clade with strong statistical support. Five isolates (23F18C–AC, 549F18C, 247F12R–AC, 765F5C–AM, and 518F1C–AM) were included in the Harzianum clade and were not conclusively characterized to the species level.
Conversely, in the Brevicompactum clade, the new taxon, T. brasiliensis, represented by four isolates (COAD2324, COAD2642, COAD2643, and COAD2644) and was clearly distinguished by strong support from the other clade species.
Of the 30 isolates, 19 were isolated from stems. They were identified as T. heveae sp. nov. (three isolates), T. acreanum sp. nov. (three isolates), T. ararianum sp. nov. (four isolates), and Trichoderma sp. (11 isolates) present in all clades. A total of nine were isolated from roots, and they were identified as T. brasiliensis sp. nov. (four isolates) and Trichoderma sp. (five isolates). Trichoderma brasiliensis sp. nov. was found exclusively in the roots of H. brasiliensis in Amazonas. In contrast, two isolates were found in the leaves of H. brasiliensis and H. guianensis and are proposed here as a new species, T. heveae sp. nov.
Taxonomy
Trichoderma acreanum V. N. Brito, J. L. Alves & M. V. Queiroz, sp. nov., Figure 3.
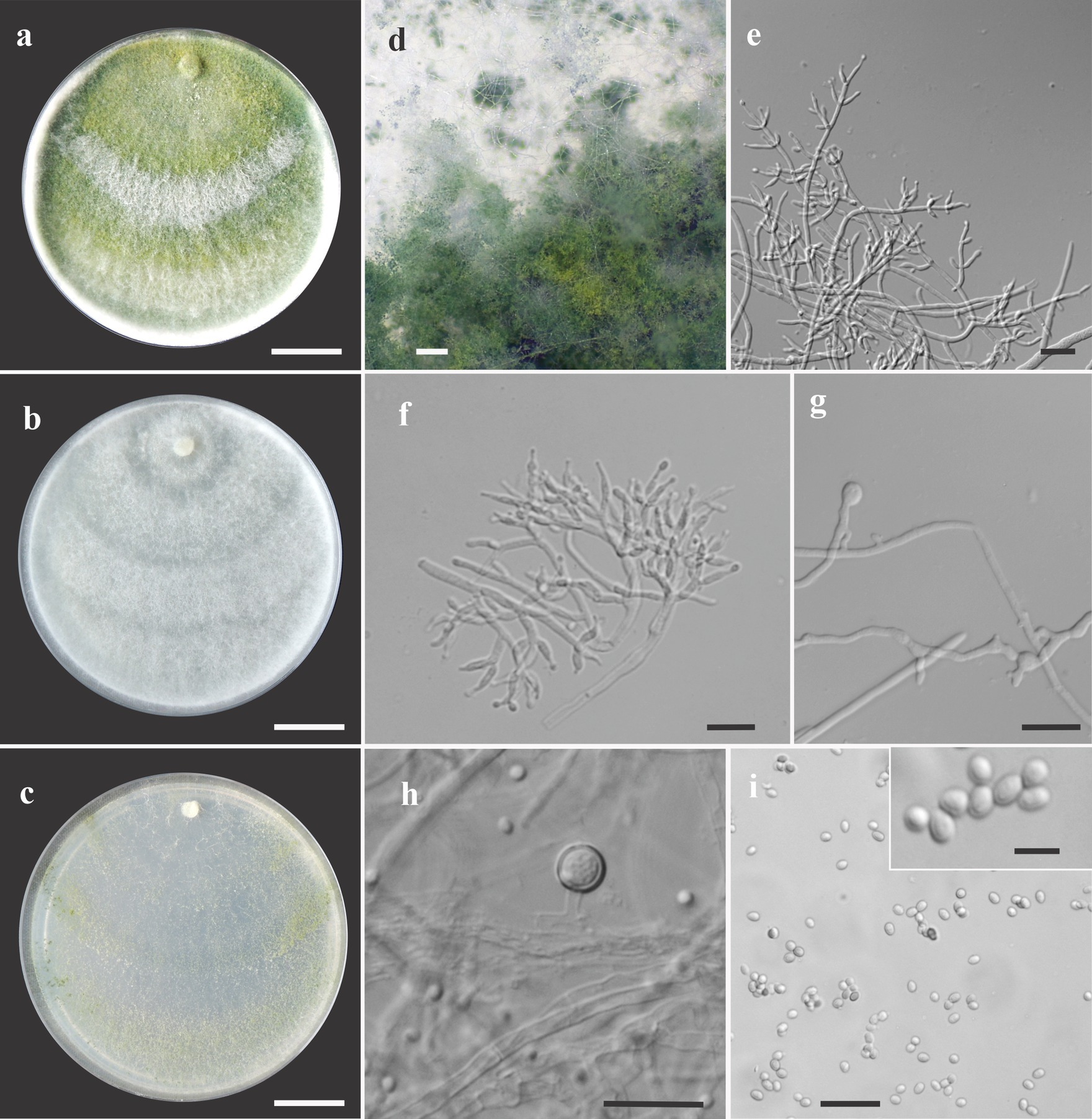
Figure 3. Asexual morph of Trichoderma acreanum sp. nov. from Hevea brasiliensis (VIC47506, holotype). (A–C) Colony appearances (PDA, CMD, and SNA; 7 days, 25°C). (D) Sporulating pustules (SNA). (E,F) Conidiophores and phialides (SNA). (G) Chlamydospore (after 48 h). (H) Chlamydospore (after 7 days). (I) Conidia. Scale bars: (A–C) 20 mm, (D) 0.2 mm, (E,F) 15 μm, (G) 10 μm, (H) 25 μm, (I) 20 μm (inside = 5 μm).
MycoBank MB830208
Etymology: “acreanum,” the specific epithet, refers to the state of Acre, Brazil.
Holotype: Brazil. State of Acre, Cruzeiro do Sul, Igarapé Campinas, coordinates 7°46′00.6″S, 72°15′03.3″W, K. S. Araújo, 26 July 2017 (holotype VIC47506). Ex-type culture of COAD3342.
Sexual morph not seen. Colonies fast growing at 25 and 30°C on all media; no growth at 35°C. No pigment or distinctive odor was observed on media PDA, SNA, and CMD. The radial growth rate on CMD was 77–79 mm at 25°C and 55–62.5 mm at 30°C, after 72 h. Colonies covering the plates after 4 days at 25°C; mycelium white, flat, and floccose in texture. After 7 days, some areas became dull green due to sporulation. Chlamydospores globose to irregular, terminal, intercalary, thick-walled, brown. Conidiophores comprising a main axis, and short fertile branches or phialides arising along the length of this axis, more or less paired with internodes, the terminal part of conidiophore sparingly branched. Phialides solitary or in diverging terminal whorls of 2–3, ampulliform, slightly inclined or curved in upper part, widest part mostly central, with a short neck, 6–11 μm × 2–3 μm (n = 30). Conidia yellow-green, smooth, globose to subglobose, 2–4 μm × 2.5–3 μm (n = 30). The radial growth rate on PDA was 78–80 mm at 25°C, 67.5–70 mm at 30°C, and 5–12 mm at 35°C, after 72 h. Colonies covered the plate after 4 days at 25°C; mycelium dense, texture floccose to velvety, mixed mycelia dark-herbage green, white, and grayish-yellow green. Chlamydospores terminal and intercalary, mostly globose, thick-walled, brown. On SNA, the radial growth rate was 42.5–45 mm at 25°C, 40–44 mm at 30°C, and 6–8 mm at 35°C, after 72 h. Colonies covered the plate after 6 days at 25°C; mycelia thin, floccose, and white, with discrete concentric zones due to sporulation, arranged in several irregular zones spreading from the center. Chlamydospores terminal and intercalary, brown, and globose.
Habitat and host range: endophytic in the stems of living wild Hevea spp. trees.
Additional specimens examined: Brazil. State of Acre, Xapuri, Reserve Chico Mendes, coordinates 10°49′59.1″S, 68°23′11.2″W, K. S. Araújo, 26 July 2017 (COAD3343, VIC47507); Brazil. State of Acre, Cruzeiro do Sul, Igarapé Campinas, coordinates 7°46′00.6″S, 72°15′03.3″W, K. S. Araújo, 26 July 2017 (COAD3344, VIC47508).
Notes: The concatenated tree with TEF1-α and RPB2 sequences clearly indicates that the three isolates belong to a new taxon (Figure 1). In addition, phylogenetic species were recognized based on two main previously accepted criteria genealogical concordance, wherein the clade (T. acreanum) was present in the two single-locus genealogies and Genealogical Non-Discordance, wherein the clade (T. acreanum) was well supported in the least one single-locus genealogy and was not contradicted in any other single-locus genealogy at the same level of support (Figure 2, Supplementary Figures 1, 2). Trichoderma acreanum sp. nov. is morphologically similar to several species of the Trichoderma viride Pers. clade (Samuels et al., 2006). In Trichoderma acreanum sp. nov., the phialides arise singly and are widely spaced directly from the main axis, which can have up to three phialides at its tip and is flask-shaped, straight, or slightly hooked; those forming at the tips of long conidiophores are often narrowly cylindrical or tapering uniformly from the base to the tip and swollen in the middle. However, in T. koningiopsis, the phialides are densely clustered with very short internodes, typically straight, hooked, or sinuous, narrowly lageniform, in whorls of 2–5, with several phialides at times arising from the same point and crowded. In addition, the conidia of the new taxon are shorter in length and subglobose compared with T. koningiopsis which has larger, ellipsoidal conidia (Table 3).
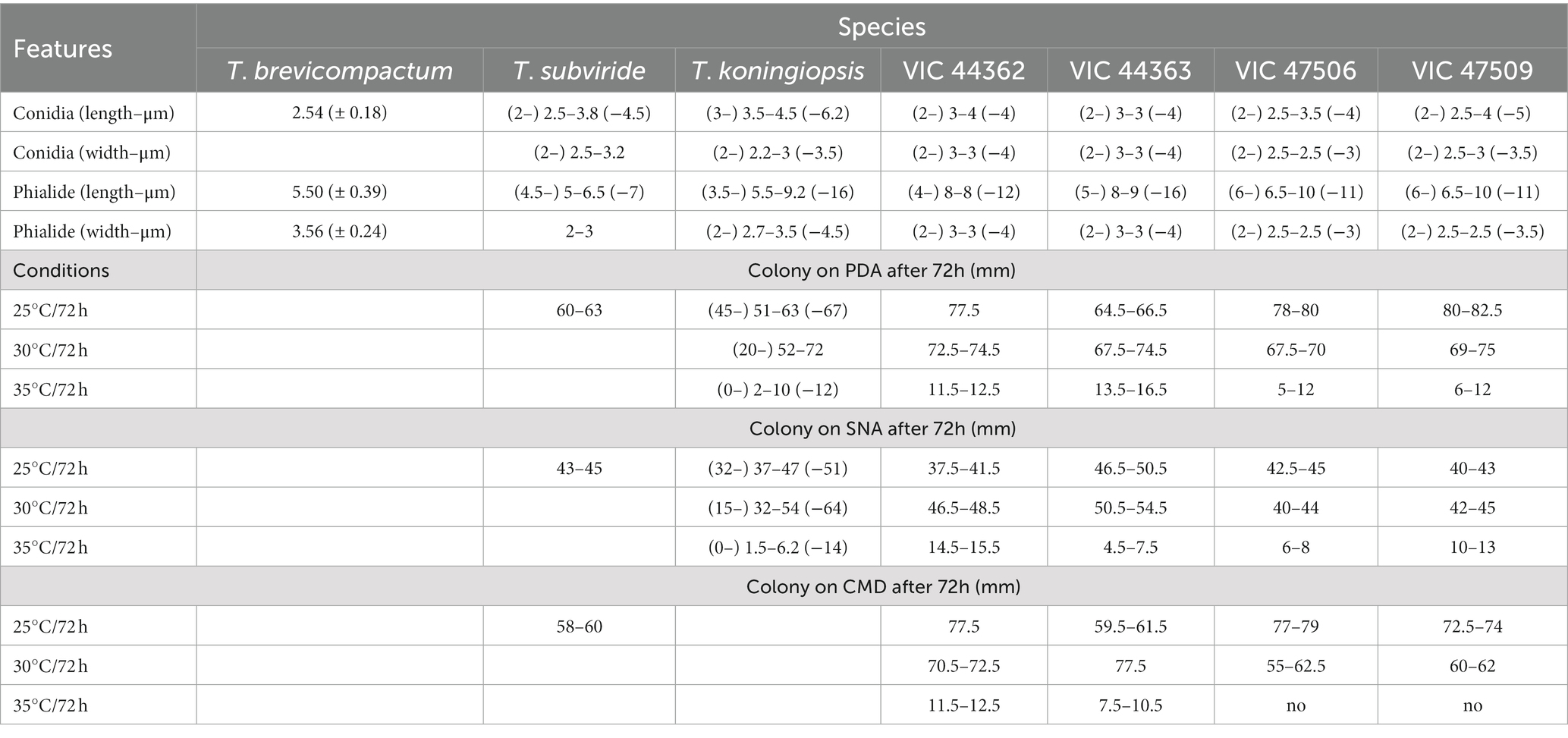
Table 3. Morphological differences relevant to the separation of new species of Trichoderma described as endophytes from Hevea spp. from the Amazon Forest.
Trichoderma ararianum V. N. Brito, J. L. Alves & M. V. Queiroz, sp. nov., Figure 4.
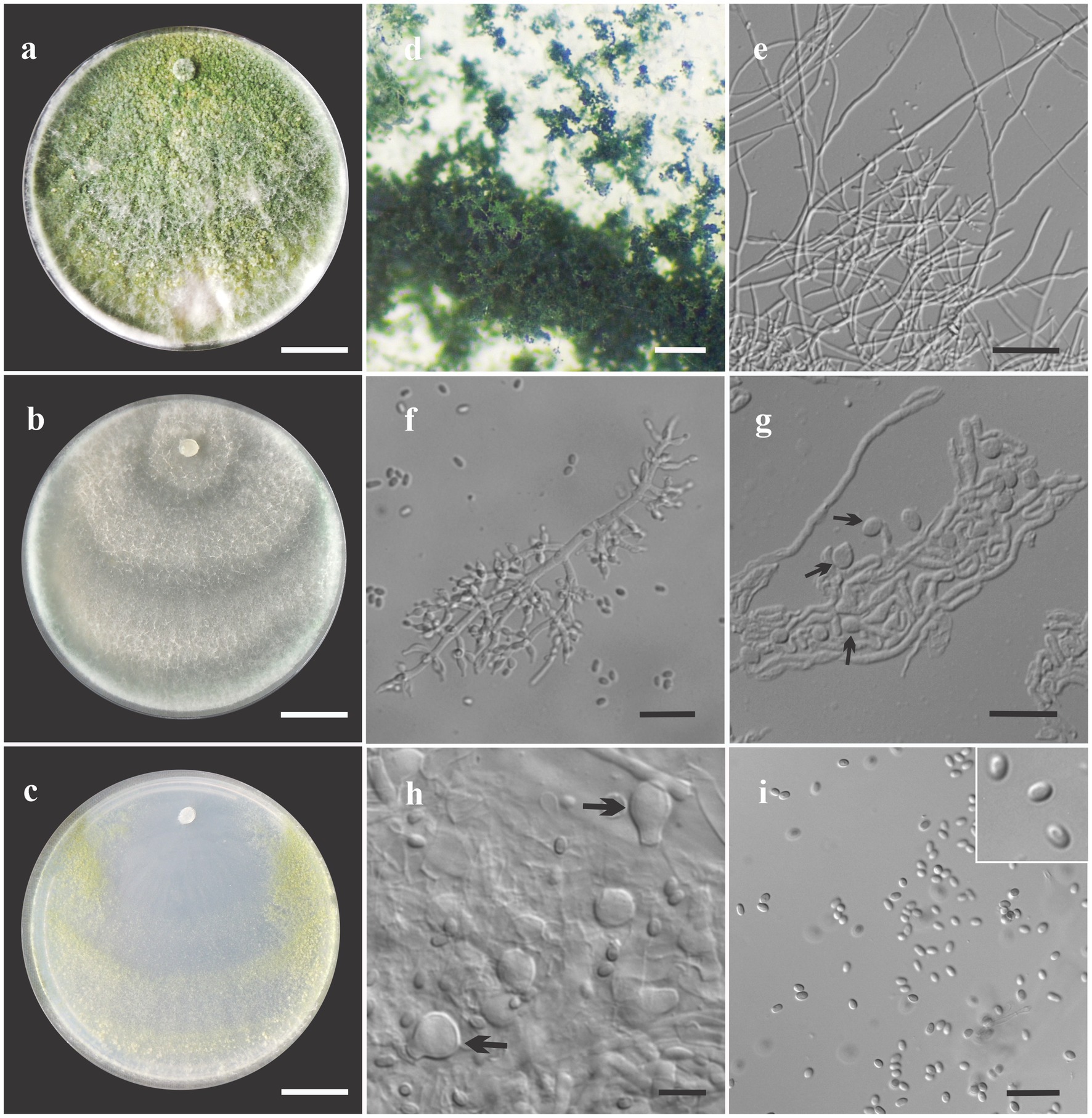
Figure 4. Asexual morph of Trichoderma ararinum sp. nov. from Hevea brasiliensis (VIC47509, holotype). (A–C) Colony appearances (PDA, CMD, and SNA; 7 days, 25°C). (D) Sporulating pustules (SNA). (E,F) Conidiophores and phialides (SNA). (G) Chlamydospore (after 48 h). (H) Chlamydospore (after 7 days). (I) Conidia. Scale bars: (A–C) 20 mm, (D) 0.2 mm, (E) 50 μm, (F,G,I) 20 μm, and (H) = 10 μm.
MycoBank MB830209
Etymology: “ararianum,” the specific epithet, refers to the river Arari from Itacoatiara, state of Manaus, Brazil.
Holotype: Brazil. State of Amazonas, Itacoatiara hydrographic basin, coordinates: 03°24′25.4″S, 058°29′38.8″W, V. N. Brito, 26 July 2017 (holotype VIC47509). Ex-type culture of COAD3345.
Sexual morph not seen. Colonies on PDA, SNA, and CMD fast growing at 25 and 30°C, with no growth at 35°C. The radial growth rate on CMD was 72.5–74 mm at 25°C and 60–62 mm at 30°C, after 72 h. Colonies covering plate after 4 days at 25°C, mycelia flat and thin, with a velvety texture, white, cottony, and raised. Chlamydospores globose to subglobose, terminal, intercalary, thick-walled, brown. Sporulation after 4 days, with dull green, fluffy tufts arranged in several concentric zones. Conidiophores with a central axis and short fertile branches or phialides growing along with it, additional branches coupled with internodes, and terminal part of conidiophore not branched. Phialides sparse, isolated; short side branches bearing terminal whorls of 2–3 phialides, slightly inclined or curved upward, 6–11 μm × 2–3.5 μm (n = 30), ampulliform, widest part mostly central, with a long neck. Conidia yellow-green, smooth, globose to subglobose, 2–5 μm × 2–3.5 μm (n = 30). On PDA, 80–82 mm at 25°C, 69–75 mm at 30°C, and 6–12 mm at 35°C, after 72 h. Colonies covering plate after 4 days at 25°C, mycelia dense and flat with floccose texture, mixed grayish yellow-green with dark herbage green hues due to sporulation; mycelia white and cottony in some areas of the colonies. The radial growth rate on SNA was 40–43 mm at 25°C, 42–45 mm at 30°C, and 10–13 mm at 35°C, after 72 h. Colonies covering the plate after 5 days at 25°C; mycelia white and thin. Sporulation unevenly spread in dull green areas. Chlamydospores globose to subglobose, terminal and intercalary, thick-walled, brown.
Habitat and host range: endophytic in the stems of living wild Hevea brasiliensis trees.
Additional specimens examined: Brazil. State of Amazonas, Itacoatiara hydrographic basin, coordinates 03°02′37,6″S, 058°30′11,8″W, V. N. Brito, 26 July 2017 (VIC47510, COAD3346); Brazil. State of Amazonas, Itacoatiara, coordinates 03°24′25.4″S, 058°29′38.8″W, V. N. Brito, 26 July 2017 (VIC47511, COAD3347); Brazil. State of Amazonas Itacoatiara, hydrographic basin, coordinates 03°24′25.4″S, 058°29′38.8″W, V. N. Brito, 26 July 2017 (VIC47512, COAD3348).
Notes: The concatenated tree with TEF1-α and RPB2 sequences clearly indicates that the four isolates belong to a new taxon (Figure 1). In addition, phylogenetic species were recognized based on two main previously accepted criteria Genealogical Concordance, wherein the clade (T. ararianum) was present in the two single-locus genealogies and Genealogical Non-Discordance, wherein the clade (T. ararianum) was well supported in the least one single-locus genealogy and was not contradicted in any other single-locus genealogy at the same level of support (Figure 2, Supplementary Figures 1, 2). Trichoderma ararianum sp. nov. is morphologically similar to several species of the T. viride clade and phylogenetically closely related to Trichoderma koningiopsis (Samuels et al., 2006). Trichoderma ararianum sp. nov. is distinct from T. koningiopsis based on; the faster colony growth rate on PDA at 25°C after 72 h (Table 3); the shorter, subglobose conidia, whereas T. koningiopsis has larger and broadly ellipsoidal conidia. In addition, the phialides are smaller, typically straight, lageniform, or slightly swollen in the middle, isolated and intercalary, or held in terminal whorls (with three); whereas T. koningiopsis has larger phialides, densely clustered with a very short internode and typically straight, though at times hooked or sinuous, narrowly lageniform held in whorls of 2–5, with several phialides at times arising from the same point and crowded (Table 3).
Trichoderma brasiliensis V. N. Brito & M. V. Queiroz, sp. nov., Figure 5.
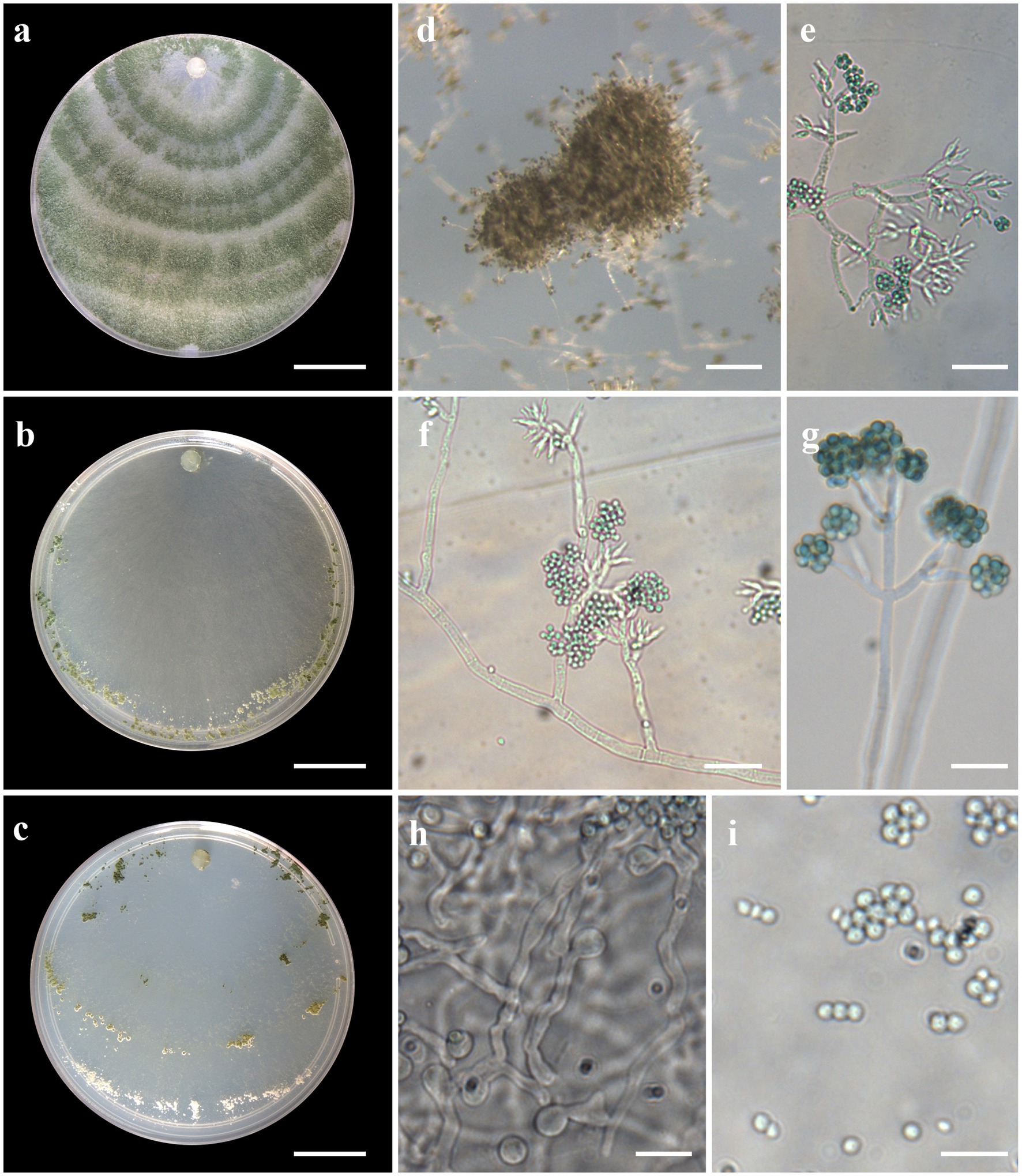
Figure 5. Asexual morph of Trichoderma brasiliensis sp. nov. from Hevea brasiliensis (VIC44363, holotype). (A–C) Colony appearances (PDA, CMD, and SNA; 7 days, 25°C). (D) Sporulating pustules (SNA). (E–G) Conidiophores and phialides (SNA). (H) Chlamydospore. (I) Conidia. Scale bars: (A–C) 20 mm, (D) 0.2 mm, (E,F,H) 20 μm, and (G,I) 10 μm.
MycoBank: MB830210
Etymology: “brasiliensis,” for its origin in Hevea brasiliensis.
Holotype: Brazil, State of Amazonas, Itacoatiara, coordinates 03°57′08.3″S, 59°05′16.8″W, V. N. Brito, 26 July 2017 (holotype VIC44363). Ex-type culture COAD2324.
Sexual morph not seen. Colonies with a fast growth rate at 30°C on all media with limited growth at 35°C. The radial growth rate on CMD was 59.5–61.5 mm at 25°C, 77.5 mm at 30°C, and 7.5–10.5 mm at 35°C, after 72 h. Colonies white, thin, with a downy surface. Chlamydospores globose, terminal, and intercalary, thick-walled, pale brown. Sporulation after 1 day, with dark green, fluffy tufts, or loose pustules arranged in several indistinctly separated, irregular concentric zones. Conidiophores consist of long main axes with pyramidal, verticillate branching in a pachybasium-type pattern (Kraus et al., 2004). Phialides 5–16 μm × 2–4 μm (n = 30), ampulliform to flask-shaped. Conidia green, smooth, globose to subglobose, 2–4 μm × 2–4 μm (n = 30). On PDA, after 72 h, 64.5–66.5 mm at 25°C, 67.5–74.5 mm at 30°C, and 13.5–16.5 mm at 35°C. Colonies covering the plate after 4 days at 25°C, mycelium dense and hairy. The radial growth rate on SNA was 46.5–50.5 mm at 25°C, 50.5–54.5 mm at 30°C, and 4.5–7.5 mm at 35°C, after 72 h. Colonies covered the plate after 6 days at 25°C; mycelium white and thin. Chlamydospores observed after 1 day; numerous, globose, grouping together in clusters, terminal and intercalary, thick-walled, brown. Sporulation starting after 2 days, arranged in several concentric zones spreading from the center and turning dark green.
Habitat and host range: In the roots of living wild Hevea brasiliensis trees.
Additional specimens examined: Brazil. State of Amazonas, Itacoatiara, coordinates 03°24′34.00″S, 058°29′18.3″W, V. N. Brito, 26 July 2017 (COAD2642); Brazil. Amazon state, Itacoatiara hydrographic basin, coordinates 03°24′34.00″S, 058°29′18.3″W, V. N. Brito, 26 July 2017 (COAD2643); Brazil. State of Amazonas, Itacoatiara, coordinates 03°24′34.00”S, 058°29′18.3”W, V. N. Brito, 26 July 2017 (COAD2644).
Notes: Phylogenetic analysis clearly indicates that the three isolates belong to a new taxon, with robust distinction in multilocus trees (Figure 1), the GCPSR concept (Figure 2), and single-locus trees (Supplementary Figures 1, 2). Trichoderma brasiliensis sp. nov. shares morphological similarities with Trichoderma brevicompactum (Kraus et al., 2004), such as ampulliform phialides arising in crowded verticils, however, the conidia of T. brevicompactum are subglobose to short ellipsoidal while those of T. brasiliensis sp. nov. are globose to subglobose.
Trichoderma heveae V. N. Brito & M. V. Queiroz, sp. nov., Figure 6.
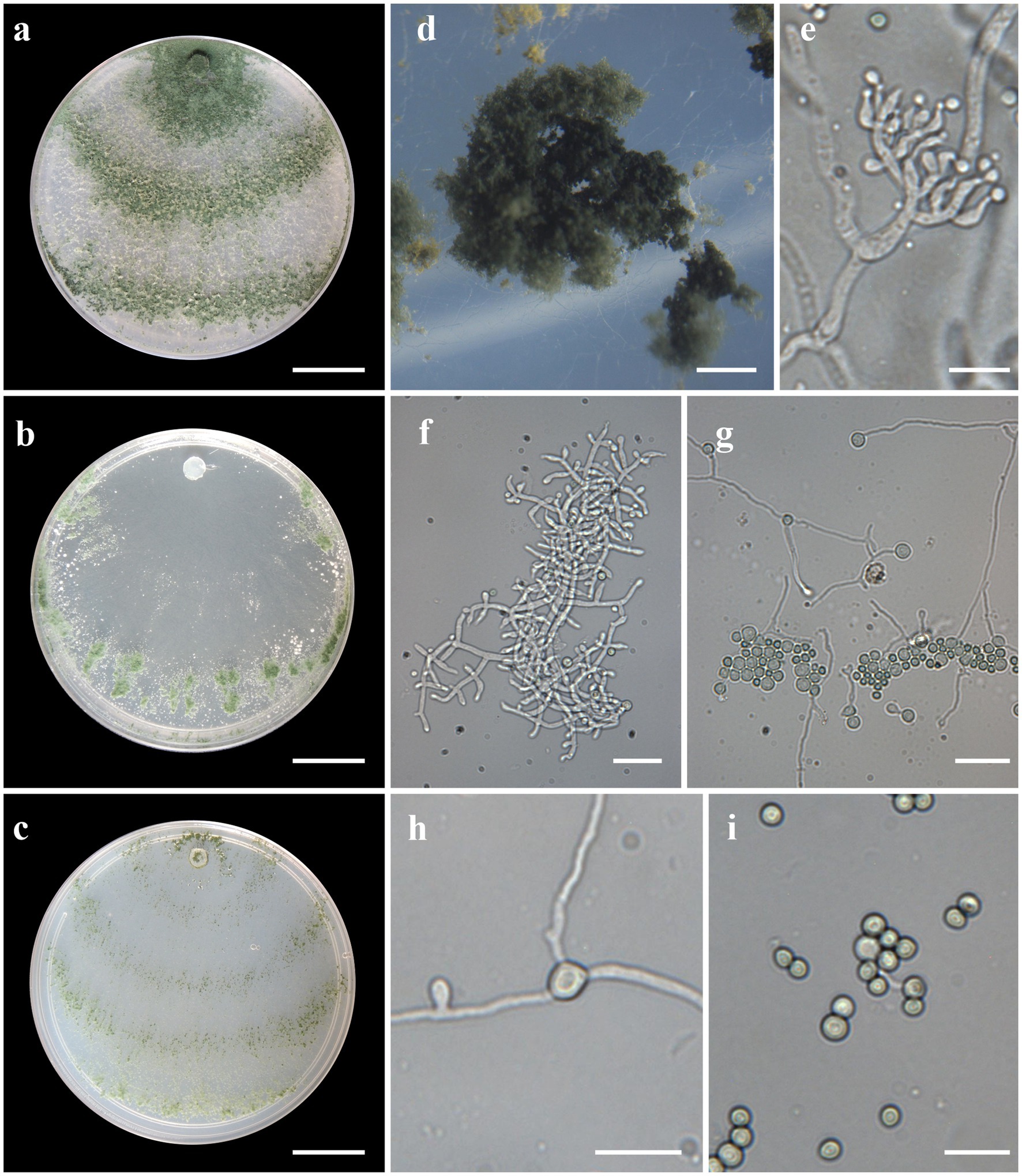
Figure 6. Asexual morph of Trichoderma heveae sp. nov. from Hevea spp. (VIC44362, holotype). (A–C) Colony appearances (PDA, CMD, and SNA; 7 days, 25°C). (D) Sporulating pustules (SNA). (E,F) Conidiophores and phialides (SNA). (G,H) Chlamydospores. (I) Conidia. Scale bars: (A–C) 20 mm, (D) 0.5 mm, (E,I) 10 μm, and (F–H) 20 μm.
MycoBank: MB830211
Etymology: Latin, heveae, refers to the host from which the fungus was isolated.
Holotype: Brazil, State of Amazonas, Itacoatiara, coordinates 7°44′04.1″S/72°49′51.2″W, V. N. Brito, 26 July 2017 (holotype VIC44362). Ex-type culture: COAD2323.
Sexual morph not seen. Colonies with a rapid growth rate at 25°C on all media with limited growth at 35°C. The radial growth rate on CMD was 77.5 mm at 25°C, 70.5–72.5 mm at 30°C, and 11.5–12.5 mm at 35°C, after 72 h. Colonies thin, surface downy, floccose or farinose, yellowish-green to green. Chlamydospores globose, terminal, and intercalary. Sporulation starting after 2 days; dark green, fluffy tufts, or loose pustules, with irregular or circular outlines, arranged in several indistinctly separated concentric zones. Conidiophores formed along with main axes, with long branches at lower levels, and sparse, widely spaced, solitary or paired phialides; or short, one-celled side branches bearing terminal whorls of 2–3 phialides, slightly inclined or curved upward, 4–12 μm × 2–4 μm, (n = 30), ampulliform, widest part mostly central. Conidia green, smooth, subglobose or oval, 2–4 μm × 2–4 μm (n = 30). The radial growth rate on PDA was 77.5 mm at 25°C, 72.5–74.5 mm at 30°C, and 11.5–12.5 mm at 35°C, after 72 h. Mycelium dense and hairy covering the plate after 3 days at 25°C. Radial growth rate on SNA was 37.5–41.5 mm at 25°C, 46.5–48.5 mm at 30°C, and 14.5–15.5 mm at 35°C, after 72 h. Mycelium covering the plate after 5 days at 25°C; colonies hyaline and thin. Chlamydospores formed after 2 days; globose, terminal, intercalary, thick-walled, brown. Sporulation starting after 2 days, arranged in several concentric zones spreading from the center and turning dark green.
Habitat and host range: In the stems (COAD2646) and leaves (COAD2645 and COAD2323) of living, wild Hevea brasiliensis trees.
Additional specimens examined: Brazil. State of Amazonas, Manaus, Coari hydrographic basin, coordinates 03°45′27.4″S, 063°24′34.6″W, V. N. Brito, 26 July 2017 (COAD2645); Brazil. State of Amazonas, Manaus, Itacoatiara, hydrographic basin, coordinates 03°02′40.4″S, 058°30′09.7″W, V. N. Brito, 26 July 2017 (COAD2646).
Notes: Phylogenetic analysis clearly indicates that the three isolates represent a new taxon, with robust distinction in multilocus trees (Figure 1), the GCPSR concept (Figure 2), and single-locus trees (Supplementary Figures 1, 2). Trichoderma heveae sp. nov. is a tropical endophytic species present in the leaves and stems of wild Hevea; belonging to the Viride clade (Figure 1). Trichoderma subviride Qin & Zhuang, a closely related species, has slower growth rates on all media (CMD 58–60 mm, PDA 60–63 mm, and SNA 43–45 mm; Qin and Zhuang, 2016b), while T. heveae sp. nov. colony showed a fast growth rate in the media (CMD 77.5 mm, PDA 77.5 mm, and SNA 37.5–41.5 mm; Table 3).
Discussion
In recent years, there has been growing interest in the isolation and identification of endophytic Trichoderma spp. and their derived products (Contreras-Cornejo et al., 2016; Katoch et al., 2019; Nuankeaw et al., 2020), and a number of new species, such as T. amazonicum (Chaverri et al., 2010), T. spirale (Chaverri et al., 2003), Trichoderma protrudens (Samuels & Chaverri; Degenkolb et al., 2008), and Trichoderma botryosum (Rodriguez et al., 2021), have been described. In the present study, 30 endophytic isolates of the genus Trichoderma were obtained from the leaves, stems, and roots of wild rubber trees in the Brazilian Amazon region, with four new species being identified, based on morphological and molecular analyses.
Cai and Druzhinina (2021) reported that at least 60% of Trichoderma species have precise and accurate molecular identification at the species level based on an analysis of three DNA barcodes (ITS, TEF1-α, and RPB2). Other studies have recently confirmed that TEF1-α and RPB2 gene sequences well support taxonomic conclusions in Trichoderma due to their suitable interspecific variations (Jaklitsch and Voglmayr, 2015; Qiao et al., 2018; Gu et al., 2020; Rodriguez et al., 2021). Furthermore, Cai and Druzhinina (2021) reported that Trichoderma species can be identified based on pairwise similarities between RPB2 and TEF1-α. In this study, single and combined TEF1-α and RPB2 datasets were used for molecular analysis to reveal the phylogenetic relationships between the species.
The Viride and Harzianum clades had the highest percentage of isolates. In addition, isolates belonging to the Virens, Strictipile, and Brevicompactum clades were identified, demonstrating that endophytic species in tropical climates are distributed over different clades of the genus Trichoderma, as demonstrated by Zhang et al. (2007) and Gazis and Chaverri (2010).
Trichoderma species were categorized into five clades, with a prevalence of rubber-tree isolates in the Viride clade, represented by 63% of isolates obtained in this study. The Viride clade is among the largest in Trichoderma, and its taxonomy has been examined in detail over the past decades, revealing a number of new species (Jaklitsch et al., 2006; Qin and Zhuang, 2016b). This is a cosmopolitan clade containing both saprotrophic and endophytic species, reportedly found in Theobroma cacao L., Theobroma gileri Cuatrec., and H. brasiliensis (Samuels et al., 2006; Gazis and Chaverri, 2010). Among the species of the Viride clade, T. ovalisporum has been reported as an endophyte in Theobroma spp. (Evans et al., 2003; Holmes et al., 2004), which has been found in both Southeast Asia and South America (Samuels et al., 2006).
Another related species within the Viride clade is T. atroviride, which occurs in Neotropical regions, as well as in Europe, colonizing tree bark or living as a mycoparasite (Hoyos-Carvajal et al., 2009; Jaklitsch, 2011). Trichoderma atroviride has also been isolated as an endophyte from the ornamental trees Cephalotaxus fortunei Hook. and Camptotheca acuminata Decne. in China (Zheng et al., 2011; Pu et al., 2013).
Trichoderma koningiopsis is a cosmopolitan species that has been recorded mainly from the Neotropics. Its Hypocrea sexual morph has been found as far north as the State of New York (Samuels et al., 2006), although the sexual form was not found in this study. Evidence suggests that T. koningiopsis is effective in plant protection, and it has been used in biological control applications. In addition, it has been reported as an endophyte of both cultivated and wild Coffea canephora Pierre ex Froehner in Cameroon (Rodriguez et al., 2021).
In this study, three new species belonging to the Viride clade were identified. Among them, two species are close to T. koningiopsis and the other is related to T. subviride. Trichoderma acreanum sp. nov. and T. ararianum sp. nov. are different from T. koningiopsis, as is evident from both phylograms (Figures 1, 2 and Supplementary Figures 1, 2). Additional morphological data show that the new taxa vary in the size and shape of the conidia and phialides, as well as in their growth rates on several media. Trichoderma heveae sp. nov. is different from T. subviride, as evidenced by both phylograms (Figures 1, 2 and Supplementary Figures 1, 2). Trichoderma subviride is a common wood-inhabiting species, while T. heveae sp. nov. has been identified as an endophyte in the leaves and stems of Hevea spp. in the Amazon Forest. Although these species have different ecological niches, their morphological characteristics are similar.
Trichoderma brasiliensis sp. nov. is most closely related to T. brevicompactum, an endophytic species from North America, South America, and Asia. However, T. brevicompactum has only been reported as an endophyte in garlic (Allium sativum L.) in China (Shentu et al., 2014) and not in tropical tree species. The Brevicompactum clade originally comprised Trichoderma arundinaceum (Zafari, Gräfenhan & Samuels), Trichoderma brevicompactum, Trichoderma protrudens, Trichoderma rodmanii (Samuels and Chaverri; Jaklitsch and Voglmayr, 2013) and Trichoderma turrialbense (Samuels, Degenkolb, Nielsen and Gräfenhan; Degenkolb et al., 2008). Trichoderma aurantioeffusum (Jaklitsch) and Trichoderma margaretense (Jaklitsch) were added later (Jaklitsch, 2011), likewise that Trichoderma grande (Qin & Zhuang) and Trichoderma limonium (Qin and Zhuang, 2016a). Samuels et al. (1998) comprehensively studied this section and emphasized the importance of asexual features and sequence data (TEF1-α and RPB2) in its taxonomy. Recently, several species have been described based on their phenotypic and genotypic differences. The species in the Brevicompactum clade are characterized by a conidiophore type known as “pachybasium,” with small and compact phialides (Bissett, 1991; Kraus et al., 2004). The new species related to this clade displayed similar characteristics, including the size, shape, and color of the conidia (Table 3). However, T. brasiliensis sp. nov. has larger and less aggregated phialides compared with T. brevicompactum.
The Harzianum clade of Trichoderma comprises species associated with a wide variety of substrates. However, this clade is characterized by a “species complex” and is morphologically heterogeneous and phylogenetically complex (Chaverri et al., 2015). Several morphologically similar species are known to be phylogenetically related to T. harzianum. In our study, five isolates grouped in the Harzianum clade, together with T. azevedoi, T. breve, and T. lentiforme. The taxonomic complex of the Harzianum clade includes combinations of different geographic origins and hosts (Rodriguez et al., 2021; Zheng et al., 2021), of ecological and economic importance, with promising prospects for use in biological control (Chaverri et al., 2015).
Trichoderma virens has a worldwide distribution, mainly in soil, and is recently reported as an endophyte in the aerial tissues of wild coffee in Africa (Rodriguez et al., 2021). In our study, one isolate (84F15C–AM) obtained from a stem of H. brasiliensis is phylogenetically close to T. virens and grouped with two isolates within the Virens clade.
Considering the significance of Trichoderma spp. in biocontrol, Baiyee et al. (2019) reported the mechanisms involved in the biological control of pathogens in lettuce (Lactuca sativa L.) by which T. spirale T76-1 inhibits the severity of leaf spot disease. In the present study, T. spirale belonging to the Strictipile clade was isolated from the stems of H. brasiliensis, and this endophytic characteristic may provide additional benefits.
In conclusion, our results indicate the presence of eight endophytic species of the genus Trichoderma previously reported in other studies (T. atroviride, T. erinaceum, T. koningiopsis, T. lentiforme, T. ovalisporum, T. sparsum, T. spirale, and T. virens), as well as four new endophytic species (T. acreanum sp. nov., T. ararianum sp. nov., T. heveae sp. nov., and T. brasiliensis sp. nov.) isolated from Hevea spp. native to the Amazon Forest. From the small sample size, it is obvious that wild rubber trees harbor an array of endophytic Trichoderma species with the potential for use in the management of rubber diseases, as well as bioprospecting for bioactive compounds.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
MQ designed the project, supervised its execution, and reviewed and edited the manuscript. VB contributed to the overall data analysis and wrote the first draft of the manuscript. VB, JL, KS, and TS contributed to the preparation of phylogenetic trees. VB and KS contributed to the collection of material, isolation, identification, and preservation of fungal isolates. VB, KS, and TS contributed to the standardization of the endophytic fungal isolation technique and sequence analysis. VB, KS, and CB contributed to DNA extraction, PCR, and gene sequencing. VB, KS, and JL contributed to morphological and molecular characterization. VB, KS, TS, OL, JL, and MQ interpreted the bioinformatics analysis. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)—Finance Code 001, the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG).
Acknowledgments
We would like to thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)—Finance Code 001, the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) for financing the project and granting the scholarships. In addition, we are grateful to Harry C. Evans (CABI Bioscience) for correcting the English grammar.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1095199/full#supplementary-material
Footnotes
References
Abramoff, M. D., Magalhães, P. J., and Ram, S. J. (2004). Image processing with ImageJ. Biophoton. Int. 11, 36–42.
Almeida, F. B. R., Cerqueira, F. M., do Nascimento, S. R., Ulhoa, C. J., and Lima, A. L. (2007). Mycoparasitism studies of Trichoderma harzianum strains against Rhizoctonia solani: evaluation of coiling and hydrolytic enzyme production. Biotechnol. Lett. 29, 1189–1193. doi: 10.1007/s10529-007-9372-z
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Araújo, K. S., Brito, V. N., Veloso, T. G. R., Leite, T. S., Alves, J. L., da Hora Júnior, B. T., et al. (2020). Diversity and distribution of endophytic fungi in different tissues of Hevea brasiliensis native to the Brazilian Amazon Forest. Mycol. Prog. 19, 1057–1068. doi: 10.1007/s11557-020-01613-4
Araújo, K. S., Brito, V. N., Veloso, T. G. R., Leite, T. S., Pereira, O. L., Mizubuti, E. S. G., et al. (2018). Diversity of culturable endophytic fungi of Hevea guianensis: a latex producer native tree from the Brazilian Amazon. Afr. J. Microbiol. Res. 12, 953–964. doi: 10.5897/AJMR2018.8980
Bae, H., Sicher, R. C., Kim, M. S., Kim, S. H., Strem, M. D., Melnick, R. L., et al. (2009). The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao. J. Exp. Bot. 60, 3279–3295. doi: 10.1093/jxb/erp165
Baiyee, B., Pornsuriya, C., Ito, S. I., and Sunpapao, A. (2019). Trichoderma spirale T76-1 displays biocontrol activity against leaf spot on lettuce (Lactuca sativa L.) caused by Corynespora cassiicola or Curvularia aeria. Biol. Control 129, 195–200. doi: 10.1016/j.biocontrol.2018.10.018
Baldoni, D. B., Antoniolli, Z. I., Mazutti, M. A., Jacques, R. J. S., Dotto, A. C., Silveira, A. D. O., et al. (2020). Chitinase production by Trichoderma koningiopsis UFSMQ40 using solid state fermentation. Braz. J. Microbiol. 51, 1897–1908. doi: 10.1007/s42770-020-00334-w
Bissett, J. (1991). A revision of the genus Trichoderma. III. Section Pachybasium. Can. J. Bot. 69, 2373–2417. doi: 10.1139/b91-298
Brankovics, B., van Dam, P., Rep, M., de Hoog, G. S., Van Der Lee, T. A., Waalwijk, C., et al. (2017). Mitochondrial genomes reveal recombination in the presumed asexual fusarium oxysporum species complex. BMC Genomics 18:735. doi: 10.1186/s12864-017-4116-5
Cai, F., and Druzhinina, I. S. (2021). In honor of John Bissett: authoritative guidelines on molecular identification of Trichoderma. Fungal Divers. 107, 1–69. doi: 10.1007/s13225-020-00464-4
Carbone, I., and Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.1080/00275514.1999.12061051
Chaverri, P., Branco-Rocha, F., Jaklitsch, W., Gazis, R., Degenkolb, T., and Samuels, G. J. (2015). Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia 107, 558–590. doi: 10.3852/14-147
Chaverri, P., Castlebury, L. A., Overton, B. E., and Samuels, G. J. (2003). Hypocrea/Trichoderma: species with conidiophore elongations and green conidia. Mycologia 95, 1100–1140. doi: 10.1080/15572536.2004.11833023
Chaverri, P., Gazis, R. O., and Samuels, G. J. (2010). Trichoderma amazonicum, a new endophytic species on Hevea brasiliensis and H. guianensis from the Amazon basin. Mycologia 103, 139–151. doi: 10.3852/10-078
Contreras-Cornejo, H. A., Rodriquez, L., Del-Val, E., and Larsen, J. (2016). Ecological functions of Trichoderma spp. and their secondary metabolites in the rhizosphere: interactions with plants. FEMS Microbiol. Ecol. 92, 1–17. doi: 10.1093/femsec/fiw036
Crozier, J., Thomas, S. E., Aime, M. C., Evans, H. C., and Holmes, K. A. (2006). Molecular characterization of fungal endophytic morphospecies isolated from stems and pods of Theobroma cacao. Plant Pathol. 55, 783–791. doi: 10.1111/j.1365-3059.2006.01446.x
De Souza, J. T., Bailey, B. A., Pomella, A. W. V., Erbe, E. F., Murphy, C. A., Bae, H., et al. (2008). Colonization of cacao seedlings by Trichoderma stromaticum, a mycoparasite of the witches’ broom pathogen, and its influence on plant growth and resistance. Biol. Control 46, 36–45. doi: 10.1016/j.biocontrol.2008.01.010
Degenkolb, T., Dieckmann, R., and Nielsen, K. F. (2008). The Trichoderma brevicompactum clade: a separate lineage with new species, new peptaibiotics, and mycotoxins. Mycol. Prog. 7, 177–219. doi: 10.1007/s11557-008-0563-3
Dettman, J. R., Jacobson, D. J., Turner, E. I., Jeremy, R., Pringle, A., and Taylor, J. W. (2003). Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution 57, 2721–2741. doi: 10.1111/j.0014-3820.2003.tb01515.x
Dey, U., Harlapur, S. I., and Dhutraj, D. N. (2013). Bioassay of fungicides, botanicals, bioagents and indigenous technology knowledge (itk) against Pyricularia oryzae Cav. Causal agent of blast of rice. J. Plant Dis. Sci. 8, 102–106. doi: 10.5897/AJAR2013.7770
Druzhinina, I. S. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 9, 749–759. doi: 10.1038/nrmicro2637
Druzhinina, I. S., Kopchinskiy, A. G., and Kubicek, C. P. (2006). The first 100 Trichoderma species characterized by molecular data. Mycoscience 47, 55–64. doi: 10.1007/S10267-006-0279-7
Evans, H. C., Holmes, K. A., and Thomas, S. E. (2003). Endophytes and mycoparasites associated with an indigenous forest tree, Theobroma gileri, in Ecuador and a preliminary assessment of their potential as biocontrol agents of cocoa diseases. Mycol. Prog. 2, 149–160. doi: 10.1007/s11557-006-0053-4
Gazis, R., and Chaverri, P. (2010). Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecol. 3, 240–254. doi: 10.1016/j.funeco.2009.12.001
Gond, S. K., Verma, V. C., Kumar, A., Kumar, V., and Kharwar, R. N. (2007). Study of endophytic fungal community from different parts of Aegle marmelos Correae (Rutaceae) from Vanasi (India). World J. Microbiol. Biotechnol. 23, 1371–1375. doi: 10.1007/s11274-007-9375-x
Gu, X., Wang, R., Sun, Q., Wu, B., and Sun, J. Z. (2020). Four new species of Trichoderma in the Harzianum clade from northern China. MycoKeys 73, 109–132. doi: 10.3897/mycokeys.73.51424
Guigón-López, C., Vargas-Albores, F., Guerrero-Prieto, V., Ruocco, M., and Lorito, M. (2015). Changes in Trichoderma asperellum enzyme expression during parasitism of the cotton root rot pathogen Phymatotrichopsis omnivora. Fungal Biol. 119, 264–273. doi: 10.1016/j.funbio.2014.12.013
Harada, M. L., Schneider, H., Schneider, M. C., Sampaio, I., Czelusniak, J., and Goodman, M. (1995). DNA evidence on the phylogenetic systematics of New World monkeys: support for the sister-grouping of Cebus and Saimiri from two unlinked nuclear genes. Mol. Phylogenet. Evol. 4, 331–349. doi: 10.1006/mpev.1995.1029
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Holmes, K. A., Schroers, H. J., Thomas, S. E., Evans, H. C., and Samuels, G. J. (2004). Taxonomy and biocontrol potential of a new species of Trichoderma from the Amazon basin of South America. Mycol. Prog. 3, 199–210. doi: 10.1007/s11557-006-0090-z
Hora Júnior, B. T. D., de Macedo, D. M., Barreto, R. W., Evans, H. C., Mattos, C. R. R., and Maffia, L. A. (2014). Erasing the past: a new identity for the dam clean pathogen causing south American leaf blight of rubber. PLoS One 9:e104750. doi: 10.1371/journal.pone.0104750
Hoyos-Carvajal, L., Orduz, S., and Bissett, J. (2009). Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropical regions. Fungal Genet. Biol. 46, 615–631. doi: 10.1016/j.fgb.2009.04.006
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jaklitsch, W. M. (2009). European species of Hypocrea part I. the green-spored species. Stud. Mycol. 63, 1–91. doi: 10.3114/sim.2009.63.01
Jaklitsch, W. M. (2011). European species of Hypocrea part II: species with hyaline ascospores. Fungal Divers. 48, 1–250. doi: 10.1007/s13225-011-0088-y
Jaklitsch, W. M., Komon, M., Kubicek, C. P., and Druzhinina, I. S. (2005). Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97, 1365–1378. doi: 10.3852/mycologia.97.6.1365
Jaklitsch, W. M., Poldmaa, K., and Samuels, G. J. (2008). Reconsideration of Protocrea (Hypocreales, Hypocreaceae). Mycologia 100, 962–984. doi: 10.3852/08-101
Jaklitsch, W. M., Samuels, G. J., Dodd, S. L., Lu, B. S., and Druzhinina, I. S. (2006). Hypocrea rufa/ Trichoderma viride: a reassessment, and description of five closely related species with and without warted conidia. Stud. Mycol. 56, 135–177. doi: 10.3114/sim.2006.56.04
Jaklitsch, W. M., and Voglmayr, H. (2013). New combinations in Trichoderma (Hypocreaceae, Hypocreales). Mycotaxon 126, 143–156. doi: 10.5248/126.143
Jaklitsch, W. M., and Voglmayr, H. (2015). Biodiversity of Trichoderma (Hypocreaceae) in southern Europe and Macaronesia. Stud. Mycol. 80, 1–87. doi: 10.1016/j.simyco.2014.11.001
Katoch, M., Singh, D., Kapoor, K. K., and Vishwakarma, R. A. (2019). Trichoderma lixii (IIIM-B4), an endophyte of Bacopa monnieri L. producing peptaibols. BMC Microbiol. 19, 98–10. doi: 10.1186/s12866-019-1477-8
Kopchinskiy, A., Komoń, M., Kubicek, C. P., and Druzhinina, I. S. (2005). TrichoBLAST: a multiloci database of phylogenetic markers for Trichoderma and Hypocrea powered by sequence diagnosis and similarity search tools. Mycol. Res. 109, 658–660. doi: 10.1017/S0953756205233397
Kraus, G. F., Druzhinina, I., Gams, W., Bissett, J., Zafari, D., Szakacs, G., et al. (2004). Trichoderma brevicompactum sp. nov. Mycologia 96, 1059–1073. doi: 10.2307/3762089
Kuswinanti, T., Syamun, E., and Masniawati, A. (2015). The potency of endophytic fungal isolates collected from local aromatic rice as indole acetic acid (IAA) producer. Procedia Food Sci. 2016, 96–103. doi: 10.1155/2016/5897486
Leite, T. S., Cnossen-Fassoni, A., Pereira, O. L., Mizubuti, E. S. G., Araújo, E. F., and Queiroz, M. V. (2013). Novel and highly diverse fungal endophytes in soybean revealed by the consortium of two different techniques. J. Microbiol. 51, 56–69. doi: 10.1007/s12275-013-2356-x
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Mbarga, J. B., Begoude, B. A. D., Ambang, Z., Meboma, M., Kuate, J., Schiffers, B., et al. (2014). A new oil-based formulation of Trichoderma asperellum for the biological control of cacao black pod disease caused by Phytophthora megakarya. Biol. Control 77, 15–22. doi: 10.1016/j.biocontrol.2014.06.004
Meenakumari, T., Meenattoor, J. R., Thirunavoukkarasu, M., Vinod, K. K., Krishan, B., Gireesh, T., et al. (2018). Dynamics of long-term adaptive responses in growth and rubber yield among Hevea brasiliensis genotypes introduced to a dry sub-humid climate of eastern India. Ind. Crop. Prod. 119, 294–303. doi: 10.1016/j.indcrop.2018.02.066
Mukherjee, P. K., Horwitz, B. A., Herrera-Estrella, A., Schmoll, M., and Kenerley, C. M. (2013). Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51, 105–129. doi: 10.1146/annurev-phyto-082712-102353
Nath, V. S., John, N. S., Anjanadevi, I. P., Hegde, V. M., Jeeva, M. L., Misra, R. S., et al. (2014). Characterization of Trichoderma spp. antagonistic to Phytophthora colocasiae associated with leaf blight of taro. Ann. Microbiol. 64, 1513–1522. doi: 10.1007/s13213-013-0794-7
Nirenberg, H. (1976). Untersuchungen uber die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitt. Biol. Bundesanst. Land. Forstwirtsch Berl Dahl 169, 1–117.
Nuankeaw, K., Chaiyosang, B., Suebrasri, T., Kanokmedhakul, S., Lumyong, S., and Boonlue, S. (2020). First report of secondary metabolites, Violaceol I and Violaceol II produced by endophytic fungus, Trichoderma polyalthiae and their antimicrobial activity. Mycoscience 61, 16–21. doi: 10.1016/j.myc.2019.10.001
Nylander, J. A. A. (2004). MrModeltestv2. Program distributed by the author. Evolutionary biology Centre, Uppsala University, Sweden.
Pereira, J. C., Marques, N. P., Rodrigues, A., Oliveira, T. B., Boscolo, M., Da Silva, R., et al. (2015). Thermophilic fungi as new sources for production of cellulases and xylanases with potential use in sugarcane bagasse saccharification. J. Appl. Microbiol. 118, 928–939. doi: 10.1111/jam.12757
Pu, X., Qu, X., Chen, F., Bao, J., Zhang, G., and Luo, Y. (2013). Camptothecin-producing endophytic fungus Trichoderma atroviride LY357: isolation, identification, and fermentation conditions optimization for camptothecin production. Appl. Microbiol. Biotechnol. 97, 9365–9375. doi: 10.1007/s00253-013-5163-8
Qiao, M., Du, X., Zhang, Z., Xu, J., and Yu, Z. (2018). Three new species of soil-inhabiting Trichoderma from southwest China. MycoKeys. 44, 63–80. doi: 10.3897/mycokeys.44.30295
Qin, W. T., and Zhuang, W. Y. (2016a). Four new species of Trichoderma with hyaline ascospores in the Brevicompactum and Longibrachiatum clades. Mycosystema 15, 811–825. doi: 10.1007/s11557-016-1211-y
Qin, W. T., and Zhuang, W. Y. (2016b). Seven wood-inhabiting new species of the genus Trichoderma (fungi, Ascomycota) in Viride clade. Sci. Rep. 6:27074. doi: 10.1038/srep27074
Rannala, B., and Yang, Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Rivano, F., Mattos, C. R., Cardoso, S. E., Martinez, M., Cevallos, V., Le Guen, V., et al. (2013). Breeding Hevea brasiliensis for yield, growth and SALB resistance for high disease environments. Ind. Crop. Prod. 44, 659–670. doi: 10.1016/j.indcrop.2012.09.005
Rivera-Méndez, W., Obregón, M., Morán-Diez, M. E., Hermosa, R., and Monte, E. (2020). Trichoderma asperellum biocontrol activity and induction of systemic defenses against Sclerotium cepivorum in onion plants under tropical climate conditions. Biol. Control 141, 104–145. doi: 10.1016/j.biocontrol.2019.104145
Rodriguez, M. C. H., Evans, H. C., de Abreu, L. M., Macedo, D. M., Ndacnou, M. K., Bekele, B. K., et al. (2021). New species and records of Trichoderma isolated as mycoparasites and endophytes from cultivated and wild coffee in Africa. Sci. Rep. 11, 5671–5630. doi: 10.1038/s41598-021-84111-1
Samuels, G. J., Dodd, S. L., Lu, B. S., Samuels, G. J., Dodd, S. L., Lu, B. S., et al. (2006). The Trichoderma koningii aggregate species. Stud. Mycol. 56, 67–133. doi: 10.3114/sim.2006.56.03
Samuels, G. J., Petrini, O., Kuhls, K., Lieckfeldt, E., and Kubicek, C. P. (1998). The Hypocrea schweinitzii complex and Trichoderma sect. Longibrachiatum. Stud. Mycol. 41.
Saravanakumar, K., Yu, C., Dou, K., Wang, M., Li, Y., and Chen, J. (2016). Synergistic effect of Trichoderma-derived antifungal metabolites and cell wall degrading enzymes on enhanced biocontrol of fusarium oxysporum f. sp. cucumerinum. Biol. Control 94, 37–46. doi: 10.1016/j.biocontrol.2015.12.001
Shentu, X., Zhan, X., Ma, Z., Yu, X., and Zhang, C. (2014). Antifungal activity of metabolites of the endophytic fungus Trichoderma brevicompactum from garlic. Braz. J. Microbiol. 45, 248–254. doi: 10.1590/S1517-83822014005000036
Singh, S. K., Singh, P. N., Maurya, D. K., and Rana, S. (2020). “Advances in systematics, taxonomy, and conservation of Trichoderma species” in Trichoderma: Agricultural Applications and Beyond. eds. C. Manoharachary, H. B. Singh, and A. Varma (Switzerland AG: Springer Nature), 1–20.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Swofford, D. L., Waddell, P. J., Huelsenbeck, J. P., Foster, P. G., Lewis, P. O., and Rogers, J. S. (2001). Bias in phylogenetic estimation and its relevance to the choice between parsimony and likelihood methods. Syst. Biol. 50, 525–539. doi: 10.1080/106351501750435086
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Taylor, J. W., Jacobson, D. J., Kroken, S., Kasuga, T., Geiser, D. M., Hibbett, D. S., et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31, 21–32. doi: 10.1006/fgbi.2000.1228
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multigene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vitti, A., Pellegrini, E., Nali, C., Lovelli, S., Sofo, A., Valerio, M., et al. (2016). Trichoderma harzianum T-22 induces systemic resistance in tomato infected by cucumber mosaic virus. Front. Plant Sci. 7:1520. doi: 10.3389/fpls.2016.01520
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR protocols: a guide to the methods and applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York: Academic Press), 315–322.
Wirsel, S. G. R., Leibinger, W., Ernst, M., Mendgen, K., and Mendgen, K. (2001). Genetic diversity of fungi closely associated with common reed. New Phytol. 149, 589–598. doi: 10.1046/j.1469-137.2001.00038.x
Zhang, C. L., Liu, S. P., Lin, F. C., Kubicek, C. P., and Druzhinina, I. S. (2007). Trichoderma taxi sp. nov., an endophytic fungus from Chinese yew Taxus mairei. FEMS Microbiol. Lett. 270, 90–96. doi: 10.1111/j.1574-6968.2007.00659.x
Zheng, H., Qiao, M., Lv, Y., Du, X., Zhang, K. Q., and Yu, Z. (2021). New species of Trichoderma isolated as endophytes and saprobes from Southwest China. J. Fungi 7:467. doi: 10.3390/jof7060467
Zheng, C. J., Sun, P. X., Jin, G. L., and Qin, L. P. (2011). Sesquiterpenoids from Trichoderma atroviride, an endophytic fungus in Cephalotaxus fortunei. Fitoterapia 82, 1035–1038. doi: 10.1016/j.fitote.2011.06.010
Keywords: DNA barcoding, GCPSR, hevea, hypocreales, phylogeny, taxonomy
Citation: Nascimento Brito V, Lana Alves J, Sírio Araújo K, de Souza Leite T, Borges de Queiroz C, Liparini Pereira O and de Queiroz MV (2023) Endophytic Trichoderma species from rubber trees native to the Brazilian Amazon, including four new species. Front. Microbiol. 14:1095199. doi: 10.3389/fmicb.2023.1095199
Edited by:
Kezia Goldmann, Helmholtz Association of German Research Centers (HZ), GermanyReviewed by:
Karin Jacobs, Stellenbosch University, South AfricaAbolfazl Narmani, University of Tabriz, Iran
Copyright © 2023 Nascimento Brito, Lana Alves, Sírio Araújo, de Souza Leite, Borges de Queiroz, Liparini Pereira and de Queiroz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marisa Vieira de Queiroz, mvqueiro@ufv.br;; mvqueiro@hotmail.com
 Vanessa Nascimento Brito1
Vanessa Nascimento Brito1 Marisa Vieira de Queiroz
Marisa Vieira de Queiroz