- 1Department of Laboratory Medicine, The Affiliated Hospital of Southwest Medical University, Luzhou, China
- 2Department of Respiratory and Critical Care Medicine, The Affiliated Hospital of Southwest Medical University, Luzhou, China
- 3Department of Laboratory Medicine, Southwest Medical University, Luzhou, China
Emergence of blaNDM–1 and blaKPC–2 co-producing Klebsiella pneumoniae strains is currently attracting widespread attention, but little information is available about their tigecycline resistance, virulence, and prevalence in Southwest China. In July 2021, an extensively drug-resistant K. pneumoniae strain AHSWKP25 whose genome contained both blaNDM–1 and blaKPC–2 genes was isolated from the blood of a patient with the malignant hematological disease in Luzhou, China. We investigated the resistance profiles of AHSWKP25 using microbroth dilution, agar dilution, modified carbapenemase inactivation (mCIM), and EDTA-modified carbapenemase inactivation methods (eCIM). The virulence of AHSWKP25 was assessed through string tests, serum killing assays, and a Galleria mellonella larval infection model. Conjugation and plasmid stability experiments were conducted to determine the horizontal transfer capacity of plasmids. And efflux pump phenotype test and real-time quantitative reverse transcription-PCR (RT-PCR) were used to determine its efflux pump activity. Sequencing of AHSWKP25 determined that AHSWKP25 belonged to ST464, which is resistant to antibiotics such as carbapenems, tetracycline, fluoroquinolones, tigecycline, and fosfomycin. The efflux pump phenotype tests and RT-PCR results demonstrated that efflux pumps were overexpressed in the AHSWKP25, which promoted the tigecycline resistance of the bacteria. AHSWKP25 also showed hypervirulence and serum resistance in vitro model. AHSWKP25 carried several different plasmids that contained blaNDM–1, blaKPC–2, and mutated tet(A) genes. Sequence alignment revealed that the plasmids carrying blaNDM–1 and blaKPC–2 underwent recombination and insertion events, respectively. We demonstrated that an X3 plasmid carrying blaNDM–1 was transferred from pSW25NDM1 to E. coli J53. We also identified missense mutations in the ramR, rcsA, lon, and csrD genes of AHSWKP25. Our results highlighted the potential of blaNDM–1 and blaKPC–2 co-producing K. pneumoniae strains to further develop antimicrobial resistance and hypervirulent phenotypes, but measures should be taken to closely monitor and control the spread of superbugs with multidrug-resistant phenotypes and hypervirulence.
Introduction
Klebsiella pneumoniae is a common Gram-negative bacteria without spores or flagella that belongs to the Bacillus genus. In clinical infections involving K. pneumoniae, the classical (cKp) and hypervirulent (hvKp) strains are the most common, and the hvkp strains are considerably more dangerous because they can infect multiple organs, leading to high mortality rates from multiorgan failure. The pathogenicity of hvKp is usually attributed to multiple virulence factors, one of the most representative of which is the capsular antigen, which not only enhances virulence but also allows the bacterium to evade the host’s immune response (Podschun and Ullmann, 1998). There are six main types of the capsular antigen that enable the invasion of the species: K1, K2, K5, K54, K20, and K57. These antigens together with other virulence factors form the hypervirulence phenotype of K. pneumoniae (Turton et al., 2010; Shon et al., 2013). K1 and K2 are the most well-described capsular serotypes and are often detected in the K. pneumoniae multilocus sequence types ST23 and ST65 (Liao et al., 2014). Virulence plasmids are a significant marker of these hvkp strains (Shon et al., 2013). The rmpA gene product regulates the synthesis of capsular polysaccharides, conferring a hypermucoviscous phenotype on these pathogens (Cheng et al., 2010). A recent study revealed that, in addition to rmpA, the presence of rmpC and rmpD genes at the rmp locus also contributed to the formation of the hypermucoviscous K. pneumoniae phenotype (Walker et al., 2020). However, not all hypervirulent strains have a mucinous phenotype or carry the rmpA/rmpA2 or iucA genes (Yan et al., 2021). One study indicated that mutations in the genes (wzi, wza, wzc, rcsAB, and lon) that encode the enzymes and regulators responsible for capsule production contributed to aberrant capsule production that promoted the pathogenicity and antiserum phagocytosis of K. pneumoniae strains. And these mutants producing hypercapsule are often associated with bloodstream infection (Ernst et al., 2020). Therefore, more studies are required to better understand the hvKp strains.
The resistance status of hvKp is not promising. Like cKp, hvKp can acquire resistance to antimicrobial agents with the acquisition of mobile elements carbapenem-resistant-hvKp is a typical example of a stain that can acquire resistance to various antimicrobial agents by obtaining exogenous plasmids carrying antimicrobial resistance genes (Yao et al., 2015). cKp can also develop into hypervirulent strains by acquiring virulence plasmids. The high pathogenicity and antimicrobial resistance of these pathogens translate to high treatment costs and poor prognoses (Shankar et al., 2018). Colistin and tigecycline represent the last line of defense against carbapenem-resistant Enterobacteriaceae bacteria, especially metallo-beta-lactamase-producing strains (Maltezou, 2009). The horizontal transfer of mcr, tet(A), and tet(X) allowed K. pneumoniae to rapidly acquire resistance to colistin and tigecycline. Also, disruptions in chromosomal genes (mgrB, pmrAB, and phoPQ), as well as resistance nodulation cell division (RND)-type efflux transporters and their regulators (marRA, ramRA, and rarA), are prominently responsible for conferring resistance of these microorganisms to antimicrobial agents (Osei Sekyere et al., 2016; Poirel et al., 2017). Recently, one study reported the emergence of blaNDM–1 and blaKPC–2 co-producing K. pneumoniae strains that could have both high-level carbapenem resistance and hypervirulent phenotypes (Liu et al., 2019; Gao et al., 2020). However, little information is available on the virulence characteristics, or tigecycline resistance of the blaNDM–1 and blaKPC–2 co-producing K. pneumoniae strains. To monitor these bacteria and prevent the development of further antimicrobial resistance, more information on these strains is required. In this study, we isolated an extensively drug-resistant K. pneumoniae isolate (AHSWKP25) from the blood of a patient with a bloodstream infection. AHSWKP25 was found to be an isolate harboring blaNDM–1, blaKPC–2, and tet(A) with an uncommon sequence type and was resistant to tigecycline and fosfomycin. In this study, we examined the virulence, genetic characteristics, and resistance mechanisms of AHSWKP25.
Materials and Methods
Source of the Isolate
In July 2021, AHSWKP25 was isolated from the blood of a patient diagnosed with acute myeloid leukemia with a bloodstream infection at the Affiliated Hospital of Southwest Medical University (Luzhou, China). The patient had previously been tread with ceftazidime, meropenem, moxifloxacin, tigecycline, and voriconazole for fungal infection and repeated fever. The clinical microbiology laboratory identified AHSWKP25 as K. pneumonia that was resistant to carbapenems, tigecycline, fluoroquinolones, and other common antimicrobial agents. The patient ultimately died of multiorgan failure. To better understand the antimicrobial resistance mechanisms and virulence characteristics of AHSWKP25, we undertook a series of experiments described below.
Antimicrobial Susceptibility Testing, Efflux Pump Phenotype Test, and DNA Amplification
The antimicrobial minimum inhibitory concentrations (MICs) of ceftazidime, cefepime, gentamicin, amikacin, chloramphenicol, ciprofloxacin, levofloxacin, tetracycline, polymyxin B, ceftazidime/avibactam, fosfomycin (agar dilution method), and tigecycline were determined according to the (Clinical and Laboratory Standards Institute (CLSI), 2020) standards. The MIC breakpoints of imipenem, ertapenem, aztreonam, levofloxacin, nitrofurantoin, and trimethoprim-sulfamethoxazole were determined using the MicroScan Walk Away System (Siemens, Germany). The breakpoint of tigecycline was interpreted according to the guidelines of the United States Food and Drug Administration (FDA1) on Antimicrobial Susceptibility Testing. The modified carbapenemase inactivation (mCIM) and EDTA-modified carbapenemase inactivation methods (eCIM) were also used following the CLSI 2020 standards.
To evaluate the efflux pump activity of AHSWKP25, we first measured the changes in the MICs of several antimicrobial agents in the presence of 1-(1-Naphthylmethyl)-piperazine (NMP, 100 mg/L). A fourfold or greater reduction in MIC was considered an indicator of overexpression in the efflux pumps (Schumacher et al., 2006). Polymerase chain reaction (PCR) was performed to detect whether the isolate carried the blaKPC or blaNDM genes, and the primers used are listed in Supplementary Table 1.
String Test, Serum Killing Assay, and Galleria mellonella Infection Model
The hypermucoviscous phenotype was determined using a string test. A single purified colony on a blood agar plate incubated at 37°C for 18-24 hours was picked with an inoculation loop for string, and the string test was considered positive if the length of the viscous string was longer than 5 mm (Yan et al., 2021). A serum killing assay was performed as described previously. In brief, 25 μL of the bacterial suspension (concentration of ∼1 × 106 CFU/mL) was added to 75 μL of healthy human serum for co-culture. After 0, 1, 2, and 3 h, the plates were inoculated with the mixture and the numbers of viable bacteria were calculated. The strains were classified as “highly sensitive,” “moderately sensitive,” or “resistant” to serum, depending on the results (Liu et al., 2017).
The pathogenicity of AHSWKP25 was assessed using a G. mellonella larvae infection model as previously described (Kim et al., 2021). A total of 15 healthy vigorous larvae were inoculated with the bacterial suspension of AHSWKP25 isolate at a dose equivalent to 106CFU. When the larvae were inactive and black, they were considered dead. The numbers of larval deaths were recorded every 12 h. K. pneumoniae NTUH-K2044 and ATCC 700603 were used as the hypervirulent control and negative control, respectively. The bacterial suspension was serially diluted and injected into G. mellonella larvae for 3 days of incubation, and the lethal dose 50 (LD50) was calculated using the probit model (Shi et al., 2018).
Whole Genome Sequencing, Identification of Mutant rcsAB, lon, csrD, and pal, Phylogenetic Reconstruction
Luria-Bertani (LB) broth was inoculated with the selected purified Klebsiella colonies, and the bacteria were cultured to log phase. Extraction of bacterial genomic DNA using a magnetic bead-based kit (Qiagen, Germany). The extracted bacterial DNA was purified and sequenced using the Illumina NovaSeq PE150 and Oxford nanopore platforms. The bacterial genome was assembled de novo using Canu (v 1.7). Prokka (v 1.10) was used to predict and annotate the coding genes, tRNAs, and rRNAs in the assembled genome. The plasmid replicon types, acquired resistance genes, sequence types, and virulence genes were determined using the online services of the Center for Genomic Epidemiology2 and VFDB (virulence factor database3). IS finder4 was used to identify the insertion sequences. The genomes were compared using the Basic Local Alignment Search Tool (BLAST5), BRIG v0.95, Mauve6, and the OAT software (Lee I. et al., 2016).
We used K. pneumoniae UCI 38 (accession number: JCMB01) as a reference strain to identify point mutations in the rcsAB and lon (Lon protease) genes of AHSWKP25 to evaluate whether AHSWKP25 was associated with hypercapsule production (Ernst et al., 2020). The rscD and pal mutants were identified using K. pneumoniae ATCC13883 (accession number: JOOW01) as the reference genome.
A goeBURST full MST analysis based on multilocus sequence typing data was performed by the Phyloviz 1.1a software to infer the phylogeny of the sequence types7. The Orthologous average nucleotide identity (OrthoANI) was calculated by comparing homologous genes between the genomes to construct a phylogenetic tree (Lee I. et al., 2016), which was visualized by iTOL8. The accession numbers of the genomes/sequences obtained from the NCBI database were listed in Supplementary Table 2.
Measurement of Efflux Pump Transcription Levels Using Quantitative Reverse Transcription-PCR
To evaluate the role of chromosomal point mutations in the antimicrobial resistance of AHSWKP25, we used RT-PCR to measure the transcriptional levels of efflux pump-encoding genes (acrA, acrB, marA, marR, rarA, and ramA) involved in conferring tigecycline resistance. In brief, single purified colonies of AHSWKP25 were inoculated into 5 mL of LB broth and the bacteria were grown to log phase. Their RNA was extracted according to the recommendations of the reagent’s manufacturer (Magen, China). The RT-PCR primers used in the present study are listed in Supplementary Table 1. The rpoB gene was used as the internal reference, and the tigecycline-susceptible K. pneumoniae strain NTUH-K2044 was used as the control strain. All experiments were performed in triplicate independently, with three biological replicates per experiment. The gene expression results were compared to the expression levels of rpoB to calculate the relative expression of the target genes (2–ΔΔCt method).
Conjugation Experiments and Plasmid Stability
Sodium azide-resistant Escherichia coli J53 was used as the recipient and AHSWKP25 as the donor in the conjugation experiments. The McFarland (McF) standard turbidity of the bacterial suspension was adjusted to 0.5 McF. The recipient bacteria (200 μL) and donor bacteria (400 μL) were inoculated into LB broth (800 μL) (Xiang et al., 2020). After 16–18 h of culturing at 35°C, MH plates containing 180 μg/mL sodium azide and antimicrobial agents [4 μg/mL meropenem; 4 μg/mL meropenem + 5 mM EDTA (final concentration); 0.25 μg/mL ciprofloxacin] were inoculated with 100 μL of the culture solution to screen for transconjugants. The transferred genes in the transconjugants were confirmed by PCR using the primers listed in Supplementary Table 1.
The stability of the self-transferred plasmids in the positive transconjugants was calculated using the plate count method, as described previously with some modifications (Nang et al., 2018). Briefly, antibiotic-free LB broth was inoculated with the positive transconjugants at a ratio of 1,000:1, and the bacteria were passaged continuously for 10 days. The mixed culture was sampled every day by inoculating aliquots (100 μL) of the mixed culture into meropenem-containing (4 μg/mL), antibiotic-free LB agar plates. The plates were incubated for 24 h to calculate the plasmid retention rate.
Results
Antimicrobial Susceptibility and General Characteristics of AHSWKP25
The antimicrobial susceptibility tests indicated that AHSWKP25 was resistant to common antibacterial agents, such as meropenem, tetracycline, ciprofloxacin, levofloxacin, and gentamicin. Notably, AHSWKP25 was also resistant to fosfomycin, tigecycline (16 μg/mL), and ceftazidime-avibactam, as shown in Table 1. The mCIM and eCIM results suggested that AHSWKP25 carries serine carbapenems. Subsequently, PCR and Sanger sequencing confirmed that AHSWKP25 was a blaNDM–1 and blaKPC–2 co-producing K. pneumonia isolate.
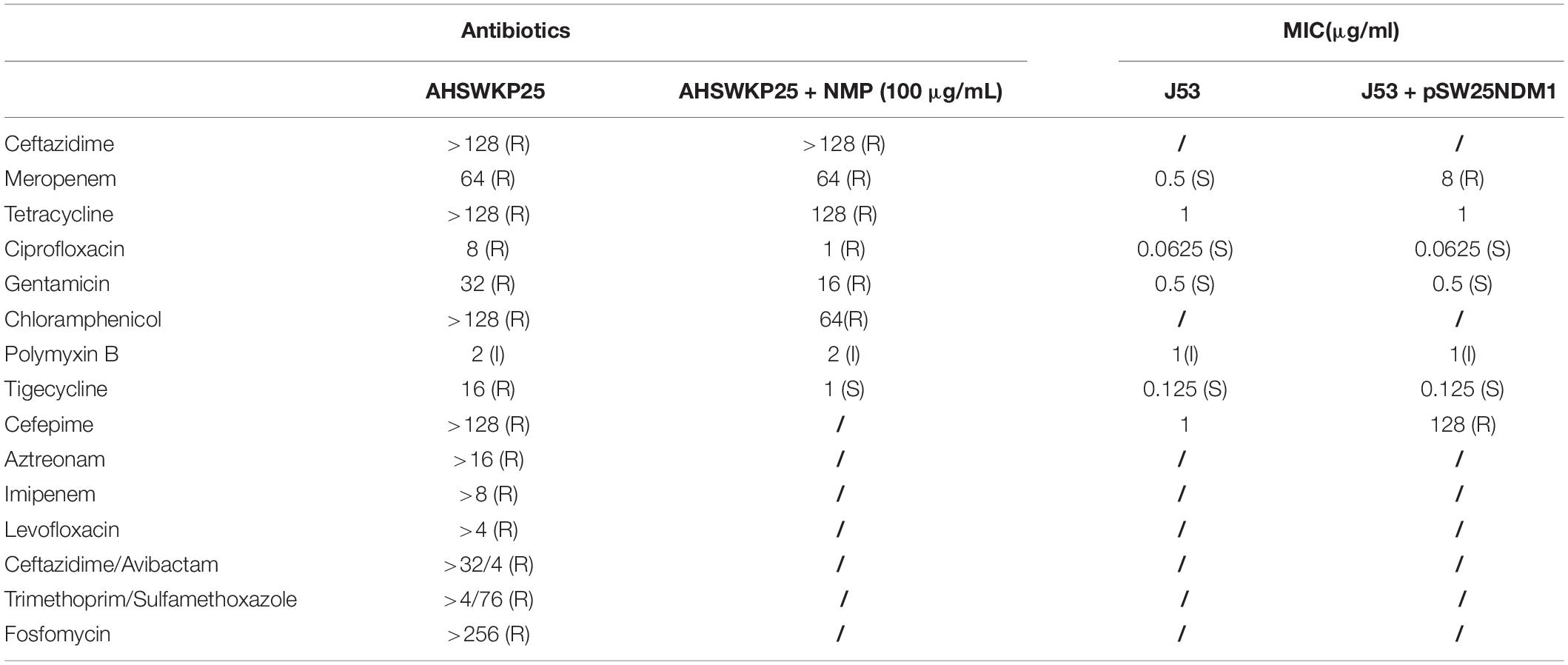
Table 1. Antibiotic susceptibility tests of AHSWKP25 and its conjugants, and efflux pump phenotype test of AHSWKP25.
The string test of AHSWKP25 was negative, as the mucoid string length was < 5 mm. AHSWKP25 was highly pathogenic in the G. mellonella infection model. Of the 15 G. mellonella larvae that were inoculated with 106 CFU of AHSWKP25, 15 died within 24 h, corresponding to a mortality rate of 100%, which was similar to the rate for the hypervirulent control strain NTUH-K2044 (P > 0.05) but different from that of K. quasipneumoniae ATCC700603 (negative control, P < 0.0001) (Figure 1B). The LD50 of AHSWKP25 was 4.44 ± 0.23 (log10 CFU), and the LD50 of the NTUH-K2044 was 4.21 ± 0.17 (log10 CFU) (P > 0.05, Supplementary Figure 1). Both NTUH-K2044 and AHSWKP25 showed resistance to the serum, whereas ATCC 700603 was highly sensitive to the serum (Figure 1A and Supplementary Table 3). To further investigate the virulence characteristics and resistance mechanisms of AHSWKP25, we sequenced its whole genome to identify mutations in the genes that regulate the resistance mechanisms.
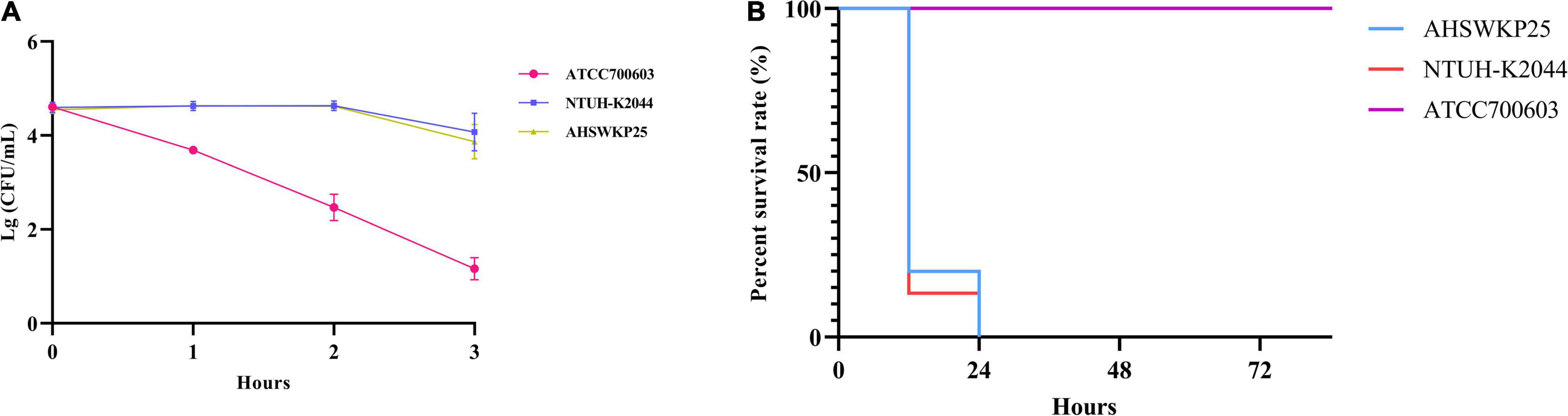
Figure 1. Serum resistance and pathogenicity of AHSWKP25.(A) The survival rate of AHSWKP25, ATCC700603, and NTUH-K2044 in healthy human serum over 3 h. The data are expressed as the mean ± SEM (standard error of the mean). (B) The survival rate of G. mellonella larvae (15 biological replicates) inoculated with AHSWKP25, NTUH-K2044, and ATCC700603, respectively. The experiments were repeated in triplicate independently.
Sequence Characteristics of the AHSWKP25 Genome
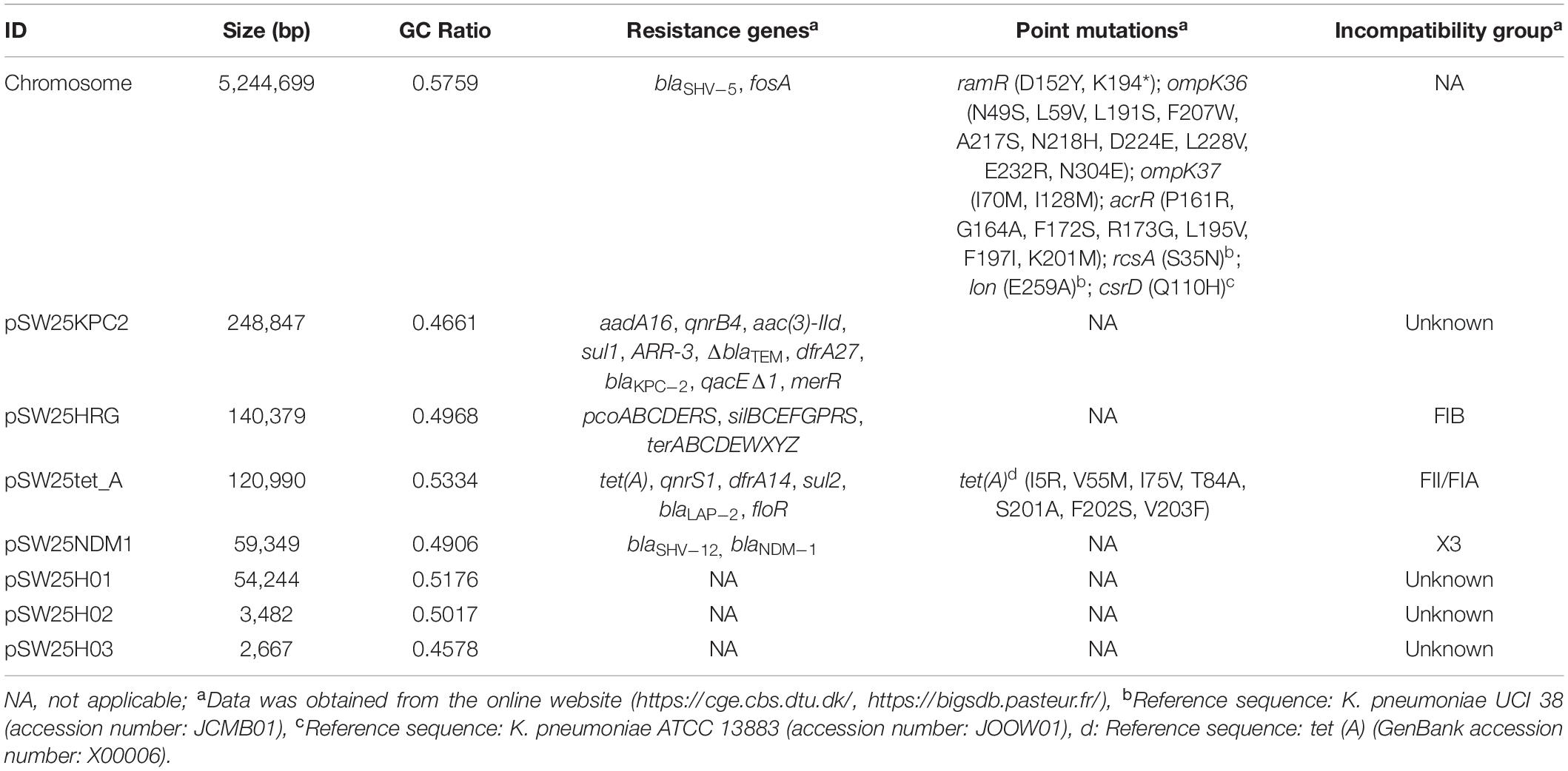
The full length of the AHSWKP25 genome was 5,874,657 bp, and it consisted of a chromosomal backbone carrying blaSHV–5 and seven circular plasmids. Of these, the plasmid incompatibility (Inc) groups for pSW25NDM1, pSW25HRG, and pSW25tet(A) were X3, FIB, and FII/FIA, respectively. The blaKPC–2 and blaNDM–1 genes were located on plasmids pSW25KPC2 and pSW25NDM1, respectively. The AHSWKP25 genome was longer than K. pneumoniae MGH78578 (5,694,894 bp, GenBank accession number: CP000647-652) and ATCC13883 (5,545,784 bp, accession number: JOOW01). The capsular serotype of AHSWKP25 was identified as K53 based on the wzc locus, which was consistent with the serotype of K. quasipneumoniae ATCC700603 (GenBank accession number: CP014696.2). The wzi allele (wzi outer membrane protein of cluster) of AHSWKP25 was most similar to allele 534 but it differed by one base pair. Therefore, we uploaded the wzi allele of AHSWKP25 into a public database9 and assigned a novel profile to it defined as wzi 725. The mrkABCDFHIJ, fimABCDEFGHIK, rcsAB, entABCDEFS, and iroE genes were all identified on the chromosomal backbone of AHSWKP25, whereas the iucA and rmpA/rmpA2 genes were not detected (Supplementary Table 4). AHSWKP25 shared an average nucleotide identity (ANI) of 98.97 and 98.96% with K. pneumonia MGH78578 and ATCC 13883 (Supplementary Figure 2), respectively. The multilocus sequence type of AHSWKP25 was ST464, which differed from ST4292 and ST2439 by only one allele, whereas ST464 differed from ST11 by five alleles (Supplementary Figure 3). The amino acid substitutions in ramR, acrR, and ompK36/ompK37 genes of AHSWKP25, as shown in Table 2. Furthermore, we also identified Ser35Asn, Glu259Ala, and Gln110His missense mutations in the rcsA, lon, and csrD genes of AHSWKP25, respectively. However, no missense mutations were found in the pal gene. A phylogenetic tree generated from orthoANI data revealed that AHSWKP25 and the K. pneumoniae strains co-producing NDM-1 and KPC-2 previously isolated from other regions of China (Liu et al., 2019; Gao et al., 2020; Xu et al., 2020) had obvious regional and temporal differences and were significantly separated phylogenetically (Figure 2).
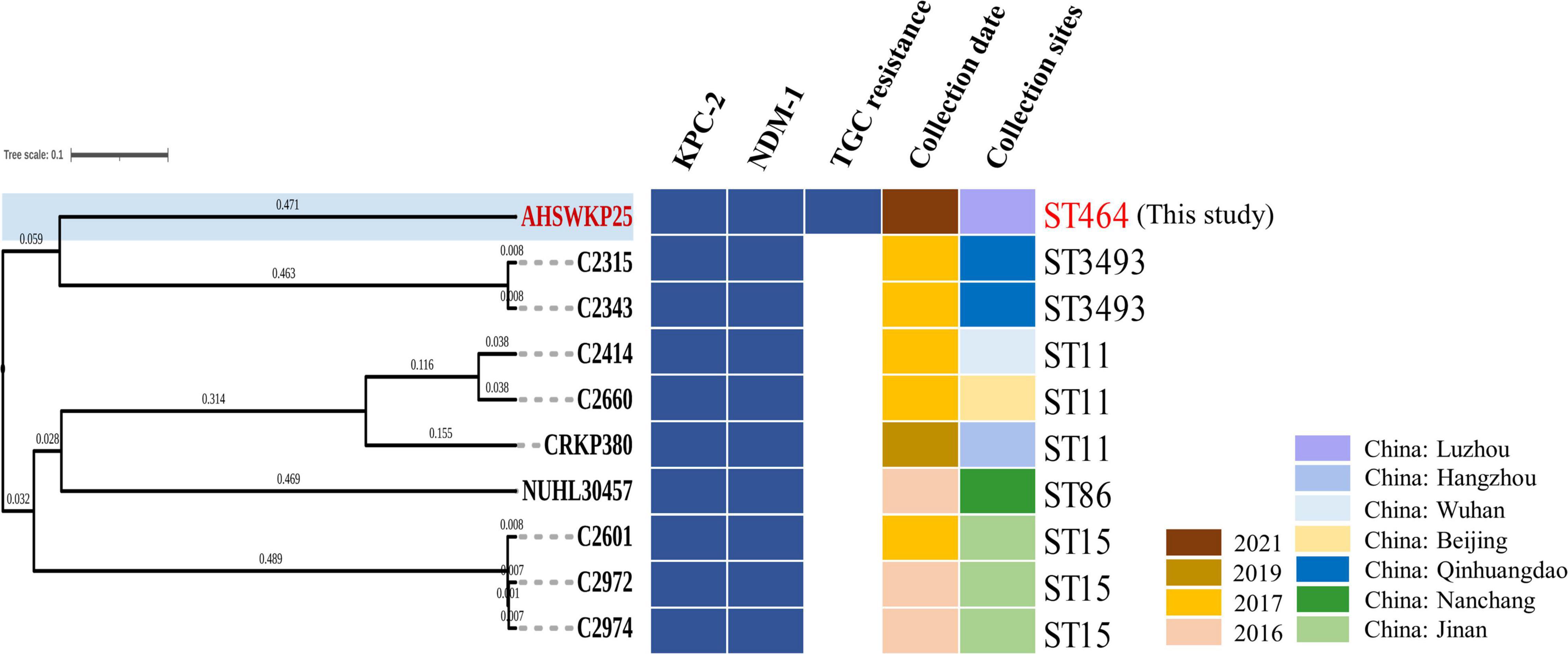
Figure 2. OrthoANI-based phylogenetic tree of AHSWKP25 and other 9 NDM-1 and KPC-2 co-producing K. pneumoniae strains previously isolated from other parts of China. The different colors, respectively represented the resistance genes, collection date, and sites, blanks represent strains that are susceptible to tigecycline.
Sequence Analysis of Plasmids Carried by AHSWKP25
Among the seven circular plasmids carried by AHSWKP25, we focused on four of them, namely pSW25KPC2, pSW25HRG, pSW25tet_A, and pSW25NDM1. pSW25KPC2 was a circular plasmid with a length of 248,847 bp, but we failed to identify its incompatibility group with Plasmid Finder 2.1. This plasmid carried multiple resistance determinants, including blaKPC–2 (beta-lactam resistance), aadA16 (aminoglycosides resistance), aac(3)-IId (aminoglycosides resistance), sul1 (sulfamethoxazole resistance), dfrA27 (trimethoprim resistance), qnrB4 (quinolones resistance), ARR-3 (rifampicin resistance), merR [Hg(II)-responsive transcriptional regulator] and qacEΔ1 (quaternary ammonium compound resistance) (Figure 3A). We searched the National Center for Biotechnology Information (NCBI) database and found that pSW25KPC2 shared a 99.97% identity (with an 92% query coverage) compared to the plasmid pYNKP001-dfrA isolated from R. ornithinolytica (GenBank accession number: KY270853.1). Sequence alignment indicated that the backbone of pSW25KPC2 was highly similar to the plasmids pKP04VIM (K. pneumoniae), pYNKP001-dfrA (R. ornithinolytica), and pRJA166a (K. pneumoniae), but there were multiple Local Colinear Blocks (LCBs) that were inverted (Figures 3A, 4A). Notably, pKP04VIM, pYNKP001-dfrA (K. pneumoniae), and pRJA166a (K. pneumoniae) did not carry the blaKPC–2 gene. Further analysis revealed that the blaKPC–2 gene of pSW25KPC2 was located in an LCB of about 6 kb that was highly homologous with pK55602_2 (GenBank accession number: CP042976.1) and pKPC2_095132 (GenBank accession number: CP028389.3) and was highly collinear with no inversions or rearrangements. The mapping of this LCB to Figure 4B revealed that pSW25KPC2, pK55602_2, and pKPC2_095132 all contained a conserved region carrying korC (encoding transcriptional repressor protein), ISKpn6, blaKPC–2, ISKpn27, Tn3, and a truncated blaTEM. Transposase TnAs1 (Tn3-like element) and IS4321 were also present on the pSW25KPC2 plasmid flanking a region encoding blaKPC–2. Additionally, a region encoding aac(3)-IId was inverted in pSW25KPC2 compared to pKPC2_095132 (Figure 4A). The Tn4401 transposon and its variants were not identified in pSW25KPC2. Lastly, pSW25KPC2 also contained a region connecting ISCR1 and intl1, that contained ARR-3-dfrA27-aadA16- qacEΔ1-sul1 genes (Figure 3A).
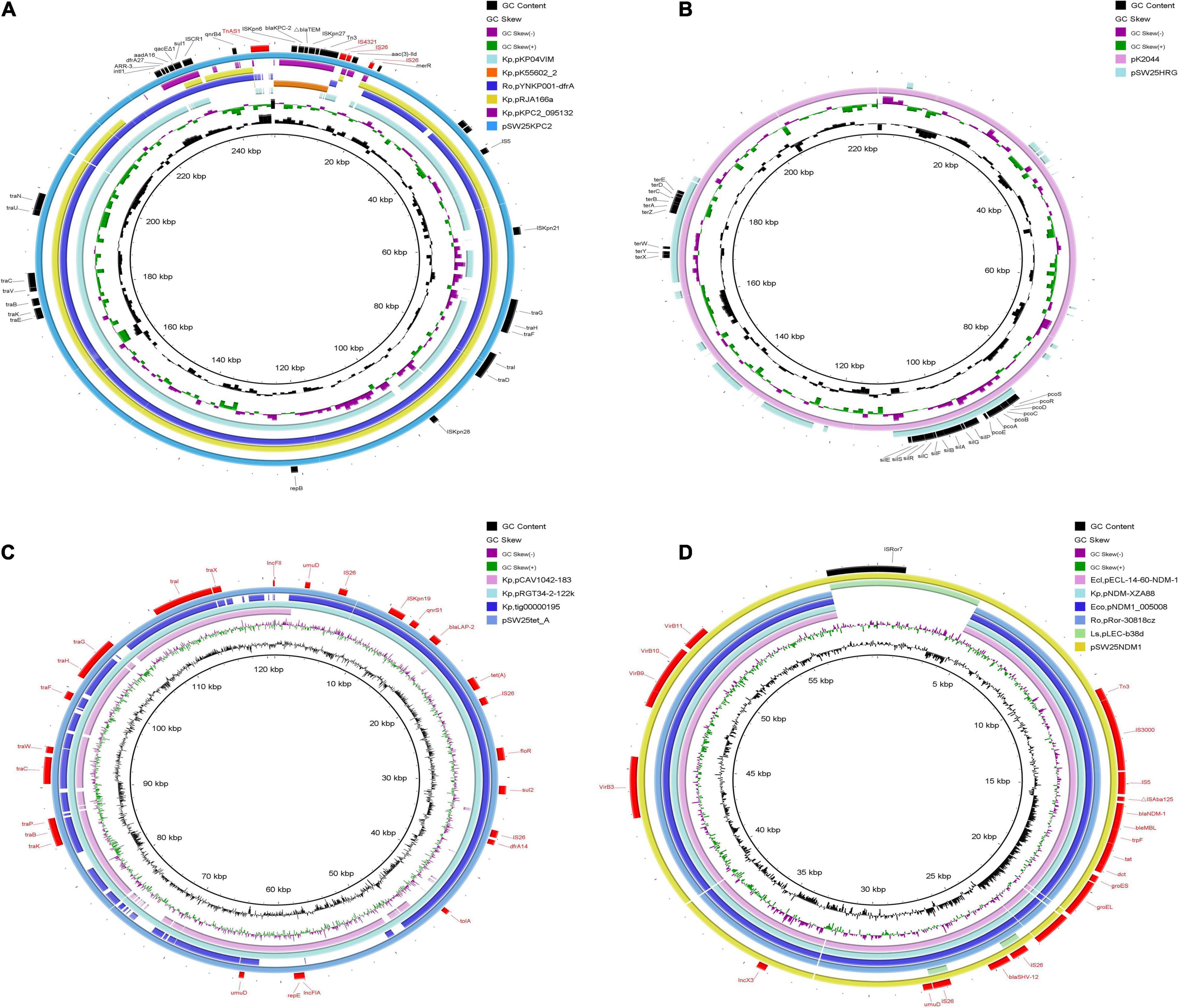
Figure 3. Comparative genomic circle of plasmids. (A) Circular comparison of pSW25KPC2 with other similar plasmids, pSW25KPC2 was used as a reference. (B) Circular comparison between pSW25HRG and virulence plasmid pK2044, pK2044 was used as a reference. (C) Circular comparison of pSW25tet_A with other similar plasmids, pSW25tet_A was used as a reference. (D) Circular comparison of pSW25NDM1 with other similar plasmids, pSW25NDM1 was used as a reference. The outermost circle annotates the genetic information, and different plasmids are assigned different colors. Blanks represent deleted regions compared to the reference plasmids. Kp: K. pneumoniae, Ro: R. ornithinolytica, Ecl: E. cloacae, Eco: E. coli, Ls: Leclercia sp.
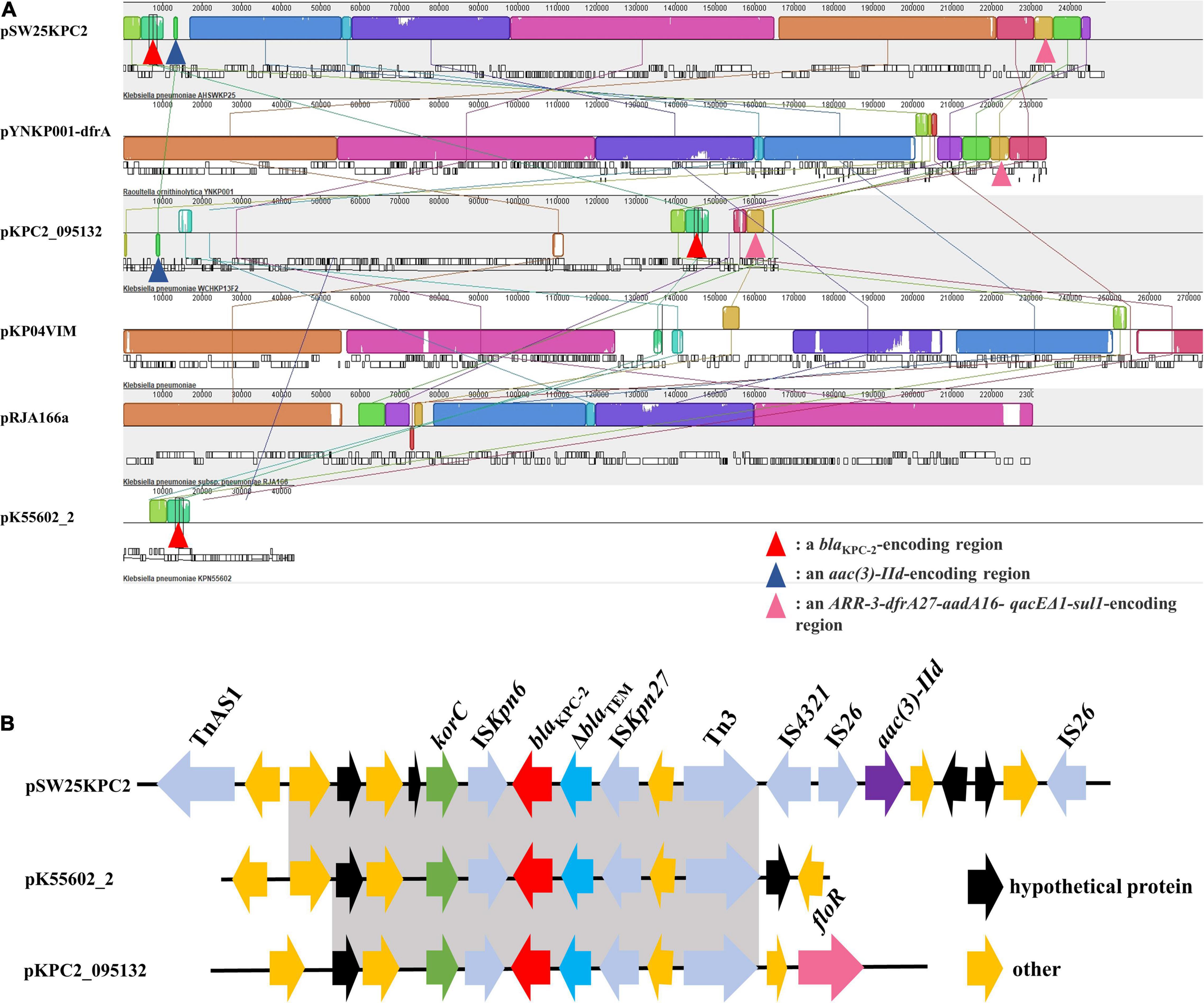
Figure 4. (A) Mauve alignment between pSW25KPC2 and other five plasmids. The grids of the same color represent collinear areas and are connected by lines. The blank area in the grid indicates that the sequences are incompletely aligned. (B) Comparison of genetic elements surrounding blaKPC–2 between pSW25KPC2 and plasmids carrying blaKPC–2. Gray areas represent homologous portions between sequences.
pSW25HRG was a FIB plasmid that did not carry any acquired antimicrobial resistance genes. There were several heavy metal resistance genes in the backbone of pSW25HRG: pcoABCDERS (copper resistance), silBCEFGPRS (silver resistance), and terABCDEWXYZ (tellurium resistance). Sequence alignments constructed with BLAST (see text footnote 5) revealed that most plasmid sequences similar to pSW25HRG have been isolated from K. pneumoniae strains. pSW25HRG shared a 99.93% identity (with an 80% query coverage) with a plasmid isolated from the K. pneumoniae strain FDAARGOS_1322 (GenBank accession number: CP070038.1) and a 99.90% identity (with an 83% query coverage) with a plasmid isolated from K. pneumoniae strain NICU_2_P7 (GenBank accession number: CP060050.1). Notably, pSW25HRG had a 23-kb homologous region compared to the well-known virulence plasmid pK2044 (GenBank accession number: NC_006625.1), wherein the above mentioned heavy-metal-resistance genes mainly occurred (Figure 3B).
The pSW25tet_A carried two replicon types, FII and FIA, and was 120,990 bp in length. Similar to pSW25KPC2, pSW25tet_A carried many genes encoding type IV conjugative transfer systems and multiple antimicrobial resistance determinants, including tet(A) (tetracycline resistance), dfrA14 (trimethoprim resistance), sul2 (sulfamethoxazole resistance), qnrS1 (quinolones resistance), and blalAP–2 (penicillins resistance). We identified that tet(A) in the pSW25tet_A was a type I mutant with a mutation profile of I5R, V55M, I75V, T84A, S201A, F202S, and V203F. This tet(A) mutant was previously demonstrated to be associated with reduced tigecycline susceptibility (Chiu et al., 2017). By searching the NCBI database, we found that pSW25tet_A was also similar to several other plasmids isolated from K. pneumoniae strains. Of these, pSW25tet_A shared a 99.97% identify (with a 100% query coverage) with a plasmid pRGT34-2-122k (GenBank accession number: CP075310.1) that was also isolated from K. pneumoniae. The homology between pSW25tet_A and pRGT34-2-122k was confirmed by comparing genomic circles (Figure 3C). pSW25tet_A shared only 28 and 29% coverage compared with pYUSHP2-2 (GenBank accession number: CP073773.1), a plasmid isolated from E. hormaechei, and pWP8-W19-CRE-01_3 (GenBank accession number: AP022271.1), a plasmid isolated from R. ornithinolytica, respectively.
Lastly, pSW25NDM1 was identified as an X3 plasmid carrying blaNDM–1 and blaSHV–12 genes. It was 59,349 bp in length and had a GC ratio of 49.06%. The genetic structure surrounding blaNDM–1 included IS3000, IS5, bleMBL, trpF, tat, dct, groES and groEL. pSW25NDM1 shared a 100% identity (with an 89% query coverage) with pECL-14-60-NDM-1 (GenBank accession number: MN061454.1), a plasmid isolated from E. cloacae. Sequence alignment revealed that pSW25NDM1 was very similar to plasmids isolated from K. pneumoniae, E. coli, E. cloacae, and R. ornithinolytica (Figure 3D). pSW25NDM1 contained an additional region of about 6 kb, which was not present in other plasmids carrying NDM-1, and it was highly homologous to a plasmid isolated from Leclercia sp. (GenBank accession number: CP026168.1) (Figure 3D). Further analysis showed that this region could be inserted by ISRo7 (marked black in Figure 3D). The blaSHV–12 was located in a 4-kb inserted region that could be inserted by IS26 at both ends.
Stable Dissemination and Expression of pSW25NDM1 Carrying blaNDM–1 Among Species
Among the three plasmids carrying the acquired antimicrobial resistance genes mentioned above, we only demonstrated the horizontal transferability of pSW25NDM1 in conjugation experiments. The pSW25NDM1 can be transferred to E. coli J53 at a transfer frequency of about 2.03 × 10–3. Recipient strain E. coli J53 showed significantly reduced susceptibility to meropenem and cefepime after the acquisition of plasmid pSW25NDM1 (Table 1). E. coli J53 carrying pSW25NDM1 displayed high plasmid stability during its continuous passage in an antibiotic-free environment for 10 days, and more than 95% of the recipient strains cells still retained pSW25NDM1 on day 10 (Supplementary Figure 4).
Efflux Pump Phenotype Test and RT-PCR
The MICs of ceftazidime and meropenem for AHSWKP25 did not change significantly after the addition of NMP (100 μg/mL). However, there were ≥ 4-fold reductions in the MICs of ciprofloxacin and chloramphenicol in the presence of NMP (100 μg/mL). And NMP restored the susceptibility of AHSWKP25 to tigecycline (Table 1). Furthermore, the RT-PCR results indicated that the expressions of acrA and acrB genes were up-regulated by 1.4-fold (1.43 ± 0.05) and 1.5-fold (1.54 ± 0.12), respectively, in AHSWKP25 compared to that in NTUH-K2044. The expressions of rarA, marA, and marR were up-regulated approximately twofold in AHSWKP25 (2.25 ± 0.24, 2.82 ± 0.43, 1.94 ± 0.19, respectively), while ramA was up-regulated 19-fold (19.18 ± 1.04) (Figure 5).
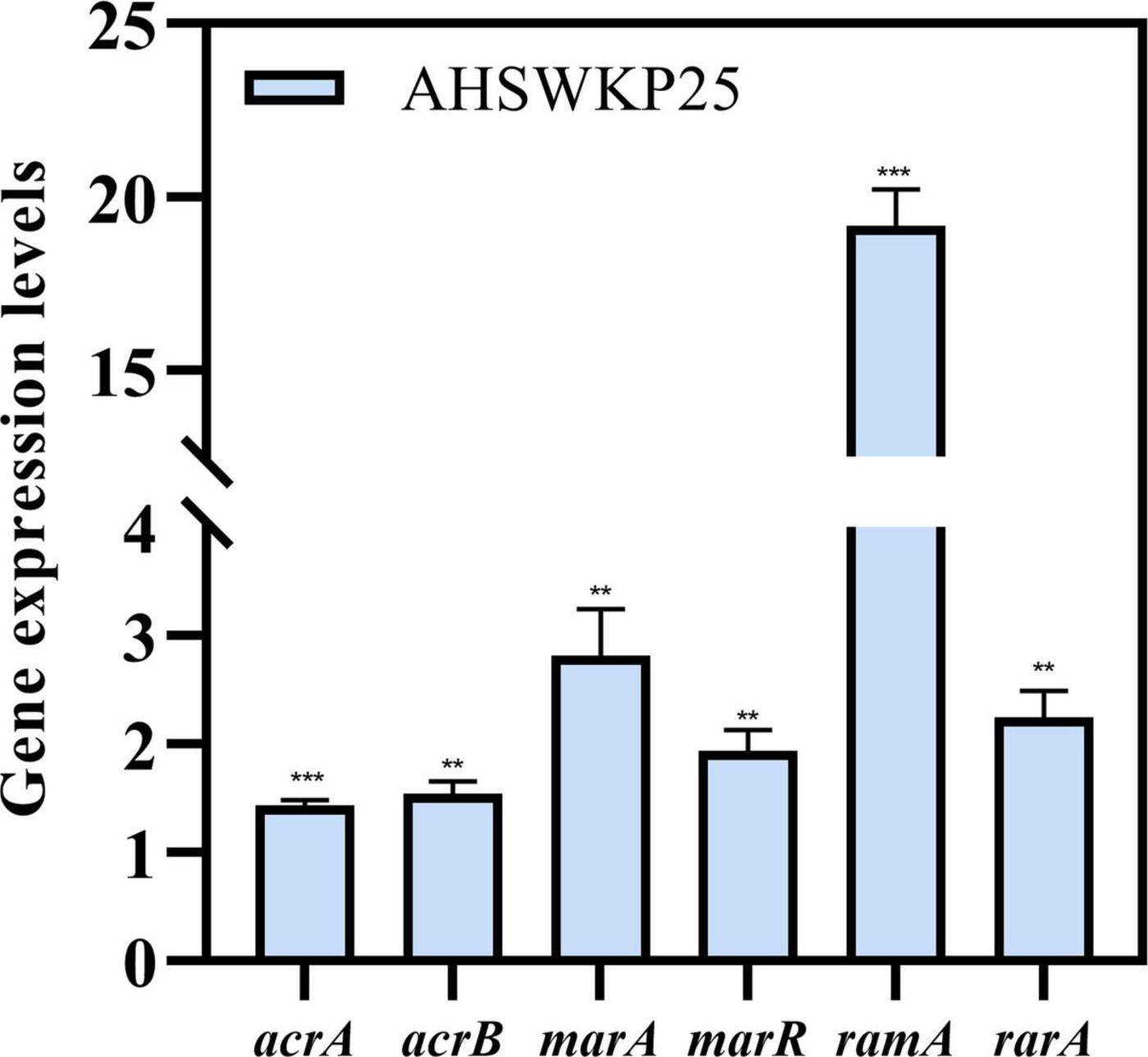
Figure 5. Relative expression levels of acrA, acrB, marA, marR, ramA, and rarA genes in AHSWKP25. Gene expression levels were normalized using the rpoB (RNA polymerase) gene. The experiment was repeated in triplicate independently, and data are expressed as the mean ± SD (standard deviation). **P < 0.01; ***P < 0.001 by Student’s t-test.
Discussion
In this study, we reported on a blaNDM–1 and blaKPC–2 co-producing K. pneumoniae isolate (AHSWKP25) that was isolated in Southwestern China. To the best of our knowledge, this is the first report of a tigecycline resistant K. pneumoniae wzi 725 strain co-producing blaNDM–1 and blaKPC–2 in China, and its resistance profiles meet the definition of extensively drug-resistant (Magiorakos et al., 2012). In particular, AHSWKP25 belonged to an uncommon sequence type ST464, that was previously identified in neonatal patients in a few countries and for which little formation is available (Rakovitsky et al., 2020; Sands et al., 2021). We searched the database and found here was a one-allele difference between ST464 and ST2439 and two-allele differences between ST464 and ST5620, respectively. ST464 is markedly different from the sequence types of other blaNDM–1 and blaKPC–2 co-producing K. pneumoniae strains that have been reported in previous studies, suggesting that the combination of blaKPC–2 and blaNDM–1 not only appeared in several common sequence types (Figure 2 and Supplementary Figure 3). Therefore, continuous monitoring of such bacteria is still necessary. To date, we have not isolated any other K. pneumoniae strains with the same resistance pattern and sequence type as AHSWKP25 from other patients and settings in the local hospital. However, the patient was dead, and we were unable to obtain further information regarding colonization. The AHSWKP25 genome contained seven circular plasmids carrying multiple mobile elements and acquired antimicrobial resistance genes, reflecting the high plasticity of the K. pneumoniae genome. In marked contrast to blaNDM–1 and blaKPC–2 co-producing K. pneumoniae strains previously isolated from other parts of China, AHSWKP25 was also resistant to fosfomycin and tigecycline (Figure 2). As a common tetracycline-resistance determinant of Gram-negative bacteria, tet(A) can increase the efflux of tetracyclines by activating the expression of the major facilitator superfamily (MFS) efflux pumps (Nguyen et al., 2014). Tigecycline belongs to the glycylcycline family of antibiotics and has a much stronger antimicrobial activity than both tetracycline and minocycline. tet(A) mutants are associated with reduced bacterial susceptibility to tigecycline (Linkevicius et al., 2016). Mutated tet(A) can act synergistically with defective ramR to significantly increase the level of tigecycline resistance in K. pneumoniae strains (Chiu et al., 2017). Because the patient in this study had previously received tigecycline therapy, we speculated that the selection pressure exerted by tigecycline might have promoted the formation of tet(A) mutants (Xu et al., 2021). Furthermore, overexpression of acrAB and ramA is another common mechanism of tigecycline resistance in Enterobacteriaceae (Zhong et al., 2014; Osei Sekyere et al., 2016). In this study, we determined the role of efflux pumps in the tigecycline resistance of AHSWKP25 using the efflux pump inhibition test and RT-PCR (Figure 4). the transcriptional regulation of acrAB and ramA was closely related to the mutation in ramR (Abouzeed et al., 2008). Previous studies showed that the A19V substitution mutation in the ramR gene was more common in tigecycline and carbapenem-resistant K. pneumoniae (Sheng et al., 2014; Chiu et al., 2017). However, we did not find the A19V mutation in the ramR of AHSWKP25, rather, we did identify a D152Y substitution mutation that was previously identified in a tigecycline-non-susceptible K. pneumoniae strain that did not carry tet(A) mutant (MIC: 4 μg/ml) (Cheng et al., 2020). Thus, we considered that the tigecycline resistance of AHSWKP25 could be manifested by the combination of the tet(A) variant and the ramR mutation. rarA encodes an AraC-type regulator that can also independently activate the AcrAB and OqxAB efflux pumps, endowing K. pneumoniae with a multidrug resistance phenotype and tigecycline resistance (Veleba et al., 2012; De Majumdar et al., 2013).
Of the three antibiotic resistance plasmids carried by AHSWKP25, only the X3 plasmid pSW25NDM1 containing the blaNDM–1 gene was shown to be horizontally transferred in the present conjugation experiments. The horizontal transfer of plasmids is often more restricted in stains carrying multiple plasmids compared to strains with only single plasmid. Distorting interactions, which can affect the horizontal transfer efficiency, have previously been observed between three plasmids carried by the same host, (Gama et al., 2017a). In addition to being inhibited by fertility inhibition systems (FIN), the co-transfer of multiple plasmids is also limited in plasmids with low conjugation rates (Gama et al., 2017b). Additionally, the horizontal transfer of these resistant plasmids may result in huge fitness costs to the recipient, thereby disturbing conjugation (San Millan and MacLean, 2017). Furthermore, the host ranges of plasmids pSW25KPC2, pSW25HRG, and pSW25tet_A could be relatively narrow (Figure 3). Therefore, when assessing the horizontal transfer capacity of multiple plasmids carried by the same host, the influence of several factors must be considered. Sequence alignment showed that although the backbones of pSW25KPC2 and pSW25NDM1 were homologous to those of other plasmids isolated from Enterobacteriaceae, the plasmids acquired other genes by mobile elements, demonstrating the evolutionary potential of these antibiotic resistance plasmids (Figure 3). Because ISCR1 can mobilize its adjacent sequences through rolling circle transposition, we speculated that the ISCR1 element was involved in the capture of multiple antimicrobial resistance genes, including ARR-3-dfrA27-aadA16-qacEΔ1-sul1 on pSW25KPC2 (Figure 3A; Cheng et al., 2016). Transposition events mediated by transposon Tn4401 were the predominant reason for the rapid spread of the blaKPC genes to different plasmids (Naas et al., 2008). While we did not identify the Tn4401 transposon and its variants in pSW25KPC2, the region on pSW25KPC2 encoding blaKPC–2 was homologous with other plasmids carrying blaKPC–2. In addition, the backbone of pSW25KPC2 was highly similar to other plasmids without blaKPC–2, suggesting that the acquisition of blaKPC–2 by pSW25KPC2 can be enabled by homologous recombination between plasmids (Figures 3A, 4). TnAs1, Tn3, and IS4321 may play a significant role in this process. Notably, our results revealed that pSW25NDM1 carrying blaNDM–1 was capable of horizontal transfer to the recipient strain E. coli J53 and a variety of other wide hosts (Figure 3D), highlighting the flexibility of plasmids carrying blaNDM–1 in the formation of blaNDM–1 and blaKPC–2 co-producing strains (Gao et al., 2020).
The virulence phenotype of AHSWKP25 was also a cause for concern (Figure 1). Our results also supported the observation that K. pneumoniae strains did not carry rmpA/rmpA2, iucA, and capsular serotype other than K1, K2, K5, K20, K54, and K57 could be had a hypervirulent phenotype (Supplementary Table 4). However, the accuracy of distinguishing cKp and hvKp strains based on the G. mellonella infection model was insufficient compared to the murine model (Russo and MacDonald, 2020). Therefore, the lack of a mammalian infection model for evaluating the pathogenicity of AHSWKP25 was a limitation of this study. In hvkp strains, the emergence of tigecycline resistance mediated predominantly by ramR mutations was accompanied by decreased mucoviscosity and serum resistance (Park et al., 2020; Di Pilato et al., 2022). This phenomenon can be explained by the fitness cost, meaning, in the absence of antibiotic selection, resistance mutations could disturb the physiological function of bacteria, resulting in reduced pathogenicity, growth rate, and competitiveness compared to susceptive strains accordingly (Andersson and Hughes, 2010). However, several recent studies have revealed that genes and their regulators responsible for capsule biosynthesis play a dominant role in the adaptive evolution of K. pneumoniae. Disruption to these genes (rsAB, lon, and csrD) can ultimately affect capsule production, especially if these disruptions promote capsule hyperproduction, which would lead to enhancements in the pathogenicity and serum resistance of K. pneumoniae (Ernst et al., 2020; Mike et al., 2021). And mutations in the pal are mainly associated with reduced virulence (Hsieh et al., 2013). As mentioned above, missense mutations were identified in the rcsA, lon, and csrD genes that regulate capsule production in AHSWKP25. In particular, the S35N missense mutation in the rcsA gene of the AHSWKP25 genome was previously identified in a hypervirulent and hypermucoviscous K. pneumoniae isolate with hypercapsule production (Morales-León et al., 2021). The hypercapsule production is not necessarily associated with hypermucoviscosity (Mike et al., 2021). Therefore, the hypervirulent phenotype of AHSWKP25 was more likely to be associated with hypercapsule production through the regulators of capsule production mutations, but the exact mechanism remains to be further studied. The Type 3 fimbriae-encoding genes mrkABCDFHIJ and the Type I fimbriae-encoding gene fimABCEFGHIK are common in ckp and hvkp strains and can promote bacterial adhesion during pathogenic infection and biofilm formation (Di Martino et al., 2003). Among the siderophores carried by AHSWKP25, enterobactin is produced in both cKp and hvKp, whereas salmonellin is found more commonly in hvKp (El Fertas-Aissani et al., 2013; Lee I. R. et al., 2016). Furthermore, the heavy metal resistance genes carried by pSW25HRG were highly homologous to the gene carried by the virulence plasmid pK2044 and might be associated with the homologous recombination in these variable regions carrying heavy metal resistance genes of virulence plasmids. Although previous studies of S. aureus and A. baumannii demonstrated a relationship between these heavy metal resistance genes and pathogenicity, their contribution to the virulence of K. pneumoniae strains is still unclear (Alquethamy et al., 2019; Lawal et al., 2021). Overall, we determined that virulence of AHSWKP25 differ from those of common hypervirulent strains, and our results suggested that the emergence of hvkp strains might not depend exclusively on the genetic backgrounds and that point mutations in chromosomal loci also contributed to the development of the hypervirulent phenotype (Ernst et al., 2020).
There were several other limitations of this study that must be addressed in subsequent studies. First, although we reported NDM-1 and KPC-2 co-producing K. pneumoniae in Southwest China for the first time, the local prevalence of such microorganisms requires further clarification. Second, as mentioned above, mammalian models are still necessary to further elucidate the virulence behavior of AHSWKP25 in the future. Third, we used only E. coli J53 as the recipient in the conjugation experiments, in subsequent studies, we will use K. pneumoniae stains as recipients to assess the horizontal transferability of these plasmids.
Conclusion
In conclusion, this study was the first to describe and sequence a hypervirulent tigecycline-resistant and serum-resistant K. pneumoniae strain containing both blaNDM–1 and blaKPC–2 in Southwestern China. The resistance phenotypes displayed by AHSWKP25 suggested that blaNDM–1 and blaKPC–2 co-producing K. pneumoniae strains can potentially develop further antimicrobial resistance. Notably, we identified missense mutations in the genes associated with hypercapsule production in AHSWKP25, which will provide valuable information for illustrating the formation mechanism of hypervirulent K. pneumoniae. Importantly, strict monitoring measures must be taken to prevent the spread of these superbugs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank, CP091048-CP091055 (K. pneumoniae AHSWKP25).
Ethics Statement
The study protocol was approved by the Institutional Review Board of the Affiliated Hospital of Southwest Medical University (Project No. KY2020043).
Author Contributions
JH, BZ, JD, and JL isolated AHSWKP25 and designed the study and experiments. JH, JD, and YW performed the assays. JH, BZ, and XX analyzed the data. JH and BZ drafted and revised the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was supported by the grants from Sichuan Science and Technology Department Program (20QYCX0056 and 2021YFH001), and Sichuan Education Department Training Program of Innovation and Entrepreneurship for College Students (S202010632250).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the team of the curators of the Institut Pasteur MLST system (Paris, France) for importing novel alleles, profiles, and/or isolates at http://bigsdb.pasteur.fr.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.868705/full#supplementary-material
Footnotes
- ^ https://www.fda.gov
- ^ http://www.genomicepidemiology.org/
- ^ http://www.mgc.ac.cn/VFs/
- ^ https://www-is.biotoul.fr/blast.php
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://sourceforge.net/projects/mauve/
- ^ http://www.phyloviz.net/
- ^ https://itol.embl.de/
- ^ http://bigsdb.pasteur.fr
References
Abouzeed, Y. M., Baucheron, S., and Cloeckaert, A. (2008). ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 52, 2428–2434. doi: 10.1128/aac.00084-08
Alquethamy, S. F., Khorvash, M., Pederick, V. G., Whittall, J. J., Paton, J. C., Paulsen, I. T., et al. (2019). The Role of the CopA Copper Efflux System in Acinetobacter baumannii Virulence. Int. J. Mol. Sci. 20:13. doi: 10.3390/ijms20030575
Andersson, D. I., and Hughes, D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271. doi: 10.1038/nrmicro2319
Cheng, C., Sun, J., Zheng, F., Lu, W., Yang, Q., and Rui, Y. (2016). New structures simultaneously harboring class 1 integron and ISCR1-linked resistance genes in multidrug-resistant Gram-negative bacteria. BMC Microbiol. 16:71. doi: 10.1186/s12866-016-0683-x
Cheng, H. Y., Chen, Y. S., Wu, C. Y., Chang, H. Y., Lai, Y. C., and Peng, H. L. (2010). RmpA Regulation of Capsular Polysaccharide Biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 192, 3144–3158. doi: 10.1128/jb.00031-10
Cheng, Y. H., Huang, T. W., Juan, C. H., Chou, S. H., Tseng, Y. Y., Chen, T. W., et al. (2020). Tigecycline-non-susceptible hypervirulent Klebsiella pneumoniae strains in Taiwan. J. Antimicrob. Chemother. 75, 309–317. doi: 10.1093/jac/dkz450
Chiu, S. K., Huang, L. Y., Chen, H., Tsai, Y. K., Liou, C. H., Lin, J. C., et al. (2017). Roles of ramR and tet(A) Mutations in Conferring Tigecycline Resistance in Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates. Antimicrob. Agents Chemother. 61, e391–e317. doi: 10.1128/aac.00391-17
Clinical and Laboratory Standards Institute. (2020). Performance Standards for Antimicrobial Susceptibility Testing, 30th Edn. Wayne: Clinical and Laboratory Standards Institute.
De Majumdar, S., Veleba, M., Finn, S., Fanning, S., and Schneiders, T. (2013). Elucidating the Regulon of Multidrug Resistance Regulator RarA in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 57, 1603–1609. doi: 10.1128/aac.01998-12
Di Martino, P., Cafferini, N., Joly, B., and Darfeuille-Michaud, A. (2003). Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 154, 9–16. doi: 10.1016/s0923-2508(02)00004-9
Di Pilato, V., Henrici De Angelis, L., Aiezza, N., Baccani, I., Niccolai, C., Parisio, E. M., et al. (2022). Resistome and virulome accretion in an NDM-1-producing ST147 sublineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: a genotypic and phenotypic characterisation. Lancet Microb. 3, e224–e234. doi: 10.1016/S2666-5247(21)00268-8
El Fertas-Aissani, R., Messai, Y., Alouache, S., and Bakour, R. (2013). Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol. Biol. 61, 209–216. doi: 10.1016/j.patbio.2012.10.004
Ernst, C. M., Braxton, J. R., Rodriguez-Osorio, C. A., Zagieboylo, A. P., Li, L., Pironti, A., et al. (2020). Adaptive evolution of virulence and persistence in carbapenem-resistant Klebsiella pneumoniae. Nat. Med. 26, 705–711. doi: 10.1038/s41591-020-0825-4
Gama, J. A., Zilhao, R., and Dionisio, F. (2017a). Multiple plasmid interference - Pledging allegiance to my enemy’s enemy. Plasmid 93, 17–23. doi: 10.1016/j.plasmid.2017.08.002
Gama, J. A., Zilhao, R., and Dionisio, F. (2017b). Co-resident plasmids travel together. Plasmid 93, 24–29. doi: 10.1016/j.plasmid.2017.08.004
Gao, H., Liu, Y. D., Wang, R. B., Wang, Q., Jin, L. Y., and Wang, H. (2020). The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. Ebiomedicine 51:11. doi: 10.1016/j.ebiom.2019.102599
Hsieh, P. F., Liu, J. Y., Pan, Y. J., Wu, M. C., Lin, T. L., Huang, Y. T., et al. (2013). Klebsiella pneumoniae peptidoglycan-associated lipoprotein and murein lipoprotein contribute to serum resistance, antiphagocytosis, and proinflammatory cytokine stimulation. J. Infect. Dis. 208, 1580–1589. doi: 10.1093/infdis/jit384
Kim, M., Park, J., and Park, W. (2021). Genomic and phenotypic analyses of multidrug-resistant Acinetobacter baumannii NCCP 16007 isolated from a patient with a urinary tract infection. Virulence 12, 150–164. doi: 10.1080/21505594.2020.1867421
Lawal, O. U., Fraqueza, M. J., Worning, P., Bouchami, O., Bartels, M. D., Goncalves, L., et al. (2021). Staphylococcus saprophyticus Causing Infections in Humans Is Associated with High Resistance to Heavy Metals. Antimicrob. Agents Chemother. 65:17. doi: 10.1128/aac.02685-20
Lee, I., Ouk Kim, Y., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Lee, I. R., Molton, J. S., Wyres, K. L., Gorrie, C., Wong, J., Hoh, C. H., et al. (2016). Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci. Rep. 6:12. doi: 10.1038/srep29316
Liao, C. H., Huang, Y. T., Chang, C. Y., Hsu, H. S., and Hsueh, P. R. (2014). Capsular serotypes and multilocus sequence types of bacteremic Klebsiella pneumoniae isolates associated with different types of infections. Eur. J. Clin. Microbiol. Infect. Dis. 33, 365–369. doi: 10.1007/s10096-013-1964-z
Linkevicius, M., Sandegren, L., and Andersson, D. I. (2016). Potential of Tetracycline Resistance Proteins To Evolve Tigecycline Resistance. Antimicrob. Agents Chemother. 60, 789–796. doi: 10.1128/aac.02465-15
Liu, Y., Liu, P. P., Wang, L. H., Wei, D. D., Wan, L. G., and Zhang, W. (2017). Capsular Polysaccharide Types and Virulence-Related Traits of Epidemic KPC-Producing Klebsiella pneumoniae Isolates in a Chinese University Hospital. Microb. Drug Resist. 23, 901–907. doi: 10.1089/mdr.2016.0222
Liu, Y., Long, D., Xiang, T. X., Du, F. L., Wei, D. D., Wan, L. G., et al. (2019). Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J. Antimicrob. Chemother. 74, 1233–1240. doi: 10.1093/jac/dkz023
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Maltezou, H. C. (2009). Metallo-beta-lactamases in Gram-negative bacteria: introducing the era of pan-resistance? Int. J. Antimicrob. Agents 33:7. doi: 10.1016/j.ijantimicag.2008.09.003
Mike, L. A., Stark, A. J., Forsyth, V. S., Vornhagen, J., Smith, S. N., Bachman, M. A., et al. (2021). A systematic analysis of hypermucoviscosity and capsule reveals distinct and overlapping genes that impact Klebsiella pneumoniae fitness. PLoS Pathog. 17:e1009376. doi: 10.1371/journal.ppat.1009376
Morales-León, F., Opazo-Capurro, A., Caro, C., Lincopan, N., Cardenas-Arias, A., Esposito, F., et al. (2021). Hypervirulent and hypermucoviscous extended-spectrum β-lactamase-producing Klebsiella pneumoniae and Klebsiella variicola in Chile. Virulence 12, 35–44. doi: 10.1080/21505594.2020.1859274
Naas, T., Cuzon, G., Villegas, M. V., Lartigue, M. F., Quinn, J. P., and Nordmann, P. (2008). Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob. Agents Chemother. 52, 1257–1263. doi: 10.1128/aac.01451-07
Nang, S. C., Morris, F. C., McDonald, M. J., Han, M. L., Wang, J. P., Strugnell, R. A., et al. (2018). Fitness cost of mcr-1-mediated polymyxin resistance in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1604–1610. doi: 10.1093/jac/dky061
Nguyen, F., Starosta, A. L., Arenz, S., Sohmen, D., Donhofer, A., and Wilson, D. N. (2014). Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 395, 559–575. doi: 10.1515/hsz-2013-0292
Osei Sekyere, J., Govinden, U., Bester, L. A., and Essack, S. Y. (2016). Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: emerging resistance mechanisms and detection methods. J. Appl. Microbiol. 121, 601–617. doi: 10.1111/jam.13169
Park, S., Lee, H., Shin, D., and Ko, K. S. (2020). Change of Hypermucoviscosity in the Development of Tigecycline Resistance in Hypervirulent Klebsiella pneumoniae Sequence Type 23 Strains. Microorganisms 8:1562. doi: 10.3390/microorganisms8101562
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/cmr.11.4.589
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/cmr.00064-16
Rakovitsky, N., Frenk, S., Kon, H., Schwartz, D., Temkin, E., Solter, E., et al. (2020). Fourier Transform Infrared Spectroscopy Is a New Option for Outbreak Investigation: a Retrospective Analysis of an Extended-Spectrum-Beta-Lactamase-Producing Klebsiella pneumoniae Outbreak in a Neonatal Intensive Care Unit. J. Clin. Microbiol. 58:8. doi: 10.1128/jcm.00098-20
Russo, T. A., and MacDonald, U. (2020). The Galleria mellonella Infection Model Does Not Accurately Differentiate between Hypervirulent and Classical Klebsiella pneumoniae. Msphere 5:7. doi: 10.1128/mSphere.00850-19
San Millan, A., and MacLean, R. C. (2017). Fitness costs of plasmids: a limit to plasmid transmission. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.MTBP-0016-2017
Sands, K., Carvalho, M. J., Portal, E., Thomson, K., Dyer, C., Akpulu, C., et al. (2021). Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nat. Microbiol. 6, 512–523. doi: 10.1038/s41564-021-00870-7
Schumacher, A., Steinke, P., Bohnert, J. A., Akova, M., Jonas, D., and Kern, W. V. (2006). Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Enterobacteriaceae other than Escherichia coli. J. Antimicrob. Chemother. 57, 344–348. doi: 10.1093/jac/dki446
Shankar, C., Nabarro, L. E., Anandan, S., Ravi, R., Babu, P., Munusamy, E., et al. (2018). Extremely High Mortality Rates in Patients with Carbapenem-resistant, Hypermucoviscous Klebsiella pneumoniae Blood Stream Infections. J. Assoc. Physicians India 66, 13–16.
Sheng, Z. K., Hu, F. P., Wang, W. X., Guo, Q. L., Chen, Z. J., Xu, X. G., et al. (2014). Mechanisms of Tigecycline Resistance among Klebsiella pneumoniae Clinical Isolates. Antimicrob. Agents Chemother. 58, 6982–6985. doi: 10.1128/aac.03808-14
Shi, Q., Lan, P., Huang, D., Hua, X., Jiang, Y., Zhou, J., et al. (2018). Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol. 18:94. doi: 10.1186/s12866-018-1236-2
Shon, A. S., Bajwa, R. P. S., and Russo, T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae A new and dangerous breed. Virulence 4, 107–118. doi: 10.4161/viru.22718
Turton, J. F., Perry, C., Elgohari, S., and Hampton, C. V. (2010). PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J. Med. Microbiol. 59, 541–547. doi: 10.1099/jmm.0.015198-0
Veleba, M., Higgins, P. G., Gonzalez, G., Seifert, H., and Schneiders, T. (2012). Characterization of RarA, a Novel AraC Family Multidrug Resistance Regulator in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 56, 4450–4458. doi: 10.1128/aac.00456-12
Walker, K. A., Treat, L. P., Sepúlveda, V. E., and Miller, V. L. (2020). The Small Protein RmpD Drives Hypermucoviscosity in Klebsiella pneumoniae. mBio 11, e1750–e1720. doi: 10.1128/mBio.01750-20
Xiang, T., Chen, C., Wen, J., Liu, Y., Zhang, Q., Cheng, N., et al. (2020). Resistance of Klebsiella pneumoniae strains carrying blaNDM–1 gene and the genetic environment of blaNDM–1. Front. Microbiol. 11:700. doi: 10.3389/fmicb.2020.00700
Xu, J., Zhao, Z., Ge, Y., and He, F. (2020). Unravelling the genome sequence of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae ST11 isolated from a bloodstream infection. J. Glob. Antimicrob. Resist. 20, 339–341. doi: 10.1016/j.jgar.2020.01.021
Xu, J., Zhu, Z. L., Chen, Y. M., Wang, W. Z., and He, F. (2021). The Plasmid-Borne tet(A) Gene Is an Important Factor Causing Tigecycline Resistance in ST11 Carbapenem-Resistant Klebsiella pneumoniae Under Selective Pressure. Front. Microbiol. 12:10. doi: 10.3389/fmicb.2021.644949
Yan, R., Lu, Y., Zhu, Y., Lan, P., Jiang, S., Lu, J., et al. (2021). A Sequence Type 23 Hypervirulent Klebsiella pneumoniae Strain Presenting Carbapenem Resistance by Acquiring an IncP1 bla (KPC-2) Plasmid. Front. Cell Infect. Microbiol. 11:641830. doi: 10.3389/fcimb.2021.641830
Yao, B., Xiao, X. M., Wang, F., Zhou, L., Zhang, X. W., and Zhang, J. (2015). Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int. J. Infect. Dis. 37, 107–112. doi: 10.1016/j.ijid.2015.06.023
Keywords: Klebsiella pneumoniae, NDM-1, KPC-2, tigecycline resistance, hypervirulent
Citation: Hao J, Zhang B, Deng J, Wei Y, Xiao X and Liu J (2022) Emergence of a Hypervirulent Tigecycline-Resistant Klebsiella pneumoniae Strain Co-producing blaNDM–1 and blaKPC–2 With an Uncommon Sequence Type ST464 in Southwestern China. Front. Microbiol. 13:868705. doi: 10.3389/fmicb.2022.868705
Received: 03 February 2022; Accepted: 21 March 2022;
Published: 29 April 2022.
Edited by:
Eun-Jeong Yoon, Korea National Institute of Health, South KoreaReviewed by:
Shangshang Qin, Zhengzhou University, ChinaVincenzo Di Pilato, University of Genoa, Italy
Copyright © 2022 Hao, Zhang, Deng, Wei, Xiao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinbo Liu, liulab2019@163.com
†These authors have contributed equally to this work
 Jingchen Hao
Jingchen Hao Bangqin Zhang1†
Bangqin Zhang1† Jiamin Deng
Jiamin Deng Xue Xiao
Xue Xiao