Erratum: Comparative analysis reveals the modular functional structure of conjugative megaplasmid pTTS12 of Pseudomonas putida S12: A paradigm for transferable traits, plasmid stability, and inheritance?
- 1Institute of Biology Leiden, Leiden University, Leiden, Netherlands
- 2Department of Fundamental Microbiology, University of Lausanne, Lausanne, Switzerland
Originating from various environmental niches, large numbers of bacterial plasmids have been found carrying heavy metal and antibiotic resistance genes, degradation pathways and specific transporter genes for organic solvents or aromatic compounds. Such genes may constitute promising candidates for novel synthetic biology applications. Our systematic analysis of gene clusters encoded on megaplasmid pTTS12 from Pseudomonas putida S12 underscores that a large portion of its genes is involved in stress response to increase survival under harsh conditions like the presence of heavy metal and organic solvent. We investigated putative roles of genes encoded on pTTS12 and further elaborated on their roles in the establishment and maintenance under several stress conditions, specifically focusing on solvent tolerance in P. putida strains. The backbone of pTTS12 was found to be closely related to that of the carbapenem-resistance plasmid pOZ176, member of the IncP-2 incompatibility group, although the carbapenem resistance cassette is absent from pTTS12. Megaplasmid pTTS12 contains multiple transposon-flanked cassettes mediating resistance to various heavy metals such as tellurite, chromate (Tn7), and mercury (Tn5053 and Tn5563). Additionally, pTTS12 also contains a P-type, Type IV secretion system (T4SS) supporting self-transfer to other P. putida strains. This study increases our understanding in the modular structure of pTTS12 as a member of IncP-2 plasmid family and several promising exchangeable gene clusters to construct robust microbial hosts for biotechnology applications.
Introduction
Bacteria may use plasmids as autonomous, self-replicating elements driving horizontal transfer of genes (HGT) that confer resistance to otherwise detrimental conditions. As such, these extrachromosomal entities often confer advantageous characteristics for the host strain (Molina et al., 2011; Xiong et al., 2013; Cazares et al., 2020). The rapid spread of resistance genes through large conjugative plasmids is further facilitated by the presence of a variety of mobile genetic elements such as transposons, integrons and insertion sequences (IS’s) (Kholodii et al., 1993; Nojiri et al., 2004; Xiong et al., 2006; Hosseini et al., 2017). The spread of multidrug resistance (MDR) via mobile genetic elements has been the subject of investigation for a number of years (Partridge et al., 2018). Despite those efforts, the role and functioning of megaplasmids is still poorly understood. Recent studies highlighted the role of megaplasmids in the spread of MDR in the opportunistic pathogen Pseudomonas aeruginosa (Xiong et al., 2013; Cazares et al., 2020). Strains of its close relative, Pseudomonas putida, have been known to harbor large conjugational plasmids, conferring resistance to environmental threats (Ramos et al., 1995; Molina et al., 2011). Plasmids of the Pseudomonas family are classified by incompatibility groups, that exhibit various modes of compatibility and transferability (Dennis, 2005; Shintani et al., 2015). We have chosen to study the recently identified megaplasmid pTTS12 of Pseudomonas putida S12 in comparison with a number of other large plasmids of the Pseudomonas family, in order to elucidate key elements governing HGT, stability and essential functions.
Pseudomonas putida S12 is a gram-negative soil bacterium which was isolated to utilize styrene as a sole carbon source (Hartmans et al., 1990). This strain shows a remarkable tolerance toward non-metabolized organic solvents (e.g., toluene) (Kieboom et al., 1998a). Such high tolerance toward organic solvents presents a beneficial trait and advantage for bioproduction of aromatics and biofuel (Mukhopadhyay, 2015; Kusumawardhani et al., 2018). Due to its solvent tolerance and versatile metabolism, P. putida S12 excels as a microbial host for production of valuable chemicals (Wierckx et al., 2005; Verhoef et al., 2007, 2009; Koopman et al., 2010; Janardhan Garikipati and Peeples, 2015). Removal of organic solvent molecules from the bacterial cell membrane is essential and carried out by SrpABC, a resistance-nodulation-cell division (RND) family efflux pump (Kieboom et al., 1998a,b). Membrane compaction and the upregulation of chaperones, general stress responses, and TCA cycle-related enzymes support a further intrinsic solvent tolerance in P. putida S12 (Volkers et al., 2006, 2015; Wijte et al., 2011; Rühl et al., 2012).
We recently found through whole-genome sequencing that the genome of P. putida S12 consists of a 5.8 Mbp chromosome and a 583 kbp single-copy megaplasmid pTTS12 (Kuepper et al., 2015). Both the SrpABC RND-efflux pump and the styrene degradation pathway which are the major distinctive features of P. putida S12, are encoded on this megaplasmid. The genome of P. putida S12 contains a large number of several types of mobile elements, spread over both chromosome and megaplasmid pTTS12 (Wery et al., 2001; Volkers et al., 2010; Kuepper et al., 2015). Some of these insertion sequences were shown to be involved in the regulation and adaptation toward stress conditions, for example during the solvent stress (Sun and Dennis, 2009; Hosseini et al., 2017). Like P. putida S12, related solvent-tolerant P. putida DOT-T1E and Pseudomonas taiwanensis VLB120 have been shown to harbor megaplasmids of 121 and 312 kb, respectively (Panke et al., 1998; Rodríguez-Herva et al., 2007; Köhler et al., 2013). Indeed, those plasmids encode RND-type efflux pumps and biodegradative pathways for aromatic compounds, similar to P. putida S12. To further characterize pTTS12, we here performed a comparative analysis against a large number of other megaplasmids from Refseq and Nuccore databases. With this analysis, we aimed at identifying the origin of the modular structure of environmental-stress related gene clusters in pTTS12.
Materials and methods
Cultivation of Pseudomonas putida
Strains and plasmids used in this report were listed in Table 1. All P. putida strains were grown in Lysogeny Broth (LB) containing 10 g L–1 tryptone, 5 g L–1 yeast extract and 5 g L–1 sodium chloride at 30°C with 200 rpm shaking. E. coli strains were cultivated in LB at 37°C with 250 rpm shaking in a horizontal shaker (Innova 4330, New Brunswick Scientific). For solid cultivation, 1.5% (w/v) agar was added to LB. M9 minimal medium used in this report was supplemented with 2 mg L–1 MgSO4 and 0.2% w/v of citrate as sole carbon source (Hartmans et al., 1990). Bacterial growth was observed by optical density measurement at 600 nm (OD600nm) using a spectrophotometer (Ultrospec 2100 pro, Amersham Biosciences). Maximum growth rate and other parameters were calculated using growthcurver R-package ver.0.3.0 (Sprouffske and Wagner, 2016). Solvent tolerance analysis was performed by growing P. putida strains in LB starting from OD600nm = 0.1 in Boston bottles with Mininert bottle caps. When required, potassium tellurite (6.75–200 mg L–1), indole (100 mg L–1), gentamicin (25 mg L–1), ampicillin (100 mg L–1), tetracycline (25 mg L–1), and kanamycin (50 mg L–1) were added to the media.
DNA methods
All PCR reactions were performed using Phire polymerase (Thermo Fischer) according to the manufacturer’s manual. All primers are listed in Table 2 and were obtained from Sigma-Aldrich. PCR reactions were visualized and analyzed by gel electrophoresis on 1% (w/v) TBE agarose gels containing 5 mg L–1 ethidium bromide in an electric field (110 V, 0.5× TBE running buffer).
Megaplasmid pTTS12 transfer into Pseudomonas putida KT2440 and Escherichia coli strains
Gentamicin resistance and GFP containing cassette were incorporated into P. putida KT2440 chromosome at the Tn7 site using pBG35 plasmid resulting in the strain P. putida KT-BG35 as previously described (Zobel et al., 2015). Correct integration of this construct was verified by observing the gentamicin resistance, GFP expression and colony PCR (glmS_Fw and glmS_Rv primers), followed by Sanger sequencing of the upstream region of glmS locus. The resulting strain is free of pTnS-1 plasmid and the helper plasmid pRK2013 by observing the absence of Ampicillin and Kanamycin resistance, respectively.
Kanamycin resistance gene was introduced into the megaplasmid pTTS12 by integrating plasmid pEMG using homologous recombination resulting in P. putida S12.1 as previously described (Martínez-García and de Lorenzo, 2011). A single homologous recombination site was obtained by PCR with 90285_Fw and 90825_Rv primer pair and this fragment was used to construct pEMG-TS plasmid. Correct integration into P. putida S12 was verified by observing kanamycin resistance and colony PCR using primers 4,963,661_Fw-4,966,726_Rv.
The transfer of pTTS12 into P. putida KT-BG35, E. coli XL1-Blue or E. coli MV1190 were performed by biparental mating between P. putida S12.1 and the recipient strain on LB agar. Both donor and recipient strains were grown overnight on LB agar at 30°C for P. putida strains and 37°C for E. coli strains. The colonies were resuspended in liquid LB media and diluted to about 2 × 108 organisms per ml. A donor and a recipient strain suspension (100 μl) were mixed and collected on a cellulose-acetate filter (Sartorius; 0.45-μm pore size; 25-mm diameter) which was incubated for 24 h at 30°C on LB agar. Cells were resuspended in 1× Phosphate-buffered saline (PBS) solution by vigorous agitation and plated at appropriate dilutions on LB agar containing Gentamicin or Tetracycline and Kanamycin to select for for transconjugants. Plasmid transfer rate was determined by comparing the event of successful plasmid transconjugant with the colony formation unit (cfu) of the recipient strain (P. putida KT-BG35, E. coli XL1-Blue or E. coli MV1190) after biparental mating (GmR/TcR).
The correct transconjugants were selected using LB agar supplemented with gentamicin and kanamycin for P. putida KT-BG35 or with tetracycline and kanamycin for E. coli strains. Additionally, transconjugants were verified using colony PCR (53,496_Fw-56,596_Rv, 200,497_Fw-203,602_Rv, and 286,448_Fw-289,462_Rv). Plasmid stability was determined by calculating the event of megaplasmid loss in P. putida S12 and P. putida KTpS12 grown in liquid media without supplementation of kanamycin as the selective pressure for pTTS12. Plasmid pSW-2 was used as a control for megaplasmid loss events (Martínez-García and de Lorenzo, 2011). P. putida S12 ΔpTTS12 (Kusumawardhani et al., 2020) was used as a control strain of plasmid-less P. putida S12.
pTTS12 sequence and comparative analyses
The pTTS12 sequence used in this analysis is the curated version of CP009975, in which we removed extra ISS12 mobile elements according to previous analysis (Hosseini et al., 2017) and corrected the disrupted genes. For this analysis, pTTS12 232 putative operons on pTTS12 were defined as sets of genes in close proximity which have the same direction in forward or reverse strand with less than 20 bps distance. The operons were clustered together based on the function of genes encoded within these clusters (Supplementary Table 1). Transposases were clustered together with the resistance genes if such transposase is known to carry the cassette.
All plasmid sequences were downloaded from the NCBI database, 22389 plasmid sequences from the Refseq database (retrieved 8 March 2020) and complemented with several plasmid sequences from NUCORE that were omitted from Refseq. The CGviewer (Stothard and Wishart, 2005) was used to generate circular plots of entire pTTS12 with standard settings and MultiGeneBlast tool (Medema et al., 2013) was used to generate synteny plots of specific regions and operons. Further basic alignments and visualization of alignments were performed using Geneious software (BioMatters), and selected sequences were aligned using MAFFT (Katoh and Standley, 2013) for DNA sequences or MUSCLE (Edgar, 2004) for protein sequences applying default settings in all cases.
Results
Comparative analysis of megaplasmid pTTS12
The 583 Kbps megaplasmid pTTS12 (GenBank accession no. CP009975) is a single-copy plasmid encoded in P. putida S12 (Kuepper et al., 2015). pTTS12 encodes 629 genes of which 583 are single copy and 15 genes are duplicated at least once. For this paper, annotations for pTTS12 were further extended (Supplementary Table 1) and the overall sequence was corrected based on our previous observations of an additional seven ISS12 mobile elements (Hosseini et al., 2017). Comparative analysis was performed with 28915 plasmids larger than 2 kb in length, acquired from the Refseq database1 combined with additional sequences from the Nuccore database.2 The top 50 plasmids encoding homologues of pTTS12 proteins are visualized in a circular plot using CGView (Stothard and Wishart, 2005) as shown on Figure 1 and listed in Table 3. The extended circular plot and list of the top 500 plasmids encoding homologues of pTTS12 can be found in Supplementary Figure 1 and Supplementary Table 2, respectively.
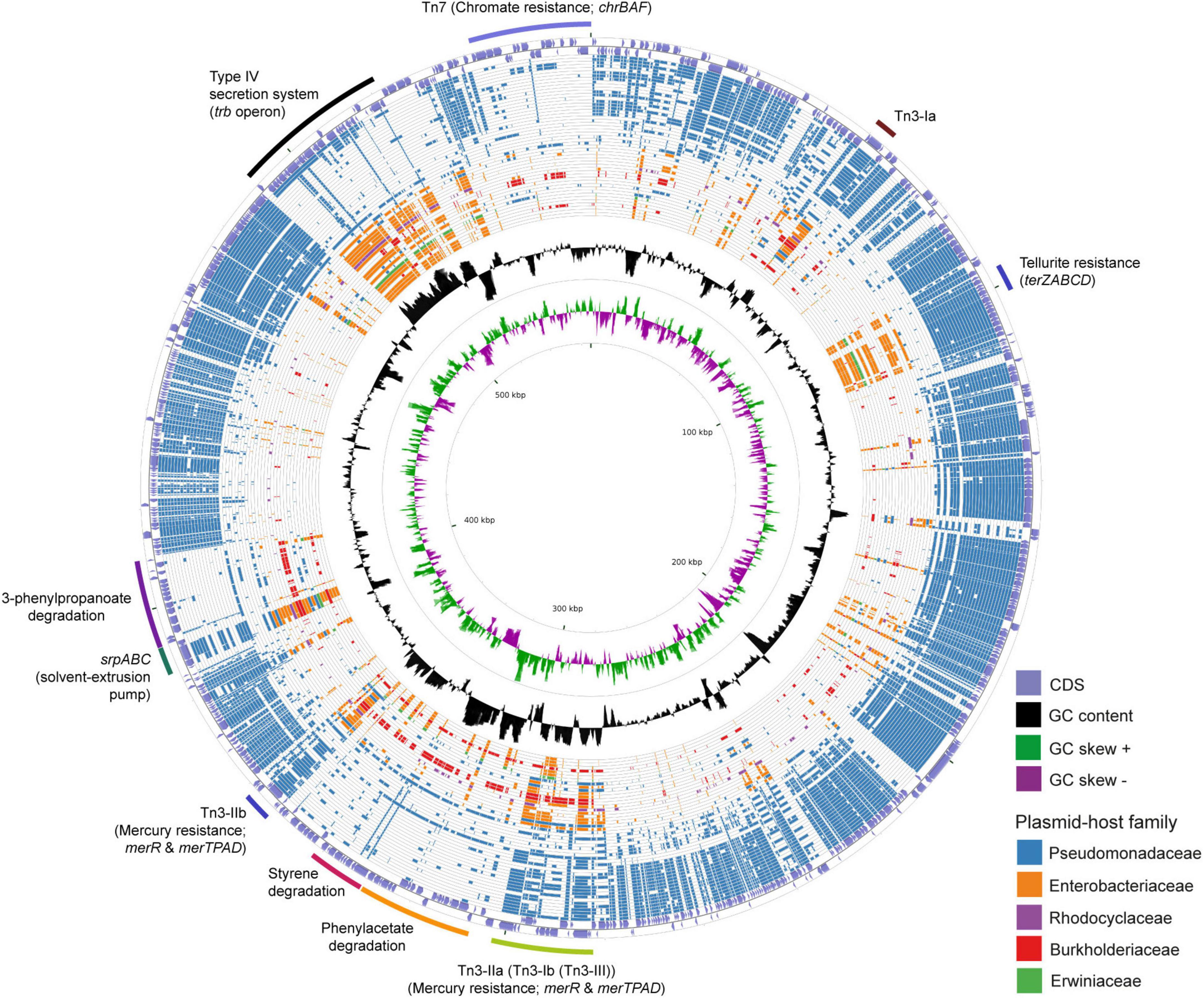
Figure 1. Circular plot of the top 50 plasmids with the highest identity scores to pTTS12. pTTS12 coding sequences (CDS) were aligned to 28915 other plasmids available at NCBI Refseq and Nuccore databases. The outer light purple ring represents the CDS of plus and minus strand of pTTS12. In the inner ring, the GC content is represented in black and the positive and negative GC skew are represented in green and purple respectively. The plasmids are ordered based on their similarity and coverage to pTTS12 CDS from outermost to innermost of the plot with each ring representing a single plasmid as listed in Table 3. In case no homologous protein was identified for a pTTS12 counterpart, that space on the circle was left blank. Ring colors represent different plasmid-host families; Pseudomonadaceae (blue), Enterobacteriaceae (orange), Rhodocyclaceae (purple), Burkholderiaceae (red), and Erwiniaceae (green). The position of the gene clusters of interest are annotated in the figure. An extended circular plot of the top 500 plasmids with highest identity scores to pTTS12 are shown in Supplementary Figure 1.
pTTS12 is highly similar with the carbapenem-resistance plasmid pOZ176 from Pseudomonas aeruginosa PA96
The majority of plasmids highly similar to pTTS12 were found in Pseudomonas genera (Table 3). The relative identity scores presented in Table 3 were calculated as a percentage of total scores of each plasmid divided by the total identity score obtained for pTTS12 itself. The plasmid most similar to pTTS12 is pOZ176 from P. aeruginosa PA96 which was previously categorized as a member of incompatibility group P-2 (IncP-2). pOZ176 and pTTS12 share more than 71% similarity of their encoded genes indicating that these plasmids share a similar IncP-2 backbone. Whereas pOZ176 encodes a carbapenem-resistance gene cluster typical for several pathogenic P. aeruginosa strains (Xiong et al., 2013), pTTS12 of P. putida S12 does not share this feature. Instead, pTTS12 contains gene clusters for styrene degradation, phenylpropionic acid degradation, and solvent efflux pump (srpABC), which are absent in pOZ176.
Other features of pTTS12 are widely distributed among other soil bacteria and enterobacteria
Between 8 to 12% of encoded proteins from pTTS12 can also be found in other genera than Pseudomonas (Table 3). Several plasmids from Salmonella, Enterobacter, Citrobacter, Leclersia, Klebsiella, Pantoea, Polaromonas, and Cupriavidus species shared homologous proteins involved in the T4SS conjugation, replication machinery, and plasmid maintenance. Like other IncP-2 plasmids, pTTS12 contains multiple transposable elements, particularly Tn3 mobile elements (Figure 1). This mobile element encodes for heavy metal resistance genes that are predominantly present in IncP-2 plasmids. Additionally, pTTS12 carries a unique Tn7-element consisting of several chromate resistance genes.
Replication and maintenance of pTTS12
Megaplasmid pTTS12 contains two replication and partitioning machineries. In a recent report, Shintani and colleagues argued that pOZ176 encodes two replication initiation protein (RIP); the primary RIP gene (pOZ176_183) and the auxiliary RIP gene (pOZ176_301) (Shintani et al., 2022). In pTTS12, we found that RPPX_27060 putatively encodes the primary RIP that shares 99% identity, 100% coverage with the primary RepA protein of pOZ176 (pOZ176_301) (Supplementary Figure 4A). In addition, the putative genes around this region show synteny and high similarity between pTTS12 and pOZ176. It must be noted, however, that there are two mobile elements present in pTTS12, disrupting the homologous regions with pOZ176_182 and pOZ176_186 (Supplementary Figure 4A).
Meanwhile, RPPX_28775 putatively encodes for the auxiliary RIP (285 amino acids) that shares 86.48% identity, 98% coverage with the auxiliary RepA protein of pOZ176 (pOZ176_301) (Supplementary Figure 4B). Downstream of RPPX_28775, pTTS12 contains three copies of direct repeats TCGTGCTATCAGGAGTA (N7) TCGTGCTATCGGGAGTA (N6) TCGTGCTATCAGGAGTA with one base mismatch compared to the direct repeats found in pOZ176 (Xiong et al., 2013). These direct repeats may constitute putative iterons at the origin of replication site. RPPX_28770 and RPPX_28765 loci, downstream of RPPX_28775 and iterons, putatively encodes for ParA-ATPase (91.04% identity, 100% coverage) and ParB (85.88% identity, 89% coverage) similar to pOZ176, respectively.
In addition to this system, pTTS12 contains another putative partitioning system encoded on RPPX_26215, RPPX_26845 and RPPX_26840 loci. RPPX_26215 putatively encodes for TrfA protein, which is essential for initiating DNA replication at the origin of repilcation. The protein encoded at RPPX_26840 locus appears to belong to the ParB/RepB/Spo0J (KorB) family partition protein. This family of partition protein is known as a regulator of replication and maintenance of the IncP plasmids by inhibiting trfA expression (Kornacki et al., 1987; Rosche et al., 2000). Typical to low copy plasmid, this putative ParA-ParB protein pair may act as an active partitioning system to ensure that a copy of the plasmid segregates to each daughter cell during cell division by targeting a nearby centromere-like element (Rosche et al., 2000).
Distinctive conjugation machinery of pTTS12
Megaplasmid pTTS12 contains a P-type T4SS (Type IV secretion system) conjugation system (RPPX_28670-RPPX_28725), sharing synteny with the prototype trb operon from A. tumefaciens pTiC58 (Figure 2A). For an operational T4SS conjugation machinery, a T4SS gene cluster, type IV coupling protein (T4CP) and a relaxase protein are required (Christie et al., 2014). An additional traG (RPPX_28650), which may serve as a T4CP, is located upstream of the T4SS cluster. Further upstream of the T4SS and T4CP, a putative virD2-like gene (RPPX_28750) and an operon consisting of parB, parA and repA (RPPX_28765-28775) are encoded on pTTS12. The putative virD2-like gene (RPPX_28750) or the ATP-dependent exonuclease RecD (RPPX_28655) may play a role as a relaxase, which is important for the transferability of pTTS12.
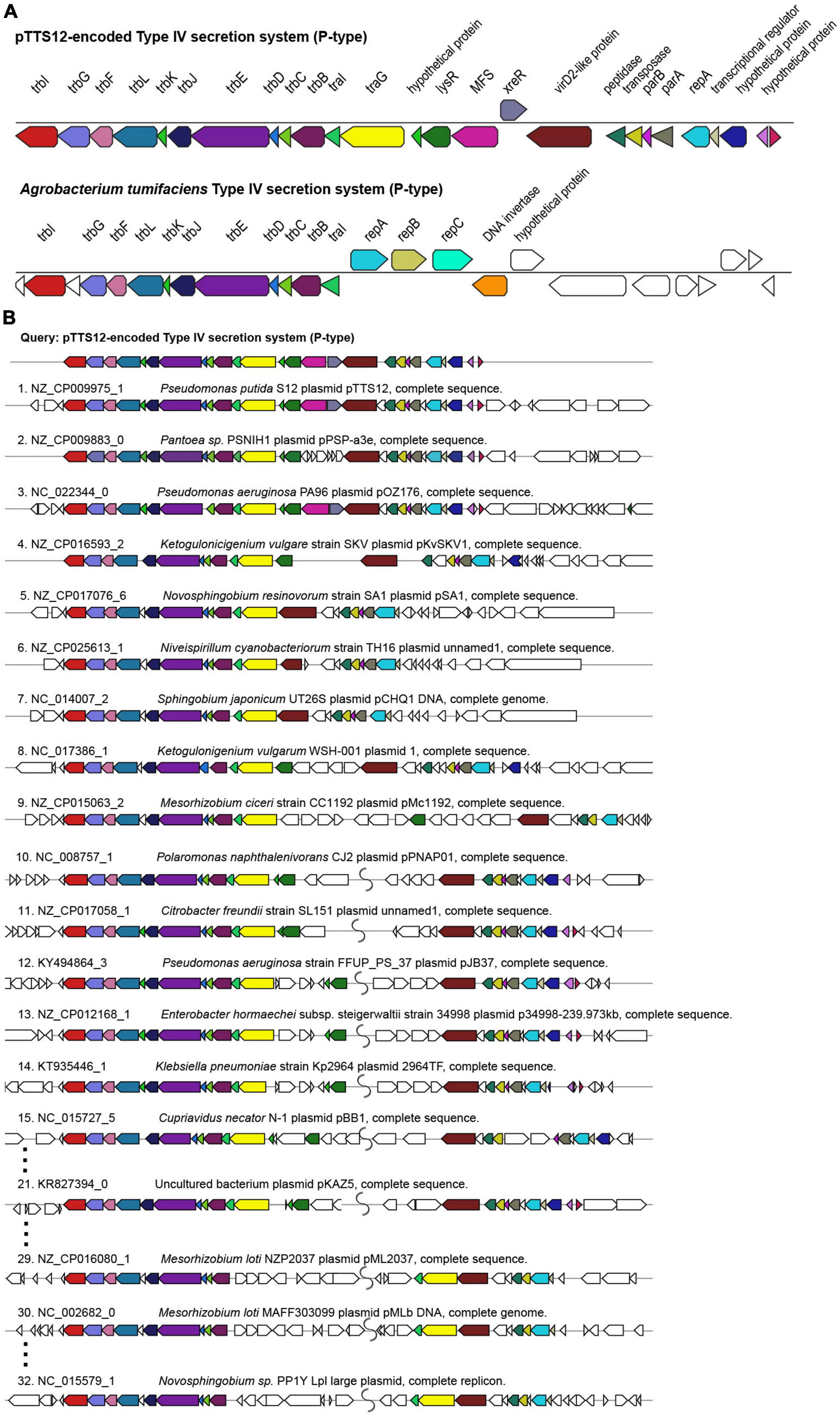
Figure 2. Structure and synteny of pTTS12 conjugation system. (A) The arrangement of the T4SS gene cluster found in pTTS12 and the prototype trb operon from Agrobacterium tumefaciens (pTiC58). The colors represent different genes in the cluster and same colors are assigned for the homologous genes. The gene names are indicated above the respective clusters. pTTS12/T4SS and the trb operon of pTiC58 share synteny for 11 genes (trbI to traI), while other parts are clearly different. (B) Synteny plot of the T4SS gene cluster of pTTS12 for plasmid conjugation, replication and partitioning compared with other plasmids. This visualization was generated using multigeneblast software (Medema et al., 2013). The numbers refer to the order of decreasing synteny. For the sake of clarity, several plots were removed from this figure, indicated by the dots. The colors represent different genes in the cluster corresponding to color-coding in panel (A). Putative coupling-protein (T4CP) traG is indicated in yellow and the putative relaxase virD2-like protein is indicated in brown.
T4SS, traG, and upstream genes involved in replication and partitioning shared synteny with the T4SS clusters on other plasmids such as Pantoea sp. PSNIH1 (pPSP-a3e), Pseudomonas aeruginosa PA96 (pOZ1760), and Ketogulonicigenium vulgare SKV (pKvSKV1) (Figure 2B). Typically, traG (T4CP) is coupled to the same operon of T4SS while the region encoding replication and partitioning may be separated (Figure 2B), in some cases relatively far. It is interesting to note that in two Mesorhizobium loti strains and Novosphingobium sp. PP1Y, the T4SS operon and traG-traI are completely separated (Figure 2B). Instead, VirD2-like protein (relaxase), traG (T4CP) and traI formed a single operon.
Most of the Pseudomonadaceae-family plasmids do not contain this trb operon (Figure 1) except for pTTS12, pOZ176 from P. aeruginosa PA96, pJB37 from P. aeruginosa FFUP_PS_37 and an unnamed plasmid from P. aeruginosa PA83. Therefore, this conjugative operon may not be common in IncP-2 plasmid family. However, this trb operon is abundant among Enterobacteriaceae-family plasmids (Figure 1).
The unique arrangement of Tn3 transposable elements is important for the dissemination of styrene degradation pathway
pTTS12 shares 21% of sequence similarity with the pSTY plasmid of P. taiwanensis VLB120. The major and only gene clusters shared between pTTS12 and pSTY are involved in styrene degradation, phenylpropionic acid degradation, and solvent efflux pump (SrpABC). The genes shared between pTTS12 and pSTY are absent in all other Pseudomonas plasmids, except for the solvent efflux pump gene cluster which is similar with TtgGHI efflux pump from the pGRT-1 plasmid of P. putida DOT-T1E. The regions encoding solvent efflux pump and phenylpropionic acid degradation are clustered together and have identical synteny in pTTS12 and pSTY (Supplementary Figure 2). However, pTTS12 and pSTY do not share any mobile genetic elements surrounding the solvent efflux pump and phenylpropionic acid degradation gene clusters that might indicate a shared mechanism of acquisition or transfer.
The unique arrangement of the three different Tn3 elements in pTTS12 is present on pSTY of P. taiwanesis VLB120, including an additional copy of Tn5563. In between these two Tn5563-elements, both megaplasmids contain the complete styrene degradation pathway, enabling both strains to grow on styrene as sole carbon source (Köhler et al., 2013; Kuepper et al., 2015). Detailed comparison of the regions between the two Tn5563 elements from pTTS12 and pSTY reveals a high similarity between Tn5563-I and Tn5563-II (Figure 3B). This sequence is around 77 kbps and 60 kbps for pTTS12 and pSTY respectively, including the IRs bracketing both sequences.
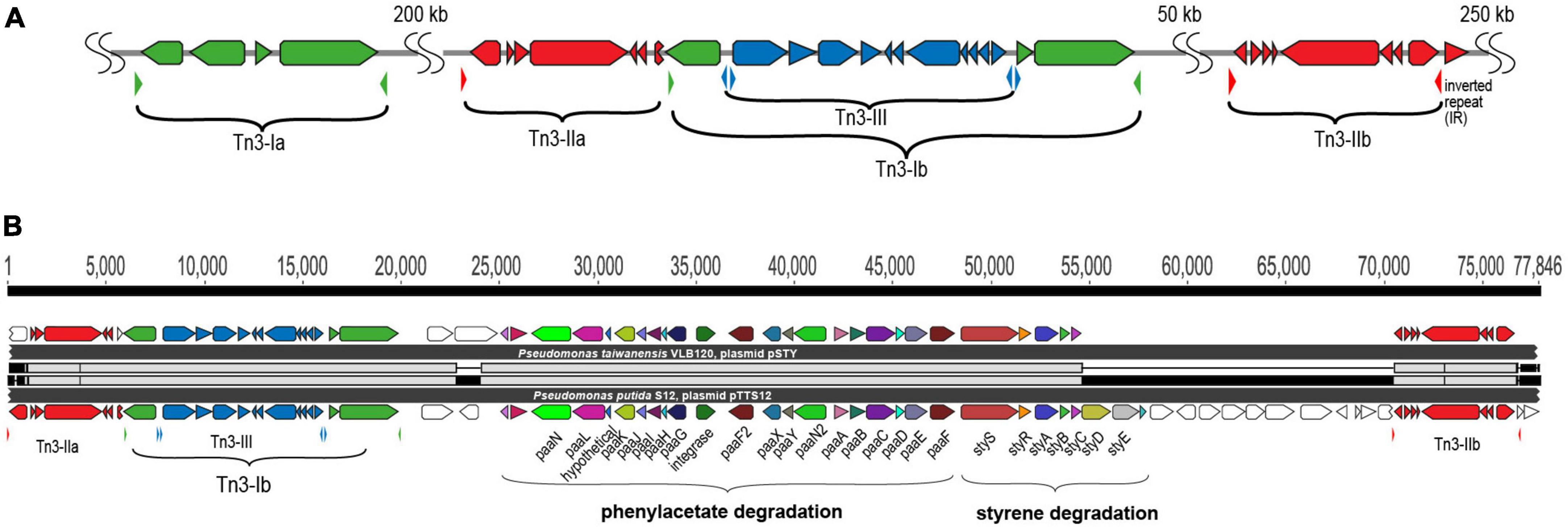
Figure 3. Tn3 transposon family elements responsible for horizontal gene transfer of styrene and phenylacetate degradation pathway. (A) Genetic organization of the three different Tn3-family transposable elements in pTTS12. Colors represent the three different Tn3 transposon families; Tn3-I with highest identity to Tn4656 (green), Tn3-IIa and b with highest identity to Tn5053 (red) and Tn3-III with highest identity to Tn5563 (blue). The inverted repeats (IRs) flanking each element are marked by small, lowered triangles in the same colors, accordingly. (B) Alignment of the Tn3-mediated horizontal gene transfer of the styrene and phenylacetate degradation cluster in pTTS12 (bottom) and pSTY from Pseudomonas taiwanensis VLB120 (top). Colors represent the different genes constituting styrene-phenylacetate degradation cluster and the three Tn3 transposable elements, corresponding to the color-coding in panel A. Gene names are indicated.
pTTS12 as well as many other highly similar plasmids contain a multitude of mobile genetic elements, such as ISS12 and Tn3-family transposases (Hall et al., 2015). pTTS12 harbors five copies of Tn3-family transposable elements; two copies of Tn3-I with highest identity to Tn4656, two copies of Tn3-II that are highly similar to Tn5053 and a single copy of Tn3-III with highest similarity to Tn5563 (Figure 3A). Tn3-I is an identical transposase (RPPX_RS27515, tnpR and RPPX_RS27515, tnpA) to Tn4656 encoded on pWW53 plasmid from P. putida MT53. Moreover, the 39-bps inverted repeat (IR) sequences found on both ends of these two elements are highly identical with only a single mismatch difference. In addition to tnpA and tnpR, this Tn3-element contains a putative methyl-accepting chemotaxis protein (mcpT-2) and an insertion sequence IS256. Tn3-II of pTTS12 is identical (99.8% similarity) to Tn5053 of Xanthomonas sp. W17 encoding a mercury resistance gene cluster merR and merTPAD (Kholodii et al., 1993). This Tn3-II has 25 bp inverted repeats, bracketing the ends and 5 bp directed repeats (DR). Tn3-III is identical to Tn5563, that is found in several other plasmids, e.g., pAMBL of P. aeruginosa, pSTY of P. taiwanensis VLB-120 and pRA2 of P. alcaligenes. The Tn3-III has identical 38 bps IRs bracketing the element, also identical to IRs found for this element in other plasmids. This element contains several genes such as merR, merTP, pilT and a gene with a PIN nuclease domain.
The Tn4656 and Tn5563 are both duplicated and rearranged on the megaplasmid together with Tn5053. This rearrangement resulted in a highly characteristic sequence, which partially resulted from insertion of Tn5053 transposition in the second copy of Tn4656 (Tn4656-II), between the mcpT-2 and tnpR loci (Figure 3). The IRs as well as DRs of Tn5053 indicate the insertion site of this element. The IRs of Tn4656-II are well preserved bracketing both elements. Other parts of this sequence consist of the second copy of Tn5563 (Tn5563-II), which is truncated at the right end by insertion of Tn4656-II in merT and thus truncated the 5’ end of merT sequence, can be identified. Due to this truncation, the IRs of Tn4656-II cannot be found on the right side of the element anymore except for the right IR of initial Tn4656, which is located 58 kbps upstream of this element.
Conjugative megaplasmid pTTS12 is highly stable in Pseudomonas putida KT2440
To characterize the conjugative transferability of pTTS12, we performed biparental mating between P. putida S12.1 and P. putida KT-BG35. P. putida KT-BG35 is a strain derived from P. putida KT2440, does not harbor a megaplasmid and carries a gentamicin resistant marker and green fluorescence protein (GFP) at its attn7 site (Zobel et al., 2015). The transfer was performed using biparental mating between P. putida S12.1 containing pTTS12 with kanamycin resistant marker and P. putida KT-BG35. Transconjugant colonies resistant to both kanamycin and gentamicin occurred at the frequency of 4.20 (±0.51) × 10–7. After appropriate selection on agar plates, the identity of transconjugant colonies was confirmed by observing GFP expression of the colonies derived from P. putida KT-BG35. Additionally, transfer of entire pTTS12 was confirmed using PCR. Amplification of several regions of pTTS12 using primer pairs 53,496_Fw-56,596_Rv, 200,497_Fw-203,602_Rv, and 286,448_Fw-289,462_Rv resulted in an expected band of 3,100, 511, and 3,015 bp in transconjugant colonies, respectively (Supplementary Figure 5). Fifteen randomly selected colonies chosen for colony PCR showed correct bands. This confirmed the transfer of entire pTTS12 into P. putida KT-BG35 and the resulting strains will further be referred to as P. putida KTpS12. Several attempts to transfer pTTS12 from P. putida S12.1 into E. coli strains represented by E. coli MV1190 and E. coli XL1-Blue by biparental mating did not result in any successful transconjugant colonies.
The stability of pTTS12 in P. putida KTpS12 and P. putida S12 was examined for 5 passages in the absence of antibiotic selective pressure (approximately 10 generations/passage step). No plasmid loss was observed from either P. putida KTpS12 or P. putida S12 growing without selective pressure while the negative control pSW-2 showed a steady plasmid loss (Figure 4A). Serendipitous deletion of pTTS12 was not found in P. putida S12 nor P. putida KTpS12. Hence, pTTS12 is a stable megaplasmid in both P. putida S12 and P. putida KT2440.
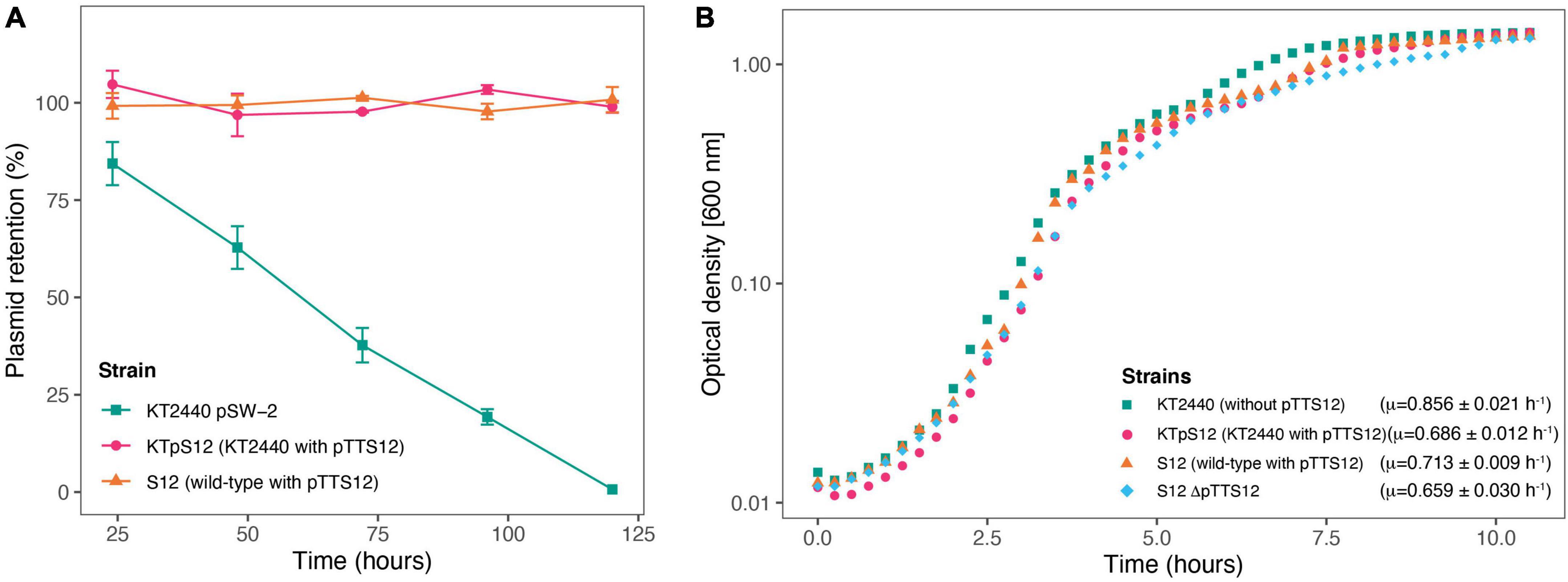
Figure 4. Megaplasmid pTTS12 is highly stable in P. putida strains and reduces maximal growth rate in P. putida KT2440. (A) Plasmid retention of pTTS12 in P. putida KT2440 and P. putida S12 on liquid LB without selection pressure for approximately 50 generations. Colors and shapes indicate the different strains with green squares indicating P. putida KT2440 pSW-2, magenta circles indicating P. putida KTpS12, and orange triangles indicating P. putida S12 (containing pTTS12). Plasmid pSW-2 in P. putida KT2440 was used as a control for loss of unstable plasmid. pTTS12 is stably maintained in both P. putida S12 and P. putida KTpS12. (B) Growth curve of P. putida KT2440, P. putida KTpTTS12 P. putida S12, and P. putida S12 ΔpTTS12. Growth was followed on liquid LB at 30°C, 200 rpm shaking. Growth curves represent data obtained from three biological replicates for each strain, starting from OD600nm = 0.05. Green squares indicate P. putida KT2440 pSW-2, magenta circles indicate P. putida KTpS12, orange triangles indicate P. putida S12 (containing pTTS12), and blue diamond indicate P. putida S12 ΔpTTS12. Maximum growth rates calculated for each strais are indicated within the figure.
The occurrence of large plasmids in bacteria may cause metabolic burden (Wein et al., 2020), which is reflected in reduced bacterial growth rate (μ). P. putida KTpS12 exhibited a lower maximum growth rate compared to P. putida KT2440, 0.686 ± 0.012 and 0.856 ± 0.021 h–1 respectively (Figure 4B). However, P. putida S12 and P. putida S12 ΔpTTS12 did not show a significant maximum growth rate difference (0.713 ± 0.009 and 0.659 ± 0.030 h–1 respectively). The length of lag-phase and biomass yield at stationary phase remained unaffected with pTTS12 present in both P. putida S12 and KTpS12. Apparently, the presence of pTTS12 imposes significant metabolic burden in P. putida KT2440 (Student’s t-test, p-value 0.0002553), but not in S12.
Phenotypic features of pTTS12 and attained solvent tolerance in Pseudomonas putida KTpS12
P. putida S12 is intrinsically tolerant to an assortment of environmental xenobiotics. The majority of these characteristic traits are encoded on pTTS12; the styrene degradation operon styABCDE, the tellurite resistance operon terZABCDE and a solvent efflux pump srpABC. To explore functionality of pTTS12 following conjugative transfer, these characteristic traits were investigated in P. putida KTpS12 (Figure 5). Activity of the styrene degradation operon in P. putida KTpS12 was tested by growing on minimal media with styrene as carbon source and inspecting transformation of indole into indigo. Similarly as observed in P. putida S12, P. putida KTpS12 was able to transform indole into indigo, whereas wild-type KT2440 was not, indicating activity of styrene monooxygenase (styA) and styrene oxide isomerase (styB) in these strains (Figure 5A). In addition, P. putida KTpS12 was able to grow on minimal media supplemented with styrene as carbon source or minimal media agar plates incubated in styrene atmosphere (data not shown).
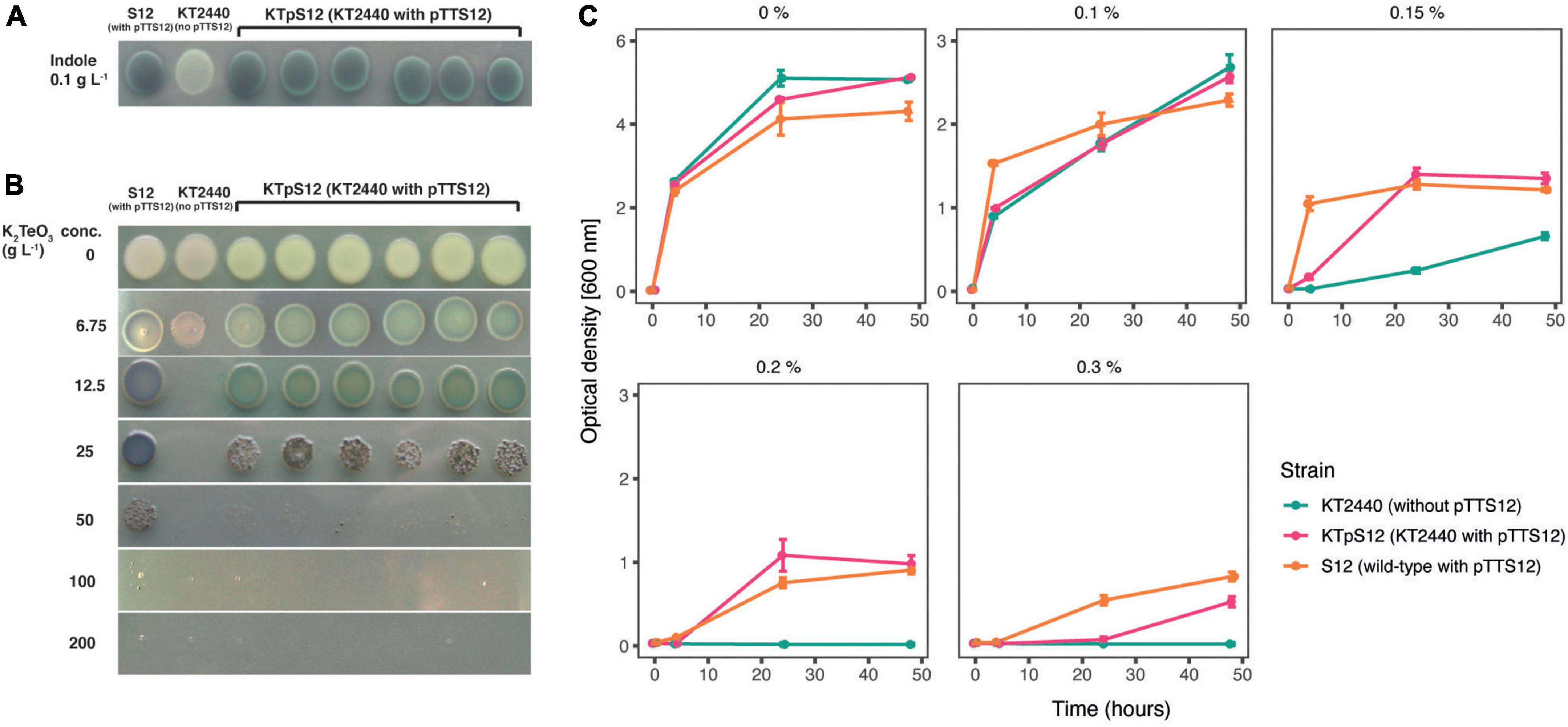
Figure 5. Transfer of pTTS12 phenotypic characteristics. (A) Production of indigo from indole indicating activity of styrene monooxygenase (StyA) and styrene oxide isomerase (StyB). Minimal medium (M9) was supplemented with indole which, in the presence of StyAB enzymes encoded on pTTS12, is converted into indigo. Positive control P. putida S12 showed indigo coloration, whereas negative control P. putida KT2440 remained white. P. putida KTpS12 showed indigo coloration indicating activity of pTTS12-encoded styAB. (B) Growth of P. putida strains in the presence of potassium tellurite (K2TeO3). Minimum inhibitory concentration (MIC) of positive control P. putida S12 was 200 g L– 1 whereas the MIC of the wild-type P. putida KT2440 was 12.5 g L– 1. The MIC of P. putida KTpS12 was 100 g L– 1 indicating the presence and activity of the ter operon on pTTS12. (C) Growth of P. putida strains on increasing concentrations of toluene. Optical density (OD600nm) of the cultures was measured at 4, 24, and 48-h time points with starting point of OD600nm = 0.1. The y-axis range is different between the first panel (0–6) and the other panels (0–3). Green dots indicate P. putida KT2440, magenta dots indicate P. putida KTpS12, and orange dots indicate P. putida S12 (containing pTTS12). P. putida KTpS12 showed an increase in solvent tolerance indicating the presence and acivity of the srp operon from pTTS12.
Potassium tellurite (K2TeO3) exhibits antimicrobial activity in bacteria. Tellurite resistance in bacteria is achieved through reduction of tellurite (TeO32–) into a less toxic metallic tellurium (Te0), causing the formation of black colonies in the presence of tellurite (Taylor, 1999). The resistance genes on pTTS12 are encoded as an operon terZABCDE (RPPX_26360-26385). To investigate whether this gene cluster is able to increase resistance toward tellurite in P. putida KT2440, the minimum inhibitory concentration (MIC) was determined for the three different stains (Figure 5B). The MIC of potassium tellurite for P. putida KT2440 was 16-fold less compared to P. putida S12, 12.5 and 200 g L–1 respectively. P. putida KTpS12 showed an 8-fold increase in tellurite resistance (MIC 100 g L–1) compared to its parental strain P. putida KT2440.
The solvent efflux pump srpABC encoded on the megaplasmid pTTS12, enables survival and growth of P. putida S12 in non-utilized organic solvents (Kieboom and de Bont, 2001). In order to investigate the effect of induced tolerance toward organic solvents due to the introduction of pTTS12 in P. putida KTpS12, a growth assay was performed in the presence of different toluene concentrations (Figure 5C). P. putida KT2440 was able to grow in LB liquid media supplemented with toluene up to a maximum of 0.15% v/v concentration, although demonstrating a significant growth reduction compared to P. putida S12. With the introduction of pTTS12, P. putida KTpS12 tolerance toward toluene increased to 0.30% v/v. Similar concentration was obtained for P. putida S12, while P. putida KTpS12 exhibited slightly slower growth in the presence of toluene. These observations indicated that the introduction of megaplasmid pTTS12 provided a full set of characteristic features to P. putida KT2440.
Discussion
Megaplasmid pTTS12 defines an environment-adapted member of IncP-2 family plasmids, carrying distinct accessory gene clusters
Megaplasmid pTTS12 of P. putida S12 is closely related to other plasmids in proteobacteria, especially within the Pseudomonas genus. In this study we demonstrated high similarity of the pTTS12 ‘backbone’ with pOZ176 from P. aeruginosa PA96 (Xiong et al., 2013) and several other IncP-2 plasmids, such as pJB37 of P. aeruginosa FFUP_PS_37 (Botelho et al., 2017). pOZ176 has been categorized within incompatibility group IncP-2 based on its replication, partitioning, and transfer machinery along with conferring tellurite resistance as a key feature of IncP-2 plasmid (Xiong et al., 2006, 2013). Indeed, pTTS12 encodes typical characteristics of IncP-2 plasmids, such as heavy metal resistance (tellurite, mercury, and chromate) and plasmid maintenance via the parA/parB/repA system (Jacoby et al., 1983; Shintani et al., 2015). Despite high similarity between pTTS12 and pOZ176, pTTS12 lacks the Tn6016 blaIMP–9-carrying class 1 integron cassette which is an important trait of pOZ176 conferring resistance to aminoglycosides and carbapenems (Xiong et al., 2006, 2013). On the other hand, pOZ176 lacks the solvent efflux pump, styrene degradation pathway, and phenylpropionic acid degradation pathway which are main characteristics of pTTS12. This demonstrates that very similar megaplasmid backbones may carry resistance to a diversity of xenobiotics, metabolic functions, or virulence gene clusters (Harrison and Brockhurst, 2012). Whereas the traits disseminated by pTTS12 and pOZ176 are highly divergent and distinctive, a similar observation of environment-adaptive traits conferred by flexible and diverse accessory gene clusters contained on IncP-2 plasmids was recently reported for P. aeruginosa plasmids pBT2436 and pBT2101, carrying multiple MDR cassettes (Cazares et al., 2020).
Convergent distribution of plasmid-encoded solvent tolerance gene clusters
pTTS12 shares its unique features of styrene and phenyl-propanoate degradation pathway with pSTY from P. taiwanensis VLB120 and the solvent extrusion pump SrpABC with TtgGHI from pSTY and pGRT-1 P. putida DOT-T1E. Hence, these plasmids isolated from different environmental sources show convergent organization of highly similar gene clusters with similar features related to environmental stresses (e.g., organic solvents) on different plasmid backbones. In pSTY, the arrangement of gene clusters encoding the styrene degradation pathway and solvent efflux pump - phenylpropionic acid degradation pathway is highly similar to pTTS12, with 99 and 80% similarity, respectively. This suggests that exchange of the styrene degradation pathway occurred more recently than exchange of the efflux pump/phenylpropionic acid degradation cluster.
The styrene degradation gene cluster is encoded within a unique arrangement of Tn3 family transposases shared by pTTS12 and pSTY (Figure 3). Due to the transposition of Tn3-Ib, Tn3-IIa lost its right flank inverted repeat (IR). Because of this arrangement, Tn3-IIa may only “jump” while carrying the entire Tn3 and styrene-phenylacetate degradation clusters using the right flank of Tn3-IIb IR. This would explain the occurrence of the styrene-phenylacetate degradation cluster distinctive arrangement shared between pTTS12 and pSTY. Although we could not find evidence of mobile genetic elements carrying the efflux pump and the phenylpropionic acid degradation gene clusters on pSTY and pTTS12, it is well possible that exchange originally occurred via such route.
Transferability of pTTS12
pTTS12 contains a P-Type type IV secretion system (T4SS) with synteny similar to the prototype trb operon of the A. tumefaciens pTiC58 system (Li et al., 1999; Christie et al., 2014) and it is self-transferable toward other P. putida strains. Interestingly, this locus contains a putative relaxase encoded by virD2 (RPPX_28750, Figure 2). This relaxase is predicted to be responsible for creating a nick prior to plasmid transfer via the conjugative bridge. However, complete deletion of virD2 did not result in a significant reduction of transfer frequency (4.32 (± 1.53) × 10–7; Student’s t-test performed, p-value 0.9078), compared to the wild-type pTTS12. This may indicate the presence of other relaxase(s) which act in-trans with this secretion system. P. putida S12 contains other T4SS within its genome which shares homology with the I-type T4SS represented by Dot/Icm from L. pneumophila (Supplementary Figure 3), although it is unclear whether Dot/Icm can support the transfer of pTTS12 (Nagai and Kubori, 2011). Further research is required to establish putative crosstalk between these secretion systems.
Stable and efficient accommodation of mega plasmid in Pseudomonas putida S12
We observed that in its original host P. putida S12 on rich media, pTTS12 did not impose an apparent metabolic burden whereas in another strain as P. putida KT2440, pTTS12 caused a 20% reduction of maximum growth rate. We previously reported on the occurrence of metabolic burden caused by pTTS12 in P. putida S12 in the presence of toluene (Kusumawardhani et al., 2020). In addition, pTTS12 was stably maintained in both P. putida S12 and P. putida KT2440 (Figure 4A). Apparently, P. putida S12 has found ways to accommodate reduced fitness cost, metabolic burden, and stability of this mega plasmid.
Conjugative transfer may impose substantial cost and burden due to the energy investment on pili formation during conjugation (Turner et al., 1998; Harrison and Brockhurst, 2012). Indeed, pTTS12 showed a substantially lower conjugation rate (4.20 × 10–7 transfer frequency after 24 h) in comparison to other IncP-2 plasmids (10–1 to 10–4 transfer frequency after 2 h) (Jacoby et al., 1983). Moreover, the expression of plasmid-encoded resistance genes, like the solvent extrusion pump, can typically be a source of metabolic burden imposed by plasmids (MacLean and San Millan, 2015). pTTS12 contains multiple types of mobile genetic elements, with ISS12 being the most abundant (Wery et al., 2001; Hosseini et al., 2017). Substantial duplication of the ISS12 mobile element has previously been reported to interrupt srpA, encoding the periplasmic subunit of the solvent efflux pump (Hosseini et al., 2017). In the prolonged absence of organic solvent, expression and maintenance of the solvent efflux pump may be costly for the bacterial cell, hence, interruption of the srp efflux pump gene cluster may further reduce the plasmid burden of pTTS12. In addition to these mechanisms, we recently described the contribution of a toxin-antitoxin module SlvT-SlvA to the stability of pTTS12 (Kusumawardhani et al., 2020).
Future outlook
IncP-2 family plasmids are widely distributed among environmental and clinical Pseudomonas isolates. These plasmids contain variable regions encoding MDR, xenobiotics extrusion pumps, degradation pathways, and heavy metal resistance cassettes. In contrast to the dynamic variable regions, the core backbone of these plasmids shows a general conservation. It is interesting to discover the minimal backbone of IncP-2, which enables Pseudomonads to scavenge gene clusters important for its survival both in environmental and clinical set-up, as a model of horizontal gene transfer. Ultimately, the IncP-2 plasmid family appears to be promising for biotechnological and bioremediation applications due to its stability and relatively low metabolic burden. On the other hand, these plasmids may exchange their traits, thus creating hybrid plasmids when they occur within the same host (Jacoby et al., 1983). The mechanisms for such exchange are still poorly understood and may well involve transposition and conjugation as suggested by the shared and type-specific characteristics of pTTS12 and pOZ176 as described in this study. Further clarification of these mechanisms is important to shed light on the rapid dissemination of bacterial tolerance and resistance to antibiotics, and chemical stresses in the environment.
Successful attempts have been made in exploiting traits from environmental plasmids for standardized components in synthetic biology (Martínez-García et al., 2015). Environmental plasmids are exchangeable between different hosts and may express their genetic features in various genomic and metabolic backgrounds. Moreover, they are an excellent source of novel biological parts such as origin of replication, metabolic pathway, resistance marker, and regulated promoters. New sequencing technologies and comparative genomics analyses support the identification of the genes enabling these features. Here, we demonstrated that pTTS12 contains promising exchangeable gene clusters and building blocks to construct robust microbial hosts for high-value biotechnology applications.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
HK, RH, and J-AV performed the data acquisition. HK and RH contributed to the data analysis and wrote the main text of the manuscript. HK, RH, and JHdW did the conceptualization, reviewing, and editing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
RH was funded by the Dutch National Organization for Scientific Research NWO, through the ERAnet-Industrial Biotechnology Program, project “Pseudomonas 2.0”. HK was supported by the Indonesia Endowment Fund for Education (LPDP) as scholarship provider from the Ministry of Finance, Indonesia. Open access funding was provided by the University of Lausanne.
Acknowledgments
We thank Erik Vijgenboom and Gerben Voshol (Leiden University) for helpful discussions and Omar Qachach for his assistance during the data acquisition process.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1001472/full#supplementary-material
Footnotes
References
Bagdasarian, M., Lurz, R., Rückert, B., Franklin, F. C. H., Bagdasarian, M. M., Frey, J., et al. (1981). Specific-purpose plasmid cloning vectors II. Broad host range, high copy number, RSF 1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16, 237–247. doi: 10.1016/0378-1119(81)90080-9
Bethesda Research Laboratories (1986). E. coli DH5 alpha competent cells. Focus Bethesda Res. Lab. 8:9.
Botelho, J., Grosso, F., Quinteira, S., Mabrouk, A., and Peixe, L. (2017). The complete nucleotide sequence of an IncP-2 megaplasmid unveils a mosaic architecture comprising a putative novel blaVIM-2-harbouring transposon in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 72, 2225–2229. doi: 10.1093/jac/dkx143
Boyer, H. W., and Roulland-dussoix, D. (1969). A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41, 459–472. doi: 10.1016/0022-2836(69)90288-5
Cazares, A., Moore, M. P., Hall, J. P. J., Wright, L. L., Grimes, M., Emond-Rhéault, J. G., et al. (2020). A megaplasmid family driving dissemination of multidrug resistance in Pseudomonas. Nat. Commun. 11:1370. doi: 10.1038/s41467-020-15081-7
Chavda, K. D., Chen, L., Fouts, D. E., Sutton, G., Brinkac, L., Jenkins, S. G., et al. (2016). Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. mBio 7:e002093-16. doi: 10.1128/mBio.02093-16
Choi, K.-H., Gaynor, J. B., White, K. G., Lopez, C., Bosio, C. M., Karkhoff-Schweizer, R. R., et al. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2, 443–448. doi: 10.1038/nmeth765
Christie, P. J., Whitaker, N., and González-Rivera, C. (2014). Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta Mol. Cell Res. 1843, 1578–1591.
Dennis, J. J. (2005). The evolution of IncP catabolic plasmids. Curr. Opin. Biotechnol. 16, 291–298.
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Figurski, D. H., and Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. doi: 10.1073/pnas.76.4.1648
Hall, J. P. J., Harrison, E., Lilley, A. K., Paterson, S., Spiers, A. J., and Brockhurst, M. A. (2015). Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects. Environ. Microbiol. 17, 5008–5022. doi: 10.1111/1462-2920.12901
Harrison, E., and Brockhurst, M. A. (2012). Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 20, 262–267. doi: 10.1016/j.tim.2012.04.003
Hartmans, S., van der Werf, M. J., and de Bont, J. A. (1990). Bacterial degradation of styrene involving a novel flavin adenine dinucleotide-dependent styrene monooxygenase. Appl. Environ. Microbiol. 56, 1347–1351. doi: 10.1128/aem.56.5.1347-1351.1990
Herrero, M., De Lorenzo, V., and Timmis, K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172, 6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990
Hosseini, R., Kuepper, J., Koebbing, S., Blank, L. M., Wierckx, N., and de Winde, J. H. (2017). Regulation of solvent tolerance in Pseudomonas putida S12 mediated by mobile elements. Microb. Biotechnol. 10, 1558–1568. doi: 10.1111/1751-7915.12495
Jacoby, G. A., Sutton, L., Knobel, L., and Mammen, P. (1983). Properties of IncP-2 plasmids of Pseudomonas spp. Antimicrob. Agents Chemother. 24, 168–175. doi: 10.1128/AAC.24.2.168
Janardhan Garikipati, S. V. B., and Peeples, T. L. (2015). Solvent resistance pumps of Pseudomonas putida S12: applications in 1-naphthol production and biocatalyst engineering. J. Biotechnol. 210, 91–99. doi: 10.1016/j.jbiotec.2015.06.419
Jiang, X., Yin, Z., Yuan, M., Cheng, Q., Hu, L., Xu, Y., et al. (2020). Plasmids of novel incompatibility group IncpRBL16 from Pseudomonas species. J. Antimicrob. Chemother. 75, 2093–2100. doi: 10.1093/jac/dkaa143
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kholodii, G. Y., Yurieva, O. V., Lomovskaya, O. L., Gorlenko, Z. M., Mindlin, S. Z., and Nikiforov, V. G. (1993). Tn5053, a mercury resistance transposon with integron’s ends. J. Mol. Biol. 230, 1103–1107.
Kieboom, J., and de Bont, J. A. M. (2001). Identification and molecular characterization of an efflux system involved in Pseudomonas putida S12 multidrug resistance. Microbiology 147, 43–51. doi: 10.1099/00221287-147-1-43
Kieboom, J., Dennis, J. J., de Bont, J. A. M., and Zylstra, G. J. (1998a). Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 273, 85–91. doi: 10.1074/jbc.273.1.85
Kieboom, J., Dennis, J. J., Zylstra, G. J., and de Bont, J. A. (1998b). Active efflux of organic solvents by Pseudomonas putida S12 is induced by solvents. J. Bacteriol. 180, 6769–6772. doi: 10.1128/JB.180.24.6769-6772.1998
Köhler, K. A. K., Rückert, C., Schatschneider, S., Vorhölter, F. J., Szczepanowski, R., Blank, L. M., et al. (2013). Complete genome sequence of Pseudomonas sp. strain VLB120 a solvent tolerant, styrene degrading bacterium, isolated from forest soil. J. Biotechnol. 168, 729–730. doi: 10.1016/j.jbiotec.2013.10.016
Koopman, F., Wierckx, N., de Winde, J. H., and Ruijssenaars, H. J. (2010). Efficient whole-cell biotransformation of 5-(hydroxymethyl)furfural into FDCA, 2,5-furandicarboxylic acid. Bioresour. Technol. 101, 6291–6296.
Kornacki, J. A., Balderes, P. J., and Figurski, D. H. (1987). Nucleotide sequence of korB, a replication control gene of broad host-range plasmid RK2. J. Mol. Biol. 198, 211–222. doi: 10.1016/0022-2836(87)90307-x
Kubasova, T., Cejkova, D., Matiasovicova, J., Sekelova, Z., Polansky, O., Medvecky, M., et al. (2016). Antibiotic resistance, core-genome and protein expression in inchi1 plasmids in Salmonella typhimurium. Genome Biol. Evol. 8, 1661–1671. doi: 10.1093/gbe/evw105
Kuepper, J., Ruijssenaars, H. J., Blank, L. M., de Winde, J. H., and Wierckx, N. (2015). Complete genome sequence of solvent-tolerant Pseudomonas putida S12 including megaplasmid pTTS12. J. Biotechnol. 200, 17–18. doi: 10.1016/j.jbiotec.2015.02.027
Kusumawardhani, H., Hosseini, R., and de Winde, J. H. (2018). Solvent tolerance in bacteria: fulfilling the promise of the biotech era? Trends Biotechnol. 36, 1025–1039. doi: 10.1016/j.tibtech.2018.04.007
Kusumawardhani, H., van Dijk, D., Hosseini, R., and de Winde, J. H. (2020). A novel toxin-antitoxin module SlvT–SlvA regulates megaplasmid stability and incites solvent tolerance in Pseudomonas putida S12. Appl. Environ. Microbiol. 86:e00686-20. doi: 10.1128/AEM.00686-20
Li, P. L., Hwang, I., Miyagi, H., True, H., and Farrand, S. K. (1999). Essential components of the Ti plasmid trb system, a type IV macromolecular transporter. J. Bacteriol. 181, 5033–5041. doi: 10.1128/JB.181.16.5033-5041.1999
MacLean, R. C., and San Millan, A. (2015). Microbial evolution: towards resolving the plasmid paradox. Curr. Biol. 25, R764–R767. doi: 10.1016/j.cub.2015.07.006
Martínez-García, E., Benedetti, I., Hueso, A., and de Lorenzo, V. (2015). Mining environmental plasmids for synthetic biology parts and devices. Microbiol. Spectr. 3:LAS–0033–2014. doi: 10.1128/microbiolspec.PLAS-0033-2014
Martínez-García, E., and de Lorenzo, V. (2011). Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 13, 2702–2716. doi: 10.1111/j.1462-2920.2011.02538.x
Medema, M. H., Takano, E., and Breitling, R. (2013). Detecting sequence homology at the gene cluster level with multigeneblast. Mol. Biol. Evol. 30, 1218–1223. doi: 10.1093/molbev/mst025
Molina, L., Duque, E., Gómez, M. J., Krell, T., Lacal, J., García Puente, A., et al. (2011). The pGRT1 plasmid of Pseudomonas putida DOT-T1E encodes functions relevant for survival under harsh conditions in the environment. Environ. Microbiol. 13, 2315–2327. doi: 10.1111/j.1462-2920.2011.02492.x
Mukhopadhyay, A. (2015). Tolerance engineering in bacteria for the production of advanced biofuels and chemicals. Trends Microbiol. 23, 498–508.
Nagai, H., and Kubori, T. (2011). Type IVB secretion systems of Legionella and other gram-negative bacteria. Front. Microbiol. 2:136. doi: 10.3389/fmicb.2011.00136
Nojiri, H., Shintani, M., and Omori, T. (2004). Divergence of mobile genetic elements involved in the distribution of xenobiotic-catabolic capacity. Appl. Microbiol. Biotechnol. 64, 154–174. doi: 10.1007/s00253-003-1509-y
Panke, S., Witholt, B., Schmid, A., and Wubbolts, M. G. (1998). Towards a biocatalyst for (S)-styrene oxide production: characterization of the styrene degradation pathway of Pseudomonas sp. strain VLB120. Appl. Environ. Microbiol. 64, 2032–2043. doi: 10.1128/AEM.64.6.2032-2043.1998
Papousek, I., Papagiannitsis, C. C., Medvecky, M., Hrabak, J., and Dolejska, M. (2017). Complete nucleotide sequences of two VIM-1-encoding plasmids from Klebsiella pneumoniae and Leclercia adecarboxylata isolates of Czech origin. Antimicrob. Agents Chemother. 61:e002648-16. doi: 10.1128/AAC.02648-16
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17.
Ramos, J. L., Duque, E., Huertas, M. J., and Haïdour, A. (1995). Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J. Bacteriol. 177, 3911–3916. doi: 10.1128/jb.177.14.3911-3916.1995
Rodríguez-Herva, J. J., García, V., Hurtado, A., Segura, A., and Ramos, J. L. (2007). The ttgGHI solvent efflux pump operon of Pseudomonas putida DOT-T1E is located on a large self-transmissible plasmid. Environ. Microbiol. 9, 1550–1561. doi: 10.1111/j.1462-2920.2007.01276.x
Rosche, T. M., Siddique, A., Larsen, M. H., and Figurski, D. H. (2000). Incompatibility protein IncC and global regulator KorB interact in active partition of promiscuous plasmid RK2. J. Bacteriol. 182, 6014–6026. doi: 10.1128/JB.182.21.6014-6026.2000
Rühl, J., Hein, E.-M., Hayen, H., Schmid, A., and Blank, L. M. (2012). The glycerophospholipid inventory of Pseudomonas putida is conserved between strains and enables growth condition-related alterations. Microb. Biotechnol. 5, 45–58. doi: 10.1111/j.1751-7915.2011.00286.x
Shintani, M., Sanchez, Z. K., and Kimbara, K. (2015). Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 6:242. doi: 10.3389/fmicb.2015.00242
Shintani, M., Suzuki, H., Nojiri, H., and Suzuki, M. (2022). Precise classification of antimicrobial resistance-associated IncP-2 megaplasmids for molecular epidemiological studies on Pseudomonas species. J. Antimicrob. Chemother. 77, 1203–1205. doi: 10.1093/jac/dkac006
Sprouffske, K., and Wagner, A. (2016). Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17:172. doi: 10.1186/s12859-016-1016-7
Stothard, P., and Wishart, D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539. doi: 10.1093/bioinformatics/bti054
Suenaga, H., Fujihara, H., Kimura, N., Hirose, J., Watanabe, T., Futagami, T., et al. (2017). Insights into the genomic plasticity of Pseudomonas putida KF715, a strain with unique biphenyl-utilizing activity and genome instability properties. Environ. Microbiol. Rep. 9, 589–598. doi: 10.1111/1758-2229.12561
Sun, F., Zhou, D., Wang, Q., Feng, J., Feng, W., Luo, W., et al. (2016). Genetic characterization of a novel blaDIM-2-carrying megaplasmid p12969-DIM from clinical Pseudomonas putida. J. Antimicrob. Chemother. 71, 909–912. doi: 10.1093/jac/dkv426
Sun, X., and Dennis, J. J. (2009). A novel insertion sequence derepresses efflux pump expression and preadapts Pseudomonas putida S12 for extreme solvent stress. J. Bacteriol. 191, 6773–6777. doi: 10.1128/JB.00832-09
Tett, A., Spiers, A. J., Crossman, L. C., Ager, D., Ciric, L., Dow, J. M., et al. (2007). Sequence-based analysis of pQBR103; a representative of a unique, transfer-proficient mega plasmid resident in the microbial community of sugar beet. ISME J. 1, 331–340. doi: 10.1038/ismej.2007.47
Turner, P. E., Cooper, V. S., and Lenski, R. E. (1998). Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution 52, 315–329. doi: 10.1111/j.1558-5646.1998.tb01634.x
Verhoef, S., Ruijssenaars, H. J., de Bont, J. A. M., and Wery, J. (2007). Bioproduction of p-hydroxybenzoate from renewable feedstock by solvent-tolerant Pseudomonas putida S12. J. Biotechnol. 132, 49–56. doi: 10.1016/j.jbiotec.2007.08.031
Verhoef, S., Wierckx, N., Westerhof, R. G. M., de Winde, J. H., and Ruijssenaars, H. J. (2009). Bioproduction of p-hydroxystyrene from glucose by the solvent-tolerant bacterium Pseudomonas putida S12 in a two-phase water-decanol fermentation. Appl. Environ. Microbiol. 75, 931–936. doi: 10.1128/AEM.02186-08
Volkers, R. J. M., Ballerstedt, H., de Winde, J. H., and Ruijssenaars, H. J. (2010). Isolation and genetic characterization of an improved benzene-tolerant mutant of Pseudomonas putida S12. Environ. Microbiol. Rep. 2, 456–460. doi: 10.1111/j.1758-2229.2010.00172.x
Volkers, R. J. M., de Jong, A. L., Hulst, A. G., van Baar, B. L. M., de Bont, J. A. M., and Wery, J. (2006). Chemostat-based proteomic analysis of toluene-affected Pseudomonas putida S12. Environ. Microbiol. 8, 1674–1679. doi: 10.1111/j.1462-2920.2006.01056.x
Volkers, R. J. M., Snoek, L. B., Ruijssenaars, H. J., and de Winde, J. H. (2015). Dynamic response of Pseudomonas putida S12 to sudden addition of toluene and the potential role of the solvent tolerance gene trgI. PLoS One 10:e0132416. doi: 10.1371/journal.pone.0132416
Wein, T., Wang, Y., Hülter, N. F., Hammerschmidt, K., and Dagan, T. (2020). Antibiotics interfere with the evolution of plasmid stability. Curr. Biol. 30, 3841–3847.
Weiser, R., Green, A. E., Bull, M. J., Cunningham-Oakes, E., Jolley, K. A., Maiden, M. C. J., et al. (2019). Not all pseudomonas aeruginosa are equal: strains from industrial sources possess uniquely large multireplicon genomes. Microb. Genomics 5:e000276. doi: 10.1099/mgen.0.000276
Wery, J., Hidayat, B., Kieboom, J., and de Bont, J. A. (2001). An insertion sequence prepares Pseudomonas putida S12 for severe solvent stress. J. Biol. Chem. 276, 5700–5706. doi: 10.1074/jbc.M007687200
Wierckx, N. J. P., Ballerstedt, H., de Bont, J. A. M., and Wery, J. (2005). Engineering of solvent-tolerant Pseudomonas putida S12 for bioproduction of phenol from glucose. Appl. Environ. Microbiol. 71, 8221–8227. doi: 10.1128/AEM.71.12.8221-8227.2005
Wijte, D., van Baar, B. L. M., Heck, A. J. R., and Altelaar, A. F. M. (2011). Probing the proteome response to toluene exposure in the solvent tolerant Pseudomonas putida S12. J. Proteome Res. 10, 394–403. doi: 10.1021/pr100401n
Xiong, J., Alexander, D. C., Ma, J. H., Déraspe, M., Low, D. E., Jamieson, F. B., et al. (2013). Complete sequence of pOZ176, a 500-kilobase IncP-2 plasmid encoding IMP-9-mediated carbapenem resistance, from outbreak isolate Pseudomonas aeruginosa 96. Antimicrob. Agents Chemother. 57, 3775–3782. doi: 10.1128/AAC.00423-13
Xiong, J., Hynes, M. F., Ye, H., Chen, H., Yang, Y., M’Zali, F., et al. (2006). blaIMP-9 and its association with large plasmids carried by Pseudomonas aeruginosa isolates from the People’s Republic of China. Antimicrob. Agents Chemother. 50, 355–358. doi: 10.1128/AAC.50.1.355-358.2006
Zhang, X., Wang, L., Li, D., Li, P., Yuan, L., Yang, F., et al. (2021). An IncP-2 plasmid sublineage associated with dissemination of bla IMP-45 among carbapenem-resistant Pseudomonas aeruginosa. Emerg. Microbes Infect. 10, 442–449. doi: 10.1080/22221751.2021.1894903
Keywords: Pseudomonas putida, genome sequence, solvent tolerance, megaplasmid, mobile genetic elements, comparative analysis
Citation: Kusumawardhani H, Hosseini R, Verschoor J-A and de Winde JH (2022) Comparative analysis reveals the modular functional structure of conjugative megaplasmid pTTS12 of Pseudomonas putida S12: A paradigm for transferable traits, plasmid stability, and inheritance? Front. Microbiol. 13:1001472. doi: 10.3389/fmicb.2022.1001472
Received: 23 July 2022; Accepted: 06 September 2022;
Published: 23 September 2022.
Edited by:
Ulrike Kappler, The University of Queensland, AustraliaReviewed by:
Rongzhen Zhang, Jiangnan University, ChinaChiho Suzuki-Minakuchi, The University of Tokyo, Japan
Copyright © 2022 Kusumawardhani, Hosseini, Verschoor and de Winde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johannes H. de Winde, j.h.de.winde@biology.leidenuniv.nl
†These authors have contributed equally to this work and share first authorship
 Hadiastri Kusumawardhani
Hadiastri Kusumawardhani Rohola Hosseini1†
Rohola Hosseini1† Jo-Anne Verschoor
Jo-Anne Verschoor Johannes H. de Winde
Johannes H. de Winde