- 1Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, MD, United States
- 2Fachbereich Biologie, Abt. Mikrobiologie, Technische Universität Kaiserlautern, Kaiserslautern, Germany
The ability to obtain purified biliverdin IX (BVIX) isomers other than the commercially available BVIXα is limited due to the low yields obtained by the chemical coupled oxidation of heme. Chemical oxidation requires toxic chemicals, has very poor BVIX yields (<0.05%), and is not conducive to scalable production. Alternative approaches utilizing recombinant E. coli BL21 expressing a cyanobacterial heme oxygenase have been employed for the production BVIXα, but yields are limited by the rate of endogenous heme biosynthesis. Furthermore, the emerging roles of BVIXβ and BVIXδ in biology and their lack of commercial availability has led to a need for an efficient and scalable method with the flexibility to produce all three physiologically relevant BVIX isomers. Herein, we have taken advantage of an optimized non-pathogenic E. coli Nissle (EcN(T7)) strain that encodes an endogenous heme transporter and an integrated T7 polymerase gene. Protein production of the Pseudomonas aeruginosa BVIXβ and BVIXδ selective heme oxygenase (HemO) or its BVIXα producing mutant (HemOα) in the EcN(T7) strain provides a scalable method to obtain all three isomers, that is not limited by the rate of endogenous heme biosynthesis, due to the natural ability of EcN(T7) to transport extracellular heme. Additionally, we have optimized our previous LC-MS/MS protocol for semi-preparative separation and validation of the BVIX isomers. Utilizing this new methodology for scalable production and separation we have increased the yields of the BVIXβ and -δ isomers >300-fold when compared to the chemical oxidation of heme.
Introduction
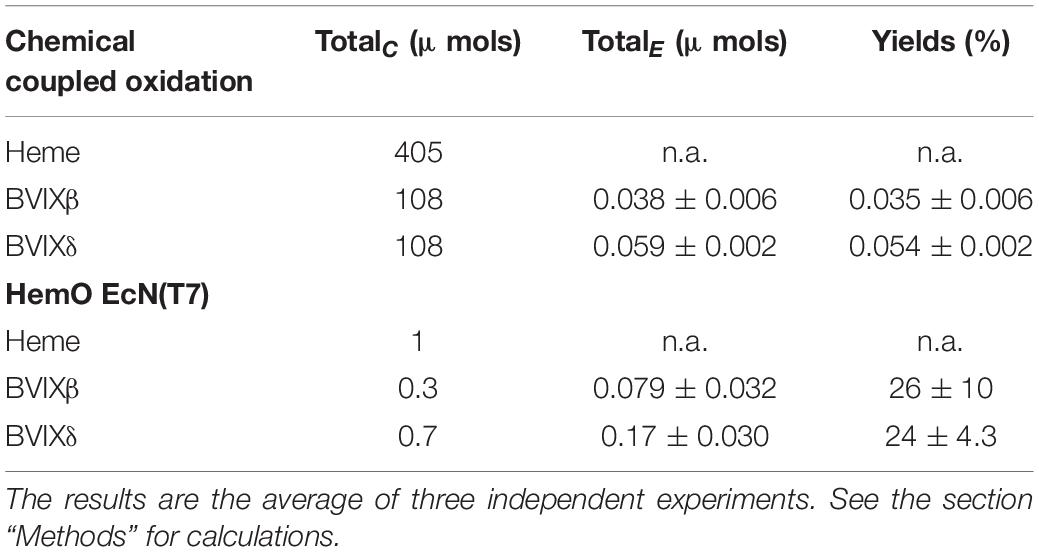
The catabolism of heme by the mammalian heme oxygenase (HO) produces biliverdin IXα (BVIXα) a metabolite well known for its anti-inflammatory and immunosuppressant functions through indirectly downregulating pro-inflammatory cytokines (TLR-4) (Bisht et al., 2014a) and upregulating anti-inflammatory cytokines (IL-10) (Bisht et al., 2014b) via biliverdin reductase (BVR) nitrosylation or phosphorylation, respectively. These modifications through subsequent signal transductions pathways lead to decreased TLR-4 expression and increased IL-10 production (Bisht et al., 2014a,b). In addition, BVIXα has anti-mutagenic (Arimoto et al., 1980; Bulmer et al., 2008) as well as anti-oxidant functions via the conversion of bilirubin IXα (BRIXα) to BVIXα and back via BVR (Stocker et al., 1987; Sedlak and Snyder, 2004; Baylor and Butler, 2019). HOs are seen across all kingdoms of life from bacteria to mammals where their main function is to break down heme into CO, BVIX metabolites, and iron for reutilization. Most HO’s have been found to produce the BVIXα isomer, except for the pathogen Pseudomonas aeruginosa whose heme oxygenase (HemO) degrades heme with the release of BVIXβ and -δ (Figure 1; Friedman et al., 2004). Recently there has been an increase in reports that other isoforms of BVIX, specifically BVIXβ and BVIXδ, play important physiological roles in eukaryotic and prokaryotic systems. In humans, BVIXβ is only seen during the first 20 weeks of gestation via the detection of bilirubin-IXβ in human fetal bile (Yamaguchi and Nakajima, 1995) and the expression of a human BVIXβ reductase (Yamaguchi et al., 1994; Pereira et al., 2001). While the degradation of heme to BVIXα by the mammalian HO’s (HO-1 and HO-2) has been well studied (Maines et al., 1986), the origin of the BVIXβ isomer is not as well understood. It is suggested that the production of BVIXβ may be the result of the switch from embryonic to fetal to adult hemoglobin during the birthing process (Pereira et al., 2001).
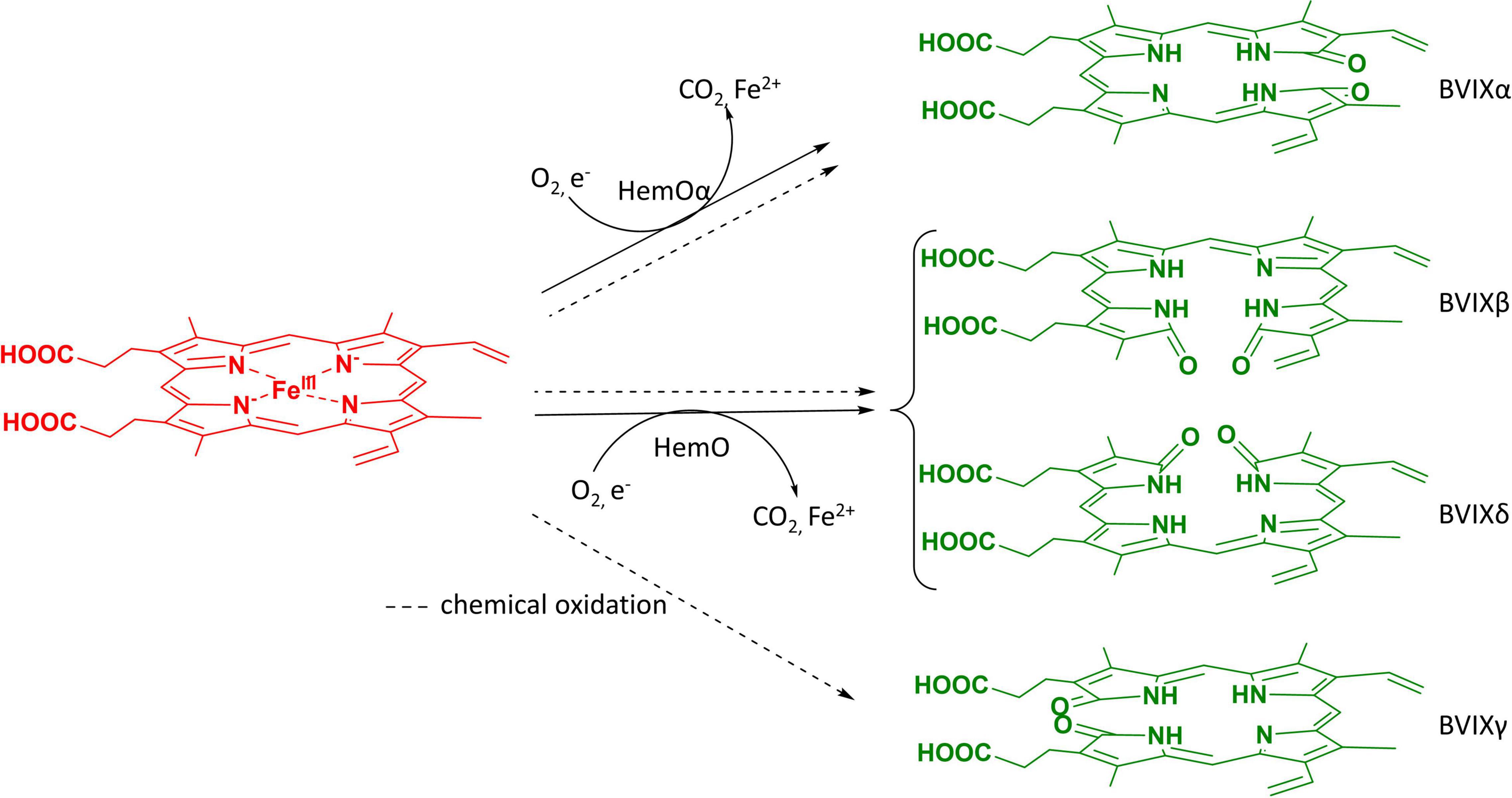
Figure 1. BVIX isomers produced by heme oxygenase or chemical oxidation. Heme oxygenases (HO’s) including mammalian and bacterial catalyze heme to BVIXα, CO2, and Fe2+. P. aeruginosa HemO degrades heme releasing the BVIXβ, and -δ isomers. All four isomers, including BVIXγ, are produced during the chemical oxidation reaction.
In the last two decades several bacterial pathogens have been shown to encode heme oxygenases and dioxygenases that degrade heme to release iron, a micronutrient required for survival and pathogenesis (Wilks and Heinzl, 2014; Wilks and Ikeda-Saito, 2014). In the opportunistic pathogen P. aeruginosa the iron regulated HemO oxidatively cleaves heme to release BVIXβ and BVIXδ (Ratliff et al., 2001). Heme uptake studies combined with LC-MS/MS analysis of the P. aeruginosa hemO deletion strain showed HemO is absolutely required to drive heme uptake into the cell (O’Neill et al., 2012). The significance of HemO in P. aeruginosa iron acquisition and its role in virulence has led to extensive study of the protein itself (Ratliff et al., 2001; Friedman et al., 2004). Additionally, recent studies by our lab have also shown the importance of HemO metabolites, BVIXδ and -β isomers, as signaling molecules in the post-transcriptional regulation of the P. aeruginosa heme sensing and acquisition pathways (Mourino et al., 2016). As heme acquisition has been shown to be the primary source of iron in P. aeruginosa chronic infection at the expense of iron siderophore systems (Marvig et al., 2014; Nguyen et al., 2014), HemO’s dual function in both sensing and uptake is clearly important for P. aeruginosa colonization and infection. Furthermore, targeting P. aeruginosa HemO has proven to be an effective therapeutic strategy by inhibiting both iron acquisition and the ability to sense extracellular heme (Liang et al., 2018; Centola et al., 2020; Robinson et al., 2021). Moreover, P. aeruginosa encodes a second BVIXα-dependent heme oxygenase, BphO, directly upstream of BphP, a sensor kinase as part of a two-component signaling system (Wegele et al., 2004; Barkovits et al., 2008). BVIXα functions as a far-red light chromophore for BphP and has been shown to be linked to the two component system KinB-AlgB which controls genes involved in biofilm formation (Mukherjee et al., 2019).
Currently, BVIXα is a commercially available compound produced by chemical oxidation of bilirubin sourced from mammalian bile (McDonagh and Palma, 1980), or by utilizing recombinant E. coli expression of HO from either cyanobacteria (Cornejo et al., 1998; Chen et al., 2012), rat (Ishikawa et al., 1991; Wilks and Ortiz de Montellano, 1993), or yeast (Michael and Pendrak, 2005). A scalable bioreactor method utilizing a cyanobacterial HO showed enhanced production and comparable purity to commercial BVIXα, however, the method is limited by the rate of endogenous heme biosynthesis (Chen et al., 2012). Furthermore, recombinant expression systems to date have been limited to the production of BVIXα. For the past 80 years, chemical oxidation of heme (McDonagh and Palma, 1980) and the oxidation of bilirubin IXα to BVIXα(McDonagh and Assisi, 1971; Elich et al., 1989) have been the main methods for producing all four BVIX isomers (Figure 1). While there has been some improvement in the production and separation of the four isomers, through preparative TLC methods of the esterified BVIX isomers (Heirwegh et al., 1991) to high-performance liquid chromatography (HPLC) methods of the BVIX isomers (Barker et al., 2012; Robinson et al., 2021), the overall yields especially those of BVIXβ and -δ are very low (Zhang et al., 2020). Recently, Zhang et al., (2020) described the scalable recombinant expression of the P. aeruginosa HemO as a means of obtaining BVIXβ and BVIXδ, however, the system is again limited by the rate of endogenous heme biosynthesis. Herein we report on a scalable BVIX production method that exploits the over expression of P. aeruginosa HemO in a recently engineered strain of the non-pathogenic probiotic (O6:K5:H1) E. coli strain Nissle 1917 (EcN) (Lodinova-Zadnikova et al., 1998; Rembacken et al., 1999; Sonnenborn, 2016). The EcN genome encodes an endogenous heme receptor, ChuA, which was shown to increase exogenous heme uptake into the cell (Fiege et al., 2018). This strain was further optimized by integrating a T7 RNA polymerase gene into the chromosome of EcN (EcN(T7)) for use with a wide array of T7-promoter expression vectors (Fiege and Frankenberg-Dinkel, 2020). The combination of an outer membrane receptor ChuA for heme uptake and a T7 RNA polymerase for over-expression of genes make this an ideal strain for the production of fully reconstituted heme proteins. Additionally, the strain can be used to express or co-express genes to produce heme metabolites such as the BVIXs or related linear tetrapyrroles. Herein, we have exploited the EcN(T7) strain for the over-expression of the P. aeruginosa HemO as a means of generating BVIXβ and BVIXδ on supplementation of the cultures with exogenous heme. Additionally, over expression of the BVIXα producing HemO N19K/K34A/ F117Y/K132A mutant (Barker et al., 2012; Heinzl et al., 2016) (here on referred to as HemOα) allowed for production of BVIXα by a similar extraction and purification method for direct comparison purposes.
Additionally, we have improved upon the biliverdin extraction and HPLC protocol for the separation of the BVIX isomers utilizing our recently optimized LC-MS/MS assay (Dent and Wilks, 2020). This method to produce the BVIXβ and -δ isomers give a >300-fold increase in yield of the individual isomers when compared to the chemical coupled oxidation of heme while avoiding the use of toxic chemicals. Furthermore, the EcN(T7) strain has several advantages over other reported expression systems, namely the ability to feed heme to the cells side stepping the rate limiting step of heme biosynthesis and the scalable nature of the system for high-volume bioreactor systems.
Methods
Reagents and Materials
All glassware and metalware were cleaned in a solution of 0.1 M NaOH and 1% SDS, rinsed with MilliQ water, autoclaved at 550°C. Precautions to avoid contaminants within the BVIX isomer extraction and HPLC separation were taken using HPLC grade reagents and solvents (Fisher Optima). Similar precautions for LC-MS/MS BVIX isomer validation using LC-MS grade (Fisher Optima and Thermo Scientific for DMSO) reagents and solvents. Specific medias, columns, and instruments are listed below where appropriate.
Biliverdin IX Isomer Production via Heme Oxygenase Over-Expression in Escherichia coli Nissle (T7)
Escherichia coli Nissle(T7) cells were obtained from the Frankenberg-Dinkel laboratory. EcN(T7) electrocompetent cell were made fresh as previously described (Dower et al., 1988) with some modifications. Briefly, 5 mL LB (Lennox) cultures inoculated with EcN(T7) were shaken at 210 rpm overnight at 37°C in 10 mL culture tubes. The overnight culture was diluted into fresh 25 mL LB cultures in a 125 ml flask to a final OD600 = 0.05 and grown with shaking (210 rpm) at 37°C until the OD600 reached 0.5–0.6 (mid-log phase). The culture was then centrifuged at 5,000 × g at 4°C for 5 min and pellets were washed with 25 mL chilled 10% glycerol and the wash repeated twice more. Following the washing steps the culture was recentrifuged and suspended in 250 μL of 10% glycerol to a final 100x lower volume from the initial culture. 30 μL aliquots of the final cell volume (250 μL) were used for electroporation or stored at −80°C for up to 3 months. Electroporation was performed as previously described (Miller and Nickoloff, 1995) with slight modifications. Briefly, 1–5 μL of pEThemO or pEThemOα (Ratliff et al., 2001; Heinzl et al., 2016; Liang et al., 2018; Robinson et al., 2021), was electroporated into electrocompetent EcN(T7) cells using prechilled 2 mm gap electroporation cuvettes and a BioRad Micropulser Electroporator on the “Bacteria: Ec1” setting (1.8 kV, 1 pulse). 1 mL SOC media at 25°C was immediately added to the electroporated culture, transferred to an ice-cold culture tube, and set to shake at 210 rpm for at least 1 h at 37°C. 250 μL of electroporated cells were then plated onto LB agar containing a final concentration of 100 μg/mL ampicillin and incubated overnight at 37°C. The resulting colonies were inoculated into 25 mL LB media containing 100 μg/mL ampicillin and shaken at 250 rpm overnight at 37°C. HemO overexpression was performed as previously described (Ratliff et al., 2001; Heinzl et al., 2016; Robinson et al., 2021) with slight modification to increase aeration and optimize HemO expression. Briefly, an overnight culture was diluted into fresh 100 mL of LB media containing 100 μg/mL ampicillin in 250 mL baffled flasks and shaken at 250 rpm for 1.5–2 h at 37°C or until they reached an OD600 = 0.4–0.5. Cultures were induced with 1 mM IPTG and shaken at 250 rpm for a further 2 h at 25°C. The cultures were then supplemented with appropriate concentrations of heme (10, 15, 20, and 25 μM) prepared in DMSO (10–100 μL) and shaken at 250 rpm overnight at 25°C. Cultures were pelleted (5,000 × g, 20 min, 4°C) in 50 mL conical tubes, and the supernatants were collected for BVIX extraction or stored at −80°C for up to 1 month. Pellets were thawed, resuspended in Milli Q water and 2 μg/mL DNase I, sonicated at 100% amplification for 5 secs on and 15 secs off for 2 min, and run on a 12% SDS PAGE gel at 200V for 30 min as previously described with slight modifications (Heinzl et al., 2016; Liang et al., 2018; Robinson et al., 2021).
Extraction of Biliverdin IX Isomers From Escherichia coli Nissle(T7) Supernatants
Biliverdin IX isomers were extracted as previously described (Mourino et al., 2016) with slight modification. Briefly, supernatants were filtered through a 0.2 μm PVDF membrane to remove any remaining particulate matter. In the dark, supernatants were acidified to pH 2.5 with 10% TFA, and loaded over a C18 Sep-Pak column (35cc, Waters) equilibrated with 20 mL each of acetonitrile (ACN), H2O, 0.1% TFA in H2O, and methanol:0.1% TFA (10:90). After sample application, the column was washed with 40 mL 0.1% TFA, 40 mL ACN:0.1% TFA (20:80), 20 mL methanol:0.1% TFA (50:50) and eluted with 15 mL methanol. Alternatively, for smaller batches the isomers were purified over a 3cc (Waters) C18 Sep-Pak column equilibrated as above scaling back the washes and elution volumes x10. The supernatant was centrifuged (14,800 rpm, 5 min, 25°C) and the remaining solution containing purified BVIX isomers was collected. Purified BVIX isomers were diluted with an equal volume of water and extracted into CHCl3 (3–5 ml) washed with ×3 with 5 ml H2O to remove any remaining acid. The CHCl3 layer was collected, and speed vacuumed dry and stored at −80 °C prior to HPLC separation.
HPLC Separation of Biliverdin IX Isomers
Dried BVIX aliquots were resuspended in 100 μL HPLC-grade methanol and centrifuged (14,800 rpm, 5 min, 25°C) to remove any particulates. The BVIX isomers were then separated by HPLC (Thermo Fisher UltimateTM 3000 HPLC) on an SynergiTM 4 μm Fusion-RP 80 Å C18 Column (250 × 10 mm) at a flow rate of 3 mL/min. The mobile phase consisted of solvent A, H2O:0.1% formic acid, and solvent B, ACN:0.1% formic acid. A linear gradient was run starting at 64% A and 36% B and ending with 0% A and 100% B over 20 or 30 mins. BVIX isomer retention times were 9.9, 10.8, and 11.6 min for BVIXα, BVIXδ, and BVIXβ isomers, respectively, for the 20 min linear gradient and 9.7, 12.1, and 13 min, respectively for the 30 min linear gradient. Separated isomers were then extracted into choloroform as described above, speed vacuumed dry and stored at −80°C. The final yield of each BVIX isomer was calculated from their respective millimolar extinction coefficients (εmM) at 376 nm and 665 nm in methanol as previously described (Heirwegh et al., 1991). BVIXα, -β, and -δ millimolar extinction coefficients at 376 nm are 50.6, 51.7, and 47.1 mM–1 cm–1, respectively, and at 665 nm are 14.6, 15.4, and 15.5 mM–1 cm–1, respectively. Yields were calculated using Eq. 1 below:
TotalC represents the total μmols of BVIX calculated to give 100% yield and TotalE represents the total μmols of BVIX obtained experimentally. For the BVIX production method by coupled oxidation TotalC for the individual BVIX isomers was calculated based on 25% of the total BVIX yield, while the HemO EcN(T7) BVIX production method, TotalC for BVIXβ, and -δ isomers were calculated based on HemO BVIX ratio of 30:70, respectively. Molar extinction coefficients of BVIXα, -β, and -δ isomers in DMSO were obtained using 380 ± 1/658 ± 3 nm, 389 ± 1/651 ± 1 nm, and 386 ± 2/645 ± 5 nm, respectively, (n = 3) based on the UV spectrum (Supplementary Figure 5). Extinction coefficients (ε) of BVIXα, -β, and -δ isomers in DMSO were calculated to be 20 ± 0.2/4.9 ± 0.2 mM–1 cm–1, 35.9 ± 12.0/9.0 ± 4.0 mM–1 cm–1, and 11.5 ± 5.3, 2.9 ± 1.4 mM–1 cm–1, respectively (Supplementary Figure 5).
LC-MS/MS Analysis of Biliverdin IX Isomers
Following HPLC separation the respective BVIX isomers were confirmed by LC-MS/MS as previously described with slight modification (Dent and Wilks, 2020). Briefly, the purified BVIX isomers were resuspended in 10 μL DMSO, diluted to 100 μL with mobile-phase ACN:H2O (50:50, v/v), and centrifuged at 14,000 rpm for 5 min at 4°C to remove particulates. BVIX isomers (2 μL) were analyzed on a Waters TQ-XS triple quadrupole mass spectrometer with AQUITY H-Class UPLC fitted with an Ascentis RP-amide 2.7-mm C18 column (10 cm × 2.1 mm) at 30oC with a flow rate of 0.4 mL/min. The mobile phase consisted of solvent A, H2O:0.1% formic acid, and solvent B, ACN:0.1% formic acid. The initial gradient was 64% A and 36% B followed by 5 min at 55% A and 45% B, 8 min at 40% A and 60% B, 8.5 min at 5% A and 95% B, and 10 min 64% A and 36% B. Using multiple-reaction monitoring (MRM), the BVIX precursor ions were detected at 583.21 and the major ions following fragmentation for BVIXα, BVIXβ, and BVIXδ isomers were 209.2/296.6, 343.1, and 402.2 m/z, with collision energies of 38, 36, and 30 V, respectively. The source temperature was set to 150°C, the capillary voltage to 3.60 kV, and the cone voltage to 43 V. Purity of the BVIX isomers was determined by the ratio of the integration of the isomer peaks to the total integration of the LC chromatograms.
Results
Optimization of Biliverdin IX Production via Heme Oxygenase Expression in Escherichia coli Nissle(T7)
Expression of HemO in EcN(T7) was performed across a range of induction temperatures, incubation times, and shaking conditions (Figure 2). The optimal temperature and incubation time for maximum production of BVIX was found to be 25 °C for 16 h with 250 rpm shaking as judged by the green pigmentation in the supernatant (Figure 2). We found increased shaking improved BVIX yields as the increased oxygenation aided HemO-dependent heme degradation. Similarly, we tested a range of heme concentrations (10, 15, 20, and 25 μM) and found 10 μM heme gave the highest BVIX yield whereas higher concentrations led to a decrease in growth rate, most likely a result of heme toxicity. The expression of HemOα in EcN(T7) was slightly lower than that of HemO as judged by SDS PAGE (Supplementary Figure 4). The reason for the decrease in expression of HemOα is not clear as this was not previously observed on expression of HemO in E. coli BL21(DE3) (Mourino et al., 2016).
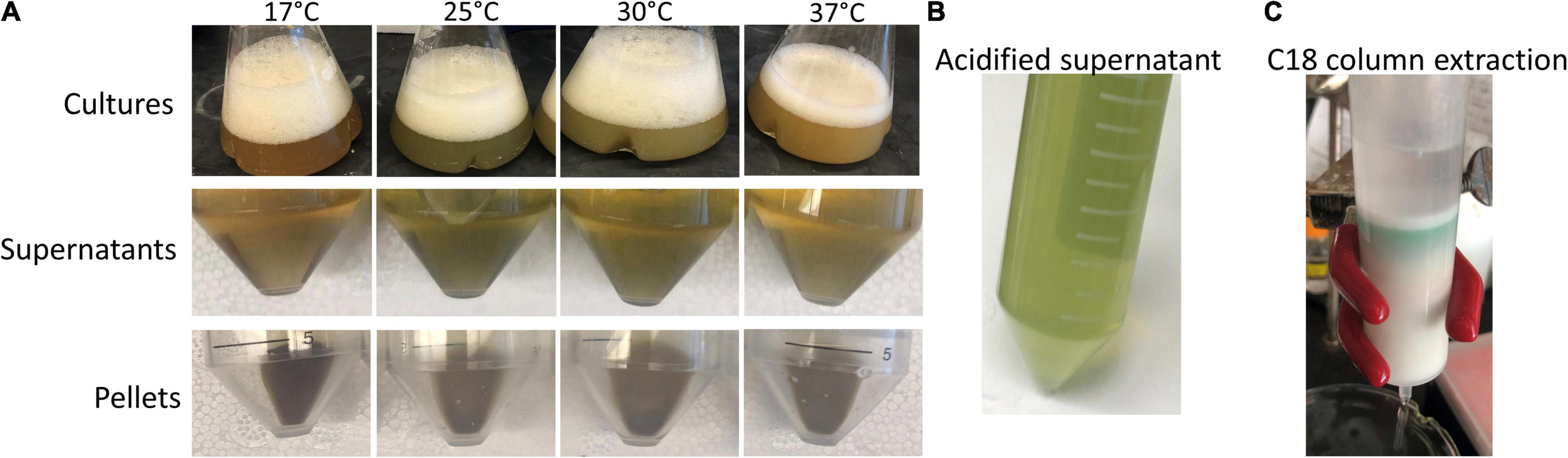
Figure 2. Induction temperature optimization of HemO over-expression in EcN(T7) cells. (A) Cultures, supernatants, and pellets after 16 h of induction at different temperatures (17, 25, 30, and 37) shaking at 250 rpm, supplemented with 10 μM heme, and induced with 1 mM IPTG. (B) Acidified supernatant. (C) Acidified and filtered supernatant applied onto a C18 Sep-Pak column (35cc, Waters) showing a green band of BVIXδ and BVIXβ isomers.
Biliverdin IX Isomers Extraction and Purification
The C18 BVIX isomer extraction was performed in semi-darkness to protect the isomers from photoisomerization. The C18 eluted BVIX fraction was centrifuged to remove any excess acidified proteins and the UV-visible spectrum of the fraction was recorded. The absorption spectrum while characteristic of BVIX with a broad absorbance at 680 nm (Heirwegh et al., 1991) did appear to contain some contaminants most likely heme, as judged by the peak at 400 nm and the shoulder at 630 nm. (Figure 3B). Following HPLC purification (Figure 3A) the separated BVIXβ and -δ isomers were analyzed by absorption spectroscopy (Figure 3B) and LC-MS/MS (Figure 4). The absorption spectrum of the purified BVIXβ, and -δ isomers showed peaks at 378 and 650 nm, and 376 and 650 nm, respectively (Figure 3B). The BVIXα from the over-expression of HemOα in EcN(T7) was similarly purified and analyzed as shown in the Supplementary Information (Supplementary Figures 2, 3). Purified BVIXα yielded an absorption spectrum with peaks at 374 and 650 nm (Supplementary Figure 2B). The resulting purified BVIX isomers were extracted into chloroform washed, speed vacuum dried and stored in the dark at −80°C.
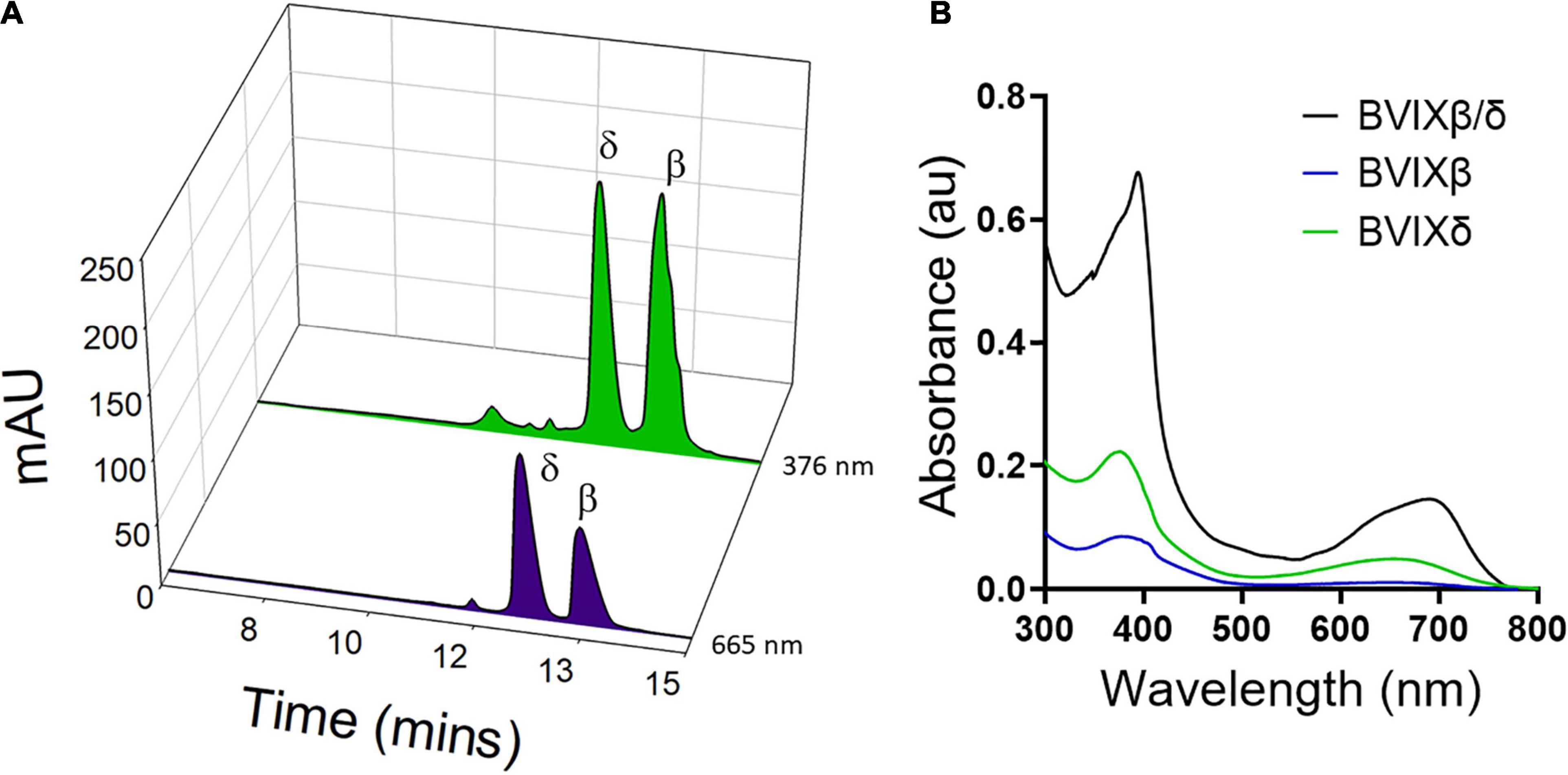
Figure 3. HPLC separation of BVIXδ, and -β isomers from supernatant. (A) HPLC separation of extracted BVIXδ and -β isomers using a linear gradient of solvent A, ACN:0.1% formic acid and B, H2O:0.1% formic acid over 30 min with retention times 12.1 min and 13.0 min, respectively. (B) UV-vis spectrum of C18 purified BVIXβ/δ (peaks at 394 and 680 nm) and HPLC purified BVIXβ (peaks at 378 and 650 nm), and -δ isomers (peaks at 376 and 650 nm) in 100% MeOH.
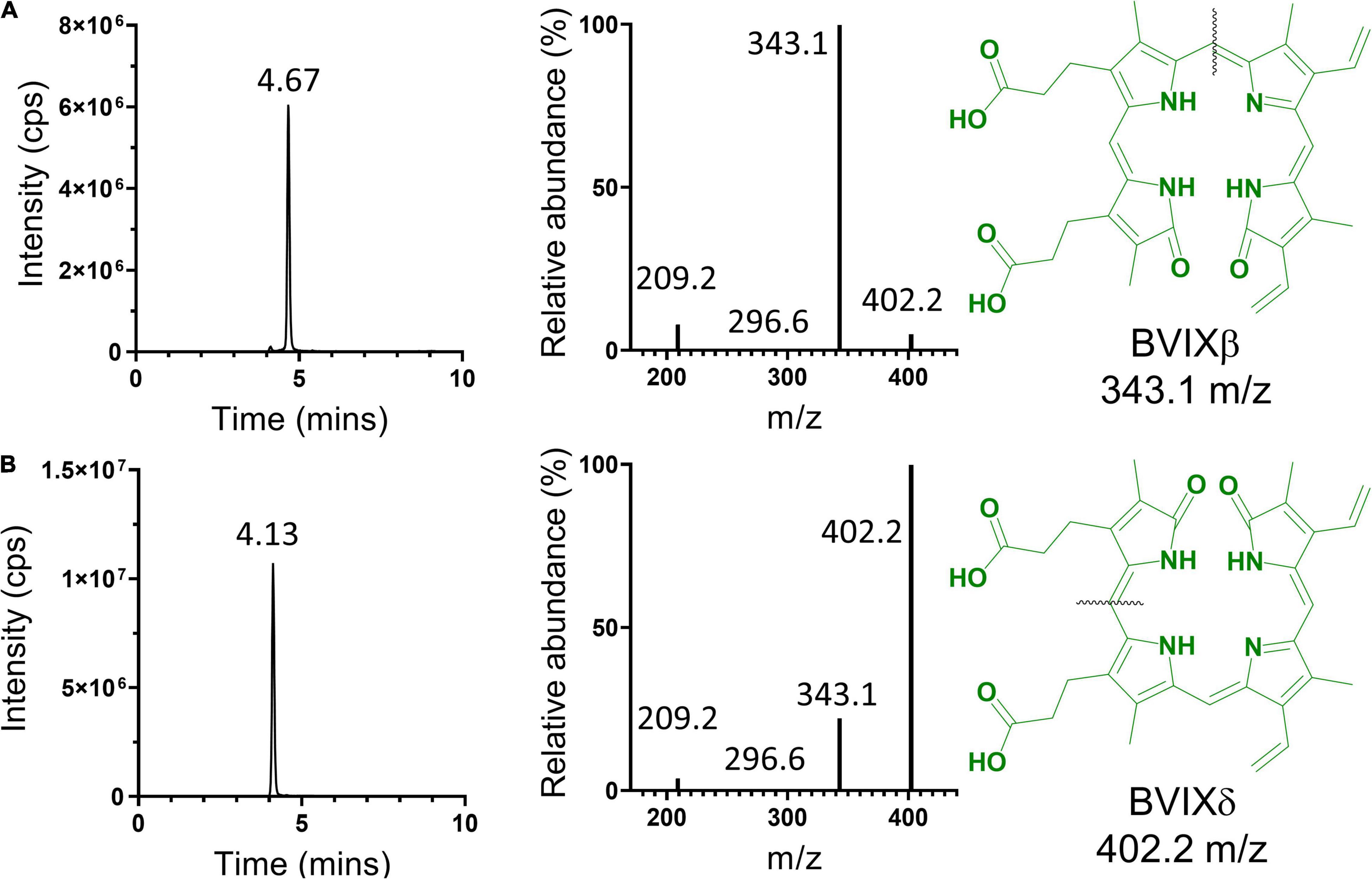
Figure 4. LC-MS/MS analysis of HPLC separated BVIX isomers. BVIXβ (A), and -δ (B) isomers with LC retention times of 4.67 and 4.13 min, respectively. BVIXβ (A), and -δ (B) isomers with majority precursor ions of 343.1, and 402.2 m/z, respectively. Black lines represent the fragmentation pattern for the respective isomers.
Our newly optimized HPLC method using a linear gradient consisting of solvent A, H2O:0.1% formic acid, and solvent B, ACN:0.1% formic acid can achieve baseline separation of the BVIXβ and -δ isomers, (Figure 3A). Comparing the yields between the BVIXβ and -δ isomer production by over expression of HemO in EcN(T7) (26 and 24%, respectively) to the chemical coupled oxidation (0.035 and 0.054%, respectively) shows a >700-fold and >400-fold increase, respectively (Table 1). The yields of BVIXα from the over expression of HemOα in EcN(T7) (1.4%) are lower than that of HemO WT but still >15-fold higher than the yield from the chemical coupled oxidation (0.09%) (Table 1).
LC-MS/MS Validation of Biliverdin IX Isomers
The HPLC purified BVIXδ and -β isomers were further validated by LC-MS/MS and shown to have >99% purity based on the fragmentation patterns (Figure 4). Fragmentation and MRM of the BVIX precursor ion (583.21 m/z) yielded ions for BVIX-δ and -β isomers at 402.2 and 343.1 m/z, respectively (Figure 4). Similarly, for BVIXα the purity was >99% as judged by the major ion at 209.2 m/z (Supplementary Figure 3).
Discussion
Previous work utilizing HemO over-expression in E. coli BL21 (DE3) has been used as a means of producing BVIXδ and BVIXβ (Zhang et al., 2020). Similarly, in our own work the E. coli BL21 (DE3) strain was employed to produce BVIXδ and BVIXβ, but the yields obtained were lower than that of the chemical coupled oxidation of heme (<0.05%), even when supplementing cultures with exogenous heme or the heme precursor δ-aminolevulinic acid to promote heme biosynthesis. Given the ability of the engineered EcN(T7) to naturally transport heme we coupled this to the over expression of HemO in an attempt to maximize BVIX yields. Interestingly, following centrifugation of the cultures, we saw no green coloration in the pellet as was observed on HemO expression in E. coli BL21 (DE3) (Wilks, 2003; Zhang et al., 2020). However, the green pigmentation in the supernatant, especially after acidification (Figures 2A,B), suggests that EcN(T7) cells may excrete the BVIX isomers more efficiently (Figure 2A and Supplementary Figure 1). We have previously shown P. aeruginosa (PAO1) also secretes BVIXα into the supernatant (Mourino et al., 2016), suggesting that bacteria such as EcN(T7) and PAO1 that acquire extracellular heme may also have a mechanism to secrete the BVIX metabolites. The secretion of the BVIX isomers proved advantageous by eliminating the necessity to lyse the cell pellet and denature HemO to release the BVIX isomers, as was necessary when expressing the protein in E. coli BL21 (DE3) strain where the majority of BVIX remains in the cell pellet (Supplementary Figure 1). The same lack of green coloration was observed in pellets following over-expression of HemOα. However, while BVIXα yields increased compared to the chemical oxidation of heme, they were significantly lower than the BVIXβ and -δ isomers produced by HemO WT, presumably due to the decreased HemOα protein levels compared to HemO (Supplementary Figure 1). Interestingly, we have not observed differences in the protein levels of HemO and HemOα in E. coli BL21 (DE3) cells (Mourino et al., 2016). Furthermore, following purification of HemO and HemOα, CD analysis showed both proteins have a similar overall structural fold and thermal stability profile (Mourino et al., 2016). Therefore, further optimization of the EcN(T7) expression system will be required for scale up of BVIXα production by either optimizing HemOα protein levels or employing an alternative bacterial or mammalian BVIXα-selective heme oxygenase. However, while the current yields on a per liter basis were lower (5–10 fold) than that previously reported for production of BVIXα in E. coli BL21(DE3), it should be noted that the authors utilized a modified cyanobacterial ho1 gene and performed the expression in a large scale Bioreactor (Chen et al., 2012). Furthermore, in the current protocol we employed an extra HPLC purification step following extraction and semi-purification by C18 chromatography. However, while the production of BVIXα in EcN(T7) systems can be further optimized the main goal of the current work was to develop a method to produce the non-commercially available BVIXβ and -δ isomers by a method other than chemical coupled oxidation. Compared to the chemical coupled oxidation of heme, this approach provides a greener alternative to obtain BVIXβ and -δ that no longer uses harsh chemicals such as pyridine and decreases the total chloroform (CHCl3) extraction steps of which both are toxic and carcinogenic (National Toxicology Program, 2000; Meek et al., 2002).
In addition to optimizing production of the BVIXδ and -β isomers, we further improved on the HPLC purification by optimizing a method that allows for baseline separation of the BVIXβ, and -δ isomers utilizing a linear gradient of ACN 0.1% formic acid and H2O 0.1% formic acid. Previous ACN-based HPLC methods for the separation of the BVIX isomers have reported baseline separation of BVIXα from the BVIXδ and -β isomers, but no baseline separation between the BVIXδ and -β isomers (Yamaguchi et al., 1979). Baseline separation of the BVIXδ and -β isomers by HPLC has been previously reported with a mobile phase consisting of 20 mM formic acid in acetone (Barker et al., 2012; Zhang et al., 2020). However, the use of such a mobile phase is not ideal given the increased polarity and volatility when compared to ACN leading to inconsistent retention times between HPLC runs.
Conclusion
Chemical oxidation heme or the oxidation of bilirubin have been the primary methods to obtain all four BVIX isomers or BVIXα, respectively. While the predominant role of BVIXα in mammalian physiology led to its commercial production, the growing awareness of the role of the BVIXδ and -β isomers particularly in bacterial systems has led to the need for alternative and greener methods to produce all three BVIX isomers. The method developed herein utilizing HemO’s unique regioselectivity coupled with the optimized EcN(T7) E. coli strain provides a cost effective and less toxic approach to obtaining semi-preparative quantities of the physiologically relevant BVIX isomers. This method increases the BVIXδ and -β isomer yields from <0.05 to >20%, a 400-fold increase. We have improved the HPLC separation process to include more widely acceptable and less harsh solvents. Furthermore, we have developed a bioreactor compatible bacterial expression system by which to produce all three physiologically relevant BVIX isomers and more importantly eliminate the use of harsh chemicals.
Data Availability Statement
The authors acknowledge that the data presented in this study must be deposited and made publicly available in an acceptable repository, prior to publication. Frontiers cannot accept a manuscript that does not adhere to our open data policies. The data presented in the study are deposited in the MetaboLights repository, accession number MTBLS3555.
Author Contributions
ER and AW were involved in the initial concept and design of the experiment as well as the acquisition and analysis of the data. ER, AW, NF-D, and FX were involved in the subsequent experimental design and data interpretation, and the drafting and revising of the manuscript for publication. All authors contributed to the article and approved the submitted version.
Funding
Funding from this work was generously provided by the NIH grants AI134886 to AW and NIH 5T32GM066706-17 to ER through the Chemistry and Biology Interface Fellowship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the University of Maryland Baltimore, School of Pharmacy Mass Spectrometry Center (SOP1841-IQB2014) for support and Center Director Professor Maureen Kane for her help and expertise.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.787609/full#supplementary-material
References
Arimoto, S., Negishi, T., and Hayatsu, H. (1980). Inhibitory effect of hemin on the mutagenic activities of carcinogens. Cancer Lett. 11, 29–33. doi: 10.1016/0304-3835(80)90125-1
Barker, K. D., Barkovits, K., and Wilks, A. (2012). Metabolic flux of extracellular heme uptake in Pseudomonas aeruginosa is driven by the iron-regulated heme oxygenase (HemO). J. Biol. Chem. 287, 18342–18350. doi: 10.1074/jbc.M112.359265
Barkovits, K., Harms, A., Benkartek, C., Smart, J. L., and Frankenberg-Dinkel, N. (2008). Expression of the phytochrome operon in Pseudomonas aeruginosa is dependent on the alternative sigma factor RpoS. FEMS Microbiol. Lett. 280, 160–168. doi: 10.1111/j.1574-6968.2007.01058.x
Baylor, J. L., and Butler, M. W. (2019). Immune challenge-induced oxidative damage may be mitigated by biliverdin. J. Exp. Biol. 222(Pt 6):jeb200055. doi: 10.1242/jeb.200055
Bisht, K., Tampe, J., Shing, C., Bakrania, B., Winearls, J., Fraser, J., et al. (2014a). Endogenous tetrapyrroles influence leukocyte responses to lipopolysaccharide in human blood: pre-clinical evidence demonstrating the anti-inflammatory potential of biliverdin. J. Clin. Cell Immunol. 5:1000218. doi: 10.4172/2155-9899.1000218
Bisht, K., Wegiel, B., Tampe, J., Neubauer, O., Wagner, K. H., Otterbein, L. E., et al. (2014b). Biliverdin modulates the expression of C5aR in response to endotoxin in part via mTOR signaling. Biochem. Biophys. Res. Commun. 449, 94–99. doi: 10.1016/j.bbrc.2014.04.150
Bulmer, A. C., Ried, K., Blanchfield, J. T., and Wagner, K. H. (2008). The anti-mutagenic properties of bile pigments. Mutat. Res. 658, 28–41. doi: 10.1016/j.mrrev.2007.05.001
Centola, G., Deredge, D. J., Hom, K., Ai, Y., Dent, A. T., Xue, F., et al. (2020). Gallium(III)-salophen as a dual inhibitor of Pseudomonas aeruginosa heme sensing and iron acquisition. ACS Infect. Dis. 6, 2073–2085. doi: 10.1021/acsinfecdis.0c00138
Chen, D., Brown, J. D., Kawasaki, Y., Bommer, J., and Takemoto, J. Y. (2012). Scalable production of biliverdin IXalpha by Escherichia coli. BMC Biotechnol. 12:89. doi: 10.1186/1472-6750-12-89
Cornejo, J., Willows, R. D., and Beale, S. I. (1998). Phytobilin biosynthesis: cloning and expression of a gene encoding soluble ferredoxin-dependent heme oxygenase from Synechocystis sp. PCC 6803. Plant J. 15, 99–107. doi: 10.1046/j.1365-313x.1998.00186.x
Dent, A. T., and Wilks, A. (2020). Contributions of the heme coordinating ligands of the Pseudomonas aeruginosa outer membrane receptor HasR to extracellular heme sensing and transport. J. Biol. Chem. 295, 10456–10467. doi: 10.1074/jbc.RA120.014081
Dower, W. J., Miller, J. F., and Ragsdale, C. W. (1988). High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16, 6127–6145. doi: 10.1093/nar/16.13.6127
Elich, T. D., Mcdonagh, A. F., Palma, L. A., and Lagarias, J. C. (1989). Phytochrome chromophore biosynthesis. Treatment of tetrapyrrole-deficient Avena explants with natural and non-natural bilatrienes leads to formation of spectrally active holoproteins. J. Biol. Chem. 264, 183–189.
Fiege, K., and Frankenberg-Dinkel, N. (2020). Construction of a new T7 promoter compatible Escherichia coli Nissle 1917 strain for recombinant production of heme-dependent proteins. Microb. Cell Fact 19:190. doi: 10.1186/s12934-020-01447-5
Fiege, K., Querebillo, C. J., Hildebrandt, P., and Frankenberg-Dinkel, N. (2018). Improved method for the incorporation of heme cofactors into recombinant proteins using Escherichia coli Nissle 1917. Biochemistry 57, 2747–2755. doi: 10.1021/acs.biochem.8b00242
Friedman, J., Lad, L., Li, H., Wilks, A., and Poulos, T. L. (2004). Structural basis for novel delta-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa. Biochemistry 43, 5239–5245. doi: 10.1021/bi049687g
Heinzl, G. A., Huang, W., Yu, W., Giardina, B. J., Zhou, Y., and Mackerell, A. D. Jr., et al. (2016). Iminoguanidines as allosteric inhibitors of the iron-regulated heme oxygenase (HemO) of Pseudomonas aeruginosa. J. Med. Chem. 59, 6929–6942. doi: 10.1021/acs.jmedchem.6b00757
Heirwegh, K. P., Blanckaert, N., and Van Hees, G. (1991). Synthesis, chromatographic purification, and analysis of isomers of biliverdin IX and bilirubin IX. Anal. Biochem. 195, 273–278. doi: 10.1016/0003-2697(91)90329-r
Ishikawa, K., Sato, M., and Yoshida, T. (1991). Expression of rat heme oxygenase in Escherichia coli as a catalytically active, full-length form that binds to bacterial membranes. Eur. J. Biochem. 202, 161–165. doi: 10.1111/j.1432-1033.1991.tb16357.x
Liang, D., Robinson, E., Hom, K., Yu, W., Nguyen, N., Li, Y., et al. (2018). Structure-based design and biological evaluation of inhibitors of the pseudomonas aeruginosa heme oxygenase (pa-HemO). Bioorg. Med. Chem. Lett. 28, 1024–1029. doi: 10.1016/j.bmcl.2018.02.027
Lodinova-Zadnikova, R., Sonnenborn, U., and Tlaskalova, H. (1998). Probiotics and E. coli infections in man. Vet. Q. 20 Suppl 3, S78–S81.
Maines, M. D., Trakshel, G. M., and Kutty, R. K. (1986). Characterization of two constitutive forms of rat liver microsomal heme oxygenase. Only one molecular species of the enzyme is inducible. J. Biol. Chem. 261, 411–419. doi: 10.1016/s0021-9258(17)42488-4
Marvig, R. L., Damkiaer, S., Khademi, S. M., Markussen, T. M., Molin, S., and Jelsbak, L. (2014). Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. mBio 5:e00966–14. doi: 10.1128/mBio.00966-14
McDonagh, A. F., and Assisi, F. (1971). Commercial bilirubin: a trinity of isomers. FEBS Lett. 18, 315–317. doi: 10.1016/0014-5793(71)80475-1
McDonagh, A. F., and Palma, L. A. (1980). Preparation and properties of crystalline biliverdin IX alpha. Simple methods for preparing isomerically homogeneous biliverdin and [14C[biliverdin by using 2,3-dichloro-5,6-dicyanobenzoquinone. Biochem. J. 189, 193–208. doi: 10.1042/bj1890193
Meek, M. E., Beauchamp, R., Long, G., Moir, D., Turner, L., and Walker, M. (2002). Chloroform: exposure estimation, hazard characterization, and exposure-response analysis. J. Toxicol. Environ. Health B Crit. Rev. 5, 283–334. doi: 10.1080/10937400290070080
Michael, L., and Pendrak, D. D. R. (2005). Methods for the Production of Biliverdin. US 2005/0209305 A1. Washington, DC: U.S. Patent and Trademark Office.
Miller, E. M., and Nickoloff, J. A. (1995). Escherichia coli electrotransformation. Methods Mol. Biol. 47, 105–113. doi: 10.1385/0-89603-310-4:105
Mourino, S., Giardina, B. J., Reyes-Caballero, H., and Wilks, A. (2016). Metabolite-driven regulation of heme uptake by the biliverdin ixbeta/delta-selective heme oxygenase (HemO) of Pseudomonas aeruginosa. J. Biol. Chem. 291, 20503–20515. doi: 10.1074/jbc.M116.728527
Mukherjee, S., Jemielita, M., Stergioula, V., Tikhonov, M., and Bassler, B. L. (2019). Photosensing and quorum sensing are integrated to control Pseudomonas aeruginosa collective behaviors. PLoS Biol. 17:e3000579. doi: 10.1371/journal.pbio.3000579
National Toxicology Program (2000). NTP toxicology and carcinogenesis studies of pyridine (CAS No. 110-86-1) in F344/N Rats, wistar rats, and B6C3F1 Mice (Drinking Water Studies). Natl. Toxicol. Program Tech. Rep. Ser. 470, 1–330.
Nguyen, A. T., O’neill, M. J., Watts, A. M., Robson, C. L., Lamont, I. L., Wilks, A., et al. (2014). Adaptation of iron homeostasis pathways by a Pseudomonas aeruginosa pyoverdine mutant in the cystic fibrosis lung. J. Bacteriol. 196, 2265–2276. doi: 10.1128/JB.01491-14
O’Neill, M. J., Bhakta, M. N., Fleming, K. G., and Wilks, A. (2012). Induced fit on heme binding to the Pseudomonas aeruginosa cytoplasmic protein (PhuS) drives interaction with heme oxygenase (HemO). Proc. Natl. Acad. Sci. U.S.A. 109, 5639–5644. doi: 10.1073/pnas.1121549109
Pereira, P. J., Macedo-Ribeiro, S., Parraga, A., Perez-Luque, R., Cunningham, O., Darcy, K., et al. (2001). Structure of human biliverdin IXbeta reductase, an early fetal bilirubin IXbeta producing enzyme. Nat. Struct. Biol. 8, 215–220. doi: 10.1038/84948
Ratliff, M., Zhu, W., Deshmukh, R., Wilks, A., and Stojiljkovic, I. (2001). Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183, 6394–6403. doi: 10.1128/JB.183.21.6394-6403.2001
Rembacken, B. J., Snelling, A. M., Hawkey, P. M., Chalmers, D. M., and Axon, A. T. (1999). Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet 354, 635–639. doi: 10.1016/s0140-6736(98)06343-0
Robinson, E. A., Wilks, A., and Xue, F. (2021). Repurposing acitretin as an antipseudomonal agent targeting the. Biochemistry 60, 689–698. doi: 10.1021/acs.biochem.0c00895
Sedlak, T. W., and Snyder, S. H. (2004). Bilirubin benefits: cellular protection by a biliverdin reductase antioxidant cycle. Pediatrics 113, 1776–1782. doi: 10.1542/peds.113.6.1776
Sonnenborn, U. (2016). Escherichia coli strain Nissle 1917-from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett. 363:fnw212. doi: 10.1093/femsle/fnw212
Stocker, R., Yamamoto, Y., Mcdonagh, A. F., Glazer, A. N., and Ames, B. N. (1987). Bilirubin is an antioxidant of possible physiological importance. Science 235, 1043–1046. doi: 10.1126/science.3029864
Wegele, R., Tasler, R., Zeng, Y., Rivera, M., and Frankenberg-Dinkel, N. (2004). The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem. 279, 45791–45802. doi: 10.1074/jbc.M408303200
Wilks, A. (2003). Purification and characterization of heme oxygenase. Curr. Protoc. Toxicol. Chapter 9:Unit 9.9.
Wilks, A., and Heinzl, G. (2014). Heme oxygenation and the widening paradigm of heme degradation. Arch. Biochem. Biophys. 544, 87–95. doi: 10.1016/j.abb.2013.10.013
Wilks, A., and Ikeda-Saito, M. (2014). Heme utilization by pathogenic bacteria: not all pathways lead to biliverdin. Acc. Chem. Res. 47, 2291–2298. doi: 10.1021/ar500028n
Wilks, A., and Ortiz de Montellano, P. R. (1993). Rat liver heme oxygenase. High level expression of a truncated soluble form and nature of the meso-hydroxylating species. J. Biol. Chem. 268, 22357–22362.
Yamaguchi, T., and Nakajima, H. (1995). Changes in the composition of bilirubin-IX isomers during human prenatal development. Eur. J. Biochem. 233, 467–472. doi: 10.1111/j.1432-1033.1995.467_2.x
Yamaguchi, T., Komoda, Y., and Nakajima, H. (1994). Biliverdin-IX alpha reductase and biliverdin-IX beta reductase from human liver. Purification and characterization. J. Biol. Chem. 269, 24343–24348.
Yamaguchi, T., Yamaguchi, N., Nakajima, H., Komoda, Y., and Ishikawa, M. (1979). Studies on bilirubin metabolism: separation and identification of the isomers of biliverdin-IX and biliverdin-IX dimethyl ester by means of high-performance liquid chromatography. Proc. Jpn. Acad. 55, 79–83.
Keywords: E. coli Nissle (EcN(T7)), Pseudomonas aeruginosa, HemO, heme, biliverdin IXβ, biliverdin IXδ, biliverdin IXα, biliverdin isomer
Citation: Robinson EA, Frankenberg-Dinkel N, Xue F and Wilks A (2021) Recombinant Production of Biliverdin IXβ and δ Isomers in the T7 Promoter Compatible Escherichia coli Nissle. Front. Microbiol. 12:787609. doi: 10.3389/fmicb.2021.787609
Received: 01 October 2021; Accepted: 11 November 2021;
Published: 08 December 2021.
Edited by:
Carlos Alberto Moreira-Filho, University of São Paulo, BrazilReviewed by:
Gunhild Layer, University of Freiburg, GermanyShin-ichi Ozaki, Yamaguchi University, Japan
Copyright © 2021 Robinson, Frankenberg-Dinkel, Xue and Wilks. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angela Wilks, awilks@rx.umaryland.edu
 Elizabeth A. Robinson
Elizabeth A. Robinson Nicole Frankenberg-Dinkel
Nicole Frankenberg-Dinkel Fengtian Xue
Fengtian Xue Angela Wilks
Angela Wilks