- 1Key Laboratory of Systems Bioengineering (Ministry of Education), Tianjin University, Tianjin, China
- 2SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), School of Chemical Engineering and Technology, Tianjin University, Tianjin, China
- 3Department of Chemistry, National University of Singapore, Singapore, Singapore
- 4Institute of Materials Research and Engineering, Singapore, Singapore
- 5Dalian Petrochemical Research Institute of Sinopec, Dalian, China
In order to improve the capability of Klebsiella pneumoniae to produce an important chemical raw material, 1,3-propanediol (1,3-PDO), a new type of K. pneumoniae x546 was obtained by glycerol acclimation and subsequently was used to produce 1,3-PDO. Under the control of pH value using Na+ pH neutralizer, the 1,3-PDO yield of K. pneumoniae x546 in a 7.5-L fermenter was 69.35 g/L, which was 1.5-fold higher than the original strain (45.91 g/L). After the addition of betaine, the yield of 1,3-PDO reached up to 74.44 g/L at 24 h, which was 40% shorter than the original fermentation time of 40 h. To study the potential mechanism of the production improvement of 1,3-PDO, the Tandem Mass Tags (TMT) technology was applied to investigate the production of 1,3-PDO in K. pneumoniae. Compared with the control group, 170 up-regulated proteins and 291 down-regulated proteins were identified. Through Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis, it was found that some proteins [such as homoserine kinase (ThrB), phosphoribosylglycinamide formyltransferase (PurT), phosphoribosylaminoimidazolesuccinocarboxamide synthase (PurC), etc.] were involved in the fermentation process, whereas some other proteins (such as ProX, ProW, ProV, etc.) played a significant role after the addition of betaine. Moreover, combined with the metabolic network of K. pneumoniae during 1,3-PDO, the proteins in the biosynthesis of 1,3-PDO [such as DhaD, DhaK, lactate dehydrogenase (LDH), BudC, etc.] were analyzed. The process of 1,3-PDO production in K. pneumoniae was explained from the perspective of proteome for the first time, which provided a theoretical basis for genetic engineering modification to improve the yield of 1,3-PDO. Because of the use of Na+ pH neutralizer in the fermentation, the subsequent environmental pollution treatment cost was greatly reduced, showing high potential for industry application in the future.
Introduction
The rise of the biodiesel industry leads to the overproduction of glycerol as a by-product, which now threatens the economic feasibility of the industry (Pan et al., 2019; Kim et al., 2020). This situation has prompted scientists to explore the utilization of glycerol as a carbon source to produce 1,3-propanediol (1,3-PDO), which is a precursor of some important commercial polymers such as polyester and polyurethane (Zhou et al., 2019b; Bao et al., 2020; Chen et al., 2020). 1,3-PDO can be produced by chemical synthesis or biosynthesis using Klebsiella pneumoniae. Because of its relatively high yield and low environmental pollution, K. pneumoniae is preferable to be used in 1,3-PDO production (Li et al., 2019; Mitrea and Vodnar, 2019; Zabed et al., 2019). Researchers have adopted several strategies such as the domestication method, genetic modification, medium optimization, and other methods to significantly increase the output of 1,3-PDO (Zhang et al., 2018; Lee et al., 2019; Ma et al., 2019; Zhou et al., 2019a). However, the industrial-scale production of 1,3-PDO using bacteria is still limited by low efficiency, which seriously hinders the competitiveness of the process (Dexter Tam et al., 2019; Guo et al., 2019; Park et al., 2019).
The proteomic analysis of protein expression patterns under experimental conditions can provide sufficient information on the function and the regulation of metabolic networks, which is important in the reasonable and purposeful exploration of genome and proteome datasets for the pathway analysis of actual biological processes in post–genome research. The Tandem Mass Tags (TMT) technology was one of the most powerful analytical methods with the highest flux, the smallest systematic error, and the most powerful function (Sogame et al., 2014). It could provide more accurate digital signals, higher detection fluxes, and wider detection ranges. A more detailed understanding of the metabolic pathway of K. pneumoniae and other species could help to provide a better way to promote the transformation of glycerol into 1,3-PDO in this system. Therefore, it is necessary to apply the TMT technology to the study of 1,3-PDO production by K. pneumoniae.
Ca2+ salt as a divalent cation can reduce the drastic changes in the activities of various intracellular dehydrogenases in the oxidation pathway, adjust and maintain the intracellular redox pressure, shift the metabolic flow to 1,3-PDO synthesis, and reduce the types of by-products caused by metabolic disorders. Therefore, Ca2+ salts have been commonly used as a pH neutralizer in industry (Nakano et al., 2012; Zhang et al., 2016; Tee et al., 2017). However, various Ca2+ salt precipitates were formed when using Ca2+ neutralizer at the bottom of the fermentation tank, which not only increases the cost of the subsequent product purification, but also causes significant environmental pollution. Considering the environmental pressure caused by the utilization of Ca2+ neutralizer, it is critical that Ca2+ pH neutralizer be replaced with a new neutralizer without compromising with production efficiency, so as to make the 1,3-PDO bioproduction more environment-friendly. Na+ pH neutralizer can reduce the solid pollutants produced after fermentation reaction. As Na2CO3 can be synthesized by a chemical method from the electrolysis of high salt wastewater containing Na+ and re-extracted for reuse (Shin et al., 2011; Simon et al., 2014), the use of Na2CO3 as a pH adjuster in fermentation would promote a new industrial recycling. However, the use of Na+ in fermentation leads to an increase in osmotic pressure, thereby restricting the yield of 1,3-PDO (Glaasker et al., 1998; Guerzoni et al., 2001), whereas betaine can slow down the effect of salt stress (Hussain et al., 2020). It can maintain the balance of osmotic pressure inside and outside, thereby maintaining the normal physiological function of the cell (Louesdon et al., 2014). Moreover, the betaine may have an effect on fermentation under Na+ conduction.
In this study, Na2CO3 was used as the pH neutralizer in fermentation, and betaine was added to alleviate the high osmotic pressure caused by excessive Na+, which would significantly enhance the yield of 1,3-PDO. The 1,3-PDO production further increased to 74.44 g/L and shortened the fermentation time from 40 to 24 h. TMT was used to study the mechanism effects of the introduction of the Na+ neutralizer and betaine on the yield of 1,3-PDO during the fermentation. This is the first comprehensive investigation of TMT analysis for the production of 1,3-PDO by K. pneumoniae x546, and the results will provide new insights on enhancing the production of 1,3-PDO (genes, proteins, and metabolites), as well as the subsequent industrial strain transformation and process optimization.
Materials and Methods
Strains, Media, and Cultivations
Klebsiella pneumoniae American Type Culture Collection (ATCC) 15380 was purchased from the ATCC. Following the previously published adaptive laboratory evolution (Willke and Vorlop, 2008; Gungormusler et al., 2011; Raghunandan et al., 2014), K. pneumoniae x546 (domesticated strain with 120–20 g/L glycerol: the strain was first domesticated with a concentration of 120 g/L glycerol and then returned to a concentration of 20 g/L glycerol for domestication) could be obtained (the details could be seen in Supplementary File 1). The seed and solid medium (pH 7.0) contained 40 g/L (60, 80, 100, 120, 140, 100–20, 120–20, and 140–20 g/L) glycerol, 4.08 g/L NH4Cl, 0.57 g/L KCl, 0.95 g/L NaH2PO4⋅2H2O, 0.28 g/L Na2SO4, 0.25 g/L MgCl2⋅6H2O, 0.38 g/L citric acid, 0.95 g/L yeast extract, 0.15 g/L Vc, and 4 mL of nutrient solution. Nutrient solution contained 0.035 g/L Na2MoO4, 0.029 g/L ZnCl2, 0.29 g/L CoCl2⋅6H2O, 0.148 g/L MgSO4⋅7H2O, 0.033 g/L NiCl2⋅6H2O, and 1.0 mL HCl. The K. pneumoniae was domesticated with 40 g/L (60, 80, 100, 120, 140, 100–20, 120–20, and 140–20 g/L), and glycerol was labeled G40, G60 G80, G120, G140, G100–20, G120–20, and G140–20 (Liang et al., 2018).
The production medium was a little different from the seed medium, which contained 40 g/L glycerol, 6.17 g/L NH4Cl, 0.86 g/L KCl, 1.40 g/L NaH2PO4⋅2H2O, 0.32 g/L Na2SO4, 0.3 g/L MgCl2⋅6H2O, 1.06 g/L citric acid, 1.15 g/L yeast extract, 0.25 g/L betaine, 0.11 g/L Vc, 0.23 g/L C5H11NO2, and 5 mL of nutrient solution. Nutrient solution contained 5.4 g/L FeCl3⋅6H2O, 0.004 g/L Na2MoO4, 0.04 g/L ZnCl2, 0.17 g/L MnCl2⋅4H2O, 0.47 g/L CoCl2⋅6H2O, 0.06 g/L H3BO4, 0.68 g/L CuSO4⋅5H2O, and 1.0 mL HCl. The betaine was added only in the production medium for 7.5 L fermenter.
The seed was cultivated in 250-mL flask containing a 100-mL seed medium at 150 revolutions/min (rpm) for 8.5 h at 37°C. The production of 1,3-PDO was carried out in a 250-mL flask with 100 mL working volume at 150 rpm for 48 h at 37°C and in a 7.5-L BioFlo 110 fermenter (New Brunswick Scientific, Edison, NJ, United States) at 400 rpm for 40 h at 37°C after adding 770 g glycerol (with a final 5.4 L working volume). Three biological replicates were used for each fermentation experiment. The pH of the seed medium and fermentation medium was adjusted to 7.0 with 3.125 M Na2CO3 solution, respectively.
Determination of 1,3-Propanediol and Glycerol Concentrations
The concentration of 1,3-PDO and glycerol was measured by an HPX-87H column (300 mm × 7.8 mm) (Bio-Rad, Palo Alto, CA, United States) with a differential refractive index detector (SFD GmbH, Schambeck, Germany); 5 mM H2SO4 was used as a mobile phase with a flow rate of 0.5 mL/min at a working temperature of 65°C.
Total Protein Extraction
The samples were ground into a powder in liquid nitrogen. Then the powder was suspended in lysis buffer (1% sodium deoxycholate, 8 M urea). The mixture was allowed to settle at 4°C for 30 min during which the sample was vortexed every 5 min and treated by ultrasound at 40 kHz and 40 W for 2 min. After centrifugation at 16,000 rpm at 4°C for 30 min, the concentration of protein supernatant was determined by bicinchoninic acid (BCA) method by BCA Protein Assay Kit (Pierce, Thermo, United States). Protein quantification was performed according to the kit protocol (Chen et al., 2021).
Protein Digestion and Tandem Mass Tags Labeling
Protein digestion was performed according to a standardized procedure, and the resulting peptide mixture was labeled using 10-plex TMT reagent (Thermo Fisher, Scientific). In brief, an aliquot of protein (100 μg) from each sample was mixed with 100 μL of the lysate. Then 10 mM TCEP was added, and the mixture was stored at 37°C for 60 min, followed by the addition of 40 mM iodoacetamide and the storage of the sample in the dark at room temperature for 40 min.
Sixfold volumes of cold acetone were added to precipitate the protein at −20°C for 4 h. After centrifuging at 10,000 rpm for 20 min at 4°C, the pellet was resuspended with 100 μL of 50 mM triethylammonium bicarbonate buffer. Trypsin was added at a trypsin-to-protein mass ratio of 1:50 and incubated at 37°C overnight. One unit of TMT reagent was thawed and reconstituted in 50 μL acetonitrile. After tagging for 2 h at room temperature, hydroxylamine was added to react with mixture for 15 min at room temperature (Dai et al., 2020).
In this work, the strains grown under control (fermentation of the strain domesticated at a glycerol concentration of 40 g/L without betaine) and optimal conditions (fermentation of the strain domesticated at a glycerol concentration of 120–20 g/L with betaine) at 10 h (each sample with two biological replicates) were collected by centrifugation (12,000 rpm, 10 min at 4°C) and frozen in liquid nitrogen, respectively. The samples were labeled as A1, A2, B1, and B2. Finally, all samples were pooled, desalted, and vacuum-dried for subsequent use. To verify the accuracy of proteomic data, quantitative real-time polymerase chain reaction (qRT-PCR) was also done (the details are shown in Supplementary File 1).
Liquid Chromatography–Tandem Mass Spectrometry Analysis
Labeled peptides were analyzed by online nano flow liquid chromatography tandem mass spectrometry (MS/MS) using the 9RKFSG2_NCS-3500R system (Thermo Fisher Scientific) connected to the Q_Exactive HF-X system (Thermo Fisher Scientific) via a nanoelectrospray ion source. Briefly, a C18-reversed phase column (75 μm × 25 cm, Thermo Fisher Scientific) was equilibrated with solvent A (A: 2% acetonitrile and 0.1% formic acid) and solvent B (B: 80% acetonitrile and 0.1% formic acid). The peptides were eluted using the following gradient: 0–2 min, 0–3% B; 2–92 min, 5–25% B; 92–102 min, 25–45% B; 102–105 min, 45–100% B; 105–120 min, 100–0% B at a flow rate of 300 μL/min. The Q_Exactive HF-X was operated in the data-dependent acquisition mode to automatically switch between full scan MS and MS/MS acquisition. The survey of full scan MS spectra (m/z 350–1,500) was acquired in the Orbitrap with 70,000 resolutions. The top 20 most intense precursor ions were selected into the collision cell for fragmentation by higher-energy collision dissociation. The MS/MS resolution was set at 35,000 (at m/z 100), with the maximum fill time of 50 ms and a dynamic exclusion of 30 s (Wang et al., 2020a,b).
Protein Identification
The RAW data files were analyzed by Proteome Discoverer 2.2 (Thermo Fisher Scientific) against the K. pneumoniae database1. The MS/MS search criteria were as follows: a mass tolerance of 20 ppm for MS and 0.02 Da for MS/MS tolerance, trypsin as the enzyme with two-missed cleavages allowed, carbamido methylation of cysteine and the TMT of the N-terminus and lysine side chains of peptides as fixed modification, and methionine oxidation as dynamic modifications, respectively. The false discovery rate for peptide identification was set at ≤0.01. A minimum of one unique peptide identification was used to support protein identification (Wang et al., 2020c).
Statistical Analyses
The thresholds of fold change (FC) (>1.2 or <0.83) and p < 0.05 were used to identify differentially expressed proteins (DEPs). Annotation of all identified proteins was performed by Gene Ontology (GO)2 and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway3 analyses. DEPs were further used for GO and KEGG enrichment analysis. Protein–protein interaction analysis was performed using the String v10.5.
Results and Discussion
Comparison of the 1,3-Propanediol Production
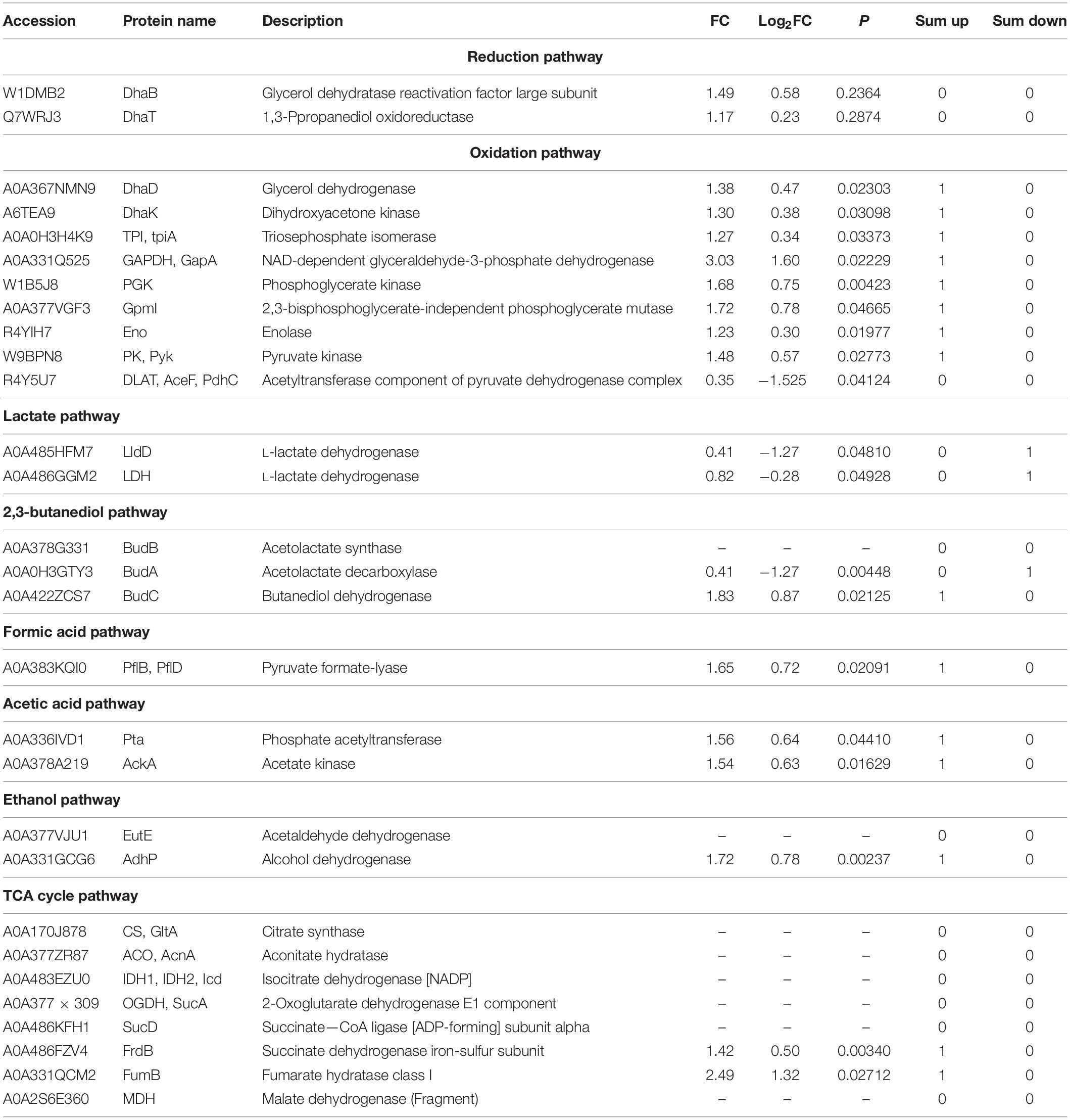
The yields of 1,3-PDO produced in 250-mL shaker by the original K. pneumoniae and the domesticated K. pneumoniae with and without betaine are summarized in Figure 1A. The 1,3-PDO production increased from G40 (13.22 g/L) to G120 (18.34 g/L), but decreased at G140. To further investigate the changes in yield during fermentation, the production of 1,3-PDO in the 7.5-L fermentation tank with glycerol domestication concentrations of 40, 60, 80, 100, 120, and 140 g/L were studied, and the results are shown in Figure 1B. The 1,3-PDO yields for glycerol domestication concentration from 40 to 120 g/L were 45.91, 49.71, 52.21, 55.02, 59.82, and 57.34 g/L, respectively. When the glycerol acclimation concentration increased to 140 g/L, the 1,3-PDO production decreased. With the increase in the domestication concentration of glycerol for the K. pneumoniae, the yield of 1,3-PDO decreased, which was consistent with the previous results (Colin et al., 2000; Yiqiang et al., 2007; Metsoviti et al., 2012; Raghunandan et al., 2014). When the concentration of glycerol increased, metabolism was inhibited, and 1,3-PDO production decreased. Therefore, G100, G120, and G140 were redomesticated under the concentration of 20 g/L glycerol. From Figure 1C, the yields of 1,3-PDO increased to 63.11, 69.35, and 66.03 g/L for G100–20, G120–20, and G140–20, respectively. After comparing G120–20 with G40, the yield of 1,3-PDO was improved by 51.06%. During the whole fermentation process, Na2CO3 was used as the pH neutralizer, which had an effect on the osmotic pressure of the fermentation liquid and the production yield of 1,3-PDO, so betaine as a fermentation medium was added (Fan et al., 2018). In Figure 1D, the 1,3-PDO yields of the G40 (+betaine) and G120–20 (+betaine) reached the 53.42 g/L and 75.92 g/L, respectively, which were higher than those without betaine, consistent with a previous report (Jantama et al., 2010). Thus, betaine could alleviate the osmotic pressure problem and ensure the activity of bacteria during fermentation. Compared to G120–20 (+betaine), which reached a yield of 74.44 g/L at 24 h, the 1,3-PDO production yield of G40 was very low (45.91 g/L) and the fermentation time was very long (40 h), further confirming that betaine could alleviate the increase in the osmotic pressure and counter-suppress the Na+ effect during fermentation. To explore the enhanced mechanism of 1,3-PDO production after the addition of Na2CO3 and betaine, G40 was used as a control group, and TMT was used to compare the differences in protein expression between the control and optimal G120–20 (+betaine).
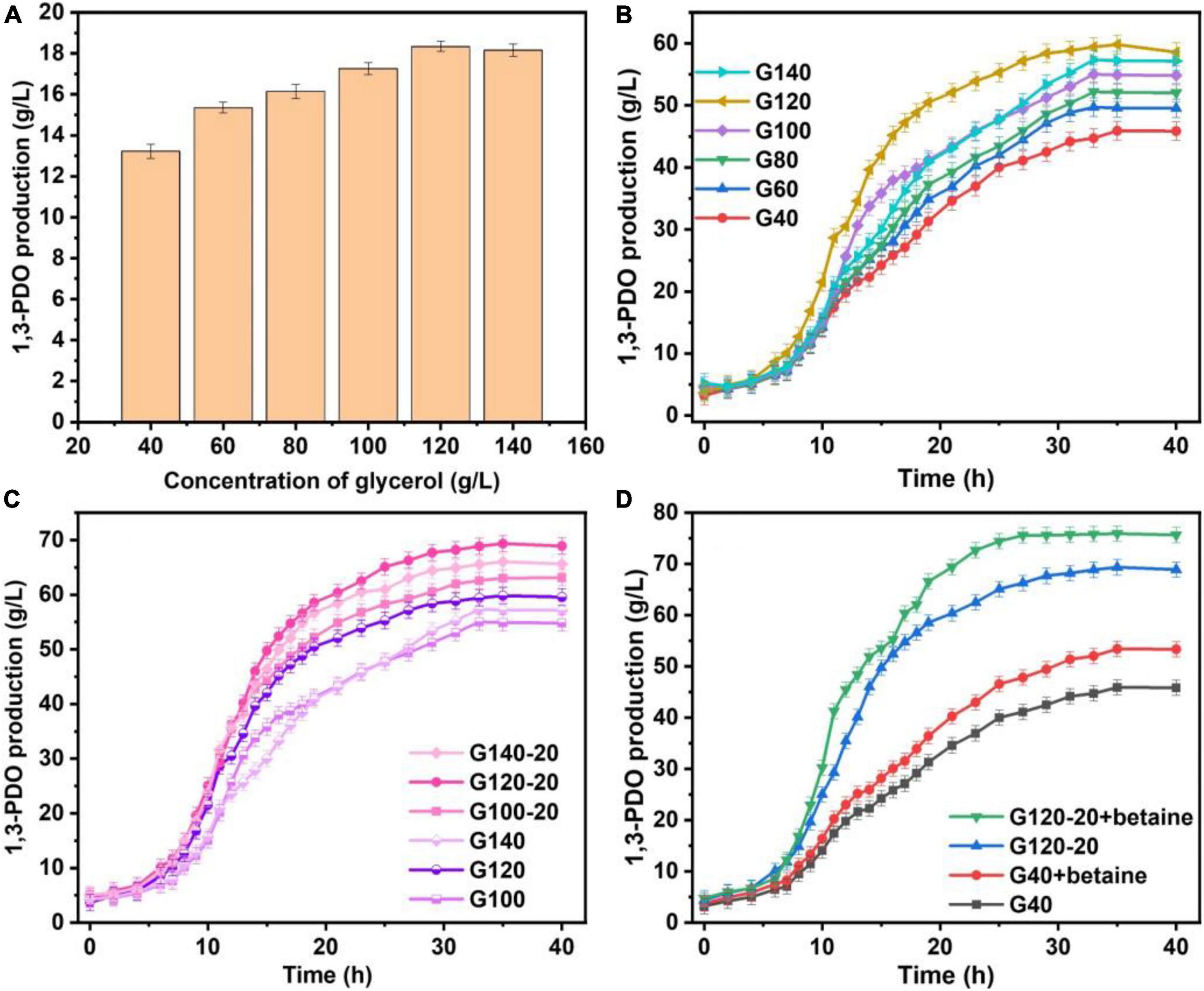
Figure 1. (A) Compa re the production of 1,3-PDO in the 250-mL shaker with glycerine domestication concentration at 40, 60, 80, 100, 120, and 140 g/L. (B) The comparison of the yield of 1,3-PDO in the 7.5-L fermentation tank with glycerine domestication concentration at 40, 60, 80, 100, 120, and 140 g/L. (C) The production 1,3-PDO in the 7.5-L fermentation tank with glycerine domestication concentration at 100, 100–20, 120, 120–20, 140, and 140–20 g/L. (D) The capacity of 1,3-PDO in the 7.5-L fermentation tank with glycerine domestication concentration at 40 g/L, 40 g/L +betaine, 120–20 and 120–20 g/L +betaine.
Protein Identification and Quantitation
After the G40 and G120–20 (+betaine) fermented broths were labeled by TMT, the primary and secondary mass spectra were analyzed statistically. With the help of the Protein Discoverer search library, a total of 3,284 proteins were identified from the four samples. Supplementary Figure 3A shows the number distribution of peptides contained in the identified proteins. For example, there were 521 proteins matched with one peptide. The length distribution of the identified peptides is shown in Supplementary Figure 3B. For instance, there were 1,860 proteins with a peptide length of eight amino acids. Most of the peptides had 5 to 20 amino acids after enzymatic hydrolysis, accounting for 83.87% of the total, which indicated that the enzymatic hydrolysis was sufficient, and the identification results were reliable. As shown in Supplementary Figure 3C, the molecular weight distribution of the identified proteins was determined, especially for these proteins with low molecular weights of less than 20 kDa. The molecular weights of most proteins were from 1 to 60 kDa, and 65 types of macromolecular proteins with molecular weights of more than 100 kDa were identified. Supplementary Figure 3D shows the coverage distribution of the identified proteins. The number of amino acids in the peptide was higher than the total number of amino acids in the protein. The identification results were more persuasive with the expansion of coverage distribution. The coverages of polypeptides with more than 10 and 20% of the identified proteins were 78.72 and 62.68%, respectively.
Usually, proteins with differences between G40 and G120–20 were determined based on the FC and the p-value. In this work, p < 0.05 indicates the difference among the groups. As shown in Figure 2, there were 3,284 proteins, including 170 up-regulated proteins (FC > 1.2) and 291 down-regulated proteins (FC < 0.83). These detected proteins were analyzed by GO term and KEGG pathway analysis to identify the biological functions of the differential proteins and the target proteins.
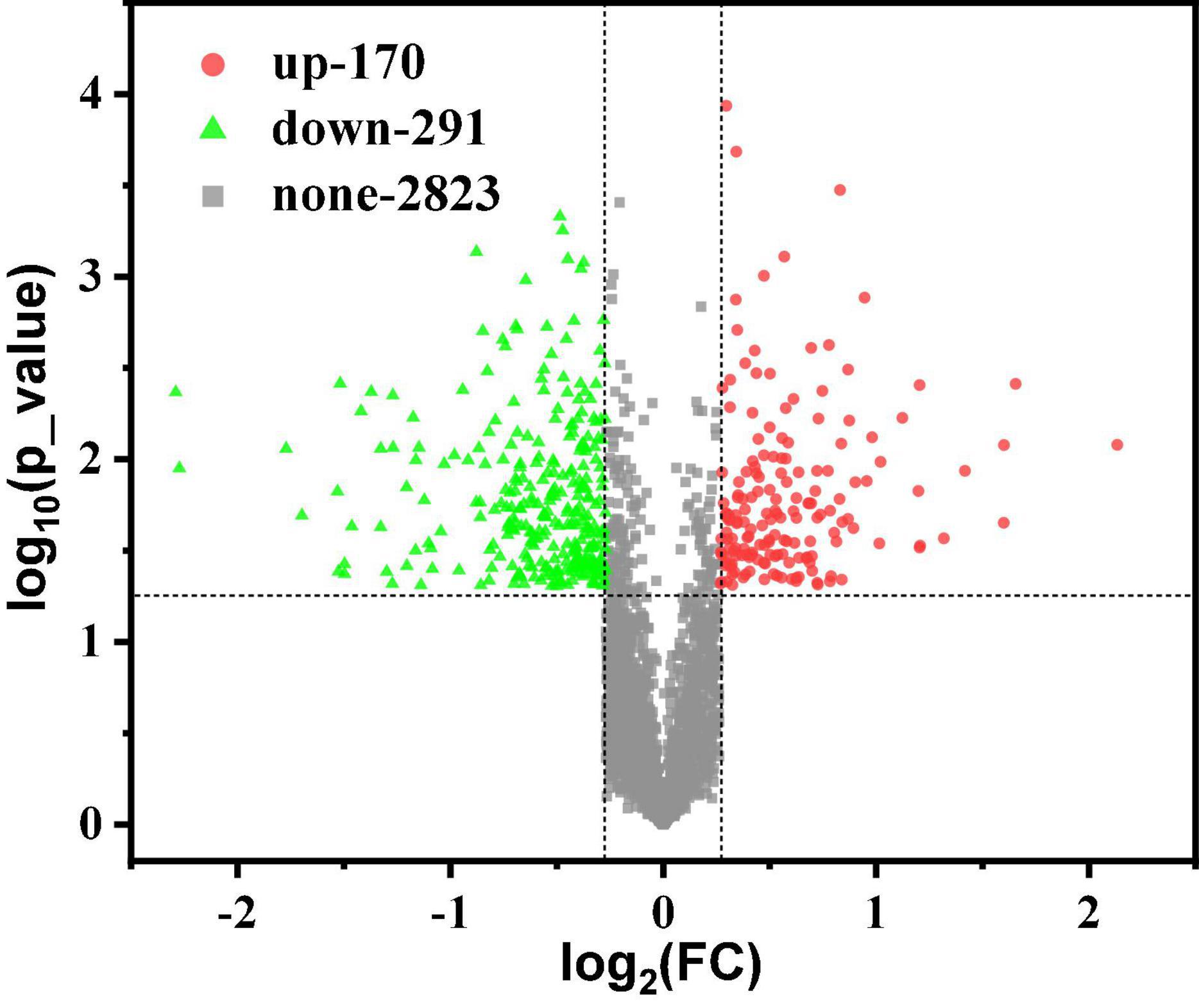
Figure 2. Scatter plot of volcano differences. A volcano plot between the G40 and the G120–20 (+betaine) whose proteins are shown. The horizontal axis is the value of log2(FC). The vertical axis is the –log10 (p-value). The red dots represent up-regulated proteins (FC > 1.2), and the green dots represent down-regulated proteins (FC < 0.83).
The Analysis for Gene Ontology Term
With the GO database, genes and gene products can be classified and annotated as follows: cellular component (CC), molecular function (MF), and Biological Process (BP). It is a bioinformatics analysis tool (Zhong et al., 2019). Figures 3A,B show the level 2 of GO classification for 3,284 proteins and 461 differential proteins, respectively. For the 3,284 proteins: 2,190, 2,029, and 1,670 proteins were detected in metabolic process, the cellular process, and single organization process of BP, respectively. There were 1,174 and 1,149 proteins detected in cell and cell part of CC, respectively. There were 2,121 and 1,639 proteins detected in catalytic activity and binding of MF, respectively. As can be seen from Figure 3B, in BP: 115 up-regulated and 189 down-regulated differential proteins were detected in metabolic process; 104 up-regulated and 172 down-regulated differential proteins were detected in the cellular process; 85 up-regulated and 154 down-regulated proteins were detected in the single organization process. In CC, 52 up-regulated and 93 down-regulated differential proteins were detected in cell; 51 up-regulated proteins and 89 down-regulated proteins were detected in the cell part. In MF, 108 up-regulated and 192 down-regulated differential proteins were detected mainly in catalytic activity; 86 up-regulated and 139 down-regulated differential proteins were detected in binding. Compared with Figures 3A,B, the main functional area was the same, which indicated that these functions played an important role in the production of 1,3-PDO by K. pneumoniae. However, because of the excessive number of proteins, the enrichment of differentially abundant proteins requires further analysis.
Figure 3. Gene Ontology (GO) classification and enrichment analysis of G40 and G120–20 (+betaine). (A) GO classification of 3,284 proteins. The ordinate on the left is –log10 (percent of proteins), and the label on the column is the number of proteins. (B) GO classification of 170-up and 291-down proteins. (C) The enrichment analysis of the BP (170-up proteins). (D) The enrichment analysis of the BP (291-down proteins). (E) The enrichment analysis of the MF (170-up proteins). (F) The enrichment analysis of the MF (291-down proteins). [The p-value (corrected) < 0.005].
Gene Ontology functional enrichment analysis can clarify the biological process, cell components, and molecular functions (Zhong et al., 2019). The enrichment of up-regulated and down-regulated proteins for BP is shown in Figure 3C and Table 1. Up-regulated proteins were related to the ribonucleoside monophosphate metabolic process, nucleoside monophosphate metabolic process, purine nucleoside monophosphate metabolic process, and so on. In MF (Figure 3E and Table 1), up-regulated proteins were related to phosphotransferase activity (alcohol group as acceptor). In BP (Figure 3D), down-regulated proteins were associated with sulfur compound metabolic process, sulfur compound biosynthetic process, sulfur amino acid biosynthetic process, sulfur amino acid metabolic process, methionine biosynthetic process, and methionine metabolic process. Finally, there were 41 up-regulated proteins and 19 down-regulated proteins after further analysis. Therefore, further analysis was needed through the chord diagram of GO term enrichment. As shown in Figure 4B, the most up-regulated proteins (1.23 ∼ 3.03-fold) were identified to be NAD-dependent glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 4-hydroxythreonine-4-phosphate dehydrogenase (PdxA), phosphoenolpyruvate-dihydroxyacetone phosphotransferase (DhaL), ThrB, PurT, phosphoglycerate kinase (PGK), 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (GpmI), PurC, and so on. PdxA, whose function is similar to the isocitrate dehydrogenase and isopropylmalate dehydrogenase, can contribute to the phosphotransacetylase (Pta) activity (Sivaraman et al., 2003; Xu et al., 2005). It has been reported that Pta plays a role in the reduction pathway of 1,3-PDO produced by K. pneumoniae. It is well known that amino acid metabolism plays an important role in the life of K. pneumoniae, which can balance the intracellular pH, generate energy, reduce power, and resist environmental pressures. With the increase in the ThrB, the more threonine is produced. The K. pneumoniae can use threonine as a nitrogen source (Reitzer, 2005), and the cavity near ADP is very suitable for homoserine binding, so it can improve the catalytic activity by stabilizing the transition state (Fan et al., 2009; Zhang et al., 2019) and contribute to the 1,3-PDO production. The ligation of amino and carboxylate groups of small molecule metabolites is catalyzed by the ATP-grasp superfamily, which is widespread across primary metabolic processes (Zhang et al., 2008). PurT is a member of the ATP-grasp superfamily, and PurC also has several structural elements in common. With the up-regulation of PurC, the expression levels of diverse proteins involved in purine and pyrimidine synthesis, carbon and energy metabolisms, iron uptake, proteolysis, protein secretion, and signal transduction can be improved. Purine can save energy from the beginning and the consumption of some amino acids (Yuan et al., 2013). As a key enzyme of glycolysis, the up-regulation of GpmI accelerates the catalysis of the interconversion between 3-phosphoglycerate and 2-phosphoglycerate, whereas enolase (Eno) catalyzes the conversion of 2-phosphoglycerate into phosphoenolpyruvate (Yin et al., 2020). GpmI also plays an important role in the carbohydrate transport and metabolism. In addition, PGK not only is a glycolytic enzyme that plays an important role in the growth of biofilm, but also contributes to the formation of surface proteins. In biofilm formation, bacterial cells are embedded in the extracellular matrix, which can protect bacteria from a variety of environmental damages (Wang et al., 2016). Therefore, the tolerance of the strain could be effectively improved during the process of glycerol acclimation, so that the related proteins in the glycolysis pathway were up-regulated, and finally, the production yield of 1,3-PDO was increased. Concurrently, the multiple of down-regulated proteins, such as methylenetetrahydrofolate reductase (MetF), phosphoadenosine phosphosulfate reductase (CysH), 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase (MetE), methionine adenosyltransferase (MetK) and so on, were down-regulated from 0.40 to 0.79. Commonly, a previous study has shown that high concentrations of homoserine are toxic to cells (Kingsbury and McCusker, 2010). In order to avoid the excessive accumulation of homoserine, decreased MetE, MetF, and MetK levels can inhibit met regulator. To prevent threonine biosynthesis, the up-regulation of ThrB can catalyze the over conversion of homoserine to o-phosphate-L-homoserine, which also can inhibit growth. This is a reversible transformation. When the concentration of homoserine decreased, o-phosphate-L-homoserine could be converted to homoserine to provide the precursor of methionine (Li et al., 2017). A dynamic balance is beneficial to the growth of cells and the synthesis of 1,3-PDO. For CysH, its down-regulation decrease cysteine (Longo et al., 2016), which reduces the effect of 1,3-PDO production.

Table 1. The enrichment analysis of the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway.
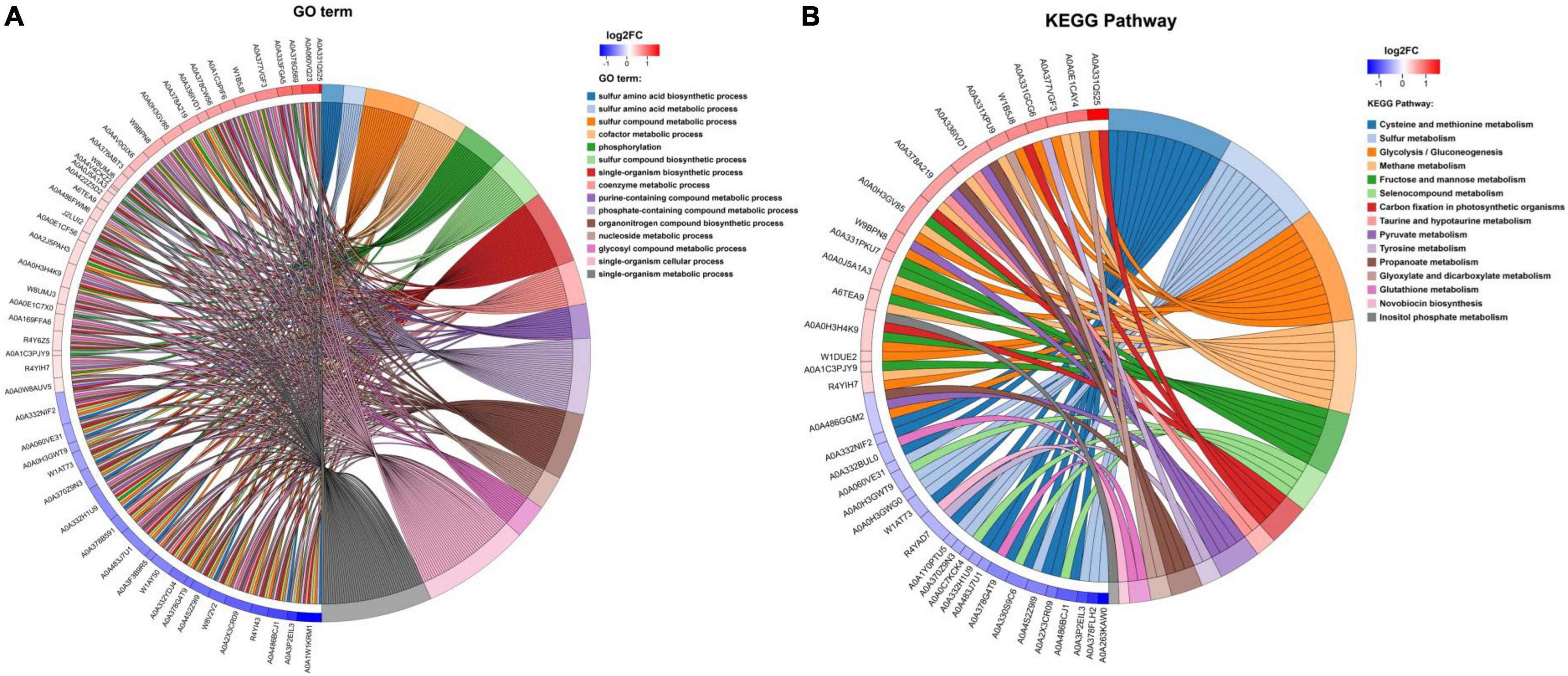
Figure 4. (A) Chord diagram of GO term enrichment. (B) Chord diagram of KEGG pathway enrichment. KEGG enrichment chord diagram shows the corresponding relationship among the target proteins, the annotation, and enrichment; the left is protein, and log2FC is displayed from top to bottom. When log2FC > 0, the larger the log2FC is, the greater the differential expression of up-regulated protein is; when log2FC < 0, the smaller the log2FC is, the greater the differential expression of down-regulated protein is. The closer the log2FC is to 0, the smaller the differential expression fold is. The right is KEGG pathway and the score of z score. When z score > 0, there are more up-regulated proteins than down-regulated proteins involved in this pathway, and this pathway is more likely to be activated. On the contrary, when z score < 0, the up-regulated proteins involved in this pathway are less than the down-regulated proteins, and this pathway is more likely to be inhibited. (The same analysis is applied to GO term).
The Analysis for Kyoto Encyclopedia of Genes and Genomes Pathway
In organisms, different gene products perform different biological functions through an orderly coordination. Therefore, the pathway information in the KEGG database helped us to understand the biological function of genes at the system level in K. pneumoniae (Jia et al., 2021). Figure 5A shows the KEGG pathway for differential proteins. The proteins were classified and annotated as follows: metabolism, genetic information processing, environmental information processing, cellular process, organismal systems, and human diseases. In this study, carbohydrate metabolism, amino acid metabolism, and energy metabolism were the most DEPs annotated in metabolism, with 65, 53, and 50 proteins, respectively. Combining with Figures 5B,C, there were 38 up-regulated proteins and 27 down-regulated proteins in carbohydrate metabolism. Simultaneously, in genetic information processing, the most DEPs were annotated in (translation), (folding, sorting, and degradation), and (replication and repair); in environmental information processing, the most DEPs were annotated in Immune system and environmental adaptation; in cellular processes, the most DEPs were annotated in cellular community–prokaryotes and cell motility. Despite the superfluous proteins, it required to investigate the KEGG pathway enrichment analysis. As shown in Figure 5D and Table 1, up-regulated proteins were linked to the methane metabolism, glycolysis/gluconeogenesis, fructose, and mannose metabolism. By contrast, the down-regulated proteins were involved in cysteine and methionine metabolism, sulfur metabolism, and selenocompound metabolism. Ultimately, there were 17 up-regulated proteins and 21 down-regulated proteins after further analysis. Therefore, further analysis was needed through the chord diagram of KEGG pathway enrichment. As can be seen from Figure 4B, the up-regulated proteins (glycolysis/gluconeogenesis, methane metabolism, fructose and mannose metabolism, carbon fixation in photosynthetic organisms, taurine and hypotaurine metabolism, pyruvate metabolism, propanoate metabolism, glyoxylate, and dicarboxylate metabolism, inositol phosphate metabolism) are more than down-regulated proteins, indicating that these KEGG pathways were activated. It suggested that the domesticated strain had an improvement in the production of 1,3-PDO compared to the predomesticated strain. As the degree of acclimation enhanced, the production of 1,3-PDO also increased, corresponding to the results in Figure 3. Therefore, the changes in the proteins of the metabolic pathway that produce 1,3-PDO needed to be further analyzed. In addition, the up-regulated and down-regulated proteins were similar to the analysis identified by GO terms, except that taurine import ATP-binding protein (TauA) and taurine-binding periplasmic protein (TauB) decreased, with the FC ranging from 0.35 to 0.43. As these two proteins were related to ABC transporters, further analysis of ABC transporters was given in Figure 6, illustrating that 17 differential proteins had changed. The CysA, TauA, TauB, ModB, PorG, MglB, ArtJ, FhuD, BtuF, ZnuA, CbiO, and LptB
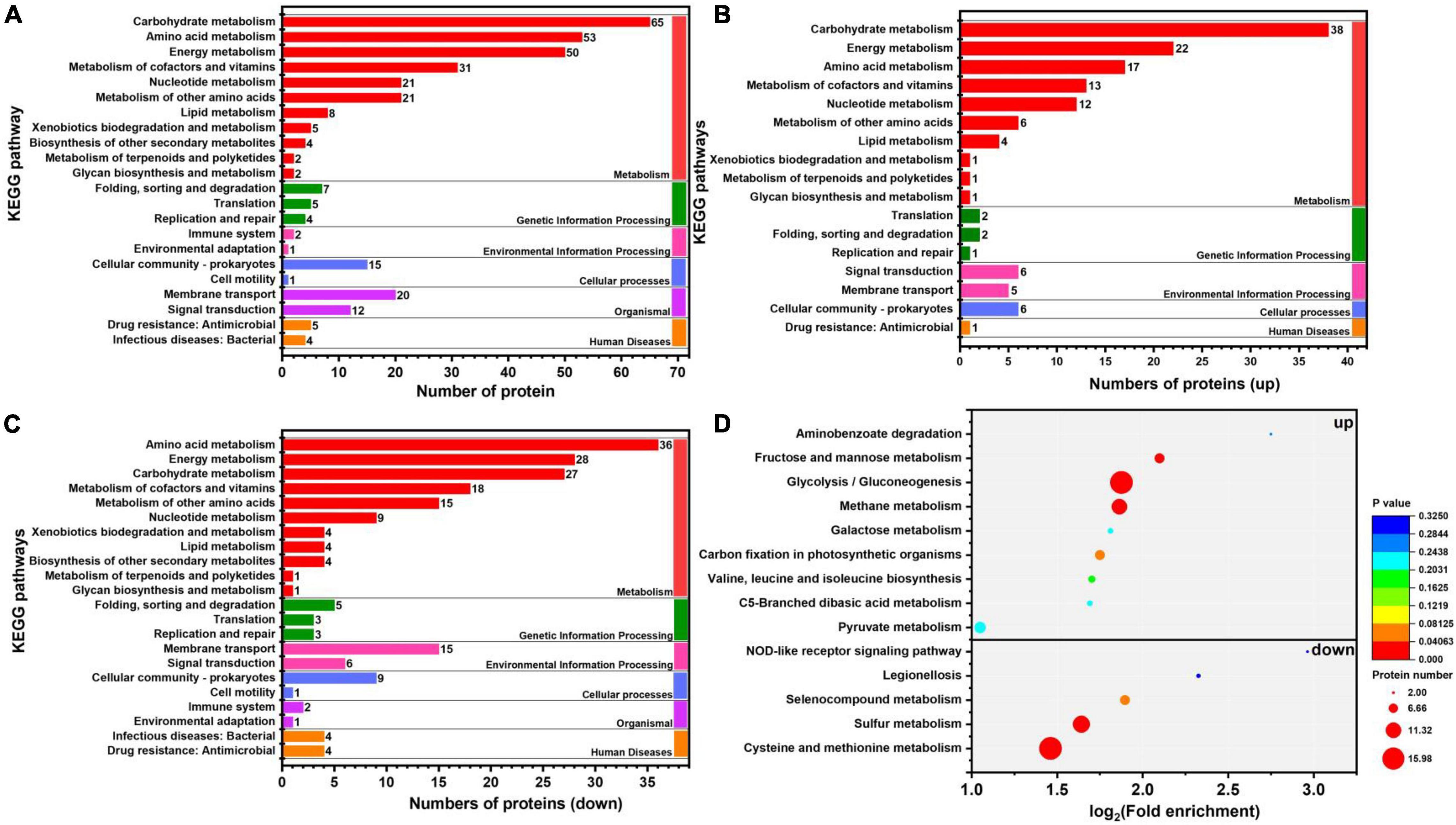
Figure 5. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and enrichment analysis of G40 and G120–20 (+betaine). (A) KEGG pathway classification of differential protein. (B) The enrichment analysis of the KEGG pathway (up proteins). (C) The enrichment analysis of the KEGG pathway (down proteins). (D) The enrichment analysis of the KEGG pathway (up and down proteins). [The p-value (corrected) < 0.05].

Figure 6. The ABC transporters of the G40 is compared with the G120–20 (+betaine) in KEGG pathway map. Proteins in orange block belong to the 3,284 proteins and in white block belonged to the undetected sample. Up-regulated proteins are marked by red, whereas down-regulated proteins are marked by green in 461 differential proteins. (The abbreviations for full name of all proteins are given in Supplementary Table 3).
were down-regulated, whereas the ProX, ProW, ProV, and Livk were up-regulated. ProV and ProW were membrane-associated proteins, and ProV had a considerable sequence identity with ATP-binding proteins from other periplasmic systems. ProX encoded the periplasmic glycine betaine-binding protein (Stirling et al., 1989; May et al., 2010). The biggest FC among them was ProX, which reached 4.39, revealing that betaine played an important role in fermentation.
When Na2CO3 was used to adjust the pH in the fermentation process, salt stress appeared with the continuous increase in Na2CO3. The osmotic pressure induced by the Na+ salt increased gradually, which resulted in the outflow of water, the loss of cell swelling pressure, and the change of solute concentration and cell volume (Fan et al., 2018). Compared with Ca(OH)2 used in commercialization as a pH-neutralizing agent (Wang et al., 2017; Liang et al., 2018), the output of 1,3-PDO was greatly affected. After the use of betaine, it was found that the yields were significantly improved as shown in Figure 1D. Betaine can be used not only as a stress protector or a stabilizer of intracellular enzymes to resist stress conditions, but also as a methyl donor for methylation. It can be accumulated at high concentrations (through transport or biosynthesis) in the cell to balance the osmotic pressure inside and outside the cell. On the other hand, it can increase the cell growth rate and improve the fermentation performance of the strain under high osmotic stress (high concentration of carbohydrate substrate or product) (Fan et al., 2018). In the 1,3-PDO fermentation process of K. pneumoniae, the stress of the Na+ salt as a neutralizer Na2CO3 and the yield of 1,3-PDO were improved by betaine, which offers an alternative way for the industrial pH neutralizer to avoid producing solid pollutants and thus greatly alleviating subsequent sewage treatment and environmental pollution.
Analysis of Several Important Proteins in Metabolic Pathway
In the above, some proteins that play a role in the production of 1,3-PDO by K. pneumoniae with the GO and KEGG pathway analysis and the important role of betaine in the fermentation process were analyzed However, protein changes of G120–20 compared with G40 in the metabolic pathway of K. pneumoniae producing 1,3-PDO still need further investigation. Combined with KEGG pathway analysis, the metabolic pathway of glycerol in K. pneumoniae is shown in Figure 7A: bacterial formation pathway, reduction pathway, and oxidation pathway. In the bacterial formation pathway, ATP was consumed, whereas ADP was produced in metabolism. In the reduction pathway: first, glycerol was converted to 3-hydroxyglyceraldehyde by DhaB and then converted to 1,3-PDO by DhaT. In G120–20 g/L (+betaine). Usually, 1,3-PDO oxidoreductase (DhaT) catalyzes the conversion of 3-hydroxypropionaldehyde (3-HPA) to 1,3-PDO, which is a key enzyme in the preparation of 1,3-PDO from glycerol. But DhaT is seriously inactivated by 3-HPA due to the reaction of 3-HPA with the sulfhydryl group of cysteine residue (Li et al., 2016). Although the DhaB and DhaT were not differential proteins in this article, they still had an effect on the production of 1,3-PDO by K. pneumoniae.
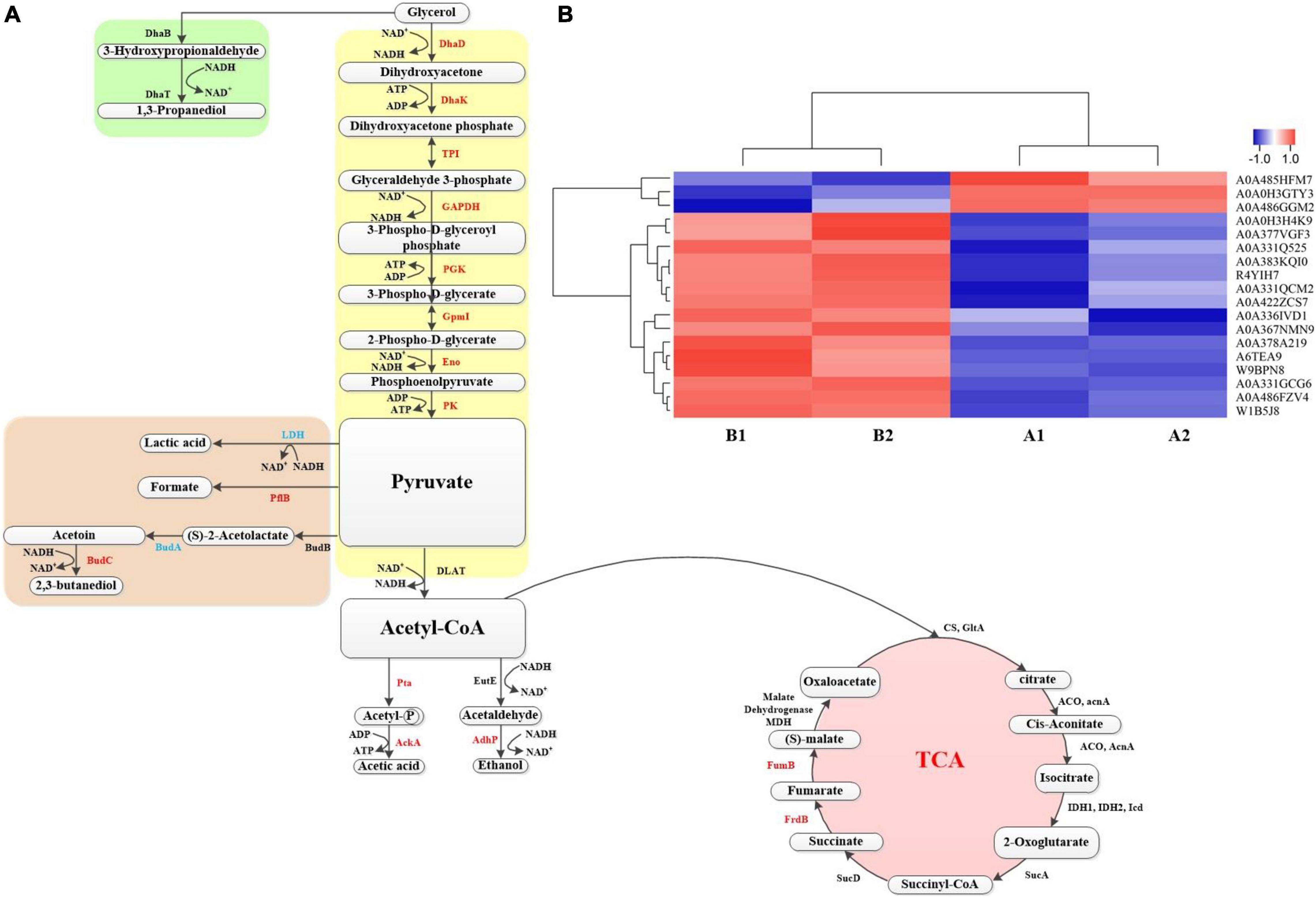
Figure 7. (A) The metabolic pathway for production of 1,3-propanediol by K. pneumoniae. The red represents the up-proteins, and the blue represents the down proteins, whereas the black represents the no change. (B) Heat map of proteins on the metabolic pathway. Each column in the figure represents a sample, and each row represents a protein. The color in the figure represents the relative expression amount of the protein in the group of samples; red represents the high expression amount of the protein in the sample, and blue represents the low expression amount. For the specific change trend of the expression amount, see the number label under the color bar at the top left.
The oxidation pathway was similar to the glycolysis pathway, in that it generated ATP and reduced NADH2, which was required for bacterial growth. NADH2 was consumed during the 3-HPA-mediated production of 1,3-PDO, which was produced in the oxidation pathway. The oxidation pathway was mainly divided into three phases.
First, glycerol is converted into dihydroxyacetone through DhaD (NAD+ is required as a coenzyme to produce NADH2). Dihydroxyacetone is phosphorylated into glycolysis under the action of Dhak. As shown in Figure 7A and Table 2, DhaD and DhaK were up-regulated, and their FCs were 1.38 and 1.30, respectively. According to the report, DhaD encodes glycerol dehydrogenase in K. pneumoniae (Tang et al., 1982), and glycerol dehydrogenase and 1,3-propylene glycol oxidoreductase are the key enzymes for the conversion of glycerol to 1,3-PDO (Zhao et al., 2009). Therefore, the up-regulation of DhaD was conducive to the production of 1,3-PDO. It has been reported (Raynaud et al., 2011) that the DhaK, DhaL, and DhaM were belong to the PEP-dependent dihydroxyacetone kinases. It can be seen from Supplementary Table 3, DhaK [A6TEA9 contains dihydroxyacetone-binding sites (Gutknecht et al., 2001)], DhaL [A0A378G569 contains ADP-binding sites (Gutknecht et al., 2001)] and DhaM [A0A486FQT7, a phosphohistidine protein that can transfer phosphoryl groups from a phosphoryl carrier protein of the phosphotransferase system (HPr or enzyme I) to the DhaL-ADP complex (Gutknecht et al., 2001; Bachler et al., 2005)] were also up-regulated, indicating that dihydroxyacetone kinases were up-regulated. Glycerol dehydratase, 1,3-PDO oxidoreductase, glycerol dehydrogenase, and dihydroxyacetone kinase are encoded by an operon named dha, and their expression was consistent (Forage and Lin, 1982). It can be seen from Figures 7A,B; both were up-regulated at the same time, which also explains that the domesticated strain favors to produce 1,3-PDO.
Second, the dihydroxyacetone phosphate was further oxidized to pyruvate. In this process, triosephosphate isomerase (TPI), GAPDH, PGK, GpmI, Eno, and pyruvate kinase (PK) were up-regulated, and their corresponding FCs were 1.27, 3.03, 1.68, 1.72, 1.23, and 1.48, respectively. TPI plays a vital role in metabolism and is the key to efficient energy production (Zheng et al., 2006). The dihydroxyacetone phosphate was transformed into glyceraldehyde 3-phosphate through TPI. Because of the up-regulation of TPI, the accumulation of dihydroxyacetone phosphate can be reduced, as its toxicity would affect cell growth and survival (Kang et al., 2014). GAPDH catalyzes the conversion of glyceraldehyde 3-phosphate to glycerol 1,3-diphosphate and reduces NAD+ to NADH. Moreover, the up-regulation of GAPDH can shorten the fermentation time and inhibit the accumulation of some harmful by-products (such as lactic acid) (Yang et al., 2013). By comparing the protein changes in the two cases, most of the proteins in the glycolysis pathway were up-regulated, which helped to provide ATP and NADH for bacteria, promote the growth of the bacteria, and finally increase the yield of 1,3-PDO.
In addition, the by-products of pyruvate metabolism were lactic acid, formic acid, and 2,3-butanediol. The strain domestication may increase in yields of both the main product 1,3-PDO and by-products, so BudC and PflB were up-regulated. AckA and AdhP were also up-regulated when acetyl CoA produced by-products, such as ethanol and acetic acid. LDH and LldD encode L-lactate dehydrogenase (Aguilera et al., 2008; Fu et al., 2016). The down-regulation of lactate dehydrogenase reduces both the consumption of NADH and the formation of the by-product lactic acid and finally improves the output of 1,3-PDO (Xu et al., 2009).
The third step was that pyruvate could be further transferred to produce acetyl-CoA, and acetyl-CoA could enter the tricarboxylic acid (TCA) cycle to produce other small molecular substances. In this process, some energy was consumed, but there was a regeneration process of force reduction. In the whole TCA cycle, FumB and FrdB provide energy for bacteria. Therefore, the up-regulation of FumB and FrdB might accelerate cell growth and increase the yield of 1,3-PDO (Tseng et al., 2001; Xue et al., 2010; Huang et al., 2013; Li et al., 2019).
For the domesticated bacteria, most of the proteins in the metabolic process were up-regulated, resulting in a significant increase in the yield of 1,3-PDO. On the other hand, the 1,3-PDO yield could be further improved by the following genetic modifications, for example, the overexpression of some up-regulated genes (such as dhaD, dhaK, tpi, gapA, etc.), blocking the by-product pathway (such as knocking out budC to reduce the competition of by-products) and weakening ldh to reduce the consumption of NADH.
Conclusion
The fermentation capabilities of 1,3-PDO by glycerol domesticated strains at the concentrations of 40, 60, 80, 100, 120, and 140 g/L were compared. It was found that the strain domesticated with 120 g/L glycerol had the highest capability to produce 1,3-PDO, reaching 59.41 g/L. To further improve the yield of 1,3-PDO, the strain domesticated with 120–20 g/L glycerol concentration (K. pneumoniae x546) was obtained, and the yield reached 69.35 g/L. In addition, in order to overcome the osmotic pressure problem caused by excessive Na+ in the fermentation system, betaine was added to the fermentation medium, making the yield further increase to 74.44 g/L and shortening the fermentation time from 40 to 24 h. Based on TMT, it was found that regulating genes, such as dhaD, dhak, budC, ldh, and so on, were able to enhance the yield of 1,3-PDO. Moreover, the introduction of Na2CO3 and betaine in the fermentation process will render the formation of 1,3-PDO more environment-friendly and facilitate industrial adoption of this technology in the future.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: PRIDE database Project Name: Klebsiella pneumoniae x546, ATCC15380, TMT Project accession: PXD028396.
Author Contributions
XW carried out the experimental work, analyzed the data, and wrote the manuscript. HC and PW performed the data analysis and participated in the manuscript editing and revise. JJ and LZ helped to partial experiment and figure processing. JX helped to edit the manuscript and involved in discussion in the manuscript preparation. JW was responsible for the experiment design and supervision. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Key Research and Development Program (No. 2018YFA0902200), the China Petrochemical Corporation (No. 2018GKF-0283), the China Scholarship Council (No. 202006250069), and the Frontiers Science Center for Synthetic Biology (Ministry of Education), Tianjin University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.770109/full#supplementary-material
Footnotes
- ^ www.uniprot.org/taxonomy/?query=ATCC15380&sort=score
- ^ http://geneontology.org/
- ^ http://www.genome.jp/kegg/
References
Aguilera, L., Campos, E., Gimenez, R., Badia, J., Aguilar, J., and Baldoma, L. (2008). Dual role of LldR in regulation of the lldPRD operon, involved in L-lactate metabolism in Escherichia coli. J. Bacteriol. 190, 2997–3005. doi: 10.1128/JB.02013-07
Bachler, C., Flukiger-Bruhwiler, K., Schneider, P., Bahler, P., and Erni, B. (2005). From ATP as substrate to ADP as coenzyme. J. Biol. Chem. 280, 18321–18325.
Bao, W. J., Wei, R. Q., Liu, X. X., Dong, S. F., Chen, T. Y., Fu, S. L., et al. (2020). Regulation of pyruvate formate lyase-deficient Klebsiella pneumoniae for efficient 1,3-propanediol bioproduction. Curr. Microbiol. 77, 55–61. doi: 10.1007/s00284-019-01795-5
Chen, W. C., Chuang, C. J., Chang, J. S., Wang, L. F., Soo, P. C., Wu, H. S., et al. (2020). Exploring dual-substrate cultivation strategy of 1,3-propanediol production using Klebsiella pneumoniae. Appl. Biochem. Biotechnol. 191, 346–359. doi: 10.1007/s12010-019-03208-6
Chen, Z., Zhong, W., Chen, S., Zhou, Y., Ji, P., Gong, Y., et al. (2021). TMT-based quantitative proteomics analyses of sterile/fertile anthers from a genic male-sterile line and its maintainer in cotton (Gossypium hirsutum L.). J. Proteomics 232:104026. doi: 10.1016/j.jprot.2020.104026
Colin, T., Bories, A., and Moulin, G. (2000). Inhibition of Clostridium butyricum by 1,3-propanediol and diols during glycerol fermentation. Appl. Microbiol. Biotechnol. 54, 201–205. doi: 10.1007/s002530000365
Dai, J., Yu, X., Han, Y., Chai, L., Liao, Y., Zhong, P., et al. (2020). TMT-labeling proteomics of papillary thyroid carcinoma reveal invasive biomarkers. J. Cancer 11, 6122–6132. doi: 10.7150/jca.47290
Dexter Tam, T. L., Ng, C. K., Lim, S. L., Yildirim, E., Ko, J., Leong, W. L., et al. (2019). Proquinoidal-conjugated polymer as an effective strategy for the enhancement of electrical conductivity and thermoelectric properties. Chem. Mater. 31, 8543–8550. doi: 10.1021/acs.chemmater.9b03684
Fan, C., Fromm, H. J., and Bobik, T. A. (2009). Kinetic and functional analysis of L-threonine kinase, the PduX enzyme of Salmonella enterica. J. Biol. Chem. 284, 20240–20248. doi: 10.1074/jbc.M109.027425
Fan, X., Zhang, T., Jie, L. I., Han, H., Gao, L., Zhang, S., et al. (2018). Betaine metabolism in microorganism and its application. Bull. Fermentation Sci. Technol. 47:3. doi: 10.16774/j.cnki.issn.1674-2214.2018.03.006
Forage, R. G., and Lin, E. C. (1982). DHA system mediating aerobic and anaerobic dissimilation of glycerol in Klebsiella pneumoniae NCIB 418. J. Bacteriol. 151, 591–599. doi: 10.1128/jb.151.2.591-599.1982
Fu, J., Huo, G., Feng, L., Mao, Y., Wang, Z., Ma, H., et al. (2016). Metabolic engineering of Bacillus subtilis for chiral pure meso-2,3-butanediol production. Biotechnol. Biofuels 9:90. doi: 10.1186/s13068-016-0502-5
Glaasker, E., Heuberger, E., Konings, W. N., and Poolman, B. (1998). Mechanism of osmotic activation of the quaternary ammonium compound transporter (QacT) of Lactobacillus plantarum. J. Bacteriol. 180, 5540–5546. doi: 10.1128/JB.180.21.5540-5546.1998
Guerzoni, M. E., Lanciotti, R., and Cocconcelli, P. S. (2001). Alteration in cellular fatty acid composition as a response to salt, acid, oxidative and thermal stresses in Lactobacillus helveticus. Microbiology 147, 2255–2264. doi: 10.1099/00221287-147-8-2255
Gungormusler, M., Gonen, C., and Azbar, N. (2011). 1,3-propanediol production potential by a locally isolated strain of Klebsiella Pneumoniae in comparison to Clostridium beijerinckii NRRL B593 from waste glycerol. J. Polym. Environ. 19, 812–817.
Guo, J., Cao, Y. J., Liu, H., Zhang, R. B., Xian, M., and Liu, H. Z. (2019). Improving the production of isoprene and 1,3-propanediol by metabolically engineered Escherichia coli through recycling redox cofactor between the dual pathways. Appl. Microbiol. Biotechnol. 103, 2597–2608. doi: 10.1007/s00253-018-09578-x
Gutknecht, R., Beutler, R., Garcia-Alles, L. F., Baumann, U., and Erni, B. (2001). The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J. 20, 2480–2486. doi: 10.1093/emboj/20.10.2480
Huang, C.-J., Wang, Z.-C., Huang, H.-Y., Huang, H.-D., and Peng, H.-L. (2013). YjcC, a c-di-GMP phosphodiesterase protein, regulates the oxidative stress response and virulence of Klebsiella pneumoniae CG43. PLoS One 8:e66740. doi: 10.1371/journal.pone.0066740
Hussain, S., Chachar, Q., Keerio, M. I., and Shirazi, M. U.-U. (2020). Physiological and biochemical response of wheat genotypes under temperature stress. Pak. J. Bot. 52, 365–374.
Jantama, K., Haupt, M. J., Svoronos, S. A., Zhang, X., and Ingram, L. O. (2010). Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng. 99, 1140–1153. doi: 10.1002/bit.21694
Jia, C., Lu, X., Gao, J., Wang, R., Sun, Q., and Huang, J. (2021). TMT-labeled quantitative proteomic analysis to identify proteins associated with the stability of peanut milk. J. Sci. Food Agric. 101, 6424–6433. doi: 10.1002/jsfa.11313
Kang, T. S., Korber, D. R., and Tanaka, T. (2014). Metabolic engineering of a glycerol-oxidative pathway in Lactobacillus panis PM1 for utilization of bioethanol thin stillage: potential to produce platform chemicals from glycerol. Appl. Environ. Microbiol. 80, 7631–7639. doi: 10.1128/AEM.01454-14
Kim, C., Lee, J. H., Baek, J., Kong, D. S., Na, J. G., Lee, J., et al. (2020). Small current but highly productive synthesis of 1,3-propanediol from glycerol by an electrode-driven metabolic shift in Klebsiella pneumoniae L17. Chemsuschem 13, 564–573. doi: 10.1002/cssc.201902928
Kingsbury, J. M., and McCusker, J. H. (2010). Homoserine toxicity in Saccharomyces cerevisiae and Candida albicans homoserine kinase (thr1 Delta) mutants. Eukaryot. Cell 9, 717–728.
Lee, J. H., Jung, H. M., Jung, M. Y., and Oh, M. K. (2019). Effects of gltA and arcA mutations on biomass and 1,3-propanediol production in Klebsiella pneumoniae. Biotechnol. Bioprocess Eng. 24, 95–102. doi: 10.1007/s12257-018-0246-0
Li, H., Wang, B. S., Li, Y. R., Zhang, L., Ding, Z. Y., Gu, Z. H., et al. (2017). Metabolic engineering of Escherichia coli W3110 for the production of l-methionine. J. Ind. Microbiol. Biotechnol. 44, 75–88. doi: 10.1007/s10295-016-1870-3
Li, X., Chen, L., Wang, X., and Tian, P. (2019). Physiological investigations of the influences of byproduct pathways on 3-hydroxypropionic acid production in Klebsiella pneumoniae. J. Basic Microbiol. 59, 1195–1207. doi: 10.1002/jobm.201800640
Li, Z., Ro, S. M., Sekar, B. S., Seol, E., Lama, S., Lee, S. G., et al. (2016). Improvement of 1,3-propanediol oxidoreductase (DhaT) stability against 3-hydroxypropionaldehyde by substitution of cysteine residues. Biotechnol. Bioprocess Eng. 21, 695–703. doi: 10.1007/s12257-016-0560-3
Liang, S., Gao, D., Liu, H., Wang, C., and Wen, J. (2018). Metabolomic and proteomic analysis of d-lactate-producing Lactobacillus delbrueckii under various fermentation conditions. J. Ind. Microbiol. Biotechnol. 45, 681–696. doi: 10.1007/s10295-018-2048-y
Longo, F., Motta, S., Mauri, P., Landini, P., and Rossi, E. (2016). Interplay of the modified nucleotide phosphoadenosine 5’-phosphosulfate (PAPS) with global regulatory proteins in Escherichia coli: modulation of cyclic AMP (cAMP)-dependent gene expression and interaction with the HupA regulatory protein. Chem. Biol. Interact. 259, 39–47. doi: 10.1016/j.cbi.2016.04.016
Louesdon, S., Charlot-Rouge, S., Juillard, V., Tourdot-Marechal, R., and Beal, C. (2014). Osmotic stress affects the stability of freeze-dried Lactobacillus buchneri R1102 as a result of intracellular betaine accumulation and membrane characteristics. J. Appl. Microbiol. 117, 196–207. doi: 10.1111/jam.12501
Ma, J. S., Jiang, H., Hector, S. B., Xiao, Z. H., Li, J. L., Liu, R. K., et al. (2019). Adaptability of Klebsiella pneumoniae 2e, a newly isolated 1,3-propanediol-producing strain, to crude glycerol as revealed by genomic profiling. Appl. Environ. Microbiol. 85:15. doi: 10.1128/AEM.00254-19
May, G., Faatz, E., Lucht, J. M., Haardt, M., and Bremer, E. (2010). Characterization of the osmoregulated Escherichia coli proU promoter and identification of ProV as a membrane-associated protein. Mol. Microbiol. 3, 1521–1531. doi: 10.1111/j.1365-2958.1989.tb00138.x
Metsoviti, M., Paraskevaidi, K., Koutinas, A., Zeng, A.-P., and Papanikolaou, S. (2012). Production of 1,3-propanediol, 2,3-butanediol and ethanol by a newly isolated Klebsiella oxytoca strain growing on biodiesel-derived glycerol based media. Process Biochem. 47, 1872–1882. doi: 10.1016/j.procbio.2012.06.011
Mitrea, L., and Vodnar, D. C. (2019). Klebsiella pneumoniae-a useful pathogenic strain for biotechnological purposes: diols biosynthesis under controlled and uncontrolled pH levels. Pathogens 8:293. doi: 10.3390/pathogens8040293
Nakano, S., Ugwu, C. U., and Tokiwa, Y. (2012). Efficient production of D-(-)-lactic acid from broken rice by Lactobacillus delbrueckii using Ca(OH)(2) as a neutralizing agent. Bioresour. Technol. 104, 791–794. doi: 10.1016/j.biortech.2011.10.017
Pan, D. T., Wang, X. D., Shi, H. Y., Yuan, D. C., and Xiu, Z. L. (2019). Ensemble optimization of microbial conversion of glycerol into 1, 3-propanediol by Klebsiella pneumoniae. J. Biotechnol. 301, 68–78. doi: 10.1016/j.jbiotec.2019.06.001
Park, Y. S., Kang, J., Chung, W. H., Lim, M. Y., Seo, M. J., Nam, Y. D., et al. (2019). Complete genome sequence of acetate-producing Klebsiella pneumoniae L5-2 isolated from infant feces. 3 Biotech 9:84. doi: 10.1007/s13205-019-1578-y
Raghunandan, K., McHunu, S., Kumar, A., Kumar, K. S., Govender, A., Permaul, K., et al. (2014). Biodegradation of glycerol using bacterial isolates from soil under aerobic conditions. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 49, 85–92. doi: 10.1080/10934529.2013.824733
Raynaud, C., Lee, J., Sarcabal, P., Croux, C., Meynial-Salles, I., and Soucaille, P. (2011). Molecular characterization of the glycerol-oxidative pathway of Clostridium butyricum VPI 1718. J. Bacteriol. 193, 3127–3134. doi: 10.1128/JB.00112-11
Reitzer, L. (2005) Catabolism of amino acids and related compounds. EcoSal Plus 1:2. doi: 10.1128/ecosalplus.3.4.7
Shin, W. S., June, C. S., Lee, Y., Park, Y., Choi, J., and Jeon, W. (2011). Synthesis of low concentration of NaOH solution using Na^+ ion in the concentrated water from membrane separation process. Korean Chem. Eng. Res. 49, 810–815. doi: 10.9713/kcer.2011.49.6.810
Simon, A., Fujioka, T., Price, W. E., and Nghiem, L. D. (2014). Sodium hydroxide production from sodium carbonate and bicarbonate solutions using membrane electrolysis: a feasibility study. Sep. Purif. Technol. 127, 70–76. doi: 10.1016/j.seppur.2014.02.020
Sivaraman, J., Li, Y. G., Banks, J., Cane, D. E., Matte, A., and Cygler, M. (2003). Crystal structure of Escherichia coli PdxA, an enzyme involved in the pyridoxal phosphate biosynthesis pathway. J. Biol. Chem. 278, 43682–43690. doi: 10.1074/jbc.M306344200
Sogame, Y., Kojima, K., Takeshita, T., Kinoshita, E., and Matsuoka, T. (2014). Identification of cAMP-dependent phosphorylated proteins involved in the formation of environment-resistant resting cysts by the terrestrial ciliate Colpoda cucullus. ISJ Invertebrate Surviv. J. 11, 213–218.
Stirling, D. A., Hulton, C. S., Waddell, L., Park, S. F., Stewart, G. S., Booth, I. R., et al. (1989). Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol. Microbiol. 3, 1025–1038. doi: 10.1111/j.1365-2958.1989.tb00253.x
Tang, J., Forage, R. G., and Lin, E. (1982). Immunochemical properties of NAD+-linked glycerol dehydrogenases from Escherichia coli and Klebsiella pneumoniae. J. Bacteriol. 152, 1169–1174. doi: 10.1128/jb.152.3.1169-1174.1982
Tee, Z. K., Jahim, J. M., Tan, J. P., and Kim, B. H. (2017). Preeminent productivity of 1,3-propanediol by Clostridium butyricum JKT37 and the role of using calcium carbonate as pH neutraliser in glycerol fermentation. Bioresour. Technol. 233, 296–304. doi: 10.1016/j.biortech.2017.02.110
Tseng, C. P., Yu, C. C., Lin, H. H., Chang, C. Y., and Kuo, J. T. (2001). Oxygen- and growth rate-dependent regulation of Escherichia coli fumarase (FumA, FumB, and FumC) activity. J. Bacteriol. 183, 461–467. doi: 10.1128/JB.183.2.461-467.2001
Wang, M., Wang, G., Zhang, T., Fan, L., and Tan, T. (2017). Multi-modular engineering of 1,3-propanediol biosynthesis system in Klebsiella pneumoniae from co-substrate. Appl. Microbiol. Biotechnol. 101, 647–657. doi: 10.1007/s00253-016-7919-4
Wang, S., Yang, Y., Zhao, Y., Zhao, H., Bai, J., Chen, J., et al. (2016). Sub-MIC tylosin inhibits Streptococcus suis biofilm formation and results in differential protein expression. Front. Microbiol. 7:384. doi: 10.3389/fmicb.2016.00384
Wang, C., Chen, L., Cai, Z. C., Chen, C., Liu, Z., Liu, X., et al. (2020a). Comparative proteomic analysis reveals the molecular mechanisms underlying the accumulation difference of bioactive constituents in Glycyrrhiza uralensis Fisch under salt stress. J. Agric. Food Chem. 68, 1480–1493. doi: 10.1021/acs.jafc.9b04887
Wang, Z., Wang, L., Zhou, J., Zou, J., and Fan, L. (2020b). New insights into the immune regulation and tissue repair of Litopenaeus vannamei during temperature fluctuation using TMT-based proteomics. Fish Shellfish Immunol. 106, 975–981. doi: 10.1016/j.fsi.2020.09.014
Wang, Z., Li, M. X., Xu, C. Z., Zhang, Y., Deng, Q., Sun, R., et al. (2020c). Comprehensive study of altered proteomic landscape in proximal renal tubular epithelial cells in response to calcium oxalate monohydrate crystals. BMC Urol. 20:136. doi: 10.1186/s12894-020-00709-z
Willke, T., and Vorlop, K. (2008). Biotransformation of glycerol into 1,3-propanediol. Eur. J. Lipid Sci. Technol. 110, 831–840. doi: 10.1002/ejlt.200800057
Xu, Q. S., Jancarik, J., Lou, Y., Kuznetsova, K., Yakunin, A. F., Yokota, H., et al. (2005). Crystal structures of a phosphotransacetylase from Bacillus subtilis and its complex with acetyl phosphate. J. Struct. Funct. Genomics 6, 269–279. doi: 10.1007/s10969-005-9001-9
Xu, Y.-Z., Guo, N.-N., Zheng, Z.-M., Ou, X.-J., Liu, H.-J., and Liu, D.-H. (2009). Metabolism in 1,3-propanediol fed-batch fermentation by a D-lactate deficient mutant of Klebsiella pneumoniae. Biotechnol. Bioeng. 104, 965–972. doi: 10.1002/bit.22455
Xue, X., Li, W., Li, Z., Xia, Y., and Ye, Q. (2010). Enhanced 1,3-propanediol production by supply of organic acids and repeated fed-batch culture. J. Ind. Microbiol. Biotechnol. 37, 681–687. doi: 10.1007/s10295-010-0711-z
Yang, T., Rao, Z., Zhang, X., Xu, M., Xu, Z., and Yang, S.-T. (2013). Improved production of 2,3-butanediol in Bacillus amyloliquefaciens by over-expression of glyceraldehyde-3-phosphate dehydrogenase and 2,3-butanediol dehydrogenase. PLoS One 8:e76149. doi: 10.1371/journal.pone.0076149
Yin, L., Luo, X., Zhang, Y., Zheng, W., Yin, F., and Fu, Y. (2020). Comparative proteomic analysis of Rhizopus oryzae hyphae displaying filamentous and pellet morphology. 3 Biotech 10:469. doi: 10.1007/s13205-020-02458-0
Yiqiang, P., Lin, L., Baishan, F., and Wenzhen, Z. (2007). The selection of 1,3-propanediol produced bacterium enduring high glycerol concentration mutate and its immobidi ferment study. Acta Laser Biol. Sin. 16, 754–758.
Yuan, Z. H., Wang, L., Sun, S. T., Wu, Y., and Qian, W. (2013). Genetic and proteomic analyses of a Xanthomonas campestris pv. campestris purC mutant deficient in purine biosynthesis and virulence. J. Genet. Genomics 40, 473–487. doi: 10.1016/j.jgg.2013.05.003
Zabed, H. M., Zhang, Y. F., Guo, Q., Yun, J. H., Yang, M. M., Zhang, G. Y., et al. (2019). Co-biosynthesis of 3-hydroxypropionic acid and 1,3-propanediol by a newly isolated Lactobacillus reuteri strain during whole cell biotransformation of glycerol. J. Clean. Prod. 226, 432–442. doi: 10.1016/j.jclepro.2019.04.071
Zhang, L., Fan, Y., Li, X., Liao, S., Wang, P., and Qiao, K. (2016). Effects of two alkaline pH regulators on the fermentation of 1,3-propanediol. Chem. Ind. Eng. Prog. 35, 2542–2546.
Zhang, L. J., Bao, W. J., Wei, R. Q., Fu, S. L., and Gong, H. (2018). Inactivating NADH:quinone oxidoreductases affects the growth and metabolism of Klebsiella pneumoniae. Biotechnol. Appl. Biochem. 65, 857–864. doi: 10.1002/bab.1684
Zhang, Y., Ma, C., Dischert, W., Soucaille, P., and Zeng, A.-P. (2019). Engineering of phosphoserine aminotransferase increases the conversion of l-homoserine to 4-hydroxy-2-ketobutyrate in a glycerol-independent pathway of 1,3-propanediol production from glucose. Biotechnol. J. 14:e1900003. doi: 10.1002/biot.201900003
Zhang, Y., Morar, M., and Ealick, S. E. (2008). Structural biology of the purine biosynthetic pathway. Cell. Mol. Life Sci. 65, 3699–3724. doi: 10.1007/s00018-008-8295-8
Zhao, L., Ma, X., Zheng, Y., Zhang, J., Wei, G., and Wei, D. (2009). Over-expression of glycerol dehydrogenase and 1,3-propanediol oxidoreductase in Klebsiella pneumoniae and their effects on conversion of glycerol into 1,3-propanediol in resting cell system. J. Chem. Technol. Biotechnol. 84, 626–632. doi: 10.1002/jctb.2092
Zheng, P., Sun, J., van den Heuvel, J., and Zeng, A.-P. (2006). Discovery and investigation of a new, second triose phosphate isomerase in Klebsiella pneumoniae. J. Biotechnol. 125, 462–473. doi: 10.1016/j.jbiotec.2006.03.034
Zhong, Q. P., Wang, B., Wang, J., Liu, Y. F., Fang, X., and Liao, Z. L. (2019). Global proteomic analysis of the resuscitation state of Vibrio parahaemolyticus compared with the normal and viable but non-culturable state. Front. Microbiol. 10:1045. doi: 10.3389/fmicb.2019.01045
Zhou, S., Lama, S., Sankaranarayanan, M., and Park, S. (2019b). Metabolic engineering of Pseudomonas denitrificans for the 1,3-propanediol production from glycerol. Bioresour. Technol. 292:121933. doi: 10.1016/j.biortech.2019.121933
Keywords: Klebsiella pneumoniae, 1, 3-propanediol production, betaine, Na+ pH neutralizer, proteomics
Citation: Wang X, Zhang L, Chen H, Wang P, Yin Y, Jin J, Xu J and Wen J (2021) Rational Proteomic Analysis of a New Domesticated Klebsiella pneumoniae x546 Producing 1,3-Propanediol. Front. Microbiol. 12:770109. doi: 10.3389/fmicb.2021.770109
Received: 07 September 2021; Accepted: 26 October 2021;
Published: 26 November 2021.
Edited by:
Roshan Kumar, Magadh University, IndiaReviewed by:
Xixian Xie, Tianjin University of Science & Technology, ChinaQuan Luo, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences (CAS), China
Copyright © 2021 Wang, Zhang, Chen, Wang, Yin, Jin, Xu and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianping Wen, jpwen@tju.edu.cn
 Xin Wang1,2,3,4
Xin Wang1,2,3,4 Jianwei Xu
Jianwei Xu Jianping Wen
Jianping Wen