- 1GSK, Siena, Italy
- 2Department of Medical Microbiology, University Medical Center Utrecht, Utrecht University, Utrecht, Netherlands
Staphylococcus aureus is the main cause of human skin and soft tissue infections. However, S. aureus pathogenicity within the skin is not fully characterized. Here, we implemented an S. aureus cutaneous infection model using human skin explants and performed a time-course infection to study the gene expression profile of a large panel of virulence-related factors of S. aureus USA300 LAC strain, by high-throughput RT-PCR. We pinpointed the genes that were differentially regulated by the bacteria in the skin tissues and identified 12 virulence factors that were upregulated at all time points assessed. Finally, using confocal microscopy, we show that the expression of alpha-hemolysin by S. aureus varies dependent on the skin niche and that the bacteria preferentially accumulates inside sweat glands and ducts. Taken together, our study gives insights about the pathogenic lifestyle of S. aureus within human skin tissues, which may contribute for the development of anti-S. aureus therapeutic strategies.
Introduction
Staphylococcus aureus is an important human pathogen that persistently colonizes 20% of human population and transiently colonizes another 60% (Kluytmans et al., 1997). These bacteria can cause different diseases that range from skin and soft tissue infections (SSTIs) as impetigo, folliculitis, abscesses and cellulitis (Olaniyi et al., 2016), to more invasive diseases like endocarditis, osteomyelitis, pneumonia and sepsis (Lowy, 1998; Salgado-Pabón and Schlievert, 2014). Available antibiotics are not sufficiently effective against multidrug resistant S. aureus strains and therefore this pathogen represents a major concern for human health. Although several S. aureus vaccines were tested in clinical trials, they failed to show efficacy in humans (Bagnoli et al., 2012). The overreliance on animal models and the incomplete understanding of S. aureus pathogenesis during human infection may explain this failure.
S. aureus is equipped with a large array of virulence factors that include adhesins, immune evasion factors, toxins and proteases (Foster, 2005; Foster et al., 2014; Thammavongsa et al., 2015; Tam and Torres, 2019). Complex regulatory networks modulate the expression of metabolic and virulence factors and enable the bacteria to adapt to different host environments (Cheung et al., 2004; Balasubramanian et al., 2017; Haag and Bagnoli, 2017; Jenul and Horswill, 2018). A number of global regulators are influenced by environmental stimuli as nutrients and oxygen availability, cell density, pH and osmolarity (Cheung et al., 2004; Balasubramanian et al., 2017; Haag and Bagnoli, 2017; Jenul and Horswill, 2018). Moreover, there is increasing evidence that host niche-specific factors also have an impact on S. aureus gene expression (Rothfork et al., 2003; Cheung et al., 2004). The best well-characterized regulator of S. aureus virulence is the accessory gene regulator (agr), a quorum-sensing system. Generally, S. aureus expresses virulence factors that facilitate adhesion to the host tissues as surface-bound proteins in the beginning of infection (or mid-exponential phase), while an activated agr system leads to the expression of secreted toxins and proteases that enable tissue invasion and dissemination later in infection (or late-exponential/stationary phase) (Cheung et al., 2004; Le and Otto, 2015).
The skin tissues are often the primary site of S. aureus infection, from which bacteria can spread to other organs to cause more invasive diseases. However, little is known about S. aureus pathogenicity during infection of human skin tissues. Because human skin differs from mouse skin in both histology and immunology (Pasparakis et al., 2014) and S. aureus produces human-specific virulence factors, it is important to study S. aureus infections in human tissues. To our knowledge, only two studies analyzed the transcriptional response of S. aureus in human skin (Loughman et al., 2009; Date et al., 2014). These studies were performed in clinical samples of cutaneous abscesses, which, although relevant, do not permit to investigate gene expression of S. aureus at initial stages of infection and also limit the comprehension about transcription kinetics while infection evolves.
In this study, we assess the virulence gene expression of S. aureus USA300 LAC strain in human skin tissues over time and highlight the factors that might be required to establish and/or maintain S. aureus cutaneous infections. Additionally, we show that S. aureus accumulates in sweat glands and demonstrate that the transcription profile of the bacteria also varies within skin niches. Overall, our study contributes for a better understanding of S. aureus pathogenicity on SSTI progression.
Materials and Methods
Ethics Statement
The studies involving human participants were reviewed and approved by French Ministry of Higher Education, Research and Innovation. The patients/participants provided their written informed consent to participate in this study.
Bacterial Strains and Culture
S. aureus USA300 LAC strains were grown overnight in tryptic soy broth (TSB) at 37°C, diluted 1:100 in fresh TSB and cultured at 37°C until mid-log phase (OD600 0.6–0.7). Cells were centrifuged, washed and resuspended in PBS to obtain 5 × 106 CFU/μL. For USA300 LAC pOS-hla-mCherry, the promoter of hla gene was fused to mCherry in a pOS plasmid backbone and transformed into USA300 LAC strain.
Process and Infect Fresh Human Skin Explants
Human skin explants were processed as described before (Olaniyi et al., 2018), with modifications (Figure 1). In short, after removal of adipose tissue, the skin sheet was stripped 30 times with a hypoallergenic tape (Transpore, 3M). Using disposable biopsy punches (Kai Medical), 8 mm biopsies in diameter were collected and subsequently submerged in culture medium (Advanced DMEM/F-12 supplemented with 4 mM L-Glutamine; Gibco). After two washes with PBS, the punches were placed in 12-well transwell plates with 0.4 μm pore size (Corning), previously filled with 1 mL of culture medium. The transwells enable culture of the skin at air-liquid interface (the surface of the epidermis layer is exposed to the air, while the other skin layers are submerged in the culture medium), as depicted in Figure 1. Finally, the punches were infected with 5 × 106 CFU USA300 LAC. To ensure that the bacteria does not contact with the culture medium, we carefully placed 1 μL of bacteria on top of the dry epidermis. After infection, the skin was cultured at air-liquid interface for 2, 3, 24, or 72 h, at 37°C, 5% CO2. For the early time points (2 and 3 h of infection), non-adherent bacteria were removed by washing the skin surface twice with PBS before RNA isolation. This protocol was performed using skin explants collected from three different donors and technical duplicates or triplicates were performed for each time point.
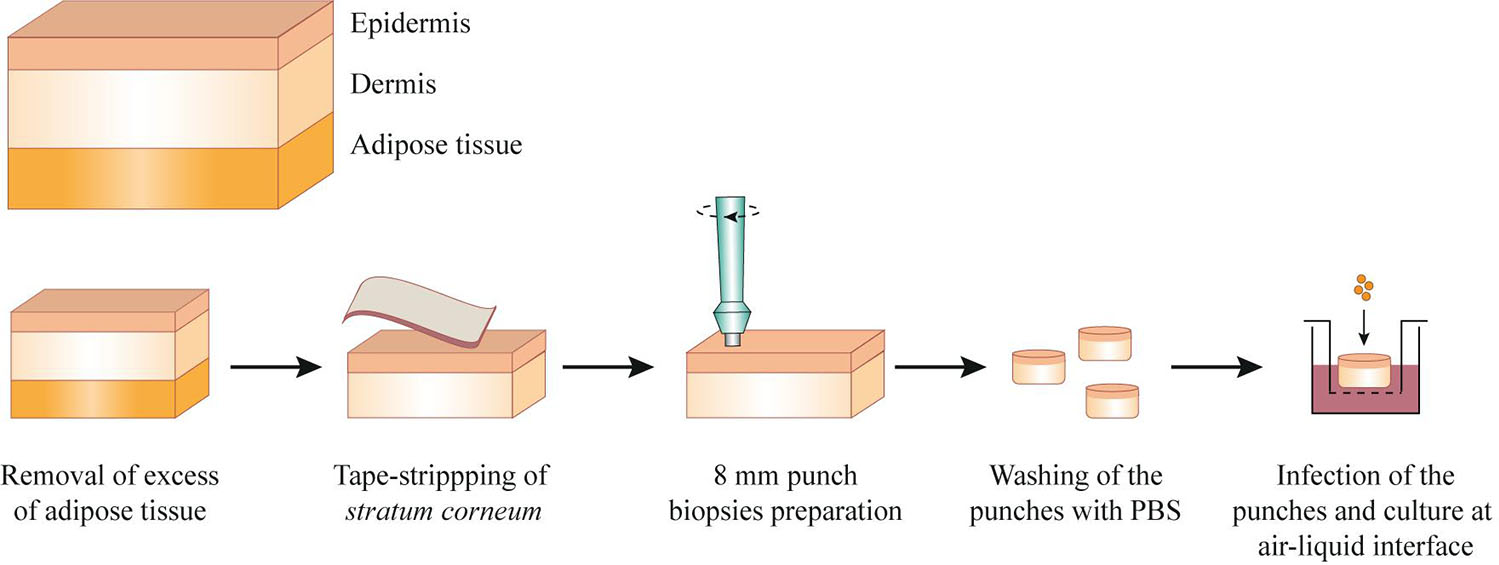
Figure 1. Schematic representation of the protocol to process and infect fresh human skin explants. After removal of the adipose tissue, the skin explants were pinned in a dissection board and the surface of epidermis was tape-stripped with an hypoallergenic tape. Skin punches of 8 mm were prepared, washed in PBS and placed in a transwell, previously filled with culture medium. The skin was infected with 5 × 106 CFU of USA300 LAC strain and cultured at air-liquid interface for 2, 3, 24, or 72 h.
Confocal Microscopy
Skin punches were fixed with 4% formaldehyde, overnight at RT. The punches were washed in PBS, embedded in tissue freezing medium (Leica Biosystems), snap-frozen in a dry ice-ethanol bath and stored at –80°C. Leica cryostat CM1950 was used to cut 8 μm-thick cryosections that were mounted onto SuperFrost Plus adhesion slides (Thermo Fisher Scientific). Cryosections were permeabilized with staining buffer (0.3% Triton-100, 3% BSA in PBS) for 1h at RT, in a humidity chamber, and stained with antibodies. The following antibodies were used: rabbit anti-wide spectrum cytokeratin (Abcam), rabbit anti-cleaved caspase-3 (Abcam), anti-Staphylococcus aureus antibody biotin (Abcam), Alexa Fluor 647 Phalloidin (Life technologies), goat anti-rabbit Alexa Fluor 568 (Life Technologies) and streptavidin Alexa Fluor 488 (Life technologies). Primary antibodies were incubated for 1h at RT and, after three washes with PBS, secondary antibodies were added for 30 min at RT, in the dark. After antibody staining, slides were mounted with ProLong Gold Antifade Reagent with DAPI (Life Technologies). Confocal images were acquired using Zeiss LSM 700 confocal microscope.
Two-Photon Microscopy
For two-photon microscopy, skin punches were infected with a GFP-expressing USA300 LAC strain for 72 h. The punches were fixed with 4% formaldehyde overnight at RT, washed in PBS and stored at –80°C until imaging.
RNA Extraction From Infected Skin Tissues, cDNA Synthesis and qPCR
Infected skin punches were incubated in RNAlater (Thermo Fisher Scientific) up to 1 week, at 4°C. Tissue disruption was performed in cold TRIzol (Invitrogen) using a Tissue Homogenizer (Omni International) and sterile OMNI Hard Tissue Tips (Omni International). The skin punches were submitted to 3 cycles of disruption of 60 s at maximum speed and incubated on ice for 2 min in between each run. After that, the samples were transferred to Lysis Matrix B tubes (Mp Biomedical) and 2 cycles of lysis of 60 s at 6.5 m s–2 were performed using MP Fastprep 24 (Mp Biomedical). In between the cycles, samples were placed on ice for 5 min. Tubes were centrifuged at 12,000 g for 10 min at 4°C and the supernatants were collected and mixed with chloroform to obtain phase separation. The upper aqueous phase containing RNA was purified and concentrated with RNA Clean&Concentrator-5 (Zymo research). DNA elimination was performed by incubation of samples with 20 U of Turbo DNase (Ambion), followed by a second cycle of RNA Clean&Concentrator-5. RNA integrity was confirmed using the Agilent RNA 6000 Pico Kit (Agilent Technologies). Reverse transcription was performed using SuperScript III First-Strand Synthesis SuperMix (Invitrogen). After pre-amplification of the cDNA samples (12 pre-amplification cycles for bacterial cDNA and 14 cycles for mixed samples), quantitative real time PCR was performed using the high-throughput qRT-PCR Fluidigm Biomark HD. The transcription profile of 65 S. aureus virulence factors was assessed using specific TaqMan assays (see Supplementary Table 1; Brignoli et al., 2019). gyrB gene was used as the endogenous control to normalize the data. All the samples were normalized to inoculated bacteria using the ΔΔct method and presented as log2 (fold-change). The gene expression data obtained from three independent experiment showed good reproducibility (Pearson’s r > 0.6 for early time points and r > 0.9 for late time points; see Supplementary Figure 2). Due to the low levels of bacterial RNA obtained from the skin punches infected over 2 and 3 h (see Supplementary Figure 3), absolute quantification of two selected transcripts (spa and hla) was assessed by droplet digital PCR (BioRad), which showed to corroborate Fluidigm data (Supplementary Figure 4).
Statistical Analysis
Statistical analysis was performed with GraphPad Prism v.8.0 software for one-way ANOVA and with SPSS v.25.0.0.2 to calculate Pearson correlation coefficients. Multiple array viewer (Mev) software was used for hierarchical clustering, using Pearson correlation as distance metric. Differences were considered significant at P < 0.05.
Results
Human Skin Explants Can Be Used to Study Staphylococcus aureus Skin Infection
To investigate colonization and infection of human skin by S. aureus, we first set up a model resembling the natural S. aureus cutaneous infections, using abdominal human skin explants. As shown in Figure 1, the uppermost layer of the epidermis (stratum corneum) was impaired by tape-stripping, to mimic skin abrasion, subsequently, the skin punches were inoculated with USA300 strain and cultured at air-liquid interface. We selected the community-acquired methicillin-resistant USA300 strain for this study because it is the predominant cause of SSTIs (King et al., 2006; Moran et al., 2006; Talan et al., 2011). To validate this model, we verified whether the non-infected skin punches could be cultured at air-liquid interface for up to 72 h without undergoing apoptosis, we compared the detection of active-caspase-3 (a marker for apoptosis) in fresh tissue, tissue cultured for over 72 h and for over 6 days, as a control. Fresh tissue and tissue cultured over 72 h did not express active-caspase-3, contrarily to tissue cultured up to 6 days (Supplementary Figure 1A), indicating that the skin punches can be incubated at air-liquid interface for at least 72 h. By comparing skin sections of non-stripped and stripped skin punches stained with anti-cytokeratin antibodies, we also demonstrated that the tape-stripping procedure effectively removed most of the stratum corneum (Supplementary Figure 1B). Finally, after infection of skin punches with GFP-expressing USA300 strain for 72 h, we performed two-photon microscopy and detected bacteria in the dermis layer, at different depths (Supplementary Figure 1C), confirming that S. aureus could invade the skin tissues from epidermis to the dermis. Overall, we optimized a human cutaneous infection model that is suitable to study S. aureus skin infections.
S. aureus Virulence Factors Group Into Four Main Clusters According to Transcription Profile During Skin Infection
To monitor S. aureus virulence in human skin, we studied the gene expression of a large panel of virulence factors after 2 and 3 h of infection for colonization, and after 24 and 72 h for established infection. We selected 65 virulence-related genes that encode adhesion, invasion and immune evasion proteins (see Supplementary Table 1), that are normally expressed by the bacteria at different stages of infection. The heatmap shows the overall transcription kinetics of the selected genes during skin colonization and infection (Figure 2A).
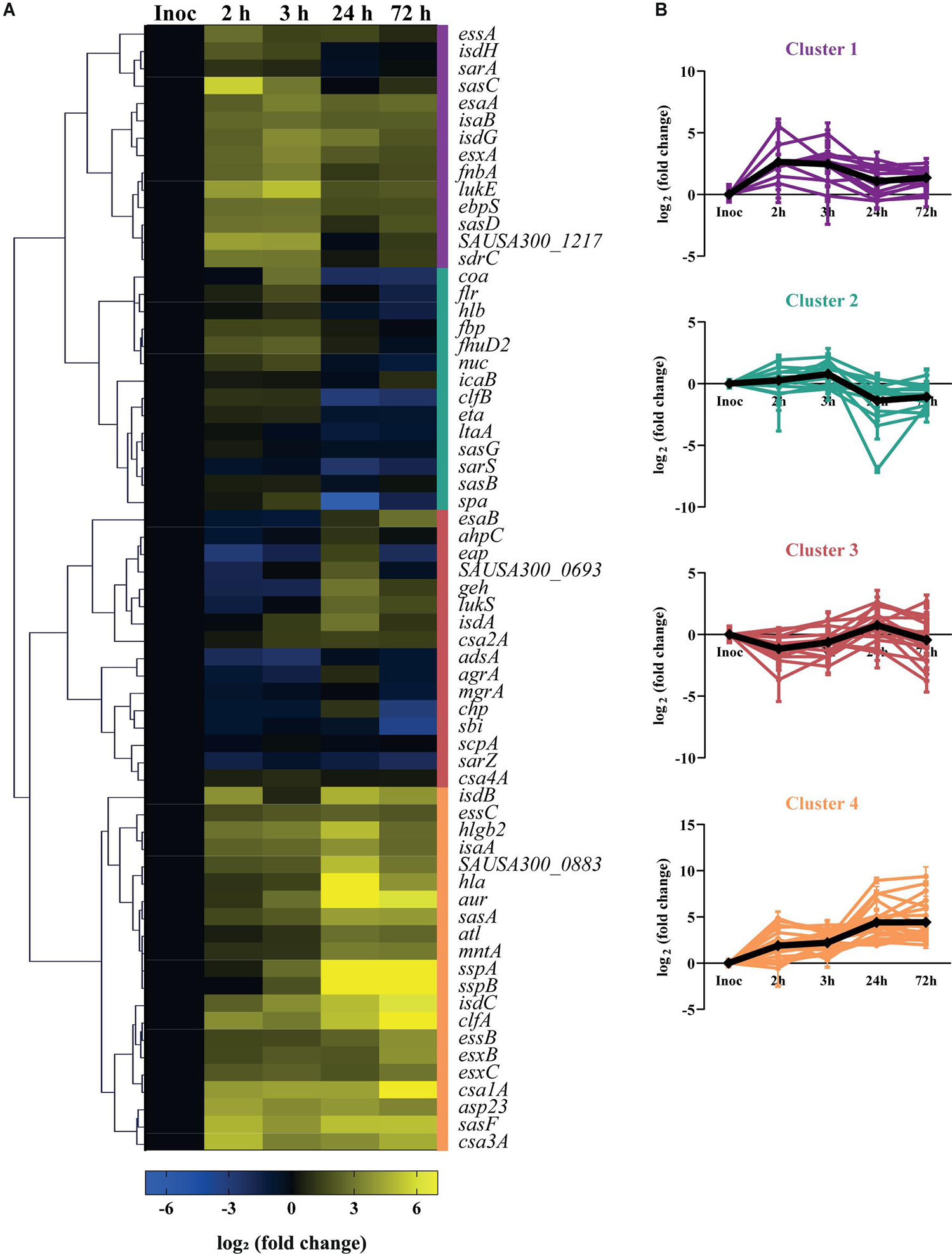
Figure 2. Virulence gene expression kinetics of USA300 during infection of human skin explants. (A) Heatmap showing the transcription profile of 65 virulence factors of S. aureus during cutaneous infection. Values represent the mean of three independent experiments, normalized to the transcription levels of the inoculated bacteria (exponential phase, OD600 = 0.6–0.7) and shown as log2 (fold change). The genes that were less or more transcribed in the skin model than in the inoculated bacteria are depicted in blue or yellow, respectively. The colored bars on the right side of the heatmap represent the four major clusters: cluster 1 (purple), cluster 2 (green), cluster 3 (red), cluster 4 (orange). (B) Transcript level variation over infection of the virulence factors grouped within the same cluster. The black line represents the mean profile of the cluster genes.
Cluster analysis subdivided the analyzed genes into four main clusters, based on their transcription profile over the time-course infection (Figures 2A,B). Cluster 1 includes genes whose transcription increases upon skin contact and slightly decreases while infection progresses, like sasC and sasD, which encode for the S. aureus surface proteins C and D, respectively (Figures 2A,B; cluster 1). Cluster 2 includes the genes whose transcripts levels are similar with those of the inoculated bacteria during colonization but that decrease in expression after 24 h of skin infection. This cluster also includes genes that encode for surface proteins such as clumping factor B (clfB), staphylococcal protein A (spa) and S. aureus surface proteins G and B (sasG, sasB) (Figures 2A,B; cluster 2). Cluster 3 represents the genes that are downregulated early in infection but increase in expression after 24 h of infection, and includes genes that encode proteins that play a role in transcription regulation like agrA, mgrA, and sarZ (Figures 2A,B; cluster 3). Finally, cluster 4 contains the genes that are highly expressed over the entire course of infection, and includes many genes that encode secreted factors such as alpha-hemolysin (hla), aureolysin (aur) and the ESAT-6 secretion system (also known as Type VII secretion system, or T7SS) extracellular protein C (esxC) (Figures 2A,B; cluster 4). Taken together, these data show the transcription kinetics of 65 virulence genes of USA300 during colonization and infection of human skin.
Bacteria Modulate Virulence Gene Expression During Skin Colonization and Infection
Having shown the transcription kinetics of the selected virulence factors within human skin tissues, we next identified the factors that were significantly up or downregulated during infection by at least twofold, relative to their expression level in inoculated bacteria (P < 0.05; Figure 3A, see Supplementary Table 2). As a strong correlation was observed between the transcription profile of S. aureus virulence factors at early time points (after 2 and 3 h of skin infection) and also the two late time points (24 and 72 h of infection) (Pearson’s r > 0.8; see Supplementary Figure 5), we considered the early time points as early skin infection/colonization and the late time points as late infection.
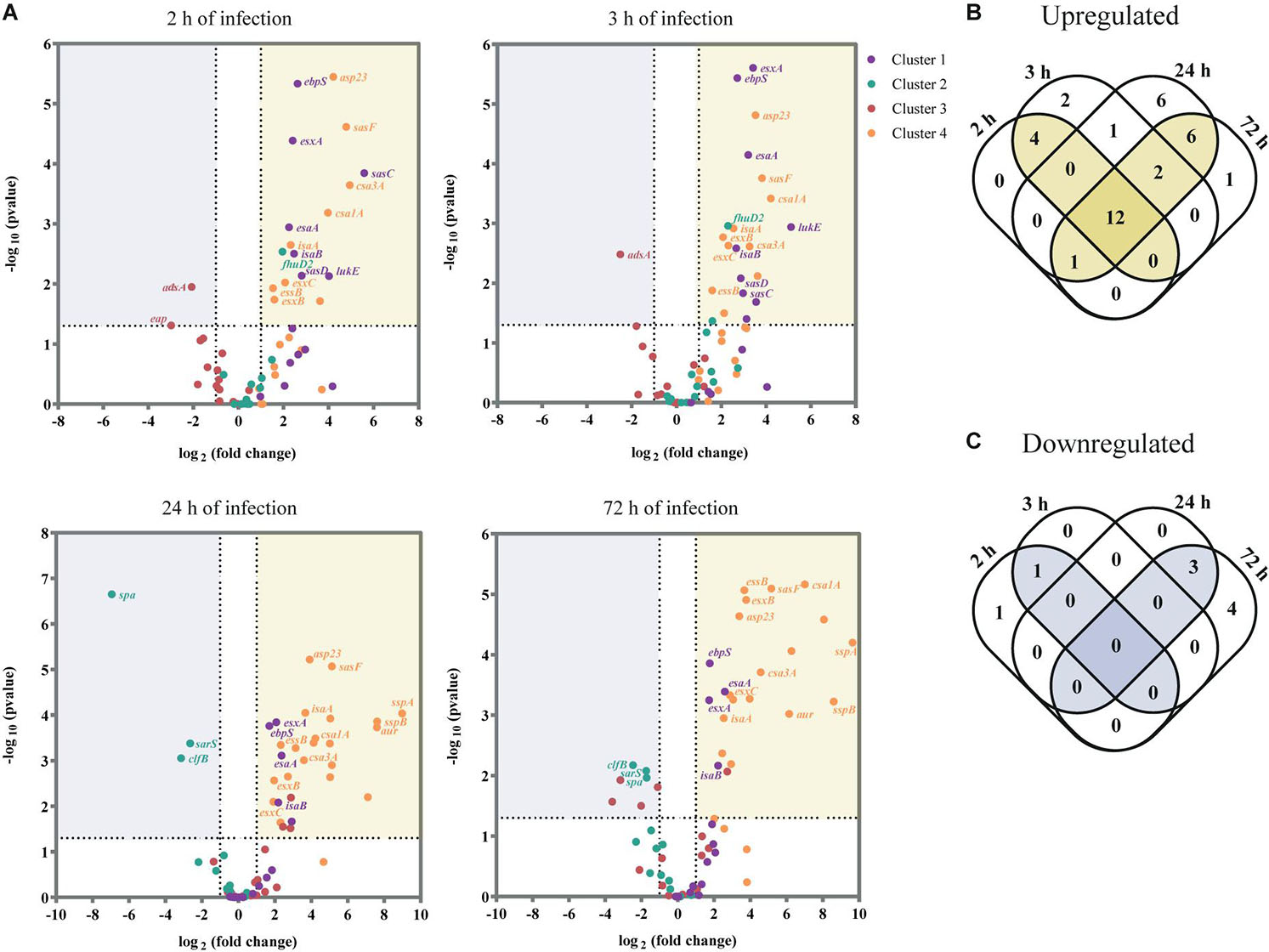
Figure 3. USA300 virulence factors differentially expressed during ex vivo skin infection when compared with inoculated bacteria. (A) Volcano plot showing the transcripts that are significantly downregulated (≥2-fold decrease, P < 0.05) or upregulated (≥2-fold increase, P < 0.05) for each time point (2, 3, 24, and 72 h of infection). Data were normalized over the inoculated bacteria and are expressed in log2 (fold change). The results are shown as the mean of three independent experiments. For each assay, statistical analysis was performed by one-way ANOVA, with Tukey’s multiple comparison tests. (B,C) Venn diagram showing the number of virulence factors that are upregulated (B) and downregulated (C) per time point.
Of the 65 virulence factors, 16 genes were significantly upregulated at early infection/colonization while 1 gene was downregulated (Figures 3B,C; see Supplementary Table 3). As anticipated, most of these genes encode for virulence factors that are associated with tissue colonization and evasion of host defenses. Namely, genes that encode surface proteins as the elastin binding protein (ebpS) and the S. aureus surface proteins C, D and F (sasC, sasD, and sasF) were upregulated upon skin contact. However, we also identified as upregulated at colonization stage the ferric hydroxamate uptake D2 lipoprotein (fhuD2) that is involved in iron uptake and in early stages of invasive S. aureus infection (Mishra et al., 2012) and Leukotoxin ED (lukE), responsible for neutrophil killing (Alonzo et al., 2012). The immune evasion protein adenosine synthase A (adsA), was the only gene identified as significantly downregulated at both 2 and 3 h of infection.
Later in infection, 21 genes were significantly upregulated and 3 were downregulated (Figures 3B,C; see Supplementary Table 3). We verified that most of the upregulated genes are secreted factors as toxins and proteases while the downregulated genes are associated to adhesion to host tissue and immune evasion. For example, V8 protease (sspA), staphopain B (sspB), and aureolysin (aur), three major secreted proteolytic enzymes of S. aureus were all significantly upregulated at this stage. On the other hand, the genes that encode the immune evasion protein SpA and the adhesion protein ClfB were significantly downregulated at late infection, as well as the transcriptional regulator sarS, which represses the expression of spa (Cheung et al., 2001).
Remarkably, 12 virulence factors were significantly upregulated over the entire course of the infection (Figure 3B and Table 1). These 12 genes encode for four proteins that are part of the ESAT-6 secretion system (esxA, esxB, esxC, esaA, and essB), two immunodominant antigens (isaA and isaB), two proteins from the family of conserved staphylococcal antigens (csa1A and csa2), two adhesion proteins (ebpS and sasF) and the alkaline shock protein 23 (asp23).
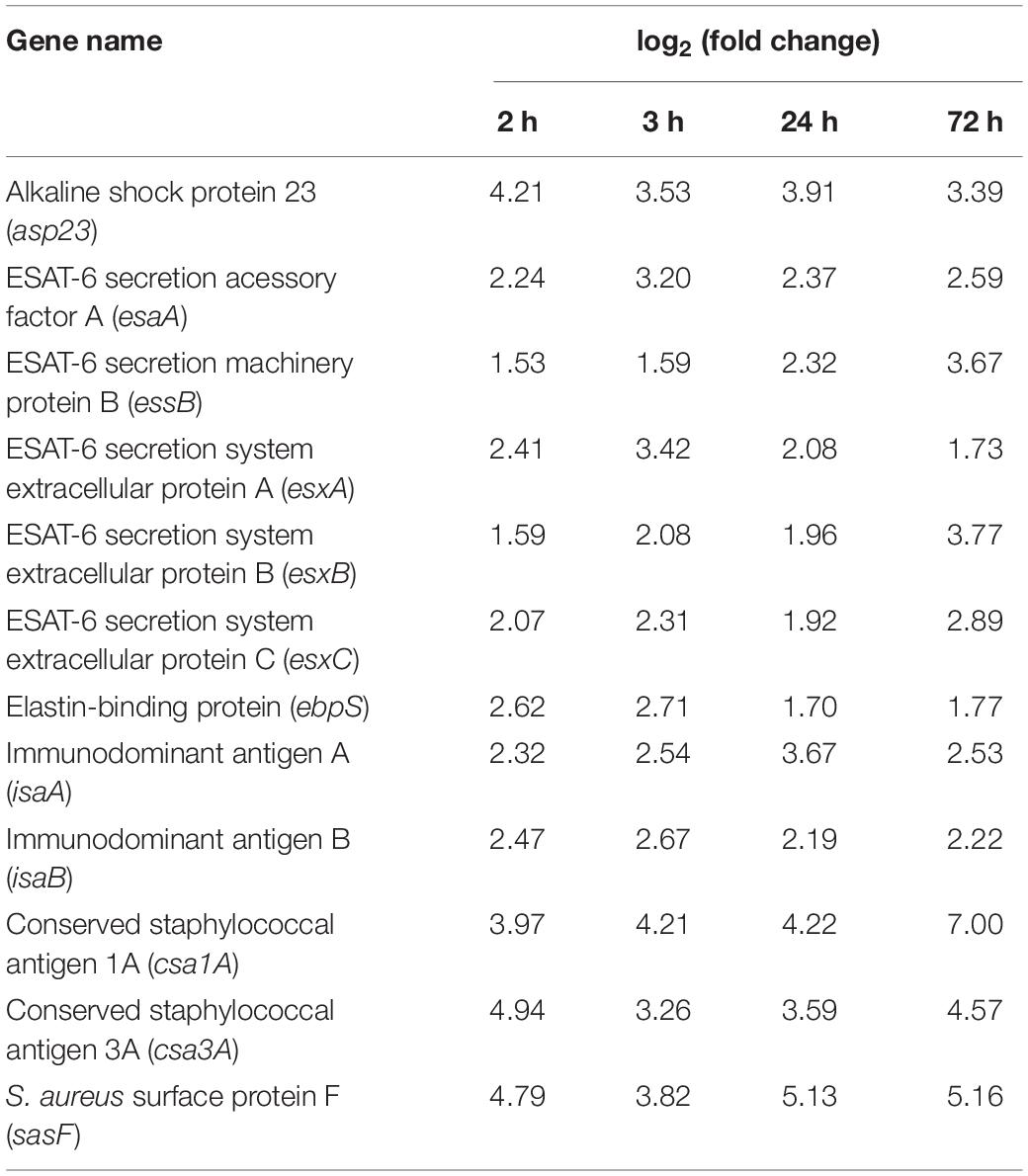
Table 1. Virulence gene expression profile of the 12 factors that were upregulated over the entire course of the infection, when compared with inoculated bacteria.
Of note, when comparing the relative gene expression profile of S. aureus within skin tissue with that of S. aureus grown in broth medium, a weak correlation was observed for most of the time points assessed (see Supplementary Figure 6). Taken together, we observed that several genes were differently regulated within the skin tissues when compared with the initial inoculum, which likely reflects their importance for colonization and infection of human skin.
S. aureus Accumulates in Sweat Glands and hla Promoter Is Activated Within This Niche
Having studied the overall virulence gene expression of bacteria within a human skin punch, we next verified whether S. aureus location in the skin tissues could influence virulence gene transcription. Following skin abrasion, skin punches were infected with a USA300 reporter fusion strain, where the promoter of alpha hemolysin gene was fused together with the mCherry gene, which enables direct visualization of differences in hla expression in situ. The infection was monitored by confocal microscopy after 2, 24, and 72 h (Figure 4). After only 2 h, S. aureus preferentially accumulated on the skin surface and inside sweat glands and ducts (Figure 4A). Remarkably, after 2 h of infection, hla promoter was activated inside the sweat glands and ducts but not on the skin surface, where the infection begun (Figure 4A). mCherry signal was also detected inside these skin appendices after 24 and 72 h of infection, but not at the skin surface (Figures 4B,C). Altogether, these data show that S. aureus accumulates inside sweat glands and ducts and suggests that S. aureus niches within the skin tissues influences its transcription profile.
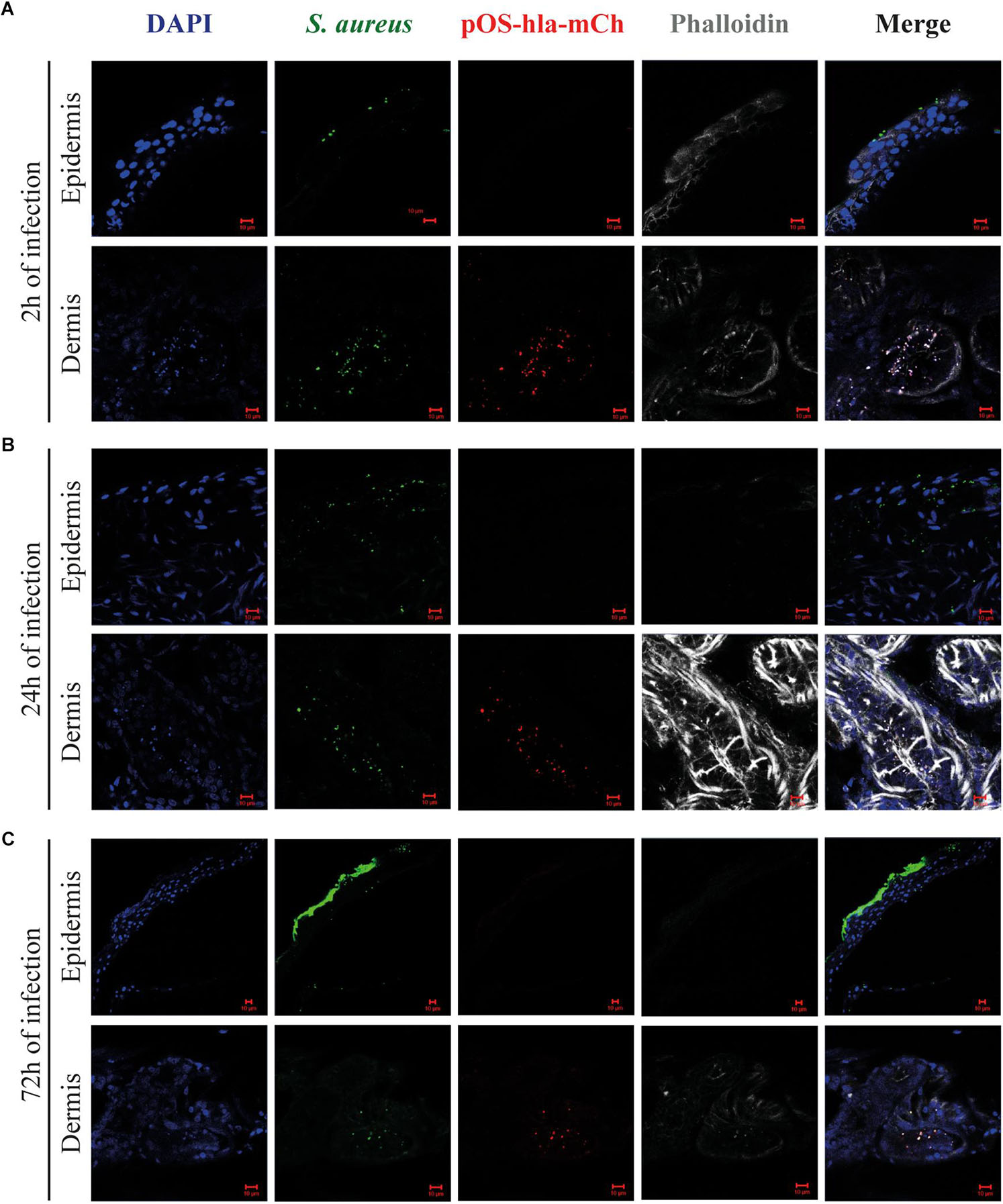
Figure 4. Alpha hemolysin reporter system activation within human skin tissues. Representative images of cryosections of skin infected with S. aureus (pOS-hla-mCherry, red), stained with anti-Staphylococcus aureus antibody (green), phalloidin (gray) and DAPI (blue) after 2 (A), 24 (B) and 72 h of infection (C). mages were taken at the epidermis (upper images) and the dermis (lower images) (scale bar, 10 μm).
Discussion
Staphylococcus aureus is the predominant cause of SSTIs (McCaig et al., 2006). Its success as a pathogen is largely dependent on its ability to adapt to different environments by modulating the expression of cell surface-associated and secreted factors that promote adherence, immune evasion and invasion of host tissues. To better understand staphylococcal pathogenesis in human tissues, it is important to determine which bacterial virulence factors are expressed in the context of the infection. Therefore, in this study we established a S. aureus cutaneous infection model using human skin explants and examined the gene expression profile of a vast array of S. aureus virulence factors at different stages of cutaneous infection.
We identified four main clusters of genes that respond similarly to environmental and host-specific signals during infection. In general, cluster 1 and 2 include the virulence factors that are more expressed by the bacteria early in infection. These clusters mostly comprise surface proteins that might be essential for human skin colonization. On the other hand, cluster 3 and 4 comprise the genes whose transcripts are increased later in infection and hence are likely important for skin invasion and long term persistence of infection. These clusters include genes that are involved in gene expression regulation and many genes that encode immune evasion proteins, toxins and secreted proteases. The fact that most adhesins are expressed in the beginning of infection and virulence-related genes are more expressed later in infection suggests that agr quorum sensing plays an important role in transcription regulation in human tissues, which is in accordance with a previous publication where the gene expression of a small panel of S. aureus virulence factors was analyzed in samples collected from human cutaneous abscesses (Loughman et al., 2009). Nevertheless, we detected only a weak correlation between the relative gene expression pattern of USA300 grown in vitro vs. in skin explants for most time points analyzed, which underlines the importance of environmental and host-specific cues for S. aureus gene expression regulation. An example is the upregulation of fhuD2 early in skin infection. This upregulation is likely a consequence of the iron-limiting conditions that S. aureus faces upon contact with human tissues, as the concentration of free iron in vivo is very limited when compared to laboratory media (Ratledge and Dover, 2000; Hazmanian et al., 2003).
In this study, we also pinpointed 12 genes that were upregulated over the entire course of the infection when comparing with inoculated bacteria. EsxA, esxB, esxC, esaA, and essB are part the Ess pathway that contributes for S. aureus virulence (Burts et al., 2005) and establishment of persistent infection (Burts et al., 2008; Anderson et al., 2011). The overexpression of Ess genes upon skin contact may be explained by bacterial interaction with host-specific fatty acids, which were shown to activate Ess operon (Lopez et al., 2017). The transcript levels of the immunodominant antigens isaA and isaB were also enhanced during skin infection. High titers of IgG against both proteins were detected in sera from sepsis patients, indicating that S. aureus expresses IsaA and IsaB during human infection (Lorenz et al., 2000). The genes that encode SasF and EbpS were also upregulated over infection. SasF is described to protect the bacteria from bactericidal effects of long-chain unsaturated fatty acids (Kenny et al., 2009) and was also shown to be involved in S. aureus virulence in mice skin abscess infections (Kwiecinski et al., 2014), therefore, its upregulation might occur as a S. aureus defense mechanism. Although the role of EbpS has not been implicated in S. aureus skin infections, by binding to elastin, EbpS may promote colonization of skin tissues that are rich in elastin (Downer et al., 2002). The gene that encodes the Asp23 was also highly expressed in the skin tissues. Asp23 is involved in cell homeostasis (Müller et al., 2014) and was identified as a CD4 T Cell antigen (Lawrence et al., 2012) but, as EbpS, it was not associated before to skin infection. Also csa1A and csa3A genes, which are members of the conserved staphylococcal antigens family that was recently identified to be involved in biofilm formation (Kavanaugh et al., 2019), were upregulated over infection. Although the effect of Csa1A and Csa3A were not studied alone in skin tissues, a four-component S. aureus vaccine (Hla, EsxA-EsxB, FhuD2, and Csa1A) showed to reduce abscess formation, CFU counts and dermonecrosis (Bagnoli et al., 2015).
Further studies are needed to clarify which stimulus result in a specific alteration in the gene expression of the bacteria and to pinpoint the factors that are essential for colonization and infection. The use of mutant strains lacking the expression of a global regulator to infect the ex vivo model presented here may aid the understanding of gene expression regulation in human tissues. In addition, it would be of interest to compare the gene expression pattern of USA300 LAC strain in human skin with other S. aureus strains. Importantly, although human skin explants contain all cells types found in skin tissues, including immune cells (Nestle et al., 2009), they lack the presence of neutrophils. Neutrophils play an important role during S. aureus skin infections, as these cells are recruited from blood to the site of infection for bacterial clearance (Miller and Cho, 2011; Kobayashi et al., 2015). Therefore, the explant model used here may not perfectly mimic the micro-environment encountered by the bacteria in human skin. Future developments in the field of microfluidic skin-on-a-chip devices may enable culture of the human skin tissues in presence of a blood flow and help to further improve the skin model used in this study.
We also observed that S. aureus accumulates inside sweat glands and ducts. The visualization of staphylococci biofilm occluding sweat glands was previously reported in atopic dermatitis skin samples (Allen et al., 2014) and also in miliaria (retention of eccrine sweat caused by blockage of sweat ducts) (Mowad et al., 1995). Additionally, although the relationship between hidradenitis suppurativa (chronic inflammatory skin disease) and bacterial infections is still controversial, S. aureus and coagulase-negative staphylococci have been reported among the most prevalent isolates detected (Ring et al., 2015; Ring and Emtestam, 2016). Therefore, skin appendices might represent a preferential niche for colonization/infection. Eccrine sweat glands are known for their function in thermoregulation but also for being part of innate immune defense of human skin against infection, due to production of the antimicrobial peptide dermcidin (DCD). DCD is specifically and constitutively expressed in eccrine sweat glands and secreted in the sweat (Schittek et al., 2001). Interestingly, DCD was shown to upregulate Staphylococcus epidermidis agr system and downregulate SarA and SaeRS regulators (Lai et al., 2007), inducing protease expression. Therefore, although the effect of DCD on S. aureus agr system was not assessed, exposure to DCD in a sweat glands and ducts might affect hla expression. Besides, the environment cues characteristic of the sweat gland, as its acidic pH (4–6.8) and high salt concentrations, can influence S. aureus virulence regulation.
Overall, our work shows for the first time the kinetics of S. aureus virulence gene expression within human skin and also suggests that sweat glands and ducts are a preferential niche for S. aureus colonization, where the virulence-related gene hla is more expressed. Increasing knowledge about gene expression profile within the target tissue is key for the design of new therapeutics to control infections caused by S. aureus.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the French Ministry of Higher Education, Research and Innovation. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AC, JS, FB, and AM were involved in designing or conceiving the study. AC performed experiments and analyzed the data. AC and AM wrote the manuscript. JS and FB acquired the funding. FB and AM supervised the project. All authors critically revised the manuscript and approved it before submission.
Funding
This work was co-sponsored by the University of Utrecht and GSK Vaccine Srl, and received funding by the European Union’s Horizon 2020 Research Programs H2020-MSCA-ITN (#675106, to JS and FB). The research was performed in the preclinical research labs of GSK Vaccine Srl.
Conflict of Interest
AC participated in a postgraduate studentship program at GSK. FB and AM are employees of GSK group of companies and FB holds shares in the GSK group of companies. FB and AM were named inventors of pending and issued patents on S. aureus vaccine formulations.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank to Tarcisio Brignoli for his help with the high-throughput RT-PCR experiments, Silvia Maccari for her help with confocal microscopy and histology staining, Alfredo Pezzicoli for his support with two-photon microscopy and Elisabetta Soldaini, Stefano Censini and Manuel Amieva for the fruitful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.692023/full#supplementary-material
References
Allen, H. B., Vaze, N. D., Choi, C., Hailu, T., Tulbert, B. H., Cusack, C. A., et al. (2014). The presence and impact of biofilm-producing staphylococci in atopic dermatitis. JAMA Dermatol. 150, 260–265. doi: 10.1001/jamadermatol.2013.8627
Alonzo, F., Benson, M. A., Chen, J., Novick, R. P., Shopsin, B., and Torres, V. J. (2012). Staphylococcus aureus leucocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo. Mol. Microbiol. 83, 423–435. doi: 10.1111/j.1365-2958.2011.07942.x
Anderson, M., Chen, Y. H., Butler, E. K., and Missiakas, D. M. (2011). EsaD, a secretion factor for the Ess pathway in Staphylococcus aureus. J. Bacteriol. 193, 1583–1589. doi: 10.1128/JB.01096-10
Bagnoli, F., Bertholet, S., and Grandi, G. (2012). Inferring reasons for the failure of Staphylococcus aureus vaccines in clinical trials. Front. Cell. Infect. Microbiol. 2:16. doi: 10.3389/fcimb.2012.00016
Bagnoli, F., Fontana, M. R., Soldaini, E., Mishra, R. P. N., Fiaschi, L., Cartocci, E., et al. (2015). Vaccine composition formulated with a novel TLR7-dependent adjuvant induces high and broad protection against Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 112, 3680–3685. doi: 10.1073/pnas.1424924112
Balasubramanian, D., Harper, L., Shopsin, B., and Torres, V. J. (2017). Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis. 75:ftx005. doi: 10.1093/femspd/ftx005
Brignoli, T., Manetti, A. G. O., Rosini, R., Haag, A. F., Scarlato, V., Bagnoli, F., et al. (2019). Absence of protein a expression is associated with higher capsule production in staphylococcal isolates. Front. Microbiol. 10:863. doi: 10.3389/fmicb.2019.00863
Burts, M. L., DeDent, A. C., and Missiakas, D. M. (2008). EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol. Microbiol. 69, 736–746. doi: 10.1111/j.1365-2958.2008.06324.x
Burts, M. L., Williams, W. A., DeBord, K., and Missiakas, D. M. (2005). EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. U.S.A. 102, 1169–1174. doi: 10.1073/pnas.0405620102
Cheung, A. L., Bayer, A. S., Zhang, G., Gresham, H., and Xiong, Y. Q. (2004). Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40, 1–9. doi: 10.1016/S0928-8244(03)00309-2
Cheung, A. L., Schmidt, K., Bateman, B., and Manna, A. C. (2001). SarS, a SarA homolog repressible by agr, is an activator of protein a synthesis in Staphylococcus aureus. Infect. Immun. 69, 2448–2455. doi: 10.1128/IAI.69.4.2448-2455.2001
Date, S. V., Modrusan, Z., Lawrence, M., Morisaki, J. H., Toy, K., Shah, I. M., et al. (2014). Global gene expression of methicillin-resistant staphylococcus aureus USA300 during human and mouse infection. J. Infect. Dis. 209, 1542–1550. doi: 10.1093/infdis/jit668
Downer, R., Roche, F., Park, P. W., Mecham, R. P., and Foster, T. J. (2002). The elastin-binding protein of Staphylococcus aureus (EbpS) is expressed at the cell surface as an integral membrane protein and not as a cell wall-associated protein. J. Biol. Chem. 277, 243–250. doi: 10.1074/jbc.M107621200
Foster, T. J. (2005). Immune evasion by staphylococci. Nat. Rev. Microbiol. 3, 948–958. doi: 10.1038/nrmicro1289
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., and Höök, M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. doi: 10.1038/nrmicro3161
Haag, A. F., and Bagnoli, F. (2017). The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Curr. Top. Microbiol. Immunol. 409, 145–198. doi: 10.1007/82_2015_5019
Hazmanian, S. K., Skaar, E. P., Gaspar, A. H., Humayun, M., Gornicki, P., Jelenska, J., et al. (2003). Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299, 906–909. doi: 10.1126/science.1081147
Jenul, C., and Horswill, A. R. (2018). Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 6, 1–21. doi: 10.1128/microbiolspec.gpp3-0031-2018
Kavanaugh, J. S., Flack, C. E., Lister, J., Ricker, E. B., Ibberson, C. B., Jenul, C., et al. (2019). Identification of extracellular DNA-binding proteins in the biofilm matrix. MBio 10:e01137-19. doi: 10.1128/mBio.01137-19
Kenny, J. G., Ward, D., Josefsson, E., Jonsson, I. M., Hinds, J., Rees, H. H., et al. (2009). The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications. PLoS One 4:e4344. doi: 10.1371/journal.pone.0004344
King, M. D., Humphrey, B. J., Wang, Y. F., Kourbatova, E. V., and Ray, S. M. (2006). Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann. Intern. Med. 144, 309–318. doi: 10.7326/0003-4819-144-5-200603070-00005
Kluytmans, J., Van Belkum, A., and Verbrugh, H. (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10, 505–520. doi: 10.1128/cmr.10.3.505
Kobayashi, S. D., Malachowa, N., and Deleo, F. R. (2015). Pathogenesis of Staphylococcus aureus abscesses. Am. J. Pathol. 185, 1518–1527. doi: 10.1016/j.ajpath.2014.11.030
Kwiecinski, J., Jin, T., and Josefsson, E. (2014). Surface proteins of Staphylococcus aureus play an important role in experimental skin infection. APMIS 122, 1240–1250. doi: 10.1111/apm.12295
Lai, Y., Villaruz, A. E., Li, M., Cha, D. J., Sturdevant, D. E., and Otto, M. (2007). The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol. Microbiol. 63, 497–506. doi: 10.1111/j.1365-2958.2006.05540.x
Lawrence, P. K., Rokbi, B., Arnaud-Barbe, N., Sutten, E. L., Norimine, J., Lahmers, K. K., et al. (2012). CD4 T cell antigens from Staphylococcus aureus newman strain identified following immunization with heat-killed bacteria. Clin. Vaccine Immunol. 19, 477–489. doi: 10.1128/CVI.05642-11
Le, K. Y., and Otto, M. (2015). Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol. 6:1174. doi: 10.3389/fmicb.2015.01174
Lopez, M. S., Tan, I. S., Yan, D., Kang, J., McCreary, M., Modrusan, Z., et al. (2017). Host-derived fatty acids activate type VII secretion in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 114, 11223–11228. doi: 10.1073/pnas.1700627114
Lorenz, U., Ohlsen, K., Karch, H., Hecker, M., Thiede, A., and Hacker, J. (2000). Human antibody response during sepsis against targets expressed by methicillin resistant Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 29, 145–153. doi: 10.1016/S0928-8244(00)00199-1
Loughman, J. A., Fritz, S. A., Storch, G. A., and Hunstad, D. A. (2009). Virulence gene expression in human community-acquired Staphylococcus aureus infection. J. Infect. Dis. 199, 294–301. doi: 10.1086/595982
Lowy, F. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
McCaig, L. F., McDonald, L. C., Mandal, S., and Jernigan, D. B. (2006). Staphylococcus aureus-associated skin and soft tissue infections in ambulatory care. Emerg. Infect. Dis. 12, 1715–1723. doi: 10.3201/eid1211.060190
Miller, L. S., and Cho, J. S. (2011). Immunity against Staphylococcus aureus cutaneous infections. Nat. Rev. Immunol. 11, 505–518. doi: 10.1038/nri3010
Mishra, R. P. N., Mariotti, P., Fiaschi, L., Nosari, S., MacCari, S., Liberatori, S., et al. (2012). Staphylococcus aureus FhuD2 is involved in the early phase of staphylococcal dissemination and generates protective immunity in mice. J. Infect. Dis. 206, 1041–1049. doi: 10.1093/infdis/jis463
Moran, G. J., Krishnadasan, A., Gorwitz, R. J., Fosheim, G. E., Mcdougal, L. K., Carey, R. B., et al. (2006). Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 335, 666–674. doi: 10.1056/NEJMoa055356
Mowad, C. M., McGinley, K. J., Foglia, A., and Leyden, J. J. (1995). The role of extracellular polysaccharide substance produced by Staphylococcus epidermidis in miliaria. J. Am. Acad. Dermatol. 33, 729–733. doi: 10.1016/0190-9622(95)91809-4
Müller, M., Reiß, S., Schlüter, R., Mäder, U., Beyer, A., Reiß, W., et al. (2014). Deletion of membrane-associated Asp23 leads to upregulation of cell wall stress genes in Staphylococcus aureus. Mol. Microbiol. 93, 1259–1268. doi: 10.1111/mmi.12733
Nestle, F. O., Di Meglio, P., Qin, J. Z., and Nickoloff, B. J. (2009). Skin immune sentinels in health and disease. Nat. Rev. Immunol. 9, 679–691. doi: 10.1038/nri2622
Olaniyi, R. O., Pancotto, L., Grimaldi, L., and Bagnoli, F. (2018). Deciphering the pathological role of staphylococcal α-Toxin and panton-valentine leukocidin using a novel ex vivo human skin model. Front. Immunol. 9:951. doi: 10.3389/fimmu.2018.00951
Olaniyi, R., Pozzi, C., Luca, G., and Bagnoli, F. (2016). Staphylococcus aureus-associated skin and soft tissue infections: anatomical localization, epidemiology, therapy and potential prophylaxis. Curr. Top. Microbiol. Immunol. 409, 199–227. doi: 10.1007/82_2016_32
Pasparakis, M., Haase, I., and Nestle, F. O. (2014). Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 14, 289–301. doi: 10.1038/nri3646
Ratledge, C., and Dover, L. G. (2000). Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54, 881–941. doi: 10.1146/annurev.micro.54.1.881
Ring, H. C., and Emtestam, L. (2016). The microbiology of hidradenitis suppurativa. Dermatol. Clin. 34, 29–35. doi: 10.1016/j.det.2015.08.010
Ring, H. C., Riis Mikkelsen, P., Miller, I. M., Jenssen, H., Fuursted, K., Saunte, D. M., et al. (2015). The bacteriology of hidradenitis suppurativa: a systematic review. Exp. Dermatol. 24, 727–731. doi: 10.1111/exd.12793
Rothfork, J. M., Dessus-Babus, S., Van Wamel, W. J. B., Cheung, A. L., and Gresham, H. D. (2003). Fibrinogen depletion attenuates staphyloccocus aureus infection by preventing density-dependent virulence gene up-regulation. J. Immunol. 171, 5389–5395. doi: 10.4049/jimmunol.171.10.5389
Salgado-Pabón, W., and Schlievert, P. M. (2014). Models matter: the search for an effective Staphylococcus aureus vaccine. Nat. Rev. Microbiol. 12, 585–591. doi: 10.1038/nrmicro3308
Schittek, B., Hipfel, R., Sauer, B., Bauer, J., Kalbacher, H., Stevanovic, S., et al. (2001). Dermcidin: a novel human antibiotic peptide secreted by sweat glands. Nat. Immunol. 2, 1133–1137. doi: 10.1038/ni732
Talan, D. A., Krishnadasan, A., Gorwitz, R. J., Fosheim, G. E., Limbago, B., Albrecht, V., et al. (2011). Comparison of Staphylococcus aureus from skin and soft-tissue infections in us emergency department patients, 2004 and 2008. Clin. Infect. Dis. 53, 144–149. doi: 10.1093/cid/cir308
Tam, K., and Torres, V. J. (2019). Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol. Spectr. 7, 640–668. doi: 10.1128/9781683670131.ch40
Keywords: Staphylococcus aureus, human skin, virulence factors, gene expression, pathogenicity, niche, sweat glands
Citation: Cruz AR, van Strijp JAG, Bagnoli F and Manetti AGO (2021) Virulence Gene Expression of Staphylococcus aureus in Human Skin. Front. Microbiol. 12:692023. doi: 10.3389/fmicb.2021.692023
Received: 07 April 2021; Accepted: 19 May 2021;
Published: 11 June 2021.
Edited by:
Lígia M. Saraiva, Universidade Nova de Lisboa, PortugalReviewed by:
Tao Jin, Sahlgrenska University Hospital, SwedenHanne Ingmer, University of Copenhagen, Denmark
Copyright © 2021 Cruz, van Strijp, Bagnoli and Manetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea G. O. Manetti, andrea.g.manetti@gsk.com
 Ana Rita Cruz
Ana Rita Cruz Jos A. G. van Strijp
Jos A. G. van Strijp Fabio Bagnoli
Fabio Bagnoli Andrea G. O. Manetti
Andrea G. O. Manetti