Potential Links between Hepadnavirus and Bornavirus Sequences in the Host Genome and Cancer
- 1IRCCS MultiMedica, Milan, Italy
- 2Medical Genetics, University of Siena, Siena, Italy
- 3Genetica Medica, Azienda Ospedaliera Universitaria Senese, Siena, Italy
A commentary on
Potential Links between Hepadnavirus and Bornavirus Sequences in the Host Genome and Cancer
by Honda, T. (2017). Front. Microbiol. 8:2537. doi: 10.3389/fmicb.2017.02537
In the paper by Honda et al. (Honda, 2017), sequences of non-retroviral RNA viruses seem to have been integrated into the host genome possibly by machineries of host retrotransposon, long interspersed nuclear element 1 (LINE-1, or L1). The integration events of viral sequences occur not only in somatic cells, but also in germ line cells and then can be inherited as EVEs (endogenous viral elements). Honda reports various possibilities for EVEs to have oncogenic potential and focuses both on HBV viruses for the occurrence of HCC and on endogenous bornavirus-like elements (EBLs).
Because EBLs are the only non-retroviral RNA virus-derived EVEs found in the human genome thus far, Honda focuses on the possible link between these elements and cancer, although the links have not been demonstrated.
Among the 7 Homo sapiens EBLNs (hsEBLN), hsEBLN-2 is most closely linked to cancer. Gene ontology analysis concerning EBLN-2 suggests that it might be a tumor suppressor gene (TSG).
Interestingly, because mitochondrial dysfunction is found in cancer, hsHBLN-2 might play important roles in mitochondrial function and then act as a TSG. Honda correctly states that “Epidemiologic studies on the links between EBL mutations and cancers are clearly required. Furthermore, the causal relationship between such EBL mutations and cancers should be demonstrated in the future.” In particular, Honda quotes extensively one of our papers: “Whole exome sequencing (WES) using two sibling pairs of non-smokers with lung adenocarcinoma reveals that a truncated mutation in hsEBLN-2 is only detected in affected siblings (Renieri et al., 2014). The authors concluded that this mutation in hsEBLN-2 might predispose an individual to lung adenocarcinoma (ADCA)” (Honda, 2017). We indeed identified a frameshift mutation, the c.250dupA, p.N83fs in EBLN2 gene (NM_018029) (Renieri et al., 2014). Interestingly, according to Honda, our report is the only study reporting a germline mutation of hsEBLN-2 associated with tumor occurrence in the clinical setting.
We would like to make some comments, namely concerning our original approach to oligogenic signatures (i.e., the germline mutation of 5–8 genes) in the occurrence of common sporadic tumors.
We have used WES, associated with a discordant sibling strategy, to show that siblings sharing 50% of the genome, could differ for 5–8 germ line mutations, likely facilitating cancer occurrence in one sibling and its absence in the other. This is in line with the modern trend toward “personalized medicine,” i.e., each subject finds his/her personal way to cancer and every individual cancer is different from the other.
Therefore, we suggest caution before extrapolating that EBNL-2, a suspected tumor suppressor gene, could play a role such as APC germ line mutations in the occurrence of familial adenomatous polyposis (FAP) (Cetta et al., 2001)—or of other TSGs as causal responsible for inherited multitumoral syndromes—, in the occurrence of lung ADCA. EBLN-2 protein may be involved in regulation of tumor-related genes, in mitochondrial function and microtubule regulation (Figure 1A). Honda suggests that this mechanism could act as a pathogenic factor for the formation of a given tumor, namely lung adenocarcinoma, maybe according to “conventional” pathogenic pathways. In our study, on the contrary germ line mutation of the EBLN gene has been found as a part of an “oligogenic signature” responsible for tumor occurrence in a discordant sibling model, that aims at detecting the congenital basis for the emergence of a “personalized” tumoral variant, according to current knowledge concerning personalized medicine (Figure 1B).
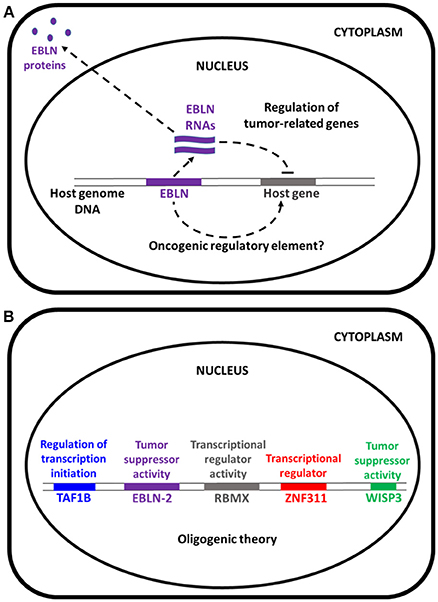
Figure 1. (A) Modified from Honda (2017). The hsEBLN-2 protein may be involved in mitochondrial function and might be a tumor suppressor. (B) EBLN-2 is involved in cancers together with others genes. For example in our study about lung adenocarcinoma susceptibility we found the involvement of TAF1b (component of RNA polymerase I core factor complex that acts as a GTF2B/TFIIB-like factor and plays a key role in multiple steps during transcription initiation), RBMX (RNA-binding protein that plays several roles in the regulation of pre- and post-transcriptional processes), ZNF311 (may be involved in transcriptional regulation), WISP3 (appears to be required for normal postnatal skeletal growth and cartilage homeostasis, and WISP3 regulates accumulation of cellular reactive oxygen species) as a proof-of-concept of the oligogenic theory (Renieri et al., 2014).
Our study is based on a philosophy that is completely different from the “gene hunting” approach of 30 years ago, aimed at detecting a single gene responsible for a single cancer. Namely, we have already found a peculiar oligogenic signature, responsible for lung ADCA in six additional discordant siblings (Cetta et al., 2017). Interestingly, a total of more than 50 different genes have been detected as “facilitating germ line mutations” in oncogenes or TSG in the entire series of eight patients with lung ADCA. They are responsible for the formation of a “personal” cancer, even if they could not include the single main determinant of tumor occurrence. Similar findings have also been detected in a non-smoker with lung squamous cell carcinoma (Baldassarri et al., 2018).
We hope that this comment could help to clarify better the significance of our findings. EBNL2 is not a gene specifically causing the formation of lung adenocarcinoma, but a germline mutation that may facilitate its occurrence as a component of a tumor predisposing oligogenic signature, at individual level. While waiting for more stringent causal relationships between EBLN-2 and various types of cancers, we think that the present clarification could prevent misleading interpretations.
Author Contributions
FC, MP, and EF wrote the manuscript. MP performed Figure 1. EF and AR made substantial contributions to the manuscript idea. All authors have given final approval of the version to be published and agree to be accountable for all aspects of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The funding was provided by Regione Toscana—Istituto Toscano Tumori (ITT) (Project “Identification of genetic bases of individual predisposition to lung cancer in non-smokers”).
References
Baldassarri, M., Fallerini, C., Cetta, F., Ghisalberti, M., Bellan, C., Furini, S., et al. (2018). Omic approach in non-smoker female with lung squamous cell carcinoma pinpoints to germline susceptibility and personalized medicine. Cancer Res. Treat 50, 356–365. doi: 10.4143/crt.2017
Cetta, F., Montalto, G., Gori, M., Curia, M. C., Cama, A., and Olschwang, S. (2001). Germline mutations of the APC gene in patients with familial adenomatous polyposis-associated thyroid carcinoma: results from a European cooperative study. J. Clin. Endocrinol. Metab. 85, 286–292. doi: 10.1210/jcem.85.1.6254
Cetta, F., Renieri, A., and Frullanti, E. (2017). Germline mutations in lung cancer and personalized medicine. Fam. Cancer 17, 429–430. doi: 10.1007/s10689-017-0044-4
Honda, T. (2017). Potential links between hepadnavirus and bornavirus sequences in the host genome and cancer. Front. Microbiol 19:2537. doi: 10.3389/fmicb.2017.02537
Keywords: cancer susceptibility, lung cancer, oligogenic inheritance, whole exome sequencing, cancer genetics
Citation: Cetta F, Palmieri M, Renieri A and Frullanti E (2018) Commentary: Potential Links between Hepadnavirus and Bornavirus Sequences in the Host Genome and Cancer. Front. Microbiol. 9:1649. doi: 10.3389/fmicb.2018.01649
Received: 14 May 2018; Accepted: 02 July 2018;
Published: 23 July 2018.
Edited by:
Gkikas Magiorkinis, National and Kapodistrian University of Athens, GreeceReviewed by:
Masayuki Horie, Kyoto University, JapanMartin Sebastian Staege, Martin Luther University of Halle-Wittenberg, Germany
Copyright © 2018 Cetta, Palmieri, Renieri and Frullanti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elisa Frullanti, elisa.frullanti@dbm.unisi.it
 Francesco Cetta1
Francesco Cetta1 Maria Palmieri
Maria Palmieri Elisa Frullanti
Elisa Frullanti