HDS screening with patient-derived primary cells guided individualized therapy for esophageal squamous cell carcinoma–in vivo and vitro
- 1Department of Gastroenterology, The 901th Hospital of Joint Logistics Support Force, Hefei, Anhui, China
- 2Anhui Province Key Laboratory of Medical Physics and Technology, Institute of Health and Medical Technology, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei, Anhui, China
Objective: To analyze and evaluate the role of the High-throughput Drug Sensitivity (HDS) screening strategy in identifying highly sensitive drugs against esophageal squamous cell carcinoma (ESCC).
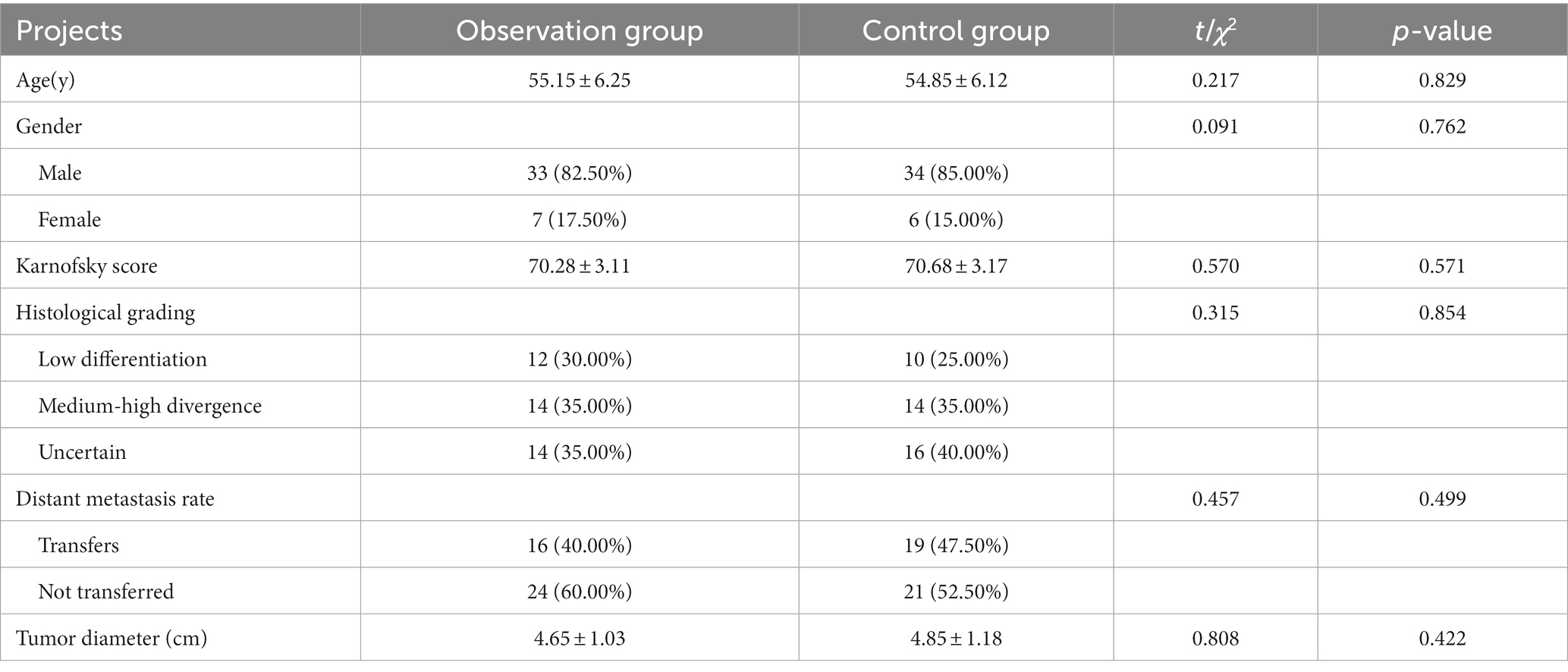
Methods: A total of 80 patients with progressive ESCC were randomly divided into the observation (40 cases) and the control groups (40 cases). In the observation group, primary ESCC cells were isolated from the tumor tissues with a gastroscope, and drug sensitivity screening was performed on cells derived from the 40 ESCC cases using the HDS method, followed by verification in a patient-derived tumor xenograft (PDX) mouse model. Finally, the differences in the therapeutic efficacy (levels of CEA, CYFRA21-1, SCCA after chemotherapy and the rates of overall survival, local progression, and distant metastasis at 12 months and 18 months time points after chemotherapy) were compared between the observation group (Screened drug-treated) and the control group (Paclitaxel combined with cisplatin regimen-treated).
Results: Forty ESCC patients were screened for nine different high-sensitive chemotherapeutics, with the majority showing sensitivity to Bortezomib. Experiments on animal models revealed that the tumor tissue mass of PDX mice treated with the HDS-screened drug was significantly lower than that of the Paclitaxel-treated mice (p < 0.05), and the therapeutic efficacy of the observation group was better than the control group (p < 0.05).
Conclusion: HDS screening technology can be beneficial in screening high-efficacy anticancer drugs for advanced-stage ESCC patients, thereby minimizing adverse drug toxicity in critically ill patients. Moreover, this study provides a new avenue for treating advanced ESCC patients with improved outcomes.
Introduction
Esophageal carcinoma (EC) is a highly prevalent and dangerous malignancy of the gastrointestinal (GI) tract (1, 2), accounting for the seventh most common cancer worldwide as well as ranking sixth for cancer-related deaths (3). EC claims approximately 450,000 lives annually (4), with a 5-year survival rate of only 18% (5). Notably, the incidence of EC is increasing rapidly in several countries, including China, in recent times (6, 7). In China, EC causes about 150,000 annual deaths, which equals 21.8% of all cancer deaths, ranking fourth in the list of deadliest carcinomas. The etiology of EC is complex, many factors such as drinking, smoking, diet, genetics, microorganisms and gender have been reported to be associated with elevated risk of EC by previous studies (5–7). Therefore, it is technically challenging and difficult to reduce the occurrence of EC. EC patients primarily undergo endoscopic diagnosis for GI symptoms. Esophagectomy is the major surgical intervention in EC patients, while other treatment modalities involve chemo alone or a combination of chemo and radiotherapy, depending on the clinical characteristic of the cancerous lesion (8, 9). Most often, patients are diagnosed with an advanced stage EC, leaving no option for radical surgery. Importantly, postoperative recurrence and metastasis are frequently detected in surgical EC patients, hence, chemotherapy is considered a potentially effective second line of treatment (10). Interestingly, there is a wide variation in EC tumor classification in different geographical regions, such that >90% of EC cases in Western countries are adenocarcinomas, while esophageal squamous cell carcinoma (ESCC) is the most prevalent (>90% of all cases) among the Chinese populations (11). Therefore, this wide range of variabilities hinders the universal application of the National Comprehensive Cancer Network (NCCN) guidelines to Chinese ESCC patients, directly affecting their treatment outcomes and tumor recurrence. Moreover, establishing an in vitro screening strategy for determining patient-specific sensitivity of chemotherapeutics may resolve the discrepancies in the generalized application of NCCN guidelines.
High-throughput drug sensitivity (HDS) screening strategy is a new method with the advantages of high efficiency, wide coverage, and potential adaptation to personalized therapy, which are promising for determining the antitumor drug sensitivity in liver cancer, head and neck cancer, and lymphomas (12–14). However, the application of this technique in antitumor drug screening of ESCC is rarely reported. Therefore, this study aimed to generate ESCC patient-derived cell models for HDS analyses and develop a patient-derived xenograft (PDX) model of human ESCC in rodents for in vivo investigation. This study provides optimized assay platform able to offer drug screening outcomes in ESCC samples with varying clinical features and guides the precision therapy for ESCC patients (see Figure 1).
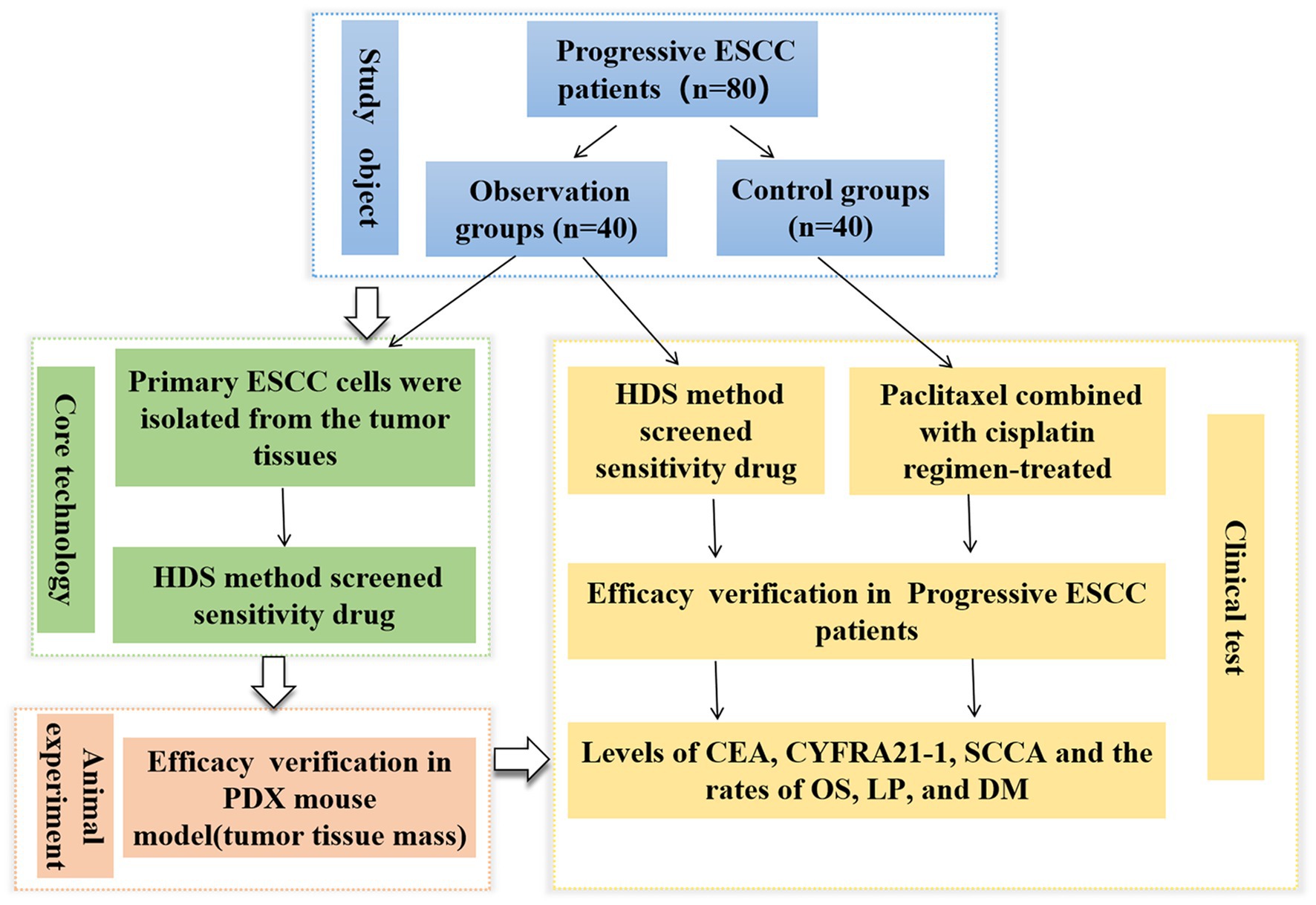
Figure 1. Flowchart showing the entire work procedure of this study including sections of study object, core technology, animal experiment and clinical test. ESCC, esophageal squamous cell carcinoma; PDX, patient-derived xenograft; HDS, High-throughput drug sensitivity.
Materials and methods
Patient selection
A total of 80 outpatient and inpatient candidates with progressive ESCC were recruited from the Department of Gastroenterology of the 901 Hospital of the Joint Logistics and Security Forces of the Chinese People’s Liberation Army from December 2018 to December 2019 and randomly assigned to either the observation group or the control group (n = 40 per group). There were no statistical differences in terms of age distribution, male-to-female ratio, tumor characteristics, and patient’s Karnofsky score (15) between the two groups (p > 0.05). Patients’ inclusion criteria were: (1) compliance with the diagnostic criteria of progressive ESCC (16); (2) availability of complete clinical data, and (3) no indication for surgery. On the other hand, patients were excluded if they had: (1) cardiovascular diseases; (2) penetrating ulcers; (3) hepatic and renal insufficiency; (4) coagulation disorder, and (5) severe immune system diseases. The study was approved by the Institutional Ethics Committee (Ethics Committee of the 901th Hospital of the Joint Support Force), and all participants provided their signed informed consent by themselves or their family members (Chinese clinical trial registration number: ChiCTR2300068566) (see Table 1).
Experimental animals
Severely immunodeficient (NCG) six-week-old female mice were purchased from Nanjing Model Animal Research Institute. The animal experiment protocol was approved by the Institutional Animal Care and Use Committee (Ethics Committee of the 901th Hospital of the Joint Support Force).
The main instruments and reagents that were used in this study are listed in Table 2.
Experimental methods
ESCC primary cell lines were derived from biopsied patient tumor tissue specimens and subsequently characterized as follows:
1. Gastroscopy and biopsy of ESCC tumors were performed using an Olympus CLV-260 electronic gastroscope. Hematoxylin and eosin (H&E) staining and immunohistochemical (IHC) analysis using anti-pCK, anti-p63, anti-Ki-67, and anti-Vim antibodies indicated that tumor tissues contained abundant ESCC cells (Figure 2A).
2. Isolation of ESCC primary cells was performed in the following steps: tissue washing, digestion, centrifugation, collection and resuspension of the cell pellet in the basal medium, centrifugation, removal of the supernatant and lysis of blood cells in appropriate buffer, and final collection of ESCC primary cells by centrifugation.
3. Culture of tumor-derived ESCC cells for (1) microscopic studies, (2) live cell counting, and (3) cell culture for downstream assays (Figure 3B).
4. Identification and characterization of cultured ESCC primary cells (Figures 3D,E).
5. The cells were washed with 200 μL of 0.25% trypsin for 1 min, aspirated, and then 500 μL of 0.05% trypsin solution was added to each well and placed in an incubator at 37°C with 5% CO2 for 10 min until complete cell detachment was observed under the microscope. After centrifugation at 1500 rpm for 4 min, the supernatant was discarded, and 500 μL of ESCC primary cell medium was added to resuspend and plate the cells (Figure 2B and Figure 3C).
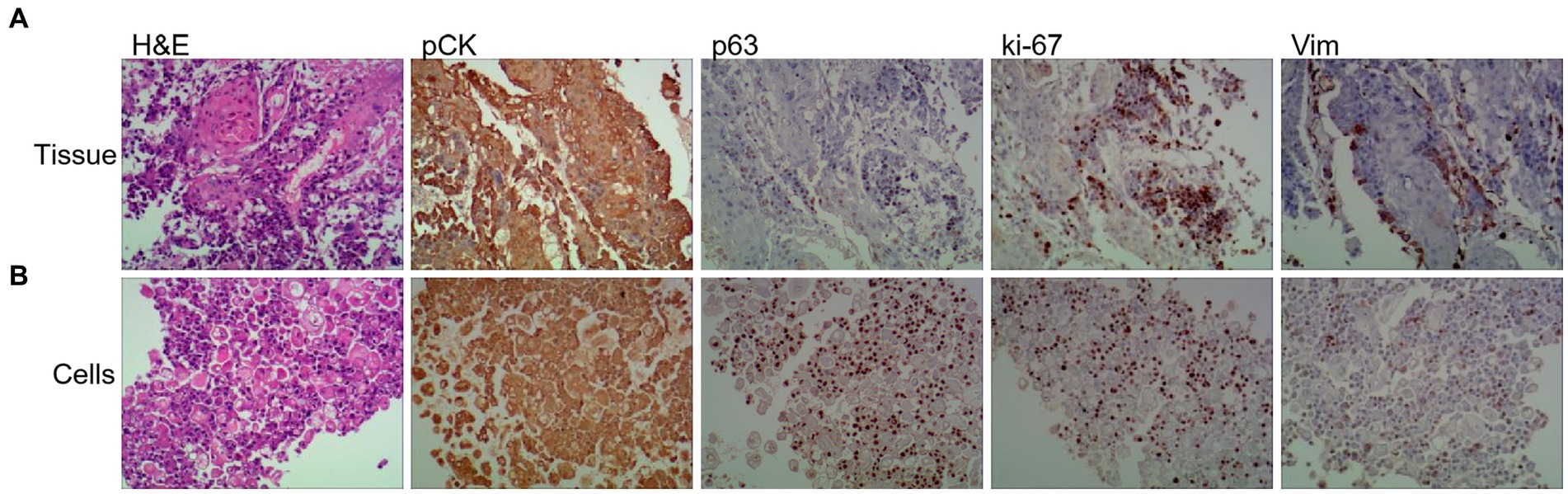
Figure 2. Tumor primary cell cultures share histopathological features with patient-matched tissue and disruption of polarity. Representative images of H&E, and IHC of Vim (Vimentin), panCK (pan-cytokeratin), ki-67 and p63 from primary tissue (A) and patient-matched primary cell (B) (20× magnifification used for primary cells and tissues).
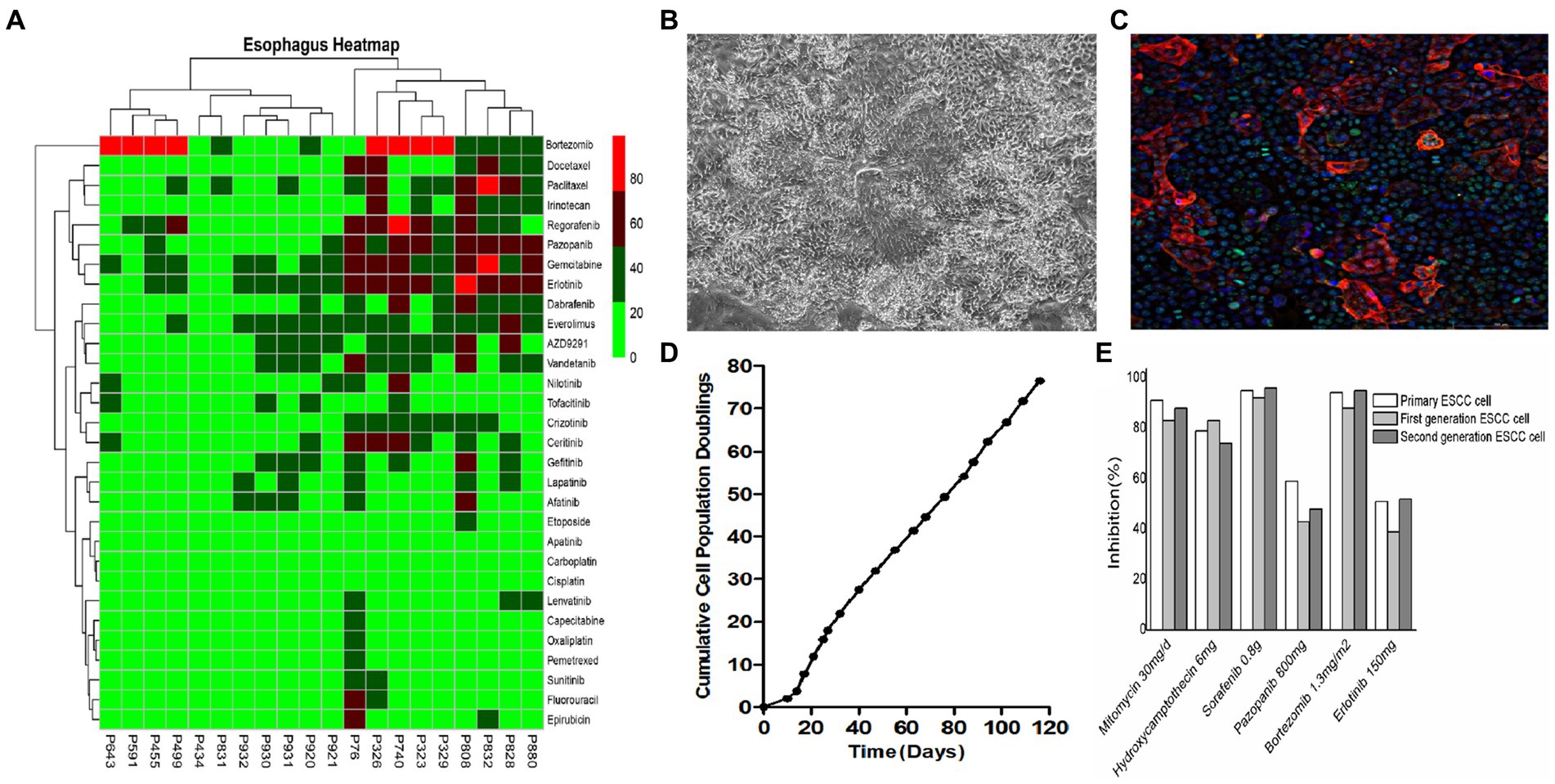
Figure 3. In vitro culture and drug sensitivity testing in ESCC cells. (A) Primary cells derived from different ESCC patients exhibited different sensitivities to different drugs. (B) Representative bright field images of primary ESCC cells in culture. (C) Primary ESCC cells characterized by immunofluorescence (IF) staining with squamous cell carcinoma marker anti-p63 antibody. (D) Cell proliferation curve of ESCC cells. (E) There was no significant difference in drug susceptibility between algebraic ESCC primary cells amplified in vitro.
HDS screening for drug sensitivity in patient-derived ESCC lines
The patient-specific primary ESCC cells were separately treated with different first-line and second-line drug regimens as well as subjected to the FDA drug library screening using the HDS method. Steps in this process include:
1. Preparation of cultured cells on petri dishes: ESCC primary cells were harvested by trypsin-digestion, centrifuged, and resuspended in DPBS buffer for cell counting by an image cytometer. The cell suspension was then diluted to 6 × 104–1 × 105 cells/ml, and seeded into 384-well plates using 50 μL of cell suspension per well.
2. Serial dilutions of drugs: the testing drug was serially diluted to different working concentrations (10 μM, 3 μM, 1 μM, 0.3 μM, 0.1 μM, 0.03 μM, and 0.01 μM).
3. Drug treatments: 24 h after seeding, cells were treated with different doses of serially diluted drug solutions or vehicle control in triplicates in 384-well plates using the automated JANUS liquid handler workstation.
4. Cell viability testing: 72 h post-treatment, the cell viability in different treatment and control groups was measured using a plate reader-based CellTiter Glo assay and calculated by the formula, Cell viability (%) = luminescence value of the spiked wells/luminescence value of the control wells × 100%. Cell inhibition (%) = 100- Cell viability (Figure 3A and Table 3).
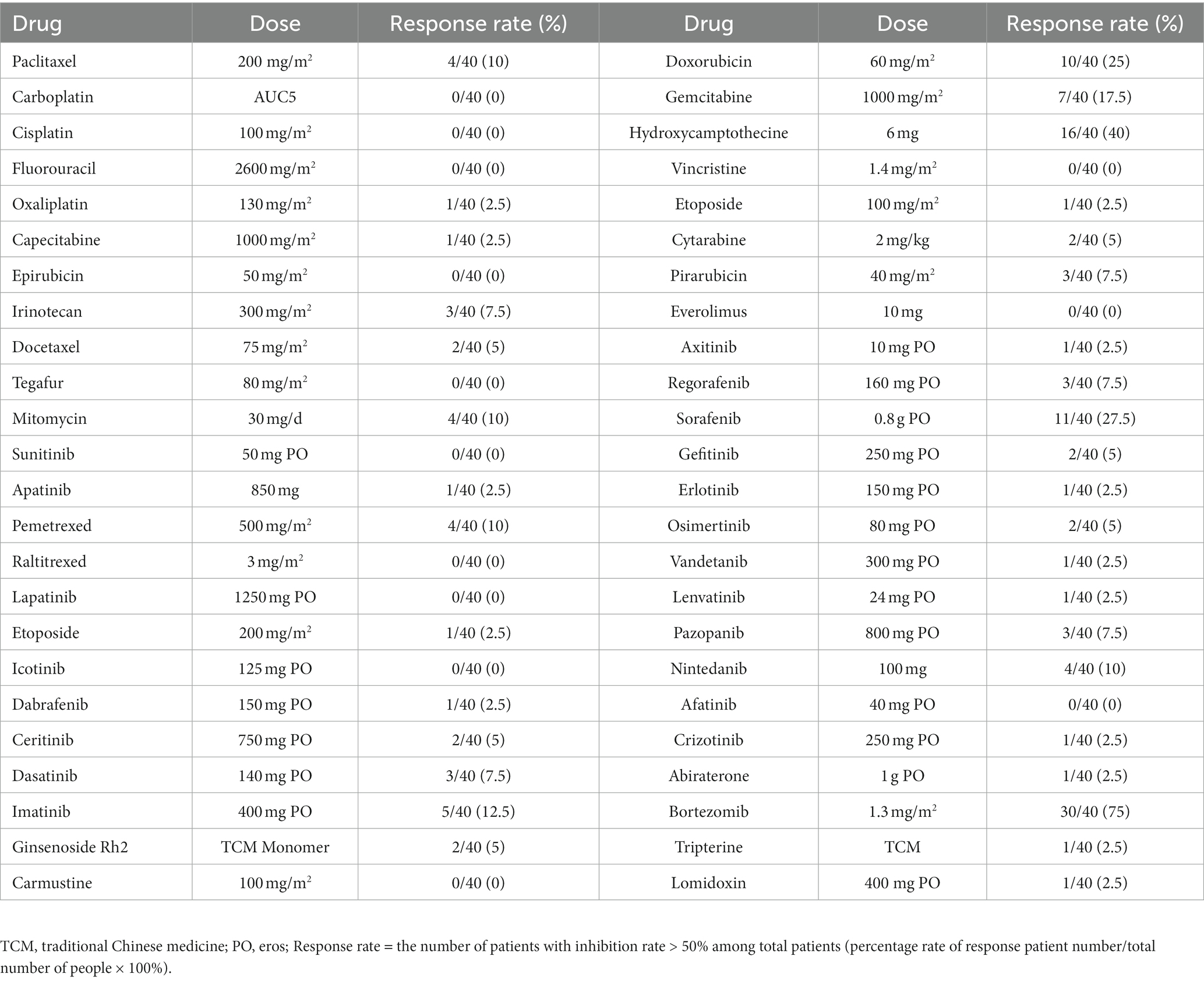
Table 3. The response rate of ESCC cells by different therapeutic drugs determined by HDS technique.
In vivo validation of drug effects in PDX mouse model
Model construction
Approximately 5 × 106 ESCC progenitor cells were injected into the ESCC fat pad and the axillary area of the right forelimb of 6-week-old female NCG mice. The volume and growth rate of ESCC tumors in mice were routinely recorded every 3 days. Tumor formation was observed at both graft sites on day 15 post-administration. Tumor proliferation was evident from day 15 to day 30, indicating a successful model establishment.
Dose optimization
The doses of the screening drugs were set at 25, 50, and 100 mg/kg/d. After the model was successfully constructed, the drug treatment group mice were administered with designated drug regimens either by oral gavage or intraperitoneal (i.p) injection for 20 days, while paclitaxel (70 mg/kg/d) (17) treated mice served as the positive control group, and vehicle (DMSO)-treated mice were included in the sham group.
Treatment protocol for ESCC subjects in the observation and control groups
Forty ESCC patients in the control group were treated with paclitaxel combined with a cisplatin regimen (18). On day 1, 135 mg/m2 of paclitaxel injection (Jiangsu Aosikang Pharma., State Drug Quantification H20083848; 2 mL: 30 mg) was added to 500 mL of glucose injection (Jiangsu Hengrui Pharma., State Drug Quantification H32022368; 250 mL: 12.5 g) and diluted into intravenous (i.v) drip for 3 h/d. On days 2–4, subjects received i.v injections of 50 mg/m2 of cisplatin (Dezhou Deyao Pharma., State Drug Quantification H20023236; 20 mg), dissolved in 500 mL of 0.9% injection-grade sodium chloride solution (Shijiazhuang IV Pharma., State Drug Quantification H13023200; 500 mL) for 2 h each time in a stretch of 3 weeks. The treatment lasted six cycles.
On the other hand, 40 ESCC patients in the observation group were treated according to screened therapeutic agents.
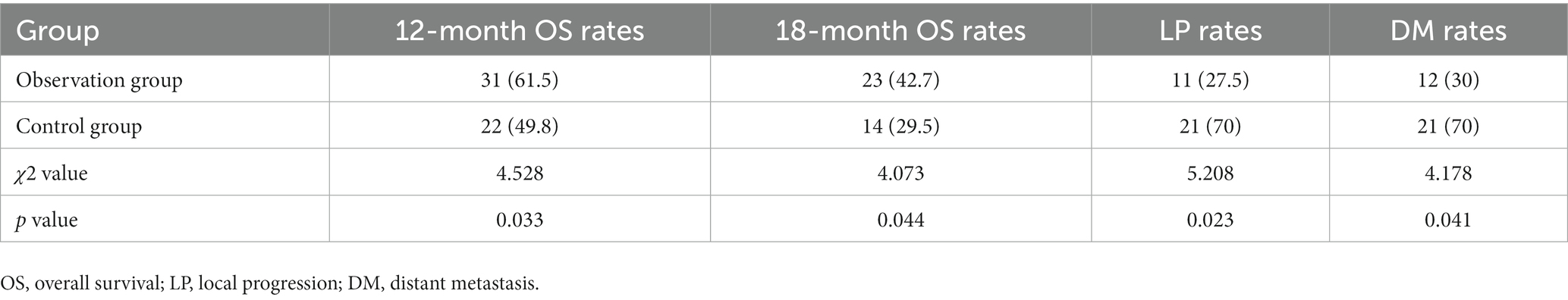
We compared several parameters between these two groups, such as the sensitivity of different anticancer drugs in ESCC cells by the HDS method, the difference in tumor growth curves before and after drug therapy in the PDX mice as well as relative expression levels of tumor markers, including carcinoembryonic antigen (CEA), serum cytokeratin protein fragment 19 (CYFRA21-1) and squamous epithelial cell carcinoma antigen (SCCA) in the ESCC patients. Furthermore, we compared the differences in overall survival (OS) rates, local progression (LP) rates, and distant metastasis (DM) rates between the two groups of patients at 12 and 18 months after chemotherapy. OS rate = number of survivors/total number × 100%. LP rate = number of local tumor diameter increase/total number × 100%. DM rate = number of new distant metastases/total number × 100%.
Statistical analysis
SPSS v22.0 software (IBM Corp., Armonk, NY, United States) was used for all statistical analyses. Measured and counted data were expressed as xs and %, respectively. The t-test was used to compare the differences in expressions of CEA, CYFRA21-1, and SCCA in ESSC patients before and after treatment with different drugs as well as tumor volumes and masses in PDX models under various treatment conditions. Least significant difference (LSD) method was used for multiple comparisons. Chi-square (χ2) test was used to compare the rate of local progression and distant metastasis between the two groups. A p-value of less than 0.05 was considered statistically significant.
Results
Identification of highly effective drugs against patient-derived ESCC cell models by HDS method
Forty ESCC patients were subjected to treatment with 9 different highly sensitive chemotherapeutic agents, with the largest number of patients exhibiting significant sensitivities to bortezomib, as described in Table 4.
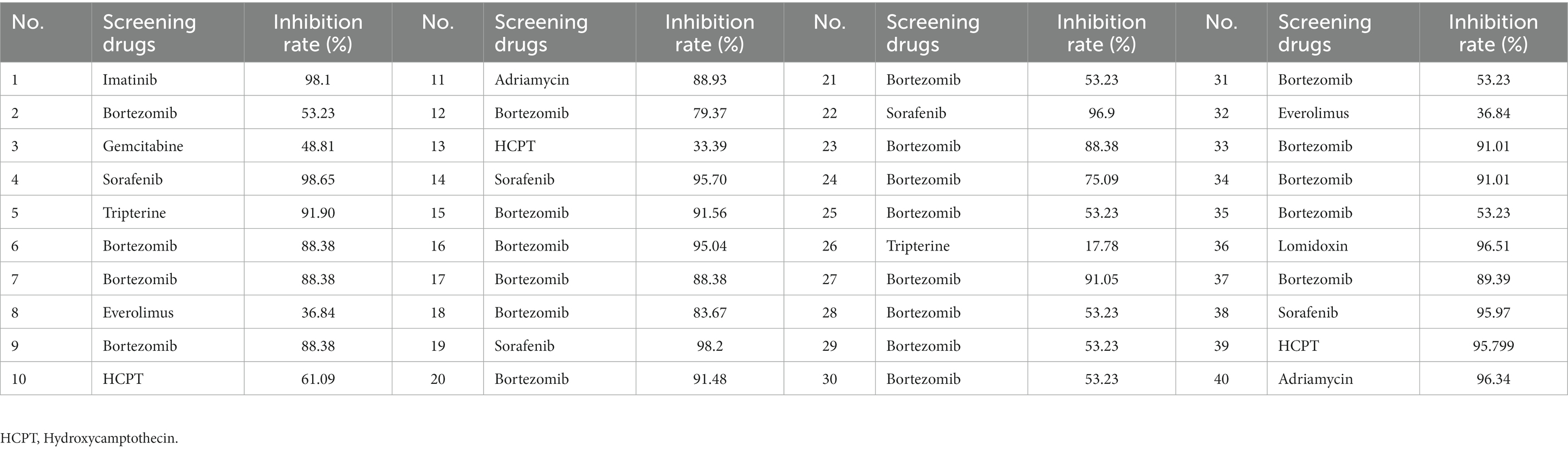
Table 4. Screening of drug sensitivity by measuring tumor cell inhibition rates (%)in 40 ESCC patients in the observation group.
HDS-screened drugs show inhibition of tumor growth in the PDX mice
We found that HDS-screened drug-treated PDX mice had significantly smaller tumor Volumes and masses compared to that in the vehicle-treated sham (p < 0.05), and Paclitaxel treatment groups, as illustrated in Figure 4.
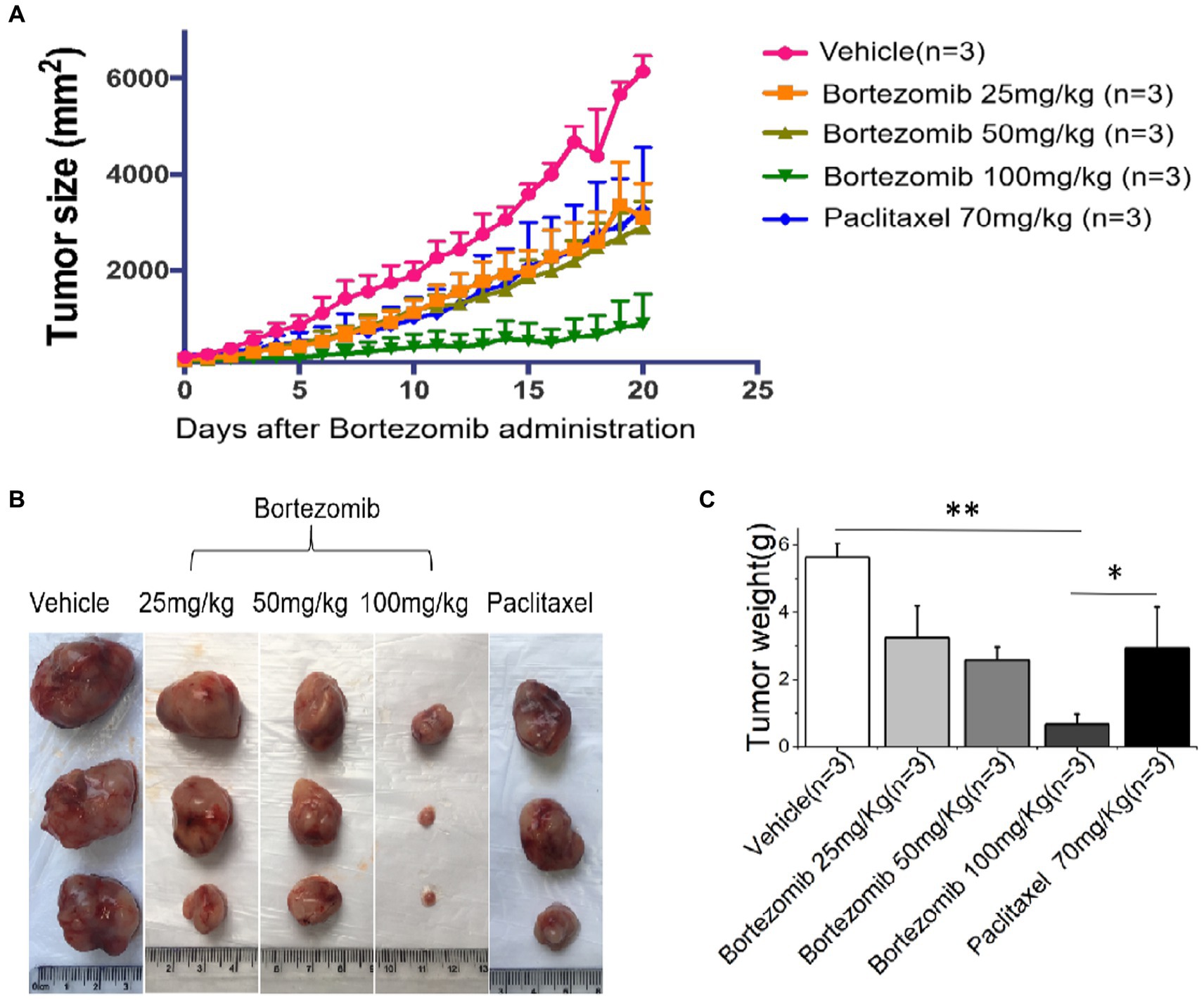
Figure 4. An improved inhibitory effect of HDS-screened drugs on tumor cell growth in PDX animal models compared to paclitaxel and vehicle. (A) The PDX mice were treated with vehicle, Bortezomib (25 mg/kg, 50 mg/kg, 100 mg/kg), or Paclitaxel (70 mg/kg) every day for 20 consecutive days. Tumor volumes were measured each day. The curves show the mean volume ± SD of tumor sizes. (B) Representative images of xenografts harvested after 20 days of treatment for comparison of tumor volumes across the groups. (C) The bar chart shows the difference in xenograft quality in different groups.*p < 0.05, **p < 0.01.
Treatment with antitumor agents alleviates the expression of tumor markers in ESCC patients
There were no statistical differences in the expression of CEA, CYFRA21-1, and SCCA between the two groups at the baseline (t = 0.327, 0.088, 0.276; p = 0.745, 0.930, 0.783), while significantly lower expression levels of these protein markers were detected in the observation group than those in the control group (t = 5.692, 5.748, 10.817; p < 0.001) after chemotherapy, as shown in Table 5.
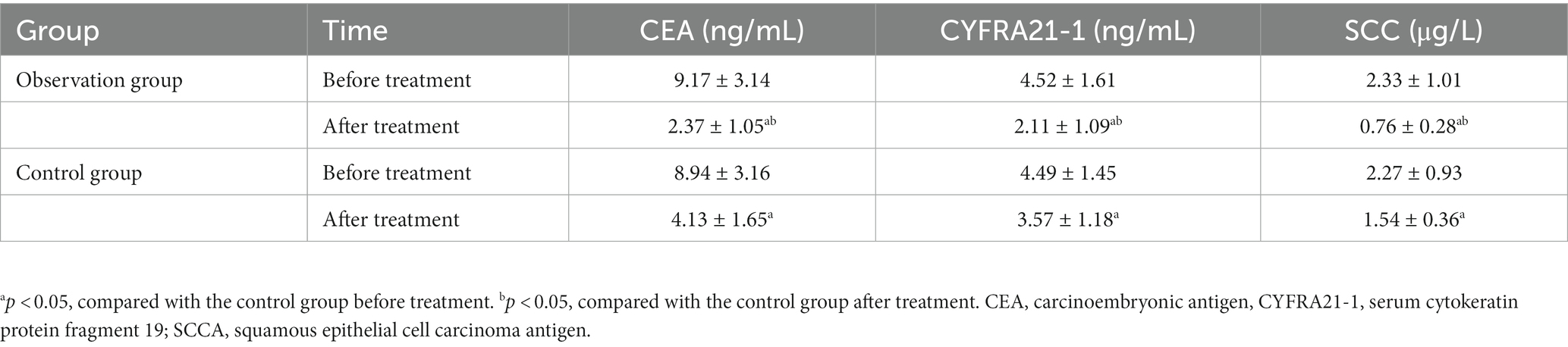
Table 5. Comparison of tumor markers’ levels before and after drug therapy between the observation and control groups.
HDS-guided treatment enhanced the clinical efficacy of chemotherapeutics
We found that the OS rates at 12 months and 18 months were significantly higher in the observation group than those in the control group (p < 0.05), while rates of LP and DM at 18 months were significantly lower in the observation group than those in the control group (p < 0.05), as shown in Table 6.
Discussion
Since the majority of patients with progressive ESCC are diagnosed at the advanced stage, most clinicians adopt palliative radiotherapy, chemotherapy, biological therapy, and traditional Chinese medicine (TCM) therapies (19–22). Notably, due to extensive heterogeneity in tumor characteristics across ESCC patients, individualized chemotherapeutic strategies are urgently needed to achieve the best therapeutic outcome and minimum drug toxicity (23, 24). Most clinical researchers exploit the patient-derived tumor cell-derived PDX rodents to assess the cell growth inhibitory effect of different regimens of anticancer drugs to overcome the problem of adverse drug toxicity in critically ill ESCC patients (25, 26). However, these in vitro testing models have their limitations, such as high cost and limited capability in accommodating large panels of therapeutic drugs. Some studies have demonstrated the broader benefits of organoid models using patient-derived cancer stem cells mimicking in vivo tumor microenvironment (TME) for drug screening (27, 28). Again, the efficacy of this model is limited by challenges in accurately modeling cancer organoids, high maintenance costs, and difficulty in simultaneously generating numerous organoids for high-throughput screening strategies.
Recently, high-throughput gene sequencing (HGS) technology has been used for the targeted therapy of ESCC. Some groups have reported the genetic landscape of human ESCCs using the whole genome and exome sequencing approaches (29–31), which has led to promising results in early studies of personalized and targeted therapies (32). However, due to an increase in the oncogenic mutational burden, differences in cellular signaling pathways, and complex interactions among tumor genes, the same tumor gene target may respond differentially to different drugs affecting the overall outcome of individualized therapy (33).
In contrast, the HDS technology used by our group extended the scope of screening with a large panel of FDA-approved drugs for determining the best possible drug regimen exerting maximum cell growth inhibitory effects. With this technically upgraded screening platform, we can rapidly identify highly sensitive and targeted chemotherapeutics for different ESCC patients, thereby solving most of the previously mentioned technical hurdles.
The primary aim of this study was to obtain high-quality primary tumor cells from ESCC patients and subsequently exploit them for in vitro drug sensitivity screening strategies as an alternative to in vivo models. We used fresh tumor tissue masses to isolate primary ESCC cells through trypsin digestion with a success rate as high as 90%, which was considerably higher than that reported in other studies (34). The HDS technique developed by our group could simultaneously measure the sensitivity of more than 300 drugs to patient-derived ESCC tumor cells, along with the screening of optimal drug concentrations to reduce the adverse drug toxicity effects. In this study, primary tumor cells from 40 ESCC patients were tested for drug sensitivity against 9 different chemotherapeutic agents, which fully illustrated the inter-patient heterogeneity of tumor cells (35). Moreover, the results of this study further signify the importance of individualized chemotherapeutic regimens in treating highly heterogeneous carcinomas like ESCC. Through this screening assay, we noticed that most ESCC patients were sensitive to Bortezomib.
Bortezomib is a 20S proteasome inhibitor that exerts its antitumor effects by interfering with cell signaling, causing cell cycle arrest, inducing apoptosis, and inhibiting angiogenesis and is currently used mainly for the treatment of multiple myeloma (36, 37). However, one study (38) reported its role in enhancing the sensitivity of ESCC to radiotherapy. Besides, there are few a studies on the application of Bortezomib in chemotherapy for advanced-stage ESCC (39). Here, we found that Bortezomib was effective in chemotherapy for esophageal squamous carcinoma by in vitro HDS assay, which might provide a new avenue for the treatment of ESCC.
To further verify the results of in vitro experiments, the previously reported PDX model (40) was used to determine the therapeutic effects of screened anticancer drugs in vivo. We exploited patient-derived primary ESCC cells for constructing the PDX mouse model to screen potential anticancer drugs. In addition, we used Paclitaxel (41) as the positive control in this animal study. The results showed that the tumor tissue mass and volume of PDX mice treated with screened high-sensitivity drugs were significantly smaller than those treated with Paclitaxel or the vehicle control (p < 0.05). Finally, we determined the optimal drug concentration to achieve the best possible tumor growth inhibition in this PDX mouse model.
After achieving encouraging results in animal experiments, we conducted individualized treatment for 40 ESCC patients with Bortezomib and compared the anticancer efficacy with 40 ESCC patients who received Paclitaxel combined with cisplatin chemotherapy in the same period of time. We first analyzed the differences in expressions of tumor markers CEA, CYFRA21-1, and SCCA between the two groups. CEA is one of the first tumor markers found to be widely present in tumor cells. CYFRA21-1 is also commonly detected in lung and esophageal tumor cells and exhibits a high sensitivity along with the diagnostic value in the evaluation of therapeutic effect against ESCC (42). SCCA is mainly present in tumor cells of the lung, pharynx, and esophagus and exhibits a high sensitivity for occurrence and progression of ESCC with high specificity (43). Some studies (44–46) have reported that the combination of these three biomarkers could be a valuable prediction tool for the precise diagnosis of EC and for predicting the disease progression rate as well. Here, we found that CEA, CYFRA21-1, and SCCA levels were significantly lower than those in the control group after chemotherapy treatment in the observation group (p < 0.05), suggesting that the HDS-screened anticancer drug was effective in controlling the ESCC tumor growth in the observation group. Furthermore, we analyzed the treatment efficacies in the two groups based on clinical symptoms and found that the 12-month and 18-month OS rates in the observation group were significantly higher than those in the control group (p < 0.05), while rates of LP and DM were significantly lower than those in the control group (p < 0.05), indicating promising efficacy of HDS-screened drugs against ESCC.
In summary, the HDS method is an efficient, reproducible, and easy-to-use drug sensitivity screening tool that can expedite the ESCC diagnosis, especially for terminally ill patients. Moreover, drugs for antitumor sensitivity on the HDS platform consistently exhibited significant inhibitory effects on ESCC tumor growth in disease animal models as well as human ESCC patients.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary materials, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of the 901th Hospital of the Joint Support Force. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Ethics Committee of the 901th Hospital of the Joint Support Force.
Author contributions
The study was conceived and designed by XnH, JT, HY, JH, and WW. XnH, HY, and JH equally contributed to performing all the required experiments, analyzing data, preparing the manuscript, and further revisions. SY and XD contributed to the data acquisition and analysis. JT, HL, JW, JL, YG, XC, and MZ provided human adipose tissue samples. JH, HY, WW, and XnH performed the HDS experiments. QG, XlH, GL, and JH performed animal studies and immunohistochemical analysis. XlH, HY, XD, HL, JW, SG, and JT were responsible for clinical studies. WW and JT provided critical suggestions for this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Anhui Key Research and Development Program (1804 h08020257).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2023.1212851/full#supplementary-material
Abbreviations
HDS, High-throughput Drug Sensitivity; EC, Esophageal Carcinoma; ESCC, Esophageal Squamous Cell Carcinoma; PDX, Patient-derived Tumor Xenograft; OS, Overall Survival; LP, Local Progression; DM, Distant Metastasis; CEA, Carcinoembryonic Antigen; CYFRA21-1, Serum Cytokeratin Protein Fragment 19; SCCA, Squamous Epithelial Cell Carcinoma Antigen; NCCN, National Comprehensive Cancer Network; FDA, Food and Drug Administration; HGS, High-throughput Gene Sequencing.
References
1. Zhou, B, Bie, F, Zang, R, Zhang, M, Song, P, Liu, L, et al. Global burden and temporal trends in incidence and mortality of oesophageal cancer. J Adv Res. (2022) S2090-1232:00236–3. doi: 10.1016/j.jare.2022.10.007
2. Fan, J, Liu, Z, Mao, X, Tong, X, Zhang, T, Suo, C, et al. Global trends in the incidence and mortality of esophageal cancer from 1990 to 2017. Cancer Med. (2020) 9:6875–87. doi: 10.1002/cam4.3338
3. Sung, H, Ferlay, J, Siegel, RL, Laversanne, M, Soerjomataram, I, Jemal, A, et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
4. Arnold, M, Abnet, CC, Neale, RE, Vignat, J, Giovannucci, EL, McGlynn, KA, et al. Global burden of 5 major types of gastrointestinal Cancer. Gastroenterology. (2020) 159:335–49.e15. doi: 10.1053/j.gastro.2020.02.068
5. Huang, FL, and Yu, SJ. Esophageal cancer: risk factors, genetic association, and treatment. Asian J Surg. (2018) 41:210–5. doi: 10.1016/j.asjsur.2016.10.005
6. Xi, Y, Shen, Y, Wu, D, Zhang, J, Lin, C, Wang, L, et al. CircBCAR3 accelerates esophageal cancer tumorigenesis and metastasis via sponging miR-27a-3p. Mol Cancer. (2022) 21:145. doi: 10.1186/s12943-022-01615-8
7. Niu, C, Liu, Y, Wang, J, Liu, Y, Zhang, S, Zhang, Y, et al. Risk factors for esophageal squamous cell carcinoma and its histological precursor lesions in China: a multicenter cross-sectional study. BMC Cancer. (2021) 21:1034. doi: 10.1186/s12885-021-08764-x
8. Bolger, JC, Donohoe, CL, Lowery, M, and Reynolds, JV. Advances in the curative management of oesophageal cancer. Br J Cancer. (2022) 126:706–17. doi: 10.1038/s41416-021-01485-9
9. Sheil, F, Donohoe, CL, King, S, O'Toole, D, Cunningham, M, Cuffe, S, et al. Outcomes for esophageal squamous cell carcinoma treated with curative intent in a Western cohort: should multimodal therapy be the gold standard? World J Surg. (2018) 42:1485–95. doi: 10.1007/s00268-017-4289-8
10. Xu, J, Li, Y, Fan, Q, Shu, Y, Yang, L, Cui, T, et al. Clinical and biomarker analyses of sintilimab versus chemotherapy as second-line therapy for advanced or metastatic esophageal squamous cell carcinoma: a randomized, open-label phase 2 study (ORIENT-2). Nat Commun. (2022) 13:857. doi: 10.1038/s41467-022-28408-3
11. Li, Y, Xu, F, Chen, F, Chen, Y, Ge, D, Zhang, S, et al. Transcriptomics based multi-dimensional characterization and drug screen in esophageal squamous cell carcinoma. EBioMedicine. (2021) 70:103510. doi: 10.1016/j.ebiom.2021.103510
12. Li, J, Xiong, X, Wang, Z, Zhao, Y, Shi, Z, Zhao, M, et al. In vitro high-throughput drug sensitivity screening with patient-derived primary cells as a guide for clinical practice in hepatocellular carcinoma-a retrospective evaluation. Clin Res Hepatol Gastroenterol. (2020) 44:699–710. doi: 10.1016/j.clinre.2020.01.003
13. Kochanek, SJ, Close, DA, and Johnston, PA. High content screening characterization of head and neck squamous cell carcinoma multicellular tumor spheroid cultures generated in 384-well ultra-low attachment plates to screen for better Cancer drug leads. Assay Drug Dev Technol. (2019) 17:17–36. doi: 10.1089/adt.2018.896
14. Xing, L, Wang, H, Liu, D, He, Q, and Li, Z. Case report: successful management of a refractory double-expressor diffuse large B-cell lymphoma patient under the guidance of in vitro high-throughput drug sensitivity test. Front Oncol. (2023) 12:1079890. doi: 10.3389/fonc.2022.1079890
15. Zhang, X, Liang, H, Li, Z, Xue, Y, Wang, Y, Zhou, Z, et al. Perioperative or postoperative adjuvant oxaliplatin with S-1 versus adjuvant oxaliplatin with capecitabine in patients with locally advanced gastric or gastro-oesophageal junction adenocarcinoma undergoing D2 gastrectomy (RESOLVE): an open-label, superiority and non-inferiority, phase 3 randomised controlled trial. Lancet Oncol. (2021) 22:1081–92. doi: 10.1016/S1470-2045(21)00297-7
16. Kato, K, Shah, MA, Enzinger, P, Bennouna, J, Shen, L, Adenis, A, et al. KEYNOTE-590: phase III study of first-line chemotherapy with or without pembrolizumab for advanced esophageal cancer. Future Oncol. (2019) 15:1057–66. doi: 10.2217/fon-2018-0609
17. Wang, J, Li, Q, Yuan, J, Wang, J, Chen, Z, Liu, Z, et al. CDK4/6 inhibitor-SHR6390 exerts potent antitumor activity in esophageal squamous cell carcinoma by inhibiting phosphorylated Rb and inducing G1 cell cycle arrest. J Transl Med. (2017) 15:127. doi: 10.1186/s12967-017-1231-7
18. Ajani, JA, D'Amico, TA, Bentrem, DJ, Cooke, D, Corvera, C, Das, P, et al. Esophageal and Esophagogastric junction cancers, version 2.2023, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. (2023) 21:393–422. doi: 10.6004/jnccn.2023.0019
19. Yang, YM, Hong, P, Xu, WW, He, QY, and Li, B. Advances in targeted therapy for esophageal cancer. Signal Transduct Target Ther. (2020) 5:229. doi: 10.1038/s41392-020-00323-3
20. He, S, Xu, J, Liu, X, and Zhen, Y. Advances and challenges in the treatment of esophageal cancer. Acta Pharm Sin B. (2021) 11:3379–92. doi: 10.1016/j.apsb.2021.03.008
21. Sheikh, M, Roshandel, G, McCormack, V, and Malekzadeh, R. Current status and future prospects for esophageal Cancer. Cancers (Basel). (2023) 15:765. doi: 10.3390/cancers15030765
22. Kakeji, Y, Oshikiri, T, Takiguchi, G, Kanaji, S, Matsuda, T, Nakamura, T, et al. Multimodality approaches to control esophageal cancer: development of chemoradiotherapy, chemotherapy, and immunotherapy. Esophagus. (2021) 18:25–32. doi: 10.1007/s10388-020-00782-1
23. Cancer Genome Atlas Research Network, Analysis Working Group: Asan University, BC Cancer Agency, Brigham and Women’s Hospital, Broad Institute, Brown University, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. (2017) 541:169–75. doi: 10.1038/nature20805
24. Zhao, W, Zhao, J, Kang, L, Li, C, Xu, Z, Li, J, et al. Fluoroscopy-guided salvage photodynamic therapy combined with nanoparticle albumin-bound paclitaxel for locally advanced esophageal Cancer after Chemoradiotherapy: a case report and literature review. Cancer Biother Radiopharm. (2022) 37:410–6. doi: 10.1089/cbr.2020.4595
25. Lan, T, Xue, X, Dunmall, LC, Miao, J, and Wang, Y. Patient-derived xenograft: a developing tool for screening biomarkers and potential therapeutic targets for human esophageal cancers. Aging (Albany NY). (2021) 13:12273–93. doi: 10.18632/aging.202934
26. Zhong, M, and Fu, L. Culture and application of conditionally reprogrammed primary tumor cells. Gastroenterol Rep (Oxf). (2020) 8:224–33. doi: 10.1093/gastro/goaa023
27. Sachdeva, UM, Shimonosono, M, Flashner, S, Cruz-Acuña, R, Gabre, JT, and Nakagawa, H. Understanding the cellular origin and progression of esophageal cancer using esophageal organoids. Cancer Lett. (2021) 509:39–52. doi: 10.1016/j.canlet.2021.03.031
28. Lau, HCH, Kranenburg, O, Xiao, H, and Yu, J. Organoid models of gastrointestinal cancers in basic and translational research. Nat Rev Gastroenterol Hepatol. (2020) 17:203–22. doi: 10.1038/s41575-019-0255-2
29. Zeng, H, Zheng, R, Guo, Y, Zhang, S, Zou, X, Wang, N, et al. Cancer survival in China, 2003-2005: a population-based study. Int J Cancer. (2015) 136:1921–30. doi: 10.1002/ijc.29227
30. Gao, YB, Chen, ZL, Li, JG, Hu, XD, Shi, XJ, Sun, ZM, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. (2014) 46:1097–102. doi: 10.1038/ng.3076
31. Lin, DC, Hao, JJ, Nagata, Y, Xu, L, Shang, L, Meng, X, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. (2014) 46:467–73. doi: 10.1038/ng.2935
32. Roychowdhury, S, Iyer, MK, Robinson, DR, Lonigro, RJ, Wu, YM, Cao, X, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. (2011) 3:111ra121. doi: 10.1126/scitranslmed.3003161
33. Kang, X, Chen, K, Li, Y, Li, J, D'Amico, TA, and Chen, X. Personalized targeted therapy for esophageal squamous cell carcinoma. World J Gastroenterol. (2015) 21:7648–58. doi: 10.3748/wjg.v21.i25.7648
34. Rockett, JC, Larkin, K, Darnton, SJ, Morris, AG, and Matthews, HR. Five newly established oesophageal carcinoma cell lines: phenotypic and immunological characterization. Br J Cancer. (1997) 75:258–63. doi: 10.1038/bjc.1997.42
35. Dinh, HQ, Pan, F, Wang, G, Huang, QF, Olingy, CE, Wu, ZY, et al. Integrated single-cell transcriptome analysis reveals heterogeneity of esophageal squamous cell carcinoma microenvironment. Nat Commun. (2021) 12:7335. doi: 10.1038/s41467-021-27599-5
36. Tan, CRC, Abdul-Majeed, S, Cael, B, and Barta, SK. Clinical pharmacokinetics and pharmacodynamics of Bortezomib. Clin Pharmacokinet. (2019) 58:157–68. doi: 10.1007/s40262-018-0679-9
37. Cengiz Seval, G, and Beksac, M. The safety of bortezomib for the treatment of multiple myeloma. Expert Opin Drug Saf. (2018) 17:953–62. doi: 10.1080/14740338.2018.1513487
38. Wang, D, Qin, Q, Jiang, QJ, and Wang, DF. Bortezomib sensitizes esophageal squamous cancer cells to radiotherapy by suppressing the expression of HIF-1α and apoptosis proteins. J Xray Sci Technol. (2016) 24:639–46. doi: 10.3233/XST-160571
39. Zhang, L, Niu, X, Bi, Y, Cui, H, Li, H, and Cheng, X. Potential role of targeting KDR and proteasome inhibitors in the therapy of esophageal squamous cell carcinoma. Technol Cancer Res Treat. (2020) 19:1533033820948060. doi: 10.1177/1533033820948060
40. Liu, J, Liu, ZX, Wu, QN, Lu, YX, Wong, CW, Miao, L, et al. Long noncoding RNA AGPG regulates PFKFB3-mediated tumor glycolytic reprogramming. Nat Commun. (2020) 11:1507. doi: 10.1038/s41467-020-15112-3
41. Shitara, K, Özgüroğlu, M, Bang, YJ, Di Bartolomeo, M, Mandalà, M, Ryu, MH, et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet. (2018) 392:123–33. doi: 10.1016/S0140-6736(18)31257-1
42. Ju, M, Ge, X, Di, X, Zhang, Y, Liang, L, and Shi, Y. Diagnostic, prognostic, and recurrence monitoring value of plasma CYFRA21-1 and NSE levels in patients with esophageal squamous cell carcinoma. Front Oncol. (2022) 11:789312. doi: 10.3389/fonc.2021.789312
43. Kitasaki, N, Hamai, Y, Emi, M, Kurokawa, T, Yoshikawa, T, Hirohata, R, et al. Prognostic factors for patients with esophageal squamous cell carcinoma after neoadjuvant chemotherapy followed by surgery. In Vivo. (2022) 36:2852–60. doi: 10.21873/invivo.13025
44. Zheng, Q, Zhang, L, Tu, M, Yin, X, Cai, L, Zhang, S, et al. Development of a panel of autoantibody against NSG1 with CEA, CYFRA21-1, and SCC-ag for the diagnosis of esophageal squamous cell carcinoma. Clin Chim Acta. (2021) 520:126–32. doi: 10.1016/j.cca.2021.06.013
45. Yajima, S, Suzuki, T, Oshima, Y, Shiratori, F, Funahashi, K, Kawai, S, et al. New assay system Elecsys anti-p 53 to detect serum anti-p 53 antibodies in esophageal Cancer patients and colorectal Cancer patients: multi-institutional study. Ann Surg Oncol. (2021) 28:4007–15. doi: 10.1245/s10434-020-09342-4
Keywords: esophageal squamous cell carcinoma, high-throughput drug sensitivity screening technology, drug sensitivity, patient-derived tumor xenografts, therapeutic efficacy
Citation: He X, Yan H, Hu J, Duan X, Zhang M, Li H, Wang J, Gao Q, Yu S, Hou X, Liao G, Guo S, Li J, Ge Y, Chen X, Wang W and Tang J (2023) HDS screening with patient-derived primary cells guided individualized therapy for esophageal squamous cell carcinoma–in vivo and vitro. Front. Med. 10:1212851. doi: 10.3389/fmed.2023.1212851
Edited by:
Behzad Mansoori, Molecular & Cellular Oncogenesis Program, United StatesReviewed by:
Kajal Gupta, Rush University, United StatesManuela Petti, Sapienza University of Rome, Italy
Daniel Gracilla, Augusta University, United States
Copyright © 2023 He, Yan, Hu, Duan, Zhang, Li, Wang, Gao, Yu, Hou, Liao, Guo, Li, Ge, Chen, Wang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenchao Wang, wwcbox@hmfl.ac.cn; Jun Tang, tangjun1974@163.com
†These authors have contributed equally to this work
 Xing He
Xing He Hezhong Yan1†
Hezhong Yan1†  Wenchao Wang
Wenchao Wang